
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet., 10 May 2022
Sec. Human and Medical Genomics
Volume 13 - 2022 | https://doi.org/10.3389/fgene.2022.897440
This article is part of the Research TopicAdoption of Artificial Intelligence in Human and Clinical GenomicsView all 11 articles
Background: To elucidate the potential biological function of hsa_circ_0062270 in the malignant process of melanoma and its potential target.
Methods: Quantitative real-time polymerase chain reaction (qRT-PCR) was conducted to examine relative level of hsa_circ_0062270 in melanoma tissues and normal skin tissues. The diagnostic and prognostic potentials of hsa_circ_0062270 in melanoma were evaluated. The regulatory effect of hsa_circ_0062270 on the expression of linear transcript Cell division cycle protein 45 (CDC45) was also examined.
Results: Hsa_circ_0062270 was up-regulated in melanoma samples and cell lines, which displayed certain diagnostic and prognostic potentials in melanoma. Inhibition of hsa_circ_0062270 attenuated the proliferative, migratory and invasive functions. Hsa_circ_0062270 could stabilize the expression of linear transcript CDC45, and thus participated in the malignant process of melanoma.
Conclusion: Hsa_circ_0062270 promotes proliferative, migratory and invasive functions of melanoma cells via stabilizing the linear transcript CDC45. Hsa_circ_0062270 can be used to diagnosis and treatment of melanoma.
Melanoma is a highly malignant skin cancer derived from melanocytes. Melanoma accounts for more than 70% of skin cancer deaths (Testori et al., 2017). More seriously, estimated numbers of new cases and death cases of melanoma are on the rise. Surgical resection combined lymph node dissection is an effective treatment for early stage melanoma (Chattopadhyay et al., 2016). Nevertheless, most of melanoma patients cannot be operated because of metastasis, leading to a poor 5-year survival (20%) (Shain and Bastian, 2016). Molecular mechanisms underlying melanoma process and metastasis remain largely unclear.
CircRNAs are novel noncoding RNAs to be widely analyzed. They are extensively involved in various fields of life sciences (Hsiao et al., 2017; Chen and Huang, 2018; Han et al., 2018). Since circRNAs do not have 3′ end, 5′ end and poly (A) tail, they can escape degradation from exonucleases. CircRNAs are more conservative and stable than linear RNAs. Moreover, they are evolutionarily conserved in different species, and specifically expressed in different tissues and developmental stages (Dong et al., 2019; Patop et al., 2019). According to the origins, circRNAs are classified to circular exonic RNAs that only contain reverse-splicing exons, circular intronic RNAs (ciRNAs) that only contain reverse-splicing introns, and exon-intron circRNAs (ElciRNAs) that introns are remained in circular exons (Salzman, 2016). The following mechanisms explain the biological functions of circRNAs: 1) CircRNAs inhibit miRNA activities through exerting the miRNA sponge effect; 2) CircRNAs interact with RNA-binding proteins as protein sponges; 3) CircRNAs regulate Pol II transcription of parent genes; 4) CircRNAs regulate linear splicing through competitively targeting splicing sites of pre-mRNAs; 5) CircRNAs have protein-encoding ability and can translate proteins (Kristensen et al., 2019). Abundant evidences have proven the vital functions of circRNAs in physiological and pathological processes (Salzman, 2016; Qu et al., 2017).
Hsa_circ_0062270 is located on chromosome 22: 19496052-19502571, and its gene symbol is CDC45. Evidence has showed that hsa_circ_0062270 is obviously up-regulated in melanoma (Hao et al., 2021). A previous study have demonstrated that downregulation of hsa_circ_0062270 can inhibit the progression of melanoma, however, the mechanism still remains unclear (Hao et al., 2021). The aim of the current research was to explore the biological effect of hsa_circ_0062270 on malignant phenotypes of melanoma and its potential target.
The normal skin tissues and melanoma tissues of 50 patients with melanoma in our hospital were selected. The ethics committee of The First People’s Hospital of Lianyungang approved our study. Signed written informed consents were obtained from all participants before the study.
