
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Genet. , 14 April 2022
Sec. Stem Cell Research
Volume 13 - 2022 | https://doi.org/10.3389/fgene.2022.891159
This article is part of the Research Topic iPS Cell Models of Human Single Gene Inheritance Diseases View all 4 articles
Human induced pluripotent stem cells (hIPSC) have been used to produce almost all types of human cells currently, which makes them into several potential applications with replicated patient-specific genotype. Furthermore, hIPSC derived cardiomyocytes assembled engineering heart tissue can be established to achieve multiple functional evaluations by tissue engineering technology. This short review summarized the current advanced applications based on the hIPSC derived heart tissue in molecular mechanisms elucidating and high throughput drug screening.
Human-induced pluripotent stem cells (hIPSC) are currently recognized as the most suitable cell source for producing almost all types of human cells. The interest in reprogramming cell fate has led to a revolutionary change in regenerative medicine, which achieves a “copy-paste” of patient-specific genotype cells from the bench to the bedside (Carvajal-Vergara et al., 2010; Itzhaki et al., 2011; Yazawa et al., 2011; Kim et al., 2013). In the past decade, researchers in the field of hIPSC have attempted to solve a series of fundamental issues, including whether hIPSC have the ability to differentiate into different types of functional cells (Ramalho-Santos, 2009; Maher, 2013; Zhao et al., 2019; Cowan et al., 2020; Lee et al., 2022); whether hIPSC-derived cells could mimic pathophysiological processes; and whether this completely “in vitro” model could help researchers unravel underlying pathogenetic mechanisms and provide clues on diagnostic and therapeutic choices (Pavez-Giani and Cyganek, 2021; Faulkner-Jones et al., 2022; Yang et al., 2022). Along with their performance in several functional tests, hIPSC-derived cells show unique advantages, although even fully mature tissue characteristics still cannot be achieved today. As more complicated syndromes have been modeled, hIPSC-based disease modeling has been praised as a simple tool to obtain essential knowledge of complicated molecular pathogenetic processes (Park et al., 2019; Liu et al., 2021). Undoubtedly, high-throughput drug testing for particular diseases based on engineering of heart tissue (EHT) using hIPSC has given birth to the transfer of in vitro models to more productive applications (Lam et al., 2019).
Investment in drug development is considered a high-risk enterprise and requires an extremely prolonged commitment. Irrespective of the chemical compounds, small peptides, microRNAs, or even gene delivery approach, drug screening methods need to be complemented by various substrates once the candidate libraries have been built (Liu et al., 2022). Traditionally, the toxicity of new drugs has been tested using different types of cell lines. Regardless of cost and efficiency, a simple cell line test supplies information to allow the selection of preliminary candidates and confirms an active serum concentration. In vitro drug screening then must advance into physiological experiments using animal models. Mice have been involved for the initial process. And transgenic mice are critical for inherit cardiovascular diseases. Then large animals are considered as the further step before clinical transition to measure specific cardiac electrophysiology. This approach can lead to the simulation of potential drug reactions in different organs throughout the body, although numerous failures have reminded researchers that interspecies differences may represent a barrier to fully transfer experimental test results to clinic practice (Pavez-Giani and Cyganek, 2021; Bourque et al., 2022; Tricot et al., 2022). Then the human based clinical trial is critical to fully validate a newly invented drugs. Commonly, this process would take more than 10 years. However, due to various genetic and epigenetic backgrounds between animal models and human, even within humans, differences in sex hormone-cardiac-electrophysiology, races, environmental exposure, failure always been obtained for final pharmacy validations leading to huge waste of costs and time. As in the Long-QT syndrome, genetic ablation of KCNQ1 in mice did not consistently lead to a cardiac phenotype similar to that of the observed in the long-QT syndrome in patients, which is due to differences in the functions of potassium channels between humans and mice; thus, it is not scientifically feasible to perform cardiotoxicity testing of drugs as there is a risk of inducing prolongation of the QT interval (Casimiro et al., 2001). Accordingly, there is an obvious gap in translational medicine research, which has raised the urgent demand for a humanized functional test model. In this context, hIPSC-derived human functional cells may perfectly fit this purpose. The marked increased in differentiation efficiency exceeds the limits placed on cell numbers (Klug et al., 1996), and the completely in vitro culture and maturation models provide the opportunity to build a heart organoid. Moreover, EHT (Kehat et al., 2001; Paik et al., 2020) and even a three-dimensional (3D) ventricle (Liang and Zhang, 2013) have been proposed to mimic cardiac tissue and heart physiological functions at various levels.
