
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol. , 15 August 2018
Sec. Nutritional Immunology
Volume 9 - 2018 | https://doi.org/10.3389/fimmu.2018.01830
This article is part of the Research Topic Human Microbiome and Personalized Nutrition View all 17 articles
 Veronica Lazar1,2†
Veronica Lazar1,2† Lia-Mara Ditu1,2
Lia-Mara Ditu1,2 Gratiela Gradisteanu Pircalabioru1,2†
Gratiela Gradisteanu Pircalabioru1,2† Irina Gheorghe1,2
Irina Gheorghe1,2 Carmen Curutiu1,2
Carmen Curutiu1,2 Alina Maria Holban1,2
Alina Maria Holban1,2 Ariana Picu1,3
Ariana Picu1,3 Laura Petcu1,3
Laura Petcu1,3 Mariana Carmen Chifiriuc1,2*†
Mariana Carmen Chifiriuc1,2*†
The microbiota consists of a dynamic multispecies community of bacteria, fungi, archaea, and protozoans, bringing to the host organism a dowry of cells and genes more numerous than its own. Among the different non-sterile cavities, the human gut harbors the most complex microbiota, with a strong impact on host homeostasis and immunostasis, being thus essential for maintaining the health condition. In this review, we outline the roles of gut microbiota in immunity, starting with the background information supporting the further presentation of the implications of gut microbiota dysbiosis in host susceptibility to infections, hypersensitivity reactions, autoimmunity, chronic inflammation, and cancer. The role of diet and antibiotics in the occurrence of dysbiosis and its pathological consequences, as well as the potential of probiotics to restore eubiosis is also discussed.
The microbiota consists of a multispecies microbial community living within a particular niche in a mutual synergy with the host organism. Besides bacteria, the microbiota includes fungi, archaea, and protozoans (1, 2), to which viruses are added, which seem to be even more numerous than microbial cells (3). The gastrointestinal tract (GIT), with its epithelial barrier with a total area of 400 m2, is a complex, open, and integrated ecosystem with the highest exposure to the external environment. The GIT contains at least 1014 microorganisms belonging to >2,000 species and 12 different phyla, the associated microbiome containing 150- to 500-fold more genes than the human DNA (1, 4–7). The GIT microbiota exhibits a huge diversity, being individually shaped by numerous and incompletely elucidated factors, such as host genetics, gender, age, immune system, antropometric parameters, health/disease condition, geographic and socio-economical factors (urban or rural, sanitary conditions), treatments, diet, etc. (8, 9). Recent metagenomic data demonstrated that the majority of component species is not present in the same time and in the same person, but, however, few species are abundant in healthy individuals, while other species are less represented (4, 7). In addition to the distribution along the digestive tract segments, the GIT microbiota of the three distinct transversal microhabitats, i.e., floating cells in the intestinal lumen, cells adherent to the mucus layer and respectively to the surface of the epithelial cells, is also different (3).
Recent findings suggest that the microbial colonization of the GIT starts before birth, as revealed by the placental microbiome profile, being composed of members of Firmicutes, Proteobacteria, Tenericutes, Bacteroidetes, and Fusobacteria groups, which were found to share some similarities with the human oral microbiome (10). Also, the meconium of full term infants is not sterile, harboring 30 genera normally found in the amniotic fluid, vagina, and the oral cavity (8, 9, 11). We can assume that the bacteria reach these sites mainly from the vaginal tract, although selective translocation is also possible. Archaea were also detected in the vaginal microbiota of pregnant women, accounting for a mother-to-child transmission (12).
Vaginally born infants have a microbiota containing species derived from the vaginal microbiota of their mothers. Conversely, in the case of cesarean section delivered babies, the microbiota is similar to the skin microbiota and is rich in Propionibacterium spp. and Staphylococcus spp. (13).
It is generally accepted that the pregnancy period and the first 1,000 days after birth are the most critical timeframes for interventions and any modulation made at this point has the potential to improve child growth and development (14). Delivery mode seems to influence immunological maturation through microbiota development. Cesarean section delivered children were found to have a higher number of antibody-secreting cells (11).
Furthermore, the human milk is involved in the GIT microbiota and immune system development. In addition to its nutritional components, this natural functional food contains numerous bioactive substances and immunological components that control the maturation of the newborn intestine and the composition of the microbial community. Numerous studies revealed that breast-feeding has a protective role in infants, conferred by a complex mixture of molecules, including lysozyme, sIgA, alpha-lactalbumin, lactoferrin, but also free oligosaccharides, complex lipids, and other glycoconjugates (14). The proteolytic processing of glycoprotein k-casein, with the release of glycomacropeptides, prevents colonization of the gut by pathogens, through competition with the receptors of the gut epithelial cells in breast-fed infants. Lactoferricin is a potent antimicrobial agent, explaining the decreased infant death rate caused by gastrointestinal and respiratory infections in breast-fed infants (14, 15). Moreover, breast milk contains ~109 bacterial cells/L (16) and prebiotic oligosaccharides (fructans) which stimulate the multiplication of Bifidobacterium spp. and Lactobacillus spp., while follow-on milk powder stimulates proliferation of enterococci and enterobacteria (17, 18). As the infant grows, solid foods are introduced, therefore the microbiota diversity increases, and the microbiota community evolves toward the adult-like state. Although some dominant enterotypes represented by Bacteroides, Prevotella, and Ruminococcus genera are recognized, however, the final composition of the adult microbiota is unique and the factors guiding this feature are still a matter of debate (19).
The very active microbial community has been shown to mutually interact with the host and to exert a lot of beneficial roles, explaining its tolerance by the host organism. The GIT microbiota is involved in energy harvest and storage, and, due to its particular metabolic pathways and enzymes, it extends the potential of the host metabolism. This property is believed to exhibit a potent evolutionary pressure toward the establishment of bacteria as human symbionts (11). The GIT microbiota influences the normal gut development, due to its ability to influence epithelial cell proliferation and apoptosis of host cells. Although the intimate interactions between microbiota and host cells are widely unknown, a major mechanism seems to involve short-chain fatty acids (SCFA), resulted from the fermentation of indigestible polysaccharides (fibers), such as butyrate, acetate, and propionate with an important anti-inflammatory role. SCFAs also support intestinal homeostasis in the normal colon, by aiding intestinal repair through the promotion of cellular proliferation and differentiation. However, SCFAs seem to inhibit the cancerous cells proliferation. Among the different SCFAs, butyrate has a paramount role in intestinal homeostasis due to its role as a primary energy source for colonocytes (20, 21). In addition, the GIT microbiota stimulates the nonspecific and specific immune system components development, just after birth and during the entire life and it acts as an antiinfectious barrier by inhibiting the pathogens’ adherence and subsequent cellular substratum colonization and by the production of bacteriocins and of other toxic metabolites. Moreover, the microbiota is predominantly composed of anaerobes which prevent the process of translocation of aerobic/facultatively anaerobic bacteria and the consecutive systemic infections in immunodeficient individuals. Importantly, some GIT microbiota representatives (Escherichia coli and Bacteroides fragilis) are involved in the synthesis of vitamins, such as B1, B2, B5, B6, B12, K, folic acid, and biotin. Also, the GIT microbiota has the ability to degrade xenobiotics, sterols and to perform biliary acids deconjugation (B. fragilis and Fusobacterium spp.) (19).
All these aforementioned effects are occurring when the microbiota community is characterized by an interspecies balance, known as eubiosis. Any perturbation of eubiosis, known as dysbiosis, could become a pivotal driver for various infectious and non-infectious diseases, each of them with specific microbiota signatures that can further trigger pathophysiologies in different organs (11).
Our aim was to review these physiological roles, focusing on one side the GIT microbiota contribution to the immune system development and education, and on the other side, to what is happening when eubiosis is replaced by the dysbiosis status; in this case the immunostasy is altered, the host becomes more susceptible to infections, both exogenous and endogenous; immunotolerance is affected and the immune system will react against the self-components (autoimmunity), or vary in intensity, being either over (allergic reactions and chronic inflammation) or less/inappropriately (immunodeficiency and cancer) activated.
The mucosal immune system is highly specialized, its functions are largely independent of the systemic immune system (15) and it undergoes major changes after bacterial colonization of the intestinal tract (22).
Commensal microorganisms are required for the maturation of the immune system, which “learns” to differentiate between commensal bacteria (which are becoming almost quasi-self and tolerated antigens) and pathogenic bacteria (23, 24). Toll-like receptors (TLRs) from the membrane of the epithelial and lymphoid cells of the small intestine are involved in this differential recognition, being responsible for the normal development of the intestinal mucosal immune system. TLRs suppress the occurrence of an inflammatory response and promote immunological tolerance to normal microbiota components. The role of TLRs is to recognize different general microbe-associated molecular patterns (MAMPs) [containing various bacterial antigens (e.g., peptidoglycan components—muramic acid, capsular polysaccharides and lipopolysaccharides, flagellin and unmethylated bacterial DNA CpG motifs)] and to trigger the innate intestinal immunity (25, 26). Following stimulation, a complex cascade of signals is initiated, leading to the release of nuclear factor kappa-light-chain-enhancer of activated B cells (NF-kB), which activates a variety of genes coding for chemokines, cytokines, acute phase proteins, and other effectors of the humoral immune response (27, 28). TLR activity decreases during the first weeks of life, potentially allowing the development of a stable gut bacterial community. Furthermore, TLR activation by antigens belonging to the normal intestinal microbiota is signaling the inhibition of inflammatory reactions, being thus essential to maintain intestinal homeostasis (29). Complementarily, NOD-like receptors (NLRs) recognize various microbial specific molecules and trigger the assembly of inflammasomes, which can act as sensors of damage-associated patterns. The NLPRP6 deficiency has been associated with an altered immune response (e.g., decreased IL-18 levels), dysbiosis, and intestinal hyperplasia (11, 30).
Gastrointestinal tract microbiota has been shown to modulate neutrophil migration and function (31) and to affect the differentiation of T cell populations into different types of helper cells (Th), respectively: Th1, Th2, and Th17 or into regulatory T cells (Tregs) (25). The Th17 cells are a subset of TCD4+ cells, which secrete multiple cytokines (IL-22, IL-17A, and IL-17F), with a significant impact on immune homeostasis and inflammation (32, 33). Unlike Th1 and Th2 cells, which have a stable secretory profile after differentiation, Th17 cells retain divergent cytokine expression profiles and functions (34). It has been shown that the administration of the purified capsular polysaccharide from the commensal bacterium B. fragilis suppresses the production of IL-17 and protects the colonic mucosa against inflammatory reactions initiated by bacterial antigens, stimulating TCD4+ lymphocytes to produce IL-10 (35). On the other side, the colonic environment also stimulates de novo differentiation and expansion of peripherally derived regulatory T cells from naïve CD4+ T cells (36). Tregs are key mediators of immune tolerance, limiting an inappropriately high inflammatory response (37), their dysfunction leading to autoimmune disorders (38).
sIgA has a crucial role in the local immune response, being considered the first line of defense against pathogens and toxins. sIgA production specific to different mucosal antigens is following their capture by Peyer’s patches M cells, transformation by underlying antigen-presenting cells [dendritic cells (DCs)], activation of T cells, and ultimately B cell class switch recombination in mesenteric lymph nodes (MLNs) and gut-associated lymphoid tissue. The commensal antigens induce the production of low amounts of sIgA through the modulation of their immunodominant epitopes, thus harboring an advantage for the colonization of the intestinal niche (11). A set of cytokines, including TGF-β, IL-4, IL-10, IL-5, and IL-6 stimulates IgA production. Some of these cytokines, notably IL-10 and TGF-β are crucial in maintaining the mucosal tolerance, therefore proving the link among sIgA production, immunity, and intestinal homeostasis (39).
In individuals with dysbiosis, immune responsiveness could be upregulated to promote the development of a more optimal status. This could be obtained through specific effects of sIgA, or less specific effects of innate immunity effectors (such as defensins) or local environment changes (i.e., diarrhea). In case of diarrhea, the host eliminates undesirable microbial communities in order to prepare niches for recolonization with more beneficial microbial populations, as a last resort to healing (14).
The host-commensal microbiota communication triggers antimicrobial responses from the epithelium including the release of several antibacterial lectins, including RegIIIc, α-defensins, and angiogenins (40, 41). These antibacterial effectors reduce the amount of potentially pathogenic microbes and provide protection against subsequent abnormal immune responses. For instance, Bacteroides thetaiotaomicron triggers the production of antimicrobial peptides which target other intestinal microbes. The microbiota of mice expressing a human enteric α-defensin, DEFA5, has no segmented filamentous bacteria (42), which are responsible for inducing IL-17-producing Th17 cells, which have been correlated with inflammatory bowel disease (IBD) and colorectal cancer.
Furthermore, aberrant microbial development during maturation of the innate immune system leads to defective immunological tolerance, which subsequently promotes exacerbated autoimmune and inflammatory diseases (e.g., allergen-induced airway hyperreactivity) (3). Microbial products may induce chronic stimulation of immune responses, leading to chronic, non-resolving inflammation and tissue damage, particularly after mucosal injury.
As also named “the last undiscovered human organ,” the intestinal microbiome has an impact on immune system development and differentiation. In addition, the microbiome holds a paramount role in the initiation and progression of infectious diseases (43).
Through the colonization of the mucosal entry sites of pathogens, microbiota could directly prevent the invasion by foreign microbes—a process known as colonization resistance (by competing with pathogenic bacteria in the gut for adhesion sites and nutrients, but also by releasing toxic molecules to counteract pathogen colonization), as well as indirectly, through the stimulation of the immune response. As stated above, the gut microbiota provides signals to stimulate the normal development of the immune system as well as the maturation of immune cells (44–46). The microbiota stimulates the secretory IgA response that is involved in inactivating rotaviruses, competes Clostridium difficile colonization, and neutralizes cholera toxin (47). Moreover, the signaling molecules released by the microbiota actively shape the host systemic immune response by regulating haematopoesis, and consequently potentiating the response to infection (48). Signals derived from the commensal microbiota trigger the development of granulocyte/monocyte progenitors in the bone marrow and hence affects tissue-resident innate immune populations which in turn promotes the early host innate response. In line with this, the absence of the microbiota derived signaling molecules cause alterations in tissue-resident myeloid populations prior to infection and leads to susceptibility to systemic infection by Staphylococcus aureus and Listeria monocytogenes (49).
The synergic interactions of the innate immune system and microbiota could be also exploited by pathogens to evade the antiinfectious mucosal barrier. A suggestive example is given by the oral bacterium Porphyromonas gingivalis, which escapes the host immune response via TLR2 signaling pathway modulation leading to dysbiosis and subsequent inflammation (50). Also, some viruses are able to interfere with the interplay between bacteria and the innate immune system (i.e., TLR4 signaling), for guaranteeing their efficient transmission (51). It has been proved that the antiviral host response is improved by antibiotic depletion of commensal microbiota. Intestinal antiviral innate immunity is the result of the induction of IL-18, interferon (IFN)-λ, or IL-22 pathways, which promote the expression of signal transducer and activator of transcription 1 (STAT1) and antiviral genes. Although IL-22 and IL-18 are both stimulated by commensal bacteria, IFN-λ expression is inhibited by the microbiota, hence enabling viral persistence. It has been also clearly demonstrated that interactions between gut epithelial cells and microbiota are crucial to maintain barrier defenses and gut homeostasis. For instance, the microbiota has a role in maintaining tight junctions’ integrity which limits Salmonella typhimurium invasion (52). On its turn, the intestinal pathogen S. typhimurium induces IL-22 production which targets commensal bacteria and liberates a colonization niche for the pathogen.