Melanoma cells (SKMEL1, A375, A2058 and A875) and normal human epidermal melanocytes (NHEM) were provided by Cell Bank of Type Culture Collection (Shanghai, China). Cells were cultivated in DMEM containing 10% fetal bovine serum (FBS), 100 U/mL penicillin and 100 μg/ml streptomycin at 5% CO2, 37°C. Cell transfection was performed using Lipofectamine 3,000 as per the protocols. Cell proliferation was determined by EdU (Beyotime, Shanghai, China).
RNAs isolation was done with TRIzol and were then reversely transcribed into cDNAs. U6 and GAPDH were used as the internal controls with the method of 2−ΔΔCt. Primers used were shown below: hsa_circ_0062270: Forward: 5′-AGGATGGCTCAGGGACAGAT-3′, reverse: 5′-AGGCCATGGTACAGCTTGTC-3′; CDC45: Forward: 5′-TTCGTGTCCGATTTCCGCAAA-3′, reverse: 5′-TGGAACCAGCGTATATTGCAC-3′; GAPDH: Forward: 5′-CGGAGTCAACGGATTTGGTCGTAT-3′, reverse: 5′-AGCCTTCTCCATGGTGGTGAAGAC-3′: U6: Forward: 5′-GCTGAGGTGACGGTCTCAAA-3′, reverse: 5′-GCCTCCCAGTTTCATGGACA-3′.
A375 cells were exposed to Actinomycin D (3 μg/ml). They were collected for isolating total RNAs. Expressions of hsa_circ_0062270 and CDC45 were detected by Quantitative real-time polymerase chain reaction. Cellular RNA (4 mg) was treated either with RNase R (10 U/μg) at 37°C for 30 min or not, followed by purification using RNeasy MinElute (Qiagen, Hilden, Germany).
Cells were seeded into the top chamber and bottom chamber. After 48-h incubation, cells in the bottom were fixed, dyed in crystal violet and captured. Migratory cells were counted in five randomly selected fields per sample. Invasion assay was conducted using transwell chamber precoated with 100 μL of Matrigel (Corning, Corning, NY, United States). In detail, Matrigel was diluted in serum-free medium at 1:3, which was coated on the top of a chamber.
Data were expressed as mean ± SD (standard deviation) and they were processed using Statistical Product and Service Solutions (SPSS) 20.0 (IBM, Armonk, NY, United States). Prognostic value of hsa_circ_0062270 in melanoma were evaluated by Kaplan-Meier and receiver operating characteristic (ROC) method, respectively. The correlation between hsa_circ_0062270 and CDC45 levels was assessed through Pearson correlation test. A significant difference was set at p < 0.05.
Firstly, we explored the expression of hsa_circ_0062270 in melanoma tissues and the normal skin tissues. Results revealed that the expression hsa_circ_0062270 in melanoma tissues was significantly higher than that in normal ones (Figure 1A). Then, receiver operating characteristic (ROC) curves depicted that the AUC of hsa_circ_0062270 was 0.6312 (Figure 1B). Kaplan-Meier analysis found that highly expressed hsa_circ_0062270 predicted a poor prognosis for melanoma patients (p = 0.0234) (Figure 1C). Hsa_circ_0062270 significantly increased in melanoma cell lines as well (Figure 1D). We furthermore examined the stability of hsa_circ_0062270 by detecting mRNA levels of hsa_circ_0062270 and CDC45 in Actinomycin D-treated A375 cells. Compared with that of CDC45 (<12 h), the half-life of hsa_circ_0062270 was over 24 h (Figure 1E). RNase R induction did not affect expression level of hsa_circ_0062270, but markedly down-regulated CDC54 (Figure 1F). Therefore, we have verified that hsa_circ_0062270 was highly stable in melanoma cells.
