- 1AI Innovation Lab, Weill Cornell Medicine-Qatar, Doha, Qatar
- 2Department of Medicine, Weill Cornell Medicine-Qatar, Doha, Qatar
Early detection and management of diabetic peripheral neuropathy (DPN) are critical to reducing associated morbidity and mortality. Corneal Confocal Microscopy (CCM) facilitates the imaging of corneal nerves to detect early and progressive nerve damage in DPN. However, its wider adoption has been limited by the subjectivity and time-intensive nature of manual nerve fiber quantification. This study investigates the diagnostic utility of state-of-the-art Vision Transformer (ViT) models for the binary classification of CCM images to distinguish between healthy controls and individuals with DPN. The ViT model's performance was also compared to ResNet50, a convolutional neural network (CNN) previously applied for DPN detection using CCM images. Using a dataset of approximately 700 CCM images, the ViT model achieved an AUC of 0.99, a sensitivity of 98%, a specificity of 92%, and an F1-score of 95%, outperforming previously reported methods. These findings highlight the potential of the ViT model as a reliable tool for CCM-based DPN diagnosis, eliminating the need for time-consuming manual image segmentation. Moreover, the results reinforce CCM's value as a non-invasive and precise imaging modality for detecting nerve damage, particularly in neuropathy-related conditions such as DPN.
1 Introduction
The Burden of Diseases, Injuries, and Risk Factors Study (GBD) estimated that, in 2021, diabetes affected 529 million people across 204 countries and territories, underscoring the high prevalence of the condition among various age groups worldwide (Ong et al., 2023). Diabetic Peripheral Neuropathy (DPN) is a neuropathic condition affecting the peripheral nerves, often presenting as a distal, symmetrical sensory or motor deficit. As a major long-term complication of diabetes, DPN can result in painful neuropathy, foot ulceration, and amputation.
Early and accurate diagnosis of DPN is essential for timely intervention and effective disease management (Ponirakis et al., 2021, 2022). Without treatment, DPN can lead to serious outcomes, including loss of sensation, falls, foot ulcers, and even limb amputations. Additionally, diabetic patients with DPN face a higher risk of mortality from any cause or cardiovascular disease compared to those without DPN (Jensen et al., 2021; Elafros et al., 2022; Eid et al., 2023).
Corneal Confocal Microscopy (CCM) is a non-invasive imaging technique that serves as a precise surrogate biomarker for small fiber neuropathy. The corneal nerves, accessible through CCM, are frequently impacted in the early stages of DPN, enabling clinicians to detect nerve damage before more severe symptoms develop. Manual analysis of CCM images is labor-intensive, subjective, and requires significant expertise, with interobserver variability that can limit diagnostic accuracy for DPN. Using Deep Learning (DL), Salahouddin et al. (2021) employed a U-Net-based model to automate the segmentation and quantification of corneal nerves in CCM images, achieving discrimination between patients with and without DPN, with an average area under the curve (AUC) of 0.93. Moving toward eliminating the need for pixel-wise annotations, Preston et al. (2022) utilized a ResNet model to diagnose peripheral neuropathy, reporting an average sensitivity of 84% in correctly identifying DPN patients on a test set of 40 images.
Following recent advancements in automated DPN diagnostics, we evaluated a state-of-the-art Vision Transformer (ViT) model for classifying DPN patients using CCM images, comparing its performance to the established ResNet architecture. Our approach, which eliminates the need for pixel-wise annotations, is the first to apply ViTs for DPN classification on CCM images, demonstrating high accuracy on a relatively large dataset. Additionally, we employed Grad-CAM to generate heatmaps, visually highlighting regions that contribute most to the classification decision and confirming a focus on corneal nerves. Figure 1 shows an overview of the transformer-based model architecture for corneal nerve classification.
