
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 18 March 2020
Sec. RNA
Volume 11 - 2020 | https://doi.org/10.3389/fgene.2020.00233
Several long non-coding RNAs (lncRNAs) have been reported regulate the expression of neighbor protein-coding genes at post-transcriptional, transcriptional and epigenetic levels. Dmp1 (Dentin matrix protein 1), encoding a non-collagenous extracellular matrix protein, plays an important role in dentin and bone mineralization. However, the transcriptional regulation of lncRNA on Dmp1 has not been reported. In this study, we identified a novel lncRNA named lnc-DMP1, which is near the Dmp1 gene region and undergoes remarkable changes during mandible development. lnc-DMP1 is co-localized and significantly expressed correlation with Dmp1 in embryonic and postnatal mouse mandibles. In MC3T3-E1 cells, lnc-DMP1 positively regulates DMP1 expression and skeletal mineralization. Furthermore, lnc-DMP1 induces the promoter activity of Dmp1 by modulating H3K27Ac enrichment in the Dmp1 promoter. In conclusion, our results indicate that lnc-DMP1 is a novel lncRNA near the Dmp1 gene region and regulates Dmp1 expression by modulating the H3K27 acetylation level of Dmp1 promoter.
Long non-coding RNAs (lncRNAs) are defined as a subgroup of non-coding RNA molecules that consist of at least 200 nucleotides and exhibit no or limited protein-coding capability (Roberts et al., 2014; Marchese et al., 2017). Since the major role of Xist in X-chromosome inactivation was first described (Brockdorff et al., 1992; Brown et al., 1992), studies have demonstrated that lncRNAs function in multiple cellular processes, such as genomic locus imprinting (Kanduri, 2016), antiviral response (Fortes and Morris, 2016) and differentiation and development (Fatica and Bozzoni, 2014). Studies have also investigated various mechanisms underlying lncRNA functions. Some nuclear lncRNAs are expressed from imprinted loci function as molecular scaffolds that recruit chromatin-modifying complexes and regulate gene expression in cis by altering the chromatin structures of target genes (Lee and Bartolomei, 2013; Melo et al., 2013). Other lncRNAs modulate gene expression in trans by interfering with transcriptional machineries or maintaining the structures of nuclear speckles (Prasanth et al., 2005; Clemson et al., 2009; Sunwoo et al., 2009). Some cytosolic lncRNAs have been suggested to regulate mRNA splicing, mRNA decay, protein translation and protein stability (Yoon et al., 2013; Quinn and Chang, 2016).
Dentin matrix protein 1 (DMP1), a highly phosphorylated acidic non-collagenous phosphoprotein, is initially identified by cDNA cloning in rat odontoblasts and highly expressed in bone tissues (George et al., 1993; D’Souza et al., 1997; Hirst et al., 1997; MacDougall et al., 1997; Feng et al., 1998). DMP1 plays a key role in the control of mineralization and phosphate homeostasis in dentin and bone (Qin et al., 2007). The deletion of Dmp1 leads to severe defects in the odontogenesis of the dentin and cartilage formation of bones in mice (Ye et al., 2004, 2005). Importantly, the loss of Dmp1 results in autosomal recessive hypophosphatemic rickets, a novel disorder in humans (Feng et al., 2006; Lorenz-Depiereux et al., 2006).
Narayanan et al. (2003) investigated the transcriptional regulation of Dmp1 by c-Fos and c-Jun, two AP-1 transcriptional factor family members, which play important roles in early osteoblast differentiation. They also demonstrated that JunB can interact with p300 and dramatically modulate the promoter activity of Dmp1 during osteoblast mineralization (Narayanan et al., 2002). The transcription factor TCF11 binds to the Dmp1 promoter and activates Dmp1 transcription in osteoblasts (Jacob et al., 2014). However, the effect of lncRNA on the transcriptional regulation of Dmp1 has yet to be reported.
Since the transcription factor JunB regulates the expression of Dmp1 gene through interaction with the transcriptional coactivator p300, which is a histone acetyltransferase (Narayanan et al., 2002). Meanwhile, lncRNAs could function as molecular scaffolds of histone-modifying enzyme to modulate the expression of target gene through specific histone modification such as methylation and acetylation (Marchese et al., 2017). So, we assumed that lncRNAs could play a certain regulatory role in the expression of Dmp1. Thus, we used lncRNA-seq technology to screen lncRNAs near the promoter of Dmp1 and differently expressed between embryonic and postnatal mouse mandibles. lnc-DMP1 was selected on the basis of the lncRNA sequence profiles. Then, the expression patterns of lnc-DMP1 and Dmp1 in the embryonic and postnatal mouse mandibles were detected through qRT-PCR and RNA-fluorescence in situ hybridization (FISH) to investigate whether lnc-DMP1 was significantly correlated with Dmp1. Subsequently, whether lnc-DMP1 could participate both mRNA and protein expression of DMP1 and mineralization were tested in osteoblast cells MC3T3-E1. Finally, luciferase assay and chromatin immunoprecipitation (ChIP) methods were performed to investigate whether lnc-DMP1 could transcriptionally regulate the Dmp1 expression in MC3T3-E1 cells by modulating the enrichment of H3K27Ac in Dmp1 promoter.
Embryonic (16, 18, and 20 days old) and postnatal (2 and 4 weeks old) C57 mice were purchased from the Animal Experiment Center of Sun Yat-sen University (Guangzhou, China). The body weight of the mice was 10–15 g, and the mice were fed at room temperature (20–22°C) (License code: SYKX: 2012-0081). The mice were anesthetized by intraperitoneally injecting 5% chloral hydrate (0.1 mL/10 g) and then fixed on a dry ice anatomy bench. The mandibles were extracted under a microscope, frozen in liquid nitrogen and stored at −80°C. After sampling was completed, the mice were sacrificed by dislocating their necks and then treated in accordance with the death treatment method for experimental animals. All animal experiments were conducted following the Ministry of Health national guidelines for housing and care of laboratory animals and performed in accordance with institutional regulations after review and approval by the Institutional Animal Care and Use Committee at the Sun Yat-sen University (IACUC-DB-13-132).
The MC3T3-E1 cell line (American Type Culture Collection, Manassas, VA, United States) was cultured in α-MEM (Gibco, New York, NY, United States) and pretreated with 10% fetal bovine serum. The cells were cultured in an incubator with humidified atmosphere and 5% CO2 at 37°C.
