
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet., 07 November 2019
Sec. Epigenomics and Epigenetics
Volume 10 - 2019 | https://doi.org/10.3389/fgene.2019.00909
This article is part of the Research TopicEpigenetic Biomarker and Personalized Precision MedicineView all 33 articles
Endometrial cancer (EC) is one of the most common gynecologic malignancies. Emerging studies had demonstrated the mutations in genes could serve as diagnostic or prognostic markers for human cancers. In this study, we screened mutated genes in EC and found that the mutations in KIAA1109, CACNA1C, BSN, AKAP13, CELSR2, and HELZ2 were correlated to the overall survival time in patients with EC. Bioinformatics analysis showed KIAA1109 was involved in regulating NIK/NF-kappaB signaling, CACNA1C was found to regulate cell migration and proliferation, BSN was found to regulate Wnt signaling pathway, CELSR2 was involved in regulating cell–cell adhesion, nuclear import, and protein folding, HELZ2 was found to regulate multiple immune related biological processes, and AKAP13 was involved in regulating translation, mRNA nonsense-mediated decay, rRNA processing, translational initiation, and mRNA splicing via spliceosome. The findings provided a novel therapeutic strategy in patients with EC.
Endometrial cancer (EC) is one of the most common gynecologic malignancies (Attarha et al., 2011). Despite the prognosis of the early stage EC is good with a 5-year survival rate of 69–88% (Gottwald et al., 2010). However, the prognosis of metastatic EC remained very poor, with a median survival of 7–12 months. Therefore, there is an urgent need to identify novel biomarkers for the prognosis of EC. Moreover, the mechanisms underlying the progression of EC remained largely unclear.
With the development of next-generation sequencing, multiple EC related mutations were identified. Emerging studies had demonstrated the mutations in genes could serve as diagnostic or prognostic markers for human cancers. For example, McConechy et al. identified a series of mutations in PTEN, CTNNB1, PIK3CA, ARID1A, ARID5B, and KRAS were associated with EC (Mcconechy et al., 2012). The mutations in FGFR2 were associated with poor outcomes in endometrioid endometrial cancer (Jeske et al., 2017). The genetic alterations in CTCF could promote EC cell survival and alter cell polarity (Marshall et al., 2017). Jing et al. found that MUC16 mutations could improve patients’ prognosis by enhancing the infiltration of cytotoxic T lymphocytes in the EC microenvironment (Jing and Jing, 2014).
The present study identified prognosis related gene mutations in EC by analyzing TCGA databases (Collins, 2007). The mutations in 6 genes were correlated to the overall survival time in patients with EC. Bioinformatics analysis was used to predict the potential functions of these genes. The purpose of this study was to evaluate the impact of somatic tumor mutation on recurrence-free survival in this patient population.
In this study, we identified the gene mutations in EC using TCGA database (https://portal.gdc.cancer.gov/). All searches were performed according to cBioPortal’s online instructions (http://www.cbioportal.org/index.do) (Jianjiong et al., 2013). The survival analysis related to gene mutations was performed on the TCGA database (https://portal.gdc.cancer.gov/).
In this study, the Pearson correlation coefficient was calculated according to the expression value between lncRNA–mRNA pair using cBioPortal’s online instructions (http://www.cbioportal.org/index.do). The top 500 co-expressing genes were selected as potential targets of mutated genes in EC.
GO and KEGG pathway enrichment analysis were performed to determine the biological significance of DEGs, using the Database for Annotation, Visualization, and Integrated Discovery (Dennis et al., 2003) (DAVID; version 6.8; http://david.ncifcrf.gov/).
Survival curves were depicted using the Kaplan-Meier method and compared with log-rank test. Cox proportional hazards regression analysis was used for univariate and multivariate analyses to explore the association of clinical features, gene mutational status, and patients’ prognosis. All the prognostic analyses were conducted by survival R package.
The two groups were compared using Student’s t‐test. Overall survival time analyses were estimated using the Kaplan-Meier product-limit estimator, and then a log-rank test was conducted to compare wildtype and mutation status. Overall survival was measured from the date of surgery to the date of last contact or death. Patients alive were censored at the date of last contact or clinic visit. Stata v14.2 (College Station, TX) was used to conduct statistical analysis.
The present study analyzed TCGA database to identify mutated genes in EC. As shown in Figure 1, the top 50 mutated genes in EC included TTN, MUC4, MUC16, PIK3CA, KMT2C, KMT2D, SYNE1, FLG, SYNE2, EP300, OBSCN, ADGRV1, RYR2, LRP1B, USH2A, MUC17, NEB, MDN1, MUC5B, CSMD1, PCLO, HUWE1, FBXW7, DMD, NSD1, NAV3, DNAH8, DST, PLEC, AHNAK2, LRP2, MKI67, DNAH2, TENM1, DNAH10, PRKDC, FAT1, TP53, HMCN1, ZFHX4, DNAH6, UBR4, NOTCH1, CREBBP, NIPBL, EYS, AHNAK, CSMD3, XIRP2, and MACF1. Among these genes, TTN, MUC4, and PIK3CA are the most frequently mutated genes. The mutation rates in TTN, MUC4, and PIK3CA from the TCGA provisional data sets were 43.25% (125/289), 31.83% (92/289), and 29.41% (85/289), respectively.
