
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Imaging, 20 June 2024
Sec. Imaging Applications
Volume 3 - 2024 | https://doi.org/10.3389/fimag.2024.1416114
This article is part of the Research TopicDeep Learning for Medical Imaging ApplicationsView all 15 articles
Introduction: Self-supervised learning (SSL) is a strategy for addressing the paucity of labelled data in medical imaging by learning representations from unlabelled images. Contrastive and non-contrastive SSL methods produce learned representations that are similar for pairs of related images. Such pairs are commonly constructed by randomly distorting the same image twice. The videographic nature of ultrasound offers flexibility for defining the similarity relationship between pairs of images.
Methods: We investigated the effect of utilizing proximal, distinct images from the same B-mode ultrasound video as pairs for SSL. Additionally, we introduced a sample weighting scheme that increases the weight of closer image pairs and demonstrated how it can be integrated into SSL objectives.
Results: Named Intra-Video Positive Pairs (IVPP), the method surpassed previous ultrasound-specific contrastive learning methods' average test accuracy on COVID-19 classification with the POCUS dataset by ≥ 1.3%. Detailed investigations of IVPP's hyperparameters revealed that some combinations of IVPP hyperparameters can lead to improved or worsened performance, depending on the downstream task.
Discussion: Guidelines for practitioners were synthesized based on the results, such as the merit of IVPP with task-specific hyperparameters, and the improved performance of contrastive methods for ultrasound compared to non-contrastive counterparts.
Medical ultrasound (US) is a modality of imaging that uses the amplitude of ultrasonic reflections from tissues to compose a pixel map. With the advent of point-of-care ultrasound devices, ultrasound has been increasingly applied in a variety of diagnostic clinical settings, such as emergency care, intensive care, oncology, and sports medicine (Yim and Corrado, 2012; Whitson and Mayo, 2016; Sood et al., 2019; Soni et al., 2020; Lau and See, 2022). It possesses several qualities that distinguish it from other radiological modalities, including its portability, lack of ionizing radiation, and affordability. Despite morphological distortion of the anatomy, ultrasound has been shown to be comparable to radiological alternatives, such as chest X-ray and CT, for several diagnostic tasks (van Randen et al., 2011; Alrajhi et al., 2012; Nazerian et al., 2015).
Deep learning has been extensively studied as a means to automate diagnostic tasks in ultrasound. As with most medical imaging tasks, the lack of open access to large datasets is a barrier to the development of such systems, since large training sets are required for deep computer vision models. Organizations that have privileged access to large datasets are also faced with the problem of labeling ultrasound data. Indeed, many point-of-care ultrasound examinations in acute care settings are not archived or documented (Hall et al., 2016; Kessler et al., 2016).
When unlabeled examinations are abundant, researchers turn to unsupervised representation learning to produce pretrained deep learning models that can be fine-tuned using labeled data. Self-supervised learning (SSL) is a broad category of methods that has been explored for problems in diagnostic ultrasound imaging. Broadly, SSL refers to the supervised pretraining of a machine learning model for a task that does not require labels for the task of interest. The pretraining task (i.e., pretext task) is a supervised learning task where the target is a quantity that is computed from unlabeled data. After optimizing the model's performance on the pretext task, the weights are recast as initial weights for a new model that is trained to solve the task of interest (referred to as the downstream task). If the pretrained model has learned to produce representations of salient information in ultrasound images, then it is likely that it can be fine-tuned to perform the downstream task more proficiently than had it been randomly initialized. Contrastive learning is a type of pretext task in SSL that involves predicting whether two inputs are related (i.e., positive pairs) or unrelated (i.e., negative pairs). In computer vision, a common way to define positive pairs is to apply two randomly defined transformations to an image, producing two distorted views of the image with similar content. Positive pairs satisfy a pairwise relationship that indicate semantic similarity. All other pairs of images are regarded as negative pairs. Non-contrastive methods disregard negative pairs, focusing only on reducing the differences between representations of positive pairs.
Unlike other forms of medical imaging, US is a dynamic modality acquired as a stream of frames, resulting in a video. Despite this, there are several US interpretation tasks that can be performed by assessing a still US image. Previous studies exploring SSL in US have exploited the temporal nature of US by defining contrastive learning tasks with intra-video positive pairs – positive pairs comprised of images derived from the same video (Chen et al., 2021; Basu et al., 2022). Recent theoretical results indicate that the pairwise relationship must align with the labels of the downstream task in order to guarantee that self-supervised pretraining leads to non-inferior performance on the downstream task (Balestriero and LeCun, 2022). For classification tasks, this means that positive pairs must have the same class label. Due to the dynamic nature of US, one cannot assume that all frames in a US video possess the same label for all downstream US interpretation tasks. As a result, it may be problematic to indiscriminately designate any pair of images originating from the same US video as a positive pair. Moreover, since US videos are often taken sequentially as a part of the same examination or from follow-up studies of the same patient, different US videos may bear a striking resemblance to each other. It follows that designating images from different US videos as negative pairs may result in negative pairs that closely resemble positive pairs.
In this study, we aimed to examine the effect of proximity and sample weighting of intra-video positive pairs for common SSL methods. We also intended to determine if non-contrastive methods are more suitable for classification tasks in ultrasound. Since non-contrastive methods do not require the specification of negative pairs, we conjectured that non-contrastive methods would alleviate the issue of cross-video similarity and yield stronger representations for downstream tasks. Our contributions and results are summarized as follows:
• A method for sampling intra-video positive pairs for joint embedding SSL with ultrasound.
• A sample weighting scheme for joint embedding SSL methods that weighs positive pairs according to the temporal or spatial distance between them in their video of origin.
• A comprehensive assessment of intra-video positive pairs integrated with SSL pretraining methods, as measured by downstream performance in B-mode and M-mode lung US classification tasks. We found that, with proper downstream task-specific hyperparameters, intra-video positive pairs can improve performance compared to the standard practice of producing two distortions of the same image.
• An comparison of contrastive and non-contrastive learning for multiple lung US classification tasks. Contrary to our initial belief, a contrastive method outperformed multiple non-contrastive methods on multiple lung US downstream tasks.
Figure 1 encapsulates the novel methods proposed in this study. To the authors' knowledge, there are no preceding studies that systematically investigate the effect of sampling multiple images from the same US video in non-contrastive learning. More generally, we believe that this study is the first to integrate sample weights into non-contrastive objectives.