Total RNA was isolated from the mandibles of embryonic (18 days old) and postnatal (2 weeks old) C57 mice with TRIzol reagent (Thermo Fisher Scientific, Carlsbad, CA, United States) in accordance with the protocol. RNA quantity and quality were evaluated with Agilent Bioanalyzer 2100. The qualified RNA samples were purified and synthesized to cDNA. After cluster amplification was conducted through PCR, a sequencing library was sequenced with Illumina HiSeqTM2000. Raw sequences were filtered to remove the joint, undetected sequences and low-quality sequences. Transcriptional transcripts were constructed from the remaining sequence, and known non-coding RNAs and protein-encoding fragments were discarded. The new non-coded sequence and the known non-coding sequence were compared and statistically analyzed to screen the differentially expressed lncRNAs in the two-time-point tissue samples.
On the basis of the Poisson distribution model, we simultaneously calculated the differential expression multiples [log2(E18D/P2W)] of lncRNA in E18D and P2W samples in accordance with the gene expression (RPKM value) and examined the differences in P-value by controlling the false discovery rate (FDR) to determine the P-value of the domain. LncRNAs with | log2(E18D/P2W) | >4 and FDR < 0.01 were considered differentially expressed in E18D and P2W samples.
The mandible samples of embryonic (16, 18, and 20 days old) and postnatal (2 and 4 weeks old) C57 mice were collected and fixed in 4% paraformaldehyde for 48 h and then decalcified in EDTA. The decalcified specimens were paraffin embedded, and 5 mm serial sections were prepared for the experiments. The slides were hybridized with lnc-DMP1 and Dmp1 probes overnight at 37°C. Afterward, the slides were washed twice with 2 × saline sodium citrate (SSC) and three times with deionized water formamide: 2 × SSC (50:50) for 20 min. DAPI solution was used to stain the cell nuclei. Fluorescence images were acquired through fluorescence microscopy.
For lnc-DMP1 overexpression, the full length of lnc-DMP1 was cloned into the lentivirus vector pCDH-CMV-MCS-EF1-copGFP (Gene, Shanghai, China). The shRNA sequence targeting lnc-DMP1 was cloned into the lentivirus vector psi-LVRU6MH (Gene) for the interference of the lnc-DMP1 expression. All of the vectors were transfected into MC3T3-E1 cells in accordance with the manufacturer’s instructions. The efficiency of transfection was confirmed through qRT-PCR. The cells were subjected to RNA extraction, Western blot assay and functional assays.
The total RNA of mandible samples and cells was extracted with TRIzol reagent in accordance with the manufacturer’s instructions (Thermo Fisher Scientific) and reverse transcribed with a high-capcity cDNA reverse transcription kit (Vazyme, Nanjing, China). SYBR Green PCR master Mix (Vazyme) and primers (Table 1) were used for qRT-PCR. The qRT-PCR program was set at the following parameters: 95°C for 2 min, followed by 40 cycles at 95°C for 15 s and 60°C for 32 s. GAPDH was used as reference genes, and the relative gene expression level was calculated by using the comparative threshold (Ct) cycle method (2–ΔΔCt).
The cultured cells were extracted by RIPA (Radio Immunoprecipitation Assay) ordinary type reagent (Vazyme), which is a traditional cell tissue lysis buffer to gain proteins for western and IP experiments. Then, 30 μg of each protein was loaded to each lane, fractionated by 10% SDS-PAGE, transferred onto PVDF membranes and probed with the primary antibody of DMP1 and DSPP (Abcam, Cambridge, United Kingdom) as a target and GAPDH (CST, Boston, MA, United States) as an internal control at 4°C overnight. The secondary antibodies were added for 2 h. The blots were visualized through enhanced chemiluminescence and autoradiography.
The lnc-DMP1 overexpression and RNAi stable cell lines were induced by osteoinductive factors (50 μg/mL ascorbic acid phosphate, 10 mM β-glycerol phosphate disodium salt and 10 nmol/L dexamethasone) for 28 days. After mineralization was induced, the medium was discarded. The cells were rinsed with PBS three times, fixed with 95% ethanol for 20 min and stained in alizarin red solution for 10 min. The calcification nodules formed in each group were compared.
The dual-luciferase reporter vector pGL3-Basic vector, which contains renilla luciferase with 35S promoter and firefly luciferase without driven promoter, was used to construct the pGL3-pDmp1 reporter vector that including both renilla luciferase with 35S promoter and firefly luciferase driven by the Dmp1 promoter (2000 bp before transcription start site). The plasmids of CMV empty vector, CMV-lnc-DMP1, U6-Sh empty vector and U6-Sh-lnc-DMP1 were co-transfected with pGL3-pDmp1 plasmid into MC3T3-E1 cell. Lipofectamine 2000 was used. After 48 h of transfection, the cells were collected, and luciferase activity was measured by using a dual luciferase reporter assay kit (Promega, Madison, United States) in accordance with the manufacturer’s protocol. The firefly luciferase activities of the Dmp1 promoter co-transfected with CMV vectors or U6-Sh vectors were firstly normalized to renilla luciferase activity of itself, then showed as a relative LUC activity to the empty vector control CMV-lnc-DMP1 or U6-Sh empty vector.
ChIP was conducted by using a ChIP kit (Qiagen, Düsseldorf, Germany) in accordance with the manufacturer’s instructions. In brief, the cells were incubated with formaldehyde, and the link between DNA and protein was built. Then, the cross-linked chromatin was sonicated into fragments. Anti-H3K27ac antibodies (Abcam) were used for the immunoprecipitation of the chromatin fragments, and IgG was used as the negative control. Subsequently, the precipitated chromatin DNA was measured through qRT-PCR.
In vitro, part of purified PCR product pDmp1 DNAs, which was 378 bp length of Dmp1 promoter before the transcriptional start site (TSS), were biotin-labeled with a Biotin DNA labeling mix (Roche, Basel, Switzerland). The Biotin-labeled and unbiotin-labeled pDmp1 DNAs were mixed with MC3T3-E1 cell protein extract in 500 μL incubation buffer (RIP buffer (100 mM NaCl, 10 mM Tris, 1 mM EDTA, 1 mM EGTA, PH7.6) with 0.2% TritonX 100 and protease inhibitor) and then incubated at 4°C overnight. Afterward, 50 μL of washed streptavidin agarose beads (Thermo Fisher Scientific) was added to each binding reaction and further incubated at RT for 1 h. The beads were washed briefly with wash buffer [RIP buffer (100 mM NaCl, 10 mM Tris, 1 mM EDTA, 1 mM EGTA, PH7.6) with 0.1% TritonX 100] five times and boiled in SDS buffer. The retrieved proteins were detected by Western blot.
