- 1Department of Biochemistry, Microbiology and Biotechnology, Kenyatta University, Nairobi, Kenya
- 2Department of Plant Sciences, Kenyatta University, Nairobi, Kenya
- 3Department of Forestry and Land Resources Management, South Eastern Kenya University, Kitui, Kenya
In agroecosystems, microbial communities play a crucial role in delivery of various ecosystem services. These microbial communities are affected by several factors such as soil physicochemical properties which contribute to the diversity of bacterial and fungal communities. In this study, we investigated the soil physicochemical parameters and the diversity and abundance of bacterial and fungal communities in rhizospheric soil collected from banana growing regions in Kisii, Nyamira and Embu Counties of Kenya. Rhizospheric soil samples from the three regions showed significant differences at (P= 0.01) with the lowest recorded pH being 4.43 in Embu County. Based on Next-generation sequencing results, there was a significant diversity and abundance of bacterial division Proteobacteria while the predominant fungal division was basidiomycota, Several genera in the fungal division such as Penicillium and Cladosporium as well as bacterial genera such as Acidobacterium and Pseudomonas sp. were those associated with soil. There were several plant pathogenic and beneficial bacteria and fungi. Based on redundancy analysis (RDA) the distribution of these microbes was affected negatively by soil parameters such as total organic carbon (TOC) and pH. In conclusion, Soil health and continuous mono-cropping systems play a significant role in the diversity and abundance of both beneficial and harmful soil microbes. Metagenomics approaches in studying microbial communities in agroecosystems is a revolutionary approach which will aid in the development of sustainable tools in agriculture that improve microbiome structures as well as overall productivity.
Introduction
Banana farming is a major economic activity in Kenya. However, the current production trends by the smallholder farmers who form 85% of the banana producers in Kenya do not meet the market demand (Okumu et al., 2011). This is because there are various factors directly affecting production such as soil health as well as pests and diseases (Köberl et al., 2017).
There has been increasing interest in soil health, specifically microbial diversity in soil habitat. This is because soil microbial communities are considered as essential in improving soil health and quality. A wide range of microorganisms are essential in soil function since microbial communities are key contributors to sustainable agriculture. Microbes are known to act as mediators in many processes that are involved in agricultural production (Lupwayi et al., 1998; Rondon et al., 2000 and Gupta et al., 2018).
It has been observed that farm practices such as conservation tillage and legume-based crop rotation supports soil microbial communities and may have direct positive effects on agricultural ecosystems (Nguyen et al., 2018). The culturing techniques of studying microorganisms are not efficient in their study since majority of the microbes are unculturable (Goel et al., 2018)). Advances in molecular application in determining microbial diversity in nature shows that there is a lot of information that was not previously accessible (Gupta et al., 2018). Therefore, understanding the factors that influence microbial communities can assist greatly in the innovation of new agricultural tools for sustainable environment (Gupta et al., 2018).
There are numerous soil borne diseases that act as limiting factors in banana production all over the world (Köberl et al., 2017). Most farmers use conventional methods in their control that mainly target pathogens using antibacterial agents, pesticides and fungicides and this has the potential of reducing the soil microbial community (Dita et al., 2018). Therefore, studying microbial communities can aid in creating tools to mitigate problems affecting essential microbial communities (Chaparro et al., 2012).
Recent reports indicate that stressors that affect soil biodiversity such as human activities, the decline in soil organic matter and degradation of land has led to the loss of aboveground and below ground biodiversity (Orgiazzi et al., 2016). The decline in soil health and fertility including biological degradation (organic matter) is a worrying trend and has the potential to lower productivity significantly. Constant monitoring of soil health status is an essential factor in processes of development of land management systems that ensure the sustainability of soil resources (Tilhou et al., 2021). In order to reclaim the damage to soil and ensure global agricultural sustainability, the identification of these stressors as well as development of sustainable methods such as bioremediation is important (Tahat et al., 2020).
The soil microbial communities in agroecosystems play a crucial role in plant growth and health. Recent studies have been focusing on microbial communities in continuous cropping systems and their roles. Understanding their roles as well as environmental factors that affect the communities is important in improving banana production (Berg et al., 2017 and Gómez-Lama et al., 2021). Studying soil microbial diversity in relation to other soil physicochemical parameters is essential. This aids greatly in identifying factors affecting beneficial microbial communities, thus highlighting the need for adoption and development of tools for sustainable agricultural practices (Gómez-Lama et al., 2021). The current study was aimed at determining microbial diversity and abundance in banana rhizospheric soil from banana producing regions in Kenya and the influence of soil physicochemical parameters on the distribution of fungal and bacterial communities.
Materials and methods
Soil sampling and characterization
Rhizospheric soils samples were collected from the farmers’ fields in Kisii, Nyamira and Embu Counties at a depth of 15-20 cm using a sterile hand shovel. The GPS coordinates were used to create a map where soil sampling was carried out (Figure 1). Soil sampling was carried out across and diagonally from 20 points per field and were mixed to form a composite sample which was packed in sterile bags for transportation to Kenyatta University, Tissue Culture Laboratory and stored in a freezer at -30 °C. Soil physicochemical parameters were determined according to the standard procedures described by Anderson and Ingram (1993) and Okalebo et al. (2002).
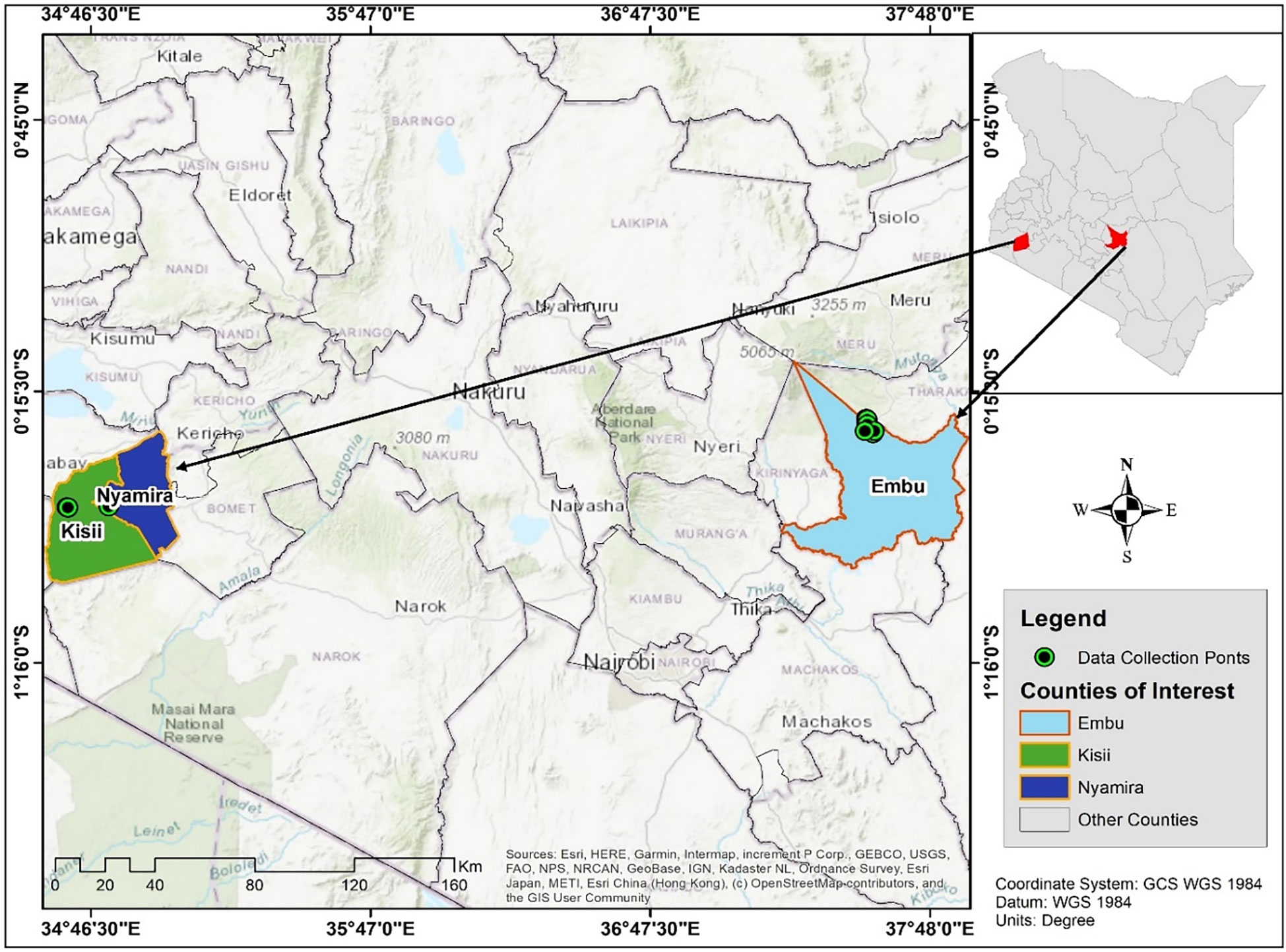
Figure 1 Map of Kenya showing the sampling sites in the study regions of Kisii, Nyamira and Embu Counties. Source: Generated using GPS co-ordinates.