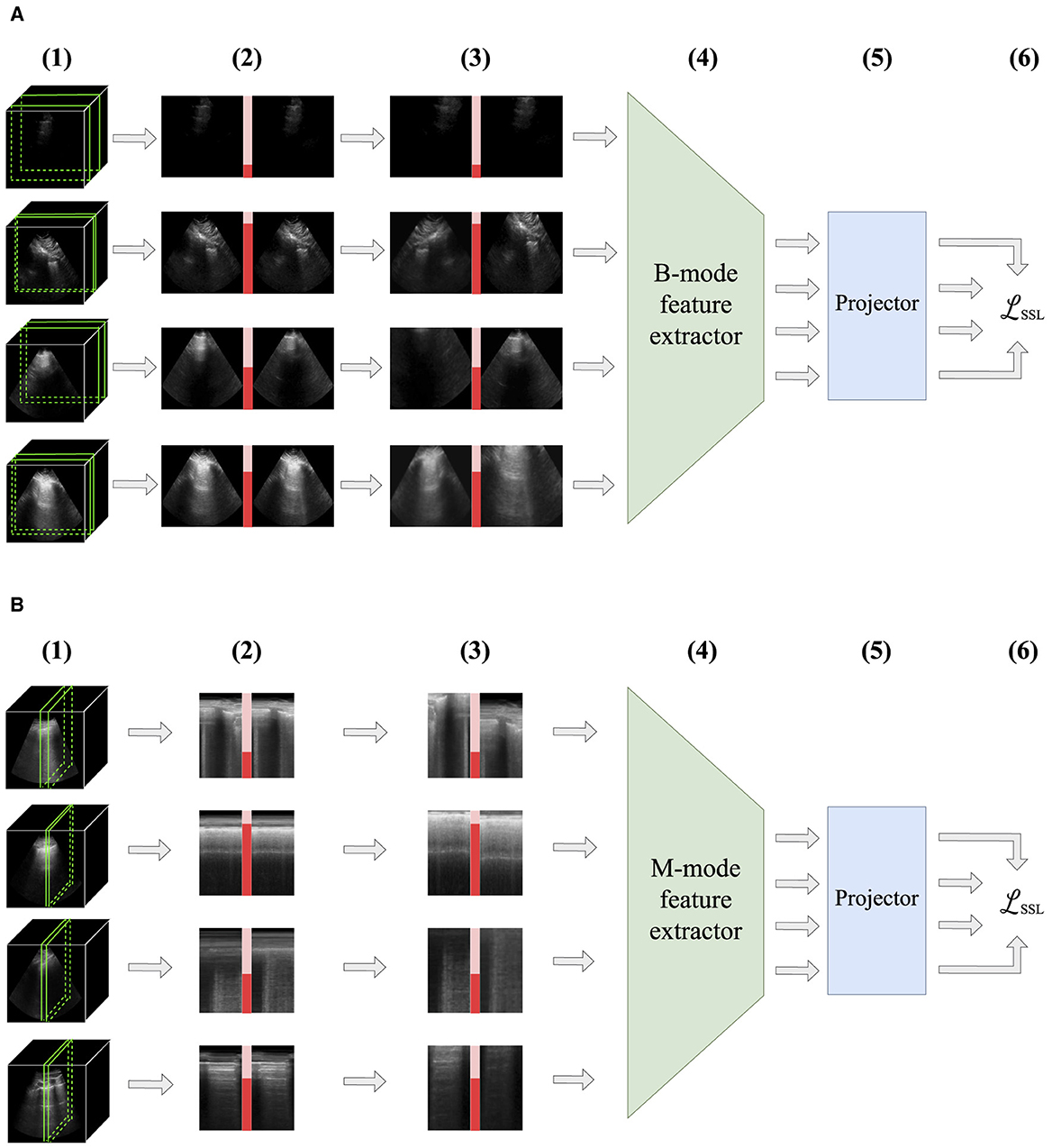
Figure 1. An overview of the methods introduced in this study. Positive pairs of images separated by no more than a threshold are sampled from the same B-mode video (1). Sample weights inversely proportional to the separation between each image (red bars) are calculated for each pair (2). Random transformations are applied to each image (3). Images are sent to a neural network consisting of a feature extractor (4) and a projector (5) connected in series. The outputs are used to calculate the objective (6). The trained feature extractor is retained for downstream supervised learning tasks. (A) For B-mode ultrasound, positive pairs are temporally separated images from the same video. (B) For M-mode ultrasound, positive pairs are spatially separated images from the same video.
Having gained popularity in recent years in multiple imaging modalities, joint embedding SSL refers to a family of methods where the pretext task is to produce output vectors (i.e., embeddings) that are close for examples satisfying a similarity pairwise relationship. Pairs of images satisfying this relationship are known as positive pairs, and they assumed to share semantic content with respect to the downstream task. For example, positive pairs could belong to the same class in a downstream supervised learning classification task. On the other hand, negative pairs are pairs of images that do not satisfy the pairwise relationship. In the label-free context of SSL, positive pairs are often constructed by sampling distorted versions of a single image (Chen et al., 2020; Grill et al., 2020; Zbontar et al., 2021; Bardes et al., 2022). The distortions are sampled from a distribution of sequentially applied transformations that are designed to preserve the semantic content of the image. Horizontal reflection is a common example of a transformation that meets this criterion in many forms of imaging.
The architecture of joint embedding models commonly consists of two modules connected in series: a feature extractor and a projector. The feature extractor is typically a convolutional neural network (CNN) or a variant of a vision transformer, while the projector is a multi-layer perceptron. After pretraining, the projector is discarded and the feature extractor is retained for weight initialization for the downstream task.
Contrastive learning and non-contrastive learning are two major categories of joint embedding methods. Contrastive methods rely on objectives that explicitly attract positive pairs and repel negative pairs in embedding space. Many of these methods adopt the InfoNCE objective (Oh Song et al., 2016), which may be viewed as cross-entropy for predicting which combination of embeddings in a batch correspond to a positive pair. In most contrastive methods, positive pairs and negative pairs are distorted versions of the same image and different images, respectively. MoCo is a contrastive method that computes pairs of embeddings using two feature extractors: a “query” encoder and a “key” encoder (He et al., 2020). The key encoder, which is an exponentially moving average of the query encoder, operates on negative examples Its output embeddings are queued to avoid recomputation of negative embeddings. SimCLR (Chen et al., 2020) is a widely used contrastive method that employs a variant of the InfoNCE objective that does not include the embedding of the positive pair in the demoninator (Oh Song et al., 2016). It does not queue negative embeddings, relying instead on large batches of negative examples.
Non-contrastive methods dispense with negative pairs altogether, limiting their focus to reducing the difference between embeddings of positive pairs. By design, they address the information collapse problem – a degenerate solution wherein all examples map to a null representation vector. Self-distillation non-contrastive methods use architectural and asymmetrical training strategies to avoid collapse [e.g., BYOL (Grill et al., 2020)]. Information maximization non-contrastive methods address collapse by employing objectives that maximize the information content of the embedding dimensions. For instance, the Barlow Twins method is a composite objective that contains a term for penalizing dimensional redundancy for batches of embeddings, in addition to a term for the distances between embeddings of individual positive pairs (Zbontar et al., 2021). VICReg introduced an additional term that explicitly maximizes variance across dimensions for batches of embeddings (Bardes et al., 2022). Despite a common belief that contrastive methods need much larger batch sizes than non-contrastive methods, recent evidence showed that hyperparameter tuning can boost the former's performance with smaller batch sizes (Bordes et al., 2023). Non-contrastive methods have been criticized for requiring embeddings with greater dimensionality than the representations outputted by the feature extractor; however, a recent study suggested that the difference may be alleviated through hyperparameter and design choices (Garrido et al., 2022).
Theoretical works have attempted to unify contrastive and non-contrastive methods. Balestriero and LeCun (2022) found that SimCLR, VICReg, and Barlow Twins are all manifestations of spectral embedding methods. Based on their results, they recommended that practitioners define a pairwise relationship that aligns with the downstream task. For example, if the downstream task is classification, then positive pairs should have the same class. Garrido et al. (2022) challenged the widely held assumptions that non-contrastive methods perform better than contrastive methods and that non-contrastive methods rely on large embedding dimensions. They showed that the methods perform comparatively on benchmark tasks after hyperparameter tuning and that VICReg can be modified to reduce the dependence on large embeddings (Garrido et al., 2022).
Ultrasound is a dynamic imaging modality that is typically captured as a sequence of images and stored as a video. As such, images originating from the same video are highly correlated and are likely to share semantic content. Accordingly, recent works have developed US-specific contrastive learning methods that construct positive pairs from the same video. The Ultrasound Contrastive Learning (USCL) method (Chen et al., 2021) is a derivative of SimCLR in which positive pairs are weighted sums of random images within the same video [i.e., the mixup operation (Zhang et al., 2017)], while negative pairs are images from different videos. They reported an improvement on the downstream task of COVID-19 classification with the POCUS dataset (Born et al., 2020). Improving on USCL, Meta-USCL concurrently trains a separate network that learns to weigh positive pairs (Chen et al., 2022). The work was inspired by the observation that the intra-video positive pairs may exhibit a wide range of semantic similarity or dissimilarity. Lastly, the HiCo method's objective is the sum of a softened InfoNCE loss calculated for the feature maps outputted by various model blocks (Zhang et al., 2022). The authors reported greatly improved performance with respect to USCL.