The primer sequences are shown in Table 1.
Data were analyzed with SPSS 20.0 and presented as mean ± SD. Differences between the two groups were assessed by using two-tailed Student’s t-test. Differences with a P < 0.05 were considered statistically significant (∗P < 0.05 and ∗∗P < 0.01).
To examine the expression profiles of lncRNAs in the development of mouse mandibles, we performed lncRNA sequencing (lncRNA-seq) analysis. RNA was collected from the mouse mandibles of embryonic18-day-old (E18D) and postnatal 2-week-old (P2W) mice. A total of 38,566 lncRNAs were identified in the two samples (Figure 1A). Through stringent classification (| log2 fold change (E18D/P2W) | >4, FDR < 0.001), 808 and 408 lncRNAs were found to be up-regulated (log2 fold change (E18D/P2W) >4) and down-regulated (log2 fold change (E18D/P2W) < −4) (Figure 1B), respectively. These results suggested that lncRNAs might be involved in the regulation of mouse mandible development.
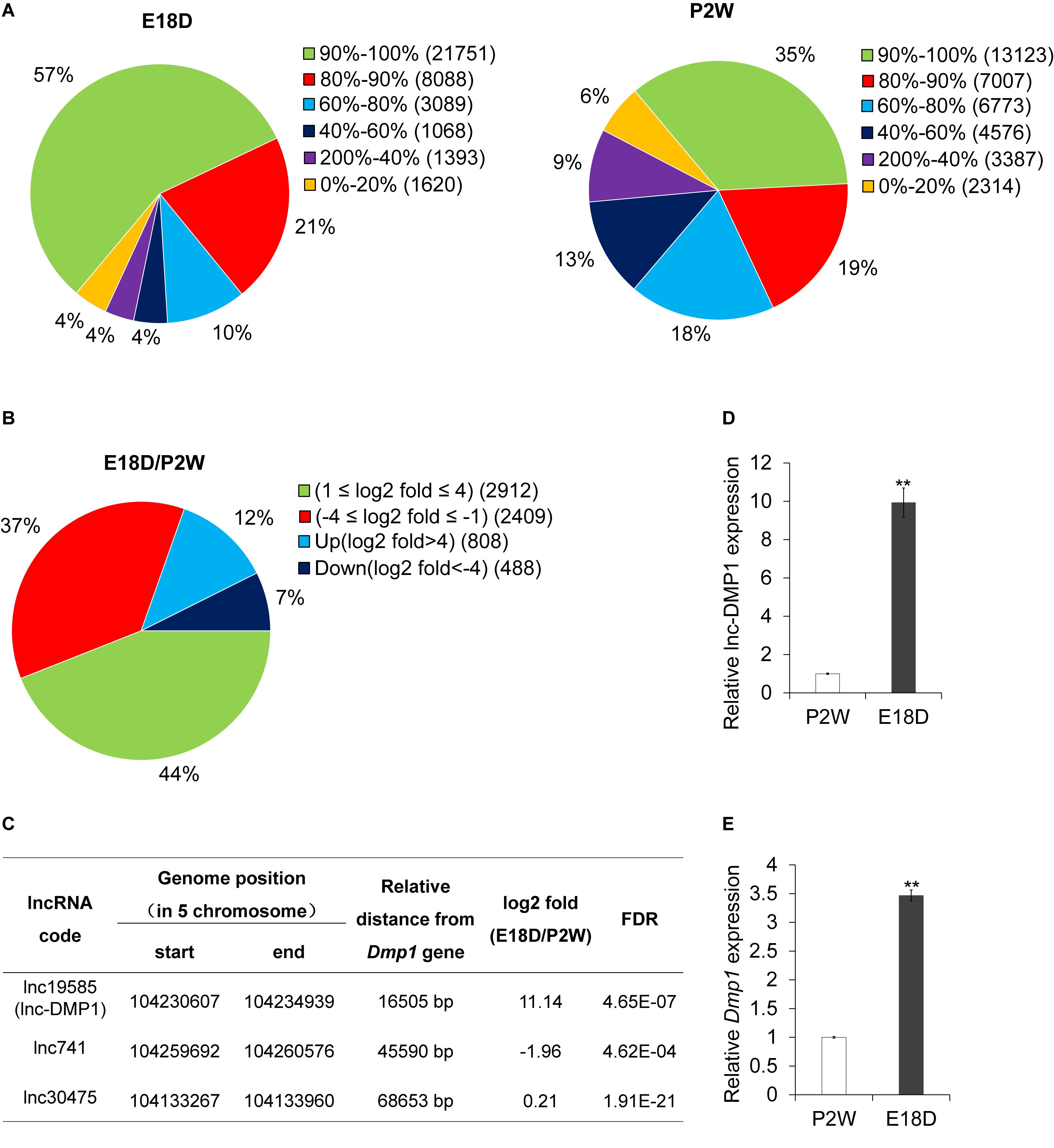
Figure 1. lnc-DMP1 is a novel lncRNA near the Dmp1 gene region, differentially expressed in E18D and P2W samples. (A) The distribution of lncRNAs’ coverage in E18D and P2W samples from lncRNA-seq data. (B) The distribution of differentially expressed lncRNAs (| log2 fold change (E18D/P2W) | ≥ 1, FDR < 0.001) in E18D and P2W samples. (C) lncRNAs located within 100 kb of Dmp1 (Chromosome 5:104202613-104214102). (D) and (E) qRT-PCR detection of lnc-DMP1 (D) and Dmp1 (E) in E18D and P2W samples. Data were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression, and P2W was set to a value of 1. **P < 0.01 by Student’s t-test. E18D, embryonic18-day-old mouse mandible; P2W, postnatal 2-week-old mouse mandible.
To investigate lncRNAs that might modulate the transcriptional expression of Dmp1 in cis, we searched lncRNAs located within 100 kb of the Dmp1 gene region (Chromosome 5:104202613-104214102) from the lncRNA-seq data. We found that lnc19585 and lnc741 were located downstream of Dmp1, with distances of 16.7 and 45.6 kb, respectively, whereas lnc30475 was located upstream of Dmp1, with distance of 68.6 kb (Figure 1C). Amongst these three lncRNAs, lnc19585 was the nearest to Dmp1 and the only one that was up-regulated in the E18D sample relative to the P2W sample. Thus, we renamed lnc19585 as lnc-DMP1.