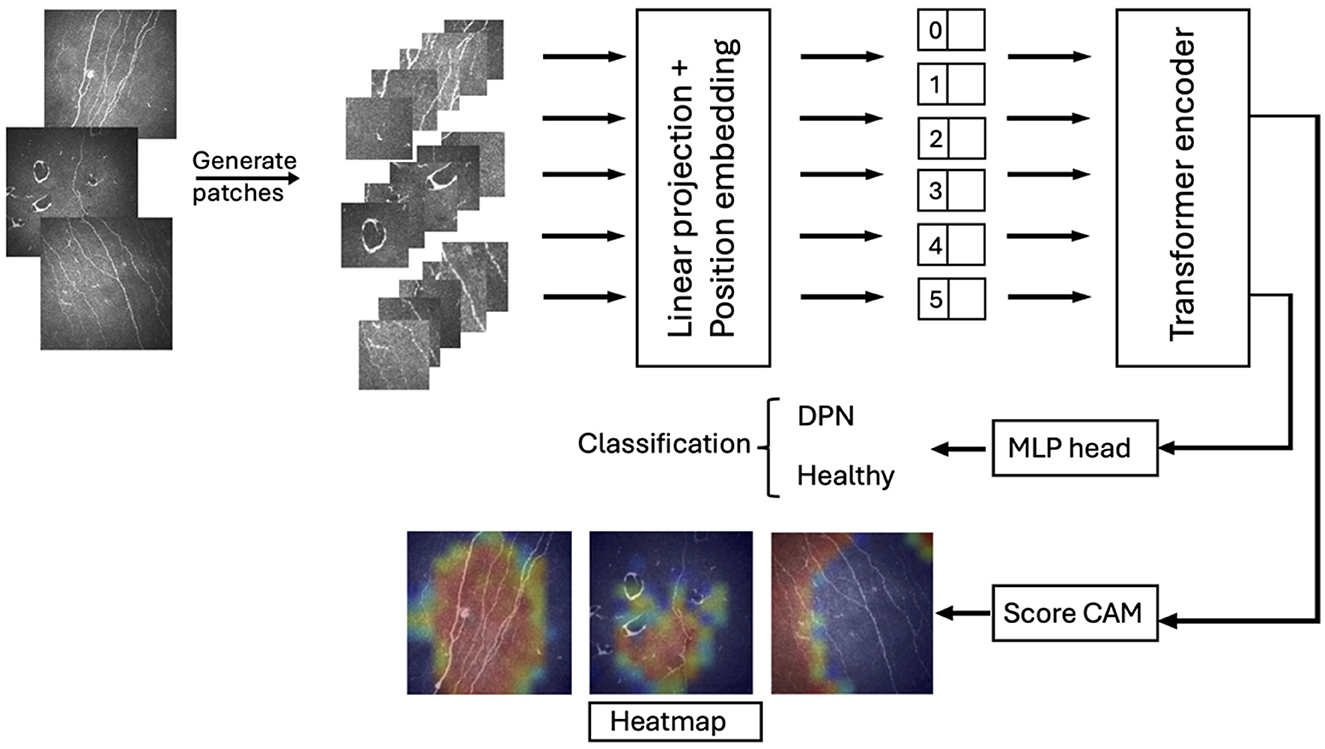
Figure 1. Overview of the transformer-based model architecture for corneal nerve classification. The input image is divided into patches, which are linearly projected and positionally embedded before being fed into the transformer encoder. The output representations are used for classification between DPN and healthy cases through an MLP head. Additionally, Score-CAM generates heatmaps highlighting relevant regions in the patches, aiding interpretability in classification.
2 Method
2.1 Dataset
The experiment was carried out on a database of 692 CCM images (358 healthy controls and 334 DPN cases) collected from 106 subjects (29 patients with DPN and 77 healthy controls), captured using the Heidelberg HRTIII corneal confocal microscope. This is a sub-analysis of the LANDMark study (Pritchard et al., 2014)—a multi-center study conducted at the University of Manchester, UK and Queensland University of Technology, Australia in 2009–2014. The LANDMark study adhered to the tenets of the Declaration of Helsinki and was approved by the relevant institutional review boards. Informed, written consent was obtained from all subjects prior to participation.
The images have a size of 384 × 384 pixels, 8-bit gray levels, and are stored in BMP format. To mitigate potential biases arising from the relatively small sample size and the varying number of images per subject, we employed a rigorous data splitting strategy. The dataset was divided into training (60%), validation (20%), and testing sets (20%). To ensure balanced representation across sets, we performed stratified splitting based on subject-level allocation. This ensured that no images from the same subject were included in more than one set, preventing potential bias arising from inter-subject variability.