Standard non-contrastive methods have been applied for various tasks in US imaging. In addition to assessing contrastive methods, Anand et al. (2022) conducted pretraining with two self-distillation non-contrastive methods [BYOL (Grill et al., 2020) and DINO (Caron et al., 2021)] on a large dataset of echocardiograms. BYOL pretraining has also been applied in anatomical tracking tasks (Liang et al., 2023). Information maximization methods have been investigated for artifact detection tasks in M-mode and B-mode lung ultrasound (VanBerlo et al., 2023a,b). To our best knowledge, no studies have trialed non-contrastive learning methods for B-mode ultrasound with intra-video positive pairs. The present study seeks to address this gap in the literature by investigating the effect of sampling positive pairs from the same video on the efficacy of non-contrastive pretraining for tasks in ultrasound.
We consider the standard joint embedding scenario where unlabeled data are provided and the goal is to maximize the similarity between embeddings of positive pairs. In contrastive learning, the goal is augmented by maximizing the dissimilarity of negative pairs. Let x1 and x2 denote a positive pair of US images. Self-supervised pretraining results in a feature extractor f(x) that outputs representation vector h. The goal of SSL is to produce a feature extractor that is a better starting point for learning the downstream task than random initialization.
In this study, we propose a simple method for sampling and weighing positive pairs in the joint embedding setting that can be adopted for any joint embedding SSL method. We adopt SimCLR (Chen et al., 2020), Barlow Twins (Zbontar et al., 2021) and VICReg (Bardes et al., 2022) for our experiments. In these methods, a MLP projector is appended to the feature extractor during pretraining. z = g(h) = g(f(x)) is the embedding vector outputted by the projector. The SSL objective is then computed in embedding space.
Recall that positive pairs are images that are semantically related. Previous work in contrastive SSL for US has explored the use of intra-video positive pairs (Chen et al., 2021, 2022; Basu et al., 2022; Zhang et al., 2022). A problem with naively sampling intra-video positive pairs is that it rests on the assumption that all images in the video are sufficiently similar. However, clinically relevant signs commonly surface and disappear within the same US video as the US probe and/or the patient move. For example, B-lines are an artifact in lung US that signify diseased lung parenchyma (Soni et al., 2020). B-lines may disappear and reappear as the patient breathes or as the sonographer moves the probe. The A-line artifact appears in the absence of B-lines, indicating normal lung parenchyma. In the absence of patient context, an image containing A-lines and an image containing B-lines from the same video convey very different impressions. While most previous methods only considered inter-video images to be negative pairs, Basu et al. (2022) argued that that temporally distant intra-video pairs of US images are more likely to be dissimilar, which inspired their method that treats such instances as negative pairs. Despite this, we argue that distant intra-video images may sometimes exhibit similar content. For example, the patient and probe may remain stationary throughout the video, or the probe may return to its original position and/or orientation. Moreover, periodic physiological processes such as the respiratory cycle may result in temporally distant yet semantically similar images. Without further knowledge of the US examinations in a dataset, we conjectured that it may be safest to only assume that positive pairs are intra-video images that are close to each other. Closer pairs are likely to contain similar semantic content, yet they harbor different noise samples that models should be invariant to. In summary, this method distinguishes itself from prior work by only considering proximal frames to be positive pairs and treating distant pairs as neither positive nor negative pairs.
For B-mode US videos, we define positive pairs as intra-video images x1 and x2 that are temporally separated by no more than δmax seconds. To accomplish this, x1 is randomly drawn from the video's images, and x2 is randomly drawn from the set of images that are within δt seconds of x1. The frame rate of the videos must be known in order to determine which images are sufficiently close to x1. Note that videos with higher frame rates will provide more candidates for positive pairs, potentially increasing the diversity of pairs with respect to naturally occurring noise.
A similar sampling scheme is applied for M-mode US images. Like previous studies, we define M-mode images as vertical slices through time of a B-mode video, taken at a specific x-coordinate in the video (Jasčǔr et al., 2021; VanBerlo et al., 2022b, 2023b). The x-axis of an M-mode image is time, and its y-axis is the vertical dimension of the B-mode video. We define positive pairs to be M-mode images whose x-coordinates differ by no more than δx pixels. To avoid resolution differences, all B-mode videos are resized to the same width and height prior to sampling M-mode images. The positive pair sampling process for B-mode and M-mode images is depicted in Figure 2.
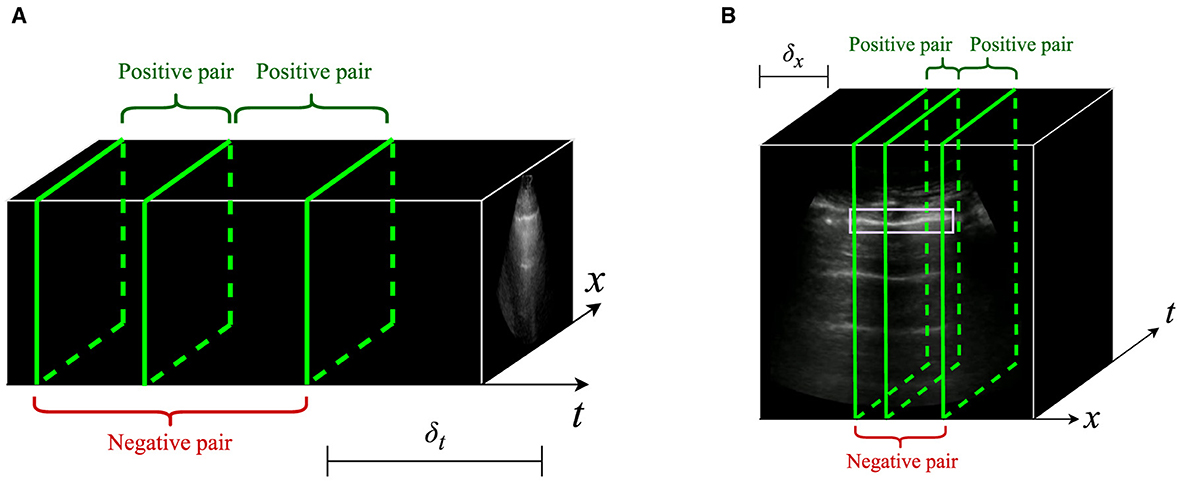
Figure 2. Illustration of intra-video positive pairs. Positive pairs are considered images that are no more than a threshold apart from each other within the same ultrasound video. (A) For B-mode ultrasound, positive pairs are frames in the same video that are within δt seconds of each other. (B) For M-mode ultrasound, positive pairs are M-mode images originating from the same B-video that are located within δx pixels from each other. In the context of lung ultrasound, M-mode images should intersect the pleural line (outlined in mauve).
As is customary in joint embedding methods, stochastic data augmentation is applied to each image, encouraging the feature extractor to become invariant to semantically insignificant differences. Any data augmentation pipeline may be adopted for this formulation of intra-video positive pairs; however, we recommend careful selection of transformations and the distributions of their parameters to ensure that the pairwise relationship continues to be consistent with the downstream US task.
The chance that intra-video images are semantically related decreases as temporal or spatial separation increases. To temper the effect of unrelated positive pairs, we apply sample weights to positive pairs in the SSL objective according to their temporal or spatial distance. Distant pairs are weighed less than closer pairs. For a positive pair of B-mode images occurring at times t1 and t2 or M-mode images occurring at positions x1 and x2, the sample weight is calculated using Equation 1:
Sample weights were incorporated into each SSL objective trialed in this study. Accordingly, we modified the objective functions for SimCLR, Barlow Twins, and VICReg in order to weigh the contribution to the loss differently based on sample weights. Appendix 1 describes the revised objective functions. To the authors' knowledge, this study is the first to propose sample weighting schemes for the aforementioned self-supervised learning methods.
