- 1College of Animal Sciences, Zhejiang University, Hangzhou, China
- 2Department of Biotechnology, Hindustan Institute of Technology and Science, Chennai, India
Numerous missense mutations have been reported in autosomal dominant polycystic kidney disease which is one of the most common renal genetic disorders. The underlying mechanism for cystogenesis is still elusive, partly due to the lack of suitable animal models. Currently, we tried to establish a porcine transgenic model overexpressing human PKD2-D511V (hPKD2-D511V), which is a dominant-negative mutation in the vertebrate in vitro models. A total of six cloned pigs were finally obtained using somatic cell nuclear transfer. However, five with functional hPKD2-D511V died shortly after birth, leaving only one with the dysfunctional transgenic event to survive. Compared with the WT pigs, the demised transgenic pigs had elevated levels of hPKD2 expression at the mRNA and protein levels. Additionally, no renal malformation was observed, indicating that hPKD2-D511V did not alter normal kidney development. RNA-seq analysis also revealed that several ADPKD-related pathways were disturbed when overexpressing hPKD2-D511V. Therefore, our study implies that hPKD2-D511V may be lethal due to the dominant-negative effect. Hence, to dissect how PKD2-D511V drives renal cystogenesis, it is better to choose in vitro or invertebrate models.
Introduction
As the most common renal inherited disease, autosomal dominant polycystic kidney disease (ADPKD) affects millions of people across the world. Progressively enlarged kidneys with numerous fluid-filled cysts are the hallmark of ADPKD, leading to end-stage renal disease in half of the patients in the sixth decade of life (Torres and Harris, 2009). ADPKD is also a systemic disorder with extrarenal manifestations, such as cysts in the liver, pancreas, and spleen, hypertension, cardiac valve malformation, and intracranial aneurysms (Gabow, 1990). Although many people are afflicted by the disease, the only treatment available besides renal replacement therapy is performed by tolvaptan which targets the vasopressin V2 receptor (Torres et al., 2012). However, tolvaptan was reported causing side effects, e.g., hepatic toxicity and nocturia, and only slow down the disease progression (Torres et al., 2017). Thus, novel therapeutic approaches must be developed to reverse renal function loss.
Extensive studies on ADPKD have been conducted for several decades. However, the precise molecular mechanisms responsible for it are still elusive. Thus far, several genes have been reported to be associated with ADPKD, of which PKD1 and PKD2 are the main causative genes (Pignatelli et al., 1992; Kimberling et al., 1993; Mochizuki et al., 1996). PKD1 and PKD2 encode multiple-transmembrane proteins, polycystin-1 (PC1) and polycystin-2 (PC2), respectively, which form a heterotetrameric protein complex mediating ion influx in the plasma membrane and primary cilium by sensing extracellular mechanical or chemical stimuli (Harris et al., 1995; Qian et al., 2003). Other than primary cilium, PC2 is abundant in the endoplasmic reticulum (ER) (Newby et al., 2002). Initial work suggested PC2 can mediate the influx of calcium, but contrarily, the recent study shows that the PC2 channel has a lower permeability to calcium than potassium and sodium (Koulen et al., 2002; Liu et al., 2018). A newly published paper by Padhy et al. further demonstrated that PC2 in the ER is a potassium channel, which mediates potassium entering the ER and drives calcium release from the ER (Padhy et al., 2022). Thus, it is essential to study the relationship between channel activity and disease phenotype.
Numerous mutations have been deposited in the mutation database of ADPKD (https://pkdb.mayo.edu/). The PKD2 c.1532A>T/p.511D>V (hereafter PKD2-D511V for gene and PC2-D511V for protein) missense mutation, which results in the replacement of an aspartic acid with valine in the bottom of S3 of PC2, is one of the most extensively studied mutations (Koulen et al., 2002). The genetic studies showed that this mutation impacts the alternative splicing, leading to aberrantly spliced PKD2 transcripts (Reynolds et al., 1999; Gonzalez-Paredes et al., 2016). The experiments using electrophysiological methods find the mutation is a dominant-negative mutation, which could abolish the channel activity of PC2 (Ma et al., 2005). In vivo zebrafish studies on PKD2-D511V transient overexpression resulted in the tail curvature, which is a reflection of zebrafish with renal cysts (Feng et al., 2008). Moreover, morpholino targeting zebrafish pkd2 could not be rescued by the PKD2-D511V (Pavel et al., 2016), demonstrating the dominant-negative effect of the mutation. To better characterize the mutation, in vivo genetic modified animal models are needed. Currently, only one fruit fly model with transgenic Amo-D627V (equivalent to human PKD2-D511V) is available to study the molecular consequence of PKD2-D511V mutation. However, no dominant-negative effect of the mutation was noticed in the invertebrate model (Köttgen et al., 2011; Hofherr et al., 2016). Although rodent models have proven critical to study the molecular basis of PKD in the past decade, there is no report on PKD2-D511V modified mouse model (Happé and Peters, 2014). These results suggest that it is necessary to establish a vertebrate model with PKD2-D511V mutation.
Since, the differences in renal anatomy (kidney size and renal function), and life span, a mouse model may not perfectly mimic human ADPKD. Thus, pig might be an ideal alternative to study the disease. Previously, our group constructed a series of pig models with overexpressed PKD2 and MYC, and PKD1 monoallelic knockout (He et al., 2013; Ye et al., 2013; He et al., 2015). This study aims to construct a transgenic pig model with the human PKD2-D511V gene to mimic the ADPKD genotype. Using somatic cell nuclear transfer (SCNT), we obtained six cloned piglets, of which only one cloned pig survived the perinatal stage. Genetic testing showed that the five demised piglets all have hPKD2-D511V transgene, while the survived contained a truncated structure. This study is novel on the PKD2-D511V vertebrate model, indicating the mutation might be lethal for vertebrates.
Materials and method
Plasmids
The plasmids pCAG-WThPKD2-3 × FLAG-floxP-neo-pH11 (hereafter pCAG-WT-hPKD2) and pCAG-muhPKD2 (c.1532A > T/p.511D > V)-3 × FLAG-floxP-neo-pH11 (hereafter pCAG-MU-hPKD2) were constructed in our previous work (Zhang et al., 2020). The pX330-pH11 plasmid, encoding Cas9 protein and gRNA targeting pH11 locus, was adapted from a previous report using primer annealing and ligation (Ruan et al., 2015). For zebrafish injection and cell transfection, these plasmids were extracted using the EndoFree Midi Plasmid kit (Tiangen, Beijing, China).
