Abstract
Tomato is one of the most important vegetable crops cultivated and consumed worldwide. Demand is increasing daily because of increase in per capita fresh fruit consumption. Despite the economic importance of tomato, it has relatively short shelf-life after ripening and experiences remarkable post-harvest losses. This study was aimed at using marker-assisted backcrossing (MABC) to transfer shelf-life gene (alc) into the genetic background of two elite Ghanaian tomato breeding lines. The MABC derived lines at BC2F3 were evaluated to confirm selections using molecular markers. All the MABC-derived lines gave significant extended shelf life compared to the checks except for Alc-LA3134, the alc donor parent, which was, however, not significantly different from one of the backcrosses (BC2F2.3-E-80-19-26). This confirms that the genetic regulation of the shelf-life trait is dependent on the alc gene. The performance of the donor check (Alc-LA3134) against the 12 MABC-derived lines indicated no significant differences for the fruit firmness (except for BC2F2.3-E-80-19-4), number of locules per fruit and shelf life (for only BC2F2.3-E-80-19-26), thus suggesting similar backgrounds of the MABC as the recurrent parents. A considerable increase in the shelf-life value was observed among the MABC-derived lines. The introgression of the alc gene into cultivated tomatoes for extended shelf-life could reduce post-harvest losses of tomato in Ghana.
1 Introduction
The demand for tomato is increasing daily because of increase in per capita fresh fruit consumption. Nonetheless as a perishable fruit vegetable, it has a relatively short-life after ripening (Zapata et al., 2008). The onset of fruit ripening involves rapid production of ethylene at the breaker stage of the fruit that moderates a series of reactions during fruit ripening. During this period, fruit tissues soften, starch is converted to sugar, and secondary metabolites influencing appearance, taste and aroma accumulate (Seymour, 1993). One of the key softening enzymes of tomato is polygalacturonase (PG). This enzyme only surfaces and accumulates in fruits during ripening. The occurrence of PG enzyme strongly associates with the onset of cell wall degradation. The natural synthesis of ethylene starts before the synthesis of PG enzyme and the exogenous ethylene that facilitate the accumulation of the enzyme in mature fruits (Grierson and Kader, 1986; Casals et al., 2021). The crop is characterized by remarkable post-harvest losses generally due to its high perishability and the poor storage conditions under which fruits are kept. Several strategies exist to reduce storage losses such as refrigeration, heat treatment, modified atmosphere packaging (MAP) and treatment with 1-methylcyclopropent and calcium chloride (Arah et al., 2016). Additionally, clustered regularly interspaced short palindromic repeats (CRISPR)-CRISPR-associated protein-9 (Cas9) also has the potential to improve many traits rapidly (Wan et al., 2021). Moreover, the process of ripening of the tomato fruit can be controlled through RNA Interference (RNAi) which shuts down gene expression of ethylene biosynthesis enzymes and cell wall degrading enzymes such as endo polygalacturonase (PG) and pectin methyl esterase (PME) (Carrari, 2007). Nevertheless, these strategies are expensive, time consuming and requires societal acceptance. Consequently, genetic enhancement of the most important fruit quality characteristics appears to be the best choice available.
Studies on the physiology and genetics of tomato have resulted in the identification and characterization of several ripening mutant genes such as alcobaca (alc), non-ripening (nor) and ripening inhibitor (rin). Whereas fruits of alc mutant gene delayed ripening and produced uniform ripening in tomato, fruits of nor and rin do not display any climacteric rise. All three mutants demonstrate little or no polygalacturonase (PG) activity during ripening. Gavrish and Korol (1991) reported that acceptable colour is developed in the heterozygous state of the alc alleles. In addition the extended shelf life is greater than the normal (Dhatt, 2001; Kitagawa et al., 2005) and develops good flavour attributes (Kopeliovitch et al., 1982; Agar et al., 1994; Seroczynska et al., 1998). Traditional breeding has utilized of alc, nor, and rin genes for the development of lines and cultivars with delayed ripening (Kopeliovitch et al., 1979) using the backcross approach. In backcross breeding, the main goal is to incorporate one or few genes from a donor into the background of an elite variety and recover the recurrent (elite) parent’s phenotype as quickly as possible (Semagn et al., 2006). In the past, this was usually achieved by conventional backcross method, but that requires many generations over several years before recovering the background of the elite cultivar. The development of genomic resources in the past two decades has allowed introgression of genes using molecular markers. The use of single nucleotide polymorphic (SNP) markers and high throughput screening in marker-assisted backcrossing (MABC) programmes has allowed more rapid recovery of the recurrent parent’s phenotype (Mammadov et al., 2012).
The MABC technology allows the tagging of regions of each chromosome so that the process of the recovery of the recurrent parent’s phenotype can be followed (Semagn et al., 2006). This approach has been developed to avoid problems associated with traditional breeding by shifting criteria for selection from phenotypes to genes that regulate the traits of interest, either directly or indirectly. Integrating molecular approaches with traditional breeding for long shelf-life could significantly increase the storage life of tomato. A marker-assisted backcross (MABC) strategy (Hospital and Charcosset, 1997) was thus chosen in order to introgress the alc gene into the genetic background of two promising elite Ghanaian tomato breeding lines.
2 Materials and methods
2.1 Location and experimental period
The experiment was conducted during the period from August 2017 to December 2018 in the greenhouse of CSIR-Crops Research Institute, Kwadaso, Ghana and the Ag-Biotech facility, Monterey, California, USA.
2.2 Plant materials
This study involved using two elite Ghanaian tomato breeding lines as recurrent parents for incorporation of long shelf-life gene derived from tomato mutant alcobaca (Alc-LA3134) as a donor parent. The two recurrent parents used were CSIR/CRI-P002 and CSIR/CRI-ATS06; these are breeding lines from the CSIR-Crops Research Institute, Kumasi, Ghana. They are popular open-pollinated tomato varieties and were developed through pure line selection method (Osei et al., 2015). The CSIR/CRI-P002 and CSIR/CRI-ATS06 varieties have semi-determinate plant growth with regular and horizontal leaf type and attitude, respectively. The fruit shape of CSIR/CRI-P002 is typically bell shaped with exterior green immature and red mature fruit colours. CSIR/CRI-ATS06 has a rounded shape with light green colour for immature fruit and red colour for mature fruit. The varieties can be planted throughout the year with average yield of 20 tons/ha. These varieties are desirable to tomato farmers because of their relative high yield, fruit quality and adaptability. However, they lack firmness and long shelf life. The donor parent, LA3134 (alc), is a ripening gene mutant of Solanum lycopersicum obtained from Tomato Genetic Resource Center (TGRC), UC-Davis, USA (accession no. LA3134). The gene ‘alc’ from the locus name alcobaca greatly delays fruit ripening processes and is noted for prolonging the shelf-life of tomatoes. The donor is also characterized by determinate plant growth habit and rounded fruit shape with big fruit size but lacks important farmer-preferred horticultural traits.
