- 1Department of Nephrology, The Seventh Affiliated Hospital, Sun Yat-Sen University, Shenzhen, China
- 2Center of Nephrology and Urology, The Seventh Affiliated Hospital, Sun Yat-Sen University, Shenzhen, China
- 3Scientific Research Center, The Seventh Affiliated Hospital, Sun Yat-Sen University, Shenzhen, China
It was previously published that single-nucleotide polymorphism rs2476601 (PTPN22 [protein tyrosine phosphatase non-receptor type 22]-C1858T) might be related to increased sensibility to Mycobacterium tuberculosis and M. leprae infection. However, the results were inconclusive despite a high degree of similarity between both parameters. Herein, we carried out this meta-analysis to systematically summarize and articulate the correlation between PTPN22-C1858T polymorphism and mycobacterial infection. The susceptibility of PTPN22-C1858T carriers with autoimmune conditions receiving immunosuppressive therapy to M. tuberculosis and M. leprae infection was determined. A systematic retrieval of studies on relevance of PTPN22-C1858T polymorphism to susceptibility of M. tuberculosis or M. leprae infection was performed in Chinese National Knowledge Infrastructure, PubMed and Embase databases. We regarded Odds ratios (ORs) and 95% confidence intervals (CIs) as the determined effect size. Finally, four and two case-control studies on tuberculosis and leprosy, respectively, were included. In all genetic models, without indicated association between PTPN22-C1858T polymorphism and tuberculosis’s susceptibility. [C versus T: OR = 0.22 (95% CI: 0.09–0.50, PH = 0.887); CT versus CC: OR = 0.21 (95% CI: 0.09–0.49, PH = 0.889); TT+CT versus CC: OR = 0.21 (95% CI: 0.09–0.49, PH = 0.889)]. A significantly increased risk of leprosy was perceived in patients with the PTPN22-C1858T polymorphism [C versus T: OR = 2.82 (95% CI: 1.02–7.81, PH = 0.108)]. While the PTPN22-C1858T polymorphism is irrelevant to higher susceptibility to the infection of M. tuberculosis in Caucasians and Asians, it is relevant to increased susceptibility to the infection of M. leprae. However, the results of M. leprae are supposed to interpreted with prudence owing to the limited quantity of studies and heterogeneity. Further well-designed studies with sufficient populations are required to verify our conclusions.
Introduction
Worldwide, tuberculosis is one of the principal lethal causes, ranking above human immunodeficiency virus infection/acquired immunodeficiency syndrome. Despite being a preventable and curable disease, the annual number of people died from tuberculosis is 1.5 million approximately. (1). Tuberculosis accounts for a great share of global burden of disease, particularly among vulnerable populations, and exerts significant impacts on 10 million people annually (2). By recent estimates, over 200,000 new leprosy cases are diagnosed annually. At the end of 2018, the prevalence rate of leprosy is 0.2/10,000 and a total of 184,212 leprosy cases has been registered (3). Leprosy can be cured using effective and affordable multidrug therapy; however, it is a major problem in resource-poor tropical and warm temperate countries and keeps endemic in several low- and middle-income countries globally (4, 5). Available evidence indicates that inferior living conditions are possible to raise the risk of leprosy (6). Moreover, the correlation between tuberculosis or leprosy and sociodemographic risk markers such as crowded living conditions, poor sanitation, and poverty has been validated across diverse geographical settings, both in ecological- and individual-level studies (4, 7).
Tuberculosis and leprosy are caused by mycobacteria (8). In humans, tuberculosis is predominantly caused by Mycobacterium tuberculosis and occasionally by other components of M. tuberculosis complex, and Mycobacterium leprae result in leprosy. M. tuberculosis and M. leprae are obligate pathogens with a similar morphology and high cell wall lipid content, which is responsible for their increased chromaticity, resistance, and pathogenicity. Moreover, these mycobacteria can cause chronic granuloma in most infected individuals.
Locating on chromosome 1p13, the protein tyrosine phosphatase non-receptor type 22 (PTPN22) gene encodes the protein lymphoid tyrosine phosphatase (LYP), which regulates the activation of protein kinases to modulate intracellular tyrosine phosphorylation incidence and induce various biological effects. PTPN22-C1858T is the most talked about single-nucleotide polymorphism (SNP) in the field of autoimmunity (9); this SNP causes R620W substitution in PTPN22 gene’s C-terminal region. PTPN22-C1858T was identified as a missense SNP in PTPN22 gene’s exon 14 by using the candidate gene approach in 2004. Moreover, genome-wide association studies showed a correlation between PTPN22-C1858T polymorphism and increased risk of autoimmune diseases (10–15) as well as bacterial infections (16). In addition to disease-causing microorganisms in the common disease spectrum, the susceptibility of individuals carrying PTPN22-C1858T SNP to tuberculosis and leprosy has been proposed in preceding studies (17–19).
Herein, we performed this meta-analysis to systematically summarize and articulate the correlation between PTPN22-C1858T polymorphism and the risk of mycobacterial infection.
Methods
Retrieval of Studies
Two investigators searched literatures from Chinese National Knowledge Infrastructure (CNKI), PubMed and Embase databases independently and systematically, which were updated on April 15, 2020. Retrieved items including “tuberculosis or (pulmonary tuberculosis) or (mycobacterium tuberculosis) or (tubercle bacillus) or (tubercle bacillus) or mtb)”, “leprosy or (leprosy bacillus) or (mycobacterium leprae) or (mycogerms leprae) or (wholemycobacterium leprae)”, “PTPN22 OR LYP”, “polymorphism OR poly-morphism” and susceptibility” were searched in the National Institutes of Health (PubMed) and European (Embase) databases without limitation. The key words “PTPN22基因多态性” and “结核” were searched in the CNKI database. The references in retrieved studies and related reviews were also under reviewing by two investigators without communications.
Inclusion Criteria and Excluding Criteria
All studies pertaining to PTPN22 polymorphisms and infection susceptibility were electronically retrieved. Thereafter, case-control studies were identified from their titles and abstracts. The full article of manuscripts fulfilled the inclusion criteria was perused. Inclusion criteria were as follows: (a) studies associated between PTPN22 polymorphisms and infection susceptibility; (b) the category of the study is case-control study among human; (c) original research with detailed explanation of the sample size; (d) with odds ratios (ORs) and its corresponding 95% confidence intervals (CIs); (e) with clear determination of genotype frequency; (f) genotype distribution in controls complying with Hardy-Weinberg equilibrium (HWE) law. Excluding criteria were as follows: (a) non-conformance to the inclusion criteria; (b) article type: abstract, review, comment, and letter; (c) studies with insufficient or no explanation of the sample size; (d) studies without genotype data; (e) repeated publications; (f) family-based studies. If several studies were with same population, the larger one will be included. According to the inclusion criteria and excluding criteria, two investigators screened out eligible literatures independently through titles, abstracts, and full-texts. We settled all dissents through discussion. The third researcher was consulted in the absence of an agreement. Finally, two studies were discussed and excluded after our discussions.
Data Withdrawal
Two investigators withdrawn the data from all included studies independently. The extracted data included the name of first author, year of publication, subject resource, ethnicity, infectious type, diagnostic criteria for infection, genotyping method, number of participants in the control and case groups, cases’ and controls’ characteristics, all subjects’ PTPN22-C1858T genotype frequency and HWE. Therein, populations from all included studies were classified into two ethnic: Caucasians and Asians. Two investigators verified the withdrawn data and achieved agreements on the conclusive data. In the case of a disagreement, the original data was re-verified and re-discussed to reach consensus. If disagreements are still subsistent, we consulted for final judgement from a third investigator. In the end, a study was discussed through third investigator.
