
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
METHODS article
Front. Genet. , 10 January 2023
Sec. Statistical Genetics and Methodology
Volume 13 - 2022 | https://doi.org/10.3389/fgene.2022.1000290
This article is part of the Research Topic Insights in Statistical Genetics and Methodology: 2022 View all 14 articles
 Huagui Wei1†
Huagui Wei1† Chunfang Wang1†
Chunfang Wang1† Weiyi Huang1
Weiyi Huang1 Liqiao He1
Liqiao He1 Yaqun Liu2
Yaqun Liu2 Huiying Huang2
Huiying Huang2 Wencheng Chen1
Wencheng Chen1 Yuzhong Zheng2
Yuzhong Zheng2 Guidan Xu1
Guidan Xu1 Liyun Lin2
Liyun Lin2 Wujun Wei1
Wujun Wei1 Weizhong Chen3
Weizhong Chen3 Liying Chen1
Liying Chen1 Junli Wang4*
Junli Wang4* Min Lin1,2,4*
Min Lin1,2,4*Objectives: Baise, a multiethnic inhabited area of southwestern China, is a historical malaria-endemic area with a high prevalence of G6PD deficiency. However, few studies of G6PD deficiency have been conducted in this region. Therefore, we performed a genetic analysis of G6PD deficiency in the Baise population from January 2020 to June 2021.
Methods: A SNPscan assay was developed to simultaneously detect 33 common Chinese G6PD mutations. 30 G6PD-deficient samples were used for the method’s validation. Then, a total of 709 suspected G6PD-deficient samples collated from the Baise population were evaluated for G6PD status, type of mutation and effect of mutations.
Results: The SNPscan test had a sensitivity of 100% [95% confidence interval (CI): 94.87%–100%] and a specificity of 100% (95% CI: 87.66%–100%) for identifying G6PD mutations. A total of fifteen mutations were identified from 76.72% (544/709) of the samples. The most common mutation was discovered to be G6PD Kaiping (24.12%), followed by G6PD Canton (17.91%), and G6PD Gaohe (11.28%). We compared the G6PD mutation spectrum among Zhuang, Han and other Southeast Asian populations, and the Zhuang population’s mutation distribution was quite similar to that in the Han population.
Conclusion: This study provided a detailed G6PD mutation spectrum in Baise of southwestern China and will be valuable for the diagnosis and research of G6PD deficiency in this area. Furthermore, the SNPscan assay could be used to quickly diagnose these G6PD mutations accurately.
Glucose-6-phosphate dehydrogenase (G6PD) deficiency is one of the most common enzymatic disorders of red blood cells, with a particularly high prevalence in tropical and subtropical regions, including southern China (Howell, 2006). According to the degree and extent of the enzyme deficiency, the World Health Organization (WHO) divided G6PD variants into four classifications in homozygous and hemizygous individuals (WHO, 2022). G6PD insufficiency manifests clinically as a range of conditions, ranging from severe enzyme deficiency to enhanced enzyme activity (Filosa et al., 1996). The most frequent clinical symptoms in patients are acute hemolysis, newborn hyperbilirubinemia, and chronic hemolysis, which are brought on by external factors including eating fava beans, taking specific medications, contracting an infection, or having a metabolic disorder (Jiang et al., 2006).
The G6PD gene (OMIM ID: 305900) spans 18 kb on the X chromosome (Xq28), contains an open reading frame of 1,545 bp, and encodes 515 amino acids (Tian et al., 2013; Wisnumurti et al., 2019). To date, approximately 217 mutations have been described worldwide (Gómez-Manzo et al., 2016). The G6PD mutation spectrum varies between different regions and ethnicities. The frequency distribution of these mutations closely correlates with populations that were exposed historically to endemic malaria (Dombrowski et al., 2017). Baise is a multiethnic inhabited area of southwestern China. The minority population accounts for 85% of the total population. It has a monsoon-influenced, humid subtropical climate and is a historical malaria-endemic area (Ji-Guang et al., 2017; Liang et al., 2020; Zheng et al., 2020). The spectrum of G6PD mutations, however, is poorly understood.
Currently, several analytical methods have been validated and developed to detect G6PD mutations, such as direct sequencing (Maloukh et al., 2021), reverse dot blot (RDB) assays (Chen et al., 2012; Duan et al., 2017; Zhang et al., 2016), high-resolution melting analysis (HRMA) (Boonyuen et al., 2021; Yang et al., 2015) and PCR-restriction fragment length polymorphism (PCR-RFLP) (Kumar et al., 2020). Although the aforementioned methods are powerful and exact, they are expensive, time-consuming and have low throughput (Zhang et al., 2016). The accuracy, sensitivity, and specificity of the SNPscan technology have been shown in numerous investigations. It is also high-throughput and cost-effective (Duan et al., 2017). Because of this, SNPscan is regarded as an acceptable method for the genetic diagnosis of G6PD deficiency.
In the present study, we established a SNPscan assay to identify 33 G6PD mutations. Combining the SNPscan assay with DNA sequence analysis for genotype detection and phenotypic screening, we studied the spectrum of G6PD mutations in Baise. Our research is essential for creating a community-based carrier screening and prevention program in the area.
A total of 709 suspected G6PD-deficient samples were enrolled from the Baise region of Guangxi Zhuang Autonomous Region between January 2020 and June 2021. These subjects included 346 males and 363 females, between the ages of 1 day old and ninety. Information on ethnic groups was collected. The Affiliated Hospital of Youjiang Medical University for Nationalities’ Ethics Committee accepted the study. Informed written consent was obtained from all adult participants or the guardians of pediatric participants. Ethylenediaminetetraacetic acid (EDTA) tubes were used to collect blood samples, which were then brought to the lab and kept in storage at 4°C.