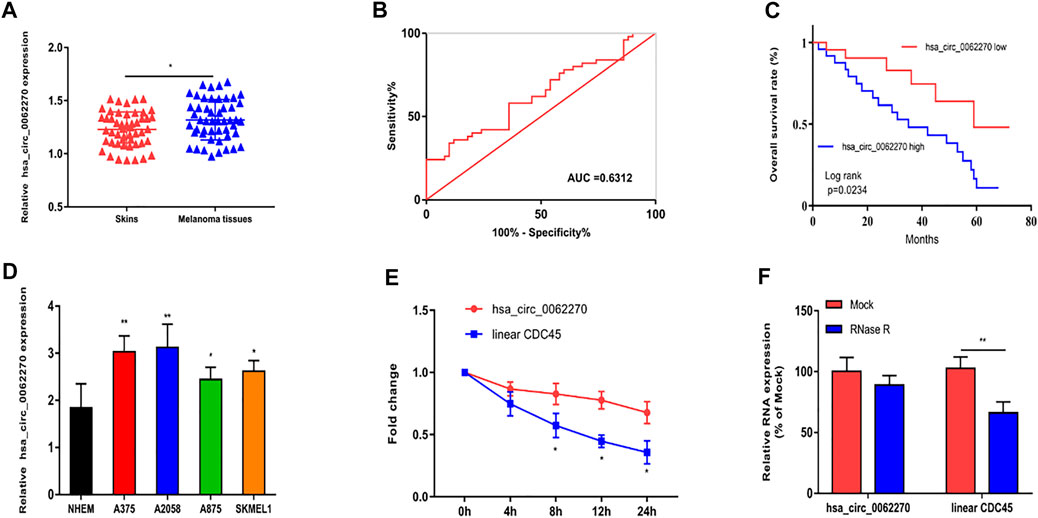
FIGURE 1. Up-regulated hsa_circ_0062270 predicted poor prognosis in melanoma. (A) Expression of hsa_circ_0062270 in melanoma and normal skin tissues; (B) Diagnostic potential of hsa_circ_0062270 in melanoma; (C) Prognostic potential of hsa_circ_0062270 in melanoma; (D) Differential level of hsa_circ_0062270 in melanoma cells and normal human epidermal melanocytes; (E) Expression changes of hsa_circ_0062270 and CDC45 in A375 cells treated with Actinomycin D; (F) Half-life of hsa_circ_0062270 and CDC45 in A375 cells treated with RNase R. *p < 0.05; **p < 0.01.
To investigate the effects of hsa_circ_0062270 on melanoma cell proliferation, migration and invasion, cells were treated with hsa_circ_0062270 siRNA. Results indicated that transfection of hsa_circ_0062270 siRNA markedly down-regulated hsa_circ_0062270 level in A375 and A2058 cells (Figure 2A). Knockdown of hsa_circ_0062270 reduced EdU-positive rate in melanoma cells (Figure 2B). In addition, hsa_circ_0062270 siRNA markedly reduced migratory and invasive rates (Figures 2C,D).
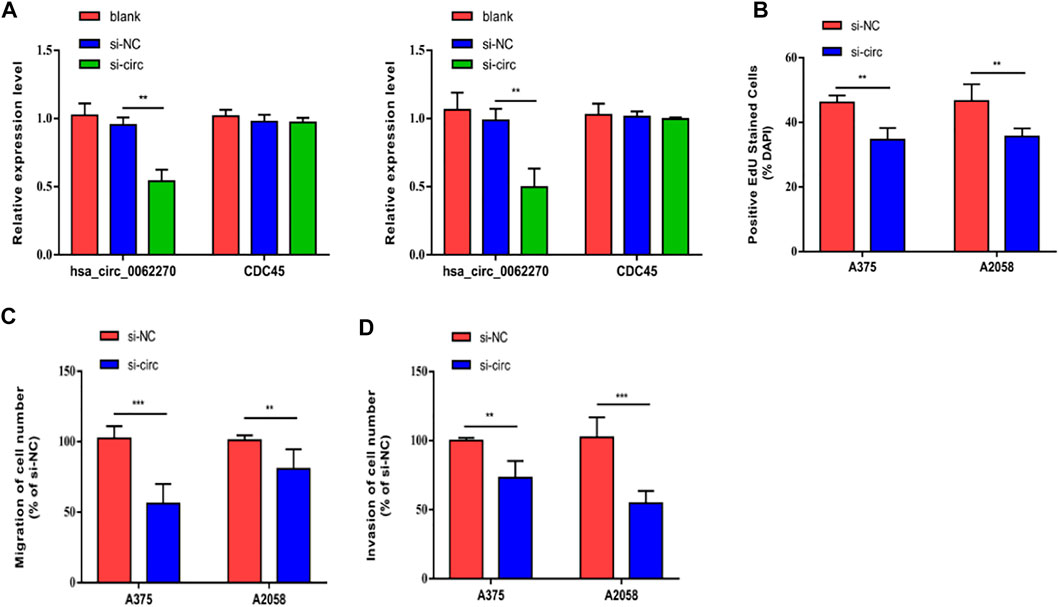
FIGURE 2. Hsa_circ_0062270 accelerated melanoma to proliferate, migrate and invade. (A) Transfection efficacy of hsa_circ_0062270 siRNA; (B) EdU-positive rate with hsa_circ_0062270 knockdown; (C) Migration in cells with hsa_circ_0062270 knockdown; (D) Invasion in cells with hsa_circ_0062270 knockdown. **p < 0.01; ***p < 0.001.
