
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
PERSPECTIVE article
Front. Hortic. , 29 September 2023
Sec. Sustainable Pest and Disease Management
Volume 2 - 2023 | https://doi.org/10.3389/fhort.2023.1263604
Australia is a major grain exporter, and this trade makes an important contribution to its economy. Fortunately, it remains free of many damaging virus diseases and virus vectors found elsewhere. However, its crop biosecurity is under increasing pressure from global ecological, climatic, and demographic challenges. Stringent biosecurity and plant health programs safeguard Australian grain production from damaging virus and virus vector incursions entering via different pathways. These programs formerly relied upon traditional testing procedures (indicator hosts, serology, PCRs) to intercept incoming virus-contaminated plant material. Recently, the integration of rapid genomic diagnostics innovation involving High Throughput Sequencing (HTS) smart tools into sample testing schedules is under exploration to improve virus testing accuracy, efficiency, and cost effectiveness under diverse circumstances. This process includes evaluating deployment of Illumina and Oxford Nanopore Technology shotgun sequencing. It also includes evaluating targeted viral genome HTS and virus vector metabarcoding approaches. In addition, using machine learning and deep learning capacities for big data analyses and remote sensing technologies will improve virus surveillance. Tracking damaging virus variants will be improved by surveillance networks which combine virus genomic-surveillance systems with an interoperable virus database. Sequencing Australian virus specimen collections will help ensure the accuracy of virus identifications based solely on genetic information. Enhancing routine diagnosis and data collection using these innovations will improve post entry virus interception and background virus and vector surveillance. This will help reduce the frequency of new incursions, improve virus management during eradication, containment and other plant health activities, and achieve more profitable Australian grain production.
Virus disease outbreaks occur globally, causing damage varying from small-scale losses to total crop failure. They diminish the growth and vigour of plants and decrease the yield and quality of their produce (Bos, 1982; Thresh, 1982; Thresh, 2004; Jones, 2006; Thresh, 2006a; Hull, 2014; Jones and Naidu, 2019). They damage all types of cultivated plants, including those grown to feed humans and their livestock, such as grains, oilseeds, roots, tubers, fruits and vegetables, and those grown for fibre, ornamental and medicinal purposes. Moreover, since they damage staple food crops of crucial importance for food-insecure world regions, such as maize, wheat, rice, potato, cassava, sweet potato and banana, minimizing the losses they cause is vital to achieving global food security (Thresh, 2004; Fargette et al., 2006; Thresh, 2006a; Loebenstein and Thottappilly, 2013; Hull, 2014; Jones and Naidu, 2019; Jones, 2020; Jones, 2021). In addition to plants grown as monocultures, virus diseases also damage plants growing amongst plant species mixtures in diverse situations. These include intercropping, mixed cropping, smallholder, market garden and subsistence farming (Thresh, 1982; Thresh, 2006b; Jones, 2009; Boudreau, 2013; Hull, 2014; Chai et al., 2021; Jones, 2022), and managed pastures (Edwardson and Christae, 1986; Barbetti et al., 1996; McLaughlin et al., 1996; Jones, 2012; Jones, 2022). They also damage wild plants growing in disturbed and undisturbed wild plant ecosystems, and at the interface between natural and managed vegetation (Bos, 1981; Thresh, 1981; Malmstrom et al., 2005a; Malmstrom et al., 2005b; Cooper and Jones, 2006; Webster et al., 2007; Alexander et al., 2014; Jones and Coutts, 2015; Malmstrom and Alexander, 2016). In managed pastures and natural vegetation, they diminish the fitness of plant species sensitive to infection. This reduces their ability to compete with non-host species and leads to alterations in the plant species balance (McLaughlin et al., 1992; Friess and Maillet, 1996; Jones and Nicholas, 1998; Maskell et al., 1999; Coutts and Jones, 2002; Malmstrom et al., 2005a; Malmstrom et al., 2005b; Cooper and Jones, 2006; Alexander et al., 2017; Jones, 2022). Wild plants also serve as an important infection reservoir from which viruses spread to crops (Bos, 1981; Thresh, 1981; Hull, 2014). In 2014, plant viruses were estimated to cause a worldwide economic impact of more than US$30 billion annually (Sastry and Zitter, 2014), which is likely to have grown considerably since then. Furthermore, since the world’s population is projected to increase to approximately 10 billion by 2050, the need to feed this burgeoning human population is paramount.
Factors driving the increasing threat posed by plant virus diseases worldwide include expansion of: (i) agricultural intensification, extensification and diversification; (ii) globalisation; and (iii) disturbance and fragmentation of natural vegetation (Thresh, 1980; Thresh, 1982; Fargette et al., 2006; Thresh, 2006a; Thresh, 2006b; Thresh, 2006c; Jones, 2009; Jones and Naidu, 2019 Jones, 2020; Jones, 2021). Furthermore, unpredictable weather conditions and global warming from climate change make plant virus diseases more difficult to control and alter their global distributions (Canto et al., 2009; Jones, 2009; Jones and Barbetti, 2012; Jones, 2016; Trebicki, 2020). Frequent reports of new and emerging viral diseases have elevated plant viruses to become an increasingly important global biosecurity challenge (Anderson et al., 2004; Fargette et al., 2006; Jones, 2009; Elena et al., 2014; Fereres, 2015; Gilbertson et al., 2015; Jones and Naidu, 2019). Agricultural diversification involving dissemination of crops away from their centres of origin and domestication has important consequences with respect to virus diseases. Firstly, due to virus infection in the seed or other planting material introduced, viruses that evolved in crop domestication centres become introduced inadvertently to new world regions, and then spread to local crops and native vegetation, neither of which have encountered them previously. Secondly, the newly introduced crops become invaded by viruses they never encountered before, which spread to them from infected crops occurring locally and native vegetation. Both scenarios lead to rapid virus evolution resulting in new viral variants or strains and host species jumps, which sometimes cause severe crop losses, major epidemics and even global pandemics (Thresh, 1980; Thresh, 2004; Jones, 2006; Thresh, 2006a; Thresh, 2006b; Jones, 2009; Dombrovsky et al., 2017; Jones and Naidu, 2019; Jones, 2020; Jones, 2021). The first scenario also damages natural ecosystems (Alexander et al., 2014; Vincent et al., 2014; Jones and Coutts, 2015). Under these circumstances, preventing incursions of damaging viruses and their virus vectors, and eradicating or containing any that establish successfully, is becoming an increasingly difficult challenge for biosecurity authorities located within different world regions and countries. Fortunately, rapid technological advances that improve the effectiveness of virus diagnosis in preborder, border and postborder situations are providing new opportunities to detect viruses earlier and more reliably (Jones, 2014; Martin et al., 2016; Kreuze, et al., 2023; Waite et al., 2022; Adams and Fox, 2016; Barrero et al., 2017; Pecman et al., 2017; Bronzato-Badial et al., 2018; FAO, 2019; Piper et al., 2019; Liefting et al., 2021; Maina et al., 2021; Whattam et al., 2021; Gauthier et al., 2022; Lelwala et al., 2022; Mackie et al., 2022; Alcalá Briseño et al., 2023).
Viruses are transmitted from virus-infected to healthy plants in diverse ways depending upon the pathosystem involved. These include transmission via insect, mite, nematode or fungal vectors, contact, vegetative propagules, seed, pollen, water, soil, and parasitic plants (Hull, 2014; Jones, 2018). This means that distinct pathways exist by which they can be introduced from one location to another. The most significant pathway for long-distance plant virus dissemination is via virus-infected vegetative planting material, fruits and seeds distributed via trade supply chains involving ship, plane, railway or road transport. However, international germplasm exchange for plant breeding purposes also contributes (Thresh, 1980; Thresh, 1982, Bos, 1992; Lapierre and Signoret, 2004; Jones, 2009; Sastry, 2013; Jones, 2020; Jones, 2021; Whattam et al., 2021; Lenzen et al., 2023). Moreover, the movement via trade supply chains of viruliferous arthropod vectors adhering to vegetative planting material and cut flowers, or viruliferous nematode vectors and resting spores of fungal vectors in infested soils, constitutes another major pathway for long-distance plant virus dissemination (Lecoq et al., 2003; Jones, 2009; Jones and Barbetti, 2012; Fereres, 2015; Wamaitha et al., 2018; Jones and Naidu, 2019). Furthermore, viruses and their vectors can also be disseminated over large distances when viruliferous arthropod vectors are transported by major wind currents including jet winds (Thresh, 1982; Irwin and Thresh, 1988; Maina et al., 2017a; Maina, 2018; Maina et al., 2018a; Maina et al., 2018b; Maina et al., 2019);. Dissemination also occurs when airline, ship or railway passengers unknowingly, or deliberately, carry virus-infected fruits, seeds, vegetative planting material or virus-infested soil in their luggage (Swain et al., 1952; Crooks et al., 1983; Alvarez Quinto et al., 2023).
Australia differs from other continents and major world regions regarding the threat virus diseases pose to its agriculture. It has a unique situation in the world because of being an island continent where the only plants cultivated during the limited agriculture practiced for thousands of years before Europeans first settled in 1788 were derived from Australian native plants (Gerritsen, 2010). Therefore, before its colonisation the continent lacked the numerous virus and virus vector introductions that occurred elsewhere in the world when seeds and vegetative propagules of cultivated plants were moved between continents (Gibbs et al., 2008; Jones, 2009). Furthermore, since the Australian Quarantine Service commenced with the Quarantine act of 1908, a national biosecurity framework has gradually been developed that minimizes the likelihood of incursions by viral pathogens that damage cultivated plants and native flora in other parts of the world (Rodoni, 2009; Burgman et al., 2014; Barrero et al., 2017; Davis et al., 2021; Jones et al., 2021c; Whattam et al., 2021). For these reasons, Australia remains free of many damaging viral diseases and virus vectors found elsewhere. Nevertheless, its quarantine barrier is sometimes breached and the ensuing post border operations may fail to eradicate or contain virus and vector incursions. For example, this occurred when two seed-borne viruses entered via infected seeds: wheat streak mosaic virus (WSMV) which arrived around 1999 (Dwyer et al., 2007; Jones et al., 2022), and cucumber green mottle mosaic virus which arrived in 2014 (Dombrovsky et al., 2017; Kehoe et al., 2022; Mackie et al., 2022). It also occurred when the insect ‘supervectors’ Franklinella occidentalis (western flower thrips) and Bemisia tabaci Biotype MEAM1 (silverleaf whitefly) arrived in 1993 and 1994, respectively (Malipatil et al., 1993; Gunning et al., 1995). Moreover, Australia’s northern coastline is near to the coastlines of its neighbouring countries of Indonesia, East Timor and Papua New Guinea (PNG). It is, therefore vulnerable to virus and vector incursions from wet season monsoonal wind currents carrying insect vectors (Maina et al., 2017a; Maina, 2018; Maina et al., 2019; Davis et al., 2021). It is also vulnerable to virus incursions via migrating birds bringing infected seeds, and vessel landings which inadvertently introduce virus-infected seeds and/or virus-infected or vector-infested vegetative propagules, fruits or soils (Gibbs et al., 2008; Horwood et al., 2018; Davis et al., 2021; Jones et al., 2021c).
The Australian continent has diverse climatic zones ranging from temperate to tropical, which allow production of a wide diversity of grain crops grown mainly for export (Henzell, 2007; Brown et al., 2020). Grain crop cultivation is a major Australian economic activity worth $16.7 billion annually (ABARES, 2022). As the world’s population continues to increase, the importance of Australia’s grain exports in helping to satisfy the ever-growing demand for food worldwide is likely to increase further. Consequently, ongoing prevention of plant virus introductions is fundamental to mitigate viral disease impacts on Australian grain crops and safeguard its economy. Here, after describing Australian plant biosecurity procedures in relation to virus and vector interceptions and incursions, we describe advancements in detection technologies, innovative tools and database analytics which can improve Australian plant virus disease diagnosis and surveillance, and highlight areas that warrant further investigation. Examples of virus biosecurity threats to Australian cereal and oilseed crops were provided in two recent reviews (Davis et al., 2021; Jones et al., 2021c). Therefore, the examples used here mostly involve biosecurity threats to the legume component of the continent’s grain crops. Our primary objective in writing this review is to enhance the effectiveness of Australian plant biosecurity at (i) intercepting damaging grain viruses when they first arrive in the country, and (ii) providing the extensive background information its post entry surveillance activities require to detect, assess and manage new virus and vector incursions in grain crops.