2.2 Vision transformer
Introduced by Dosovitskiy et al. (2020), Vision Transformers (ViTs) have quickly gained prominence in classification tasks, often outperforming traditional methods (Bazi et al., 2021; Ding et al., 2023; Long et al., 2024). The ViT model includes an embedding layer, a transformer encoder, and an MLP head. The input image is divided into non-overlapping patches, each treated as a token, with position embeddings added to retain spatial information. These embeddings are processed by the encoder, which consists of stacked layers with multiheaded self-attention (capturing relationships across image regions), an MLP block (refining extracted information), and a normalization layer (ensuring data stability). Finally, the MLP head translates encoded information into the predicted class.
Our work leverages the capabilities of ViTs to construct a robust and scalable system, while addressing technical complexities associated with data preprocessing and model development. To enhance efficiency and potentially improve performance, we optimized the original ViT architecture (Dosovitskiy et al., 2020) by reducing the number of Transformer layers, thereby streamlining the model and overfitting. Furthermore, we decreased the MLP size, leading to a substantial decrease in model parameters and computational cost. We modified the input patch size. This trade-off increases the effective sequence length for the Transformer while simultaneously reducing computational complexity, as the number of patches decreases quadratically with the increase in patch size. These modifications resulted in a dramatic reduction in model parameters from 86M to 6M, making our model significantly more compact and potentially easier to deploy on resource-constrained devices.
To enhance model performance and stability, we incorporated a batch normalization layer after the Transformer block. Unlike the original model's layer normalization, which normalized across all features within a sample, our batch normalization normalizes each feature independently across the mini-batch. This modification aims to improve training stability and potentially enhance generalization. To further mitigate overfitting, we integrated Dropout throughout the model architecture. Dropout randomly deactivates a fraction of neurons during training, preventing excessive reliance on specific features and encouraging weight sharing across the network, ultimately leading to more robust and generalizable models.
2.3 Model training
We trained our ViT model using Python 3.7.10 and TensorFlow with Keras on a GPU P100 for 150 epochs. Images were resized to 256 × 256 pixels and divided by the ViT into 144 patches of 20 × 20 pixels each. During training, we applied a combination of feature normalization and data augmentation techniques on each patch, including horizontal flipping, zooming (height and width factor 0.2), and slight rotation (factor 0.02), to enhance model robustness. Optimizing ViT's complex structure is challenging, so we used the AdamW optimizer with Decoupled Weight Decay Regularization, with specific parameters listed in Table 1, carefully selected for a balance of accuracy and efficiency (https://github.com/serag-ai/ViT-CCM).
3 Evaluation metrics
We assessed our model's performance using several key metrics: Area Under the Receiver Operating Characteristic Curve (AUC), Specificity, Sensitivity, and F1-score.
AUC is a threshold-independent metric that evaluates the performance of a classification model. It represents the probability that the model will rank a randomly chosen positive instance higher than a randomly chosen negative instance. The AUC ranges from 0 to 1, where a value closer to 1 indicates superior discriminative ability. An AUC of 0.5 suggests no discriminative power, equivalent to random guessing.
Sensitivity, also known as recall, measures the proportion of true positives (TP) correctly identified out of all actual positives. It is calculated as:
Specificity measures the proportion of true negatives (TN) correctly identified out of all actual negatives. It is calculated as:
The F1-score is a harmonic mean of Precision (Pre) and Recall (Rec), combining them into a single metric. It is calculated as:
Recall (Rec) is defined as in Equation (1), while Precision (Pre) is defined as the proportion of true positives out of all positive predictions:
In these formulas, TP (True Positives) refers to instances correctly classified as positive, while FP (False Positives) denotes negative instances that are incorrectly classified as positive. Similarly, FN (False Negatives) represents positive instances that are incorrectly classified as negative, and TN (True Negatives) refers to instances correctly classified as negative.
3.1 Statistical analysis
We also performed a statistical analysis to test the differences between classification results. A t-test was conducted, and a P-value > 0.05 was interpreted as indicating insufficient evidence to conclude a significant difference between the classification results.
4 Results
4.1 Model performance
The trained ViT model demonstrated outstanding performance in this binary classification task, achieving an AUC of 0.99, which underscores the effectiveness of ViT architectures in extracting discriminative features from CCM images. Specifically, the model correctly classified 75 out of 81 healthy controls, with only one misclassification among DPN cases, resulting in a sensitivity of 98%, specificity of 92%, and a high F1-score of 95%.