2.3 Population development
Each of the recurrent parents, CSIR/CRI-P002 and CSIR/CRI-ATS06, was crossed with the donor parent LA3134 (alc) (Figure 1) during the major season of June 2017 at CSIR-Crops Research Institute, Kwadaso to produce F1s. The F1 seeds were later grown and tested for hybridity using SNP markers from Ag-Biotech. This was done at the early stages of plant growth to detect the presence of the marker allele and eliminate false hybrids.
Figure 1
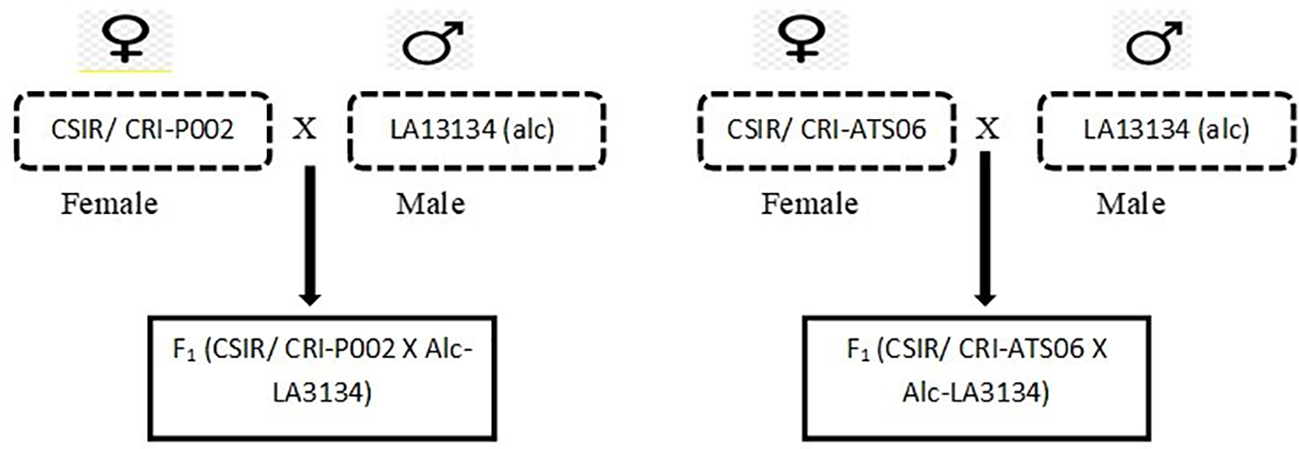
Scheme of F1 development between two elite CSIR/CRI tomato breeding lines and alc (Donor) line (LA3134).
The true F1 plants were then crossed back to the recurrent parents (CSIR/CRI-P002 and CSIR/CRI-ATS06) during the minor season of August 2017 to produce 88 and 86 BC1F1 plants respectively (The F1 plants were used as females while the recurrent parents were males) (Figure 2). Individual plants of the BC1F1 population were screened for the marker (alc) at early growth stages in April 2018. Plants that carried the desired marker allele and had the most SNPs from the recurrent parent were crossed back to the recurrent parents to create BC2F1 populations in July 2018. For this study, the final backcrossing population ended at BC2F1. Individual plants of the BC2F1 population (88 plants) were likewise screened with the SNP marker for the target trait (alc) and 19 other polymorphic SNP markers. Plants with the alc SNP marker and the most SNP markers for the recurrent parent were then selfed and harvested to produce BC2F2 generations. Progenies of this backcross-selfing (BC2F2) were screened in December 2018 to detect the markers, and the seeds were harvested from individuals carrying the homozygous donor parent marker alleles of the target trait in March 2019. Fresh leaf tissues were harvested from individual plants from selected backcross populations (BC1 & BC2 generations) including selfed individuals from BC2 population of the final generation developed from CSIR/CRI-P002 and CSIR/CRI-ATS06 tomato breeding lines. Each recurrent parent (CSIR/CRI-P002; CSIR/CRI-ATS06) comprised of 86, 88 and 86 plants of backcross populations for BC1F1, BC2F1 and BC2F2, respectively. These were placed in known positions of 96-deep-well plate and shipped to Ag-Biotech, Inc. via USDA-APHIS office in the USA. Leaf samples of parents were also included in each plate as controls. Twenty-two SNPs that were polymorphic in the original 140 SNPs were used to test these BC1 plants.
Figure 2
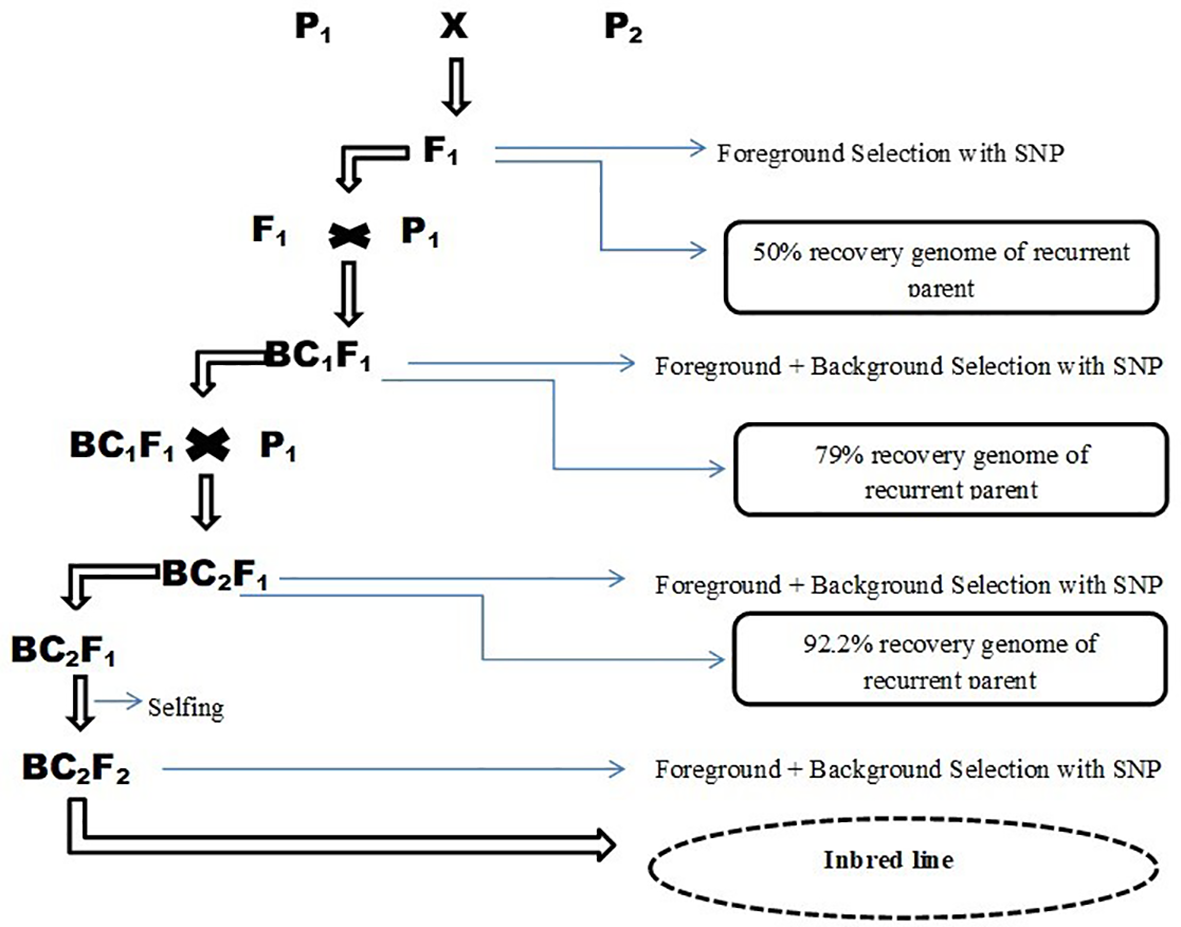
Scheme of backcross population development between two elite Ghanaian breeding lines and donor line with alc gene. Source modified from Babu et al. (2005).