The statutory plant health term ‘plant biosecurity’ used in Australia is referred to as ‘plant quarantine’ by some countries (Burgman et al., 2014). The generic term ‘pest’ refers to ‘all pathogenic agents that are injurious to plants or plant products’, and includes damaging plant viruses (FAO, 2010). The plant biosecurity term ‘risk’ refers to ‘the probability of an event occurring and the consequence of that event’ (IPPC, 1997). The terms ‘introduction’ and ‘incursion’ refer to ‘the entry of a pest resulting in its establishment’, and ‘establishment’ refers to the ‘perpetuation for the foreseeable future of a pest within an area after entry’. ‘Pest risk analysis’ (PRA) refers to ‘the process of evaluating biological or other scientific and economic evidence to determine whether an organism is a ‘pest’, whether it should be regulated, and the strength of any phytosanitary measures to be taken against it’ (FAO, 2010; Burgman et al., 2014).
In Australia, a PRA framework is in place by which the risks associated with the introduction of each pest is identified and assessed. Its role is to deter the incursion and establishment of new pests, including viruses (and virus vectors) that cause diseases likely to have a damaging impact (Burgman et al., 2014; Anderson et al., 2017). This framework consists of four components, ‘integrated biosecurity and planning’, ‘preparedness for rapid response’, deploying ‘innovative detection and surveillance tools’ and ‘innovative disease management approaches’ (Figure 1). All damaging viruses or vectors intercepted during border surveillance and monitoring of places of greatest risk (seaport, airport and mail centres) are destroyed. Post border eradication programs commence when these procedures fail to detect and prevent their entry and PRAs deem them to be significant economic threats to Australian agriculture, forestry or endangered native ecosystems. Such programs involve widespread surveillance and the destruction of all virus infection and vector infestation foci found. Should they fail and their establishment result, PRAs determine whether the economic threat justifies the expense of undertaking containment programs or no further biosecurity measures are warranted (Ebbels, 2003; Rodoni, 2009; Jeggo, 2012; Burgman et al., 2014; Davis et al., 2021; Jones et al., 2021c). In this context, the innovative detection and surveillance tools discussed below in this review are of paramount importance.
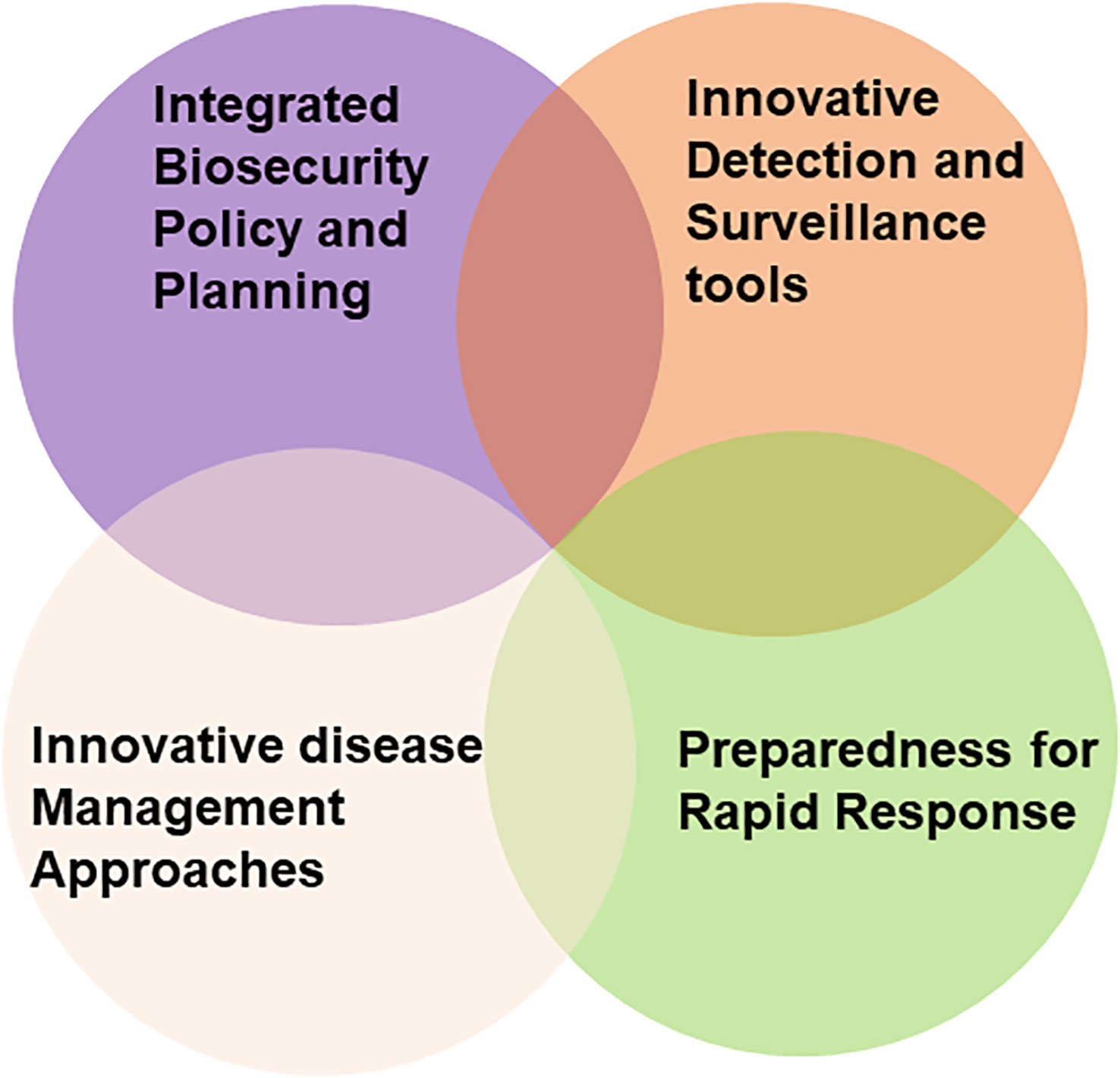
Figure 1 Venn diagram representing current and future integrated grains biosecurity strategies, required to mitigate virus associated grain yield losses in Australia.
When damaging plant viruses or their vectors gain a foothold within Australia, achieving effective eradication or containment often requires very substantial control measures that can be extremely costly. They typically include destroying all the infected crops and plant material found in each affected region, and applying strict quarantine restrictions over movement of plant materials, machinery, vehicles and humans onto and off each quarantined farm/property. Moreover, the owner of each quarantined farm/property may require financial compensation for loss of income. Table 1 provides an example of the often drastic actions required for each quarantined farm/property. Similar types of control measures are required for quarantined protected cropping but need to be adjusted to reflect the more intensive production systems involved. The integrated disease management approaches currently used to manage established virus diseases in Australian crops include a diverse range of phytosanitary, cultural, chemical, host resistance, and sometimes biological control measures (Jones, 2004; Jones, 2006; Jones and Naidu, 2019). They include both selective and nonselective control measures that, when used in combination, target not only external and internal virus sources but also the early and late phases of virus spread, and operate in as many different ways as possible, e.g., when used to suppress the spread of virus diseases of grain legumes (Jones, 2001; Jones, 2004; Jones, 2006; Makkouk et al., 2014; Makkouk, 2020). The integrated disease management approaches devised for managing viruses in high value situations, such as plant breeding plots and seed crops of newly released cultivars, are the most comprehensive and effective at supressing virus spread (e.g., Jones, 2001; Jones, 2004; Jones and Naidu, 2019). However, they are also the most expensive to deploy. Although they are sometimes suitable for use by virus containment programs that don’t warrant a more expensive approach, their stringency is inadequate for effective virus eradication.

Table 1 Example of a typical Australian biosecurity plan for virus eradication from an infested farm - annual crop version.
The Northern Australia Quarantine Strategy (NAQS) is an additional biosecurity program that commenced in 1990 (NAQS, 2020). Instead of focussing on initial border protection at seaports airports and postal facilities, it conducts regular surveillance across Northern Australia for pest incursions arriving via unregulated pathways from Australia’s northern neighbours (Indonesia, East Timor and PNG). As outlined briefly in the Introduction, virus and virus vector incursions can occur in three ways: (i) virus vectors with or without viruses being blown to the south by wet-season monsoonal winds; (ii) migrating birds with virus-infected seeds in their intestines flying, or being blown, southward over the sea; or (iii) virus-infected seeds or discarded plant material, vegetative propagules, fruits or soils being left behind after legal or illegal vessel visits by traders, fishermen or drug traffickers from Australia’s northern neighbours (e.g., Gibbs et al., 2008; Jones, 2009; Maina et al., 2017a; Horwood et al., 2018; Maina, 2018; Maina et al., 2019; Davis et al., 2021). The early detection of new virus or vector incursions arriving like this from the north means they can be intercepted and removed promptly, preventing them from spreading further south in the continent and infecting cultivated plants or threatened ecosystems there. NAQS is necessary because Australia’s northern island neighbours have a much longer history of growing locally domesticated and introduced crops (>5,000 years) than Australia where crops were first introduced after 1788. Therefore, they already have many pests, including viruses and vectors, not yet present in Australia (e.g., Maina et al., 2017a; Maina et al., 2017b; Horwood et al., 2018; Maina et al., 2018a; Maina et al., 2018b; Maina et al., 2019; Davis et al., 2021). The NAQS remit also includes preborder surveillance involving intergovernmental collaborative surveys with Indonesia, East Timor and PNG to determine which damaging pests, including viruses and vectors, are present but have not yet reached Australia (Davis et al., 2021). Furthermore, collaborative surveys conducted within these countries outside the NAQS system have not only found potentially damaging viruses that are absent from Northern Australia, but also revealed evidence of past virus incursions. This evidence was obtained by demonstrating ‘genetic connectivity’ between virus populations from crops in East Timor or PNG with virus populations from the same crops growing in Northern Australia (Maina et al., 2017a; Maina et al., 2017b; Maina et al., 2017c; Maina et al., 2018a; Maina et al., 2018b; Maina et al., 2019). Indeed, mediated by climate change and an increase in illegal vessel visits, it seems likely that the frequency of new virus and virus vector incursion from Australia’s neighbours will accelerate in the future (Jones and Barbetti, 2012; Eagles et al., 2013; Jones, 2016; Firth et al., 2017; Trebicki, 2020).
Australian government-mandated biosecurity import condition guidelines apply to all plant imports (BICON, 2023). However, they still remain a potential pathway for the inadvertent introduction of damaging viruses and vectors, e.g. via virus-infected vegetative plant propagules or seeds. Australia’s World Trade Organization (WTO) membership requires application of its Sanitary and Phytosanitary Measures (SPS) agreement. Under this agreement, Australia applies a stringent biosecurity protection policy designed to minimize risk without diminishing this to zero. Its aim is to provide fast but still reliable access to the new genetic material Australia’s plant industries require (Whattam et al., 2021). The import arrangements depend upon the exporting country, and the species and type of plant material being imported. Before entry, a phytosanitary certificate stating the imported plant material is free from damaging pests and diseases is required. BICON (2023) provide a detailed description of the requirements for importation of live plants and seeds. After their arrival, they are inspected for pest or disease presence by biosecurity personnel, and either released, treated or directed to a post-entry quarantine (PEQ) facility for screening and/or testing, including for virus diseases and virus vectors. This testing for plant viruses includes traditional (symptom inspections, bioassays, serology, electron microscopy) and currently used PCR and real-time PCR diagnostics (Whattam et al., 2021). Labour saving procedures like high throughput automated RNA/DNA extraction platforms and liquid handling robotics have made these currently used molecular procedures more efficient. However, employing such a wide range of test assays across a very disparate range of target species remains a very labour intensive process (Whattam et al., 2021). Also, these tests can be very specific and unable to identify diverse strains, variants or new viruses (Villamor et al., 2019). In the future, innovative molecular diagnostic technologies that are less resource demanding without compromising accuracy, need to be used to update and improve Australian PEQ and other Australian biosecurity and plant health diagnostics (Maina et al., 2021; Whattam et al., 2021).
