- 1College of Chemistry and Life Sciences, Zhejiang Normal University, Jinhua, China
- 2Guangxi Academy of Agricultural Sciences, Nanning, China
- 3State Key Laboratory of Rice Biology, China National Rice Research Institute, Hangzhou, China
Plant lesion mimics refer to necrotic spots spontaneously produced by the plant without mechanical damage, pathogen invasion, and adversity stress. Here, we isolated and characterized two rice (Oryza sativa L) mutants, namely, spl88-1 (spotted leaf88-1) and spl88-2 (spotted leaf88-2), which were identified from an ethyl methanesulfonate-mutagenized japonica cultivar Xiushui 11 population. Physiological and biochemical experiments indicated that more ROS accumulated in spl88-1 and spl88-2 than in wild type. spl88-1 and spl88-2 displayed spontaneous cell death and enhanced their resistance to bacterial blight by affecting the expression of defense-related genes. We isolated SPL88 by map-based cloning, which encoded a highly conserved Cullin protein. A single base deletion was detected in spl88-1 and spl88-2, in which the 132nd base C of SPL88-1 and the 381th base T of SPL88-2 were deleted, causing premature termination of protein translation. SPL88 was expressed in root, stem, leaf, leaf sheath, and panicle. The Cullin protein was localized in the cytoplasm and nucleus. The aforementioned results indicate that SPL88 regulates the growth and development of rice by affecting the expression of defense-related genes.
Introduction
Plant lesion mimics are the spontaneous formation of necrotic spots on leaves and sheath (even stem and seed) in abiotic or biotic stress (Cai et al., 2021). This process involves programmed cell death (PCD) (Huang et al., 2010; McGrann et al., 2015), and the spot area can reflect the degree of cell death (Shirasu and Pual, 2000). According to the developmental process of the lesion mimics, it can be divided into the whole life lesion mimics, such as lrd32, lrd35, and lrd40.; vegetative initiation lesion mimics, such as lrd31, lrd41, and lrd44; and reproductive initiation lesion mimics, such as lrd27, lrd28 and lrd39 (Wang et al., 2004).
The mechanism underlying the generation of lesion mimics is very complex, mainly due to resistance-related gene mutations, chlorophyll metabolism disorders, programmed cell death defects, and changes in signal pathways (Takahashi et al., 1999; Wu et al., 2008). The mutation of the NLS1 gene encoding CC-NB-LRR protein blocks the independent defense signaling pathway between salicylic acid (SA) and NPR1, causing a large accumulation of SA and hydrogen peroxide (H2O2), resulting in lesion mimics on rice leaf sheaths (Tang et al., 2011). Les22 encodes a key enzyme in the chlorophyll biosynthesis pathway, uroporphyrinogen decarboxylase (UROD). After mutation, it leads to the accumulation of its substrate uroporphyrin and induces lesion mimics on maize leaves (Hu et al., 1998). LSD1P encodes a zinc finger protein that negatively regulates programmed cell death depending on the accumulation of SA and the expression of NIM1/NPR1 (Aviv et al., 2002). Copines are a calcium-dependent phospholipid binding protein Arabidopsis mutant cpn1-1 that inhibits the production of lesion mimics in high humidity, indicating that the primary function of CPN1 may be the regulation of plant responses to low humidity (Jambunathan et al., 2001).
The current study shows that since not all lesion mimic mutants are caused by mutations in disease resistance–related genes, most of them have significantly increased disease resistance (Mizobuchi et al., 2002). In 1965, the lesion mimic mutant spl1 was first discovered in the rice cultivar Asahi (Liu et al., 2004), which enhanced blast resistance and was accompanied with a high expression of defense-related genes. Wu et al. (Wu et al., 2008) obtained 21 lesion mimic mutants through four different mutagenesis methods and found spl21 and spl24 had bacterial blight enhanced resistance and provided essential materials for dissection of the disease resistance pathways in rice. To further explore the relationship between PCD and disease resistance, Yin et al. (Yin et al., 2000) found that spl11 leaf lesion mimics were caused by apoptosis through trypan blue staining, and spl11 had non-race-specific resistance. It means not only resistance to bacterial blight but also against blast, providing clues for developing broad-spectrum resistance to multiple pathogens.
The Cullin protein family has a conserved CULLIN homology domain, which can bind itself as a molecular scaffold to the RING E3 ubiquitin ligase to form the Cullin-RING E3 ubiquitin ligase complex (CRL); the ubiquitin/proteasome pathway plays an important role in growth and development (Zeng et al., 2004; Figueroa et al., 2005) and defense responses (Gao et al., 2020). In the mouse model, Cullin3 plays an indispensable role in early embryogenesis and cell division cycle (Singer et al., 1999). In Arabidopsis thaliana, Cullin3a and Cullin3b are functionally redundant, but the absence of Cullin3a and Cullin3b affects endosperm development and leads to embryo death (Thomann et al., 2005). Jin et al. (Zhao et al., 2014) found that DWARF3, a nuclear-localized protein of F-box, was rich in leucine repeats, interacts with OsCcullin1 to form an SCF complex (comprising Skip, Cullin, and F-box), and binds DWARF14 to inhibit branching of rice shoots. OsCUL3a interacts with OsRBX1a and OsRBX1b in rice to form a CUL loop-like E3 ubiquitin ligase complex, which depends on the 26s proteasome to degrade OsNPR1 and negatively regulate plant immunity and cell death (Liu et al., 2017).
Here, we described two rice lesion mimic mutants that exhibited programmed cell death and increased resistance to bacterial blight. On the basis of map-based cloning and complementation research studies, we demonstrated that the phenotype of the mutants was caused by a mutation in SPL88.
Materials and Methods
Plant Material and Population Construction
The spotted leaf mutants spl88-1 and spl88-2 were isolated from a methanesulfonate (EMS)-induced mutant library of Xiushui 11 rice (wild type, WT). Two mutants were hybridized with NJ06 as the male parent, and the F1 offspring and the F2 population were grown in the rice experimental field of Zhejiang Normal University, Jinhua City, Zhejiang Province, China, during the summer of 2020 and 2021. The F2 populations of both spl88-1/NJ06 and spl88-2/NJ06 were used for genetic analysis, and the F2 recessive individuals of spl88-1/NJ06 and spl88-2/NJ06 were used for gene mapping. The agronomic traits of WT, spl88-1, and spl88-2 were statistically analyzed, including plant height, tiller number, number of branches, grain number per panicle, 1000-grain weight, and seed setting rate. These results were analyzed based on the average of 10 replicates. The relevant data were statistically analyzed by SPSS 20.0 software, and graphs were plotted by GraphPad Prism 7 software.
Shading Assay
At the tillering stage, when the mutants just appeared the lesion mimicked the phenotype. WT, spl88-1, and spl88-2 were subjected to shading experiments and used about 2 cm of tin foil to shade the leaves where no lesion mimics appeared. Observations, photographs, and comparisons were made after 7 days.
Measuring Photosynthetic Parameters and Chlorophyll Content
At noon in summer, ten WT, spl88-1, and spl88-2 with relatively uniform growth vigor were selected to measure with an LI-6400XT portable photosynthesis tester at the tillering stage. Three biological replicates were performed for each measurement.
Flag leaves of wild type, spl88-1, and spl88-2 were sampled separately for chlorophyll determination. The leaves were weighed, and 0.2 g of leaf tissue was cut into pieces, soaked in 95% alcohol, wrapped in tin foil and stored at 4°C for 24 h, and inverted up and down every 8 h until the green of the leaves was completely faded. The absorbance values at 470 nm, 645, and 663 nm were measured by a spectrophotometer, and the photosynthetic pigment content was calculated by the method of Lichtenthaler (Lichtenthaler, 1987). Three biological replicates were performed for each group and statistically analyzed by Student’s t-test.
Electron Microscope Observation and Histochemical Analysis
At the tillering stage, WT and mutant plants with relatively uniform growth vigor were selected for scanning electron microscope (SEM) and transmission electron microscope (TEM) observation. The experimental method was referred to Rao (Rao, 2016). The content of hydrogen peroxide (H2O2), reactive oxygen species (ROS) and malondialdehyde (MDA) and the enzymatic activity of superoxide dismutase (SOD) and peroxidase (POD) were analyzed under the instructions of the manufacturer. (Nanjing Jiancheng Bioengineering Institute, Nanjing, China). TdT-mediated dUTP Nick-End Labeling (TUNEL) was used to detect DNA fragmentation and determine the degree of apoptosis. At the tillering stage, the leaves of the WT, spl88-1, and spl88-2 were taken and placed in a centrifuge tube filled with 2 ml of FAA fixative (Liang and Zhou, 2018) and the centrifuge tube was pumped into a vacuum until the tissue sample was completely sunk to the bottom of the tube. An apoptosis detection kit was used to measure apoptosis in the samples (Inada et al., 1998).
