- 1IRCCS Centro Neurolesi “Bonino-Pulejo”, Messina, Italy
- 2Department of Medical, Oral and Biotechnological Sciences, University “G. d’Annunzio” Chieti and Pescara, Chieti, Italy
- 3ASL02 Lanciano-Vasto-Chieti, “Ss. Annunziata” Hospital, Chieti, Italy
The human Periodontal Ligament Stem Cells (hPDLSCs) exhibit self-renewal capacity and clonogenicity potential. The Extracellular Vesicles (EVs) secreted by hPDLSCs are particles containing lipids, proteins, mRNAs, and non-coding RNAs, among which microRNAs, that are important in intercellular communication. The purpose of this study was the analysis of the non-coding RNAs contained in the EVs derived from hPDLSCs using Next Generation Sequencing. Moreover, our data were enriched using bioinformatic tools. The analysis highlighted the presence of non-coding RNAs and five microRNAs: MIR24-2, MIR142, MIR335, MIR490, and MIR296. Our results show that these miRNAs target the genes classified in two terms of the Gene Ontology: “Ras protein signal transduction” and “Actin/microtubule cytoskeleton organization.” Noteworthy, the in-deep analysis of our EVs highlights that the miRNAs could be implicated in the silencing of proto-oncogenes involved in 12 different types of tumors.
Introduction
The human Periodontal Ligament Stem Cells (hPDLSCs) are mesenchymal stem cells that can be easily harvested from periodontal tissue. Non-invasive surgery during standard dental scaling does not entail any additional risk to the donor (Chiricosta et al., 2019). The hPDLSCs showed self-renewal capacity, differentiation and immunomodulatory proprieties (Eleuterio et al., 2013). The Extracellular Vesicles (EVs) are particles delimited by a lipid bilayer capable of crossing biological barriers and being internalized in the target cells so that they play an essential role in intercellular communication (Valadi et al., 2007; Pizzicannella et al., 2019b). EVs contain different macromolecules such as lipids, proteins, DNA, mRNA and non-coding RNAs (ncRNAs) among which microRNAs (or miRNAs) (Zhang et al., 2019). The dynamic light-scattering analysis, previously demonstrated from our research group, highlighted the presence of two different populations of EVs derived from hPDLSCs. The average diameter of the EVs populations is between 90 ± 20 nm and 1,200 ± 400 nm and they have the same ζ-potential of −10.7 ± 0.9 mV. The tapping-mode topographic 3D atomic force-microscopy measurements showed that EVs derived from hPDLSCs have a globular shape with a central depression and a relatively smooth surface (Diomede et al., 2018).
The ncRNAs are circulating RNAs that represent the 99% of total RNAs and can be classified according with length (small 18–200 nucleotides and long >200), with function (ribosomal RNAs and transfer RNAs), or with regulation [miRNAs and long non-coding RNAs (lncRNAs)] (Dozmorov et al., 2013; Palazzo and Lee, 2015). The ncRNAs are involved in gene regulation by RNA interference, RNA modification or spliceosomal cycle. Specifically, miRNAs are small ncRNAs (18–25 nucleotides in length) that prevent the translation of mRNAs and consequently contribute to define the amount of proteins inside the cell (Bartel, 2004). miRNAs play an important role in physiological functions such as cell-cell communication, cell proliferation and vascularization. The ncRNAs and miRNAs are implicated in various diseases including cancer, in which they play an important biological role (Slaby et al., 2017).
Therefore, in this study, using Next Generation Sequencing (NGS), we want to analyze the ncRNAs and miRNAs contained in EVs derived from hPDLSCs in the early stages of stemness. Particularly, we investigated the possible implications of ncRNAs in the biological processes and in the proto-oncogenes modulation.
Materials and Methods
Culture and Extraction of the hPDLSCs
All the subjects that were involved in the study gave their informed consent. The protocol executed for this study was approved by the Medical Ethics Committee at the Medical School, “G. d’Annunzio” University of Chieti-Pescara, Chieti, Italy (n°266 17 April 2014) and it is in accordance with the Declaration of Helsinki. Five patients in healthy general conditions were chosen for tooth removal for orthodontic purposes. Subsequently, the cells were cultured in the Mesenchymal Stem Cell Growth Medium Chemically Defined (MSCGM-CD) Bulletin medium (Lonza, Basel, Switzerland). In order to minimize exposure to the non-human substances and facilitate the growth of human MSCs, the MSCGM-CD was changed twice a week. Cells isolation and characterization were performed as previously described (Rajan et al., 2016; Trubiani et al., 2016). The hPDLSCs were collected and washed with PBS (Lonza) several times. Finally, they were cultured at 37°C with 5% of CO2 with the MSCGM-CD medium.
Isolation of the hPDLSCs-Derived EVs
The hPDLSCs, isolated from each patient, were collected from conditioned medium (CM; 10 mL) at passage 2 after 48 h of incubation. For 15 min, the CM was centrifuged at 3,000 × g in order to remove debris and suspended cells. EVs were extracted using the commercial agglutinant ExoQuick TC (System Biosciences, Euroclone SpA, Milan, Italy). Specifically, 10 mL of CM of the hPDLSCs were mixed with 2 mL ExoQuick TC and the whole was incubated at 4°C overnight without rotation. Subsequently, in order to deposit the EVs, a centrifugation step was carried out for 30 min at 1,500 × g. Finally, the pellets were resuspended in 200 μL of PBS (Ca2+ and Mg2+). The EVs, obtained from hPDLSCs of each patient, were used for transcriptomic analyses.
RNA Extraction and Non-coding Analysis
The RNA extraction and processing were conducted as previously reported (Silvestro et al., 2020). Briefly, the Total Exosome RNA and Protein Isolation Kit (catalog #4478545; Thermo Fisher Scientific, Rockford, IL, United States) was used to isolate the RNA following the manufacturer’s protocol and 30 μL of RNA solution were collected from each sample (Pizzicannella et al., 2019a).
The manufacturer instructions and the TruSeq RNA Exome protocol (Illumina, San Diego, CA, United States) were followed in order to prepare the library. The normalized libraries were analyzed using the MiSeq instrument (Illumina) in a single read mode. For the NGS analysis, the RNA extracted from the EVs of each patient was repeated in triplicate.
The software fastQC (Babraham Institute, Cambridge, United Kingdom) was used to perform the quality check of the reads. Then, the adapters and the low-quality bases were identified and removed by the software Trimmomatic (Usadel Lab, Aachen, Germany) (Bolger et al., 2014). The “Homo Sapiens” reference genome, available from the University of California Santa Cruz website1, was used to align the reads taking advantage of the software Spliced Transcripts Alignment to a Reference RNA-seq aligner (STAR) (Dobin et al., 2013). Finally, the Cufflinks software (Trapnell Lab, Washington, DC, United States), version 2.2.1, was used to assign the gene symbol to each transcript (Trapnell et al., 2013). In order to study the transcripts, the whole set was classified with HUGO Gene Nomenclature Committee website2 (Stelzer et al., 2016) using the “non-coding RNAs” group (475). Specifically, the genes that are targeted by the identified miRNAs were analyzed with TargetScanHuman (version 7.2) (Agarwal et al., 2015). Moreover, the target genes were enriched with the “Biological Process” terms included in the “Gene Ontology” using PANTHER. Finally, the Human microRNA Disease Database (HMDD) v3.2 was used in order to associate the genes target of our miRNAs to the disease in which their expression is deregulated. All the plots were depicted with the software R.
