
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet., 08 June 2022
Sec. Evolutionary and Population Genetics
Volume 13 - 2022 | https://doi.org/10.3389/fgene.2022.922634
Complete mitochondrial genomes (mitogenomes) can provide valuable information for phylogenetic relationships, gene rearrangement, and molecular evolution. Here, we report the mitochondrial whole genomes of three Garra species and explore the mechanisms of rearrangements that occur in their mitochondrial genomes. The lengths of the mitogenomes’ sequences of Garra dengba, Garra tibetana, and Garra yajiangensis were 16,876, 16,861, and 16,835, respectively. They contained 13 protein-coding genes, two ribosomal RNAs, 22 transfer RNA genes, and two identical control regions (CRs). The mitochondrial genomes of three Garra species were rearranged compared to other fish mitochondrial genomes. The tRNA-Thr, tRNA-Pro and CR (T-P-CR) genes undergo replication followed by random loss of the tRNA-Thr and tRNA-Pro genes to form tRNA-Thr, CR1, tRNA-Pro and CR2 (T-CR-P-CR). Tandem duplication and random loss best explain this mitochondrial gene rearrangement. These results provide a foundation for future characterization of the mitochondrial gene arrangement of Labeoninae and further phylogenetic studies.
The subfamily Labeoninae is wildly distributed in Asia and Africa as one of the most diverse freshwater fishes in the family Cyprinidae (Cypriniformes). About 26 genera are distributed in China, 12 of which are endemic, with a great diversity of genera and species (Pei-Qi and Een-Duan, 1984). Once, Howes et al. (1991) and Winfield et al. (2012) proposed that the tribe Labeonini is essentially equivalent to the subfamily Labeoninae in Cyprininae (Howes et al., 1991; Winfield and Nelson, 2012). Because of the great diversity of Labeoninae orofacial morphology, it is often used as a key feature for identifying the included genera and for recovering the phylogenetic history hypothesis suite. However, since the morphological diversity and the difficulty of distinguishing the ambiguous morphological characteristics, the taxonomy of the Labeoninae has been constantly subject to revision, and the phylogenetic relationships within them remain amid change. Although some researchers erected a few new genera (e.g., Hongshuia, Qianlabeo, and Stenorhinchus) (Zhang and Chen, 2004; Zhang and Fang, 2005; LI et al., 2006; Zhang et al., 2008), and rearranged the generic assignment of some species based on the oromandibular morphology (Yuan et al., 2008). There was no input from molecule-based phylogenetic evidence. Furthermore, Yang and Mayden (2010) constructed the most comprehensive molecular phylogeny just including one species of each of 11 genera, supporting the monophyly of Labeoninae and proposing a model for the relationships of the major sublineages within Labeoninae (Yang and Mayden, 2010). However, limited by the number of mitogenomes, the monophyly of many genera and the validity of the nomenclature are unresolved. Therefore, re-establishing the Labeoninae phylogenetic relationship using suitable species is necessary.
Garra is a genus of fish in the carp family of the subfamily Labeoninae, distributed in southern China, across Southeast Asia, India, and the Middle East to northern and central Africa (STIASSNY and GETAHUN, 2007). Garra is usually found in swift-flowing streams, where they hold on to rocks with their highly modified mouths like suckers, including bottom-dwelling fishes. They feed on plant debris in open-water habitats and on periphyton in bottom-surface habitats (Kullander and Fang, 2004; Nebeshwar et al., 2009). Currently, 20 different Garra species have been identified from the Brahmaputra River basin, three of which in China, namely, G. dengba, G. Tibetan, and G. yajiangensis caught our attention due to the rearrangement in their mitochondrial genome (Deng et al., 2018; Gong, 2018; Gong et al., 2018; Zhang et al., 2020). They were all collected from high altitudes. Among them, G. dengba is derived from the Chayu-Qu, a stream that flows into the Brahmaputra River in eastern Tibet, China. It shares with the five Garra fishes (G. arupi, G. elongata, G. gravelyi, G. kalpangi, and G. rotundinasus) an incipient proboscis on the nose, but differs from them in its less divergent dorsal and anal fins and more perforated lateral line scales (Deng et al., 2018). Meanwhile, due to the similarity of morphological characters, G. tibetana was for a long time mistakenly identified as G. kempi (Galtier et al., 2009). G. yajiangensis, a member of the long-nosed species group, is distinguished from other members of the group mainly by the presence of a prominent, quadrangular, slightly bilobed proboscis. Apart from G. tibetana has been reported as a complete mitochondrial sequence, current researches on these three Garra fishes are mainly stagnant in morphology while their corresponding molecular evolutions are still limited (Zhang et al., 2020). In addition, Zhang et al. (2020) identified two control regions in the mitochondrial genome of G. tibetana (Zhang et al., 2020). Also, Li et al. (2016) reported that the tRNA-Pro gene of G. kempi is positioned between two control regions, and a 246 bp repeat unit is identified in the second control region, which is different from the typical mitochondrial genome organization in vertebrate (Li et al., 2016). However, He et al. (2016) reported that the complete mitochondrial genome of G. imberba is a classic structure (He et al., 2016). A different mitochondrial genome composition is present in Garra fish, and no one has yet identified the reason for this phenomenon. Therefore, this study reports novel gene rearrangement in the mitochondrial genome of G. dengba, G. tibetana, and G. yajiangensis, which could provide important genomic information for studying the evolutionary relationships and population genetics of the genus Garra.
The mitochondrial genome (Mitogenome) houses its DNA and encodes many key proteins for the assembling and activation of the mitochondrial respiratory complex (Yan et al., 2019). Mitogenomes as a molecule marker were used for species classification, population genetics, molecular systematic geography, molecular ecology, and other areas (Groves and Shields, 1996; Nielsen et al., 2010; Cheng et al., 2012; Zhang et al., 2018). Vertebrate mitogenomes are generally 16–20 kb in length (Boore, 1999), compared to plant mitochondrial genomes, which are shorter. The structure of the vertebrate mitogenome typically consists of 13 protein-coding genes (PCGs), 22 tRNA genes (tRNAs), and two rRNA genes (rRNAs) (Boore, 1999). In the vertebrate mitogenome, the order of these genes is conserved and generally unaltered (Bibb et al., 1981; Roe et al., 1985). The mitochondrial genome is inherited maternally in most animals and therefore has a very low rate of recombination (Brown et al., 1979). However, variants of the gene sequence have now been identified in a variety of vertebrates, including amphibians, reptiles, birds, marsupials, and fish (Pääbo et al., 1991; Macey et al., 1997; Eberhard and Wright, 2015; Liu et al., 2019a; Lü et al., 2019). In recent years, with the advancement of sequencing techniques, more and more mitochondrial rearrangements are being identified.
In general, the structure of the fish mitochondrial genome is extremely conserved (Gong et al., 2013). With the gradual increase in mitochondrial DNA sequence data for fish, reports of mitogenome rearrangements continue to appear (Miya et al., 2001; Inoue et al., 2003; Zhang et al., 2021a). To date, a few models have been employed for mitochondrial gene rearrangement. These models include the tandem duplication/random loss (TDRL) (Moritz and Brown, 1987), recombination model (Lunt and Hyman, 1997), tRNA mismatch model (Cantatore et al., 1987), tandem duplication/nonrandom loss model (RDNL) (Lavrov et al., 2000) and double replication/random loss model (DRRL) (Shi et al., 2014). The TDRL model occurs by tandem duplication of certain genes that undergo rearrangement, followed by random deletion of repetitive sequences. The model has been widely applied to explain the translocation of genes encoded on the same strand (Shi et al., 2015; Wang et al., 2018). The RDNL model differs from TDRL in non-random loss and is dependent on the transcriptional polarity and location of the gene.
In this study, three mitogenomes of Garra species (G. dengba, G. tibetana, and G. yajiangensis) were sequenced and annotated. We have also characterized their mitochondrial genome structure and phylogenetic analysis within their subfamily Labeoninae. In addition to this, we have explored the mechanisms by which their mitochondrial genes undergo rearrangement. Based on the above analysis, we hope that these results will provide a better understanding of the mechanisms by which rearrangements occur in the mitochondrial genomes of Garra species, as well as their evolution.
The sample of G. dengba was collected in Chayu City, Tibet, China (28°66′ N, 97°46′ E). Samples of G. tibetana and G. yajiangensis were collected in Motuo City, Tibet, China (29°32′ N, 95°33′ E). Total genomic DNA was extracted from muscle tissue using the Qiagen QIAamp tissue kit according to the manufacturer’s protocol. The DNA sediment was solubilized in double-distilled water, stored at 4°C, and then quantified in concentration. The G. tibetana mitogenome was amplified with ten pairs of universal primer by general PCR. The general PCR cycle requirement for DNA amplification is 5 min at 94°C, [30 s at 94°C, 30 s at 55–56°C, 1 min at 72°C] X 35 cycles, and 10 min at 72°C. Sequences were sequenced using an ABI Genetic Analyzer (Applied Biosystems, China). Complete mitogenome sequencing of G. dengba and G. yajiangensis was performed on an Illumina HiSeq X Ten platform.
