
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Immunol., 23 July 2020
Sec. B Cell Biology
Volume 11 - 2020 | https://doi.org/10.3389/fimmu.2020.01564
This article is part of the Research TopicMouse Models of B Cell MalignanciesView all 14 articles
Chromosomal translocations linking various oncogenes to transcriptional enhancers of the immunoglobulin heavy chain (IgH) locus are often implicated as the cause of B-cell malignancies. Two major IgH transcriptional enhancers have been reported so far. The Eμ enhancer located upstream of the Cμ gene controls early events in B-cell maturation such as VDJ recombination. The 3' regulatory region (3'RR) located downstream from the Cα gene controls late events in B-cell maturation such as IgH transcription, somatic hypermutation, and class switch recombination. Convincing demonstrations of the essential contributions of both Eμ and 3'RR in B-cell lymphomagenesis have been provided by transgenic and knock-in animal models which bring the oncogene c-myc under Eμ/3'RR transcriptional control. This short review summarizes the different mouse models so far available and their interests/limitations for progress in our understanding of human c-myc-induced B-cell lymphomagenesis.
RAG-induced recombination, AID-induced DNA breaks and mutations throughout B-cell development make the IgH locus a hotspot for translocations (1) (Figures 1A,B). Bcl-2 translocation, the typical hallmark of follicular lymphomas (FL), occurs during RAG-induced VDJ recombination. Cyclin D1 translocation, associated with mantle cell lymphomas (MCL), occurs either during AID-induced somatic hypermutation (SHM) or AID-induced class switch recombination (CSR). C-myc translocation, the typical hallmark of Burkitt lymphoma (BL), takes place during AID-induced SHM and CSR. Finally, several translocations (such as c-myc, c-maf, cyclin D1/D3) found in myelomas are also related to AID-induced CSR. During CSR, AID-induced DNA double strand breaks (DSB) appear in the switch (S) donor region (usually Sμ) and in the S acceptor region (for example Sγ1 and Sα for CSR toward IgG1 and IgA, respectively). S regions are of various lengths (for example 3.5 and 10 kb long for Sμ and Sγ1, respectively) and are unusually G-rich. AID deaminates C into U at preferential AID hotspot motifs located throughout S regions. The AID-introduced U in S region DNA is removed by UNG to generate an abasic site that is recognized by the endonuclease APE1 generating a nick. A closely spaced, similarly created nick on the opposite strand induces a staggered DSB. Translocation of the DNA fragment encompassing c-myc is due to an off target AID effect on the chromosome bearing c-myc. Since AID transforms C to U all along S donor/acceptor regions, there is no common breakpoint identified in S regions for mature B-cell lymphomas. It is the same AID effect for SHM where AID targets the VDJ rearranged segments (and up to several kB in 3') and can induce DNA DSB for c-myc translocation. Similarly to CSR, there is no common breakpoint established in VDJ regions for mature B-cell lymphomas. During VDJ recombination RAG binds to recombination signal sequences adjacent to V, D, and J coding segments and induces DNA DSB. C-myc translocation could take place during this process. Similarly to CSR/SHM, there is no common breakpoint singled out in VDJ regions for B-cell lymphomas. The common point for all these c-myc translocations is the occurrence of DSB in the IgH locus during its remodeling required for B-cell repertoire formation and B-cell maturation. All remodeling events of the IgH locus (VDJ recombination, SHM, and CSR) require transcription to occur (2). Transcriptional control and remodeling of the IgH locus are under the control of several cis-regulatory elements located throughout the IgH locus. In the murine IgH locus seven regions of interest can be defined including cis-regulatory elements, matrix attachment regions (MARs), and hypersensitivity (hs) sites with potential transcriptional enhancer or insulator activity: 4 hs sites located 5' of the first V segments, 6 hs sites in the V–D intergenic region, the DQ52 promoter–enhancer, the Eμ enhancer (between JH and Cμ) and its flanking MARs, the γ1 enhancer element, the 3' regulatory region (3'RR) downstream from Cα with its four enhancers (hs3a, hs1,2, hs3b, and 4) and the 3'CBE insulator region (hs5, 6, 7, 8) as the 3' boundary of the locus (Figure 1A). Two potent transcriptional enhancers act during B-cell maturation: Eμ (during early B-cell maturation stages) and 3'RR (during late B-cell maturation stages) (Figure 1A). These elements obviously intervene in oncogene-induced B-cell lymphomagenesis as reported by several transgenic mouse models (using both transgene and knock-in (KI) strategies) developed in order to mimic human mature B-cell lymphomagenesis. Since c-myc is a key regulator of cell growth through its action on cell cycle progression, metabolism, differentiation, death receptor signaling, and DNA damage recovery, the vast majority of available models use c-myc as a deregulated oncogene (3). This short review describes how Eμ and 3'RR enhancers might play a critical role in c-myc deregulation during c-myc-induced mature B-cell lymphomas, why these models are not silver bullets to totally mimic human B-cell lymphomagenesis and why it is possible that targeting the 3'RR would be an interesting strategy in human B-cell lymphomagenesis.