4.2 Comparison against other methods
We further compared our method against ResNet50 pretrained on ImageNet (Deng et al., 2009), which has previously been used for detecting DPN in CCM images (Preston et al., 2022; Meng et al., 2023). Table 2 presents the AUC, sensitivity, specificity, and F1-scores for both methods. Our proposed ViT model outperformed ResNet50, achieving a higher AUC compared to ResNet50. Although ResNet50 exhibited a slightly higher F1-score than the ViT model, the difference was not significant (P = 0.397).
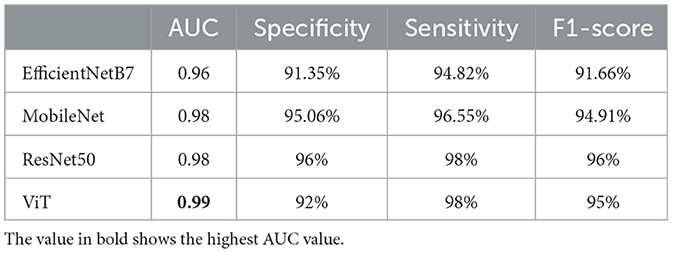
Table 2. Comparison of AUC, sensitivity, specificity, and F1-score between the ViT model and other methods for the binary classification task.
Besides ResNet50, we have compared our results to well-known DL models including the EfficientNetB7 (Tan and Le, 2019), and MobileNet (Howard, 2017), chosen for their exceptional performance in tasks such as feature extraction and image classification, particularly their capability to detect anomalies within images. In Table 2, we reported a remakrbale AUC for MobileNet of 0.98. However, our ViT beats all these models in term of AUC and F1-score.
To enhance the interpretability of our model's predictions on test images and provide clinicians with greater insight, we employed Grad-CAM (Selvaraju et al., 2017). This attribution method uses the gradients flowing into the final convolutional layer to generate a coarse “attribution map,” visually highlighting the regions of the image with the strongest influence on the classification outcome. In essence, the map reveals which parts of the image were most significant in the model's decision-making process. Figure 2 illustrates original and Grad-CAM images from healthy controls (Figure 2A) and patients with DPN (Figure 2B). This clearly identifies areas where corneal nerves are located as providing the most influence to identify DPN.
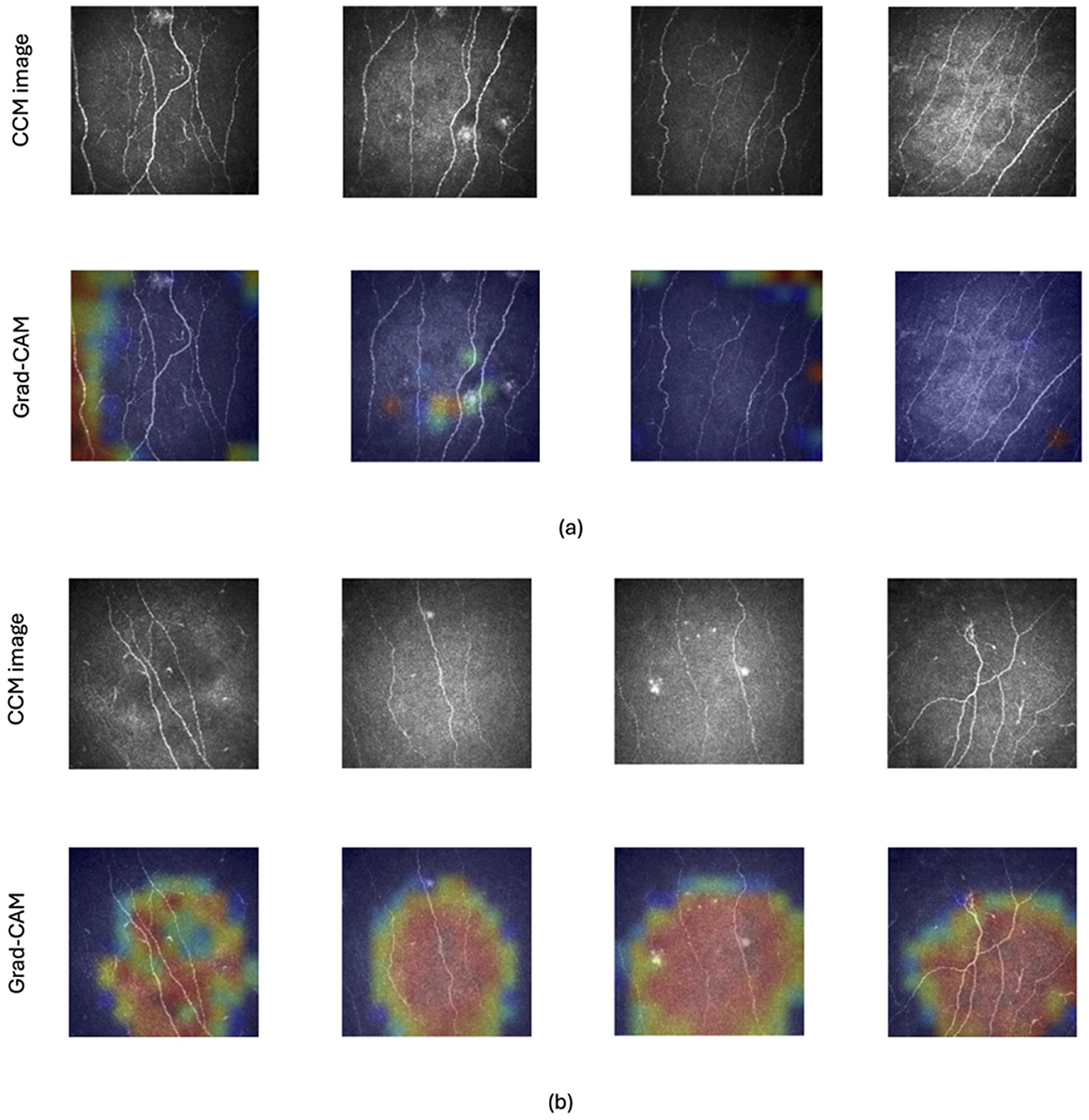
Figure 2. Example of CCM images of healthy controls and patients with and without DPN along with their corresponding Grad-CAM images. Grad-CAM creates a heatmap where hotter colors (red) indicate the image regions that had the strongest impact on the model's classification decision. Cooler colors (orange, yellow, and green) represent progressively less influential areas, with shades of blue highlighting the regions with the weakest influence. The top two rows (A) display images of healthy control subjects and the bottom two rows (B) present images of patients with DPN.