Plasmids injection of zebrafish embryos
The plasmids were diluted to 100 ng/μl with 10% phenol red before injecting into the yolk of fertilized one-cell stage zebrafish eggs. Mock-injected was performed using an equal volume (1 nL) of phenol red solution. The survived fertilized eggs were counted daily, and the dead embryos were discarded. At 5 dpf, the morphological characteristics of zebrafish seedlings were observed under the microscope. The seedlings with typical characteristics were picked for photography. Three biological replicates were performed. The total number of survived zebrafish with mock-injection, and injection of pCAG-WT-hPKD2 and pCAG-MU-hPKD2 were 179, 251, and 82, respectively.
Generation of transgenic pigs
A million primary CEMP embryonic fibroblast cells were cultured in DMEM (Gibco, Thermo Fisher, Shanghai, China) with 10% fetal bovine serum (Gibco, Thermo Fisher, Australia) at 37°C in a humidified incubator. On the day of transfection, the fibroblasts were digested and mixed with the 3 μg pX330-pH11 and 3 μg pCAG-MU-hPKD2. The transfection was performed using the Lonza Nucleofector (V4XP-3032, Lonza, Germany) with the CA-137 program. Then, the transfected cells were recovered for 24 h in a 6-well-plate before seeding in the 96-well plates at a density of 200 cells/well. 500 ng/μl Geneticin (Thermo fisher, Shanghai, China) was added to each well, and positively selected cells were subcultured to the 24-well-plates after 7–10 days. After that, the cells were split into two aliquots, which were then subjected to PCR identification and cryopreservation. The primers for the identification are detailed in the Supplementary Table S1. Somatic cell nuclear transfer was conducted using the pooled positive cells according to the previously published method (Supplementary Data S1) (Wei et al., 2008). A total of 7 surrogate sows were chosen and received an average of 300 reconstructed embryos. After 121–123 days, the cloned piglets were delivered naturally.
Genotyping of cloned piglets
Genotyping of cloned pigs was carried out using the same primers to identify positive transgenic cells, except that all the PCR amplified fragments were subjected to sanger sequencing.
Quantitative real-time polymerase chain reaction and western blotting
Total RNA was isolated from the skin and kidney tissues using the TRIzol Reagent (Tsingke, Beijing, China) according to the manufacturer’s instructions. The RNA was reverse-transcribed (Vazyme, Nanjing, China) following the manufacturer’s instructions, using the HiScript III Reverse Transcriptase (Vazyme, Nanjing, China) to perform the quantitative real-time polymerase chain reactions (qRT-PCR). The primers used in our study were listed in the Supplementary Table S1.
Membrane protein was extracted from the kidney tissues by the membrane and cytosol protein extraction kit (Beyotime, Shanghai, China) for western blotting. Membrane protein was subjected to 8% SDS-polyacrylamide gel electrophoresis. The separated proteins were transferred to PVDF membranes. The membranes were blocked with 5% skimmed milk at 37°C for 2 h, and incubated overnight with diluted primary antibodies against Polycystin-2 (D-3) (PC-2) (1:1000, E20, Santa Cruz Biotechnology, Dallas, TX, United States), DYKDDDDK Tag (FLAG) (1:1000, M2, Sigma-Aldrich, Shanghai, China), followed by goat anti-rabbit IgG (H + L)-HRP (1:5000, BIOKER, Hangzhou, China) and goat anti-mouse IgG (H + L)-HRP (1:5000, BIOKER, Hangzhou, China). An image vertical overflow drain (VCD) gel imaging system was used to determine the optical density (OD) of the protein bands. Finally, the relative abundance was calculated as the ratio of the OD of each protein to the OD of Ponceau S (Beyotime, Shanghai, China) stain.
Histological examination of cloned piglets
The sample of kidney tissues was fixed with 4% paraformaldehyde, dehydrated, and immersed in transparent wax. Next, the sections were sliced from the prepared paraffin blocks. Then, these sections were stained with hematoxylin and eosin. Antibodies for PC-2 (1:100, E20, Santa Cruz Biotechnology, Dallas, TX, United States) and FLAG (1:100, M2, Sigma-Aldrich, Shanghai, China) were used for immunohistochemistry.
RNA sequencing
RNA-seq experiments were carried out by Novogene (Beijing, China) using RNA extracted from the porcine kidneys. Following the manufacturer’s recommendations, sequencing libraries were built using the NEBNext UltraTM RNA Library Prep Kit for Illumina (NEB, United States), and index codes were added to attribute sequences to each sample. According to the manufacturer’s instructions, TruSeq PE Cluster Kit v3-cBot-HS (Illumina, United States) was used to cluster the index-coded samples on a cBot Cluster Generation System. The library preparations were sequenced on an Illumina Novaseq 6000 platform, and 150 bp paired-end reads were generated. Next, the cDNA sequence of human PKD2 was inserted into the Sus scrofa reference genome (http://ftp.ensembl.org/pub/release-107/fasta/sus_scrofa/cdna/) before indexing the reference. Salmon was used to align reads to the reference genome and quantify gene expression (Patro et al., 2017). DEGs were screened using the “DESeq2” package (Love et al., 2014) in R software (version 4.1.2) with the cutoff |Log2 fold change| > 1 and adjusted p-value < 0.05.
The principal component analysis (PCA) was performed by the bioladder cloud platform (https://www.bioladder.cn/) using the whole genome expression profile. The volcano and bubble charts were exported by the “ggplot2″ package (Kolde, 2015; Gómez-Rubio, 2017). The heatmap was exported by the “pheatmap” package (Kolde, 2015).
Gene ontology functional enrichment analyses
Gene ontology (GO) analyses were performed using the identified DEGs by the Database for Annotation, Visualization and Integrated Discovery (DAVID) with default parameters (https://david.ncifcrf.gov/) (Jiao et al., 2012).