Then we focused on the potential target of hsa_circ_0062270 in the regulation of phenotypes of melanoma. CircRNAs are involved in pathological process via mediating expression levels of their linear transcripts. CDC45 was the linear transcript of hsa_circ_0062270 (Figure 3A) and positively correlated to hsa_circ_0062270 level (Figure 3B). Identically, CDC45 was highly expressed in melanoma cell lines (Figure 3C). Knockdown of hsa_circ_0062270 could downregulate CDC45 and as expected, CDC45 was up-regulated in A375 and A2058 cells overexpressing hsa_circ_0062270 (Figures 3D,E). It is concluded that hsa_circ_0062270 could stabilize the expression of CDC45.
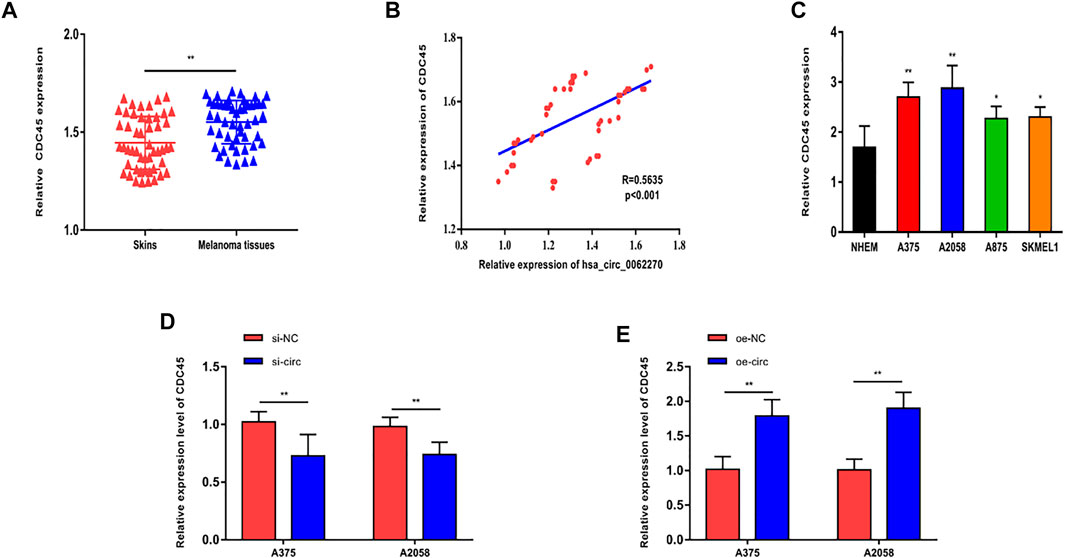
FIGURE 3. hsa_circ_0062270 stabilized CDC45 expression. (A) Differential level of CDC45 in melanoma and normal skin tissues; (B) A positive correlation between hsa_circ_0062270 and CDC45 levels in melanoma tissues; (C) Differential level of CDC45 in melanoma cells and normal human epidermal melanocytes; (D) CDC45 in cells with hsa_circ_0062270 knockdown; (E) CDC45 in cells overexpressing hsa_circ_0062270. *p < 0.05; **p < 0.01.
