- 1Guangdong Provincial Key Laboratory of Pathogenic Biology and Epidemiology for Aquatic Economic Animal, Key Laboratory of Control for Disease of Aquatic Animals of Guangdong Higher Education Institutes, Southern Marine Science and Engineering Guangdong Laboratory, College of Fishery, Guangdong Ocean University, Zhanjiang, China
- 2Laboratory for Marine Biology and Biotechnology, Qingdao National Laboratory for Marine Science and Technology, Qingdao, China
- 3Guangdong Provincial Engineering Research Center for Aquatic Animal Health Assessment, Shenzhen, China
- 4Guangxi Key Lab for Marine Natural Products and Combinational Biosynthesis Chemistry, Guangxi Beibu Gulf Marine Research Center, Guangxi Academy of Sciences, Nanning, China
Galectin-8 is a member of the galectin family that is involved in immune response against pathogens. However, the roles of fish galectin-8 during pathogen infection require comprehensive studies. In this study, a galectin-8 homolog (OnGal8-like, OnGal8-L) was characterized from Nile tilapia (Oreochromis niloticus), and its roles in response to bacterial infection were analyzed. The OnGal8-L contains an open reading frame of 891 bp, encoding a peptide of 296 amino acids with two CRD regions of tandem-repeat galectin and two carbohydrate recognition sites. The OnGal8-L protein shares 46.42% identities with reported Oreochromis niloticus galectin-8 protein. Transcriptional expression analysis revealed that OnGal8-L was constitutively expressed in all examined tissues and was highly expressed in spleen. The transcript levels of OnGal8-L were up-regulated in the spleen, head kidney, and brain, following Streptococcus agalactiae (S. agalactiae) challenge. Further in vitro analysis indicated that the recombinant protein of OnGal8-L (rOnGal8L) could agglutinate erythrocyte, S. agalactiae, and A. hydrophila and bind S. agalactiae, A. hydrophila, and various PAMPs (lipopolysaccharides, lipoteichoic acid, poly I:C, peptidoglycan, galactose, mannose, and maltose). Also, rOnGal8L could regulate inflammatory-related gene expression, phagocytosis, and a respiratory burst of monocytes/macrophages. Moreover, in vivo analysis showed that OnGal8-L overexpression could protect O. niloticus from S. agalactiae infection through modulating serum antibacterial activity (AKP, ACP, and LZM), antioxidant capacity (CAT, POD, and SOD), and monocyte/macrophage proliferation and cytokine expression, as well as reducing bacterial burden and decreasing tissue damage. Our results collectively indicate that OnGal8-L plays important regulatory roles in immune response against bacterial infection.
Introduction
Galectins are a family of proteins that possess high affinity for β-galactosides, which play a variety of roles in cell–cell adhesion, cell–matrix interaction, and transmembrane signaling (1, 2). Galectins can be found in cytoplasm, nucleus, and extracellular space (3). Presently, galectins serve as pattern recognition receptors (PRR) that regulate the innate immune processes triggered by pathogen-associated molecular patterns (4, 5). On the other hand, the intracellular galectins are involved in cellular functional processes including pre-mRNA splicing, cell growth regulation, and cell cycle processes (6, 7). Until now, 15 galectins have been discovered in mammals, which are subdivided into three subfamilies based on their conserved carbohydrate recognition domain (CRD): “prototype,” “chimera,” and “tandem repeat” (8).
Galectin-8 belongs to the tandem-repeat subfamily, consists of two CRDs linked by a peptide. Human galectin-8 has six isoforms encoded by 14 alternatively spliced transcripts (9). As a secreted protein, the immobilized galectin-8 can promote cell adhesion via attaching and aggregating integrin receptors on the cell surface, which subsequently trigger integrin-mediated signaling cascades (10). Furthermore, the involvement of galectin-8 in host defenses against bacterial infection has been well-described in mammals. The mammalian galectin-8 can detect the bacterial invasion by monitoring the integrity of endosomes and lysosomes, and then activate antibacterial autophagy to protect cells from bacterial infection (11).
To date, galectin-8 homologs have been identified in many species, but the functions of teleost galectin-8 are not well-understood yet (12). Nile tilapia (Oreochromis niloticus) is one of the most economically important fish all around the world (13). In recent years, the outbreak of bacterial disease has resulted in massive losses of tilapia culture (14). Although the involvement of galectin-8 in bacterial infection has been recorded in Nile tilapia (Oreochromis niloticus) (15), the exact roles of this molecular during bacterial infection still need to be clarified. In this study, a new tandem-repeat galectin-8 isoform (OnGal8-L) was identified and characterized from O. niloticus. The regulatory roles of OnGal8-L in immune response against bacterial infection were also investigated in vitro and in vivo. Our data will provide new insights into the function of fish galectin-8 against bacterial infection.
Materials and Methods
Fish Preparation and Bacterial Challenge
Nile tilapia (50 ± 10 g) were acquired from a local fish farm in Zhanjiang, Guangdong, China. The fish were cultured in a 1,000-L tank with aerated freshwater under 28 ± 2°C for 3 weeks (16). All experiments were conducted according to the principles and procedures of Guangdong Province laboratory animal management regulations (17).
To explore the tissue distribution of OnGal8-L in healthy tilapia, the tissues including brain, head kidney, gill, spleen, thymus, heart, liver, muscle, skin, and intestine were collected and frozen quickly by liquid nitrogen and stored at −80°C until use.
S. agalactiae (ZQ1901) used in the experiment was isolated from Nile tilapia and kept in our laboratory (18). The S. agalactiae was dissolved in phosphate-buffered saline (PBS) with a final concentration of 1 × 107 cells/mL. The stimulation groups were injected with 100 μL S. agalactiae. The samples including spleen, head kidney, and brain were collected at seven time points (0, 4, 12, 24, 48, 72, and 96 h), and then stored at −80°C until use.
Stimulation of S. agalactiae on Head Kidney-Derived Monocytes/Macrophages
Monocytes/macrophages were prepared as previous studies (18, 19). Briefly, healthy tilapia was anesthetized by MS222. The head kidney was carefully excised and transferred through a 40-μm stainless nylon mesh (Greiner Bio-OneGmbH, Germany). The cell suspension was suspended in Dulbecco's Modified Eagle Medium (DMEM) (Gibco, US) supplemented with 100 U/mL penicillin, 100 μg/mL streptomycin, and 25 U/mL heparin (Gibco). The cell suspension was slowly added into a 51%/34% percoll (GE Healthcare) density gradient, centrifuged at 400 g for 40 min, and the cell layer of the interface was carefully aspirated. The harvested cells were washed with PBS and collected again through centrifugation at 500 g for 10 min. Cell viability was measured using a Trypan Blue Staining kit (Sangon Biotech). The cells were cultured at 25°C for 24 h and then removed the non-adherent cells.
Challenge experiments were carried out according to the method of Mu et al. (20). The monocytes/macrophages were stimulated by formalin-inactivated S. agalactiae (1 × 107 cells/mL). The cells were collected and lysed by Trizol Reagent (Sangon Biotech) at 0, 3, 6, 12, 24, and 48 h post-challenge, respectively.
Cloning and Sequence Analysis of OnGal8-L
In order to obtain the open reading frame of OnGal8-L, total RNA from spleen was extracted by using EasyPure RNA Kit (TransGen, China) according to the manual protocol. The first-stand cDNA was synthesized from total RNA by using EasyScript One-Step gDNA Removal and cDNA Synthesis SuperMix (TransGen, China). The specific primers (OnGal8-L-F, OnGal8-L-R) were designed based on Nile tilapia transcriptome data (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA244908) and the prediction of NCBI Basic Local Alignment Search Tool. All the primers used in this study were designed by Primer 6.0 and showed in Table 1.
The potential open reading frame of OnGal8-L was analyzed with ORF finder program (https://www.ncbi.nlm.nih.gov/orffinder/). Molecular weight, theoretical pI, amino acid composition of OnGal8-L were predicted by ProtParam tool (https://web.expasy.org/protparam/). Multiple sequence alignment of OnGal8-L protein sequences was performed by Clustal W program (http://www.clustal.org/clustal2/). The similarity analyses of the determined amino acid sequence were performed by UniProt programs (https://www.uniprot.org/). Phylogenic trees were constructed by the neighbor-joining method using MEGA X software with 1,000 bootstrap replications. The evolutionary distances were computed using the Poisson correction method (21) and are in the units of the number of amino acid substitutions per site.
Quantitative Real-Time PCR of OnGal8-L
The RNA from 10 different tissues (heart, brain, spleen, liver, thymus, head kidney, intestine, muscle, gill, and skin) of healthy tilapia were extracted and then reverse-transcribed into cDNA, as described in section Stimulation of S. agalactiae on Head Kidney-Derived Monocytes/Macrophages. The β-actin (housekeeping gene) was chosen as the internal reference gene. Quantitative RT-PCR (qRT-PCR) was carried out on Roche LC384 LightcyclerTM (Roche, Switzerland) with a PCR reaction volume of 10 μL containing 5 μL of FastStart Essential DNA Green Master (Roche, Switzerland), 0.5 μL of each primer (2 μM), and 3.5 μL of nuclease-free water. The program was performed as follows: 1 cycle of 10 min at 95°C, 40 cycles of 10 s at 95°C, 15 s at 55°C, and 15 s at 72°C. The relative expression level of OnGal8-L mRNA were calculated using 2−ΔΔCt method (22).
Preparation of OnGal8-L Recombinant Protein (rOnGal8L)
The specific primers (EOnGal8-L-F and EOnGal8-L-R) with restriction sites (BamH I and Xho I) were designed (Table 1) to amplify OnGal8-L ORF. The PCR product was purified and ligated into the pMD18-T vector. The recombinant plasmids pMD-18T-OnGal8-L and pGEX-4T-1 were digested with BamH I and Xho I. The digested products were ligated and transformed into E. coli BL21 (DE3) (TransGen, China). The positive clone was verified by PCR and DNA sequencing. Then, the positive clone was picked to culture in fresh LB liquid medium containing ampicillin (100 μg/mL). When OD600 reached 0.4–0.6, isopropyl-β-Dthiogalactopyranpside (IPTG) was added into cultured bacteria with a final concentration of 1 mmol/L. The IPTG added cultures were continuously induced at 37°C for 5 h. The bacteria were collected and washed three times with PBS. Lysozyme was added into bacterial solution with a final concentration of 1 mg/mL, and placed on ice for 30 min, then centrifuged at 4°C for 10 min. The supernatant was purified using a GST-tag protein purification kit (Beyotime, China), desalted and concentrated using an Amicon Ultra Centrifugal Filter (Amicon, USA). The purified protein was analyzed by 10% reducing SDS-PAGE and Western blot. The empty vector (pGEX-4T-1)-expressed GST-tagged protein was prepared as described above and used as control in further analysis.
