- 1School of Medicine, Southeast University, Nanjing, Jiangsu, China
- 2Department of Orthopaedics, Zhongda Hospital, School of Medicine, Southeast University, Nanjing, Jiangsu, China
Osteoarthritis (OA) is the most common joint disease globally, and its progression is irreversible. The mechanism of osteoarthritis is not fully understood. Research on the molecular biological mechanism of OA is deepening, among which epigenetics, especially noncoding RNA, is an emerging hotspot. CircRNA is a unique circular noncoding RNA not degraded by RNase R, so it is a possible clinical target and biomarker. Many studies have found that circRNAs play an essential role in the progression of OA, including extracellular matrix metabolism, autophagy, apoptosis, the proliferation of chondrocytes, inflammation, oxidative stress, cartilage development, and chondrogenic differentiation. Differential expression of circRNAs was also observed in the synovium and subchondral bone in the OA joint. In terms of mechanism, existing studies have mainly found that circRNA adsorbs miRNA through the ceRNA mechanism, and a few studies have found that circRNA can serve as a scaffold for protein reactions. In terms of clinical transformation, circRNAs are considered promising biomarkers, but no large cohort has tested their diagnostic value. Meanwhile, some studies have used circRNAs loaded in extracellular vesicles for OA precision medicine. However, there are still many problems to be solved in the research, such as the role of circRNA in different OA stages or OA subtypes, the construction of animal models of circRNA knockout, and more research on the mechanism of circRNA. In general, circRNAs have a regulatory role in OA and have particular clinical potential, but further studies are needed in the future.
1 Introduction
Osteoarthritis (OA) is a classic degenerative chronic disease with significant symptoms, including pain, morning stiffness, and joint instability, leading to disability and ultimately impairing quality of life (Martel-Pelletier et al., 2016). The incidence of osteoarthritis is still high worldwide, with approximately 303.1 million hip and knee osteoarthritis cases, according to the Global Burden of Disease Project (GBD). As presented by Safari et al.'s analysis of GBD data up to 2017, the incidence of osteoarthritis has increased by approximately 8%–10% since 1990, which is based only on hip and knee osteoarthritis (Peat and Thomas, 2021). As a chronic disease, osteoarthritis impacts patients’ quality of life and burdens the country and society long-term. Although, at present, the subtypes of OA, risk factors, or etiologic factors, the mechanism of development cannot be revealed entirely clearly. The corresponding treatment methods are also under study. Osteoarthritis is characterized by cartilage degeneration, osteophyte formation, damage and remodeling of the cartilage, and varying degrees of synovitis and other joint structural damage, including ligaments and menisci. Identifying the molecular biological mechanisms of osteoarthritis development is essential to the treatment of osteoarthritis (Katz et al., 2021). The current mechanism of osteoarthritis development mainly focuses on changes in the structure of tissues and their functions in the joints, with the participation of various molecular biological processes and a variety of cells and cytokines (van den Bosch, 2021). The understanding of the molecular biological mechanism is constantly improving, including epigenetics as a new hot research direction in recent years. The osteoarthritis research process is increasingly often mentioned, of which noncoding RNA research is also increasingly in-depth, especially circRNA research (Ratneswaran and Kapoor, 2021). Osteoarthritis is an inflammatory disease produced by various factors, and circRNAs also participate in and regulate its progression (Zhang et al., 2021a). Circular RNA is currently one of the heated topics in research, and the purpose of this paper is to review the progress of its research in osteoarthritis and discuss its significance, breakthroughs, and deficiencies in current research and future research directions.
2 Introduction to CircRNA
A wide variety of noncoding RNAs are involved in osteoarthritis, including microRNAs, lncRNAs, and circRNAs. CircRNA is a long-chained and closed-loop RNA with better stability and a longer half-life due to its unique circular structure, which is more resistant to RNase R (Jeck and Sharpless, 2014), making it a potential candidate for diagnostic biomarkers and therapeutic targets. CircRNAs form by reverse splicing, wherein the 3′end of the exon is connected to its own or upstream exon’s 5′end through a 3'- 5′phosphate bond, forming a closed-loop structure with a reverse splicing connection site (Chen and Yang, 2015). They participate in a variety of physiological or pathological processes through a variety of mechanisms.
In terms of mechanism, the most common studies have focused on its molecular sponge as a microRNA, competitively inhibiting its host gene by affecting its intervention transcription function, also known as the competing endogenous RNAs (ceRNA) mechanism (Panda, 2018). An increasing number of studies have shown that its mechanisms and physiological functions are diverse (Chen, 2020a), such as regulating transcription and binding with its host gene to form an R-loop structure and upregulating the transcription process of skipping exons or intercepts (Conn et al., 2017), cooperating with the U1 snRNP junction for Pol II regulation of nuclear transcription (Li et al., 2015). The translation function of circRNA has received increasing attention in recent years, and circRNA with IRES structure can be used as a template for translation, translating the corresponding biologically functional peptide segment (Legnini et al., 2017). Moreover, circRNAs with ORFs are translated in a rolling circle manner, even up to a hundredfold linear translation, due to the property of their circularity (Abe et al., 2013). However, more studies have also found that its recognition by YTHDF3 after N6-methyladenosine (m6A) methylation, which recruits eIF4G2, can also enable circRNA translation (Di Timoteo et al., 2020). CircRNAs can also affect protein function by interacting with DNA or RNA-binding proteins (Du et al., 2017; Luo et al., 2019; Zhou et al., 2020a; Huang et al., 2020) or affect protein-to-protein interactions (Zhou et al., 2020a). (Figure 1)
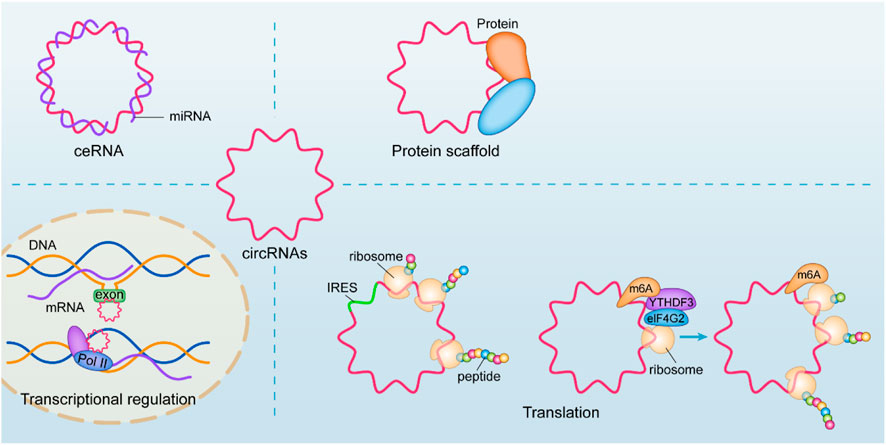
FIGURE 1. Biological functions of circRNAs. An increasing number of studies have shown that its mechanisms and physiological functions are diverse, such as regulating transcription and binding with its host gene to form an R-loop structure and upregulating the transcription process of skipping exons or intercepts, cooperating with the U1 snRNP junction for Pol II regulation of nuclear transcription. CircRNA with IRES structure can be used as a template for translation. More studies have also found that its recognition by YTHDF3 after m6A methylation, which recruits eIF4G2, can also enable circRNA translation. CircRNAs can also affect protein function by interacting with DNA or RNA-binding proteins or affect protein-to-protein interactions.
The metabolic mechanisms of circRNAs have also been clarified in recent years, including their upstream regulation and downstream metabolism (Xiao et al., 2020). CircRNAs are regulated by cis-acting elements and transcription factors (Ashwal-Fluss et al., 2014). The reverse repeated Alu sequence in the flank inclusion promotes exon circularization (Zhang et al., 2014). According to recent literature, N6-methyladenosine (m6A) controls the biogenesis of circRNA. Methyltransferase-like 3 (METTL3) or YTH domain 1 (YTHDC1) is reported to regulate circRNA (Di Timoteo et al., 2020). It has also been reported to be regulated by m6A concerning its transport out of the nucleus and degradation, methylated circRNA translocated into the cytosol through YTHDC1 recognition (Chen et al., 2019a), and its degradation through YTHDF2 (Park et al., 2019). Other degradation mechanisms, including RNase L, have also been reported to degrade circRNAs (Liu et al., 2019).
The physiopathological functions of circRNAs have been elucidated in a significant fraction of diseases as regulatory roles, involving almost all physiological processes in all organisms and regulating the function of cells and organs. Its clinical transformation has also been documented, and circRNAs can be used as biomarkers of liquid biopsies for the early diagnosis of diseases such as tumors and the evaluation of disease progression (Li et al., 2021a; Li et al., 2021b; Gui et al., 2021; Kuo et al., 2021; Zhang et al., 2022a; Kristensen et al., 2022).
3 CircRNAs in OA chondrocytes
Cartilage degeneration is a significant event in the development and progression of OA, and in this study, we summarize the various pathological processes in which circRNAs participate in cartilage degeneration.
3.1 CircRNAs in cartilage development and differentiation
CircRNAs have significantly different expression levels at different stages of developmental processes and promote the differentiation of bone marrow-derived stem cells or adipose-derived stem cells (Zhou et al., 2021a). In recent years, research on OA has also focused on bone and cartilage development and differentiation. In terms of noncoding RNAs, it has been well documented that miRNAs act as important regulators involved in cartilage development and differentiation. Especially in recent years, there has been a blowout increase in the study of miRNAs in cartilage differentiation, and many miRNAs have been identified to play a regulatory role in cartilage differentiation. miRNAs can also regulate mesenchymal stem cell differentiation by targeting the transcription of growth-related genes such as IHH, SOX5/6, and Sox9. (Iaquinta et al., 2021).
As a result, circRNAs, as noncoding RNAs closely related to miRNAs, are also likely to play a regulatory role in the development and differentiation of cartilage. CircPSM3 has been proven to regulate cartilage differentiation in cartilage and is upregulated in OA, targeting miRNA-296-5p. After detecting BMP2, BMP4, BMP6, and Runx2 at the mRNA and protein levels, it was found that high expression of miRNA-296-5p could effectively promote OA chondrocyte differentiation, while a miRNA-296-5p inhibitor could reverse the differentiation of OA chondrocytes promoted by si-circPSM3 (Ni et al., 2020). The recent study also showed that the circATRNL1/Sox9 pathway presented a clear high expression and a positive correlation with increased chondrogenesis in adipose mesenchymal stem cells and was regulated by miR-145-5p (Zhu et al., 2021a). CircNFIX/miR758-3p/KDM6A axis has also been reported as a possible target for regulating chondrogenesis (Liao et al., 2022). Furthermore, differentiation into proliferative chondrocytes may effectively increase the likelihood of cartilage repair, and studies on the roles of circRNAs in chondrogenesis may be necessary.
3.2 circRNA is involved in cartilage degeneration
Cartilage degeneration and loss as typical osteoarthritis and explicit pathophysiological changes are most studied in osteoarthritis research. Many studies have shown that circRNAs are involved in a variety of microRNA-regulated cartilage loss through ceRNA mechanisms, including apoptosis, proliferative function change, autophagic function change, inflammatory status, and degradation of the extracellular matrix of chondrocytes (Mao et al., 2021a). Although a considerable number of current studies confirm it, most of them appear too similar, making it quite difficult and necessary to identify critical circRNAs and pathways with regulatory functions. Nevertheless, an increasing number of studies have verified multifaceted functions, such as validating the simultaneous regulation of cartilage inflammation, chondrocyte apoptosis, and cartilage extracellular matrix degradation by circRNAs, which reflects that inflammation, apoptosis, and extracellular matrix degradation may be common in the terminal state of OA cartilage (Figure 2).
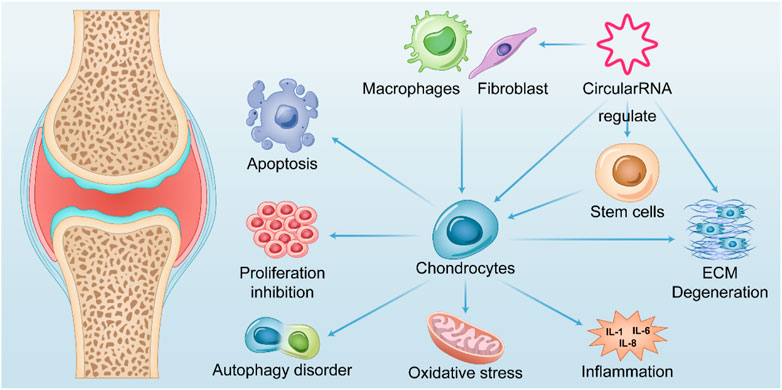
FIGURE 2. Summary of the role of CircRNAs in OA. CircularRNAs function in multiple tissues within the OA joint and their upregulation or downregulation in chondrocytes is associated with apoptosis, abnormal autophagy, impaired proliferation, oxidative stress, and cellular inflammatory status of chondrocytes, thereby mediating catabolism of extracellular matrix. Further exacerbates chondrocyte degeneration after losing the extracellular matrix environment. It also mediates fibroblast proliferation and polarization of macrophages in synoviocytes, leading to their release of inflammatory factors that exacerbate chondrocyte degeneration.