Results
Evaluation of Non-coding Transcripts of EVs Derived From hPDLSCs
The analysis of EVs derived from hPDLSCs reveals 955 non-coding transcripts (Supplementary Table S1). The HUGO website characterizes 212 transcripts distributed in 11 groups as showed in Figure 1. The most of the non-coding transcripts of EVs belong to the “Antisense RNAs” (109), to the “Long intergenic non-protein coding RNAs” (40) and to the “Long non-coding RNAs with non-systematic symbols” (25). The remaining transcripts are included in smaller categories. Our EVs contain also 5 miRNAs: MIR24-2, MIR142, MIR296, MIR335, MIR490. We studied the biological roles of our miRNAs using TargetScanHuman (Agarwal et al., 2015) that associates each miRNA to its target genes. We used the database PANTHER (Thomas et al., 2003) to visualize all the “Biological Process” terms in which the target genes are involved (Figure 2). The deepest terms that better characterize our transcripts are related to the cytoskeletal organization (“Actin/microtubule cytoskeleton organization”) and the signal transduction mediated by Ras protein (“Ras protein signal transduction”). PANTHER was also used to associate the genes with the pathway in which they are involved. In particular, the most characterized pathways include the genes CDC42, RAC1, RHOC, RHOA, RHOJ, SPRY3, PFN2, WASL, TUBB, TUBB4B, SSH3, SSH1, ARPC5L, PARVA, RHOQ. The first five genes are in common to “Actin/microtubule cytoskeleton organization” and “Ras protein signal transduction.” SPRY3 is found only in the pathway related to Ras protein while the other genes are exclusively related to the cytoskeletal organization. Moreover, we inspected the amount of genes that are simultaneously targeted by our miRNAs as represented in the Venn Diagram in Figure 3. There is no gene that is targeted by all the miRNAs. Nevertheless, we observed that one gene (GABRB1) is not targeted just by MIR142, one gene (ONECUT2) just by MIR24, one gene (CNNM2) just by MIR490. Furthermore, MIR-24 targets the most of the genes (1,500) immediately followed by MIR142 (1,250). MIR490, MIR335, and MIR296 target exclusively less than 200 genes each. Furthermore, using HMDD (Huang et al., 2019) we reported in Table 1 the associations found among our miRNAs and the target genes deregulated in specific diseases.
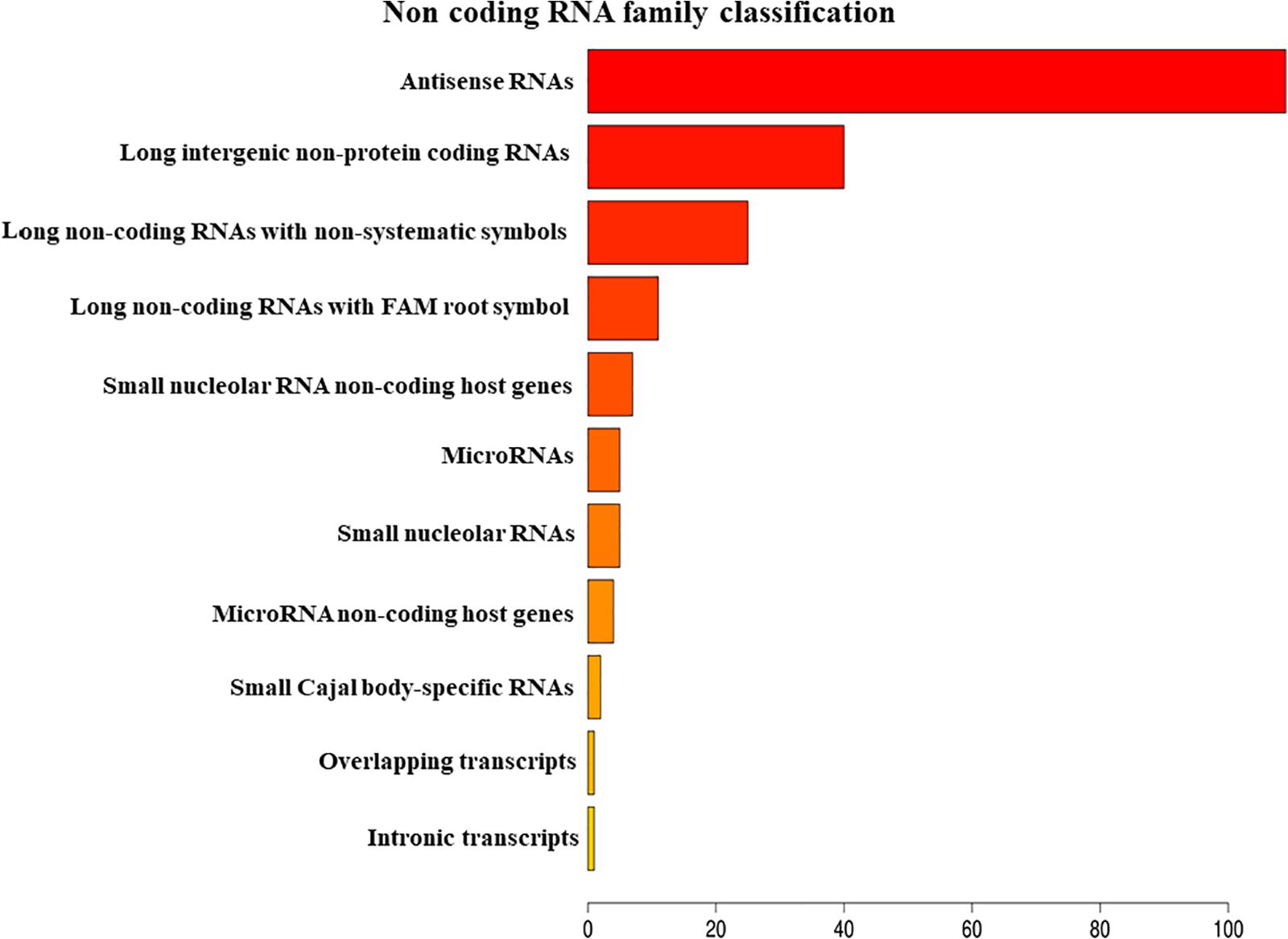
Figure 1. Classification of the non-coding RNAs families contained in EVs belong using HUGO database.