Australia’s grain breeding programs rely on germplasm imported as seeds to enhance the genetic diversity of new grain cultivars. This includes introducing traits to improve yield, and protection not only from pest and disease threats but also from threats arising from increasing climate instability and changes in farming systems. As mentioned in the previous paragraph, these disease threats include damaging seed-borne viruses. Grain crop germplasm imported as seed requires rigorous biosecurity procedures to prevent their introduction, including inspection of seeds on-arrival and mandatory growth in a PEQ facility where seedlings are grown out and tested for seed-borne viruses. Once all the biosecurity measures have been met, seed lines can be released. As mentioned in the introduction, an example of where PEQ procedures employed in the past failed to prevent an important seed-borne virus of a grain crop from entering via infected seed, and then becoming established across Australia, is WSMV. It arrived first around the year 1999 in infected seed of wheat breeding lines (Dwyer et al., 2007; Jones et al., 2022). High plains wheat mosaic virus (HPWMV), which has the same eriophyid mite vector as WSMV, and often occurs in mixed infection with it, probably arrived in Australia via maize (or wheat) seed in the same way and around the same time (Jones et al., 2023). These failures likely occurred because WSMV and HPWMV were not known to be seed-borne in wheat or maize, respectively, until soon afterwards (Forster et al., 2001; Jones et al., 2005).
An increasing problem for Australian biosecurity is the likelihood of entry of significant viral pathogens from overseas via illegal seed imports purchased from on-line retailers, non-conforming legally imported seed sources or smuggled seeds brought in by returning travellers intending to sow them in their back gardens (e.g. Constable et al., 2021). An important reason why this problem is growing is the rise in immigration involving people from different parts of the world, which is fuelling an increasing demand for ‘country-of-origin’ sourced cultural heritage grains intended for planting or consumption in Australia. Similar considerations apply to the likelihood of entry of significant viral pathogens from overseas via illegal imports of vegetative propagules, such as tubers, tuberous roots, bulbs and corms that end up being planted in Australia. Examples of such virus introductions include infected tubers from South America entering Europe via the internet trade or in the luggage of returning travellers (Fox et al., 2019; Fuentes et al., 2021; Alvarez Quinto et al., 2023). Also, when tubers of heritage (=gourmet) potato cultivars were tested in Australia an unusual strain of potato virus Y (PVYD) was found (Jones and Vincent, 2018; Jones et al., 2021a) which, although not of biosecurity significance, illustrates how viruses of biosecurity concern can become established via ‘country-of-origin’ sourced cultural heritage introductions. To address the threat of virus entry from these pathways, greater biosecurity vigilance at mail centres, airports and seaports combined with more effective yet labour saving diagnostics is required. Indeed, the escalating plant biosecurity risk posed by entry of damaging viral pathogens via these diverse pathways constitutes an increasing cause for concern for Australia. This is because the viral diseases pose a serious threat to the economic and ecological wellbeing of the continent’s agricultural industries and natural ecosystems. Moreover, since Australia is a major food exporter, any threat to its crops is also a cause for concern elsewhere, especially for the world’s food insecure regions.
Currently more than 1.8 million hectares of grain legumes are grown in Australia, an average 2.2 million metric tonnes being produced annually (AEGIC, 2020). The principal grain legumes grown are chickpea (Cicer arietinum), field pea (Pisum sativum), faba bean (Vicia faba), lentil (Lens culinaris), lupin (Lupinus spp.), mungbean (Vigna radiata), common bean (Phaseolus vulgaris), cowpea (Vigna unguiculata), soybean (Glycine max), and peanut (Arachis hypogea) (AEGIC, 2020). The main grain legume crops in tropical northern Australia are chickpea, mungbean, soybean, peanut, cowpea, common bean and cowpea. Those grown within Australia’s subtropical northern grainbelt are mungbean, soybean, peanut, lupin, field pea, chickpea and lentil, while those grown in the Mediterranean and temperate climates of its southern grainbelt are lupin, chickpea, faba bean, lentil and field pea (AEGIC, 2020; Brown et al., 2020; Jones et al., 2021c).
Examples of more important grain legume viruses absent from Australia are shown in Table 2, which lists information for each of them on the main crops they affect, their principal vectors, whether they are seed-borne, and their global distributions, disease impacts and entry risks. Figures 2A–H illustrate the disease symptoms some of them cause in grain legume foliage and seeds. The losses these viral pathogens engender within grain legume crops growing elsewhere in the world vary from devastating, e.g., groundnut rosette disease in sub-Saharan Africa (Figures 2A, B; Naidu et al., 1999) and faba bean necrotic yellows disease in the Middle East and North Africa (Figures 2C, D; Makkouk, 2020), to major losses only occurring sporadically (e.g. bean dwarf mosaic disease in South America and pea early browning disease in Europe) (Table 2). Nine of the virus examples in Table 2 are seed-borne so these are therefore of greater concern for Australian biosecurity authorities than those only likely to enter Australia via vegetative propagule imports. Moreover, three of the viruses in Table 2, bean common mosaic necrosis virus (BCMNV), groundnut bud necrosis virus (GBNV) and mungbean yellow mosaic India virus (MBYMIV), have been found in the neighbouring countries of Indonesia or East Timor closest to Australia’s northern coastline. Amongst these, BCMNV is both found in East Timor (Maina et al., 2016) and seed-borne so likely to enter Australia via birds or insect vectors crossing the ocean or directly via introduction of virus-contaminated seed (see previous Section). Since they are not seed-borne, GBNV and MBYMIV are more likely to arrive in northern Australia from Indonesia (Damayanti and Naidu, 2009; Hema et al., 2014) via insect vectors crossing the ocean. In the 1970s, the seed-borne viruses broad bean stain virus (BSV; Figure 2H) and broad bean true mosaic virus were found infecting faba bean seeds sourced in the Australian Capital Territory and the state of South Australia (Moghal and Francki, 1974; Randles and Dube, 1977; Buchen-Osmond et al., 1988). However, there have been no further detections in Australia, so both are deemed no longer present and, therefore, still considered to be biosecurity threats (Table 2).
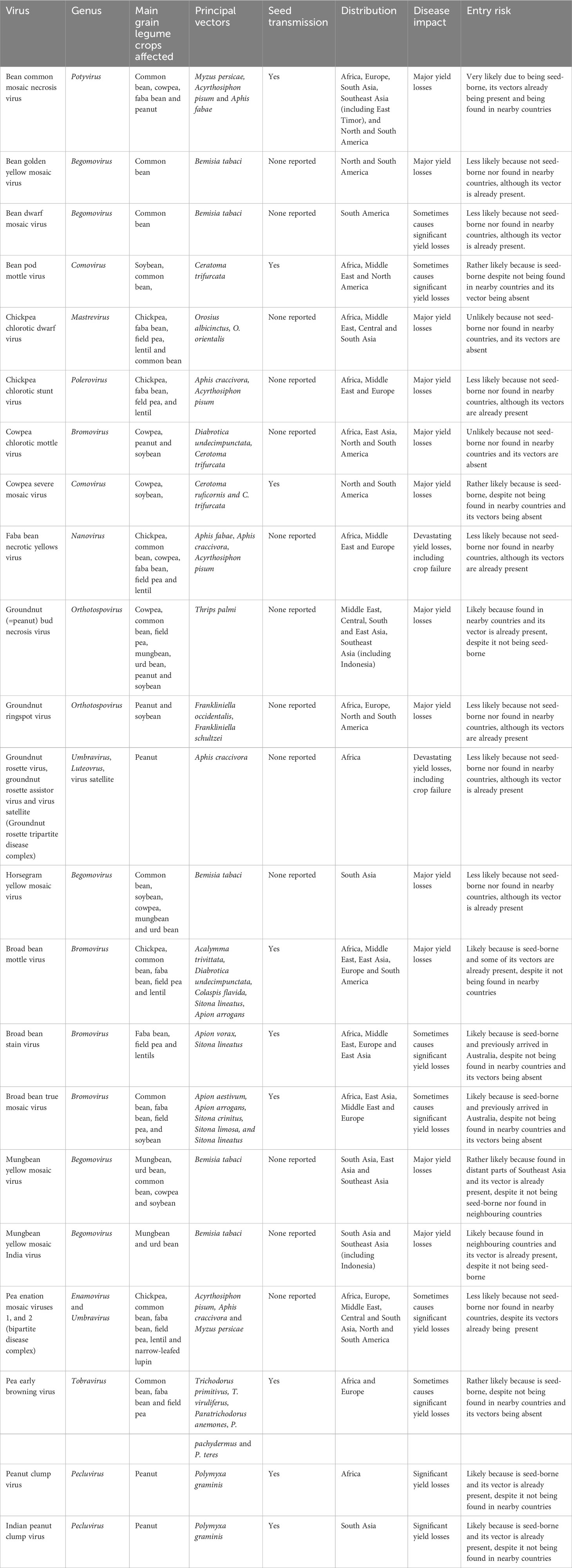
Table 2 Examples of viruses that cause disease in grain legume crops in other continents but not yet in Australia.
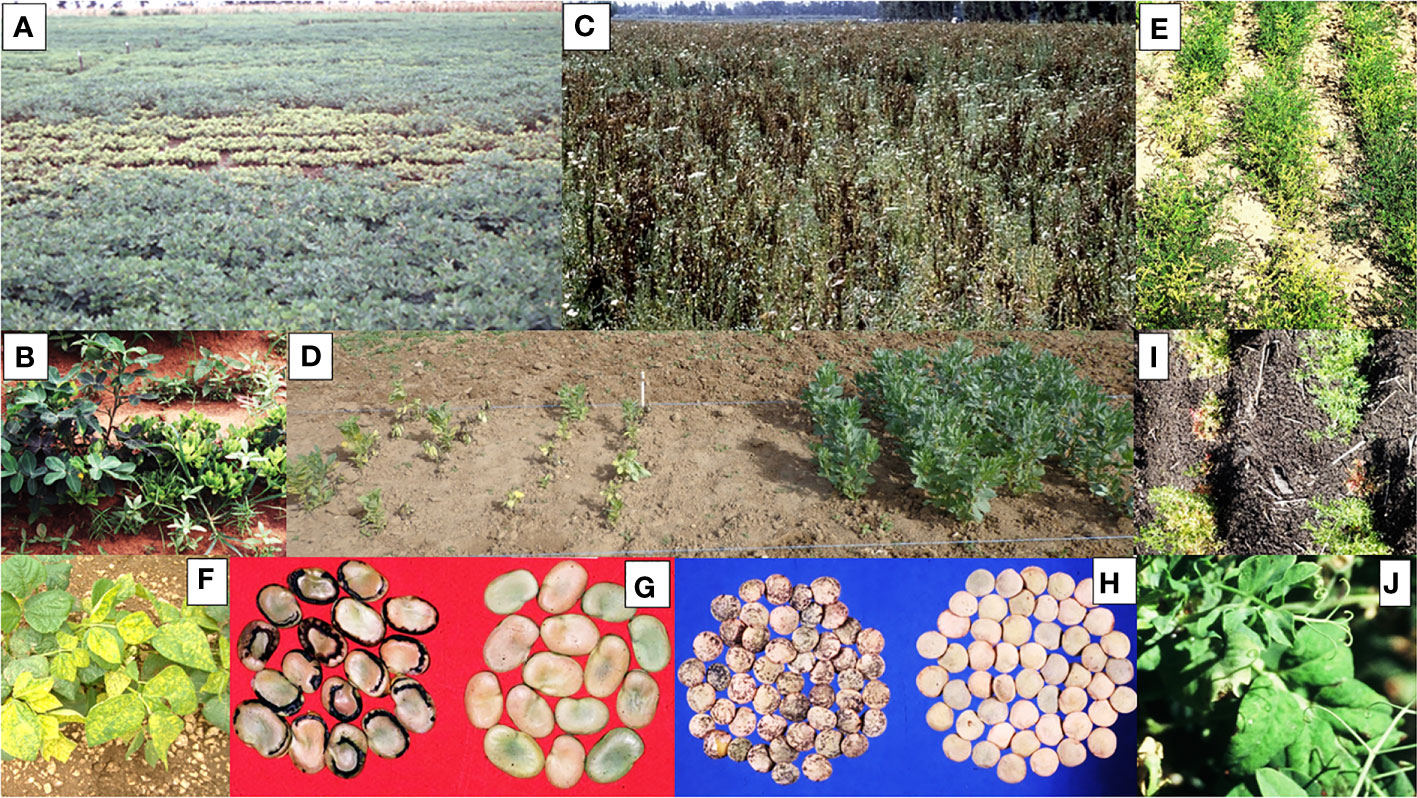
Figure 2 Examples of damage caused to grain legume foliage and seeds by virus diseases of biosecurity concern for Australia. (A) A field of peanut growing in East Africa with a large central area of chlorotic severely dwarfed plants caused by infection with groundnut rosette disease (image credit@Washington State University/Rayapati Naidu). (B) A row of peanut plants growing in East Africa showing foliage bushiness and severe plant stunting caused by infection with groundnut rosette disease (right), healthy plant (left) (image credit @Washington State University/Rayapati Naidu). (C) A field of faba bean peanut growing in Egypt showing dead plants killed by infection with faba bean necrotic yellows virus (image credit@International Center for Agricultural Research in the Dry Areas/Safaa Kumari). (D) Faba bean field experiment in Tunisia, plot on left contains dying plants with chlorotic and necrotic foliar symptoms caused by infection with faba bean necrotic yellows virus, insecticide protected control plot on right contains healthy plants (image credit@International Center for Agricultural Research in the Dry Areas/Safaa Kumari). (E) Rows of chickpea plants growing in Pakistan showing a mixture of stunted plants with chlorotic foliage caused by chickpea chlorotic dwarf virus and healthy plants. (F) A row of common bean plants growing in Florida showing bright yellow leaf mosaic symptoms caused by infection with bean golden mosaic virus. (G) Seeds harvested from faba bean plants growing in the Middle East: marginal necrosis, malformation and size reduction in seeds from a plant infected with broad bean mottle virus (left), seeds from healthy plant (right) (image credit@International Center for Agricultural Research in the Dry Areas/Khaled Makkouk). (H) Seeds harvested from lentil plants growing in the Middle East: necrotic rings and line patterns, malformation and size reduction in seeds from a plant infected with broad bean stain virus (left), seeds from healthy plant (right) (image credit@International Center for Agricultural Research in the Dry Areas/Khaled Makkouk). (I) Rows of lentil plants growing in Ethiopia showing symptoms of leaf chlorosis, reddening and size reduction and severe plant stunting caused by infection with the lentil strain of pea seed-borne mosaic virus (image credit@New South Wales Department of Primary Industries/Joop van Leur). (J) Foliage of field pea plant growing in south-west Australia (field breeding plots, Medina Research Station, 1997) showing leaf symptoms of severe mottle, deformation and size reduction due to infection with a severe strain of pea seed-borne mosaic virus.
