
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Immunol. , 18 October 2022
Sec. Viral Immunology
Volume 13 - 2022 | https://doi.org/10.3389/fimmu.2022.1008285
Since immune system and internal environment in vivo are large and complex, the interpretation of the observed immune effect from the perspective of a single immune cell or antibody seems a little feeble. Many studies have shown that specific antibodies against “ former” viruses have a reduced ability to neutralize “new” mutant strains. However, there is no comprehensive and clear view of whether there will be Antibody-dependent enhancement (ADE). We review the latest relevant studies, hoping to explain the ADE of SARS-CoV-2 infection sometimes observed in some patients.
Antibody-dependent enhancement (ADE) has been observed in many coronaviruses, such as Feline infectious peritonitis (FIP) virus, SARS-CoV and Middle East Respiratory Syndrome Coronavirus (MERS-CoV) (1), which raises concerns about a possible aggravation of SARS-CoV-2 infection due to preexisting antibodies (2). Most of the studies on the mechanism of ADE of SARS-CoV-2 are related to the severity of infection. There is increasing evidence that ADE of SARS-CoV-2 does exist although rarely observed. At present, there is a lack of comprehensive and in-depth research on its specific mechanism. Results of different researches are even controversial. Different results are observed in different tissues of the body (3, 4). The results observed in vivo or in vitro also differ (5). Different results were observed even for the same cells (6). We review some recent researches on the mechanism of ADE of SARS-CoV-2, and summarize relevant viewpoints and countermeasures, as well as multiple roles of macrophages in SARS-CoV-2 infection, in order to benefit the research on ADE of SARS-CoV-2.
Several previous studies (2–8) showed that ADE of SARS-CoV-2 infection had at least two mechanisms. The Receptor binding domain (RBD)-specific ADE antibodies enhance infection depending on Fc receptors. The N-terminal domain (NTD)-specific ADE antibodies are independent of Fc receptors. Instead, they affect the binding of S protein to Angiotensin-converting enzyme 2 (ACE2) receptor by changing the conformation of S protein. Neutralizing antibodies to the RBD of SARS-CoV-2 or the NTD of SARS-CoV-2 both mediate ADE effects, as shown in Figure 1. And some nonspecific antibodies also mediate ADE effects.