Statistics
Data were presented as mean ± standard errors of the mean (SEMs). For comparing two groups, either t-test or Mann-Whitney test was used. Pearson’s chi-square test was utilized for comparing category data, and the resulting p-values were adjusted with the FDR approach. The type I error was set as 0.05.
Results and discussion
By accessing the PKD mutation database (https://pkdb.mayo.edu/), there are currently 2600 germline mutation records. However, no mutation hotspot has been identified in both PKD genes, indicating that ADPKD mutation is quite family-specific (Rossetti et al., 2001; Gout et al., 2007). Thus, it is unrealistic to establish a representative animal model to study the underline molecular mechanisms. Numerous rodent models have been generated to mimic ADPKD; however, most are null mutation models (Happé and Peters, 2014). Additionally, pig and monkey ADPKD models have been reported using the same strategy by knocking out PKD1 (He et al., 2015; Tsukiyama et al., 2019; Watanabe et al., 2022). Moreover, approximately 1/3 of the mutations deposited in the database are missense mutations, which might be crucial to understand the disease’s etiology. One of the established missense mutation models is the PKD1-R3277C mutation, which has been widely used as the slow progression model of ADPKD (Hopp et al., 2012). Here, we intended to construct a mini-pig model to study the consequence of another missense mutation PKD2-D511V.
Overexpression of hPKD2-D511V resulted in tail curvature of zebrafish
Since there is currently no published PKD2-D511V vertebrate model to our knowledge. In addition, in vivo study demonstrated that PKD2-D511V is a dominant-negative mutation related to ion influx. Therefore, generating a porcine model expressing PKD2-D511V is risky. The overexpression of the plasmids pCAG-WT-hPKD2 and pCAG-MU-hPKD2 resulted in the enrichment of ADPKD-related pathways in a pig kidney cell line (Zhang et al., 2020). To further prove that hPKD2-D511V would be functional in vivo and test the consequence of overexpressing the mutant form, the plasmids are injected into the one-cell stage of zebrafish. Unlike other reports, plasmids, not mRNA, were used in our experiment to extend the expression period of hPKD2. Similar to previous publications (Feng et al., 2008; Pavel et al., 2016), hPKD2-D511V resulted in a more severe tail curvature percentage than WT hPKD2 injection (Figure 1). Another feature observed is that the hPKD2-D511V injected zebrafish had a higher ratio of pericardial edema, implying that heart development was compromised (Figures 1B,C). Zebrafish with induction of acute kidney injury or disturbance of genes related to cilia displayed pericardial edema and ventrally curved tail (Rbaibi et al., 2012; Lee et al., 2015; Wen et al., 2018; Xie et al., 2019). Thus, our zebrafish experiments indicated that the constructed pCAG-MU-hPKD2 vector might be functional in vivo.
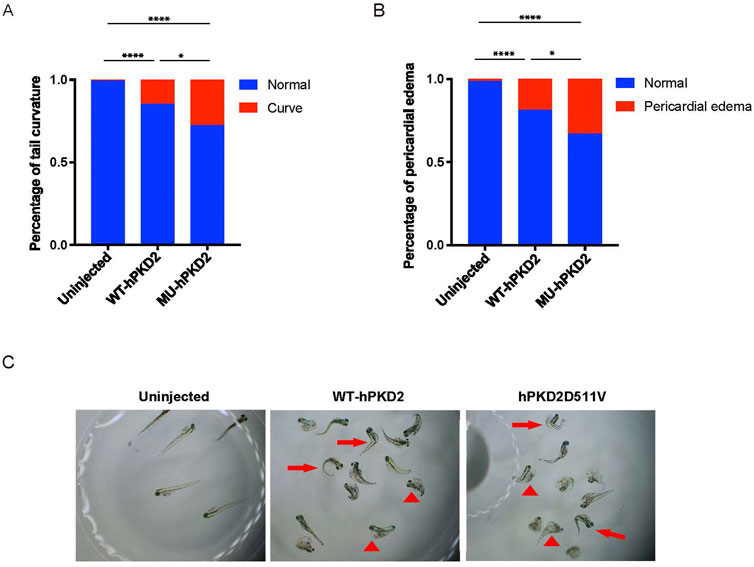
FIGURE 1. Overexpression of hPKD2-D511V resulted in tail curvature of zebrafish. (A) Statistical analysis of the percentage of tail curvature zebrafish shows a more severe phenotype in zebrafish injected with the pCAG-MU-hPKD2. (B) Statistical analysis of the percentage of pericardial edema zebrafish. (C) The morphological observation of zebrafish with pCAG-MU-hPKD2 and pCAG-WT-hPKD2 injection. The arrows indicate tail curvature, and the arrowheads indicate pericardial edema.
Efficient generation of transgenic pigs for hPKD2-D511V
To precisely analyze the in vivo function of PKD2-D511V, transgenic pigs were created with hPKD2-D511V. To avoid position-effect in transgenic animals, the non-homologous end joining mediated targeted insertion was adapted to insert the transgene vector into the pH11 safe harbor (Ruan et al., 2015; Zheng et al., 2017). Using this method, Cas9 protein could cut the targets both in the plasmid and genomic DNA. Theoretically, the linearized plasmid could integrate into the pH11 site of the pig genome. Nevertheless, the linearized plasmid could also integrate into other genomic locations akin to conventional transgenesis (Ju et al., 2015). To preclude the random insertion events, several pairs of primers were designed. Two pairs of upstream and downstream primers could amplify the intended targeted insertion sequence, as shown in Figure 2A. If the vector was inserted reversely, a different combination of these primers could also detect the event (Supplementary Table S1).
