
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet., 04 August 2020
Sec. RNA
Volume 11 - 2020 | https://doi.org/10.3389/fgene.2020.00853
 Yun Liu1,2†
Yun Liu1,2† Zeyao Zhu3†
Zeyao Zhu3† Idy H. T. Ho1,4
Idy H. T. Ho1,4 Yujian Shi1
Yujian Shi1 Jianzhen Li5
Jianzhen Li5 Xia Wang3
Xia Wang3 Matthew T. V. Chan4
Matthew T. V. Chan4 Christopher H. K. Cheng1*
Christopher H. K. Cheng1*MiR-430 is considered an important regulator during embryonic development, but genetic loss-of-function study is still lacking. Here we demonstrated that genetic deletion of the miR-430 cluster resulted in developmental defects in cell movement, germ layer specification, axis patterning and organ progenitor formation in zebrafish. Transcriptome analysis indicated that the maternally provided transcripts were not properly degraded whereas the zygotic genome expressed genes were not fully activated in the miR-430 mutants. We further found that a reciprocal regulatory loop exists between miR-430 and maternally provided transcripts: the maternally provided transcripts (Nanog, Dicer1, Dgcr8, and AGOs) are required for miR-430 biogenesis and function, whereas miR-430 is required for the clearance of these maternally provided transcripts. These data provide the first genetic evidence that miR-430 is required for maternal-zygotic transition and subsequent establishment of embryonic body plan.
MicroRNAs (miRNAs) are ∼22-nucleotide (nt) non-coding RNAs that repress mRNA expression post-transcriptionally (Bartel, 2009). Transcribed by RNA polymerase II, the primary miRNA transcripts are processed by Dgcr8 and RNase III enzyme Drosha into 70-nt precursors which are further processed into 21–23-nt mature microRNAs by Dicer (Kim et al., 2009). One strand of the mature microRNAs is loaded into argonaute (AGO) protein to recognize target mRNAs by pairing with miRNA binding sites in the 3′ untranslated region (UTR) (Kim et al., 2009). AGO proteins further recruit other factors that could induce translational repression, mRNA deadenylation and mRNA decay (Fabian et al., 2010). One miRNA can modulate hundreds of mRNA targets and more than 60% of human protein-coding genes are considered targets for miRNAs (Friedman et al., 2009).
In mice, zygotic deletion of Dgcr8 or Dicer leads to embryonic arrest shortly after implantation (Bernstein et al., 2003; Wang et al., 2007). In zebrafish, Dicer1 transcripts are maternally provided and maternal-zygotic dicer (MZdicer) mutant exhibits cell movement defects at the onset of gastrulation (Wienholds et al., 2003; Giraldez et al., 2005, 2006). Recently, we have successfully generated zebrafish maternal-zygotic dgcr8 (MZdgcr8) mutant and found that MZdgcr8 and MZdicer share similar phenotypes (Liu et al., 2017; Zhu et al., 2020), suggesting that the canonical miRNAs play important roles in early development. Interestingly, miR-430 could rescue some developmental defects of MZdicer and MZdgcr8 mutants, suggesting that suggesting that miR-430 might be the key effector of the miRNA pathway in early embryogenesis (Giraldez et al., 2005, 2006; Liu et al., 2017).
The maternally provided transcripts play central roles in the regulation of early embryonic development (Lee et al., 2013; Leichsenring et al., 2013). In zebrafish, the maternally provided transcripts Pou5f1, Nanog, and Sox19b are required for zygotic genome activation (ZGA) (Lee et al., 2013; Leichsenring et al., 2013). MiR-430 is among the first-wave of zygotic genome expressed genes activated by Nanog (Lee et al., 2013). Using MZdicer mutants, miR-430 was suggested to destabilize hundreds of maternal transcripts after ZGA (Giraldez et al., 2006). Moreover, miR-430 was also found to regulate the nodal pathway and other developmental pathways during early development (Mishima et al., 2006; Choi et al., 2007; Rosa et al., 2009; Staton et al., 2011; Wylie et al., 2014; van Boxtel et al., 2015; Takacs and Giraldez, 2016). However, genetic loss-of-function study of miR-430 is still lacking. In this study, we generated miR-430-deficient zebrafish and found that miR-430 is required for maternal-zygotic transition (MZT) and establishment of embryonic body plan.
AB zebrafish were maintained at 28°C in the zebrafish facility of Sun Yat-sen University and the Chinese University of Hong Kong. All animal experiments were conducted in accordance with the guidelines and approval of the respective Animal Research and Ethics Committees.
The method for deleting the miR-430 gene cluster was reported in previous studies (Lei et al., 2012; Liu et al., 2013, 2014). Germline transmission of miR-430 deletion was reported in our previous study (Liu et al., 2013). To obtain homozygous mutants, heterozygous mutants of the same mutation were obtained and self-crossed.
To analyze the phenotypic consequences of miR-430 loss-of-function, embryos from incrosses of miR-430 heterozygote adults were collected and genotyped by PCR (Supplementary Figure S1). Because the miR-430–/– mutants showed developmental delay, embryos of the same developmental stages or at same time points were photographed.
To perform rescue experiments, miR-430 mimics were synthesized as described (Giraldez et al., 2005, 2006). For phenotypic rescue, miR-430 mimics (10 pg/embryo) or mismatched miR-430b mimics were injected into one-cell stage embryos and the phenotypic changes were recorded on a stereomicroscope (Olympus). For miRNA rescue efficiency analysis, marker gene analysis and transcriptome sequencing, miR-430–/– embryos produced by incrosses of the rescued miR-430–/– adult fish were used.
The antisense probes of marker genes were prepared using the DIG RNA Labeling Kit (Roche, United States). The probe information was provided in the Supplementary Table S1. Whole mount in situ hybridization was performed as described (Li et al., 2014). The embryos of similar developmental stages were collected and the ratios of the affected embryos were calculated.
Total RNA was isolated from shield stage embryos of WT, miR-430–/– and rescued groups with the RNeasy Mini Kit (QIAGEN). Deep-sequencing of total RNA (ribosomal RNA minus) was performed on an Illumina HiSeq2000 platform at BGI (Shenzhen, China). Approximately 5 Gb of raw reads were generated for each sample. The fragments per kilobase million (FPKM) values of the mapped genes were obtained using the Cufflink software (Trapnell et al., 2012). The differentially expressed genes were identified by the defined thresholds of fold changes and p value. The miR-430 binding sites were predicted by TargetScanFish 6.01 (Ulitsky et al., 2012). Gene ontology (GO) analysis was performed by the DAVID software2 (Huang et al., 2009). The reported transcriptome data in a previous study (Yang et al., 2013) were used to examine gene expression profiles during early embryonic development in zebrafish.
Total RNAs were isolated from zebrafish embryos of shield stage using the RNeasy Mini Kit (QIAGEN). Complementary DNA was synthesized using the PrimeScript RT Reagent Kit (TAKARA). Real-time Q-PCR was performed on an ABI PRISM 7900 Sequence Detection System (Applied Biosystems) using the SYBR Green I Kit (Applied Biosystem). The mRNA transcript levels were normalized against the ef1α transcript level. Primers used in this study are listed in Supplementary Table S1.
The MO sequence for lft2 knockdown was designed as reported (Feldman et al., 2002). The lft2 MO or control MO (8 ng/embryo) were injected into the miR-430–/– mutants. The embryos were collected at 75% epiboly and 36 h post fertilization (36 hpf) for marker gene expression analysis.