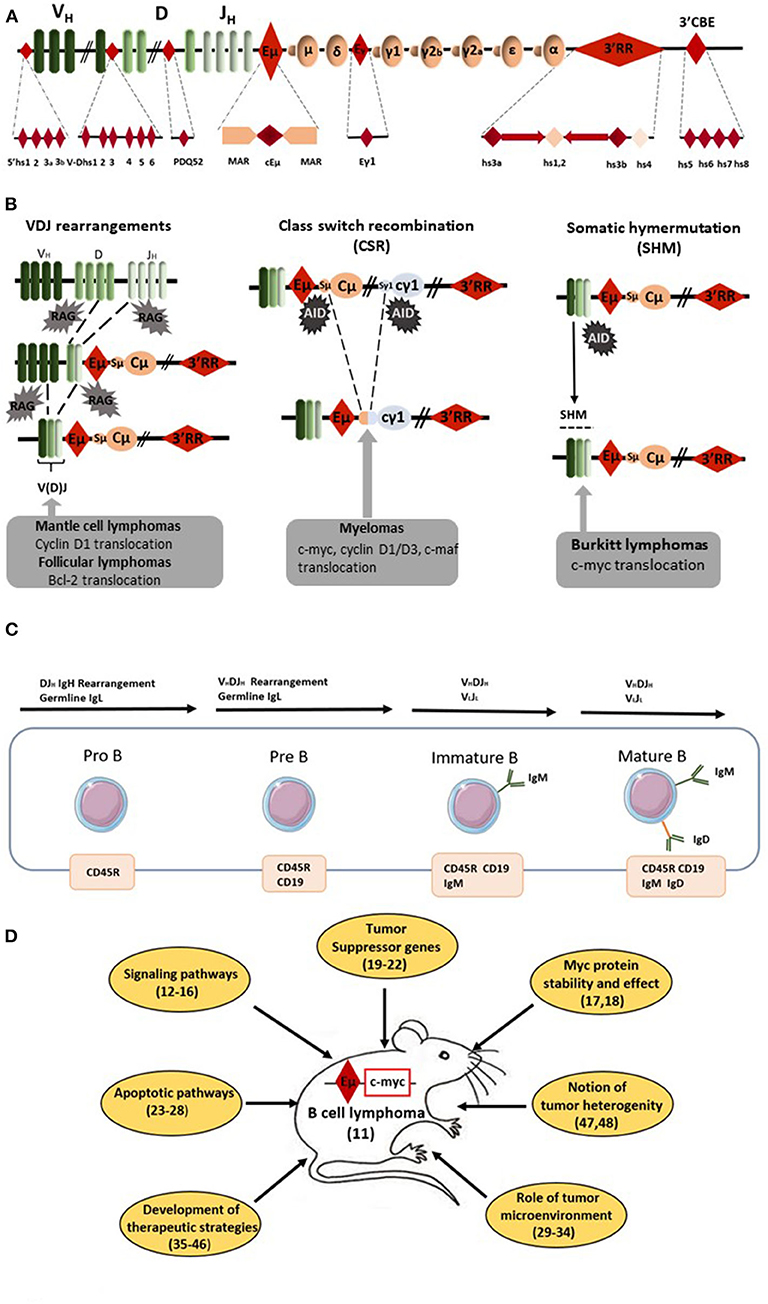
Figure 1. Eμ-Myc mice as a model of B-cell lymphomagenesis. (A) Schematic diagrams of the mouse IgH locus. Locations of the various IgH cis-regulatory elements with enhancer or insulator activity are reported: four hs sites located 5' of the first V segments, six hs sites in the V–D intergenic region, the DQ52 promoter-enhancer, the Eμ enhancer (the core region (cEμ) and its flanking MARs), the γ1 enhancer, the 3' regulatory region (3'RR) [four enhancers (namely hs3a, hs1,2, hs3b, and 4) with flanking inverted repeats] and the 3'CBE insulator region (hs5, 6, 7, and 8) as the 3' boundary of the locus. (B) Schematic representation of oncogene translocation affecting the IgH locus during VDJ recombination, CSR and SHM. Arrows indicate the site of oncogene translocation found during follicular lymphomas, mantle cell lymphomas, myelomas, and Burkitt lymphomas. (C) Schematic representation of B-cell development from pro-B to mature B-cells. Lymphomas from Eμ-Myc mice are from the pre-B to the mature B-cell stages. The immature B-cell stage is characterized by the expression of membrane IgM whereas membrane IgD occurs at the mature B-cell stage. (D) Schematic representation of the various field of research developed with Eμ-Myc mice. Bibliographic references are reported (number in parenthesis).
Forty years ago, Eμ was the first discovered IgH cis-transcriptional enhancer (4–6). It is located upstream of the Cμ gene (Figure 1A). Eμ-deficient mice revealed its role in controlling IgH locus access at immature B-cell stages and thus its key role for efficient VDJ recombination (7, 8). In contrast, Eμ is dispensable for late B-cell maturation events such as IgH locus transcription for Ig synthesis and CSR (9, 10). In 1985, transgenic mice bearing c-myc coupled to the Eμ enhancer were reported to consistently develop immature (pre-B) and sometimes mature B-cell lymphomas (11). Our entire knowledge of Eμ involvement in c-myc oncogenic deregulation for B-cell lymphoma development was built from this model. Since 1985, 183 papers with “Eμ-Myc mice” in their abstract have been referenced. Of note, 153 have been published in the last 15 years showing the great interest of the scientific community for this transgenic mouse model of B-cell lymphoma. It is thus impossible in this short review to reference them all. Therefore, the authors apologize in advance for the numerous interesting manuscripts which have not been cited in the present review. Lymphomas from Eμ-Myc mice range from the pre-B to the mature B-cell stages (Figure 1C). They are usually all positive for the CD45R (B220), CD19 and CD93 (AA4.1) B-cell specific markers and negative for the CD3 T-cell marker. Tumors of pre-B-cell type are characterized by the lack of membrane IgM and no Ig light chain (IgL) rearrangements. Tumors of immature B-cell types are more mature and express membrane IgM after efficient IgL rearrangements. Tumors of mature B-cell types are even more mature and express both membrane IgM and IgD. The majority of lymphomas in Eμ-Myc mice are at the pre-B and immature B-cell stages. In their original study, Adams et al. (11) stated that “these myc mice should aid study of lymphoma development, B-cell ontogeny and Ig regulation.” Clearly 35 years later this is the case. Creation of these mice resulted in the dissection of many mechanisms implicated in B-cell lymphomagenesis (Figure 1D). They have highlighted the importance of several signaling pathways (such as Ras/Mapk, mTOR, and Akt) (12–14), several cell cycle check-points (such as Mdm2/p53/p73) (15, 16) and processes that affect c-myc stability and action (17, 18). Using these mice clearly demonstrated the importance of numerous (new and well-known) tumor suppressor genes (such as FoxO3, CDK4, Mtap, and Smchd1) (19–22). This model reinforced our knowledge concerning the signaling/regulation of the B-cell apoptotic program (members of the Bcl-2 family of apoptosis regulator) and deficiencies in apoptotic pathways leading to B-cell lymphomagenesis (23–28). To our knowledge the influence of genetic background in the development of B-cell lymphomas in Eμ-Myc mice has not been documented. The Eμ-Myc model has also opened a new area of research concerning the role of tumor microenvironment via release of angiocrine/chemokine factors (29–31) and the importance of cells from the vascular niche for NK cell surveillance, senescence, and homing of B-cell lymphomas (32–34). Perhaps most importantly, this model is at the origin of a wide number of publications investigating new therapeutic treatments or combinations of drugs in order to affect (among various targets) DNA synthesis (cytarabine, doxyrubicin, cyclophosphamide), mTOR signaling (rapamycin analogs), microtubule formation (vincristine), c-myc (decursin), apoptosis (venetoclax and BET inhibitors), protein synthesis (silvestrol), or B-cell receptor (BCR)-induced, or chemokine-mediated signaling (ibrutinib) (35–42). The rapid occurrence of lymphoma in Eμ-Myc mice and its high penetrance make this mouse model an accurate, reliable, easy, and fast