Generally, the antiinfectious barrier is efficient when the microbiota is complex and stable, in a eubiotic status. On the contrary, when dysbiosis occurs (due to different causes, e.g., poor colonization, antibiotherapy or an unbalanced, unhealthy diet, different pathological conditions leading to secondary immunodeficiencies), the microbiota loses its antiinfectious barrier potency and the host can be easily infected with different pathogenic microorganisms from the environment. In addition, some species of microbiota, enriched in the new condition of dysbiosis, can manifest their pathogenic potential by producing opportunistic infections. For example, antibiotics can be used for treating certain pathological GIT diseases, but the induced alteration of the intestinal microbiota could lead to metabolic disturbances, such as increased intestinal permeability, and may also increase susceptibility to infections [e.g., fungal and Clostridium difficile infections (CDIs)].
Recent findings proved a clear correlation between microbiome composition and risk of infectious diseases. For example, microbiota composition represents an infection risk for Plasmodium falciparum infection, and also a key factor for diverse vaccine responses (43).
Recent studies aiming to investigate the specific role of gut microbiota and immune system interactions in infectious diseases focused mainly on microbiome manipulation. This was achieved either by probiotics administration or fecal microbiota transplantation. Serious conditions which are prevalent in children, such as necrotizing and acute infectious diarrhea, but also antibiotic-associated diarrhea, CDIs and ventilator-associated pneumonia could be treated more efficiently by microbiota manipulation, with a better outcome, reduced mortality, and faster recovery rates. Since microbiota manipulation could control the balance between health and infectious disease, intestinal microbiota alteration by a pathogen or a pathobiont can lead to chronic diseases. In vivo studies demonstrated that the colonization of adherent-invasive Escherichia coli (AIEC, an E. coli pathovar involved in Crohn’s disease pathogenesis) during microbiota acquisition drove chronic colitis in mice (53). It seems that AIEC, Yersinia enterocolitica and probably other pathobionts, may promote chronic inflammation in susceptible hosts by producing gut microbiota alterations which lead to a higher capacity in activating innate immunity/pro-inflammatory gene expression (54). A recent study by Inoue et al. shed light on the impact of hepatitis C virus (HCV) infection on the gut microbiota. Unlike healthy individuals, HCV infected patients showed dysbiosis characterized by a decrease in Clostridiales and enrichment in Streptococcus and Lactobacillus genera. Microbiota alterations were present even in patients with mild liver disease, as revealed by the transient increase in Bacteroides and Enterobacteriaceae (55).
Gastrointestinal tract microbiota members can translocate from the digestive mucosa and reach the general circulation, indirectly by stimulating IL-12 production by splenic macrophages, DCs, which, in turn, regulates the Th1/Th2 balance toward a cell-mediated Th1 response (56). Studies have shown that the soluble products of Lactobacillus fermentum DSMZ 20052 determine the decrease of IL-8 levels by inhibiting the NF-kB pathway, thus alleviating the pro-inflammatory effect induced by Yersinia enterocolitica infection (57). Other studies support the activation of NF-kB signaling pathway with the subsequent activation of inflammatory genes by some probiotics. One of the hallmarks of NF-kB activation is the production of IL-6 (56). It has been shown that the colonization of the digestive tract of germ-free rats with Bifidobacterium lactis BB 12 strain stimulates the IL-6 synthesis (58).
All bacteria are able to communicate with each other by signaling molecules, which allow the bacterial cells to sense the environment, monitor population density and to adjust accordingly their gene expression. Through this type of communication, bacteria acquire an advantage crucial for dissemination and survival in highly competitive environments, which harbor hundreds of coexisting species (e.g., the oral cavity, the intestine). Depending on the involved members, intercellular communication is divided into two categories, based on the quorum-sensing (QS) mechanism. QS is a density-dependent molecular language responsible for the regulation of cellular phenotype/behavior as a response to environmental changes. The first type is the intraspecific cell-to-cell communication though specific QS molecules and the second mechanism consists of the interspecific communication based on an universal chemical “language,” which provides interspecific signaling between bacteria and eukaryotic/host cells. QS is orchestrated by small molecules, usually considered hormone-like organic molecules called autoinducers (AIs). AIs are represented by diffusible molecules called homoserine-lactones (Acyl-HSL) in Gram-negative bacteria and not diffusible peptidic molecules (AIP) in Gram-positive ones. A universal interspecies signal (“cross talk”) which contains AIs common for both Gram-positive and Gram-negative bacteria has been identified in 55 pathogens so far. These compounds depend on the microbial cellular density and hold a paramount role in various niches, especially in highly colonized sites, such as the gut and the oral cavity (59). This mechanism of communication regulates the expression of virulence genes in pathogens, with an important role in infection. For example, a relatively low virulence factors production by a limited population of bacteria may promote a robust host response that neutralizes these molecules, while the coordinated virulence factors gene expression by high-density bacterial populations can lead to higher secretion of extracellular factors (60, 61). The produced molecules have also an immunomodulatory effect, controlling the inflammatory response which can induce severe damaging of host tissues (62). Recent studies reported they may have also a therapeutic potential, for autoimmune diseases as immunosuppressive drugs (63). The QS mechanism allows bacteria to regulate the host colonization by commensal bacteria and to modulate the host response (64–66). Although the specific mechanism(s) through which AIs influence mammalian cells is unclear, a modified immune response was observed. For example, the 3-oxododecanoyl homoserine lactone (HSL-C12) induces apoptosis and Ca2+ release from endoplasmic reticulum stores. HSL-C12 has also been reported to modulate the inflammatory signaling (67), being immunosuppressive at or below 10 µM concentrations, but pro-inflammatory and proapoptotic at 20 µM and above (68). HSL-C12 acts through TLR- and Nod/Ipaf/caterpillar-independent signaling and activates multiple NF-κB-associated pro-inflammatory genes including IL-1α, IL-6, IL-8, Cox2, mPGES, PGE2, and MUC5AC in different cell types. The pro-inflammatory effects may be achieved through activation of MAPKs, extracellular-signal-regulated kinases, inhibition of peroxisome proliferator-activated receptor γ, or Ca2+ (69). In the presence of pro-inflammatory molecules, such as lipopolysaccharides (LPS) or TNFα, HSL-C12 may inhibit NF-κB signaling and expression of pro-inflammatory cytokines in macrophages and epithelial cells (69). In vivo experiments proved that direct injection of HSL-C12 in C57BL/6 mice lead to the expression of macrophage inflammatory protein-2 (MIP-2) (the mouse analog of the human cytokine IL-8) and also other cytokines. Significantly, higher concentration of MIP-2 was found in mice infected with QS active microbial strains than those inoculated with the QS-deficient bacteria (70).
Quorum-sensing is also used by microbiota members in order to detect the presence of other similar microbes (71); their well-known antiinfectious barrier effect is to the result of the antagonistic relationships with pathogens; is well known that probiotic strains are able to produce antimicrobial molecules as well as small QSIs which are interfering with the QS mechanism and virulence expression of the pathogens (72–74). It seems that the antimicrobial eosinophil-derived neurotoxin, cathelicidins, defensins, AI2 signaling molecules hold paramount functions in intra- and interspecies communication.
Certain intestinal mammalian hormones mimic the action of bacterial signaling molecules, thus increasing the complexity level of the bidirectional communication between bacteria and the host (75). In this context, a particular field of the exchange of molecular information between the many microorganisms and the host (76) is represented by microbial endocrinology, defined by the ability of GIT microbiota to orchestrate a bidirectional communication with the central nervous system by producing and sensing neurochemicals that are derived either within the microorganisms themselves or within their host (77). Steroid hormones (adrenaline and noradrenaline), due to their ability to pass through the plasmatic membrane are involved in the inter-kingdom communication between microorganisms and their mammalian host (78, 79). Although bacteria do not express adrenergic receptors, some studies indicate that bacterial cells are responsive to adrenaline and/or noradrenaline (NA) and recent studies suggest they have an important impact in maintaining the homeostasis of gut microbiota (80). The existing data sustain that NA may work as a siderophore (81). It is believed that NA is involved in overexpression of enterobactin and in the iron chelating mechanism in E. coli, subsequently increasing the bacterial growth rate. On the other hand, gut microbiota can produce neurochemicals with hormonal activities that could extend beyond the gut, being involved in the modulation of anxiety, depression, cognition, pain, inflammatory, autoimmune, and metabolic diseases (82–87).
The alteration of the complexity and eubiotic state of microbiota might promote intestinal and extraintestinal autoimmune and inflammatory disorders (type I diabetes, rheumatoid arthritis, ankylopsing spondilosis, IBD, pulmonary disease, atopy, non-alcoholic fatty liver disease, obesity, atherosclerosis, carcinogenesis, etc.) although the mechanisms involved are not well understood (3). Many researchers reported an opposite connection between the incidence of immune disorders and the infectious process. Within this line of thought, children under the age of 5 years living in developed countries are not exposed to many of the microbes, compounds and antigens they would have encountered a century ago. This lack of early immune stimulation by biotic factors that humans and their ancestors have evolved with may hinder the functioning of the immune system later in life and lead to hypersensitivity, autoimmune, or inflammatory diseases (88).
Initially called juvenile-onset diabetes, type 1 DM (T1DM) is a chronic illness associated with high morbidity and premature mortality. This disease is caused by the patient’s inability to secrete insulin as a result of the autoimmune destruction of the pancreatic beta cells (89). Usually, T1DM occurs early in life, but recent studies reported that up to 50% of new-onset T1DM patients are older than 20 years (90). The major factor in the pathophysiology of T1DM is represented by autoimmunity. The genetically susceptible individuals (around 95% of patients with T1DM) harbor either human leukocyte antigen DR3-DQ2 or DR4-DQ8 haplotypes, or have the UBASH3A mutation, also known as STS2, located on chromosome 21, which are linked also with other autoimmune diseases, such as celiac disease (91, 92), viral infections (mumps, enterovirus, coxsackie virus B4, and rubella), but also toxic chemicals, exposure to cytotoxins or cow’s milk in infancy and may stimulate the production of antibodies against antigenically similar beta cell molecules. T1DM is associated with a low diversity of microbiota and with the expansion of distinct groups of bacteria (93, 94). However, human studies have not yet elucidated the causal relationship between the gut microbiome and pathogenesis of T1DM. Some models have linked the gut microbiome with the development of T1DM, respectively, the Hygiene Hypothesis, the Leaky Gut Hypothesis, the Perfect Storm Hypothesis, and the Old Friends Hypothesis. Based on the Leaky Gut Hypothesis, the increased permeability of the intestinal epithelium develops from loss of tight barrier function (95). Macromolecules derived from diet and microbial antigens are able to pass through the epithelial barrier and consequently trigger intestinal inflammation that could lead to pancreatic beta cell attack (95). The Old Friends Hypothesis sustains the role of commensal microbes which have evolved together with their host and highlights that loss of these commensal microbes may impact the host’s immune response regulation and homeostasis (96). On the other hand, the Perfect Storm Hypothesis reunites aspects from the Leaky Gut Hypothesis and the Old Friends Hypothesis advocating that a combination of both increased intestinal permeability an altered microbiota composition, and an impaired intestinal immune responsiveness interact together culminating in anti-islet autoimmunity (97). The Hygiene Hypothesis was formulated by David Strachan (1989) who, trying to explain the actual high incidence of allergic and autoimmune diseases, postulated that increasing T1DM incidences is the result of a diminished or a lack of contact with infectious agents due to elevated hygienic conditions (98). In a recent study by Maffeis et al. on children at risk of developing T1DM, increased intestinal permeability was correlated with microbiota alterations. Unlike healthy controls, children with T1DM risk exhibited high levels of Globicatella sanguinis, Dialister invisus, and Bifidobacterium longum (99). In addition, it was also reported that the Bacteroidaceae family is enriched in children with T1DM. Moreover, T1DM children exhibited a decrease of Bifidobacterium pseudocatenulatum and Bifidobacterium adolescentis (100). A subsequent study revealed that the microbiota of genetically predisposed infants from 3 months to 3 years old was characterized by an enrichment of Rikenellaceae, Ruminococcus, Streptococcus, and Blautia as well as by reduced alpha diversity (101).
Recent studies found a correlation between rheumatoid arthritis, the enrichment of Prevotella copri and colitis susceptibility, suggesting that the inflammatory component of autoimmune diseases might be modulated by an impaired communication between the host and the microbiota (102). These data are also sustained by a recent study which characterized the gut microbiota of DBA1 mice after collagen induction arthritis (CIA) and found altered distribution of the microbiota. Mice susceptible to CIA harbored Lactobacillus as the dominant genus prior to the onset of arthritis. During disease progression, the operational taxonomic units (OTUs) of the Lachnospiraceae, Bacteroidaceae, and S24-7 families were significantly elevated in CIA-susceptible mice. Also, germ-free mice receiving microbiota harvested from CIA-susceptible mice presented an elevated induction of arthritis compared to those receiving microbiota from CIA-resistant mice (103).
Modification of the normal gut microbiota may have a role in the onset and/or progression of celiac disease. Species such as Staphylococcus epidermidis, Staphylococcus pasteuri, and Klebsiella oxytoca were enriched in duodenal biopsies harvested from patients diagnosed with active celiac disease. Species such as Streptococcus mutans and Streptococcus anginosus were reduced in patients with celiac disease compared to healthy people, independently of the inflammatory status. Fucosyltransferase 2 (FUT2) gene regulates the expression of ABH blood group antigens in mucus as well as other body secretions and also influences the composition of mucosa-associated bacteria. A mutation in FUT2 gene lead to decreased bacterial heterogeneity and abundance, including a lower quantity of Bifidobacterium spp., in the human gut. Fut2-deficient mice presented more susceptibility to Candida albicans colonization comparing to wild-type mice and Candida albicans infection is a culprit in the onset of celiac disease. Bifidobacterium spp. were shown to have a protective role against C. albicans colonization. Therefore, alteration of the microbiota due to mutation of FUT2 gene decreases colonization resistance and has a role in the pathogenesis of celiac disease (104).