The G6PD enzyme activity was measured by a commercial G6PD Detection kit (Korfang Biotechnology Co., Guangzhou, Guangdong, China) according to the rate method (Zhong et al., 2018), which was approved by the China Food and Drug Administration (CFDA) (reg. no. CFDA (P) 20193400771). According to the National Inspection Operational Regulations, 1 mL solution (Korfang Biotechnology Co., Guangzhou, Guangdong, China) was added to a small cup, and then 20 μL of erythrocyte was accurately absorbed into the solution without the plasma layer. The activity of G6PD was detected by the rate method on Hitachi 7170A automatic biochemical analyzer (HITACHI, Japan), and the concentration of hemoglobin in hemolysis was detected by the HiCN method. This method can detect NADPH production in fixed time, which reflect G6PD activity in red blood cells. In each test run, the accuracy of the test findings was checked by calibration and the use of controls offered by KOFA Medical. The reference range of adults with values below 1.30 KU/L (1.30–3.60) and infants with values below 1.70 KU/L (1.70–4.00).
According to the manufacturer’s recommendations, genomic DNA was extracted from all samples using a QIAamp DNA Blood Mini kit (Qiagen, Hilden, Germany). The DNA concentration was measured using a Thermo Scientific Nanodrop™ 2000 spectrophotometer and subsequently adjusted to 50 ng/L.
A multiplex SNPscan assays were designed to detect 33 G6PD mutations reported in Chinese population (Wang et al., 2021) as follow: G6PD Gaohe (c.95A>G), G6PD Songklanagarind (c.196T>A), G6PD Asahi (c.202G>A), G6PD Chinese-4 (c.392G>T), G6PD Valladolid (c.406C>T), G6PD Liuzhou (c.442G>A), G6PD Shenzhe (c.473G>A), G6PD Mahidol (c.487G>A), G6PD Taipei (c.493A>G), G6PD Nankang (c.517T>C), G6PD Miaoli (c.519C>T/G), G6PD Mediterranean (c.563C>T), G6PD Shunde (c.592C>T), G6PD Nanning (c.703C>T), G6PD Haikou (c.835A>G/T), G6PD Viangchan (c.871G>A), G6PD Fushan (c.1004C>A/T), G6PD Chinese-5 (c.1024C>T), G6PD Beverly Hills (c.1160G>A), G6PD Santiago de Cuba (c.1339G>A), G6PD Jiangxi (c.1340G>T), G6PD Union (c.1360C>T), G6PD Canton (c.1376G>T), G6PD Yannan (c.1381G>A), G6PD Kamiube (c.1387C>T), G6PD Kaiping (c.1388G>A), G6PD Laibin (c.1414A>C), and four unnamed mutations (c.274C>T, c.371A>G, c.691G>C and c.1225C>T) and two Silent mutation (c.1311C>T, c.1365-13T>C). The 33 G6PD mutation sites in the G6PD gene are shown in Figure 1A.
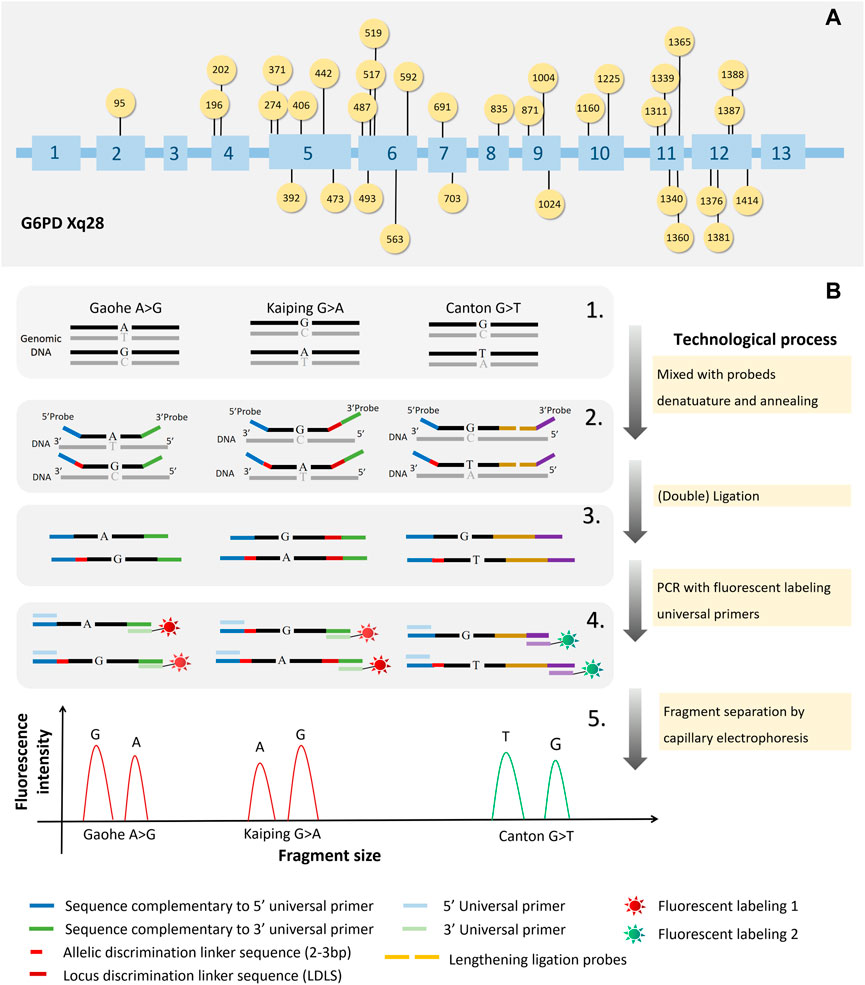
FIGURE 1. The workings of SNPscan technology and the locations of 33 G6PD gene mutations. The locations of the 33 mutations in the G6PD gene are shown in (A). The principles of SNPscan technology are shown in (B).