Early in 1996, human beating cells were first observed following spontaneous Emory body (EB) differentiation (Klug et al., 1996). Unfortunately, the differentiation efficiency was really low, which was around 5–10%. Subsequently, more than 20 years were spent improving cardiomyocyte differentiation protocols. Since Yamanaka’s research team reported the convinced method to improve the production of hIPSC, several studies devoted to establish a stable protocol to simulate the differential efficiency. Particular regulation of several signaling pathways, which were involved in cardiomyocytes development including bone morphogenetic protein (BMP), wingless/integrated (Wnt) and transforming growth factor-β (TGF- β), had been indicated to promote efficacy of hIPSC derived cardiomyocytes (hIPSC-CMs), which could reach more than 90% purity of CMs. Currently, cardiomyocytes differentiation efficiency is now one of the most robust among all the differentiation lineages. Although, the evaluation and qualification of hIPSC-CMs did not achieve cell cluster contractility, it can be applied to cell-based assays, including differentiation efficiency, cardiomyocytes structure (Kehat et al., 2001), single cell contraction, and electrophysiology. The development of an efficient, stable, and low-cost differentiation model stimulated an unprecedented boost in subsequent research activity. However, it is important to be noted that the hIPSC-CMs demonstrated phenotypic plasticity. And current differential protocol could only derived CMs into neonatal or under-maturated stages, which could not be fully identified as matured CMs. So that, the cultural conditions and duration are critical to make hIPSC-CMs function. Thus, it should be build a specific protocol for differentiation according to genotypes and applications of hIPSC-CMs. Specific gene expressions had been used to determine which stages the hIPSC-CMs remaining, such as mesoderm formation (MIXL1 and BRY), cardiogenic mesoderm (ISL1 and MESP1), cardiac-specific progenitors (GATA4, TBX5 and NKX2.5) and relative mature CMs (TNNT2 and MYH7). Besides, ventricular and atrial specific differentiation could be achieved recently, which provided more platforms to assess the drugs targeting different cellular types respectively. A series of studies have evaluated whether the methods could be designed to improve cardiomyocyte maturation in vitro (both at the cell level and at the micro-tissue level). EHT constructs greatly expanded the potential and breadth of functional testing, presenting an in vitro functional assay a model with a phenotype that may truly mimic clinical symptoms. Successful cardiomyocyte modeling shifted the hIPSC-CM-based drug screening model from a traditional target driven assay to a functional model, which expands the strategies for drug validation and measurement of pharmacodynamic effects (Paik et al., 2020). So that, if EHT had been used to test the adverse effects on cardiac development, it should to make the hIPSC-CMs remained in specific developmental phases. While exploring the therapeutic effects on mature CMs, purification and advanced protocol should be applied to make EHT functional contraction and electrical activity. As shown in a recent study, four types of calcium channel blockers initially showed therapeutic efficacy, but the mechanism of action and transcriptional activity differed in hIPSC models, and partly explained the origins of drug resistance and unexpected side effects (Lam et al., 2019).
One of the most well-known advantages of hIPSC is the generation of personalized genotype cells. Although more than 99.9% of the genome sequence is exactly the same across populations, only a 0.1% difference is responsible for the diversity in the response to different drugs. From this standpoint, hIPSC-based drug testing will represent a simple and effective model to verify personalized-based outcomes. So that, to establish a personal genotype based EHT and determine the best drug applications according to a specific genetic background could be considered as the personalization strategy, which would reduce the differences between population results and individual application. In a previous study (Lam et al., 2019), based on only three different cell lines, four drugs showed different response combinations. Although we could explain the variability in gene transcription in cell lines due to genetic variants and not to drugs, another explanation is also due to the variability of differently expressed differentiation characteristics within the same genotype (Liang and Zhang, 2013). Accordingly, another important question has been raised: is there a standard mechanism involved in cardiac differentiation and EHT assembly? Since 2001, cardiomyocyte differentiation has undergone extensive research to identify how to produce mature cells. From the “beating” phenotype to the detailed subcellular structure (Chung et al., 2007; Qi et al., 2022), even atrial or ventricular cardiomyocytes exhibit a specific physiology, which has gradually become a new standard in the field (Kattman et al., 2011). At this point, the development of universal quality standards for EHT will promote the scientific advancement among different research groups and will direct the development of specifically targeted drugs. In addition, Zhang et al. (2013). identified an engineered tissue patch function was associated with cardiomyocyte purity within the tissue. Interestingly, contractility showed the highest score in 65–75% of cardiomyocytes and the conduction velocity was positively associated with purity. This finding suggests that it is possible to establish a specific functionally-enhanced tissue model not only via a spatial organization pattern but also by establishing a subtype ratio.