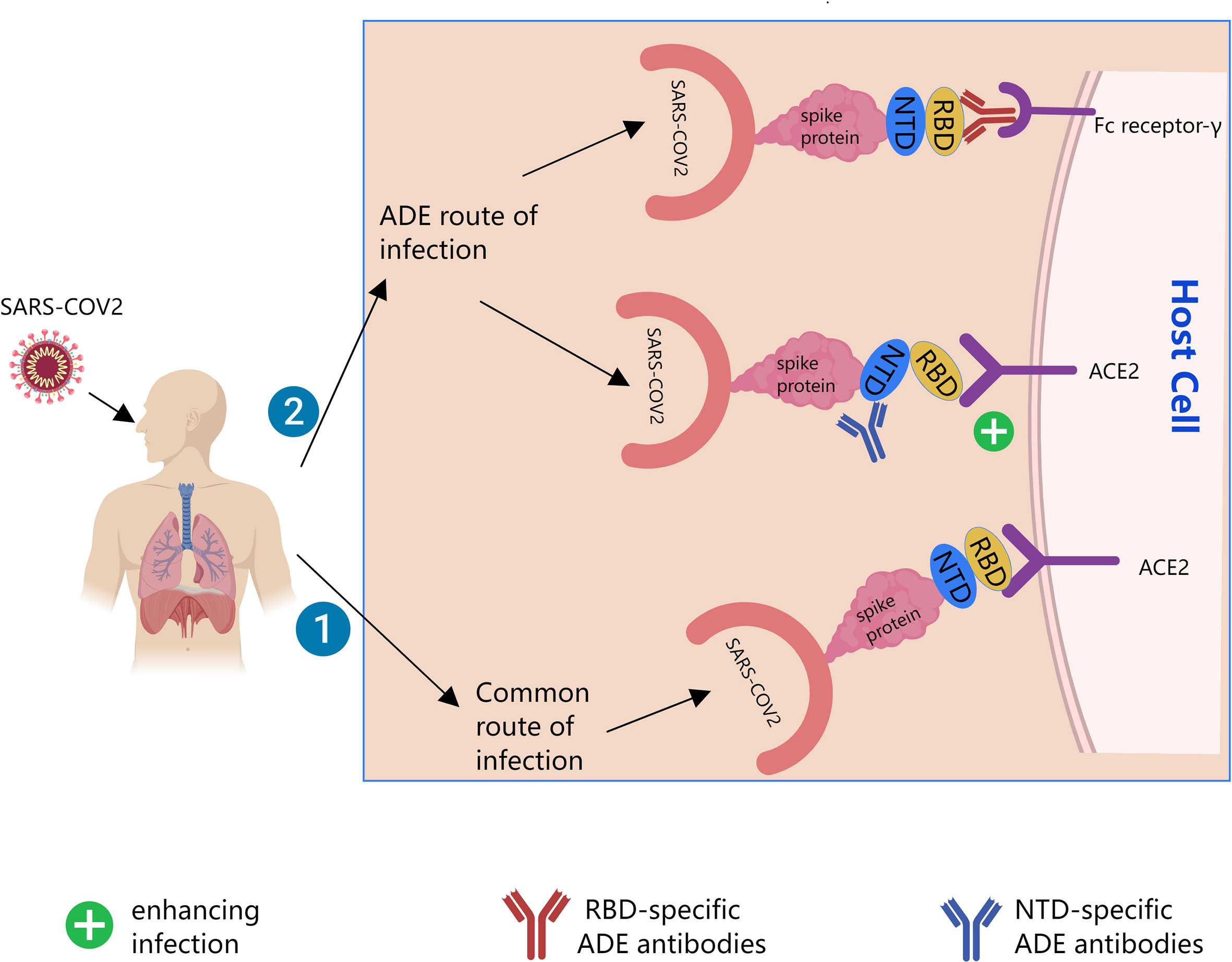
Figure 1 Possible mechanisms of Antibody-dependent enhancement (ADE) of SARS-CoV-2. The black line is the path direction of the infection. Pathway 1 is common pathway by which SARS-CoV-2 infect cells. Pathway 2 is possible ADE pathway by which SARS-CoV-2 infect cells. ACE2, angiotensin-converting enzyme 2; RBD, receptor binding domain; NTD, the N-terminal domain.
In 2020, Wang et al. (5) reported a monoclonal antibody (mAb)-MW05, which had SARS-CoV-2 neutralizing activity by disrupting the interaction of RBD with ACE2 receptor. ADE mediated by cross-linking of the Fc of MW05 with FcγRIIB provided the first evidence that SARS-CoV-2 monoclonal antibodies had an ADE effect in vitro, which could be eliminated by introducing LALA mutations into the Fc region (MW05/LALA). Their latest study in 2022 (4) reported that two neutralizing monoclonal antibodies, MW01 and MW05, could enhance the infection of FcγRIIB-expressing B cells by SARS-CoV-2 pseudovirus. This identified a novel ADE mechanism that FcγRIIB mediated the uptake of bivalent virus-antibody complexes for the SARS-CoV-2 pseudovirus in vitro. FcγRIIB (CD32B) is mainly expressed on myeloid cells, such as leukocyte lineages (Raji, THP-1 and K562), B cells (the only FcR on the surface of B cells), and plays an important role in the negative regulation of B cell function.
Some researchers studied the infection mechanism of ADE of SARS-CoV-2 using convalescent plasma from Coronavirus disease 2019 (COVID-19) patients and found that the ADE was mainly mediated by two types of FcγRs, FcγRIIA and FcγRIIIA (3). FcγRIIA (CD32A) is mainly expressed on the surface of neutrophils, monocytes, platelets and DC cells. It has been shown that the IgG lacking core fucosylation could initiate enhanced antibody-dependent cytotoxicity by increasing affinity for the Fc receptor FcRIIIA (9). FcγRIIIA (CD16A) is expressed on almost all leukocytes, mainly natural killer cells (NK) and macrophages.