Inoculation Test
Xanthomonas oryzae pv. Oryzae, Xoo (the causal agent of bacterial blight) was inoculated onto the fresh leaf, free of disease spots, and senescence flag leaves of WT, spl88-1, and spl88-2 at the tillering stage. Specifically, the tip of each green and healthy rice leaf (∼1.5 cm) was cut with scissors that was dipped into bacterial blight solution before making each cut. The phenotype was observed 15 days after inoculation. Moreover, the length of the lesion was photographed and measured.
Quantitative Reverse-Transcription PCR Analysis
Root, stem, leaf, leaf sheath, and panicle samples were collected from WT and mutant plants at each stage of development. Total RNA of rice was extracted from the samples by an RNAprep pure Plant Kit (Cat No. DP432, Tiangen Biotech, Beijing, China), amplified using a ReverTra Ace® qPCR RT Master with a+ gDNA Remover kit (Code: FSQ-301, TOYOBO, Japan) and backup for post-reverse transcription. Reverse-transcription PCR (qRT-PCR) was used to detect the expression of defense-related genes in mutant plants and the expression of SPL88 in tissues, with the OsActin gene used as an internal reference (GenBank accession number: NM001058705). The reaction mixtures contained 1 μl cDNA template, 5 μl 2 × SYBR qPCR mix, 1 μl each of forward and reverse primers, and ddH2O to a final volume of 10 μl. The reaction program was 98°C for 30 s; 98°C for 5 s, 55°C for 10 s; and 72°C for 5 s for 40 cycles (Jozefczuk and Adjaye, 2011). Each reaction was performed in triplicate, using the 2−ΔΔCt method for calculations. RT-PCR was performed using a quantitative fluorescence gene amplification instrument (qTOWER3G; Jena, Germany). Data were analyzed by Excel and SPSS 20.0 software. Student’s t-test was used to analyze the significance of the differences. The primers used for qRT-PCR are shown in Supplementary Table S1.
Vector Construction
The complete genomic DNA fragment (including the promoter) of WT SPL88 was amplified by PCR with primers SPL88-1300-F/SPL88-1300-R and then ligated with the digested binary vector pCAMBIA1300 by the Clontech In-Fusion PCR (TaKaRa) kit to construct complementary vectors of mutants spl88-1 and spl88-2. The primer pair SPL881305.1-F/SPL88-1305.1-R was used to amplify the promoter sequence of the SPL88 gene, and the sequence was ligated with the binary vector pCAMBIA1305.1 to obtain the GUS vectors, investigating the tissue expression of SPL88 genes. The full-length SPL88 open-reading frame (ORF) was amplified with the primer pair SPL88-1132-F/SPL88-1132-R, and the coding sequence of SPL88 was inserted into the binary vector pYBA1132 containing the 35S promoter (p35S::SPL88) for subcellular localization analysis. The primers used for vector construction are shown in Supplementary Table S2.
Transcriptome Sequencing and data Analysis
At the tillering stage, flag leaves of wild-type, spl88-1, and spl88-2 were sampled, and RNA extraction was performed with an RNAprep pure Plant Kit (Cat No. DP432, Tiangen Biotech, Beijing, China). Illumina HiSeq (Illumina, San Diego, CA, United States) library construction was performed according to the manufacturer’s instructions. Differentially expressed genes (DEGs) were detected by DESeq (http://www-huber.embl.de/users/anders/DESeq/) programs. Gene function was analyzed with the KEGG analysis tool (http://www.genome.jp/kegg/). Sequencing and bioinformatics services were completed by Shanghai Paisenuo Biotechnology Co., Ltd (China).
Result
Phenotype of the Lesion Mimic Mutants spl88-1 and spl88-2
Under normal planting conditions in the summer field, the leaves of spl88-1 and spl88-2 were not significantly different from those of WT (Figure 1A) before the tillering stage. During the tillering stage, the necrotic spots appeared from the apex of the leaf, and from the tillering stage to the heading stage, the necrotic spots became more serious and spread to the whole leaf (Figure 1B). The phenotype of most rice lesion formation is induced by light (Livak and Schmittgen, 2001). During the tillering stage, spl88-1 and spl88-2 were subjected to shading experiments, and some newly emerged leaves were covered with 2–3 cm of aluminum foil and their phenotype was observed after 7 days. It was found that no lesions were produced in the area covered by tinfoil, but the lesions spread on the uncovered leaves (Figure 1C). These results indicate that spl88-1 and spl88-2 developed lesions in a light-dependent manner.
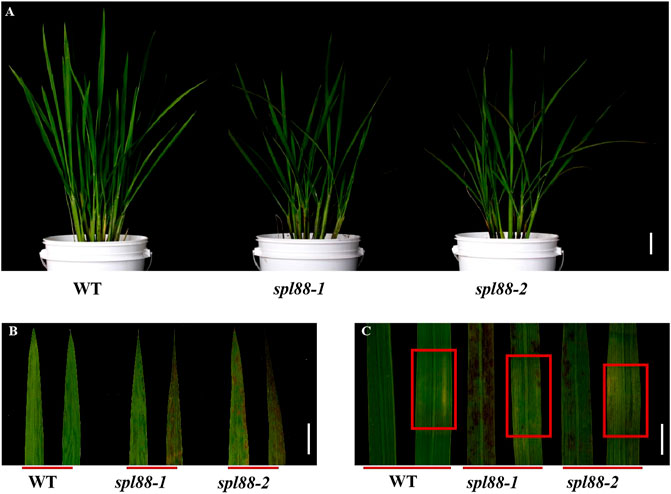
FIGURE 1. Phenotypic analysis of the wild type and mutant plants. (A) Phenotype diagram of wild type and mutant plants; Bar = 15 cm. (B) Leaf phenotype of wild type and mutant plants; Bar = 3 cm. (C) Phenotype of wild type and mutant plants before and after shading; Bar = 2 cm.
Agronomic Traits of the Lesion Mimic Mutants spl88-1 and spl88-2
The main agronomic traits of WT, spl88-1, and spl88-2 were statistically analyzed at the booting and maturity stages. Compared with those of WT, plant height, tiller number, number of branches, number of grains, 1000-grain weight, and seed setting rate of spl88-1 and spl88-2 were significantly decreased (Figures 2A–F).
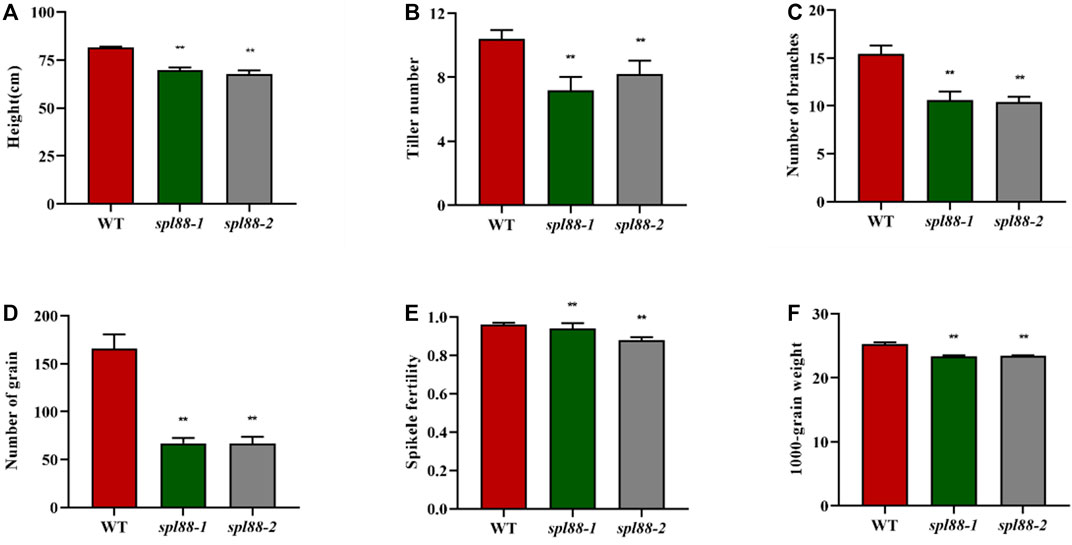
FIGURE 2. Main agronomic traits of wild-type and mutant plants. (A): plant height; (B): tiller number; (C): number of branches; (D): number of grain; (E): spikele fertility; (F): 1000-grain weight; values are means ± SD (n = 10); * indicates significance at p ≤ 0.05 by Student’s t test; ** indicates significance at p ≤ 0.01 by Student’s t test.