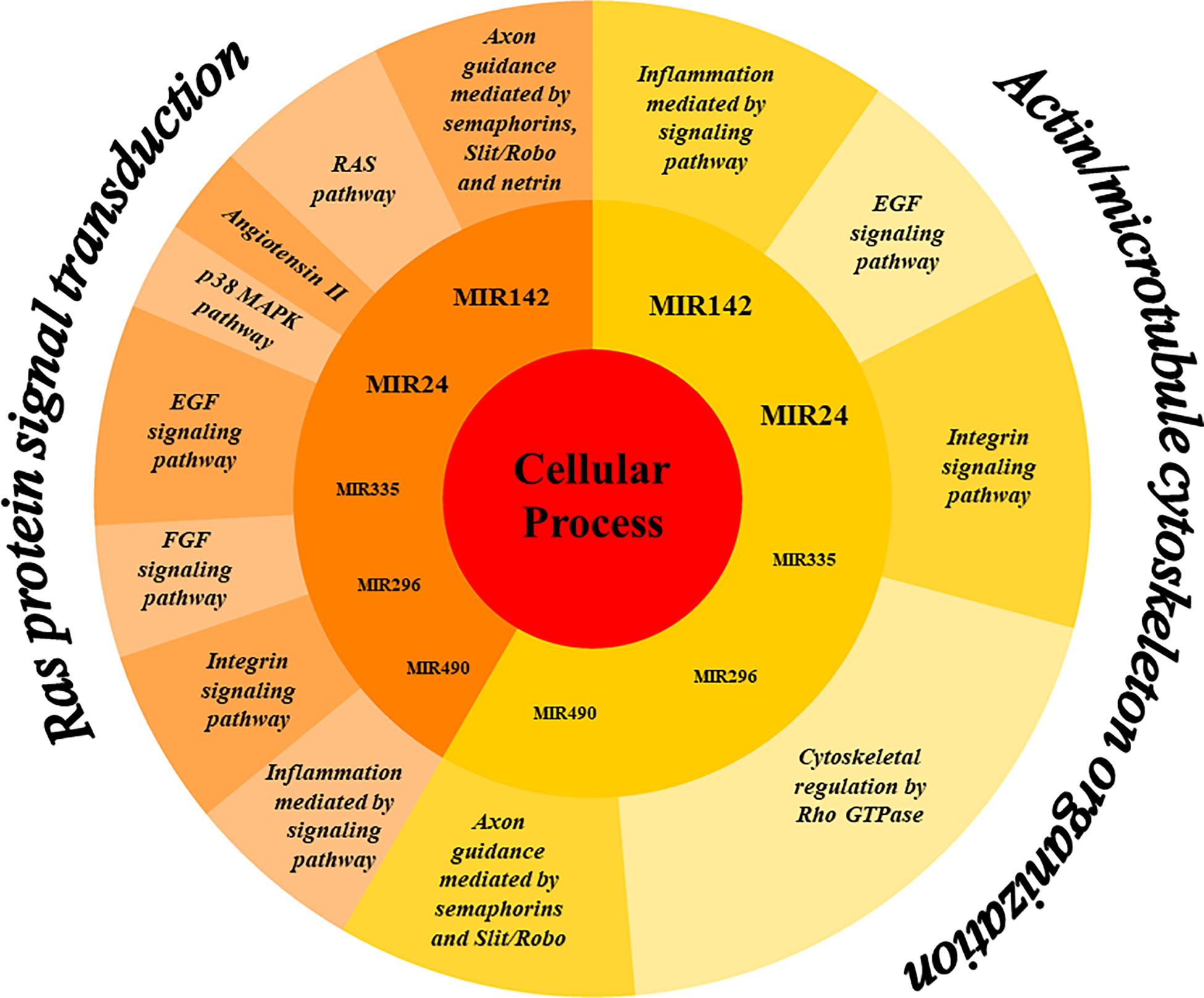
Figure 2. Representation of the biological processes in which the genes targeted by the miRNAs in EVs are mainly involved. The miRNAs MIR24, MIR142, MIR335, MIR296, and MIR490 mainly target genes belonging to the “Cellular processes” category of the Gene Ontology. More specifically, the genes fall into two sub-categories “Ras protein signal transduction” and “Actin/microtubule cytoskeleton organization.” The genes involved in these subprocesses are characteristic of the pathways represented in the outer ring.
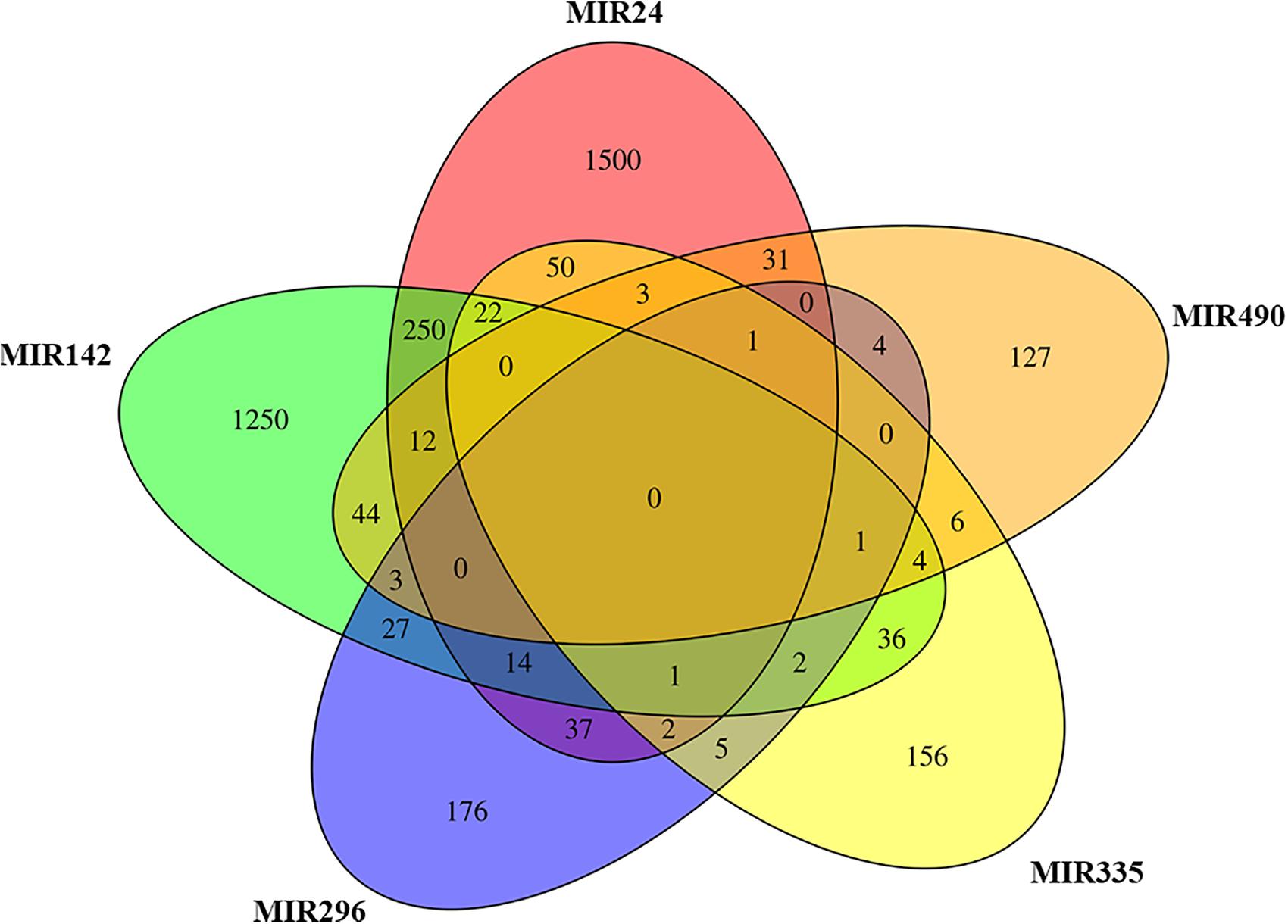
Figure 3. Venn diagram. The plot represents for each miRNA in EVs the genes that inhibits in comparison to the others. MIR24 and MIR142 target exclusively the highest number of genes.