The acquired sequence PCR fragments were processed through CodonCode Aligner 9.0.1 (CodonCode Corporation, Dedham, MA) and the complete mitochondrial genome was assembled. Clean data without sequencing adapters were assembled de novo using NOVOPlasty software (Dierckxsens et al., 2016). The new mitogenome was annotated with the vertebrate mitochondrial code using the MITOS (Donath et al., 2019). The structures of the 22 tRNAs were determined using tRNA-scan 2.0 (Chan et al., 2021) and then the tRNA structures were mapped by online web software Forna (Kerpedjiev et al., 2015). The circular map of the mitochondrial genome was produced by using the OGDRAW v1.3.1 (Greiner et al., 2019). Base composition and relative synonymous codon usage (RSCU) for 13 PCGs of Garra were computed and sorted using MEGA X (Kumar et al., 2018). Composition skew values were calculated by the following formulas: AT skew = (A—T)/(A + T); GC skew = (G—C)/(G + C) (Perna and Kocher, 1995). The putative origin of L-strand replication (OL) was identified by the Mfold Web Server (Zuker, 2003).
Rapid identification of 12 PCGs in the mitochondrial genome was performed using DAMBE v7.2.3 software (Xia, 2018). 13 PCGs were used excluding ND6 owing to its heterogeneous base composition and poor consistent phylogenetic performance (Miya et al., 2003). Sequences were compared using the default parameters of Clustal X 2.0 (Larkin et al., 2007). The ambiguous sequences were eliminated by Gblock (Talavera and Castresana, 2007). Subsequently, the results of the multiple sequence comparisons were then used to construct phylogenetic trees based on maximum likelihood (ML) and Bayesian inference (BI) analyses. The ML tree was built in PhyML 3.0 (Guindon et al., 2010), using the best model (TVM + F + R8) selected in ModelFinder with 1000 nonparametric bootstrap replications (Kalyaanamoorthy et al., 2017). Bayesian inference (BI) methods were used with the program MrBayes 3.2 (Ronquist et al., 2012). Based on the Akaike information criteria (AIC), MrModeltest 2.2 (Nylander et al., 2004) was performed to determine the best evolutionary model among 24 models for BI analysis and pointed out that GTR + G + I was the analytical data set for the best-fit substitution model. BI analysis was performed using Markov chain Monte Carlo (MCMC), sampled every 1,000 generations each with three heated chains and one cold chain run for 6,000,000 generations, and the first 25% of the burns were discarded. Visualization of the tree was realized using online web iTOL v5 (https://itol.embl.de) (Letunic and Bork, 2021).
The complete mitochondrial genomes of three Garra (G. dengba, accession no. OL826794; G. tibetana accession no. NC_045032 and G. yajiangensis, accession no. OL826795) in GenBank were 16,876, 16,861 and 16,835 bp in length, respectively (Table 1). The structure of the mitochondrial genomes of these three fish species is extremely similar. The mitochondrial genomes of all three species are very similar in structure, containing 13 protein-coding genes (PCGs), two rRNAs, 22 tRNAs, a light-stranded replication initiation region (OL), and two control regions (CR). Compared to the structure of the mitochondrial genomes of most teleost fishes, the mitochondrial genomes of the three fishes in this study have an extra CR. The main reason for this is the rearrangement of mitochondrial genes in these three fish species (Figure 5). The presence of replicating CR was considered a special feature in the vertebrate mitochondrial genome (Jiang et al., 2007; Shi et al., 2013). The nucleotide composition of G dengba, G tibetana, and G yajiangensis mitogenomes had a higher A + T bias of 57.42, 58.44, and 58.44%, respectively, and both showed positive AT-skew and negative GC-skew (Table 2).
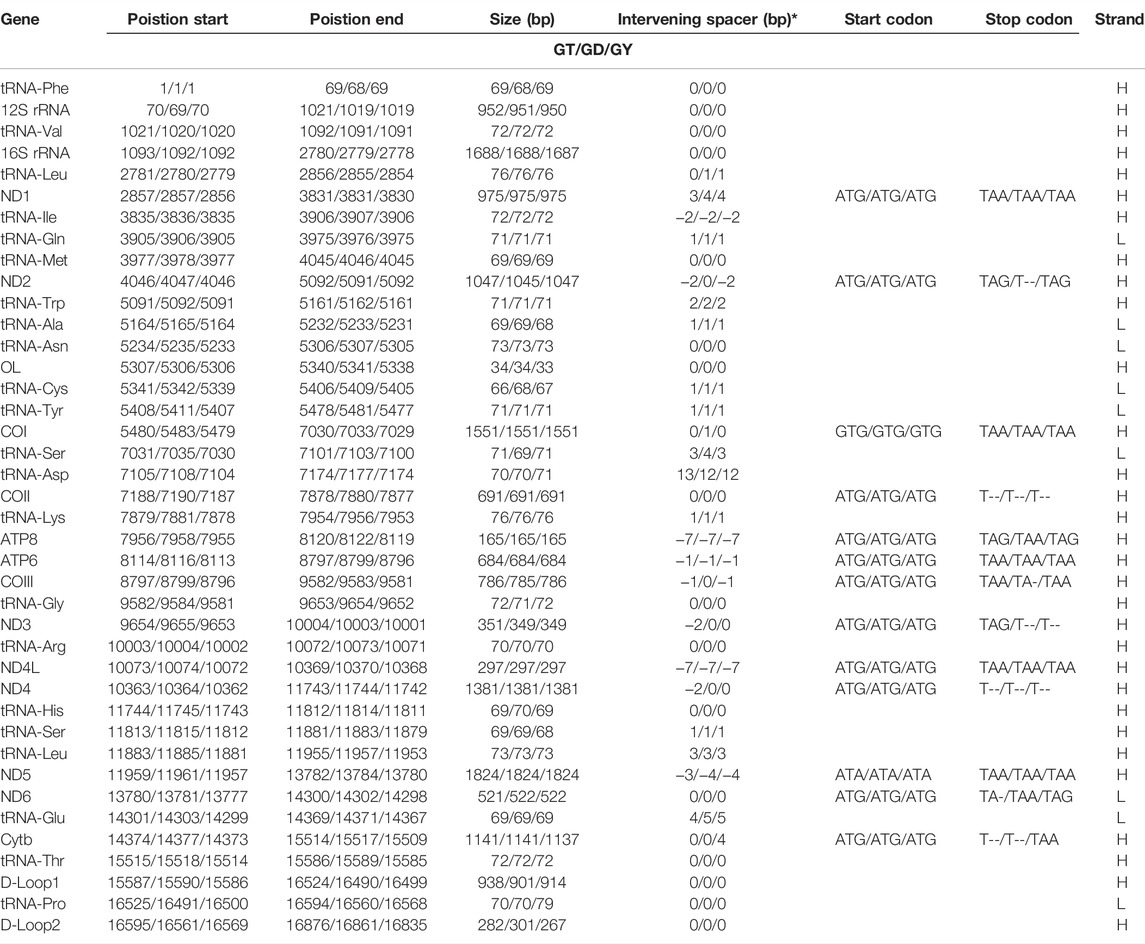
TABLE 1. Summary of gene/element feature of Garra dengba (GD), Garra tibetana (GT) and Garra yajiangensis (GY).
Like the typical mitochondrial genome of vertebrates (Zhang et al., 2021b; Liu et al., 2019b), the 13 PCGs consist of one cytochrome b (Cyt b), two ATPases (ATP6 and ATP8), and three cytochrome c oxidases (COI-COIII), and seven NADH dehydrogenases (ND1-ND6 and ND4L). Twelve PCGs (ND1, ND2, COI, COII, ATP8, ATP6, COIII, ND3, ND4L, ND4, ND5, and Cytb) were coded on the heavy strand (H-strand) and the remaining ND6 gene was coded on the light strand (L-strand) (Table 1). All PCGs use the start codon ATG except for COI, which uses GTG. Most of the 13 PCGs ended with TAA or TAG, whereas four other PCGs (COI, ND4, ND6, and Cytb) use a single T-- or TA- as the stop codon (Table 1). The incomplete stop codon has also been found in all other Garra species (Su et al., 2015; Xiong et al., 2016). The presence of incomplete stop codons is a common phenomenon in invertebrate and vertebrate mitochondrial genes (Lin et al., 2020; Zhang et al., 2021c). All 12 PCGs genes (ND1-ND3, COI-COIII, ATP8, ATP6, ND4L, ND4, ND5) in the three Garra mitochondria had negative GC-skew values and they were all encoded on the H-strand, in addition, the ND6 gene encoded on the L-strand had a positive GC-skew value (Table 2; Supplementary Table S1, S2).