experimental model not only to test new therapeutic approaches but also combinatory associations. This model is also unique by providing the possibility to monitor the assay of new NK therapeutic vaccination strategies (43, 44), to stimulate immune defenses for tumor rejection (45) and to test protocols for monoclonal antibody therapies (46). Eμ-Myc mice have thus proven their great potential as a model to study human B-cell lymphomagenesis during the past decade. Moreover, arising lymphomas are heterogeneous (47, 48) mirroring genomic differences observed between human BL, germinal center B-cell lymphomas (GCBCL), activated B-cell lymphomas (ABCL), and diffuse large B-cell lymphomas (DLBCL). The different genomic signatures (toward specific proliferative and/or apoptotic pathways) of B-cell lymphomas in Eμ-Myc mice might be used as biomarkers of response against specific therapeutic strategies. Thus, and especially with the development of transcriptomic tools, Eμ-Myc mice can serve as relevant model for human B-cell lymphoma subtype experimental or associated treatments. The only but nevertheless major drawback of Eμ-Myc mice relates to the window of activity for Eμ which has been clearly demonstrated to occur at the immature pro-B/pre-B B-cell stages (49, 50). Eμ is not implicated in IgH hypertranscription occurring at the mature/plasma cell stages. Eμ is also not implicated in DNA breaks occurring during SHM/CSR and thus clearly not implicated in oncogenic translocation induced by off target AID action occurring during CSR or SHM in the majority of human mature B-cell lymphoma subtypes. As confirmation of this fact, the great majority of lymphomas from Eμ-Myc mice have a pre-B/immature B-cell stage.
The second transcriptional enhancer located in the IgH locus is the 3'RR (Figure 1). The 3'RR is a complex element with four transcriptional enhancers (namely hs3a, hs1,2, hs3b, and hs4) encompassed in a unique and functional 3D palindromic architecture (51). The 3'RR controls μ transcription (7), CSR (52, 53), and SHM (54) in mature B-cells. The transcriptional activity of the 3'RR occurs from pre-B to mature B-cell stages (55) and thus has a much larger window of activity than the Eμ enhancer. In 1994, Madisen and Groudine reported (in stable transfection assays in plasmacytomas and BL cells) that the 3'RR was efficient and sufficient to deregulate c-myc transcription (56). Convincing demonstration of 3'RR involvement in lymphomagenesis has been produced by a transgenic 3'RR-deficient model of B-cell lymphomas with IgH-c-myc translocations (57). The integrity of the 3'RR (deletion of hs3b to hs4) has been shown to be dispensable for development of pro-B-cell lymphomas with V(D)J recombination-initiated translocations suggesting the key role of Eμ. In contrast, 3'RR integrity (for its optimal transcriptional activity) is required for B-cell lymphomas with CSR-associated translocations (57). In another study modeling murine plasmacytomas with T (12, 15) translocations, the same hs3b-hs4 deletion of the 3'RR in Bcl-xL transgenic mice was without effect for Myc deregulation and mouse plasmacytoma generation (58). However, total 3'RR deletion in these plasmacytomas lowered Myc expression and cell growth confirming 3'RR involvement for myc deregulation by T (12, 15). Nevertheless, these models are not sufficient to monitor in detail and to modulate signaling pathways for B-cell lymphoma development. The same comments can be made for the transgenic mouse model of Wang and Boxer (59) which develops mature B-cell lymphomas (CD19+B220+IgM+IgDlow) after the KI of a 3'RR cassette upstream of the endogenous c-myc gene (this model is the reverse of natural c-myc translocation into the human IgH locus) (Figure 2). More than 15 years after the development of transgenic Eμ-Myc mice, transgenic Myc-3'RR mice were generated and were shown to develop BL-like proliferations and diffuse anaplastic B-cell lymphomas (60). All these lymphomas exhibited a mature B-cell phenotype (CD19+B220+IgM+IgD+) but differed by their Ki67 status (low and high for diffuse anaplastic B-cell lymphomas and BL lymphomas, respectively). This model was used to study the role of second hits such as p53 deficiency, Cdk4 mutation, and change of class-specific B cell receptor (BCR) tonic signals. Results clearly demonstrated that a second hit affects the phenotype of B-cell lymphomas, their aggressiveness and transcriptomic signatures differently (61–64). This model was, however, progressively abandoned due to its medium B-cell lymphoma penetrance (compared to Eμ-Myc mice), long delay for B-cell lymphoma development (compared to Eμ-Myc mice), key differences with human B-cell lymphomas (such as mutations lacking for the p53-ARF-Mdm2 apoptotic pathways in numerous cases) and the description that the occurrence of B-cell lymphomas was much too sensitive to genetic background [C57Bl/6 mice developed BL-like lymphomas while none occurred in a Balb/c background (65)]. All these points argued against the use of Myc-3'RR mice as an accurate experimental model to test new pharmacologic or vaccination strategies.