Hemagglutination Assay
The hemagglutination assay was performed according to the method of Madusanka et al. (12). Briefly, the blood of Nile tilapia was collected in a centrifuge tube containing 1% sodium heparin. After centrifuging at 2,000 g for 15 min, the supernatant was removed and the collected erythrocytes were washed four times with sterile TBS. The harvested erythrocytes were adjusted to 1 × 105 cells/mL with sterile TBS. Then, a 500-μL erythrocyte suspension was incubated with 10 μL of rOnGal8L (1 mg/mL) at room temperature for 10 min. Both PBS and the GST-tag protein were used as control. The incubated erythrocytes were transferred onto slide, and the agglutination was observed by fluorescence microscope.
Bacterial Agglutination Assay
Briefly, S. agalactiae and A. hydrophila were cultured until the OD600 reached 0.4–0.6; the bacteria were collected and washed three times with PBS. The collected bacteria were resuspended in 0.1 M Na2CO3. Then FITC (Solaribo, China) was added into resuspended bacterial solution with a final concentration of 0.1 mmol/L. The FITC-added solutions were incubated at 37°C for another 30 min. After incubation, the bacteria were centrifuged three times to completely remove the FITC and incubated with 10 μL of rOnGal8L (1 mg/mL) at room temperature for 1 h. The incubated bacteria were transferred onto slide and observed with a fluorescence microscope.
Binding Assay of rOnGal8L to Bacterial Pathogens and Carbohydrates
The binding abilities of rOnGal8L to Gram-positive bacteria (S. agalactiae) and Gram-negative bacteria (A. hydrophila) were detected by the method of Bai et al. (23). Briefly, the bacteria were cultured until the OD600 reached 0.4–0.6. Ten microliters of rOnGal8L (1 mg/mL) was added into 500 μL of bacterial solution and incubated at 37°C for 1 h. The incubated bacterial were collected and washed three times with PBS, then lysed the bacteria with 7% SDS for 1 min and centrifuged at 12,000 rpm for 10 min. The supernatant was collected for Western blot analysis. The anti-GST mouse monoclonal antibody (1:1,000; 1 mg/mL) (Sangon Biotech, Shanghai) was used as the primary antibody, and the HRP-goat α-mouse antibody (1: 5,000; 1 mg/mL) (Sangon Biotech, Shanghai) was used as the secondary antibody. The bacteria incubated with PBS or pGEX-4T-1 were used as control. Previous study shows that galectins bind to bacteria through the carbohydrates on bacterial cell wall, which can be competitively inhibited by another carbohydrates (24). So several saccharides including lipopolysaccharide (LPS), lipoteichoic acid (25), galactose, mannose, and maltose were chosen to explore the inhibitory capacities on the binding activity of rOnGal8L to bacterial through competitive binding assay. The rOnGal8L or GST-tag protein (1 mg/mL, 10 μL) was incubated with each saccharide (1 mg/mL, 50 μL) and 500 μL of S. agalactiae and A. hydrophila at room temperature for 1 h and incubated mixtures were examined by western blot as described above.
To further understand the binding activity of rOnGal8L to carbohydrates, enzyme-linked immunosorbent assay (ELISA) was performed according to the method of Zhang et al. (26). Briefly, LTA, LPS, Poly I:C, PGN, galactose, mannose, and maltose were chosen and adjust to a concentration of 100 μg/mL with 20× Coating Buffer (Sangon Biotech, Shanghai). The 10-fold serial dilutions of carbohydrates were made. Then the diluted carbohydrates solutions (100 μL/well) were added into 96-well-plates for coating at 4°C overnight and blocked at 37°C for 2 h in the next day. The empty wells were used as blank control. Plates were washed with TBST three times between each step, and rOnGal8L (100 μg/mL, 100 μL) was added to the wells and incubated at 37°C for 2 h after final wash. Following incubation, the anti-GST mouse monoclonal antibody (1: 1,000, 1 mg/mL) (Sangon Biotech, Shanghai) was used as the primary antibody, and the HRP-goat α-mouse antibody (1: 5,000, 1 mg/mL) (Sangon Biotech, Shanghai) was used as the secondary antibody. One-hundred milliliters of TMB mixed reaction solution was added to each well and incubated at 37°C for 15 min in the dark, then used H2SO4 (2 M, 50 μL) to terminate the reaction. The optical density (O.D.) at 405 nm was measured using a Microplate Reader (Thermo, USA).
Bacterial Growth Inhibition Assay
The bacterial growth inhibition assay of rOnGal8L was performed as described in a previous study (27). In brief, S. agalactiae and A. hydrophila were cultured overnight, the bacterial pellet was collected by centrifugation, and then adjusted to an initial concentration of 1 × 105 cells/mL with culture medium. One-hundred microliters of diluted bacterial solution was added into a 96-well-microtiter plate, the rOnGal8L, GST-tag protein, or PBS was added with a final concentration of 50 μg/mL at the same time. The plates were incubated at 37°C and the O.D. value of 600 nm were measured every 2 h. The antibacterial activities were evaluated by the bacterial growth density (O.D. value).
Analysis of Inflammatory-Related Gene Expression in Monocytes/Macrophages After rOnGal8L Stimulation
The experiment was performed according to the method of Crinier et al. (28). Briefly, the monocytes/macrophages were stimulated with 50 μL of rOnGal8L (1 mg/mL), GST-tag protein (1 mg/mL), and PBS, respectively. The cells were collected, and the cDNA were synthesized, as described in section Cloning and Sequence Analysis of OnGal8-L. The expression levels of IL-6, IL-10, IL-8, and MIF (Table 1) in monocytes/macrophages were measured by qRT-PCR. Similar with the method described in section Quantitative Real-Time PCR of OnGal8-L, the qRT-PCR was performed on Roche LC384 LightcyclerTM with a reaction volume of 10 μL. The β-actin was applied as the internal reference gene. The relative expression levels of inflammatory-related genes were calculated using 2−ΔΔCt method.
Phagocytosis Assay
Phagocytosis assay was performed as the method of Bai et al. (29). Briefly, 200 μL of FITC-labeled S. agalactiae and A. hydrophila suspension was mixed with 180 μL of monocytes/macrophages and 20 μL of rOnGal8L (1 mg/mL) and incubated in the dark for 1 h with shaking every 5 min. In the control group, 20 μL of PBS or GST-tag protein (1 mg/mL) was used instead of rOnGal8L. Then centrifuge at 500 g for 10 min to completely remove the non-ingested bacteria. The results were analyzed using flow cytometer. The fluorescence data for this experiment is limited to a gate to ensure the accuracy of the analysis. Each sample measurement was repeated in triplicate.
Respiratory Burst Assay
The effect of rOnGal8L on the respiratory burst activity of monocytes/macrophages was performed by the methods described in the References (30–32). In brief, PMA was used to stimulate monocytes/macrophages to produce reactive oxides that induced the oxidation of dihydrorhodamine to rhodamine 123, which was capable of emitting fluorescence. According to the protocol of PMN Oxidative Burst Quantitative Assay Kit (Absin Bioscience, Shanghai), 300 μL of monocytes/macrophages cells (2 × 107 cells/mL) were mixed with 10 μL of rOnGal8L (1 mg/mL) and incubated at 25°C for 1 h. Then 50 μl of PMA was added and the mixture was incubated at 37°C for 15 min. Subsequently, 25 μL of dihydrorhodamine was added to the mixture and incubated at 37°C for 5 min in the dark. One milliliter of diluted hemolysin was added at room temperature and hemolyzed for 15 min. The mixture was washed twice with PBS, centrifuged at 1,500 rpm for 5 min and the supernatant was discarded. Then the cells were resuspended in 0.5 mL PBS for flow cytometry analysis. The experimental procedures of control groups (GST-tag protein or PBS) were the same as described above.
Plasmid Construction and Overexpression in vivo
Per the method described in section Preparation of OnGal8-L Recombinant Protein (rOnGal8L), pEGFP-C1 eukaryotic expression vector was constructed using specific primers (Table 1). The recombinant plasmid pEGFP-Gal8-L and pEGFP-C1 were extracted using E.Z.N.A. Endo-free Plasmid Mini Kit (Promega, USA) in compliance with manufacturer's instructions and then used for further analysis.
According to the in vivo overexpression protocol described in Huang et al. (33), the plasmids pEGFP-Gal8-L and pEGFP-C1 were adjusted to a final concentration of 200 μg/mL and 50 μl of each diluted plasmid was injected intramuscularly into fish. PBS group was set as a control. After 7 days post-injection, the spleen of each group was collected and fixed using standard histological techniques. The fluorescence signal in collected spleens were detected by fluorescence microscope (LEICA, CTR6000, Germany).
Analysis of the Regulatory Roles of OnGal8-L Overexpression on Immune Response in vivo
Fish blood samples from each group were collected from caudal vein with a sterile syringe at day 7 after plasmid (pEGFP-Gal8-L and pEGFP-C1) and PBS administration. The serum was isolated for the detection of non-specific immune parameters, including alkaline phosphatase activity (AKP), acid phosphatase (ACP), lysozyme (LZM), catalase (CAT), peroxidase (POD), and superoxide dismutase (SOD) using the corresponding protease detection kit (Nanjing Jiancheng, Bioengineering Institute, China). At day 7 after plasmids and PBS administration, the fish were infected intramuscularly with 100 μl of S. agalactiae resuspended in PBS (1 × 107 cells/mL). After bacterial stimulation, three tissues samples including head kidney, spleen, and liver were obtained from each group fish at 3, 6, 12, 24, 48, 72, and 96 hour post injection (hpi), respectively. The qRT-PCR was conducted per section Cloning and Sequence Analysis of OnGal8-L to test the express level of immune-related genes, including IL-10, IL-1β, and TNF-α (Table 1).
Determination of the Effects of OnGal8-L Overexpression on Bacterial Resistance in vivo
As the challenge experiment design in section Analysis of the Regulatory Roles of OnGal8-L Overexpression on Immune Response in vivo, the bacterial burden in tilapia tissues of different treatment groups were evaluated after bacterial infection. The bacterial counts were carried out as the method described in the References (34). Briefly, the spleen, head kidney, and liver were collected from the experimental fish at 24 hpi. After weighing, the collected tissues were homogenized in 1 mL of sterile PBS. The homogenized tissues were serially diluted and spread on brain–heart infusion agar plates and cultured at 37°C for 24 h. Single colonies were identified by PCR using S. agalactiae-specific primers. Finally, colony-forming units (CFU) in all plates were counted and multiplied by the dilution factor. For histological observation, various tissues including the head kidney, spleen, and liver were collected at 24 h after bacterial injection. The histological changes of these samples were observed under microscope.
The survival assay was performed according to the method described in our previous study (33). Briefly, 160 fish were divided into four groups and injected with plasmids (pEGFP-Gal8-L or pEGFP-C1) and PBS. At day 7 post injection (p.i.), fish in each group were challenged with activated S. agalactiae at 1 × 108 colony-forming units (CFU)/fish by intraperitoneal injection. Daily statistical morbidity lasted for 10 days, and the survival rate (SR) was calculated as SR = (surviving fish÷40) × 100%.