The DNA sequence of Dicer-3′UTR-WT or Dicer-3′UTR-mut were amplified by PCR and cloned into pCS2-3XIPT-miR-430 plasmid by XhoI and XbaI (Giraldez et al., 2005). GFP sensor mRNAs were transcribed using the mMESSAGE mMACHINE SP6 kit (Ambion) and purified using the RNeasy Mini Kit (QIAGEN). One hundred nanogram of mRNA was injected into a miR-430–/– mutant or WT embryo at one cell stage. The GFP signal was analyzed on a fluorescent microscope (Olympus) at 6 hpf.
The Q-PCR data are expressed as the mean values ± SEM. Statistical analyses were performed using t-test or one-way ANOVA followed by Tukey’s Multiple Comparison Test (GraphPad). The enrichment analyses were performed using Chi-squared test (Microsoft Excel software). Results were considered statistically significant at P < 0.05.
To investigate the functional roles of miR-430 during early embryonic development, we assembled two pairs of TALENs to delete a ∼80 kb genomic fragment containing the zebrafish miR-430 cluster (Figure 1A) (Liu et al., 2013). This deleted region mainly contains repeated sequences with no recognizable functional genes other than miR-430. The miR-430 cluster is the largest identified miRNA cluster which contains 57 miRNAs with the same miRNA seed sequence in zebrafish (Figure 1A). In this study, we have characterized the phenotypes of miR-430 homozygous mutants. Whole mount in situ hybridization analysis indicated that the primary miR-430 transcript was abundantly expressed in the WT (4 hpf) but not in the miR-430 mutants (Figure 1B). Moreover, the mature miR-430s were the most abundantly expressed miRNAs in the 6-somite WT embryos (92% of total miRNA reads) and decreased about 4,000 folds in the miR-430–/– mutants (Figure 1C). These data indicated that we have successfully established the loss-of-function model of the miR-430 cluster in zebrafish.
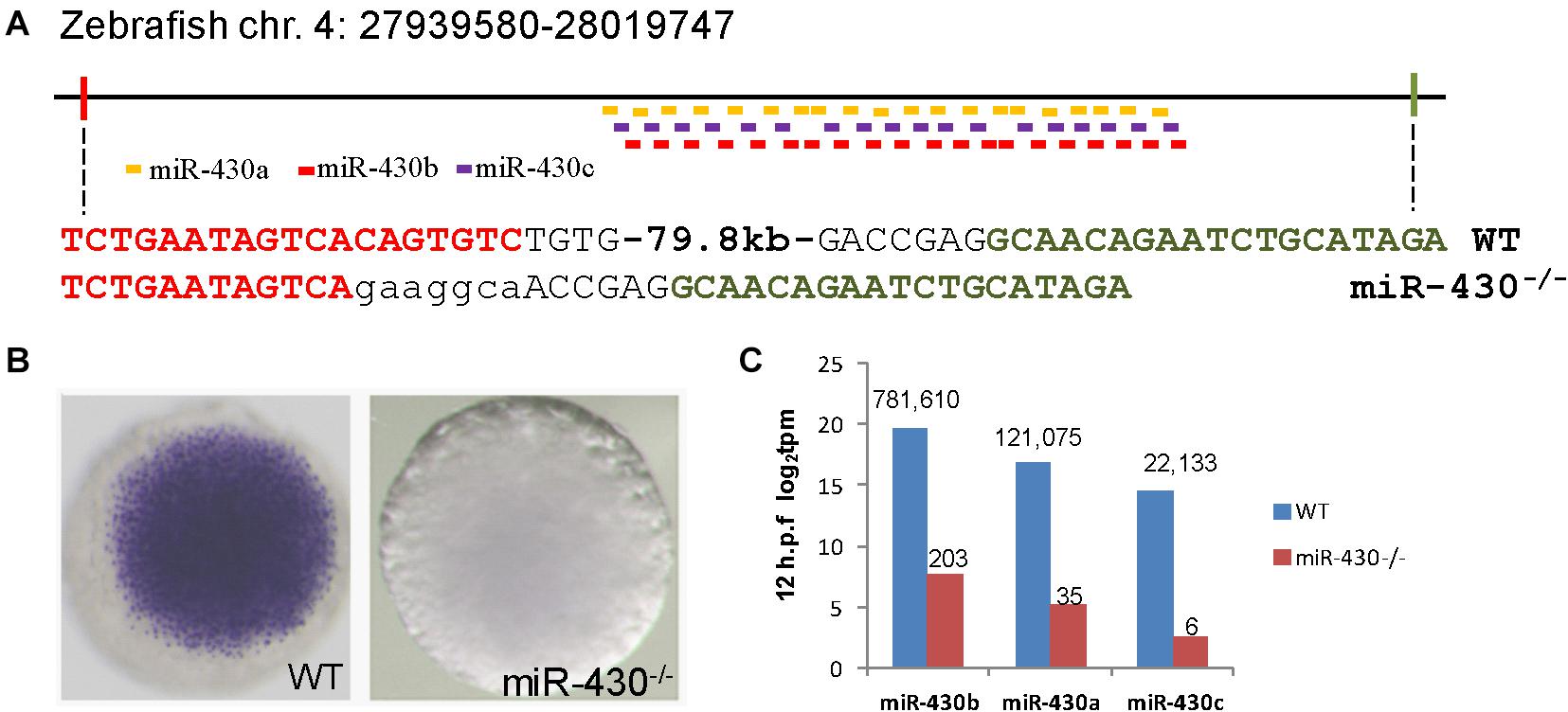
Figure 1. Targeted deletion of miR-430 in zebrafish. (A) Genetic deletion of zebrafish miR-430 cluster using TALENs. A 79.8 kb genomic fragment containing the zebrafish miR-430 was deleted using two pairs of TALENs. The TALEN binding sites were shown in colors and the inserted nucleotides were shown in lower case letters. MiR-430 isoforms were indicated by colored short lines. (B) In situ hybridization detection of pri-mir-430 expression in the WT and miR-430–/– mutant embryos at 4 hpf. (C) Expression profiles of mature miR-430 isoforms in the WT and miR-430–/– mutant embryos (12 hpf) by small RNA deep sequencing. The numbers of normalized reads were indicated. TPM, transcripts per kilobase million.
We then analyzed the phenotypic consequences of miR-430 deletion. Compared to WT embryos, the miR-430–/– mutants exhibited developmental delay since the germ ring stage (5.7 hpf; Figures 2A,F). The formation of shield at the dorsal site was reduced at 6 hpf (Figures 2B,G). The mutant embryos exhibited a longer animal-vegetal axis and the yolk was not completely covered by cell sheet at the end of epiboly cell movements (Figures 2C,H). The extension of the anterior-posterior body axis was reduced in the mutant (Figures 2D,E,I,J). The regionalization of the brain was disrupted at 24 hpf (Figures 2K,M). No heart beat or circulation can be observed but tail blister was developed in the mutant at 48 hpf (Figures 2L,N). All mutant died at around 5 days post fertilization (dpf) with a swollen body cavity (n = 500) (Supplementary Figure S2).