Next, we screened somatic mutations associated with overall survival time in patients with EC. As shown in Figure 2, Log-rank test showed that mutations in KIAA1109, CACNA1C, BSN, AKAP13, and HELZ2 were significantly associated with the longer overall survival time in EC patients, however, mutations in CELSR2 were significantly associated with the shorter overall survival time in EC patients.
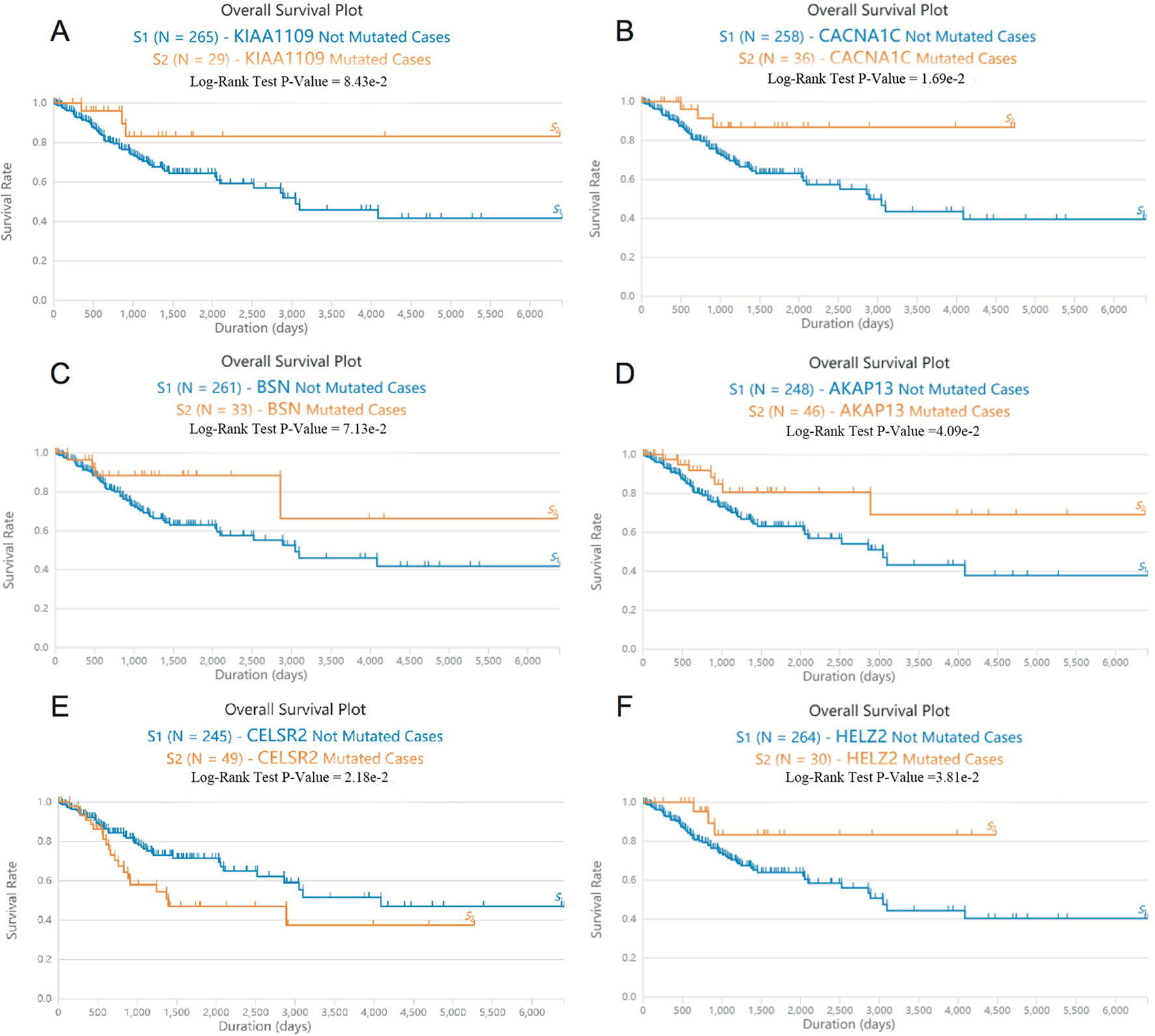
Figure 2 The somatic mutations of KIAA1109, CACNA1C, BSN, AKAP13, CELSR2, and HELZ2 were correlated to overall survival time in patients with EC. (A–F) Log-rank test showed that mutations in KIAA1109 (A), CACNA1C (B), BSN (C), AKAP13 (D), CELSR2 (E) and HELZ2 (F) were associated with the overall survival time in EC patients.