Amongst some of the grain legume viruses already present in Australia, there are virulent strains which have not yet arrived but pose a threat to Australia’s grain legume crops. For example, two virulent strains of bean common mosaic virus (BCMV) fit this category. These are its peanut stripe (BCMV-PSt) and blackeye cowpea mosaic virus (BCMV-BICMV) strains, both of which were formerly considered distinct viruses. Moreover, BCMV-BICMV already occurs in Indonesia, and BCMV-PSt in both Indonesia and PNG (Green et al., 1988; Akin, 1997; Davis et al., 2002; Davis et al., 2021). Therefore, both are likely to enter Australia via birds or insect vectors crossing the ocean or directly via introduction of virus-contaminated seed. In 2013, an eastern Australian study reported finding a seed-borne PSbMV strain in seed of lentil germplasm imported from the USA and demonstrated that it belonged to PSbMV pathotype 2 (Figure 2I; Van Leur et al., 2013). This report considered this PSbMV strain to be a major biosecurity threat to the Australian lentil industry and ways of avoiding its establishment in commercial lentil crops were proposed. However, in 2001 an earlier study in south-western Australia had reported finding PSbMV-infected plants with atypically severe foliage symptoms growing within plots of field pea breeding lines (Figure 2J; Latham and Jones, 2001). This report suggested that these plants were infected with a severe PSbMV strain introduced via field pea germplasm from overseas. Soon afterwards, virus isolate W1 obtained from these plants was found to belong to PSbMV pathotype 2 to which the virulent lentil strain of PSbMV, sometimes called the lentil strain (L-1, Alconero et al., 1986), also belongs (Coutts et al., 2008; Wylie et al., 2011). Moreover, in a field experiment PSbMV isolate W1 infection decreased lentil seed yield by 96% and was seed-borne in 6% of seedlings grown from harvested lentil seed (Coutts et al., 2008). Therefore, the PSbMV variants found first in field pea breeding plots in south-western Australia and later in imported lentil seed in eastern Australia might both be representatives of the same virulent seed-borne lentil strain, which has apparently not yet spread within commercial crops belonging to the Australian lentil industry.
Recent advances in high-throughput sequencing (HTS) and bioinformatic analysis have provided considerable opportunities to upgrade virus surveillance for plant health services and virus discovery, so this subject has recently received considerable worldwide attention (Massart et al., 2014; Adams and Fox, 2016; Massart et al., 2017; Pecman et al., 2017; Adams et al., 2018; Villamor et al., 2019; EPPO, 2022; Lebas et al., 2022; Alcalá Briseño et al., 2023; Fontdevila Pareta et al., 2023), including within Australia (Barrero et al., 2017; Piper et al., 2019; Maina et al., 2021; Whattam et al., 2021; Gauthier et al., 2022; Lelwala et al., 2022; Mackie et al., 2022; Maina et al., 2022). Moreover, the cost of HTS is declining at a significant rate due to advances in sequencing technology, so its application as a routine diagnostic tool is becoming increasing attractive (Villamor et al., 2019; Lebas et al., 2022). Indeed, it has been endorsed as a robust testing tool for screening viral pathogens in inter-country commodity trading by both the World Trade Organisation and Food and the Agriculture Organisation of the United Nations (FAO, 2019). Furthermore, its integration is recommended for critical sample testing within healthy stock, seed and crop certification programs around the world to enhance the availability of virus-tested plant material in a timely manner (Villamor et al., 2019). However, despite these benefits its main drawback remains that, in addition to known viral pathogens, it also reveals presence of potential viral pathogens that exist only as sequences lacking any biological data (Massart et al., 2017; Jones et al., 2021b; Whattam et al., 2021). In 2017 in Europe, a collective framework was devised to provide guidelines designed to help both plant inspection agencies and biosecurity and plant health authorities with their collection of the biological data needed to establish whether such potential viral pathogens pose any actual economic risks to the agricultural sector (Massart et al., 2017). However, the biological data obtained from following these guidelines was insufficient for meaningful biosecurity PRAs to be performed on each potential viral pathogen. This was because they required comprehensive information for each virus about its incidence, distribution, symptomatology, pathogenicity, vectors, host range, genetic diversity and epidemiology. Therefore, this framework was expanded to make it sufficiently comprehensive to guide these agencies and authorities toward assembling all the biological data necessary for their PRAs to be effective (Adams et al., 2018; Olmos et al., 2018; Fontdevila Pareta et al., 2023). Figure 2 of Fontdevila Pareta et al. (2023) provides an up-to-date, detailed, explanatory diagram of the most recent version of the European PRA framework, which illustrates all its different components and how they relate to the other. This revised framework will help greatly with the collection of biological data needed to strengthen the effectiveness of biosecurity PRAs employed in Australia (see the Australian Plant Biosecurity Section above).
In the pre-sequencing era, there was far greater focus on the provision of extensive biological data about plant viruses than currently. Sequencing virus isolates preserved in historical plant virus collections, and establishing whether the sequences obtained match those of recently found potential viral pathogens existing only as sequences, can, in instances where there is a match, help provide access to otherwise missing but extensive biological baseline data needed for biosecurity PRAs (Jones et al., 2021b). It will also help establish when apparently newly found viruses have been provided inadvertently with incorrect names, because they had actually been studied and named in the past. This scenario is also important for biosecurity and other plant health authorities to be aware of when conducting their PRAs. Nevertheless, over the last ten years, the demand for HTS in global plant health diagnostics has remained high. Notably, whole genome sequencing can be costly when used for routine massive parallel sequencing. Therefore, over the last five years, considerable efforts have been underway in Australia to explore the possible integration of rapid diagnostics innovation involving HTS genome based smart tools into plant health and biosecurity programs (Barrero et al., 2017; Piper et al., 2019; Maina et al., 2021; Whattam et al., 2021; Gautier et al., 2022; Lelwala et al., 2022; Mackie et al., 2022). The following Sections describe examples of the most promising, cost-effective approaches suitable for use by plant biosecurity and other plant health authorities.
Whole genome sequencing platforms, such as Illumina, remain the most widely used procedures due to their high throughput and low error rate. Illumina can deliver up to 20 billion reads per run with paired read lengths of 150 bp. It uses technology based on reversible terminator–based methods to detect single bases as they are added into growing DNA strands. Such procedures significantly decrease errors and missed base calls associated with homopolymers which makes them extremely viable for biosecurity diagnoses (Maina et al., 2021; Pecman et al., 2022). However, Illumina produces shorter read lengths and involves a longer turn around time before results become available than third-generation platforms such as Oxford Nanopore Technology (ONT) (Pecman et al., 2022). Also, ONT provides a convenient means of identifying plant viruses (Bronzato-Badial et al., 2018; Ben Chehida et al., 2021; Liefting et al., 2021; Maina et al., 2022; Pecman et al., 2022; Waite et al., 2022). Its ONT direct RNA sequencing approach involves sequencing RNA without the need for prior amplification and it directly reads RNA without the need for reverse transcription (Phannareth et al., 2020). This avoids the complexity of some transcriptome assembly steps required with short reads, especially in highly variable regions. Its library preparation protocol (e.g SQK-RNA002) is straightforward and quick providing long reads lacking PCR bias (Garalde et al., 2018). Moreover, although ONT's high error rates were a drawback initially (ONT, 2020; Wongsurawat et al., 2019), during its last few years, its chemistry has progressed rapidly (Phannareth et al., 2020; Sereika et al., 2022). Indeed, its level of accuracy was reported to have improved from 85% to >99.9% when used to test macrofaunal reef samples (Chang et al., 2020; ONT, 2020). In addition, the ONT approach directly sequences RNA without modification of their native form (Wongsurawat et al., 2019; Cozzuto et al., 2020). Therefore, since RNA virus genomes undergo high rates of recombination and are notoriously variable, ONT direct RNA long read sequencing serves as an attractive option for detecting RNA viruses within a metagenomic sample. It not only negates PCR amplification biases but also helps achieve success in sequencing problematic viral genome sections. The absence of an RNA to cDNA reverse transcribing step means that ONT can provide results rapidly. Moreover, its use of long read, single molecule sequencing mitigates the issue of low sequence coverage and depth, making it suitable for use as a virus diagnostic metagenomic approach to sample testing within plant health and biosecurity virus screening programs. Indeed, when virus sample and library preparation procedures being used with it were optimised, Nanopore MinION sequencing proved as effective as Illumina sequencing at testing virus samples (Pecman et al., 2022). Overall, the Nanopore MinION is better suited to situations where few samples need to be tested rapidly, whereas the low error rates and higher throughput of other widely used platforms, such as Illumina, make them better suited to handling large numbers of samples (Villamor et al., 2019; Pecman et al., 2022). In New Zealand, deploying a combination of the MinION portable sequencer with the ONT Flongle platform increased both the efficiency and the accuracy of routine PEQ testing and biosecurity surveillance (Liefting et al., 2021). Notably, it was highly effective at detecting new host associations, mixed viral infections and previously undescribed viruses.
Innovative modifications could render ONT HTS a more cost-effective genomic diagnostic tool, offering a paradigm shift in PEQ biosecurity screening. Such modifications include taking advantage of the LAMP diagnostic method, which involves targeted isothermal amplification and colorimetric detection (Notomi et al., 2000). It is portable and faster than conventional PCR/qPCR, making it suitable for near-field/in-field applications and it can generate many copies of the target region in around 30 minutes at 65°C. Its successful amplification is often inferred from a proxy measurement, including high turbidity and colour fluorescence changes. However, these can be affected by contaminating substances present in a plant sample, e.g., colour change in no-template controls, arising from primer artifact amplification, can lead to a false positive detections. Instead of relying on the principle of proxy measurements, combining LAMP with ONT chemistry (LamPORE) can offer a raw sequence readout (Schmid-Burgk et al., 2020; Thompson and Lei, 2020). LamPORE uses a combined multiplexed barcoded loop-mediated amplification, transposase library preparation and real-time nanopore sequencing. This approach can be improved further and adopted to screen large numbers of samples for the presence or absence of viruses. Its target amplification profile sequences bind to the primers and can be identified without ambiguity by sequence analysis (Peto et al., 2021). LamPORE sequencing provides the additional opportunity to amplify and detect multiple targets in a single tube which can be adopted for virus biosecurity and other plant health surveillance in the field. The real time read generation of ONT combined with fast library preparation, reduces virus diagnostic costs and shortens decision-making timelines. This means that innovations like LamPORE offer a feasible solution in helping to improve grain biosecurity. It is also likely to be useful on a large scale as a portable plant health virus identification tool suitable for on farm virus genomic surveillance. In the future, further innovations and research within both Illumina and ONT will include real-time high-quality score data generation combined with faster robust library preparation procedures. This will lead to further reductions in costs associated with virus biosecurity diagnosis and shorten decision-making timelines. Such efforts, will enhance biosecurity diagnostics, surveillance, and decision-making, thereby contributing towards higher grains industry productivity.