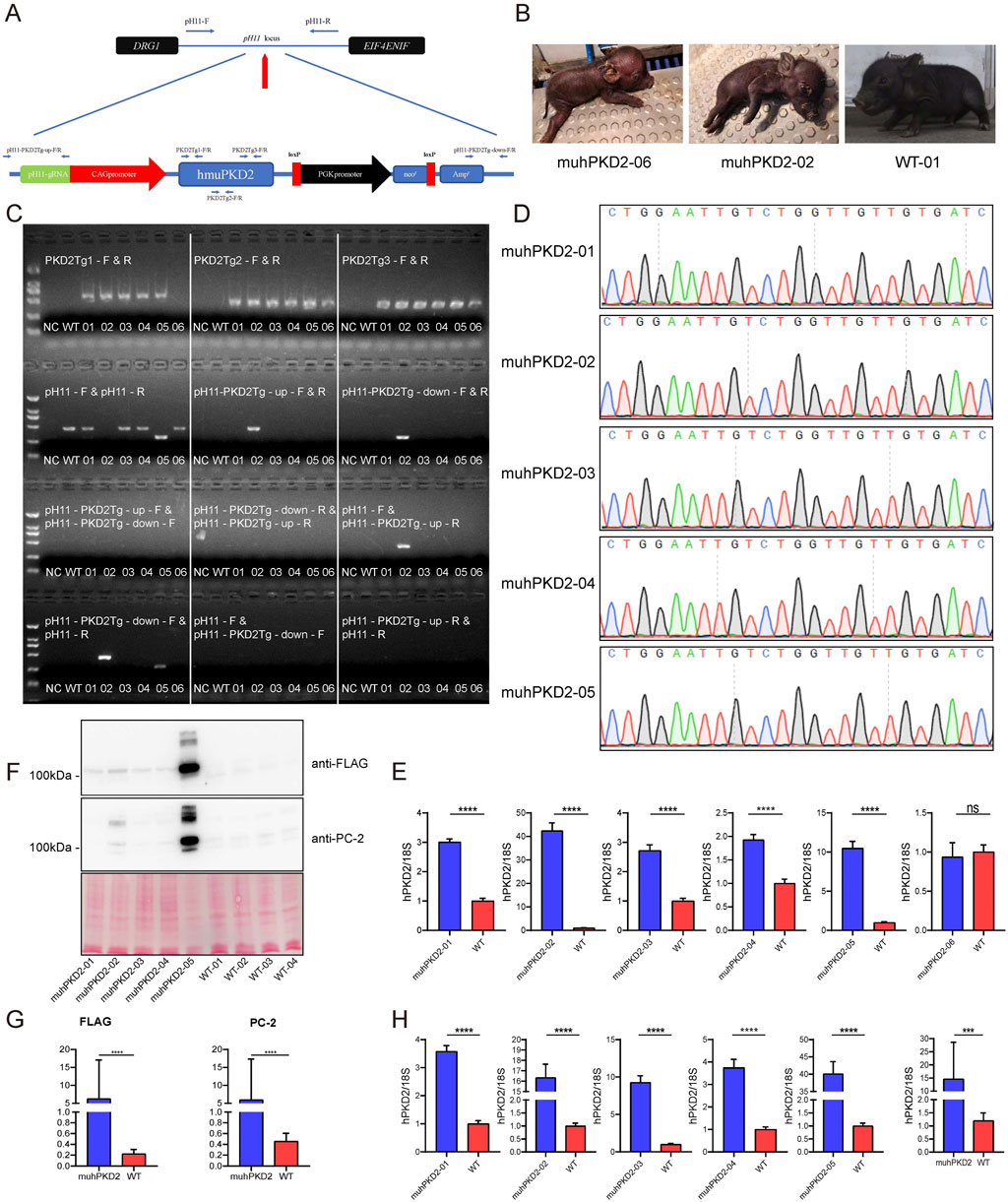
FIGURE 2. Overexpression of hPKD2-D511V resulted in postnatal deaths of the pigs. (A) The schematic representation of insertion of the transgene vector to pH11 safe harbor by the non-homologous end joining mediated targeted insertion. (B) Photos of newborn cloned pigs. (C) The results of PCR identification. (D) Sequencing analyses of cloned pigs using primers PKD2Tg1-F/R. (E) The mRNA levels of hPKD2 in the skin tissues of different cloned pigs. (F,G) Western blot analysis of FLAG and PC-2 in the cloned pig kidney tissues. Ponceau S staining was used as a loading control in the measurements. (H) The mRNA levels of hPKD2 in the kidney tissues of different cloned pigs. The relative mRNA expression and membrane protein expression levels from the WT group were used as the reference values, and * indicates that there were significant differences between the two groups. Results were expressed as mean ± SEM (n = 3).
The limited dilution method was first utilized for screening the transgenic cells. In this case, if the colonies were formed, all the cells should be derived from the same single cell with an identical genotype. However, due to the status of the available CEMP primary fibroblasts, no cell colonies were formed in our initial screen. Thus, 200 cells/well are seeded to circumvent the problems of obtaining transgenic cells, which would inevitably contain cells with different genotypes. After screening, 16 colonies were secured. PCR identification showed that they all contained transgenic vectors (Supplementary Figure S1). Colonies #3 and #5 were chosen for subsequent SCNT, because they had the intended targeted insertion event, which is presented in Figure 2A. Three batches of embryo transfer were conducted serially; however, only the first batch of embryo transfer resulted in the pregnancy of 2 surrogate sows, which finally delivered 4 and 2 piglets (labeled from MU-hPKD2-01 to -06), respectively. We found that MU-hPKD2-01, 03, 04, and 05 died immediately after birth, while MU-hPKD2-02 died at P1 (Figure 2B). Autopsies showed that the cloned piglets had hepatic congestion, splenomegaly, enlarged pancreas, and lung congestion (Supplementary Figure S2). Only MU-hPKD2-06 survived to date. PCR and sequencing analyses revealed that of the 6 cloned piglets, only MU-hPKD2-02 contained targeted insertion of the transgene in the pH11 locus, while the others were randomly transgenic events (Figures 2C,D). Interestingly, the survived MU-hPKD2-06 contained a 5′ truncated hPKD2, which was validated by qRT-PCR using skin tissues with comparable hPKD2 levels with the WT piglets (Figure 2E). Thus, the transgenic hPKD2 might be disrupted in the survived piglet.
Next, the kidneys of the demised pigs were harvested. qRT-PCR and western blotting were then performed to see whether hPKD2 was elevated in these transgenic pigs. As shown in Figures 2F–H, the transgenic vector could encode the hPKD2-D511V in the kidneys at both the mRNA and protein levels.