To further elucidate the effects of CDC45 on melanoma phenotypes, we established the overexpression models of CDC45. Transfection of overexpressed plasmid of CDC45 effectively up-regulated CDC45 in A375 and A2058 cells (Figure 4A). In melanoma cells overexpressing CDC45, EdU-positive rate increased (Figure 4B). Moreover, migratory and invasive potentials of melanoma were promoted by overexpressed CDC45 (Figures 4C,D). Therefore, CDC45 could stimulate the malignant process of melanoma.
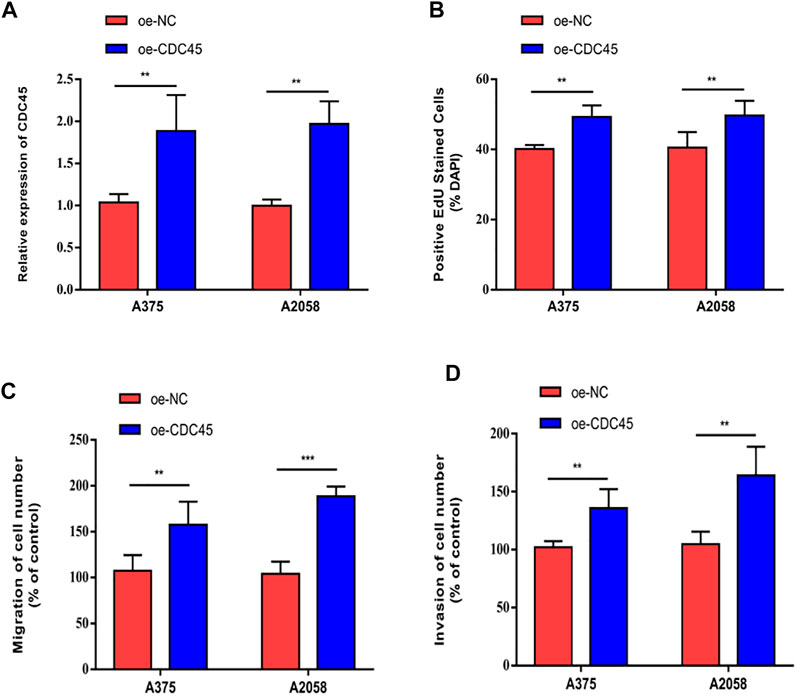
FIGURE 4. CDC45 promoted melanoma to proliferate, migrate and invade. (A) Transfection efficacy of overexpressed plasmid of CDC45; (B) EdU-positive rate in cells overexpressing CDC45; (C) Migration in cells overexpressing CDC45; (D) Invasion in cells overexpressing CDC45. **p < 0.01; ***p < 0.001.
To uncover the co-regulation of hsa_circ_0062270 and CDC45, cells were co-transfected using si-CDC45 and overexpressed plasmid of hsa_circ_0062270 (Figure 5A). Overexpression of hsa_circ_0062270 could enhance the down-regulated CDC45 in melanoma cells transfected with si-CDC45. Knockdown of CDC45 reduced EdU-positive rate, migratory cell number and invasive cell number, which were reversed by overexpressed hsa_circ_0062270 (Figures 5B–D). All above indicated that hsa_circ_0062270 may play a carcinogenic role in melanoma by stabilizing its linear transcription CDC45.