As was done in previous studies on on US-specific joint embedding methods (Chen et al., 2021, 2022; Basu et al., 2022; Zhang et al., 2022), we evaluate IVPP on the public POCUS lung US dataset (Born et al., 2020). This dataset contains 140 publicly sourced US videos (2116 images) labeled for three classes: COVID-19 pneumonia, non-COVID-19 pneumonia, and normal lung.1 When evaluating on POCUS, we pretrain on the public Butterfly dataset, which contains 22 unlabeled lung ultrasound videos (Butterfly Network, 2020).2
A-lines and B-lines are two cardinal artifact in B-mode lung US that can provide quick information on the status of a patient's lung tissue. A-lines are reverberation artifacts that are indicative of normal, clear lung parenchyma (Soni et al., 2020). On lung US, they as horizontal lines deep to the pleural line. Conversely, B-lines are indicative of diseased lung tissue (Soni et al., 2020). Generally, the two are mutually exclusive. We evaluate on the binary classification task of A-lines versus B-lines on lung US, as was done in previous work benchmarking joint embedding SSL methods for lung US tasks (VanBerlo et al., 2023a).
We use a private dataset of 25917 parenchymal lung US videos (5.9e6 images), hereafter referred to as ParenchymalLUS. It is a subset of a larger database of de-identified lung US videos that was partially labeled for previous work (Arntfield et al., 2021; VanBerlo et al., 2022b). Access to this database was permitted via ethical approval by Western University (REB 116838). Before experimentation, we split the labeled portion of ParenchymalLUS by anonymous patient identifier into training, validation, and test sets. The unlabeled portion of ParenchymalLUS was assembled by gathering 20000 videos from the unlabeled pool of videos in the database that were predicted to contain a parenchymal view of the lungs by a previously trained lung US view classifier (VanBerlo et al., 2022a). All videos from the same patient were in either the labeled or the unlabeled subset. Table 1 provides further information on the membership of ParenchymalLUS.
Lung sliding is a dynamic artifact that, when observed on a parenchymal lung US view, rules out the possibility of a pneumothorax at the site of the probe (Lichtenstein and Menu, 1995). The absence of lung sliding is suggestive of pneumothorax, warranting further investigation. On B-mode US, lung sliding manifests as a shimmering of the pleural line (Lichtenstein and Menu, 1995). The presence or absence of lung sliding is also appreciable on M-mode lung US images that intersect the pleural line (Lichtenstein et al., 2005; Lichtenstein, 2010). We evaluate on the binary lung sliding classification task, where positive pairs are M-mode images originating from the same B-mode video.
ParenchymalLUS is adopted for the lung sliding classification task. We use the same train/validation/test partition as described above. Following prior studies, we estimate the horizontal bounds of the pleural line using a previously trained object detection model (VanBerlo et al., 2022b) and use the top half of qualifying M-mode images, in decreasing order of total pixel intensity (VanBerlo et al., 2023b).
Unless otherwise stated, all feature extractors are initialized with ImageNet-pretrained weights. Similar studies concentrating on medical imaging have observed that this practice improves downstream performance when compared to random initialization (Azizi et al., 2021; VanBerlo et al., 2023b). Moreover, we designate fully supervised classifiers initialized with ImageNet-pretrained weights as a baseline against which to compare models pretrained with SSL.
Evaluation on POCUS follows a similar protocol employed in prior works (Chen et al., 2021; Basu et al., 2022). Feature extractors with the ResNet18 architecture (He et al., 2016) are pretrained on the Butterfly dataset. Prior to training on the POCUS dataset, a 3-node fully connected layer with softmax activation was appended to the pretrained feature extractor. Five-fold cross validation is conducted with POCUS by fine-tuning the final three layers of the pretrained feature extractor. Unlike prior works, we adopt the average across-folds validation accuracy, instead of taking the accuracy of the combined set of validation set predictions across folds. Presenting the results in this manner revealed the high variance of model performance across folds, which may be due to the benchmark dataset's small video sample size.
All experiments with ParenchymalLUS utilize the MobileNetV3-Small architecture as the feature extractor, which outputs a 576-dimensional representation vector (Howard et al., 2019). Feature extractors are pretrained on the union of the unlabeled videos and labeled training set videos in ParenchymalLUS. Performance is assessed via test set classification metrics. Prior to training on the downstream task, a single-node fully connected layer with sigmoid activation was appended to the pretrained feature extractor. We report the performance of linear classifiers trained on the frozen feature extractor's representations, along with classifiers that are fine-tuned end-to-end.
For each joint embedding method, the projectors were multilayer perceptrons with two 768-node layers, outputting 768-dimensional embeddings. Pretraining is conducted for 500 epochs using the LARS optimizer (You et al., 2019) with a batch size of 384 and a learning rate schedule with warmup and cosine decay as in Bardes et al. (2022).
The pretraining and training data augmentation pipelines consist of random transformations, including random cropping, horizontal reflection, brightness jitter, contrast jitter, and Gaussian blurring. Additional data preprocessing details are available in Appendix 2.
Source code will be made available upon publication.3
The two main proposed features of IVPP are intra-video positive pairs and distance-based sample weights. Accordingly, we assess the performance of IVPP across multiple assignments of the maximum image separation. Separate trials were conducted for SimCLR, Barlow Twins, and VICReg pretraining. For the COVID and AB tasks, we explored the values δt∈{0, 0.5, 1, 1.5} seconds. The LS task is defined for M-mode US, and so we explored δx∈{0, 5, 10, 15} pixels. The standardized width of B-mode US videos should be considered when determining an appropriate range for δx. Note that when δ = 0, sample weights are all 1 and therefore do not modify any of the SSL objectives investigated in this study.
Figure 3 summarizes the performance of IVPP on the public POCUS dataset after pretraining on the Butterfly dataset, which is measured by average test accuracy in 5-fold cross validation. In most cases, pretrained models obtained equal or greater average accuracy than the ImageNet-pretrained baseline, with the exception of Barlow Twins with δt = 0 and δt = 0.5. The performance of models pretrained with SimCLR, Barlow Twins, and VICReg peaked at different nonzero values of δt (0.5, 1, and 1.5 respectively), indicating a possible benefit of selecting temporally close yet distinct intra-video positive pairs. It was also observed across all three pretraining methods that the inclusion of sample weights resulted in worsened test AUC when δ = 0.5, but improved test AUC when δ = 1.0 and δ = 1.5.
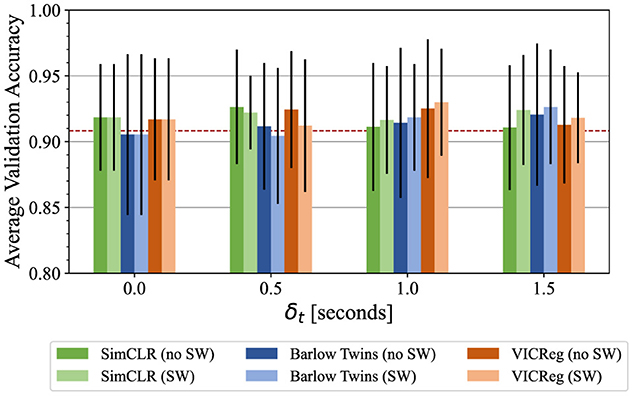
Figure 3. Average test accuracy across 5-fold cross validation on the POCUS dataset. Models were pretrained with a variety of intra-video positive pair thresholds with and without sample weights. Error bars indicate the standard deviation across folds. The dashed line indicates initialization with ImageNet-pretrained weights.
Similar experiments were conducted with ParenchymalLUS for the AB task and LS task, using B-mode and M-mode images respectively as input. ParenchymalLUS represents a scenario where there is an abundance of unlabeled data, which differs greatly from the preceding evaluation on public, yet small, datasets. The unlabeled and labeled portions of ParenchymalLUS contained at least an order of magnitude more videos than either the public Butterfly and POCUS datasets. B-mode and M-mode feature extractors were pretrained on the union of the unlabeled and training portions of ParenchymalLUS—one for each value of δ, with and without sample weights. For these evaluations, we use all training examples that have been assigned a label for the downstream task. Figure 4 provides a visual comparison of the test AUC obtained by linear feature representation classifiers and fine-tuned models for the AB and LS tasks. An immediate trend across both tasks and evaluation types is that SimCLR consistently outperformed Barlow Twins and VICReg, which are both non-contrastive methods. Furthermore, pretraining with non-contrastive methods often resulted in worse test AUC compared to initialization with ImageNet-pretrained weights. Another observation across all experiments was that there was no discernible trend for the effect of sample weights that was consistent for any task, pretraining method, δt, or δx.