Soil DNA extraction and next generation sequencing (NGS) of soil metagenome targeting 16S rDNA and ITS regions
DNA for metagenomic analysis was extracted from 200 mg composite soil samples using soil DNA extraction kit (Invitrogen by Thermofisher Scientific) according to the manufacturer’s instructions. The DNA pellets were washed with 70% ethanol and stained using SYBR Green dye and subjected to gel electrophoresis in 1.2% agarose gel in 0.5X TBE buffer at 80 V for 20 minutes. A 100 bp gene ruler was used to estimate the band sizes (Gupta et al., 2018).
The 16S rRNA primer pair, 515F GTGYCAGCMGCCGCGGTAA/806R GGACTACNVGGGTWTCTAAT for bacterial communities and the internal transcribed spacer (ITS) primer pair, ITS1F CTTGGTCATTTAGAGGAAGTAA/ITS4R TCCTCCGCTTATTGATATGC for fungal communities were used to evaluate microbial ecology of each sample on the Illumina NovaSeq with methods via the bTEFAP® DNA analysis service. Prokaryotic and eukaryotic libraries were constructed and each sample underwent a single-step 35 cycle PCR using HotStarTaq Plus Master Mix Kit (Qiagen, Valencia, CA). The PCR amplification conditions were as follows: initial denaturation at 95°C for 10 minutes, followed by 35 cycles (denaturation at 95°C for 30 seconds; annealing at 53°C for 40 seconds and elongation at 72°C for 1 minute) and a final elongation step at 72°C for 10 minutes. Following PCR, all amplicon products from different samples were purified using SPRI beads (Soliman et al., 2017). Samples were sequenced utilizing the Illumina NovaSeq chemistry following manufacturer’s instruction (Gastauer et al., 2019).
Statistical analysis
The open source package DADA 2 running under R was used to process the raw reads. The Q25 sequence data derived from the sequencing process was processed using the MR DNA ribosomal and functional gene analysis pipeline (www.mrdnalab.com, MR DNA, Shallowater, TX). The unique sequences were deionized and identified with illumina sequencing.PCR point errors were removed, followed by chimera removal, thereby providing a or zOTU. Final sequence operational taxonomic units (zOTUs) were taxonomically classified using BLAST against a curated database derived from NCBI (www.ncbi.nlm.nih.gov) and compiled into each taxonomic level into both counts and percentages. Counts files contain the actual number of sequences while the percent files contain the relative (proportion) percentage of sequences within each sample that map to the designated taxonomic classification. Statistical analysis was performed using a variety of computer packages including XLstat (Dowd et al., 2008a), NCSS 2007 (Dowd et al., 2008b), “R” (Tasnim et al., 2017); and NCSS 2010 (Eren et al., 2011 and Swanson et al., 2011). Alpha and beta diversity analysis was conducted as described previously using Qiime 2 (Bolyen et al., 2018). Based on the analysis, percentages, counts and diversity indices with a P value < 0.05 were recorded as significant.
Results
Soil composition
Soil samples collected from Kisii, Nyamira and Embu counties had varied physicochemical parameters. The samples obtained from the three counties varied in soil organic carbon levels with majority of the soil samples from Embu and Nyamira containing moderate organic carbon levels (Table 1). The study showed the average pH recorded in the soils obtained from the Counties was 4.94. Additionally, there was a significant difference in the pH between the three Counties (P= 0.01).
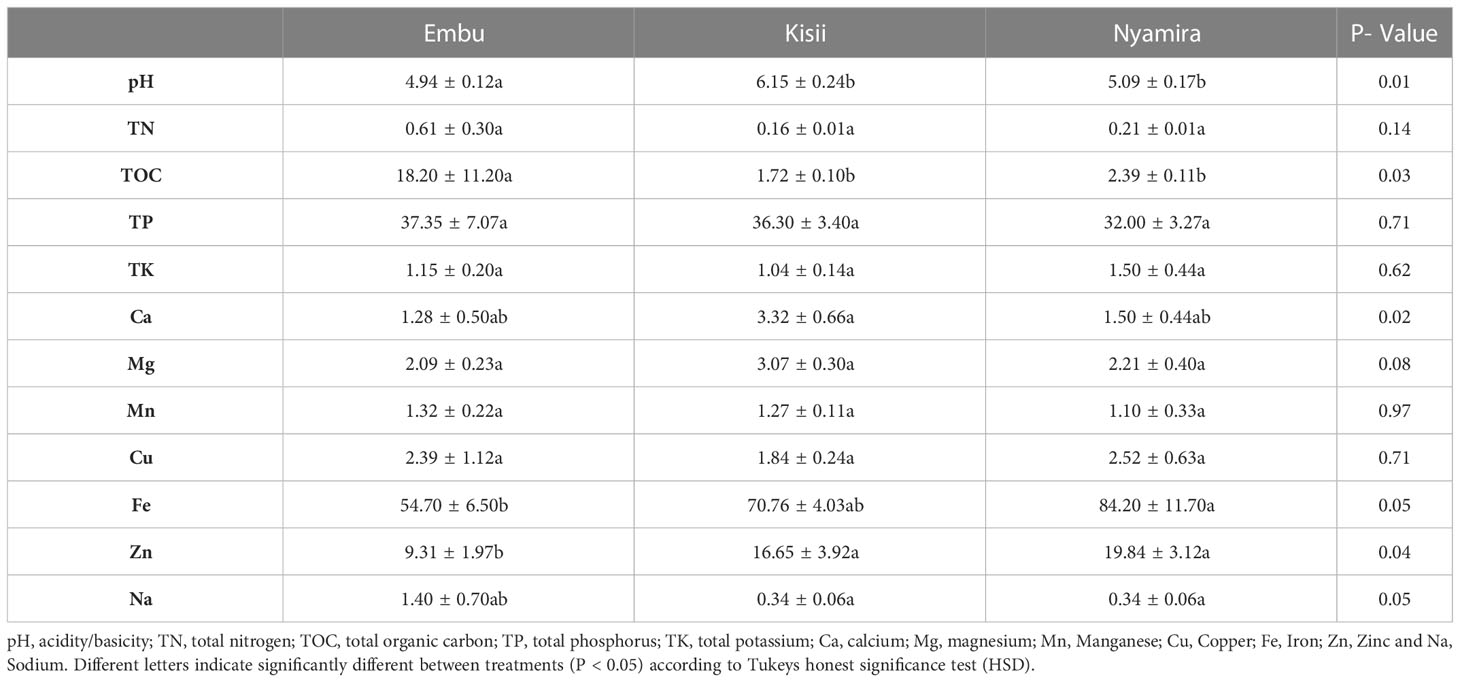
Table 1 Soil physico- chemical properties of rhizospheric soil obtained from Kisii, Nyamira and Embu Counties.
Microbial distribution
The bacterial communities varied in the different rhizospheric soil samples obtained from the three Counties. A total of 420000 sequences were parsed and 335089 were then mapped to (operational taxonomic units (OTUs), 324697 sequences identified within the Bacteria and Archaea domains. The dominant bacterial phyla were Actinobacteria, Proteobacteria, Acidobacteria (Figure 2).