The specific functionally enhanced tissue model could be used directly to model a target disease dysfunction, or different functional characteristics of the same disease. Most importantly, the mimicking mini hearts share the same genetic background, which could validate novel therapeutic strategies. Beyond the applications of drug tests on various genetic backgrounds, hIPSC-CM-EHTs have been used for gene mutation induced inherit cardiovascular diseases. Over the last decade, several cell lines of genetically inherited diseases have been developed to test the efficacy of old and newly invented drugs, including LMNA (Lee et al., 2019), TTN (Hinson et al., 2015), MYH7 (Vander Roest et al., 2021), TNNT2 (Pettinato et al., 2020), MYBPC3 (Seeger et al., 2019), PKP2 (Kim et al., 2013), DSP (Xia et al., 2020), TAFAZZIN (Wang et al., 2014), PTPN11 (Carvajal-Vergara et al., 2010)-induced cardiomyopathy, and RYR2 (Park et al., 2019), and KCNQ1 (Casimiro et al., 2001) induced arrhythmia. Furthermore, multiple genetic variants have also been evaluated using iPSC-based models, which has provided a further understanding on the complex genetic background (Gifford et al., 2019). Thus, the in vitro heart may represent an optimal approach for cardiac research both in terms of the mechanisms of inherited cardiovascular disease and screening of new drugs.
Accordingly, we could easy hypothesize that the application of the hIPSC-derived cardiomyocyte-assembled EHT (hIPSC-CM-EHT) model will define a novel drug screening model system. In this system, the variable genetic background of the micro-tissue will play the role of a tiny drug tester, for different genotypes, thus, different types of functional micro-tissue models can be used to mimic different clinical phenotypes. According to current strategies, there are two kinds of common aspects of EHT as 3D architecture and monolayer. Usually, monolayer model is more suitable for high throughput screening. And this kind of monolayer model is great for evaluation for toxicity, proliferation, development, and even for calcium transit. While 3D structural model is more difficult to be established and reduce the capability for large size screening. But it presents the advantages in determining contractile activity and interaction between various cells and extracellular matrix. Thus, to select the optimal model based on the propose of research is also crucial for drug screening. Due to the different strategies to build EHT, each type of EHTs presents different capabilities in determining contractile parameters (Table 1). Generally. There are five approved strategies of EHT establishment. Organoid has been used to mimic the function of whole heart with several approaches (Lewis-Israeli et al., 2021). Although it could be recorded contraction, but it still doesn’t fully address the cardiac function with very limited parameters could be underlined. Then thin film has been widely used to measure part of contractile activities (Wang et al., 2014). But it could not be loaded mechanical stress and still a kind of 2D EHT. EHTs based on muscle ring/network/bundle/biowire satisfied most of contractile parameters measurements (Zhang et al., 2013; Zhao et al., 2019; Li et al., 2020; Bliley et al., 2021). However such models required more hIPSC-CMs and complex steps, which would reduce the capability of high throughput. Similarly, in vitro cardiomyocyte maturation methods, for example, electrostimulation (Sun and Nunes, 2016; Ronaldson-Bouchard et al., 2018) and mechanical rocking (Zhang et al., 2013; Shadrin et al., 2017), will represent additional options to mimic a differentiated adult-like phenotype of cardiac tissue function. Although the engineered “maturation” of cardiomyocytes has always been difficult, it is also difficult to derive equivalent EHT at a specific stage of development. Nonetheless, the hIPSC-CM-EHT model is an easy-to-harvest artificial heart tissue, compared with in vivo human heart tissue specimens, which presents similar functional properties (Eschenhagen et al., 2015).
Considering the advantages of such a model system, it may be possible to mimic the chronic side effects of prolonged treatment (years) with specific agents only after a few weeks and rare phenotypes may be rapidly identified in vitro. Furthermore, the process of cardiomyocyte differentiation and cardiac tissue assembly provides an extended treatment window able to mimic almost all periods of heart development. A recent study by Theodoris et al. (2021) established a network-based screening strategy using iPSC-derived cells to identify therapeutic candidates for heart valve disease, which was made feasible by human iPSC technology, network analysis, and machine learning and represents an effective path for drug discovery. In particular, due to the limits posed by ethical issues, this model system provides the valuable opportunity to focus on the “leopard spot” transformation of cardiomyocytes specificity, proliferation, and maturation directly form the fetal cell’s black box. So that, the application of hIPSC-CM-EHT could help to accelerate the drug screening by the following steps (Figure 1). First, hIPSC should be obtained from a specific group or individual patients and healthy population. Then the basic hIPSC-CM could take over the initial candidate molecules screening. After purification of CMs, “heart in dish” could be established and involved in tissue screening, and benefit drug development, impacting the therapeutic strategy for the donors representing population. Then, specific genotype hIPSC-CMs would be participated in building “disease in disease”, which had been used in functional measurement, providing information for target digging and molecular design.
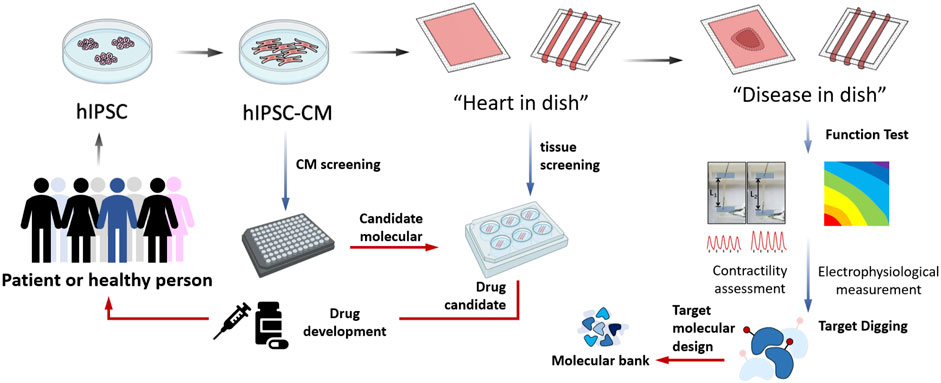
FIGURE 1. The applications of hIPSC derived cardiomyocytes assembled engineering heart tissue model in cardiovascular researches.