Literature Assessment
The Newcastle-Ottawa Scale (NOS) was applied to assess the quality of six eligible studies in meta-analysis by two independent investigators (20) (Methods were shown in Supplement 2). All study were assessed by using a “star system” on the basis of three categories: selection of case and control, subjects’ comparability, and exposure’s ascertainment. The maximum score is 9 on the NOS scale. If a study with a score ≥ 7, it is considered as a high-quality study. The details of assessment has been shown in Table S3.
Statistical Methods
The PRISMA checklist was followed throughout the course of this meta-analysis strictly. (21). The HWE was used to assess genotyping errors in control groups by Chi-squared test in all studies (P < 0.05 was significant). ORs and its corresponding 95% CI is was applied to determine the extent of correlation between PTPN22-C1858T polymorphism and susceptibility to mycobacterial infection. We calculated the pooled ORs in allelic comparison (PTPN22: C versus T), the heterozygote model (PTPN22: CT versus CC), and the dominant model (PTPN22: CT+TT versus CC). P < 0.05 was significant according to Z-test. Heterogeneity was assessed by Q test (P < 0.1 was significant) and I-squared statistic (I2: 0%–25%, without heterogeneity; I2: 25%–50%, moderate heterogeneity; I2: 50%–75%, large heterogeneity; I2: 75%–100% extreme heterogeneity; I2 > 50% represented significant inconsistency in our meta-analysis) (22). We applied a fixed-effect or a random-effects model to pool effect sizes depending on data heterogeneity (23). Heterogeneity was analyzed by using meta-regression model (P < 0.1 was considered significant) with pathogen (M. tuberculosis and M. leprae) and ethnicity and subgroup analysis stratified by pathogen and ethnicity. No deviation from HWE was shown among controls in all included studies, which are used for further meta-analysis. Additionally, we assessed publication bias by Begg’s funnel plots (24). An asymmetric plot with P < 0.05 was identified a significant publication bias. Besides, we also performed sensitivity analysis to estimate the pooled ORs of PTPN22-C1858T polymorphism in each study. The results of the meta-analysis were recalculated after excluding each studies. All statistical analyses were performed by Stata 14.0 software (StataCorp, College Station, TX, USA). Except for certain conditions defined a specific P value, a two-tailed P value < 0.05 was regarded as significant.
Result
Study Retrieval
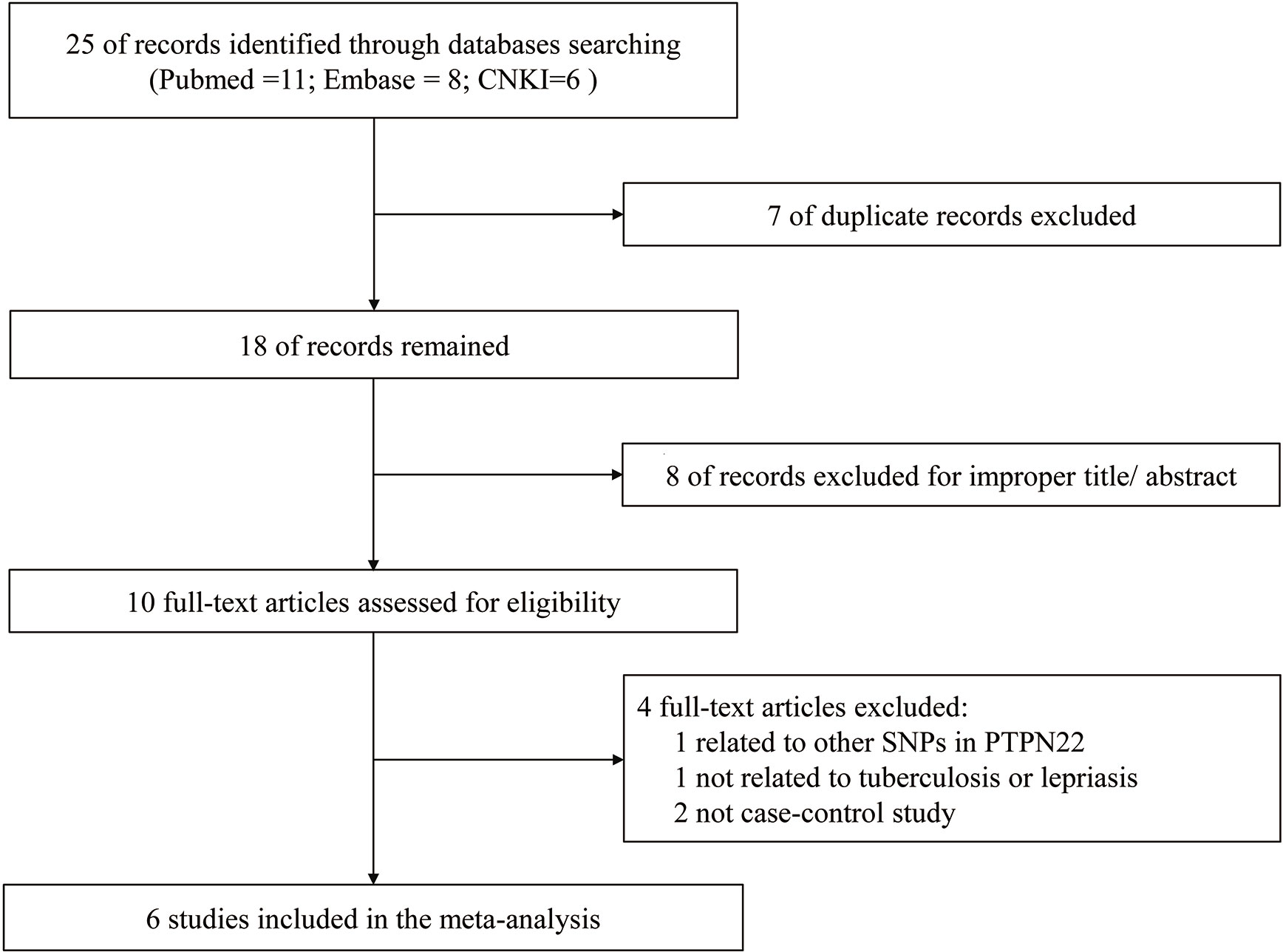
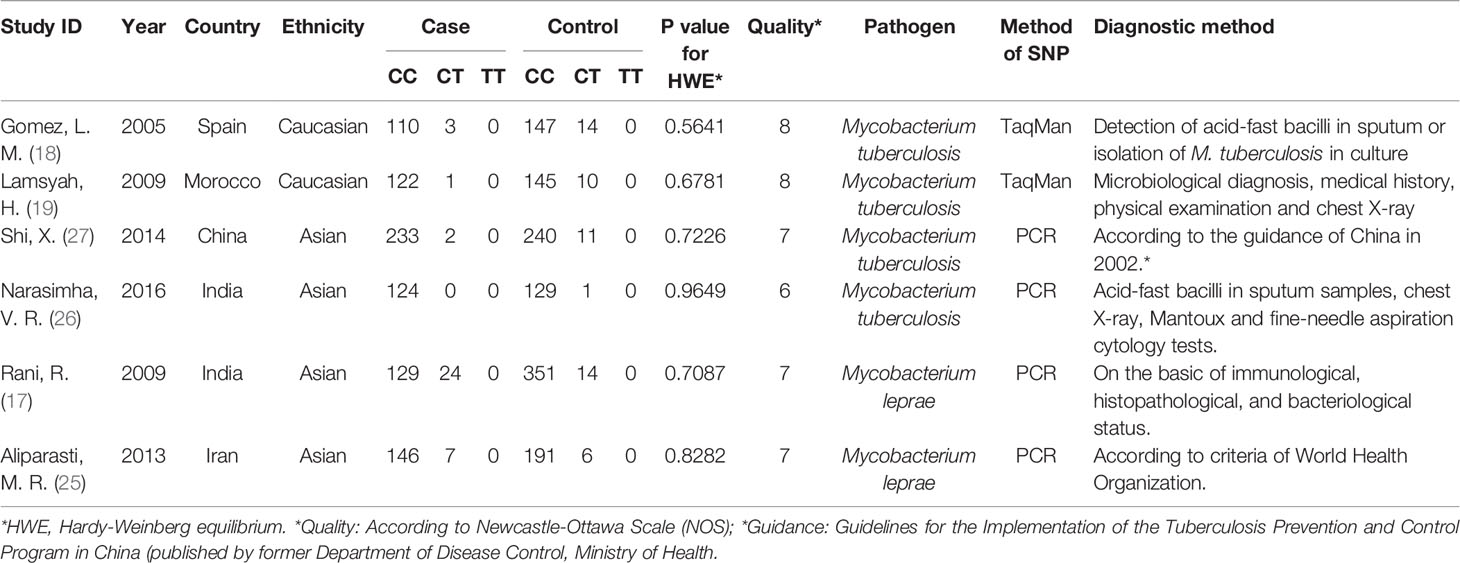
The study selection process during meta-analysis of association of rs2476601(PTPN22-C1858T) polymorphism with M. tuberculosis and M. leprae infection is shown in Figure 1. Following the initial retrieval of 25 publications through a database search (eleven from PubMed, eight from Embase, and six from CNKI), 18 records were selected after the removal of seven duplicates. Moreover, after careful review of the title and abstract, eight publications were rejected because of their irrelevance to this meta-analysis. The remaining ten publications were full-article reviewed; of these, four were excluded. One of them was related to other SNPs in PTPN22, one was not related to tuberculosis or lepriasis, and two were not case-control studies. Finally, six case-control studies (17–19, 25–27) consisting of 2,160 participants (cases = 901; controls = 1,259) were included in meta-analysis. General characteristics of the six studies are in Table 1 while the genotype distribution of subjects is shown in Table 2.