Contrary to the readily detectable hPKD2 expression, no malformation was noticed in the kidney sections, indicating at least that hPKD2-D511V did not affect normal kidney development (Figures 3A,B). Furthermore, immunohistochemical analyses showed increased PC2 in the epithelia of renal tubules when compared to age- and gender-matched WT pigs (Figures 3C,D). These results indicated that the hPKD2-D511V overexpression did not elicit the cystic phenotype in newborn piglets. Similar phenomena were also uncovered in our transgenic pigs overexpressing MYC and pig PKD2, in addition to some rodent models (He et al., 2013; Ye et al., 2013; Cai et al., 2014; Li et al., 2015), making it still divisive whether overexpression model is an explanation to ADPKD (Nagao et al., 2012).
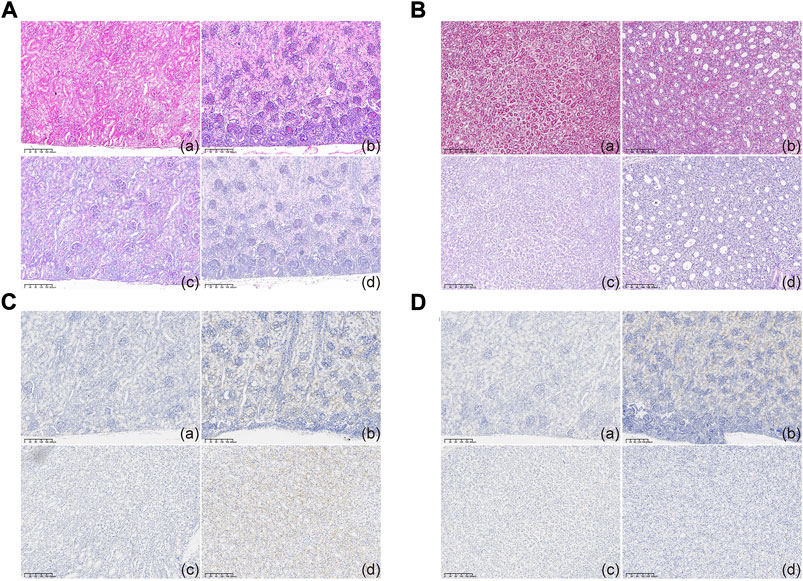
FIGURE 3. Histological examination of cloned piglets (A,B) Optical microscopy observation of the kidney tissues in the MU-hPKD2-02 and WT pigs. (C,D) Immunohistochemical characterization of the kidney tissues in the MU-hPKD2-02 and WT pigs. (a and c) MU-hPKD2-02. (b and d) WT pigs. (a,b) renal cortex. (c,d) medulla.
RNA sequencing reveals the enrichment of ADPKD-related pathways
The kidneys were subjected to RNA-seq due to the lack of renal cystic phenotype to see whether disease-related pathways were disturbed. Four age- and gender-matched WT CEMP piglets were sacrificed, and kidneys were harvested for RNA-seq. The principal component analysis (PCA) showed that overexpression of hPKD2-D511V significantly changed the transcriptome compared with the WT group (Figure 4A), which was further confirmed by the heatmap and clustering analysis (Figure 4B). Meanwhile, the volcano plot visualized many differentially expressed genes (DEGs), including 449 upregulated genes and 389 downregulated genes (Figure 4C). GO analysis shows several ADPKD-related biological processes were enriched in our transgenic pigs, e.g., Wnt, MAPK, EGF, JAK-STAT, cell proliferation, apoptosis, and migration (Figures 4D,E) (Bhunia et al., 2002; Wei and Liu, 2002; Ma et al., 2005; Lal et al., 2008). The EGF-mediated ion flux could be activated when overexpression of WT hPKD2 in a cell line (Ma et al., 2005). Moreover, this observation could not be replicated when overexpression hPKD2-D511V. Our analysis discovered that seven genes were significantly upregulated in hPKD2-D511V pig kidneys (Supplementary Table S3), validating the relationship between PKD2-D511V and the EGF pathway. One of the pathological hallmarks of ADPKD is massive transcriptomic dysregulation, including EGFR pathway. Some of the major signaling pathways initiated by EGFR activation, and then dependent on the recruitment and binding of specific signaling proteins to the phosphorylated tyrosine residues on the carboxyl termini of EGFR receptor molecules. Further regulate cell function including cell growth, proliferation, migration, differentiation, and apoptosis (He et al., 2013). Besides, the heart development term is also enriched, which might be used to explain the pericardial edema in the zebrafish injected with MU-hPKD2 (Figure 1B). In accordance with qRT-PCR results, the RNA-seq also validated elevated exogenous hPKD2 transcripts and unaltered endogenous pPKD2 levels in the transgenic pigs (Figure 4F).