Depending on the codon degeneration pattern, the amino acids serine and leucine are encoded by six synonymous codons, and the remaining amino acids are encoded by four or two codons. A count of the translated amino acid content of 13 PCGs was found. The top utilized amino acids are Leu, Ile, and Ala, and the few top utilized amino acids are Asp, Arg, and Cys (Figure S1). This phenomenon is also commonly seen in the mitochondrial genomes of other fish (Su et al., 2015; Zhang et al., 2021b). We measured the RSCU of the three Garra species mitogenomes (Figure 1) and the results showed that the usage of NNA and NNC (N for A, T, C, G) was more frequent than NNT and NNG in the G. dengba, G. tibetana, and G. yajiangensis.
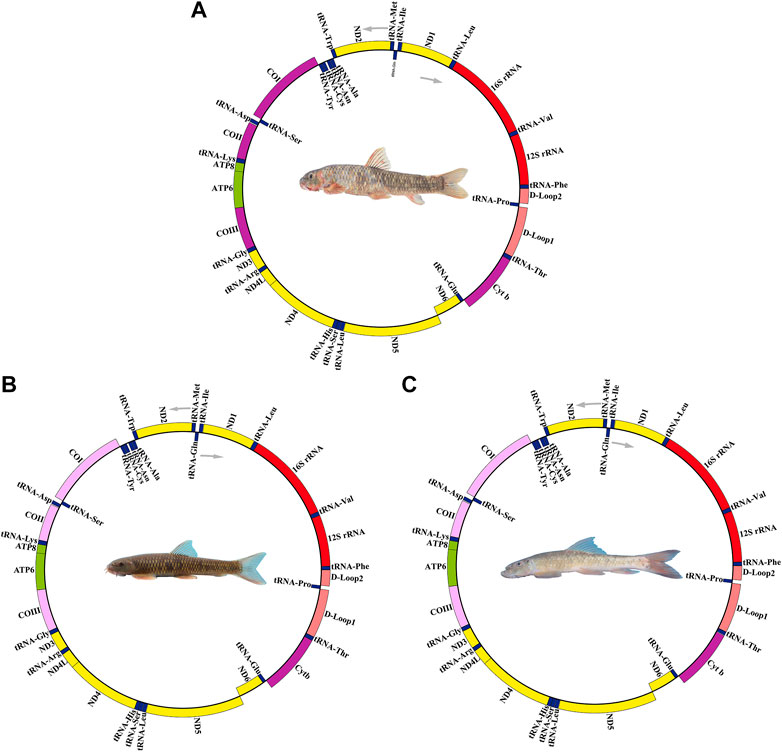
FIGURE 1. The RSCU of the mitogenome in (A) Garra dengba, (B) Garra tibetana and (C) Garra yajiangensis. The RSCU of the three Garra species mitogenomes show that the usage of NNA and NNC (N for A, T, C, G) was more frequent than NNT and NNG in the G. dengba, G. tibetana, and G. yajiangensis.
Like the typical set of tRNA genes in fish mitogenomes, all three Garra species mitogenomes included 22 tRNAs, a 12S rRNA, and 16S rRNA. Of these 22 tRNAs, 14 tRNAs were encoded on the H-strand and the remaining eight (tRNA-Gln, tRNA-Ala, tRNA-Asn, tRNA-Cys, tRNA-Tyr, tRNA-Ser, tRNA-Glu, and tRNA-Pro) were encoded on the L-strand (Figure 2). All of them had two kinds of tRNA-Leu and tRNA-Ser (Table 1). Secondary clover structure of tRNA genes identified in the mitogenome of G. dengba, G. tibetana, and G. yajiangensis were shown in Supplementary Figures S2–4. Out of 22 tRNAs, all have a typical cloverleaf structure except for tRNA-Ser (GCT), which lacks the entire dihydrouridine arm. The 12S rRNA gene was positioned between tRNA-Phe and tRNA-Val, and the 16S rRNA gene was located between tRNA-Val and tRNA-Leu (UUR). The lengths of 12S rRNA were 952, 951, and 950 in the mitochondrial genomes of G. dengba, G. tibetana, and G. yajiangensis, respectively. The size of the 16S rRNA in G. dengba and G. tibetana were 1,688 bp, both a little longer than in G. yajiangensis (1,687bp). Positive AT-skew values for both tRNA and rRNA in the mitochondrial genomes of G. dengba, G. tibetana, and G. yajiangensis indicate that the As content is higher than the Ts content (Table 2, Supplementary Tables S1, S2).
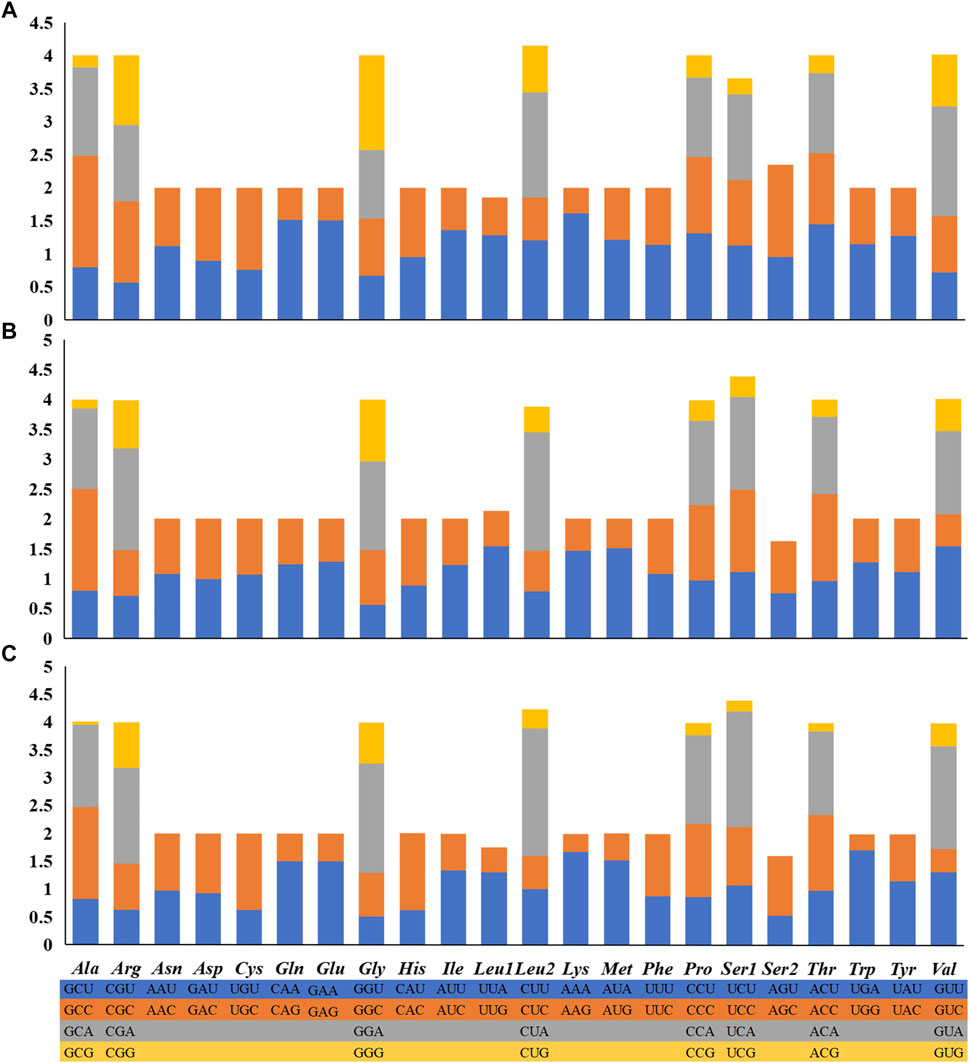
FIGURE 2. Mitochondrial genome maps of the three Garra in this study. (A) Garra dengba mitochondrial genome visualization ring diagram. (B) Garra tibetana mitochondrial genome visualization ring diagram. (C) Garra yajiangensis mitochondrial genome visualization ring diagram. Genes encoded on the heavy or light strands are shown at the outside or inside of the circular gene map, respectively. The mitochondrial genome visualization ring diagram of the three species show the same structure and all have two control regions.