Aberrant immune responses against commensal bacteria may promote the development of IBDs, such as ulcerative colitis and Crohn’s disease, providing experimental models for studying different aspects of the immune system-microbiota crosstalk, such as oxidative stress, microbial sensing, and antigen processing. Some alleles of the genes encoding for innate immunity mechanisms, i.e., ATG16L1, which is involved in autophagy; NOD2, which is connected to the activation of the immune system by peptidoglycans; and CLEC7A, linked with the recognition of fungi by DCs have been shown to predispose to IBD (105). Dysbiosis controls the pathogenesis of IBD, affecting over one million people in the United States and one-quarter million in the UK (106). The IBD pathogenesis involves the bacterial adherence to the gut mucosa and invasion into mucosal epithelial cells leading to the occurrence of an inflammatory response, mediated by the production of TNF-α by monocytes/macrophages. This chronic bowel inflammation affects the epithelial cell tolerance to intestinal bacteria leading to changes in intestinal microbiota composition with an increase in aerobic bacteria accompanied by a significant decrease in the fecal levels of butyric and propionic acid in IBD patients. However, despite the generally accepted involvement of LPS in triggering an inflammatory effect (2), the main species adhering to the mucosa surrounding the colon mucus layer are Bifidobacterium spp. and Clostridium coccoides, suggesting that IBD is not triggered by a microbial species, but by an unbalanced microbiota. The hydrogen peroxide-producing colonic bacteria have been also suggested as causative agents of IBD in young adults (107). The studies performed on a mouse model of colitis (dextran sodium sulfate-induced colitis) showed that the introduction of anaerobic, noncultivatable segmented filamentous bacteria stimulates Th17 development, while commensals such as Bacteroides fragilis or Clostridium species, facilitate the differentiation of regulatory T-cell and IL-10 production in the gut. In most cases of spontaneous colitis models, including the IL-10−/− mouse, antibiotics or a germ-free state have been shown to prevent the development of colitis (24). The presence of Gram-positive bacteria, such as Lachnospirillaceae seems to be necessary for the infiltration of colitogenic macropahages and monocytes into the colon through induction of C–C chemokine receptor type-2 ligands, as revealed by the decrease of inflammatory reaction in mice treated with vancomycin (24). In humans, Faecalibacterium prausnitzii is one of the most abundant colonic bacteria found within the fecal mass but is also present in the adjacent mucosa, representing 5–20% of the total fecal microbiota of healthy individuals. Low counts of Faecalibacterium prausnitzii have been linked to several pathological disorders including Crohn’s disease (108).
Allergic diseases affect more than half a billion people worldwide. The development of allergy is clearly associated with some genetic and molecular factors but environmental factors including the gut microbiota are also involved. Indeed, reduced microbial diversity in infancy was correlated with an increased risk for allergies later in life. The commensal microbiota was reported to provide protection against allergic airway inflammation and food allergy (109) since mice treated with antibiotics and germ-free mice developed an exacerbated disease. TLR2- or TLR4-deficient mice develop pulmonary damage after chronic intake of a high-fat diet, a feature that can be transmitted to wild-type mice by fecal transplantation (110).
Recent data proved that lower prevalence of bacteria such as Akkermansia, Faecalibacterium, and Bifidobacterium, along with higher abundance of fungi such as Rhodotorula and Candida in neonates may lead to allergy susceptibility by modulating T-cell differentiation (48).
Systemic lupus erythematosus is a systemic autoimmune disease with unknown etiology characterized by the presence of hyperactive and aberrant antibody response to nuclear and cytoplasmic antigens (111). The dysbiosys observed in SLE is characterized by an increase of the Bacteroides phyla and a decrease in the Firmicutes (112). Despite the fact that the role of microbiota in the development of SLE is poorly understood, it is suggested that the dysbiosis observed in the SLE patients could be related to this disease. In line with this, mouse models of lupus exhibited an accelerated development of the disease that was linked to increased levels of Lachnospiraceae and low levels of Lactobacillaceae (113).
Skin autoimmune diseases have also been linked to microbiome shifts. For instance, a recent study by Scher et al. compared the composition of gut microbiota in patients with psoriatic arthritis or with psoriasis to that of healthy controls (114). The gut microbiota signature of psoriatic arthritis and psoriasis groups exhibited decreased bacterial diversity and a reduced relative abundance of Ruminococcus, Pseudobutyrivibrio, and Akkermansia. Importantly, the microbiota profile of psoriatic arthritis was similar to that of IBD patients, therefore suggesting a link between the gut microbiota and this skin disease (114). In case of scleroderma, most patients suffer from GIT symptoms that may be due to changes in intestinal microbiota composition. Recently, Volkmann et al. (115) revealed that Firmicutes were highly abundant in systemic sclerosis patients compared to healthy controls, whereas Bacteroidetes were lower in one of the cohorts, compared to healthy controls (115).
In the last decade, more studies proved that the gut microbiota has an impact on brain development and function. The studies compared germ-free and conventional laboratory rodents and the results suggest that the absence of microbiota alters anxiety-like behavior and also enhances the hypothalamic pituitary adrenal system stress reactivity. This abnormal behavior observed in germ-free animals was eradicated if the intestinal microbiota was restored in early life but not in the adulthood stage, suggesting the existence of a critical period of time for microbiota imprinting on stress responsiveness. The mechanism of action is not completely understood. Neuroactive bacterial metabolites are transported through the bloodstream to the brain and stimulate entero-endocrine cells or the vagus nerve, or modulate the immune system and, subsequently the inflammatory status, proving that dysbiosis could impact anxiety-related disorders of the treatment of anxiety-prone rodents with antibiotics or probiotics exhibited an anxiolytic-like activity (116). For example, there are studies involving the gut microbiota in host cognition or Alzheimer’s disease (AD)-related pathogenesis. Species from gut microbiota can generate large amounts of amyloids and LPS, which may modulate various signaling pathways and the synthesis of pro-inflammatory cytokines involved in AD pathogenesis. Furthermore, imbalances in the gut microbiota can lead to inflammation that is associated with AD (117). Microbial dysbiosis was also seen in the gut of multiple sclerosis (MS) patients and significant differences in microbiota composition between patients with MS and healthy controls were observed. Patients with active disease exhibited reduced species richness, whereas the microbiota of patients in remission was similar to that of the healthy controls. Comparing treated and untreated MS patients, certain genera such as Sutterella (Proteobacteria) and Prevotella (Bacteroidetes) were found to be reduced in untreated patients but restored after treatment. However, in treated patients Sarcina spp. was also reduced, a fact that proves the potential of MS therapies to alter the gut microbiome. As in case of MS, neuromyelitis optica (NMO) is driven by pathogenic Th17 cells reactive against self-proteins, in this case aquaporin-4 (AQP4) which is a channel protein transporting water across cell membranes. AQP4 is expressed by brain astrocytes and shows sequence homology with an ATP-binding cassette transporter permease from Clostridium perfringens. AQP4-specific T cells cross-react with Clostridium perfringens. Significant compositional differences were observed between the microbiome of patients with NMO compared to healthy controls: Clostridium perfringens and Fibrobacteres were enriched in NMO patients and together may have an impact on disease progression (118).
Chronic inflammation could drive carcinogenesis (e.g., colorectal cancer in patients with untreated IBD, hepatocellular carcinomas following chronic hepatitis). Through epithelial injury and inflammation, chronic infections (viruses, Helicobacter pylori and other Helicobacter spp., Bacteroides fragilis, Bacteroides vulgatus, Escherichia coli, Citrobacter rodentium, Citrobacter freundii, and protozoa) are linked to carcinogenesis with approximately 18% of the worldwide cancer burden (119). However, cancer, as well as other diseases, is not attributable to a single pathogen but to overall microbiome changes. Dysbiosis of gut microbiota leads to an increase in bacterial populations that stimulate tumorigenesis and the loss of protective ones. Inflammation might augment community-level alterations in the microbiota and aid the bacterial translocation into the neoplastic tissue, which in turn promotes the expression of inflammatory cytokines subsequently leading to tumor growth. The colonic microbiota may also promote colorectal cancer by stimulating exaggerated immune responses (i.e., via Th17 cells) (120). The dysbiosis caused by a deficiency of the NLRP6 inflammasome promotes cancer development via IL-6-induced epithelial proliferation (121, 122).
Oral cancer, particularly oral squamous cell carcinoma (OSCC) which evolves from the lining mucosae of the lips and the mouth is a multifactorial disease caused environmental factors (tobacco, human papillomavirus, and alcohol consumption) and host genetics (123). The microbiota shifts linked to OSCC have been analyzed in several studies. The culture-based analysis of surface swabs revealed that the levels of Fusobacterium spp. and Porphyromonas spp. were significantly higher in the OSCC tissue compared with the adjacent healthy mucosa (124). Subsequently, it was reported that sections of gingival squamous cell carcinoma harbored higher levels of Porphyromonas ginigivalis (125). A study targeting the differences in bacterial counts in 45 OSCC samples and 229 healthy controls revealed that cancer samples exhibited elevated Capnocytophaga gingivalis, Streptococcus mitis, and Prevotella melaninogenica (126). Another bacteria, Streptococcus anginosus has been linked to all head and neck squamous cell carcinoma, including OSCC (127). Subsequent studies by Sasaki et al. and Morita et al. detected Streptococcus anginosus in 45 and 13% OSCC samples, respectively (128, 129). Importantly, recent studies have revealed that Streptococcus anginosus is found in non-tumorous oral tissue even in higher levels compared with tumoral tissue, thus implying that Streptococcus anginosus is a normal inhabitant of the oral microbiota (130). Hooper et al. used culture methods as well as 16S rRNA Sanger sequencing to isolate 108 bacterial species from within the tissue of OSCC biopsies including Ralstonia insidiosa, Fusobacterium naviforme, Peptostreptococcus micros, Clavibacter michiganensis subspp. tessellarius, Fusobacterium naviforme, Micrococcus luteus, Prevotella melaninogenica, Staplylococcus aureus, Exiguobacterium oxidotolerans, and Veillonella parvula (131). These studies revealed that bacteria have tumor specificity, since several species were found in either the non-cancerous or cancerous sites. Pushalkar et al. analyzed 10 specimens of OSCC by 16S rRNA Sanger sequencing and detected 35 novel species. Nevertheless, this study identified in the tumors a totally different panel of bacteria including Eubacterium infirmum, Eubacterium brachy, Gemella haemolysans, Streptococcus gordonii, Peptostreptococcus stomatis, Gemella morbillorum, Streptococcus parasanguinis, Johnsonella ignava, Streptococcus salivarius, and Gemella sanguinis (130). These differences were likely caused by the fact that conventional culture methods and Sanger sequencing have low depth of analysis; thus, these techniques cannot guarantee robust identification of possibly relevant low abundant species. The advent of next-generation sequencing (NGS) has overcome this caveat. Several studies have used NGS to investigate the OSCC-associated microbiome. Pushalkar et al. analyzed salivary samples from three OSCC cases and two healthy controls and identified 860 OTUs among which 244 were exclusively present in the OSCC. Thus, the genera Gemella, Peptostreptococcus, Porphyromonas, Micromonas, Streptococcus, Rothia, and Lactobacillus had a higher abundance in the OSCC samples, while Capnocytophaga, Leptotrichia, Actinobacillus, Oribacterium Prevotella, and Neisseria were prevalent in samples without cancer (132). Schmidt et al. investigated swabs of lesion surface and normal mucosa from 8 pre-cancer cases, 18 OSCC samples, and 9 cancer free controls and observed that the Bacteroidetes phylum was significantly enriched in cancerous and normal tissues of OSCC patients compared to pre-cancer and healthy controls. This suggests that elevated colonization with bacteria from this phylum may be considered a possible biomarker for OSCC risk. In addition, tumor samples were associated with significantly higher levels of Fusobacterium and reduced abundance of Rothia and Streptococcus (133). A subsequent study by Al-hebshi et al. employed additional bioinformatics analysis and showed that OSCC samples contained 228 bacterial species, among which B. fragilis (134). Recently, Guerrero-Preston et al. compared the saliva microbiota in DNA isolated from Oropharyngeal (OPSCC), Oral OCSCC patients, and normal epithelium controls. The authors characterized the HNSCC saliva microbiota before and after surgical resection and revealed a predominance of Bacteroidetes, Proteobacteria, and Firmicutes with low abundance of Actinobacteria and Fusobacteria before surgery. The most enriched genera were Veillonella, Haemophilus, Streptococcus, Lactobacillus, and Prevotella with lower levels of Neisseraceae and Citrobacter. HNSCC patients exhibited a significant loss in microbiota diversity and richness (135). A novel study by Zhao et al. revealed that a group of periodontitis-correlated taxa, including Dialister, Fusobacterium, Peptococcus, Filifactor, Catonella, Parvimonas, and Peptostreptococcus was more abundant in OSCC samples (136).
Gastric cancer is one of the most common malignancies worldwide, gastrointestinal malignancies being responsible for about one-third of global cancer (137). The pathogenesis of this disease is a multi-stage process of affected by multiple factors still not clearly elucidated. Environmental factors, Helicobacter pylori infection and genetic factors are all involved in gastric cancer pathogenesis. Helicobacter pylori are closely linked to gastric cancer and whether other intragastric microbes participate in gastric cancer development requires further investigation (138). Emerging evidence suggests that other microorganisms exhibit a role in the pathophysiology of gastric cancer. This so called non-Helicobacter pylori bacteria that are enriched in a hypoacidic environment could potentially trigger carcinogenesis via different mechanisms, such as producing toxic metabolites, promoting inflammation, modifying stem cell dynamics, and stimulating cell proliferation (139). A recent study of the stomach microbiota in patients with gastric cancer revealed similarities with the stomach microbiota of patients with dyspepsia and a normal gastric mucosa (140). An analysis performed on two human populations with high and low gastric cancer risk in Columbia revealed two significantly more abundant OTUs, Veillonella spp. and Leptotrichia wadei in the high-risk area and 16 OTUs, including Staphylococcus spp. were more frequent in the low-incidence region (139). In addition, Ferreira et al. showed that gastric carcinoma is associated with a significant decrease in Helicobacter, and an increase in Lactobacillus, Citrobacter, Achromobacter, Clostridium, and Rhodococcus genera abundance. Interestingly, Phyllobacterium, a bacteria commonly found in plant roots, was enriched in gastric carcinoma samples (141).
Colorectal carcinogenesis is caused by a combination of host- and microbiota-dependent mechanisms. In this equation, the most common environmental factors are lifestyle choices and diet. Unhealthy diets high in fat, red meat, alcohol, and low in fiber are associated with an increased risk of adenomas and CRC (142). In addition, smoking, gender, ethnicity, and lifestyle, obesity all impact CRC development (143, 144). Certain bacteria promote carcinogenesis directly via the secretion of substances that lead to DNA damage. Several outstanding examples include the release of reactive oxygen species by Enterococcus faecalis, the excessive production of nitric oxide from immune cells triggered by Helicobacter hepaticus, as well as enterotoxin secretion by enterotoxigenic Bacteroides fragilis (ETBF), which can activate the c-MYC oncogene. Also, the ETBF fragilysin toxin activates signaling pathways such as Wnt/β-catenin and NF-κB to induce excessive cell proliferation and inflammation (145). In addition, BFT could potentially generate a multi-step pro-tumorigenic signaling requiring NF-κB, IL-17R, and STAT3 in colonic epithelial cells leading to myeloid-cell-dependent distal colon tumorigenesis (146). Other species including Parvimonas micra, Solobacterium moorei, and Peptostreptococcus anaerobius were also significantly correlated with CRC. In a recent study by Tsoi et al., P. anaerobius was significantly elevated in biopsies from tumor lesions as well as in stool samples from CRC patients compared to healthy controls (147). The accumulation of hydrogen sulfide generated by sulfate reducing bacteria in response to a diet rich in meat, promotes chronic inflammation and the release of mutagenic compounds or genotoxins (such as CDTs produced by Salmonella enterica serovar typhi and Escherichia coli and colibactin, a secondary metabolite, a hybrid polyketide/nonribosomal peptide) (148, 149) (Figure 1).