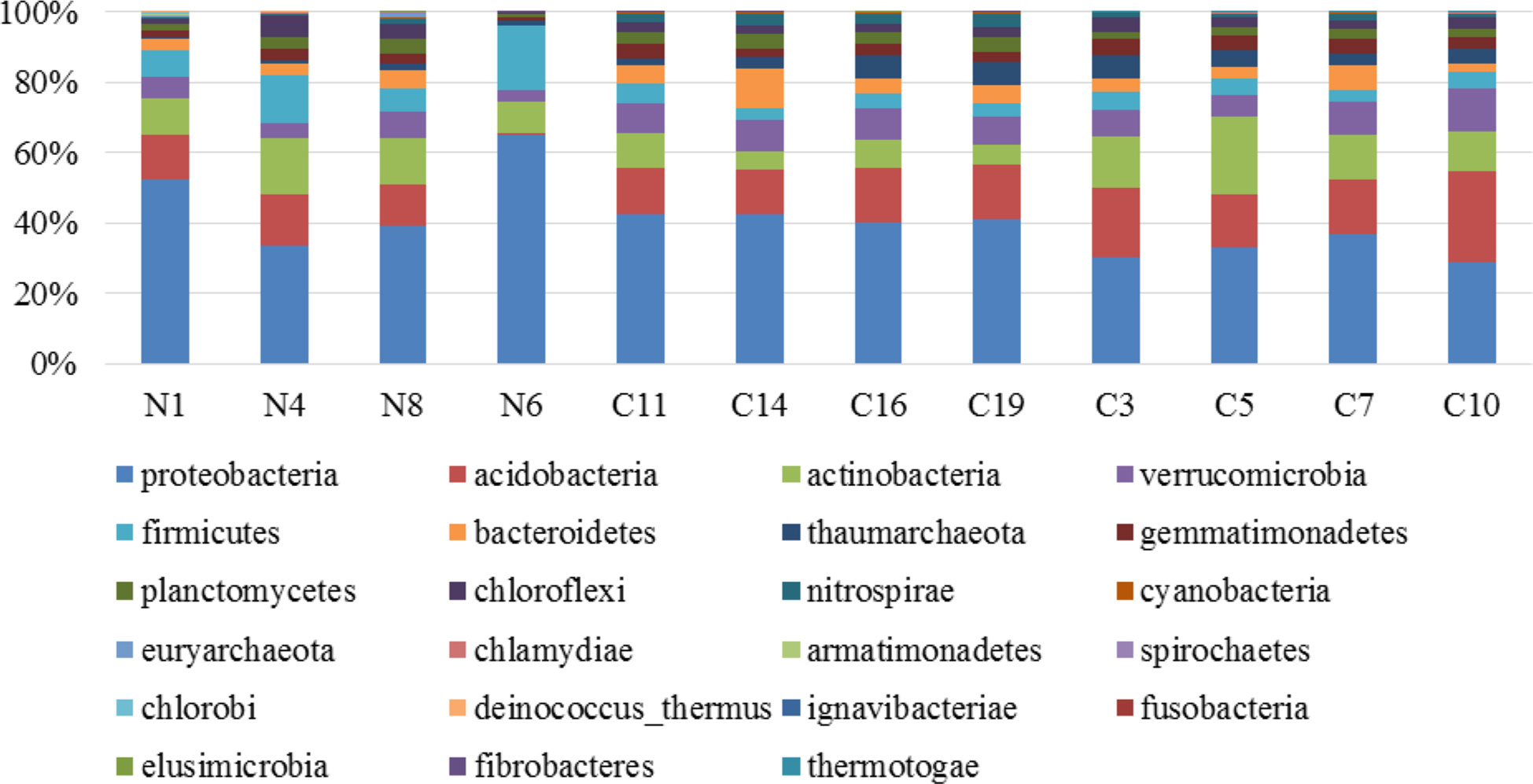
Figure 2 Relative abundance of total observed bacteria and archea domain phyla in banana soil rhizosphere samples from Nyamira County (N1, N4, N6, N8), Kisii County (C11, C14, C16, C19) and Embu County (C3, C5, C7, C10).
A total of 391220 sequences were parsed and 314531 sequences identified within the Fungi kingdom and were the basis of identification of fungal phyla (Figure 3). Fungal diversity was higher in soil samples obtained from Kisii County based on Shannon diversity metrics.
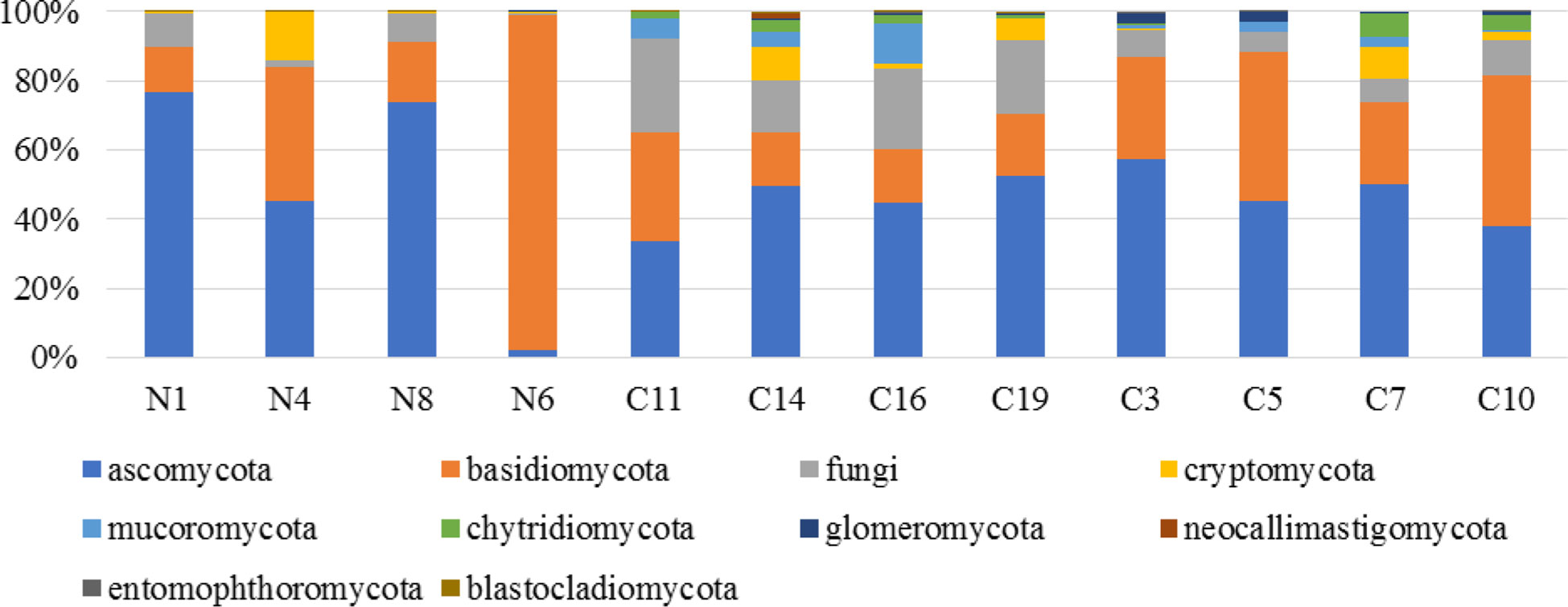
Figure 3 Relative abundance of total observed fungi kingdom phyla in banana rhizosphere soil samples from Nyamira County (N1, N4, N6, N8), Kisii County (C11, C14, C16, C19) and Embu County (C3, C5, C7, C10).
Based on the dual hierarchal dendrogram, there was a lack of distinct clustering between the presumed sample groups N and C (Figure 4). There was no clear evidence of a significant difference between sample groups. The heatmap obtained from the dual hierarchal classification of the predominant genera enabled the visualization and analysis of multi-dimensional datasets such as data obtained from this study.