During the worldwide outbreak of COVID-19, the hIPSC-CM-EHT model has contributed to several types of investigations that: 1) mimicked myocardial injuries caused by SARS-CoV-2 infection (Li et al., 2021; Perez-Bermejo et al., 2021; Salerno et al., 2021; Xie et al., 2022); 2) evaluated the safety of different types of vaccines including mRNA and adenovirus vaccines, especially across different ethnic populations with different genomic backgrounds (Bojkova et al., 2020; Lu et al., 2020; Davidson et al., 2021); and 3) assessed the potential side effects of anti-COVID-19 developed treatments on cardiomyocytes (Charrez et al., 2021; Deguchi et al., 2021; Dimai et al., 2021). Research involving the hIPSC-CM-EHT model in COVID-19 has rapidly extended the application of hIPSC, making it a more common conceptual and powerful investigative tool.
The application of hIPSC-CM-EHT as a systematic, standardized, and high throughput tool will be the objective next step, and much more research will stem from here.
SL, YH and DZ conceived the presented idea. LW, SX, YL, YW and YQ summarized the reference and draft the manuscript. YL organized the figure. SL, YH and YL supervised the project and contributed equally to the final version of the manuscript.
All phase of this study was supported by a Hubei Province Science Fund for Distinguished Young Scholars (2019CFA092), Key R&D Program of Sichuan Province of China (2020YFS0102) and Natural Science Foundation of China (81700360, 31871496). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.891159/full#supplementary-material
Bliley, J. M., Vermeer, M. C. S. C., Duffy, R. M., Batalov, I., Kramer, D., Tashman, J. W., et al. (2021). Dynamic Loading of Human Engineered Heart Tissue Enhances Contractile Function and Drives a Desmosome-Linked Disease Phenotype. Sci. Transl. Med. 13 (603), 13. eng. Epub 2021/07/23. Cited in: Pubmed; PMID 34290054. doi:10.1126/scitranslmed.abd1817
Bojkova, D., Wagner, J. U. G., Shumliakivska, M., Aslan, G. S., Saleem, U., Hansen, A., et al. (2020). SARS-CoV-2 Infects and Induces Cytotoxic Effects in Human Cardiomyocytes. Cardiovasc. Res. 116 (14), 2207–2215. eng. Epub 2020/09/24. Cited in: Pubmed; PMID 32966582. doi:10.1093/cvr/cvaa267
Bourque, K., Jones-Tabah, J., and Hébert, T. E. (2022). Tools for Drug Discovery and Disease Modeling- the Future Is upon Us. Methods. eng. Epub 2022/02/14. Cited in: Pubmed; PMID 35151888. doi:10.1016/j.ymeth.2022.02.003
Carvajal-Vergara, X., Sevilla, A., D’Souza, S. L., Ang, Y.-S., Schaniel, C., Lee, D.-F., et al. (2010). Patient-specific Induced Pluripotent Stem-Cell-Derived Models of LEOPARD Syndrome. Nature 465 (7299), 808–812. eng. Epub 2010/06/11. Cited in: Pubmed; PMID 20535210. doi:10.1038/nature09005
Casimiro, M. C., Knollmann, B. C., Ebert, S. N., Vary, J. C., Greene, A. E., Franz, M. R., et al. (2001). Targeted Disruption of the Kcnq1 Gene Produces a Mouse Model of Jervell and Lange- Nielsen Syndrome. Proc. Natl. Acad. Sci. U.S.A. 98 (5), 2526–2531. eng. Epub 2001/02/28. Cited in: Pubmed; PMID 11226272. doi:10.1073/pnas.041398998
Charrez, B., Charwat, V., Siemons, B., Finsberg, H., Miller, E. W., Edwards, A. G., et al. (2021). In Vitro safety "clinical Trial" of the Cardiac Liability of Drug Polytherapy. Clin. Transl Sci. 14 (3), 1155–1165. eng. Epub 2021/04/01. Cited in: Pubmed; PMID 33786981. doi:10.1111/cts.13038
Chung, S., Dzeja, P. P., Faustino, R. S., Perez-Terzic, C., Behfar, A., and Terzic, A. (2007). Mitochondrial Oxidative Metabolism Is Required for the Cardiac Differentiation of Stem Cells. Nat. Rev. Cardiol. 4 (Suppl. 1), S60–S67. eng. Epub 2007/01/19. Cited in: Pubmed; PMID 17230217. doi:10.1038/ncpcardio0766