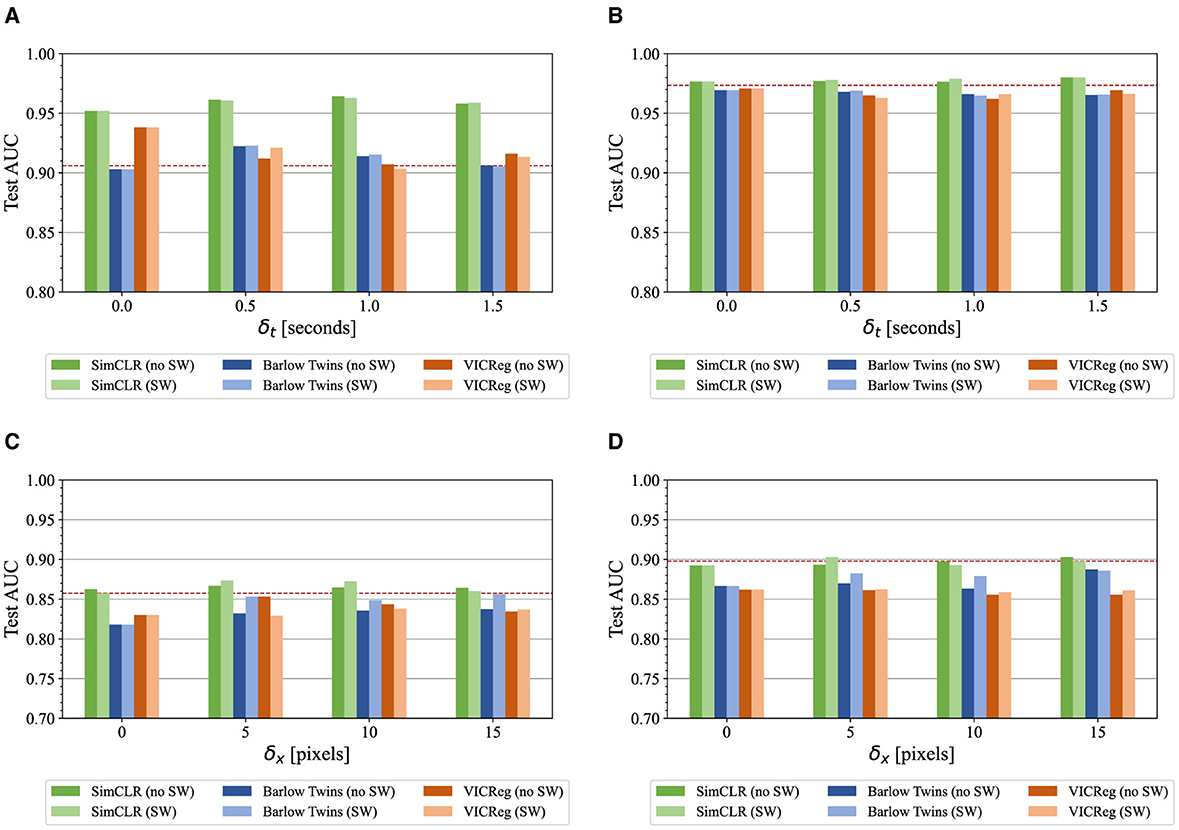
Figure 4. ParenchymalLUS test set AUC for the AB and LS binary classification tasks, calculated for models pretrained with a selection of contrastive and non-contrastive learning methods and employing a variety of intra-video positive pair thresholds with and without sample weights (SW). The dashed line indicates initialization with ImageNet-pretrained weights. (A) Linear classifiers for the AB task. (B) Fine-tuned classifiers for the AB task. (C) Linear classifiers for the LS task. (D) Fine-tuned classifiers for the LS task.
Focusing on AB, linear classifiers achieved the greatest performance when δt>0, with the exception of VICReg (Figure 4A). The use of SimCLR compared to the other pretraining methods appeared to be responsible for the greatest difference in test performance. As shown in Figure 4A, SimCLR-pretrained models outperformed non-contrastive methods, and were the only models that outperformed ImageNet-pretrained weights. The use of a nonzero δt resulted in slight improvement in combination with SimCLR pretraining, but degraded performance of non-contrastive methods.
Similar results were observed for the LS M-mode classification task. Models pretrained with SimCLR were the only ones that matched or surpassed fully supervised models. Nonzero δx generally improved the performance of linear classifiers, with δx = 5 pixels corresponding to the greatest test AUC for SimCLR and VICReg, and δx = 15 for Barlow Twins. Inclusion of sample weights appreciably improved the performance of Barlow Twins-pretrained models. Fine-tuned models pretrained with SimCLR performed similarly to fully supervised models, while non-contrastive methods resulted in degradation of test AUC.
Table 2 compares the top-performing IVPP-pretrained models for each SSL method with two prior US-specific contrastive learning methods— USCL (Chen et al., 2021) and US UCL (Basu et al., 2022). Of note is that all three self-supervised methods pretrained with IVPP outperformed ImageNet-pretrained initialization for POCUS, a task where very little pretraining and training data were utilized. For the B-mode and M-mode tasks assessed with ParenchymalLUS, a contrastive method (including the baseline) outperformed non-contrastive methods. Appendix 4 provides additional results that exhibit a similar trend with different pretraining batch sizes. Overall, the most salient result from the above experiments is that SimCLR, a contrastive method, outperformed both non-contrastive methods when unlabeled data is abundant.
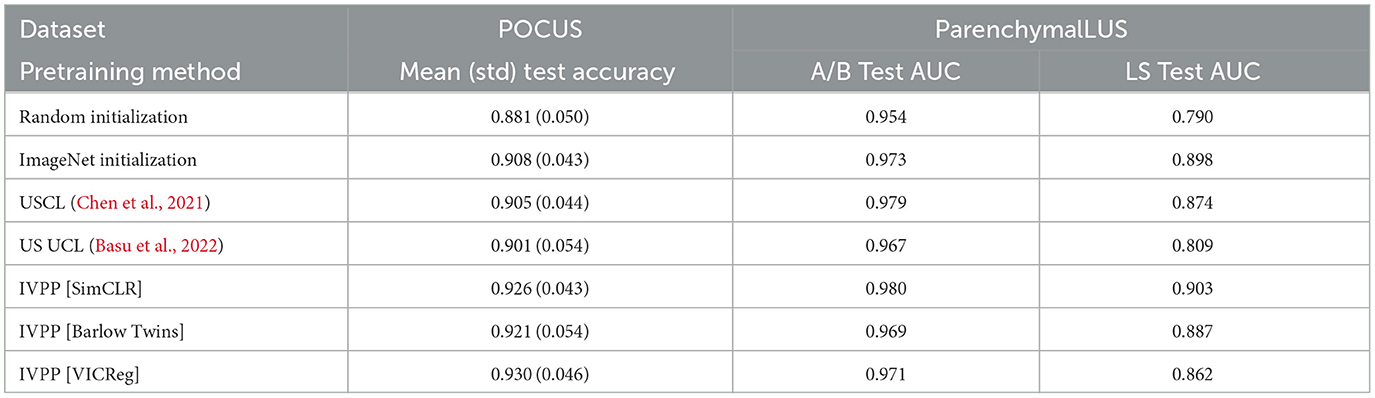
Table 2. Performance of fine-tuned models pretrained using IVPP compared to US-specific contrastive learning methods, USCL, and UCL, and to baseline random and ImageNet initializations.
ParenchymalLUS is much larger than public ultrasound datasets for machine learning. Although the majority of its videos are unlabeled, it contains a large number of labeled examples. To simulate a scenario where the fraction of examples that are labeled is much smaller, we investigated the downstream performance of models that were pretrained on all the unlabeled and training ParenchymalLUS examples and then fine-tuned on a very small subset of the training set.