The Relevance Between PTPN22-C1858T Polymorphism and the Susceptibility to Mycobacterial Infection
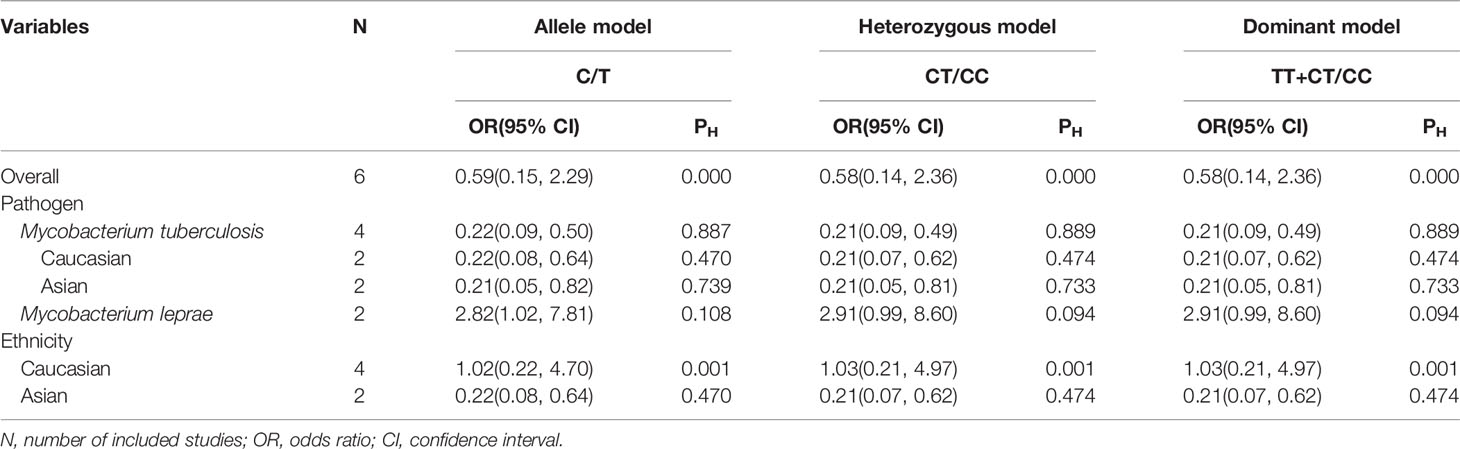
We explored that PTPN22-C1858T polymorphism associates susceptibility to mycobacterial infection. After Q-test and I-squared statistics in various genetic models, it indicated significant heterogeneity. Therefore, we used the random-effects model in meta-analysis. No genetic model showed correlation between PTPN22-C1858T polymorphism and increased susceptibility to mycobacterial infection [C versus G: OR = 0.59 (95% CI: 0.15–2.29, PH = 0.000) (Figure 2); CT versus CC: OR = 0.58 (95% CI: 0.14–2.36, PH = 0.000); CT+TT versus CC: OR = 0.58, (95% CI: 0.14–2.36, PH = 0.000)] (Table 2). Then, we further performed meta-analysis in the subgroups with different ethnicities. In Asians, no genetic model revealed correlation between PTPN22-C1858T polymorphism and increased susceptibility to mycobacterial infection [C versus G: OR = 0.22 (95% CI: 0.08–0.64, PH = 0.470); CT versus CC: OR = 1.03 (95% CI: 0.21–4.97, PH = 0.001); CT+TT versus CC: OR = 1.03 (95% CI: 0.21–4.97, PH = 0.001)] (Table 2). In Caucasians, the result indicated that PTPN22-C1858T polymorphism correlates the susceptibility to mycobacterial infection [C versus G: OR = 1.02 (95% CI: 0.22–4.70, PH = 0.001); CT versus CC: OR = 1.03 (95% CI: 0.21–4.97, PH = 0.001); CT+TT versus CC: OR = 1.03 (95% CI: 0.21–4.97, PH = 0.001)] (Table 2).