5 Discussion
In our research, e investigated the potential of the Vision Transformer (ViT) model for classifying corneal confocal microscopy (CCM) images. By splitting images into patches and processing them within a transformer-based architecture, the ViT model effectively captures both local and global features, making it particularly well-suited for tasks requiring a comprehensive view of image content. To our knowledge, this is the first study to apply a ViT model for analyzing and classifying CCM images, achieving a high AUC of 0.99, which surpasses results reported in previous studies (Silva et al., 2015; Salahouddin et al., 2021; Alam et al., 2022; Preston et al., 2022; Meng et al., 2023). These classification results underscore the effectiveness of ViT in distinguishing between healthy controls and individuals with DPN in this context.
To enhance model interpretability and provide clinicians with insights into the ViT model's predictions, we employed Grad-CAM as an explainability tool. Recognizing that Grad-CAM is traditionally designed for CNNs with their hierarchical convolutional layers, we adapted this technique for our ViT architecture. Instead of relying on convolutional feature maps, we leveraged the attention maps generated by the Transformer encoder. By analyzing the attention weights assigned to different image patches, we effectively identified the regions within the CCM images that most significantly influenced the model's predictions. The generated heatmaps, qualitatively validated for their effectiveness, highlighted regions within images that are clinically relevant for diagnosing DPN, such as corneal nerves. This approach not only provides valuable insights into the model's decision-making process but also enhances clinician trust and confidence in its predictions, thereby facilitating potential adoption in clinical settings. To address this, one of the co-authors (RAM), a pioneer of corneal nerve analysis undertook visual inspection of the Grad-CAM heatmaps and confirmed that the highlighted regions were identifying corneal nerve fiber loss, a hallmark of DPN.