As shown in Figure 1B, previously mentioned, the double ligation and multiplex fluorescence PCR serves as the foundation for the SNPscan test. (Wei et al., 2013). The primers and probes are listed in Supplementary Table S1. For each SNPscan assay, 12 µL of ligation mixture was first prepared to contain 2 μL of 10 × ligase buffer, 1 μL of 1 × probe mix, .5 μL of ligase, 7 μL of ddH2O and 1 μL of 30–250 ng of DNA sample. The ligation reaction was performed on an ABI 2720 thermal cycler with the following cycling program: 98°C for 2 min; 5 cycles of 95°C for 1 min, 58°C for 3 h; 94°C for 2 min, hold at 72°C. Fluorescence in multiplex After that, PCRs were run on each ligation product. Every PCR mixture was made in 20 μL containing 2× PCR Buffer, 1 μL of primer mix, 8 μL of ddH2O, and 1 μL of ligation product. The PCR program was as follows: 95°C for 2 min; 9 cycles of 94°C for 20 s, 62°C–.5°C/cycle for 40 s, and 72°C for 1.5 min; 26 cycles of 94°C for 20 s, 58°C for 40 s, and 72°C for 1.5 min; 60°C for 1 h; and hold at 4°C. Using a capillary electrophoresis system and an ABI 3730XL sequencer, PCR products were separated and identified. Raw data were analysed with GeneMapper 4.1 software (Applied Biosystems, United States), and the genotypes of each locus were determined.
In order to confirm the SNPscan assay results, PCR amplification and DNA sequencing of the entire G6PD coding region was performed as described in our earlier research (Pan et al., 2013; Zheng et al., 2020). Purification and sequencing of PCR products were done by Shanghai Vebery Biotechnology (Shanghai, China). All primers are in Supplementary Table S4 (Pan et al., 2013).
The bioinformatics software used in this work was used to analyze each G6PD mutation identified. Moreover, the pI of G6PD variants (i.e., monomers) was determined using Kozlowski’s protein isoelectric point (IP) calculator (http://isoelectric.org/). Utilizing ConSurf (http://bental.tau.ac.il/new_ConSurfDB/), we looked at the evolutionary conservation of mutant amino acid residues. The pathogenicity of these potential variants was assessed by PolyPhen-2 (Polymorphism Phenotyping v2) (http://genetics.bwh.harvard.edu/pph2/) and Sorting Intolerant from Tolerant (SIFT) web server (http://sift.jcvi.org) prediction models.
The data are collated in Excel. All data were statistical using SPSS 22.0. Descriptive statistics were used to estimate the accuracy.
A SNPscan assay was developed to detect 33 G6PD mutations reported in Chinese individuals. As shown in Figure 1, it could precisely distinguish heterozygous mutations and homozygous/hemizygous mutations by capillary electrophoresis (Figure 2). To confirm the accuracy of the SNPscan assay, 30 samples were blindly analysed using PCR amplification and DNA sequencing (Supplementary Table S2). Comparatively speaking to direct DNA sequencing, the SNPscan assay was 100% sensitive [95% confidence interval (CI): 94.87–100%] and 100% specific (95% CI: 87.66–100%), without any cross-reactivity for the identification of G6PD mutations. Additionally, the SNPscan assay could precisely distinguish double mutations, such as Canton/Viangchan, Gaohe/Kaiping and Canton/Kaiping. The created approach is dependable for identifying G6PD mutations, according to all of the evidence, detailed data are shown in Supplementary Table S3.
Fifteen G6PD mutations were identified by the SNPcan assay in the Baise population (Table 1). Among the 709 G6PD-deficient people, 544 (277 females and 267 males) had at least one mutation in the G6PD gene. Among the 277 females with mutated G6PD deficiency, we identified 22 (3.10%) homozygotes and 255 heterozygotes, including 73 compound heterozygotes. The mutations of G6PD Kaiping, G6PD Canton and G6PD Gaohe were the three dominant mutations with an overall frequency of higher than 79.06%, followed by G6PD Chinese-5, G6PD Viangchan and G6PD Valladolid, with a frequency of 2.11% as a minimum, respectively. The number and frequency of various mutations are presented in Table 1.
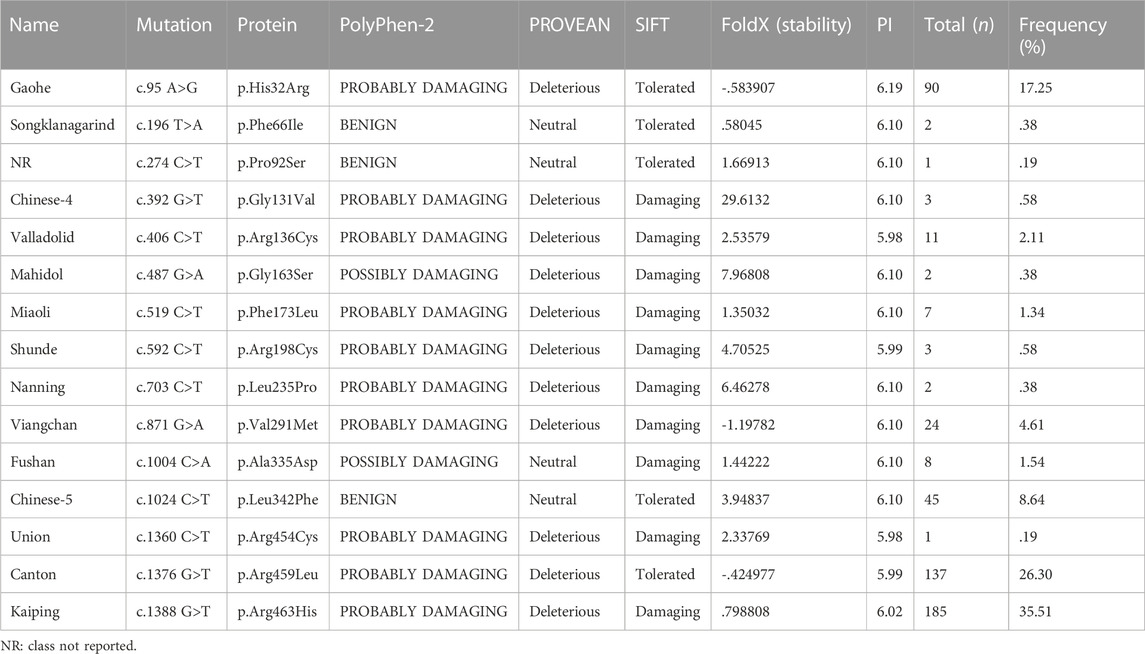
TABLE 1. Frequency of all G6PD-positive mutations and predicted consequences before and after amino acid changes.