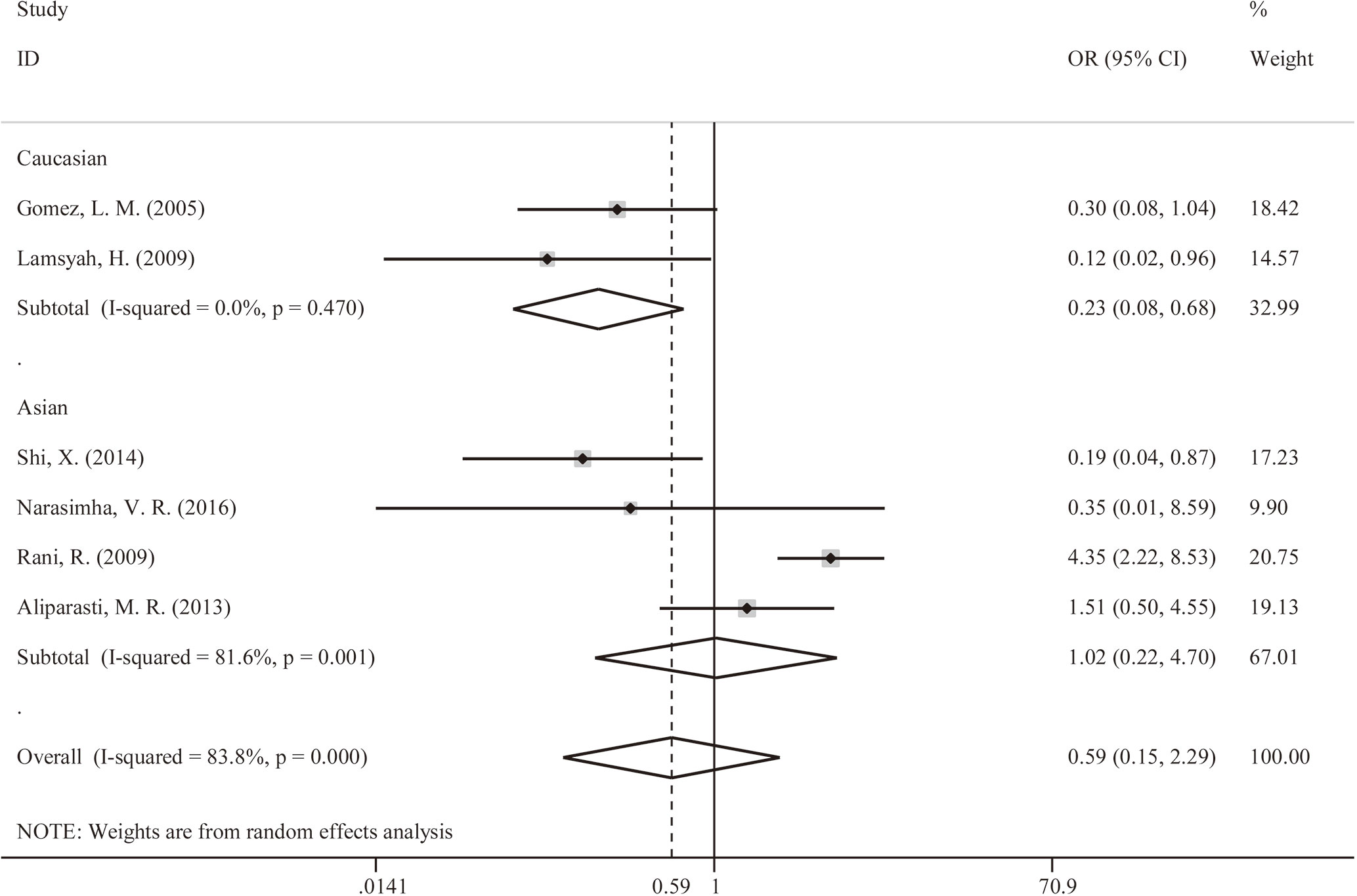
Figure 2 Forest plot for meta-analysis of association of rs2476601 (PTPN22-C1858T) polymorphism with increased susceptibility to Mycobacterium tuberculosis or M. leprae infection in Caucasians and Asians (C versus T). The size of blocks or diamonds is on behalf of the weight and the length of the straight line is on behalf of 95% confidence interval’s width.
PTPN22-C1858T Polymorphism Uncorrelated With Susceptibility to M. tuberculosis Infection
Initially, we analyzed the correlation between PTPN22-C1858T polymorphism and M. tuberculosis infection in our included studies of M. tuberculosis infection. There was no genetic model supported significant correlation between PTPN22-C1858T polymorphism and increased possibility to M. tuberculosis infection [C versus T: OR = 0.22 (95% CI: 0.09–0.50, PH = 0.887) (Figure 3A); CT versus CC: OR = 0.21 (95% CI: 0.09–0.49, PH = 0.889); TT+CT versus CC: OR = 0.21 (95% CI: 0.09–0.49, PH = 0.889)] (Table 2).
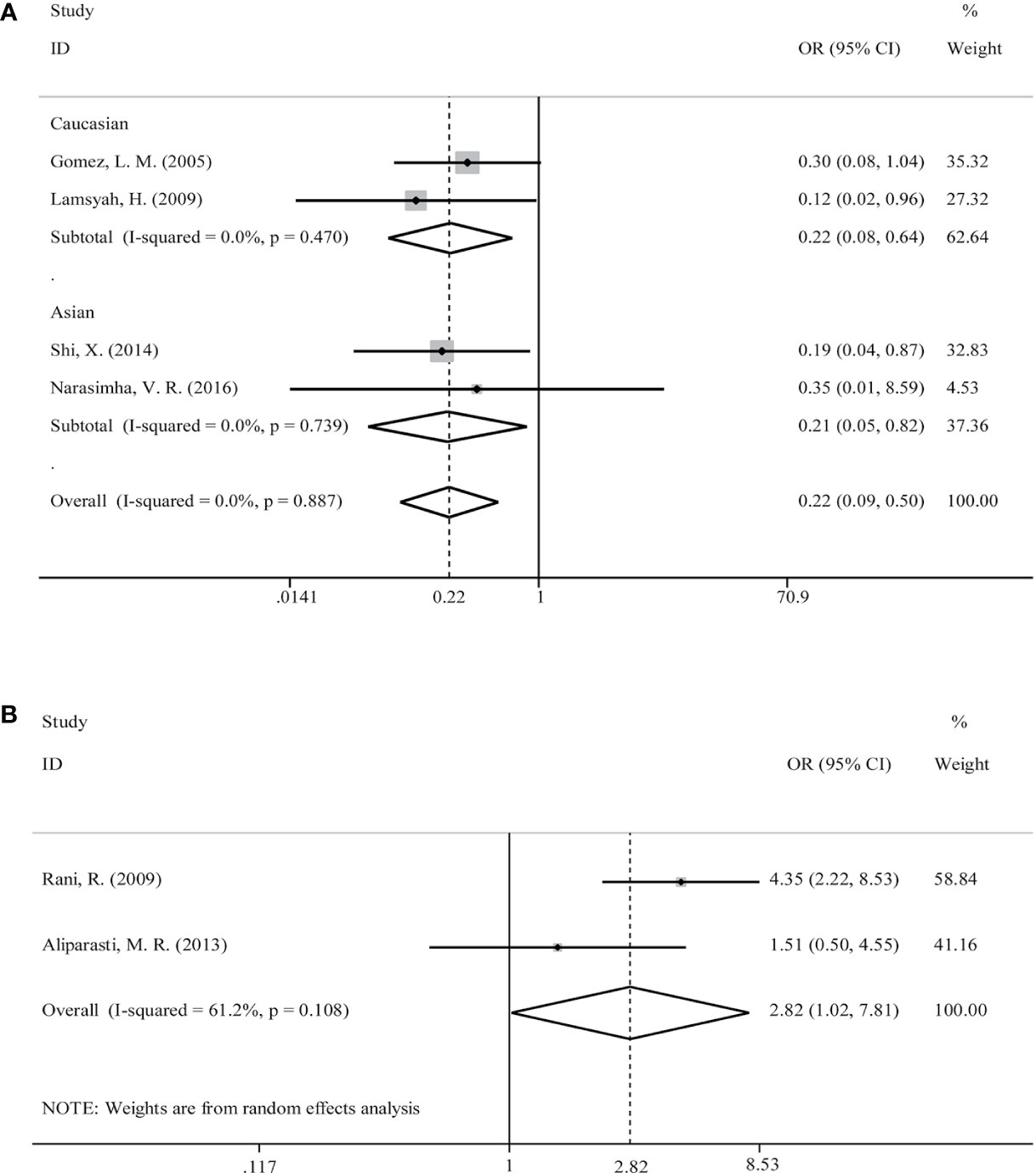
Figure 3 Forest plot for meta-analysis of association of rs2476601 (PTPN22-C1858T) polymorphism with increased susceptibility to Mycobacterium tuberculosis (A) infection in Caucasians and Asians and M. leprae (B) (C versus T). The size of blocks or diamonds is on behalf of the weight and the length of the straight line is on behalf of 95% confidence interval’s width.