Targeted HTS such as TG-Seq has proved successful in detecting multiple plant viruses simultaneously in a single reaction (e.g., Maina et al., 2021; Mackie et al., 2022). This approach targets conserved genome regions found within multiple viruses (pan-genomes). These are amplified by employing multiplexed oligonucleotide panels followed by using bead-linked transposomes to enrich the libraries for viral detection (Maina et al., 2021). Before TG-Seq was developed, studies that demonstrated a targeted HTS enrichment approach towards detecting a broad spectrum of plant viruses simultaneously were lacking, although this had been done previously in samples containing vertebrate viruses in clinical samples (e.g., Briese et al., 2015). The TG-Seq findings enabled a systematic understanding to be obtained of how multiple plant virus specific genome sections can be targeted and sequenced simultaneously to generate homology information (Maina et al., 2021). Such approaches maximise the sensitivity and specificity of the assay for the viruses targeted which reduces the sequencing costs associated with the detection assay and simplifies the downstream bioinformatic analyses. However, as yet, it remains to be determined how many target primers from diverse variants of RNA and DNA viruses can be multiplexed together in a single reaction. Other detection strategies include viral genome skimming sequencing (VgSkim-Seq) which also provides an effective and low-cost tool for routine viral genome sequencing and homology analysis (Bohmann et al., 2020). More importantly, these approaches can evaluate the incorporation of unique dual indices within the libraries derived to maximise the multiplexing capacity and reduce the risk of any indexing crossover.
Metabarcoding is also a targeted HTS approach. It can detect multiple insect virus vector species within a single reaction using the cytochrome C oxidase I (COI) mitochondrial gene, thus enabling massive parallel identification of diverse individuals from diverse insect taxonomic groups (e.g., Piper et al., 2019; Batovska et al., 2021). This approach is applicable to surveillance for incursions of vector insects that transmit viruses of grain crops, such as the recently arrived Diuraphis noxia (Russian wheat aphid), which still remains the subject of active Australian biosecurity surveillance and containment programs (Damsteegt et al., 1992; Ward et al., 2020). Deploying a targeted HTS platform such as the NovaSeq system(s), that can deliver up to 20 billion reads per run, would provide an efficient and very cost effective virus vector identification procedure, particularly in surveillance for virus and vector incursions in the field. This would contribute to virus management, surveillance and associated biosecurity decision-making.
Machine learning and deep learning algorithms for big data analyses, and remote sensing technologies, such as hyperspectral, multispectral, thermal, and other types of optical sensing, hold considerable promise toward achieving more effective virus disease and virus vector surveillance (Jones, 2014; Jones and Naidu, 2019; Oerke, 2020; Ahmad Loti et al., 2021; Khan et al., 2022; Gollapudi et al., 2023; Sarkar et al., 2023). Moreover, the combined use of ground with aerial remote sensing increases the accuracy and precision of virus disease or vector detection (Jones, 2014; Neupane and Baysal-Gurel, 2021; Rhodes et al., 2022; Wang et al., 2022). Furthermore, these innovations are becoming increasingly successful at distinguishing virus-infected or insect vector-infested plants from healthy plants (Griffel et al., 2018; Nansen et al., 2019; Abd El-Ghany et al., 2020; Terentev et al., 2022; Wang et al., 2022). During searches for new virus and vector incursions, adopting such technologies can strengthen the effectiveness and efficiency of different kinds of surveillance activities. This applies not only to NAQS activities in northern Australia, but also to post entry biosecurity eradication and containment programs in affected regions throughout Australia. It also applies to other plant health activities such as healthy stock and seed schemes and large-scale seed crop certification inspections. Such imaging would be applied on different scales depending on what it is being used for, ranging from very intensive during eradication programs to moderate or infrequent, as appropriate, during NAQS or containment activities.
Database surveillance networks could serve as an effective way of sharing datasets to help track viral genome mutations and variants of concern as they enter Australia. For this, a prospective virus genomic-surveillance system would be combined with an interoperable virus database adhering to standard practices. These practices would guide the process of creating and submitting sequence metadata records for standardization across different Australian biosecurity and plant health agencies. A dedicated high-performance computing system would be built in partnership with computing system administrators with extensive computing expertise, and both virology and virus vector researchers with strong virus biology and bioinformatics linkages. Database and surveillance systems would facilitate tracking viruses of biosecurity or plant health concern using sequence predictive tools to identify lineages and the origins of different incursions (Figure 1). Nextstrain (Hadfield et al., 2018) proved a successful approach with which real-time phylogenetic information was used to track variants within SARS-2 (COVID-19) populations. By revealing virus evolutionary trends, geographical origins and diversity, the development of such phylogenetic inference tools would help with tracking Australian grain virus incursions. This information would ensure more effective surveillance thereby enabling more effect virus management during eradication and containment programs.
Notably, as explained in the ‘High Throughput Sequencing’ Section above, historical virus specimen collections can help with understanding more recent virus incursions and their subsequent spread (Jones et al., 2021b). Likewise, building and enhancing Australian virus specimen collections, would provide a significant baseline data contribution to future virus biosecurity and plant health programs. These collections need to be sequenced, and the datasets obtained linked back to surveillance databases. Doing this would not only support the accurate identification of damaging viral variants linked to new incursions, but also help to ensure that all virus species identifications and classifications based on sequences are correct.
Here we explain the critical importance of grain industry exports to the Australian economy, the reasons why the continent lacks many of the virus diseases that damage grain crops in other continents and the importance of having a stringent biosecurity program in place to prevent their introduction and establishment. We also specify the different pathways by which damaging grain viruses and their vectors can reach Australia, and the measures currently in place to prevent their introduction and establishment. Next, we explain how Australian biosecurity programs currently test seeds, vegetative propagules or other plant imports after their arrival for the presence of viruses of concern. When such viruses are found the consignment is destroyed. When these efforts fail to detect threatening viruses, and an affected consignment is inadvertently released, an incursion occurs. In these circumstances, PRAs are done to provide an estimate of the risk of economic loss likely to occur if no further action is taken. In this context, we recommend using the most recent European framework providing comprehensive guidelines to help biosecurity and plant health authorities assemble all the biological data required to conduct effective PRAs. When the estimated risk level justifies the cost of taking action, an eradication campaign is authorised. Although such campaigns normally involve taking drastic measures, they sometimes fail to eliminate the incursion. When this transpires, a decision is made to continue to localise the distribution of the viral pathogen with a containment exercise or accept its presence and manage its epidemics in the same way as those of established viruses. Similar procedures to those used with virus threat arrivals also apply to incoming virus vectors.
Fortunately, exciting recent advances in technological innovation provide major opportunities to improve the effectiveness and reduce the costs of Australian biosecurity detection, diagnosis and surveillance procedures that target threatening grain crop viruses and their vectors. Where appropriate, they also provide opportunities to improve other Australian plant health activities. These include the effectiveness of future virus surveillance during healthy stock and seed programs, and crop certification. We recommend increasing their adoption, and continuous investment in future research innovations in Australian grains biosecurity to enhance their effectiveness and reduce the costs of virus testing and surveillance activities. In particular, we advocate more widespread integration of HTS genome based smart tools and research on their application into PEQ testing and other biosecurity testing of imported seeds, vegetative propagules, and further planting material. Additionally, these technologies offer major advantages for testing NAQS surveillance samples collected in the field. Where appropriate, we also suggest their use to ensure accurate diagnosis of viruses present in critical surveillance samples collected during eradication and containment activities, healthy stock and seed program inspections and seed crop health certification. This is because conventional plant biosecurity techniques are less adaptable to the detection of distinct viruses than HTS. This challenge underscores the necessity of undertaking future innovation focused on developing effective broad-spectrum HTS detection methods for use in detecting the growing number of virus biosecurity threats to the Australian grain industry. Globally, HTS has revolutionized the identification and characterization of plant viruses. Its high robustness has enabled the discovery of many viruses which have been fully or partially characterized subsequently. This has improved diagnostic procedures, disease diagnosis, and the understanding of virus diseases of unknown etiology. HTS has superseded conventional detection methods, due to its ability to identify both known and unknown viruses without prior knowledge of their presence, and in a reduced timeframe. The greater comprehensiveness of HTS compared to conventional detection methods means that it can profile multiple viruses present in a single biological sample, including viruses co-existing within a synergistic complex.
Illumina technology significantly reduces errors and missed base calls associated with homopolymers and other experimental stochasticity. It provides high quality short reads and has a massive multiplexing capability. These are all desirable cost-effective features especially suited for screening large number of samples for targets of low abundance viruses. However, future research innovation should focus on developing faster and shorter robust library preparation procedures and the capability to stop/pause the run as soon as enough data has accumulated to allow initial analysis. Further, future on board platforms should have the ability to allow analysis of base called datasets in real time sequencing. This could dramatically reduce grain virus biosecurity diagnostics associated costs and shorten critical biosecurity decision-making timelines. The ONT MinION portable sequencer is ideally suited to circumstances where few samples need to be tested rapidly by a cheaper method. Combining real-time nanopore sequencing with multiplexed barcoded loop-mediated amplification and transposase library preparation (LamPORE) increases accuracy, reduces virus diagnostic costs and shortens decision-making timelines. ONT future innovation should strive to develop robust library preparation procedures which incorporate both rRNA depletion and reverse transcription, for example in direct RNA-Seq and cDNA-PCR approaches, and higher quality score sequencing procedures which lead to a considerable increase in obtaining accurate results in virus detection. Such a portable tool would be ideally suited to on farm virus genomic surveillance.
TG-Seq is a targeted HTS approach that maximises the sensitivity and specificity of virus assays, decreases sequencing costs and simplifies downstream bioinformatic analyses. Likewise, VgSkim-Seq offers an effective and low-cost tool for use in routine viral genome sequencing. Moreover, inclusion of metabarcoding in an HTS platform such as the NovaSeq system makes it efficient and cost effective for use in surveillance for new virus vector incursions in the field. To further optimise the effectiveness of virus surveillance activities, we recommend widespread adoption of machine learning and deep learning algorithms for big data analyses, and both surface and aerial remote sensing technologies. Combining in-field machine learning high throughput phenotyping and real-time genotyping, will help track vector arrivals, viral genome mutations and virus variants of concern as they enter Australia. We also recommend using these technologies along with database surveillance networks that combine virus genomic-surveillance systems with an interoperable virus database. Finally, we recommend, enhancing and HTS of Australian virus specimen collections to help ensure all virus species identifications based on sequences are correct and viral variants linked to new incursions are identified accurately. These recommendations will help improve Australian grain biosecurity and plant health policy planning and grain virus and vector research. They will also help towards bettering both virus diagnosis and virus and virus vector data collection, thereby upgrading not only post entry virus interceptions but also background virus and vector surveillance information. In turn, this will reduce the frequency of new incursions, improve virus management during eradication, containment and other plant health activities, and help towards safeguarding the Australian grains industry and achieving more profitable Australian grain production.
The original contributions presented in the study are included in the article. Further inquiries can be directed to the corresponding author.
SM: Conceptualization, Funding acquisition, Investigation, Project administration, Writing – original draft, Writing – review & editing. RJ: Conceptualization, Investigation, Supervision, Visualization, Writing – original draft, Writing – review & editing.
The first author declares financial support was received for the research, authorship, and/or publication of this article. We thank the Grains Research and Development Corporation for funding the New South Wales Department of Primary Industries through the National Grains Diagnostics and Surveillance Initiative (DPI2305-010RTX). The funder was not involved in the study design, collection, analysis, interpretation, writing of this review or the decision to submit it for publication
We thank Rayapati Naidu, Khaled Makkouk, Safaa Kumari and Joop van Leur for permission to use their images, Will Cuddy, Katie Robinson, Anil Raghavendra, Paul Arthur and Krista Plett, New South Wales Department of Primary Industries, for their comments on the manuscript and Brendan Rodoni for useful discussions. The first author acknowledges receiving a Scholarship from the Cooperative Research Centre for Plant Biosecurity during his PhD studies at the University Western Australia. The authors acknowledge gaining plant virus biosecurity experience during previous employment at Agriculture Victoria Research (SM), the Western Australian Department of Primary Industries and Regional Development (RACJ), and the former UK Ministry of Agriculture Fisheries and Food (RACJ).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Author SM declared that he was an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
ABARES (2022). Ag commodities statistics. Available at: https://www.agriculture.gov.au/abares/research-topics/agricultural-outlook/data#_2022 (Accessed 12/9/22).