In this review, we summarize the existing research on the regulatory mechanisms of circRNAs in OA cartilage and present the results in Table 1. The mechanisms, pathways, biological functions, and animal models used in the research of circRNAs in OA progression are summarized. Considering that these studies all experimentally validated circRNAs and their targets for expression levels at the nucleic acid or protein level, we pooled and analyzed these circRNAs and their targets. We found that 42 circRNAs were downregulated and 61 upregulated in OA cartilage. After performing an analysis of circRNAs differentially expressed in OA, we found that the CircRNAs play a significant role in OA mainly through the following pathways and genes, including the classic Ras/MAPK pathway, PI3K/AKT pathway, TGF-β/SMAD path, JAK/STAT path, and FGF/ERK signal path. In addition, circRNAs in OA regulate SOX9 related cartilage differentiation pathway, BMPR2 represented osteoblastic differentiation pathway, CCDN1 represented cell cycle pathway, PON2 related oxidative stress pathway or directly regulate genes related to extracellular matrix metabolism such as TIMP3, MMP13 and ADAMTS5 related matrix metalloproteinase. Meanwhile, circRNAs also participate in the regulation of the expression of a considerable number of transcription factors like FOXO1 and KLF5 as well as ubiquitination related genes like FBXO21 and FBXW7, to affect the expression of downstream genes (Table 1).
3.2.1 Regulation of chondrocyte proliferation, apoptosis, and autophagy
In studies of cartilage degeneration loss, the status of chondrocytes is undoubtedly crucial. Involvement of circRNAs has been found in apoptotic pathways, changes in chondrocyte proliferation function, and changes in autophagy function (Jiang et al., 2020). A large number of studies have shown that circRNAs play a regulatory role in the apoptosis of chondrocytes (Table 1).
Almost all of the existing studies focus on the ceRNA mechanism. CircRNA regulates the expression of target genes through the competitive binding of miRNA, thus regulating the apoptosis of chondrocytes. Therefore, chondrocyte apoptosis and the weakening of proliferation should be considered the final outcome of cell fate. Targets for therapy should focus on more upstream pathways.
Reasonable autophagy often protects joint chondrocytes from further damage (Valenti et al., 2021). In osteoarthritic cartilage, however, chondrocyte autophagy disorders are also a significant cause of cartilage degeneration. CircRNAs significantly regulate autophagy in chondrocytes, and the ciRS-7/miR-7/PI3K/AKT/mTOR pathway (Zhou et al., 2020b), Hsa_circ_0005567/miR-495/ATG14 pathway (Zhang et al., 2020a), circRNA-MSR/miR-761/FBXO21 (Jia et al., 2022)and CircPan3/miR-667-5p/ghrelin (Zeng et al., 2021) have been shown to have regulatory effects in OA.
3.2.2 CircRNAs regulate the inflammatory state of chondrocytes
Osteoarthritis is an inflammatory disease. The inflammatory state of chondrocytes is naturally also considered in the study, and circRNA-mediated inflammatory processes also play an essential role. CircRNAs have a regulatory role in several inflammatory processes (Saaoud et al., 2021). In osteoarthritis models, upregulated or downregulated circRNAs have also been observed to impact the production and degradation of inflammatory factors. The main inflammatory factors regulated by circRNAs were IL-6/IL-8/TNF-α/IL-17, and IL-1β and TNF-α also induced most osteoarthritis in vitro cell models. In addition, macrophages within the joint environment have also been implicated in the inflammation of OA cartilage. CircRNAs have also been found to play a regulatory role in inducing macrophage polarization. One study showed that the expression of hsa_circ_0005567 in OA synovium is downregulated. Overexpression of hsa_circ_0005567 inhibits M1-type macrophage polarization and promotes M2-type macrophage polarization. After being treated with the supernatant of LPS-induced THP-1 macrophages, the proliferation of chondrocytes was significantly reduced, while the apoptosis rate was significantly increased. Hsa_circ_0005567 overexpression reversed this phenomenon. Mechanistically, hsa_circ_0005567 acts through the miR-492/SOCS2 axis to suppress M1 macrophage polarization and thereby mitigate chondrocyte apoptosis in OA cartilage (Zhang et al., 2021b). A large number of studies have shown that circRNAs play a regulatory role in the inflammation of chondrocytes (Table 1). These studies demonstrate that circRNAs can influence the fate of chondrocytes and their extracellular matrix through the involvement of miRNAs in the regulation of inflammatory factors.
3.2.3 Changes in the extracellular matrix of chondrocytes
Degradation of the extracellular matrix is an essential mechanism for the development of osteoarthritis and is also thought to be a characteristic phenotype of osteoarthritis. MMP1, MMP3, MMP13, aggrecanase, ADAMTS4, ADAMTS5, and cathepsins have been proven to be specific markers of matrix degradation. Currently, many studies have confirmed that circRNAs, by competitively inhibiting miRNAs, affect the function of their target genes, causing the composition of the extracellular mechanism to change, eventually leading to the occurrence and progression of osteoarthritis.
3.2.4 circRNA is involved in the oxidation stress process
Oxidative stress regulated by circRNAs has also been observed to be involved in several other mechanisms that cause damage to chondrocytes. YANG Y et al. found that circRSU1 in human chondrocytes affects downstream oxidative stress processes in osteoarthritis models through the miR-93-5p/MAP3K8 axis. Abnormally high expression of circRSU1 leads to the production of more ROS and the loss of cartilage extracellular matrix, leading to the occurrence and development of osteoarthritis (Yang et al., 2021). The role of circRNA in oxidative stress deserves attention, and Zhao found that in NASH (nonalcoholic fatty liver disease), mitochondrial circRNA-circSCAR reduces mitochondrial oxidative stress processes affecting the interaction between CypD and ATP5B. In the case of lipid exposure, circSCAR is regulated by endothelial mesh stress and PCG-1. After overexpression of circSCAR in mitochondria through mitochondrial-targeted nanograms in mice, oxidative stress in the NASH liver was significantly reversed, and liver function was improved (Zhao et al., 2020). This study reveals that circRNAs also play an essential regulatory role in metabolic diseases and mitochondrial function. At the same time, a significant proportion of patients with OA have significant metabolic syndrome, considering the possibility of metabolic osteoarthritis (Kuusalo et al., 2021). On the other hand, mitochondria also play an essential role in aging-induced degenerative diseases such as OA (Sun et al., 2020). CircRNAs have also been reported to play an essential role in mitochondrial stabilization in OA. SIRT3, an essential gene for mitochondrial function, significantly decreased expression in OA chondrocytes and was regulated by miR-505-3p. Overexpression of miR-505-3p can cause ROS to rise, and apoptosis of chondrocytes increases, while overexpression of CircFAM160A2 plays a therapeutic role. Both in vivo and in vitro experiments have demonstrated that CircFAM160A2 regulates chondrocyte mitochondrial stability and chondrocyte apoptosis through the miR-505-3p/SIRT3 axis (Bao et al., 2021). Recent studies have also demonstrated that CircHIPK3, through the mir-30a-3p/PON2 axis, regulates mitochondrial function, which in turn affects chondrocyte apoptosis (Shang et al., 2022). These findings suggest that our circRNA may also regulate mitochondria in OA, providing new research ideas.
3.2.5 circRNAs act as protein scaffolds in OA chondrocytes
It is clear that circRNAs’ rich mechanisms of action have been increasingly recognized in recent years, such as translation function and interactions with proteins, and in OA studies, little is known about mechanisms other than ceRNA. In a recent study, Shen et al. found that the reduction in circPDE4B in cartilage in OA patients was regulated by upstream FUS (an RNA-binding protein). The downregulation of circPDE4B resulted in the degradation of the extracellular matrix and decreased viability of chondrocytes. At the same time, AGO2 RIP was found to not function through the ceRNA mechanism. After cloning and amplification of its ORF sequence, no protein translated by circPDE4B was identified. Therefore, after RPD-MS and qRT–PCR verification, the authors found that RIC8 guanine-nucleotide exchange factor A (RIC8A) interacted with circPDE4B. Mass spectrometry has identified MID1 as an E3 ligase interacting with circPDE4B. After identifying the downstream pathway, the author screened out the p38/MAPK pathway. In medial meniscal enucleation models (DMM) model mice, overexpressed circPDE4B has also been proven to inhibit activating the RIC8A and p38/MAPK pathways and reverse the OA phenotype in mice, indicating that circPDE4B can be an effective molecular target drug for OA (Shen et al., 2021). Recent studies have shown that circNFKB1 regulates the expression of its host gene NFKB1 by interacting with the ENO1 protein (Tang et al., 2022a). A novel study found that circFOXO3 regulated the downstream PI3K/Akt pathway and affected chondrocyte autophagy through interaction with its parental gene FOXO3 (Zhao et al., 2022). In the future, more circRNA functions and mechanisms of action may be revealed in OA, which also suggests the possibility of circRNA as a molecularly targeted drug.
4 CircRNA is involved in the regulation of the environment within the joint
As a multi-component organization, osteoarthritis is a complex mechanism and an unclear chronic inflammatory disease, and changes in the joint environment are also essential in its development. Current studies of osteoarthritis also focus on other components of the joint, including synovium and subchondral bone, and abnormal vascular and neural factors in osteoarthritis (Ching et al., 2021; Zhang and Wen, 2021).
4.1 CircRNA is abnormally expressed in the synovium or infrapatellar fat pad of patients with osteoarthritis
The role of the synovium is not negligible in the development of osteoarthritis. Synovitis is often observed in the OA joint. Both synovial hyperplasia and the secretion of proinflammatory factors drive the progression of osteoarthritis (van den Bosch et al., 2020). MRI and ultrasound have identified a positive correlation between the risk of structural progression of synovial and osteoarthritis and joint symptoms (Roemer et al., 2011). Shuai et al. performed circRNA sequencing of OA synovial samples and controls and identified 122 circRNAs differentially expressed in osteoarthritic synovium. Using GO analysis and KEGG enrichment analysis, differentially expressed circRNAs were rich in adhesion molecules, tumor pathways, TGF-β, and some osteoarthritis pathways, such as Hippo and WNT pathways. This article also establishes the circRNA-miRNA network, exploring possible molecular regulation mechanisms for specific expression of circRNAs, and the miR-20, miR-29, and miR-136 families have all been reported in previous OA studies and interact with several differentially expressed circRNAs (Xiang et al., 2019). On the other hand, the infrapatellar fat pad and the surrounding synovium are also essential tissues involved in regulating the intra-articular environment. Circular RNA expression profiles of the infrapatellar fat pad/synovium unit reveal that hsa_circ_0005265 was down-expressed in both OA synovium and IPFP, targeting hsa-miR-6769b-5p and hsa-miR-1249-5p (Jiang et al., 2021a). However, the molecular mechanisms of differentially expressed circRNAs in OA synovium or infrapatellar fat pad in the progression of OA have not been thoroughly investigate.
4.2 Abnormal expression of circRNA in osteoclasts of osteoarthritis
Changes in subchondral bone, especially the imbalance between rupture and remodeling, are also essential mechanisms for the development of osteoarthritis (Fan et al., 2021). In such a process, osteoclasts play a central role (Zhu et al., 2020). Dou C and others first identified the differential expression of circRNAs in abnormally activated osteoclasts. Many differentially expressed circRNAs were identified in osteoclasts and activated osteoclasts, suggesting that circRNA may also be extensively involved in remodeling subchondral bone. In another study, the expression of circRNA in bone marrow stromal cells during RANKL- and CSF1-stimulated osteoclast formation was sequenced, and their difference was analysed. This study focuses on the role of circRNA-28313 in bone marrow osteoclast differentiation. The results showed that circRNA-28313 knockout inhibited the differentiation of bone marrow mesenchymal stem cells and RANKL-induced osteoclasts and partially prevented the bone loss induced by ovariectomy (OVX). Further downstream experiments further found that circRNA-28313 alleviated miR-195a-mediated inhibition of CSF1 through a ceRNA mechanism, thereby regulating osteoclast differentiation (Chen et al., 2019b). The destruction of subchondral bone in OA is related to bone destruction between OA and osteoporosis, and RANKL was also identified in OA (Kovacs et al., 2019). These studies also indicate that circRNA is more likely to participate in the pathogenesis of osteoarthritis.
4.3 Abnormal expression of circRNA in osteoarthritis meniscus
Meniscal degeneration and wear are also prevalent in the knee tissue of OA patients, and a large number of OA model animals have used medial meniscal enucleation models to mimic OA (Zaki et al., 2021). Few studies have focused on the mechanism of meniscal changes in OA, but the meniscus plays an essential role in the stability of joints and the protection of articular cartilage in the function of the knee joint. Bin Wang et al. performed bioinformatics analysis and prediction using public databases (GEOs) and revealed 360 differentially expressed genes in the OA meniscus. It is expected that hsa_circ_0025119, hsa_circ_0025113, hsa_circ_0009897, and hsa_circ_0002447 are the most critical circRNAs. The article failed to verify and identify the expression and function of circRNA in subsequent experiments, it also suggested that circRNA might also play a regulatory role in the OA meniscus (Wang et al., 2020a). Meanwhile, The hsa_ circ_ 0018069/mir-147b-3p/tjp2 axis was also found to play a regulatory role in the OA meniscus (Jiang et al., 2021b).