2.4 Introgression of alc gene (MABC scheme)
2.4.1 Foreground selection
Target gene linked marker (Sly10-14) and selected SolCap SNPs were initially tested for polymorphism between donor and recurrent parents to establish their usage in the MABC programme using 19 SNP markers located across the chromosomes. Information on the primers is a trade secret of Ag-Biotech. These markers were associated with chromosomes 2 to 12. Most of the markers were designed from SolCap SNPs (M. Massoudi, Personal communication, 2017). The A/T SNP linked with the shelf life mutant gene, alc was used to design the SNP marker, Sly10-14 (Casals et al., 2011; Osei et al., 2019). For each backcross one generation, the F1s were tested for hybridity for the introduced gene using the foreground marker. This was to validate that the selected plants carried the target gene. The successful F1 plants, which showed the target marker, were used for further backcrossing and for the final selfing. The backcross populations were genotyped using SNP marker Sly10-14 for foreground selection to detect heterozygous plants in these populations. At BC2F2 generation, the foreground marker was used for selecting homozygous plants that carried the marker allele from the donor parent.
2.4.2 Background selection
An optimized subset of 140 SNPs markers that were derived from the 7,725 SNP array developed by the Solanaceae Coordinated Agricultural Project (SolCap) were used (Panthee et al., 2017). The polymorphism rates between the tomato lines aided in the selection of the markers. The tomato genotypes or lines were screened using the 140 SNPs with a minimum of 10 SNPs for each tomato chromosome. One hundred and forty (140) SolCap SNPs were converted to allele-specific primers by Ag-Biotech, Inc. Information from Yogendra and Ramanjini (2013) and Vrebalov et al. (2002) was used to design KASP primer sets by Ag-Biotech, Inc. A total of 19 SNPs that were unlinked to the target gene were used for background selection. Here individuals with genetic background closest to the recipient line in each case were selected. The recovery was calculated according to Van (2008) and Douglas Maxwell (D. Maxwell 2018, personal communication). The progeny with the greatest proportion of recurrent parent genome (RPG) SNP markers and the alc SNP were selected.
2.5 Evaluation and validation of MABC derived lines of BC2F3
The seeds from the homozygous BC2F2 for the donor parent marker alleles were evaluated on field in June 2019 to validate the shelf life (alc) gene. Sixteen tomato genotypes comprising three parents and 12 BC2F2.3 lines selected based on the SNP markers and one farmers’ variety as check (popular local variety) were used for field evaluation. The experiment was conducted at Wenchi (Latitude =07044’53.2” N; Longitude = 002004’25.4” W) in the Transition agro-ecological zone of Ghana. The tomato seeds were nursed and transplanted in June 2019 for planting. The soil was sandy loam. Wenchi has bimodal rainfall pattern but gave irregular rains from June, 2019 to September 2019 during the period this experiment was conducted. The experiment was laid out in a randomized complete block design with three replications. Each experimental unit consisted of two rows of 24 plants at 60 cm x 50 cm inter and intra row distances respectively. The experiment was conducted under a rain-fed condition with supplemental drip irrigation. Standard agronomic and plant protection measures were adopted to grow a healthy crop. Data collected include days to 50% flowering, plant height (cm), number of fruits per plant, yield per plant (g), yield per plot (kg), fruit firmness (N/cm2), number of locules, pericarp thickness (mm), total soluble solids (TSS) and shelf-life (days).
The Recurrent Parent Genome (RPG) recovery was calculated according to Van (2008) and Douglas Maxell (personal communication, 2018). Data from the field evaluation were subjected to analysis of variance (ANOVA) using R statistical software version 3.6 and mean separation using LSD at p<0.05.
3 Results
3.1 Parental polymorphism survey between recipient parents and the donor parent
Nineteen (19) SNP markers revealed polymorphism between CSIR/CRI-ATS06 and Alc-LA3134 whereas seven SNPs were polymorphic between CSIR/CRI-P002 and Alc-LA3134 (Tables 1 and 2). These SNP markers were employed for the confirmation and selection during the introgression programme.
Table 1
| SNPs Entry | Sly02-9 | Sly03-8 | Sly04-7 | Sly05-5 | Sly05-9 | Sly06-1 | Sly06-6 | Sly06-7 | Sly07-1 | Sly08-1 | Sly08-8 | Sly08-9 | Sly09-1 | Sly10-1 | Sly10-alc | Sly10-11 | Sly11-13 | Sly11-Rx4 | Sly12-10 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Alc-LA3134 | X:X | X:X | X:X | X:X | Y:Y | Y:Y | Y:Y | X:X | Y:Y | X:X | X:Y | X:X | Y:Y | Y:Y | X:X | X:X | Y:Y | X:X | X:X |
| CSIR/CRI-ATS06 | Y:Y | Y:Y | Y:Y | Y:Y | X:X | X:X | X:X | Y:Y | X:X | Y:Y | X:X | Y:Y | X:X | X:X | Y:Y | Y:Y | X:X | Y:Y | Y:Y |
| Remarks | PM | PM | PM | PM | PM | PM | PM | PM | PM | PM | PM | PM | PM | PM | PM | PM | PM | PM | PM |
Parental polymorphism survey between CSIR/CRI-ATS06 (recipient) and Alc-LA3134 (donor).