While ResNet, a widely adopted CNN architecture, demonstrated competitive results, it has certain limitations. ResNet requires a fixed input size, which can be restrictive when working with images of varying dimensions (Salehi et al., 2023), and it struggles to capture long-range dependencies, which are often essential for identifying complex patterns. In contrast, ViT models, in principle, can process images of different dimensions due to their inherent self-attention mechanisms and the fact of processing images with patches. Practical implementations often necessitate training with a specific input resolution for computational efficiency. In our case, the input to our ViT model consists of CCM images with their original size of 384 × 384 pixels. However, during the internal image augmentation process within the model, these images are resized to 256 × 256 pixels. This choice was made to optimize training efficiency by enabling efficient batch processing and optimized memory usage, leading to faster training times. This approach, while introducing a degree of constraint, does not inherently limit the model's generalizability to images of different dimensions. ViT's architecture, with its self-attention mechanisms, allows it to flexibly handle varying input sizes while capturing long-range dependencies, making it a more adaptable and powerful choice for tasks that demand a deep understanding of image-wide context. In real-world applications, this approach, combined with the inherent flexibility of the ViT architecture, allows for a degree of adaptability to varying input dimensions.
Furthermore, ViTs are renowned for their scalability (Pan et al., 2021; Chen et al., 2022; Dehghani et al., 2023), as their performance typically improves with larger datasets and increased model complexity. This scalability is particularly advantageous for medical applications, where large datasets and robust models are often essential for achieving high diagnostic accuracy. Building on this scalability, our research demonstrates that ViT models can effectively detect DPN using CCM images without requiring complex pre-processing steps, segmentation, or adaptive feature extraction techniques.
This study, while demonstrating promising results, acknowledges several limitations. Firstly, the relatively small sample size (692 images) may limit the generalizability of the findings.
Secondly, the integration of this AI model into clinical practice presents several challenges. The computational demands of ViT models, while mitigated through optimizations employed in this study, may still pose challenges in resource-limited clinical settings.
Furthermore, the use of AI in healthcare raises important ethical considerations, including data privacy, algorithmic bias, and the potential for unintended consequences. Ensuring responsible and equitable AI development and deployment is paramount. To safeguard patient privacy while advancing AI models in healthcare, two promising approaches are federated learning and synthetic data generation. Federated learning enables model evaluation and refinement without transferring sensitive patient data, while synthetic data generation creates artificial data that mimics real data without containing any actual patient information. These innovative solutions offer a balance between model improvement and robust privacy protection.
These findings suggest that ViT models may offer a more efficient and accurate approach to DPN diagnosis compared to traditional methods. To fully harness the potential of ViTs, future research should focus on developing training sets encompassing a broader range of normal and abnormal pathologies, exploring the practical implementation of this algorithm in clinical workflows, and comparing its performance to existing diabetic neuropathy screening techniques. This will be crucial for translating this technology into real-world healthcare solutions.
This study serves as a foundation for future research that will address the identified shortcomings. Further research with larger, more diverse cohorts is warranted to confirm these initial observations. Moreover, incorporating other medical image modalities can be used to assess the robustness of the model in peripheral neuropathies classification.
In conclusion, this study presents a novel application of AI for the automated classification of CCM images, enabling rapid and objective detection of DPN. Our vision transformer-based model demonstrated remarkable accuracy in distinguishing patients with DPN from healthy controls. By eliminating the subjectivity and time-intensive processes of manual image segmentation and interpretation, this approach offers a faster and more consistent analysis. The integration of this AI-driven tool into clinical workflows has the potential to revolutionize DPN diagnosis by enabling quicker decision-making, facilitating timely interventions, and ultimately improving patient outcomes. While the results are promising, further research is needed to refine the model and extend its applicability. Future studies should utilize larger datasets, including diabetic patients with diverse comorbidities, to enhance model interpretability and provide clinicians with more actionable insights. This research highlights the transformative potential of AI in medical diagnostics. By automating complex tasks and improving diagnostic accuracy, AI-driven solutions can advance patient care and contribute to the effective management of diabetes-related neuropathies.
Data availability statement
The data analyzed in this study is subject to the following licenses/restrictions: the dataset contains de-identified patient data and is subject to ethical and confidentiality requirements. Access is limited to authorized researchers for academic and non-commercial purposes only. Approval from the institutional review board is required before access is granted. All requests for data access should be submitted to RM. along with a detailed research proposal and signed data use agreement. Requests to access these datasets should be directed to cmFtMjA0NUBxYXRhci1tZWQuY29ybmVsbC5lZHU=.
Ethics statement
The study was approved by the North Manchester Research Ethics Committee. Informed consent: All study participants provided written informed consent. Registry and the registration no. of the study/trial: (Ethical approval number: #09/H1006/38). The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation was not required from the participants or the participants' legal guardians/next of kin in accordance with the national legislation and institutional requirements.