Statistical Analysis
All data in this study were displayed as means ±standard deviation (SDs). Statistical analysis was performed by the least significant difference (LSD) test using SPSS 17.0 software. Difference between two groups was considered as significant at *p < 0.05 or highly significant at **p < 0.01.
Results
Sequence Analysis of OnGal8-L
The ORF of OnGal8-L was 891 bp in length and encoded 296 amino acids. The predicted molecular mass of OnGal8-L was 33.14 KDa and the theoretical pI was 5.03. It had two CRD domains (N- and C-CRD domains) and a carbohydrate recognition site. Multiple sequence alignment of OnGal8-L and other reported galectin-8 found that OnGal8-L possessed conservative sequences of the galectin family such as V-N, H-N-RL, and W-E-R in two CRDs, which shared higher identities with known galectin-8. While the linker peptide between two CRDs in OnGal8-L was much shorter than that in galectin-8 (Figure 1A). Phylogenetic analysis showed that OnGal8-L located within fish galectin-8 subgroup and was clustered close to Oreochromis niloticus galectin-8 and Maylandia zebra galectin-8 (Figure 1B).
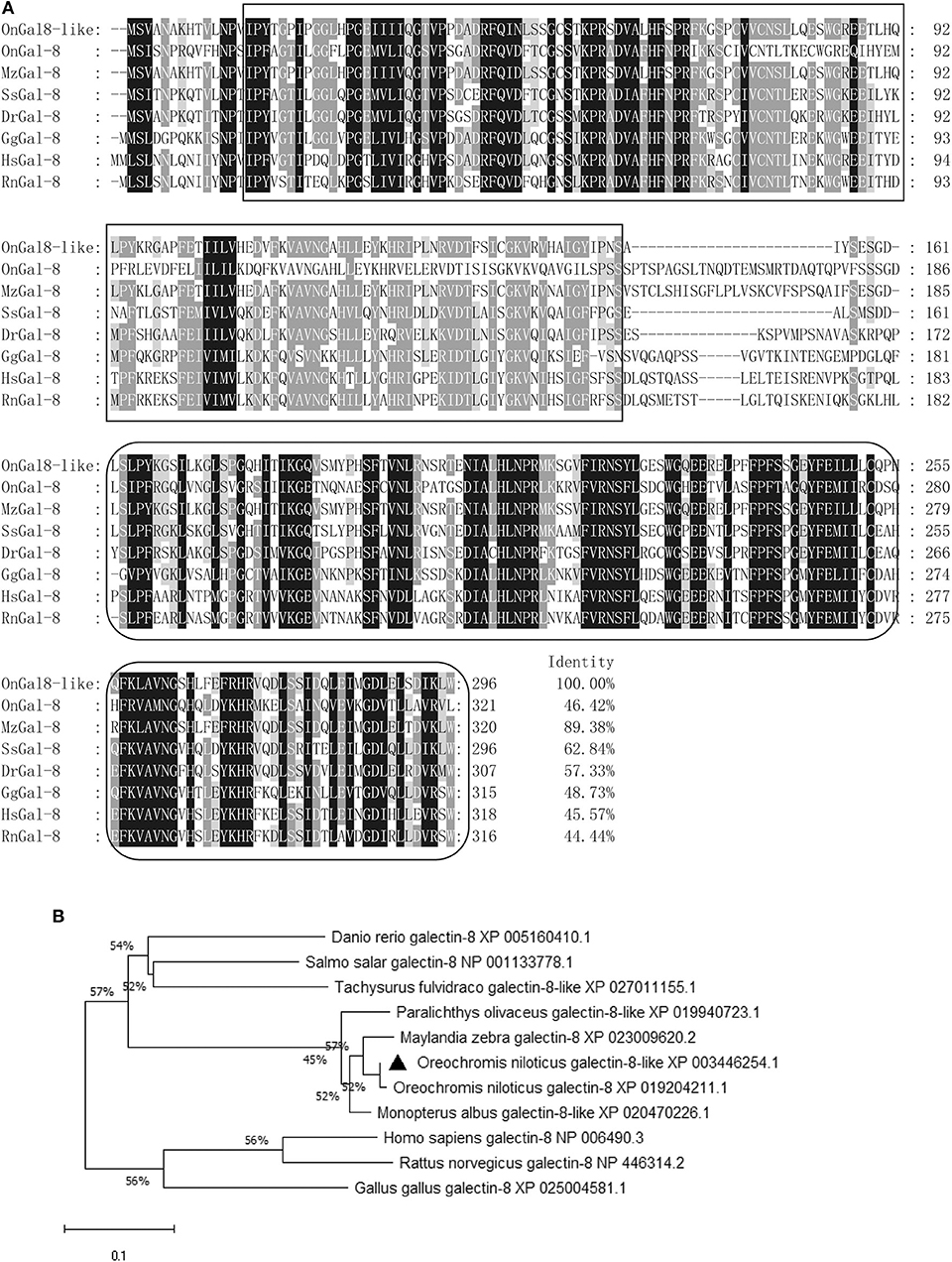
Figure 1. (A) Multiple sequence alignment of galectin-8(like) from various species. Conserved amino acid residues are shaded dark gray, and similar amino acids are shaded light gray. The CRDs are in two black boxes. Conserved residues (H-N-R, V-N, and W-E-R) for carbohydrate recognition and binding are shown above the alignment sequence. The GenBank accession numbers of genes involved are as below: M. zebra galectin-8, XP_023009620.2; S. salar galectin-8, NP_001133778.1; D. rerio galectin-8, XP_005160410.1; G. gallus galectin-8, NP_001010843.1; O. niloticus galectin-8, XP_019204211.1; H. sapiens galectin-8, NP_006490.3; and R. norvegicus galectin-8, NP_446314.2; (B) Phylogenetic tree of OnGal8-like family members constructed using the NJ method by MEGA X program based on the alignment of 10 members of the Gal-8 group. The numbers at each branch indicates the bootstrap values (%).
The Expression Profiles of OnGal8-L in vivo and in vitro
The tissue distribution of OnGal8-L was investigated by qRT-PCR analysis. The results showed that the relatively higher transcriptional levels of OnGal8-L were observed in the spleen, skin, brain, and intestine; followed by muscle, liver, gill, heart, and head kidney; and the lowest level in thymus (Figure 2A).
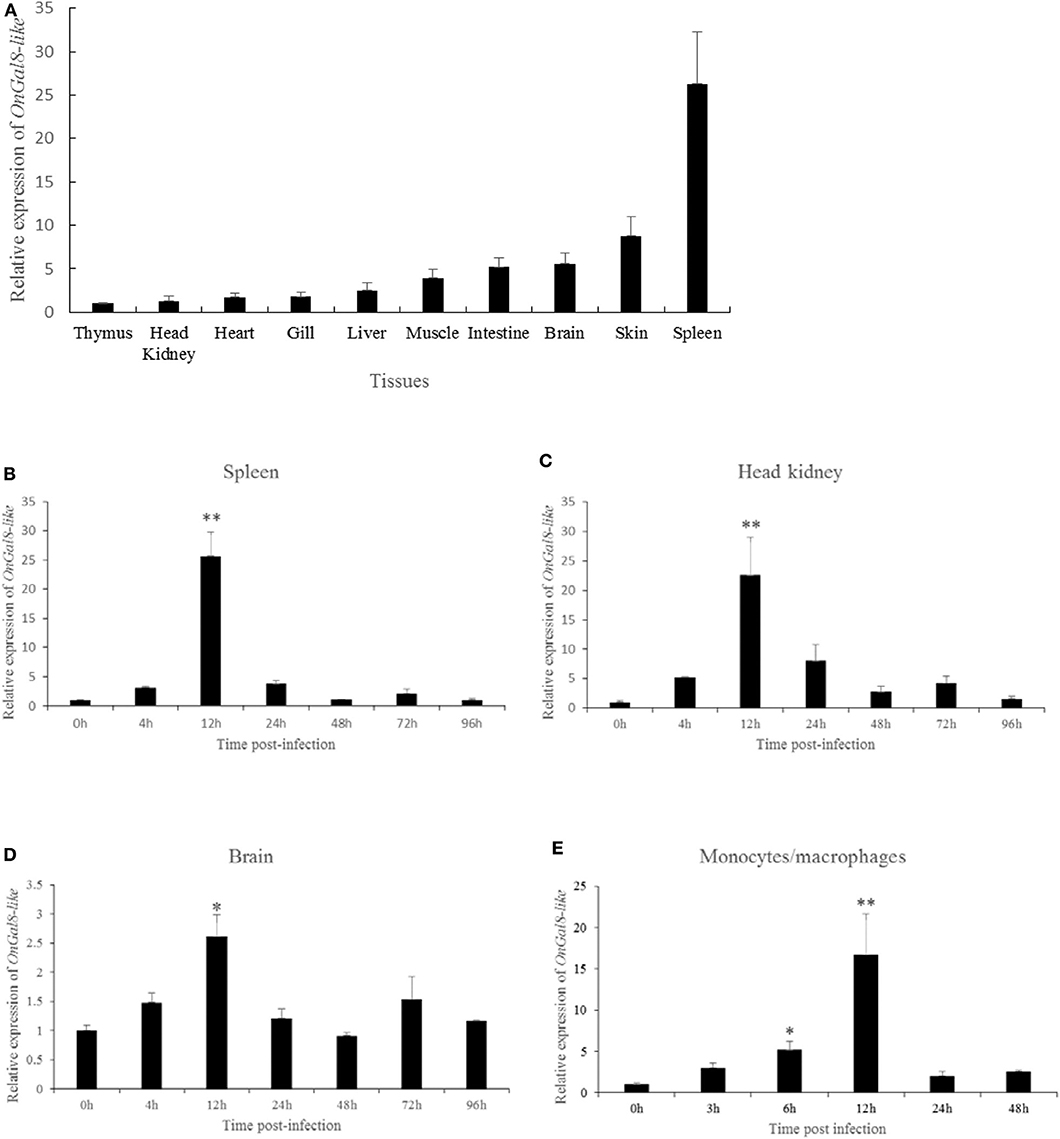
Figure 2. (A) Tissue distribution of OnGal8-L mRNA in healthy Nile tilapia. The ratio refers to the gene expression in different tissues relative to that in thymus, and target gene expression is normalized to β-actin. The results were mean ± SD of three replicate measurements. (B–E) Expression analysis of OnGal8-like in spleen (B), head kidney (C), brain (D), and monocytes/macrophages (E) after S. agalactiae challenge was performed by qRT-PCR. Data are shown as mean ± SD from three replicate measurements. Significant difference was indicated by asterisks as *0.01 < P < 0.05 and **P < 0.01.