M, Monomorphic; PM, Polymorphic.
Table 2
| SNPs Entry | Sly02-9 | Sly03-8 | Sly04-7 | Sly05-5 | Sly05-9 | Sly06-1 | Sly06-6 | Sly06-7 | Sly07-1 | Sly08-1 | Sly08-8 | Sly08-9 | Sly09-1 | Sly10-1 | Sly10-alc | Sly10-11 | Sly11-13 | Sly11-Rx4 | Sly12-10 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Alc-LA3134 | X:X | X:X | X:X | X:X | Y:Y | Y:Y | Y:Y | X:X | X:X | X:X | Y:Y | X:X | Y:Y | Y:Y | X:X | X:X | Y:Y | X:X | X:X |
| CSIR/CRI-P002 | X:X | Y:Y | X:X | X:X | Y:Y | Y:Y | Y:Y | X:X | X:X | X:X | X:X | Y:Y | X:X | X:X | Y:Y | Y:Y | Y:Y | X:X | X:X |
| Remarks | M | PM | M | M | M | M | M | M | M | M | PM | PM | PM | PM | PM | PM | M | M | M |
Parental polymorphism survey between CSIR/CRI-P002 (recipient) and Alc-LA3134 (donor).
M, Monomorphic; PM, Polymorphic.
3.2 Confirmation of alc gene in F1, BC1F1, BC2F1 and BC2F2 generations of two elite lines
Nineteen polymorphic SNPs were identified for the cross, CSIR/CRI-P002 x Alc-LA3134 and 18 polymorphic SNPs displayed successful F1s. For the cross, CSIR/CRI-ATS06 x Alc LA3134, 22 SNPs were polymorphic and 20 SNPs indicated successful F1’s. Of the 88 and 86 backcross individuals (BC1F1) produced from CSIR/CRI-ATS06 x Alc-LA3134 and CSIR/CRI-P002 x Alc-LA3134, only 23 and 39 plants were positive for the alc gene in the backcross populations, respectively (Table 3). Of the 23 BC1F1 individuals from CSIR/CRI-ATS06 that were positive for the alc gene, six plants produced SNP score of more than 30 with the highest SNP score of 34 in BC line (E1 x E)-79. Similarly, in the CSIR/CRI-P002 BC1 population, 39 BC1F1 individuals were positive for the alc gene. The highest SNP score was 10 and was associated with seven plants.
Table 3
| Name of BC1F1 crosses | No. of BC1F1 plants produced | No. of positive BC1F1 plants for alc gene |
|---|---|---|
| BC1=F1 x CSIR/CRI-ATS06 | 88 | 23 |
| BC1=F1 x CSIR/CRI-P002 | 86 | 39 |
| Name of BC2F1 crosses | No. of BC2F1 plants produced | No. of positive BC2F1 plants for alc gene |
| BC2=BC1 x CSIR/CRI-ATS06-80 | 88 | 38 |
| BC2=BC1 x CSIR/CRI-ATS06-22 | 88 | 6 |
| BC2=BC1 x CSIR/CRI-P002-80 | 88 | 42 |
| BC2=BC1 x CSIR/CRI-P002-13 | 88 | 43 |
List of BC1F1 and BC2F1 produced and the number of true F1 at each generation positive for the alc gene.
Of the total of 38 BC2F1 individuals from CSIR/CRI-ATS06 (line BC2-E-80) that were positive for the alc gene, only four plants produced SNP score of recurrent genome recovery (RGR) of more than 19 with the highest SNP score of 21 for BC lines (BC2- E-80-1, BC2-E-80-4, BC2-E-80-34) (Table 3). Similarly, in line BC2-E-22 population of CSIR/CRI-ATS06 where only six plants showed positive for the alc gene, only three plants produced more than a 16 SNP score for RGR with the highest SNP score of 20 for BC2-E-22-27 (Table 3). A total of 42 BC2F1 plants (line BC2-A-80) that were identified to be positive for the alc gene produced more than a 10 SNP score of RGR for 18 plants. The highest score of 12 was identified for BC2-A-80-36 (Table 3). In addition, of the total of 43 BC2F1 individuals from CSIR/CRI-P002 (line BC2-A-13) genotyped, 13 lines had SNP scores of more than 10 for RGR with the highest RGR of 12 for BC2-A-13-45 (Table 3).
Of the 86 BC2F2 plants representing CSIR/CRI-ATS06 BC population for plant numbers E-80-19 and E-22-27, only 15 and 22 plants were identified as homozygous for the alc gene, respectively (Table 4). Likewise, in the CSIR/CRI-P002 BC population, out of the 86 BC2F2 plants generated from BC2F1 (plant numbers A-80-70 and A-13-39), 19 plants from each population were identified to be homozygous for the alc gene (Table 4).
Table 4
| BC2F1 lines selected for selfing | No. of BC2F2 plants produced | No. of positive BC2F2 plants for alc gene |
|---|---|---|
| BC2-E-80- 19[BC2= (E xE1) x E-80-19] | 86 | 15 |
| BC2-E-22-27 [BC2= (E xE1) x E-22-27] | 86 | 22 |
| BC2-A-80-70 [BC2= (A xA1) x A-80-70] | 86 | 19 |
| BC2-A-13-39 [BC2= (A xA1) x A-13-39] | 86 | 19 |
List of BC2F1 lines selected for selfing.
3.3 Variability of measured traits in MABC-derived lines and their field performance
There were significant differences among the parents and MABC-derived lines for all the studied characters (Table 5).
Table 5
| Source | Df | D50F | PHT | NFPPLT | FYPLOT | FF | PT | TSS | NOL | SL | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Rep | 2 | 0.14 | 0.21 | 0.43 | 5.3* | 0.03 | 0.02 | 0.09 | 1.18** | 1.4 | |
| Entry | 15 | 12.13*** | 51.11*** | 16.13*** | 137.88*** | 1.49*** | 0.20*** | 2.25*** | 1.18*** | 275.91*** | |
| Total | 30 | 0.32 | 0.44 | 16.13 | 1.08 | 0.01 | 0.02 | 0.08 | 0.18 | 2.42 | |
| CV | 2.38 | 1.01 | 5.94 | 1.86 | 0.25 | 2.97 | 4.91 | 9.49 | 3.83 | ||
Mean squares of studied traits in parents and MABC derived lines.