Label efficiency investigations are typically conducted by fitting a model for the downstream task using progressively smaller fractions of training data to gauge how well self-supervised models fare in low-label scenarios. The results of these experiments may be unique to the particular training subset that is randomly selected. We designed an experiment to determine if the choice of δt, δx, or the introduction of sample weights influenced downstream performance in low-label settings. To reduce the chance of biased training subset sampling, we divided the training set into 20 subsets and repeatedly performed fine-tuning experiments on each subset for each pretraining method and δ value, with and without sample weights. To ensure independence among the subsets, we split the subsets by patient. Inspection of the central moments and boxplots from each distribution indicated that the normality and equal variance assumptions for ANOVA were not violated. For each pretraining method, a two-way repeated-measures analysis of variance (ANOVA) was performed to determine whether the mean test AUC scores across values of δ and sample weight usage were different. The independent variables were δ and the presence of sample weights, while the dependent variable was test AUC. Whenever the null hypothesis of the ANOVA was rejected, post-hoc paired t-tests were performed to compare the following:
• Pretraining with nonzero δ against standard positive pair selection (δ = 0).
• For the same nonzero δ value, sample weights against no sample weights.
For each group of post-hoc tests, the Bonferroni correction was applied to establish a family-wise error rate of α = 0.05. To ensure that each training subset was independent, we split the dataset by anonymous patient identifier. This was a necessary step because intra-video images are highly correlated, along with videos from the same patient. As a result, the task became substantially more difficult than naively sampling 5% of training images because the volume and heterogeneity of training examples was reduced by training on a small fraction of examples from a small set of patients.
The fine-tuning procedure was identical to that described in Section 4.1, with the exception that the model's weights at the end of training were retained for evaluation, instead of restoring the best-performing weights on the validation set. Figure 5 provides boxplots for all trials that indicate the distributions of test AUC under the varying conditions for both the AB and LS tasks. Again, SimCLR performance appeared to be substantially higher than both non-contrastive methods.
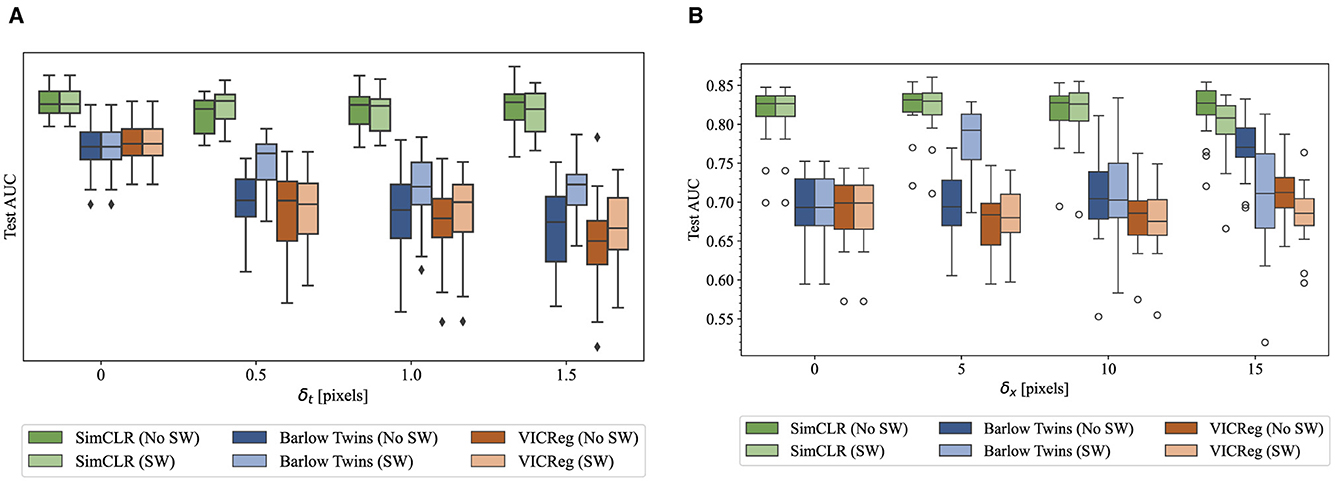
Figure 5. Boxplots conveying the quartile ranges of test AUC distributions for each pretraining method and assignment to δ, with and without sample weights. Each experiment is repeated 20 times on disjoint subsets of the training set, each containing all images from a group of patients. (A) AB task. (B) LS task.
Table 3 gives the mean and standard deviation of each set of trials, for each hyperparameter combination. For each task and each pretraining method, the ANOVA revealed significant interaction effects (p ≤ 0.05). Accordingly, all intended post-hoc t-tests were performed to ascertain (1) which combinations of hyperparameters were different from the baseline setting of augmenting the same frame twice (δ = 0) and (2) values of δ where the addition of sample weights changes the outcome. First, we note that SimCLR was the only pretraining method that consistently outperformed full supervision with ImageNet-pretrained weights. Barlow Twins and VICReg pretraining – both non-contrastive methods – resulted in worse performance.

Table 3. ParenchymalLUS test AUC for the the AB and LS tasks when trained using examples from 5% of the patients in the training set.
For the AB task, no combination of intra-video positive pairs or sample weights resulted in statistically significant improvements compared to dual distortion of the same image (δt = 0). For Barlow Twins and VICReg, several nonzero δt resulted in significantly worse mean test AUC. Sample weights consistently made a difference in Barlow Twins across δt values, but only improved mean test AUC for δt = 1 and δt = 1.5.
Different trends were observed for the LS task. SimCLR with δx = 5 and no sample weights improved mean test AUC compared to the baseline where δx = 0. No other combination of hyperparameters resulted in a significant improvement. For Barlow Twins, multiple IVPP hyparameter combinations resulted in improved mean test AUC over the baseline. No IVPP hyperparameter combinations significantly improved the performance of VICReg.
Insights were derived to guide practitioners working with deep learning for ultrasound interpretation. First, SimCLR was observed to achieve the greatest performance consistently across multiple tasks. With the exception of the data-scarce COVID-19 classification task, SimCLR decisively outperformed Barlow Twins and VICReg on the A/B and LS tasks. The results provide evidence toward favoring contrastive learning over non-contrastive learning for problems in ultrasound. It could be that the non-contrastive methods studied may be less effective for lung ultrasound examinations. We suspect that the lack of diversity in parenchymal lung ultrasound and the fine-grained nature of the classification tasks is problematic for non-contrastive methods, as the objectives are attractive and focus on maximizing embedding information. Perhaps explicit samples of negative pairs may be needed to learn a meaningful embedding manifold for fine-grained downstream tasks. Future work assessing non-contrastive methods for tasks in different ultrasound examinations or alternative imaging modalities altogether would shed light on the utility of non-contrastive methods outside the typical evaluation setting of photographic images.
While the experimental results do not support the existence of overarching trends for hyperparameter assignments for intra-video positive pairs across pretraining methods, it was observed that some combinations improved performance on particular downstream tasks. For example, each pretraining method's downstream performance on COVID-19 classification was improved by a nonzero value of δt. Overall, the results indicated that the optimal assignment for IVPP hyperparameters may be problem-specific. Clinically, IVPP may improve performance on downstream ultrasound interpretation tasks; however, practitioners are advised to include a range of values of δ with and without sample weights in their hyperparameter search.
The methods and experiments conducted in this study were not without limitations. As is common in medical imaging datasets, the ParenchymalLUS dataset was imbalanced. The image-wise representation for the positive class was 30.0% for the AB task and 11.7% for the lung sliding task. Although some evidence exists in support for self-supervised pretraining for alleviating the ill effects of class imbalance in photographic images (Yang and Xu, 2020; Liu et al., 2021), computed tomography, and fundoscopy images (Zhang et al., 2023), we found no such evidence for tasks in medical ultrasound.