SPL88 Regulates Plant Growth and Development
We speculated that the lesion mimic phenotype of spl88-1 and spl88-2 affected the growth and development of plants. The chlorophyll content of WT, spl88-1, and spl88-2 was determined at the tillering stage. The results showed that the carotenoid content of spl88-1 and spl88-2 did not change significantly, while chlorophyll a and chlorophyll b were significantly lower than those in WT, and the chlorophyll a of spl88-1 decreased more significantly (Figure 3A). Compared with WT, the maximum photosynthetic rates of spl88-1 and spl88-2 were significantly decreased (Figure 3B). In addition, we observed the ultrastructure of chloroplasts by transmission electron microscope and found that the chloroplasts of spl88-1 and spl88-2 were shrunken and the internal lamella structure was disordered. More obviously, the osmium granules in the chloroplasts of spl88-1 and spl88-2 were obviously increased (Figures 3C–H). Scanning electron microscopy (SEM) results showed that the stomatal pore sizes of spl88-1 and spl88-2 were significantly larger than those of WT (Figure 3I–N). The aforementioned results indicate that the changes in chloroplast structure of spl88-1 and spl88-2 affect the growth and development of plants.
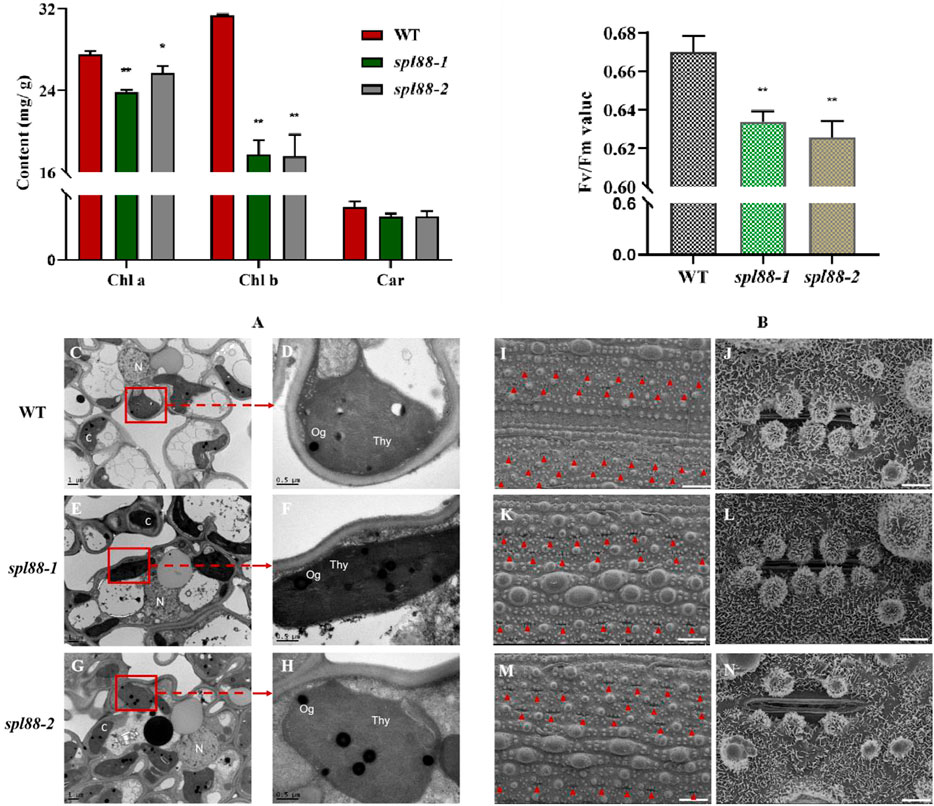
FIGURE 3. Chloroplast development, maximum photosynthetic rate, and the observation of mesophyll cell electron microscope in wild-type and mutant plants. (A) Chlorophyll content in flag leaves of wild-type and mutant plants. (B) Maximum photosynthetic rate of wild-type and mutant plants. (C–H) Ultrastructural observation of chloroplast of wild type and mutant plants; (N): nucleus; Thy: chloroplast; Og: osmium granules; Bar = 1 μm. (I–N): observation of stomatal pore diameter of wild-type and mutant plant leaves; I, (K,M): Bar = 300 μm; (J,L,N): Bar = 30 μm * indicates significance at p ≤ 0.05 by Student’s t-test; ** indicates significance at p ≤ 0.01 by Student’s t-test.
SPL88 Regulates ROS Accumulation and Cell Death in Rice
The TUNEL assay is a marker of programmed cell death (Kim et al., 2009; Wang et al., 2017), which is used to detect DNA fragmentation. To explore whether this type of lesion mimic phenotype could cause the PCD pathway of mutants spl88-1 and spl88-2, we performed TUNEL signal detection. A weaker TUNEL signal was observed in WT, while the TUNEL signals of spl88-1 and spl88-2 were significantly enhanced, and the fluorescence signal distribution was random (Figures 4A–F). When plants are under stress or senescence, high concentrations of ROS will accumulate, leading to formation of H2O2, which directly or indirectly oxidizes biological macromolecules such as nucleic acids, proteins, and lipids, thereby destroying the cell membrane structure and causing programmed cell death (Chen et al., 2018; Hoang et al., 2019). POD can catalyze the direct oxidation of H2O2 into phenolic or amine compounds to eliminate the toxic effect of H2O2 (Rechcigl and Evans, 1963); SOD can eliminate superoxide anion-free radicals and plays a crucial role in anti-oxidation (Sharma et al., 20122012; Yuchun et al., 2021). Therefore, we measured H2O2 content, POD activity, and SOD activity in the plants and found that a large amount of H2O2 accumulated in spl88-1 and spl88-2, while POD and SOD activities were significantly reduced, and the soluble protein content also decreased significantly. When plants accumulate large amounts of H2O2, lipid peroxidation occurs, producing large amounts of MDA (Sun et al., 2017; Tu et al., 2020). The MDA content in spl88-1 and spl88-2 was significantly higher than that in WT (Figures 4G–K).
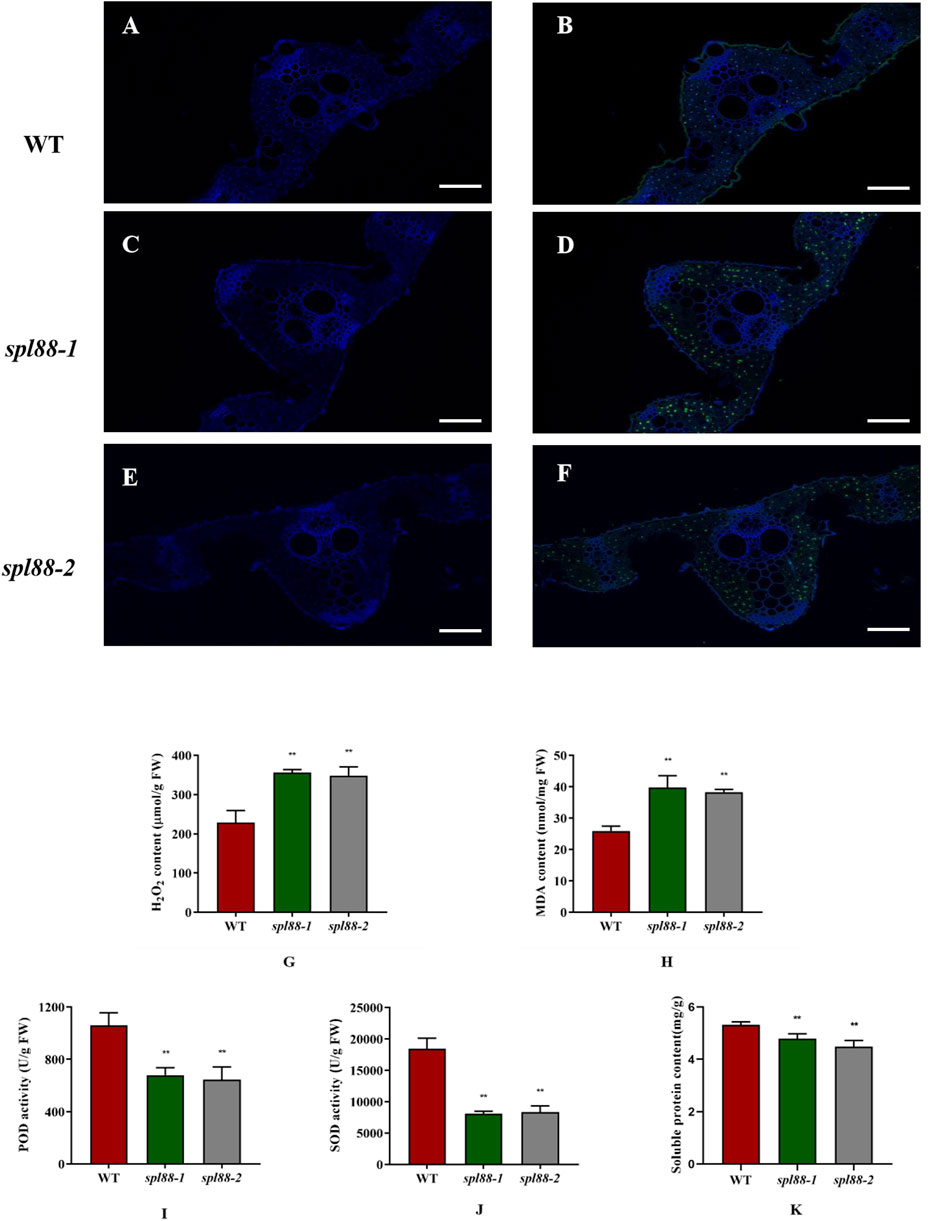
FIGURE 4. Physiological and biochemical analysis of wild-type and mutant plants at the tillering stage. (A–F): TUNEL assay of DNA fragmentation in mesophyll cells; Bar: 200 μm. (G–K): H2O2, MDA, and soluble protein contents of leaves, POD, and SOD activities in the leaves. *indicates significance at p ≤ 0.05 by Student’s t-test, ** indicates significance at p ≤ 0.01 by Student’s t-test.