*, **, *** =Significant at P = 0.05, 0.01, 0.001 probability levels respectively; df, degree of freedom; D50F, Days to 50% flowering; PHT, Plant height; NFPPLT, Number of fruits per plant; FYPLOT, Fruit yield per plot; FF, Fruit firmness; PT, Pericarp thickness; TSS, Total soluble solids; NOL, Number of locules per fruit; SL, Shelf life; CV, coefficient of variation.
3.3.1 Field performance of MABC-derived lines
The field performance of the four checks (local, CSIR/CRI-P002, CSIR/CRI-ATS06, Alc-LA3134) against the 12 MABC-derived hybrids are presented in Table 6. Among the 12 MABC-derived hybrids, a subset of six hybrids of the BC2F2.3-A lines were not significantly different from CSIR/CRI-P002 (parent check) for days to 50% flowering, plant height, number of fruits per plant, fruit yield per plot and TSS. Similarly, all six of the MABC-derived hybrids of BC2F2.3-E lines were not significantly different from the parent check (CSIR/CRI-ATS06) for the 50% flowering, plant height, number of fruits per plant, fruit yield per plot and brix. From the result, it is clear that the Alc-LA3134 (donor check) was significantly different from MABC-derived lines for days to 50% flowering, plant height, number of fruits per plant and TSS. For the fruit yield per plot, the donor check (Alc-LA3134) was however, not significantly different from only the BC2F2.3-E lines. Field performance of the donor check (Alc-LA3134) against the 12 MABC derived lines indicated no significant differences for the fruit firmness, pericarp thickness, number of locules and shelf-life. Nonetheless, there was significant differences between the 12 MABC derived lines and the other parent checks (CSIR/CRI-P002, CSIR/CRI-ATS06) including the local check for the fruit quality and the shelf-life traits. Within the 12 MABC-derived, lines were also significant differences for fruit quality and shelf -life traits.
Table 6
| Tomato genotypes | D50F(days) | PHT(cm) | NFPPLT | FYPLOT(kg) | FF(N/cm2) | PT(mm) | BX(%) | NOL | SL(days) |
|---|---|---|---|---|---|---|---|---|---|
| BC2F2.3-A-13-39-30 | 22.33 | 64.00 | 16.66 | 52.00 | 54.83 | 5.15 | 6.66 | 4.33 | 39.66 |
| BC2F2.3-A-13-39-33 | 22.00 | 65.00 | 17.33 | 52.00 | 54.83 | 5.15 | 6.83 | 4.33 | 41.00 |
| BC2F2.3.-A-13-39-65 | 22.33 | 64.57 | 18.00 | 51.94 | 54.83 | 5.13 | 6.76 | 4.00 | 40.00 |
| BC2F2.3-A-80-70-23 | 22.00 | 64.88 | 16.66 | 51.66 | 54.80 | 5.17 | 6.66 | 4.33 | 45.00 |
| BC2F2.3-A-80-70-4 | 22.33 | 65.76 | 17.33 | 51.66 | 54.80 | 5.16 | 6.00 | 4.33 | 45.33 |
| BC2F2.3-A-80-70-8 | 22.66 | 65.66 | 17.33 | 52.00 | 54.80 | 5.17 | 6.33 | 4.33 | 45.00 |
| BC2F2.3-E-22-27-1 | 25.00 | 68.29 | 13.66 | 62.00 | 54.67 | 5.25 | 5.16 | 4.33 | 47.66 |
| BC2F2.3-E-22-27-23 | 25.00 | 68.40 | 14.66 | 62.29 | 54.67 | 5.24 | 5.00 | 4.66 | 46.66 |
| BC2F2.3-E-22-27-8 | 25.33 | 68.00 | 14.66 | 61.66 | 54.67 | 5.26 | 5.16 | 4.33 | 45.00 |
| BC2F2.3-E-80-19-26 | 25.66 | 68.33 | 15.00 | 61.33 | 54.70 | 5.19 | 5.33 | 4.33 | 50.33 |
| BC2F2.3-E-80-19-30 | 25.00 | 68.04 | 14.00 | 60.84 | 54.70 | 5.20 | 5.00 | 4.66 | 42.33 |
| BC2F2.3-E-80-19-4 | 25.33 | 68.68 | 13.66 | 62.12 | 54.47 | 5.22 | 5.16 | 4.33 | 41.33 |
| CSIR/CRI-ATS06 (check) | 25.33 | 68.41 | 14.00 | 62.18 | 54.31 | 4.74 | 5.10 | 5.00 | 28.66 |
| CSIR/CRI-P002 (check) | 22.00 | 64.33 | 18.00 | 52.04 | 54.53 | 4.90 | 6.93 | 5.00 | 23.66 |
| ALC-LA3134 (check) | 28.33 | 51.93 | 9.66 | 61.18 | 54.82 | 5.54 | 4.06 | 4.00 | 50.33 |
| Local (check) | 21.00 | 69.76 | 12.33 | 38.40 | 51.94 | 4.37 | 5.93 | 6.66 | 16.33 |
| Mean | 23.85 | 65.88 | 15.18 | 55.95 | 54.52 | 5.11 | 5.75 | 4.56 | 40.52 |
| CV | 2.38 | 1.01 | 5.94 | 1.86 | 0.25 | 2.97 | 4.91 | 9.49 | 3.83 |
| LSD | 0.95 | 1.07 | 1.51 | 1.74 | 0.23 | 0.25 | 0.47 | 0.72 | 2.59 |
| MSE | 0.32 | 0.44 | 0.81 | 1.08 | 0.01 | 0.02 | 0.08 | 0.18 | 2.41 |
Mean performance of parents and MABC derived lines of BC2F2.3.
D50F ,Days to 50% flowering; PHT, Plant height; NFPPLT, Number of fruits per plant; FYPLOT, Fruit yield per plot; FF, Fruit firmness; PT, Pericarp thickness; TSS, Total soluble solids; NOL, Number of locules per fruit; SL, Shelf life; CV, coefficient of variation; LSD, Least significant difference; MSE, Mean squared error.