The mutation rates in KIAA1109, CACNA1C, BSN, AKAP13, CELSR2, and HELZ2 from the TCGA provisional data sets were 6.92% (20/289), 7.27% (21/289), 7.96% (23/289), 7.61% (22/289), 6.92% (20/289), and 7.27% (21/289), respectively in Figure 3. A, majority of mutations identified were missense and nonsense resulting in amino acid, changes and a truncation of these proteins. However, there was no evidence of a mutational hotspot in KIAA1109, CACNA1C, BSN, AKAP13, CELSR2, and HELZ2 in EC patients (Figure 4).
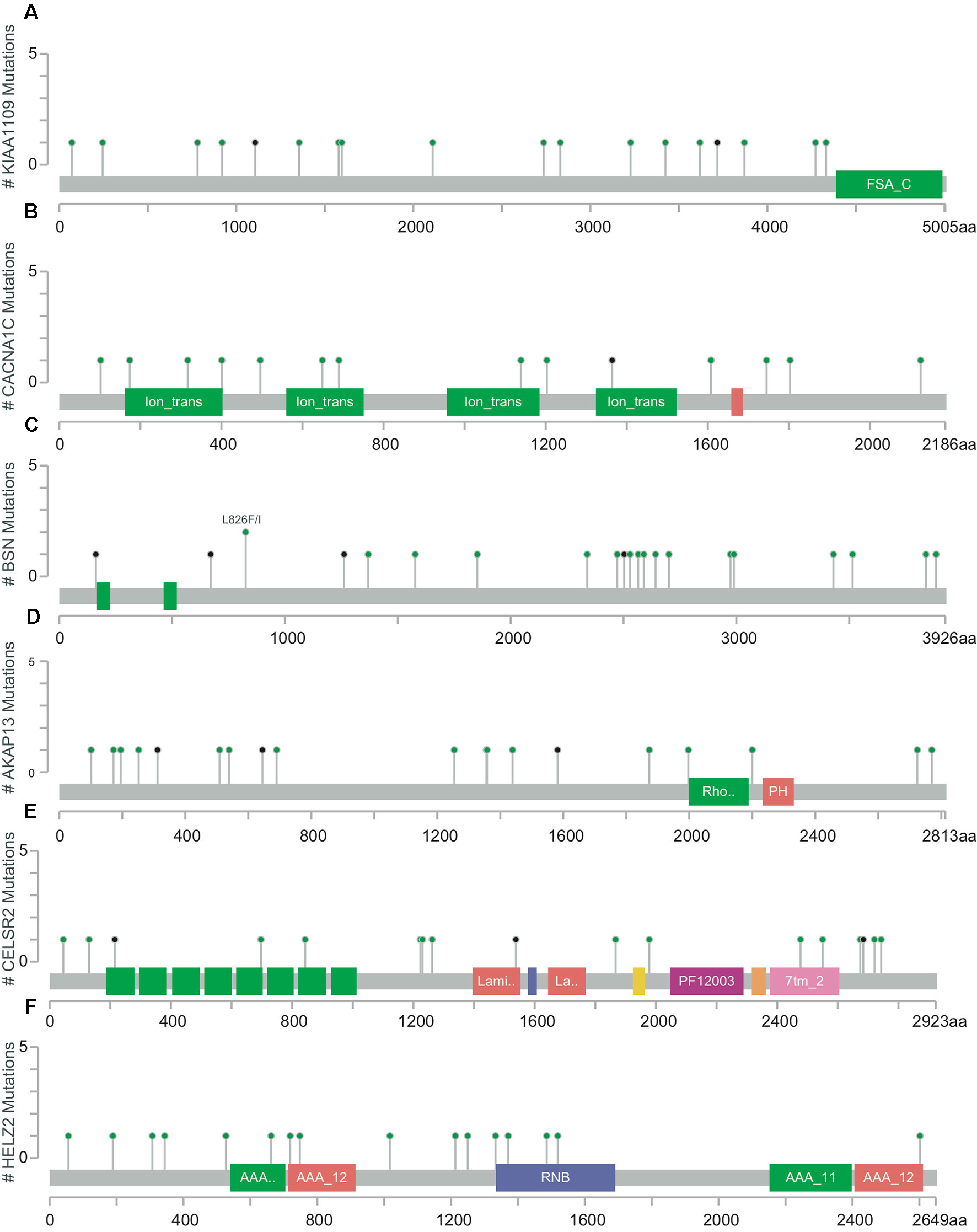
Figure 4 Detailed mutation maps of KIAA1109 (A), CACNA1C (B), BSN (C), AKAP13 (D), CELSR2 (E) and HELZ2 (F) found in patients with EC. Each dot above the protein molecule represents a mutation, which spreads across the entire encoded protein of these genes.