After S. agalactiae infection, OnGal8-L transcripts increased in a time-dependent pattern and reached its peak at 12 h (Figures 2B–D) in head kidney, spleen, and brain. Additionally, the transcriptional levels in head kidney and spleen were much higher than that in brain. Similarly, the expression of OnGal8-L in monocytes/macrophages was also induced by bacterial stimulation and reached the peak at 12 h (16.35-fold) post-stimulation (Figure 2E).
Purification and Western Blotting Analysis of rOnGal8-L
The prokaryotic recombinant vector pGEX-4T-1-OnGal8-L was transformed into E. coli and induced to produce the OnGal8-L recombinant protein (rOnGal8-L). As predicted, an OnGal8-L recombinant protein of 59 KDa was obtained (Figure 3A, lane 2 and 3). Western blot showed that the protein could be specifically recognized by GST-tagged proteins (Figure 3A, lane 4), indicating that the recombinant protein was successfully expressed.
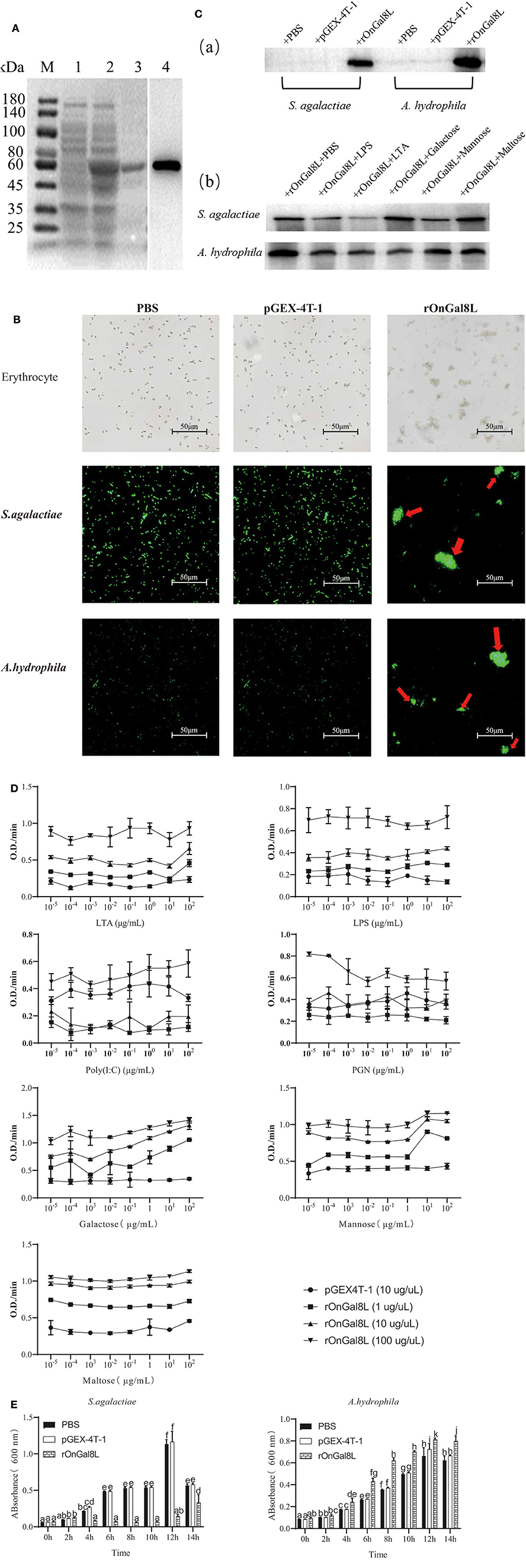
Figure 3. (A) SDS-PAGE and Western blot of rOnGal8L. Lane M: markers (25–180 kDa); lane 1: bacteria liquid before IPTG induction; lane 2: bacteria liquid after IPTG induction; lane 3: purified rOnGal8L; lane 4: Western blot analysis of rOnGal8L. (B) Agglutinating activity of rOnGal8L against fish erythrocytes, FITC-labeled S. agalactiae and A. hydrophila. PBS or pGEX-4T-1 was incubated with bacteria as a negative control. The agglutination was presented with red arrows; (C) a: Western blot analysis of rOnGal8L binding to S. agalactiae and A. hydrophila; b: Western blot analysis of the inhibitory effect of carbohydrate on the binding of rOnGal8L to bacteria. (D) ELISA analysis of rOnGal8L incubated with LTA, LPS, poly I:C PGN, galactose, mannose, and maltose in different concentration, respectively. The 10-fold serial dilutions of carbohydrates were made and the diluted carbohydrates solutions (100 μL/well) were added into 96-well-plate. The empty wells were applied as blank control. (E) The bacteriostatic activity of rOnGal8L. The growth curve of S. agalactiae and A. hydrophila. The bacteria were incubated with rOnGal8L, pGEX-4T-1, and PBS, respectively. OD600 was measured at different time points. Data are shown as mean ± SD. The error bars represent standard deviation (n = 3), and different letters (a–c) depict statistical significance between different groups (p < 0.05).
Agglutinating Activity of rOnGal8L to Erythrocytes and Bacterial Pathogens
To clarify the agglutination ability, rOnGal8L was used to incubate with erythrocytes, S. agalactiae and A. hydrophila. The strong agglutination activities of rOnGal8L on these three cells were seen, while no agglutinations were observed in the pGEX-4T-1 group and the PBS group (Figure 3B).
Binding Assay of rOnGal8L to Bacterial Pathogens and Carbohydrates
For the determination of the binding ability of rOnGal8L to bacteria, the S. agalactiae and A. hydrophila were incubated with rOnGal8L and then lysed with 7% SDS as described above (23). The supernatant after centrifugation was collected for Western blotting analysis. The results showed that rOnGal8L could strongly bind to both chosen bacteria (Figure 3Ca). Then, different polysaccharides were added to perform competitive binding experiments on bacteria. The results indicated that LTA reduce the binding of rOnGal8L to bacteria, while no obvious inhibitory effects were found on the other sugars (Figure 3Cb).
The results described above showed that different polysaccharides could affect the binding capacity of rOnGal8L, so we used ELISA to detect the binding capacities of rOnGal8L to carbohydrates. As shown in Figure 3D, only the highest concentration rOnGal8L (100 μg/μL) exhibited stable binding activity to carbohydrates of each concentration, which showed strongest binding activity to galactose, followed by maltose, mannose, LTA, LPS, PGN, and poly I:C. In addition, the binding activities of highest concentration of rOnGal8L to polysaccharides increased with the increasing carbohydrates concentration except that to PGN.
Inhibitory Effect of rOnGal8L on Bacterial Growth
After incubating with rOnGal8L, the growth of the S. agalactiae and A. hydrophila were monitored for 14 h. The results showed that rOnGal8L significantly suppressed the growth of S. agalactiae, but not A. hydrophila (Figure 3E).
Effects of rOnGal8L on the Activity of Monocytes/Macrophages
To determine the modulatory effects of rOnGal8L on the activity of monocytes/macrophages, several inflammatory-related genes, including IL-6, IL-10, IL-8, and MIF, were chosen for qRT-PCR analysis. After rOnGal8L incubation, the chosen genes in rOnGal8L-treated group were significantly up-regulated. Compared to PBS-treated group, the IL-6 and IL-10 of rOnGal8L-treated group reached their peak at 12 h and showed higher expression levels (IL-6, 36.2- vs. 5.5-fold, 12 h; IL-10, 19.8- vs. 6.8-fold, 12 h), while IL-8 and MIF reached their peak at 24 h. Moreover, rOnGal8L could significantly enhance the phagocytosis of macrophages to S. agalactiae and A. hydrophila (Figure 4B). The enhancement of rOnGal8L on respiratory burst of monocytes/macrophages was also seen (Figure 4C).
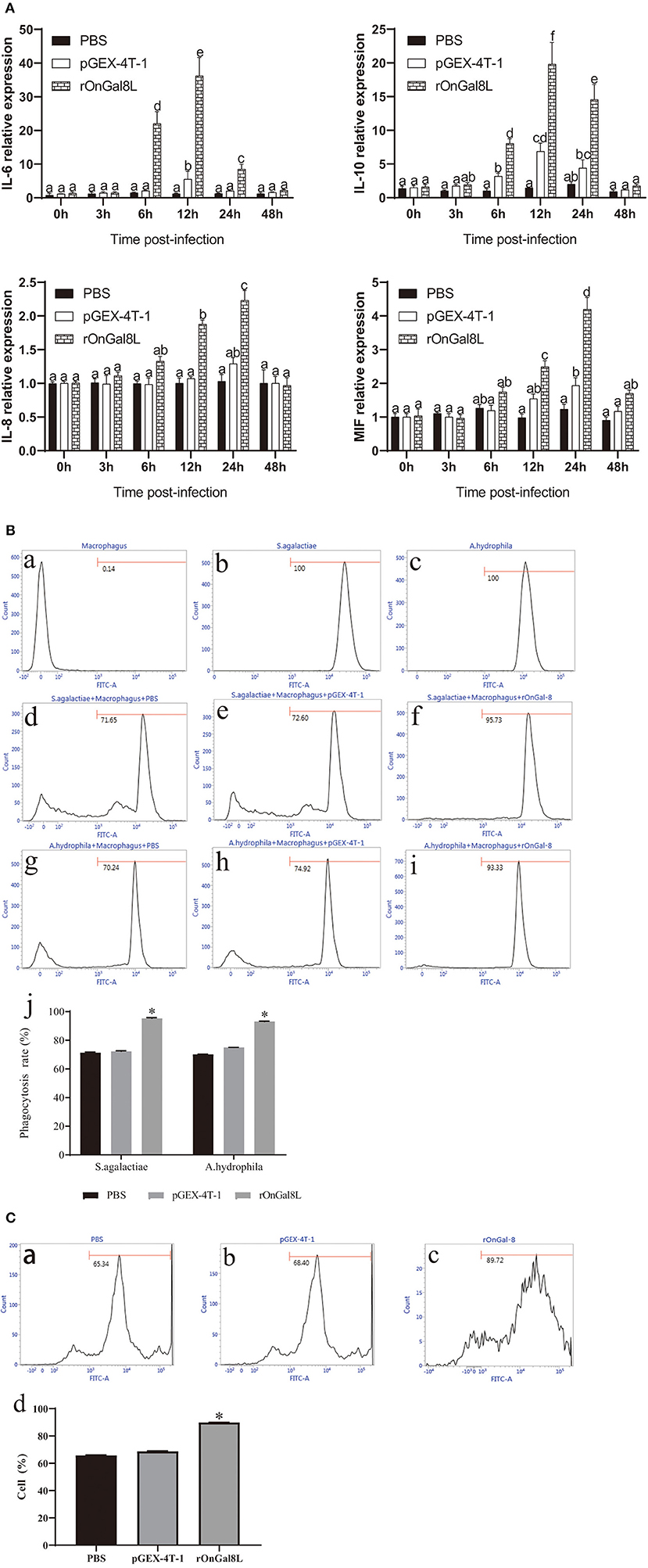
Figure 4. (A) The mRNA expressions of IL-6, IL-10, IL-8, and MIF in the head kidney-derived monocytes/macrophages. The monocytes/macrophages were treated with PBS, pGEX-4T-1 (50 μg/mL), and rOnGal8L (50 μg/mL). The mRNA levels of IL-6, IL-8, IL-10, and MIF genes were normalized to that of β-actin gene. (B) a–c: The effects of rOnGal8L on phagocytosis activity of head kidney-derived macrophages. d–i: The histogram of macrophages, FITC-labeled S. agalactiae, and A. hydrophila. The histogram of flow cytometric analyses of the macrophages phagocytosing S. agalactiae and A. hydrophila after pre-incubated with PBS, pGEX-4T-1, and rOnGal8L, respectively. j: The histogram of the phagocytosis rates. The average standard deviation was obtained from three replicate tests. Values were shown as mean ± SD (n = 3; *p < 0.05). (C) a–c: Flow cytometric analysis of the respiratory burst of head kidney-derived monocytes/macrophages. The histogram of the respiratory burst activities of monocytes/macrophages after pre-incubated with PBS (a), pGEX-4T-1 (b), or rOnGal8L (c) (10 μg/mL). (d): The histogram of the positive cell rates. The average standard deviation was obtained from three replicate tests. Values were shown as mean ± SD (n = 3; *p < 0.05).