4 Discussion
A successful marker-assisted backcrossing strategy requires the availability of polymorphic markers closely linked to genes controlling traits desired in the recurrent parent (Hasan et al., 2015). These polymorphic markers are very useful for MABC scheme in each backcross generation. A marker which is monomorphic bears no value in selection work because this type of marker cannot differentiate the two parental genotypes. With the recent development in the field of molecular marker analysis, single nucleotide polymorphic (SNP) markers have enormous potential to improve efficiency and precision of conventional plant breeding via Marker Assisted Selection (MAS) (Gregorio et al., 2002; Bundo et al., 2022). The results from this study indicated patterns of genetic variation at different loci on chromosomes within parental materials. This agrees with (Sim et al., 2012) who in a similar study revealed patterns of genetic variation through the use of high-design SNPs for genotyping of tomato. The selected polymorphic SNP markers distinguished the parents and consequently justified the choice of the markers for selection of the generated populations. Thus, these SNP markers could be used to select for the recurrent parent.
A successful backcrossing programme is also dependent on the ability to select for the donor gene at each backcross generation and the ability to recover the recurrent parent’s phenotype. The application of DNA-based molecular markers offers a chance to carry out indirect selection of target traits at any stage of the plant growth and or selection for recurrent parent background or against unwanted donor parent genome. In the present study, while screening for plants possessing the alc gene among the BC1F1 plants, a total of 23 and 39 plants were observed for CSIR/CRI-ATS06 and CSIR/CRI-P002, respectively, to be positive as BC1F1. When the confirmed positive F1 plants are backcrossed with the respective recurrent parent, one out of every two progeny BC1F1 plants is expected to have the entire shelf-life gene. Nonetheless as noticed earlier, in the present study, some of the number of positives generated from the BC1F1 cross from CSIR/CRI-ATS06 deviated from the typical Mendelian pattern of segregation, probably due to selfed seeds, which might have resulted in fewer number of positive plants in that backcross. According to Sayed et al. (2002) even though markers will generally segregate in a Mendelian manner, distorted segregation ratios may be encountered. Identification of positives for alc gene in BC1F1 generation through phenotypic screening is very difficult and is time consuming. Phenotypically, it will require harvesting and holding of fruits on the shelf until they are unmarketable or wrinkled to estimate the shelf life and confirm presence of alc gene based on fruit colour and length of shelf life. Molecular markers can thus, be used as supplement for phenotyping, since they allow selection at the early stage of the plant (seedling stage) and even during the off-season, thus making breeding more cost-effective by growing more generations per year and reducing the number of breeding lines that are needed to be tested. This facilitates elimination of undesirable lines at early generations (Ribaut and Hoisington, 1998). Like other genetic markers, the SNP marker used in the present study is gene specific to alc gene. The foreground marker used in this study, Sly10-14-SNP, has been validated in earlier study (Osei et al., 2019). Hence, this marker can be used to complement classical breeding techniques in order to select segregating plants that have the recessive alc gene at early stage rather than waiting to observe the phenotypic characters (storage life). It must be noted that even though the phenotype of Alc/alc vs. Alc/Alc is detectable, the difference may not always be very great so one might not be absolutely certain of the Alc/alc genotypes. Selfing every plant is another option, but it would take a long time to get the fruit and to check if you had any alc/alc plants. In the analysis, it was figured that the alc gene for shelf life was located on chromosome 10 which is consistent with the literature (Mutschler et al., 1988). Based on Chi-square analysis to validate the alc gene on F2 generation data, the segregation ratio obtained from the cross was in good agreement with the theoretical ratio of 1:2:1. Clearly, this was expected, given that Sly10-14-SNP used in the present studies was a functional marker for shelf-life trait that has been validated (Osei et. al., 2019).
Application of marker-assisted backcrossing enables the successful transfer of recessive alleles, which is difficult to do when using conventional approaches. This selection involved the use of markers tightly linked to long shelf-life in tomato to screen BC1F1 progenies for the presence of alc gene. They were able to successfully identify individuals that carried homozygous loci from the heterozygote ones though they were phenotypically indistinguishable. The selected BC2F1 plant had a very high similarity to the recurrent parents for most of the plant characters, despite the fact that the selected BC1F1 plant from the cross displayed a low resemblance. Even though there is no apparent reason to explain this higher percentage of recurrent parent genome recovery, it can be assumed that this could be because of some unknown mechanisms which might have resulted in transfer of some chromosomal segments from recurrent parent genome to be transmitted as such transferred to the progenies in the second cycle of backcrossing. Also, it must be noted that the SNP marker recover was not as complete as would be desirable; it would be best to have at least 10 polymorphic markers per chromosome or even more. Chen et al. (2002) reported similar results where they report recovery of a higher percentage of recurrent parent genome at each backcross generation as compared to expected values. In general, RPG recovery can be accelerated by using markers for background selection (Servin and Hospital, 2002; Sowjanya and Sridevi, 2021). The results agreed with many other MAS studies in tomato (Menda et al., 2013) demonstrating that continued selection in a self-pollinated BC2 generation with the aid of molecular markers would lead to a higher recovery in RPG. Nonetheless, some studies have reported otherwise (Jairin et al., 2009), and this could be because background screening was not done in the early backcross generations. The background selection is paramount in eliminating such deleterious genome regions of the donor parents that may negatively affect the final product. This is extremely useful because the recurrent parent recovery can be greatly accelerated. Traditional backcrossing takes a minimum of six backcross generations to recover the genome of the recurrent parent, with some fragments of the donor genome still lingering on. However, the genome of the recurrent parent can be achieved at the BC2, BC3 or BC4, with background selection thus shortening the process by two of the four backcross generations when markers are employed for background selection. In this study, the phenotype recovery of the recurrent parent was achieved at BC2. The MABC approach in the present study has clearly demonstrated the ability to accelerate the breeding process of high-yielding tomato lines with long shelf-life fruits. Within two backcross generations, a considerable increase in shelf life was observed among the selected lines derived. Through MAS, we advanced two backcross generations within one year. When conventional breeding strategies are applied, the advancement of two backcross generations with selection for the long shelf-life character would take two years or more, and it might be very difficult to differentiate reliably between individuals heterozygous for more than one of the genomic regions contributing to the trait. Identification of homozygous BC2F2 lines is very important because if the selected BC2F2 lines contain one or more of the target genes in heterozygous condition, they will segregate in the next generation. In the present study, only homozygous lines with the desirable gene combinations were selected for further advancement and evaluation, thus, ensuring homozygosity of the material with respect to shelf life. It has been observed that the recovery of plants with alc gene was more frequent and more in number, this may be due to the fact that the marker was closely linked to the gene on chromosome 10. The use of gene specific marker Sly10-14 will be of prime importance for the selection and tagging of alc gene associated with shelf life. Identification of homozygous plants from the segregating populations is of extreme importance in MAS. This helps in the stabilization of the genotypes in shortest possible time. This is evident from the results observed. Selected F1s were carefully genotyped for presence of the alc gene with the respective linked markers. The homozygous F2s carrying the alc gene were selected and backcrossed with the respective recurrent parents. The positive BC2F1 of CSIR/CRI-ATS06 and CSIR/CRI-P002 plants were then selected, selfed and advanced for further generations for agro-morphological traits, yield, fruit quality and shelf life. Background selection was thus achieved to select the individuals with the genetic background closest to the recipient line in each case.