These 709 samples were classified by ethnicity. There were 412 allele mutations and 15 variants in the Zhuang nationality, most of which were G6PD Kaiping, G6PD Canton and G6PD Gaohe, accounting for 79.08% (Table 2). However, there were only 87 (16.70%) allele mutations in the Han nationality. In addition, in order to examine the association of the major G6PD-deficient alleles in Chinese people and Southern Asian populations, data from our research or other studies were further analysed (Liu et al., 2020; Zheng et al., 2020). As shown in Figure 3, the frequencies of different G6PD-deficient alleles in different regions were plotted on a heatmap. The color of each block varies with the corresponding frequency. Purple represented the lowest allele frequency on the color scale, which went up to red for the greatest allele frequency. Obviously, Four G6PD-deficient alleles (Canton, Kaiping, Gaohe and Chinese-5) were present in relatively high frequencies in Chinese people, whereas G6PD Viangchan and G6PD Kaiping were prevalent in Southern Asian populations (Vietnam populations).
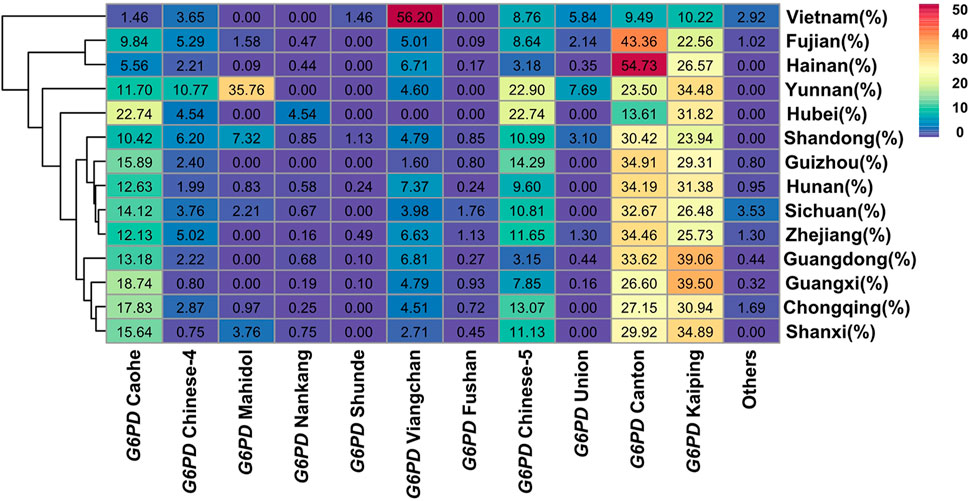
FIGURE 3. Heatmap of G6PD-deficient allele frequency distributions for Baise populations and others cities populations. Red represents the highest G6PD-deficient allele frequency, while purple represents the lowest.
Tools from bioinformatics were used to forecast how changing an amino acid might affect how a protein function (Table 1). According to PolyPhen2.0, all variants were identified as potentially damaging (prediction score close to 1) except for G6PD Songklanagarind, G6PD c.274C>T and G6PD Chinese-5), similar to the results predicted by PROVEAN (except G6PD Fushan). However, SIFT predicted that five missense mutations (G6PD Gaohe, G6PD Songklanagarind, G6PD c.274C>T, G6PD Chinese-5, and G6PD Canton) could be tolerated, and the rest were damaging. FoldX was used to predict changes in the protein stability of G6PD (Table 1), and three variants (G6PD Gaohe, G6PD Viangchan and G6PD Canton) were found to increase the stability of the G6PD protein, while other missense variants were predicted to destabilize the G6PD protein. Additionally, Table 1 provides an overview of the expected pI values for each of the 15 G6PD variations. The changes in protein structure and polar bonds before and after G6PD mutation are shown in Figure 4.
In this study, we looked studied the distribution of different G6PD gene variants, the prevalence of G6PD deficiency, and the relationship between genotypes and phenotypes related to enzyme function in Baise, Guangxi Zhuang Autonomous Region. The results showed that six of the most prevalent mutations were G6PD Kaiping, G6PD Canton, G6PD Gaohe, G6PD Chinese-4, G6PD Viangchan and G6PD Chinese-5, accounting for more than 60% of G6PD-deficient alleles. This result is consistent with LinZou’s research (Liu et al., 2020). The sexes and different sorts of mutation patterns affected how G6PD activities were distributed (Driscoll and Migeon, 1990). These findings present a more precise and thorough characterization of G6PD deficiency in Baise, Guangxi.