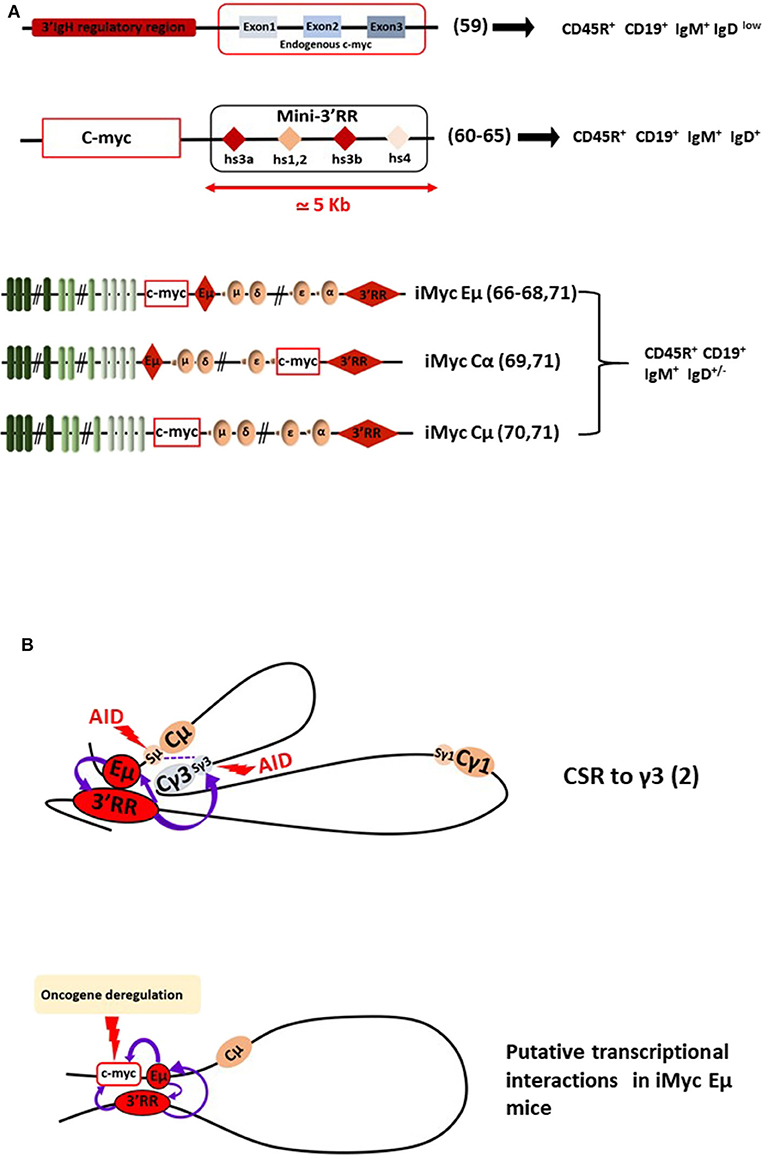
Figure 2. The 3'RR and B-cell lymphomagenesis. (A) Schematic representation of several transgenic mouse models reporting c-myc 3'RR-driven deregulation leading to B-cell lymphomagenesis. B-cell lymphoma phenotypes are reported. Bibliographic references are reported (number in parenthesis). The “Mini-3'RR” contains the four transcriptional enhancers hs3a, hs1,2, hs3b, and hs4 but not the 3'RR palindromic sequences flanking hs1,2 and the DNA sequence between hs3a and hs4. (B) Long-range loop interactions between chromatin segments of the IgH locus comprise the mechanism of normal gene transcription regulation by the Eμ and 3'RR transcriptional enhancers. The example of the IgG3 CSR process is schematized. Putative long-range interactions leading to c-myc oncogene deregulation in iMycEμ mice are schematized.
As reported above, a transgenic model with IgH-c-myc translocations in response to pristine demonstrated the involvement of IgH cis-transcriptional enhancers in B-cell lymphomagenesis (57). In another manner, this study confirmed results obtained with three transgenic mouse models with a c-myc KI in various locations in the IgH locus (i.e., under the dependence of both Eμ and 3'RR elements) (Figure 2). These models provided the most convincing data for the essential roles of both Eμ and 3'RR in c-myc B-cell lymphomagenesis. The KI of c-myc in the mouse IgH locus just 5′ to Eμ (namely iMycEμ mice), thus modeling human endemic BL, induced, as expected, B-cell lymphoma development with alterations in the p19Arf-Mdm2-p53 tumor suppressor axis (66) and NF?B/STAT3/PI3K signaling (67). In this model, c-myc is under the control of both Eμ and 3'RR at immature and mature B-cell stages, respectively. iMycEμ mice also mimic T (12, 15) mouse plasmacytoma translocation and thus also lead to plasmacytomas (68). KI of c-myc directly into Cα just 5' to the 3'RR (iMycCα mice) produced B-cell lymphomas with low kinetics which were increased after overexpression of the anti-apoptotic Bcl-XL gene (69). In this model, c-myc is located in a site where Eμ has no transcriptional influence, c-myc transcription being only under the dependence of 3'RR at mature B-cell stages. c-myc KI in the mouse IgH locus just 5′ to Cμ with Eμ deletion (namely iMycCμ mice), thus modeling human sporadic BL, confirmed that 3'RR alone is sufficient to deregulate c-myc in the B-cell lineage and to induce B-cell lymphoma development (70). Taken altogether, these KI models carrying c-myc at the IgH locus are prone to B-cell lymphomas of various penetrance, kinetics, and fate as recently reported in a study comparing the three mouse models (71). The lymphoma signatures are also heterogeneous even comparing lymphomas from a specific KI, mirroring the genomic differences observed between the various subtypes of human mature B-cell lymphomas and those previously reported with the model of transgenic Eμ-Myc mice. In our opinion, these transgenic mouse models represent the “most physiological” experimental mouse models by mimicking the direct effect of c-myc in the context of the endogenous IgH locus. However, the main drawbacks of these various KI mice (and similarly to Myc-3'RR mice) remain their low lymphoma penetrance and their low kinetics of B-cell lymphoma development arguing against their use as efficient and easy experimental models to test new experimental therapeutic approaches. The low kinetics of B-cell lymphoma development compared with 3'RR-Myc mice would be related to the 3'CBE insulator region at the 3' boundary of the endogenous IgH locus (72, 73). This region is not present in the transgenic mouse model of 3'RR-induced c-myc deregulation. The 3'CBE insulator region contains a high density of binding sites for CCCTC-binding factor (CTCF), a protein associated with mammalian insulator activity. Deletion of the 3'CBE insulator region resulted in significant effects on VDJ rearrangement, IgH locus compaction, and IgH locus insulation. Furthermore, physical interactions occur in B-cells between 3'CBE and 3'RR enhancers suggesting that the entire 3' region (3'RR enhancers + 3'CBE insulators) works as a physical unit. The lack of 3'CBE in 3'RR-Myc mice could induced stronger and longer c-myc deregulation (and thus faster lymphoma emergence) than that obtained when c-myc is inserted into the IgH locus under the control of the entire (enhancer + insulator) region.