Cowan, C. S., Renner, M., De Gennaro, M., Gross-Scherf, B., Goldblum, D., Hou, Y., et al. (2020). Cell Types of the Human Retina and its Organoids at Single-Cell Resolution. Cell 182 (6), 1623–1640.e34. eng. Epub 2020/09/19. Cited in: Pubmed; PMID 32946783. doi:10.1016/j.cell.2020.08.013
Davidson, S. M., Padró, T., Bollini, S., Vilahur, G., Duncker, D. J., Evans, P. C., et al. (2021). Progress in Cardiac Research: from Rebooting Cardiac Regeneration to a Complete Cell Atlas of the Heart. Cardiovasc. Res. 117 (10), 2161–2174. eng. Epub 2021/06/12. Cited in: Pubmed; PMID 34114614. doi:10.1093/cvr/cvab200
Deguchi, S., Serrano-Aroca, Á., Tambuwala, M. M., Uhal, B. D., Brufsky, A. M., and Takayama, K. (2021). SARS-CoV-2 Research Using Human Pluripotent Stem Cells and Organoids. Stem Cell Transl Med 10 (11), 1491–1499. eng. Epub 2021/07/25. Cited in: Pubmed; PMID 34302450. doi:10.1002/sctm.21-0183
Dimai, S., Semmler, L., Prabhu, A., Stachelscheid, H., Huettemeister, J., Klaucke, S. C., et al. (2021). COVID19-associated Cardiomyocyte Dysfunction, Arrhythmias and the Effect of Canakinumab. PLoS One 16 (8), e0255976. eng. Epub 2021/08/20. Cited in: Pubmed; PMID 34411149. doi:10.1371/journal.pone.0255976
Eschenhagen, T., Mummery, C., and Knollmann, B. C. (2015). Modelling Sarcomeric Cardiomyopathies in the Dish: from Human Heart Samples to iPSC Cardiomyocytes. Cardiovasc. Res. 105 (4), 424–438. eng. Epub 2015/01/27. Cited in: Pubmed; PMID 25618410. doi:10.1093/cvr/cvv017
Faulkner-Jones, A., Zamora, V., Hortigon-Vinagre, M. P., Wang, W., Ardron, M., Smith, G. L., et al. (2022). A Bioprinted Heart-On-A-Chip with Human Pluripotent Stem Cell-Derived Cardiomyocytes for Drug Evaluation. Bioengineering 9 (1), 32. eng. Epub 2022/01/21. Cited in: Pubmed; PMID 35049741. doi:10.3390/bioengineering9010032)
Gifford, C. A., Ranade, S. S., Samarakoon, R., Salunga, H. T., de Soysa, T. Y., Huang, Y., et al. (2019). Oligogenic Inheritance of a Human Heart Disease Involving a Genetic Modifier. Science 364 (6443), 865–870. eng. Epub 2019/05/31. Cited in: Pubmed; PMID 31147515. doi:10.1126/science.aat5056
Hinson, J. T., Chopra, A., Nafissi, N., Polacheck, W. J., Benson, C. C., Swist, S., et al. (2015). Titin Mutations in iPS Cells Define Sarcomere Insufficiency as a Cause of Dilated Cardiomyopathy. Science 349 (6251), 982–986. eng. Epub 2015/09/01. Cited in: Pubmed; PMID 26315439. doi:10.1126/science.aaa5458
Itzhaki, I., Maizels, L., Huber, I., Zwi-Dantsis, L., Caspi, O., Winterstern, A., et al. (2011). Modelling the Long QT Syndrome with Induced Pluripotent Stem Cells. Nature 471 (7337), 225–229. eng. Epub 2011/01/18. Cited in: Pubmed; PMID 21240260. doi:10.1038/nature09747
Kattman, S. J., Witty, A. D., Gagliardi, M., Dubois, N. C., Niapour, M., Hotta, A., et al. (2011). Stage-specific Optimization of Activin/nodal and BMP Signaling Promotes Cardiac Differentiation of Mouse and Human Pluripotent Stem Cell Lines. Cell Stem Cell 8 (2), 228–240. eng. Epub 2011/02/08. Cited in: Pubmed; PMID 21295278. doi:10.1016/j.stem.2010.12.008
Kehat, I., Kenyagin-Karsenti, D., Snir, M., Segev, H., Amit, M., Gepstein, A., et al. (2001). Human Embryonic Stem Cells Can Differentiate into Myocytes with Structural and Functional Properties of Cardiomyocytes. J. Clin. Invest. 108 (3), 407–414. eng. Epub 2001/08/08. Cited in: Pubmed; PMID 11489934. doi:10.1172/jci200112131
Kim, C., Wong, J., Wen, J., Wang, S., Wang, C., Spiering, S., et al. (2013). Studying Arrhythmogenic Right Ventricular Dysplasia with Patient-specific iPSCs. Nature 494 (7435), 105–110. eng. Epub 2013/01/29. Cited in: Pubmed; PMID 23354045. doi:10.1038/nature11799