5 Upstream regulation and downstream metabolism of circRNAs in OA
The reason for the change in circRNA in OA is nothing more than the increase and decrease in metabolism. The metabolism and upstream regulation of circRNAs have also been hot topics of research, with clear evidence that circRNAs are regulated by transcription factors. In OA studies, there has not been much research on the upstream regulatory factors of circRNAs in osteoarthritis. Wang et al. found that LEF1, as a transcription regulator, affects the expression changes of downstream circRNF121, regulates the miR-665/MYD88/NF-Кb pathway, and ultimately regulates the apoptosis and proliferation of chondrocytes and the metabolism of extracellular matrix (Wang et al., 2020b). RNA binding proteins regulate back splicing mainly by directly bridging distal splice sites and by binding to intronic complementary sequence (ICS) (Chen, 2020a). RNA binding proteins including QKI, HNRNPL, Mbl, SLU7, NF110, NF110, DHX9, ADAR1 were reported to potentially regulate back splicing of circRNAs (Ashwal-Fluss et al., 2014; Conn et al., 2015; Ivanov et al., 2015; Aktas et al., 2017; Li et al., 2017a). Some of them, such as QKI and DHX9, were reported to be differentially expressed in osteoarthritic tissues as well (Li et al., 2016; Tang et al., 2022b), which may also be one of the upstream mechanisms regulating expression of circRNAs in osteoarthritis. However, another possible change in the expression of circRNA in OA is its downstream metabolic changes. Changes in downstream metabolic clearance have rarely been mentioned in OA studies. With a further understanding of the circRNA metabolic pathway, this may be one of the follow-up research directions.
6 The role of circRNA in OA of different etiologies and stages
6.1 circRNAs in different subtypes of OA
In fact, from a clinical point of view, some risk factors for osteoarthritis should also be taken into account. OA is a highly heterogeneous disease, and different drivers tend to shape different OA phenotypes (Van Spil et al., 2019). In existing circRNA-related studies, different types of osteoarthritis caused by different factors have not yet been considered by most researchers. However, some factors were considered, such as the specimen selection of load-bearing stress areas in osteoarthritis cartilage compared with non-load-bearing stress areas. Some studies selected chondrocytes from load-bearing versus non-load-bearing areas to identify differential expression. The etiology of osteoarthritis in the clinic is diverse and complex, and it has been established as a risk factor for several conditions: obesity, physical activity, structural factors, and genetics (Martel-Pelletier et al., 2016). Patients with OA of different etiologies or etiological factors should be treated separately. Therefore, studies should be more refined regarding OA of different etiologies, and corresponding studies aimed at different etiological subtypes of OA, such as RNA sequencing and identification of differential expression in patients with distinct metabolic profiles or those with distinct stress factors, may also yield different results. The construction of different OA models usually simulates different OA initiating factors, especially the design of animal models (Zaki et al., 2021). Most research on OA circRNAs is anterior cruciate ligament transection (ACLT) and medial meniscal enucleation models (DMM) models in rats and mice. These two models can simulate the initiating factors of traumatic osteoarthritis. More pathogenetic factors for OA should be considered. Designing different animal models may be a solution to this problem. Additionally, molecular subtype of OA has been mentioned by more and more researchers. Julia Steinberg et al. conducted a cluster analysis of mRNA sequencing data obtained from cartilage and synovium samples of OA patients. They discovered two subgroups in the synovium, one related to inflammation and the other related to extracellular matrix metabolism. High-grade inflammation of cartilage was also linked to female gender and proton pump inhibitor use (Steinberg et al., 2021). In another study, researchers from China sequenced cartilage samples from 131 OA patients and carried out a cluster analysis of their expression profiles. The patients were divided into four subtypes: a subtype characterized by glycosaminoglycan metabolism disorder, a subtype marked by collagen metabolism disorder, a subtype with sensory neuron activation, and an inflammation subtype (Yuan et al., 2020). Biochemical markers have also been employed to cluster and classify patients into three groups, C1, C2, and C3. C1 is associated with low tissue turnover, including low repair and turnover of articular cartilage and subchondral bone. C2 is characterized by structural damage, such as high bone formation and resorption and cartilage degeneration. C3 is linked to systemic inflammation, joint tissue degeneration, and cartilage degeneration. In the FNIH/OAI cohort, C1 had the highest proportion of progressors, C2 was linked to the progression of mechanical structure, and C3 was associated with pain, which was consistent with molecular typing (Angelini et al., 2022). Future research can construct circRNA expression profiles of patients with different OA subtypes and conduct in-depth exploration of the relevant mechanisms.
6.2 circRNAs in different stages of OA
Another important aspect is that OA, as a chronic disease, has a significantly long pathological process, and different stages of OA have different characteristics. Sun et al. (2011) measured the miRNA expression profiles of rat articular cartilage at different developmental stages and sequenced femoral head cartilage on zeroth days, twenty-first days, and forty-second days after birth. The authors observed in the results that, on the one hand, some miRNA clusters were continuously expressed at three stages, which may illustrate the existence of specific conserved sequences of miRNAs during development. On the other hand, the expression of some miRNAs changed obviously at different stages, and in the sequencing results combined with PCR validation, it could be found that some miRNAs showed early high expression. In contrast, some showed high expression at later stages of developmen. This study reveals the phased change in the miRNA expression profile at different stages of development. S. A. Ali et al. classified 91 OA patients into early OA (n = 41) and late OA groups (n = 50) using K-L grading of standard X-ray, performed next-generation sequencing on plasma samples from patients, and identified that hsa-mir-335-3p, hsa-mir-199a-5p, hsa-mir-671-3p, hsa-mir-1260b, hsa-mir-191-3p, hsa-mir-335-5p, and hsa-mir-543 were more highly expressed in early OA compared to late OA, while hsa-mir-193b-5p, hsa-mir-193a-5p, and hsa-mir-455-5p were more highly expressed in late OA. Meanwhile, the authors also identified some novel miRNAs, some of which showed high expression in early OA, some of which showed high expression in late OA, and others that showed high expression in both early and advanced stages (Ali et al., 2020). This result illustrated that the expression of miRNAs might change continuously with the progression of OA. Therefore, we can infer that in the different stages of the disease, such as early and late OA, the characteristic circRNA expression profile in the determination stage may bring different results. Determination of circRNA expression profiles based on the patient’s symptoms, signs, functional assessment such as WOMAC or KOOS score of the knee (Roos and Toksvig-Larsen, 2003), and imaging grading such as Kellgren-Lawrence grade to stratify different patients may be considered, which may help us to understand the role of circRNAs in OA more deeply.
7 Clinical application and potential of circRNA
7.1 circRNA is the marker of a liquid biopsy
CircRNAs are less susceptible to degradation by RNase R due to their lack of 3′and 5′ends, which confers a longer half-life and stability than other noncoding RNAs or mRNAs, suggesting the possibility of circRNAs as indicators for clinical testing. CircRNA as a liquid biopsy indicator has been frequently mentioned in tumor studies (Li et al., 2022a; Wang et al., 2022a; Ruan et al., 2022; Xue et al., 2022). CircRNA was detected in cartilage, synovial fluid, and serum in OA patients and was different from the control group, suggesting that it is an indicator of early diagnosis of osteoarthritis. Chen C et al. measured hsa_circ_101178 levels in serum and fluid and control group serum and fluid in OA patients and found that group OA was significantly higher than the control group. Meanwhile, there was a positive correlation between circ-101178 levels in serum and synovial fluid. In addition, serum hsa_circ_101178 was positively correlated with OA’s KL score and WOMAC pain score (Chen, 2020b). Fangyu et al. identified five circRNAs in the synovial fluid of OA patients who were significantly elevated compared with healthy controls. AUC analysis of diagnostic value found that hsa_circ_0104873, hsa_circ_0104595, and hsa_circ_0101251 can effectively distinguish between OA patients and healthy controls. However, it was also found that three circRNAs were positively correlated with the radiological grade and symptom severity of OA patients (Yu et al., 2018). Similarly, Ying Wang et al. also identified that circ-0032131 levels in the peripheral blood of OA patients were significantly different from those in the healthy population (Wang et al., 2019). Additional studies have also reported that plasma circRNA-016901 can effectively distinguish osteoarthritis from rheumatoid arthritis and is correlated with disease severity (Du et al., 2022). Although these studies failed to carry out clinical trials to prove the diagnostic value of circRNA, they also suggested, to some extent, the possibility of circRNA as a molecular marker for the diagnosis of osteoarthritis.
7.2 Clinical transformation potential of circRNA
The study of circRNAs is constantly moving from basic research to clinical translation, and its clinical potential has been explored as it has become more aware of its mechanisms and functions. For example, several animal trials have demonstrated that intraarticular injection of AVV or AV carried shRNA or plasmid targeting circRNAs, and silencing or overexpressing osteoarthritis-associated circRNAs in articular cartilage can effectively alleviate the progression of osteoarthritis in animal models. Treatment with intra-articular injection of sodium hyaluronate and glucocorticoids is widely used in the clinic (Yuan et al., 2022). However, it remains controversial. The development of precision medicine approaches targeting specific nucleic acid drugs within OA joints, combined with individual sequencing results, may lead to better outcomes and personalized treatment options. While circRNAs are stable and are not easily degraded by RNase, the potential for translation and interaction with proteins may make them suitable carriers for nucleic acid drugs (Figure 3).
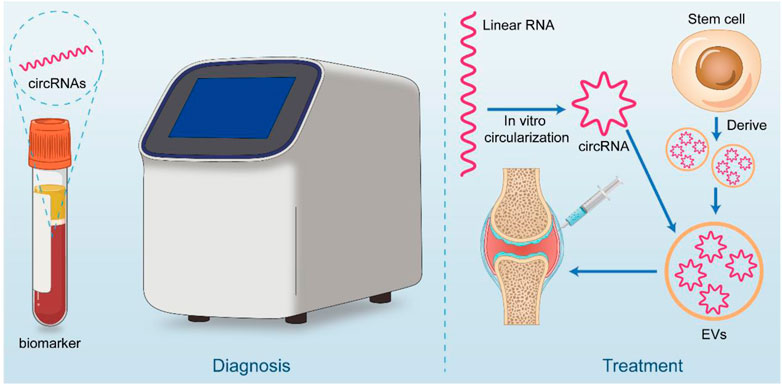
FIGURE 3. Clinical application potential of circRNA. CircRNA has great potential for both diagnostic and therapeutic applications. In diagnosis, it can be used as a liquid biopsy tool for early screening of OA patients. On the therapeutic side, circRNA is considered to be good for treatment by in vitro synthesis (linear RNA cyclization) or stem cell-derived circRNA piggybacked by materials such as extracellular vesicles and injected through the joint cavity.
7.2.1 Extracellular vesicles loaded with circRNA
Extracellular vesicles are also a significant research hotspot, and extracellular vesicles have been suggested to be promising drug carriers, particularly nucleic acid drugs. Extracellular vesicles (EVs) are small vesicles released from different cells into the extracellular matrix, classified by origin and size, and include three subtypes: 1. Apoptotic bodies (500 nm—5 μm); 2. Microvesicles (150–500 nm); 3. Exosomes (40–150 nm), which can participate in intercellular communication (Wu et al., 2022). Exosomes are a subset of EVs secreted by most cells and have good biocompatibility, low toxicity and immunogenicity as well as great designability. They have received extensive attention over the past decades as therapeutic carriers and diagnostic markers. Circular RNAs naturally carried by exosomes have been widely used in the treatment of cancer, cardiovascular, and metabolic diseases (Zhang et al., 2022b).
In the treatment of OA, several studies have proposed that extracellular vesicles may be used as carriers to loading on specific circRNAs and thereby exert their therapeutic effects on OA. As an example, Songlong Li et al. isolated extracellular vesicles from MSCs and observed an increase in COL2A1, Sox9, and aggrecan expression after coculturing circHIPK3-overexpressing extracellular vesicles with chondrocytes, along with inhibited expression of OA-related markers such as MMP-13 and Runx2. Meanwhile, functional experiments also found that CircHIPK3 could alleviate IL-1β-induced inhibition of chondrocyte apoptosis, proliferation, and migration (Li et al., 2021c). Shi Cong Tao et al. found that circRNA3503 was significantly increased after melatonin (MT)-induced cellular sleep. Mechanistically, circRNA3503 acted as a sponge for hsa-mir-181c-3p and hsa-let-7b-3p. Prepared and isolated circRNA3503-loaded extracellular vesicles (circRNA3503-OE-SEVs) from SMSCs. The feasibility of circRNA3503-OE-SEVs in preventing OA progression was validated by in vivo and in vitro experiments (Tao et al., 2021). In addition, circRNAs transported by exosomes (circ_0001236) suppresses IL-1 β via the miR-3677-3p/Sox9 pathway in TGF-β induced cartilage degeneration, promoting chondrocyte proliferation (Mao et al., 2021b).