Knock-out mice models have clarified the functions of Eμ and 3'RR enhancers as essential for DNA remodeling and IgH locus transcription at specific stages of B-cell development and maturation. Thus, these enhancers have a major potential to be oncogene deregulators for IgH-translocated oncogenes, even when the breakpoints lie several 100 kb away from them. All these models contribute different but interesting data to our understanding of human B-cell lymphoma development and treatments especially with regards to the great functional and structural similarities found between human and mouse IgH loci (74). However, we must keep in mind that these mice are experimental models that do not reflect 100% of what happens in humans. For example, if the vast majority of human mature B-cell lymphomas are mutated in their VDJ region (highlighting their post-germinal center status) it is not the case in mice where mature B-cell lymphomas are unmutated (highlighting their pre-germinal center status) (75). Long-range loop interactions between chromatin segments of the IgH locus comprise the mechanism of normal and abnormal gene transcription regulation by the 3'RR (2, 76) (Figure 2B). Therefore, it is possible to suggest that targeted inhibition of the 3'RR would be a therapeutic approach for the treatment of some mature B-cell lymphomas. Finally, it is also of importance to mention that the c-myc oncogene driven by Ig light chain enhancers also induces B-cell lymphoid malignancy in transgenic mice (11, 77). These models highlight not only the importance of all Ig enhancers for B-cell lymphoma development but also that a 3'RR targeting strategy (if any) would not be a silver bullet to treat all B-cell lymphomas but at best some mature B-cell subtypes.
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
This work was supported by grants from Equipe Labellisée LIGUE 2018 and ANR Episwitch 2016. MF was supported by a grant from Université de Limoges and Région Nouvelle Aquitaine. NG and HI were supported by ANR Episwitch 2016.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
1. Lieber MR. Mechanisms of human lymphoid chromosomal translocations. Nat Rev Cancer. (2016) 16:387–98. doi: 10.1038/nrc.2016.40
2. Pinaud E, Marquet M, Fiancette R, Peron S, Vincent-Fabert C, Denizot Y, et al. The IgH locus 3' regulatory region: pulling the strings from behind. Adv Immunol. (2011) 110:27–70. doi: 10.1016/B978-0-12-387663-8.00002-8
3. Kuzyk A, Mai S. C-MYC induced genomic instability. Cold Spring Harb Perspect Med. (2014) 4:a014373. doi: 10.1101/cshperspect.a014373
4. Alt FW, Rosenberg N, Casanova RJ, Thomas E, Baltimore D. Immunoglobulin heavy-chain expression and class switching in a murine leukaemia cell line. Nature. (1982) 296:325–331. doi: 10.1038/296325a0
5. Gillies SD, Morrison SL, Oi VT, Tonegawa S. A tissue-specific transcription. enhancer element is located in the major intron of a rearranged immunoglobulin heavy chain gene. Cell. (1983) 33:717–28. doi: 10.1016/0092-8674(83)90014-4
6. Banerji J, Olson L, Schaffner W. A lymphocyte-specific cellular enhancer is located downstream of the joining region in immunoglobulin heavy chain genes. Cell. (1983) 33:729–40. doi: 10.1016/0092-8674(83)90015-6
7. Perlot T, Alt FW, Bassing CH, Heikyung S, Pinaud E. Elucidation of IgH intronic enhancer functions via germ-line deletion. Proc Natl Acad Sci USA. (2005) 102:14362–7. doi: 10.1073/pnas.0507090102
8. Marquet M, Garot A, Bender S, Carrion C, Rouaud P, Lecardeur S, et al. The Eμ enhancer region influences H chain expression and B cell fate without impacting IgVH repertoire and immune response in vivo. J Immunol. (2014) 193:1171–83. doi: 10.4049/jimmunol.1302868
9. Saintamand A, Rouaud P, Garot A, Carrion C, Oblet C, Cogné M, et al. The IgH 3' regulatory region governs μ chain transcription in mature B lymphocytes and the B cell fate. Oncotarget. (2015) 6:4845–52. doi: 10.18632/oncotarget.3010
10. Issaoui H, Ghazzaui N, Ferrad M, Boyer F, Denizo Y. Class switch recombination junctions are not affected by the absence of the immunoglobulin heavy chain Eμ enhancer. Cell Mol Immunol. (2019) 16:671–3. doi: 10.1038/s41423-019-0229-x
11. Adams JM, Harris AW, Pinkert CA, Corcoran LM, Alexander W, Cory S, et al. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature. (1985) 318:533–8. doi: 10.1038/318533a0
12. Hussain S, Bedekovics T, Liu Q, Hu W, Jeon H, Johnson H, et al. UCH-L1 bypasses mTOR to promote protein biosynthesis and is required for MYC-driven lymphomagenesis in mice. Blood. (2018) 132:2564–74. doi: 10.1182/blood-2018-05-848515
13. Hussain S, Foreman O, Perkins SL, Witzig TE, Miles RR, van Deursen J, et al. The de-ubiquitinase UCH-L1 is an oncogene that drives the development of lymphoma in vivo by deregulating PHLPP1 and Akt signaling. Leukemia. (2010) 24:1641–55. doi: 10.1038/leu.2010.138
14. Gramling MW, Eischen CM. Suppression of Ras/Mapk pathway signaling inhibits Myc-induced lymphomagenesis. Cell Death Differ. (2012) 19:1220–7. doi: 10.1038/cdd.2012.1
