
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
CORRECTION article
Front. Genet., 29 August 2022
Sec. Evolutionary and Genomic Microbiology
Volume 13 - 2022 | https://doi.org/10.3389/fgene.2022.1009773
This article is part of the Research TopicLinkage Between Membrane Composition, Cellular Functions and Membrane- Mediated StressView all 5 articles
This article is a correction to:
Re-Expression of Tafazzin Isoforms in TAZ-Deficient C6 Glioma Cells Restores Cardiolipin Composition but Not Proliferation Rate and Alterations in Gene Expression
 Gayatri Jagirdar1
Gayatri Jagirdar1 Matthias Elsner2
Matthias Elsner2 Christian Scharf3
Christian Scharf3 Stefan Simm4
Stefan Simm4 Katrin Borucki5
Katrin Borucki5 Daniela Peter5
Daniela Peter5 Michael Lalk6
Michael Lalk6 Karen Methling6
Karen Methling6 Michael Linnebacher7
Michael Linnebacher7 Mathias Krohn7
Mathias Krohn7 Carmen Wolke1
Carmen Wolke1 Uwe Lendeckel1*
Uwe Lendeckel1*by Jagirdar G, Elsner M, Scharf C, Simm S, Borucki K, Peter D, Lalk M, Methling K, Linnebacher M, Krohn M, Wolke C and Lendeckel U (2022). Front. Genet. 13:931017. doi: 10.3389/fgene.2022.931017
In the published article, there was an error in “Figures 1–5 and Table 1” as published. The figures were not in correct order and some human primers shown in Table 1 were not used in the study. The corrected figures and tables appear below.
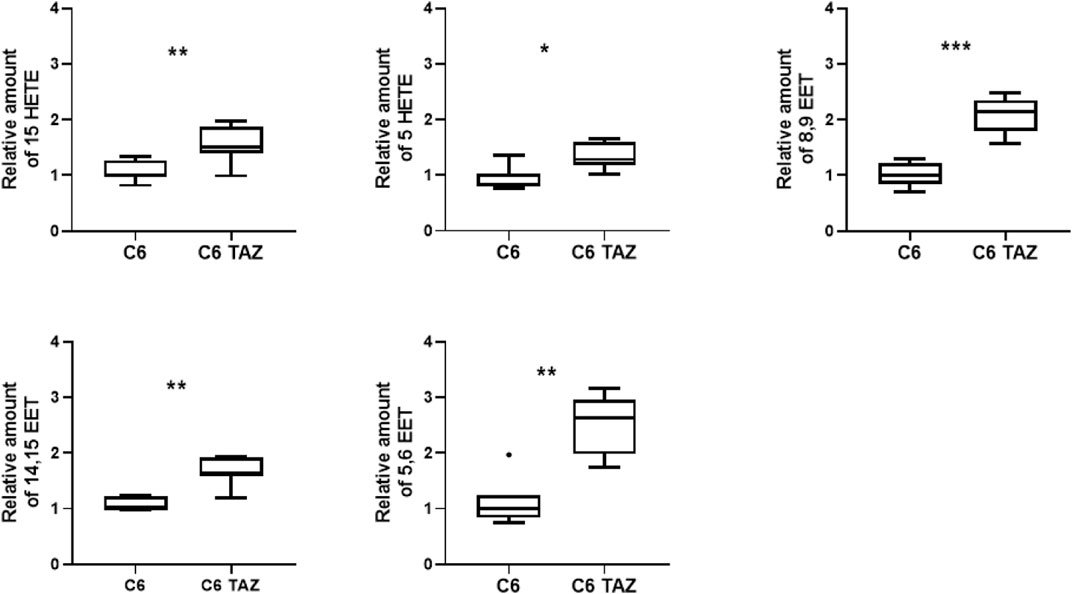
FIGURE 1. Tafazzin deficiency alters oxylipin levels in C6 cells: Effect of tafazzin knockout on cellular lipids, oxylipins was determined by HPLC-MS/MS. The Data analysis was performed with Agilent Mass Hunter Qualitative and Quantitative Analysis software (version B.08.00 for both). For all detected oxylipins, a relative quantification was done by normalizing the area of the metabolite signal to the area of the signal of the internal standard compound (relative amount). Data is represented as median,quartile and interquartile range for n = 4.

FIGURE 2. Tafazzin deficiency alters gene expression in C6 cells: Quantification levels of indicated genes in C6 and C6 TAZ cells was preformed and the amount of mRNA was determined by using RT-qPCR and normalized to RPL13a. Data is given as boxplot with median, quartile, and interquartile range (*p < 0.05; **p < 0.01; ***p < 0.001). Outliers are indicated using Tukey method. n = 8.