The origin of light chain replication (OL) was usually found within a WANCY group and can fold into a stem-loop ring secondary structure (Zhang et al., 2021a). The OL of G. dengba and G. tibetana were located between tRNA-Asn and tRNA-Cys with the same length of 34 bp, and 1 bp longer than G. yajiangensis (33 bp) (Table 1). The two-structure prediction of the OL region revealed that the stem lengths in the OL regions of G. dengba, G. tibetana, and G. yajiangensis were 10, 5, and 9 bp, respectively, and the loops were 11, 15, and 10nt, respectively (Figure 3). In these OL structures, the utilization of stem codons showed a clear asymmetry, with more pyrimidines at the 5′ end of the sequence. In contrast to other fish studies (Catanese et al., 2010), the conserved sequence 5′-GCCGG-3′ was not present in the OL region in this study, but rather a segment of the 5′-GGCGG-3′ sequence was present.
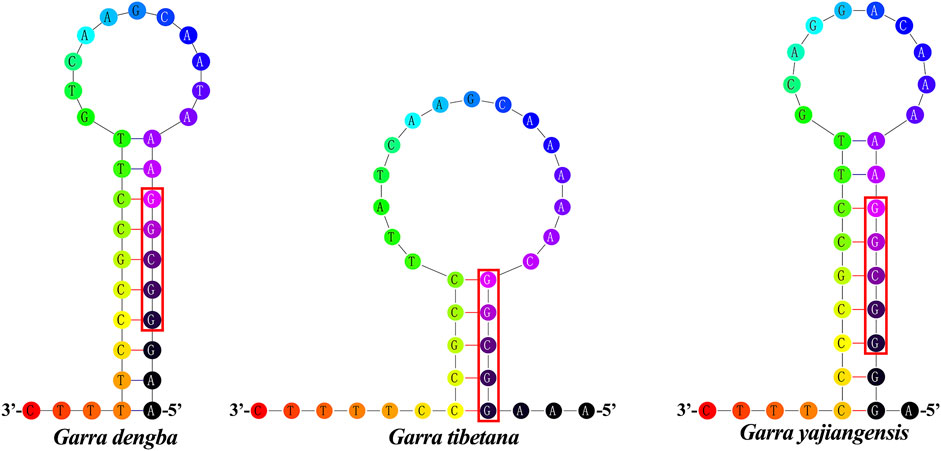
FIGURE 3. Inferred secondary structures of the OL of Garra mitogenome, (A) Garra dengba, (B) Garra tibetana, and (C) Garra yajiangensis. The OLs of the three Garra mitogenomes were located between tRNA-Asn and tRNA-Cys, and G. dengba and G. tibetana were of the same length (34 bp) and longer than G. yajiangensis (33 bp).
The CR is the greatest non-coding region in the fish mitogenome which is analogous to the A + T enriched region of the insect mitogenome (Liu et al., 2013). The CR1 and CR2 of Garra mitogenomes were located between tRNA-Thr and tRNA-Pro, tRNA-Pro, and tRNA-Phe, respectively (Figure 1). The lengths of CR1 in the three Garra mitogenomes were 938, 901, and 914 bp (Table 1), and the ratio of AT was 66.10, 66.37, and 65.75%, respectively (Table 2, Supplementary Tables S1, S2). The length of CR2 in the three Garra mitogenomes was 282, 301, and 267 bp (Table 1), and the ratio of AT was 73.76, 71.43, and 73.41%, respectively (Table 2, Supplementary Tables S1, S2). The palindromic sequence motifs “TACAT” and “ATGTA” related to heavy chain replication termination were found in both CRs. The “TACAT” and “ATGTA” motifs are generally referred to as the terminator region because of their ability to form a stable hairpin structure (Figure 4, in the red box). Six Garra CRs were compared to probe the conservation of sequences. The results revealed 12 conserved blocks in Figure 4 (In the yellow box). As far as we know, this is the first report of a study reporting conserved blocks of the CR in Garra mitogenomes.

FIGURE 4. Compositional features of the first control region of the rearranged Garra mitochondrial genome. Palindromic motif sequences “TACAT” and “ATGTA” are marked in red boxes. Conservative sequences are marked using yellow boxes. DBCR1: The first control region of Garra dengba. XZCR1: The first control region of Garra tibetana. YJCR1: The first control region of Garra yajiangensis. GKCR1: The first control region of Garra kempi. GQCR1: The first control region of Garra qiaojiensis. GSCR1: The first control region of Garra salweenica.
By comparing the structure of the mitochondrial genomes of other vertebrates (Kim et al., 2004), we found that the mitochondrial genomes of the three Garra species had undergone partial recombination of genes. The two genes that changed position in the mitochondrial genome of these three Garra species were tRNA-Pro and CR. In general, the tRNA-Pro was positioned between the tRNA-Thr and CR, and only one CR region was positioned at the end of the mitogenome. However, the position of tRNA-Pro in the mitochondrial genome of these three Garra species changed, and a CR was copied (Figure 5). In this study, the gene sequences were identical to those of the vertebrate mitochondrial genome, except for the tRNA-Pro translocation and CR repeats.
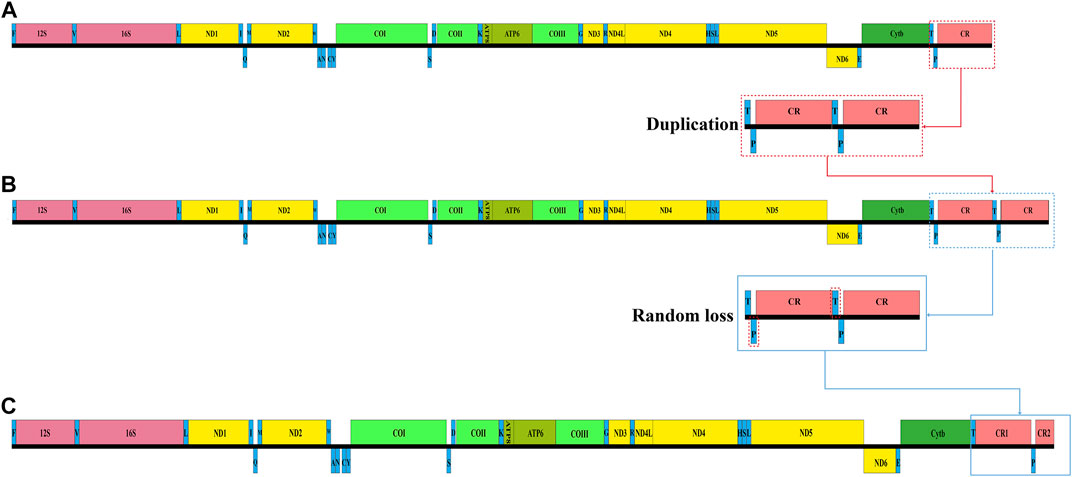
FIGURE 5. Analysis of Garra mitochondrial gene rearrangement. (A) The typical mitochondrial genome organization in Garra. (B) The normal mitochondrial gene blocks (T-P-CR) form gene blocks (T-P-CR-T-P-CR) after complete duplication. (C) The rearrangement mitochondrial genome organization of Garra species. The replicated CR is left behind in some genes (T and P) that undergo random loss events. F: tRNA-Phe; V: tRNA-Val; L: tRNA-Leu; I: tRNA-Ile; Q: tRNA-Gln; M: tRNA-Met; W: tRNA-Trp; A: tRNA-Ala; N: tRNA-Asn; C: tRNA-Cys; Y: tRNA-Tyr; S: tRNA-Ser; D: tRNA-Asp; K: tRNA-Lys; G: tRNA-Gly; R: tRNA-Arg; H: tRNA-His; S: tRNA-Ser; L: tRNA-Leu; E: tRNA-Glu; T: tRNA-Thr; P: tRNA-Pro.
To better characterize the mechanics of gene rearrangement that occur in the Garra mitochondrial genome, we have explored this in-depth. Several models have been proposed for the mechanism of rearrangement of the mitochondrial genome (Poulton et al., 1993; Arndt and Smith, 1998; Lavrov et al., 2002; Shi et al., 2014). The recombination model is only appropriate for the exchange and inversion of small fragments (Dowton and Campbell, 2001; Kong et al., 2009); thus, this recombination model was not suited to explaining the mitochondrial gene rearrangement of Garra. Regarding the two models, TDNL and DRRL, they are often used to explain mitochondrial gene rearrangements, but with genes aggregated in the same pole (encoded by the L- or H-strand) and with no change in relative order. Therefore, neither model can be used to explain the rearrangement mechanism that occurred in this study. The TDRL model was due to incomplete deletion of duplicated genes resulting in intergenic spacer regions or pseudogenes (Shi et al., 2015; Gong et al., 2019). This TDRL phenotype explains well the gene rearrangements with redundant genes. Evidence for the TDRL model was the presence of pseudogenes or duplicate genes and the location of gene spacer regions. Therefore, in the present study, the TDRL model was recommended to study rearrangement events, as gene rearrangements with duplicate CRs were observed in the mitochondrial genome of Garra.