Author contributions
CBR: Conceptualization, Methodology, Project administration, Writing – original draft. INP: Data curation, Writing – review & editing. RAM: Data curation, Validation, Visualization, Writing – review & editing. AS: Conceptualization, Methodology, Supervision, Validation, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Acknowledgments
The authors thank all volunteers for their participation.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The author(s) declare that no Gen AI was used in the creation of this manuscript.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Alam, U., Anson, M., Meng, Y., Preston, F., Kirthi, V., Jackson, T. L., et al. (2022). Artificial intelligence and corneal confocal microscopy: the start of a beautiful relationship. J. Clin. Med. 11:6199. doi: 10.3390/jcm11206199
Bazi, Y., Bashmal, L., Rahhal, M. M. A., Dayil, R. A., and Ajlan, N. A. (2021). Vision transformers for remote sensing image classification. Rem. Sens. 13:516. doi: 10.3390/rs13030516
Chen, S., Ge, C., Tong, Z., Wang, J., Song, Y., Wang, J., et al. (2022). “Adaptformer: adapting vision transformers for scalable visual recognition,” in Advances in Neural Information Processing Systems, 16664–16678.
Dehghani, M., Djolonga, J., Mustafa, B., Padlewski, P., Heek, J., Gilmer, J., et al. (2023). “Scaling vision transformers to 22 billion parameters,” in International Conference on Machine Learning (PMLR), 7480–7512.
Deng, J., Dong, W., Socher, R., Li, L.-J., Li, K., and Fei-Fei, L. (2009). “ImageNet: a large-scale hierarchical image database,” in 2009 IEEE Conference on Computer Vision and Pattern Recognition (IEEE), 248–255. doi: 10.1109/CVPR.2009.5206848
Ding, M., Qu, A., Zhong, H., Lai, Z., Xiao, S., and He, P. (2023). An enhanced vision transformer with wavelet position embedding for histopathological image classification. Patt. Recognit. 140:109532. doi: 10.1016/j.patcog.2023.109532
Dosovitskiy, A., Beyer, L., Kolesnikov, A., Weissenborn, D., Zhai, X., Unterthiner, T., et al. (2020). An image is worth 16x16 words: transformers for image recognition at scale. arXiv preprint arXiv:2010.11929.
Eid, S. A., Rumora, A. E., Beirowski, B., Bennett, D. L., Hur, J., Savelieff, M. G., et al. (2023). New perspectives in diabetic neuropathy. Neuron 111, 2623–2641. doi: 10.1016/j.neuron.2023.05.003
Elafros, M. A., Andersen, H., Bennett, D. L., Savelieff, M. G., Viswanathan, V., Callaghan, B. C., et al. (2022). Towards prevention of diabetic peripheral neuropathy: clinical presentation, pathogenesis, and new treatments. Lancet Neurol. 21, 922–936. doi: 10.1016/S1474-4422(22)00188-0
Howard, A. G. (2017). MobileNets: efficient convolutional neural networks for mobile vision applications. arXiv preprint arXiv:1704.04861.
Jensen, T. S., Karlsson, P., Gylfadottir, S. S., Andersen, S. T., Bennett, D. L., Tankisi, H., et al. (2021). Painful and non-painful diabetic neuropathy, diagnostic challenges and implications for future management. Brain 144, 1632–1645. doi: 10.1093/brain/awab079
Long, Z., McCreadie, R., and Imran, M. (2024). CrisisViT: a robust vision transformer for crisis image classification. arXiv preprint arXiv:2401.02838.