The study of Liu et al. (7) screened some anti-spike monoclonal antibodies from patients with COVID-19. Mutational analysis revealed that all antibodies enhancing infection recognized a specific site on the NTD. Structural analysis showed that all infectivity-enhancing antibodies bound NTDs in a similar manner. And by inducing the open conformation of RBD, the binding ability of the spike protein to ACE2 and the infectivity of SARS-CoV-2 were enhanced. Not only neutralizing antibodies but also ADE-enhancing antibodies are produced during SARS-CoV-2 infection.
Macrophages play multiple roles in viral infection. First, they can remove the virus, infected cells or debris. Second, although they do not directly damage the lymphoid organs, they can act as a SARS-CoV-2 Trojan horse, facilitating the hiding and spreading of the virus (10–12), thereby promoting the formation of syncytia, which could target the lymphocytes for internalization and cell-in-cell (CIC) mediated death, contributing to lymphopenia (13). Third, they can enhance the recruitment of NK and Cytotoxic T lymphocyte (CTL) at the site of infection, which may kill the infected autologous tissue cells, damage the lymphoid organs. In addition, they can also recruit a variety of lymphocytes and maintain their activity.
Alveolar macrophages are sentinels for immune surveillance of foreign viruses (14). However, macrophages are diverse and are mainly divided into two categories according to their origin, alveolar macrophages (AM) and recruited monocyte-derived macrophages (MDM). Their interaction mechanisms with viruses are not exactly the same. Among them, AM (12) are polarized to M1AM and M2AM, and AM in humans seems to be biased towards M2AM. M1AM promote infection (ADE) and enhance inflammation. M2AM limit viral spread, and suppress inflammation, and enhance viral clearance. Recruited MDM are polarized into M1 and M2 (15). Some studies have found that both M1 and M2 inhibit viral infection. However, some studies have claimed that they have both pro-infection and scavenging effects. M1 destroy pathogens by producing large amounts of pro-inflammatory cytokines. M2 exhibit anti-inflammatory properties and higher phagocytic activity against pathogens. In the early stages of infection, monocyte-derived macrophages recruited to the lung clear infected cells and cellular debris (16). While tissue-resident alveolar macrophages (M1AM) promoted early infection in the lungs. Research has found that human ACE2-overexpressing or knockdown AMs don’t have a major impact on uptake of SARS-CoV-2, suggesting that ACE2 is dispensable for M1AMs to efficiently take up SARS-CoV-2. Mechanical softness could be used to reflect deformability, with greater the deformability comes greater ability to absorb extracellular particles. M1 are much softer than M2 AMs, which favors M1 AMs to efficiently take up SARS-CoV-2. On the other hand, M1 AMs have a lower endosomal pH, favoring membrane fusion and allowing the entry of viral RNA from the endosomes into the cytoplasm, where the virus achieves replication and is packaged to be released.
As the infection progresses, alveolar macrophages are largely depleted. Recruited monocyte-derived macrophages gradually surpass and replace alveolar macrophages. And Lungs have more and more activated M1 macrophages, M1 release a large number of pro-inflammatory factors, which are both beneficial to fight against SARS-CoV-2, and aggravate the inflammatory response, causing severe COVID-19 inflammation. This provides one explanation for the increasing of severe morbidity and mortality in people who are older or have underlying medical conditions, such as hypertension and diabetes (15, 17).
SARS-CoV-2 infects resident CD169+ macrophages in the spleen and lymph nodes via the ACE2 receptor, potentially leading to spleen and lymph node damage, lymphopenia (18). Compared with the normal healthy control group, the total lymphocyte counts were significantly lower in sections from viruses infected spleens, which were was mainly necrotic and apoptotic lymphocytes. This result is also consistent with the observation by Diao et al. (19) that T cell count in COVID-19 patients were significantly reduced, and that surviving T cells were functionally exhausted. There may be several reasons for this. First, the resident macrophages aggravated the infection by promoting the spread of the virus. Second, phagocytosis and clearance of macrophages at the site of infection promoted the recruitment of various cells, such as NK and CTL, which might attack infected lymphoid organs. Furthermore, SARS-CoV-2 could induce lymphocyte apoptosis by enhancing Fas signaling, and could also trigger macrophage secretion of IL-6 and promote lymphopenia.