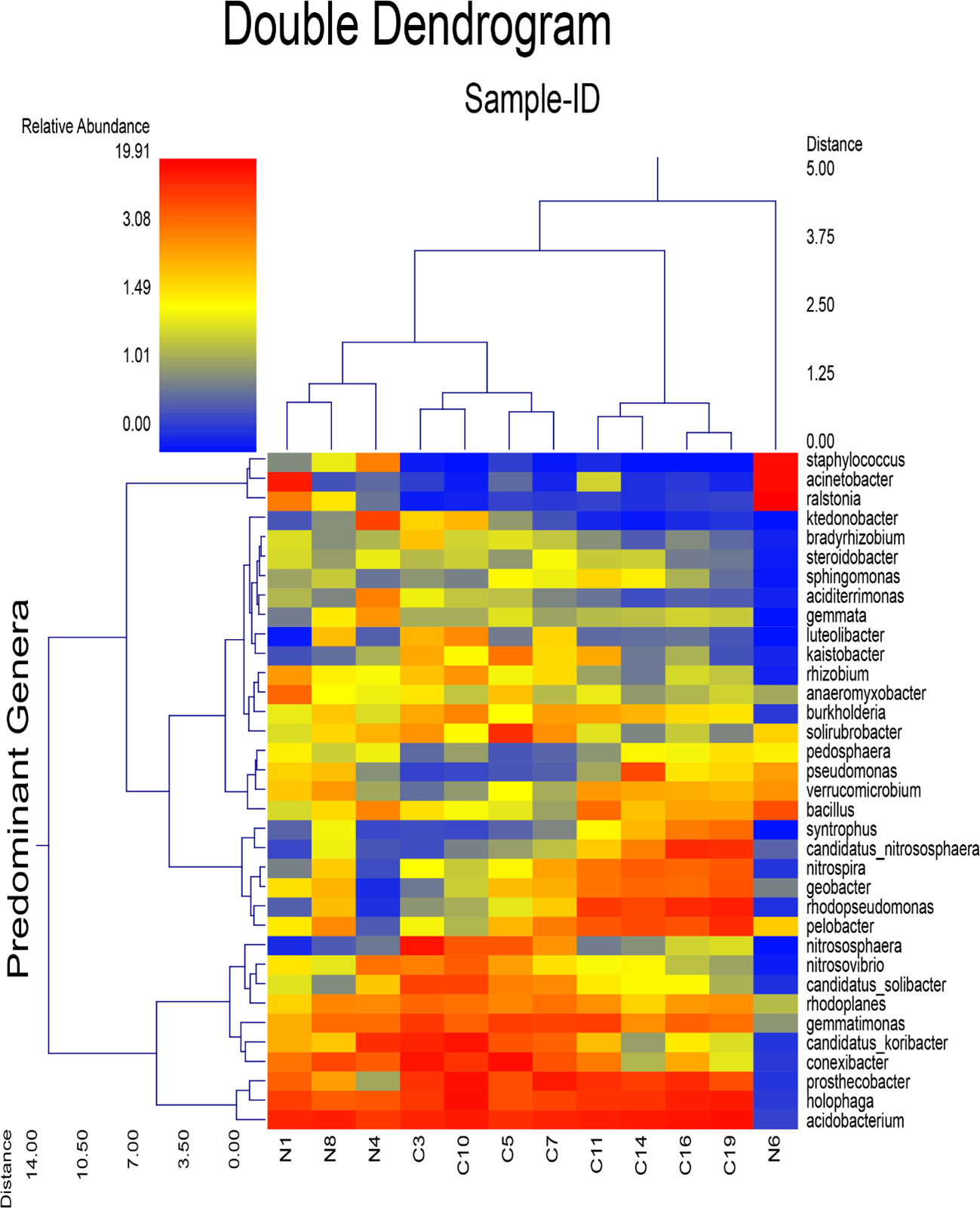
Figure 4 Dual hierarchal dendrogram evaluation of the taxonomic classification data of bacterial genera, with each sample clustered on the X-axis labeled based soil sample from Nyamira County (N1, N4, N6, N8), Embu County (C3, C5, C7, C10) and Kisii County (C11, C14, C16, C19). The heatmap represents the relative percentages of each genus. The predominant genera are represented along the Y-axis (right).
A dual hierarchal dendrogram displayed the predominant genera with clustering related to the different groups. Based on the lack of distinct clustering between the presumed sample groups N and C (Figure 5), there is no clear evidence of a significant difference between sample groups. Based on the results, Fusarium sp., a commonly known plant pathogenic genera was significantly abundant in soil samples from Nyamira County (Figure 5).
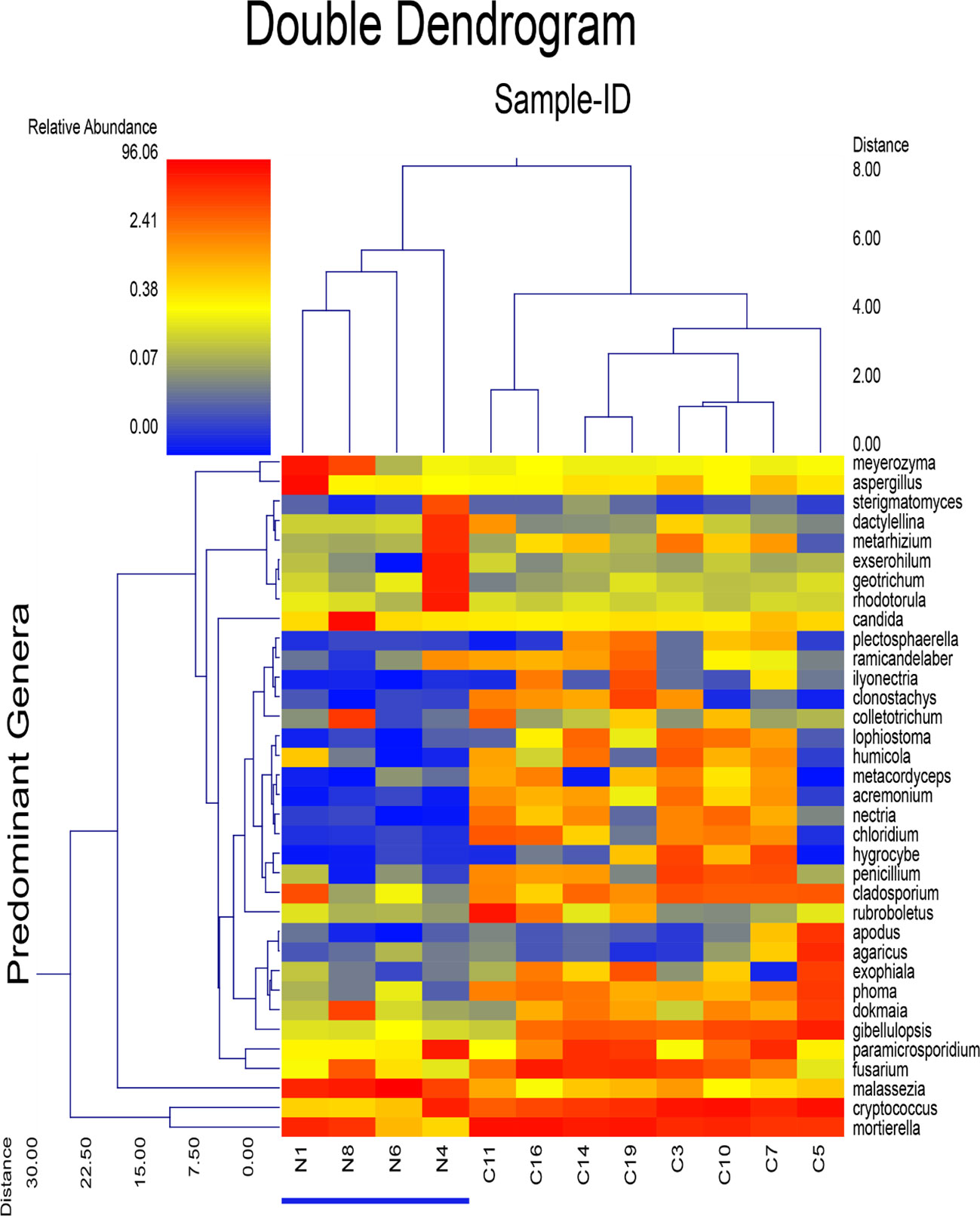
Figure 5 Dual Hierarchal dendrogram evaluation of the taxonomic classification data of fungal genera, with each sample clustered on the X-axis labeled based soil sample from Nyamira County (N1, N4, N6, N8), Embu County (C3, C5, C7, C10) and Kisii County (C11, C14, C16, C19). The heatmap represents the relative percentages of each genus. The predominant genera are represented along the Y-axis (right).