The significant difference observed between the MABC derived lines and recurrent parents for shelf life, fruit firmness, pericarp thickness and number of locules confirm that the genetic regulation for these traits are dependent on the alc gene. The non-significance between the recurrent parents and the MABC derived lines for days to 50% flowering, plant height, number of fruits per plant, number of fruits per plot and TSS suggest similar backgrounds of the MABC as the recurrent parents. These results demonstrate that genetic gains to improve shelf-life have been achieved through the use of molecular markers following some recovery of the recurrent background into the MABC-derived lines. This agrees with Ribaut and Hoisington, (1998) who used MABC to improve drought tolerance in maize. The use of MAS to improve shelf-life, therefore, appears very promising. The significant difference observed between the MABC derived lines and recurrent parents for shelf-life, fruit firmness, pericarp thickness and number of locules confirm that the genetic regulation for these traits are dependent on the alc gene. The non-significant difference between the recurrent parents and the MABC-derived lines for days to 50% flowering, plant height, number of fruits per plant, number of fruits per plot and TSS suggest similar backgrounds of the MABC as the recurrent parents. These results demonstrate that genetic gains to improve shelf life have been achieved through the use of molecular markers following some recovery of the recurrent background into the MABC-derived lines. This agrees with Ribaut and Hoisington (1998) who used MABC to improve drought tolerance in maize. The use of MAS to improve shelf-life, therefore, appears very promising.
5 Conclusion
Using marker-assisted selection, it was possible to introgress genomic regions from ripening mutant donor parent Alc-LA3134 into the genetic backgrounds of two elite recurrent parents (CSIR/CRI-ATS06 and CSIR/CRI-P002) over two generations. The use of markers allowed rapid recovery of the backgrounds of the CSIR/CRI adapted/improved tomato lines. The findings clearly reflect the reduced accuracy of phenotypic selection compared to background selection using markers. The field performance of the MABC-derived lines has confirmed that the genetic regulation for shelf-life is dependent on the alc gene.
Statements
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author contributions
MK, AD, and HA-D: did literature search, conceptualize the concept, and wrote manuscript. ED, EB, MM and DM: provided guideline and edited manuscript. All authors contributed to the article and approved the submitted version.
Acknowledgments
The authors thank the staff of Ag-Biotech for their enormous assistance regarding DNA extraction and SNP marker analysis. This work was supported by Ag-Biotech Lab., Monterrey, CA, USA, and Syngenta.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fhort.2022.1024042/full#supplementary-material
References
1
AgarI. T.AbakK.YarsiG. (1994). Effect of different maturity stages on the keeping quality of nor (non-ripening), rin (ripening-inhibitor) and normal type tomatoes. Acta Horticulture368, 742–753. doi: 10.17660/ActaHortic.1994.368.88
2
ArahI. K.AhorboG. K.AnkuE. K.KumahE. K.AmagloH. (2016). Postharvest handling practices and treatment methods for tomato handlers in developing countries: A mini review. Adv. Agric.1–8. doi: 10.1155/2016/6436945
3
BabuR.NairS. K.KumarA.VenkateshS.SekharJ. C.SinghN. N. (2005). Two-generation marker aided backcrossing for rapid conversion of normal maize lines to quality protein maize (QPM). Theor. Appl. Genet.111, 888–897. doi: 10.1007/s00122-005-0011-6
4
BundoM.Martin-CardosoH.PesentiM.Gomez-ArizaJ.CastilloL.FrouinJ.et al. (2022). Integrative approach for precise genotyping and transcriptomics of salt tolerant introgression rice lines. Front. Plant Sci.12, 797141 doi: 10.3389/fpls.2021.797141
5
CarrariF. (2007). The metabolic shifts underlying tomato fruit development. Plant Biotechnol.24 (1), 45–55. doi: 10.5511/plantbiotechnology.24.45
6
CasalsJ.Cebolla-CornejoJ.RoselloS.BeltranJ.CasanasF.NuezF. (2011). Long-term postharvest aroma evolution of tomatoes with the alcobaca (alc) mutation. Eur. Food Res. Technol.233 (2), 331–342. doi: 10.1007/s00217-011-1517-6
7
CasalsJ.RullA.Giné-BordonabaJ. (2021). Changes in ripening-related quality traits of long shelf life tomatoes as influenced by water deficit and short-term postharvest storage. Agronomy11, 2304. doi: 10.3390/agronomy11112304
8
ChenM.PrestingG.BarbazukW. B.GoicoecheaJ. L.BlackmonB.FangG.et al. (2002). An integrated physical and genetic map of the rice genome. Plant Cell14 (3), 537–545.
9
DhattA. S. (2001). Evaluation of F1 hybrids incorporating nor, rin and alc for yield, quality and shelf life of tomato (Lycopersicon esculentum mill).” ph. d, thesis (Ludhiana: Punjab Agricultural University).
10
GavrishS. F.KorolV. G. (1991). Some biological features of F1 tomato hybrids carrying the nor gene. Izvestiya Timiryazevskoi Sel’skhozyaistvennoi Akademii1, 118–132.
11
GregorioG.SenadhiraD.MendozaR.ManigbasN.RoxasJ. P.GuertaC. Q. (2002). Progress in breeding for salinity tolerance and associated abiotic stresses in rice. Field Crops Res.76, 91–101. doi: 10.1016/S0378-4290(02)00031-X
12
GriersonD.KaderA. A. (1986). “Fruit ripening and quality,” in The tomato crop: A scientific basis for improvement. Eds. AthertonJ. G.RudichJ. (London: Chapman and Hall), 241–280.