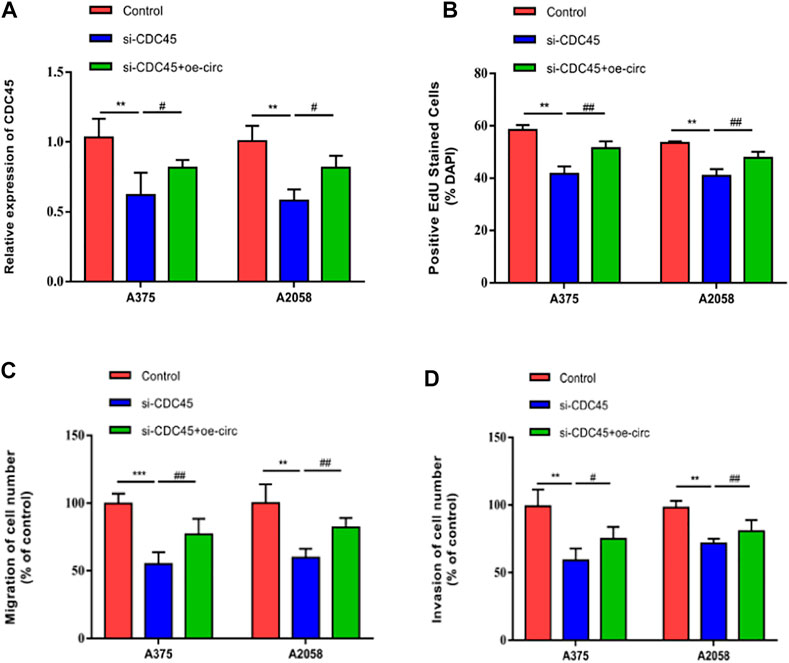
FIGURE 5. hsa_circ_0062270 stimulated the malignant process of melanoma by stabilizing CDC45. (A) Co-transfection of si-CDC34 and overexpressed plasmid of hsa_circ_0062270; (B) EdU-positive rate co-regulated by hsa_circ_0062270 and CDC45; (C) Migrative ability regulated by hsa_circ_0062270 and CDC45; (D) Invasion in A375 and A2058 cells co-regulated by hsa_circ_0062270 and CDC45. **p < 0.01; ***p < 0.001; #p < 0.05; ##p < 0.01.
The incidence of melanoma is not high, covering 4–5% of malignant tumors. Family history, multiple atypical moles and dysplastic moles are risk factors that trigger the carcinogenesis of melanoma (Pavri et al., 2016). In addition, ultraviolet rays can induce melanoma by damaging DNA repair genes.
CircRNAs, as a type of emerging noncoding RNAs, have been well concerned because of their unique structure and vital functions (Meng et al., 2017; Chen and Huang, 2018). They can be utilized as molecular biomarkers for diagnosing tumors and evaluating their prognosis (Bian et al., 2018; Zhou et al., 2018; Chen et al., 2020). The involvement of circRNAs in melanoma has been reported through literature review.
We previously found that the expression of hsa_circ_0062270 in melanoma was up-regulated. The diagnostic and prognostic potentials of hsa_circ_0062270 in melanoma were verified through depicting ROC and Kaplan-Meier curves, respectively. In vitro experiment results illustrated that knockdown of hsa_circ_0062270 remarkably suppressed proliferative, migratory and invasive functions of melanoma cells.
Recent studies have demonstrated that circRNAs have an important role in disease progression by mediating expressions of their linear transcripts (Wu et al., 2020; Tang et al., 2021; Mecozzi et al., 2021; Peng and Wang, 2021). Through qRT-PCR and rescue experiments, we found that hsa_circ_0062270 was able to stabilize the expression of its linear transcript CDC45. Knockdown of CDC45 blocked proliferative, migratory and invasive functions of melanoma cells, which could be reversed by overexpression of hsa_circ_0062270. The binding of CDC45 to chromatins regulated by the convergent effect of CDKs and DDK coincides with the time point of the start of DNA replication, suggesting that CDC45 is crucial in regulating the initiation of DNA replication (Köhler et al., 2016; Szambowska et al., 2017; Can et al., 2019).