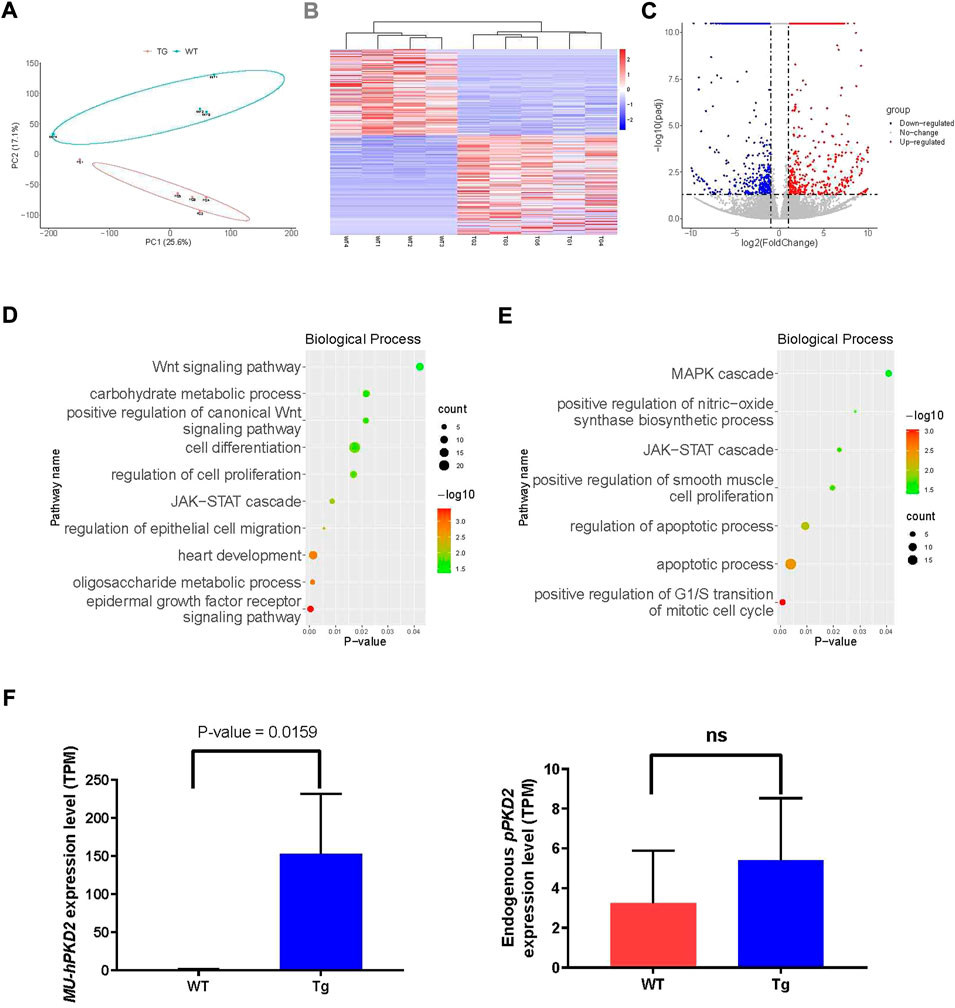
FIGURE 4. RNA-seq analysis of pig kidneys. (A) Principal component analysis (PCA) of merged RNA-seq data indicated the separation of the hPKD2-D511V group (MU-hPKD2-01-05) and the WT group (WT-hPKD2-01-04). (B) The Heatmap of RNA-seq data with the hPKD2-D511V group (MU-hPKD2-01-05) or the WT group based on the Log2 intensity. (C) Volcano plots showing the adjusted p-values and the log2 fold change (FC) values of genes in the hPKD2-D511V group (MU-hPKD2-01-05) versus the WT group (WT-hPKD2-01-04). DEGs are indicated by the red or blue dots. (D,E) Bubble plots of significant gene ontology (GO) (biological processes) terms enriched in genes from the DEG sets (upregulated DEG for D and downregulated DEG for E) of the two genotypes. (F) The TPM of hPKD2 and pPKD2 in hPKD2-D511V group (MU-hPKD2-01-05) and WT group.
Conclusion
The current study showed that hPKD2-D511V resulted in more tail curvature and pericardial edema in zebrafish experiment, and the hPKD2-D511V as a dominant-negative mutation might be lethal in a transgenic pig model. Out of the six cloned piglets, only one survived; moreover, the survived one contained a truncated form of the transgene that would not be expressed at the mRNA level (Figure 2E). Therefore, to study the molecular mechanism of PKD2-D511V driving cystogenesis in human patients, other approaches, such as transient expression in zebrafish embryos or the construction of non-invertebrate models, should be considered.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI SRA, PRJNA881011.
Ethics statement
The animal study was reviewed and approved by Zebrafish AB wildtype (WT) strain was used in this study. Fertilized eggs were placed in a petri-dish containing egg water and allowed to grow to 5 dpf at 28°C. The zebrafish procedures were adopted per the requirement of the “Regulation for the Use of Experimental Animals in Zhejiang Province”. This work is approved by the Animal Welfare Committee in the School of Medicine, Zhejiang University (No. ZJU 2011-1-11-009Y). The Chinese experimental mini-pigs (CEMP) and surrogate large white pigs were purchased from the China Agricultural University, which approved our use of the pigs (No. AW31059102-3). The subsequent procedures conducted on the pigs were also approved by the Animal Welfare Committee of Zhejiang University (No. 11834).
Author contributions
JH conceived the study and designed the experiment. YZ, SX, and HW performed the molecular biology and pig experiments. SX, JL, and CG conducted the experiments on zebrafish. QJ and SJ analyzed the data. JH wrote the manuscript, which was further edited by SJ and LZ. All authors have read the manuscript and confirmed its submission and publication.
Funding
This study was supported by the Natural Science Foundation of Zhejiang Province (No. LY20C170001), the National Natural Science Foundation of China (81941003 and 32270884), the Key Research and Development Program of Zhejiang Province (No. 2021C02060), and the open research fund of Wenzhou Engineering Research Center of Pet (WP03).
Acknowledgments
We are grateful to Jinrong Peng and Li Jan Lo of the Department of Animal Science and Technology for providing the zebrafish facilities. We also thank Fu-shan Shi of the Department of Veterinary Medicine for the pathological assistance.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.1059682/full#supplementary-material
References
Bhunia, A. K., Piontek, K., Boletta, A., Liu, L., Qian, F., Xu, P. N., et al. (2002). PKD1 induces p21waf1 and regulation of the cell cycle via direct activation of the JAK-STAT signaling pathway in a process requiring PKD2. Cell 109, 157–168. doi:10.1016/s0092-8674(02)00716-x
Cai, Y., Fedeles, S. v., Dong, K., Anyatonwu, G., Onoe, T., Mitobe, M., et al. (2014). Altered trafficking and stability of polycystins underlie polycystic kidney disease. J. Clin. Invest. 124, 5129–5144. doi:10.1172/JCI67273
Feng, S., Okenka, G. M., Bai, C. X., Streets, A. J., Newby, L. J., DeChant, B. T., et al. (2008). Identification and functional characterization of an N-terminal oligomerization domain for polycystin-2. J. Biol. Chem. 283, 28471–28479. doi:10.1074/jbc.M803834200
Gabow, P. A. (1990). Autosomal dominant polycystic kidney disease-more than a renal disease. Am. J. Kidney Dis. 16, 403–413. doi:10.1016/S0272-6386(12)80051-5
Gómez-Rubio, V. (2017). ggplot2 - elegant graphics for data analysis (2nd edition). J. Stat. Softw. 77, b02. doi:10.18637/jss.v077.b022nd Edition
Gonzalez-Paredes, F. J., Ramos-Trujillo, E., and Claverie-Martin, F. (2016). Three exonic mutations in polycystic kidney disease-2 gene (PKD2) alter splicing of its pre-mRNA in a minigene system. Gene 578, 117–123. doi:10.1016/j.gene.2015.12.019