Then, a further meta-analysis was performed in M. tuberculosis infection on the four included studies according to ethnicity (18, 19, 26, 27). All three genetic models showed significant homogeneity among Caucasians; therefore, a fixed-effects model was applied. However, all of involved genetic models didn’t illustrate an association with significance between PTPN22-C1858T polymorphism and increased susceptibility to mycobacterial infection in Caucasians [C versus T: OR = 0.22 (95% CI: 0.08–0.64, PH = 0.470) (Figure 3A); CT versus CC: OR = 0.21 (95% CI: 0.07–0.62, PH = 0.474); TT+CT versus CC: OR = 0.21 (95% CI: 0.07–0.62, PH = 0.474)] (Table 2). What’s more, all three genetic models showed heterogeneity among Asians and we applied a random-effects model in Asian subgroup. No association with significance between PTPN22-C1858T polymorphism and probability to M. tuberculosis infection in Asians [C versus T: OR = 0.21 (95% CI: 0.05–0.82, PH = 0.739) (Figure 3A); CT versus CC: OR = 0.21 (95% CI: 0.05–0.81, PH = 0.733); TT+CT versus CC: OR = 0.21 (95% CI: 0.05–0.81, PH = 0.733)] (Table 2).
PTPN22-C1858T Polymorphism Correlated With Increased Susceptibility to M. leprae Infection
The relevance between PTPN22-C1858T polymorphism and increased possibility to M. leprae infection in Caucasian and Asian populations was investigated in two studies (17, 25). Owing to limited quantity of studies and heterogeneity (P = 0.108 and I-squared = 61.2%) (Figure 3B), among the study populations, we reviewed carefully with the results and adopted random-effects model in allele (C versus T), heterozygote (CT versus CC), and dominant (TT versus CT+CC) models for meta-analysis. A significantly increased risk of M. leprae infection was observed in the allele model [C versus T: OR = 2.82 (95% CI: 1.02–7.81, PH = 0.108)] (Figure 3B) (Table 2). An elevated risk of leprosy was perceived in patients with PTPN22-C1858T polymorphism in heterozygote models [CT versus CC: OR = 2.91 (95% CI: 0.99–8.60, PH = 0.094)] and dominant models [TT/CT versus CC: OR = 2.91 (95% CI: 0.99–8.60, PH = 0.094)] (Table 2).
Meta-Regression Was Applied to Analyzing Heterogeneity
Ethnicity and pathogen were included in meta-regression model for heterogeneity analysis. The coefficient of ethnicity is -0.060 (P = 0.955) and the coefficient of pathogen is 2.560 (P = 0.060) in all patients with Mycobacterial infection (Table 3), which indicated that pathogen (M. tuberculosis/M. leprae) was able to account for the heterogeneity of the associations between mycobacterial infections and PTPN22-C1858T polymorphism.

Table 3 Multivariate meta-regression analysis of PTPN22-C1858T in patients with mycobacterium infection.
Publication Bias
No publication bias on relavance between PTPN22-C1858T polymorphism and possibility to tuberculosis (P = 0.734) or leprosy (P = 1.000) was reflected in Begg’s funnel plot. Meanwhile, we obtained a symmetrical funnel plots (Figure 4).
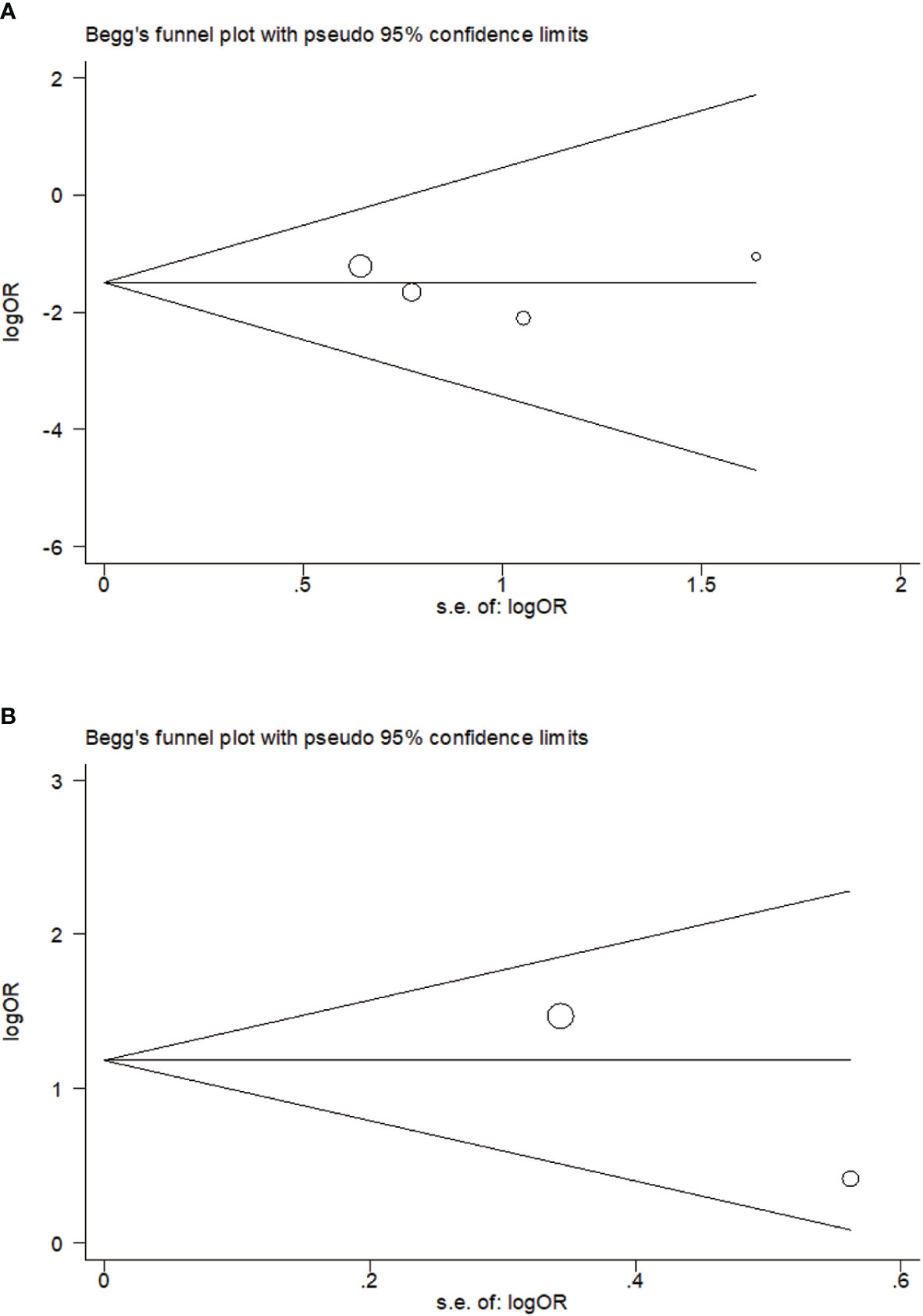
Figure 4 Begg’s funnel plot of rs2476601 (PTPN22-C1858T) polymorphism in individuals with Mycobacterium tuberculosis (A) and M. leprae (B) infection (C versus T).
Sensitivity Analysis
In order to determine impact from each study on pooled ORs for PTPN22-C1858T polymorphism, we performed sensitivity analysis by the means of deleting one of single study each time in every genetic model. The pooled ORs of both four studies related to M. tuberculosis infection (Figure S1A) and two studies related to M. leprae infection (Figure S1B) showed no significant association with PTPN22-C1858T polymorphism in above mentioned genetic models.