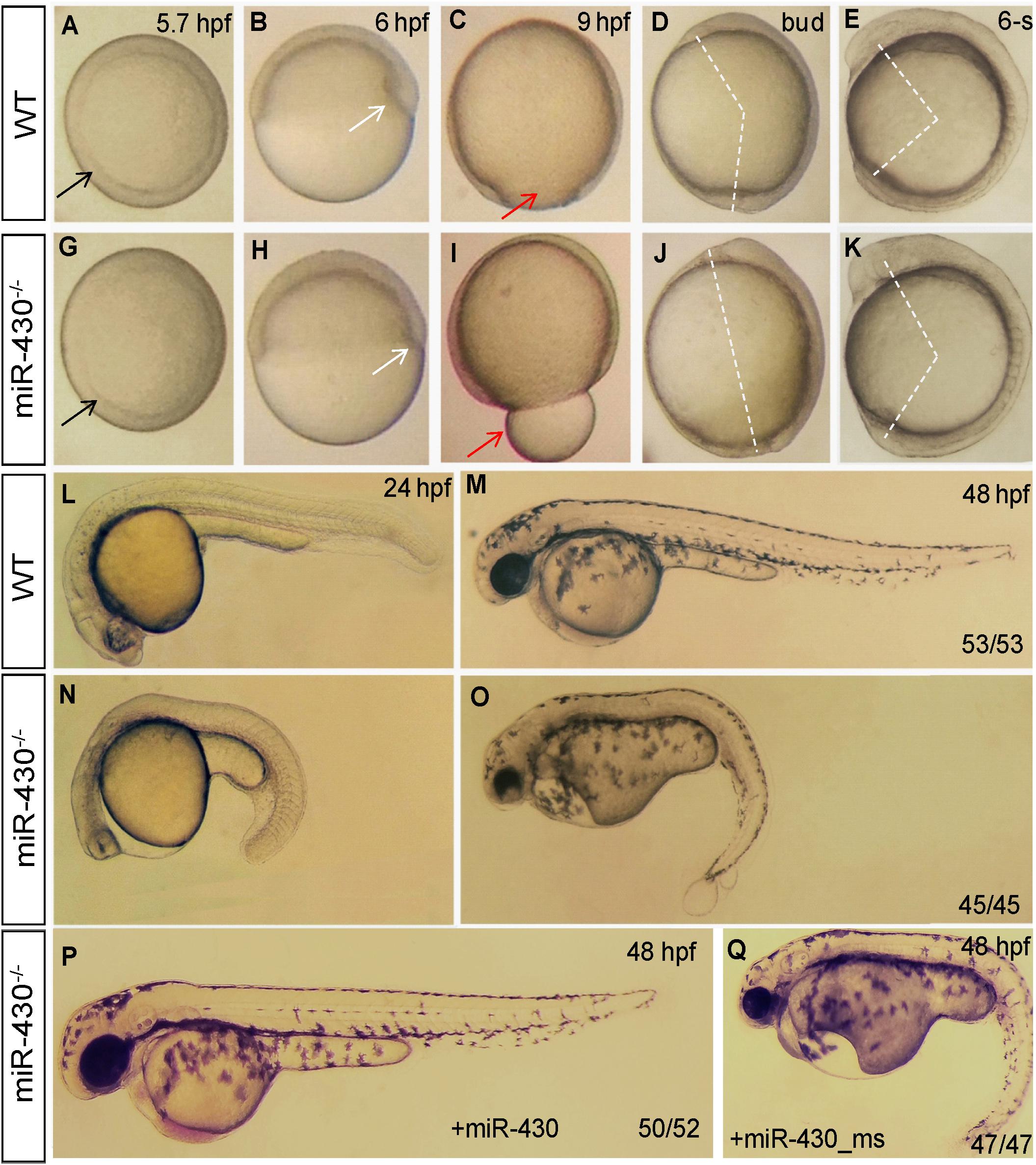
Figure 2. Morphological defects in the miR-430-deficient embryos. (A–O) The morphology of WT and miR-430-deficient embryos. The phenotypes are fully penetrant. Formation of the germ ring (black arrows) and shield (white arrows) were delayed in the miR-430–/– mutants (A,B,F,G). The yolk was not enclosed by epiboly in the mutant at 9 hpf (C,H). The extension of the anterior-posterior body axis was reduced in the miR-430 mutants (D,E,I,J). The miR-430 mutants had no brain vesicles and lacked heart tissue and circulation (K,M). Tail blisters were observed in the mutant at 48 hpf (L,N). The miR-430 mimics but not the mismatched miR-430 with 2-nucleotide alterations could rescue the defects observed in the miR-430 mutants (O,P). The ratios of observed phenotypes were indicated.
To test whether the observed phenotypes were due to the loss of function of miR-430, we then performed rescue experiments using miR-430 duplexes (8, 9). Injection of miR-430 duplexes but not the mismatched mimics efficiently rescued the mutant phenotypes (Figures 2O,P). Some of rescued embryos even survived to adulthood. We further crossed the rescued miR-430–/– mutant adult fish to produce the MZmiR-430 mutants. The MZmiR-430 embryos had the same phenotypes as the ZmiR-430 mutants. Further rescue of the MZmiR-430 embryos indicated that eight of the 52 rescued embryos survived to adulthood. These data indicated that miR-430 is mainly functional during early embryonic development.
We next analyzed the miR-430–/– phenotypes using marker genes. In the mutant embryos, expression of the dorsal mesoderm markers (chd and gsc) were reduced and expression domain of the ventral markers (eve1 and bmp4) were expanded (Figure 3A). The mesoderm (snai1a and mixer1) and endoderm markers (sox17 and sox32) were down-regulated (Figure 3B). The erythroid progenitor marker (gata1) was expanded but the myeloid progenitor marker (pu.1) was absent (Figure 3C). The cardiac progenitors (nkx2.5) were reduced and the cardiocytes (cmlc2) failed to migrate to the middle line, resulted in cardiac bifida (Figures 3C,E). The angioblast marker fli-1 was specified but the vascular system was disrupted at later stages (Figure 3C and Supplementary Figure S3). In the neuronal system, the expression of the mid-hind boundary domain markers (eng2 and pax2a) was decreased. The forebrain region was reduced (six3) and the optic placode was expanded (pax2a) (Figure 3D). Consistent with the morphological observations, the anterior neural plate was reduced anterior-posteriorly but expanded medio-laterally (Figure 3D). These results indicated that miR-430 is required for axis patterning, germ layer specification, organ progenitor formation and cell movements, thus contributing to the establishment of embryonic body plan (Schier and Talbot, 2005).