Plant beneficial fungi (Trichoderma, Talaromyces and arbuscular mycorrhiza fungal species such as Gigaspora) commonly used as bioenhancers were detected in some samples but at low abundance levels.
Microbial diversity and richness
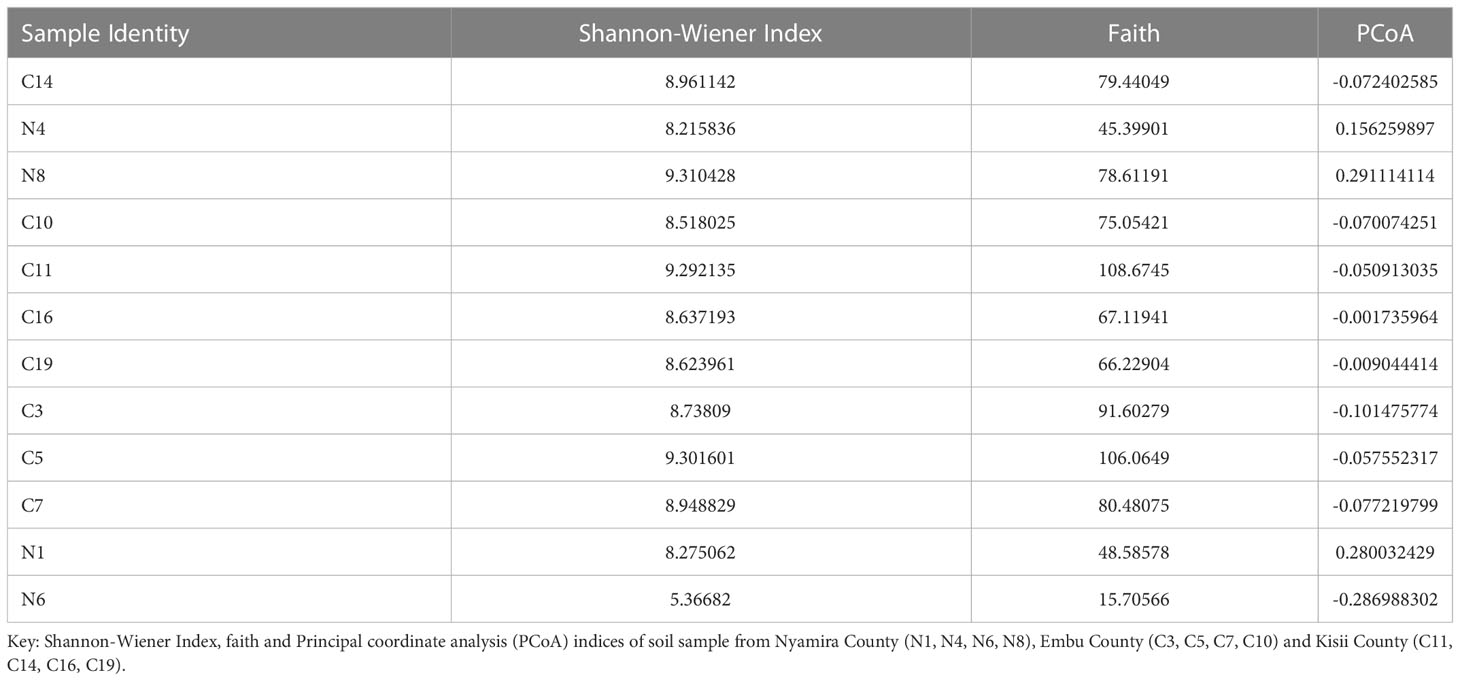
To determine the alpha diversity, the Shannon-Wiener Index at 20,000 reads were sufficient. This indicated that all the samples were equally and sufficiently sampled and the sequence depth was sufficient to capture all the fungal community diversity. Based on Shannon diversity metrics, the highest bacterial diversity was recorded in a soil sample obtained from Embu County (Table 2).
The α- and β-diversity of the bacterial and fungal communities in the sampled farms showed no significant differences. The Shannon diversity metrics of fungal communities were low especially in samples obtained from Nyamira County (N1, N4, N6 and N8). However, bacterial community diversity metrics were higher with the lowest diversity recorded in soil sample N6 obtained from Nyamira County (Table 3).
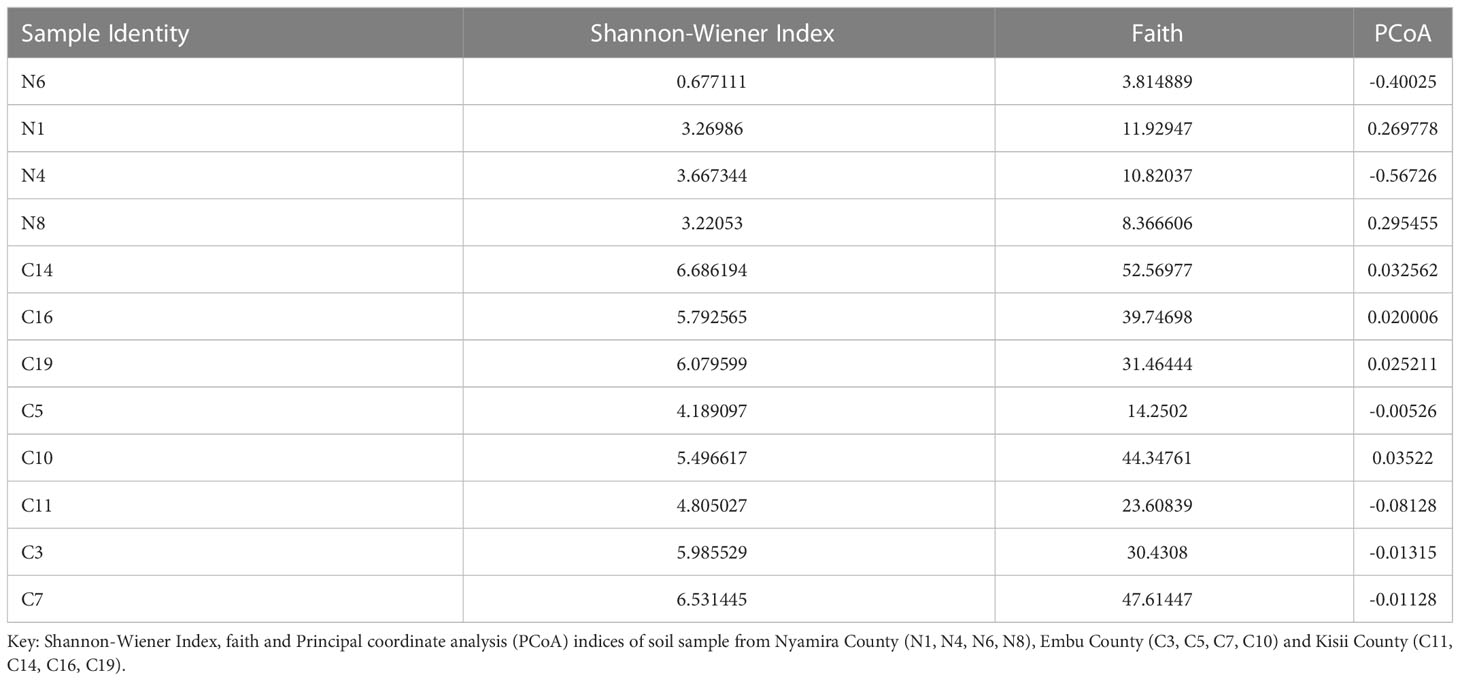
Table 3 Alpha and Beta diversity metrics of the fungal communities’ analysis of banana rhizospere soil samples.
A Bray-Curtis dissimilarity matrix using principal coordinate analysis (PCoA) of the microbial communities showed that there appears to be phylogenetic grouping amongst sample group N1, N4, N6, sample group C3, C5, C7, C10 that is distinct from sample group C11, C14, C16, C19. Primary vector explains 45.1% of the variation between the samples. Beta diversity analysis showed the microbial community structure of the different soil samples. A principal coordinate analysis allowed for visualization and comparison of the communities of microbes as a whole taking into account differences in the samples and phylogenetic relatedness.
There were variations in bacterial and fungal communities which closely correlated with the soil parameters based on Canonical correspondence analysis. These included soil pH, total organic carbon (TOC), and total nitrogen which were the main drivers of microbial community distribution (Figure 6).