Discussion
EVs are phospholipids membrane-enclosed organizations that carry proteins, lipids and nucleic acids including mRNAs and ncRNAs (Trubiani et al., 2019).
In this study, we analyzed the ncRNAs contained in EVs derived from hPDLSCs at early stage of stemness with a focus on miRNAs.
Our results show that the ncRNAs contained in EVs are classified in 11 families (Figure 1). The most represented class of ncRNAs is the “Antisense RNAs,” small molecules that regulate gene’s expression by binding the complementary mRNAs (Xu et al., 2018). Our results show also the presence of lncRNAs, an important class of transcripts that indirectly regulates the transcription recruiting transcription factors and affecting mRNA stability (Fatima and Nawaz, 2017). EVs transport lncRNAs from one cell to another and can induce epigenetic modifications in the receiving cells (Fatima and Nawaz, 2017). Other classes of ncRNAs represented in our EVs are the small nucleolar RNAs and miRNAs. The small nucleolar RNAs are involved in the chemical alteration of different RNAs and in the regulation of the alternative splicing in pre-mRNAs (Decatur and Fournier, 2002; Falaleeva et al., 2016; Sharma et al., 2016).
Among ncRNAs, miRNAs are the most studied since they can regulate the expression of 60% of the human genes. Specifically, miRNAs are post-transcriptional regulators that bind complementary mRNA and consequently reduce protein expression. miRNAs, released from the EVs, can determine the cell fate through their participation in differentiation and in the regulation of reprogramming processes (Friedman et al., 2009; Fatima and Nawaz, 2017; Mens and Ghanbari, 2018). In the last two decades, many studies contextualized the role of miRNAs in cancer development (Rupaimoole and Slack, 2017). Interestingly, in our EVs, we found five miRNAs: MIR24-2, MIR142, MIR335, MIR490, and MIR296.
Our bioinformatic analysis, represented in Figure 3, highlights the amount of target genes that are modulated by our miRNAs. Indeed, both MIR24-2 and MIR142 are able to influence more than 1,000 genes while MIR335, MIR490, and MIR296 less than 200 each. The representation with the Venn Diagram shows that each miRNA can regulate several genes and each gene can be regulated by more than one miRNA. This evidence suggests that MIR24-2 and MIR142 are ones most involved.
The MIR24-2, MIR142, MIR335, MIR490, and MIR296 found in our EVs target the genes mainly involved in “Ras protein signal transduction” and “Actin/microtubule cytoskeleton organization” (Figure 2).
The Ras proteins are GTPases that act as switches recruiting either the Map Kinases or the RHO/RAC family resulting in downstream signaling for transcription, cell cycle or cytoskeletal organization (Bustelo et al., 2007).
The analysis performed using PANTHER database shows that our miRNAs target CDC42, RAC1, RHOA, RHOC, and RHOJ that simultaneously participate in the regulation of “Actin/microtubule cytoskeleton organization” and “Ras protein signal transduction.” These genes encode for smalls GTPases/Rho family that are active when bound to Guanosine Triphosphate (GTP) or inactive when bound to Guanosine Diphosphate (GDP). The CDC42, RAC1, RHOC, and RHOA genes play an important role in cytoskeletal rearrangement, morphology regulation, cell motility, cell adhesion and cell cycle regulation (Aspenström, 2019; Thomas et al., 2019). RHOJ is a gene that encodes for an endothelial member of the Cdc42 subfamily involved in endothelial motility, tubulogenesis and microtubule lumen formation (Leszczynska et al., 2011). Indeed, miRNAs are very important regulators of these GTPase Rho family (Liu et al., 2012).
Additionally, as regards the Gene Ontology term “Ras protein signal transduction,” the miRNAs inside our EVs target the SPRY3 gene, which encodes for the Protein sprouty homolog 3 (Spry3), an intracellular negative regulators of Receptor tyrosine kinase signaling and of the Fibroblast Growth Factor (FGF) pathways (Hacohen et al., 1998). Furthermore, Panagiotaki et al. (2010) showed that Spry3 negatively regulates the release of calcium downstream of brain-derived neurotrophic factor signaling, and consequently cause the inhibition of the axons branching in cultured cortical neurons.
Moreover, the miRNAs present in our EVs regulate the “Actin/microtubule cytoskeleton organization” process by targeting different genes such as TUBB, TUBB4B, PFN2, WASL, SSH3, SSH1, ARPC5L, PARVA, and RHOQ. In detail, TUBB and TUBB4B encode respectively for a beta-tubulin protein and for the beta-4B tubulin chain, both representing structural components of the microtubules. RHOQ encodes for a small GTPase involved in the regulation of the cell shape through the organization of the actin cytoskeleton. SSH1 and SSH3 encode for protein phosphatases that play a role in the regulation of the dynamics of the actin filament. In addition, SSH1 and SSH3 activate the actin-binding protein family ADF/cofilin that promotes the disassembling of the actin filaments (Kurita et al., 2007). ARPC5L gene, part of Arp2/3 complex, encodes for the Actin Related Protein 2/3 Complex Subunit 5 Like. This gene regulates the actin polymerization in the cytoskeleton and generates an actin filament network (Abella et al., 2016). PARVA encodes for the α-parvin protein that is involved in the regulation of the actin cytoskeleton dynamism (Sepulveda and Wu, 2006). PFN2 is an actin-binding protein that regulates the architecture of the synapse and the cytoskeleton (Jeong et al., 2018). WASL, which encodes for Neural Wiskott-Aldrich syndrome protein, appears to be involved in the polymerization of actin. In addition, PFN2 and WASL are highly expressed in neural tissues and participate to the axon guidance in the Slit/Robo pathway. These results show that our EVs contain miRNAs involved in axon guidance and consequently in neural differentiation.
Moreover, we evaluated (using HMDD) how much our miRNAs deregulate oncogenes involved in different kinds of cancers. Noteworthy, our miRNAs are involved at least in 12 different cancers (Table 1) and specifically in their development and progression (Calin and Croce, 2006).