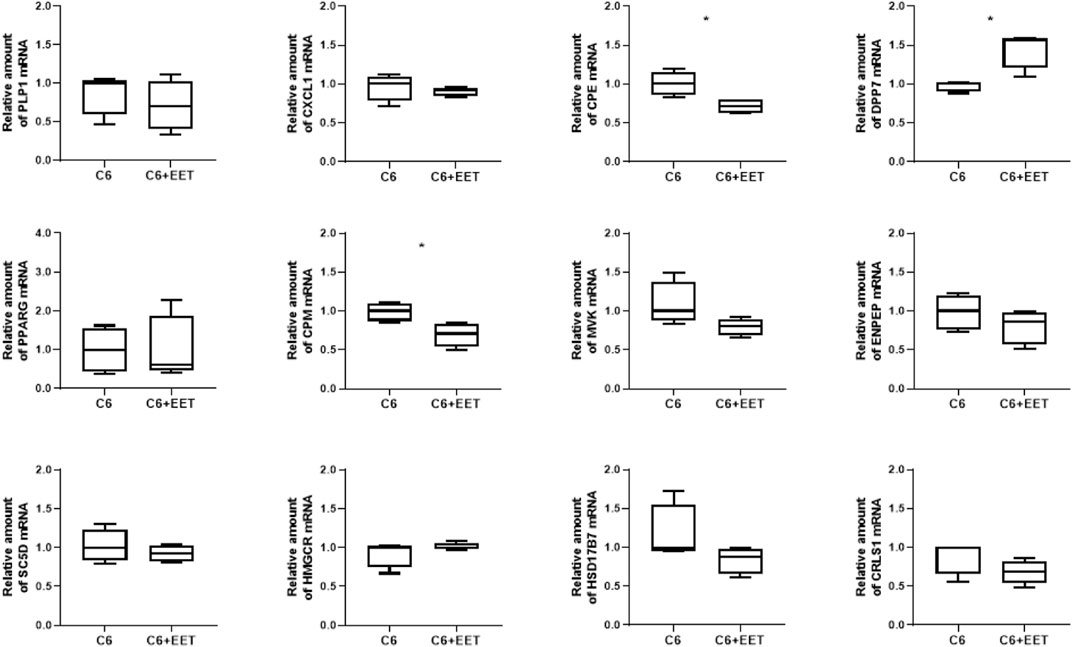
FIGURE 3. Oxylipins regulates TAZ dependent gene expression in C6 cells: Relative gene expression of C6 cells treated with ethanol (control) and C6 cells with 3 μM EET incubated at 37°C for 24 h. The amount of Mrna is determined by qpcr that is normalized to RPL13a. Data is represented as boxplot with median, quartile and interquartile range. Asterisks represent significant change.
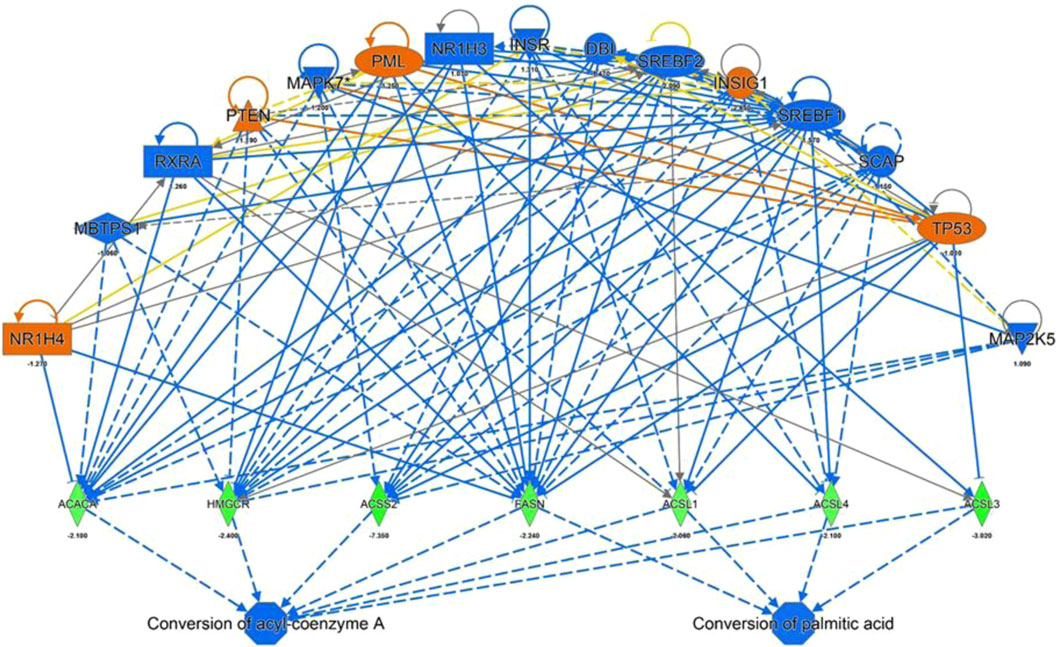
FIGURE 4. IPA Upstream Regulators Analysis for identification of upstream regulators that may be responsible for changes in gene expression observed for ACACA, HMGCR, ACSS2, FASN, ACSL 1,3 and 4 (predicted activation (orange) predicted inhibition (blue) of the regulators leads to a decreased expression (green)).