To confirm the results of the lnc-DMP1 expression from lncRNA-seq, we also conducted qRT-qPCR to detect the expression levels of lnc-DMP1 and Dmp1 in the mandibles derived from E18D and P2W mice. The results showed that the level of lnc-DMP1 was remarkably up-regulated in E18D compared with that in P2W (Figure 1D), and this expression pattern was consistent with that of Dmp1 (Figure 1E). These results indicated that lnc-DMP1 was a novel lncRNA near the Dmp1 gene region, which was differentially expressed in the development of mouse mandibles.
To test whether lnc-DMP1 was co-localized with Dmp1, we performed RNA-FISH to detect the expression of lnc-DMP1 and Dmp1 in the mandibles derived from embryonic 16-, 18-, and 20-day-old mice and postnatal 2- and 4-week-old mice. In Figure 2, the expression levels of Dmp1 (green fluorescence) and lnc-Dmp1 (red fluorescence) were highly co-localized (yellow fluorescence) in all of the developmental processes of mouse mandibles, and the co-localization was distributed in the region, including the nuclei (blue fluorescence) and the cytoplasm. We found that the expression levels of lnc-DMP1 and Dmp1 decreased in the postnatal mouse mandibles (Figures 2D,E) compared with those in the embryonic mouse mandibles (Figures 2A–C). These data suggested that the expression of lnc-DMP1 was significantly correlated with that of Dmp1 in different developmental processes of mouse mandibles.
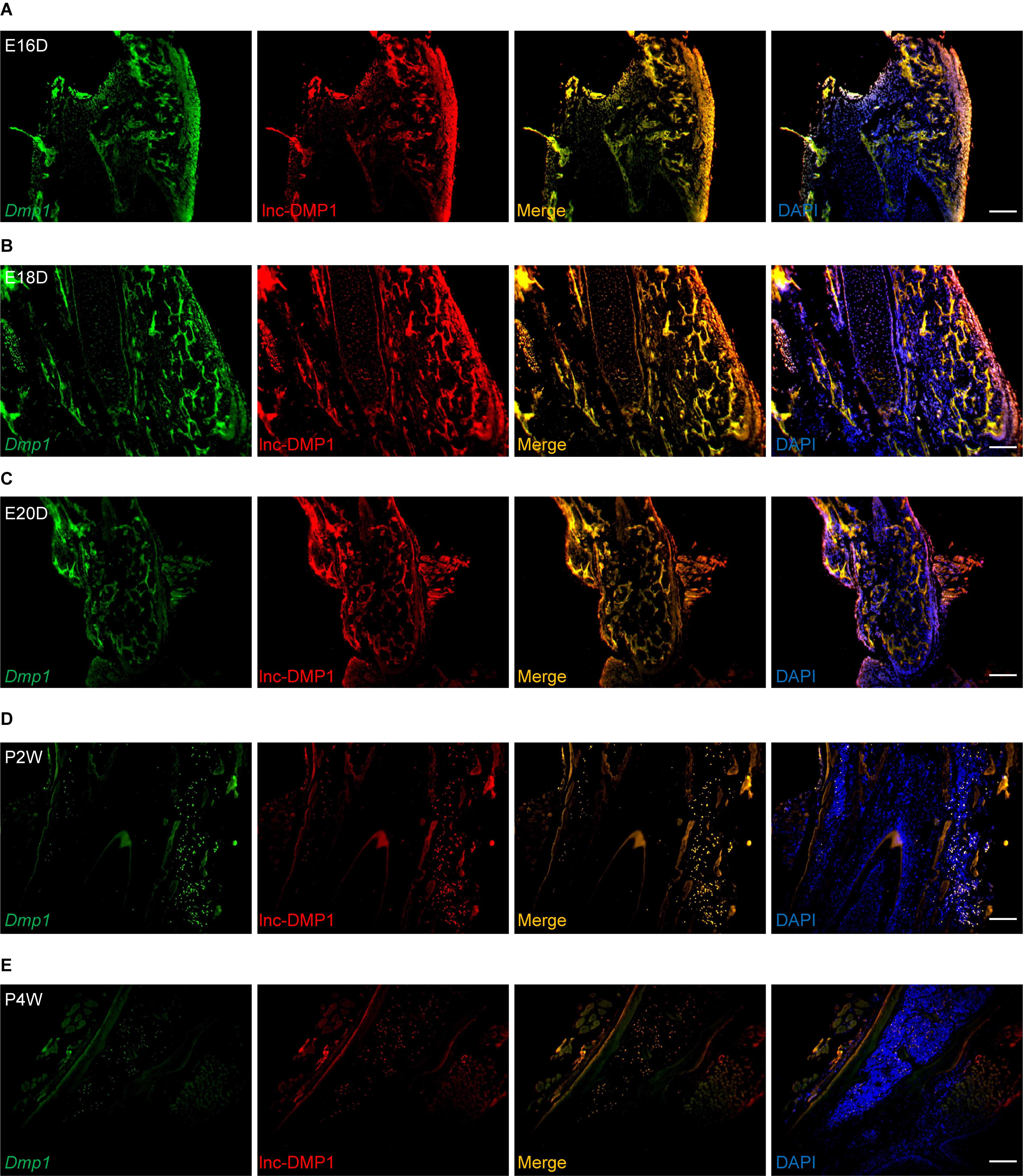
Figure 2. RNA-FISH assay of lnc-DMP1 and Dmp1 in the development of mouse mandibles. Representative images of lnc-DMP1 (green) and Dmp1 (red) expression in embryonic 16- (A), 18- (B) and 20-day (C) mouse mandibles and in postnatal 2- (D) and 4-week (E) mouse mandibles. Yellow denoted the co-localization of lnc-DMP1 and Dmp1. Nuclei were stained with DAPI (blue). Scale bar represented 500 μm.
To test the role of lnc-DMP1 on the modulation of Dmp1 expression, we selected MC3T3-E1 cells to construct stable OE and RNAi lines of lnc-DMP1, which were, respectively, transduced with the expression vectors of pCDH-CMV-lnc-DMP1 and psi-LVRU6MH-sh-lnc-DMP1, and the empty vectors were transduced as control lines (Figure 3A). The mRNA expression level of Dmp1 was detected in stable lnc-DMP1 OE and RNAi lines, and qRT-PCR results showed that the Dmp1 expression increased in the lnc-DMP1 OE line and decreased in the lnc-DMP1 RNAi line relative to the control lines (Figure 3B). Dspp was a downstream gene of Dmp1, whose expression was regulated by Dmp1. We also detected the mRNA expression level of Dspp in lnc-DMP1 OE and RNAi lines, and the results showed the same expression pattern of Dspp as Dmp1 in these stable lines (Figure 3C). We further tested the alteration on the protein level through an immunoblot assay. The results showed that the protein levels of DMP1 and DSPP increased in the lnc-DMP1 OE line and decreased in the lnc-DMP1 RNAi line (Figure 3D). These data suggested that lnc-DMP1 acted as an upstream regulator promoted both mRNA and protein expression of DMP1.