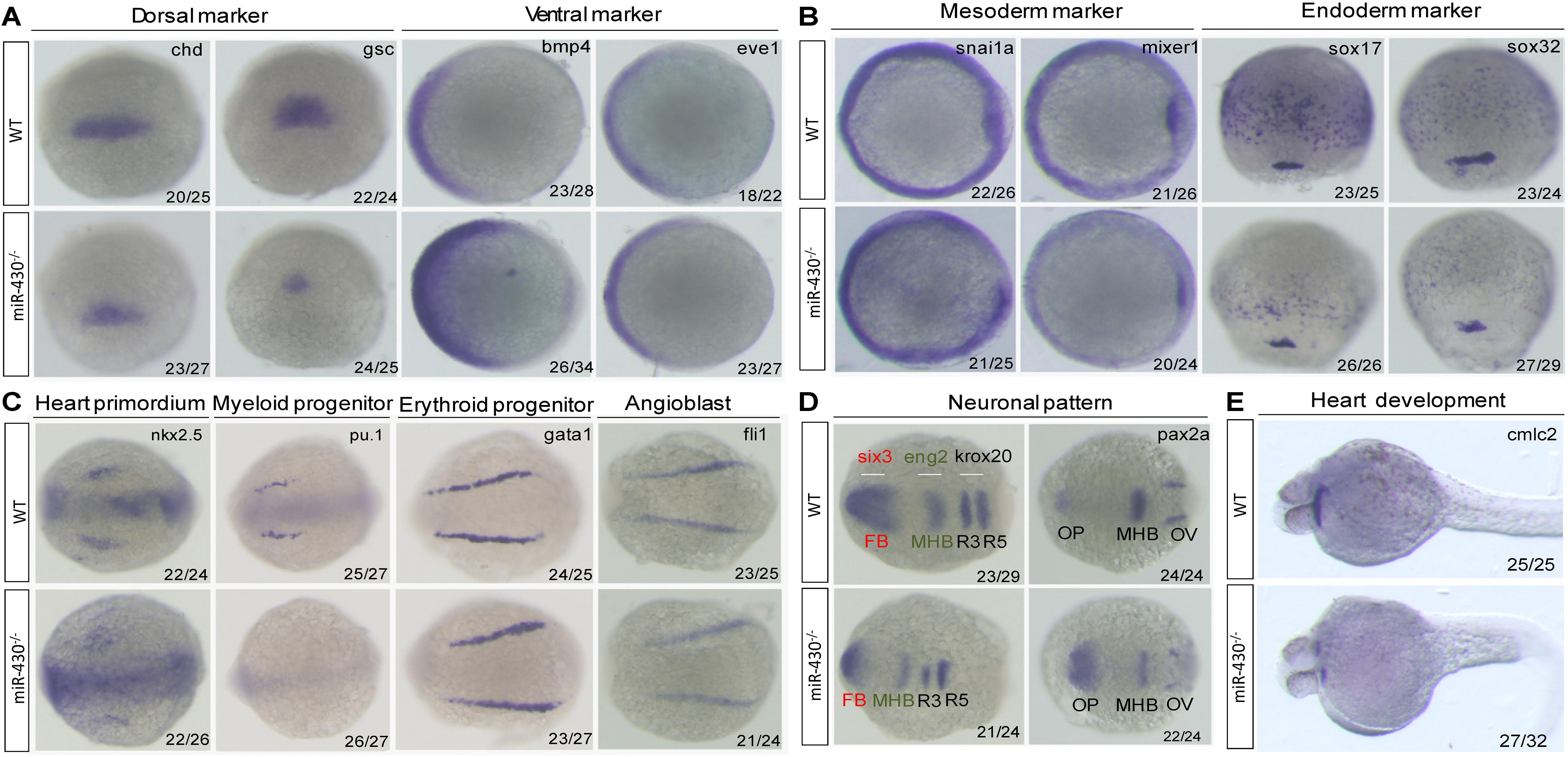
Figure 3. Marker gene analysis. Marker gene expression of the WT and miR-430–/– mutant was analyzed at shield stage (A), 75% epiboly (B), 6-somite (C,D), and 36 hpf (E). Marker genes of axis formation (A), germ layer formation (B), organ progenitor formation, (C) neuronal pattern (D), and heart formation (E) were analyzed in the miR-430–/– mutants. The ratios of the observed phenotypes were indicated. FB, forebrain; MHB, midbrain hindbrain boundary; OV, otic vesicle; OP, optic placode; R3, rhombomere 3; R5, rhombomere 5.
To identify miR-430 targets, we collected the WT, miR-430–/– and rescued embryos at shield stage and performed transcriptome sequencing (Supplementary Table S2). The transcriptome data were validated by Q-PCR (Supplementary Figure S4). We reasoned that the miR-430 regulated genes would be up-regulated in the miR-430–/– mutants but down-regulated in the rescued embryos (miR-430–/– + miR-430 mimics) and vice versa. We have identified 803 up-regulated genes and 421 down-regulated genes (miR-430–/– VS WT, fold change > 1.5, p < 0.05; rescued VS miR-430–/–, fold change < 0.8, p < 0.05) (Figure 4A and Supplementary Table S2). About half of the up-regulated genes possess canonical miR-430 binding sites (8er, 7er-m8, and 7er-1A) in their 3′UTR (1) (Figure 4B and Supplementary Table S2). The miR-430 recognizing sites were enriched in the 3′UTR of up-regulated genes but depleted in the down-regulated genes (Figure 4B).
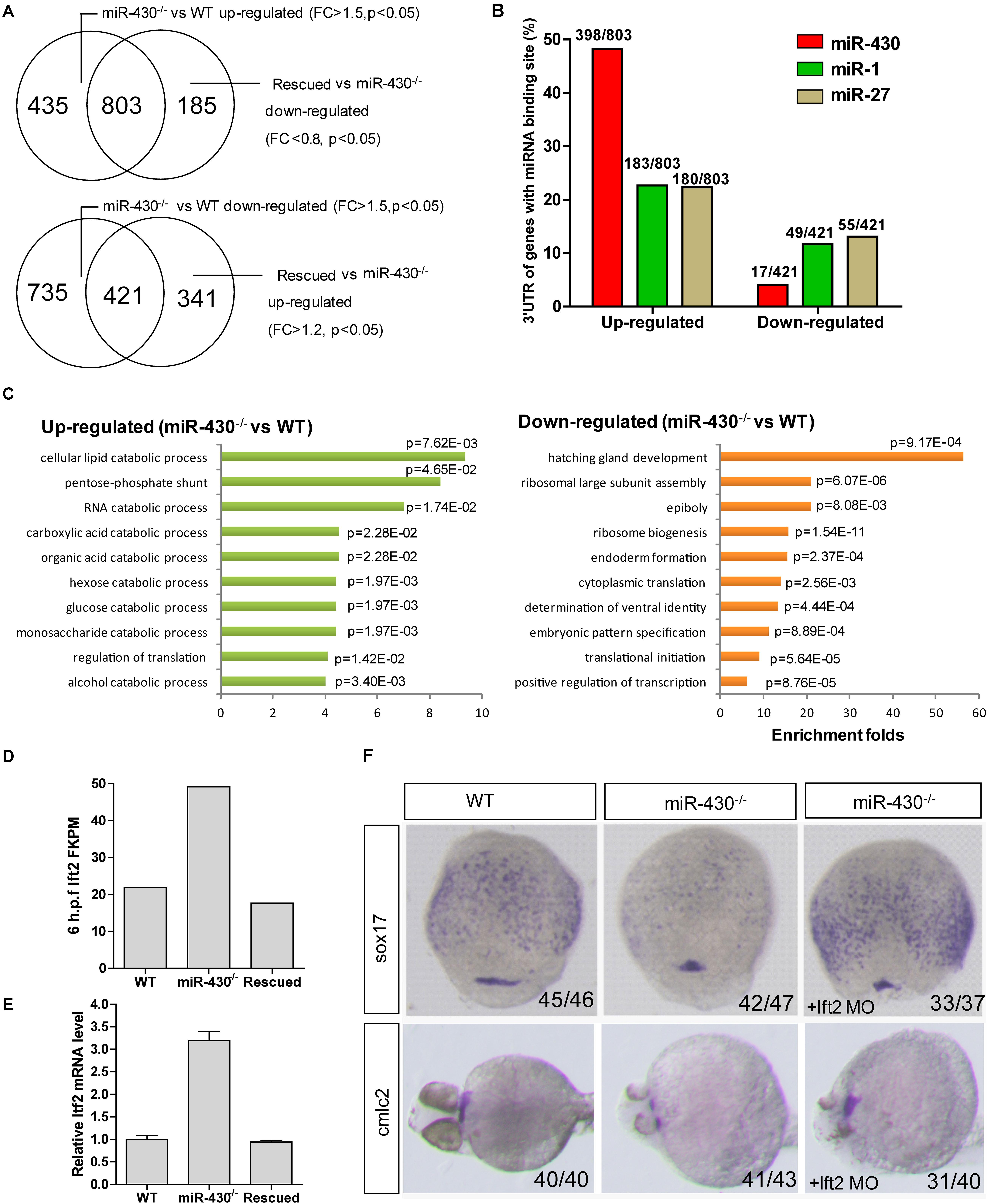
Figure 4. Transcriptome analysis. Total mRNAs of embryos from WT, miR-430–/– mutant and rescued groups were collected at shield stage for transcriptome sequencing. (A) Identification of miR-430-regulated genes. (B) MiR-430 binding site analysis. The ratios indicate the numbers of genes with canonical sites of the indicated miRNA in their 3′UTR/total number of analyzed genes. The miRNA binding sites were predicted by TargetScanFish 6.2. (C) GO analysis of the miR-430 regulated genes, performed by the DAVID system. (D) The expression of lft2 in the transcriptome of the WT, miR-430–/– and rescued embryos. (E) Q-PCR analysis of lft2 mRNA expression levels in the WT, miR-430–/– and rescued group. Data are expressed as mean values ± S.E.M (n = 4). (F) MO knockdown of lft2 partially rescued the miR-430-deficient phenotypes. The embryos were collected at 75% epiboly and 36 hpf for WISH analysis of marker gene expression.