The MIR24-2 encodes miR-24 that plays an important role as a suppressor of the genes E2F2 and MYC. Moreover, by binding the complementary 3′-UTR mRNA of these genes, miR24-2 is involved in the regulation of the cell cycle (Lal et al., 2009). Guo et al. (2012) also showed that the up-regulation of MIR24 leads to morphological changes, low cell proliferation and enhancement of cell invasion potential in laryngeal squamous cell carcinoma by inhibiting transcription of the S100A8 gene. Additionally, the overexpression of MIR24 inhibits osteosarcoma cell proliferation by blocking the transcription of AGPAT2 gene encoding for Lysophosphatidic Acid Acyltransferase β, an enzyme involved in osteosarcoma cell proliferation (Song et al., 2013).
The MIR142 appears to be a potential proto-oncogene suppressor. Indeed, through the transcriptional block of RAC1, a gene that encodes for a GTPase that regulates cell growth and migration and promotes the activation of protein kinases. Wu et al. (2011) showed that MIR142 has also a role in suppressing migration and invasion of hepatocellular carcinoma cells. In addition, MIR142 inhibits the proliferation of pancreatic cancer cells by decreasing the expression of heat shock protein 70 (Mackenzie et al., 2013). It inhibits the growth of cells responsible for colon cancer, hindering the transcription of PROM1 (CD133), ABCG2, and LRG5 genes (Shen et al., 2013). Moreover, by blocking the transcription of the HMGA1 gene, an important gene involved in promoting cancer and increasing invasiveness, Xu et al. (2014) reveals that MIR142 can act as a suppressor in osteosarcoma. Furthermore, as reported by Dickman et al. (2017) the MIR142 is secreted by oral squamous cell carcinoma cells, promoting the growth of the tumor. The release of miR-142-3p also influences the tumor microenvironment, promoting angiogenesis (Dickman et al., 2017). Indeed, in oral squamous cell carcinoma, MIR142 was up-regulated, demonstrating how the differential expression of miRNAs could be useful in the diagnosis of oral cancer (Xu et al., 2019).
HMGA1 gene can be also targeted by MIR296, another miRNA presents in our EVs, supporting the knowledge by which different miRNAs can interact with the same mRNA. Negative regulation of this gene by MIR296 appears to reduce invasiveness in prostate cancer cells (Wei et al., 2011). In addition, MIR296 targeting PIN1, another gene involved in tumor development, suppresses cell proliferation and growth in prostate cancer cells (Lee et al., 2014). In hepatocellular carcinoma, MIR296 inhibits the proliferative capacity and progression of the cell cycle and induces apoptosis blocking the transcription of the FGFR1 proto-oncogene (Wang et al., 2016). Other targets of MIR296 are the proto-oncogenes AKT2 in pancreatic cancer (Li et al., 2017), PLK1 in non-small cell lung cancer (Xu et al., 2016), SP1 in cervical cancer (Lv and Wang, 2018) and CDX1 in gastric cancer (Li et al., 2014). Through the negative regulation of the expression of these genes, MIR296 exerts its action as a tumor suppressor. Therefore, EVs derived from hPDLSCs, through the secretion of MIR296, could act as suppressors of the malignant progression by attenuating the transcription of oncogenic targets. The MIR296, as reported by Severino et al. (2015) was also detected in the in metastases of patients with oral squamous cell carcinoma.
In the transcriptome of our EVs, we found also MIR490 that seems to have the role of tumor suppressor in lung cancer cells A549 and ovarian cancer by blocking the transcription of CCND1 and CDK1 respectively, two important genes involved in cell cycle progression (Gu et al., 2014; Chen et al., 2015). Additionally, as showed by Li et al. (2013), the MIR490 seems to be a suppressor of bladder cancer cell proliferation by blocking the transcription of FOS. Moreover, MIR490 inhibits the expression of HMGA2 and its downregulation appears to affect the development potential of the osteosarcoma (Liu et al., 2015).
MIR335, present in our EVs, also appears to play the role of proto-oncogene suppressor in a wide variety of cancers. It is known that MIR335 negatively regulates the metastasis in gastric cancer by targeting BCL2L2 and SP1 (Xu et al., 2012). Likewise targeting the same BCL2L2 gene, it is capable of inhibiting invasiveness in ovarian cancer (Cao et al., 2013). In neuroblastoma cells, MIR335 downregulates the ROCK1 and MAPK1 genes, involved in the non-canonical TGF-β pathways. This modulation implicates a reduced invasiveness of the neuroblastoma cells. In addition, MIR335 inhibits the mRNA transcribed by LRG1 gene reducing the migration of neuroblastoma cells (Lynch et al., 2012). Similarly, it negatively regulates at the post-transcriptional level the ROCK1 inhibiting tumor cell invasion and migration in osteosarcoma (Wang et al., 2013). Conversely, Kabir et al. (2016) showed that MIR335 is up-regulated in fibroblasts associated with senescent cancer. These cells present in the tumor environment of oral neoplasms. Furthermore, the authors reported that MIR335 is also involved in the development of a secretory phenotype associated with senescence. In this way, MIR335 contributing to the progression of cancer (Kabir et al., 2016).
Interestingly, in our previous manuscript, we have already demonstrated in an analysis of the oral stem cell transcriptome that, following the in vitro expansion, many oncogenes were absent or poorly expressed (Gugliandolo et al., 2017).
Conclusion
The EVs derived from hPDLSCs contain several ncRNAs among which the five miRNAs: MIR24-2, MIR142, MIR296, MIR335, and MIR490. Specifically, these miRNAs regulate genes that are involved in “Ras protein signal transduction” and “Actin/microtubule cytoskeleton organization” processes that regulate cell growth and differentiation during cytokinesis. Moreover, our results demonstrate that MIR24 and MIR142 are the most relevant miRNAs because they target more than 1,000 genes each. Furthermore, all the miRNAs detected in our EVs could have a potential role as proto-oncogenes suppressors. These findings indicated that the EVs obtained from hPDLSCs can be considered as anticancer therapeutic agents. This evidence supports the role of mesenchymal stem cell derivatives in the deregulatory functioning of human carcinogenesis in addition to their use in regenerative medicine.
Data Availability Statement
We uploaded our data to the repository Sequence Read Archive (accession number PRJNA630492).
Ethics Statement
The study involving human participants was approved by the Medical Ethics Committee at the Medical School, “G. d’Annunzio” University of Chieti-Pescara, Chieti, Italy (n°266 17 April 2014) and it is in accordance with the Declaration of Helsinki. All the subjects that were involved in the study gave their informed consent.
Author Contributions
JP and GM performed the hPDLSCs culturing and the isolation of the EVs. AG performed the RNA extraction and library preparation. LC performed the computational and statistical analysis. LC and SS wrote the manuscript. PB, OT, and EM designed the study and revised the manuscript.