SPL88 Regulates Defense Responses in Rice
Most rice lesion mimic mutants show resistance to fungal and bacterial pathogens. To investigate whether spl88-1 and spl88-2 also have this resistance, we used the leaf clipping method (Zang et al., 2000) to inoculate WT, spl88-1, and spl88-2 with the bacterial blight strain HM73. After 15 days of inoculation, the phenotype was observed and the length of the lesions was measured. The length of bacterial leaf blight lesions of WT was significantly longer than that of spl88-1 and spl88-2 (Figures 5A,B). This indicated resistance to the Xanthomonas oryzae pv. Oryzae, Xoo was significantly enhanced after the emergence of disease spots in spl88-1 and spl88-2. In addition, we detected the expression of defense-related genes in WT, spl88-1, and spl88-2 by qRT-PCR. The expression levels of defense-related genes MAPK12, AOS2, LYP6, PR2, ASP90, and PR1a in spl88-1 and spl88-2 were significantly increased (Figure 5C). Among them, the expression of AOS2 increased most obviously, and SPL88 may respond to the damage caused by the lesion mimics by inducing the synthesis of JA (Kemal and Manners, 2011; Wakuta et al., 2011). These results show that the loss of function of the SPL88-encoded protein triggers a defense response in rice, leading to enhanced resistance to pathogens in spl88-1 and spl88-2.
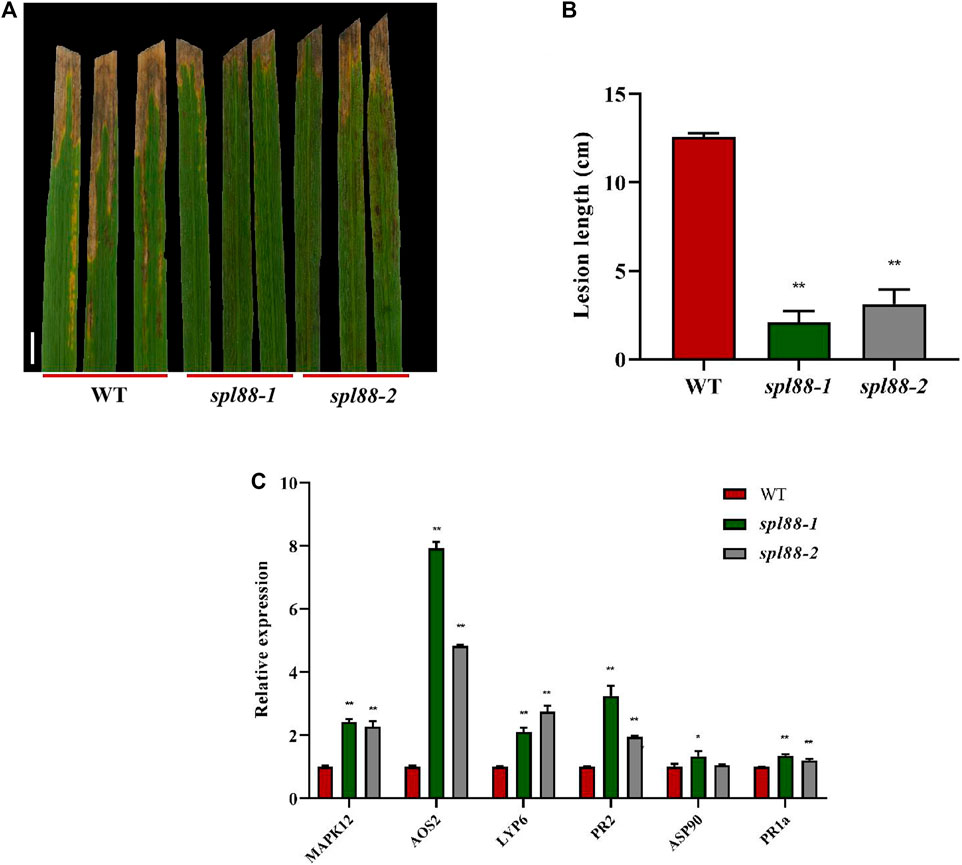
FIGURE 5. SPL88 regulates defense responses in rice. (A) Phenotype of wild-type and mutant plant leaves at 15 days after inoculation with the bacterial blight pathogen HM73, Bar = 3 cm; (B) statistical analysis of the length of bacterial leaf blight lesions; (C) relative expression of defense-related genes in plants; *indicates significance at p ≤ 0.05 by Student’s t test, **indicates significance at p ≤ 0.01 by Student’s t test.
Genetic Analysis and Map-Based Cloning of SPL88
We used spl88-1 and spl88-2 to cross with the japonica cultivar Nanjing 06 to get F1. The F1 plants did not show lesion mimics. F2 generation plants appeared similar in phenotype to spl88-1 and spl88-2. The segregation ratio of lesion mimic phenotype to the normal phenotype in the F2 population was essentially in compliance with a 1:3 ratio (Supplementary Table S4, S5), indicating that the spl88-1 and spl88-2 phenotype was caused by a mutation in a single recessive nuclear gene.
We used 238 pairs of SSR molecular markers to map the mutant gene in 21 F2 individuals with lesion mimic phenotype and preliminarily located the mutation site between the two molecular markers M1 and M10 on the long arm of chromosome 2. Further through the indica–japonica difference, we developed four newly polymorphic tags and ultimately mapped SPL88 to an 84-kb region between markers M5 and M6. It was predicted that this region contains 14 open-reading frames (ORFs) by http://rice.uga.edu/. Sequencing found that the gene LOC_Os02g51180 was mutated, in which the 132nd base C in the coding region of the SPL88-1 gene was deleted, and the 381st base T in the coding region of the SPL88-2 gene was deleted, resulting in early termination (Figures 6A–F). Therefore, LOC_Os02g51180 is the candidate gene for SPL88.