When COVID-19 occurs, within the alveolar space, SARS-CoV-2 infects alveolar macrophages. And the infected alveolar macrophages promote chemotactic recruitment of T cells and monocytes (10). The recruited T cells produce γ-interferon, which in turn promotes the secretion of pro-inflammatory factors from alveolar macrophages and promotes further activation of T cells. A positive feedback loop exists between SARS-CoV-2-infected alveolar macrophages and T cells, causing persistent alveolar inflammatory response.
Studies have pointed out that anti-recombinant native full-length S protein trimer (triSpike) antibody could mediate SARS-CoV entry into B cells through FcγRII in vitro (20). And the recombinant trimeric S protein was also able to elicit potent protective immune response in vivo. Li et al (8). isolated neutralizing antibodies (NAbs) from individuals with a history of acute or convalescent SARS-CoV-2 or SARS-CoV-1 infection. These antibodies raised against two different crucial domain of S glycoprotein, RBD and NTD, had neutralizing activity, protecting against SARS-CoV-2 infection. They also isolated some non-neutralizing antibodies targeting RBD and NTD, which could enhance viral infection mediated by Fc receptors. Five non-neutralizing NTD antibodies could enhance FcγR-independent infection in vitro. While, two of them that enhanced infection in vitro inhibited SARS-CoV-2 replication in both monkeys and mice. These studies suggest that although antibodies can enhance infection in vitro, they do not necessarily predict enhanced infection in vivo. The possible reason is that non-neutralized NTD Abs can enhance S binding to ACE2 to enhance infection. As the environment is single and few influencing factors in vitro, the enhancement of infection at the level of individual cells is the main manifestations. However, in vivo, non-neutralized NTD Abs not only mediate ADE, but may also mediate Antibody-dependent cell-medicated cytotoxicity (ADCC), Complement dependent cytotoxicity (CDC), and promote the phagocytosis and clearance of dendritic cell (DC)and macrophages. After the virus enters the body, the immune system is activated. The result at this time depends on a net balance of infection enhancement and virus clearance level. Therefore, ADE in SARS-CoV-2 infection and vaccination may be less easily observed. Only in severe infection or immunocompromised conditions, it may be observed.
Theoretically, SARS-CoV-2 has the possibility of ADE. Although there is clear evidence that antibodies against the original viruses have reduced neutralization capacity against mutant strains, there is lack of robust data for a clear ADE effect. Macrophages, which are well studied in ADE, are also found to have multiple roles.
Macrophages are scavengers and Trojan horse in the early, middle and late stages of SARS-CoV-2 infection, which do not directly contribute to ADE, but can be mediated by antibodies to facilitate viral transmission and enhance infection. Macrophages from different sources promote ADE in different stages, but also have a clearing function. In the inflammatory environment of infection with SARS-CoV-2, immune cells such as macrophages, NK, T, and B, non-immune cells such as alveolar cells and epithelial cells, various antibodies and various cytokines affect and interact with each other. The extensive expression of receptors such as pattern recognition receptor, Fc receptors, and the crosstalk between them form a complex dynamic immune network. This network affects the balance between virus clearance, ADE effect, and the extent of the inflammatory response, which determines the treatment outcome of infected patients.
Some studies have proposed some possible ways to reduce the risk of ADE of SARS-CoV-2. For example, the use of a biomimetic shell avoids the risk of ADE of SARS-CoV-2. Biodegradable calcium phosphate-encapsulated viral particles evade recognition by pre-existing antibodies extracellularly, thereby eliminating the ADE of viral infection (21). Two leucine-alanine substitutions (LALA mutations) were introduced at residues 234 and 235 of the Fc part to reduce Fc-mediated ADE (5, 22). Fucosylated anti-SARS-CoV-2 antibodies may reduce affinity for FcRIIIa receptors, thereby attenuating ADE (9). Novel activators are used to induce or deliver potent neutralizing antibodies. When infecting a mutant virus, it promotes the presentation of specific antigens against the mutant virus, restarts the immune system, and produces high-quality specific neutralizing antibodies.