15. Nemajerova A, Petrenko O, Trümper L, Palacios G, Moll UM. Loss of p73 promotes dissemination of Myc-induced B cell lymphomas in mice. J Clin Invest. (2010) 120:2070–80. doi: 10.1172/JCI40331
16. Odvody J, Vincent T, Arrate M, Pgrieb B, Wang S, Garriga J, et al. A deficiency in Mdm2 binding protein inhibits Myc-induced B-cell proliferation and lymphomagenesis. Oncogene. (2010) 29:3287–96. doi: 10.1038/onc.2010.82
17. Chen J, Shin JH, Zhao R, Phan L, Wang H, Xue Y, et al. CSN6 drives carcinogenesis by positively regulating Myc stability. Nat Commun. (2014) 5:5384. doi: 10.1038/ncomms6384
18. Wong DM, Li L, Jurado S, King A, Bamford R, Wall M, et al. The transcription factor ASCIZ and its target DYNLL1 are essential for the development and expansion of MYC-driven B cell lymphoma. Cell Rep. (2016) 14:1488–99. doi: 10.1016/j.celrep.2016.01.012
19. Kadariya Y, Tang B, Wang L, Al-Saleem T, Hayakawa K, Slifker MJ, et al. Germline mutations in Mtap cooperate with Myc to accelerate tumorigenesis in mice. PLoS ONE. (2013) 8:e67635. doi: 10.1371/journal.pone.0067635
20. Leong HS, Kelan C, Hilton DJ, Blewitt ME, Yifang HU, Lee S, et al. Epigenetic regulator Smchd1 functions as a tumor suppressor. Cancer Res. (2012) 73:1591–9. doi: 10.1158/0008-5472.CAN-12-3019
21. Lu Y, Wu Y, Feng X, Shen R, Wang JH, Fallahi M, et al. CDK4 deficiency promotes genomic instability and enhances Myc-driven lymphomagenesis. J Clin Invest. (2014) 124:1672–84. doi: 10.1172/JCI63139
22. Vandenberg CJ, Motoyama N, Cory S. FoxO3 suppresses Myc driven lymphomagenesis. Cell Death Dis. (2016) 7:e2046. doi: 10.1038/cddis.2015.396
23. Olive V, Sabio E, Bennett MJ, De Jong CS, Biton A, McGann JC, et al. A component of the mir-17-92 polycistronic oncomir promotes oncogene-dependent apoptosis. Elife. (2013) 2:e00822. doi: 10.7554/eLife.00822
24. Chu B, Kon N, Chen D, Li T, Liu T, Jiang L, et al. ALOX12 is required for p53-mediated tumour suppression through a distinct ferroptosis pathway. Nat Cell Biol. (2019) 21:579–91. doi: 10.1038/s41556-019-0305-6
25. Takahashi Y, Young MM, Serfass JM, Hori T, Wang HG. Sh3glb1/Bif-1 and mitophagy. Autophagy. (2013) 9:1107–9. doi: 10.4161/auto.24817
26. Tuzlak S, Haschka MD, Mokina AM, Rülicke T, Cory S, Labi V, et al. Differential effects of Vav-promoter-driven overexpression of BCLX and BFL1 on lymphocyte survival and B cell lymphomagenesis. FEBS J. (2018) 285:1403–18. doi: 10.1111/febs.14426
27. Sochalska M, Schuler F, Weiss JG, Prchal-Murphy M, Sexl V, Villunger A. MYC selects against reduced BCL2A1/A1 protein expression during B-cell lymphomagenesis. Oncogene. (2017) 36:2066–73. doi: 10.1038/onc.2016.362
28. Nguyen HV, Vandenberg CJ, Ng AP, Robati MR, Anstee NS, Rimes J, et al. Development and survival of MYC-driven lymphomas require the Myc antagonist MNT to curb MYC-induced apoptosis. Blood. (2020) 135:1019–31. doi: 10.1182/blood.2019003014
29. Vecchio E, Fiume G, Mignogna C, Iaccino E, Mimmi S, Maisano D, et al. IBTK haploinsufficiency affects the tumor microenvironment of Myc-driven lymphoma in E-myc mice. Int J Mol Sci. (2020) 21:885. doi: 10.3390/ijms21030885
30. Rehm A, Mensen A, Schradi K, Gerlach K, Wittstock S, Winter S, et al. Cooperative function of CCR7 and lymphotoxin in the formation of a lymphoma-permissive niche within murine secondary lymphoid organs. Blood. (2011) 118:1020–33. doi: 10.1182/blood-2010-11-321265
31. Cao Z, Ding BS, Guo P, Lee SB, Butler JM, Casey C, et al. Angiocrine factors deployed by tumor vascular niche induce B cell lymphoma invasiveness and chemotresistance. Cancer Cell. (2014) 25:350–65. doi: 10.1016/j.ccr.2014.02.005
32. Reimann M, Lee S, Loddenkemper C, Dorr JR, Tabor V, Aichele P, et al. Tumor stroma-derived TGF-b limits myc-driven lymphomagenesis via Suv39h1-dependent senescence. Cancer Cell. (2010) 17:262–72. doi: 10.1016/j.ccr.2009.12.043
33. Sauer M, Schuldner M, Hoffmann N, Cetintas A, Reiners KS, Shatnyeva O, et al. CBP/p300 acetyltransferases regulate the expression of NKG2D ligands on tumor cells. Oncogene. (2017) 36:933–41. doi: 10.1038/onc.2016.259
34. Rehm A, Gätjen M, Gerlach K, Scholz F, Mensen A, Gloger M, et al. Dendritic cell-mediated survival signals in Eμ-Myc B-cell lymphoma depend on the transcription factor C/EBPμ. Nat Comm. (2014) 5:5057. doi: 10.1038/ncomms6057
35. Lasorsa E, Smonksey M, Kirk JS, Rosario S, Hernandez-Ilizaliturri FJ, Ellis L, et al. Mitochondrial protection impairs BET bromodomain inhibitor-mediated cell death and provides rationale for combination therapeutic strategies. Cell Death Dis. (2015) 6:e2014. doi: 10.1038/cddis.2015.352
36. Hogg SJ, Newbold A, Vervoort SJ, Cluse LA, Martin BP, Gregory P, et al. BET inhibition induces apoptosis in aggressive B-cell lymphoma via epigenetic regulation of BCL-2 family members. Mol Cancer Ther. (2016) 15:2030–41. doi: 10.1158/1535-7163.MCT-15-0924
37. Lin CJ, Robert F, Sukarieh R, Michnick S, Pelletier J. The antidepressant sertraline inhibits translation initiation by curtailing mammalian target of rapamycin signaling. Cancer Res. (2010) 70:3199–208. doi: 10.1158/0008-5472.CAN-09-4072