7.2.2 mRNA therapeutics
mRNA could theoretically produce any protein on demand inside the cell, enabling treatment of disease. However, linear mRNAs also face many challenges due to their own defects. CircRNAs do not contain a 5′- end cap and a 3'- end PolyA tail and form a covalently closed circular structure by back splicing, protecting circRNAs from degradation by exonucleases. At the same time, circRNAs also need no complex modification when they are synthesized in vitro, so circRNAs have the advantages of high stability, low immunogenicity, and long-lasting expression compared to linear mRNA (Santer et al., 2019). Recent studies identified nearly a thousand endogenous circular RNAs that are translatable, half of which can synthesize large molecular weight proteins by rolling circle translation. The authors explore factors that influence translation of circular RNAs and, by optimizing relevant conditions, increase circular RNA protein production several hundred fold, providing a more abundant and long-lasting translated protein product under in vitro and in vivo conditions (Fan et al., 2022).
In vitro synthesis of circRNAs is the basis of drug development, and there have been studies that successfully prepared circRNAs in vitro and elucidated their functions. There are currently two major routes known for the in vitro synthesis of circular RNAs: direct intramolecular ligation into circles based on catalysis by T4 RNA ligase, and self splicing into circles based on type I intronic ribozymes (T4 bacteriophage or Anabaena) (Chen et al., 2017; Wesselhoeft et al., 2019; Rausch et al., 2021; Qu et al., 2022). There are studies showing that circular RNA synthesized by T4 RNA ligase does not elicit an intracellular innate immune response (Liu et al., 2022a). It provides an important foundation for the further application of circular RNAs synthesized in vitro and also holds promising prospects for the further development of nucleic acid aptamers and gene therapy fields based on circular RNA technology. Artificially manufactured circular RNA technology has also been successfully applied in drug development. CircINSR was screened for significant underexpression in heart tissue from patients with heart failure and mice with left ventricular pressure overload induced cardiac remodeling. The authors explored the therapeutic effect using two modalities: AAV loading overexpression plasmid and in vitro transcribed CircINSR, and found that in vitro transcribed CircINSR could achieve superior therapeutic and protective effects against doxorubicin mediated cardiomyocyte death (Lu et al., 2022).
8 Prospect of research
At present, there are a considerable number of studies on the function of circRNAs in osteoarthritis, which fully shows that circRNAs play a regulatory role in OA. However, there are still many deficiencies in the existing research. For example, mechanistic research is limited to acting as a sponge combined with miRNA, which fails to consider the characteristics of diseases, such as OA of different types and stages. However, the potential of circRNAs has been confirmed in many other areas, giving us some directions for studying the role of circRNAs in OA.
Many studies have confirmed the critical role of miRNA in osteoarthritis, and miRNA as a molecular target for precision therapy has also been proven feasible (Ji et al., 2020). CircRNA is an essential competitive inhibitor of miRNA, and its high degree of conservatism and stability makes it possible to fight OA effectively. However, the richness of the mechanism of action of circRNA has been described earlier. The mechanism of circRNA should not be limited to ceRNA. In comparison, only a few articles illustrate its function in OA through interactions with proteins. With the continuous progress of sequencing technology, many studies have shown that circRNAs have the potential for translation and can regulate transcription. Breakthrough progress has been made in the study of circRNA translation products as disease-specific molecular targets in cancer. In triple-negative breast carcinoma, circ-HER2 is expressed in approximately 30% of triple-negative breast carcinoma cells. The translated peptide HER2-103 promotes the proliferation and invasion of triple-negative breast cancer cells, and HER2-103 can also be used as a target molecule for the anti-HER2 targeted drug pertuzumab (Li et al., 2020). In addition, many studies have confirmed the critical role of circRNA translation products in disease development (Zhou et al., 2021b). In addition, a database on circRNA translation has been established. However, up to now, no study of osteoarthritis has found a translational function of circRNA, which may be due to the previous researchers’ lack of in-depth understanding of circRNA, and there are good reasons why we can assume that circRNA in OA may also have the translational function to regulate the progress of the disease.
At present, research on circRNAs in OA has also been performed in a considerable number of experimental animals. As shown in Table 1, animal models used extensively in existing studies are rats, mice, and rabbits. The knockout or overexpression of circRNA was achieved by intra-articular injection of adenovirus (AV), adeno-associated virus (AAV), or lentivirus. However, because of the unique and complex structure of the articular cavity, intra-articular injections can be well absorbed or unknown by articular cartilage and surrounding tissues. Therefore, the effect of such knockout or overexpression in animal models has not been effectively verified, setting obstacles to the development of drug targets and drug safety. All of these findings demonstrate the importance of establishing a circRNA knockout model. Methods to knock out circRNA have always been a challenge. In the past, such knockdowns may have been limited by technical reasons, but the emergence and advancement of new technologies have made it possible to achieve a specific knockout of circRNA (He et al., 2021). Benyu Liu et al. used CRISPR-CAS9 to build circKcnt2 knockout mice to study the role of circRNA in intestinal inflammation. The knockout site was selected as a intronic complementary sequences mediated by flanking introns, which is thought to be the critical sequence for back splicing of circRNAs. circKcnt2 knockout was constructed by deleting the intronic complementary sequences of the genome. Analysis of the expression of circKcnt2 and its host genes in knockout models proved that circKnct2 could be knocked out specifically, while the expression of its host genes was not affected (Liu et al., 2020). The experiment successfully constructed the circRNA knockout model and demonstrated the function of circRNA. Furthermore, a circRNA knockout model was established in OA to improve the current understanding.
In conclusion, circRNA may function as a regulator in OA. A growing body of evidence suggests that circRNAs can regulate chondrocyte proliferation, apoptosis, differentiation, and autophagy, regulate extracellular matrix degradation, and regulate oxidative stress processes and inflammatory processes in chondrocytes. On the other hand, circRNAs can modulate the intra-articular environment, such as the synovium, meniscus, and subchondral bone, and can serve as biomarkers for liquid biopsy. Although the number of studies available is already significant, many deficiencies remain regarding mechanisms, animal model construction, and disease heterogeneity.
Author contributions
JL supervised and was responsible for the whole study. ZL was responsible for the literature search and data analysis and wrote the first draft., drawing figures and tables. All authors revised the paper critically for intellectual content and approved the final version.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abe, N., Hiroshima, M., Maruyama, H., Nakashima, Y., Nakano, Y., Matsuda, A., et al. (2013). Rolling circle amplification in a prokaryotic translation system using small circular RNA. Angew. Chem. Int. Ed. Engl. 52, 7004–7008. doi:10.1002/anie.201302044
Aktas, T., Avsar Ilik, I., Maticzka, D., Bhardwaj, V., Pessoa Rodrigues, C., Mittler, G., et al. (2017). DHX9 suppresses RNA processing defects originating from the Alu invasion of the human genome. Nature 544, 115–119. doi:10.1038/nature21715
Ali, S. A., Gandhi, R., Potla, P., Keshavarzi, S., Espin-Garcia, O., Shestopaloff, K., et al. (2020). Sequencing identifies a distinct signature of circulating microRNAs in early radiographic knee osteoarthritis. Osteoarthr. Cartil. 28, 1471–1481. doi:10.1016/j.joca.2020.07.003
Angelini, F., Widera, P., Mobasheri, A., Blair, J., Struglics, A., Uebelhoer, M., et al. (2022). Osteoarthritis endotype discovery via clustering of biochemical marker data. Ann. Rheum. Dis. 81, 666–675. doi:10.1136/annrheumdis-2021-221763
Apizi, X., Talifujiang, D., Kasimu, A., Zhang, X., Yiming, A., Ma, X., et al. (2021). Circular RNA mmu_circ_0001598 contributes to IL-1β-induced osteoarthritis progression by regulating miR-127-3p. J. Healthc. Eng. 2021, 2793379. doi:10.1155/2021/2793379
Ashwal-Fluss, R., Meyer, M., Pamudurti, N. R., Ivanov, A., Bartok, O., Hanan, M., et al. (2014). circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell 56, 55–66. doi:10.1016/j.molcel.2014.08.019
Bai, Z. M., Kang, M. M., Zhou, X. F., and Wang, D. (2020). CircTMBIM6 promotes osteoarthritis-induced chondrocyte extracellular matrix degradation via miR-27a/MMP13 axis. Eur. Rev. Med. Pharmacol. Sci. 24, 7927–7936. doi:10.26355/eurrev_202008_22475
Bao, J., Lin, C., Zhou, X., Ma, D., Ge, L., Xu, K., et al. (2021). circFAM160A2 promotes mitochondrial stabilization and apoptosis reduction in osteoarthritis chondrocytes by targeting miR-505-3p and SIRT3. Oxid. Med. Cell Longev. 2021, 5712280. doi:10.1155/2021/5712280
Chen, L. L., and Yang, L. (2015). Regulation of circRNA biogenesis. RNA Biol. 12, 381–388. doi:10.1080/15476286.2015.1020271
Chen, Y. G., Kim, M. V., Chen, X., Batista, P. J., Aoyama, S., Wilusz, J. E., et al. (2017). Sensing self and foreign circular RNAs by intron identity. Mol. Cell 67, 228–238. doi:10.1016/j.molcel.2017.05.022
Chen, R. X., Chen, X., Xia, L. P., Zhang, J. X., Pan, Z. Z., Ma, X. D., et al. (2019). N(6)-methyladenosine modification of circNSUN2 facilitates cytoplasmic export and stabilizes HMGA2 to promote colorectal liver metastasis. Nat. Commun. 10, 4695. doi:10.1038/s41467-019-12651-2
Chen, X., Ouyang, Z., Shen, Y., Liu, B., Zhang, Q., Wan, L., et al. (2019). CircRNA_28313/miR-195a/CSF1 axis modulates osteoclast differentiation to affect OVX-induced bone absorption in mice. RNA Biol. 16, 1249–1262. doi:10.1080/15476286.2019.1624470