Funding
This study was supported by a Current Research Fund 2019, Ministry of Health, Italy.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank the Ministry of Health, Italy.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2020.00582/full#supplementary-material
Abbreviations
EVs, Extracellular Vesicles; FGF, Fibroblast Growth Factor; GDP, Guanosine Diphosphate; GTP, Guanosine Triphosphate; hPDLSCs, human Periodontal Ligament Stem Cells; lncRNAs, long non-coding RNAs; miRNAs, microRNAs; MSCGM-CD, Mesenchymal Stem Cell Growth Medium Chemically Defined; ncRNAs, non-coding RNAs; NGS, Next Generation Sequencing; Spry3, protein sprout homolog 3; STAR, Spliced Transcripts Alignment to a Reference RNA-seq aligner; HMDD, Human microRNA Disease Database.
Footnotes
- ^ http://labshare.cshl.edu/shares/gingeraslab/www-data/dobin/STAR/STARgenomes/Old/ENSEMBL/homo_sapiens/ENSEMBL.homo_sapiens.release-75/
- ^ https://www.genenames.org/
References
Abella, J. V., Galloni, C., Pernier, J., Barry, D. J., Kjaer, S., Carlier, M. F., et al. (2016). Isoform diversity in the Arp2/3 complex determines actin filament dynamics. Nat. Cell Biol. 18, 76–86. doi: 10.1038/ncb3286
Agarwal, V., Bell, G. W., Nam, J. W., and Bartel, D. P. (2015). Predicting effective microRNA target sites in mammalian mRNAs. eLife 4:e05005.
Aspenström, P. (2019). The intrinsic gdp/gtp exchange activities of Cdc42 and rac1 are critical determinants for their specific effects on mobilization of the actin filament system. Cells 8:759. doi: 10.3390/cells8070759
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bustelo, X. R., Sauzeau, V., and Berenjeno, I. M. (2007). GTP−binding proteins of the Rho/Rac family: regulation, effectors and functions in vivo. Bioessays 29, 356–370. doi: 10.1002/bies.20558
Calin, G. A., and Croce, C. M. (2006). MicroRNA signatures in human cancers. Nat. Rev. Cancer 6, 857–866. doi: 10.1038/nrc1997
Cao, J., Cai, J., Huang, D., Han, Q., Yang, Q., Li, T., et al. (2013). miR-335 represents an invasion suppressor gene in ovarian cancer by targeting Bcl-w. Oncol. Rep. 30, 701–706. doi: 10.3892/or.2013.2482
Chen, S., Chen, X., Xiu, Y.-L., Sun, K.-X., and Zhao, Y. (2015). MicroRNA-490-3P targets CDK1 and inhibits ovarian epithelial carcinoma tumorigenesis and progression. Cancer Lett. 362, 122–130. doi: 10.1016/j.canlet.2015.03.029
Chiricosta, L., Gugliandolo, A., Diomede, F., Pizzicannella, J., Trubiani, O., Iori, R., et al. (2019). Moringin pretreatment inhibits the expression of genes involved in mitophagy in the stem cell of the human periodontal ligament. Molecules 24:3217. doi: 10.3390/molecules24183217
Decatur, W. A., and Fournier, M. J. (2002). rRNA modifications and ribosome function. Trends Biochem. Sci. 27, 344–351. doi: 10.1016/s0968-0004(02)02109-6
Dickman, C. T., Lawson, J., Jabalee, J., Maclellan, S. A., Lepard, N. E., Bennewith, K. L., et al. (2017). Selective extracellular vesicle exclusion of miR-142-3p by oral cancer cells promotes both internal and extracellular malignant phenotypes. Oncotarget 8, 15252–15266. doi: 10.18632/oncotarget.14862
Diomede, F., D’aurora, M., Gugliandolo, A., Merciaro, I., Ettorre, V., Bramanti, A., et al. (2018). A novel role in skeletal segment regeneration of extracellular vesicles released from periodontal-ligament stem cells. Int. J. Nanomed. 13:3805. doi: 10.2147/IJN.S162836
Dobin, A., Davis, C. A., Schlesinger, F., Drenkow, J., Zaleski, C., Jha, S., et al. (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. doi: 10.1093/bioinformatics/bts635
Dozmorov, M. G., Giles, C. B., Koelsch, K. A., and Wren, J. D. (2013). Systematic classification of non-coding RNAs by epigenomic similarity. BMC Bioinformatics 14(Suppl. 14):S2. doi: 10.1186/1471-2105-14-S14-S2
Eleuterio, E., Trubiani, O., Sulpizio, M., Di Giuseppe, F., Pierdomenico, L., Marchisio, M., et al. (2013). Proteome of human stem cells from periodontal ligament and dental pulp. PLoS One 8:e71101. doi: 10.1371/journal.pone.0071101
Falaleeva, M., Pages, A., Matuszek, Z., Hidmi, S., Agranat-Tamir, L., Korotkov, K., et al. (2016). Dual function of C/D box small nucleolar RNAs in rRNA modification and alternative pre-mRNA splicing. Proc. Natl. Acad. Sci. U.S.A. 113, E1625–E1634. doi: 10.1073/pnas.1519292113
Fatima, F., and Nawaz, M. (2017). Vesiculated long non-coding RNAs: offshore packages deciphering trans-regulation between cells, cancer progression and resistance to therapies. Non Coding RNA 3:10. doi: 10.3390/ncrna3010010
Friedman, R. C., Farh, K. K., Burge, C. B., and Bartel, D. P. (2009). Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 19, 92–105. doi: 10.1101/gr.082701.108
Gu, H., Yang, T., Fu, S., Chen, X., Guo, L., and Ni, Y. (2014). MicroRNA-490-3p inhibits proliferation of A549 lung cancer cells by targeting CCND1. Biochem. Biophys. Res. Commun. 444, 104–108. doi: 10.1016/j.bbrc.2014.01.020
Gugliandolo, A., Rajan, T. S., Scionti, D., Diomede, F., Bramanti, P., Mazzon, E., et al. (2017). Reprogramming of oncogene expression in gingival mesenchymal stem cells following long-term culture in vitro. Cell Reprogram 19, 159–170. doi: 10.1089/cell.2016.0056
Guo, Y., Fu, W., Chen, H., Shang, C., and Zhong, M. (2012). miR-24 functions as a tumor suppressor in Hep2 laryngeal carcinoma cells partly through down-regulation of the S100A8 protein. Oncol. Rep. 27, 1097–1103. doi: 10.3892/or.2011.1571
Hacohen, N., Kramer, S., Sutherland, D., Hiromi, Y., and Krasnow, M. A. (1998). sprouty encodes a novel antagonist of FGF signaling that patterns apical branching of the Drosophila airways. Cell 92, 253–263. doi: 10.1016/s0092-8674(00)80919-8