Collectively, our present study was the first attempt to reveal that hsa_circ_0062270 was up-regulated in melanoma specimens and correlated to its prognosis. Hsa_circ_0062270 stimulated malignant process of melanoma by stabilizing its linear transcript CDC45. Our findings provide a new aspect for developing diagnostic and therapeutic strategies for melanoma. Several limitations of our study should be pointed out. First of all, in vivo role of hsa_circ_0062270 in melanoma is not explored. Secondly, how hsa_circ_0062270 regulates CDC45 remains unclear. Thirdly, other cell phenotypes of melanoma, including apoptosis, epithelial-mesenchymal transition and cell cycle progression affected by hsa_circ_0062270 are not clear.
Hsa_circ_0062270 promotes proliferative, migrative and invasive functions in melanoma cells via stabilizing the linear transcript CDC45. These findings provided strong evidence that hsa_circ_0062270 could be a novel promising therapeutic target used to diagnosis and treatment of melanoma.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
The studies involving human participants were reviewed and approved by the ethics committee of the First People’s Hospital of Lianyungang. The patients/participants provided their written informed consent to participate in this study.
CW, WS, and XD designed the study and performed the experiments, CL and FM collected the data, CL, FM, and LS analyzed the data, CW, WS, and XD prepared the manuscript. All authors read and approved the final manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Bian, D., Wu, Y., and Song, G. (2018). Novel Circular RNA, Hsa_circ_0025039 Promotes Cell Growth, Invasion and Glucose Metabolism in Malignant Melanoma via the miR-198/CDK4 axis. Biomed. Pharmacother. 108, 165–176. doi:10.1016/j.biopha.2018.08.152
Can, G., Kauerhof, A. C., Macak, D., and Zegerman, P. (2019). Helicase Subunit Cdc45 Targets the Checkpoint Kinase Rad53 to Both Replication Initiation and Elongation Complexes after Fork Stalling. Mol. Cell 73 (3), 562–573. doi:10.1016/j.molcel.2018.11.025
Chattopadhyay, C., Kim, D. W., Gombos, D. S., Oba, J., Qin, Y., Williams, M. D., et al. (2016). Uveal Melanoma: From Diagnosis to Treatment and the Science in between. Cancer 122 (15), 2299–2312. doi:10.1002/cncr.29727
Chen, B., and Huang, S. (2018). Circular RNA: An Emerging Non-coding RNA as a Regulator and Biomarker in Cancer. Cancer Lett. 418, 41–50. doi:10.1016/j.canlet.2018.01.011
Chen, Z., Chen, J., Wa, Q., He, M., Wang, X., Zhou, J., et al. (2020). Knockdown of Circ_0084043 Suppresses the Development of Human Melanoma Cells through miR-429/tribbles Homolog 2 axis and Wnt/β-Catenin Pathway. Life Sci. 243, 117323. doi:10.1016/j.lfs.2020.117323
Dong, W., Dai, Z.-h., Liu, F.-c., Guo, X.-g., Ge, C.-m., Ding, J., et al. (2019). The RNA-Binding Protein RBM3 Promotes Cell Proliferation in Hepatocellular Carcinoma by Regulating Circular RNA SCD-circRNA 2 Production. Ebiomedicine 45, 155–167. doi:10.1016/j.ebiom.2019.06.030
Han, B., Chao, J., and Yao, H. (2018). Circular RNA and its Mechanisms in Disease: From the Bench to the Clinic. Pharmacol. Ther. 187, 31–44. doi:10.1016/j.pharmthera.2018.01.010
Hao, T., Yang, Y., He, J., Bai, J., Zheng, Y., and Luo, Z. (2021). Knockdown of Circular RNA Hsa_circ_0062270 Suppresses the Progression of Melanoma via Downregulation of CDC45. Histol. Histopathol., 18412. doi:10.14670/HH-18-412