Chen, C., Yin, P., Hu, S., Sun, X., and Li, B. (2020). Circular RNA-9119 protects IL-1β-treated chondrocytes from apoptosis in an osteoarthritis cell model by intercepting the microRNA-26a/PTEN axis. Life Sci. 256, 117924. doi:10.1016/j.lfs.2020.117924
Chen, G., Liu, T., Yu, B., Wang, B., and Peng, Q. (2020). CircRNA-UBE2G1 regulates LPS-induced osteoarthritis through miR-373/HIF-1a axis. Cell Cycle 19, 1696–1705. doi:10.1080/15384101.2020.1772545
Chen, L. L. (2020). The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat. Rev. Mol. Cell Biol. 21, 475–490. doi:10.1038/s41580-020-0243-y
Chen, C. (2020). Serum hsa_circ_101178 as a potential biomarker for early prediction of osteoarthritis. Clin. Lab. 66. doi:10.7754/Clin.Lab.2020.191251
Cheng, S., Nie, Z., Cao, J., and Peng, H. (2022). Circ_0136474 promotes the progression of osteoarthritis by sponging mir-140-3p and upregulating MECP2. J. Mol. Histol. 10, 1–12. doi:10.1007/s10735-022-10100-x
Ching, K., Houard, X., Berenbaum, F., and Wen, C. (2021). Hypertension meets osteoarthritis - revisiting the vascular aetiology hypothesis. Nat. Rev. Rheumatol. 17, 533–549. doi:10.1038/s41584-021-00650-x
Conn, S. J., Pillman, K. A., Toubia, J., Conn, V. M., Salmanidis, M., Phillips, C. A., et al. (2015). The RNA binding protein quaking regulates formation of circRNAs. Cell 160, 1125–1134. doi:10.1016/j.cell.2015.02.014
Conn, V. M., Hugouvieux, V., Nayak, A., Conos, S. A., Capovilla, G., Cildir, G., et al. (2017). A circRNA from SEPALLATA3 regulates splicing of its cognate mRNA through R-loop formation. Nat. Plants 3, 17053. doi:10.1038/nplants.2017.53
Di Timoteo, G., Dattilo, D., Centron-Broco, A., Colantoni, A., Guarnacci, M., Rossi, F., et al. (2020). Modulation of circRNA metabolism by m(6)A modification. Cell Rep. 31, 107641. doi:10.1016/j.celrep.2020.107641
Ding, R., Zhou, J., Xu, J., Lu, H., Zhang, T., Xiang, X., et al. (2021). Circ_0045714/miR-331-3p interaction affects IL-1β-evoked human articular chondrocyte injury through regulating PIK3R3 in a ceRNA regulatory cascade. J. Orthop. Surg. Res. 16, 595. doi:10.1186/s13018-021-02738-2
Du, W. W., Zhang, C., Yang, W., Yong, T., Awan, F. M., and Yang, B. B. (2017). Identifying and characterizing circRNA-protein interaction. Theranostics 7, 4183–4191. doi:10.7150/thno.21299
Du, M., Fan, S., Liu, Y., Hao, Y., and Guo, J. (2022). The application of circRNA-016901 in improving the diagnostic accuracy of osteoarthritis. Biomed. Res. Int. 2022, 1158562. doi:10.1155/2022/1158562
Fan, X., Wu, X., Crawford, R., Xiao, Y., and Prasadam, I. (2021). Macro, micro, and molecular. Changes of the osteochondral interface in osteoarthritis development. Front. Cell Dev. Biol. 9, 659654. doi:10.3389/fcell.2021.659654
Fan, X., Yang, Y., Chen, C., and Wang, Z. (2022). Pervasive translation of circular RNAs driven by short IRES-like elements. Nat. Commun. 13, 3751. doi:10.1038/s41467-022-31327-y
Feng, M., Jing, L., Cheng, J., An, S., Huang, J., and Yan, Q. (2021). Circ_0020093 ameliorates IL-1β-induced apoptosis and extracellular matrix degradation of human chondrocytes by upregulating SPRY1 via targeting miR-23b. Mol. Cell Biochem. 476, 3623–3633. doi:10.1007/s11010-021-04186-2
Fu, Q., Li, L., Wang, B., Wu, J., Li, H., Han, Y., et al. (2021). CircADAMTS6/miR-431-5p axis regulate interleukin-1β induced chondrocyte apoptosis. J. Gene Med. 23, e3304. doi:10.1002/jgm.3304
Fu, G., Yin, F., and Zhao, J. (2022). Depletion of circ_0128846 ameliorates interleukin-1β-induced human chondrocyte apoptosis and inflammation through the miR-940/PTPN12 pathway. Int. Immunopharmacol. 110, 108996. doi:10.1016/j.intimp.2022.108996
Gao, L., Wang, X., Xiong, J., and Ma, Y. (2022). Circular RNA from phosphodiesterase 4D can attenuate chondrocyte apoptosis and matrix degradation under OA milieu induced by IL-1β via circPDE4D/miR-4306/SOX9 Cascade. Immunopharmacol. Immunotoxicol. 44, 682–692. doi:10.1080/08923973.2022.2077215
Gui, C. P., Liao, B., Luo, C. G., Chen, Y. H., Tan, L., Tang, Y. M., et al. (2021). circCHST15 is a novel prognostic biomarker that promotes clear cell renal cell carcinoma cell proliferation and metastasis through the miR-125a-5p/EIF4EBP1 axis. Mol. Cancer 20, 169. doi:10.1186/s12943-021-01449-w
Guo, Z., Wang, H., Zhao, F., Liu, M., Wang, F., Kang, M., et al. (2021). Exosomal circ-BRWD1 contributes to osteoarthritis development through the modulation of miR-1277/TRAF6 axis. Arthritis Res. Ther. 23, 159. doi:10.1186/s13075-021-02541-8
He, A. T., Liu, J., Li, F., and Yang, B. B. (2021). Targeting circular RNAs as a therapeutic approach: Current strategies and challenges. Signal Transduct. Target Ther. 6, 185. doi:10.1038/s41392-021-00569-5
He, Y., Zhou, F., and Cheng, X. (2022). CircKMT2E participates in osteoarthritis through promotes apoptosis of chondrocytes via sponging miR-140-5p to activate TLR4. Evid. Based Complement. Altern. Med. 2022, 2957844. doi:10.1155/2022/2957844
He, M., Jia, Z., Wen, Y., and Chen, X. (2022). Circ_0043947 contributes to interleukin 1β-induced injury in chondrocytes by sponging miR-671-5p to up-regulate RTN3 expression in osteoarthritis pathology. J. Orthop. Surg. Res. 17, 177. doi:10.1186/s13018-022-02970-4
Huang, A., Zheng, H., Wu, Z., Chen, M., and Huang, Y. (2020). Circular RNA-protein interactions: Functions, mechanisms, and identification. Theranostics 10, 3503–3517. doi:10.7150/thno.42174
Huang, Z., Ma, W., Xiao, J., Dai, X., and Ling, W. (2021). CircRNA_0092516 regulates chondrocyte proliferation and apoptosis in osteoarthritis through the miR-337-3p/PTEN axis. J. Biochem. 169, 467–475. doi:10.1093/jb/mvaa119
Iaquinta, M. R., Lanzillotti, C., Mazziotta, C., Bononi, I., Frontini, F., Mazzoni, E., et al. (2021). The role of microRNAs in the osteogenic and chondrogenic differentiation of mesenchymal stem cells and bone pathologies. Theranostics 11, 6573–6591. doi:10.7150/thno.55664
Ivanov, A., Memczak, S., Wyler, E., Torti, F., Porath, H. T., Orejuela, M. R., et al. (2015). Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. 10, 170–177. doi:10.1016/j.celrep.2014.12.019
Jeck, W. R., and Sharpless, N. E. (2014). Detecting and characterizing circular RNAs. Nat. Biotechnol. 32, 453–461. doi:10.1038/nbt.2890
Ji, M. L., Jiang, H., Wu, F., Geng, R., Ya, L. K., Lin, Y. C., et al. (2020). Precise targeting of miR-141/200c cluster in chondrocytes attenuates osteoarthritis development. Ann. Rheum. Dis. 80, 356–366. doi:10.1136/annrheumdis-2020-218469
Jia, Z., and Wei, Q. J. (2021). CircRNA-MSR regulates LPS-induced C28/I2 chondrocyte injury through miR-643/MAP2K6 signaling pathway. Cartilage 13, 785S. doi:10.1177/19476035211044826
Jia, Z., Liu, J., and Wang, J. (2022). circRNA-MSR regulates the expression of FBXO21 to inhibit chondrocyte autophagy by targeting miR-761 in osteoarthritis. Kaohsiung J. Med. Sci. 10, 1168–1177. doi:10.1002/kjm2.12604
Jiang, S., Liu, Y., Xu, B., Zhang, Y., and Yang, M. (2020). Noncoding RNAs: New regulatory code in chondrocyte apoptosis and autophagy. Wiley Interdiscip. Rev. RNA 11, e1584. doi:10.1002/wrna.1584
Jiang, T., Lu, Y., Chen, Z., Lin, X., Zhang, J., Shan, J., et al. (2021). RNA sequencing reveals the circular RNA expression profiles of the infrapatellar fat pad/synovium unit. Ann. Transl. Med. 9, 1685. doi:10.21037/atm-21-5739
Jiang, Z., Du, X., Wen, X., Li, H., Zeng, A., Sun, H., et al. (2021). Whole-Transcriptome sequence of degenerative meniscus cells unveiling diagnostic markers and therapeutic targets for osteoarthritis. Front. Genet. 12, 754421. doi:10.3389/fgene.2021.754421
Jiang, H., Dai, J., Zhang, C., Sun, H., and Tang, X. (2021). Circ_0045714 alleviates TNF-α-induced chondrocyte injury and extracellular matrix degradation through miR-218-5p/HRAS axis. J. Bioenerg. Biomembr. 53, 97–107. doi:10.1007/s10863-020-09868-y
Jiang, R., Gao, H., Cong, F., Zhang, W., Song, T., and Yu, Z. (2021). Circ_DHRS3 positively regulates GREM1 expression by competitively targeting miR-183-5p to modulate IL-1β-administered chondrocyte proliferation, apoptosis and ECM degradation. Int. Immunopharmacol. 91, 107293. doi:10.1016/j.intimp.2020.107293
Katz, J. N., Arant, K. R., and Loeser, R. F. (2021). Diagnosis and treatment of hip and knee osteoarthritis: A review. JAMA 325, 568–578. doi:10.1001/jama.2020.22171
Kovacs, B., Vajda, E., and Nagy, E. E. (2019). Regulatory effects and interactions of the wnt and OPG-RANKL-RANK signaling at the bone-cartilage interface in osteoarthritis. Int. J. Mol. Sci. 20, 4653. doi:10.3390/ijms20184653
Kristensen, L. S., Jakobsen, T., Hager, H., and Kjems, J. (2022). The emerging roles of circRNAs in cancer and oncology. Nat. Rev. Clin. Oncol. 19, 188–206. doi:10.1038/s41571-021-00585-y
Kuo, M. C., Liu, S. C., Hsu, Y. F., and Wu, R. M. (2021). The role of noncoding RNAs in Parkinson's disease: Biomarkers and associations with pathogenic pathways. J. Biomed. Sci. 28, 78. doi:10.1186/s12929-021-00775-x
Kuusalo, L., Felson, D. T., Wang, N., Lewis, C. E., Torner, J., Nevitt, M. C., et al. (2021). Metabolic osteoarthritis - relation of diabetes and cardiovascular disease with knee osteoarthritis. Osteoarthr. Cartil. 29, 230–234. doi:10.1016/j.joca.2020.09.010
Lai, X., Song, Y., and Tian, J. (2022). CircCDK14 ameliorates interleukin-1β-induced chondrocyte damage by the miR-1183/KLF5 pathway in osteoarthritis. Autoimmunity 55, 408–417. doi:10.1080/08916934.2022.2081843
Legnini, I., Di Timoteo, G., Rossi, F., Morlando, M., Briganti, F., Sthandier, O., et al. (2017). Circ-ZNF609 is a circular RNA that can Be translated and functions in myogenesis. Mol. Cell 66, 22. doi:10.1016/j.molcel.2017.02.017
Li, Z., Huang, C., Bao, C., Chen, L., Lin, M., Wang, X., et al. (2015). Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 22, 256–264. doi:10.1038/nsmb.2959
Li, Y. H., Tavallaee, G., Tokar, T., Nakamura, A., Sundararajan, K., Weston, A., et al. (2016). Identification of synovial fluid microRNA signature in knee osteoarthritis: Differentiating early- and late-stage knee osteoarthritis. Osteoarthr. Cartil. 24, 1577–1586. doi:10.1016/j.joca.2016.04.019
Li, X., Liu, C. X., Xue, W., Zhang, Y., Jiang, S., Yin, Q. F., et al. (2017). Coordinated circRNA biogenesis and function with NF90/NF110 in viral infection. Mol. Cell 67, 214–227. doi:10.1016/j.molcel.2017.05.023
Li, B. F., Zhang, Y., Xiao, J., Wang, F., Li, M., Guo, X. Z., et al. (2017). Hsa_circ_0045714 regulates chondrocyte proliferation, apoptosis and extracellular matrix synthesis by promoting the expression of miR-193b target gene IGF1R. Hum. Cell 30, 311–318. doi:10.1007/s13577-017-0177-7
Li, Z., Yuan, B., Pei, Z., Zhang, K., Ding, Z., Zhu, S., et al. (2019). Circ_0136474 and MMP-13 suppressed cell proliferation by competitive binding to miR-127-5p in osteoarthritis. J. Cell Mol. Med. 23, 6554–6564. doi:10.1111/jcmm.14400