OnGal8-L Overexpression Improved Serum Non-specific Immunological Parameters
To confirm the overexpression of OnGal8-L in vivo, the spleen sections were observed at 7 days after plasmid injection. The results showed that green fluorescence appeared in both pEGFP-Gal8-L group and pEGFP-C1 group, while no fluorescence was observed in the PBS group (Figure 5A). Furthermore, a series of non-specific immunological parameters were examined. The results showed that the activities of parameters including AKP, ACP, LZM, CAT, POD, and SOD in pEGFP-Gal8-L group were significantly higher than pEGFP-C1 and PBS group (Figure 5B).
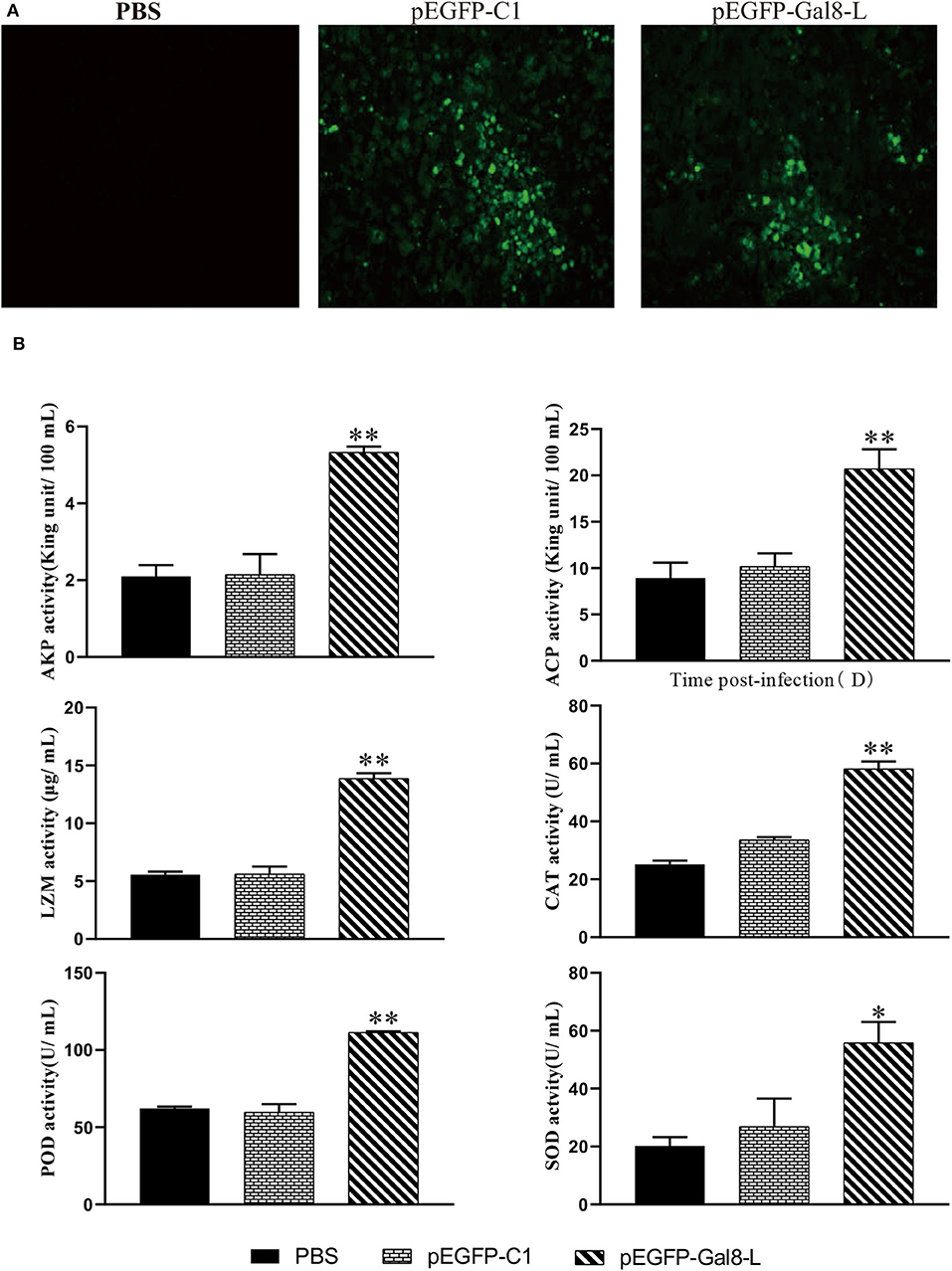
Figure 5. (A) Determination of fluorescence signals in pEGFP-Gal8-L- or pEGFP-C1-administered tilapia. Fish were injected with PBS, pEGFP-C1, and pEGFP-Gal8-L. Spleen tissue was obtained at day 7 after plasmid administration and examined with a fluorescence microscope. (B) Changes in antioxidant activity and non-specific immune response of administered tilapia. The average standard deviation was obtained from three replicate samples (*p < 0.05; **p < 0.01).
OnGal8-L Overexpression Reduced the Bacterial Burden in S. agalactiae-Infected Fish
The plate-counting method was applied to calculate the bacterial burden in the spleen, head kidney, and liver. The amounts of bacteria per milligram of tissues were represented by CFU. The PBS-injected group was set as the negative control, and no S. agalactiae was seen in the spleen and head kidney. After being challenged with S. agalactiae, the number of bacteria in head kidney of pEGFP-Gal8-L-injected group (2.16 × 104 CFU per mg) was much lower than that in the PBS-injected group (5.12 × 104 CFU/mg) and the pEGFP-C1-injected group (4.6 × 104 CFU/mg). Similarly, onGal8-L overexpression significantly reduced the bacterial burden in spleen (1.28 × 104 CFU/mg) compared with the PBS-injected group (3.68 × 104 CFU/mg) and the pEGFP-C1-injected group (3.96 × 104 CFU/mg). In liver, the amounts of bacteria in PBS-injected group was 1.6 × 105 CFU/mg, the pEGFP-C1-treated group was 9.6 × 104 CFU/mg, while the pEGFP-Gal8-L-injected group was significantly lower (3.24 × 104 CFU/mg; Figure 6A).
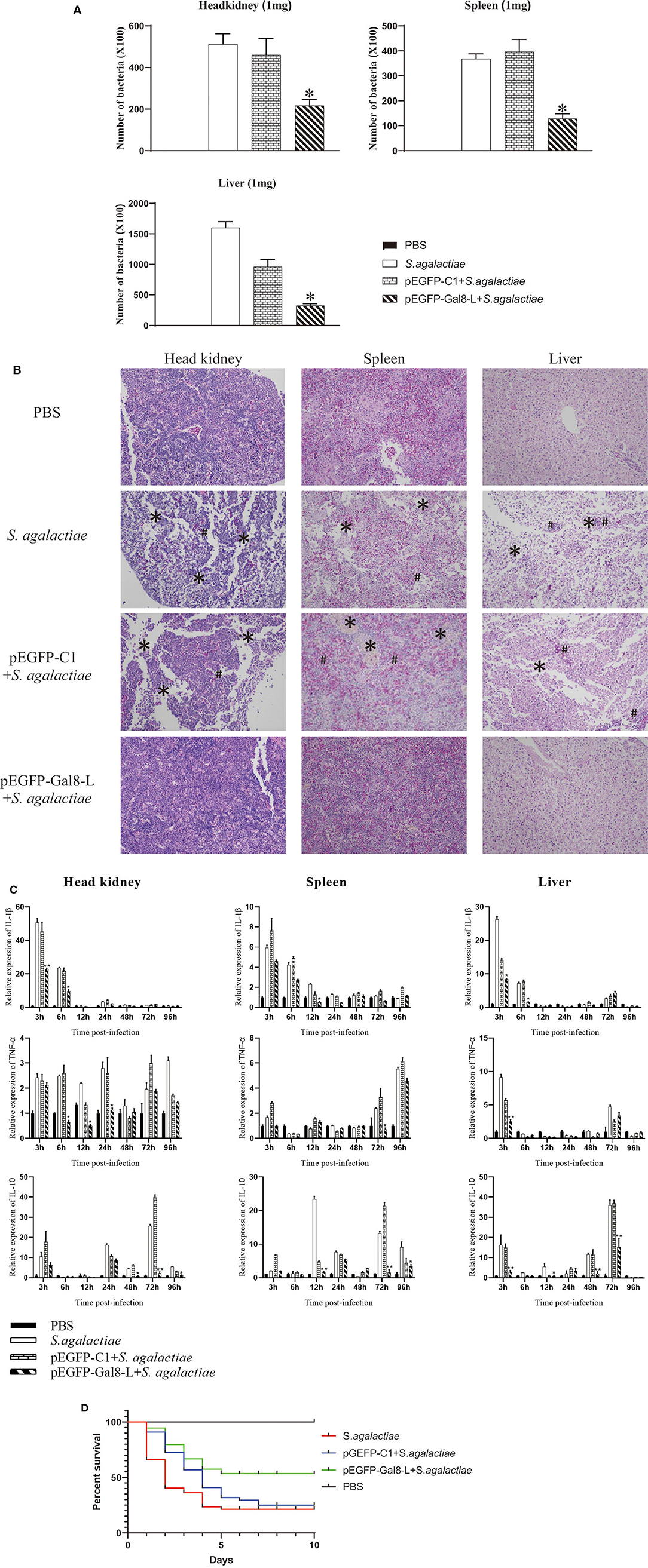
Figure 6. (A) The bacterial burden in administered tilapia after challenge with live S. agalactiae. Nile tilapia were administered with PBS, pEGFP-C1, or pEGFP-Gal8-L and then infected with S. agalactiae. Bacterial numbers in the spleen were determined at 24 hpi. Values are shown as means ± SDs (n = 3). **p < 0.01. (B) Histological observation of the head-kidney (left lane), spleen (middle lane), and liver (right lane) in administered fish after S. agalactiae infection. *markers, necrotic foci; #markers, severe hemorrhage. N = 5 each group. All sections were stained with HE and photographed under an optical microscope (×400 magnification). (C) qRT-PCR analysis of cytokine transcripts in head kidney, spleen, and liver of administered tilapia following S. agalactiae infection. Five fish/group were sampled at 3, 6, 12, 24, 48, 72, and 96 hpi, respectively. Cytokine expressions were normalized to β-actin. Data are shown as mean ± SD. Significant difference of pEGFP-Gal8-L-injected group to PBS-injected group was indicated by asterisks as *0.01 < p < 0.05 and **p < 0.01. (D) Survival rates of administered tilapia after challenge with live S. agalactiae. The fish were injected with PBS, pEGFP-C1, or pEGFP-Gal8-L. Fish were challenged with S. agalactiae (108 CFU/fish), daily statistical morbidity lasted for 10 days, and the survival rate (SR) was calculated as: SR = (surviving fish÷40) × 100%; n= 40 each group.