Based on the putative model described above, we inferred the regions of mitochondrial gene rearrangement in three Garra species. First, normal mitochondrial gene blocks (T-P-CR) form gene blocks (T-P-CR-T-P-CR) after complete duplication (Figure 5A). Subsequently, after successive replications, some genes (T and P) undergo random loss events, leaving only an incompletely replicated CR (Figure 5B). Thus, after this replication and random loss, both CRs are preserved (Figure 5C).
To further study the evolutionary status of G. dengba, G. tibetana, and G. yajiangensis in the Labeoninae, we selected 58 closely related subfamilies and two outgroups (Sahyadria chalakkudiensis and Sahyadria denisonii) to construct evolutionary trees (BI and ML) to analyze phylogenetic relationships (Table 3). The results showed that the ML tree and the BI tree have the same topological structure. Thus, only one topology (BI) is displayed. In addition, the support values of BI tree are higher than ML tree (Figure 6). The phylogenetic trees showed that the subfamily Labeoninae is divided into three major clades and 13 genera (Cirrhinus, Labeo, Henicorhynchus, Crossocheilus, Thynnichthys, Osteochilus, Labiobarbus, Garra, Bangana, Discogobio Parasinilabeo, Semilabeo, Ptychidio). First, the clade1, genera Cirrhinus, and Labeo show a sister relationship, both of them were once classified in the genus Labeonini (Yang et al., 2012). Then, the clade2, including Henicorhynchus, Crossocheilus, Thynnichthys, Osteochilus, and Labiobarbus, which belonged to “Osteochilini” (Saenjundaeng et al., 2020). The clade3 aggregates all mitochondrial genes with rearrangements in the genus Garra. Except for Bangana and Garra2, all other species of the fourth clade used to belong to the genus Semilabeonini (Stout et al., 2016). The genus Bangana belonged to the cyprinid tribe Labeonini too (Zhang and Chen, 2006; Kottelat, 2017).
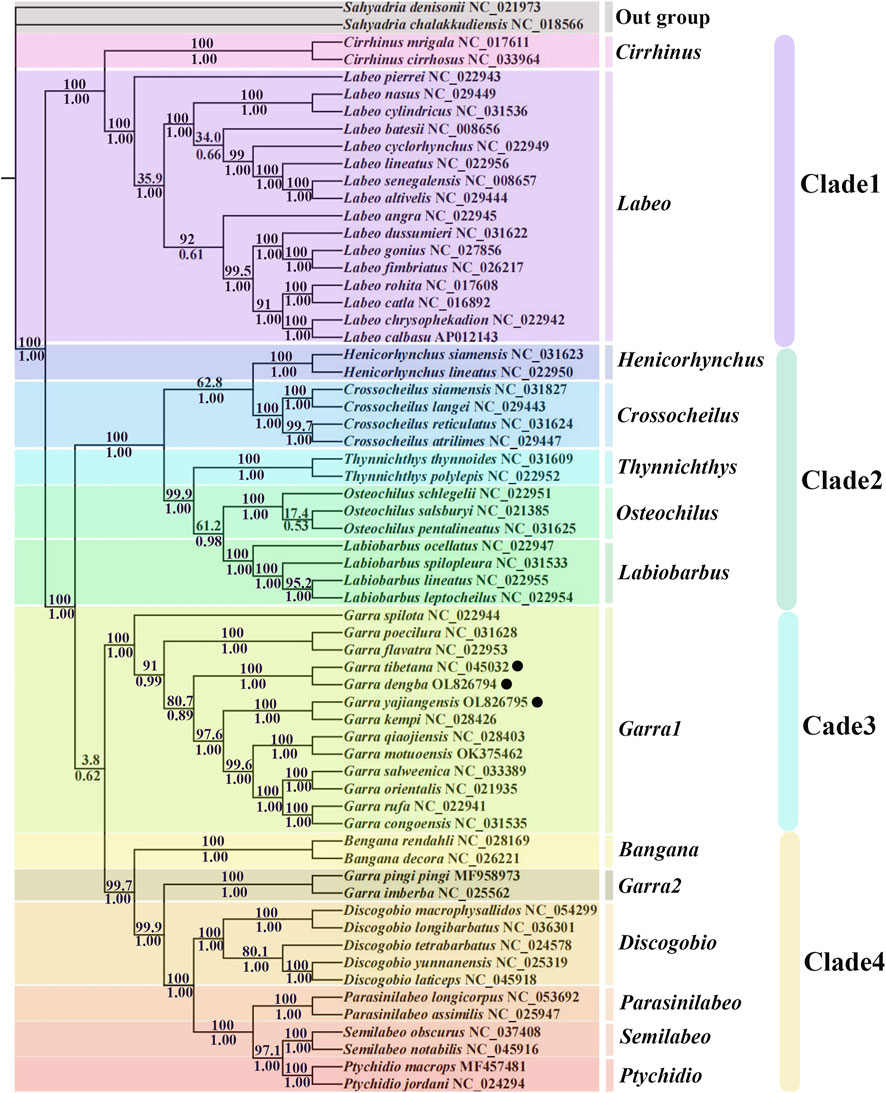
FIGURE 6. Phylogenetic analysis is based on the nucleotide sequences of the 12 PCGs in the mitogenome. The numbers beside the nodes are posterior probabilities (BI, bottom) and bootstrap (ML, top). Black point: gene rearrangement occurs.
In addition, four clades in this phylogenetic tree show high support values except for the value between clade3 and clade4 (posterior probabilities = 0.62; bootstrap = 3.8; Figure 6), indicating that these two clades in Labeoninae are not highly differentiated. All mitochondrial sequences of genus Garra in clade3 have two control regions (Figure 5C) (Li et al., 2016; Su et al., 2015; Xiong et al., 2016; Li et al., 2016), while G. pingi pingi and G. imberba have only one control region and clusters with other Labeoninae species in clade4 without rearrangements, which is consistent with previous studies (Figure 5A) (Zou et al., 2018). Furthermore, each of the 13 different Garra fishes in clade3 clustered into six pairs of sister branches, with only G. spilota forming a monophyletic branch. The three species from Tibet described in this study (species marked by dots in the tree), G. tibetana and G. dengba, form sister branches, showing the close relationship between them. G. yajiangensis and G. kempi cluster together, while G. qiaojiensis showed a close relationship with G. kempi in Zou et al. (2018)’ s study, which may be limited by the number of mitochondria in Garra (Zou et al., 2018). It is certain that Garra1 and Garra2 are distantly related due to the rearrangement of mitochondria in Garra1 species, whereas Garra2 species are not rearranged. Thus, we also found that the species of Garra2 are all from Sichuan (He et al., 2016; Zou et al., 2018), and we speculated that none mitochondria of Garra in this region have two control regions or other rearrangements. However, due to the lack of Garra samples from Sichuan, further sample collection is needed to confirm this conjecture.
With the development of genetic investigations, the reliance on molecular info is gradually surpassing morphological data, and molecular assays have become the most used method in the study of biological system development. Therefore, in this study, we sequenced and assembled the complete mitochondrial gene of G. dengba, G. tibetana, and G. yajiangensis and determined its characteristics, which contains 37 genes and two control regions. Upon comparison with typical vertebrate mitochondrial genes, we discovered a clear rearrangement of mitochondrial genes in G. dengba, G. tibetana, and G. yajiangensis, the position of tRNA-Pro in the mitochondrial genome of these three Garra species changed, accompanied by CR repeat. The TDRL model is most suitable to explain the gene rearrangements with redundant genes in this study. The two phylogenetic trees (BI and ML) were generated based on the mitochondrial genomes of 61 Labeoninae. Both phylogenetic trees strongly favor the non-monophyly of Garra, which were divided into two different clades (rearrangement and no rearrangement), providing a more advanced classification of Labeoninae. Further, our findings provide a theoretical ground for a deeper understanding of the mechanism and evolution of genus Garra gene rearrangement and phylogenetic studies of Labeoninae.
The raw data is publicly accessible at GenBank under Bioproject accession number PRJNA837076 and the annotation sequences were submitted to NCBI (G. dengba, accession no. OL826794; G. tibetana accession no. NC_045032 and G. yajiangensis, accession no. OL826795).
The animal study was reviewed and approved by the Zhejiang Ocean University.
CZ, KZ, and BL conceived and designed research. CZ, KZ, YP, JZ,YL, and BL conducted experiments, analyzed data, and wrote the manuscript. The authors critically reviewed and approved the manuscript.