We next analyzed the differentially expressed genes by GO enrichment analysis. The up-regulated genes were enriched in the catabolic processes whereas the down-regulated genes were enriched in the regulation of development (Figure 4C). Genes involved in the epiboly (pou3f1), embryonic pattern (chd, gsc, flh, zic3, spl5, and tcf7l1) and germ layer specification (gata5, snail1a, bon, sox17, and sox32) were down-regulated whereas genes involved in the regulation of cell movements (sdf-1 and cap1) were increased in the miR-430–/– mutants.
Nodal signaling plays a critical role in the induction of mesendoderm cell fate specification (Schier and Talbot, 2005). Consistent with the reported functional roles of miR-430 in the regulation of nodal signaling, we found that the down-stream genes of the nodal pathway (e.g., chd and gata5) were decreased and mesendoderm development was disrupted in the mutant. The expression level of the nodal antagonist, lft2, was markedly increased in the miR-430–/– mutants (Figures 4D,E). We tested whether knockdown of lft2 could rescue some phenotypes of the mutants. Lft2 knockdown rescued endoderm development and the cardiac bifida (Figure 4F). These data indicate that hundreds of targets are regulated by miR-430 and regulation of nodal signaling by miR-430 is required for endoderm development.
Using the MZdicer embryos, previous studies suggested that miR-430 is required for clearance of maternally provided transcripts (Giraldez et al., 2006; Lee et al., 2013). We further explored this hypothesis using the transcriptome data (Supplementary Table S2). We grouped the miR-430-regulated genes into three categories: (i) maternally provided transcripts that are rapidly degraded after ZGA; (ii) the steadily expressed genes during early development; and (iii) the up-regulated genes after ZGA (Figure 5A). Compared to the control data set (genes not significantly changed in the miR-430–/– mutants), the up-regulated genes in the miR-430–/– mutants were significantly enriched in category (i) (p = 3.0147E-41, Chi-squared test) but depleted in category (iii) (p = 2.6503E-17, Chi-squared test) (Figures 5B,C), supporting that miR-430 is required for the clearance of maternally provided transcripts. Conversely, the down-regulated genes in the miR-430–/– mutants were significantly enriched in the category (iii) (p = 7.1046E-18, Chi-squared test) but depleted in category (i) (p = 3.8758E-28, Chi-squared test) (Figure 5D), demonstrating that the zygotic genome expressed genes were not fully activated in the miR-430–/– mutants. These data indicate that hundreds of maternally provided transcripts were not timely eliminated and ZGA was impeded upon loss-of-function of miR-430, demonstrating a critical role for miR-430 during MZT (Supplementary Figure S5).
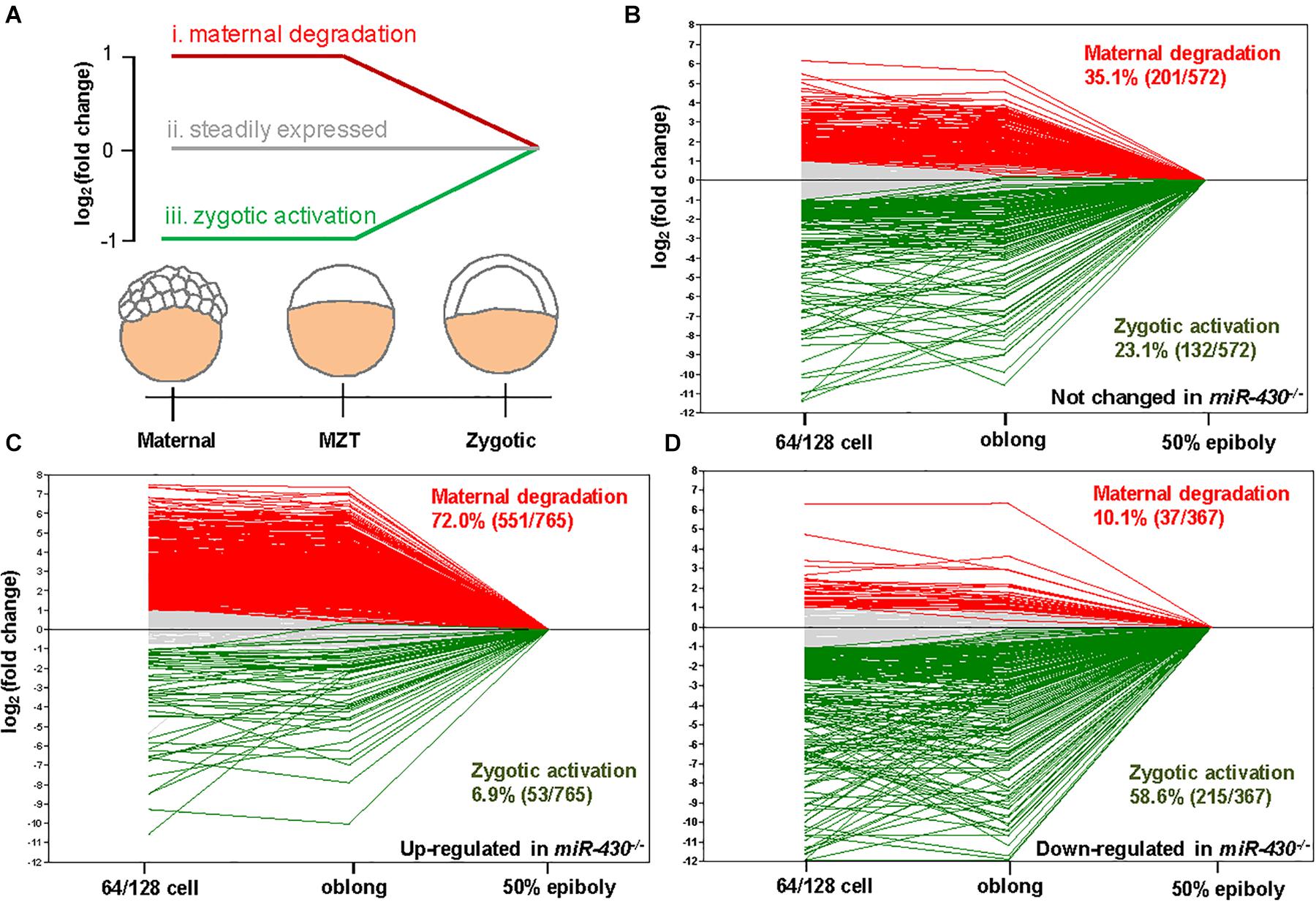
Figure 5. miR-430 is required for the degradation of maternally provided transcripts and activation of zygotic gene expression. (A) Schematic representation of the expression profiles of miR-430-regulated genes during MZT (64/128-cell stages, oblong and 50 epiboly). The miR-430-regulated genes were grouped into three categories based on their expression profiles. The expression profiles of genes in the control set (B), up-regulated set (C), and down-regulated set (D) of the miR-430 mutants during MZT.