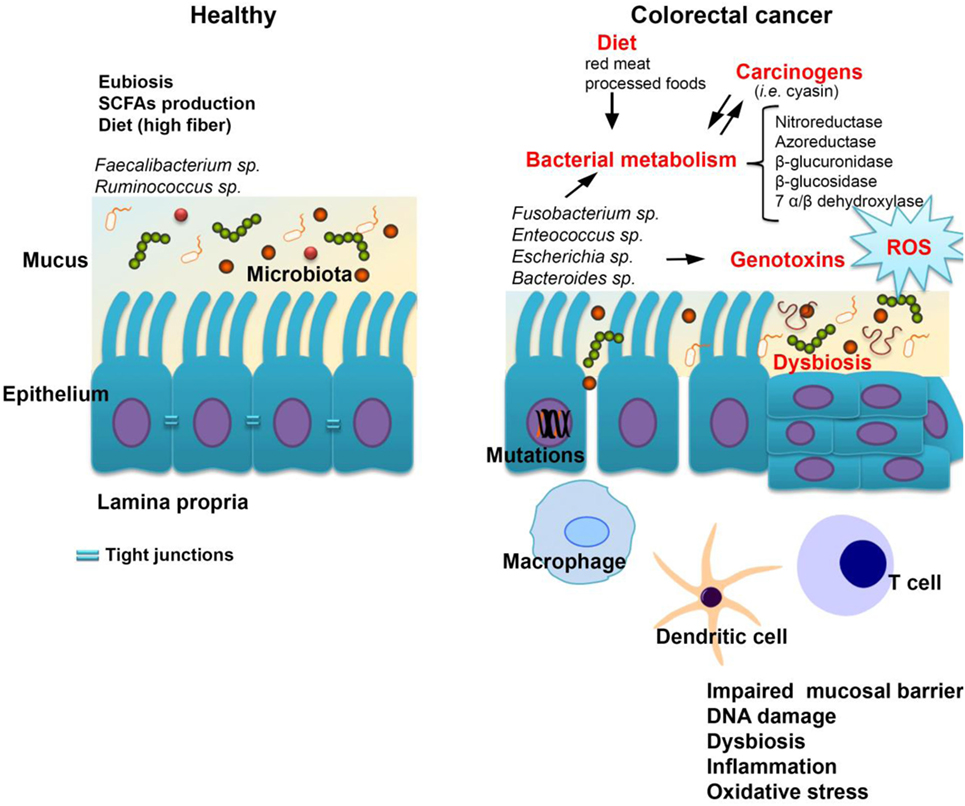
Figure 1. The host-microbiome interplay in colorectal cancer. Some bacterial species including Fusobacterium spp., Enterococcus spp., Escherichia coli, and Bacteroides spp. are most commonly associated with colorectal cancer. Changes in microbiota composition (dysbiosis) impair the gut barrier function of epithelial tight junctions and the mucus layer. Consequently, this increases the exposure of the epithelium to bacteria and their metabolites which may have carcinogenic potential. Bacterial translocation also leads to increased inflammation associated with the production of procarcinogens or toxic chemicals such as reactive oxygen species (ROS), bacterial genotoxins (colibactin), and hydrogen sulfide (H2S). Altogether, these changes trigger oxidative stress which leads to DNA damage. A series of pathways independent of the inflammatory response, but related to bacterial enzymes which could generate carcinogenic compounds, are cited in the literature by which the gastrointestinal microbiome may be involved in the genesis and evolution of neoplastic processes.
In experimental models of CRC precancerous lesions were characterized by elevated levels of Allobaculum spp. and Ruminococcus obeum. These data suggest that modifications in the composition of adherent microbial populations may exhibit a role in adenoma development (150), by inducing a pro-inflammatory response (e.g., Fusobacterium nucleatum through its virulence factor FadA) (151). Novel studies identified additional virulence factors in Fusobacterium nucleatum (Fap2, LPS) that may act as triggers in the evolution from healthy epithelial cells to tumor cells (152).
It seems that microbial status may have an effect on tumorigenesis without direct influence on the inflammatory status. In line with this, up to 80% of IBD patients with long-standing disease (<30 years) do not develop colitis-associated CRC, hence highlighting that inflammation is not enough to trigger cancer (153).
It has been suggested that a higher microbial density in a sterile organ is correlated with a higher incidence of cancer, due to exposure to microbe-associated molecular patterns (MAMPs) and bacterial metabolites, while a decreasing microbiota in mice by antibiotics reduced the liver and colon cancer frequency (23).
However, microbiota can also exert anti-tumor effects in patients with sarcomas, by converting the tumor tolerance in an anti-tumor immune response, involving TLR agonists and NLR (154).
Microbiota can synthesize large amounts of folic acid, which plays a crucial role in regulating DNA synthesis, and other antioxidant substances such as selenium, vitamin C, and vitamin A, which help prevent DNA lesions. Some anaerobic species (Clostridium orbiscindens and Eubacterium ramulus) are involved in the process of colon and breast cancerogenesis, by degradation of some food glicozides and flavonoids with protective, antineoplasic effect (155). Methylazoxymethanol, a bioactive carcinogenic compound, the downstream active metabolite of plant glycoside cyasin is generated under the action of the bacterial β-glucosidase enzyme (156). This could explain the anti-tumoral effect of probiotic lactic acid bacteria including Lactobacillus acidophilus and Lactobacillus casei which express a decreased activity of azoreductase, β-glucuronidase, and nitroreductase (157). Butyrate producing bacteria also have anti-tumor effects in colon cancer cells by promoting cancerous cells apoptosis (2, 158). Butyrate induced apoptosis in colon cancer cell lines in vitro is correlated with its role as a histone deacetylase inhibitor, achieved through the modulation of different molecular pathways (159–162).
Since the gut microbiota is an important modulator of host immunity, it is natural to think that it could have an impact on the response to cancer therapy. As initially revealed by animal studies and further confirmed by human studies, the gut microbiome is an important factor in mediating the host response and toxicity to various anticancer therapies (e.g., immunotherapy with CpG oligonucleotides or chemotherapy). Commensal Bifidobacterium spp. enhances the tumor control in a similar way to programmed cell death protein 1 ligand 1-specific antibody therapy, through the augmentation of dendritic-cell function (163).
Patients with hematologic malignancies undergoing hematopoietic stem cell transplant (HSCT) are often treated with broad-spectrum antibiotics, immunosuppressants, and even total body irradiation thus dysbiosis is fairly common in these individuals (164, 165). After HSCT, dysbiosis is characterized by a loss of bacterial diversity and stability, reduced levels of Faecalibacterium and Ruminococcus and an enrichment of Enterococcus, Streptococcus, and Proteobacteria (166).
An improved overall survival after HSCT treatment was correlated with certain microbiota signatures characterized by higher levels of the genus Blautia (167). In addition, a higher density of Eubacterium limosum was linked to reduced relapse risk (168).
Conventional chemotherapy is also interacting with the immune system and the microbiota. For instance, during treatment with cyclophosphamide, the translocation of Enterococcus hirae and Lactobacillus johnsonii into MLNs can facilitate Th17 and Th1 responses. In addition, the effects of cyclophosphamide as well as other chemotherapy drugs were absent in antibiotic-treated or germ-free mice (169, 170). The impact of the microbiota was analyzed in case of treatment with immune checkpoint inhibitors targeting the immunomodulatory molecules found on the surface of T cells. Recent studies try to find means to control therapeutic resistance by identifying the predictors of the host response to immune checkpoint blockade (171, 172). The microbiota may impact the response to immune checkpoint inhibitors by targeting the programmed death receptor 1 (PD-1) and the cytotoxic T lymphocyte antigen 4 (CTLA-4) (163, 173).
Gopalakrishnan et al. reported that patients responsive to anti-PD-1 therapy harbored a higher microbial diversity characterized by higher Ruminococcaceae, Faecalibacterium, and Clostridiales relative abundance (174, 175). By contrast, nonresponders exhibited significantly lower bacterial diversity and a higher levels of Bacteroidales. In addition, the comparison between the composition of the gut microbiota with the immune profile in the tumor microniche showed that patients hosting a favorable gut microbiota exhibited enhanced antigen processing and presentation and elevated expression of cytolytic T cell markers compared to patients with unfavorable dysbiotic microbiota (166).
Toxicity scores were reported to be improved in anti-CTLA-4-treated mice after oral gavage with Bacteroides fragilis and Burckholderia cepacia (173). The impact of the microbiota on toxicity was also investigated in human cohorts (176–178). Anti-CTLA-4-treated melanoma patients without colitis showed enhanced levels of Bacteroidetes as opposed to those who did develop colitis (177). Additional studies revealed that patients with low abundance of Bacteroidetes and elevated Faecalibacterium prausnitzii and other related Firmicutes harbored an elevated risk of colitis in anti-CTLA-4 therapy (176, 178). Taxa within the Bacteroidales order (Bacteroidetes phylum) were linked to lack of responsiveness to immune checkpoint blockade, whereas their elevated abundance was correlated with a lower toxicity incidence (166, 174, 176–178). Nevertheless, at lower taxonomic levels several taxa within Firmicutes such as Roseburia and Streptococcus were linked with a lack of response (175, 178) whereas some taxa within Bacteroidetes (Porphyromonas and Alistipes) were correlated with response (164, 174, 175). In addition, other taxa were correlated with response (such as Bifidobacterium longum, Collinsella aerofaciens, Akkermansia muciniphila, and Bifidobacterium adolescentis) and non-response (such as Gardnerella vaginalis and Actinomyces viscosus) (164, 166, 174, 175, 178).
These studies might open up an appealing route of investigation for cancer prevention as well as to develop cancer therapeutics through microbiome manipulation.
Time series data reveal that microbiota composition is relatively stable in case of healthy adult individuals over time. Two of the most important factors triggering changes in the microbiota are the dietary intake and the overuse of antimicrobials (179).
A recent study by Rothschild et al. investigated the interplay between environmental factors, host genetics and gut microbiota, and revealed that the gut microbiota community structure was mostly influenced by environmental factors rather than single nucleotide polymorphisms or genetic ancestry. In addition, only 1.9% of the gut microbiome is estimated to be heritable, whereas more than 20% of the microbiome β-diversity is shaped by the environment (diet, lifestyle, etc.) (180).
It was demonstrated that shifting to a high-sugar high-fat, “Western” diet from a plant polysaccharide-rich, low-fat or from a low-fiber/high-fat diet to a high-fiber/low-fat diet can change the mouse microbiota within a day (181) the monitoring of temporal dynamics of microbial communities within an individual through time could predict the disease states and help develop strategies to correct dysbiosis. Diet is also correlated with the gut enterotype, as proven by the fact that individuals eating a diet high in animal fat exhibit a Bacteroides-enterotype, whereas a carbohydrate-rich diet leads to a Prevotella-dominated enterotype (182). Several studies revealed that a diet rich in non-digestible carbohydrates is enriched in probiotic bacteria including bifidobacteria and lactic acid bacteria. In addition, diets that were rich in wheat bran and whole grain promoted an increase in intestinal bifidobacteria and lactobacilli (183, 184). A significant influence is held by non-digestible polysaccharides, but microbiota-accessible carbohydrates (MAC), which are not present in adequate amounts in the Western diet, which generally is based on heavily processed foods, rich in sugar, protein, fat, and different additives, and very low in fibers and micronutrients. For example, individuals on the Western diet only consume half of the recommended daily intake of dietary fiber. Recent reports demonstrate that the prevalence of these non-communicable diseases has increased dramatically in Western lifestyle countries, some of them doubling (e.g., asthma and MS) or even tripling (Crohn’s disease) in different Western European countries (155). In a multigenerational study performed on mice, the consumption of a Western-style diet aggravated the loss of microbiota diversity with a predicted loss in diversity of glycoside hydrolases, compared with a diet that was rich in MACs (155). A large study performed on a cohort of 168,999 women and 219,123 men showed that, in both genders, dietary fiber intake was significantly linked to a 22% decrease in mortality rates from infectious, cardiovascular, and respiratory diseases (185). Low-MAC content food, such as food desserts were associated with an increased incidence of asthma in children (155). Disinfectants and antibiotics generally induce a long-term decrease in bacterial diversity and, in some individuals, can affect certain particular taxa, which do not recover even months after treatment. The affected microbiota will have a decreased colonization resistance, hence allowing foreign microbes to cause permanent changes in the microbiome and varying states of disease. The outgrowth of opportunistic pathogens (pathobionts) may favor their translocation from mucosa to the extraintestinal compartment where they can initiate infectious processes (186). The majority of the antibiotics available at this moment have a broad spectrum of activity and they act not only on pathogens but also on beneficial members of the gut microbiota (187). It was reported that overexposure to antibiotics may promote the development of antibiotic resistance genes (ARGs)—studied by genomic and metagenomic approaches in the commensal microbiota and to their potential transfer to pathogenic species; they enrich phage-encoded genes and enhance the ARGs exchange between phages and bacteria (188). The repeated use of antibiotics in humans augments the reservoir of ARGs in the host microbiome, as revealed by the increased incidence of multidrug resistant (MDR) E. coli or vancomycin resistant enterococci (VRE) in children receiving antibiotics for respiratory infections (11). At the same time, it was revealed that limited exposure to antibiotics select the resistant pathogens (189). Recently it was shown that antibiotics change the functions of the normal gut microbiota after destroying its structure (190) by altering the diversity of microbial associated molecular patterns. After antibiotic treatment, the gut microbiota manifests resiliency and is able to present a similar composition after a long period (191). ARGs can be horizontally transferred through transformation, conjugation, and phage transduction, the mobilization promoted by mobile genetic elements such insertion sequences, integrons, and transposons (192). Several researchers have demonstrated that ARGs are present not only the microbiota of adults also in that of children and infants (193). Several antibiotic classes lead to different patterns of microbiota alterations because of their different spectrum of activity. For example, Gibson et al. demonstrated that ticarcillin-clavulanate, meropenem, and cefotaxime treatments were correlated with decreased microbiota species richness in children (194). It was also demonstrated that β-lactam combinations of beta-lactamase inhibitors and penicillins or cephalosporins determined an increase in Proteobacteria (specially Enterobacteriaceae) and Bacteroidetes and a decrease in Firmicutes as well as to a reduced microbial richness (195). By contrast, it was demonstrated that penicillin V and amoxicillin did not correlate to significant microbiota changes (196). Broad-spectrum antibiotics, like clindamycin, which are active against anaerobic species (197) were demonstrated to decrease the abundance of lactobacilli and bifidobacteria (198); macrolides have been shown to increase Bacteroides spp. and Proteobacteria levels (196) and decrease the abundance of Actinobacteria and Firmicutes taxa. Ciprofloxacin has been shown to diminish the Gram-negative facultative anaerobes, increase the abundance of the Gram-positive aerobes, and reduce the microbiota diversity; levofloxacin decreased the number of Gram-positive anaerobic microbes, including Bifidobacteria (197). Jakobsson et al. reported that clarithromycin decreased the number of Actinobacteria intrinsically resistant to metronidazole (199). Nitrofurantoin (active against Gram-negative and Gram-positive species) lead to a temporary increase in the number of Bifidobacteria in the gut microbiota of patients treated for uncomplicated urinary tract infections (200). Along with the multiple benefits of the uses of antibiotics in public health, agriculture, and medicine, they induce dysbiosis with negative effects on health which can remain for long periods of time after cessation of treatment. Antibiotic treatment increases the opportunities for horizontal gene transfer with crucial implications for the emergence of resistance.