In addition, ADE is not easy to occur, which may require many conditions. Such as the subtype of the antibody, the quality, specificity, titer, affinity of the antibody, and antibodies against “former” viruses have reduced neutralization capacity against mutant strains etc (23). In addition, race, genetic diversity, age, gender, vaccination/infection history, underlying health conditions, etc. may all have an impact. At present, vaccination is the best way to prevent SARS-CoV-2 infection, which can reduce the severe case fatality rate. But special populations (elders, people with underlying diseases or chronic diseases, etc.) need to be vigilant against ADE of SARS-CoV-2.
YY completed writing of the paper. FX designed the paper and guided writing. All authors contributed to the article and approved the submitted version.
This work is supported by the National Natural Science Foundations of China [82171814]. The funders have no roles in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Karthik K, Senthilkumar TMA, Udhayavel S, Raj GD. Role of antibody-dependent enhancement (ADE) in the virulence of SARS-CoV-2 and its mitigation strategies for the development of vaccines and immunotherapies to counter COVID-19. Hum Vaccin Immunother (2020) 16(12):3055–60. doi: 10.1080/21645515.2020.1796425
2. Lee WS, Wheatley AK, Kent SJ, DeKosky BJ. Antibody-dependent enhancement and SARS-CoV-2 vaccines and therapies. Nat Microbiol (2020) 5(10):1185–91. doi: 10.1038/s41564-020-00789-5
3. Maemura T, Kuroda M, Armbrust T, Yamayoshi S, Halfmann PJ, Kawaoka Y. Antibody-dependent enhancement of SARS-CoV-2 infection is mediated by the IgG receptors FcγRIIA and FcγRIIIA but does not contribute to aberrant cytokine production by macrophages. mBio (2021) 12(5):e0198721. doi: 10.1128/mBio.01987-21
4. Wang S, Wang J, Yu X, Jiang W, Chen S, Wang R, et al. Antibody-dependent enhancement (ADE) of SARS-CoV-2 pseudoviral infection requires FcγRIIB and virus-antibody complex with bivalent interaction. Commun Biol (2022) 5(1):262. doi: 10.1038/s42003-022-03207-0
5. Wang S, Peng Y, Wang R, Jiao S, Wang M, Huang W, et al. Characterization of neutralizing antibody with prophylactic and therapeutic efficacy against SARS-CoV-2 in rhesus monkeys. Nat Commun (2020) 11(1):5752. doi: 10.1038/s41467-020-19568-1
6. Zhou Y, Liu Z, Li S, Xu W, Zhang Q, Silva IT, et al. Enhancement versus neutralization by SARS-CoV-2 antibodies from a convalescent donor associates with distinct epitopes on the RBD. Cell Rep (2021) 34(5):108699. doi: 10.1016/j.celrep.2021.108699
7. Liu Y, Soh WT, Kishikawa JI, Hirose M, Nakayama EE, Li S, et al. An infectivity-enhancing site on the SARS-CoV-2 spike protein targeted by antibodies. Cell (2021) 184(13):3452–66.e18. doi: 10.1016/j.cell.2021.05.032
8. Li D, Edwards RJ, Manne K, Martinez DR, Schäfer A, Alam SM, et al. The functions of SARS-CoV-2 neutralizing and infection-enhancing antibodies in vitro and in mice and nonhuman primates. bioRxiv (2021). doi: 10.1101/2020.12.31.424729
9. Larsen MD, de Graaf EL, Sonneveld ME, Plomp HR, Nouta J, Hoepel W, et al. Afucosylated IgG characterizes enveloped viral responses and correlates with COVID-19 severity. Sci (NY) (2021) 371(6532). doi: 10.1126/science.abc8378
10. Grant RA, Morales-Nebreda L, Markov NS, Swaminathan S, Querrey M, Guzman ER, et al. Circuits between infected macrophages and T cells in SARS-CoV-2 pneumonia. Nature (2021) 590(7847):635–41. doi: 10.1038/s41586-020-03148-w