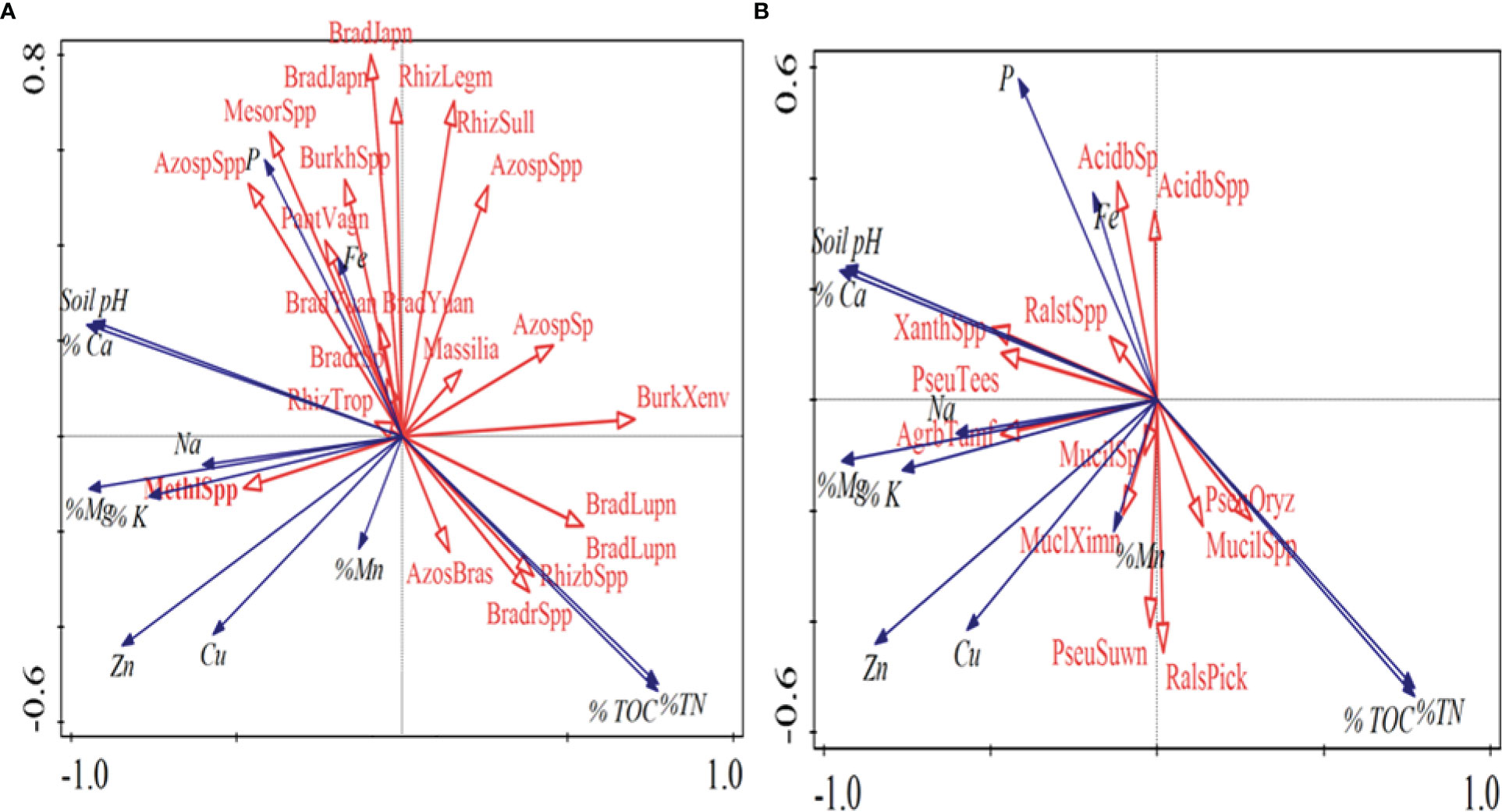
Figure 6 Canonical correspondence analysis (A) Beneficial and (B) Pathogenic bacterial species and soil chemical parameters for banana rhizospheric soil. Red arrows represent fungal species while the blue arrows represent soil physico-chemical parameters.
Plant beneficial bacteria species such as Bradyrhizobium, Azotobacter and Rhizobium species distribution was mainly affected by soil pH, soil P (Phosphorus), Total Nitrogen (TN) and TOC. In plant pathogenic bacterial species TN, TOC and soil pH were the main drivers, in which soils with high pH, adequate TN and TOC had low incidences of pathogenic bacterial species such as Xanthomonas and Ralstonia.
The distribution and abundance of beneficial fungal species such as arbuscular mycorrhiza fungi (AMF) which includes Gigaspora sp was affected by soil pH with low counts observed in highly acidic soil. The abundance of fungal genera such as Acaulospora were affected by total nitrogen (TN) and total organic carbon (TOC) (Figure 7). Population of plant pathogenic species such as Colletotrichum and Verticillium were TOC and TN while majority of Fusarium species were affected by soil pH and P.
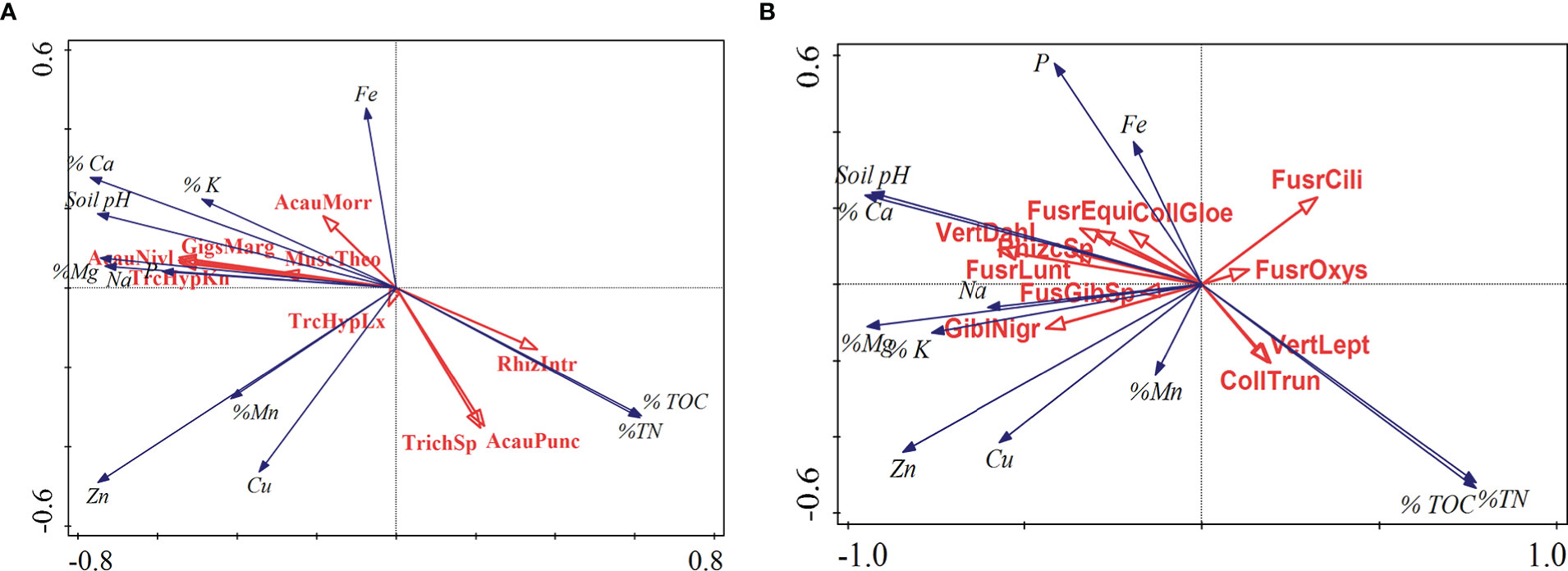
Figure 7 Canonical correspondence analysis of the (A) Beneficial and (B) Pathogenic fungal species and soil chemical parameters for banana rhizospheric soil. Red arrows represent fungal species while the blue arrows represent soil physico-chemical parameters.