Lactic acid-producing bacteria including Enterococcus spp., Lactobacillus spp., Bifidobacterium spp., but also several Bacillus spp., E. coli, and Streptococcus spp. strains are considered the most beneficial microbial species isolated from the human microbiota and are proposed as probiotics. Probiotics can be used to correct the antibiotics-induced disbiosis specifically in critically ill patients. The intrinsic resistance of different probiotic strains to current antimicrobials facilitates their concomitant use with specific antibacterial treatments. For example, Lactobacillus rhamnosus GG is constitutively resistant to metronidazole and vancomycin and is routinely used for the treatment of pseudomembranous colitis and antibiotic-associated diarrhea, while Lactobacillus fermentum ME-3 (DSM14241) can be used in association with ofloxacin for the treatment of Salmonella enterica serovar typhimurium infections (201, 202). Manges et al. employed a comparative metagenomic approach and reported that the development of CDI in humans is linked to an increase in Firmicutes and a depletion of Bacteroidetes phylum. It was shown that human probiotic infusion corrects the dysbiosis in CDI by replacing the depleted bacterial species and re-establishing colonization resistance (203, 204). Also, in their study, Khodaii et al. evaluated the effects of cell-free supernatants of cultures belonging to 16 strains of lactobacilli and bifidobacteria on the invasive capability of enteroinvasive Escherichia coli (EIEC) strain. Thus, the treatment of the pathogen with cell-free supernatants prevented the EIEC strains invasion of CaCo-2 and T84 cells. They suggested that probiotics prevent invasion of EIEC into the small and large intestine not by competing with adhesins receptors, but by producing some metabolites that changes the environment, cell barrier, or gene expression (205). Lactic acid bacteria produce bacteriocins in a cell density-dependent manner and utilize a molecular QS regulatory mechanism. Bacteriocin production is an inducible mechanism and requires the extracellular accumulation of certain chemical messengers (AI1 and AI2) (206). So, for specific purposes either probiotic products (live cells/dead) or probiotic soluble molecules (antimicrobials, QSI molecules) can be used. Not only microorganisms are producing QS molecules and QSIs, but also plants are able to interfere with bacterial communication and processes controlled by QS mechanism, as an expression of their antiinfectious defense mechanisms and these inhibitory molecules with demonstrated in vitro activity and can be used alone or in combinations as an alternative/complementary antiinfectious strategy (60). Probiotics confer health benefits through the modulation of pro- and anti-inflammatory responses. Studies reported that cell surface molecules of Lactobacillus strains exhibit TNF-α-inducing activities in macrophages via TLR2 signaling (179). In vitro and in vivo studies suggest that Propionibacterium species can be used as probiotics, with many potential health benefits, modulating gut microbiota composition and gut activities (207, 208). Dairy propionic bacteria can impact the gut microbiota by favoring the growth of symbiotic bacteria such as Bifidobacteria, or by inhibiting the in vitro adherence of some pathogens such as H. pylori (209), Salmonella enterica serovar Enteritidis and enteropathogenic Esherichia coli strains to different cell lines (210, 211). Also, clinical studies reported the beneficial outcome of combining dairy propionibacteria with other probiotic bacteria in order to modulate the host immune system (212). Thus, two probiotic bacteria, Propionibacterium freudenreichii spp. shermanii (PJS) and Lactobacillus rhamnosus GG (GG) were tested for immunomodulatory response in the mouse intestine, using fat-fed ApoE*3Leiden mice. It was shown that mice receiving PJS and GG harbored significantly lower intestinal mast cells compared to the control. Also, GG increased intestinal IL-10, whereas PJS lowered the intestinal immunoreactivity of TNF-α. Probiotics represent also a very efficient preventive therapy against necrotizing enterocolitis (NEC), the potential mechanisms of this effect being experimentally demonstrated. Thus, oral administration of Bifidobacterium bifidum cells decreased the levels of ileal claudin-3 and occludin in neonatal rats with NEC (213). Also, bacteria-free conditioned media harvested from probiotics such as Bifidobacterium infantis, Lactobacillus plantarum, and Lactobacillus acidophilus administered in single or in multiple combinations, may confer protection against NEC, by their anti-inflammatory and cytoprotective properties, and by improving intestinal barrier function (214, 215). A new approach in the probiotic therapy is represented by the bioengineered probiotics. Probiotic strains can be employed as vehicles for expressing foreign genes. In line with this, Culligan et al. analyzed the advantages of recombinant probiotics in treating enteric infection (216) but such probiotic strains are regarded as genetically modified organisms, raising ethical issues related to their use (217).
The application of macroecological concepts to the gut microbiota indicates that the microbiota biodiversity can serve as an important measure of eubiosis status.
The easy access and possibility to modulate the microbiota makes it a good target for both establishing a link between certain patterns of gut microbiota and the physiological/pathological status. Due to the complexity and inter-individual differences of human microbiota, the identification of microbial colonization profiles specifically associated with certain disorders, as well as the characterization of microbial metabolic pathways related to health and disease state still remains a challenge.
The human microbiota interacts at multiple levels with the immune system and the alteration of this crosstalk could be involved in the pathophysiological mechanisms of the host and can further be exploited to develop clinical therapies for some immunological disorders, such as inflammatory and autoimmune diseases, allergies, cancer, dysbiosis, and opportunistic infections. This could lead to the development of potential biomarkers allowing to implement personalized healthcare strategies and to identify new tools for prevention, screening, and treatment.
However, the very diverse actors of these complex interactions (bacterial species, microbial products, host receptors, signaling molecules, and molecular pathways) as well as diet influence are still to be uncovered. Related to diet, it has long been seen as an adjuvant of medication, but recent research data are offering arguments for the use of food to efficiently modulate the microbiota and to develop microbiota-based interventional therapies or personalized diets, tailored in accordance with the host genetic background, microbiome, metabolome, as well as nutrient intake and habitual food consumption.
In perspective, we can target a homeostatic status of microbiota and host health, by designing microbiota-based therapeutics, but also by reducing antimicrobials consumption and assuring a diverse and balanced diet for the health of both host metabolism and its microbiota. In order to achieve this desideratum, an educational component, assuring the proper understanding of microbiota structure and roles in host health and disease, is absolutely needed to convince people about the necessity to introduce some long-term changes in their lifestyle, instead of turning to short time therapeutic interventions in emergency situations.
VL and GP drafted the manuscript. L-MD, GP, CC, IG, AH, AP and LP wrote the manuscript. GP made the figures; MC wrote and corrected the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This work was supported by the Research Grant PN III - P2-2.1-BG-2016-0369-116/2016, funded by the Executive Unit for Financing Higher Education, Research, Development and Innovation (UEFISCDI).
1. Neish AS. Microbes in gastrointestinal health and disease. Gastroenterology (2009) 136:65–80. doi:10.1053/j.gastro.2008.10.080
2. Sonnenburg JL, Bäckhed F. Diet–microbiota interactions as moderators of human metabolism. Nature (2016) 535:56–64. doi:10.1038/nature18846
3. Sekirov I, Russell SL, Antunes LC, Finlay BB. Gut microbiota in health and disease. Physiol Rev (2010) 90(3):859–904. doi:10.1152/physrev.00045.2009
4. Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature (2010) 464:59–65. doi:10.1038/nature08821
5. Hugon P, Dufour JC, Colson P, Fournier PE, Sallah K, Raoult D. A comprehensive repertoire of prokaryotic species identified in human beings. Lancet Infect Dis (2015) 15(10):1211–9. doi:10.1016/S1473-3099(15)00293-5
6. Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A (2007) 104(34):13780–5. doi:10.1073/pnas.0706625104
7. Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature (2012) 486:207–14. doi:10.1038/nature11234
8. Ardissone AN, de la Cruz DM, Davis-Richardson AG, Rechcigl KT, Li N, Drew JC, et al. Meconium microbiome analysis identifies bacteria correlated with premature birth. PLoS One (2014) 9(3):e90784. doi:10.1371/journal.pone.0090784
9. De Martino SJ, Mahoudeau I, Brettes JP, Piemont Y, Monteil H, Jaulhac B. Peripartum bacteremias due to Leptotrichia amnionii and Sneathia sanguinegens, Rare Causes of Fever during and after Delivery. J Clin Microbiol (2004) 42(12):5940–3. doi:10.1128/JCM.42.12.5940-5943.2004
10. Aagaard K, Ma J, Antony KM, Ganu R, Petrosino J, Versalovic J. The placenta harbors a unique microbiome. Sci Transl Med (2014) 6:237ra65. doi:10.1126/scitranslmed.3008599
11. Clemente JC, Ursell LK, Parfrey LW, Knight R. The impact of the gut microbiota on human health: an integrative view. Cell (2012) 148(6):1258–70. doi:10.1016/j.cell.2012.01.035
12. Lagier J, Armougom F, Million M, Hugon P, Pagnier I, Robert C, et al. Microbial culturomics: paradigm shift in the human gut microbiome study. Clin Microbiol Infect (2012) 18(12):1185–93. doi:10.1111/1469-0691.12023
13. Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A (2010) 107:11971–5. doi:10.1073/pnas.1002601107
14. Gordon JI, Dewey KG, Mills DA, Medzhitov RM. The human gut microbiota and undernutrition. Sci Transl Med (2012) 6(4):137. doi:10.1126/scitranslmed.3004347
15. Woof JM, Kerr MA. The function of immunoglobulin A in immunity. J Pathol (2006) 208(2):270–82. doi:10.1002/path.1877
16. Endesfelder D, Castell WZ, Ardissone A, Davis-Richardson AG, Achenbach P, Hagen M, et al. Compromised gut microbiota networks in children with anti-islet cell autoimmunity. Diabetes (2014) 63(6):2006–14. doi:10.2337/db13-1676
17. Marcobal A, Barboza M, Sonnenburg ED, Pudlo N, Martens EC, Desai P, et al. Bacteroides in the infant gut consume milk oligosaccharides via mucus-utilization pathways. Cell Host Microbe (2011) 10(5):507–14. doi:10.1016/j.chom.2011.10.007
18. Guaraldi F, Salvatori G. Effect of breast and formula feeding on gut microbiota shaping in newborns. Front Cell Infect Microbiol (2012) 2:94. doi:10.3389/fcimb.2012.00094
19. Philippe G. Metabolism of cholesterol and bile acids by the gut microbiota. Pathogens (2014) 3(1):14–24. doi:10.3390/pathogens3010014
20. Scheppach W. Effects of short chain fatty acids on gut morphology and function. Gut (1994) 35:S35–8. doi:10.1136/gut.35.1_Suppl.S35
21. Heerdt BG, Houston MA, Augenlicht LH. Short-chain fatty acid-initiated cell cycle arrest and apoptosis of colonic epithelial cells is linked to mitochondrial function. Cell Growth Differ (1997) 8:523–32.