Li, J., Ma, M., Yang, X., Zhang, M., Luo, J., Zhou, H., et al. (2020). Circular HER2 RNA positive triple negative breast cancer is sensitive to Pertuzumab. Mol. Cancer 19, 142. doi:10.1186/s12943-020-01259-6
Li, C., Ni, Y. Q., Xu, H., Xiang, Q. Y., Zhao, Y., Zhan, J. K., et al. (2021). Roles and mechanisms of exosomal non-coding RNAs in human health and diseases. Signal Transduct. Target Ther. 6, 383. doi:10.1038/s41392-021-00779-x
Li, Z., Cheng, Y., Fu, K., Lin, Q., Zhao, T., Tang, W., et al. (2021). Circ-PTPDC1 promotes the progression of gastric cancer through sponging mir-139-3p by regulating ELK1 and functions as a prognostic biomarker. Int. J. Biol. Sci. 17, 4285–4304. doi:10.7150/ijbs.62732
Li, S., Liu, J., Liu, S., Jiao, W., and Wang, X. (2021). Mesenchymal stem cell-derived extracellular vesicles prevent the development of osteoarthritis via the circHIPK3/miR-124-3p/MYH9 axis. J. Nanobiotechnology 19, 194. doi:10.1186/s12951-021-00940-2
Li, H., Liu, Z., Guo, X., and Zhang, M.(2021) Circ_0128846/miR-140-3p/JAK2 network in osteoarthritis development. Immunol. Invest. 1-19, 1529–1547. doi:10.1080/08820139.2021.1981930
Li, J., Ye, Z., Hu, X., Hou, S., and Hang, Q. (2022). Prognostic, diagnostic, and clinicopathological significance of circular RNAs in pancreatic cancer: A systematic review and meta-analysis. Cancers (Basel) 14, 6187. doi:10.3390/cancers14246187
Li, N., Wang, Y., and Wu, X. (2022). Knockdown of Circ_0037658 alleviates IL-1β-induced osteoarthritis progression by serving as a sponge of miR-665 to regulate ADAMTS5. Front. Genet. 13, 886898. doi:10.3389/fgene.2022.886898
Li, G., Luo, H., Ding, Z., Liang, H., Lai, Z., Chen, S., et al. (2022). Silencing of circ_0000205 mitigates interleukin-1β-induced apoptosis and extracellular matrix degradation in chondrocytes via targeting miR-766-3p/ADAMTS5 axis. Innate Immun. 28, 79–90. doi:10.1177/17534259221077078
Li, X., Xie, C., Xiao, F., Su, H., Li, Z., Weng, J., et al. (2022). Circular RNA circ_0000423 regulates cartilage ECM synthesis via circ_0000423/miRNA-27b-3p/MMP-13 axis in osteoarthritis. Aging (Albany NY) 14, 3400–3415. doi:10.18632/aging.204018
Li, Z., Meng, D., Liu, Y., Bi, F., Tian, K., Xu, J., et al. (2022). Circular RNA VMA21 ameliorates IL-1β-engendered chondrocyte injury through the miR-495-3p/FBWX7 signaling axis. Clin. Immunol. 238, 108995. doi:10.1016/j.clim.2022.108995
Liao, H. X., Zhang, Z. H., Chen, H. L., Huang, Y. M., Liu, Z. L., and Huang, J. (2021). CircHYBID regulates hyaluronan metabolism in chondrocytes via hsa-miR-29b-3p/TGF-β1 axis. Mol. Med. 27, 56. doi:10.1186/s10020-021-00319-x
Liao, H., Tu, Q., Kang, Y., Mao, G., Li, Z., Hu, S., et al. (2022). CircNFIX regulates chondrogenesis and cartilage homeostasis by targeting the miR758-3p/KDM6A axis. Cell Prolif. 55, e13302. doi:10.1111/cpr.13302
Lin, S., Li, H., Wu, B., Shang, J., Jiang, N., Peng, R., et al. (2022). TGF-β1 regulates chondrocyte proliferation and extracellular matrix synthesis via circPhf21a-Vegfa axis in osteoarthritis. Cell Commun. Signal 20, 75. doi:10.1186/s12964-022-00881-9
Liu, Q., Zhang, X., Hu, X., Dai, L., Fu, X., Zhang, J., et al. (2016). Circular RNA related to the chondrocyte ECM regulates MMP13 expression by functioning as a MiR-136 'sponge' in human cartilage degradation. Sci. Rep. 6, 22572. doi:10.1038/srep22572
Liu, C. X., Li, X., Nan, F., Jiang, S., Gao, X., Guo, S. K., et al. (2019). Structure and degradation of circular RNAs regulate PKR activation in innate immunity. Cell 177, 865–880. doi:10.1016/j.cell.2019.03.046
Liu, B., Ye, B., Zhu, X., Yang, L., Li, H., Liu, N., et al. (2020). An inducible circular RNA circKcnt2 inhibits ILC3 activation to facilitate colitis resolution. Nat. Commun. 11, 4076. doi:10.1038/s41467-020-17944-5
Liu, Y., Li, Q., Gao, Z., Lei, F., and Gao, X. (2021). Circ-SPG11 knockdown hampers IL-1β-induced osteoarthritis progression via targeting miR-337-3p/ADAMTS5. J. Orthop. Surg. Res. 16, 392. doi:10.1186/s13018-021-02526-y
Liu, C., Cheng, P., Liang, J., Zhao, X., and Du, W. (2021). Circular RNA circ_0128846 promotes the progression of osteoarthritis by regulating miR-127-5p/NAMPT axis. J. Orthop. Surg. Res. 16, 307. doi:10.1186/s13018-021-02428-z
Liu, C. X., Guo, S. K., Nan, F., Xu, Y. F., Yang, L., and Chen, L. L. (2022). RNA circles with minimized immunogenicity as potent PKR inhibitors. Mol. Cell 82, 420–434.e6. doi:10.1016/j.molcel.2021.11.019
Liu, K., Fan, X. E., Zhang, L., Yang, Y., and Zhou, X. L. (2022). Circ-NCX1 inhibits LPS-induced chondrocyte apoptosis by regulating the miR-133a/SIRT1 axis. Kaohsiung J. Med. Sci. 38, 992–1000. doi:10.1002/kjm2.12564
Liu, Y., Yang, Y., Lin, Y., Wei, B., Hu, X., Xu, L., et al. (2022). N(6) -methyladenosine-modified circRNA RERE modulates osteoarthritis by regulating beta-catenin ubiquitination and degradation. Cell Prolif. 56, e13297. doi:10.1111/cpr.13297
Liu, Z., Cao, J., Zhang, L., Li, J., Yan, T., Zhou, P., et al. (2022). Knockdown of circ-PRKCH alleviates IL-1β-treated chondrocyte cell phenotypic changes through modulating miR-502-5p/ADAMTS5 axis. Autoimmunity 55, 179–191. doi:10.1080/08916934.2022.2027918
Lu, D., Chatterjee, S., Xiao, K., Riedel, I., Huang, C. K., Costa, A., et al. (2022). A circular RNA derived from the insulin receptor locus protects against doxorubicin-induced cardiotoxicity. Eur. Heart J. 43, 4496–4511. doi:10.1093/eurheartj/ehac337
Luo, J., Liu, H., Luan, S., and Li, Z. (2019). Guidance of circular RNAs to proteins' behavior as binding partners. Cell Mol. Life Sci. 76, 4233–4243. doi:10.1007/s00018-019-03216-z
Luobu, Z., Wang, L., Jiang, D., Liao, T., Luobu, C., and Qunpei, L. (2022). CircSCAPER contributes to IL-1β-induced osteoarthritis in vitro via miR-140-3p/EZH2 axis. Bone Jt. Res. 11, 61–72. R2. doi:10.1302/2046-3758.112.bjr-2020-0482.r2
Ma, H. R., Mu, W. B., Zhang, K. Y., Zhou, H. K., Jiang, R. D., and Cao, L. (2021). CircVCAN regulates the proliferation and apoptosis of osteoarthritis chondrocyte through NF-κB signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 25, 3152. doi:10.26355/eurrev_202104_25720
Man, G., Yang, H., Shen, K., Zhang, D., Zhang, J., Wu, H., et al. (2022). Circular RNA RHOT1 regulates miR-142-5p/CCND1 to participate in chondrocyte autophagy and proliferation in osteoarthritis. J. Immunol. Res. 2022, 4370873. doi:10.1155/2022/4370873
Mao, X., Cao, Y., Guo, Z., Wang, L., and Xiang, C. (2021). Biological roles and therapeutic potential of circular RNAs in osteoarthritis. Mol. Ther. Nucleic Acids 24, 856–867. doi:10.1016/j.omtn.2021.04.006
Mao, G., Xu, Y., Long, D., Sun, H., Li, H., Xin, R., et al. (2021). Exosome-transported circRNA_0001236 enhances chondrogenesis and suppress cartilage degradation via the miR-3677-3p/Sox9 axis. Stem Cell Res. Ther. 12, 389. doi:10.1186/s13287-021-02431-5
Martel-Pelletier, J., Barr, A. J., Cicuttini, F. M., Conaghan, P. G., Cooper, C., Goldring, M. B., et al. (2016). Osteoarthritis. Nat. Rev. Dis. Prim. 2, 16072. doi:10.1038/nrdp.2016.72
Ni, J. L., Dang, X. Q., and Shi, Z. B. (2020). CircPSM3 inhibits the proliferation and differentiation of OA chondrocytes by targeting miRNA-296-5p. Eur. Rev. Med. Pharmacol. Sci. 24, 3467–3475. doi:10.26355/eurrev_202004_20805
Ni, W., Jiang, C., Wu, Y., Zhang, H., Wang, L., Yik, J. H. N., et al. (2021). CircSLC7A2 protects against osteoarthritis through inhibition of the miR-4498/TIMP3 axis. Cell Prolif. 54, e13047. doi:10.1111/cpr.13047
Ou, L., Huang, W., Zhang, T., Xu, D., Kong, D., and Meng, Y. (2022). Circular RNA circ_0114876 regulates osteoarthritis through upregulating ADAM10 via targeting miR-1227-3p. Transpl. Immunol. 77, 101747. doi:10.1016/j.trim.2022.101747
Ouyang, X., Ding, Y., Yu, L., Xin, F., Yang, X., Liu, X., et al. (2021). Circ_SPG11 plays contributing effects on IL-1β-induced chondrocyte apoptosis and ECM degradation via miR-665 inhibition-mediated GREM1 upregulation. Clin. Immunol. 233, 108889. doi:10.1016/j.clim.2021.108889
Pan, F., Li, Z., Luo, Y., Ma, Z., Feng, S., and Xu, N. (2022). Circ_0136474 contributes to the IL-1β-induced chondrocyte injury by binding to miR-665 to induce the FGFR1 upregulation. Transpl. Immunol. 2022. 101615. doi:10.1016/j.trim.2022.101615
Panda, A. C. (2018). Circular RNAs act as miRNA sponges. Adv. Exp. Med. Biol. 1087, 67–79. doi:10.1007/978-981-13-1426-1_6
Park, O. H., Ha, H., Lee, Y., Boo, S. H., Kwon, D. H., Song, H. K., et al. (2019). Endoribonucleolytic cleavage of m(6)a-containing RNAs by RNase P/MRP complex. Mol. Cell 74, 494–507. doi:10.1016/j.molcel.2019.02.034
Peat, G., and Thomas, M. J. (2021). Osteoarthritis year in review 2020: Epidemiology & therapy. Osteoarthr. Cartil. 29, 180–189. doi:10.1016/j.joca.2020.10.007
Qian, L., Yu, B., Chen, T., Chen, K., Ma, Z., Wang, Y., et al. (2022). Circ_0022383 alleviates IL-1β-induced apoptosis, inflammation and extracellular matrix degeneration in osteoarthritis cell model by miR-3619-5p/SIRT1 axis. Int. Immunopharmacol. 112, 109289. doi:10.1016/j.intimp.2022.109289
Qu, L., Yi, Z., Shen, Y., Lin, L., Chen, F., Xu, Y., et al. (2022). Circular RNA vaccines against SARS-CoV-2 and emerging variants. Cell 185, 1728–1744.e16. doi:10.1016/j.cell.2022.03.044
Que, W., Liu, H., and Yang, Q. (2022). CircPRKCH modulates extracellular matrix formation and metabolism by regulating the miR-145/HGF axis in osteoarthritis. Arthritis Res. Ther. 24, 216. doi:10.1186/s13075-022-02893-9
Ratneswaran, A., and Kapoor, M. (2021). Osteoarthritis year in review: Genetics, genomics, epigenetics. Osteoarthr. Cartil. 29, 151–160. doi:10.1016/j.joca.2020.11.003
Rausch, J. W., Heinz, W. F., Payea, M. J., Sherpa, C., Gorospe, M., and Le Grice, S. F. J. (2021). Characterizing and circumventing sequence restrictions for synthesis of circular RNA in vitro. Nucleic Acids Res. 49, e35. doi:10.1093/nar/gkaa1256
Roemer, F. W., Guermazi, A., Felson, D. T., Niu, J., Nevitt, M. C., Crema, M. D., et al. (2011). Presence of MRI-detected joint effusion and synovitis increases the risk of cartilage loss in knees without osteoarthritis at 30-month follow-up: The MOST study. Ann. Rheum. Dis. 70, 1804–1809. doi:10.1136/ard.2011.150243
Roos, E. M., and Toksvig-Larsen, S. (2003). Knee injury and Osteoarthritis Outcome Score (KOOS) - validation and comparison to the WOMAC in total knee replacement. Health Qual. Life Outcomes 1, 17. doi:10.1186/1477-7525-1-17
Ruan, Y., Li, Z., Xie, Y., Sun, W., and Guo, J. (2022). Detecting plasma hsa_circ_0061276 in patients with gastric cancer by reverse transcription-digital polymerase chain reaction. Front. Oncol. 12, 1042248. doi:10.3389/fonc.2022.1042248