Discussion
Soil health plays a critical role in ensuring the stability of microbial communities. Soil organic carbon levels are a good indicator of the health status of soil and positively correlate to the yield of crops (Bennett et al., 2010). Highly acidic conditions were detected in the majority of soil samples collected from Embu and Nyamira which is worrying. Delgado and Gómez (2016) has reported that a pH of below 5.5 is harmful to agroecosystems since it negatively influences plant and environment interaction, indicating that the soil pH in Embu County poses a great danger to agroecosystems.
It has been reported that rhizospheric soil that is closest to the root system and is essential in studying microbial communities (Berendsen et al., 2012). Sui et al. (2019) and Chaudhary et al. (2021) Zhu et al. (2018), reported that Actinobacteria are among the most identified rhizospheric bacteria phyla in banana plants. The abundance of Proteobacteria and Actinobacteria has been previously linked to long periods of production (Pang et al., 2021). However, Actinobacteria have been reported to produce antibiotics as well as having an important role in organic matter decomposition (Zhang et al., 2017). Berendsen et al. (2012) reported that microbial community composition in rhizosperic soil are linked to soil quality as well as the health of the crop grown. Microbiota associated with soil has great influence on how plants are able to adapt and respond to changes in the environments such as emergence of stressors such as salinity and drought (Zhang et al., 2017). High abundance were noted in the phyla Basidiomycota and Ascomycota similar to results by Sommermann et al. (2018) and Gómez-Lama et al. (2021).
Rossmann et al. (2012) reported the presence of potential plant, human and insect pathogens similar to the results obtained in this study. Gu et al. (2016) utilized high-throughput gene expression to visualize hierarchal classification and species richness, similar to this study. Plant beneficial bacteria genera diversity such as Rhizobium and Azotobacter differed with that of probable plant pathogenic bacteria genera such as Ralstonia in all the soil samples similar to results reported by Kepler et al (Kepler et al., 2020). The presence of Rhizoctonia, Colletotrichium and Verticillium was an indicator of disease causing fungal pathogens in some of the soil samples similar to results by Sommermann et al. (2018).
Investigation of abundance of plant beneficial fungi such as those used as biocontrol agents in agroecosystems is a field that has not been extensively studied (Harman et al., 2004). The variations in bacterial genera indicate differences in land use strategies resulting in changes in microbial community structure and diversity (Lynn et al., 2017). Fan et al. (2020) reported variations in bacterial and fungal communities in relation to soil parameters. It has been reported that many soil variables such as soil organic matter, salinity and pH have a lot of impact on soil microbial populations (Gans et al., 2005). Additionally, these microbial communities cannot recover to their natural state even after many years (Pellegrino et al., 2014). Based on previous studies pH impacts soil microbial communities therefore defines taxonomic composition of microbial communities (Delgado and Gómez, 2016).
It has been established that soil composition and function are associated with plant soil borne disease outbreaks in which healthy fields have different microbial communities from the infected ones (Zhang et al., 2017). Frac et al. (2018) reported that the diversity and fungi activity is regulated by biotic and abiotic factors such as soil pH, structure and some nutrients.
Conclusion
Based on the results obtained from this study, there were variations in soil composition and microbial populations in the different regions. The soil physicochemical properties played a significant role in the rhizosphere soil microbial communities. It has been established that plants modify the rhizosphere through various modes such as changes in microbiome composition and function. Studying the interaction between plants, microorganisms and soil physicochemical conditions enables one to understand how each plays a role in ecosystem balance and well as productivity in agroecosystems. The results show that studying soil microbiome in monocropping systems is a great tool in determining ideal microbial community structures for the crops. In addition, application of metagenomics techniques in studying microbial communities in agroecosystems will aid in designing banana production systems as well as targeted approaches in disease management.
Data availability statement
The data presented in the study are deposited on Figshare (https://doi.org/10.6084/m9.figshare.21939977).
Author contributions
JM, OO, EN, and JK conceived and designed the research and data collection tools and participated in drafting the manuscript. CW collected the data, participated in the data analyses, and wrote the manuscript. MM was involved in molecular work and data analyses. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by The National Research fund (NRF), Kenya.
Acknowledgments
We are grateful to the farmers in Kisii, Nyamira and Embu Counties for working with us and Mr DNA Shallowater, TX for their contribution in Next Generation Sequencing.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Anderson J. M., Ingram J. S. I. (Eds.) (1993). Tropical soil biology and fertility: A handbook of methods. 2nd ed (Wallingford: CAB International).
Bennett L. T., Mele P. M., Annett S., Kase l.S. (2010). Examining links between soil management, soil health, and public benefits in agricultural landscapes: an Australian perspective. Agric. Ecosyst. Environ. 139, 1–12. doi: 10.1016/j.agee.2010.06.017
Berendsen R. L., Pieterse C. M., Bakker P. A. (2012). The rhizosphere microbiome and plant health. Trends Plant Sci. 17, 478–486. doi: 10.1016/j.tplants.2012.04.001
Berg G., Köberl M., Rybakova D., Muller H., Grosch R., Smalla K. (2017). Plant microbial diversity is suggested as the key to future biocontrol and health trends. FEMS. Microbiol. Ecol. 93, 5. doi: 10.1093/femsec/fix050
Bolyen E., Rideout J. R., Dillon M. R., Bokulich N. A., Abnet C., Al-Ghalith G. A., et al. (2018). QIIME 2: Reproducible, interactive, scalable, and extensible microbiome data science. Peer J. 37 (8), 852–857. doi: 10.7287/peerj.preprints.27295v1
Chaparro J., Sheflin A., Manter D., Vivanco J. (2012). Manipulating the soil microbiome to increase soil health and plant fertility. Biol. Fertil. Soils. doi: 10.1007/s00374-012-0691-4
Chaudhary P., Khati P., Chaudhary A., Maithani D., Kumar G., Sharma A. (2021). Cultivable and metagenomic approach to study the combined impact of nanogypsum and On maize plant health and its rhizospheric microbiome. PloS One 16 (4), e0250574. doi: 10.1371/journal.pone.0250574
Delgado A., Gómez J. A. (2016). “The soil physical, chemical and biological properties,” in Principles of agronomy for sustainable agriculture. Eds. Villalobos F., Fereres E. (Cham: Springer). doi: 10.1007/978-3-319-46116-8_2
Dita M., Barquero M., Heck D., Mizubuti E., Staver C. (2018). Fusarium wilt of banana: Current knowledge on epidemiology and research needs toward sustainable disease management. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.01468
Dowd S. E., Callaway T. R., Wolcott D., Sun Y., McKeehan T., Hageeroot G., et al. (2008a). Evaluation of the bacterial diversity in the feces of cattle using 16S rDNA bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP).". BMC Microbiol. 8, 125. doi: 10.1186/1471-2180-8-125
Dowd S. E., Sun Y., Secor R., Rhoads D., Wolcott M., James A., et al. (2008b). Bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP) for microbiome studies: bacterial diversity in the ileum of newly weaned salmonella-infected pigs. Foodborne Pathog. Dis. 5 (4), 459–472. doi: 10.1089/fpd.2008.0107
Eren A. M., Zozaya M., Taylor C. M., Dowd S. E., Martin D. H., Fems M. J., et al. (2011). Exploring the diversity of gardnerella vaginalis in the genitourinary tract microbiota of monogamous couples through subtle nucleotide variation. PloS One 6 (10), e26732. doi: 10.1371/journal.pone.0026732