Discussion
In this meta-analysis, we analyzed four eligible case-control studies including 595 subjects with tuberculosis and 697 controls and two studies including 306 subjects with leprosy and 562 controls. Our results indicate that PTPN22-C1858T polymorphism is uncorrelated with increased susceptibility to M. tuberculosis infection both among Caucasian and Asian populations in all genetic models but is related to increased susceptibility to M. leprae infection. However, further studies are required to validate the relevance between PTPN22-C1858T polymorphism and leprosy because of the limited quantity of studies and indicated heterogeneity among studies.
In addition to mycobacteria, a number of studies have focused on the relevance of PTPN22-C1858T polymorphism with raised susceptibility to infections by other bacteria (16, 28, 29). First, it was confirmed that individuals with PTPN22-C1858T polymorphism are predisposed to invasive infections with Streptococcus pneumoniae (16). Second, allogeneic hematopoietic stem cell transplant recipients have been found to have a significantly lower risk of post-transplantation bacterial infections after accepting corresponding allografts from PTPN22-C1858T polymorphic donors (28). Third, PTPN22-C1858T polymorphic patients with chronic mucocutaneous candidiasis showed a higher incidence of bacterial pulmonary infections than normal controls (29). Moreover, other PTPN22 SNPs such as rs33996649 (PTPN22-G788A) might be related to raised susceptibility to tuberculosis. One study showed 788A allele was with higher frequency of in Moroccan patients with tuberculosis than in healthy donors [3.65% versus 0.65%, respectively; OR = 5.85 (95% CI:1.17–39.55, P = 0.01)] (19).
Detrimental effect of PTPN22-C1858T polymorphism-induced immune dysregulation on delayed-type hypersensitivity of tuberculosis and leprosy has been extensively studied. Herein, we discussed the relevance of PTPN22-C1858T polymorphism and increased risk of tuberculosis and leprosy in humans (17–19, 25–27). The PTPN22 gene encodes for LYP, which render an impact on negative regulation to immunity (30). Moreover, PTPN22-C1858T carriers have variable immune patterns (31). Under physiological conditions, LYP selectively inhibits positive selection during a self-tolerant and immunological T-cell repertoire’s formation (32, 33), dephosphorylates the suppressive tyrosine located at Src family kinases’ C-terminus, and downregulate T-cell receptor (TCR) signaling by combining with growth factor receptor-bound protein 2 adapter (34–37). Patients with PTPN22-C1858T polymorphism show decreased calcium mobilization that is TCR-induced (38), reduced TCR-CD3ζ, Zap-70 and Lck’s phosphorylation (39), and decreased interleukin-2 production (39). However, some of these studies have yielded inconsistent conclusions. The absolute expression of PTPN22 and proportion of T-cell subsets should be considered during assessment of the overall effect of PTPN22-C1858T polymorphism in T-cell-mediated immunity (40). Furthermore, increased susceptibility to mycobacterial infections is closely correlated with the immune status and genetic milieu of hosts. Most healthy individuals with a normal functioning immune system are resistant to M. tuberculosis or M. leprae and do not develop clinical tuberculosis or leprosy (41, 42). Host genetics accounts for a large proportion of inherited mycobacterial diseases; epidemiological investigations and functional genetic studies have identified several genes involved in mycobacterial infection. These studies provide information about the inherited predisposition to these infections (43–45). Hence, we conducted this meta-analysis to determine and articulate the relevance between PTPN22-C1858T polymorphism and risk of tuberculosis and leprosy.
Increased intracellular expression of LYP in patients with mycobacterial infection (46, 47), the involvement of PTPN22 in downregulation of T-cell function (30), and the participation of PTPN22 in invasive bacterial infection (16) have been considered as evidence to conclude that PTPN22-C1858T polymorphism increases the risk of mycobacterial disease onset (17). Moreover, PTPN22 status has been shown to cause lymphoproliferative diseases in animal models. Splenomegaly and lymphadenectasis accompanied by initiative germinal center formation and higher-level antibodies are subtle immune changes observed in PTPN22-knockout mice (32). In addition, PTPN22 promotes K63-linked polyubiquitination of the Toll-like receptor signal central promoter TRAF3 to upregulate type I interferons (IFNs) and promote type I IFN-dependent biological effects, causing immune cells to trigger a host defense response. In contrast, PTPN22-C1858T carriers show decreased TRAF3 K63-linked polyubiquitination and type I IFN production (48).
Roles of PTPN22-C1858T in infection and prevention mainly are beneficial to the patients with autoimmune diseases. PTPN22-C1858T is more often discussed in autoimmune disease. With the indispensable treatment with glucocorticoids, patient with autoimmune disease are suffering the from the risk of various infection. Additionally, we tend to claim that population with PTPN22-C1858T is more likely to gain autoimmune disease and the diseases itself carry a high risk of various infection. To this analysis, we are expecting our conslusions are able to provide useful informations for human to evade the potential infection and suggestion for the usage of immunosuppressors, especially for patients with autoimmune disease. To be more concrete, patients with PTPN22-C1858T are recommand a lower dose of immunosuppressors after diagnosis of autoimmne disease and they also are advised to notice all possiablity of infection.
We would like to emphasize a couple of strengths in our meta-analysis. First, we applied subgroup analysis to demonstrate the differences in the correlation between PTPN22-C1858T polymorphism and increased risk of mycobacterial infection. What’s more, the conclusion was verified in meta-regression model. Additionally, two independent investigators searched literatures without limitations so that we well controlled the selection bias. Besides, the result from Begg’s funnel plot revealed no evident publication bias, which render reliability to our conclusions. Finally, most of each studies in our meta-analysis attained a score representing high quality with regards to NOS quality assessment.
Nevertheless, we also acknowledge several limitations in this meta-analysis. In the first place, other factors contributing to mycobacterial infection were not considered due to the availability of limited information. Second, the results of M. leprae were obtained from two studies. Third, with a low allele frequency less than 1%, PTPN22-C1858T is a rare SNP in Asian population. Therefore, PTPN22-C1858T polymorphism would be rarely detected in most populations except Caucasians of Northern European descent (49). Fourth, the absence of cases and controls with TT in the six included studies may be attributed to their low survival rate for gene mutations. Fifth, tuberculosis and leprosy are considered curable diseases worldwide, especially in developed regions. Moreover, the annual incidence rate of these disease is declining. Furthermore, the diagnosis of leprosy is often not considered outside leprosy-endemic areas (50). Finally, the occurrence of a latency period following infection (42) and lengthy incubation periods of M. leprae (from a month to over 40 years) (50) might have excluded individuals with recessive infection or no infected (in the incubation period) to be registered as positive cases or were registered as controls.
In conclusion, we manifested that this PTPN22-C1858T polymorphism was uncorrelated with raised susceptibility to M. tuberculosis in Caucasians and Asians. In contrast, PTPN22-C1858T polymorphism related to increased susceptibility to M. leprae infection. However, the results of M. leprae are supposed to interpreted with. Further well-designed studies with sufficient populations are required to verify our conclusions.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author Contributions
YC and SL conceived the topic and analyzed data. SL wrote this manuscript and YC modified it. JY provided statistical suggestions. SL and YZ evaluated the quality of the six included studies. SL, YZ, XW, and TC searched references.Y, ZJZ, and ZZ provided the overall direction of series studies and funding support. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the 100 Top Talents Program of Sun Yat-sen University, National Natural Science Foundation of China (NSFC, Grant No. 31871400), the Shenzhen Science and Technology Innovation Committee of Guangdong Province of China (Grant No. JSGG20180703155802047), and the Shenzhen Science and Technology Innovation Committee of Guangdong Province of China (Grant No. JCYJ20180307150634856).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2021.592841/full#supplementary-material
Abbreviations
CI, confidence interval; CNKI, Chinese National Knowledge Infrastructure; HWE, Hardy-Weinberg equilibrium; IFN, interferon; NOS, Newcastle-Ottawa Scale; OR, odds ratio; PTPN22, protein tyrosine phosphatase non-receptor type 22; SNP, single-nucleotide polymorphism; TCR, T-cell receptor.