Gout, A. M., Martin, N. C., Brown, A. F., and Ravine, D. (2007). Pkdb: Polycystic kidney disease mutation database - a gene variant database for autosomal dominant polycystic kidney disease. Hum. Mutat. 28, 654–659. doi:10.1002/humu.20474
Happé, H., and Peters, D. J. M. (2014). Translational research in ADPKD: Lessons from animal models. Nat. Rev. Nephrol. 10, 587–601. doi:10.1038/nrneph.2014.137
Harris, P. C., Ward, C. J., Peral, B., and Hughes, J. (1995). Polycystic kidney disease 1: Identification and analysis of the primary defect. J. Am. Soc. Nephrol. 6, 1125–1133. doi:10.1681/asn.v641125
He, J., Li, Q., Fang, S., Guo, Y., Liu, T., Ye, J., et al. (2015). Pkd1 mono-allelic knockout is sufficient to trigger renal cystogenesis in a mini-pig model. Int. J. Biol. Sci. 11, 361–369. doi:10.7150/ijbs.10858
He, J., Ye, J., Li, Q., Feng, Y., Bai, X., Chen, X., et al. (2013). Construction of a transgenic pig model overexpressing polycystic kidney disease 2 (PKD2) gene. Transgenic Res. 22, 861–867. doi:10.1007/s11248-012-9686-z
Hofherr, A., Wagner, C. J., Watnick, T., and Köttgen, M. (2016). Targeted rescue of a polycystic kidney disease mutation by lysosomal inhibition. Kidney Int. 89, 949–955. doi:10.1016/j.kint.2015.11.015
Hopp, K., Ward, C. J., Hommerding, C. J., Nasr, S. H., Tuan, H. F., Gainullin, V. G., et al. (2012). Functional polycystin-1 dosage governs autosomal dominant polycystic kidney disease severity. J. Clin. Invest. 122, 4257–4273. doi:10.1172/JCI64313
Jiao, X., Sherman, B. T., Huang, D. W., Stephens, R., Baseler, M. W., Lane, H. C., et al. (2012). DAVID-WS: A stateful web service to facilitate gene/protein list analysis. Bioinformatics 28, 1805–1806. doi:10.1093/bioinformatics/bts251
Ju, H., Zhang, J., Bai, L., Mu, Y., Du, Y., Yang, W., et al. (2015). The transgenic cloned pig population with integrated and controllable GH expression that has higher feed efficiency and meat production. Sci. Rep. 5, 10152. doi:10.1038/srep10152
Kimberling, W. J., Kumar, S., Gabow, P. A., Kenyon, J. B., Connolly, C. J., and Somlo, S. (1993). Autosomal dominant polycystic kidney disease: Localization of the second gene to chromosome 4q13–q23. Genomics 18, 467–472. doi:10.1016/S0888-7543(11)80001-7
Köttgen, M., Hofherr, A., Li, W., Chu, K., Cook, S., Montell, C., et al. (2011). Drosophila sperm swim backwards in the female reproductive tract and are activated via TRPP2 ion channels. PLoS One 6, e20031. doi:10.1371/journal.pone.0020031
Koulen, P., Cai, Y., Geng, L., Maeda, Y., Nishimura, S., Witzgall, R., et al. (2002). Polycystin-2 is an intracellular calcium release channel. Nat. Cell Biol. 4, 191–197. doi:10.1038/ncb754
Lal, M., Song, X., Pluznick, J. L., di Giovanni, V., Merrick, D. M., Rosenblum, N. D., et al. (2008). Polycystin-1 C-terminal tail associates with β-catenin and inhibits canonical Wnt signaling. Hum. Mol. Genet. 17, 3105–3117. doi:10.1093/hmg/ddn208
Lee, M. S., Hwang, K. S., Oh, H. W., Ji-Ae, K., Kim, H. T., Cho, H. S., et al. (2015). IFT46 plays an essential role in cilia development. Dev. Biol. 400, 248–257. doi:10.1016/j.ydbio.2015.02.009
Li, A., Tian, X., Zhang, X., Huang, S., Ma, Y., Wu, D., et al. (2015). Human polycystin-2 transgene dose-dependently rescues ADPKD phenotypes in Pkd2 mutant mice. Am. J. Pathol. 185, 2843–2860. doi:10.1016/j.ajpath.2015.06.014
Liu, X., Vien, T., Duan, J., Sheu, S. H., DeCaen, P. G., and Clapham, D. E. (2018). Polycystin-2 is an essential ion channel subunit in the primary cilium of the renal collecting duct epithelium. Elife 7, e33183. doi:10.7554/eLife.33183
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. doi:10.1186/s13059-014-0550-8
Ma, R., Li, W.-P., Rundle, D., Kong, J., Akbarali, H. I., and Tsiokas, L. (2005). PKD2 functions as an epidermal growth factor-activated plasma membrane channel. Mol. Cell. Biol. 25, 8285–8298. doi:10.1128/mcb.25.18.8285-8298.2005
Mochizuki, T., Wu, G., Hayashi, T., Xenophontos, S. L., Veldhuisen, B., Saris, J. J., et al. (1996). PKD2, a gene for polycystic kidney disease that encodes an integral membrane protein. Science 272, 1339–1342. doi:10.1126/science.272.5266.1339
Nagao, S., Kugita, M., Yoshihara, D., and Yamaguchi, T. (2012). Animal models for human polycystic kidney disease. Exp. Anim. 61, 477–488. doi:10.1538/expanim.61.477
Newby, L. J., Streets, A. J., Zhao, Y., Harris, P. C., Ward, C. J., and Ong, A. C. M. (2002). Identification, characterization, and localization of a novel kidney polycystin-1-polycystin-2 complex. J. Biol. Chem. 277, 20763–20773. doi:10.1074/jbc.M107788200
Padhy, B., Xie, J., Wang, R., Lin, F., and Huang, C.-L. (2022). Channel function of polycystin-2 in the endoplasmic reticulum protects against autosomal dominant polycystic kidney disease. J. Am. Soc. Nephrol. 33, 1501–1516. doi:10.1681/ASN.2022010053