Huang, Z., Shi, J., Gao, Y., Cui, C., Zhang, S., Li, J., et al. (2019). HMDD v3.0: a database for experimentally supported human microRNA-disease associations. Nucleic Acids Res. 47, D1013–D1017. doi: 10.1093/nar/gky1010
Jeong, D. H., Choi, Y. N., Seo, T. W., Lee, J. S., and Yoo, S. J. (2018). Ubiquitin-proteasome dependent regulation of Profilin2 (Pfn2) by a cellular inhibitor of apoptotic protein 1 (cIAP1). Biochem. Biophys. Res. Commun. 506, 423–428. doi: 10.1016/j.bbrc.2018.10.115
Kabir, T. D., Leigh, R. J., Tasena, H., Mellone, M., Coletta, R. D., Parkinson, E. K., et al. (2016). A miR-335/COX-2/PTEN axis regulates the secretory phenotype of senescent cancer-associated fibroblasts. Aging (Albany NY) 8, 1608–1635. doi: 10.18632/aging.100987
Kurita, S., Gunji, E., Ohashi, K., and Mizuno, K. (2007). Actin filaments-stabilizing and -bundling activities of cofilin-phosphatase Slingshot-1. Genes Cells 12, 663–676. doi: 10.1111/j.1365-2443.2007.01078.x
Lal, A., Navarro, F., Maher, C. A., Maliszewski, L. E., Yan, N., O’day, E., et al. (2009). miR-24 Inhibits cell proliferation by targeting E2F2, MYC, and other cell-cycle genes via binding to “seedless” 3’ UTR microRNA recognition elements. Molecular Cell 35, 610–625. doi: 10.1016/j.molcel.2009.08.020
Lee, K.-H., Lin, F.-C., Hsu, T.-I., Lin, J.-T., Guo, J.-H., Tsai, C.-H., et al. (2014). MicroRNA-296-5p (miR-296-5p) functions as a tumor suppressor in prostate cancer by directly targeting Pin1. Biochim. Biophys. Acta Mol. Cell Res. 1843, 2055–2066. doi: 10.1016/j.bbamcr.2014.06.001
Leszczynska, K., Kaur, S., Wilson, E., Bicknell, R., and Heath, V. L. (2011). The role of RhoJ in endothelial cell biology and angiogenesis. Biochem. Soc. Trans. 39, 1606–1611. doi: 10.1042/bst20110702
Li, H., Li, J., Shi, B., and Chen, F. (2017). MicroRNA-296 targets AKT2 in pancreatic cancer and functions as a potential tumor suppressor. Mol. Med. Rep. 16, 466–472. doi: 10.3892/mmr.2017.6602
Li, S., Xu, X., Xu, X., Hu, Z., Wu, J., Zhu, Y., et al. (2013). MicroRNA-490-5p inhibits proliferation of bladder cancer by targeting c-Fos. Biochem. Biophys. Res. Commun. 441, 976–981. doi: 10.1016/j.bbrc.2013.11.006
Li, T., Lu, Y., Zhao, X., Guo, H., Liu, C., Li, H., et al. (2014). MicroRNA-296-5p increases proliferation in gastric cancer through repression of Caudal-related homeobox 1. Oncogene 33, 783–793. doi: 10.1038/onc.2012.637
Liu, M., Bi, F., Zhou, X., and Zheng, Y. (2012). Rho GTPase regulation by miRNAs and covalent modifications. Trends Cell Biol. 22, 365–373. doi: 10.1016/j.tcb.2012.04.004
Liu, W., Xu, G., Liu, H., and Li, T. (2015). MicroRNA−490−3p regulates cell proliferation and apoptosis by targeting HMGA2 in osteosarcoma. FEBS Lett. 589, 3148–3153. doi: 10.1016/j.febslet.2015.08.034
Lv, L., and Wang, X. (2018). MicroRNA-296 targets specificity protein 1 to suppress cell proliferation and invasion in cervical cancer. Oncol. Res. Featuring Preclin. Clin. Cancer Ther. 26, 775–783. doi: 10.3727/096504017X15132494420120
Lynch, J., Fay, J., Meehan, M., Bryan, K., Watters, K. M., Murphy, D. M., et al. (2012). MiRNA-335 suppresses neuroblastoma cell invasiveness by direct targeting of multiple genes from the non-canonical TGF-β signalling pathway. Carcinogenesis 33, 976–985. doi: 10.1093/carcin/bgs114
Mackenzie, T. N., Mujumdar, N., Banerjee, S., Sangwan, V., Sarver, A., Vickers, S., et al. (2013). Triptolide induces the expression of miR-142-3p: a negative regulator of heat shock protein 70 and pancreatic cancer cell proliferation. Mol. Cancer Ther. 12, 1266–1275. doi: 10.1158/1535-7163.MCT-12-1231
Mens, M. M. J., and Ghanbari, M. (2018). Cell cycle regulation of stem cells by MicroRNAs. Stem Cell Rev. Rep. 14, 309–322. doi: 10.1007/s12015-018-9808-y
Palazzo, A. F., and Lee, E. S. (2015). Non-coding RNA: what is functional and what is junk? Front. Genet. 6:2. doi: 10.3389/fgene.2015.00002
Panagiotaki, N., Dajas-Bailador, F., Amaya, E., Papalopulu, N., and Dorey, K. (2010). Characterisation of a new regulator of BDNF signalling, Sprouty3, involved in axonal morphogenesis in vivo. Development 137, 4005–4015. doi: 10.1242/dev.053173
Pizzicannella, J., Diomede, F., Gugliandolo, A., Chiricosta, L., Bramanti, P., Merciaro, I., et al. (2019a). 3D Printing PLA/Gingival Stem Cells/EVs Upregulate miR-2861 and -210 during Osteoangiogenesis commitment. Int. J. Mol. Sci. 20:3256. doi: 10.3390/ijms20133256
Pizzicannella, J., Gugliandolo, A., Orsini, T., Fontana, A., Ventrella, A., Mazzon, E., et al. (2019b). Engineered extracellular vesicles from human periodontal-ligament stem cells increase VEGF/VEGFR2 expression during bone regeneration. Front. Physiol. 10:512. doi: 10.3389/fphys.2019.00512
Rajan, T. S., Giacoppo, S., Diomede, F., Ballerini, P., Paolantonio, M., Marchisio, M., et al. (2016). The secretome of periodontal ligament stem cells from MS patients protects against EAE. Sci. Rep. 6:38743. doi: 10.1038/srep38743
Rupaimoole, R., and Slack, F. J. (2017). MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 16, 203–222. doi: 10.1038/nrd.2016.246
Sepulveda, J. L., and Wu, C. (2006). The parvins. Cell. Mol. Life Sci. 63, 25–35. doi: 10.1007/s00018-005-5355-1
Severino, P., Oliveira, L. S., Andreghetto, F. M., Torres, N., Curioni, O., Cury, P. M., et al. (2015). Small RNAs in metastatic and non-metastatic oral squamous cell carcinoma. BMC Med. Genomics 8:31. doi: 10.1186/s12920-015-0102-4
Sharma, E., Sterne-Weiler, T., O’hanlon, D., and Blencowe, B. J. (2016). Global mapping of human RNA-RNA interactions. Mol. Cell 62, 618–626. doi: 10.1016/j.molcel.2016.04.030