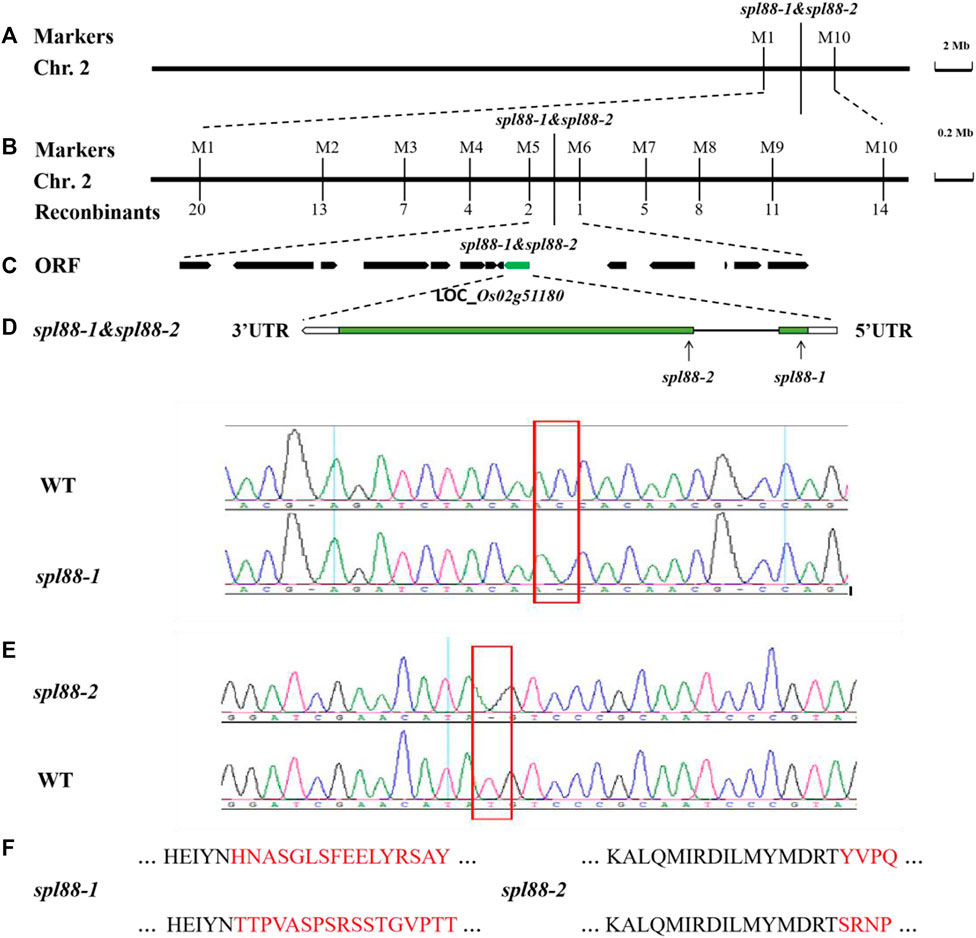
FIGURE 6. Genetic and physical maps of the SPL88. (A) SPL88 gene was localized to chromosome 2 between InDel markers M1 and M10; (B) SPL88 gene was delimited to the M5 to M6 interval on chromosome 2; (C) 14 putative ORFs located in the ∼84 kb region of this locus; (D) gene structure of LOC_Os02g51180; (E) sequence analysis of C-base deletion mutation sites in wild-type and spl88-1; sequence analysis of T-base deletion mutation sites in wild-type and spl88-2; (F) mutation causes premature termination of protein translation.
To verify that the phenotype of mutants spl88-1 and spl88-2 was caused by the deletion of the coding region of the gene LOC_Os02g51180, we constructed complementary vectors pGSPL88-1 and pGSPL88-2, respectively. The two vectors contained genomic DNA fragments including the promoter of the SPL88 gene in WT Xiushui 11 and were introduced into spl88-1 and spl88-2 by Agrobacterium tumefaciens–mediated transformation (Zhang and Chen, 2007; WangShen et al., 2011). Among the transformed T0 generation plants, the positive plants were consistent with the phenotype of WT and the negative plants were consistent with the lesion mimic phenotype of mutants spl88-1 and spl88-2 (Supplementary Figure S1). The aforementioned results verify the accuracy of the candidate gene LOC_Os02g51180 and also prove that the lesion phenotype of spl88-1 and spl88-2 is caused by the single-base deletion of the gene LOC_Os02g51180.
Expression Analysis of SPL88
Using http://bar.utoronto.ca/eplant_rice/to predict the expression patterns of SPL88 in various organs, we found that SPL88 was mainly expressed in root, leaves, and panicle, especially high in leaves and panicle (Figure 7A). To verify this prediction, we constructed the vector pSPL88::GUS, which contained promoter fragments of the SPL88 and introduced it into WT by Agrobacterium tumefaciens–mediated transformation. We stained various organs of the transgene-positive plants and observed GUS signals in various tissues. In addition, we used qRT-PCR to analyze the expression of SPL88 in various organs and found that the results were consistent with the results of GUS staining, with the highest expression in panicle and the lowest expression in the stem.
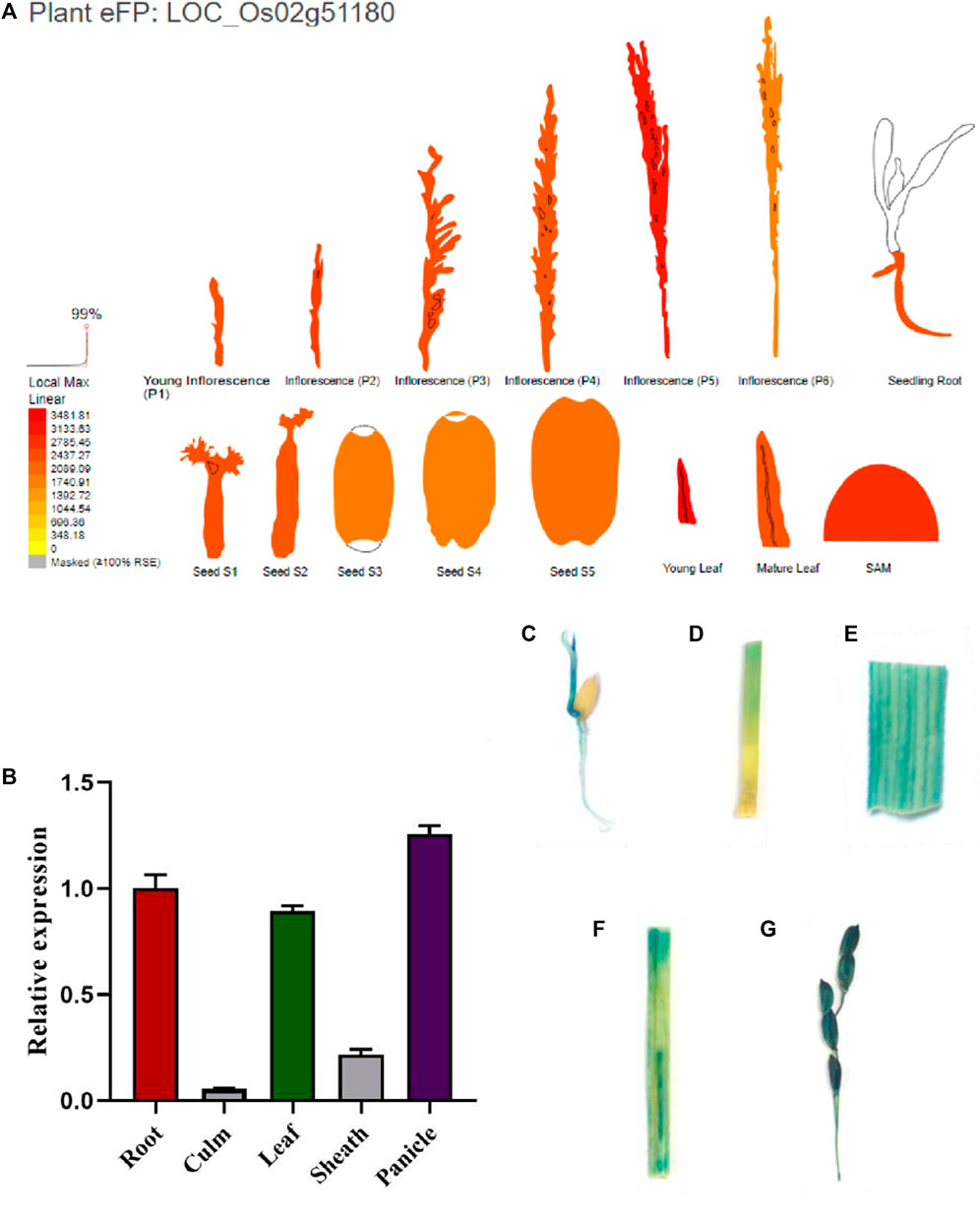
FIGURE 7. Expression analysis of SPL88. (A) Site predicts the expression pattern of SPL88; (B) expression of SPL88 in various organs of wild-type and mutant plants analyzed by qRT-PCR; (C–G) histochemical signals from the SPL88 promoter-GUS reporter gene. GUS signals were detected in root C, stem D, leaf E, leaf sheath F, and panicle (G)
Subcellular Localization of SPL88
To explore the localization pattern of SPL88 in cells, we constructed the pVBA1132::SPL88::eGFP vector, which contained the SPL88 coding sequence of WT and was transiently expressed in tobacco epidermal cells as observed by laser scanning confocal microscopy (CLSM). There are strong green fluorescence signals in the cytoplasm and nucleus, indicating that SPL88 is expressed in the cytoplasm and nucleus (Figures 8A–F).