The prevalence of G6PD deficiency varies widely by region in China, with northern China having a relatively lower prevalence than southern China. G6PD deficiency was present in 2.1% of China’s population overall (He et al., 2020), and over 35 different G6PD gene mutations were known, with G6PD Kaiping and G6PD Canton predominating in earlier investigations (Liu et al., 2020; Jiang et al., 2006). Africa, Asia, southern Europe, the Middle East, Southeast Asia, and Mediterranean nations have the highest prevalence rates, according to reports (He et al., 2020; Liu et al., 2020). In India, in various population groups, it was discovered that 8.5% of people have G6PD deficiencies (Saravu et al., 2016). The prevalence rate varies between the tribal groupings, ranging from 2.3% to 27.0%, with an overall incidence of 7.7% (Mukherjee et al., 2015). In contrast to southern India, where it is continuously low except in the states of Andhra Pradesh and Tamil Nadu, the frequency of the G6PD-deficient allele is higher in northern and western India (Devendra et al., 2020). In Indian caste groupings, G6PD Mediterranean was discovered to be the most prevalent variation (Devendra et al., 2020; Sukumar et al., 2004). However, G6PD Kaiping was found to be the most common variant in China (Lin et al., 2018; Liu et al., 2020; Yan et al., 2010). In Southeast Asia, G6PD deficiency is diverse, as previously demonstrated by epidemiological and molecular research (Louicharoen and Nuchprayoon, 2005). In Thais, Laotians, Cambodians, and Malaysian Malays, G6PD Viangchan appears to be the most prevalent form (Ainoon et al., 2003; Iwai et al., 2001; Louicharoen and Nuchprayoon, 2005; Nuchprayoon et al., 2002), while the most prevalent form of G6PD in the population of Myanmar is Mahidol (Matsuoka et al., 2004). In the current research, a total of 15 harmful mutations were found, which were dominated by G6PD Kaiping and G6PD Canton, accounting for approximately 42% of all G6PD-deficient alleles. However, it is lower than previous research results (84.1%, 75.3%) in the Guangxi population (Fu et al., 2018; Yan et al., 2006). This can be because we only collected a small number of samples or because geographical disparities exist. In addition, we are a region with a high prevalence of thalassemia, moreover, hemolysis and anemia are common (Lin et al., 2015). Medication, hemolysis, and anemia can affect the detection of G6PD activity (Nuinoon et al., 2022; Pfeffer et al., 2022). In our study, these may be one of the reasons that the 165 samples with no detection of any of the 33 common mutations. However, they were detected with G6PD activity deficiency. Certainly, the other reason is that they may have rare mutations (besides 33 common mutations).
G6PD was first described by Carson in 1956 (ALVING et al., 1956). Its clinical manifestations include fulminant hemolysis, severe hyperbilirubinemia, and kernicterus, which contribute to neonatal neurological injury and risk of death (He et al., 2020; Kaplan et al., 2015; Liu et al., 2020). This condition may be brought on by infections, specific foods (such as fava beans), oxidizing medicines, and/or specific herbal therapies (Liu et al., 2020). To date, after a newborn’s screening results in a positive result, the most effective treatment for this illness is to prevent hemolysis by avoiding some oxidative stressors (Liu et al., 2020). Therefore, the general survey of G6PD deficiency, early detection and early prevention are important measures to prevent and treat the disease. There are three common measures to prevent the disease, the most important is to avoid accidental ingestion of fava beans (Reading et al., 2016); secondly, avoid taking anti-malarial drugs (primaquine, chloroquine, malaria quinine, pentaquine and adipine), sulfones (thiazole sulfone, aminophene sulfone), sulfonamides (sulfamethoxazole, sulfadimethoxine, sulfapyridine and salazosulfapyridine) and antipyretics (acetazolamide and acetanilide) and so on (http://www.g6pd.org) (Chu and Freedom, 2019; Reading et al., 2016). Finally, when the patient has an infection (viral hepatitis, influenza, pneumonia, typhoid), which should immediately seek medical help to avoid hemolysis.
Today’s G6PD deficiency diagnosis primarily uses the enzyme activity detection assay, and the main diagnosis used to avoid oxidative hemolysis cannot be other than a phenotypic test, especially in women; however, there is an added value in G6PD genotyping, different sorts of mutations can result in various classes of variations and exhibit various symptoms (Beutler et al., 2002; WHO, 2022). So, to establish a certain diagnosis of G6PD insufficiency, genotyping of G6PD mutations is beneficial (Jiang et al., 2006). In addition, the analysis of G6PD genotypes contributes to the study of molecular biology and genetic characterization of human populations (Hamali, 2021; Lee et al., 2022). Aside from this, the genotyping of G6PD deficiency also has a significant impact on the field’s understanding of the disorder (Li et al., 2008). The SNPscan assay used in the study covered 33 common mutations in the Chinese population and could identify more than 95% of G6PD deficiencies. Based on the detection of SNP loci, SNPscan technology can simultaneously type multiple SNP loci in one detection process (Yu et al., 2021). Numerous investigations have shown that it has good accuracy, sensitivity, and specificity and is cost-effective and high-throughput (Du et al., 2014; Yin et al., 2014; Zhang et al., 2016). Compared with the direct sequencing method, it saves more tedious operations in the experimental process, can detect multiple sites in multiple samples at the same time, and reduces the cost (Zhang et al., 2016). Compared with the gene chip method, SNPscan technology has more detection sites, so it can be flexibly designed for known target gene mutation sites and achieves high throughput (Chen et al., 2012; Duan et al., 2017; Hu et al., 2015; Zhang et al., 2016). In addition, we investigated a general comparison of costs associated with these different techniques and found that the SNPscan technique has the lowest cost (SNPscan technology: $14.26/sample, direct sequencing method: $20.97/sample, gene chip method: $69.93/sample). Therefore, a trustworthy, quick, and affordable method for identifying G6PD point mutations would be beneficial to patients, their families, the doctors who treat them, and the testing labs.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
This protocol was approved by the ethics committee of the Affiliated Hospital of Youjiang Medical University for Nationalities, and written informed consent was obtained from all individuals.
All authors contributed to the study’s conception and design. ML, JW, and CW designed the study. WC, LC, and LH collected the samples and entered the data. WW, GX, and WH analysed and interpreted the data. HW, HH, and WH conducted the laboratory work (G6PD genotyping and analysis of G6PD enzyme activity). LL, WC, and YZ make the figures and tables. ML, HW, and YL wrote the paper. All authors critically reviewed the paper and approved the final version of the paper for submission.
This work was partially supported by Key Scientific Research Projects of Guangdong Provincial Department of Education, Grant/Award Number: 2018A030307074; Natural Science Foundation of Guangdong Province, Grant/Award Number: 2018A030307074; National Natural Science Foundation of China, Contract/Grant Number: 81760615; Scientific Research Project of Chaozhou Health Bureau, Grant/Award Number: 2021013.