Abd El-Ghany N. M., Abd El-Aziz S. E., Marei S. S. (2020). A review: application of remote sensing as a promising strategy for insect pests and diseases management. Env. Sci. pollut. Res. 7, 33503–33515.
Adams I. P., Fox A. (2016). “Diagnosis of plant viruses using next-generation sequencing and metagenomic analysis,” in Current research topics in plant virology’. Eds. Wang A., Zhou X. (Springer, Cham, New York, USA), 323–335. doi: 10.1007/978-3-319-32919-2_14
Adams I. P., Fox A., Boonham N., Massart S., De Jonghe K. (2018). The impact of high throughput sequencing on plant health diagnostics. Eur. J. Plant Pathol. 152, 909–919. doi: 10.1007/s10658-018-1570-0
AEGIC (2020). What grows where? In: Australian grain production – a snapshot (Sydney and Perth, Australia: Australian Export Grains Innovation Centre). Available at: https://www.aegic.org.au/Australian-grain-production-a-snapshot (Accessed 12/9/22).
Ahmad Loti N. N., Mohd Noor M. R., Chang S. W. (2021). Integrated analysis of machine learning and deep learning in chili pest and disease identification. J. Sci. Food Agric. 101, 3582–3594.
Akin H. M. (1997). Characterisation of peanut stripe virus (PStV) isolates originating from various provinces in Indonesia. Indones. J. Trop. Agric. 8, 13–20.
Alcalá Briseño R. I., Batuman O., Brawner J., Cuellar W. J., Delaquis E., Etherton B. A., et al. (2023). Translating virome analyses to support biosecurity, on-farm management, and crop breeding. Front. Plant Sci. 14, 1056603.
Alconero R., Provvidenti R., Gonsalves D. (1986). Three pea seedborne mosaic virus pathotypes from pea and lentil germ plasm. Plant Dis. 70, 783–786.
Alexander H. M., Bruns E., Schebor H., Malmstrom C. M. (2017). Crop associated virus infection in a native perennial grass: reduction in plant fitness and dynamic patterns of virus detection. J. Ecol. 105, 1021–1031.
Alexander H. M., Mauck K. E., Whitfield A. E., Garrett K. A., Malmstrom C. M. (2014). Plant-virus interactions and the agro-ecological interface. Eur. J. Plant Pathol. 138, 529–547.
Alvarez Quinto R. A., Amao M., Muller G., Fuentes S., Grinstead S., Fuentes-Bueno I., et al. (2023). Evidence that an unnamed isometric virus associated with potato rugose disease in Peru is a new species of torradovirus. Phytopathology. doi: 10.1094/PHYTO/-11-22-0449-V
Anderson P. K., Cunningham A. A., Patel N. G., Morales F. J., Epstein P. R., Daszak P. (2004). Emerging infectious diseases of plants: pathogen pollution, climate change and agrotechnology drivers. Trends Ecol. Evol. 19, 535–544.
Anderson C., Low-Choy S., Whittle P., Taylor S., Gambley C., Smith L., et al. (2017). Australian plant biosecurity surveillance systems. Crop Prot. 100, 8–20.
Barbetti M. J., Jones R. A. C., Riley I. T. (1996). “Problems and progress in assessing direct and indirect yield losses caused by pathogens in pasture species,” in Pasture and forage crop pathology. Eds. Chackraborty S., Leath K. T., Skipp R. A., Pederson G. A., Bray R., Latch G. C. M., Nutter F. W. (Madison, WI: American Society of Agronomy), 63–91.
Barrero R. A., Napier K. R., Cunnington J., Liefting L., Keenan S., Frampton R. A., et al. (2017). An internet-based bioinformatics toolkit for plant biosecurity diagnosis and surveillance of viruses and viroids. BMC Bioinf. 18, 26.
Batovska J., Piper A. M., Valenzuela I., Cunningham J. P., Blacket M. J. (2021). Developing a non-destructive metabarcoding protocol for detection of pest insects in bulk trap catches. Sci. Rep. 11 (1), 7946.
Ben Chehida S., Filloux D., Fernandez E., Moubset O., Hoareau M., Julian C., et al. (2021). Nanopore sequencing is a credible alternative to recover complete genomes of Geminiviruses. Microorganisms 9 (5), 903.
BICON (2023). Australian biosecurity import conditions. Available at: https://bicon.agriculture.gov.au/BiconWeb4.0.
Bohmann K., Mirarab S., Bafna V., Gilbert M. T. P. (2020). Beyond DNA barcoding: The unrealized potential of genome skim data in sample identification. Mol. Ecol. 29, 2521–2534.
Bos L. (1981). “Wild plants in the ecology of virus diseases,” in Plant diseases and vectors: ecology and epidemiology. Eds. Maramorosch K., Harris K. F. (New York: NJ: Academic Press), 1–33.
Bos L. (1992). New plant virus problems in developing countries. A corollary to agricultural modernization. Adv. Virus Res. 41, 349–407.
Briese T., Kapoor A., Mishra N., Jain K., Kumar A., Jabado O. J., et al. (2015). Virome capture sequencing enables sensitive viral diagnosis and comprehensive virome analysis. mBio 6 (5), e01491–e01415. doi: 10.1128/mBio.01491-15
Bronzato-Badial A. B., Sherman D., Stone A., Gopakumar A., Wilson V., Schneider W., et al. (2018). Nanopore sequencing as a surveillance tool for plant pathogens in plant and insect tissues. Plant Dis. 102, 1648–1652.
Brown A., De Costa C., Guo F. (2020) Our food future: trends and opportunities (Canberra, ACT, Australia: Research Report 20.1, Australian Bureau of Agricultural and Resource Economics and Sciences) (Accessed 12/9/21).
Buchen-Osmond C., Crabtree K., Gibbs A. J., McLean G. D. (1988). Viruses of plants in AUSTRALIA (Australian National University Printing Service: Canberra, ACT).
Burgman M., Roberts B., Sansford C., Griffin R., Mengersen K. (2014). “The role of pest risk analysis in plant biosecurity,” in The handbook of plant biosecurity: principles and practices for the identification, containment and control of organisms that threaten agriculture and the environment globally (Springer: Dordecht, The Netherlands), 235–267.
Canto T., Aranda M. A., Fereres A. (2009). Climate change effects on physiology and population processes of hosts and vectors that influence the spread of hemipteran-borne plant viruses. Global Change Biol. 15, 1884–1894.
Chai Q., Nemecek T., Liang C., Zhao C., Yu A., Coulter J. A., et al. (2021). Integrated farming with intercropping increases food production while reducing environmental footprint. Proc. Nat. Acad. Sci. 118 (38), e2106382118.
Chang J. J. M., Ip Y. C. A., Bauman A. G., Huang D. (2020). MinION-in-ARMS: nanopore sequencing to expedite barcoding of specimenrich macrofaunal samples from autonomous reef monitoring structures. Front. Mar. Sci. 7, 448.
Constable F., Kelly G., Dall D. (2021). Viruses in cucurbit seeds from on-line mail-order providers. Australas. Plant Dis. Notes 16, 1–4.
Cooper J. I., Jones R. A. C. (2006). Wild plants and viruses: under-investigated ecosystems. Adv. Virus Res. 67, 1–46.
Coutts B. A., Jones R. A. C. (2002). Temporal dynamics of spread of four viruses within mixed species perennial pastures. Ann. Appl. Biol. 140, 37–52.
Coutts B. A., Prince R. T., Jones R. A. C. (2008). Further studies on pea seed-borne mosaic virus in cool-season crop legumes: Responses to infection and seed quality defects. Aust. J. Agric. Res. 59, 1130–1145.
Cozzuto L., Liu H., Pryszcz L. P., Pulido T. H., Delgado-Tejedor A., Ponomarenko J., et al. (2020). MasterOfPores: a workflow for the analysis of oxford nanopore direct RNA sequencing datasets. Front. Genet. 11, 211.
Crooks E., Havel K., Shannon M., Snyder G., Wallenmaier T. (1983). “Stopping pest introductions, pp. 239-259,” in Exotic plant pests and north american agriculture. Eds. Wilson C. L., Graham C. L. (New York, USA: Academic Press).
Damayanti T. A., Naidu R. A. (2009). Identification of Peanut bud necrosis virus and Tomato spotted wilt virus in Indonesia for the first time. Plant Pathol. 58, 782.
Damsteegt V. D., Gildow F. E., Hewings A. D., Carroll T. W. (1992). A clone of the Russian wheat aphid (Diuraphis noxia) as a vector of barley yellow dwarf, barley stripe mosaic, and brome mosaic viruses. Plant Dis. l76, 1155–1160.
Davis R. I., Jones L. M., Pease B., Perkins S. L., Vala H. R., Kokoa P., et al. (2021). Plant virus and virus-like disease threats to Australia’s north targeted by the northern Australia quarantine strategy. Plants 10 (10), p.2175.
Davis R. I., Thomas J. E., McMichae L. A., Dietzgen R. G., Callaghan B., James A. P., et al. (2002). Plant virus surveys on the island of New Guinea and adjacent regions of northern Australia. Australas. Plant Pathol. 31, 385–390.
Dombrovsky A., Tran-Nguyen L. T., Jones R. A. C. (2017). Cucumber green mottle mosaic virus: rapidly increasing global distribution, etiology, epidemiology, and management. Annu. Rev. Phytopathol. 55, 231–256.
Dwyer G. I., Gibbs M. J., Gibbs A. J., Jones R. A. C. (2007). Wheat streak mosaic virus in Australia: Relationship to isolates from the Pacific Northwest of the USA and its dispersion via seed transmission. Plant Dis. 91, 164–170.
Eagles D., Walker P. J., Zalucki M. P., Durr P. A. (2013). Modelling spatio-temporal patterns of long-distance Culicoides dispersal into northern Australia. Prev. Vet. Med. 110, 312–322. doi: 10.1016/j
Edwardson J. R., Christae R. G. (1986). Viruses infecting forage legumes Vol. Vol III (Gainsville, FL: University of Florida), 742.
Elena S. F., Fraile A., García-Arenal F. (2014). Evolution and emergence of plant viruses. Adv. Virus Res. 88, 161–191.
EPPO (2022). Considerations for the use of high throughput sequencing in plant health diagnostics. EPPO Bull. 52, 619–642. doi: 10.1111/epp.12884
FAO (2010). International standards for phytosanitary measures publication no. 5. Glossary of phytosanitary terms. (Rome: Secretariat of the International Plant Protection Convention, Food and Agriculture Organisation of the United Nations). Available at: https://www.fao.org/fileadmin/user_upload/faoterm/PDF/ISPM_05_2016_En_2017-05-25_PostCPM12_InkAm.pdf (Accessed 15/9/22).
FAO (2019). Recommendation on: Preparing to use high-throughput sequencing (HTS) technologies as a diagnostic tool for phytosanitary purposes. (Rome: Commission on Phytosanitary Measures Recommendation No. 8. Secretariat of the International Plant Protection Convention, Food and Agriculture Organisation of the United Nations). Available at: https://www.ippc.int/static/media/files/publication/en/2019/05/R_08_En_2019-05-06_Post-CPM-14.pdf (Accessed 15/9/22).
Fargette D., Konate G., Fauquet C., Muller E., PeterscHmitt M., Thresh J. M. (2006). Molecular ecology and emergence of tropical plant viruses. Annu. Rev. Phytopathol. 44, 235–260.
Fereres A. (2015). Insect vectors as drivers of plant virus emergence. Curr. Opin. Virol. 10, 42–46.
Firth C., Blasdell K. R., Amos-Ritchie R., Sendow I., Agnihotri K., Boyle D. B., et al. (2017). Genomic analysis of bluetongue virus episystems in Australia and Indonesia. Vet. Res. 48, 82. doi: 10.1186/s13567-017-0488-4
Fontdevila Pareta N., Khalili M., Maachi A., Rivarez M. P. S., Rollin J., Salavert F., et al. (2023). Managing the deluge of newly discovered plant viruses and viroids: an optimized scientific and regulatory framework for their characterization and risk analysis. Front. Microbiol. 14. doi: 10.3389/fmicb.2023.1181562
Forster R. L., Seifers D. L., Strausbaugh C. A., Jensen S. G., Ball E. M., Harvey T. L. (2001). Seed transmission of the High Plains virus in sweet corn. Plant Dis. 85, 696–699.