The nature of these miR-430-regulated maternally provided transcripts was less explored. We found that Nanog, Dicer1, Dgcr8, and AGO transcripts are maternally provided transcripts that were degraded rapidly after ZGA (Figures 6A,C). Interestingly, these maternally provided transcripts were significantly increased in the miR-430–/– mutants (Figures 6B,C and Supplementary Figure S6), suggesting that miR-430 is required for the clearance of these maternally provided transcripts which were actually required for the activation, processing and function of miR-430. MiR-430 binding sites were identified in the 3′UTR of these genes except for Nanog (Supplementary Table S3), suggesting that miR-430 may directly regulate these transcripts through these binding sites. We have tested one of the targets, Dicer1, which possesses two miR-430 sites in its 3′UTR (Figure 6D and Supplementary Figure S7). We found that fluorescence signal from the GFP sensor with WT UTR (GFP-Dicer1-3′UTR-WT) was suppressed in the WT embryos but not in the mutant embryos (Figure 6D). Moreover, mutation of the miR-430 binding sites abolished the miR-430 mediated Dicer1 suppression (Figure 6D). These data indicate that the existence of negative regulatory loop between the maternally provided transcripts and miR-430: that the maternally provided transcripts are required for miR-430 biogenesis, whereas miR-430 is required for the clearance of these maternally provided transcripts (Figure 6E).
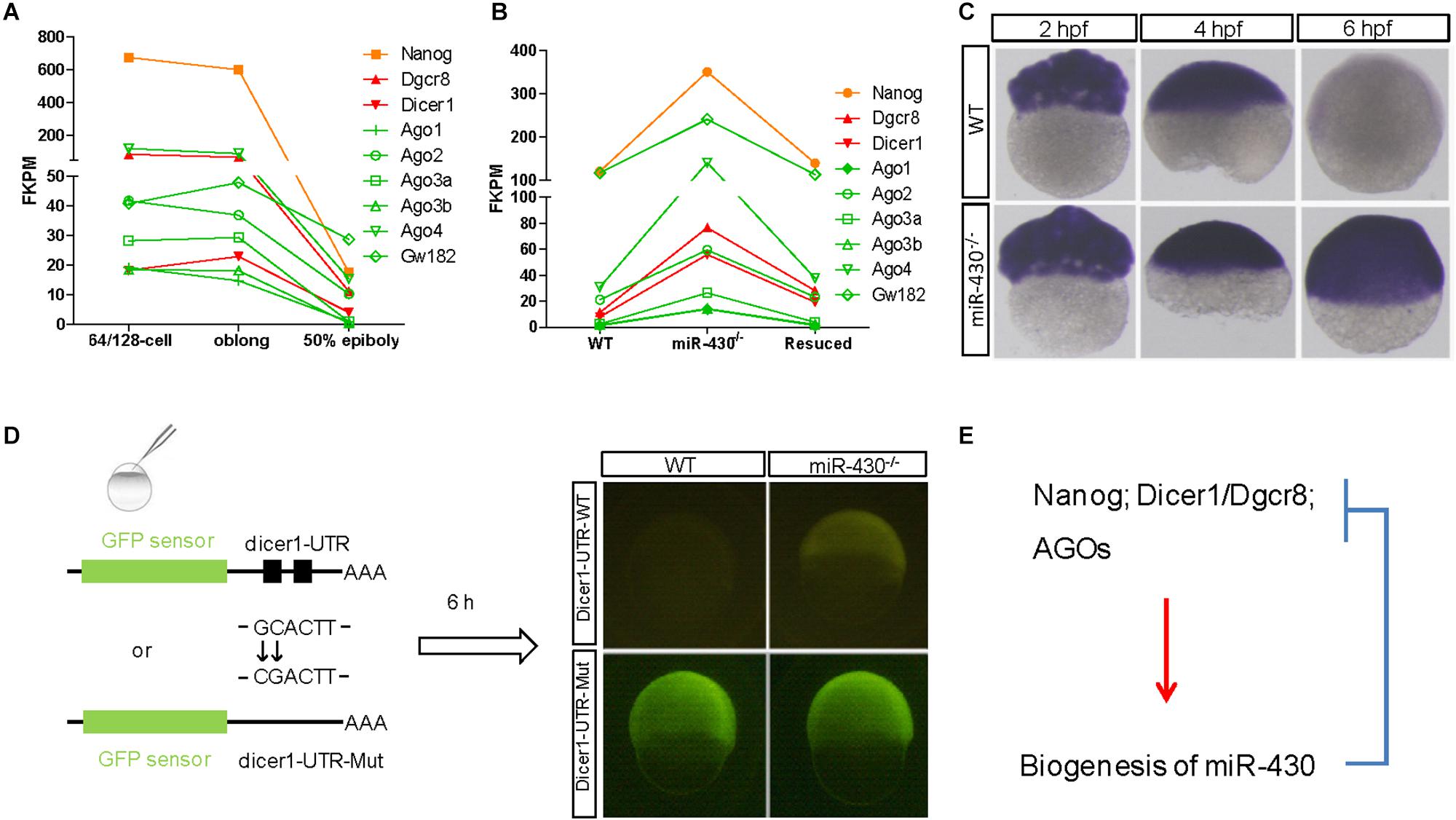
Figure 6. A reciprocal regulatory loop for miR-430 in the clearance of maternal transcript. (A) The transcripts of Nanog, Dicer1, Dgcr8, and AGOs were maternally provided. (B) The maternally provided transcripts Nanog, Dicer1, Dgcr8, and AGOs were significantly increased in the miR-430–/– mutants and down-regulated in the rescued embryos at shield stage. (C) WISH detection of Dicer1 transcripts in the WT and miR-430–/– mutants. The Dicer1 transcripts were maternally provided and degraded at shield stage in the WT but not miR-430-deficient embryos. (D) Repression of Dicer1 by miR-430 thorough miR-430 binding sites in their 3′UTR. Two miR-430 binding sites (GCACTT) were identified in the 3′UTR of Dicer1. GFP sensors with Dicer-3′UTR-WT and Dicer-3′UTR-Mut (two-nucleotide mutations in the miR-430 binding sites) were injected into embryos and fluorescence was examined at 6 hpf. (E) A proposed reciprocal regulatory loop between maternally provided transcripts and miR-430.
In contrast to transcription factors, most gene knockout studies of miRNA produced subtle phenotypes (Alvarez-Saavedra and Horvitz, 2010). In this study, we demonstrated that genetic deletion of the miR-430 cluster leads to embryonic lethal phenotypes with severe developmental defects. Although such a large deletion may disrupt genomic topological associated domains, but the consequence of these disruptions are not clear. The mutated phenotype can be well-rescued by miR-430, indicating that the large deletion event may not directly contribute to the observed phenotypes. Some rescued embryos could even survive to adulthood, indicating that miR-430 is mainly functional during early embryonic development. Moreover, most cell types are specified in the miR-430–/– mutants, but the myeloid progenitor cells failed to specify, suggesting that miR-430 is essential for specific cell type emergence.