We would like to thank all teammates for contributing to this work.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.1000290/full#supplementary-material
Ainoon, O., Yu, Y. H., Amir, M. A., Boo, N. Y., Cheong, S. K., and Hamidah, N. H. (2003). Glucose-6-phosphate dehydrogenase (G6PD) variants in Malaysian Malays. Hum. Mutat. 21 (1), 101. doi:10.1002/humu.9103
Alving, A. S., Carson, P. E., Flanagan, C. L., and Ickes, C. E. (1956). Enzymatic deficiency in primaquine-sensitive erythrocytes. Science 124 (3220), 484–485. doi:10.1126/science.124.3220.484-a
Beutler, E., Gelbart, T., and Miller, W. (2002). Severe jaundice in a patient with a previously undescribed glucose-6-phosphate dehydrogenase (g6pd) mutation and gilbert syndrome. Blood Cells Mol. Dis. 28 (2), 104–107.
Boonyuen, U., Songdej, D., Tanyaratsrisakul, S., Phuanukoonnon, S., Chamchoy, K., Praoparotai, A., et al. (2021). Glucose-6-phosphate dehydrogenase mutations in malaria endemic area of Thailand by multiplexed high-resolution melting curve analysis. Malar. J. 20 (1), 194. doi:10.1186/s12936-021-03731-0
Chen, X., Li, S., Yang, Y., Yang, X., Liu, Y., Liu, Y., et al. (2012). Genome-wide association study validation identifies novel loci for atherosclerotic cardiovascular disease. J. Thromb. Haemost. 10 (8), 1508–1514. doi:10.1111/j.1538-7836.2012.04815.x
Chu, C. S., and Freedman, D. O. (2019). Tafenoquine and G6PD: A primer for clinicians. J. Travel Med. 26 (4), taz023. doi:10.1093/jtm/taz023
Devendra, R., Gupta, V., Biradar, S. S., Bhat, P., Hegde, S., Hoti, S. L., et al. (2020). G6PD A-is the major cause of G6PD deficiency among the Siddis of Karnataka, India. Ann. Hum. Biol. 47 (1), 55–58. doi:10.1080/03014460.2019.1699954
Dombrowski, J. G., Souza, R. M., Curry, J., Hinton, L., Silva, N., Grignard, L., et al. (2017). G6PD deficiency alleles in a malaria-endemic region in the Western Brazilian Amazon. Malar. J. 16 (1), 253. doi:10.1186/s12936-017-1889-6
Driscoll, D. J., and Migeon, B. R. (1990). Sex difference in methylation of single-copy genes in human meiotic germ cells: implications for X chromosome inactivation, parental imprinting, and origin of CpG mutations. Somat. Cell. Mol. Genet. 16 (3), 267–282. doi:10.1007/BF01233363
Du, W., Cheng, J., Ding, H., Jiang, Z., Guo, Y., and Yuan, H. (2014). A rapid method for simultaneous multi-gene mutation screening in children with nonsyndromic hearing loss. Genomics 104 (4), 264–270. doi:10.1016/j.ygeno.2014.07.009
Duan, S. H., Ma, J. L., Yang, X. L., and Guo, Y. F. (2017). Simultaneous multi‑gene mutation screening using SNPscan in patients from ethnic minorities with nonsyndromic hearing‑impairment in Northwest China. Mol. Med. Rep. 16 (5), 6722–6728. doi:10.3892/mmr.2017.7431
Filosa, S., Giacometti, N., Wangwei, C., De Mattia, D., Pagnini, D., Alfinito, F., et al. (1996). Somatic-cell selection is a major determinant of the blood-cell phenotype in heterozygotes for glucose-6-phosphate dehydrogenase mutations causing severe enzyme deficiency. Am. J. Hum. Genet. 59 (4), 887–895.
Fu, C., Luo, S., Li, Q., Xie, B., Yang, Q., Geng, G., et al. (2018). Newborn screening of glucose-6-phosphate dehydrogenase deficiency in Guangxi, China: Determination of optimal cutoff value to identify heterozygous female neonates. Sci. Rep. 8 (1), 833. doi:10.1038/s41598-017-17667-6
Gómez-Manzo, S., Marcial-Quino, J., Vanoye-Carlo, A., Serrano-Posada, H., Ortega-Cuellar, D., González-Valdez, A., et al. (2016). Glucose-6-phosphate dehydrogenase: Update and analysis of new mutations around the world. Int. J. Mol. Sci. 17 (12), 2069. doi:10.3390/ijms17122069
Hamali, H. A. (2021). Glucose-6-phosphate dehydrogenase deficiency: An overview of the prevalence and genetic variants in Saudi Arabia. Hemoglobin 45 (5), 287–295. doi:10.1080/03630269.2022.2034644
He, Y., Zhang, Y., Chen, X., Wang, Q., Ling, L., and Xu, Y. (2020). Glucose-6-phosphate dehydrogenase deficiency in the han Chinese population: Molecular characterization and genotype-phenotype association throughout an activity distribution. Sci. Rep. 10 (1), 17106. doi:10.1038/s41598-020-74200-y
Howell, R. R. (2006). Advisory committee on heritable disorders and genetic diseases in newborns and children. Ment. Retard. Dev. Disabil. Res. Rev. 12 (4), 313–315. doi:10.1002/mrdd.20126
Hu, R., Lin, M., Ye, J., Zheng, B. P., Jiang, L. X., Zhu, J. J., et al. (2015). Molecular epidemiological investigation of G6PD deficiency by a gene chip among Chinese Hakka of southern Jiangxi province. Int. J. Clin. Exp. Pathol. 8 (11), 15013. doi:10.1097/01.hs9.0000566368.48943.78
Iwai, K., Hirono, A., Matsuoka, H., Kawamoto, F., Horie, T., Lin, K., et al. (2001). Distribution of glucose-6-phosphate dehydrogenase mutations in Southeast Asia. Hum. Genet. 108 (6), 445–449. doi:10.1007/s004390100527