22. Kabat AM, Srinivasan N, Maloy KJ. Modulation of immune development and function by intestinal microbiota. Trends Immunol (2014) 35(11):507–17. doi:10.1016/j.it.2014.07.010
23. Thaiss CA, Zmora N, Levy M, Elinav E. The microbiome and innate immunity. Nature (2016) 535:65–74. doi:10.1038/nature18847
24. Nakanishi Y, Sato T, Ohteki T. Commensal Gram-positive bacteria initiates colitis by inducing monocyte/macrophage mobilization. Mucosal Immunol (2015) 8:152–60. doi:10.1038/mi.2014.53
25. Francino MP. Early development of the gut microbiota and immune health. Pathogens (2014) 4:769–90. doi:10.3390/pathogens3030769
26. Shanahan F. Physiological basis for novel drug therapies to treat the inflammatory bowel diseases: I. Pathophysiological basis and prospects for probiotic therapy in inflammatory bowel disease. Am J Physiol Gastrointest Liver Physiol (2005) 288:G417–21. doi:10.1152/ajpgi.00421.2004
27. Thomas CM, Versalovic J. Probiotics-host communication modulation of signaling pathways in the intestine. Gut Microbes (2010) 1(3):1–16. doi:10.4161/gmic.1.3.11712
28. Belkaid Y. Role of the microbiota in immunity and inflammation. Cell (2014) 157(1):121–41. doi:10.1016/j.cell.2014.03.011
29. Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell (2004) 118:229–41. doi:10.1016/j.cell.2004.07.002
30. Elinav E, Strowig T, Kau AL, Henao-mejia J, Thaiss CA, Booth CJ, et al. NLRP6 inflammasome is a regulator of colonic microbial ecology and risk for colitis. Cell (2011) 145:745–57. doi:10.1016/j.cell.2011.04.022
31. Owaga E, Hsieh RH, Mugendi B, Masuku S, Shih CK, Chang JS. Th17 cells as potential probiotic therapeutic targets in inflammatory bowel diseases. Int J Mol Sci (2015) 16:20841–58. doi:10.3390/ijms160920841
32. Rossi M, Bot A. The Th17 cell population and the immune homeostasis of the gastrointestinal tract. Int Rev Immunol (2013) 32(5–6):471–4. doi:10.3109/08830185.2013.843983
33. Sonnenberg GF, Fouser LA, Artis D. Border patrol: regulation of immunity, inflammation and tissue homeostasis at barrier surfaces by IL-22. Nat Immunol (2011) 12(5):383–90. doi:10.1038/ni.2025
34. Weaver CT, Elson CO, Fouser LA, Kolls JK. The Th17 pathway and inflammatory diseases of the intestines, lungs, and skin. Annu Rev Pathol (2013) 8:477–512. doi:10.1146/annurev-pathol-011110-130318
35. Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature (2008) 453(7195):620–5. doi:10.1038/nature07008
36. Tanoue T, Atarashi K, Honda K. Development and maintenance of intestinal regulatory T cells. Nat Rev Immunol (2016) 16:295–309. doi:10.1038/nri.2016.36
37. Collins CB, Aherne CM, Kominsky D, McNamee EN, Lebsack MDP, Eltzschig H, et al. Retinoic acid attenuates ileitis by restoring the balance between T-helper 17 and T regulatory cells. Gastroenterology (2011) 141(5):1821–31. doi:10.1053/j.gastro.2011.05.049
38. Wu HJ, Wu E. The role of gut microbiota in immune homeostasis and autoimmunity. Gut Microbes (2012) 3(1):4–14. doi:10.4161/gmic.19320
39. Mantis NJ, Rol N, Corthésy B. Secretory IgA’s complex roles in immunity and mucosal homeostasis in the gut. Mucosal Immunol (2011) 4(6):603–11. doi:10.1038/mi.2011.41
40. Hooper LV, Stappenbeck TS, Hong CV, Gordon JI. Angiogenins: a new class of microbicidal proteins involved in innate immunity. Nat Immunol (2003) 4:269–73. doi:10.1038/ni888
41. Cash HL, Whitham CV, Behrendt CL, Hooper LV. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science (2006) 313(5790):1126–30. doi:10.1126/science.1127119
42. Salzman NH, Hung K, Haribhai D, Chu H, Karlsson-Sjoberg J, Amir E, et al. Enteric defensins are essential regulators of intestinal microbial ecology. Nat Immunol (2010) 11:76–83. doi:10.1038/ni.1825
43. Harris VC, Haak BW, van Hensbroek MB, Wiersinga WJ. The intestinal microbiome in infectious diseases: the clinical relevance of a rapidly emerging field. Open Forum Infect Dis (2017) 4(3):ofx144. doi:10.1093/ofid/ofx144
44. Louis P, Hold GL, Flint HJ. The gut microbiota, bacterial metabolites and colorectal cancer. Nat Rev Microbiol (2014) 12(10):661–72. doi:10.1038/nrmicro3344
45. LeBlanc JG, Milani C, de Giori GS, Sesma F, van Sinderen D, Ventura M. Bacteria as vitamin suppliers to their host: a gut microbiota perspective. Curr Opin Biotechnol (2013) 24(2):160–8. doi:10.1016/j.copbio.2012.08.005
46. Kosiewicz MM, Zirnheld AL, Alard P. Gut microbiota, immunity, and disease: a complex relationship. Front Microbiol (2011) 2:180. doi:10.3389/fmicb.2011.00180
47. Rolhion N, Chassaing B. When pathogenic bacteria meet the intestinal microbiota. Philos Trans R Soc Lond B Biol Sci (2016) 371(1707):20150504. doi:10.1098/rstb.2015.0504
48. Van Den Elsen LWJ, Poyntz HC, Weyrich LS, Young W, Forbes-Blom EE. Embracing the gut microbiota: the new frontier for inflammatory and infectious diseases. Clin Transl Immunology (2017) 6(1):e125–9. doi:10.1038/cti.2016.91
49. Khosravi A, Yáñez A, Price JG, Chow A, Merad M, Goodridge HS, et al. Gut microbiota promote hematopoiesis to control bacterial infection. Cell Host Microbe (2014) 15:374–81. doi:10.1016/j.chom.2014.02.006
50. Olsen I, Lambris JD. Porphyromonas gingivalis disturbs host–commensal homeostasis by changing complement function. J Oral Microbiol (2017) 9(1):1340085. doi:10.1080/20002297.2017.1340085
51. Wilks J, Lien E, Jacobson AN, Fischbach MA, Qureshi N, Chervonsky AV, et al. Mammalian lipopolysaccharide receptors incorporated into the retroviral envelope augment virus transmission. Cell Host Microbe (2015) 18(4):456–62. doi:10.1016/j.chom.2015.09.005
52. Hallstrom K, McCormick BA. Salmonella interaction with and passage through the intestinal mucosa: through the lens of the organism. Front Microbiol (2011) 2:88. doi:10.3389/fmicb.2011.00088
53. Carvalho FA, Koren O, Goodrich JK, Johansson MEV, Aitken JD, Su Y, et al. Transient Inability to manage proteobacteria promotes chronic gut inflammation in TLR5-deficient mice. Cell Host Microbe (2012) 12(2):139–52. doi:10.1016/j.chom.2012.07.004
54. Vayssier-Taussat M, Albina E, Citti C, Cosson J-F, Jacques M-A, Lebrun M-H, et al. Shifting the paradigm from pathogens to pathobiome: new concepts in the light of meta-omics. Front Cell Infect Microbiol (2014) 4:29. doi:10.3389/fcimb.2014.00029
55. Inoue T, NaInoue T, Nakayama J, Moriya K, Kawaratani H, Momoda R, et al. Gut dysbiosis associated with hepatitis C virus infection. Clin Infect Dis (2018). doi:10.1093/cid/ciy205
56. Lievin-Le Moal V, Servin Alain L. The front line of enteric host defense against unwelcome intrusion of harmful microorganisms: mucins, antimicrobial peptides, and microbiota. Clin Microbiol Rev (2006) 19(2):315–37. doi:10.1128/CMR.19.2.315-337.2006
57. Schenk Frick JS, Quitadamo K, Kahl M, Köberle F, Bohn M, Aepfelbacher E, et al. Lactobacillus fermentum attenuates the proinflammatory effect of Yersinia enterocolitica on human epithelial cells. Inflamm Bowel Dis (2007) 13(1):83–90. doi:10.1002/ibd.20009
58. Ruiz PA, Hoffmann M, Szcesny S, Blaut M, Haller D. Innate mechanisms for Bifidobacterium lactisto activate transient pro-inflammatory host responses in intestinal epithelial cells after the colonization of germ-free rats. Immunology (2005) 115:441–50. doi:10.1111/j.1365-2567.2005.02176.x
59. Jayaraman A, Wood TK. Bacterial quorum sensing: signals, circuits, and implications for biofilms and disease. Annu Rev Biomed Eng (2008) 10:145–67. doi:10.1146/annurev.bioeng.10.061807.160536
60. Lazar V. Quorum sensing in biofilms – how to destroy the bacterial citadels or their cohesion/power? Anaerobe (2011) 17(6):280–5. doi:10.1016/j.anaerobe.2011.03.023
61. Lazăr V, Chifiriuc MC. Medical significance and new therapeutical strategies for biofilm associated infections. Roum Arch Microbiol Immunol (2010) 69(3):125–38.
62. Hentzer M. Pharmacological inhibition of quorum sensing for the treatment of chronic bacterial infections. J Clin Invest (2003) 112:1300–7. doi:10.1172/JCI20074
63. Thomas GL, Bohner CM, Williams HE, Walsh CM, Ladlow M, Welch M, et al. Immunomodulatory effects of Pseudomonas aeruginosa quorum sensing smallmolecule probes on mammalian macrophages. Mol Biosyst (2006) 2:132–7. doi:10.1039/B517248A
64. Davey ME, O’toole GA. Microbial biofilms: from ecology to molecular genetics. Microbiol Mol Biol Rev (2000) 64:847–67. doi:10.1128/MMBR.64.4.847-867.2000
65. Tlaskalova-Hogenova H, Tuckova L, Mestecky J, Kolinska J, Rossman P, Stepankova R, et al. Interaction of mucosal microbiota with the innate immune system. Scand J Immunol (2005) 62:106–13. doi:10.1111/j.1365-3083.2005.01618.x
66. Rutherford ST, Bassler BL. Bacterial quorum sensing: its role in virulence and possibilities for its control. Cold Spring Harb Perspect Med (2012) 2(11):a012427. doi:10.1101/cshperspect.a012427
67. Grabiner MA, Fu Z, Wu T, Barry KC, Schwarzer C, Machen TE. Pseudomonas aeruginosa quorum-sensing molecule homoserine lactone modulates inflammatory signaling through PERK and eI-F2 α. J Immunol (2014) 193(3):1459–67. doi:10.4049/jimmunol.1303437
68. Atkinson S, Williams P. Quorum sensing and social networking in the microbial world. J R Soc Interface (2009) 6:959–78. doi:10.1098/rsif.2009.0203
69. Schwarzer C, Wong S, Shi J, Matthes E, Illek B, Ianowski JP, et al. Pseudomonas aeruginosa homoserine lactone activates store-operated cAMP and cystic fibrosis transmembrane regulator-dependent Cl secretion by human airway. J Biol Chem (2010) 285(45):34850–63. doi:10.1074/jbc.M110.167668
70. Curutiu C, Chifiriuc MC, Mihaela Mitache M. Pseudomonas aeruginosa-eukaryotic cell crosstalk: mediators, mechanisms and implications for the antimicrobial therapy. Curr Org Chem (2013) 17(2):149–54.
71. Falcao JP, Sharp F, Sperandio V. Cell-to-cell signaling in intestinal pathogens. Curr Issues Intest Microbiol (2004) 5:9–17.
72. Moslehi-Jenabian S, Gori K, Jespersen L. AI-2 signaling is induced by acidic shock in probiotic strains of Lactobacillus spp. Int J Food Microb (2009) 135:295–302. doi:10.1016/j.ijfoodmicro.2009.08.011
73. Chifiriuc C, Lazar V, Dracea O, Ditu LM, Smarandache D, Bucur M, et al. Drastic attenuation of Pseudomonas aeruginosa pathogenicity in a holoxenic mouse experimental model induced by subinhibitory concentrations of phenyllactic acid (PLA) produced by LAB. Int J Mol Sci (2007) 8:583–92. doi:10.3390/i8070583
74. Chifiriuc MC, Bleotu C, Ditu LM, Banu O, Bleotu C, Dracea O, et al. Subinhibitory concentrations of phenyl lactic acid interfere with the expression of virulence factors in S. aureus and Ps. aeruginosa clinical strains. Roum Arch Microbiol Immunol (2009) 68(1):27–33.
75. Sperandio V, Torres AG, Kaper JB. Quorum sensing Escherichia coli regulators B and C (QseBC): a novel two-component regulatory system involved in the regulation of flagella and motility by quorum sensing in E. coli. Mol Microbiol (2002) 43(3):809–21. doi:10.1046/j.1365-2958.2002.02803.x
76. Holban MK, Holban A. Nanosized drug delivery systems in gastrointestinal targeting: interactions with microbiota. Pharmaceuticals (Basel) (2016) 9(4):62. doi:10.3390/ph9040062
77. Lyte M. Microbial endocrinology and the microbiota-gut-brain axis. In: Lyte M, Cryan JF, editors. Microbial Endocrinology: The Microbiota-Gut-Brain Axis in Health and Disease Advances in Experimental Medicine and Biology. (Vol. 817), New York: Springer (2014) 73–113.
78. Lazăr V, Holban A. Inter-kingdom cross-talk: the example of prokaryotes – eukaryotes communication. Biointerface Res Appl Chem (2011) 1(3):095–110.
79. Hughes DT, Sperandio V. Inter-kingdom signalling: communication between bacteria and their hosts. Nat Rev Microbiol (2008) 6(2):111–20. doi:10.1038/nrmicro1836
80. Burton CL, Chhabra SR, Swift S, Baldwin TJ, Withers H, Hill SJ, et al. The growth response of Escherichia coli to neurotransmitters and related catecholamine drugs requires a functional enterobactin biosynthesis and uptake system. Infect Immun (2002) 70:5913–23. doi:10.1128/IAI.70.11.5913-5923.2002
81. Freestone PP, Lyte M, Neal CP, Maggs AF, Haigh R, Williams PH. The mammalian neuroendocrine hormone norepinephrine supplies iron for bacterial growth in the presence of transferrin or lactoferrin. J Bacteriol (2000) 182:6091–8. doi:10.1128/JB.182.21.6091-6098.2000
82. Grenham S, Clarke G, Cryan JF, Dinan TG. Brain-gut-microbe communication in health and disease. Front Physiol (2011) 2:94. doi:10.3389/fphys.2011.00094
83. Lin L, Zhang J. Role of intestinal microbiota and metabolites on gut homeostasis and human diseases. BMC Immunol (2017) 18:2. doi:10.1186/s12865-016-0187-3
84. Haemer MA, Hyang TT, Daniels SR. The effect of neurohormonal factors, epigenetic factors, and gut microbiota on risk of obesity. Prev Chronic Dis (2009) 6(3):A96.
85. Bercik P, Denou E, Collins J, Jackson W, Lu J, Jury J, et al. The intestinal microbiota affect central levels of brain-derived neurotropic factor and behavior in mice. Gastroenterology (2011) 141(2):599–609. doi:10.1053/j.gastro.2011.04.052
86. Ochoa-Repáraz J, Mielcarz DW, Begum-Haque S, Kasper LH. Gut, bugs, and brain: role of commensal bacteria in the control of central nervous system disease. Ann Neurol (2011) 69(2):240–7. doi:10.1002/ana.22344
87. Erny D, Hrabě de Angelis AL, Jaitin D, Wieghofer P, Staszewski O, David E, et al. Host microbiota constantly control maturation and function of microglia in the CNS. Nat Neurosci (2015) 18(7):965–77. doi:10.1038/nn.4030
88. Unit 15: adaptive, specific host defenses. The Hygiene Hypothesis and Hypersensitivity. Available from: https://courses.lumenlearning.com/suny-mcc-microbiology/chapter/hypersensitivity (Accessed: April 23, 2018).
89. Aathira R, Jain V. Advances in management of type 1 diabetes mellitus. World J Diabetes (2014) 5(5):689–96. doi:10.4239/wjd.v5.i5.689
90. Centers for Disease Control and Prevention. National Diabetes Fact Sheet: National Estimates and General Information on Diabetes and Prediabetes in the United States. Atlanta, GA: U.S. Department of Health and Human Services, Centers for Disease Control and Prevention (2011).
91. Kantarova D, Buc M. Genetic susceptibility to type 1 diabetes mellitus in humans. Physiol Res (2007) 56:255–66.
92. Polychronakos C, Li Q. Understanding type 1 diabetes through genetics: advances and prospects. Nat Rev Genet (2011) 12(11):781–92. doi:10.1038/nrg3069
93. Alkanani AK, Hara N, Gottlieb PA, Ir D, Robertson CE, Wagner BD, et al. Alterations in intestinal microbiota correlate with susceptibility to type 1 diabetes. Diabetes (2015) 64(10):3510–20. doi:10.2337/db14-1847
94. Brown CT, Davis-Richardson AG, Giongo A, Gano KA, Crabb DB, Mukherjee N, et al. Gut microbiome metagenomics analysis suggests a functional model for the development of autoimmunity for type 1 diabetes. PLoS One (2011) 6(10):e25792. doi:10.1371/journal.pone.0025792
95. Vaarala O. Is the origin of type 1 diabetes in the gut? Immunol Cell Biol (2012) 90:271–6. doi:10.1038/icb.2011.115
96. Rook GAW, Brunet LR. Microbes, immunoregulation, and the gut. Gut (2005) 54(3):317–20. doi:10.1136/gut.2004.053785
97. Vaarala O, Atkinson MA. The “perfect storm” for type 1 diabetes: the complex interplay between intestinal microbiota, gut permeability, and mucosal immunity. Diabetes (2008) 57(10):2555–62. doi:10.2337/db08-0331
98. Bach JF, Chatenoud L. The hygiene hypothesis: an explanation for the increased frequency of insulin-dependent diabetes. Cold Spring Harb Perspect Med (2012) 2(2):a007799. doi:10.1101/cshperspect.a007799
99. Maffeis C, Martina A, Corradi M, Quarella S, Nori N, Torriani S, et al. Association between intestinal permeability and faecal microbiota composition in Italian children with beta cell autoimmunity at risk for type 1 diabetes. Diabetes Metab Res Rev (2016) 32:700–9. doi:10.1002/dmrr.2790
100. De Goffau MC, Luopajärvi K, Knip M, Ilonen J, Ruohtula T, Härkönen T, et al. Fecal microbiota composition differs between children with β-cell autoimmunity and those without. Diabetes (2013) 62:1238–44. doi:10.2337/db12-0526
101. Kostic AD, Gevers D, Siljander H, Vatanen T, Hyötyläinen T, Hämäläinen A-M, et al. The dynamics of the human infant gut microbiome in development and in progression toward type 1 diabetes. Cell Host Microbe (2015) 17:260–73. doi:10.1016/j.chom.2015.01.001
102. Scher JU, Sczesnak A, Longman RS, Segata N, Ubeda C, Bielski C, et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. Elife (2013) 2:e01202. doi:10.7554/eLife.01202
103. Liu X, Zeng B, Zhang J, Li W, Mou F, Wang H, et al. Role of the gut microbiome in modulating arthritis progression in mice. Sci Rep (2016) 6:30594. doi:10.1038/srep30594
104. Nagao-Kitamoto H, Kitamoto S, Kuffa P, Kamada N. Pathogenic role of the gut microbiota in gastrointestinal diseases. Intest Res (2016) 14(2):127. doi:10.5217/ir.2016.14.2.127
105. Biasi F, Leonarduzzi G, Oteiza PI. Inflammatory bowel disease: mechanisms, redox considerations, and therapeutic targets. Antioxid Redox Signal (2013) 19(14):1711–47. doi:10.1089/ars.2012.4530
106. M’Koma AE. inflammatory bowel disease: an expanding global health problem. Clin Med Insights Gastroenterol (2013) 6:33–47. doi:10.4137/CGast.S12731
107. Strus M, Gosiewski T, Fyderek K, Wedrychowicz A, Kowalska-Duplaga K, Kochan P, et al. A role of hydrogen peroxide producing commensal bacteria present in colon of adolescents with inflammatory bowel disease in perpetuation of the inflammatory process. J Physiol Pharmacol (2009) 60(6):46–54.