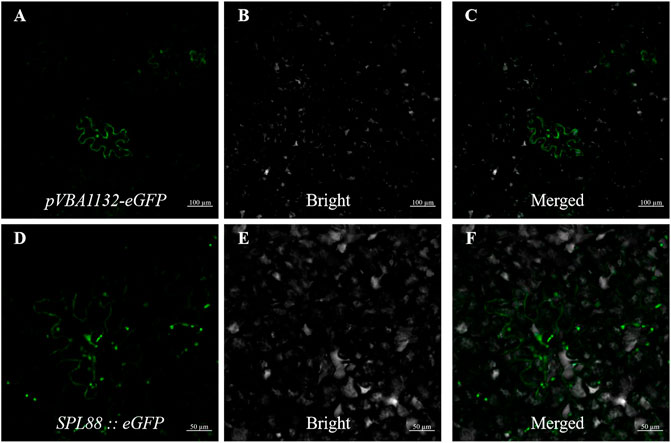
FIGURE 8. Subcellular localization of SPL88; (A–C): transient expression of pVBA1132-eGFP in tobacco, Bar: 100 μm; (D–F): transient expression of pVBA1132:SPL88::eGFP in tobacco, Bar: 50 μm.
Transcriptome Analysis Suggested That SPL88 Regulates Defense Responses in Rice
To investigate the potential regulatory mechanisms of lesion mimics in the spl88-1 and spl88-2, we sampled flag leaves from WT, spl88-1, and spl88-2 at the tillering stage with the most necrotic spots and performed transcriptome sequencing to compare global gene expression changes between spl88-1, spl88-2, and WT. The result revealed that a total of 1564 differentially expressed genes were found in spl88-1, of which 788 genes were upregulated (log2 ratio ≥ 1) and 766 were downregulated (log2 ratio ≤ −1). 531 genes were upregulated and 998 were downregulated in spl88-2. According to the Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis, the differentially expressed genes were mainly involved in signaling pathways and physiological metabolic pathways, including indole alkaloid biosynthesis, purine metabolism, diterpenoid biosynthesis, phenylpropanoid biosynthesis, taurine, and hypotaurine metabolism and ribosome synthesis (Figures 9A,B). Based on the RNA-seq results, the defense-related genes and growth and development-related genes including OsJMT1, OsPAL06, OsMADS26, OsMT2b, OsWAK14, OsWRKY6, OsTPS24, OsEXPB2, Osrboh3, OsPIN5b, OsPuf4, OsBISAMT1, OsAG O 18, OsAOS2, OsPR1b, OsGR3, OsRACK1A, NLS1, OsPR10a, OsCATA were significantly differentially expressed between spl88-1, spl88-2, and WT (Figure 9C). These results suggest that SPL88 plays an important role in the defense response in rice.
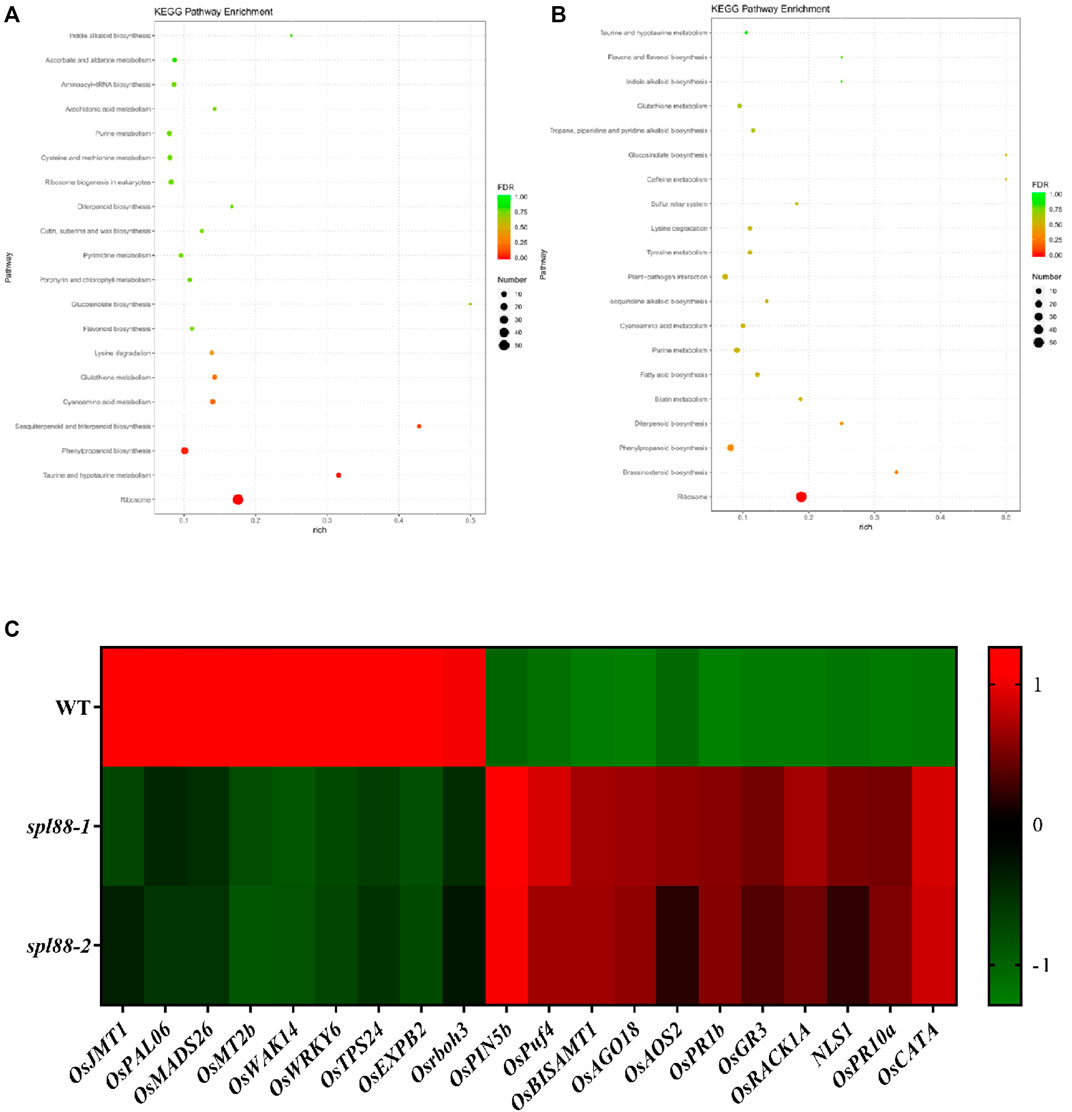
FIGURE 9. Transcriptome profiling analysis of spl88-1 and spl88-2. (A), (B) KEGG enrichment analysis of differentially expressed genes compared to wild type, spl88-1, and spl88-2; (C) expression heat map of differentially expressed genes associated with defense and growth and development.
Discussion
Lesion mimics often affect normal growth and development in plants, resulting in weakened photosynthesis and reduced yield. Therefore, the research on rice lesion mimic mutants is of great significance for elucidating the mechanism of plant growth and development. In this study, we obtained two lesion mimic mutants spl88-1 and spl88-2. At the tillering stage, the lesion mimic phenotype appeared in the leaf apex of spl88-1 and spl88-2. With the growth of the plants, the necrotic spots gradually spread throughout the leaves. A transmission electron microscope showed that the chloroplast ultrastructure of mutants spl88-1 and spl88-2 changed, which affected the growth and development of plants. Furthermore, through a series of physiological and biochemical analyses, we found that the appearance of lesion mimic phenotype led to the accumulation and burst of ROS in mutants spl88-1 and spl88-2, resulting in cell death. The gene LOC_Os02g51180 was confirmed to be the candidate gene of SPL88 by map-based cloning and genetic complementation experiments, in which the 132nd base C in the coding region of the SPL88-1 gene was deleted, and the 381st base in the coding region of the SPL88-2 gene had a T deletion, resulting in premature termination of SPL88 translation and formation of three distinct proteins (Supplementary Figure S2). We used the website (http://smart.embl-heidelberg.de/) to analyze the protein sequence encoded by SPL88 and found that the protein contained two domains of CULLIN and Cullin Nedd8 (Supplementary Figure S3). The CULLIN domain consists of 147 (407-554) amino acids and the Cullin Nedd8 domain consists of 67 (663-730) amino acids. According to the website (https://services.healthtech.dtu.dk/service.php?TMHMM-2.0), it was predicted that the 736 amino acids of the protein were located on the surface of the cell membrane and did not form a typical transmembrane helical region, which did not belong to transmembrane protein (Supplementary Figure S4). To explore whether SPL88 is involved in the pathways related to adversity stress, we conducted a hydroponic salt stress experiment on the wild type, spl88-1, and spl88-2. We treated the seedlings in the trefoil stage with 150 mM/L NaCl for 4 days and found that the phenotype of wild-type and mutant plants did not change obviously before and after the treatment, indicating that SPL88 did not involve in the response to salt stress (Yan et al., 2021) (Supplementary Figure S5).