Ji-Guang, D., Shui-Lan, Y. U., Nong-Zhi, , and Yi-Chao, Y. (2017). Analysis of inspection certification results on malaria elimination in Baise City. Zhongguo Xue Xi Chong Bing Fang. Zhi Za Zhi 29 (4), 512–514. doi:10.16250/j.32.1374.2017017
Jiang, W., Yu, G., Liu, P., Geng, Q., Chen, L., Lin, Q., et al. (2006). Structure and function of glucose-6-phosphate dehydrogenase-deficient variants in Chinese population. Hum. Genet. 119 (5), 463–478. doi:10.1007/s00439-005-0126-5
Kaplan, M., Hammerman, C., and Bhutani, V. K. (2015). Parental education and the WHO neonatal G-6-PD screening program: A quarter century later. J. Perinatol. 35 (10), 779–784. doi:10.1038/jp.2015.77
Kumar, R., Singh, M., Mahapatra, S., Chaurasia, S., Tripathi, M. K., Oommen, J., et al. (2020). Fine mapping of glucose 6 phosphate dehydrogenase (G6PD) deficiency in a rural malaria area of south west odisha using the clinical, hematological and molecular approach. Med. J. Hematol. Infect. Dis. 12 (1), e2020015. doi:10.4084/MJHID.2020.015
Lee, H. Y., Ithnin, A., Azma, R. Z., Othman, A., Salvador, A., and Cheah, F. C. (2022). Glucose-6-Phosphate dehydrogenase deficiency and neonatal hyperbilirubinemia: Insights on pathophysiology, diagnosis, and gene variants in disease heterogeneity. Front. Pediatr. 10, 875877. doi:10.3389/fped.2022.875877
Li, L., Zhou, Y. Q., Xiao, Q. Z., Yan, T. Z., and Xu, X. M. (2008). Development and evaluation of a reverse dot blot assay for the simultaneous detection of six common Chinese G6PD mutations and one polymorphism. Blood Cells Mol. Dis. 41 (1), 17–21. doi:10.1016/j.bcmd.2008.01.007
Liang, X. Y., Chen, J. T., Ma, Y. B., Huang, H. Y., Xie, D. D., Monte-Nguba, S. M., et al. (2020). Evidence of positively selected G6PD A-allele reduces risk of Plasmodium falciparum infection in African population on Bioko Island. Mol. Genet. Genom. Med. 8 (2), e1061. doi:10.1002/mgg3.1061
Lin, F., Lou, Z., Xing, S., Zhang, L., and Yang, L. (2018). The gene spectrum of glucose-6-phosphate dehydrogenase (g6pd) deficiency in guangdong province, China. Gene 678, 312–317. doi:10.1016/j.gene.2018.07.068
Lin, M., Yang, L. Y., Xie, D. D., Chen, J. T., Nguba, S. M., Ehapo, C. S., et al. (2015). G6PD deficiency and hemoglobinopathies: Molecular epidemiological characteristics and healthy effects on malaria endemic bioko island, Equatorial Guinea. PLoS One 10 (4), e0123991. doi:10.1371/journal.pone.0123991
Liu, Z., Yu, C., Li, Q., Cai, R., Qu, Y., Wang, W., et al. (2020). Chinese newborn screening for the incidence of G6PD deficiency and variant of G6PD gene from 2013 to 2017. Hum. Mutat. 41 (1), 212–221. doi:10.1002/humu.23911
Louicharoen, C., and Nuchprayoon, I. (2005). G6PD Viangchan (871G>A) is the most common G6PD-deficient variant in the Cambodian population. J. Hum. Genet. 50 (9), 448–452. doi:10.1007/s10038-005-0276-2
Maloukh, L., Kumarappan, A., El-Din, E. H., Al-Kamali, F., Gomma, F., Akhondi, A., et al. (2021). Development of allelic discrimination assay to detect Mediterranean G6PD mutation and its linked inheritance with normal vision and/colorblindness loci for 4 generations among Egyptian and Emirati families. Saudi J. Biol. Sci. 28 (9), 5028–5033. doi:10.1016/j.sjbs.2021.05.014
Matsuoka, H., Wang, J., Hirai, M., Arai, M., Yoshida, S., Kobayashi, T., et al. (2004). Glucose-6-phosphate dehydrogenase (g6pd) mutations in Myanmar: g6pd mahidol (487g> a is the most common variant in the Myanmar population. J. Hum. Genet. 49 (10), 544–547. doi:10.1007/s10038-004-0187-7
Mukherjee, M. B., Colah, R. B., Martin, S., and Ghosh, K. (2015). Glucose-6-phosphate dehydrogenase (G6PD) deficiency among tribal populations of India - country scenario. Indian J. Med. Res. 141 (5), 516–520. doi:10.4103/0971-5916.159499
Nuchprayoon, I., Sanpavat, S., and Nuchprayoon, S. (2002). Glucose-6-phosphate dehydrogenase (g6pd) mutations in Thailand: g6pd viangchan (871g> is the most common deficiency variant in the Thai population. Hum. Mutat. 19 (2), 185. doi:10.1002/humu.9010
Nuinoon, M., Krithong, R., Pramtong, S., Sasuk, P., Ngeaiad, C., Chaimusik, S., et al. (2022). Prevalence of g6pd deficiency and g6pd variants amongst the southern Thai population. PeerJ 10, e14208. doi:10.7717/peerj.14208
Pan, M., Lin, M., Yang, L., Wu, J., Zhan, X., Zhao, Y., et al. (2013). Glucose-6-phosphate dehydrogenase (G6PD) gene mutations detection by improved high-resolution DNA melting assay. Mol. Biol. Rep. 40 (4), 3073–3082. doi:10.1007/s11033-012-2381-6
Pfeffer, D. A., Satyagraha, A. W., Sadhewa, A., Alam, M. S., Bancone, G., Boum, Y. N., et al. (2022). Genetic variants of glucose-6-phosphate dehydrogenase and their associated enzyme activity: A systematic review and meta-analysis. Pathogens 11 (9), 1045. doi:10.3390/pathogens11091045