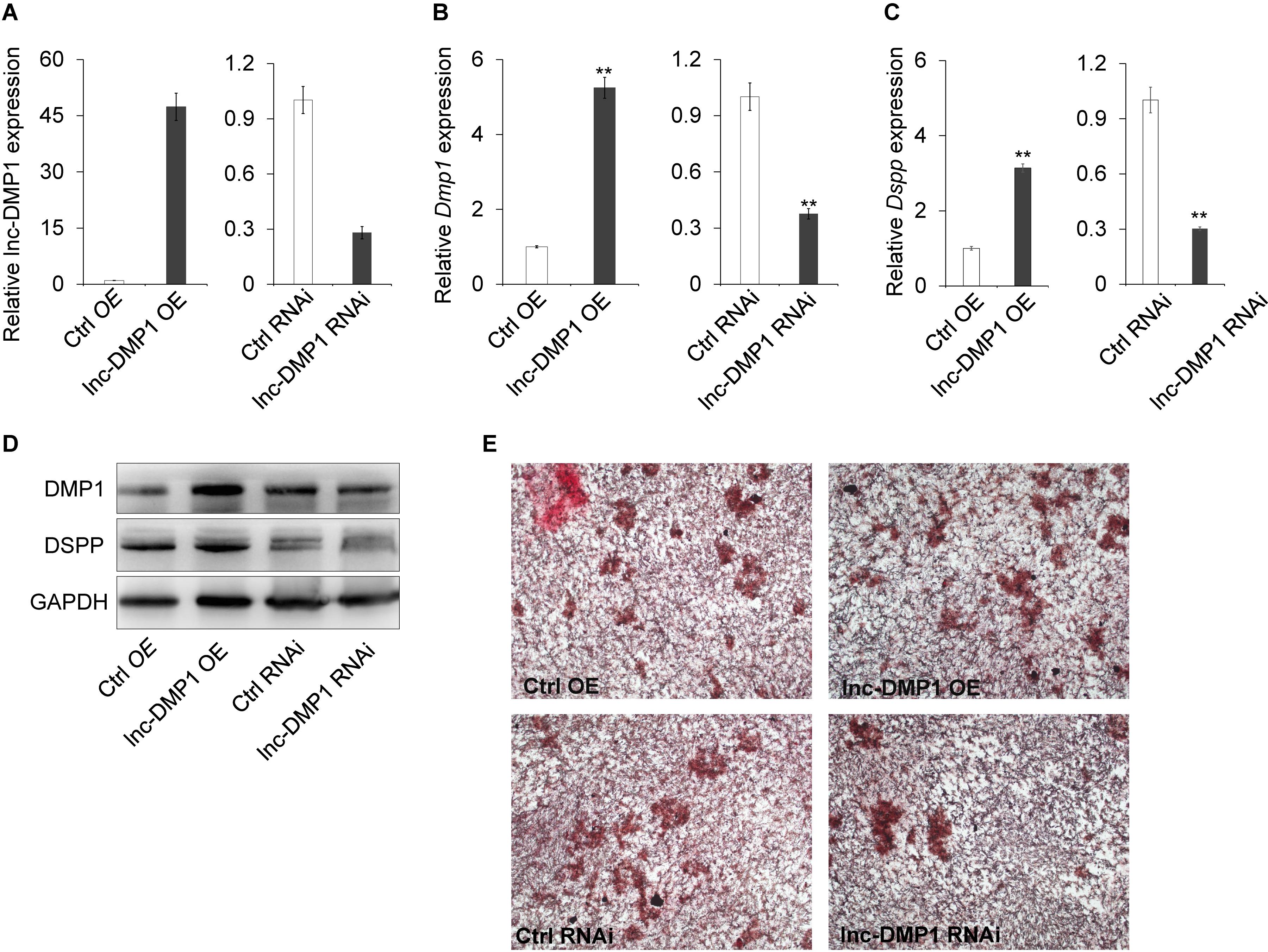
Figure 3. lnc-DMP1 controls mineralization by increasing the DMP1 expression in MC3T3-E1 cells. (A) The transformation efficiency of lnc-DMP1 OE and RNAi stable lines in MC3T3-E1 cells was determined through qRT-PCR. Data were normalised to GAPDH expression, and the empty vector control was set to a value of 1. (B,C) The alteration of the mRNA levels of Dmp1 (B) and Dspp (C) in lnc-DMP1 OE and RNAi cells. (D) The alteration of the protein levels of DMP1 and DSPP in lnc-DMP1 OE and RNAi cells. (E) Mineralization was detected by subjecting lnc-DMP1 OE and RNAi cells to alizarin red staining. **P < 0.01 by Student’s t-test.
Given that Dmp1 plays an important role in the control of osteoblast cell mineralization (Toyosawa et al., 2001; Ling et al., 2005), we determined whether lnc-DMP1 participated in the skeletal mineralization of osteoblast cells. The results of the cell mineralization assays performed after the cells were treated with osteo-inductive factors for 28 days showed that more mineralized nodules were formed in the lnc-DMP1 OE line compared with those in the control lines, whereas the lnc-DMP1 RNAi line showed obviously inhibited mineralized nodules formation compared with that in the control lines (Figure 3E). These results suggested that lnc-DMP1 controlled mineralization by increasing the Dmp1 expression in MC3T3-E1 cells.
To test whether lnc-DMP1 functions in the transcriptional regulation of Dmp1 expression, we made a construct expressing a luciferase reporter gene under the control of the Dmp1 promoter. While the pDmp1:LUC reporter gene was co-transfected with CMV-lnc-DMP1 plasmid into MC3T3-E1 cells, the expression of luciferase significantly increased relative to that of the control. The co-transfected of U6-sh-lnc-DMP1 plasmid reduced the reporter gene expression (Figure 4A). These results suggested that lnc-DMP1 induced the promoter activity of Dmp1.