13
HasanM. M.RafiiM. Y.IsmailM. R.MahmoodM.RahimH. A.AlamM. A.et al. (2015). Marker-assisted backcrossing: a useful method for rice improvement. Biotechnol. Biotechnol. Equip. 29 (2), 237–254. doi: 10.1080/13102818.2014.995920
14
HospitalF.CharcossetA. (1997). Marker assisted introgression of quantitative trait loci. Genetics147, 1469–1485. doi: 10.1093/genetics/147.3.1469
15
JairinJ.TeangdeerithS.KothcharerkJ.SansenK.YiM.VanavichitA.et al. (2009). Development of rice introgression lines with brown planthopper resistance and KDML105 grain quality characteristics through marker-assisted selection. Field Crops Res.110, 263–271. doi: 10.1016/j.fcr.2008.09.009
16
KitagawaM.ItoH.ShiinaT.NakamuranI. T.KasumiT.IshiguroY.et al. (2005). Characterization of tomato fruit ripening and analysis of gene expression in F1 hybrids of the ripening inhibitor (rin) mutant. Physiologia Plantarum123, 331–338. doi: 10.1111/j.1399-3054.2005.00460.x
17
KopeliovitchE.MizrahiY.RabinowitchH. D.KedarN. (1982). Effect of the fruit-ripening mutant genes rin and nor on the flavour of tomato fruit. J. Am. Soc. Horticulture Sci.107, 361–364. doi: 10.21273/JASHS.107.3.361
18
KopeliovitchE.RabinowitchH. D.MizrahiY.KedarN. (1979). The potential of ripening mutants for extending the storage life of the tomato fruit. Euphytica28, 99–104. doi: 10.1007/BF00029179
19
MammadovJ. A.AggarwalR.BuyyarapuR.KumpatlaS. (2012). “SNP markers and their impact on plant breeding,” in Book: The role of bioinformatics in agriculture. (New York: Taylor and Francis group). doi: 10.1201/b16568-18
20
MendaN.StricklerS. R.MuellerL. A. (2013). Advances in tomato research in the post- genome era. Plant Biotechnol.30 (3), 243. doi: 10.5511/plantbiotechnology.13.0904a
21
MutschlerM.GuttieriM. J.KinzerS.GriersonD.TuckerG. A. (1988). Changes in ripening-related processes in tomato conditioned by the alc mutant. Theor. Appl. Genet.76 (2), 285–292. doi: 10.1007/BF00257857
22
OseiM. K.BonsuK. O.Adu-GyamfiK.FrimpongM. (2015). Development of high yielding and uniform tomato fruits using pure line selection. Direct Res. J. Agric. Food Sci.3 (1), 10–16.
23
OseiM. K.DanquahE.DanquahA.MassoudiM.MaxwellD.Adu-DapaahH.et al. (2019). Validation of SNP marker linked to alc gene for long shelf life of tomato. J. Crop Improvement33 (5), 669–682. doi: 10.1080/15427528.2019.1657216
24
PantheeD. R.PiotrowskiA.IbrahimR. (2017). Mapping quantitative trait loci (QTL) for resistance to late blight in tomato. Int. J. Mol. Sci.18 (7), 1589. doi: 10.3390/ijms18071589
25
RibautJ. M.HoisingtonD. (1998). Marker-assisted selection: new tools and strategies. Trends Plant Sci.3, 236–239. doi: 10.1016/S1360-1385(98)01240-0
26
SayedH.KayyalH.RamseyL.CeccarelliS.BaumM. (2002). Segregation distortion in doubled haploid lines of barley (Hordeum vulgare l.) detected by simple sequence repeat markers. Euphytica225, 265–272. doi: 10.1023/A:1015861610226
27
SemagnK.BjornstadA.NdjiondjopM. N. (2006). An overview of molecular marker methods for plants. Afr. J. Biotechnol.l.5 (25), 2540–2568.
28
SeroczynskaA.NiemirowiczS. K.Korrenicwskl. A. (1998). Utilization of three non-ripening mutants in tomato breeding for prolonged shelf life. Folia Hortic.1, 3–14.
29
ServinB.HospitalF. (2002). Optimal positioning of markers to control genetic background in marker-assisted backcrossing. J. Heredity93, 214–217. doi: 10.1093/jhered/93.3.214
30
SeymourG. B. (1993). Biochemistry of fruit ripening (London; New York: Chapman & Hall).
31
SimS. C.Van-DeynzeA.StoffelK.DouchesD. S.ZarkaD.GanalM. W.et al. (2012). High-Density SNP Genotyping of 1478 Tomato (Solanum lycopersicum L.) reveals patterns of genetic variation due to breeding. PLoS One7, 9. doi: 10.1371/journal.pone.0045520
32
SowjanyaB. A.SrideviO. (2021). Introgression of tomato leaf curl virus (ToLCV) resistant gene into two cultivated tomato (Solanum lycopersicum l.) varieties through marker assisted backcross breeding. Biol. Forum – Int. J.13 (1), 211–226.
33
VanB. R. (2008). GGT 2.0: versatile software for visualization and analysis of genetic data. J. Heredity99, 232–236. doi: 10.1093/jhered/esm109
34
VrebalovJ.RuezinskyD.PadmanabhanV.WhiteR.MedranoD.DrakeR.et al. (2002). MADS-box gene necessary for fruit ripening at the tomato ripening-inhibitor (rin) locus. Science296, 343–346. doi: 10.1126/science.1068181
35
WanL.WangZ.TangM.HongD.SunY.RenJ.et al. (2021). CRISPR-Cas9 gene editing for fruit and vegetable crops: Strategies and prospects. Horticulturae7, 193. doi: 10.3390/horticulturae7070193
36
YogendraK. N.RamanjiniG. P. H. (2013). Phenotypic and molecular characterization of a tomato F2 population segregation for improving shelf life. Genet. Mol. Res.12 (1), 506–518. doi: 10.4238/2013.January.9.4
37
ZapataP. J.GuillenF.Martinez-RomeroD.CastilloS.ValeroD.SerranoM. (2008). Use of alginate or zein as edible coatings to delay postharvest ripening process and to maintain tomato quality. J. Sci. Food Agric.88 (7), 1287–1293. doi: 10.1002/jsfa.3220
Summary
Keywords
alc gene, tomato, shelf-life, backcrosses, introgression
Citation
Kwabena Osei M, Danquah A, Adu-Dapaah H, Danquah E, Blay E, Massoudi M and Maxwell D (2022) Marker assisted backcrossing of alcobaca gene into two elite tomato breeding lines. Front. Hortic. 1:1024042. doi: 10.3389/fhort.2022.1024042
Received
20 August 2022
Accepted
25 November 2022
Published
22 December 2022
Volume
1 - 2022
Edited by
Manuel Jamilena, University of Almeria, Spain
Reviewed by
Jorge Lora, Spanish National Research Council (CSIC), Spain; Pritam Kalia, Indian Agricultural Research Institute (ICAR), India; Shabir Hussain Wani, Sher-e-Kashmir University of Agricultural Sciences and Technology, India
Updates
Copyright
© 2022 Kwabena Osei, Danquah, Adu-Dapaah, Danquah, Blay, Massoudi and Maxwell.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michael Kwabena Osei, oranigh@gmail.com
This article was submitted to Breeding and Genetics, a section of the journal Frontiers in Horticulture
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.