Although the lesion mimic phenotypes of spl88-1 and spl88-2 were both caused by deletion of the same gene and the series of positive and negative effects brought about by the lesion mimic phenotype were generally consistent, there were still some subtle differences. In the chlorophyll assay results, the chlorophyll a of spl88-1 decreased more and the chloroplasts were more severely shrunken, while the chloroplasts of spl88-2 were more inclined to chloroplast disorder and the expression levels of defense-related genes AOS2 and ASP90 in spl88-1 were also significantly higher than those in spl88-2. According to these results, we speculate that the aforementioned results are because the SPL88-1 base is deleted in the first exon and the SPL88-2 base is deleted in the second exon, but not within the CULLIN and Cullin Nedd8 domains, and both of the two domains do not express.
We confirmed that the candidate gene SPL88 indeed controlled the phenotype of mutants spl88-1 and spl88-2 by map-based cloning and complementation experiments, indicating that SPL88 was a new allelic variant gene of cloned OsCUL3a, which interacts with OsRBX1a and OsRBX1b to form RING E3 ubiquitin ligase (CRL3) involved in plant immune responses (Liu et al., 2017; Gao et al., 2020). Like oscul3a, spl88-1 and spl88-2 exhibited enhanced defense responses and reduced seed setting. The difference was that we found SPL88 not only damaged the chloroplast structure but also hindered chlorophyll biosynthesis. We also detected many apoptotic cells in spl88-1 and spl88-2 by TUNEL assay. In addition, we demonstrated SPL88 was expressed in all organs and expressed the most in roots and leaves. Based on the aforementioned analysis, we speculate SPL88 is a different mutant form from the reported allele of OsCUL3a.
Mutants are important materials for studying gene function. For the study of the gene regulatory network of lesion mimics, it is necessary to strengthen the screening and gene function identification of different types of lesion mimic mutants in the future. The research objects spl88-1 and spl88-2 are of great significance for mining and analyzing the regulation mechanism of lesion mimics and improving the regulation network of lesion mimics.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
ZC, WY, YR, YW, and TL conceived and designed the experiment. HY and XL performed data collection. ZC, WY, and ML produced the data analysis and figures. ZC and TL wrote the original manuscript with input from YR, SL, YW, GD, and PD. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Key R&D Program of China (Grant No: 2021YFA1300703), the Zhejiang Provincial Natural Science Outstanding Youth Fund (Grant No: LR20C130001), the National Natural Science Foundation of China (31971921, U20A2030), and the State Key Laboratory of Rice Biology, China (20200102).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.918973/full#supplementary-material
References
Aviv, D. H., Rustérucci, C., Iii, B. F. H., Dietrich, R. A., Parker, J. E., and Dangl, J. L. (2002). Runaway Cell Death, but Not Basal Disease Resistance, Inlsd1is SA- andNIM1/NPR1-dependent. Plant J. 29 (3), 381–391. doi:10.1046/j.0960-7412.2001.01225.x
Cai, L., Yan, M., Yun, H., Tan, J., Du, D., Sun, H., et al. (2021). Identification and Fine Mapping of Lesion Mimic Mutant Spl36 in Rice (Oryza Sativa L.). Breed. Sci. 71 (5), 510–519. doi:10.1270/jsbbs.20160
Chen, Z., Chen, T., Sathe, A., He, Y., Zhang, X.-b., and Wu, J.-l. (2018). Identification of a Novel Semi-dominant Spotted-Leaf Mutant with Enhanced Resistance to Xanthomonas Oryzae Pv. Oryzae in Rice. Ijms 19, 3766. doi:10.3390/ijms19123766
Figueroa, P., Gusmaroli, G., Serino, G., Habashi, J., Ma, L., Shen, Y., et al. (2005). Arabidopsis Has Two Redundant Cullin3 Proteins that Are Essential for Embryo Development and that Interact with RBX1 and BTB Proteins to Form Multisubunit E3 Ubiquitin Ligase Complexes In Vivo. Plant Cell 17 (4), 1180–1195. doi:10.1105/tpc.105.031989
Gao, Z., Liu, Q., Zhang, Y., Chen, D., Zhan, X., Deng, C., et al. (2020). OsCUL3a-associated Molecular Switches Have Functions in Cell Metabolism, Cell Death, and Disease Resistance. J. Agric. Food Chem. 68 (19), 5471–5482. doi:10.1021/acs.jafc.9b07426
Hoang, T. V., Vo, K. T. X., Rahman, M. M., Choi, S.-H., and Jeon, J.-S. (2019). Heat Stress Transcription Factor OsSPL7 Plays a Critical Role in Reactive Oxygen Species Balance and Stress Responses in Rice. Plant Sci. 289, 110273. doi:10.1016/j.plantsci.11027310.1016/j.plantsci.2019.110273
Hu, G., Yalpani, N., Briggs, S. P., and Johal, G. S. (1998). A Porphyrin Pathway Impairment Is Responsible for the Phenotype of a Dominant Disease Lesion Mimic Mutant of Maize. Plant Cell 10 (7), 1095–1105. doi:10.1105/tpc.10.7.1095
Huang, Q. N., Yang, Y., Shi, Y. F., Chen, J., and Wu, J. L. (2010). Spotted-leaf Mutants of Rice (Oryza Sativa L.). Rice Sci. 17 (004), 247–256. doi:10.1016/S1672-6308(09)60024-X
Inada, N., Sakai, A., Kuroiwa, H., and Kuroiwa, T. (1998). Three-dimensional Analysis of the Senescence Program in Rice ( Oryza Sativa L.) Coleoptiles. Planta 205 (2), 153–164. doi:10.1007/s004250050307
Jambunathan, N., Siani, J. M., and Mcnellis, T. W. (2001). A Humidity-Sensitive Arabidopsis Copine Mutant Exhibits Precocious Cell Death and Increased Disease Resistance. Plant Cell 13 (10), 2225–2240. doi:10.1105/tpc.010226
Jozefczuk, J., and Adjaye, J. (2011). Quantitative Real-Time PCR-Based Analysis of Gene Expression. Methods Enzymol. 500, 99–109. doi:10.1016/B978-0-12-385118-5.00006-2
Kemal, K., and Manners, J. M. (2011). The Interplay between Light and Jasmonate Signalling during Defence and Development. J. Exp. Bot. 62 (12), 4087–4100.
Kim, J.-A., Cho, K., Singh, R., Jung, Y.-H., Jeong, S.-H., Kim, S.-H., et al. (2009). Rice OsACDR1 (Oryza Sativa Accelerated Cell Death and Resistance 1) Is a Potential Positive Regulator of Fungal Disease Resistance. Mol. Cells 28 (5), 431–439. doi:10.1007/s10059-009-0161-5
Liang, X. X., and Zhou, J. M. (2018). The Secret of Fertilization in Flowering Plants Unveiled. Sci. Bull. 63 (07), 18–20. doi:10.1016/j.scib.2018.02.010
Lichtenthaler, H. K. (1987). [34] Chlorophylls and Carotenoids: Pigments of Photosynthetic Biomembranes. Methods Enzymol. 148 (1), 350–382. doi:10.1016/0076-6879(87)48036-1
Liu, G., Wang, L., Zhou, Z., Leung, H., Wang, G.-L., and He, C. (2004). Physical Mapping of a Rice Lesion Mimic Gene, Spl1 , to a 70-kb Segment of Rice Chromosome 12. Mol. Genet. Genomics 272 (1), 108–115. doi:10.1007/s00438-004-1040-6
Liu, Q., Ning, Y., Zhang, Y., Yu, N., Zhao, C., Zhan, X., et al. (2017). OsCUL3a Negatively Regulates Cell Death and Immunity by Degrading OsNPR1 in Rice. Plant Cell 29 (2), 345–359. doi:10.1105/tpc.16.00650
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 25 (4), 402–408. doi:10.1006/meth.2001.1262
McGrann, G. R. D., Steed, A., Burt, C., Nicholson, P., and Brown, J. K. M. (2015). Differential Effects of Lesion Mimic Mutants in Barley on Disease Development by Facultative Pathogens. J. Exp. Bot. 66 (11), 3417–3428. doi:10.1093/jxb/erv154
Mizobuchi, R., Hirabayashi, H., Kaji, R., Nishizawa, Y., Satoh, H., Ogawa, T., et al. (2002). Differential Expression of Disease Resistance in Rice Lesion-Mimic Mutants. Plant Cell Rep. 21 (4), 390–396. doi:10.1007/s00299-002-0525-1
Rao, Y. C. (2016). Early Senescence Gene ES1 Affects Water Loss in Rice. Beijing: Chinese Academy of Agricultural Sciences.