Patro, R., Duggal, G., Love, M. I., Irizarry, R. A., and Kingsford, C. (2017). Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419. doi:10.1038/nmeth.4197
Pavel, M. A., Lv, C., Ng, C., Yang, L., Kashyap, P., Lam, C., et al. (2016). Function and regulation of TRPP2 ion channel revealed by a gain-of-function mutant. Proc. Natl. Acad. Sci. U. S. A. 113, E2363–E2372. doi:10.1073/pnas.1517066113
Pignatelli, P. M., Pound, S. E., Carothers, A. D., Macnicol, A. M., Allan, P. L., Watson, M. L., et al. (1992). Multipoint mapping of adult onset polycystic kidney disease (PKD1) on chromosome 16. J. Med. Genet. 29, 638–641. doi:10.1136/jmg.29.9.638
Rbaibi, Y., Cui, S., Mo, D., Carattino, M., Rohatgi, R., Satlin, L. M., et al. (2012). OCRL1 modulates cilia length in renal epithelial cells. Traffic 13, 1295–1305. doi:10.1111/j.1600-0854.2012.01387.x
Reynolds, D. M., Hayashi, T., Cai, Y., Veldhuisen, B., Watnick, T. J., Lens, X. M., et al. (1999). Aberrant splicing in the PKD2 gene as a cause of polycystic kidney disease. J. Am. Soc. Nephrol. 10, 2342–2351. doi:10.1681/asn.v10112342
Rossetti, S., Strmecki, L., Gamble, V., Burton, S., Sneddon, V., Peral, B., et al. (2001). Mutation analysis of the entire PKD1 gene: Genetic and diagnostic implications. Am. J. Hum. Genet. 68, 46–63. doi:10.1086/316939
Ruan, J., Li, H., Xu, K., Wu, T., Wei, J., Zhou, R., et al. (2015). Highly efficient CRISPR/Cas9-mediated transgene knockin at the H11 locus in pigs. Sci. Rep. 5, 14253. doi:10.1038/srep14253
Torres, V. E., Chapman, A. B., Devuyst, O., Gansevoort, R. T., Grantham, J. J., Higashihara, E., et al. (2012). Tolvaptan in patients with autosomal dominant polycystic kidney disease. N. Engl. J. Med. 367, 2407–2418. doi:10.1056/nejmoa1205511
Torres, V. E., Chapman, A. B., Devuyst, O., Gansevoort, R. T., Perrone, R. D., Koch, G., et al. (2017). Tolvaptan in later-stage autosomal dominant polycystic kidney disease. N. Engl. J. Med. 377, 1930–1942. doi:10.1056/nejmoa1710030
Torres, V. E., and Harris, P. C. (2009). Autosomal dominant polycystic kidney disease: The last 3 years. Kidney Int. 76, 149–168. doi:10.1038/ki.2009.128
Tsukiyama, T., Kobayashi, K., Nakaya, M., Iwatani, C., Seita, Y., Tsuchiya, H., et al. (2019). Monkeys mutant for PKD1 recapitulate human autosomal dominant polycystic kidney disease. Nat. Commun. 10, 5517. doi:10.1038/s41467-019-13398-6
Watanabe, M., Umeyama, K., Nakano, K., Matsunari, H., Fukuda, T., Matsumoto, K., et al. (2022). Generation of heterozygous PKD1 mutant pigs exhibiting early-onset renal cyst formation. Lab. Invest. 102, 560–569. doi:10.1038/s41374-021-00717-z
Wei, H., Li, Q., Li, J., Li, Y., Dai, Y., Ma, Y., et al. (2008). Effect of leptin on oocyte maturation and subsequent pregnancy rate of cloned embryos reconstructed by somatic cell nuclear transfer in pigs. Prog. Nat. Sci. 18, 1583–1587. doi:10.1016/j.pnsc.2008.05.018
Wei, Z., and Liu, H. T. (2002). MAPK signal pathways in the regulation of cell proliferation in mammalian cells. Cell Res. 12, 9–18. doi:10.1038/sj.cr.7290105
Wen, X., Cui, L., Morrisroe, S., Maberry, D., Emlet, D., Watkins, S., et al. (2018). A zebrafish model of infection-associated acute kidney injury. Am. J. Physiol. Ren. Physiol. 315, F291–F299. doi:10.1152/ajprenal.00328.2017
Xie, S., Jin, J., Xu, Z., Huang, Y., Zhang, W., Zhao, L., et al. (2019). Centrosomal protein FOR20 is essential for cilia-dependent development in zebrafish embryos. FASEB J. 33, 3613–3622. doi:10.1096/fj.201801235RR
Ye, J., He, J., Li, Q., Feng, Y., Bai, X., Chen, X., et al. (2013). Generation of c-Myc transgenic pigs for autosomal dominant polycystic kidney disease. Transgenic Res. 22, 1231–1239. doi:10.1007/s11248-013-9707-6
Zhang, Z., Dang, Y., Wang, Z., Wang, H., Pan, Y., and He, J. (2020). Identification of ADPKD-related genes and pathways in cells overexpressing PKD2. Genes (Basel) 11, E122. doi:10.3390/genes11020122
Keywords: ADPKD, disease model, PKD2, transgenic pig, kidney
Citation: Zhang Y, Xu S, Jin Q, Luo J, Gao C, Jayaprakash S, Wang H, Zhuang L and He J (2022) Establishment of transgenic pigs overexpressing human PKD2-D511V mutant. Front. Genet. 13:1059682. doi: 10.3389/fgene.2022.1059682
Received: 01 October 2022; Accepted: 01 November 2022;
Published: 14 November 2022.
Edited by:
Stephen J. Bush, University of Oxford, United KingdomReviewed by:
Chungang Feng, Nanjing Agricultural University, ChinaCheng Xue, Shanghai Changzheng Hospital, China
Xuguang Du, China Agricultural University, China
Yu Dawei, Institute of Animal Sciences (CAAS), China
Copyright © 2022 Zhang, Xu, Jin, Luo, Gao, Jayaprakash, Wang, Zhuang and He. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jin He, aGVqaW5Aemp1LmVkdS5jbg==
 Yuan Zhang
Yuan Zhang Saifei Xu1
Saifei Xu1 Ce Gao
Ce Gao Huanan Wang
Huanan Wang Lenan Zhuang
Lenan Zhuang Jin He
Jin He