11. Yao XH, He ZC, Li TY, Zhang HR, Wang Y, Mou H, et al. Pathological evidence for residual SARS-CoV-2 in pulmonary tissues of a ready-for-discharge patient. Cell Res (2020) 30(6):541–3. doi: 10.1038/s41422-020-0318-5
12. Lv J, Wang Z, Qu Y, Zhu H, Zhu Q, Tong W, et al. Distinct uptake, amplification, and release of SARS-CoV-2 by M1 and M2 alveolar macrophages. Cell Discov (2021) 7(1):24. doi: 10.1038/s41421-021-00258-1
13. Zhang Z, Zheng Y, Niu Z, Zhang B, Wang C, Yao X, et al. SARS-CoV-2 spike protein dictates syncytium-mediated lymphocyte elimination. Cell Death Differ (2021) 28(9):2765–77. doi: 10.1038/s41418-021-00782-3
14. Allard B, Panariti A, Martin JG. Alveolar macrophages in the resolution of inflammation, tissue repair, and tolerance to infection. Front Immunol (2018) 9:1777. doi: 10.3389/fimmu.2018.01777
15. Lian Q, Zhang K, Zhang Z, Duan F, Guo L, Luo W, et al. Differential effects of macrophage subtypes on SARS-CoV-2 infection in a human pluripotent stem cell-derived model. Nat Commun (2022) 13(1):2028. doi: 10.1038/s41467-022-29731-5
16. Speranza E, Williamson BN, Feldmann F, Sturdevant GL, Pérez-Pérez L, Meade-White K, et al. Single-cell RNA sequencing reveals SARS-CoV-2 infection dynamics in lungs of African green monkeys. Sci Trans Med (2021) 13(578). doi: 10.1126/scitranslmed.abe8146
17. Li F, Piattini F, Pohlmeier L, Feng Q, Rehrauer H, Kopf M. Monocyte-derived alveolar macrophages autonomously determine severe outcome of respiratory viral infection. Sci Immunol (2022) 7(73):eabj5761. doi: 10.1126/sciimmunol.abj5761
18. Feng Z, Diao B, Wang R, Wang G, Wang C, Tan Y, et al. The novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) direstcly decimates human spleens and lymph nodes. (2020). doi: 10.1101/2020.03.27.20045427
19. Diao B, Wang C, Tan Y, Chen X, Liu Y, Ning L, et al. Reduction and functional exhaustion of T cells in patients with coronavirus disease 2019 (COVID-19). Front Immunol (2020) 11:827. doi: 10.3389/fimmu.2020.00827
20. Kam YW, Kien F, Roberts A, Cheung YC, Lamirande EW, Vogel L, et al. Antibodies against trimeric s glycoprotein protect hamsters against SARS-CoV challenge despite their capacity to mediate FcgammaRII-dependent entry into b cells in vitro. Vaccine (2007) 25(4):729–40. doi: 10.1016/j.vaccine.2006.08.011
21. Wang X, Deng YQ, Yang D, Xiao Y, Zhao H, Nian QG, et al. Biomimetic inorganic camouflage circumvents antibody-dependent enhancement of infection. Chem Sci (2017) 8(12):8240–6. doi: 10.1039/c7sc03868b
22. Shi R, Shan C, Duan X, Chen Z, Liu P, Song J, et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature (2020) 584(7819):120–4. doi: 10.1038/s41586-020-2381-y
Keywords: SARS-CoV-2, antibody-dependent enhancement (ADE), mechanism, receptor-mediated, multiple roles of macrophages, countermeasures
Citation: Yang Y and Xu F (2022) Evolving understanding of antibody-dependent enhancement (ADE) of SARS-CoV-2. Front. Immunol. 13:1008285. doi: 10.3389/fimmu.2022.1008285
Received: 31 July 2022; Accepted: 04 October 2022;
Published: 18 October 2022.
Edited by:
Elsa Anes, Universidade de Lisboa, PortugalReviewed by:
Miguel Azevedo-Pereira, University of Lisbon, PortugalCopyright © 2022 Yang and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fenghua Xu, eHVmaEAzMDFob3NwaXRhbC5jb20uY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.