Klug, M. G., Soonpaa, M. H., Koh, G. Y., and Field, L. J. (1996). Genetically Selected Cardiomyocytes from Differentiating Embronic Stem Cells Form Stable Intracardiac Grafts. J. Clin. Invest. 98 (1), 216–224. eng. Epub 1996/07/01. Cited in: Pubmed; PMID 8690796. doi:10.1172/jci118769
Lam, C. K., Tian, L., Belbachir, N., Wnorowski, A., Shrestha, R., Ma, N., et al. (2019). Identifying the Transcriptome Signatures of Calcium Channel Blockers in Human Induced Pluripotent Stem Cell-Derived Cardiomyocytes. Circ. Res. 125 (2), 212–222. eng. Epub 2019/05/14. Cited in: Pubmed; PMID 31079550. doi:10.1161/circresaha.118.314202
Lee, J., Termglinchan, V., Diecke, S., Itzhaki, I., Lam, C. K., Garg, P., et al. (2019). Activation of PDGF Pathway Links LMNA Mutation to Dilated Cardiomyopathy. Nature 572 (7769), 335–340. eng. Epub 2019/07/19. Cited in: Pubmed; PMID 31316208. doi:10.1038/s41586-019-1406-x
Lee, K. Y., Park, S.-J., Matthews, D. G., Kim, S. L., Marquez, C. A., Zimmerman, J. F., et al. (2022). An Autonomously Swimming Biohybrid Fish Designed with Human Cardiac Biophysics. Science 375 (6581), 639–647. eng. Epub 2022/02/11. Cited in: Pubmed; PMID 35143298. doi:10.1126/science.abh0474
Lewis-Israeli, Y. R., Wasserman, A. H., Gabalski, M. A., Volmert, B. D., Ming, Y., Ball, K. A., et al. (2021). Self-assembling Human Heart Organoids for the Modeling of Cardiac Development and Congenital Heart Disease. Nat. Commun. 12 (1), 5142. eng. Epub 2021/08/28. Cited in: Pubmed; PMID 34446706. doi:10.1038/s41467-021-25329-5
Li, Y., Renner, D. M., Comar, C. E., Whelan, J. N., Reyes, H. M., Cardenas-Diaz, F. L., et al. (2021). SARS-CoV-2 Induces Double-Stranded RNA-Mediated Innate Immune Responses in Respiratory Epithelial-Derived Cells and Cardiomyocytes. Proc. Natl. Acad. Sci. U.S.A. 118 (16), 118. eng. Epub 2021/04/04. Cited in: Pubmed; PMID 33811184. doi:10.1073/pnas.2022643118
Li, Y., Song, D., Mao, L., Abraham, D. M., and Bursac, N. (2020). Lack of Thy1 Defines a Pathogenic Fraction of Cardiac Fibroblasts in Heart Failure. Biomaterials 236, 119824. eng. Epub 2020/02/07. Cited in: Pubmed; PMID 32028169. doi:10.1016/j.biomaterials.2020.119824
Liang, G., and Zhang, Y. (2013). Genetic and Epigenetic Variations in iPSCs: Potential Causes and Implications for Application. Cell Stem Cell 13 (2), 149–159. eng. Epub 2013/08/06. Cited in: Pubmed; PMID 23910082. doi:10.1016/j.stem.2013.07.001
Liu, L., Yue, P., Zhang, Y., Hua, Y., Bi, W., Yan, H., et al. (2022). Non-cell-autonomous Manner of AAV Administration to Attenuate Cardiomyocyte Hypertrophy by Targeting Paracrine Signaling on ECM to Reduce Viral Dosage. Sig Transduct Target. Ther. 7 (1), 2. eng. Epub 2022/01/05. Cited in: Pubmed; PMID 34980885. doi:10.1038/s41392-021-00715-z
Liu, X., Wang, S., Guo, X., Li, Y., Ogurlu, R., Lu, F., et al. (2021). Increased Reactive Oxygen Species-Mediated Ca 2+ /Calmodulin-dependent Protein Kinase II Activation Contributes to Calcium Handling Abnormalities and Impaired Contraction in Barth Syndrome. Circulation 143 (19), 1894–1911. eng. Epub 2021/04/02. Cited in: Pubmed; PMID 33793303. doi:10.1161/circulationaha.120.048698
Lu, D., Chatterjee, S., Xiao, K., Riedel, I., Wang, Y., Foo, R., et al. (2020). MicroRNAs Targeting the SARS-CoV-2 Entry Receptor ACE2 in Cardiomyocytes. J. Mol. Cell Cardiol. 148, 46–49. eng. Epub 2020/09/07. Cited in: Pubmed; PMID 32891636. doi:10.1016/j.yjmcc.2020.08.017
Maher, B. (2013). Tissue Engineering: How to Build a Heart. Nature 499 (7456), 20–22. eng. Epub 2013/07/05. Cited in: Pubmed; PMID 23823778. doi:10.1038/499020a