Saaoud, F., Drummer, I. V. C., Shao, Y., Sun, Y., Lu, Y., Xu, K., et al. (2021). Circular RNAs are a novel type of non-coding RNAs in ROS regulation, cardiovascular metabolic inflammations and cancers. Pharmacol. Ther. 220, 107715. doi:10.1016/j.pharmthera.2020.107715
Santer, L., Bar, C., and Thum, T. (2019). Circular RNAs: A novel class of functional RNA molecules with a therapeutic perspective. Mol. Ther. 27, 1350–1363. doi:10.1016/j.ymthe.2019.07.001
Shang, J., Li, H., Wu, B., Jiang, N., Wang, B., Wang, D., et al. (2022). CircHIPK3 prevents chondrocyte apoptosis and cartilage degradation by sponging miR-30a-3p and promoting PON2. Cell Prolif. 55, e13285. doi:10.1111/cpr.13285
Shen, S., Wu, Y., Chen, J., Xie, Z., Huang, K., Wang, G., et al. (2019). CircSERPINE2 protects against osteoarthritis by targeting miR-1271 and ETS-related gene. Ann. Rheum. Dis. 78, 826–836. doi:10.1136/annrheumdis-2018-214786
Shen, P., Yang, Y., Liu, G., Chen, W., Chen, J., Wang, Q., et al. (2020). CircCDK14 protects against Osteoarthritis by sponging miR-125a-5p and promoting the expression of Smad2. Theranostics 10, 9113–9131. doi:10.7150/thno.45993
Shen, S., Yang, Y., Shen, P., Ma, J., Fang, B., Wang, Q., et al. (2021). circPDE4B prevents articular cartilage degeneration and promotes repair by acting as a scaffold for RIC8A and MID1. Ann. Rheum. Dis. 80, 1209–1219. doi:10.1136/annrheumdis-2021-219969
Shen, L., Ji, C., Lin, J., and Yang, H. (2022). Regulation of circADAMTS6-miR-324-5p-PIK3R3 ceRNA pathway may be a novel mechanism of IL-1β-induced osteoarthritic chondrocytes. J. Bone Min. Metab. 40, 389–401. doi:10.1007/s00774-021-01308-0
Shi, L., Zhang, H., Sun, J., Gao, X., and Liu, C. (2021). CircSEC24A promotes IL-1β-induced apoptosis and inflammation in chondrocytes by regulating miR-142-5p/SOX5 axis. Biotechnol. Appl. Biochem. 69, 701–713. doi:10.1002/bab.2145
Shuai, S., Cai, Q., and Ou, Y. (2022). Circular RNA circ_0008365 regulates SOX9 by targeting miR-338-3p to inhibit IL-1β-induced chondrocyte apoptosis and extracellular matrix degradation. J. Orthop. Surg. Res. 17, 452. doi:10.1186/s13018-022-03240-z
Steinberg, J., Southam, L., Fontalis, A., Clark, M. J., Jayasuriya, R. L., Swift, D., et al. (2021). Linking chondrocyte and synovial transcriptional profile to clinical phenotype in osteoarthritis. Ann. Rheum. Dis. 10, 1070–1074. doi:10.1136/annrheumdis-2020-219760
Sun, J., Zhong, N., Li, Q., Min, Z., Zhao, W., Sun, Q., et al. (2011). MicroRNAs of rat articular cartilage at different developmental stages identified by Solexa sequencing. Osteoarthr. Cartil. 19, 1237–1245. doi:10.1016/j.joca.2011.07.002
Sun, K., Jing, X., Guo, J., Yao, X., and Guo, F. (2020). Mitophagy in degenerative joint diseases. Autophagy 1, 2082–2092. doi:10.1080/15548627.2020.1822097
Sun, M., Yang, J., Jiang, D., and Bao, G. (2021). Overexpression of hsa_circ_0094742 inhibits IL-1β-induced decline in CHON-001 cell viability by targeting microRNA-127-5p. Histol. Histopathol. 36, 207–216. doi:10.14670/HH-18-325
Tang, S., Nie, X., Ruan, J., Cao, Y., Kang, J., and Ding, C. (2022). Circular RNA circNFKB1 promotes osteoarthritis progression through interacting with ENO1 and sustaining NF-κB signaling. Cell Death Dis. 13, 695. doi:10.1038/s41419-022-05148-2
Tang, S., Cao, Y., Cai, Z., Nie, X., Ruan, J., Zhou, Z., et al. (2022). The lncRNA PILA promotes NF-κB signaling in osteoarthritis by stimulating the activity of the protein arginine methyltransferase PRMT1. Sci. Signal 15, eabm6265. doi:10.1126/scisignal.abm6265
Tao, S. C., Huang, J. Y., Gao, Y., Li, Z. X., Wei, Z. Y., Dawes, H., et al. (2021). Small extracellular vesicles in combination with sleep-related circRNA3503: A targeted therapeutic agent with injectable thermosensitive hydrogel to prevent osteoarthritis. Bioact. Mater 6, 4455–4469. doi:10.1016/j.bioactmat.2021.04.031
Valenti, M. T., Dalle Carbonare, L., Zipeto, D., and Mottes, M. (2021). Control of the autophagy pathway in osteoarthritis: Key regulators, therapeutic targets and therapeutic strategies. Int. J. Mol. Sci. 22, 2700. doi:10.3390/ijms22052700
van den Bosch, M. H. J., van Lent, P., and van der Kraan, P. M. (2020). Identifying effector molecules, cells, and cytokines of innate immunity in OA. Osteoarthr. Cartil. 28, 532–543. doi:10.1016/j.joca.2020.01.016
van den Bosch, M. H. J. (2021). Osteoarthritis year in review 2020: Biology. Osteoarthr. Cartil. 29, 143–150. doi:10.1016/j.joca.2020.10.006
Van Spil, W. E., Kubassova, O., Boesen, M., Bay-Jensen, A. C., and Mobasheri, A. (2019). Osteoarthritis phenotypes and novel therapeutic targets. Biochem. Pharmacol. 165, 41–48. doi:10.1016/j.bcp.2019.02.037
Wahafu, P., Xu, A., Zhao, B., Tuo, Y., and Yang, J. (2022). Circ_0005526 contributes to interleukin-1β-induced chondrocyte injury in osteoarthritis via upregulating transcription factor 4 by interacting with miR-142-5p. Bioengineered 13, 8407–8418. doi:10.1080/21655979.2022.2048773
Wang, Y., Wu, C., Yang, Y., Ren, Z., Lammi, M. J., and Guo, X. (2019). Preliminary exploration of hsa_circ_0032131 levels in peripheral blood as a potential diagnostic biomarker of osteoarthritis. Genet. Test. Mol. Biomarkers 23, 717–721. doi:10.1089/gtmb.2019.0036
Wang, B., Zhong, J. L., Xu, X. H., Wu, B., Shang, J., Jiang, N., et al. (2020). Gene expression profiling analysis to identify key genes and underlying mechanisms in meniscus of osteoarthritis patients. Comb. Chem. High. Throughput Screen 10, 1151–1167. doi:10.2174/1386207323666200902140656
Wang, T., Hao, Z., Liu, C., Yuan, L., Li, L., Yin, M., et al. (2020). LEF1 mediates osteoarthritis progression through circRNF121/miR-665/MYD88 axis via NF-кB signaling pathway. Cell Death Dis. 11, 598. doi:10.1038/s41419-020-02769-3
Wang, K. F., Shi, Z. W., and Dong, D. M. (2021). CircATRNL1 protects against osteoarthritis by targeting miR-153-3p and KLF5. Int. Immunopharmacol. 96, 107704. doi:10.1016/j.intimp.2021.107704
Wang, Q., Luo, S., Yang, J., Li, J., Huan, S., She, G., et al. (2021). Circ_0114876 promoted IL-1β-induced chondrocyte injury by targeting miR-671/TRAF2 axis. Biotechnol. Lett. 43, 791–802. doi:10.1007/s10529-020-03070-1
Wang, X., Zhang, C., Song, H., Yuan, J., Zhang, L., and He, J. (2022). CircCCDC66: Emerging roles and potential clinical values in malignant tumors. Front. Oncol. 12, 1061007. doi:10.3389/fonc.2022.1061007
Wang, R. Z. Z., Wang, X., Jiang, C., and Yang, Y. (2022). circPhc3 sponging microRNA-93-3p is involved in the regulation of chondrocyte function by mechanical instability in osteoarthritis. Int. J. Mol. Med. 49, 6. doi:10.3892/ijmm.2021.5061
Wesselhoeft, R. A., Kowalski, P. S., Parker-Hale, F. C., Huang, Y., Bisaria, N., and Anderson, D. G. (2019). RNA circularization diminishes immunogenicity and can extend translation duration in vivo. Mol. Cell 74, 508–520. doi:10.1016/j.molcel.2019.02.015
Wu, Y., Zhang, Y., Zhang, Y., and Wang, J. J. (2017). CircRNA hsa_circ_0005105 upregulates NAMPT expression and promotes chondrocyte extracellular matrix degradation by sponging miR-26a. Cell Biol. Int. 41, 1283–1289. doi:10.1002/cbin.10761
Wu, Q., Yuan, Z. H., Ma, X. B., and Tang, X. H. (2020). Low expression of CircRNA HIPK3 promotes osteoarthritis chondrocyte apoptosis by serving as a sponge of miR-124 to regulate SOX8. Eur. Rev. Med. Pharmacol. Sci. 24, 7937–7945. doi:10.26355/eurrev_202008_22476
Wu, R., Zhang, F., Cai, Y., Long, Z., Duan, Z., Wu, D., et al. (2021). Circ_0134111 knockdown relieves IL-1β-induced apoptosis, inflammation and extracellular matrix degradation in human chondrocytes through the circ_0134111-miR-515-5p-SOCS1 network. Int. Immunopharmacol. 95, 107495. doi:10.1016/j.intimp.2021.107495
Wu, Y., Hong, Z., Xu, W., Chen, J., Wang, Q., Chen, J., et al. (2021). Circular RNA circPDE4D protects against osteoarthritis by binding to miR-103a-3p and regulating FGF18. Mol. Ther. 29, 308–323. doi:10.1016/j.ymthe.2020.09.002
Wu, Q., Fu, S., Xiao, H., Du, J., Cheng, F., Wan, S., et al. (2022). Advances in extracellular vesicle nanotechnology for precision theranostics. Adv. Sci. (Weinh) 10, e2204814. doi:10.1002/advs.202204814
Xi, P., Zhang, C. L., Wu, S. Y., Liu, L., Li, W. J., and Li, Y. M. (2021). CircRNA circ-IQGAP1 knockdown alleviates interleukin-1β-induced osteoarthritis progression via targeting miR-671-5p/TCF4. Orthop. Surg. 13, 1036–1046. doi:10.1111/os.12923
Xiang, S., Li, Z., Bian, Y., and Weng, X. (2019). RNA sequencing reveals the circular RNA expression profiles of osteoarthritic synovium. J. Cell Biochem. 120, 18031–18040. doi:10.1002/jcb.29106
Xiao, M. S., Ai, Y., and Wilusz, J. E. (2020). Biogenesis and functions of circular RNAs come into focus. Trends Cell Biol. 30, 226–240. doi:10.1016/j.tcb.2019.12.004
Xie, W., Jiang, L., Huang, X., You, W., and Sun, W. (2022). hsa_circ_00046621 accelerates the progression of osteoarthritis via the microRNA-424-5p/VEGFA axis. Curr. Mol. Med. 10, 2174. doi:10.2174/1566524023666221103161203
Xu, J., and Ma, X. (2021). Hsa_circ_0032131 knockdown inhibits osteoarthritis progression via the miR-502-5p/PRDX3 axis. Aging (Albany NY) 13, 15100–15113. doi:10.18632/aging.203073
Xu, Y., Mao, G., Long, D., Deng, Z., Xin, R., Zhang, Z., et al. (2022). Circular RNA CREBBP modulates cartilage degradation by activating the Smad1/5 pathway through the TGFβ2/ALK1 axis. Exp. Mol. Med. 54, 1727–1740. doi:10.1038/s12276-022-00865-2
Xue, C., Li, G., Zheng, Q., Gu, X., Bao, Z., Lu, J., et al. (2022). The functional roles of the circRNA/Wnt axis in cancer. Mol. Cancer 21, 108. doi:10.1186/s12943-022-01582-0
Yan, W., Yu, H., Liu, B., Jiang, Z., Jin, H., Li, Z., et al. (2022). Andrographolide suppresses osteoarthritis progression by regulating circ_Rapgef1/miR-383-3p/NLRP3 signaling axis. Transpl. Immunol. 71, 101548. doi:10.1016/j.trim.2022.101548
Yang, Y., Shen, P., Yao, T., Ma, J., Chen, Z., Zhu, J., et al. (2021). Novel role of circRSU1 in the progression of osteoarthritis by adjusting oxidative stress. Theranostics 11, 1877–1900. doi:10.7150/thno.53307
Yang, J., Li, Q., Wang, T., and Lv, K. (2022). Circular RNA triple functional domain promotes osteoarthritis' development by modulating the microRNA-136-5p/Nicotinamide phosphoribosyltransferase axis. Bioengineered 13, 6070–6079. doi:10.1080/21655979.2021.2018095
Yao, T., Yang, Y., Xie, Z., Xu, Y., Huang, Y., Gao, J., et al. (2020). Circ0083429 regulates osteoarthritis progression via the mir-346/SMAD3 Axis. Front. Cell Dev. Biol. 8, 579945. doi:10.3389/fcell.2020.579945
Yu, F., Xie, C., Sun, J., Feng, H., and Huang, X. (2018). Circular RNA expression profiles in synovial fluid: A promising new class of diagnostic biomarkers for osteoarthritis. Int. J. Clin. Exp. Pathol. 11, 1338–1346.