OnGal8-L Overexpression Impaired Tissue Damage Caused by S. agalactiae Infection
After S. agalactiae infection, series of marked pathological changes were seen in S. agalactiae-injected group and pEGFP-C1-injected group. For examples, in head kidney, there were a large amount of tissue bleeds and shed necrosis to form necrotic foci; in spleen, diffuse hyperemia and cell necrosis was found, and the integrity of the plasma membrane was disrupted; in liver, the cells were sparsely arranged, and many necrotic foci were present in the liver parenchyma, and inflammatory hyperemia appeared locally. While the similar phenomenon was not exhibited in PBS-injected group and pEGFP-Gal8-L-injected group (Figure 6B).
Regulatory Roles of OnGal8-L Overexpression on Inflammatory-Related Gene Expression During S. agalactiae Infection in vivo
qRT-PCR was carried out to detect the expression of inflammatory-related genes in head kidney, spleen, and liver from fish in different treated groups following S. agalactiae infection (Figure 6C). Compared to the PBS-injected group (without S. agalactiae infection), the bacterial infection markedly induced the transcriptional levels of IL-1β (head kidney: 50.7-fold at 3 hpi; spleen: 5.9-fold at 3 hpi; liver: 26.4-fold at 3 hpi), TNFα (head kidney: 2.4-fold at 3 hpi; spleen: 5.5-fold at 96 hpi; liver: 9.1-fold at 3 hpi), and IL-10 (head kidney: 10.5-fold at 3 hpi; spleen: 23.4-fold at 12 hpi; liver: 16.3-fold at 3 hpi) in S. agalactiae-treated group. Similar results were observed in pEGFP-C1+S. agalactiae-treated group and pEGFP-Gal8-L+S. agalactiae-treated group. Moreover, relatively weak increases of the examined cytokines were observed in the pEGFP-Gal8-L+S. agalactiae-treated group in comparison with pEGFP-C1+S. agalactiae-treated group. For instance, IL-1β was reduced at 3 and 6 hpi in head kidney, 12 hpi in spleen, and 3 and 6 hpi in liver; TNFα was down-regulated at 6, 12, and 24 hpi in head kidney, 72 hpi in spleen, and 3 hpi in liver; and IL-10 was decreased at 48, 72, and 96 hpi in head kidney, 12, 72, and 96 hpi in spleen, and 3, 12, 48, and 72 hpi in liver.
OnGal8-L Overexpression Improved the Survival Rates of Fish After S. agalactiae Infection
At day 7 after plasmid injection, the injected fish were challenged with S. agalactiae and the survival rates were evaluated. The results showed that the survival rate of the S. agalactiae-injected group was 20%, and the survival rate of the pEGFP-C1-injected group was 22.5%, while the survival rate of the pEGFP-Gal8-L-injected group was 52.5% (Figure 6D).
Discussion
As a family of evolutionarily conserved carbohydrate-binding proteins, galectins can interact with glycoconjugates of cell surface and extracellular matrix, thereby participating extensively in innate and adaptive immune processes (35, 36). In this study, a new tandem-repeat galectin (OnGal8-L) from Nile tilapia (Oreochromis niloticus) was identified, and its roles in antibacterial immune response also were investigated. OnGal8-L encoded a 296-amino acid peptide that shared 46.42 and 45.57% identities with reported Oreochromis niloticus galectin-8 and Homo sapiens galectin-8, respectively. Further comparison of OnGal8-L with reported galectin-8 revealed that OnGal8-L protein has two characteristic CRDs with different linker length. Given that a difference in linker length was also found in two human galectin-8 isoforms produced by the alternatively spliced of LGALS8 (37), we assume that OnGal8-L and reported Oreochromis niloticus galectin-8 may be two alternatively spliced isoforms of galectin-8. This assumption needs to be clarified in future studies. Moreover, given CRD is vital to mediate cell–cell or cell–intercellular matrix interactions in developmental processes and immunity, the co-existence of two different CRDs increases the diversity of ligand recognition by tandem repeat galectin (38). The high sequence similarities in both the N- and C-CRD regions of OnGal8-L indicate that it may exerts similar functions as reported galectin-8.
The current data exhibits that the OnGal8-L transcripts are present in all examined tissues with highest level in spleen. Fish spleen is one of the major peripheral lymphoid organs in fish (39), this result indicates OnGal8-L may play roles in the lymphatic system. Additionally, the high transcriptional levels of OnGal8-L are observed in skin, brain, and intestines, which is similar to the findings of OnGal-2 and OnGal-9 in tilapia (17, 18). As organs contact the external environment, the skin and intestine of fish are considered to be the first line of defense against pathogens (40, 41), and the blood–brain barrier in brain can resist the penetration by S. agalactiae (42). The abundance of OnGal8-L in these tissues indicates that it may be associated with immune defense against pathogens. To confirm whether OnGal8-L is involved in immune response against bacterial infection, the temporal transcription expressions of OnGal8-L in different tissues (spleen, head kidney, and brain) are investigated by qRT-PCR. Our results show that OnGal8-L is up-regulated in a time-dependent manner and reached to peak at 12 h in spleen, head kidney, and brain after S. agalactiae infection, indicating that OnGal8-L plays a role in the anti-bacterial immune response of tilapia. Similar results are also observed in the studies of OnCL-K1 and OnGal-2 from Nile tilapia (17, 27). It has been proven that S. agalactiae can invade the brain microvascular endothelial cell (BMEC) of fish, leading to meningitis (43). The increased expression of OnGal8-L in brain implies its contribution in protecting self-tissue. Compared with the transcriptional level in brain, a stronger increase of OnGal8-L was seen in spleen and head kidney at 12 hpi. Spleen and head kidney are the two main immune organs in teleost (44), which are armored with various immune cell types, including B cells, macrophages, granulocytes, and T cells. These results suggest that OnGal8-L may be involved in the regulation of cell-mediated immune responses during bacterial infection. Moreover, monocyte/macrophages are vital leukocytes that modulate innate immune responses through antigen presentation and cytokine production (45, 46). In our present data, we also observed the up-regulation of OnGal8-L in monocyte/macrophages after S. agalactiae stimulation, which is in line with the result of mannose-binding lectin in monocyte/macrophages (47). The in vivo and in vitro data indicate an involvement of OnGal8-L in response to bacterial infection.
To explore the exact function of OnGal8-L, the recombinant protein (rOnGal8L) was prepared for in vitro study. rOnGal8L exhibits the hemagglutination activity toward fish erythrocyte and bacterial, which is consistent with the research of Madusanka et al. (12), and the binding of rOnGal8L to S. agalactiae is impaired by competitive binding of LTA. Also, rOnGal8L can bind to LTA, LPS, poly I:C, PGN, galactose, mannose, and maltose, suggesting its utility in recognizing a wide range of pathogens. Besides the agglutination and binding activities, antibacterial activities are observed in a few galectin family members as well (48). Interestingly, our data showed that rOnGal8L significantly suppressed the growth of S. agalactiae but not A. hydrophila, which could be attributed to the different cell wall compositions of S. agalactiae (Gram-positive) and A. hydrophila (Gram-negative). These data indicate that rOnGal8-L acts as a PRR or an antimicrobial molecule in the immune defense against bacteria.
Considering that galectins recognize molecular patterns on the surface of microbes and initiate the downstream immune signaling pathways for bacterial clearance (49, 50), we further examined the impacts of rOnGal8L on phagocytosis and cytokine production of monocyte/macrophages. As expected, rOnGal8L can remarkably enhance the phagocytosis of macrophage to both Gram-positive and Gram-negative bacteria, which is in line with the finding of galectin-2 in tilapia (17). Respiratory burst is crucial for degrading internalized particles and bacteria in phagocytes. It has been recorded that fish lectins, including C-type lectin (27), galectin (18), and rhamnose-binding lectin (51), can enhance the respiratory burst activity of monocyte/macrophages. Consistently, stronger respiratory burst of macrophage was detected after rOnGal8L treatment in this study, indicating a role of OnGal8-L as opsonin during pathogen invasion. Apart from phagocytosis, monocytes/macrophages can mediate immune response by producing a variety of cytokines such as interleukin 6 (IL-6), interleukin 10 (IL-10), interleukin 8 (IL-8), and macrophage migration inhibitory factor (MIF) (52, 53). Similar to the finding of Mu et al. (54), our present study found that IL-6 and IL-10 rose significantly and peaked at 12 h after rOnGal8L incubation (Figure 4Aa,b). IL-6 is a pleiotropic pro-inflammatory cytokine that can be rapidly expressed and secreted in the immune regulation process (55), while IL-10 inhibits the pro-inflammatory response and limits unnecessary tissue destruction caused by inflammation (56). The increase of these two cytokines in the early stages of immune response suggests that OnGal8-L can activate inflammatory response of monocyte/macrophages. In addition, relatively delayed response patterns of IL-8 and MIF were observed in rOnGal8L-treated group (Figure 4Ac,d), which slowly increased and reached to the highest level at 24 h. The results were consistent with our previous finding of OnGal-3 (32). As a neutrophil chemokine, IL-8 can recruit different immune cells including macrophages, mast cells, neutrophils, and other granulocytes to the infection sites (57). MIF, a macrophage migration inhibitory factor, which is necessary for leukocytes to transport to infection site (58). The observation implies that OnGal8-L may not only stimulate the inflammatory factors expression, but also induce the chemokines production for recruiting leukocytes to the infection site during antibacterial defense.