References
1. World Health Organization. Global tuberculosis report 2019. World Health Organization (2019). WHO/CDSTB/2018.20. Available at: https://www.who.int/teams/global-tuberculosis-programme/tb-reports/global-report-2019.
2. Janse Van Rensburg A, Dube A, Curran R, Ambaw F, Murdoch J, Bachmann M, et al. Comorbidities between tuberculosis and common mental disorders: a scoping review of epidemiological patterns and person-centred care interventions from low-to-middle income and BRICS countries. Infect Dis Poverty (2020) 9:4. doi: 10.1186/s40249-019-0619-4
3. World Health Organization. Global Leprosy Strategy 2016–2020: Accelerating towards a leprosy-free world. India: World Health Organization (2016).
4. Pescarini JM, Strina A, Nery JS, Skalinski LM, Andrade KVF, Penna MLF, et al. Socioeconomic risk markers of leprosy in high-burden countries: A systematic review and meta-analysis. PloS Negl Trop Dis (2018) 12:e0006622. doi: 10.1371/journal.pntd.0006622
6. Hotez PJ, Fenwick A, Savioli L, Molyneux DH. Rescuing the bottom billion through control of neglected tropical diseases. Lancet (2009) 373:1570–5. doi: 10.1016/S0140-6736(09)60233-6
7. Carrasco-Escobar G, Schwalb A, Tello-Lizarraga K, Vega-Guerovich P, Ugarte-Gil C. Spatio-temporal co-occurrence of hotspots of tuberculosis, poverty and air pollution in Lima, Peru. Infect Dis Poverty (2020) 9:32. doi: 10.1186/s40249-020-00647-w
8. Rees RJ, Meade TW. Comparison of the modes of spread and the incidence of tuberculosis and leprosy. Lancet (1974) 1:47–8. doi: 10.1016/S0140-6736(74)93043-8
9. Ferreira RC, Castro Dopico X, Oliveira JJ, Rainbow DB, Yang JH, Trzupek D, et al. Chronic Immune Activation in Systemic Lupus Erythematosus and the Autoimmune PTPN22 Trp(620) Risk Allele Drive the Expansion of FOXP3(+) Regulatory T Cells and PD-1 Expression. Front Immunol (2019) 10:2606. doi: 10.3389/fimmu.2019.02606
10. Bottini N, Musumeci L, Alonso A, Rahmouni S, Nika K, Rostamkhani M, et al. A functional variant of lymphoid tyrosine phosphatase is associated with type I diabetes. Nat Genet (2004) 36:337–8. doi: 10.1038/ng1323
11. Kyogoku C, Langefeld CD, Ortmann WA, Lee A, Selby S, Carlton VE, et al. Genetic association of the R620W polymorphism of protein tyrosine phosphatase PTPN22 with human SLE. Am J Hum Genet (2004) 75:504–7. doi: 10.1086/423790
12. Harley JB, Alarcon-Riquelme ME, Criswell LA, Jacob CO, Kimberly RP, Moser KL, et al. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat Genet (2008) 40:204–10. doi: 10.1038/ng.81
13. Jin Y, Birlea SA, Fain PR, Gowan K, Riccardi SL, Holland PJ, et al. Variant of TYR and autoimmunity susceptibility loci in generalized vitiligo. N Engl J Med (2010) 362:1686–97. doi: 10.1056/NEJMoa0908547
14. Diaz-Gallo LM, Ramskold D, Shchetynsky K, Folkersen L, Chemin K, Brynedal B, et al. Systematic approach demonstrates enrichment of multiple interactions between non-HLA risk variants and HLA-DRB1 risk alleles in rheumatoid arthritis. Ann Rheum Dis (2018) 77:1454–62. doi: 10.1136/annrheumdis-2018-213412
15. Krischer JP, Lynch KF, Lernmark A, Hagopian WA, Rewers MJ, She JX, et al. Genetic and Environmental Interactions Modify the Risk of Diabetes-Related Autoimmunity by 6 Years of Age: The TEDDY Study. Diabetes Care (2017) 40:1194–202. doi: 10.2337/dc17-0238
16. Chapman SJ, Khor CC, Vannberg FO, Maskell NA, Davies CW, Hedley EL, et al. PTPN22 and invasive bacterial disease. Nat Genet (2006) 38:499–500. doi: 10.1038/ng0506-499
17. Rani R, Singh A, Israni N, Singh A, Sharma P, Kar HK. The role of polymorphic protein tyrosine phosphatase non-receptor type 22 in leprosy. J Invest Dermatol (2009) 129:2726–8. doi: 10.1038/jid.2009.140
18. Gomez LM, Anaya JM, Martin J. Genetic influence of PTPN22 R620W polymorphism in tuberculosis. Hum Immunol (2005) 66:1242–7. doi: 10.1016/j.humimm.2005.11.008
19. Lamsyah H, Rueda B, Baassi L, Elaouad R, Bottini N, Sadki K, et al. Association of PTPN22 gene functional variants with development of pulmonary tuberculosis in Moroccan population. Tissue Antigens (2009) 74:228–32. doi: 10.1111/j.1399-0039.2009.01304.x
20. Wells GA, Shea B, O’Connell D, Peterson J, Welch V, Losos M, et al. The Newcastle-Ottawa Scale (NOS) for assessing the quality of nonrandomised studies in meta-analyses (2009). Available at: http://www.ohri.ca/programs/clinical_epidemiology/oxford.htm (Accessed 9 May 2010).
21. Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. J Clin Epidemiol (2009) 62:1006–12. doi: 10.1016/j.jclinepi.2009.06.005
22. Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med (2002) 21:1539–58. doi: 10.1002/sim.1186
23. Dersimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials (1986) 7:177–88. doi: 10.1016/0197-2456(86)90046-2
24. Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics (1994) 50:1088–101. doi: 10.2307/2533446
25. Aliparasti MR, Almasi S, Majidi J, Zamani F, Khoramifar AR, Azari AR. Protein tyrosine phosphatase non-receptor type 22 gene polymorphism C1858T is not associated with leprosy in Azerbaijan, Northwest Iran. Indian J Hum Genet (2013) 19:403–7. doi: 10.4103/0971-6866.124365
26. Narasimha VR, Panati K, Reddy MG, Narala VR. Protein tyrosine phosphatase nonreceptor type 22 (PTPN22) gene polymorphism in pulmonary tuberculosis in the Indian population. Int J Mycobacteriol (2016) 5:346–50. doi: 10.1016/j.ijmyco.2016.06.014
27. Shi X, Yang B, Wang Z, Lin D, Wang R. Correlation between polymorphism of PTPN22 gene and pulmonary tuberculosis: a case-control study. Chin J Microbiol Immunol (China) (2014) 34:908–12. doi: 10.3760/cma.j.issn.0254-5101.2014.12.004
28. Azarian M, Busson M, Rocha V, Ribaud P, Peffault De Latour R, Bleux H, et al. The PTPN22 R620W polymorphism is associated with severe bacterial infections after human leukocyte antigen geno-identical haematopoietic stem-cell transplantations. Transplantation (2008) 85:1859–62. doi: 10.1097/TP.0b013e31817729c4
29. Nahum A, Bates A, Sharfe N, Roifman CM. Association of the lymphoid protein tyrosine phosphatase, R620W variant, with chronic mucocutaneous candidiasis. J Allergy Clin Immunol (2008) 122:1220–2. doi: 10.1016/j.jaci.2008.10.027
30. Cloutier JF, Veillette A. Cooperative inhibition of T-cell antigen receptor signaling by a complex between a kinase and a phosphatase. J Exp Med (1999) 189:111–21. doi: 10.1084/jem.189.1.111
31. Bray C, Wright D, Haupt S, Thomas S, Stauss H, Zamoyska R. Crispr/Cas Mediated Deletion of PTPN22 in Jurkat T Cells Enhances TCR Signaling and Production of IL-2. Front Immunol (2018) 9:2595. doi: 10.3389/fimmu.2018.02595
32. Hasegawa K, Martin F, Huang G, Tumas D, Diehl L, Chan AC. PEST domain-enriched tyrosine phosphatase (PEP) regulation of effector/memory T cells. Science (2004) 303:685–9. doi: 10.1126/science.1092138
33. Moran AE, Hogquist KA. T-cell receptor affinity in thymic development. Immunology (2012) 135:261–7. doi: 10.1111/j.1365-2567.2011.03547.x
34. Cohen S, Dadi H, Shaoul E, Sharfe N, Roifman CM. Cloning and characterization of a lymphoid-specific, inducible human protein tyrosine phosphatase, Lyp. Blood (1999) 93:2013–24. doi: 10.1182/blood.V93.6.2013.406k25_2013_2024
35. Gjorloff-Wingren A, Saxena M, Williams S, Hammi D, Mustelin T. Characterization of TCR-induced receptor-proximal signaling events negatively regulated by the protein tyrosine phosphatase PEP. Eur J Immunol (1999) 29:3845–54. doi: 10.1002/(SICI)1521-4141(199912)29:12<3845::AID-IMMU3845>3.0.CO;2-U
36. Schmedt C, Saijo K, Niidome T, Kuhn R, Aizawa S, Tarakhovsky A. Csk controls antigen receptor-mediated development and selection of T-lineage cells. Nature (1998) 394:901–4. doi: 10.1038/29802
37. Hill RJ, Zozulya S, Lu YL, Ward K, Gishizky M, Jallal B. The lymphoid protein tyrosine phosphatase Lyp interacts with the adaptor molecule Grb2 and functions as a negative regulator of T-cell activation. Exp Hematol (2002) 30:237–44. doi: 10.1016/S0301-472X(01)00794-9
38. Rieck M, Arechiga A, Onengut-Gumuscu S, Greenbaum C, Concannon P, Buckner JH. Genetic variation in PTPN22 corresponds to altered function of T and B lymphocytes. J Immunol (2007) 179:4704–10. doi: 10.4049/jimmunol.179.7.4704
39. Vang T, Congia M, Macis MD, Musumeci L, Orru V, Zavattari P, et al. Autoimmune-associated lymphoid tyrosine phosphatase is a gain-of-function variant. Nat Genet (2005) 37:1317–9. doi: 10.1038/ng1673
40. Bottini N, Peterson EJ. Tyrosine phosphatase PTPN22: multifunctional regulator of immune signaling, development, and disease. Annu Rev Immunol (2014) 32:83–119. doi: 10.1146/annurev-immunol-032713-120249
41. Dornelles LN, Pereira-Ferrari L, Messias-Reason I. Mannan-binding lectin plasma levels in leprosy: deficiency confers protection against the lepromatous but not the tuberculoid forms. Clin Exp Immunol (2006) 145:463–8. doi: 10.1111/j.1365-2249.2006.03161.x
42. Moller M, Kinnear CJ, Orlova M, Kroon EE, Van Helden PD, Schurr E, et al. Genetic Resistance to Mycobacterium tuberculosis Infection and Disease. Front Immunol (2018) 9:2219. doi: 10.3389/fimmu.2018.02219
43. Orlova M, Schurr E. Human Genomics of Mycobacterium tuberculosis Infection and Disease. Curr Genet Med Rep (2017) 5:125–31. doi: 10.1007/s40142-017-0124-7
44. Uaska Sartori PV, Penna GO, Buhrer-Sekula S, Pontes MAA, Goncalves HS, Cruz R, et al. Human Genetic Susceptibility of Leprosy Recurrence. Sci Rep (2020) 10:1284. doi: 10.1038/s41598-020-58079-3
45. Sauer ME, Salomao H, Ramos GB, D’espindula HR, Rodrigues RS, Macedo WC, et al. Genetics of leprosy: expected and unexpected developments and perspectives. Clin Dermatol (2015) 33:99–107. doi: 10.1016/j.clindermatol.2014.10.001
46. Bonecini-Almeida Mda G, Werneck-Barroso E, Carvalho PB, De Moura CP, Andrade EF, Hafner A, et al. Functional activity of alveolar and peripheral cells in patients with human acquired immunodeficiency syndrome and pulmonary tuberculosis. Cell Immunol (1998) 190:112–20. doi: 10.1006/cimm.1998.1399
47. Begovich AB, Carlton VE, Honigberg LA, Schrodi SJ, Chokkalingam AP, Alexander HC, et al. A missense single-nucleotide polymorphism in a gene encoding a protein tyrosine phosphatase (PTPN22) is associated with rheumatoid arthritis. Am J Hum Genet (2004) 75:330–7. doi: 10.1086/422827
48. Wang Y, Shaked I, Stanford SM, Zhou W, Curtsinger JM, Mikulski Z, et al. The autoimmunity-associated gene PTPN22 potentiates toll-like receptor-driven, type 1 interferon-dependent immunity. Immunity (2013) 39:111–22. doi: 10.1016/j.immuni.2013.06.013
49. Zheng J, Ibrahim S, Petersen F, Yu X. Meta-analysis reveals an association of PTPN22 C1858T with autoimmune diseases, which depends on the localization of the affected tissue. Genes Immun (2012) 13:641–52. doi: 10.1038/gene.2012.46
Keywords: PTPN22-C1858T, single-nucleotide polymorphism, tuberculosis, leprosy, Mycobacterium tuberculosis, Mycobacterium leprae
Citation: Li S, Wang X, Zhao Y, Yang J, Cui T, Zhao ZJ, Chen Y and Zheng Z (2021) Association of PTPN22-C1858T Polymorphism With Susceptibility to Mycobacterium tuberculosis and Mycobacterium leprae Infection: A Meta-Analysis. Front. Immunol. 12:592841. doi: 10.3389/fimmu.2021.592841
Received: 03 September 2020; Accepted: 13 January 2021;
Published: 25 February 2021.
Edited by:
Stéphane Ranque, Aix-Marseille Université, FranceReviewed by:
Raju Kumar Mandal, Jazan University, Saudi ArabiaSajad Ahmad Dar, Jazan University, Saudi Arabia
Copyright © 2021 Li, Wang, Zhao, Yang, Cui, Zhao, Chen and Zheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhihua Zheng, emh6aGlodWFAbWFpbC5zeXN1LmVkdS5jbg==; Yun Chen, Y2hlbnk2NTNAbWFpbC5zeXN1LmVkdS5jbg==
 Shuping Li
Shuping Li Xiaohua Wang1,2
Xiaohua Wang1,2 Yun Chen
Yun Chen