Yu, J., Qin, Y., and Zhou, N. (2021). Knockdown of Circ_SLC39A8 protects against the progression of osteoarthritis by regulating miR-591/IRAK3 axis. J. Orthop. Surg. Res. 16, 170. doi:10.1186/s13018-021-02323-7
Yu, Z., Cong, F., Zhang, W., Song, T., Zhang, S., and Jiang, R. (2022). Circular RNA circ_0020014 contributes to osteoarthritis progression via miR-613/ADAMTS5 axis. Bosn. J. Basic Med. Sci. 22, 716–727. doi:10.17305/bjbms.2021.6668
Yu, L., Zhang, X., Liu, X., Li, G., Chen, M., Liu, Z., et al. (2022). CircTMOD3 promotes lipopolysaccharide-induced chondrocyte apoptosis in osteoarthritis by sponging miR-27a. J. Bone Min. Metab. 40, 415–421. doi:10.1007/s00774-022-01310-0
Yuan, C., Pan, Z., Zhao, K., Li, J., Sheng, Z., Yao, X., et al. (2020). Classification of four distinct osteoarthritis subtypes with a knee joint tissue transcriptome atlas. Bone Res. 8, 38. doi:10.1038/s41413-020-00109-x
Yuan, X., Zhang, Y., Cai, C., Liu, C., Xie, J., and Yi, C. (2021). Circular RNA circZNF652 is overexpressed in osteoarthritis and positively regulates LPS-induced apoptosis of chondrocytes by upregulating PTEN. Autoimmunity 54, 415–421. doi:10.1080/08916934.2021.1951716
Yuan, H., Mears, L. L. E., Wang, Y., Su, R., Qi, W., He, Z., et al. (2022). Lubricants for osteoarthritis treatment: From natural to bioinspired and alternative strategies. Adv. Colloid Interface Sci. 311, 102814. doi:10.1016/j.cis.2022.102814
Zaki, S., Blaker, C. L., and Little, C. B. (2021). OA foundations – experimental models of osteoarthritis. Osteoarthr. Cartil. 30, 357–380. doi:10.1016/j.joca.2021.03.024
Zeng, J., Zhang, Z., Liao, Q., Lu, Q., Liu, J., Yuan, L., et al. (2021). CircPan3 promotes the ghrelin system and chondrocyte autophagy by sponging miR-667-5p during rat osteoarthritis pathogenesis. Front. Cell Dev. Biol. 9, 719898. doi:10.3389/fcell.2021.719898
Zhang, L., and Wen, C. (2021). Osteocyte dysfunction in joint homeostasis and osteoarthritis. Int. J. Mol. Sci. 22, 6522. doi:10.3390/ijms22126522
Zhang, X. O., Wang, H. B., Zhang, Y., Lu, X., Chen, L. L., and Yang, L. (2014). Complementary sequence-mediated exon circularization. Cell 159, 134–147. doi:10.1016/j.cell.2014.09.001
Zhang, J., Cheng, F., Rong, G., Tang, Z., and Gui, B. (2020). Hsa_circ_0005567 activates autophagy and suppresses IL-1β-induced chondrocyte apoptosis by regulating miR-495. Front. Mol. Biosci. 7, 216. doi:10.3389/fmolb.2020.00216
Zhang, Q., Qiao, X., and Xia, W. (2020). CircSERPINE2 weakens IL-1β-caused apoptosis and extracellular matrix degradation of chondrocytes by regulating miR-495/TGFBR2 axis. Biosci. Rep. 40. doi:10.1042/BSR20201601
Zhang, W., Zhang, C., Hu, C., Luo, C., Zhong, B., and Yu, X. (2020). Circular RNA-CDR1as acts as the sponge of microRNA-641 to promote osteoarthritis progression. J. Inflamm. (Lond) 17, 8. doi:10.1186/s12950-020-0234-y
Zhang, W., Qi, L., Chen, R., He, J., Liu, Z., Wang, W., et al. (2021). Circular RNAs in osteoarthritis: Indispensable regulators and novel strategies in clinical implications. Arthritis Res. Ther. 23, 23. doi:10.1186/s13075-021-02420-2
Zhang, J., Cheng, F., Rong, G., Tang, Z., and Gui, B. (2021). Circular RNA hsa_circ_0005567 overexpression promotes M2 type macrophage polarization through miR-492/SOCS2 axis to inhibit osteoarthritis progression. Bioengineered 12, 8920–8930. doi:10.1080/21655979.2021.1989999
Zhang, Z., Yang, B., Zhou, S., and Wu, J. (2021). CircRNA circ_SEC24A upregulates DNMT3A expression by sponging miR-26b-5p to aggravate osteoarthritis progression. Int. Immunopharmacol. 99, 107957. doi:10.1016/j.intimp.2021.107957
Zhang, L., Sui, C., Zhang, Y., Wang, G., and Yin, Z. (2021). Knockdown of hsa_circ_0134111 alleviates the symptom of osteoarthritis via sponging microRNA-224-5p. Cell Cycle 20, 1052–1066. doi:10.1080/15384101.2021.1919838
Zhang, H., Ge, J., and Lu, X. (2021). CircFADS2 is downregulated in osteoarthritis and suppresses LPS-induced apoptosis of chondrocytes by regulating miR-195-5p methylation. Arch. Gerontol. Geriatr. 96, 104477. doi:10.1016/j.archger.2021.104477
Zhang, M., Mou, L., Liu, S., Sun, F., and Gong, M. (2021). Circ_0001103 alleviates IL-1β-induced chondrocyte cell injuries by upregulating SIRT1 via targeting miR-375. Clin. Immunol. 227, 108718. doi:10.1016/j.clim.2021.108718
Zhang, P., Sun, H., Wen, P., Wang, Y., Cui, Y., and Wu, J. (2022). circRNA circMED27 acts as a prognostic factor and mediator to promote lenvatinib resistance of hepatocellular carcinoma. Mol. Ther. Nucleic Acids 27, 293–303. doi:10.1016/j.omtn.2021.12.001
Zhang, Y., Liu, Q., Zhang, X., Huang, H., Tang, S., Chai, Y., et al. (2022). Recent advances in exosome-mediated nucleic acid delivery for cancer therapy. J. Nanobiotechnology 20, 279. doi:10.1186/s12951-022-01472-z
Zhang, Y., Zhao, P., Li, S., Mu, X., and Wang, H. (2022). CircSCAPER knockdown attenuates IL-1β-induced chondrocyte injury by miR-127-5p/TLR4 axis in osteoarthritis. Autoimmunity 55, 577–586. doi:10.1080/08916934.2022.2103798
Zhang, Y., Lu, R., Huang, X., Yin, E., Yang, Y., Yi, C., et al. (2022). Circular RNA MELK promotes chondrocyte apoptosis and inhibits autophagy in osteoarthritis by regulating MYD88/NF-κB signaling Axis through MicroRNA-497-5p. Contrast Media Mol. Imaging 2022, 7614497. doi:10.1155/2022/7614497
Zhang, S., Luo, J., and Zeng, S. (2022). Circ-LRP1B functions as a competing endogenous RNA to regulate proliferation, apoptosis and oxidative stress of LPS-induced human C28/I2 chondrocytes. J. Bioenerg. Biomembr. 54, 93–108. doi:10.1007/s10863-022-09932-9
Zhang, Y., Zheng, H., and Li, B. (2022). Circ_0110251 overexpression alleviates IL-1β-induced chondrocyte apoptosis and extracellular matrix degradation by regulating miR-3189-3p/SPRY1 axis in osteoarthritis. Autoimmunity 55, 168–178. doi:10.1080/08916934.2022.2027917
Zhang, Z., Zhao, T., Xu, H., and Wu, X. (2022). Circ_0008365 suppresses apoptosis, inflammation and extracellular matrix degradation of IL-1β-treated chondrocytes in osteoarthritis by regulating miR-324-5p/BMPR2/NF-κB signaling Axis. Immunol. Invest. 51, 1598–1611. doi:10.1080/08820139.2021.2001496
Zhang, C., Lu, Y., Yuan, F., and Jiang, S. (2022). Circular RNA CCDC66 regulates osteoarthritis progression by targeting miR-3622b-5p. Gerontology 68, 431–441. doi:10.1159/000520325
Zhao, Q., Liu, J., Deng, H., Ma, R., Liao, J. Y., Liang, H., et al. (2020). Targeting mitochondria-located circRNA SCAR alleviates NASH via reducing mROS output. Cell 183, 76–93. doi:10.1016/j.cell.2020.08.009
Zhao, J., Li, T., and Luo, W. (2021). Silencing of circ-PRKCH protects against lipopolysaccharide (LPS)-evoked chondrocyte damage and extracellular matrix loss by the miR-140-3p/ADAM10 axis. Gen. Physiol. Biophys. 40, 89–101. doi:10.4149/gpb_2021001
Zhao, C., Li, X., Sun, G., Liu, P., Kong, K., Chen, X., et al. (2022). CircFOXO3 protects against osteoarthritis by targeting its parental gene FOXO3 and activating PI3K/AKT-mediated autophagy. Cell Death Dis. 13, 932. doi:10.1038/s41419-022-05390-8
Zheng, W., Hou, G., and Li, Y. (2021). Circ_0116061 regulated the proliferation, apoptosis, and inflammation of osteoarthritis chondrocytes through regulating the miR-200b-3p/SMURF2 axis. J. Orthop. Surg. Res. 16, 253. doi:10.1186/s13018-021-02391-9
Zhou, Z. B., Du, D., Huang, G. X., Chen, A., and Zhu, L. (2018). Circular RNA Atp9b, a competing endogenous RNA, regulates the progression of osteoarthritis by targeting miR-138-5p. Gene 646, 203–209. doi:10.1016/j.gene.2017.12.064
Zhou, X., Jiang, L., Fan, G., Yang, H., Wu, L., Huang, Y., et al. (2019). Role of the ciRS-7/miR-7 axis in the regulation of proliferation, apoptosis and inflammation of chondrocytes induced by IL-1β. Int. Immunopharmacol. 71, 233–240. doi:10.1016/j.intimp.2019.03.037
Zhou, Z. B., Huang, G. X., Fu, Q., Han, B., Lu, J. J., Chen, A. M., et al. (2019). circRNA.33186 contributes to the pathogenesis of osteoarthritis by sponging miR-127-5p. Mol. Ther. 27, 531–541. doi:10.1016/j.ymthe.2019.01.006
Zhou, W. Y., Cai, Z. R., Liu, J., Wang, D. S., Ju, H. Q., and Xu, R. H. (2020). Circular RNA: Metabolism, functions and interactions with proteins. Mol. Cancer 19, 172. doi:10.1186/s12943-020-01286-3
Zhou, X., Li, J., Zhou, Y., Yang, Z., Yang, H., Li, D., et al. (2020). Down-regulated ciRS-7/up-regulated miR-7 axis aggravated cartilage degradation and autophagy defection by PI3K/AKT/mTOR activation mediated by IL-17A in osteoarthritis. Aging (Albany NY) 12, 20163–20183. doi:10.18632/aging.103731
Zhou, J., Qiu, C., Fan, Z., Liu, T., and Liu, T. (2021). Circular RNAs in stem cell differentiation: A sponge-like role for miRNAs. Int. J. Med. Sci. 18, 2438–2448. doi:10.7150/ijms.56457
Zhou, B., Yang, H., Yang, C., Bao, Y. L., Yang, S. M., Liu, J., et al. (2021). Translation of noncoding RNAs and cancer. Cancer Lett. 497, 89–99. doi:10.1016/j.canlet.2020.10.002
Zhou, J. L., Deng, S., Fang, H. S., Du, X. J., Peng, H., and Hu, Q. J. (2021). Circular RNA circANKRD36 regulates Casz1 by targeting miR-599 to prevent osteoarthritis chondrocyte apoptosis and inflammation. J. Cell Mol. Med. 25, 120–131. doi:10.1111/jcmm.15884
Zhou, Z., Ma, J., Lu, J., Chen, A., and Zhu, L. (2021). Circular RNA CircCDH13 contributes to the pathogenesis of osteoarthritis via CircCDH13/miR-296-3p/PTEN axis. J. Cell Physiol. 236, 3521–3535. doi:10.1002/jcp.30091
Zhou, J. L., Deng, S., Fang, H. S., Peng, H., and Hu, Q. J. (2022). CircSPI1_005 ameliorates osteoarthritis by sponging miR-370-3p to regulate the expression of MAP3K9. Int. Immunopharmacol. 110, 109064. doi:10.1016/j.intimp.2022.109064
Zhu, J., and Guo, Y. (2022). Circ_0020093 overexpression alleviates interleukin-1 beta-induced inflammation, apoptosis and extracellular matrix degradation in human chondrocytes by targeting the miR-181a-5p/ERG pathway. Immunol. Invest. 51, 1660–1677. doi:10.1080/08820139.2021.2021938
Zhu, X., Chan, Y. T., Yung, P. S. H., Tuan, R. S., and Jiang, Y. (2020). Subchondral bone remodeling: A therapeutic target for osteoarthritis. Front. Cell Dev. Biol. 8, 607764. doi:10.3389/fcell.2020.607764
Zhu, J., Fu, Q., Shao, J., Jinhui, P., Qian, Q., Zhou, Y., et al. (2021). Regulating effect of Circ_ATRNL1 on the promotion of SOX9 expression to promote chondrogenic differentiation of hAMSCs mediated by MiR-145-5p. J. Tissue Eng. Regen. Med. 15, 487–502. doi:10.1002/term.3189
Keywords: circRNA, osteoarthritis, cartilage, molecular mechanism, synovium
Citation: Li Z and Lu J (2023) CircRNAs in osteoarthritis: research status and prospect. Front. Genet. 14:1173812. doi: 10.3389/fgene.2023.1173812
Received: 25 February 2023; Accepted: 18 April 2023;
Published: 09 May 2023.
Edited by:
Gary S. Stein, University of Vermont, United StatesReviewed by:
Yu-Hang Xing, Massachusetts General Hospital Cancer Center, United StatesYaming Li, Renmin Hospital of Wuhan University, China
Copyright © 2023 Li and Lu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jun Lu, junlusuper@163.com
 Zhuang Li
Zhuang Li Jun Lu
Jun Lu