In fish, non-specific immunity is required for maintaining dynamic balance, preventing microbial invasion, eliminating various pathogens, and activating adaptive immune responses (59). AKP functions as a multi-functional enzyme participates in antimicrobial activity (60). ACP is a typical lysosome enzyme involved in killing and digesting pathogens (61). LZM also has potent non-specific antimicrobial activity (62). Moreover, as a component of antioxidant defenses, enzymatic antioxidants can protect fish against oxidative damage (63, 64). CAT, SOD, and POD are the primary antioxidant enzymes that represent neutrophil antimicrobial activity (65, 66). SOD can convert oxygen radicals (O2−) into H2O2, which is subsequently disproportionated or decomposed by CAT and POD (67, 68). The present study found that OnGal8-L overexpression enhanced the activities of AKP, ACP, LZM, CAT, POD, and SOD in serum, which clearly suggest that OnGal8-L can improve non-specific immunity and antioxidant activity in tilapia. The subsequent challenge experiment showed that overexpression of OnGal8-L also decreased bacterial burden and tissue damage in head kidney, spleen, and liver after S. agalactiae infection. Additionally, OnGal8-L overexpression reduced the expression of IL-1β, TNF-α caused by bacterial infection. While a significant up-regulation of IL-10 was seen in the late stage of bacterial infection, indicating OnGal8-L can regulate inflammation response in vivo during bacterial infection. After S. agalactiae infection, the pEGFP-Gal8-L injected group had a better survival rate than the other groups, which suggests that OnGal8-L may be promising for applying as a molecular adjuvant in the development of vaccine against bacterial infection.
In summary, a novel tandem-repeat fish galectin-8 homolog (OnGal8-L) and its involvement in immune response during bacterial infection were identified in Nile tilapia. The OnGal8-L was widely distributed in various tissues and was induced by bacterial infection in vivo or in vitro. OnGal8-L could protect tilapia against bacterial infection via stimulating immune response, inhibiting bacterial proliferation and impairing tissue damages. This study lays a foundation for further study of fish galectin-8 in modulating immunity and sheds light on its potential application as vaccine adjuvant.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.
Ethics Statement
The animal study was reviewed and approved by Guangdong Province Laboratory Animal Management Regulations.
Author Contributions
JC and YH designed the experiments. JN performed experiments, analyzed data, and wrote the manuscript. XL and FW performed Nile tilapia farming and cell isolation. BW and JT contributed to the graphing. JJ and YL reviewed the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by National Natural Science Foundation of China (grant nos. 31302226 and 31572651), National Key R&D Program of China (2018YFD0900501), Technology Planning Project of Guangdong Province of China (grant nos. 2015A020209181 and 2017A030307033), the Fund of Southern Marine Science and Engineering Guangdong Laboratory (Zhanjiang) (ZJW-2019-05), and Special foundation for achieving the first class of Guangdong Province (231419013).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Liu FT, Rabinovich GA. Galectins: regulators of acute and chronic inflammation. Ann N Y Acad Sci. (2010) 1183:158–82. doi: 10.1111/j.1749-6632.2009.05131.x
2. Sharma UC, Pokharel S, van Brakel TJ, van Berlo JH, Cleutjens JP, Schroen B, et al. Galectin-3 marks activated macrophages in failure-prone hypertrophied hearts and contributes to cardiac dysfunction. Circulation. (2004) 110:3121–8. doi: 10.1161/01.CIR.0000147181.65298.4D
3. Leffler H, Carlsson S, Hedlund M, Qian Y, Poirier F. Introduction to galectins. Glycoconj J. (2002) 19:433–40. doi: 10.1023/B:GLYC.0000014072.34840.04
4. Sato S, St-Pierre C, Bhaumik P, Nieminen J. Galectins in innate immunity: dual functions of host soluble β-galactoside-binding lectins as damage-associated molecular patterns (DAMPs) and as receptors for pathogen-associated molecular patterns (PAMPs). Immunol Rev. (2009) 230:172–87. doi: 10.1111/j.1600-065X.2009.00790.x
5. Vasta GR. Roles of galectins in infection. Nat Rev Microbiol. (2009) 7:424–38. doi: 10.1038/nrmicro2146
6. Haudek KC, Patterson RJ, Wang JL. SR proteins and galectins: what's in a name? Glycobiology. (2010) 20:1199–207. doi: 10.1093/glycob/cwq097
7. Rabinovich GA, Gruppi A. Galectins as immunoregulators during infectious processes: from microbial invasion to the resolution of the disease. Parasite Immunol. (2005) 27:103–14. doi: 10.1111/j.1365-3024.2005.00749.x
8. Bai Z, Zhao L, Chen X, Li Q, Li J. A galectin contributes to the innate immune recognition and elimination of pathogens in the freshwater mussel Hyriopsis cumingii. Dev Comp Immunol. (2017) 73:36–45. doi: 10.1016/j.dci.2017.03.008
9. Bidon N, Brichory F, Bourguet P, Le Pennec JP, Dazord L. Galectin-8: a complex sub-family of galectins (Review). Int J Mol Med. (2001) 8:245–50. doi: 10.3892/ijmm.8.3.245
10. Zick Y, Eisenstein M, Goren RA, Hadari YR, Levy Y, Ronen D. Role of galectin-8 as a modulator of cell adhesion and cell growth. Glycoconj J. (2002) 19:517–26. doi: 10.1023/B:GLYC.0000014081.55445.af
11. Thurston TL, Wandel MP, von Muhlinen N, Foeglein A, Randow F. Galectin 8 targets damaged vesicles for autophagy to defend cells against bacterial invasion. Nature. (2012) 482:414–8. doi: 10.1038/nature10744
12. Madusanka RK, Priyathilaka TT, Janson ND, Kasthuriarachchi TDW, Jung S, Tharuka MDN, et al. Molecular, transcriptional and functional delineation of Galectin-8 from black rockfish (Sebastes schlegelii) and its potential immunological role. Fish Shellfish Immunol. (2019) 93:449–62. doi: 10.1016/j.fsi.2019.07.072
13. Sifa L, Chenhong L, Dey M, Gagalac F, Dunham R. Cold tolerance of three strains of Nile tilapia, Oreochromis niloticus, in China. Aquaculture. (2002) 213:123–9. doi: 10.1016/S0044-8486(02)00068-6
14. Suanyuk N, Kanghear H, Khongpradit R, Supamattaya K. Streptococcus agalactiae infection in tilapia (Oreochromis niloticus). Songklanakarin J Sci Technol. (2005) 27(Suppl 1).
15. Unajak S, Pholmanee N, Songtawee N, Srikulnath K, Srisapoome P, Kiataramkul A, et al. Molecular characterization of Galectin-8 from Nile tilapia (Oreochromis niloticus Linn.) and its response to bacterial infection. Mol Immunol. (2015) 68:585–96. doi: 10.1016/j.molimm.2015.09.012
16. Huang Y, Wang Z, Zheng Q, Tang J, Cai J, Lu Y, et al. Conservation of structural and interactional features of CD28 and CD80/86 molecules from Nile tilapia (Oreochromis niloticus). Fish Shellfish Immunol. (2018) 72:95–103. doi: 10.1016/j.fsi.2017.10.008
17. Niu J, Huang Y, Niu J, Wang Z, Tang J, Wang B, et al. Characterization of Galectin-2 from Nile tilapia (Oreochromis niloticus) involved in the immune response to bacterial infection. Fish Shellfish Immunol. (2019) 87:737–43. doi: 10.1016/j.fsi.2019.02.026
18. Niu J, Huang Y, Li Y, Wang Z, Tang J, Wang B, et al. Characterization of a tandem-repeat galectin-9 from Nile tilapia (Oreochromis niloticus) involved in the immune response against bacterial infection. Fish Shellfish Immunol. (2019) 92:216–23. doi: 10.1016/j.fsi.2019.05.061
19. Mu L, Wu H, Han K, Wu L, Bian X, Li B, et al. Molecular and functional characterization of a mannose-binding lectin/ficolin-associated protein (MAp44) from Nile tilapia (Oreochromis niloticus) involved in the immune response to bacterial infection. Dev Comp Immunol. (2019) 101:103438. doi: 10.1016/j.dci.2019.103438
20. Mu L, Yin X, Xiao Y, Bian X, Yang Y, Wu L, et al. A C-type lectin (CL11X1-like) from Nile tilapia (Oreochromis niloticus) is involved in host defense against bacterial infection. Dev Comp Immunol. (2018) 84:230–40. doi: 10.1016/j.dci.2018.02.015
21. Meunier E, Broz P. Evolutionary convergence and divergence in NLR function and structure. Trends Immunol. (2017) 38:744–57. doi: 10.1016/j.it.2017.04.005
22. Livak K, Methods TSJ. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt Method. (2000) 25:402–8. doi: 10.1006/meth.2001.1262
23. Bai Y, Niu D, Bai Y, Li Y, Lan T, Peng M, et al. Identification of a novel galectin in Sinonovacula constricta and its role in recognition of Gram-negative bacteria. Fish Shellfish Immunol. (2018) 80:1–9. doi: 10.1016/j.fsi.2018.05.041
24. Zhang XW, Xu WT, Wang XW, Mu Y, Zhao XF, Yu XQ, et al. A novel C-type lectin with two CRD domains from Chinese shrimp Fenneropenaeus chinensis functions as a pattern recognition protein. Mol Immunol. (2009) 46:1626–37. doi: 10.1016/j.molimm.2009.02.029
25. Pech MF, Fong LE, Villalta JE, Chan LJ, Kharbanda S, O'Brien JJ, et al. Systematic identification of cancer cell vulnerabilities to natural killer cell-mediated immune surveillance. Elife. (2019) 8:e47362. doi: 10.7554/eLife.47362
26. Zhang DL, Lv CH, Yu DH, Wang ZY. Characterization and functional analysis of a tandem-repeat galectin-9 in large yellow croaker Larimichthys crocea. Fish Shellfish Immunol. (2016) 52:167–78. doi: 10.1016/j.fsi.2016.03.032
27. Mu L, Yin X, Bian X, Wu L, Yang Y, Wei X, et al. Expression and functional characterization of collection-K1 from Nile tilapia (Oreochromis niloticus) in host innate immune defense. Mol Immunol. (2018) 103:21–34. doi: 10.1016/j.molimm.2018.08.012