38. Rava M, D'Andrea A, Nicoli P, Gritti I, Donati G, Doni M, et al. Therapeutic synergy between tigecycline and venetoclax in a preclinical model of Myc/BCL2 double-hit B cell lymphoma. Sci Transl Med. (2018) 10:eaan8723. doi: 10.1126/scitranslmed.aan8723
39. Ross J, Rashkovan M, Fraszczak J, Beauparlant CJ, Vadnais C, Winkler R, et al. C and B-ALL/lymphoma mouse models. Cancer Res. 79:4184–95. doi: 10.1158/0008-5472.CAN-18-3038
40. Reiff SD, Mantel R, Smith LL, Greene JT, Muhowski EM, Fabian A, et al. The BTK inhibitor ARQ531 targets ibrutinib-resistant CLL and richter transformation. Cancer Discov. (2018) 8:1300–15. doi: 10.1158/2159-8290.CD-17-1409
41. Wall M, Poortinga G, Stanley KL, Lindemann RK, Bots M, Chan J, et al. The mTORC1 inhibitor everolimus prevents and treats em-Myc lymphoma by restoring oncogene-induced senescence. Cancer Discov. (2013) 3:82–95. doi: 10.1158/2159-8290.CD-12-0404
42. Rodrigo M, Cencic R, Roche SP, Pelletier J, Porco JA Jr. Synthesis of rocaglamide hydroxamates and related compounds as eukaryotic translation inhibitors: synthetic and biological studies. J Med Chem. (2012) 55:558–62. doi: 10.1021/jm201263k
43. Mattarollo SR, West AC, Steegh K, Duret H, Paget C, Martin B, et al. NKT cell adjuvant-based tumor vaccine for treatment of myc oncogene-driven mouse B-cell lymphoma. Blood. (2012) 120:3019–29. doi: 10.1182/blood-2012-04-426643
44. Kobayashi T, Doff BL, Rearden RC, Leggatt GR, Mattarollo SR. NKT cell-targeted vaccination plus anti-4-1BB antibody generates persistent CD8 T cell immunity against B cell lymphoma. OncoImmunology. (2015) 4:e990793. doi: 10.4161/2162402X.2014.990793
45. Croxford JL, Tang ML, Pan MF, Huang CW, Kamran N, Phua M, et al. ATM-dependent spontaneous regression of early Eμ-myc-induced murine B-cell leukemia depends on natural killer and T cells. Blood. (2013) 121:2512–21. doi: 10.1182/blood-2012-08-449025
46. Westwood JA, Matthews GM, Shortt J, Faulkner D, Pegram HJ, Duong P, et al. Combination anti-CD137 and anti-CD40 antibody therapy in murine myc-driven hematological cancers. Leukemia Res. (2014) 38:948–54. doi: 10.1016/j.leukres.2014.05.010
47. Johnston HE, Carter MJ, Cox KL, Dunscombe M, Manousopoulou A, Townsend A, et al. Integrated cellular and plasma proteomics of contrasting B-cell cancers reveals common unique and systemic signatures. Mol Cell Proteomics. (2017) 16:386–406. doi: 10.1074/mcp.M116.063511
48. Rempel RE, Jiang X, Fullerton P, Tan TZ, Ye J, Lau A, et al. Utilization of the Eμ-Myc mouse to model heterogeneity of therapeutic response. Mol Cancer Ther. (2014) 13:3219–29. doi: 10.1158/1535-7163.MCT-13-0044
49. Guglielmi L, Truffinet V, Carrion C, Le Bert M, Cogné N, Cogné M, et al. The 5'HS4 insulator element is an efficient tool to analyse the transient expression of an Eμ-GFP vector in a transgenic mouse model. Trans Res. (2005) 14:361–4. doi: 10.1007/s11248-005-3239-7
50. Ghazzaui N, Issaoui H, Boyer F, Martin OA, Saintamand A, Denizot Y. 3'RR and 5'Eμ immunoglobulin heavy chain enhancers are independent engines of locus remodeling. Cell Mol Immunol. (2019) 16:198–200. doi: 10.1038/s41423-018-0171-3
51. Saintamand A, Vincent-Fabert C, Garot A, Rouaud P, Oruc Z, Magnone V, et al. Deciphering the importance of the palindromic architecture of the immunoglobulin heavy chain 3' regulatory region. Nat Commun. (2016) 7:10730. doi: 10.1038/ncomms10730
52. Vincent-Fabert C, Fiancette R, Pinaud E, Truffinet V, Cogné N, Cogné M, et al. Genomic deletion of the whole IgH 3' regulatory region (hs3a, hs1,2, hs3b, hs4) dramatically affects class switch recombination and Ig secretion to all isotypes. Blood. (2010) 116:1895–98. doi: 10.1182/blood-2010-01-264689
53. Saintamand A, Rouaud P, Saad F, Rios G, Cogné M, Denizot Y. Elucidation of IgH 3' region regulatory role during class switch recombination via germline deletion. Nat Commun. (2015) 6:7084. doi: 10.1038/ncomms8084
54. Rouaud P, Vincent-Fabert C, Saintamand A, Fiancette R, Marquet M, Robert I, et al. The IgH 3' regulatory region controls AID-induced somatic hypermutation in germinal centre B-cells in mice. J Exp Med. (2013) 210:1501–7. doi: 10.1084/jem.20130072
55. Guglielmi L, Le Bert M, Truffinet V, Cogné M, Denizot Y. Insulators to improve expression of a 3'IgH LCR-driven reporter gene in transgenic mouse models. Biochem Biophys Res Commun. (2003) 307:466–71. doi: 10.1016/S0006-291X(03)01185-9
56. Madisen L, Groudine M. Identification of a locus control region in the immunoglobulin heavy-chain locus that deregulates C-Myc expression in plasmacytoma and Burkitt's lymphoma cells. Genes Dev. (1994) 8:2212–26. doi: 10.1101/gad.8.18.2212
57. Gostissa M, Yan CT, Bianco JM, Cogné M, Pinaud E, Alt FW. Long-range oncogenic activation of Igh-c-myc translocations by the Igh 3' regulatory region. Nature. (2009) 462:803–8. doi: 10.1038/nature08633
58. Kovalchuk AL, Sakai T, Qi CF, Du Bois W, Dunnick WA, Congé M, et al. (2018) 3'Igh enhancers hs3b/hs4 are dispensable for Myc deregulation in mouse plasmacytomas with T(12;15) translocations. Oncotarget. 9:34528–42. doi: 10.18632/oncotarget.26160