Consistent with the fact that miR-430 could partially rescue MZdgcr8 and MZdicer phenotypes, genetic deletion of miR-430 generates similar phenotypes as the MZdgcr8 and MZdicer in several aspects: developmental delay, affected cardiovascular and neural systems as well as germ layer specification and cell movement defects. Cardiac bifida and tail blister was observed in both the miR-430–/– and MZdgcr8 mutants but not in the MZdicer1 mutants. These phenotypes were also observed in other mutants with disrupted endoderm development and sphingosine-1-phosphate (S1P) signaling pathway (Reiter et al., 2001; Kawahara et al., 2009). In zebrafish, mutations in the S1P transporter (spns2), the S1P receptor (slpr2) and Gα13 disrupt myocardial migration, leading to cardiac bifida (Kupperman et al., 2000; Kawahara et al., 2009; Ye and Lin, 2013). Interestingly, miR-430 binding sites were predicted in the 3′UTR of both spns2 and slpr2. Whether miR-430 could directly regulate these targets awaits further investigation.
miRNAs have been proposed to be required for maternal RNA clearance across species (Giraldez et al., 2006; Bushati et al., 2008; Lund et al., 2009). We tested this hypothesis using the miR-430–/– mutants in this study. Hundreds of maternal mRNAs were not timely degraded in the miR-430 deficient embryos. Moreover, many zygotic genome-expressed genes are not fully activated in the miR-430–/– mutants. The prolonged presence of maternally provided transcripts may prevent full ZGA. A similar phenomenon was reported when the m6A-dependent RNA decay pathway was disrupted in zebrafish embryos, in which the maternally provided transcripts were not timely degraded and ZGA was impeded (Zhao et al., 2017). These data demonstrate that miR-430 is not only required for the clearance of maternal RNA but also contribute to ZGA.
In this study, we found that hundreds of maternally provided transcripts are regulated by miR-430. One interesting observation is that Nanog, Dicer1, Dgcr8, and AGOs, which are required for transcription, processing and targeting of miR-430 were regulated by miR-430, suggesting that a reciprocal regulatory loop existed between the maternal transcripts and miR-430. By analysis of gene enrichment in a particular pathway, we further found that many of the miR-430 targeted maternally provided transcripts are involved in the catabolic processes (Figure 4C), suggesting that these maternally provided transcripts may play important roles in controlling their metabolism in the embryo. After ZGA, the miR-430 was produced and these maternally provided transcripts were degraded, thus miR-430 may facilitate a shift in metabolic state during MZT.
In summary, the translation of maternally provided transcripts promotes miR-430 biogenesis shortly after fertilization and the synthesized miR-430 in turn promotes termination of the maternal program by inducing degradation of the maternally provided transcripts. Moreover, miR-430s also promote the zygotic program by regulating developmental pathways that are required for cell movement, germ layer specification, axis patterning and organ progenitor formation in the embryos. These results demonstrate that miR-430 is required for proper degradation of maternally provided transcripts and impact the later embryonic development.
NCBI Sequence Read Archive (SRA) reference number: SRX5707650 is the reference series for our publication: https://www.ncbi.nlm.nih.gov/sra/SRX5707650.
The animal study was reviewed and approved by Animal Experimentation Ethics Committee of the Chinese University of Hong Kong.
YL, IH, and ZZ analyzed the genotypes and phenotypes. YL, YS, and ZZ carried out marker gene analysis. YL, JL, and XW performed transcriptome experiments. YL, MC, and CC conceived the research and designed the experiments and wrote the manuscript. All authors contributed to the article and approved the submitted version.
This work was funded by the General Research Fund (CUHK 14113119) of the Hong Kong Research Grants Council, the Guangdong Provincial Natural Science Foundation (2017A030313109 and 2018A0303130262), the National Natural Science Foundation of China (31801213), the Shenzhen Science and Technology Innovation Commission (JCYJ20170818101216838 and JCYJ20170818141120342), and the Natural Science Research Project of Shenzhen University (2018016).
YL was employed by the company Shenzhen Zelong Biological Technology Limited Cooperation.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Ms Kathy W. Y. Sham for technical assistance and the Core Laboratories in the School of Biomedical Sciences at the Chinese University of Hong Kong for the provision of equipment and technical support.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2020.00853/full#supplementary-material
FPKM, fragments per kilobase million; hpf, hours post fertilization; lft2, lefty2; MZdgcr8, maternal zygotic dgcr8; MZdicer, maternal-zygotic dicer; MZT, maternal zygotic transition; UTR, untranslated region; ZGA, zygotic genome activation.
Alvarez-Saavedra, E., and Horvitz, H. R. (2010). Many families of C. elegans microRNAs are not essential for development or viability. Curr. Biol. 20, 367–373. doi: 10.1016/j.cub.2009.12.051
Bartel, D. P. (2009). MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233. doi: 10.1016/j.cell.2009.01.002
Bernstein, E., Kim, S. Y., Carmell, M. A., Murchison, E. P., Alcorn, H., Li, M. Z., et al. (2003). Dicer is essential for mouse development. Nat. Genet. 35, 215–217. doi: 10.1038/ng1253
Bushati, N., Stark, A., Brennecke, J., and Cohen, S. M. (2008). Temporal reciprocity of miRNAs and their targets during the maternal-to-zygotic transition in Drosophila. Curr. Biol. 18, 501–506. doi: 10.1016/j.cub.2008.02.081
Choi, W. Y., Giraldez, A. J., and Schier, A. F. (2007). Target protectors reveal dampening and balancing of nodal agonist and antagonist by miR-430. Science 318, 271–274. doi: 10.1126/science.1147535
Fabian, M. R., Sonenberg, N., and Filipowicz, W. (2010). Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 79, 351–379. doi: 10.1146/annurev-biochem-060308-103103
Feldman, B., Concha, M. L., Saude, L., Parsons, M. J., Adams, R. J., Wilson, S. W., et al. (2002). Lefty antagonism of squint is essential for normal gastrulation. Curr. Biol. 12, 2129–2135. doi: 10.1016/s0960-9822(02)01361-1
Friedman, R. C., Farh, K. K., Burge, C. B., and Bartel, D. P. (2009). Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 19, 92–105. doi: 10.1101/gr.082701.108
Giraldez, A. J., Cinalli, R. M., Glasner, M. E., Enright, A. J., Thomson, J. M., Baskerville, S., et al. (2005). MicroRNAs regulate brain morphogenesis in zebrafish. Science 308, 833–838. doi: 10.1126/science.1109020