Fan S., Zuo J., Dong H. (2020). Changes in soil properties and bacterial community composition with biochar amendment after six years. MDPI Agronomy 10, 5, 746. doi: 10.3390/agronomy10050746
Frac M., Hannula S. E., Bełka M. J., ˛Edryczka M. (2018). Fungal biodiversity and their role in soil health. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.00707
Gans J., Wolinsky M., Dunbar J. (2005). Computational improvements reveal great bacterial diversity and high metal toxicity in soil. Science 309, 1387–1390. doi: 10.1126/science.1112665
Gastauer M., Vera M., de Souza K. P., Pires E. S., Alves R., Caldeira C. F., et al. (2019). A metagenomic survey of soil microbial communities along a rehabilitation chronosequence after iron ore mining. Sci. Data 6, 190008. doi: 10.1038/sdata.2019.8
Goel R., Kumar V., Suyal D., Narayan, Soni R. (2018). “Toward the unculturable microbes for sustainable agricultural production,” in Role of rhizospheric microbes in soil. Ed. Meena V. (Singapore: Springer). doi: 10.1007/978-981-10-8402-7_4
Gómez-Lama C., Fernández-González A., Cardoni M., Valverde-Corredor A., López-Cepero J., Fernández-López M., et al. (2021). The banana root endophytome: Differences between mother plants and suckers and evaluation of selected bacteria to control Fusarium oxysporum f.sp. cubense. J. Fungi 7, 194. doi: 10.3390/jof7030194
Gu Z., Eils R., Schlesner M. (2016). Complex heat maps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32 (18), 2847–2849. doi: 10.1093/bioinformatics/btw313
Gupta N., Vats S., Bhargava P. (2018). “Sustainable agriculture: Role of metagenomics and metabolomics in exploring the soil microbiota,” in Insilico approach for sustain. agric. Springer. 183–199. doi: 10.1007/978-981-13-0347-0_11
Harman G. E., Howell C. R., Viterbo A., Chet I., Lorito M. (2004). Trichoderma species opportunistic, a virulent plant symbionts. Nat. Rev. Microbiol. 2, 43–56. doi: 10.1038/nrmicro797
Kepler R. M., Epp- Schmidt D. J., Yarwood S. A., Cavigelli M. A., Reddy K. N., Duke S. O., et al. (2020). Soil microbial communities in diverse agroecosystems exposed to the herbicide glyphosate. Appl. Environ. Microbiol. 86, e01744-19. doi: 10.1128/AEM.01744-19
Köberl M., Dita M., Martinuz A., Staver C., Berg G. (2017). Members of gammaproteobacteria as indicator species of healthy banana plants on fusarium wilt-infested fields in central America. Sci. Rep. 7, 45318. doi: 10.1038/srep45318
Lupwayi N., Rice W., Clayton G. (1998). Soil microbial diversity and community structure under wheat as influenced by tillage and crop rotation. Soil Biol. Biochem. 30 (13), 1733–1741. doi: 10.1016/S0038-0717(98)00025-X
Lynn T. M., Liu Q., Hu Y., Yuan H., Wu X., Khai A. A., et al. (2017). Influence of land use on bacterial and archaeal diversity and community structures in three natural ecosystems and one agricultural soil. Arch. Microbiol. 199 (5), 711–721. doi: 10.1007/s00203-017-1347-4
Nguyen L., Osanai Y., Lai K., Anderson I., Bange M., Tissue D., et al. (2018). Responses of the soil microbial community to nitrogen fertilizer regimes and historical exposure to extreme weather events: Flooding or prolonged-drought. Soil Biol. Biochem. 118, 227–236. doi: 10.1016/j.soilbio.2017.12.016
Okalebo J. R., Gathua K. W., Woomer P. L. (2002). Laboratory methods for soil and plant analysis: A working manual (Nairobi: TSBF).
Okumu M., Van Asten P., Kahangi E., Okech S. H., Jefwa J., Vanlauwe B. (2011). Production gradients in smallholder banana (cv. giant Cavendish) farms in central Kenya. Sci. Hortic. 127 (4), 475–481. doi: 10.1016/j.scienta.2010.11.005
Orgiazzi A., Bardgett R., Barrios E., Behan-Pelletier V., Briones M., Chotte J., et al. (2016) Global soil biodiversity atlas (Luxembourg: European Commission, Publications Office of the European Union). Available at: https://esdac.jrc.ec.europa.eu/content/global-soil-biodiversity-maps-0 (Accessed 11 June 2021).
Pang Z., Dong F., Liu Q., Lin W., Hu C., Yuan Z. (2021). Soil metagenomics reveals effects of continuous sugarcane cropping on the structure and functional pathway of rhizospheric microbial community. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.627569
Pellegrino E., Bosco S., Cicollini V., Chiara P., Sabbatini T., Silvestri N., et al. (2014). Agricultural abandonment in Mediterranean reclaimed peaty soils: long-term effects on soil chemical properties, arbuscular mycorrhizas and CO2 flux. Agric. Ecosyst. Environ. 199, 164–175. doi: 10.1128/aem.66.6.2541-2547.2000
Rondon M. R., August P. R., Bettermann A. D., Brady S. F., Grossman T. H., Liles M. R., et al. (2000). Cloning the metagenome: a strategy for accessing the genetic and functional diversity of uncultured microorganisms. Appl. Environ. Microbiol. 66, 2541–2547. 10.1128/aem.66.6.2541-2547.2000. doi: 10.1128/AEM.66.6.2541-2547.2000
Rossmann B., Muller H., Smalla K., Mpiira S., Tumuhairwe J. B., Staver C., et al. (2012). Banana associated microbial communities in Uganda are highly diverse but dominated by enterobacteriaceae. Appl. Environ. Microbiol. 78, 4933–4941. doi: 10.1128/AEM.00772-12
Soliman T., Yang S., Yamazaki T., Jenke-Kodama H. (2017). Profiling soil microbial communities with next-generation sequencing: the influence of DNA kit selection and technician technical expertise. Peer J. 5, e4178. doi: 10.7717/peerj.4178
Sommermann L., Geistlinger J., Wibberg D., Deubel A., Zwanzig J., Babin D., et al. (2018). Fungal community profiles in agricultural soils of a long-term field trial under different tillage, fertilization and crop rotation conditions analyzed by high-throughput ITS-amplicon sequencing. PloS One 13 (4), e0195345. doi: 10.1371/journal.pone.0195345
Sui X., Zhang H., Frey R., Yang L., Li M., Ni H. (2019). Land use change effects on diversity of soil bacterial, acidobacterial and fungal communities in wetlands of the sanjiang plain, northeastern China. Nat. Res. Sci. Rep. 9, 18535. doi: 10.1038/s41598-019-55063-4
Swanson K. S., Dowd S. E., Suchodolski J. S., Meddelbos I. S., Vester B. M., Barry K. A., et al. (2011). Phylogenetic and gene-centric metagenomics of the canine intestinal microbiome reveals similarities with humans and mice. ISME J. 5 (4), 639–649. doi: 10.1038/ismej.2010.162
Tahat M., Monther M., Kholoud A., Yahia I. (2020). Soil health and sustainable. Agri.Sustain. 12 (12), 4859. doi: 10.3390/su12124859
Tasnim N., Abulizi N., Pither J., Hart M. M., Gibson D. L. (2017). Linking the gut microbial ecosystem with the environment: does gut health depend on where we live? Front. Microbiol. 8. doi: 10.3389/fmicb.2017.01935
Tilhou N., Renata L., Nave S., Neal E., Travis M. (2021). Forage species and summer management impacts on soil carbon and nitrogen in winter stockpiled grazing systems. Agrosyst. Geosci. Environ. 4 (1), e20132. doi: 10.1002/agg2.20132
Zhang H., Wang R., Chen S., Qi G., He Z., Zhao X. (2017). Microbial taxa and functional genes shift in degraded soil with bacterial wilt. Sci. Rep. 7, 39911. doi: 10.1038/srep39911
Keywords: microbiome, metagenomics, banana rhizosphere, soil parameters, diversity
Citation: Wahome CN, Maingi JM, Ombori O, Njeru EM, Muthini M and Kimiti JM (2023) Diversity and abundance of bacterial and fungal communities in rhizospheric soil from smallholder banana producing agroecosystems in Kenya. Front. Hortic. 2:1061456. doi: 10.3389/fhort.2023.1061456
Received: 04 October 2022; Accepted: 18 January 2023;
Published: 08 February 2023.
Edited by:
Metin Turan, Yeditepe University, TürkiyeReviewed by:
Ees Ahmad, National Bureau of Agriculturally Important Microorganisms (ICAR), IndiaHayssam M. Ali, King Saud University, Saudi Arabia
Copyright © 2023 Wahome, Maingi, Ombori, Njeru, Muthini and Kimiti. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: John M. Maingi, bWFpbmdpam9obkBnbWFpbC5jb20=
 Caroline N. Wahome
Caroline N. Wahome John M. Maingi
John M. Maingi Omwoyo Ombori
Omwoyo Ombori Ezekiel Mugendi Njeru
Ezekiel Mugendi Njeru Morris Muthini1
Morris Muthini1 Jacinta Malia Kimiti
Jacinta Malia Kimiti