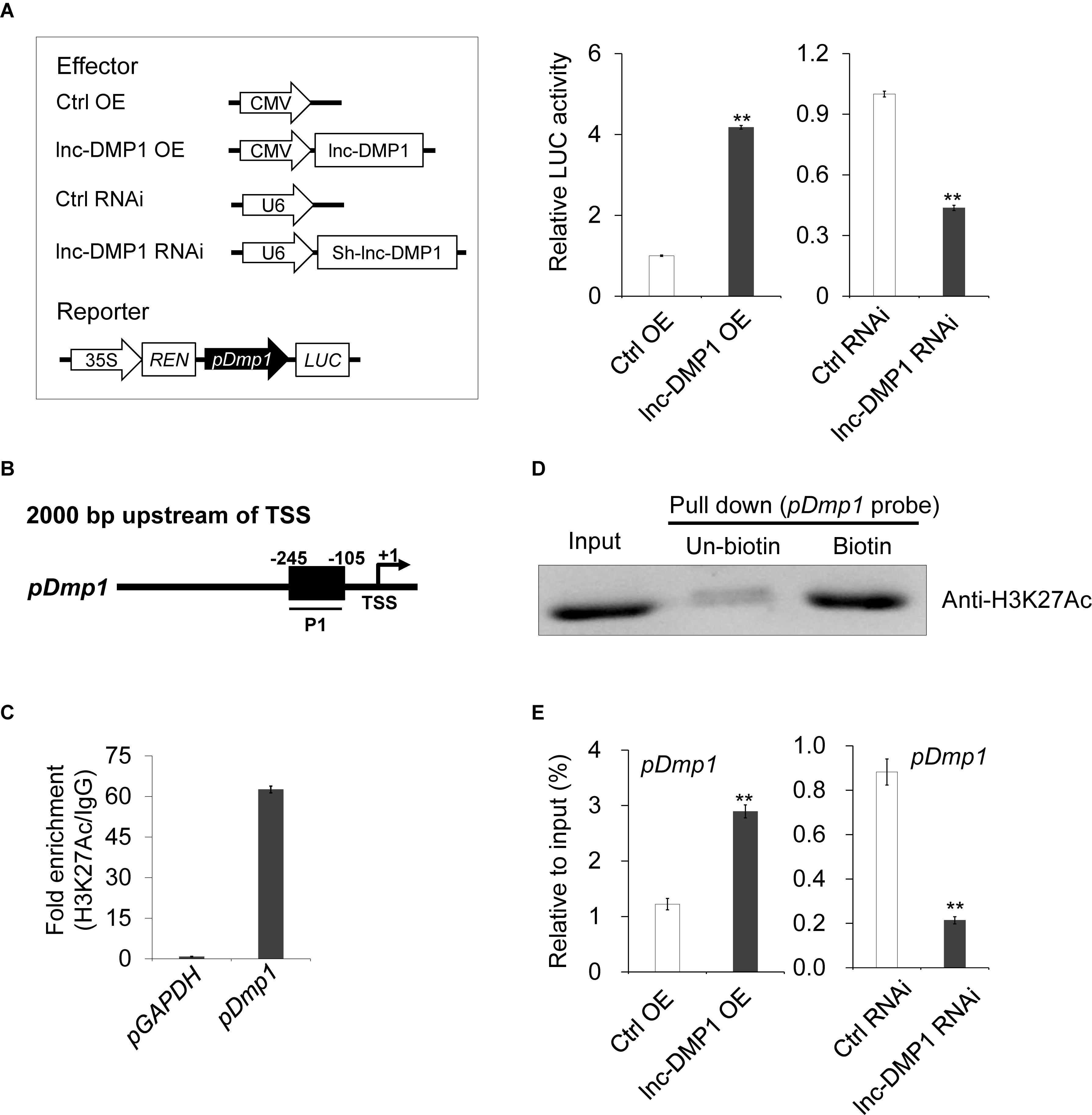
Figure 4. lnc-DMP1 induces the promoter activity by modulating the H3K27Ac enrichment of the Dmp1 promoter region in MC3T3-E1 cells. (A) Luciferase (LUC) reporter assay: the Dmp1 promoter region [2000 bp before transcription start site (TSS)] was cloned upstream of the firefly luciferase coding region. Their luciferase activities were tested in MC3T3-E1 co-transfected with CMV:Lnc-DMP1 (lnc-DMP1 OE), U6:sh-lnc-DMP1 (lnc-DMP1 RNAi) or empty vectors (Ctrl OE and Ctrl RNAi). The empty vector control was set to a value of 1. (B) Schematic of the potential H3K27Ac binding region in the promoter sequence of Dmp1. The numbers indicated the nucleotide positions relative to their transcription start site (TSS), which was shown as + 1. (C) qRT-PCR detection of the indicated DNAs retrieved by H3K27Ac-specific antibody compared with immunoglobulin G (IgG) in the CHIP assay within MC3T3-E1. (D) DNA pull down experiment with MC3T3-E1 extract. Specific bands were identified by immunoblotting H3K27Ac. (E) The alteration of the enrichment of H3K27Ac in the promoter region of Dmp1 in lnc-DMP1 OE and RNAi cells. **P < 0.01 by Student’s t-test.
lncRNA can bind adaptor protein and target transferase to drive histone modification and active gene transcription (Marchese et al., 2017). According to the ENCODE (Encyclopedia of DNA Elements) project data, we could not find any histone modification information of Dmp1 in mouse cells. Nonetheless, histone H3-lysine-27 acetylation (H3K27Ac) was enriched in the promoter region of Dmp1 in human H1-hESC cells. Hence, we conducted a ChIP assay to detect the enrichment of H3K27Ac in the promoter of Dmp1 in MC3T3-E1 cells. The results showed that the acetylation of H3K27 in the promoter of Dmp1 was enriched (Figures 4B,C). The DNA pull down assay also confirmed the binding of Dmp1 promoter to the H3K27Ac protein (Figure 4D).