108. Khan TM, Duncan SH, Stams AJ, Van Dijl JM, Flint HJ, Harmsen HJ. The gut anaerobe Faecalibacterium prausnitzii uses an extracellular electron shuttle to grow at oxic–anoxic interphases. ISME J (2012) 6(5):1578–85. doi:10.1038/ismej.2012.5
109. Cao S, Feehley TJ, Nagler CR. The role of commensal bacteria in the regulation of sensitization to food allergens. FEBS Lett (2014) 588(22):4258–66. doi:10.1016/j.febslet.2014.04.026
110. Ji Y, Sun S, Goodrich JK, Kim H, Poole AC, Duhamel GE, et al. Diet-induced alterations in gut microflora contribute to lethal pulmonary damage in TLR2/TLR4 deficient mice. Cell Rep (2014) 8(1):137–49. doi:10.1016/j.celrep.2014.05.040
111. La Paglia GMC, Leone MC, Lepri G, Vagelli R, Valentini E, Alunno A, et al. One year in review 2017: systemic lupus erythematosus. Clin Exp Rheumatol (2017) 35:551–61.
112. Hevia A, Milani C, Lopez P, Cuervo A, Arboleya S, Duranti S, et al. Intestinal dysbiosis associated with systemic lupus erythematosus. MBio (2014) 5:e01548–14. doi:10.1128/mBio.01548-14
113. Zhang H, Liao X, Sparks JB, Luo XM. Dynamics of gut microbiota in autoimmune lupus. Appl Environ Microbiol (2014) 80:7551–60. doi:10.1128/AEM.02676-14
114. Scher JU, Ubeda C, Artacho A, Attur M, Isaac S, Reddy SM, et al. Decreased bacterial diversity characterizes the altered gut microbiota in patients with psoriatic arthritis, resembling dysbiosis in inflammatory bowel disease. Arthritis Rheumatol (2015) 67:128–39. doi:10.1002/art.38892
115. Volkmann ER, Hoffmann-Vold AM, Chang YL, Jacobs JP, Tillisch K, Mayer EA, et al. Systemic sclerosis is associated with specific alterations in gastrointestinal microbiota in two independent cohorts. BMJ Open Gastroenterol (2017) 4:e000134. doi:10.1136/bmjgast-2017-000134
116. Rabot S, Jaglin M, Daugé V, Naudon L. Impact of the gut microbiota on the neuroendocrine and behavioural responses to stress in rodents. Ocl (2016) 23(1):D116. doi:10.1051/ocl/2015036
117. Jiang C, Li G, Huang P, Liu Z. The gut microbiota and Alzheimer’s disease. Alzheimers Dis (2017) 58(1):1–15. doi:10.3233/JAD-161141
118. Colpitts SL, Kasper LH. Influence of the gut microbiome on autoimmunity in the central nervous system. J Immunol (2017) 198(2):596–604. doi:10.4049/jimmunol.1601438
119. Mbulaiteye SM, Hisada ME, El-Omar EM. Helicobacter Pylori associated global gastric cancer burden. Front Biosci J Virtual Libr (2009) 14:1490–504. doi:10.2741/3320
120. Wu S, Rhee KJ, Albesiano E, Rabizadeh S, Wu X, Yen HR, et al. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat Med (2009) 15:1016–22. doi:10.1038/nm.2015
121. Chen GY. Role of Nlrp6 and Nlrp12 in the maintenance of intestinal homeostasis. Eur J Immunol (2014) 44:321–7. doi:10.1002/eji.201344135
122. Hu B, Elinav E, Huber S, Strowig T, Hao L, Hafemann A, et al. Microbiota-induced activation of epithelial IL-6 signaling links inflammasome-driven inflammation with transmissible cancer. Proc Natl Acad Sci U S A (2013) 110(24):9862–7. doi:10.1073/pnas.1307575110
123. Ng JH, Iyer NG, Tan MH, Edgren G. Changing epidemiology of oral squamous cell carcinoma of the tongue: a global study. Head Neck (2017) 39:297–304. doi:10.1002/hed.24589
124. Nagy K, Sonkodi I, Szoke I, Nagy E, Newman HN. The microflora associated with human oral carcinomas. Oral Oncol (1998) 34:304–8. doi:10.1016/S1368-8375(98)80012-2
125. Katz J, Onate MD, Pauley KM, Bhattacharyya I, Cha S. Presence of Porphyromonas gingivalis in gingival squamous cell carcinoma. Int J Oral Sci (2011) 3:209–15. doi:10.4248/IJOS11075
126. Mager DL, Haffajee AD, Devlin PM, Norris CM, Posner MR, Goodson JM. The salivary microbiota as a diagnostic indicator of oral cancer: a descriptive, non-randomized study of cancer-free and oral squamous cell carcinoma subjects. J Transl Med (2005) 3:27. doi:10.1186/1479-5876-3-27
127. Tateda M, Shiga K, Saijo S, Sone M, Hori T, Yokoyama J, et al. Streptococcus anginosus in head and neck squamous cell carcinoma: implication in carcinogenesis. Int J Mol Med (2000) 6:699–1402. doi:10.3892/ijmm.6.6.699
128. Morita E, Narikiyo M, Yano A, Nishimura E, Igaki H, Sasaki H, et al. Different frequencies of Streptococcus anginosus infection in oral cancer and esophageal cancer. Cancer Sci (2003) 94:492–6. doi:10.1111/j.1349-7006.2003.tb01471.x
129. Sasaki M, Yamaura C, Ohara-Nemoto Y, Tajika S, Kodama Y, Ohya T, et al. Streptococcus anginosus infection in oral cancer and its infection route. Oral Dis (2005) 11:151–6. doi:10.1111/j.1601-0825.2005.01051.x
130. Pushalkar S, Ji X, Li Y, Estilo C, Yegnanarayana R, Singh B, et al. Comparison of oral microbiota in tumor and non-tumor tissues of patients with oral squamous cell carcinoma. BMC Microbiol (2012) 12:144. doi:10.1186/1471-2180-12-144
131. Hooper SJ, Crean SJ, Fardy MJ, Lewis MA, Spratt DA, Wade WG, et al. A molecular analysis of the bacteria present within oral squamous cell carcinoma. J Med Microbiol (2007) 56:1651–9. doi:10.1099/jmm.0.46918-0
132. Pushalkar S, Mane SP, Ji X, Li Y, Evans C, Crasta OR, et al. Microbial diversity in saliva of oral squamous cell carcinoma. FEMS Immunol Med Microbiol (2011) 61:269–77. doi:10.1111/j.1574-695X.2010.00773.x
133. Schmidt BL, Kuczynski J, Bhattacharya A, Huey B, Corby PM, Queiroz EL, et al. Changes in abundance of oral microbiota associated with oral cancer. PLoS One (2014) 9:e98741. doi:10.1371/journal.pone.0098741
134. Al-Hebshi NN, Nasher AT, Idris AM, Chen T. Robust species taxonomy assignment algorithm for 16S rRNA NGS reads: application to oral carcinoma samples. J Oral Microbiol (2015) 7:28934. doi:10.3402/jom.v7.28934
135. Guerrero-Preston R, Godoy-Vitorino A, Jedlicka A, Rodriguez H, Gonzalez H, Sidransky D, et al. 16S rRNA amplicon sequencing identifies microbiota associated with oral cancer, Human Papilloma Virus infection and surgical treatment. Oncotarget (2016) 7(32):1–15. doi:10.18632/oncotarget.9710
136. Zhao H, Chu M, Huang Z, Yang X, Ran S, Hu B, et al. Variations in oral microbiota associated with oral cancer. Sci Rep (2017) 7(1):1–10. doi:10.1038/s41598-017-11779-9
137. Jemal A, Siegel R, Xu J. Cancer statistics. CA Cancer J Clin (2010) 60:277–300. doi:10.3322/caac.20073
138. Ilie M, Dascãlu L, Chifiriuc C, Popa M, Constantinescu G, Tãnãsescu C, et al. Correlation of anti-Helicobacter pylori Cag A IgG antibodies with resistance to first line treatment, bleeding gastroduodenal ulcers and gastric cancer. Rom Arch Microbiol Immunol (2011) 70(3):101–5.
139. Petra CV, Rus A, DumitraŞcu DL. Gastric microbiota: tracing the culprit. Clujul Med (2017) 90(4):369–76. doi:10.15386/cjmed-854
140. Dicksved J, Lindberg M, Rosenquist M, Enroth H, Jansson JK, Engstrand L. Molecular characterization of the stomach microbiota in patients with gastric cancer and in controls. J Med Microbiol (2009) 58:509–16. doi:10.1099/jmm.0.007302-0
141. Ferreira RM, Pereira-Marques J, Pinto-Ribeiro I, Costa JL, Carneiro F, Machado JC, et al. Gastric microbial community profiling reveals a dysbiotic cancer-associated microbiota. Gut (2018) 67(2):226–36. doi:10.1136/gutjnl-2017-314205
142. Willett WC, Stampfer MJ, Colditz GA, Rosner BA, Speizer FE. Relation of meat, fat, and fiber intake to the risk of colon cancer in a prospective study among women. N Engl J Med (1990) 323:1664–72. doi:10.1056/NEJM199012133232404
143. Huxley RR, Ansary-Moghaddam A, Clifton P, Czernichow S, Parr CL, Woodward M. The impact of dietary and lifestyle risk factors on risk of colorectal cancer: a quantitative overview of the epidemiological evidence. Int J Cancer (2009) 125:171–80. doi:10.1002/ijc.24343
144. Pibiri F, Kittles RA, Sandler RS, Keku TO, Kupfer SS, Xicola RM, et al. Genetic variation in vitamin D-related genes and risk of colorectal cancer in African Americans. Cancer Causes Control (2014) 25(5):561–70. doi:10.1007/s10552-014-0361-y
145. Shiryaev SA, Remacle AG, Chernov AV, Golubkov VS, Muranaka N, Dambacher CM, et al. Substrate cleavage profiling suggests a distinct function of Bacteroides fragilis metalloproteinases (fragilysin and metalloproteinase II) at the microbiome-inflammationcancer interface. J Biol Chem (2013) 288:34956–67. doi:10.1074/jbc.M113.516153
146. Chung L, Orberg ET, Geis AL, Chan JL, Fu K, Shields CED, et al. Bacteroides fragilis toxin coordinates a pro-carcinogenic inflammatory cascade via targeting of colonic epithelial cells. Cell Host Microbe (2018) 23:203–14. doi:10.1016/j.chom.2018.01.007
147. Tsoi H, Chu ESH, Zhang X, Sheng JQ, Nakatsu G, Ng SC, et al. Peptostreptococcus anaerobius induces intracellular cholesterol biosynthesis in colon cells to induce proliferation and causes dysplasia in mice. Gastroenterology (2017) 152:1419–33. doi:10.1053/j.gastro.2017.01.009
148. Taieb F, Petit C, Nougayrède JP, Oswald E. The enterobacterial genotoxins: cytolethal distending toxin and colibactin. EcoSal Plus (2016) 7(1):1–14. doi:10.1128/ecosalplus.ESP-0008-2016
149. Takeda K, Akira S. Toll-like receptors in innate immunity. Int Immunol (2005) 17(1):1–14. doi:10.1093/intimm/dxh186
150. Wei H, Dong L, Wang T, Zhang M, Hua W, Zhang C, et al. Structural shifts of gut microbiota as surrogate endpoints for monitoring host health changes induced by carcinogen exposure. FEMS Microbiol Ecol (2010) 73:577–86. doi:10.1111/j.1574-6941.2010.00924.x
151. Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin. Cell Host Microbe (2013) 14:195–206. doi:10.1016/j.chom.2013.07.012
152. Gholizadeh P, Eslami H, Kafil HS. Carcinogenesis mechanisms of Fusobacterium nucleatum. Biomed Pharmacother (2017) 89:918–25. doi:10.1016/j.biopha.2017.02.102
153. Eaden JA, Abrams KR, Mayberry JF. The risk of colorectal cancer in ulcerative colitis: a meta-analysis. Gut (2001) 48:526–35. doi:10.1136/gut.48.4.526
155. Schoefer L, Mohan R, Schwiertz A, Braune A, Blaut M. Anaerobic degradation of flavonoids by Clostridium orbiscindens. Appl Environ Microbiol (2003) 69(10):5849–54. doi:10.1128/AEM.69.10.5849-5854.2003
156. Kim DH, Jin YH. Intestinal bacterial beta-glucuronidase activity of patients with colon cancer. Arch Pharm Res (2001) 24:564–7. doi:10.1007/BF02975166
157. Goldin BR, Swenson L, Dwyer J, Sexton M, Gorbach SL. Effect of diet and Lactobacillus acidophilus supplements on human fecal bacterial enzymes. J Natl Cancer Inst (1980) 64(2):255–61. doi:10.1093/jnci/64.2.255
158. Daïen CI, Pinget GV, Tan JK, Macia L. Detrimental impact of microbiota-accessible carbohydrate-deprived diet on gut and immune homeostasis: an overview. Front Immunol (2017) 8:548. doi:10.3389/fimmu.2017.00548
159. Bordonaro M, Lazarova DL. Hyperinduction of Wnt activity: a new paradigm for the treatment of colorectal cancer? Oncol Res (2008) 317:1–9. doi:10.3727/096504008784046108
160. Hague A, Elder DJ, Hicks DJ, Paraskeva C. Apoptosis in colorectal tumour cells: induction by the short chain fatty acids butyrate, propionate and acetate and by the bile salt deoxycholate. Int J Cancer (1995) 60:400–6. doi:10.1002/ijc.2910600322
161. Ruemmele FM, Schwartz S, Seidman EG, Dionne S, Levy E, Lentze MJ. Butyrate induced Caco-2 cell apoptosis is mediated via the mitochondrial pathway. Gut (2003) 52:94–100. doi:10.1136/gut.52.1.94
162. Bolden JE, Peart MJ, Johnstone RW. Anticancer activities of histone deacetylase inhibitors. Nat Rev Drug Discov (2006) 5:769–84. doi:10.1038/nrd2133
163. Sivan A, Corrales L, Hubert N, Williams JB, Aquino-Michaels K, Earley ZM, et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science (2015) 350(6264):1084–9. doi:10.1126/science.aac4255
164. Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillere R, et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science (2018) 359:91–7. doi:10.1126/science.aan3706
165. Shono Y, Docampo MD, Peled JU, Perobelli SM, Velardi E, Tsai JJ, et al. Increased GVHD-related mortality with broad-spectrum antibiotic use after allogeneic hematopoietic stem cell transplantation in human patients and mice. Sci Transl Med (2016) 8(8):339ra71. doi:10.1126/scitranslmed.aaf2311