As outlined in the background, the pretraining objectives employed in this study have been shown to improve downstream performance when the pairwise relationship aligns with the downstream task (Balestriero and LeCun, 2022). These guarantees compare to the baseline case of random weight initialization. While it was observed that all pretraining methods outperformed full supervision with randomly initialized weights, ImageNet-pretrained weights outperformed non-contrastive methods in several of the experiments. ImageNet-pretrained weights are a strong and meaningful baseline for medical imaging tasks, as they have been shown to boost performance in several supervised learning tasks across medical imaging modalities (Azizi et al., 2021). It is possible that some extreme data augmentation transformations and intra-positive pairs could jeopardize the class agreement of positive pairs (as is likely in most pragmatic cases); however, near-consistent alignment was achieved through data augmentation design and small ranges of δ. Although there exists evidence that VICReg and SimCLR can achieve similar performance on ImageNet with judicious selection of hyperparameters (e.g., temperature, loss term weights, learning rate) (Garrido et al., 2022), we used default hyperparameters. Due to limited computational resources, we avoided expansion of the hyperparameter space by only studying IVPP hyperparameters.
Lastly, M-mode images were designated by selecting x-coordinates in B-mode videos that intersect a pleural line region of interest, as predicted by an object detection model utilized in previous work (VanBerlo et al., 2022b, 2023b). LUS M-mode images must intersect the pleural line in order to appreciate the lung sliding artifact. While we mitigated potential inaccuracies in localization by limiting training and evaluation data to the brightest half of eligible x-coordinates, it is possible that a small fraction of M-mode images were utilized that did not intersect the pleural line.
Intra-video positive pairs have been proposed as a means of improving the downsteam performance of ultrasound classifiers pretrained with joint embedding self supervised learning. In this study, we suggested a scheme for integrating such positive pairs into common contrastive and non-contrastive SSL methods. Applicable to both B-mode and M-mode ultrasound, the proposed method (IVPP) consists of sampling positive pairs that are separated temporally or spatially by no more than a threshold, optionally applying sample weights to each pair in the objective depending on the distance. Investigations revealed that using nearby images from the same video for positive pairs can lead to improved performance when compared to composing positive pairs from the same image, but that IVPP hyperparameter assignments yielding benefits may vary by the downstream task. Another salient result was the persistent top performance of SimCLR for key tasks in B-mode and M-mode lung ultrasound, indicating that contrastive learning may be more suitable than non-contrastive learning methods for ultrasound imaging.
Future work could investigate IVPP for other types of medical ultrasound exams. IVPP could also be integrated into other SSL objectives. The sample weights formulation proposed in this study could also be applied to SSL for non-US videos. Given the high performance of SimCLR, subsequent work should perform a comprehensive comparison contrastive and non-contrastive SSL methods for tasks in medical US. Lastly, future work could evaluate US-specific data augmentation transformations that preserve semantic content. As a natural source of differences between positive pairs, IVPP could be studied in tandem with US-specific augmentations.
Publicly available datasets were analyzed in this study. This data can be found here: https://github.com/983632847/USCL. Some of the datasets generated and/or analyzed during the current study are available in via online repositories. The Butterfly dataset and the 5-fold splits of the POCUS dataset can be found in the above USCL repository.
The studies involving humans were approved by Lawson Research Institute, Western University. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation was not required from the participants or the participants' legal guardians/next of kin in accordance with the national legislation and institutional requirements.
BV: Conceptualization, Formal analysis, Funding acquisition, Investigation, Methodology, Software, Visualization, Writing original draft, Writing – review & editing. AW: Methodology, Supervision, Writing – review & editing. JH: Methodology, Supervision, Writing – review & editing. RA: Data curation, Resources, Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was funded by the Natural Sciences and Engineering Research Council of Canada, as BV was a Vanier Scholar (FRN 186945). The funder played no role in study design, data collection, analysis and interpretation of data, or the writing of this manuscript.
Computational resource support was also provided by Compute Ontario (computeontario.ca) and the Digital Research Alliance of Canada (alliance.can.ca).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimag.2024.1416114/full#supplementary-material
1. ^See dataset details at the public POCUS repository (Born et al., 2020): https://github.com/jannisborn/covid19_ultrasound.
2. ^Accessed via a URL available at the public USCL repository (Chen et al., 2021): https://github.com/983632847/USCL.
Alrajhi, K., Woo, M. Y., and Vaillancourt, C. (2012). Test characteristics of ultrasonography for the detection of pneumothorax: a systematic review and meta-analysis. Chest 141, 703–708. doi: 10.1378/chest.11-0131
Anand, D., Annangi, P., and Sudhakar, P. (2022). “Benchmarking self-supervised representation learning from a million cardiac ultrasound images,” in 2022 44th Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC) (Glasgow: IEEE), 529–532.
Arntfield, R., Wu, D., Tschirhart, J., VanBerlo, B., Ford, A., Ho, J., et al. (2021). Automation of lung ultrasound interpretation via deep learning for the classification of normal versus abnormal lung parenchyma: a multicenter study. Diagnostics 11:2049. doi: 10.3390/diagnostics11112049
Azizi, S., Mustafa, B., Ryan, F., Beaver, Z., Freyberg, J., Deaton, J., et al. (2021). “Big self-supervised models advance medical image classification,” in Proceedings of the IEEE/CVF International Conference on Computer Vision, 3478–3488.
Balestriero, R., and LeCun, Y. (2022). “Contrastive and non-contrastive self-supervised learning recover global and local spectral embedding methods,” in Advances in Neural Information Processing Systems, eds. S. Koyejo, S. Mohamed, A. Agarwal, D. Belgrave, K. Cho, and A. Oh (New York: Curran Associates, Inc), 26671–26685.
Bardes, A., Ponce, J., and LeCun, Y. (2022). “VICReg: Variance-Invariance-Covariance Regularization for Self-Supervised Learning,” in International Conference on Learning Representations.
Basu, S., Singla, S., Gupta, M., Rana, P., Gupta, P., and Arora, C. (2022). “Unsupervised contrastive learning of image representations from ultrasound videos with hard negative mining,” in International Conference on Medical Image Computing and Computer-Assisted Intervention (Cham: Springer), 423–433.
Bordes, F., Balestriero, R., and Vincent, P. (2023). Towards democratizing joint-embedding self-supervised learning. arXiv[preprint] arXiv:2303.01986. doi: 10.48550/arXiv.2303.01986
Born, J., Brändle, G., Cossio, M., Disdier, M., Goulet, J., Roulin, J., et al. (2020). POCOVID-net: automatic detection of COVID-19 from a new lung ultrasound imaging dataset (POCUS). arXiv[preprint] arXiv:2004.12084. doi: 10.48550/arXiv.2004.12084
Butterfly Network (2020). Covid-19 Ultrasound Gallery. Available online at: https://www.butterflynetwork.com/covid19/covid-19-ultrasound-gallery (accessed September 20, 2020).
Caron, M., Touvron, H., Misra, I., Jégou, H., Mairal, J., Bojanowski, P., et al. (2021). “Emerging properties in self-supervised vision transformers,” in Proceedings of the IEEE/CVF International Conference on Computer Vision (Montreal, QC: IEEE), 9650–9660.
Chen, T., Kornblith, S., Norouzi, M., and Hinton, G. (2020). “A simple framework for contrastive learning of visual representations,” in International Conference on Machine Learning (New York: PMLR), 1597–1607.
Chen, Y., Zhang, C., Ding, C. H., and Liu, L. (2022). Generating and weighting semantically consistent sample pairs for ultrasound contrastive learning. IEEE Trans. Med. Imag. 42, 1388–1400. doi: 10.1109/TMI.2022.3228254
Chen, Y., Zhang, C., Liu, L., Feng, C., Dong, C., Luo, Y., et al. (2021). “USCL: pretraining deep ultrasound image diagnosis model through video contrastive representation learning,” in Medical Image Computing and Computer Assisted Intervention-MICCAI 2021: 24th International Conference (Strasbourg: Springer), 627–637.