Given that the enhanced H3K27Ac histone modification is usually an important agonist for transcriptional regulation, we hypothesized that lnc-DMP1 participated in the acetylation of H3K27 in the promoter of Dmp1. To confirm this hypothesis, we performed ChIP assay to detect the enrichment of H3K27Ac in the promoter of Dmp1 in lnc-DMP1 OE and lnc-DMP1 RNAi stable lines. As shown in Figure 4E, the overexpression of lnc-DMP1 increased the enrichment of H3K27Ac of the Dmp1 promoter, whereas the RNAi of lnc-DMP1 decreased the enrichment of H3K27Ac of the Dmp1 promoter. These results suggested that lnc-DMP1 induced the promoter activity by modulating the H3K27Ac enrichment of Dmp1 promoter.
lncRNAs play important regulatory roles during multiple biological processes and in various diseases (Wang et al., 2015; Ding et al., 2017). Histone modification through methylation and acetylation is the main regulatory mechanism by which lncRNAs regulate the expression of their neighbor protein-coding genes. lncRNAs modulate the expression of target genes by promoting the acetylation of H3K27 in the promoter regions of genes. Dmp1 is highly expressed in bone and dentin tissues and plays a key role in mineralization, but the mechanism of its regulation needs to further investigated (Narayanan et al., 2002; Jacob et al., 2014; Wang et al., 2014). In this study, we attempted to investigate the potential regulatory pattern in lncRNAs and Dmp1.
lnc-DMP1, a novel lncRNA validated by the lncRNA sequence in our study, was near the Dmp1 gene region and exhibited the most remarkable changes during mandible development. The RNA-FISH assay demonstrated that lnc-DMP1 and Dmp1 were expressed in the same areas in the mandibles and co-localized in the cytoplasm and the nuclei. Moreover, the significant correlation between the expression levels of lnc-DMP1 and Dmp1 at different stages of mandible development implied the presence of a regulatory mechanism between them. After constructing the stable lines of lnc-DMP1 overexpression and RNAi in osteoblast cell lineMC3T-E1, we found that overexpressing lnc-DMP1 promoted both mRNA and protein expression of DMP1 and skeletal mineralization in MC3T3-E1 cells. These results indicated that lnc-DMP1 positively regulated the expression of DMP1 to control the mineralization. The luciferase results implied that lnc-DMP1 induced the promoter activity of Dmp1. Given that the acetylation level of H3K27 was positively correlated with the transcriptional activity, the results of the ChIP assays confirmed our hypothesis that lnc-DMP1 enhanced Dmp1 expression by enriching H3K27Ac in Dmp1 promoter.
In summary, our study identified a novel lnc-DMP1 positively related to the Dmp1 during the development of mouse mandibles. The lnc-DMP1 regulated Dmp1 expression and mineralization by modulating the H3K27ac enrichment of Dmp1 promoter in osteoblast cells. In our future studies, we will further investigate chromatin-modifying complexes, which interact with lnc-DMP1 to modulate Dmp1 expression. Collectively, our findings will serve as a basis for exploring the mechanism of lncRNAs in the regulation of bone regeneration, including periodontal tissue reconstruction, and for tissue engineering and clinical practice.
The lnc-RNA sequencing data mentioned in our manuscript has been submitted to the SRA in NCBI, the SubmissionID is BioProject ID is PRJNA540617 and BioSample accessions are SAMN11549457 and SAMN11549458.
XX, YR, BL, YY, XK, and PZ performed the experiments and/or analyzed the data. HW designed the research. YR participated the design of some experiments. XX and HW wrote the manuscript. All co-authors corrected the manuscript.
This study was supported by the National Natural Science Foundation of China (81200825 to HW) and National Natural Science Foundation of Guangdong Province, China (2017A030313501 to YR and 2018A0303130106 to PZ).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Jinsong Li (Sun Yat-sen Memorial Hospital, China) for sending MC3T3-E1 cells, and Dong Yin (Sun Yat-sen Memorial Hospital, China) and Research center of the Sun Yat-sen Memorial Hospital for excellent technical help, and Anxun Wang (The First Affiliated Hospital, Sun Yat-sen University, China), Jun Shen (Sun Yat-sen Memorial Hospital, China) and Shilong Zhong (Guangdong provincial People’s hospital, China) for research and manuscript help.
Brockdorff, N., Ashworth, A., Kay, G. F., McCabe, V. M., Norris, D. P., Cooper, P. J., et al. (1992). The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell 71, 515–526. doi: 10.1016/0092-8674(92)90519-I
Brown, C. J., Hendrich, B. D., Rupert, J. L., Lafreniere, R. G., Xing, Y., Lawrence, J., et al. (1992). The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell 71, 527–542. doi: 10.1016/0092-8674(92)90520-M
Clemson, C. M., Hutchinson, J. N., Sara, S. A., Ensminger, A. W., Fox, A. H., Chess, A., et al. (2009). An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol. Cell 33, 717–726. doi: 10.1016/j.molcel.2009.01.026
Ding, G., Li, W., Liu, J., Zeng, Y., Mao, C., Kang, Y., et al. (2017). LncRNA GHET1 activated by H3K27 acetylation promotes cell tumorigenesis through regulating ATF1 in hepatocellular carcinoma. Biomed. Pharmacother. 94, 326–331. doi: 10.1016/j.biopha.2017.07.046
D’Souza, R. N., Cavender, A., Sunavala, G., Alvarez, J., Ohshima, T., Kulkarni, A. B., et al. (1997). Gene expression patterns of murine dentin matrix protein 1 (Dmp1) and dentin sialophosphoprotein (DSPP) suggest distinct developmental functions in vivo. J. Bone Miner. Res. 12, 2040–2049. doi: 10.1359/jbmr.1997.12.12.2040
Fatica, A., and Bozzoni, I. (2014). Long non-coding RNAs: new players in cell differentiation and development. Nat. Rev. Genet. 15, 7. doi: 10.1038/nrg3606
Feng, J. Q., Luan, X., Wallace, J., Jing, D., Ohshima, T., Kulkarni, A. B., et al. (1998). Genomic organization, chromosomal mapping, and promoter analysis of the mouse dentin sialophosphoprotein (Dspp) gene, which codes for both dentin sialoprotein and dentin phosphoprotein. J. Biol. Chem. 273, 9457–9464. doi: 10.1074/jbc.273.16.9457
Feng, J. Q., Ward, L. M., Liu, S., Lu, Y., Xie, Y., Yuan, B., et al. (2006). Loss of DMP1 causes rickets and osteomalacia and identifies a role for osteocytes in mineral metabolism. Nat. Genet. 38:1310. doi: 10.1038/ng1905
Fortes, P., and Morris, K. V. (2016). Long noncoding RNAs in viral infections. Virus Res. 212, 1–11. doi: 10.1016/j.virusres.2015.10.002
George, A., Sabsay, B., Simonian, P. A., and Veis, A. (1993). Characterization of a novel dentin matrix acidic phosphoprotein. Implications for induction of biomineralization. J. Biol. Chem. 268, 12624–12630.