Fox A., Fowkes A. R., Skelton A., Harju V., Buxton-Kirk A., Kelly M., et al. (2019). Using high-throughput sequencing in support of a plant health outbreak reveals novel viruses in Ullucus tuberosus (Basellaceae). Plant Pathol. 68, 576–587.
Friess N., Maillet J. (1996). Influence of cucumber mosaic virus infection on the intraspecific competitive ability and fitness of purslane (Portulaca oleracea). New Phytol. 132, 103–111.
Fuentes S., Gibbs A. J., Adams I. P., Wilson C., Botermans M., Fox A., et al. (2021). Potato virus A isolates from three continents: Their biological properties, phylogenetics, and prehistory. Phytopathology 111, 217–226.
Garalde D. R., Snell E. A., Jachimowicz D., Sipos B., Lloyd J. H., Bruce M., et al. (2018). Highly parallel direct RNA sequencing on an array of nanopores. Nat. Methods 15, 201–206. doi: 10.1038/nmeth.4577
Gauthier M. E. A., Lelwala R. V., Elliot C. E., Windell C., Fiorito S., Dinsdale A., et al. (2022). Side-by-side comparison of post-entry quarantine and high througput sequencing methods for virus and viroid testing. Biology 11 (2), 263.
Gibbs A. J., Mackenzie A. M., Wei K. J., Gibbs M. J. (2008). The potyviruses of AUSTRALIA. Arch. Virol. 153, 1411–1420.
Gilbertson R. L., Batuman O., Webster C. G., Adkins S. (2015). Role of the insect supervectors Bemisia tabaci and Frankliniella occidentalis in the emergence and global spread of plant viruses. Ann. Rev. Virol. 2, 67–93.
Gollapudi K., Varigonda P., Jancy S., PushpaLatha S., Helen S. (2023)Detection of plant disease using machine learning and deep learning algorithms (Accessed 15/5/23).
Green S. K., Lee D. R., Horne N. M. (1988). Survey for viruses of soybean, mung bean and peanut in Java, Indonesia. Plant Dis. 72, 994.
Griffel L. M., Delparte D., Edwards J. (2018). Using Support Vector Machines classification to differentiate spectral signatures of potato plants infected with potato Virus Y. Comput. Electron. Agric. 153, 318–324.
Gunning R. V., Bryne F. J., Condé B. D., Connelly M. I., Hergstrom K., Devonshire A. L. (1995). First report of B-Biotype Bemisia tabaci (Hemiptera: Aleyrodidae) in Australia. Aust. J. Entomol. 34, 116. doi: 10.1111/j.1440-6055.1995.tb01298.x
Hadfield J., Megill C., Bell S. M., Huddleston J., Potter B., Callender C., et al. (2018). Nextstrain: real-time tracking of pathogen evolution. Bioinformatics 34, 4121–4123.
Hema M., Sreenivasulu P., Patil B. L., Kumar P. L., Reddy D. V. (2014). Tropical food legumes: virus diseases of economic importance and their control. Adv. Virus Res. 90, 431–505.
Henzell T. (2007). Australian agriculture: its history and challenges (CSIRO Publishing: Melbourne, Australia).
Horwood P. F., McBryde E. S., Peniyamina D., Ritchie S. A. (2018). The Indo-Papuan conduit: a biosecurity challenge for Northern Australia. Aust. New Z. J. Public Health 42, 434–436.
IPPC (1997). International plant protection convention, new revised text (Rome, Italy: Approved by the FAO Conference at its 29th Session). Available at: https://assets.ippc.int/static/media/files/publication/en/2016/10/New_Revised_Text_of_the_IPPC.pdf (Accessed 15/5/23).
Irwin M. E., Thresh J. M. (1988). Long-range aerial dispersal of cereal aphids as virus vectors in North America. Philos. Trans. R. Soc. Ser. B Biol. Sci. 321, 421–446.
Jeggo M. (2012). “The Australian perspective, the biosecurity continuum from preborder, to border and postborder,” in Improving food safety through a one health approach, workshop summary (Washington, DC, USA: National Academies Press), 198–206.
Jones R. A. C. (2001). Developing integrated disease management strategies against non-persistently aphid-borne viruses: a model programme. Int. Pest Manage. Rev. 6, 15–46.
Jones R. A. C. (2004). Using epidemiological information to develop effective integrated virus disease management strategies. Virus Res. 100, 5–30.
Jones R. A. C. (2009). Plant virus emergence and evolution: origins, new encounter scenarios, factors driving emergence, effects of changing world conditions, and prospects for control. Virus Res. 141, 113–130.
Jones R. A. C. (2012). Virus diseases of annual pasture legumes: incidences, losses, epidemiology and management. Crop Pasture Sci. 63, 399–418.
Jones R. A. C. (2014). Trends in plant virus epidemiology: opportunities from new or improved technologies. Virus Res. 186, 3–19.
Jones R. A. C. (2016). Future scenarios for plant virus pathogens as climate change progresses. Adv. Virus Res. 95, 87–147.
Jones R. A. C. (2018). Plant and insect viruses in managed and natural environments: Novel and neglected transmission pathways. Adv. Virus Res. 101, 149–187.
Jones R. A. C. (2020). Disease pandemics and major epidemics arising from new encounters between indigenous viruses and introduced crops. Viruses 12, 1388.
Jones R. A. C. (2021). Global plant virus disease pandemics and epidemics. Plants 10 (2), 233. doi: 10.3390/plants10020233
Jones R. A. C. (2022). Alteration of plant species mixtures by virus infection: Managed pastures the forgotten dimension. Plant Pathol. 71, 1255–1281.
Jones R. A. C., Barbetti M. J. (2012). Influence of climate change on plant disease infections and epidemics caused by viruses and bacteria. Plant Sci. Rev. 22, 1–31.
Jones R. A. C., Barbetti M. J., Fox A., Adams I. P. (2021a). Potato virus Y biological strain group YD: Hypersensitive resistance genes elicited and phylogenetic placement. Plant Dis. 105, 3600–3609.
Jones R. A. C., Boonham N., Adams I. P., Fox A. (2021b). Historical virus isolate collections: An invaluable resource connecting plant virology’s pre-sequencing and post-sequencing eras. Plant Pathol. 70, 235–248.
Jones R. A. C., Coutts B. A. (2015). Spread of introduced viruses to new plants in natural ecosystems and the threat this poses to plant biodiversity. Mol. Plant Pathol. 16, 541–545.
Jones R. A. C., Coutts B. A., Mackie A. E., Dwyer G. I. (2005). Seed transmission of wheat streak mosaic virus shown unequivocally in wheat. Plant Dis. 89, 1048–1050.
Jones R. A. C., Naidu R. A. (2019). Global dimensions of plant virus diseases: Current status and future perspectives. Annu. Rev. Virol. 6, 387–409.
Jones R. A. C., Nicholas D. A. (1998). Impact of an insidious virus disease in the legume component on the species balance within self-regenerating annual pasture. Aust. J. Agric. Sci. 131, 155–170.
Jones R. A. C., Sharman M., Trębicki P., Maina S., Congdon B. S. (2021c). Virus diseases of cereal and oilseed crops in Australia: current position and future challenges. Viruses 13 (10), 2051.
Jones R. A. C., Vazquez-Iglesias I., Hajizadeh M., McGreig S., Fox A., Gibbs A. J. (2022). Phylogenetics and evolution of wheat streak mosaic virus: Its global origin and the source of the Australian epidemic. Plant Pathol. 71, 1660–1673.
Jones R. A. C., Vazquez-Iglesias I., McGreig S., Fox A., Gibbs A. J. (2023). Genomic High Plains wheat mosaic virus sequences from Australia: Their phylogenetics and evidence for emaravirus recombination and reassortment. Viruses 15 (2), 401.
Jones R. A. C., Vincent S. J. (2018). Strain-specific hypersensitive and extreme resistance phenotypes elicited by potato virus Y among 39 potato cultivars released in three world regions over a 117-year period. Plant Dis. 102, 185–196.
Kehoe M. A., Webster C., Wang C., Jones R. A. C., Coutts B. A. (2022). Occurrence of cucumber green mottle mosaic virus in Western Australia. Australas. Plant Pathol. 51, 1–8.
Khan K., Khan R. U., Albattah W., Qamar A. M. (2022). End-to-end semantic leaf segmentation framework for plants disease classification. Complexity 2022, 1–11. doi: 10.1155/2022/1168700
Kreuze J., Cuellar W., Kumar L., Prasanna B. M., Omondi A. (2023). New technologies provide innovative opportunities to enhance understanding of major virus diseases threatening global food security. Phytopathol. doi: 10.1094/PHYTO-12-22-0457-V
Lapierre H., Signoret P. A. (2004). Viruses and virus diseases of poaceae (Gramineae) (France: Institut National de la Recherche Agronomique (INRA).
Latham L. J., Jones R. A. C. (2001). Incidence of virus infection in experimental plots, commercial crops, and seed stocks of cool season crop legumes. Aust. J. Agric. Res. 52, 397–413.
Lebas B., Adams I., Al Rwahnih M., Baeyen S., Bilodeau G. J., Blouin A. G., et al. (2022). Facilitating the adoption of high-throughput sequencing technologies as a plant pest diagnostic test in laboratories: A step-by-step description. EPPO Bull. 52, 394–418.
Lecoq H., Desbiez C., Wipf-Scheibel C., Girard M. (2003). Potential involvement of melon fruit in the long distance dissemination of cucurbit potyviruses. Plant Dis. 87, 955–959.
Lelwala R. V., LeBlanc Z., Gauthier M. E. A., Elliot. C. E., Constable F. E., Murphy G., et al. (2022). Implemenation of GA-VirReport, a web-based bioinformatics toolkit for post-entry quarantine screening of virus and viroids in plants. Viruses 14, 1480.
Lenzen M., Tzeng M., Floerl O., Zaiko A. (2023). Application of multi-region input-output analysis to examine biosecurity risks associated with the global shipping network. Sci. Total Environ. 854, 158758.
Liefting L. W., Waite D. W., Thompson J. R. (2021). Application of Oxford Nanopore technology to plant virus detection. Viruses 13 (8), 1424.
Loebenstein G., Thottappilly G. (2013). Virus and virus-like diseases of major crops in developing countries (Dordrecht, The Netherlands: Springer).
Mackie J., Kinoti W. M., Chahal S. I., Lovelock D. A., Campbell P. R., Tran-Nguyen L. T. T., et al. (2022). Targeted Whole Genome Sequencing (TWG-Seq) of cucumber green mottle mosaic virus using tiled amplicon multiplex PCR and Nanopore sequencing. Plants 11, 2716.
Maina S. (2018) Application of viral genomics to study the connectivity between crop viruses from northern Australia and neighbouring countries (The University of Western Australia: Doctor of Philosophy) (Accessed 15/5/23).
Maina S., Barbetti M. J., Edwards O. R., de Almeida L., Ximenes A., Jones R. A. C. (2018a). Sweet potato feathery mottle virus and Sweet potato virus C from East Timorese and Australian Sweet potato: Biological and Molecular Properties, and Biosecurity Implications. Plant Dis. 102, 589–599.
Maina S., Barbetti M. J., Edwards O. R., Minemba D., Areke M. W., Jones R. A. C. (2019). Genetic connectivity between Papaya ringspot virus genomes from Papua New Guinea and northern Australia, and new recombination insights. Plant Dis. 103, 737–747.
Maina S., Barbetti M. J., Martin D. P., Edwards O. R., Jones R. A. C. (2018b). New isolates of Sweet potato feathery mottle virus and Sweet potato virus C: biological and molecular properties, and recombination analysis based on complete genomes. Plant Dis. 102, 1899–1914.
Maina S., Coutts B. A., Edwards O. R., de Almeida L., Kehoe M. A., Ximenes A., et al. (2017a). Zucchini yellow mosaic virus populations from East Timorese and northern Australian cucurbit crops: Molecular properties, genetic connectivity and biosecurity implications. Plant Dis. 101, 1236–1245.
Maina S., Coutts B. A., Edwards O. R., de Almeida L., Ximenes A., Jones R. A. C. (2017c). Papaya ringspot virus populations from east timorese and northern australian cucurbit crops: biological and molecular properties, and absence of genetic connectivity. Plant Dis. 101, 985–993.
Maina S., Edwards O. R., de Almeida L., Ximenes A., Jones R. A. C. (2016). First complete genome sequence of bean common mosaic necrosis virus from East Timor. Genome Announc. 4 (5), e01049–e01016.
Maina S., Edwards O. R., de Almeida L., Ximenes A., Jones R. A. C. (2017b). First complete Squash leaf curl China virus genomic segment DNA-A sequence from East Timor. Genome Announc 5, e00483–e00417.