Shen, W.-W., Zeng, Z., Zhu, W.-X., and Fu, G.-H. (2013). MiR-142-3p functions as a tumor suppressor by targeting CD133, ABCG2, and Lgr5 in colon cancer cells. J. Mol. Med. 91, 989–1000. doi: 10.1007/s00109-013-1037-x
Silvestro, S., Chiricosta, L., Gugliandolo, A., Pizzicannella, J., Diomede, F., Bramanti, P., et al. (2020). Extracellular vesicles derived from human gingival mesenchymal stem cells: a transcriptomic analysis. Genes (Basel) 11:118. doi: 10.3390/genes11020118
Slaby, O., Laga, R., and Sedlacek, O. (2017). Therapeutic targeting of non-coding RNAs in cancer. Biochem. J. 474, 4219–4251. doi: 10.1042/BCJ20170079
Song, L., Yang, J., Duan, P., Xu, J., Luo, X., Luo, F., et al. (2013). MicroRNA-24 inhibits osteosarcoma cell proliferation both in vitro and in vivo by targeting LPAATβ. Arch. Biochem. Biophys. 535, 128–135. doi: 10.1016/j.abb.2013.04.001
Stelzer, G., Rosen, N., Plaschkes, I., Zimmerman, S., Twik, M., Fishilevich, S., et al. (2016). The GeneCards suite: from gene data mining to disease genome sequence analyses. Curr. Protoc. Bioinformatics 54, 1.30.1–1.30.33. doi: 10.1002/cpbi.5
Thomas, P., Pranatharthi, A., Ross, C., and Srivastava, S. (2019). RhoC: a fascinating journey from a cytoskeletal organizer to a cancer stem cell therapeutic target. J. Exp. Clin. Cancer Res. 38:328. doi: 10.1186/s13046-019-1327-4
Thomas, P. D., Campbell, M. J., Kejariwal, A., Mi, H., Karlak, B., Daverman, R., et al. (2003). PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 13, 2129–2141. doi: 10.1101/gr.772403
Trapnell, C., Hendrickson, D. G., Sauvageau, M., Goff, L., Rinn, J. L., and Pachter, L. (2013). Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 31:46. doi: 10.1038/nbt.2450
Trubiani, O., Giacoppo, S., Ballerini, P., Diomede, F., Piattelli, A., Bramanti, P., et al. (2016). Alternative source of stem cells derived from human periodontal ligament: a new treatment for experimental autoimmune encephalomyelitis. Stem Cell Res. Ther. 7:1. doi: 10.1186/s13287-015-0253-4
Trubiani, O., Marconi, G. D., Pierdomenico, S. D., Piattelli, A., Diomede, F., and Pizzicannella, J. (2019). Human oral stem cells, biomaterials and extracellular vesicles: a promising tool in bone tissue repair. Int. J. Mol. Sci. 20:4987. doi: 10.3390/ijms20204987
Valadi, H., Ekström, K., Bossios, A., Sjöstrand, M., Lee, J. J., and Lötvall, J. O. (2007). Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 9, 654–659. doi: 10.1038/ncb1596
Wang, L., Bo, X., Zheng, Q., Xiao, X., Wu, L., and Li, B. (2016). miR−296 inhibits proliferation and induces apoptosis by targeting FGFR 1 in human hepatocellular carcinoma. FEBS Lett. 590, 4252–4262. doi: 10.1002/1873-3468.12442
Wang, Y., Zhao, W., and Fu, Q. (2013). miR-335 suppresses migration and invasion by targeting ROCK1 in osteosarcoma cells. Mol. Cell. Biochem. 384, 105–111. doi: 10.1007/s11010-013-1786-4
Wei, J.-J., Wu, X., Peng, Y., Shi, G., Olca, B., Yang, X., et al. (2011). Regulation of HMGA1 expression by microRNA-296 affects prostate cancer growth and invasion. Clin. Cancer Res. 17, 1297–1305. doi: 10.1158/1078-0432.ccr-10-0993
Wu, L., Cai, C., Wang, X., Liu, M., Li, X., and Tang, H. (2011). MicroRNA−142−3p, a new regulator of RAC1, suppresses the migration and invasion of hepatocellular carcinoma cells. FEBS Lett. 585, 1322–1330. doi: 10.1016/j.febslet.2011.03.067
Xu, C., Li, S., Chen, T., Hu, H., Ding, C., Xu, Z., et al. (2016). miR-296-5p suppresses cell viability by directly targeting PLK1 in non-small cell lung cancer. Oncol. Rep. 35, 497–503. doi: 10.3892/or.2015.4392
Xu, G., Wang, J., Jia, Y., Shen, F., Han, W., and Kang, Y. (2014). MiR-142-3p functions as a potential tumor suppressor in human osteosarcoma by targeting HMGA1. Cell. Physiol. Biochem. 33, 1329–1339. doi: 10.1159/000358700
Xu, G. Q., Li, L. H., Wei, J. N., Xiao, L. F., Wang, X. T., Pang, W. B., et al. (2019). Identification and profiling of microRNAs expressed in oral buccal mucosa squamous cell carcinoma of Chinese hamster. Sci. Rep. 9:15616. doi: 10.1038/s41598-019-52197-3
Xu, J.-Z., Zhang, J.-L., and Zhang, W.-G. (2018). Antisense RNA: the new favorite in genetic research. J. Zhejiang Univ. Sci. B 19, 739–749. doi: 10.1631/jzus.B1700594
Xu, Y., Zhao, F., Wang, Z., Song, Y., Luo, Y., Zhang, X., et al. (2012). MicroRNA-335 acts as a metastasis suppressor in gastric cancer by targeting Bcl-w and specificity protein 1. Oncogene 31, 1398–1407. doi: 10.1038/onc.2011.340
Keywords: human periodontal ligament stem cells, extracellular vesicles, next generation sequencing, non-coding RNAs, microRNAs, cytokinesis
Citation: Chiricosta L, Silvestro S, Gugliandolo A, Marconi GD, Pizzicannella J, Bramanti P, Trubiani O and Mazzon E (2020) Extracellular Vesicles of Human Periodontal Ligament Stem Cells Contain MicroRNAs Associated to Proto-Oncogenes: Implications in Cytokinesis. Front. Genet. 11:582. doi: 10.3389/fgene.2020.00582
Received: 05 March 2020; Accepted: 12 May 2020;
Published: 04 June 2020.
Edited by:
Saverio Bellusci, University of Giessen, GermanyReviewed by:
Federica Papaccio, INCLIVA Health Research Institute, SpainRoberto Di Primio, Marche Polytechnic University, Italy
Copyright © 2020 Chiricosta, Silvestro, Gugliandolo, Marconi, Pizzicannella, Bramanti, Trubiani and Mazzon. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Emanuela Mazzon, ZW1hbnVlbGEubWF6em9uQGlyY2NzbWUuaXQ=
†These authors have contributed equally to this work
 Luigi Chiricosta
Luigi Chiricosta Serena Silvestro
Serena Silvestro Agnese Gugliandolo
Agnese Gugliandolo Guya Diletta Marconi
Guya Diletta Marconi Jacopo Pizzicannella
Jacopo Pizzicannella Placido Bramanti
Placido Bramanti Oriana Trubiani
Oriana Trubiani Emanuela Mazzon
Emanuela Mazzon