Paik, D. T., Chandy, M., and Wu, J. C. (2020). Patient and Disease-specific Induced Pluripotent Stem Cells for Discovery of Personalized Cardiovascular Drugs and Therapeutics. Pharmacol. Rev. 72 (1), 320–342. eng. Epub 2019/12/25. Cited in: Pubmed; PMID 31871214. doi:10.1124/pr.116.013003
Park, S.-J., Zhang, D., Qi, Y., Li, Y., Lee, K. Y., Bezzerides, V. J., et al. (2019). Insights into the Pathogenesis of Catecholaminergic Polymorphic Ventricular Tachycardia from Engineered Human Heart Tissue. Circulation 140 (5), 390–404. eng. Epub 2019/07/18. Cited in: Pubmed; PMID 31311300. doi:10.1161/circulationaha.119.039711
Pavez-Giani, M. G., and Cyganek, L. (2021). Recent Advances in Modeling Mitochondrial Cardiomyopathy Using Human Induced Pluripotent Stem Cells. Front. Cel Dev. Biol. 9, 800529. eng. Epub 2022/01/28. Cited in: Pubmed; PMID 35083221. doi:10.3389/fcell.2021.800529
Perez-Bermejo, J. A., Kang, S., Rockwood, S. J., Simoneau, C. R., Joy, D. A., Silva, A. C., et al. (2021). SARS-CoV-2 Infection of Human iPSC-Derived Cardiac Cells Reflects Cytopathic Features in Hearts of Patients with COVID-19. Sci. Transl. Med. 13 (590). eng. Epub 2021/03/17. Cited in: Pubmed; PMID 33723017. doi:10.1126/scitranslmed.abf7872
Pettinato, A. M., Ladha, F. A., Mellert, D. J., Legere, N., Cohn, R., Romano, R., et al. (2020). Development of a Cardiac Sarcomere Functional Genomics Platform to Enable Scalable Interrogation of Human TNNT2 Variants. Circulation 142 (23), 2262–2275. eng. Epub 2020/10/08. Cited in: Pubmed; PMID 33025817. doi:10.1161/circulationaha.120.047999
Qi, Y., Ye, Y., Wang, R., Yu, S., Zhang, Y., Lv, J., et al. (2022). Mitochondrial Dysfunction by TFAM Depletion Disrupts Self-Renewal and Lineage Differentiation of Human PSCs by Affecting Cell Proliferation and YAP Response. Redox Biol. 50, 102248. eng. Epub 2022/01/30. Cited in: Pubmed; PMID 35091324. doi:10.1016/j.redox.2022.102248
Ramalho-Santos, M. (2009). iPS Cells: Insights into Basic Biology. Cell 138 (4), 616–618. eng. Epub 2009/08/26. Cited in: Pubmed; PMID 19703387. doi:10.1016/j.cell.2009.08.012
Ronaldson-Bouchard, K., Ma, S. P., Yeager, K., Chen, T., Song, L., Sirabella, D., et al. (2018). Advanced Maturation of Human Cardiac Tissue Grown from Pluripotent Stem Cells. Nature 556 (7700), 239–243. eng. Epub 2018/04/06. Cited in: Pubmed; PMID 29618819. doi:10.1038/s41586-018-0016-3
Salerno, J. A., Torquato, T., Temerozo, J. R., Goto-Silva, L., Karmirian, K., Mendes, M. A., et al. (2021). Inhibition of SARS-CoV-2 Infection in Human iPSC-Derived Cardiomyocytes by Targeting the Sigma-1 Receptor Disrupts Cytoarchitecture and Beating. PeerJ 9, e12595. eng. Epub 2022/01/18. Cited in: Pubmed; PMID 35036128. doi:10.7717/peerj.12595
Seeger, T., Shrestha, R., Lam, C. K., Chen, C., McKeithan, W. L., Lau, E., et al. (2019). A Premature Termination Codon Mutation in MYBPC3 Causes Hypertrophic Cardiomyopathy via Chronic Activation of Nonsense-Mediated Decay. Circulation 139 (6), 799–811. eng. Epub 2018/12/28. Cited in: Pubmed; PMID 30586709. doi:10.1161/circulationaha.118.034624
Shadrin, I. Y., Allen, B. W., Qian, Y., Jackman, C. P., Carlson, A. L., Juhas, M. E., et al. (2017). Cardiopatch Platform Enables Maturation and Scale-Up of Human Pluripotent Stem Cell-Derived Engineered Heart Tissues. Nat. Commun. 8 (1), 1825. eng. Epub 2017/12/01. Cited in: Pubmed; PMID 29184059. doi:10.1038/s41467-017-01946-x
Sun, X., and Nunes, S. S. (2016). Biowire Platform for Maturation of Human Pluripotent Stem Cell-Derived Cardiomyocytes. Methods 101, 21–26. eng. Epub 2015/11/08. Cited in: Pubmed; PMID 26546730. doi:10.1016/j.ymeth.2015.11.005