Meng, Y., Preston, F. G., Ferdousi, M., Azmi, S., Petropoulos, I. N., Kaye, S., et al. (2023). Artificial intelligence based analysis of corneal confocal microscopy images for diagnosing peripheral neuropathy: a binary classification model. J. Clin. Med. 12:1284. doi: 10.3390/jcm12041284
Ong, K. L., Stafford, L. K., McLaughlin, S. A., Boyko, E. J., Vollset, S. E., Smith, A. E., et al. (2023). Global, regional, and national burden of diabetes from 1990 to 2021, with projections of prevalence to 2050: a systematic analysis for the global burden of disease study 2021. Lancet 402, 203–234. doi: 10.1016/S0140-6736(23)01301-6
Pan, Z., Zhuang, B., Liu, J., He, H., and Cai, J. (2021). “Scalable vision transformers with hierarchical pooling,” in Proceedings of the IEEE/CVF International Conference on Computer Vision, 377–386. doi: 10.1109/ICCV48922.2021.00043
Ponirakis, G., Elhadd, T., Al Ozairi, E., Brema, I., Chinnaiyan, S., Taghadom, E., et al. (2022). Prevalence and risk factors for diabetic peripheral neuropathy, neuropathic pain and foot ulceration in the Arabian gulf region. J. Diab. Investig. 13, 1551–1559. doi: 10.1111/jdi.13815
Ponirakis, G., Elhadd, T., Chinnaiyan, S., Hamza, A. H., Sheik, S., Kalathingal, M. A., et al. (2021). Prevalence and risk factors for diabetic neuropathy and painful diabetic neuropathy in primary and secondary healthcare in Qatar. J. Diab. Investig. 12, 592–600. doi: 10.1111/jdi.13388
Preston, F. G., Meng, Y., Burgess, J., Ferdousi, M., Azmi, S., Petropoulos, I. N., et al. (2022). Artificial intelligence utilising corneal confocal microscopy for the diagnosis of peripheral neuropathy in diabetes mellitus and prediabetes. Diabetologia 65, 457–466. doi: 10.1007/s00125-021-05617-x
Pritchard, N., Edwards, K., Dehghani, C., Fadavi, H., Jeziorska, M., Marshall, A., et al. (2014). Longitudinal assessment of neuropathy in type 1 diabetes using novel ophthalmic markers (landmark): study design and baseline characteristics. Diabetes Res. Clin. Pract. 104, 248–256. doi: 10.1016/j.diabres.2014.02.011
Salahouddin, T., Petropoulos, I. N., Ferdousi, M., Ponirakis, G., Asghar, O., Alam, U., et al. (2021). Artificial intelligence-based classification of diabetic peripheral neuropathy from corneal confocal microscopy images. Diabetes Care 44:e151. doi: 10.2337/dc20-2012
Salehi, A. W., Khan, S., Gupta, G., Alabduallah, B. I., Almjally, A., Alsolai, H., et al. (2023). A study of CNN and transfer learning in medical imaging: advantages, challenges, future scope. Sustainability 15:5930. doi: 10.3390/su15075930
Selvaraju, R. R., Cogswell, M., Das, A., Vedantam, R., Parikh, D., and Batra, D. (2017). “Grad-CAM: visual explanations from deep networks via gradient-based localization,” in Proceedings of the IEEE International Conference on Computer Vision, 618–626. doi: 10.1109/ICCV.2017.74
Silva, S. F., Gouveia, S., Gomes, L., Negrão, L., Quadrado, M. J., Domingues, J. P., et al. (2015). “Diabetic peripheral neuropathy assessment through texture based analysis of corneal nerve images,” in Journal of Physics: Conference Series (IOP Publishing), 012002. doi: 10.1088/1742-6596/616/1/012002
Keywords: artificial intelligence, diabetic neuropathy, corneal confocal microscopy, image classification, disease diagnosis
Citation: Ben Rabah C, Petropoulos IN, Malik RA and Serag A (2025) Vision transformers for automated detection of diabetic peripheral neuropathy in corneal confocal microscopy images. Front. Imaging 4:1542128. doi: 10.3389/fimag.2025.1542128
Received: 09 December 2024; Accepted: 08 January 2025;
Published: 03 February 2025.
Edited by:
Simone Bonechi, University of Siena, ItalyReviewed by:
Minhyeok Lee, Chung-Ang University, Republic of KoreaFusong Jiang, Shanghai Jiao Tong University, China
Copyright © 2025 Ben Rabah, Petropoulos, Malik and Serag. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ahmed Serag, YWZzNDAwMkBxYXRhci1tZWQuY29ybmVsbC5lZHU=; Chaima Ben Rabah, Y2hiNDAzNkBxYXRhci1tZWQuY29ybmVsbC5lZHU=
 Chaima Ben Rabah
Chaima Ben Rabah Ioannis N. Petropoulos
Ioannis N. Petropoulos Rayaz A. Malik
Rayaz A. Malik Ahmed Serag
Ahmed Serag