Hsiao, K.-Y., Sun, H. S., and Tsai, S.-J. (2017). Circular RNA - New Member of Noncoding RNA with Novel Functions. Exp. Biol. Med. (Maywood) 242 (11), 1136–1141. doi:10.1177/1535370217708978
Köhler, C., Koalick, D., Fabricius, A., Parplys, A. C., Borgmann, K., Pospiech, H., et al. (2016). Cdc45 is Limiting for Replication Initiation in Humans. Cell Cycle 15 (7), 974–985. doi:10.1080/15384101.2016.1152424
Kristensen, L. S., Andersen, M. S., Stagsted, L. V. W., Ebbesen, K. K., Hansen, T. B., and Kjems, J. (2019). The Biogenesis, Biology and Characterization of Circular RNAs. Nat. Rev. Genet. 20 (11), 675–691. doi:10.1038/s41576-019-0158-7
Mecozzi, N., Vera, O., and Karreth, F. A. (2021). Squaring the Circle: circRNAs in Melanoma. Oncogene 40 (37), 5559–5566. doi:10.1038/s41388-021-01977-1
Meng, X., Li, X., Zhang, P., Wang, J., Zhou, Y., and Chen, M. (2017). Circular RNA: An Emerging Key Player in RNA World. Brief. Bioinform 18 (4), bbw045–57. doi:10.1093/bib/bbw045
Patop, I. L., Wüst, S., and Kadener, S. (2019). Past, Present, and Future of Circ RNA S. Embo J. 38 (16), e100836. doi:10.15252/embj.2018100836
Pavri, S. N., Clune, J., Ariyan, S., and Narayan, D. (2016). Malignant Melanoma: Beyond the Basics. Plastic Reconstr. Surg. 138 (2), 330e–340e. doi:10.1097/PRS.0000000000002367
Peng, Q., and Wang, J. (2021). Non-coding RNAs in Melanoma: Biological Functions and Potential Clinical Applications. Mol. Ther. Oncol. 22, 219–231. doi:10.1016/j.omto.2021.05.012
Qu, S., Zhong, Y., Shang, R., Zhang, X., Song, W., Kjems, J., et al. (2017). The Emerging Landscape of Circular RNA in Life Processes. RNA Biol. 14 (8), 992–999. doi:10.1080/15476286.2016.1220473
Salzman, J. (2016). Circular RNA Expression: Its Potential Regulation and Function. Trends Genet. 32 (5), 309–316. doi:10.1016/j.tig.2016.03.002
Shain, A. H., and Bastian, B. C. (2016). From Melanocytes to Melanomas. Nat. Rev. Cancer 16 (6), 345–358. doi:10.1038/nrc.2016.37
Szambowska, A., Tessmer, I., Prus, P., Schlott, B., Pospiech, H., and Grosse, F. (2017). Cdc45-induced Loading of Human RPA onto Single-Stranded DNA. Nucleic Acids Res. 45 (6), gkw1364. doi:10.1093/nar/gkw1364
Tang, K., Zhang, H., Li, Y., Sun, Q., and Jin, H. (2021). Circular RNA as a Potential Biomarker for Melanoma: A Systematic Review. Front. Cell Dev. Biol. 9, 638548. doi:10.3389/fcell.2021.638548
Testori, A., Ribero, S., and Bataille, V. (2017). Diagnosis and Treatment of In-Transit Melanoma Metastases. Eur. J. Surg. Oncol. 43 (3), 544–560. doi:10.1016/j.ejso.2016.10.005
Wu, X., Xiao, Y., Ma, J., and Wang, A. (2020). Circular RNA: A Novel Potential Biomarker for Skin Diseases. Pharmacol. Res. 158, 104841. doi:10.1016/j.phrs.2020.104841
Keywords: hsa_circ_0062270, melanoma, Cdc45, proliferation, metastasis
Citation: Wei C, Sun W, Liu C, Meng F, Sun L and Ding X (2022) Hsa_circ_0062270 Promotes Tumorigenesis of Melanoma by Stabilizing the Linear Transcript Cell Division Cycle Protein 45. Front. Genet. 13:897440. doi: 10.3389/fgene.2022.897440
Received: 16 March 2022; Accepted: 25 April 2022;
Published: 10 May 2022.
Edited by:
Deepak Kumar Jain, Chongqing University of Posts and Telecommunications, ChinaReviewed by:
Jian Tang, Nanjing Medical University, ChinaCopyright © 2022 Wei, Sun, Liu, Meng, Sun and Ding. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiangsheng Ding, ZGluZzg4ODM2NDAzNTE0MEAxMjYuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.