Reading, N. S., Sirdah, M. M., Shubair, M. E., Nelson, B. E., Al-Kahlout, M. S., Al-Tayeb, J. M., et al. (2016). Favism, the commonest form of severe hemolytic anemia in Palestinian children, varies in severity with three different variants of G6PD deficiency within the same community. Blood Cells Mol. Dis. 60, 58–64. doi:10.1016/j.bcmd.2016.07.001
Saravu, K., Kumar, R., Ashok, H., Kundapura, P., Kamath, V., Kamath, A., et al. (2016). Therapeutic assessment of chloroquine-primaquine combined regimen in adult cohort of plasmodium vivax malaria from primary care centres in southwestern India. PLoS One 11 (6), e0157666. doi:10.1371/journal.pone.0157666
Sukumar, S., Mukherjee, M. B., Colah, R. B., and Mohanty, D. (2004). Molecular basis of G6PD deficiency in India. Blood Cells Mol. Dis. 33 (2), 141–145. doi:10.1016/j.bcmd.2004.06.003
Tian, P. L., Zhou, B. Y., Zhao, W. Z., Zheng, L. X., Ye, J. L., Wang, B. X., et al. (2013). Identification of glucose-6-phosphate dehydrogenase gene variants in Guangdong populations. Zhonghua Xue Ye Xue Za Zhi 34 (8), 719–721. doi:10.3760/cma.j.issn.0253-2727.2013.08.017
Wei, J., Zheng, L., Liu, S., Yin, J., Wang, L., Wang, X., et al. (2013). MiR-196a2 rs11614913 T > C polymorphism and risk of esophageal cancer in a Chinese population. Hum. Immunol. 74 (9), 1199–1205. doi:10.1016/j.humimm.2013.06.012
Who, (2022). Technical consultation to review the classification of glucose-6-phosphate dehydrogenase (g6pd) Geneva: World Health Organ.
Wisnumurti, D. A., Sribudiani, Y., Porsch, R. M., Maskoen, A. M., Rahayuningsih, S. E., Asni, E. K., et al. (2019). G6PD genetic variations in neonatal Hyperbilirubinemia in Indonesian Deutromalay population. BMC Pediatr. 19 (1), 506. doi:10.1186/s12887-019-1882-z
Yan, J. B., Xu, H. P., Xiong, C., Ren, Z. R., Tian, G. L., Zeng, F., et al. (2010). Rapid and reliable detection of glucose-6-phosphate dehydrogenase (G6PD) gene mutations in Han Chinese using high-resolution melting analysis. J. Mol. Diagn. 12 (3), 305–311. doi:10.2353/jmoldx.2010.090104
Yan, T., Cai, R., Mo, O., Zhu, D., Ouyang, H., Huang, L., et al. (2006). Incidence and complete molecular characterization of glucose-6-phosphate dehydrogenase deficiency in the Guangxi Zhuang autonomous region of southern China: Description of four novel mutations. Haematologica 91 (10), 1321. doi:10.3389/fgene.2022.994015
Yang, H., Wang, Q., Zheng, L., Zhan, X. F., Lin, M., Lin, F., et al. (2015). Incidence and molecular characterization of Glucose-6-Phosphate Dehydrogenase deficiency among neonates for newborn screening in Chaozhou, China. Int. J. Lab. Hematol. 37 (3), 410–419. doi:10.1111/ijlh.12303
Yin, J., Wang, L., Shi, Y., Shao, A., Tang, W., Wang, X., et al. (2014). Interleukin 17A rs4711998 A>G polymorphism was associated with a decreased risk of esophageal cancer in a Chinese population. Dis. Esophagus. 27 (1), 87–92. doi:10.1111/dote.12045
Yu, H., Hu, W., Lin, C., Xu, L., Liu, H., Luo, L., et al. (2021). Polymorphisms analysis for association between ADIPO signaling pathway and genetic susceptibility to T2DM in Chinese han population. Adipocyte 10 (1), 463–474. doi:10.1080/21623945.2021.1978728
Zhang, F., Xiao, Y., Xu, L., Zhang, X., Zhang, G., Li, J., et al. (2016). Mutation analysis of the common deafness genes in patients with nonsyndromic hearing loss in linyi by SNPscan assay. Biomed. Res. Int. 2016, 1302914. doi:10.1155/2016/1302914
Zheng, Y., Wang, J., Liang, X., Huang, H., Ma, Y., Lin, L., et al. (2020). Epidemiology, evolutionary origin, and malaria-induced positive selection effects of G6PD-deficient alleles in Chinese populations. Mol. Genet. Genom. Med. 8 (12), e1540. doi:10.1002/mgg3.1540
Keywords: G6PD deficiency, mutation spectrum, southwestern China, SNPscan assay, G6PD genotype
Citation: Wei H, Wang C, Huang W, He L, Liu Y, Huang H, Chen W, Zheng Y, Xu G, Lin L, Wei W, Chen W, Chen L, Wang J and Lin M (2023) Simultaneous detection of G6PD mutations using SNPscan in a multiethnic minority area of Southwestern China. Front. Genet. 13:1000290. doi: 10.3389/fgene.2022.1000290
Received: 22 July 2022; Accepted: 21 December 2022;
Published: 10 January 2023.
Edited by:
Simon Charles Heath, Center for Genomic Regulation (CRG), SpainReviewed by:
Germana Bancone, Centre for Tropical Medicine and Global Health, University of Oxford, United KingdomCopyright © 2023 Wei, Wang, Huang, He, Liu, Huang, Chen, Zheng, Xu, Lin, Wei, Chen, Chen, Wang and Lin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Junli Wang, YmFpc2V3YW5nanVubGlAMTYzLmNvbQ==; Min Lin, a29uZnV0ZWFAaG90bWFpbC5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.