The study was supported by the Foreign Experts scientific research project in Tibet of China [2020WZ007]; Science and Technology Project of Zhoushan (2020C21016); Zhejiang Provincial Natural Science Foundation of China (LY22D060001&LY20C190008); National Natural Science Foundation of China (NSFC) (NO.41806156); Fund of Guangdong Provincial Key Laboratory of Fishery Ecology and Environment (FEEL-2021-8); Open Foundation from Marine Sciences in the First-Class Subjects of Zhejiang (20200201, 20200202).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.922634/full#supplementary-material
Arndt, A., and Smith, M. J. (1998). Mitochondrial Gene Rearrangement in the Sea Cucumber Genus Cucumaria. Mol. Biol. Evol. 15 (8), 1009–1016. doi:10.1093/oxfordjournals.molbev.a025999
Bibb, M. J., Van Etten, R. A., Wright, C. T., Walberg, M. W., and Clayton, D. A. (1981). Sequence and Gene Organization of Mouse Mitochondrial DNA. Cell. 26, 167–180. doi:10.1016/0092-8674(81)90300-7
Boore, J. L. (1999). Animal Mitochondrial Genomes. Nucleic Acids Res. 27 (8), 1767–1780. doi:10.1093/nar/27.8.1767
Brown, W. M., George, M., and Wilson, A. C. (1979). Rapid Evolution of Animal Mitochondrial DNA. Proc. Natl. Acad. Sci. U.S.A. 76, 1967–1971. doi:10.1073/pnas.76.4.1967
Cantatore, P., Gadaleta, M. N., Roberti, M., Saccone, C., and Wilson, A. C. (1987). Duplication and Remoulding of tRNA Genes during the Evolutionary Rearrangement of Mitochondrial Genomes. Nature 329 (6142), 853–855. doi:10.1038/329853a0
Catanese, G., Manchado, M., and Infante, C. (2010). Evolutionary Relatedness of Mackerels of the Genus Scomber Based on Complete Mitochondrial Genomes: Strong Support to the Recognition of Atlantic Scomber Colias and Pacific Scomber Japonicus as Distinct Species. Gene 452 (1), 35–43. doi:10.1016/j.gene.2009.12.004
Chan, P. P., Lin, B. Y., Mak, A. J., and Lowe, T. M. (2021). tRNAscan-SE 2.0: Improved Detection and Functional Classification of Transfer RNA Genes. Nucleic Acids Res. 49 (16), 9077–9096. doi:10.1093/nar/gkab688
Cheng, J., Ma, G.-q., Miao, Z.-q., Shui, B.-n., and Gao, T.-x. (2012). Complete Mitochondrial Genome Sequence of the Spinyhead Croaker Collichthys Lucidus (Perciformes, Sciaenidae) with Phylogenetic Considerations. Mol. Biol. Rep. 39 (4), 4249–4259. doi:10.1007/s11033-011-1211-6
Deng, S.-Q., Cao, L., and Zhang, E. (2018). Garra Dengba, a New Species of Cyprinid Fish (Pisces: Teleostei) from Eastern Tibet, China. Zootaxa 4476 (1), 94–108. doi:10.11646/zootaxa.4476.1.9
Dierckxsens, N., Mardulyn, P., and Smits, G. (2016). NOVOPlasty: De Novo Assembly of Organelle Genomes from Whole Genome Data. Nucleic Acids Res. 45 (4), e18. doi:10.1093/nar/gkw955
Donath, A., Jühling, F., Al-Arab, M., Bernhart, S. H., Reinhardt, F., Stadler, P. F., et al. (2019). Improved Annotation of Protein-Coding Genes Boundaries in Metazoan Mitochondrial Genomes. Nucleic Acids Res. 47 (20), 10543–10552. doi:10.1093/nar/gkz833
Dowton, M., and Campbell, N. J. H. (2001). Intramitochondrial Recombination - Is it Why Some Mitochondrial Genes Sleep Around? Trends Ecol. Evol. 16 (6), 269–271. doi:10.1016/s0169-5347(01)02182-6
Eberhard, J. R., and Wright, T. F. (2015). Rearrangement and Evolution of Mitochondrial Genomes in Parrots. Mol. Phylogenet Evol. 94 (Pt), 34–46. doi:10.1016/j.ympev.2015.08.011
Galtier, N., Nabholz, B., Glémin, S., and Hurst, G. D. D. (2009). Mitochondrial DNA as a Marker of Molecular Diversity: a Reappraisal. Mol. Ecol. 18 (22), 4541–4550. doi:10.1111/j.1365-294x.2009.04380.x
Gong, L., Shi, W., Si, L. Z., and Kong, X. Y. (2013). Rearrangement of Mitochondrial Genome in Fishes. Dongwuxue Yanjiu 34 (6), 666–673. doi:10.11813/j.issn.0254-5853.2013.6.0666
Gong, L., Jiang, H., Zhu, K., Lu, X., Liu, L., Liu, B., et al. (2019). Large-scale Mitochondrial Gene Rearrangements in the Hermit Crab Pagurus Nigrofascia and Phylogenetic Analysis of the Anomura. Gene 695, 75–83. doi:10.1016/j.gene.2019.01.035
Gong, Z. (2018). A New Species of Genus Garra (Cypriniformes: Cyprinidae) from the Lower Yarlung Tsangpo River Raffinage in Southeastern Tibet, China. Chin. J. Zool. 53, 857–867. doi:10.13859/j.cjz.201806004
Gong, Z., Freyhof, J., Wang, J., Liu, M., Liu, F., Lin, P., et al. (2018). Two New Species of Garra (Cypriniformes: Cyprinidae) from the Lower Yarlung Tsangpo River Drainage in Southeastern Tibet, China. Zootaxa 4532 (3), 367–384. doi:10.11646/zootaxa.4532.3.3
Greiner, S., Lehwark, P., and Bock, R. (2019). OrganellarGenomeDRAW (OGDRAW) Version 1.3.1: Expanded Toolkit for the Graphical Visualization of Organellar Genomes. Nucleic Acids Res. 47 (W1), W59–W64. doi:10.1093/nar/gkz238
Groves, P., and Shields, G. F. (1996). Phylogenetics of the Caprinae Based on Cytochrome B Sequence. Mol. Phylogenetics Evol. 5 (3), 467–476. doi:10.1006/mpev.1996.0043
Guindon, S., Dufayard, J.-F., Lefort, V., Anisimova, M., Hordijk, W., and Gascuel, O. (2010). New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0. Syst. Biol. 59 (3), 307–321. doi:10.1093/sysbio/syq010
He, B., Lai, J., Liu, Y., Zhou, J., Chen, Y., and Liu, G. (2016). The Complete Mitochondrial Genome of Garra Imberba (Teleostei, Cyprinidae, Garra). Mitochondrial DNA A DNA Mapp. Seq. Anal. 27 (3), 1646–1647. doi:10.3109/19401736.2014.958710
Howes, G., Winfield, I., and Nelson, J. (1991). Cyprinid Fishes: Systematics, Biology and Exploitation. New Dehli: Chapman & Hall.
Inoue, J. G., Miya, M., Tsukamoto, K., and Nishida, M. (2003). Evolution of the Deep-Sea Gulper Eel Mitochondrial Genomes: Large-Scale Gene Rearrangements Originated within the Eels. Mol. Biol. Evol. (11), 11. doi:10.1093/molbev/msg206
Jiang, Z. J., Castoe, T. A., Austin, C. C., Burbrink, F. T., Herron, M. D., McGuire, J. A., et al. (2007). Comparative Mitochondrial Genomics of Snakes: Extraordinary Substitution Rate Dynamics and Functionality of the Duplicate Control Region. BMC Evol. Biol. 7 (1), 123. doi:10.1186/1471-2148-7-123
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., von Haeseler, A., and Jermiin, L. S. (2017). ModelFinder: Fast Model Selection for Accurate Phylogenetic Estimates. Nat. Methods 14 (6), 587–589. doi:10.1038/nmeth.4285
Kerpedjiev, P., Hammer, S., and Hofacker, I. L. (2015). Forna (Force-directed RNA): Simple and Effective Online RNA Secondary Structure Diagrams. Bioinformatics 31 (20), 3377–3379. doi:10.1093/bioinformatics/btv372
Kim, I.-C., Kweon, H.-S., Kim, Y. J., Kim, C.-B., Gye, M. C., Lee, W.-O., et al. (2004). The complete mitochondrial genome of the javeline goby Acanthogobius hasta (Perciformes, Gobiidae) and phylogenetic considerations. Gene 336 (2), 147–153. doi:10.1016/j.gene.2004.04.009
Kong, X., Dong, X., Zhang, Y., Shi, W., Wang, Z., and Yu, Z. (2009). A Novel Rearrangement in the Mitochondrial Genome of Tongue Sole,Cynoglossus Semilaevis: Control Region Translocation and a tRNA Gene Inversion. Genome 52 (12), 975–984. doi:10.1139/g09-069
Kottelat, M. (2017). Speolabeo, a New Genus Name for the Cave Fish Bangana Musaei (Teleostei: Cyprinidae). Zootaxa 4254 (4), 493–499. doi:10.11646/zootaxa.4254.4.6
Kullander, S. O., and Fang, F. (2004). Seven New Species of Garra (Cyprinidae: Cyprininae) from the Rakhine Yoma, Southern Myanmar. Ichthyological Explor. Freshwaters 15, 257–278.