28. Crinier A, Milpied P, Escaliere B, Piperoglou C, Galluso J, Balsamo A, et al. High-dimensional single-cell analysis identifies organ-specific signatures and conserved NK cell subsets in humans and mice. Immunity. (2018) 49:971–86.e5. doi: 10.1016/j.immuni.2018.09.009
29. Bai Z, Zhao L, Chen X, Li Q, Li J. A galectin from Hyriopsis cumingii involved in the innate immune response against to pathogenic microorganism and its expression profiling during pearl sac formation. Fish Shellfish Immunol. (2016) 56:127–35. doi: 10.1016/j.fsi.2016.07.006
30. Cha GH, Liu Y, Peng T, Huang MZ, Xie CY, Xiao YC, et al. Molecular cloning, expression of a galectin gene in Pacific white shrimp Litopenaeus vannamei and the antibacterial activity of its recombinant protein. Mol Immunol. (2015) 67:325–40. doi: 10.1016/j.molimm.2015.06.014
31. Sha Z, Wang L, Sun L, Chen Y, Zheng Y, Xin M, et al. Isolation and characterization of monocyte/macrophage from peripheral blood of half smooth tongue sole (Cynoglossus semilaevis). Fish Shellfish Immunol. (2017) 65:256–66. doi: 10.1016/j.fsi.2017.04.015
32. Niu J, Huang Y, Liu X, Luo G, Tang J, Wang B, et al. Functional characterization of galectin-3 from Nile tilapia (Oreochromis niloticus) and its regulatory role on monocytes/macrophages. Fish Shellfish Immunol. (2019) 95:268–76. doi: 10.1016/j.fsi.2019.10.043
33. Huang Y, Zheng Q, Niu J, Tang J, Wang B, Abarike ED, et al. NK-lysin from Oreochromis niloticus improves antimicrobial defence against bacterial pathogens. Fish Shellfish Immunol. (2018) 72:259–65. doi: 10.1016/j.fsi.2017.11.002
34. Chen J, Chen Q, Lu XJ, Li CH. LECT2 improves the outcomes in ayu with Vibrio anguillarum infection via monocytes/macrophages. Fish Shellfish Immunol. (2014) 41:586–92. doi: 10.1016/j.fsi.2014.10.012
35. Liu FT, Rabinovich GA. Galectins as modulators of tumour progression. Nat Rev Cancer. (2005) 5:29–41. doi: 10.1038/nrc1527
36. Rabinovich GA, Liu FT, Hirashima M, Anderson A. An emerging role for galectins in tuning the immune response: lessons from experimental models of inflammatory disease, autoimmunity and cancer. Scand J Immunol. (2007) 66:143–58. doi: 10.1111/j.1365-3083.2007.01986.x
37. Weinmann D, Kenn M, Schmidt S, Schmidt K, Walzer SM, Kubista B, et al. Galectin-8 induces functional disease markers in human osteoarthritis and cooperates with galectins-1 and−3. Cell Mol Life. (2018) 75:4187–205. doi: 10.1007/s00018-018-2856-2
38. Vasta GR, Nita-Lazar M, Giomarelli B, Ahmed H, Du S, Cammarata M, et al. Structural and functional diversity of the lectin repertoire in teleost fish: relevance to innate and adaptive immunity. Dev Comp Immunol. (2011) 35:1388–99. doi: 10.1016/j.dci.2011.08.011
39. Fänge R, Nilsson S. The fish spleen: structure and function. Experientia. (1985) 41:152–58. doi: 10.1007/BF02002607
40. Salinas I. The mucosal immune system of teleost fish. Biology (Basel). (2015) 4:525–39. doi: 10.3390/biology4030525
41. Ellis AE. Innate host defense mechanisms of fish against viruses and bacteria. Dev Comp Immunol. (2001) 25:827–39. doi: 10.1016/S0145-305X(01)00038-6
42. Gan Z, Wang B, Lu Y, Cai S, Cai J, Jian J, et al. Molecular characterization and expression of CD2BP2 in Nile tilapia (Oreochromis niloticus) in response to Streptococcus agalactiae stimulus. Gene. (2014) 548:126–33. doi: 10.1016/j.gene.2014.07.032
43. Mian G, Godoy D, Leal C, Yuhara T, Costa G, Figueiredo HCP. Aspects of the natural history virulence of S. agalactiae infection in Nile tilapia. (2009) 136:180-183. doi: 10.1016/j.vetmic.2008.10.016
44. Stenvik J, Schroder MB, Olsen K, Zapata A, Jorgensen TO. Expression of immunoglobulin heavy chain transcripts (VH-families, IgM, and IgD) in head kidney and spleen of the Atlantic cod (Gadus morhua L.). Dev Comp Immunol. (2001) 25:291–302. doi: 10.1016/S0145-305X(00)00056-2
45. Swirski FK, Nahrendorf M, Etzrodt M, Wildgruber M, Cortez-Retamozo V, Panizzi P, et al. Identification of splenic reservoir monocytes and their deployment to inflammatory sites. Science. (2009) 325:612–6. doi: 10.1126/science.1175202
46. Sioud M, Pettersen S, Ailte I, Fløisand Y. Targeted killing of monocytes/macrophages and myeloid leukemia cells with pro-apoptotic peptides. Cancers (Basel). (2019) 11:1088. doi: 10.3390/cancers11081088
47. Mu L, Yin X, Yang Y, Wu L, Wu H, Li B, et al. Functional characterization of a mannose-binding lectin (MBL) from Nile tilapia (Oreochromis niloticus) in non-specific cell immunity and apoptosis in monocytes/macrophages. Fish Shellfish Immunol. (2019) 87:265–74. doi: 10.1016/j.fsi.2019.01.019
48. Arthur CM, Cummings RD, Stowell SR. Evaluation of the bactericidal activity of galectins. Methods Mol Biol. (2015) 1207:421–30. doi: 10.1007/978-1-4939-1396-1_27
49. Kamhawi S, Ramalho-Ortigao M, Pham VM, Kumar S, Lawyer PG, Turco SJ, et al. A role for insect galectins in parasite survival. Cell. (2004) 119:329–41. doi: 10.1016/j.cell.2004.10.009
50. Song X, Zhang H, Wang L, Zhao J, Mu C, Song L, et al. A galectin with quadruple-domain from bay scallop Argopecten irradians is involved in innate immune response. Dev Comp Immunol. (2011) 35:592–602. doi: 10.1016/j.dci.2011.01.006
51. Watanabe Y, Chang Y-H, Nakamura O, Naganuma T, Ogawa T, Muramoto K. Rhamnose-binding lectins induce respiratory burst activity in macrophage cells from rainbow trout. Fish Sci. (2013) 79:513–9. doi: 10.1007/s12562-013-0624-7
52. Lee EH, Rikihisa Y. Absence of tumor necrosis factor alpha, interleukin-6 (IL-6), and granulocyte-macrophage colony-stimulating factor expression but presence of IL-1beta, IL-8, and IL-10 expression in human monocytes exposed to viable or killed Ehrlichia chaffeensis. Infect Immun. (1996) 64:4211–9. doi: 10.1128/IAI.64.10.4211-4219.1996
53. Calandra T, Roger T. Macrophage migration inhibitory factor: a regulator of innate immunity. Nat Rev Immunol. (2003) 3:791–800. doi: 10.1038/nri1200
54. Mu L, Yin X, Wu H, Han K, Wu L, Ding M, et al. Expression and functional characterization of a mannose-binding lectin-associated serine protease-1 (MASP-1) from Nile tilapia (Oreochromis niloticus) in host defense against bacterial infection. Fish Shellfish Immunol. (2019) 91:68–77. doi: 10.1016/j.fsi.2019.05.014
55. Kimura A, Kishimoto T. IL-6: regulator of Treg/Th17 balance. Eur J Immunol. (2010) 40:1830–5. doi: 10.1002/eji.201040391
56. Ouyang W, Rutz S, Crellin NK, Valdez PA, Hymowitz SG. Regulation and functions of the IL-10 family of cytokines in inflammation and disease. Annu Rev Immunol. (2011) 29:71–109. doi: 10.1146/annurev-immunol-031210-101312
57. Itoh Y, Joh T, Tanida S, Sasaki M, Kataoka H, Itoh K, et al. IL-8 promotes cell proliferation and migration through metalloproteinase-cleavage proHB-EGF in human colon carcinoma cells. Cytokine. (2005) 29:275–82. doi: 10.1016/j.cyto.2004.11.005
58. Morand EF, Leech M, Bernhagen J. MIF: a new cytokine link between rheumatoid arthritis and atherosclerosis. Nat Rev Drug Dis. (2006) 5:399–410. doi: 10.1038/nrd2029
59. Riera Romo M, Pérez-Martínez D, Castillo Ferrer C. Innate immunity in vertebrates: an overview. Immunology. (2016) 148:125–39. doi: 10.1111/imm.12597
60. Xing J, Zhan WB, Zhou L. Endoenzymes associated with haemocyte types in the scallop (Chlamys farreri). Fish Shellfish Immunol. (2002) 13:271–8. doi: 10.1006/fsim.2001.0402
61. Cheng TC, Dougherty WJ. Ultrastructural evidence for the destruction of Schistosoma mansoni sporocysts associated with elevated lysosomal enzyme levels in Biomphalaria glabrata. J Parasitol. (1989) 75:928–41. doi: 10.2307/3282873
62. Cai S, Zhang Y, Wu F, Wu R, Yang S, Li Y, et al. Identification and functional characterization of a c-type lysozyme from Fenneropenaeus penicillatus. Fish Shellfish Immunol. (2019) 88:161–9. doi: 10.1016/j.fsi.2019.02.043
63. Martínez-Álvarez RM, Morales AE, Sanz A. Antioxidant defenses in fish: biotic and abiotic factors. Rev Fish Biol Fish. (2005) 15:75–88. doi: 10.1007/s11160-005-7846-4
64. Ahmad S. Oxidative Stress and Antioxidant Defenses in Biology. Dordrecht: Springer Science & Business Media (2012).
65. Jin Y, Zhang X, Shu L, Chen L, Sun L, Qian H, et al. Oxidative stress response and gene expression with atrazine exposure in adult female zebrafish (Danio rerio). Chemosphere. (2010) 78:846–52. doi: 10.1016/j.chemosphere.2009.11.044
66. Slaninova A, Smutna M, Modra H, Svobodova Z. A review: oxidative stress in fish induced by pesticides. Neuro Endocrinol Lett. (2009) 30(Suppl 1):2–12.
67. Wang X, Martinez MA, Dai M, Chen D, Ares I, Romero A, et al. Permethrin-induced oxidative stress and toxicity and metabolism. A review. Environ Res. (2016) 149:86–104. doi: 10.1016/j.envres.2016.05.003
Keywords: galectin-8, Nile tilapia, monocytes/macrophages, Streptococcus agalactiae, immune response
Citation: Niu J, Huang Y, Liu X, Wu F, Tang J, Wang B, Lu Y, Cai J and Jian J (2020) Fish Galectin8-Like Exerts Positive Regulation on Immune Response Against Bacterial Infection. Front. Immunol. 11:1140. doi: 10.3389/fimmu.2020.01140
Received: 04 February 2020; Accepted: 11 May 2020;
Published: 26 June 2020.
Edited by:
Tiehui Wang, University of Aberdeen, United KingdomCopyright © 2020 Niu, Huang, Liu, Wu, Tang, Wang, Lu, Cai and Jian. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jia Cai, bWF0cml4OTI0QGZveG1haWwuY29t; Jichang Jian, amlhbmpjQGdkb3UuZWR1LmNu
 Jinzhong Niu
Jinzhong Niu Yu Huang1,2,3
Yu Huang1,2,3 Jia Cai
Jia Cai