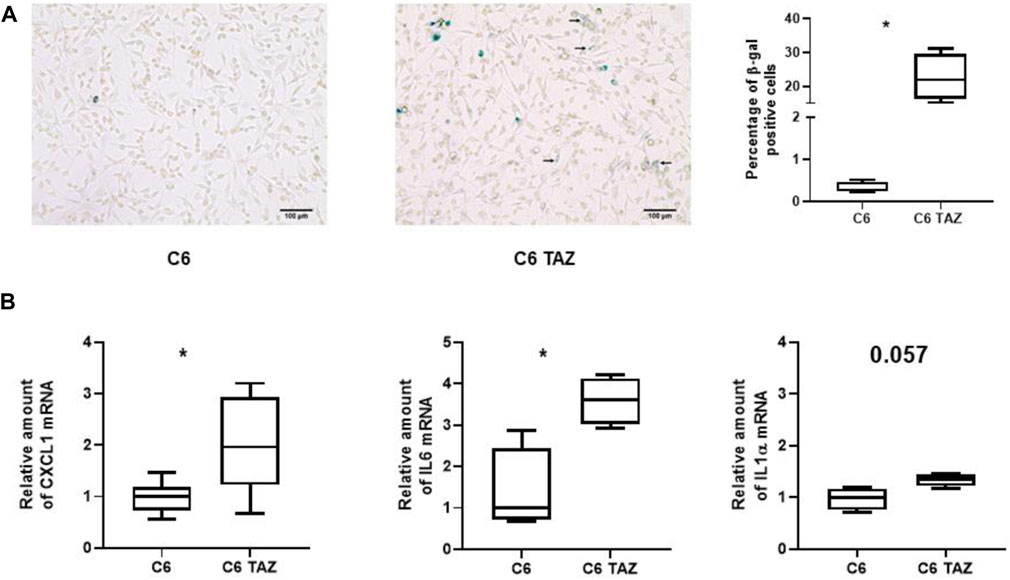
FIGURE 5. (A) Left: Microscopic images of β-gal positive cells (blue color) in C6 and C6 TAZ cells. Right: Percentage of β-gal positive cells is given as boxplot (* indicates p < 0.05). (B) Upregulation of SASP genes: Amounts of mrnas of CXCL1, IL6, and IL1α are elevated in C6 TAZ cells (mrna expression normalized to RPL13a; n = 4, p = 0.029).
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Keywords: Barth syndrome, cardiolipin, cellular proliferation, gene expression, tafazzin, Barth syndrome (BTHS)
Citation: Jagirdar G, Elsner M, Scharf C, Simm S, Borucki K, Peter D, Lalk M, Methling K, Linnebacher M, Krohn M, Wolke C and Lendeckel U (2022) Corrigendum: Re-expression of tafazzin isoforms in TAZ-deficient C6 glioma cells restores cardiolipin composition but not proliferation rate and alterations in gene expression. Front. Genet. 13:1009773. doi: 10.3389/fgene.2022.1009773
Received: 02 August 2022; Accepted: 03 August 2022;
Published: 29 August 2022.
Approved by:
Frontiers Editorial Office, Frontiers Media SA, SwitzerlandCopyright © 2022 Jagirdar, Elsner, Scharf, Simm, Borucki, Peter, Lalk, Methling, Linnebacher, Krohn, Wolke and Lendeckel. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Uwe Lendeckel, dXdlLmxlbmRlY2tlbEBtZWQudW5pLWdyZWlmc3dhbGQuZGU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.