59. Wang J, Boxer LM. Regulatory elements in the immunoglobulin heavy chain gene 3'-enhancers induce c-myc deregulation and lymphomagenesis in murine B cells. J Biol Chem. (2006) 280:12766–73. doi: 10.1074/jbc.M412446200
60. Truffinet V, Pinaud E, Cogné N, Petit B, Guglielmi L, Cogné M, et al. The 3' IgH locus control region is sufficient to deregulate a c-myc transgene and promote mature B cell malignancies with a predominant Burkitt-like phenotype. J Immunol. (2007) 179:6033–42. doi: 10.4049/jimmunol.179.9.6033
61. Fiancette R, Rouaud P, Vincent-Fabert C, Laffleur B, Magnone V, Cogné M, et al. A p53 defect sensitizes various stages of B cell development to lymphomagenesis in mice carrying an IgH 3' regulatory region-driven c-myc transgene. J Immunol. (2011) 187:5772–82. doi: 10.4049/jimmunol.1102059
62. Vincent-Fabert C, Fiancette R, Rouaud P, Baudet C, Truffinet V, Magnone V, et al. A defect of the INK4-Cdk4 checkpoint and c-myc collaborate in blastoid mantle cell lymphoma (MCL)-like lymphoma formation in mice. Am J Pathol. (2012) 180:1688–701. doi: 10.1016/j.ajpath.2012.01.004
63. Rouaud P, Fiancette R, Vincent-Fabert C, Magnone V, Cogné M, Dubus P, et al. Mantle cell lymphoma-like lymphomas in c-myc-3'RR/p53+/- mice and c-myc-3'RR/Cdk4R24C mice: differential oncogenic mechanisms but similar cellular origin. Oncotarget. (2012) 3:586–93. doi: 10.18632/oncotarget.474
64. Amin R, Marfak A, Pangault C, Oblet C, Chanut A, Tarte K, et al. The class-specific BCR tonic signal modulates lymphomagenesis in a c-myc deregulation transgenic model. Oncotarget. (2014) 15:8995–9006. doi: 10.18632/oncotarget.2297
65. Vincent-Fabert C, Fiancette R, Truffinet V, Cogné N, Cogné M, Denizot Y. Genetic background modulates susceptibility to oncogen-driven proliferation and lymphoma occurrence in mice carrying a deregulated c-myc transgene. Leuk Res. (2009) 33:e203–6. doi: 10.1016/j.leukres.2009.05.018
66. Park SS, Kim JS, Tessarollo L, Owens JD, Peng L, Su Han S, et al. Insertion of c-Myc into Igh induces B-cell and plasma-cell neoplasms in mice. Cancer Res. (2005) 65:1306–15. doi: 10.1158/0008-5472.CAN-04-0268
67. Han SS, Yun H, Son DJ, Tompkins VS, Peng L, Chung T, et al. NF-kappaB/STAT3/PI3K signaling crosstalk in iMyc E mu B lymphoma. Mol Cancer. (2010) 9:97. doi: 10.1186/1476-4598-9-97
68. Park SS, Shaffer AL, Kim JS, Dubois W, Potter M, Staudt M, et al. Insertion of Myc into Igh accelerates peritoneal plasmacytomas in mice. Cancer Res. (2005) 65:7644–52. doi: 10.1158/0008-5472.CAN-05-1222
69. Cheung WC, Kim JS, Linden M, Peng L, Van Ness B. Novel targeted deregulation of c-myc cooperates with Bcl-XL to cause plasma cell neoplasms in mice. J Clin Invest. (2004) 113:1763–73. doi: 10.1172/JCI200420369
70. Rosean TR, Holman CJ, Tompkins VS, Jing X, Krasowski MD, Rose-John S, et al. KSHV-encoded vIL6 collaborates with deregulated c-MYC to drive plasmablastic neoplasm in mice. Blood Cancer J. (2016) 6:e398. doi: 10.1038/bcj.2016.6
71. Ghazzaui N, Issaoui H, Ferrad M, Carrion C, Cook-Moreau J, Denizot Y, et al. Eμ and 3'RR transcriptional enhancers of the IgH locus cooperate to promote c-myc-induced mature B-cell lymphomas. Blood Adv. (2020) 4:28–39. doi: 10.1182/bloodadvances.2019000845
72. Volpi SA, Verma-Gaur J, Hassan R, Ju Z, Roa S, Chatterjee S, et al. Germline deletion of Igh 3′ regulatory region elements hs 5, 6, 7 (hs5–7) affects B cell-specific regulation, rearrangement, and insulation of the Igh locus. J Immunol. (2012) 188: 2556–66. doi: 10.4049/jimmunol.1102763
73. Birshtein BK. Epigenetic regulation of individual modules of the immunoglobulin heavy chain locus 3' regulatory region. Front Immunol. (2014) 5:163. doi: 10.3389/fimmu.2014.00163
74. D'addabbo P, Scascitelli M, Giambra V, Rocchi M, Frezza D. Position and sequence conservation in amniota of polymorphic enhancer HS1,2 within the palindrome of IgH 3' regulatory region. BMC Evol Biol. (2011) 11:71. doi: 10.1186/1471-2148-11-71
75. Saintamand A, Garot A, Saad F, Moulinas R, Denizot Y. Pre-germinal center origin for mature mouse B cell lymphomas: a major discrepancy with human mature lymphomas. Cell Cycle. (2015) 14:3656–8. doi: 10.1080/15384101.2015.1093708
76. Duan H, Xiang H, Ma L, Boxer LM. Functionals long-range interactions of the IgH 3' enhancers with the bcl-2 promoter region in t(14;18) lymphoma cells. Oncogene. (2008) 27:6720–8. doi: 10.1038/onc.2008.286
Keywords: MYC, B-cell lymphoma, transgenic mouse models, IgH locus, IgH transcriptional enhancers
Citation: Ferrad M, Ghazzaui N, Issaoui H, Cook-Moreau J and Denizot Y (2020) Mouse Models of c-myc Deregulation Driven by IgH Locus Enhancers as Models of B-Cell Lymphomagenesis. Front. Immunol. 11:1564. doi: 10.3389/fimmu.2020.01564
Received: 29 April 2020; Accepted: 15 June 2020;
Published: 23 July 2020.
Edited by:
Gema Perez-Chacon, Spanish National Cancer Research Center (CNIO), SpainReviewed by:
Ignacio Moreno De Alborán, Consejo Superior de Investigaciones Científicas (CSIC), SpainCopyright © 2020 Ferrad, Ghazzaui, Issaoui, Cook-Moreau and Denizot. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yves Denizot, eXZlcy5kZW5pem90QHVuaWxpbS5mcg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.