Giraldez, A. J., Mishima, Y., Rihel, J., Grocock, R. J., Van Dongen, S., Inoue, K., et al. (2006). Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science 312, 75–79. doi: 10.1126/science.1122689
Huang, D. W., Sherman, B. T., and Lempicki, R. A. (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57. doi: 10.1038/nprot.2008.211
Kawahara, A., Nishi, T., Hisano, Y., Fukui, H., Yamaguchi, A., and Mochizuki, N. (2009). The sphingolipid transporter spns2 functions in migration of zebrafish myocardial precursors. Science 323, 524–527. doi: 10.1126/science.1167449
Kim, V. N., Han, J., and Siomi, M. C. (2009). Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 10, 126–139. doi: 10.1038/nrm2632
Kupperman, E., An, S., Osborne, N., Waldron, S., and Stainier, D. Y. (2000). A sphingosine-1-phosphate receptor regulates cell migration during vertebrate heart development. Nature 406, 192–195. doi: 10.1038/35018092
Lee, M. T., Bonneau, A. R., Takacs, C. M., Bazzini, A. A., Divito, K. R., Fleming, E. S., et al. (2013). Nanog, Pou5f1 and SoxB1 activate zygotic gene expression during the maternal-to-zygotic transition. Nature 503, 360–364. doi: 10.1038/nature12632
Lei, Y., Guo, X. G., Liu, Y., Cao, Y., Deng, Y., Chen, X. F., et al. (2012). Efficient targeted gene disruption in Xenopus embryos using engineered transcription activator-like effector nucleases (TALENs). Proc. Natl. Acad. Sci. U.S.A. 109, 17484–17489. doi: 10.1073/pnas.1215421109
Leichsenring, M., Maes, J., Mossner, R., Driever, W., and Onichtchouk, D. (2013). Pou5f1 Transcription factor controls zygotic gene activation in vertebrates. Science 341, 1005–1009. doi: 10.1126/science.1242527
Li, J. Z., Wu, P., Liu, Y., Wang, D. S., and Cheng, C. H. K. (2014). Temporal and spatial expression of the four Igf ligands and two Igf type 1 receptors in zebrafish during early embryonic development. Gene Express. Patt. 15, 104–111. doi: 10.1016/j.gep.2014.05.006
Liu, Y., Luo, D. J., Lei, Y., Hu, W., Zhao, H., and Cheng, C. H. K. (2014). A highly effective TALEN-mediated approach for targeted gene disruption in Xenopus tropicalis and zebrafish. Methods 69, 58–66. doi: 10.1016/j.ymeth.2014.02.011
Liu, Y., Luo, D. J., Zhao, H., Zhu, Z. Y., Hu, W., and Cheng, C. H. K. (2013). Inheritable and precise large genomic deletions of non-coding RNA genes in zebrafish using TALENs. PLoS One 8:e0076387. doi: 10.1371/journal.pone.0076387
Liu, Y., Zhu, Z. Y., Ho, I. H. T., Shi, Y. J., Xie, Y. X., Li, J. Z., et al. (2017). Germline-specific dgcr8 knockout in zebrafish using a BACK approach. Cell. Mol. Life Sci. 74, 2503–2511. doi: 10.1007/s00018-017-2471-7
Lund, E., Liu, M. Z., Hartley, R. S., Sheets, M. D., and Dahlberg, J. E. (2009). Deadenylation of maternal mRNAs mediated by miR-427 in Xenopus laevis embryos. RNA 15, 2351–2363. doi: 10.1261/rna.1882009
Mishima, Y., Giraldez, A. J., Takeda, Y., Fujiwara, T., Sakamoto, H., Schier, A. F., et al. (2006). Differential regulation of germline mRNAs in soma and germ cells by zebrafish miR-430. Curr. Biol. 16, 2135–2142. doi: 10.1016/j.cub.2006.08.086
Reiter, J. F., Kikuchi, Y., and Stainier, D. Y. (2001). Multiple roles for Gata5 in zebrafish endoderm formation. Development 128, 125–135.
Rosa, A., Spagnoli, F. M., and Brivanlou, A. H. (2009). The miR-430/427/302 family controls mesendodermal fate specification via species-specific target selection. Dev. Cell 16, 517–527. doi: 10.1016/j.devcel.2009.02.007
Schier, A. F., and Talbot, W. S. (2005). Molecular genetics of axis formation in zebrafish. Annu. Rev. Genet. 39, 561–613. doi: 10.1146/annurev.genet.37.110801.143752
Staton, A. A., Knaut, H., and Giraldez, A. J. (2011). miRNA regulation of Sdf1 chemokine signaling provides genetic robustness to germ cell migration. Nat. Genet. 43, 204–211. doi: 10.1038/ng.758
Takacs, C. M., and Giraldez, A. J. (2016). miR-430 regulates oriented cell division during neural tube development in zebrafish. Dev. Biol. 409, 442–450. doi: 10.1016/j.ydbio.2015.11.016
Trapnell, C., Roberts, A., Goff, L., Pertea, G., Kim, D., Kelley, D. R., et al. (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578. doi: 10.1038/nprot.2012.016
Ulitsky, I., Shkumatava, A., Jan, C. H., Subtelny, A. O., Koppstein, D., Bell, G. W., et al. (2012). Extensive alternative polyadenylation during zebrafish development. Genome Res. 22, 2054–2066. doi: 10.1101/gr.139733.112
van Boxtel, A. L., Chesebro, J. E., Heliot, C., Ramel, M. C., Stone, R. K., and Hill, C. S. (2015). A temporal window for signal activation dictates the dimensions of a nodal signaling domain. Dev. Cell 35, 175–185. doi: 10.1016/j.devcel.2015.09.014
Wang, Y., Medvid, R., Melton, C., Jaenisch, R., and Blelloch, R. (2007). DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat. Genet. 39, 380–385. doi: 10.1038/ng1969
Wienholds, E., Koudijs, M. J., Van Eeden, F. J. M., Cuppen, E., and Plasterk, R. H. A. (2003). The microRNA-producing enzyme Dicer1 is essential for zebrafish development. Nat. Genet. 35, 217–218. doi: 10.1038/ng1251
Wylie, A. D., Fleming, J. A. G. W., Whitener, A. E., and Lekven, A. C. (2014). Post-transcriptional regulation of wnt8a is essential to zebrafish axis development. Dev. Biol. 386, 53–63. doi: 10.1016/j.ydbio.2013.12.003
Yang, H. X., Zhou, Y., Gu, J. L., Xie, S. Y., Xu, Y., Zhu, G. F., et al. (2013). Deep mRNA sequencing analysis to capture the transcriptome landscape of zebrafish embryos and larvae. PLoS One 8:e0064058. doi: 10.1371/journal.pone.0064058
Ye, D., and Lin, F. (2013). S1pr2/Galpha13 signaling controls myocardial migration by regulating endoderm convergence. Development 140, 789–799. doi: 10.1242/dev.085340
Zhao, B. X. S., Wang, X., Beadell, A. V., Lu, Z. K., Shi, H. L., Kuuspalu, A., et al. (2017). m(6)A-dependent maternal mRNA clearance facilitates zebrafish maternal-to-zygotic transition. Nature 542, 475–478. doi: 10.1038/nature21355
Keywords: small regulatory RNAs, miR-430, genome editing, gastrulation, cell fate
Citation: Liu Y, Zhu Z, Ho IHT, Shi Y, Li J, Wang X, Chan MTV and Cheng CHK (2020) Genetic Deletion of miR-430 Disrupts Maternal-Zygotic Transition and Embryonic Body Plan. Front. Genet. 11:853. doi: 10.3389/fgene.2020.00853
Received: 02 June 2020; Accepted: 13 July 2020;
Published: 04 August 2020.
Edited by:
Sandeep Kumar, Emory University, United StatesReviewed by:
Vladimir Korzh, International Institute of Molecular and Cell Biology in Warsaw (IIMCB), PolandCopyright © 2020 Liu, Zhu, Ho, Shi, Li, Wang, Chan and Cheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Christopher H. K. Cheng, Y2hrY2hlbmdAY3Voay5lZHUuaGs=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.