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 35 (6), 1547–1549. doi:10.1093/molbev/msy096
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., et al. (2007). Clustal W and Clustal X Version 2.0. Bioinformatics 23 (21), 2947–2948. doi:10.1093/bioinformatics/btm404
Lavrov, D. V., Boore, J. L., and Brown, W. M. (2002). Complete mtDNA Sequences of Two Millipedes Suggest a New Model for Mitochondrial Gene Rearrangements: Duplication and Nonrandom Loss. Mol. Biol. Evol. 19 (2), 163–169. doi:10.1093/oxfordjournals.molbev.a004068
Lavrov, D. V., Brown, W. M., and Boore, J. L. (2000). A Novel Type of RNA Editing Occurs in the Mitochondrial tRNAs of the Centipede Lithobius forficatus. Proc. Natl. Acad. Sci. U.S.A. 97 (25), 13738–13742. doi:10.1073/pnas.250402997
Letunic, I., and Bork, P. (2021). Interactive Tree of Life (iTOL) V5: an Online Tool for Phylogenetic Tree Display and Annotation. Nucleic acids Res. 49 (W1), W293–W296. doi:10.1093/nar/gkab301
Li, H., Liu, Y., Cao, H., Chen, Y., and Du, J. (2016). The Complete Mitochondrial Genome of Garra Kempi (Teleostei, Cyprinidae, Garra). Mitochondrial DNA Part A 27 (6), 4631–4632. doi:10.3109/19401736.2015.1101590
Li, W.-x., Ran, J.-c., and Chen, H.-m. (2006). A New Genus and Species of Labeoninae from Guizhou, 26. China: Journal of Zhanjiang Ocean University, 1–2.
Lin, J.-P., Tsai, M.-H., Kroh, A., Trautman, A., Machado, D. J., Chang, L.-Y., et al. (2020). The First Complete Mitochondrial Genome of the Sand Dollar Sinaechinocyamus Mai (Echinoidea: Clypeasteroida). Genomics 112 (2), 1686–1693. doi:10.1016/j.ygeno.2019.10.007
Liu, J., Yu, J., Zhou, M., and Yang, J. (2019). Complete Mitochondrial Genome of Japalura Flaviceps: Deep Insights into the Phylogeny and Gene Rearrangements of Agamidae Species. Int. J. Biol. Macromol. 125, 423–431. doi:10.1016/j.ijbiomac.2018.12.068
Liu, Q.-N., Zhu, B.-J., Dai, L.-S., and Liu, C.-L. (2013). The Complete Mitogenome of Bombyx mori Strain Dazao (Lepidoptera: Bombycidae) and Comparison with Other Lepidopteran Insects. Genomics 101 (1), 64–73. doi:10.1016/j.ygeno.2012.10.002
Liu, Y., Wu, P.-D., Zhang, D.-Z., Zhang, H.-B., Tang, B.-P., Liu, Q.-N., et al. (2019). Mitochondrial Genome of the Yellow Catfish Pelteobagrus fulvidraco and Insights into Bagridae Phylogenetics. Genomics 111 (6), 1258–1265. doi:10.1016/j.ygeno.2018.08.005
Lü, Z., Zhu, K., Jiang, H., Lu, X., Lu, B., Ye, Y., et al. (2019). Complete Mitochondrial Genome of Ophichthus Brevicaudatus Reveals Novel Gene Order and Phylogenetic Relationships of Anguilliformes. Int. J. Biol. Macromol. 135, 609–618. doi:10.1016/j.ijbiomac.2019.05.139
Lunt, D. H., and Hyman, B. C. (1997). Animal Mitochondrial DNA Recombination. Nature 387 (6630), 247. doi:10.1038/387247a0
Macey, J. R., Larson, A., Ananjeva, N. B., Fang, Z., and Papenfuss, T. J. (1997). Two Novel Gene Orders and the Role of Light-Strand Replication in Rearrangement of the Vertebrate Mitochondrial Genome. Mol. Biol. Evol. 14 (1), 91–104. doi:10.1093/oxfordjournals.molbev.a025706
Miya, M., Kawaguchi, A., and Nishida, M. (2001). Mitogenomic Exploration of Higher Teleostean Phylogenies: A Case Study for Moderate-Scale Evolutionary Genomics with 38 Newly Determined Complete Mitochondrial DNA Sequences. Mol. Biol. Evol. 18 (11), 1993–2009. doi:10.1093/oxfordjournals.molbev.a003741
Miya, M., Takeshima, H., Endo, H., Ishiguro, N. B., Inoue, J. G., Mukai, T., et al. (2003). Major Patterns of Higher Teleostean Phylogenies: a New Perspective Based on 100 Complete Mitochondrial DNA Sequences. Mol. Phylogenetics Evol. 26 (1), 121–138. doi:10.1016/s1055-7903(02)00332-9
Moritz, C., and Brown, W. M. (1987). Tandem Duplications in Animal Mitochondrial DNAs: Variation in Incidence and Gene Content Among Lizards. Proc. Natl. Acad. Sci. U.S.A. 84 (20), 7183–7187. doi:10.1073/pnas.84.20.7183
Nebeshwar, K., Vishwanath, W., and Das, D. N. (2009). Garra Arupi, a New Cyprinid Fish Species (Cypriniformes: Cyprinidae) from Upper Brahmaputra Basin in Arunachal Pradesh, India. J. Threat. Taxa 1 (4), 197–202. doi:10.11609/jott.o1842.197-202
Nielsen, J. L., Graziano, S. L., and Seitz, A. C. (2010). Fine-scale Population Genetic Structure in Alaskan Pacific Halibut (Hippoglossus stenolepis). Conserv. Genet. 11 (3), 999–1012. doi:10.1007/s10592-009-9943-8
Nylander, J. A. A., Ronquist, F., Huelsenbeck, J. P., and Nieves-Aldrey, J. (2004). Bayesian Phylogenetic Analysis of Combined Data. Syst. Biol. 53 (1), 47–67. doi:10.1080/10635150490264699
Pääbo, S., Kelley Thomas, W., Whitfield, K. M., Kumazawa, Y., and Wilson, A. C. (1991). Rearrangements of Mitochondrial Transfer RNA Genes in Marsupials. J. Mol. Evol. 33 (5), 426–430. doi:10.1007/bf02103134
Pei-Qi, C. X.-L. Y., and Een-Duan, Lin. (1984). Major Groups within the Family Cyprinidae and Their Phylogenetic Relationships. Acta zootaxonomica sin.