Garrido, Q., Chen, Y., Bardes, A., Najman, L., and Lecun, Y. (2022). On the duality between contrastive and non-contrastive self-supervised learning. arXiv[preprint] arXiv:2206.02574. doi: 10.48550/arXiv.2206.02574
Grill, J.-B., Strub, F., Altché, F., Tallec, C., Richemond, P., Buchatskaya, E., et al. (2020). Bootstrap your own latent-a new approach to self-supervised learning. Adv. Neural Inform. Proc. Syst. 33, 21271–21284. doi: 10.5555/3495724.3497510
Hall, M. K., Hall, J., Gross, C. P., Harish, N. J., Liu, R., Maroongroge, S., et al. (2016). Use of point-of-care ultrasound in the emergency department: insights from the 2012 medicare national payment data set. J. Ultrasound Med. 35, 2467–2474. doi: 10.7863/ultra.16.01041
He, K., Fan, H., Wu, Y., Xie, S., and Girshick, R. (2020). “Momentum contrast for unsupervised visual representation learning,” in Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (Seattle, WA: IEEE), 9729–9738.
He, K., Zhang, X., Ren, S., and Sun, J. (2016). “Deep residual learning for image recognition,” in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 770–778.
Howard, A., Sandler, M., Chu, G., Chen, L.-C., Chen, B, Tan, M., et al. (2019). “Searching for MobileNetV3,” in Proceedings of the IEEE/CVF International Conference on Computer Vision, 1314–1324. doi: 10.1109/ICCV.2019.00140
Jasčǔr, M., Bundzel, M., Malík, M., Dzian, A., Ferenčík, N., and Babič, F. (2021). Detecting the absence of lung sliding in lung ultrasounds using deep learning. Appl. Sci. 11:6976. doi: 10.3390/app11156976
Kessler, R., Stowell, J. R., Vogel, J. A., Liao, M. M., and Kendall, J. L. (2016). Effect of interventional program on the utilization of pacs in point-of-care ultrasound. J. Digi. Imag. 29, 701–705. doi: 10.1007/s10278-016-9893-x
Lau, Y. H., and See, K. C. (2022). Point-of-care ultrasound for critically-ill patients: a mini-review of key diagnostic features and protocols. World J. Criti. Care Med. 11:70. doi: 10.5492/wjccm.v11.i2.70
Liang, H., Ning, G., Zhang, X., and Liao, H. (2023). “Semi-supervised anatomy tracking with contrastive representation learning in ultrasound sequences,” in 2023 IEEE 20th International Symposium on Biomedical Imaging (ISBI).
Lichtenstein, D. A. (2010). Whole Body Ultrasonography in the Critically Ill. Cham: Springer Science & Business Media.
Lichtenstein, D. A., and Menu, Y. (1995). A bedside ultrasound sign ruling out pneumothorax in the critically iii: lung sliding. Chest 108, 1345–1348. doi: 10.1378/chest.108.5.1345
Lichtenstein, D. A., Mezière, G., Lascols, N., Biderman, P., Courret, J.-P., Gepner, A., et al. (2005). Ultrasound diagnosis of occult pneumothorax. Criti. Care Med. 33, 1231–1238. doi: 10.1097/01.CCM.0000164542.86954.B4
Liu, H., HaoChen, J. Z., Gaidon, A., and Ma, T. (2021). Self-supervised learning is more robust to dataset imbalance. arXiv[preprint] arXiv:2110.05025. doi: 10.48550/arXiv.2110.05025
Nazerian, P., Volpicelli, G., Vanni, S., Gigli, C., Betti, L., Bartolucci, M., et al. (2015). Accuracy of lung ultrasound for the diagnosis of consolidations when compared to chest computed tomography. Am. J. Emerg. Med. 33, 620–625. doi: 10.1016/j.ajem.2015.01.035
Oh Song, H., Xiang, Y., Jegelka, S., and Savarese, S. (2016). “Deep metric learning via lifted structured feature embedding,” in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 4004–4012.
Sood, R., Rositch, A. F., Shakoor, D., Ambinder, E., Pool, K.-L., Pollack, E., et al. (2019). Ultrasound for breast cancer detection globally: a systematic review and meta-analysis. J. Global Oncol. 5, 1–17. doi: 10.1200/JGO.19.00127
van Randen, A., Laméris, W., van Es, H. W., van Heesewijk, H. P., van Ramshorst, B., Ten Hove, W., et al. (2011). A comparison of the accuracy of ultrasound and computed tomography in common diagnoses causing acute abdominal pain. Eur. Radiol. 21, 1535–1545. doi: 10.1007/s00330-011-2087-5
VanBerlo, B., Li, B., Hoey, J., and Wong, A. (2023a). Self-supervised pretraining improves performance and inference efficiency in multiple lung ultrasound interpretation tasks. arXiv[preprint] arXiv:2309.02596. doi: 10.1109/ACCESS.2023.3337398
VanBerlo, B., Li, B., Wong, A., Hoey, J., and Arntfield, R. (2023b). “Exploring the utility of self-supervised pretraining strategies for the detection of absent lung sliding in m-mode lung ultrasound,” in Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (Vancouver, BC: IEEE), 3076–3085.
VanBerlo, B., Smith, D., Tschirhart, J., VanBerlo, B, Wu, D., Ford, A., et al. (2022a). Enhancing annotation efficiency with machine learning: Automated partitioning of a lung ultrasound dataset by view. Diagnostics 12:2351. doi: 10.3390/diagnostics12102351
VanBerlo, B., Wu, D., Li, B., Rahman, M. A., Hogg, G, VanBerlo, B., et al. (2022b). Accurate assessment of the lung sliding artefact on lung ultrasonography using a deep learning approach. Comp. Biol. Med. 148:105953. doi: 10.1016/j.compbiomed.2022.105953
Whitson, M. R., and Mayo, P. H. (2016). Ultrasonography in the emergency department. Crit. Care 20:1–8. doi: 10.1186/s13054-016-1399-x
Yang, Y., and Xu, Z. (2020). Rethinking the value of labels for improving class-imbalanced learning. Adv. Neural Informat. Proc. Syst. 33, 19290–19301. doi: 10.5555/3495724.3497342
Yim, E. S., and Corrado, G. (2012). Ultrasound in sports medicine: relevance of emerging techniques to clinical care of athletes. Sports Med. 42, 665–680. doi: 10.1007/BF03262287
You, Y., Li, J., Reddi, S., Hseu, J., Kumar, S., Bhojanapalli, S., et al. (2019). Large batch optimization for deep learning: training bert in 76 minutes. arXiv[preprint] arXiv:1904.00962. doi: 10.48550/arXiv.1904.00962
Zbontar, J., Jing, L., Misra, I., LeCun, Y., and Deny, S. (2021). “Barlow twins: self-supervised learning via redundancy reduction,” in International Conference on Machine Learning, 12310–12320.
Zhang, C., Chen, Y., Liu, L., Liu, Q., and Zhou, X. (2022). “Hico: hierarchical contrastive learning for ultrasound video model pretraining,” in Proceedings of the Asian Conference on Computer Vision, 229–246.
Zhang, C., Zheng, H., and Gu, Y. (2023). Dive into the details of self-supervised learning for medical image analysis. Med. Image Analy. 89:102879. doi: 10.1016/j.media.2023.102879
Keywords: self-supervised learning, ultrasound, contrastive learning, non-contrastive learning, representation learning
Citation: VanBerlo B, Wong A, Hoey J and Arntfield R (2024) Intra-video positive pairs in self-supervised learning for ultrasound. Front. Imaging. 3:1416114. doi: 10.3389/fimag.2024.1416114
Received: 11 April 2024; Accepted: 04 June 2024;
Published: 20 June 2024.
Edited by:
Sandeep Kumar Mishra, Yale University, United StatesReviewed by:
Aili Wang, Harbin University of Science and Technology, ChinaCopyright © 2024 VanBerlo, Wong, Hoey and Arntfield. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Blake VanBerlo, YnZhbmJlcmxAdXdhdGVybG9vLmNh
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.