Maina S., Norton S. L., McQueen V. L., King S., Rodoni B. (2022). Oxford Nanopore Sequencing Reveals an Exotic Broad bean mottle virus Genome within Australian Grains Post-Entry Quarantine. Microbiol. Resource Announcements 11 (7), e00211–e00222.
Maina S., Zheng L., Rodoni B. C. (2021). Targeted genome sequencing (TG-Seq) approaches to detect plant viruses. Viruses 13 (4), 583.
Makkouk K. M. (2020). Plant pathogens which threaten food security: Viruses of chickpea and other cool season legumes in West Asia and North Africa. Food Secur. 12, 495–502.
Makkouk K. M., Kumari S. G., van Leur J. A., Jones R. A. C. (2014). Control of plant virus diseases in cool-season grain legume crops. Adv. Virus Res. 90, 207–253).
Malipatil M. B., Postle A. C., Osmelak J. A., Hill M., Moran J. (1993). First record of Frankliniella occidentals (pergande) in Australia (Thysanoptera: Thripidae). Aust. J. Entomol. 32, 378–378.
Malmstrom C. M., Alexander H. M. (2016). Effects of crop viruses on wild plants. Curr. Opin. Virol. 19, 30–36.
Malmstrom C. M., Hughes C. C., Newton L. A., Stoner C. J. (2005a). Virus infection in remnant native bunchgrasses from invaded California grasslands. New Phytol. 168, 217–230.
Malmstrom C. M., McCullough A. J., Johnson H. A., Newton L. A., Borer E. T. (2005b). Invasive annual grasses indirectly increase virus incidence in California native perennial bunchgrasses. Oecologia 145, 153–164.
Martin R. R., Constable F., Tzanetakis I. E. (2016). Quarantine regulations and the impact of modern detection methods. Annu. Rev. Phytopathol. 54, 189–205.
Maskell L. C., Raybould A. F., Cooper J. I., Edwards M. L., Gray A. J. (1999). Effects of turnip mosaic virus and turnip yellow mosaic virus on the survival, growth and reproduction of wild cabbage (Brassica oleracea). Ann. Appl.Biol. 135, 401–407.
Massart S., Candresse T., Gil J., Lacomme C., Predajna L., Ravnikar M., et al. (2017). A framework for the evaluation of biosecurity, commercial, regulatory, and scientific impacts of plant viruses and viroids identified by NGS technologies. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.00045
Massart S., Olmos A., Jijakli H., Candresse T. (2014). Current impact and future directions of high throughput sequencing in plant virus diagnostics. Virus Res. 188, 90–96.
McLaughlin M. R., Larsen R. C., Trevathan L. E., Eastman C. E., Hewings A. D. (1996). “Virus diseases of American pasture and forage crops,” in Pasture and forage crop pathology. Eds. Chakraborty S., Leath K. T. L., Skipp R. A., Pederson G. A., Bray R. A., Latch G. C. M., Nutter F. W. (Madison, MN, USA: American Society of Agronomy), 323–361.
McLaughlin M. R., Pederson G. A., Evans R. R., Ivy R. L. (1992). Virus diseases and stand decline in a white clover pasture. Plant Dis. 76, 158–162.
Moghal S. M., Francki R. I. B. (1974). Occurrence and properties of broad bean stain virus in South Australia. Aust. J. Biol. Sci. 27, 341–348.
Naidu R. A., Kimmins F. M., Deom C. M., Subrahmanyam P., Chiyembekeza A. J., van der Merwe P. J. A. (1999). Groundnut rossette: a virus disease affecting groundnut production in sub-Saharan Africa. Plant Dis. 83, 700–709.
Nansen C., Stewart A. N., Gutierrez T. A. M., Wintermantel W. M., McRoberts N., Gilbertson R. L. (2019). Proximal remote sensing to differentiate nonviruliferous and viruliferous insect vectors–proof of concept and importance of input data robustness. Plant Pathol. 68, 746–754.
NAQS (2020). Safeguarding our home and way of life: 30 years of co-operative biosecurity in Northern Australia (Canberra, ACT, Australia: Northern Australia Quarantine Strategy, Department of Agriculture, Water and the Envionment, Australian Federal Government). Available at: https://www.agriculture.gov.au/sites/default/files/documents/naqs_30years_brochure_2020.pdf (Accessed 28th May 2023).
Neupane K., Baysal-Gurel F. (2021). Automatic identification and monitoring of plant diseases using unmanned aerial vehicles: A review. Remote Sens. 13 (19), p.3841.
Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N., et al. (2000). Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 28 (12), e63–e63.
Olmos A., Boonham N., Candresse T., Gentit P., Giovani B., Kutnjak D., et al. (2018). High-throughput sequencing technologies for plant pest diagnosis: challenges and opportunities. EPPO Bull. 48, 219–224. doi: 10.1111/epp.12472
ONT (2020). New research algorithms yield accuracy gains for nanopore sequencing. Oxford nanopore sequencing. Available at: https://nanoporetech.com/about-us/news/new-research-algorithms-yield-accuracy-gains-nanopore-sequencing (Accessed 16th May 2023).
Pecman A., Adams I., Gutiérrez-Aguirre I., Fox A., Boonham N., Ravnikar M., et al. (2022). Systematic comparison of nanopore and Illumina sequencing for the detection of plant viruses and viroids using total RNA sequencing approach. Front. Microbiol. 13, 883921.
Pecman A., Kutnjak D., Gutiérrez-Aguirre I., Adams I., Fox A., Boonham N., et al. (2017). Next generation sequencing for detection and discovery of plant viruses and viroids: Comparison of two approaches. Front. Microbiol. 8, p.1998.
Peto L., Rodger G., Carter D. P., Osman K. L., Yavuz M., Johnson K., et al. (2021). Diagnosis of SARS-CoV-2 infection with LamPORE, a high-throughput platform combining loop-mediated isothermal amplification and nanopore sequencing. J. Clin. Microbiol. 59 (6), e03271–e03220.
Phannareth T., Nunziata S. O., Stulberg M. J., Galvez M. E., Rivera Y. (2020). Comparison of nanopore sequencing protocols and real-time analysis for phytopathogen diagnostics. Plant Health Prog. 22, 31–36.
Piper A. M., Batovska J., Cogan N. O., Weiss J., Cunningham J. P., Rodoni B. C., et al. (2019). Prospects and challenges of implementing DNA metabarcoding for high-throughput insect surveillance. GigaScience 8 (8), giz092.
Randles J. W., Dube A. J. (1977). Three seedborne pathogens isolated from Vicia faba seed imported from the United Kingdom. Australas. Plant Pathol. Soc Newslett. 6, 37–38. doi: 10.1071/APP9770037
Rhodes M. W., Bennie J. J., Spalding A., Ffrench-Constant R. H., Maclean I. M. (2022). Recent advances in the remote sensing of insects. Biol. Rev. 97 (1), 343–360.
Rodoni B. (2009). The role of plant biosecurity in preventing and controlling emerging plant virus disease epidemics. Virus Res. 141, 150–157.
Sarkar C., Gupta D., Gupta U., Hazarika B. B. (2023). Leaf disease detection using machine learning and deep learning: Review and challenges. Appl. Soft Comput. 22, 110534.
Sastry K. S., Zitter T. A. (2014). “Management of virus and viroid diseases of crops in the tropics,” in Plant virus and viroid diseases in the tropics, epidemiology and management, vol. 2 . Eds. Sastry S. K., Zitter T. A. (Dordrecht, The Netherlands: Springer), 149–480.
Schmid-Burgk J. L., Schmithausen R. M., Li D., Hollstein R., Ben-Shmuel A., Israeli O., et al. (2020). LAMP-Seq: population-scale COVID-19 diagnostics using combinatorial barcoding. biorxiv 2020–2024.
Sereika M., Kirkegaard R. H., Karst S. M., Michaelsen T. Y., Sørensen E. A., Wollenberg R. D., et al. (2022). Oxford Nanopore R10.4 long-read sequencing enables the generation of near-finished bacterial genomes from pure cultures and metagenomes without short-read or reference polishing. Nat. Methods 19, 823–826.
Swain R. B., et al. (1952). How insects gain entry. In: Insects: the yearbook of agriculture (USDA, USA). Available at: https://naldc.nal.usda.gov/download/IND43894226/PDF (Accessed 15/5/2023).
Terentev A., Dolzhenko V., Fedotov A., Eremenko D. (2022). Current state of hyperspectral remote sensing for early plant disease detection: A review. Sensors 22 (3), 757.
Thompson D., Lei Y. (2020). Mini review: Recent progress in RT-LAMP enabled COVID-19 detection. Sensors Actuators Rep. 2 (1), 100017.
Thresh J. M. (1980). The origins and epidemiology of some important plant virus diseases. Appl. Biol. 5, 2–65.
Thresh J. M. (2004). “The impact of plant virus diseases in developing countries,” in Virus and virus-like diseases of major crops in developing countries. Eds. Loebenstein G., Thottappilly G. (Dordrecht, The Netherlands: Springer), 1–30.
Thresh J. M. (2006a). “Crop viruses and virus diseases: A global perspective,” in Virus diseases and crop biosecurity. Eds. Cooper J. I., Kuehne T., Polischuk V. P. (Dordrecht, The Netherlands: Springer), 9–32.
Thresh J. M. (2006c). Plant virus epidemiology: the concept of host genetic vulnerability. Adv. Virus Res. 67, 89–125.
Van Leur J. A. G., Freeman A. E., Aftab M., Spackman M., Redden B., Materne M. (2013). Identification of seed-borne pea seed-borne mosaic virus in lentil (Lens culinaris) germplasm and strategies to avoid its introduction in commercial Australian lentil fields. Australas. Plant Dis. Notes 8, 75–77.
Villamor D. E. V., Ho T., Al Rwahnih M., Martin R. R., Tzanetakis I. E. (2019). High throughput sequencing for plant virus detection and discovery. Phytopathology 109, 716–725.
Vincent S. J., Coutts B. A., Jones R. A. C. (2014). Effects of introduced and indigenous viruses on native plants: exploring their disease causing potential at the agro-ecological interface. PloS One 9 (3), 91224.
Waite D. W., Liefting L., Delmiglio C., Chernyavtseva A., Ha H. J., Thompson J. R. (2022). Development and validation of a bioinformatic workflow for the rapid detection of viruses in biosecurity. Viruses 14 (10), 2163.
Wamaitha M. J., Nigam D., Maina S., Stomeo F., Wangai A., Njuguna J. N., et al. (2018). Metagenomic analysis of viruses associated with maize lethal necrosis in Kenya. Virology 15, 90.
Wang Y. M., Ostendorf B., Gautam D., Habili N., Pagay V. (2022). Plant viral disease detection: From molecular diagnosis to optical sensing technology—A multidisciplinary review. Remote Sens. 14 (7), 1542.
Ward S., van Helden M., Heddle T., Ridland P. M., Pirtle E., Umina P. A. (2020). Biology, ecology and management of Diuraphis noxia (Hemiptera: Aphididae) in Australia. Aust. Entomol. 59, 238–252.
Webster C. G., Coutts B. A., Jones R. A. C., Jones M. G. K., Wylie S. J. (2007). Virus impact at the interface of an ancient ecosystem and a recent agroecosystem: studies on three legume-infecting potyviruses in the southwest Australian floristic region. Plant Pathol. 56, 729–742.
Whattam M., Dinsdale A., Elliott C. E. (2021). Evolution of plant virus diagnostics used in Australian Post Entry Quarantine. Plants 10, 1430. doi: 10.3390/plants10071430
Wongsurawat T., Jenjaroenpun P., Taylor M. K., Lee J., Tolardo A. L., Parvathareddy J., et al. (2019). Rapid sequencing of multiple RNA viruses in their native form. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.00260
Keywords: viruses, disease, grains, biosecurity, plant health, diagnosis, genomics
Citation: Maina S and Jones RAC (2023) Enhancing biosecurity against virus disease threats to Australian grain crops: current situation and future prospects. Front. Hortic. 2:1263604. doi: 10.3389/fhort.2023.1263604
Received: 20 July 2023; Accepted: 24 August 2023;
Published: 29 September 2023.
Edited by:
Francis Onono Wamonje, National Institute of Agricultural Botany (NIAB), United KingdomReviewed by:
Netsai Mhlanga, National Institute of Agricultural Botany (NIAB), United KingdomCopyright © 2023 Maina and Jones. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Solomon Maina, c29sb21vbi5tYWluYUBkcGkubnN3Lmdvdi5hdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.