Perna, N. T., and Kocher, T. D. (1995). Patterns of Nucleotide Composition at Fourfold Degenerate Sites of Animal Mitochondrial Genomes. J. Mol. Evol. 41 (3), 353–358. doi:10.1007/bf01215182
Poulton, J., Deadman, M. E., Bindoff, L., Morten, K., Land, J., and Brown, G. (1993). Families of mtDNA Re-arrangements Can Be Detected in Patients with mtDNA Deletions: Duplications May Be a Transient Intermediate Form. Hum. Mol. Genet. 2 (1), 23–30. doi:10.1093/hmg/2.1.23
Roe, B. A., Ma, D. P., Wilson, R. K., and Wong, J. F. (1985). The Complete Nucleotide Sequence of the Xenopus laevis Mitochondrial Genome. J. Biol. Chem. 260 (17), 9759–9774. doi:10.1016/s0021-9258(17)39303-1
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D. L., Darling, A., Höhna, S., et al. (2012). MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice across a Large Model Space. Syst. Biol. 61 (3), 539–542. doi:10.1093/sysbio/sys029
Saenjundaeng, P., Supiwong, W., Sassi, F. M. C., Bertollo, L. A. C., Rab, P., Kretschmer, R., et al. (2020). Chromosomes of Asian Cyprinid Fishes: Variable Karyotype Patterns and Evolutionary Trends in the Genus Osteochilus (Cyprinidae, Labeoninae, "Osteochilini"). Genet. Mol. Biol. 43, e20200195. doi:10.1590/1678-4685-GMB-2020-0195
Shi, W., Dong, X.-L., Wang, Z.-M., Miao, X.-G., Wang, S.-Y., and Kong, X.-Y. (2013). Complete Mitogenome Sequences of Four Flatfishes (Pleuronectiformes) Reveal a Novel Gene Arrangement of L-Strand Coding Genes. BMC Evol. Biol. 13 (1), 173. doi:10.1186/1471-2148-13-173
Shi, W., Gong, L., Wang, S.-Y., Miao, X.-G., and Kong, X.-Y. (2015). Tandem Duplication and Random Loss for Mitogenome Rearrangement in Symphurus (Teleost: Pleuronectiformes). BMC Genomics 16 (1), 355. doi:10.1186/s12864-015-1581-6
Shi, W., Miao, X.-G., and Kong, X.-Y. (2014). A Novel Model of Double Replications and Random Loss Accounts for Rearrangements in the Mitogenome of Samariscus Latus (Teleostei: Pleuronectiformes). BMC Genomics 15 (1), 352. doi:10.1186/1471-2164-15-352
Stiassny, M. L. J., and Getahun, A. (2007). An Overview of Labeonin Relationships and the Phylogenetic Placement of the Afro-Asian Genus Garra Hamilton, 1922 (Teleostei: Cyprinidae), with the Description of Five New Species of Garra from Ethiopia, and a Key to All African Species. Zoological J. Linn. Soc. 150 (1), 41–83. doi:10.1111/j.1096-3642.2007.00281.x
Stout, C. C., Tan, M., Lemmon, A. R., Lemmon, E. M., and Armbruster, J. W. (2016). Resolving Cypriniformes Relationships Using an Anchored Enrichment Approach. BMC Evol. Biol. 16 (1), 244–257. doi:10.1186/s12862-016-0819-5
Su, L.-W., Liu, Z.-Z., Ueng, Y.-T., Tang, W.-Q., Bao, B.-L., Liu, D., et al. (2015). The Complete Mitogenome of the Oriental Sucking Barb,Garra orientalis(Cypriniformes, Cyprinidae): Repetitive Sequences in the Control Region. Mitochondrial DNA 26 (2), 272–273. doi:10.3109/19401736.2013.823178
Talavera, G., and Castresana, J. (2007). Improvement of Phylogenies after Removing Divergent and Ambiguously Aligned Blocks from Protein Sequence Alignments. Syst. Biol. 56 (4), 564–577. doi:10.1080/10635150701472164
Wang, Z., Wang, Z., Shi, X., Wu, Q., Tao, Y., Guo, H., et al. (2018). Complete Mitochondrial Genome of Parasesarma Affine (Brachyura: Sesarmidae): Gene Rearrangements in Sesarmidae and Phylogenetic Analysis of the Brachyura. Int. J. Biol. Macromol. 118, 31–40. doi:10.1016/j.ijbiomac.2018.06.056
Winfield, I. J., and Nelson, J. S. (2012). Cyprinid Fishes: Systematics, Biology and Exploitation, 3. Springer Science & Business Media.
Xia, X. (2018). DAMBE7: New and Improved Tools for Data Analysis in Molecular Biology and Evolution. Mol. Biol. Evol. 35 (6), 1550–1552. doi:10.1093/molbev/msy073
Xiong, H., Chen, W., Gan, X., and Wang, X. (2016). Complete Sequence and Rearrangement of the Mitochondrial Genome ofGarra qiaojiensis(Cypriniformes: Cyprinidae). Mitochondrial DNA 27 (1), 36–37. doi:10.3109/19401736.2013.869682
Yan, C., Duanmu, fnm., Zeng, fnm., Liu, fnm., and Song, fnm. (2019). Mitochondrial DNA: Distribution, Mutations, and Elimination. Cells 8 (4), 379. doi:10.3390/cells8040379
Yang, L., Arunachalam, M., Sado, T., Levin, B. A., Golubtsov, A. S., Freyhof, J., et al. (2012). Molecular Phylogeny of the Cyprinid Tribe Labeonini (Teleostei: Cypriniformes). Mol. Phylogenetics Evol. 65 (2), 362–379. doi:10.1016/j.ympev.2012.06.007
Yang, L., and Mayden, R. L. (2010). Phylogenetic Relationships, Subdivision, and Biogeography of the Cyprinid Tribe Labeonini (Sensu Rainboth, 1991) (Teleostei: Cypriniformes), with Comments on the Implications of Lips and Associated Structures in the Labeonin Classification. Mol. Phylogenetics Evol. 54 (1), 254–265. doi:10.1016/j.ympev.2009.09.027
Yuan, L.-Y., Zhang, E., and Huang, Y.-F. (2008). Revision of the Labeonine Genus Sinocrossocheilus (Teleostei: Cyprinidae) from South China. Zootaxa 1809 (1), 36–48. doi:10.11646/zootaxa.1809.1.2
Zhang, C., Wang, W., Zhou, J., Chen, M., and Han, X. (2020). The Complete Mitochondrial Genome of the Garra Tibetana (Cypriniformes: Cyprinidae). Mitochondrial DNA Part B 5 (2), 1737–1738. doi:10.1080/23802359.2020.1748537
Zhang, E., and Chen, Y.-Y. (2004). Qianlabeo Striatus, a New Genus and Species of Labeoninae from Guizhou Province, China (Teleostei: Cyprinidae). Hydrobiologia 527 (1), 25–33. doi:10.1023/b:hydr.0000043315.64357.da
Zhang, E., and Chen, Y.-Y. (2006). Revised Diagnosis of the Genus Bangana Hamilton, 1822 (Pisces: Cyprinidae), with Taxonomic and Nomenclatural Notes on the Chinese Species. Zootaxa 1281 (1), 41–54. doi:10.11646/zootaxa.1281.1.3
Zhang, E., and Fang, F. (2005). Linichthys: a New Genus of Chinese Cyprinid Fishes (Teleostei: Cypriniformes). Copeia 2005 (1), 61–67. doi:10.1643/ci-03-256r2
Zhang, E., Qiang, X., and Lan, J.-H. (2008). Description of a New Genus and Two New Species of Labeonine Fishes from South China (Teleostei: Cyprinidae). Zootaxa 1682 (1), 33–44. doi:10.11646/zootaxa.1682.1.3
Zhang, K., Liu, Y., Chen, J., Zhang, H., Gong, L., Jiang, L., et al. (2021). Characterization of the Complete Mitochondrial Genome of Macrotocinclus Affinis (Siluriformes; Loricariidae) and Phylogenetic Studies of Siluriformes. Mol. Biol. Rep. 48 (1), 677–689. doi:10.1007/s11033-020-06120-z
Zhang, K., Zhu, K., Liu, Y., Zhang, H., Gong, L., Jiang, L., et al. (2021). Novel Gene Rearrangement in the Mitochondrial Genome of Muraenesox cinereus and the Phylogenetic Relationship of Anguilliformes. Sci. Rep. 11 (1), 2411. doi:10.1038/s41598-021-81622-9
Zhang, L.-P., Cai, Y.-Y., Yu, D.-N., Storey, K. B., and Zhang, J.-Y. (2018). Gene Characteristics of the Complete Mitochondrial Genomes of Paratoxodera Polyacantha and Toxodera Hauseri (Mantodea: Toxoderidae). PeerJ 6, e4595. doi:10.7717/peerj.4595
Zhang, Y., Gao, Y., Gong, L., Lu, X., Jiang, L., Liu, B., et al. (2021). Mitochondrial Genome of Episesarma Lafondii (Brachyura: Sesarmidae) and Comparison with Other Sesarmid Crabs. J. Ocean. Univ. China 20 (6), 1545–1556. doi:10.1007/s11802-021-4779-z
Zou, Y.-C., Chen, M., Qin, C.-J., Wang, Y.-M., Li, R., Qi, Z.-M., et al. (2018). Identification of the Complete Mitochondrial Genome of Garra Pingi Pingi (Cypriniformes, Cyprinidae). Conserv. Genet. Resour. 10 (4), 693–696. doi:10.1007/s12686-017-0903-1
Keywords: Garra, gene rearrangement, complete mtDNA sequence, Labeoninae, phylogeny
Citation: Zhang C, Zhang K, Peng Y, Zhou J, Liu Y and Liu B (2022) Novel Gene Rearrangement in the Mitochondrial Genome of Three Garra and Insights Into the Phylogenetic Relationships of Labeoninae. Front. Genet. 13:922634. doi: 10.3389/fgene.2022.922634
Received: 18 April 2022; Accepted: 16 May 2022;
Published: 08 June 2022.
Edited by:
Genlou Sun, Saint Mary’s University, CanadaReviewed by:
Zhong-duo Wang, Guangdong Ocean University, ChinaCopyright © 2022 Zhang, Zhang, Peng, Zhou, Liu and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bingjian Liu, YmpldGJqQDE2My5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.