Theodoris, C. V., Zhou, P., Liu, L., Zhang, Y., Nishino, T., Huang, Y., et al. (2021). Network-based Screen in iPSC-Derived Cells Reveals Therapeutic Candidate for Heart Valve Disease. Science 371 (6530), 371. eng. Epub 2020/12/12. Cited in: Pubmed; PMID 33303684. doi:10.1126/science.abd0724
Tricot, T., Verfaillie, C. M., and Kumar, M. (2022). Current Status and Challenges of Human Induced Pluripotent Stem Cell-Derived Liver Models in Drug Discovery. Cells 11 (3), 442. eng. Epub 2022/02/16. Cited in: Pubmed; PMID 35159250. doi:10.3390/cells11030442
Vander Roest, A. S., Liu, C., Morck, M. M., Kooiker, K. B., Jung, G., Song, D., et al. Hypertrophic Cardiomyopathy β-cardiac Myosin Mutation (P710R) Leads to Hypercontractility by Disrupting Super Relaxed State. Proc. Natl. Acad. Sci. U.S.A. 2021;118(24). eng. Epub 2021/06/13. Cited in: Pubmed; PMID 34117120. doi:10.1073/pnas.2025030118
Wang, G., McCain, M. L., Yang, L., He, A., Pasqualini, F. S., Agarwal, A., et al. (2014). Modeling the Mitochondrial Cardiomyopathy of Barth Syndrome with Induced Pluripotent Stem Cell and Heart-On-Chip Technologies. Nat. Med. 20 (6), 616–623. eng. Epub 2014/05/13. Cited in: Pubmed; PMID 24813252. doi:10.1038/nm.3545
Xia, S., Wang, X., Yue, P., Li, Y., and Zhang, D. (2020). Establishment of Induced Pluripotent Stem Cell Lines from a Family of an ARVC Patient Receiving Heart Transplantation in Infant Age Carrying Compound Heterozygous Mutations in DSP Gene. Stem Cel Res. 48, 101977. eng. Epub 2020/09/18. Cited in: Pubmed; PMID 32942234. doi:10.1016/j.scr.2020.101977
Xie, L.-M., Huang, Y.-F., Liu, Y.-L., Liang, J.-Q., Deng, W., Lin, G.-L., et al. (2022). Identification of the Hub Genes and the Signaling Pathways in Human iPSC-Cardiomyocytes Infected by SARS-CoV-2. Biochem. Genet. 2, 1–17. eng. Epub 2022/03/03. Cited in: Pubmed; PMID 35235083. doi:10.1007/s10528-022-10206-7
Yang, M., Fu, J.-D., Zou, J., Sridharan, D., Zhao, M.-T., Singh, H., et al. (2022). Assessment of Mitophagy in Human iPSC-Derived Cardiomyocytes. Autophagy, 1–14. eng. Epub 2022/03/01. Cited in: Pubmed; PMID 35220905. doi:10.1080/15548627.2022.2037920
Yazawa, M., Hsueh, B., Jia, X., Pasca, A. M., Bernstein, J. A., Hallmayer, J., et al. (2011). Using Induced Pluripotent Stem Cells to Investigate Cardiac Phenotypes in Timothy Syndrome. Nature 471 (7337), 230–234. eng. Epub 2011/02/11. Cited in: Pubmed; PMID 21307850. doi:10.1038/nature09855
Zhang, D., Shadrin, I. Y., Lam, J., Xian, H.-Q., Snodgrass, H. R., and Bursac, N. (2013). Tissue-engineered Cardiac Patch for Advanced Functional Maturation of Human ESC-Derived Cardiomyocytes. Biomaterials 34 (23), 5813–5820. eng. Epub 2013/05/07. Cited in: Pubmed; PMID 23642535. doi:10.1016/j.biomaterials.2013.04.026
Keywords: IPSC, disease model, tissue engeneering, heart in dish, drug testing
Citation: Wei L, Xia S, Li Y, Qi Y, Wang Y, Zhang D, Hua Y and Luo S (2022) Application of hiPSC as a Drug Tester Via Mimicking a Personalized Mini Heart. Front. Genet. 13:891159. doi: 10.3389/fgene.2022.891159
Received: 07 March 2022; Accepted: 30 March 2022;
Published: 14 April 2022.
Edited by:
Xiaoling Guo, Wenzhou Medical University, ChinaReviewed by:
Bo Pan, Children’s Hospital of Chongqing Medical University, ChinaCopyright © 2022 Wei, Xia, Li, Qi, Wang, Zhang, Hua and Luo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Donghui Zhang, ZG9uZ2guemhhbmdAaHVidS5lZHUuY24=; Yimin Hua, bmF0aGFuX2h1YUAxNjMuY29t; Shuhua Luo, bHVvc2h1aHVhQHNjdS5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.