Hirst, K. L., Ibaraki-O’Connor, K., Young, M. F., and Dixon, M. J. (1997). Cloning and expression analysis of the bovine dentin matrix acidic phosphoprotein gene. J. Dent. Res. 76, 754–760. doi: 10.1177/00220345970760030701
Jacob, A., Zhang, Y., and George, A. (2014). Transcriptional regulation of dentin matrix protein 1 (DMP1) in odontoblasts and osteoblasts. Connect. Tissue. Res. 55, 107–112. doi: 10.3109/03008207.2014.923850
Kanduri C. (2016). Long noncoding RNAs: lessons from genomic imprinting. BBA-Gene Regul. Mech. 1859, 102–111. doi: 10.1016/j.bbagrm.2015.05.006
Lee, J. T., and Bartolomei, M. S. (2013). X-inactivation, imprinting, and long noncoding RNAs in health and disease. Cell 152, 1308–1323. doi: 10.1016/j.cell.2013.02.016
Ling, Y., Rios, H. F., Myers, E. R., Lu, Y., Feng, J. Q., and Boskey, A. L. (2005). DMP1 depletion decreases bone mineralization in vivo: an FTIR imaging analysis. J. Bone and Miner. Res. 20, 2169–2177. doi: 10.1359/JBMR.050815
Lorenz-Depiereux, B., Bastepe, M., Benet-Pagès, A., Amyere, M., Wagenstaller, J., Müller-Barth, U., et al. (2006). DMP1 mutations in autosomal recessive hypophosphatemia implicate a bone matrix protein in the regulation of phosphate homeostasis. Nat. Genet. 38, 1248–1250. doi: 10.1038/ng1868
MacDougall, M., Simmons, D., Luan, X., Nydegger, J., Feng, J., and Gu, T. T. (1997). Dentin phosphoprotein and dentin sialoprotein are cleavage products expressed from a single transcript coded by a gene on human chromosome 4 Dentin phosphoprotein DNA sequence determination. J. Biol. Chem. 272, 835–842. doi: 10.1074/jbc.272.2.835
Marchese, F. P., Raimondi, I., and Huarte, M. (2017). The multidimensional mechanisms of long noncoding RNA function. Genome Bio. 18, 206–218. doi: 10.1186/s13059-017-1348-2
Melo, C. A., Léveillé, N., and Agami, R. (2013). eRNAs reach the heart of transcription. Cell Res. 23, 1151–1152. doi: 10.1038/cr.2013.97
Narayanan, K., Ramachandran, A., Hao, J., and George, A. (2002). Transcriptional regulation of dentin matrix protein 1 (DMP1) by AP-1 (c-fos/c-jun) factors. Connect. Tissue Res. 43, 365–371. doi: 10.1080/03008200290000592
Narayanan, K., Ramachandran, A., Hao, J., He, G., Park, K. W., Cho, M., et al. (2003). Dual functional roles of dentin matrix protein 1 Implications in biomineralization and gene transcription by activation of intracellular Ca2+ store. J. Biol. Chem. 278, 17500–17508. doi: 10.1074/jbc.M212700200
Prasanth, K. V., Prasanth, S. G., Xuan, Z., Hearn, S., Freier, S. M., Bennett, C. F., et al. (2005). Regulating gene expression through RNA nuclear retention. Cell 123, 249–263. doi: 10.1016/j.cell.2005.08.033
Qin, C., D’Souza, R., and Feng, J. Q. (2007). Dentin matrix protein 1 (DMP1): new and important roles for biomineralization and phosphate homeostasis. J. Dent. Res. 86, 1134–1141. doi: 10.1177/154405910708601202
Quinn, J. J., and Chang, H. Y. (2016). Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 17, 47–62. doi: 10.1038/nrg.2015.10
Roberts, T. C., Morris, K. V., and Weinberg, M. S. (2014). Perspectives on the mechanism of transcriptional regulation by long non-coding RNAs. Epigenetics 9, 13–20. doi: 10.4161/epi.26700
Sunwoo, H., Dinger, M. E., Wilusz, J. E., Amaral, P. P., Mattick, J. S., and Spector, D. L. (2009). MEN ε/β nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res. 19, 347–359. doi: 10.1101/gr.087775.108
Toyosawa, S., Shintani, S., Fujiwara, T., Ooshima, T., Sato, A., Ijuhin, N., et al. (2001). Dentin matrix protein 1 is predominantly expressed in chicken and rat osteocytes but not in osteoblasts. J. Bone Miner. Res. 16, 2017–2026. doi: 10.1359/jbmr.2001.16.11.2017
Wang, T., Liu, H., Ning, Y., and Xu, Q. (2014). The histone acetyltransferase p300 regulates the expression of pluripotency factors and odontogenic differentiation of human dental pulp cells. PLoS One 9:e102117. doi: 10.1371/journal.pone.0102117
Wang, Y., Zhong, H., Xie, X., Chen, C. Y., Huang, D., Shen, L., et al. (2015). Long noncoding RNA derived from CD244 signaling epigenetically controls CD8+ T-cell immune responses in tuberculosis infection. Proc. Natl. Acad. Sci. U.S.A. 112, E3883–E3892. doi: 10.1073/pnas.1501662112
Ye, L., MacDougall, M., Zhang, S., Xie, Y., Zhang, J., Li, Z., et al. (2004). Deletion of dentin matrix protein-1 leads to a partial failure of maturation of predentin into dentin, hypomineralization and expanded cavities of pulp and root canal during postnatal tooth development. J. Biol. Chem. 279, 19141–19148. doi: 10.1074/jbc.M400490200
Ye, L., Mishina, Y., Chen, D., Huang, H., Dallas, S. L., Dallas, M. R., et al. (2005). Dmp1-deficient mice display severe defects in cartilage formation responsible for a chondrodysplasia-like phenotype. J. Biol. Chem. 280, 6197–6203. doi: 10.1074/jbc.M412911200
Keywords: long non-coding RNA, Dentin matrix protein 1, transcriptional regulation, mice mandible, MC3T3-E1 cell, H3K27Ac
Citation: Xia X, Ruan Y, Li B, Yu Y, Kong X, Zhuang P and Wu H (2020) The Long Non-coding RNA lnc-DMP1 Regulates Dmp1 Expression Through H3K27Ac Modification. Front. Genet. 11:233. doi: 10.3389/fgene.2020.00233
Received: 15 May 2019; Accepted: 27 February 2020;
Published: 18 March 2020.
Edited by:
Graziano Pesole, Institute of Biomembranes, Bioenergetics and Molecular Biotechnologies, National Research Council, ItalyCopyright © 2020 Xia, Ruan, Li, Yu, Kong, Zhuang and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hong Wu, Y2Fycmlld2hAaG90bWFpbC5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.