Rechcigl, M., and Evans, W. H. (1963). Role of Catalase and Peroxidase in the Metabolism of Leucocytes. Nature 199, 1001–1002. doi:10.1038/1991001b0
Sharma, P., Jha, A. B., Dubey, R. S., and Pessarakli, M. (2012). Reactive Oxygen Species, Oxidative Damage, and Antioxidative Defense Mechanism in Plants under Stressful Conditions. J. Bot. 2012, 1–26. doi:10.1155/201221703710.1155/2012/217037
Shirasu, K., and Pual, S. L. (2000). Regulators of Cell Death in Disease Resistance. Plant Mol. Biol. 44, 371–385. doi:10.1023/a:1026552827716
Singer, J. D., Gurian-West, M., Clurman, B., and Roberts, J. M. (1999). Cullin-3 Targets Cyclin E for Ubiquitination and Controls S Phase in Mammalian Cells. Genes & Dev. 13 (18), 2375–2387. doi:10.1101/gad.13.18.2375
Sun, L., Wang, Y., Liu, L.-l., Wang, C., Gan, T., Zhang, Z., et al. (2017). Isolation and Characterization of a Spotted Leaf 32 Mutant with Early Leaf Senescence and Enhanced Defense Response in Rice. Sci. Rep. 7, 41846. doi:10.1038/srep41846
Takahashi, A., Kawasaki, T., Henmi, K., Shii, K., Kodama, O., Satoh, H., et al. (1999). Lesion Mimic Mutants of Rice with Alterations in Early Signaling Events of Defense. Plant J. 17 (5), 535–545. doi:10.1046/j.1365-313x.1999.00405.x
Tang, J., Zhu, X., Wang, Y., Liu, L., Xu, B., Li, F., et al. (2011). Semi-dominant Mutations in the CC-NB-LRR-type R Gene, NLS1, Lead to Constitutive Activation of Defense Responses in Rice. Plant J. 66 (6), 996–1007. doi:10.1111/j.1365-313X.2011.04557.x
Thomann, A., Brukhin, V., Dieterle, M., Gheyeselinck, J., Vantard, M., Grossniklaus, U., et al. (2005). Arabidopsis CUL3A and CUL3B Genes Are Essential for Normal Embryogenesis. Plant J. 43 (3), 437–448. doi:10.1111/j.1365-313X.2005.02467.x
Tu, R., Wang, H., Liu, Q., Wang, D., ZhouXu, X. P., Xu, P., et al. (2020). Characterization and Genetic Analysis of the Oshpl3 Rice Lesion Mimic Mutant Showing Spontaneous Cell Death and Enhanced Bacterial Blight Resistance. Plant Physiology Biochem. 154, 94–104. doi:10.1016/j.plaphy.2020.05.001
Wakuta, S., Suzuki, E., Saburi, W., Matsuura, H., Nabeta, K., Imai, R., et al. (2011). OsJAR1 and OsJAR2 Are Jasmonyl-L-Isoleucine Synthases Involved in Wound- and Pathogen-Induced Jasmonic Acid Signalling. Biochem. Biophysical Res. Commun. 409 (4), 634–639. doi:10.1016/j.bbrc.2011.05.055
Wang, J. J., Zhu, X. D., Wang, L. Y., Zhang, L. H., Xue, Q. Z., and He, Z. H. (2004). Physiological and Genetic Analysis of Lesion Resembling Disease Mutants (Lrd) of Oryza Sativa L. Zhi Wu Sheng Li Yu Fen Zi Sheng Wu Xue Xue Bao 30 (3), 331–338. doi:10.1016/j.jco.2003.08.015
Wang, S., Lei, C., Wang, J., Ma, J., Tang, S., Wang, C., et al. (2017). SPL33, Encoding an eEF1A-like Protein, Negatively Regulates Cell Death and Defense Responses in Rice. J. Exp. Bot. 68 (5), 899–913. doi:10.1093/jxb/erx001
WangShen, X. Q. X., He, Y. M., Ren, T. N., Wu, W. T., and Xi, T. (2011). Optimization of Freeze-Thaw Transformation Conditions for Agrobacterium Tumefaciens EHA105 and LBA4404. Pharm. Biotechnol. 18 (5), 5.
Wu, C., Bordeos, A., Madamba, M. R. S., Baraoidan, M., Ramos, M., Wang, G.-l., et al. (2008). Rice Lesion Mimic Mutants with Enhanced Resistance to Diseases. Mol. Genet. Genomics 279 (6), 605–619. doi:10.1007/s00438-008-0337-2
Yan, F., Zhang, J., Li, W., Ding, Y., Zhong, Q., Xu, X., et al. (2021). Exogenous Melatonin Alleviates Salt Stress by Improving Leaf Photosynthesis in Rice Seedlings. Plant Physiology Biochem. 163, 367–375. doi:10.1016/j.plaphy.2021.03.058
Yin, Z., Chen, J., Zeng, L., Goh, M., Leung, H., Khush, G. S., et al. (2000). Characterizing Rice Lesion Mimic Mutants and Identifying a Mutant with Broad-Spectrum Resistance to Rice Blast and Bacterial Blight. Mpmi 13 (8), 869–876. doi:10.1094/MPMI.2000.13.8.869
Yuchun, R. A. O., Ran, J. I. A. O., Sheng, W. A. N. G., Xianmei, W. U., Hanfei, Y. E., Chenyang, P. A. N., et al. (2021). SPL36 Encodes a Receptor-like Protein Kinase that Regulates Programmed Cell Death and Defense Responses in Rice. Rice 14 (1), 1–14. doi:10.1186/s12284-021-00475-y
Zang, Q., Zhao, B. Y., Zhao, K. J., Wang, W. C., Lin, S. M., Que, G. S., et al. (2000). Identification and Molecular Marker Mapping of a Novel Rice Bacterial Blight Resistance Gene Xa-23(t) in Oryza Sativa L. J. Crops 26 (005), 536–542.
Zeng, L.-R., Qu, S., Bordeos, A., Yang, C., Baraoidan, M., Yan, H., et al. (2004). Spotted Leaf11, a Negative Regulator of Plant Cell Death and Defense, Encodes a U-Box/Armadillo Repeat Protein Endowed with E3 Ubiquitin Ligase Activity. Plant Cell 16 (10), 2795–2808. doi:10.1105/tpc.104.025171
Zhang, B. J., and Chen, Q. Z. (2007). Study on Freeze-Thaw Method of Plasmids Introduced into Different Species of Agrobacterium. Hubei Agric. Sci. 46 (3), 329–331. doi:10.3969/j.issn.0439-8114.2007.03.003
Keywords: lesion mimics, cullin protein, SPL88, growth and development, rice
Citation: Chen Z, Yin W, Li X, Lu T, Ye H, Dai G, Mao Y, Li S, Duan P, Lu M, Rao Y and Wang Y (2022) OsSPL88 Encodes a Cullin Protein that Regulates Rice Growth and Development. Front. Genet. 13:918973. doi: 10.3389/fgene.2022.918973
Received: 13 April 2022; Accepted: 07 June 2022;
Published: 11 July 2022.
Edited by:
Christian Meyer, INRA UMR1318 Institut Jean Pierre Bourgin, FranceReviewed by:
Hai Zhou, South China Agricultural University, ChinaAnuj Kumar, Dalhousie University, Canada
Copyright © 2022 Chen, Yin, Li, Lu, Ye, Dai, Mao, Li, Duan, Lu, Rao and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuexing Wang, wangyuexing@caas.cn; Yuchun Rao, ryc@zjnu.cn
 Zhengai Chen
Zhengai Chen