166. Gopalakrishnan V, Helmink BA, Spencer CN, Reuben A, Wargo JA. The influence of the gut microbiome on cancer, immunity, and cancer immunotherapy. Cancer Cell (2018) 33(4):570–80. doi:10.1016/j.ccell.2018.03.015
167. Jenq RR, Taur Y, Devlin SM, Ponce DM, Goldberg JD, Ahr KF, et al. Intestinal Blautia is associated with reduced death from graft-versus-host disease. Biol Blood Marrow Transplant (2015) 21:1373–83. doi:10.1016/j.bbmt.2015.04.016
168. Peled JU, Devlin SM, Staffas A, Lumish M, Khanin R, Littmann ER, et al. Intestinal microbiota and relapse after hematopoietic-cell transplantation. J Clin Oncol (2017) 35:1650–9. doi:10.1200/JCO.2016.70.3348
169. Viaud S, Saccheri F, Mignot G, Yamazaki T, Daillere R, Hannani D, et al. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science (2013) 342:971–6. doi:10.1126/science.1240537
170. Iida N, Dzutsev A, Stewart CA, Smith L, Bouladoux N, Weingarten RA, et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science (2013) 342:967–70. doi:10.1126/science.1240527
171. Cogdill AP, Andrews MC, Wargo JA. Hallmarks of response to immune checkpoint blockade. Br J Cancer (2017) 117:1–7. doi:10.1038/bjc.2017.136
172. Sharma P, Hu-Lieskovan S, Wargo JA, Ribas A. Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell (2017) 168:707–23. doi:10.1016/j.cell.2017.01.017
173. Vetizou M, Pitt JM, Daillere R, Lepage P, Waldschmitt N, Flament C, et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science (2015) 350:1079–84. doi:10.1126/science.aad1329
174. Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science (2018) 359:97–103. doi:10.1126/science.aan4236
175. Matson V, Fessler J, Bao R, Chongsuwat T, Zha Y, Alegre M-L, et al. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science (2018) 359:104–8. doi:10.1126/science.aao3290
176. Chaput N, Lepage P, Coutzac C, Soularue E, Le Roux K, Monot C, et al. Baseline gut microbiota predicts clinical response and colitis in metastatic melanoma patients treated with ipilimumab. Ann Oncol (2017) 28:1368–79. doi:10.1093/annonc/mdx108
177. Dubin K, Callahan MK, Ren B, Khanin R, Viale A, Ling L, et al. Intestinal microbiome analyses identify melanoma patients at risk for checkpoint-blockade-induced colitis. Nat Commun (2016) 7:10391. doi:10.1038/ncomms10391
178. Frankel AE, Coughlin LA, Kim J, Froehlich TW, Xie Y, Frenkel EPA, et al. Metagenomic shotgun sequencing and unbiased metabolomic profiling identify specific human gut microbiota and metabolites associated with immune checkpoint therapy efficacy in melanoma patients. Neoplasia (2017) 19:848–55. doi:10.1016/j.neo.2017.08.004
179. Mikelsaar M, Lazar V, Onderdonk AB, Donnelli G. Do probiotic preparations for humans really have efficacy? Microb Ecol Heal Dis (2011) 22(1):10128. doi:10.3402/mehd.v22i0.10128
180. Rothschild D, Weissbrod O, Barkan E, Kurilshikov A, Korem TZ, Costea PI, et al. Environment dominates over host genetics in shaping human gut microbiota. Nature (2018) 555:210–5. doi:10.1038/nature25973
181. Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med (2009) 1(6):6ra14. doi:10.1126/scitranslmed.3000322
182. Wu GD, Chen J, Hoffmann C, Bittinger K, Chen Y, Sue A, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science (2012) 334(6052):105–8. doi:10.1126/science.1208344
183. Hald S, Schioldan AG, Moore ME, Dige A, Laerke HN, Agnholt J, et al. Effects of arabinoxylan and resistant starch on intestinal microbiota and short-chain fatty acids in subjects with metabolic syndrome: a randomised crossover study. PLoS One (2016) 11(7):e0159223. doi:10.1371/journal.pone.0159223
184. Wainstein J, Ganz T, Boaz M, Bar Dayan Y, Dolev E, Kerem Z, et al. Olive leaf extract as a hypoglycemic agent in both human diabetic subjects and in rats. J Med Food (2012) 15:605–10. doi:10.1089/jmf.2011.0243
185. Park Y, Subar AF, Hollenbeck A, Schatzkin A. Dietary fiber intake and mortality in the NIH-AARP diet and health study. Arch Intern Med (2011) 171(12):1061–8. doi:10.1001/archinternmed.2011.18
186. Otto GP, Sossdorf M, Claus RA, Rodel J, Menge K, Reinhart K, et al. The late phase of sepsis is characterized by an increased microbiological burden and death rate. Crit Care (2011) 15:R183. doi:10.1186/cc10332
187. MJ B. Antibiotic use and its consequences for the normal microbiome. Science (2016) 352:544–5. doi:10.1126/science.aad9358
188. Karami N, Hannoun C, Adlerberth I, Dell’Amico E, Roselli M, Strohmeyer M, et al. Colonization dynamics of ampicillin-resistant Escherichia coli in the infantile colonic microbiota. J Antimicrob Chemother (2008) 62:703–8. doi:10.1093/jac/dkn263
189. Bartoloni A, Bartalesi F, Mantella A, Dell’Amico E, Roselli M, Strohmeyer M, et al. High prevalence of acquired antimicrobial resistance unrelated to heavy antimicrobial consumption. J Infect Dis (2004) 189(7):1291–4. doi:10.1086/382191
190. Ponziani FR, Scaldaferri F, Petito V, Paroni Sterbini F, Pecere S, Lopetuso LR, et al. The role of antibiotics in gut microbiota modulation: the eubiotic effects of rifaximin. Dig Dis (2016) 34:269–78. doi:10.1159/000443361
191. Pilar Francino M, Moy A. Effects of antibiotic use on the microbiota of the gut and associated alterations of immunity and metabolism. Eur Med J Gastroenterol (2013) 2(1):74–80.
192. Modi SR, Collins JJ, Relman DA. Antibiotics and the gut microbiota. J Clin Invest (2014) 124(10):4212–8. doi:10.1172/JCI72333
193. Gosalbes MJ, Valles Y, Jimenez-Hernandez N, Balle C, Riva P, Miravet-Verde S, et al. High frequencies of antibiotic resistance genes in infants’ meconium and early fecal samples. J Dev Orig Health Dis (2015) 7(1):35–44. doi:10.1017/S2040174415001506
194. Gibson MK, Wang B, Ahmadi S, Burnham CA, Tarr PI, Warner BB, et al. Developmental dynamics of the preterm infant gut microbiota and antibiotic resistome. Nat Microbiol (2016) 1:16024. doi:10.1038/nmicrobiol.2016.24
195. Knecht H, Neulinger SC, Heinsen FA, Knecht C, Schilhabel A, Schmitz RA, et al. Effects of β-lactam antibiotics and fluoroquinolones on human gut microbiota in relation to Clostridium difficile associated diarrhea. PLoS One (2014) 9:e89417. doi:10.1371/journal.pone.0089417
196. Korpela K, Salonen A, Virta LJ, Kekkonen RA, Forslund K, Bork P, et al. Intestinal microbiome is related to lifetime antibiotic use in Finnish pre-school children. Nat Commun (2016) 7:10410. doi:10.1038/ncomms10410
197. Ianiro G, Tilg H, Gasbarrini A. Antibiotics as deep modulators of gut microbiota: between good and evil. Gut (2016) 65(11):1906–15. doi:10.1136/gutjnl-2016-312297
198. Rashid MU, Zaura E, Buijs MJ, Keijser BJ, Crielaard W, Nord CE, et al. Determining the long-term effect of antibiotic administration on the human normal intestinal microbiota using culture and pyrosequencing methods. Clin Infect Dis (2015) 60(Suppl 2):S77–84. doi:10.1093/cid/civ137
199. Jakobsson HE, Jernberg C, Andersson AF, Sjölund-Karlsson M, Jansson JK, Engstrand L. Short-term antibiotic treatment has differing long-term impacts on the human throat and gut microbiome. PLoS One (2010) 5:e9836. doi:10.1371/journal.pone.0009836
200. Vervoort J, Xavier BB, Stewardson A, Coenen S, Godycki-Cwirko M, Adriaenssens N, et al. Metagenomic analysis of the impact of nitrofurantoin treatment on the human faecal microbiota. J Antimicrob Chemother (2015) 70:1989–92. doi:10.1093/jac/dkv062
201. Salminen MK, Rautelin H, Tynkkynen S, Poussa T, Saxelin M, Valtonen V, et al. Lactobacillus bacteremia, species identification, and antimicrobial susceptibility of 85 blood isolates. Clin Infect Dis (2006) 42:e35–44. doi:10.1086/500214
202. Truusalu K, Mikelsaar RH, Naaber P, Karki T, Kullisaar T, Zilmer M, et al. Eradication of Salmonella typhimurium infection in a murine model of typhoid fever with the combination of probiotic Lactobacillus fermentum ME-3 and ofloxacin. BMC Microbiol (2008) 8:132. doi:10.1186/1471-2180-8-132
203. Adamu BO, Lawley TD. Bacteriotherapy for the treatment of intestinal dysbiosis caused by Clostridium difficile infection. Curr Opin Microbiol (2013) 16(5):596–601. doi:10.1016/j.mib.2013.06.009
204. Manges AR, Labbe A, Loo VG, Atherton JK, Behr MA. Comparative metagenomic study of alterations to the intestinal microbiota and risk of nosocomial Clostridium difficile-associated disease. J Infect Dis (2010) 202:1877–84. doi:10.1086/657319
205. Khodaii Z, Ghaderian SMH, Natanzi MM. Probiotic bacteria and their supernatants protect enterocyte cell lines from enteroinvasive Escherichia coli (EIEC) invasion. Int J Mol Cell Med (2017) 1:0–6. doi:10.22088/acadpub.BUMS.6.3.183
206. Quadri LEN. Regulation of class II bacteriocin production by cell-cell signaling. J Microbiol (2003) 41:175–82.
207. Rabah H, Rosa do Carmo F, Jan G. Dairy propionibacteria: versatile probiotics. Microorganisms (2017) 5(2):E24. doi:10.3390/microorganisms5020024
208. Altieri C. Dairy propionibacteria as probiotics: recent evidences. World J Microbiol Biotechnol (2016) 32:172. doi:10.1007/s11274-016-2118-0
209. Myllyluoma E, Ahonen AM, Korpela R, Vapaatalo H, Kankuri E. Effects of multispecies probiotic combination on Helicobacter pylori infection in vitro. Clin Vaccine Immunol (2008) 15:1472–82. doi:10.1128/CVI.00080-08
210. Nair DVT, Kollanoor-Johny A. Effect of Propionibacterium freudenreichii on Salmonella multiplication, motility, and association with avian epithelial cells. Poult Sci (2017) 96(5):1376–86. doi:10.3382/ps/pew367
211. Campaniello D, Bevilacqua A, Sinigaglia M, Altieri C. Screening of Propionibacterium spp for potential probiotic properties. Anaerobe (2015) 34:169–73. doi:10.1016/j.anaerobe.2015.06.003
212. Oksaharju A, Kooistra T, Kleemann R, van Duyvenvoorde W, Miettinen M, Lappalainen J, et al. Effects of probiotic Lactobacillus rhamnosus GG and Propionibacterium freudenreichii spp. shermanii JS supplementation on intestinal and systemic markers of inflammation in ApoE*3Leiden mice consuming a high-fat diet. Br J Nutr (2013) 110(1):77–85. doi:10.1017/S0007114512004801
213. Khailova L, Dvorak K, Arganbright KM, Halpern MD, Kinouchi T, Yajima M, et al. Bifidobacterium bifidum improves intestinal integrity in a rat model of necrotizing enterocolitis. Am J Physiol Gastrointest Liver Physiol (2009) 297(5):G940–9. doi:10.1152/ajpgi.00141.2009
214. Shiou SR, Yu Y, Guo Y, He SM, Mziray-Andrew CH, Hoenig J, et al. Synergistic protection of combined probiotic conditioned media against neonatal necrotizing enterocolitis-like intestinal injury. PLoS One (2013) 8(5):e65108. doi:10.1371/journal.pone.0065108
215. Singh RK, Chang H-W, Yan D, Lee KM, Ucmak D, Wong K, et al. Influence of diet on the gut microbiome and implications for human health. J Transl Med (2017) 15(1):73. doi:10.1186/s12967-017-1175-y
216. Culligan EP, Hill C, Sleator RD. Probiotics and gastrointestinal disease: successes, problems and future prospects. Gut Pathog (2009) 1(1):1–12. doi:10.1186/1757-4749-1-19
Keywords: gut microbiota, opportunistic infections, autoimmunity, chronic inflammation, cancer, antibiotics, probiotics, diet
Citation: Lazar V, Ditu L-M, Pircalabioru GG, Gheorghe I, Curutiu C, Holban AM, Picu A, Petcu L and Chifiriuc MC (2018) Aspects of Gut Microbiota and Immune System Interactions in Infectious Diseases, Immunopathology, and Cancer. Front. Immunol. 9:1830. doi: 10.3389/fimmu.2018.01830
Received: 22 January 2018; Accepted: 24 July 2018;
Published: 15 August 2018
Edited by:
Nadiya V. Boyko, Uzhhorod National University, UkraineReviewed by:
Claudio Nicoletti, Università degli Studi di Firenze, ItalyCopyright: © 2018 Lazar, Ditu, Pircalabioru, Gheorghe, Curutiu, Holban, Picu, Petcu and Chifiriuc. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mariana Carmen Chifiriuc, Y2FybWVuX2JhbG90ZXNjdUB5YWhvby5jb20=
†These authors have contributed equally to this work.
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.