Furthermore, we detected the effect of mutations in KIAA1109, CACNA1C, BSN, AKAP13, CELSR2, and HELZ2 on mRNA expression based on the RNA-Seq data. As shown in Figure 5, we found the mutations in CACNA1C, BSN, CELSR2, and HELZ2 did not result in a significant alteration of their mRNA levels. However, we found that the mRNA levels in KIAA1109 and AKAP13 mutated EC samples were lower than that in KIAA1109 and AKAP13 wild type EC samples.
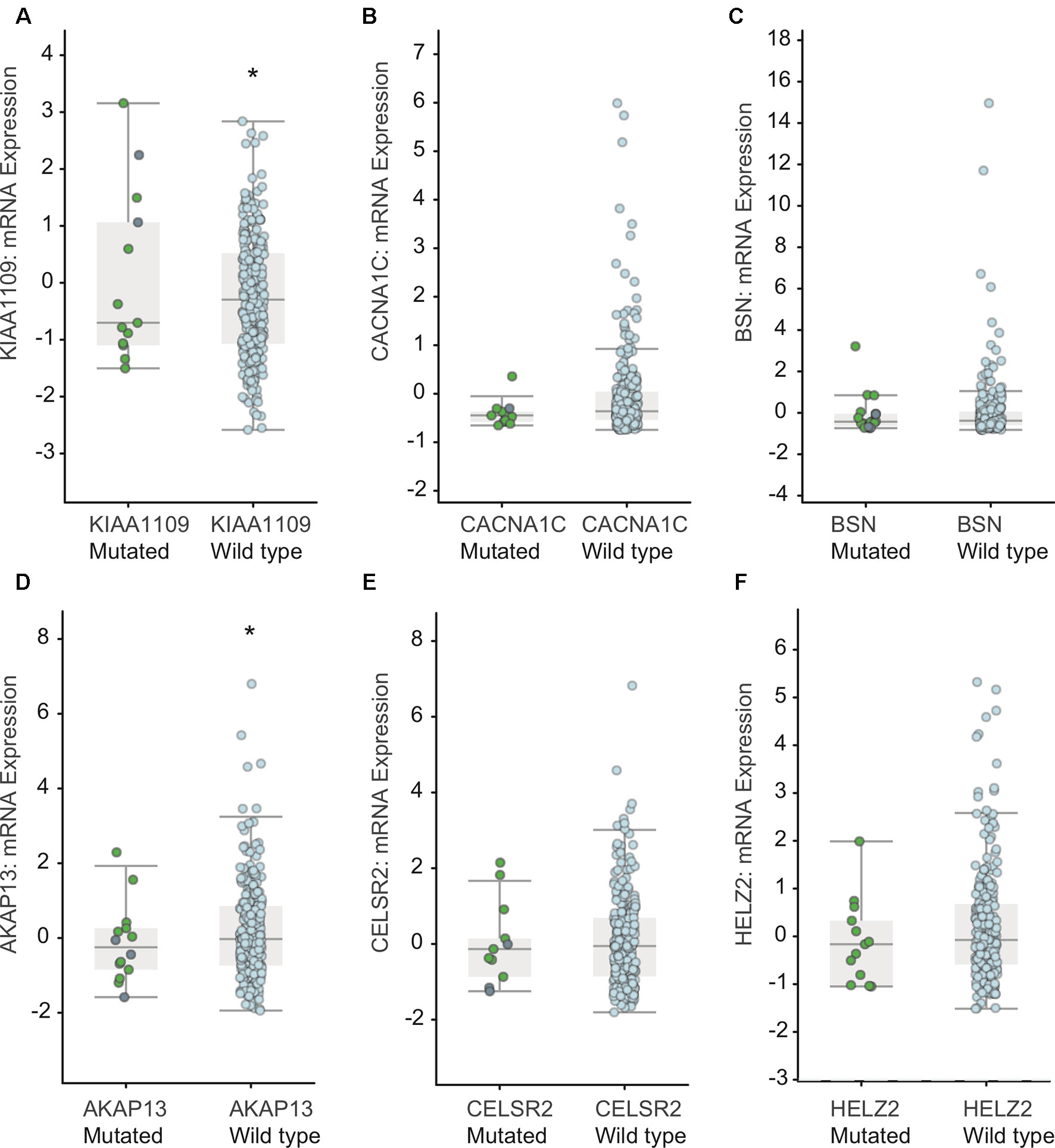
Figure 5 Association between mutations in KIAA1109 (A), CACNA1C (B), BSN (C), AKAP13 (D), CELSR2 (E) and HELZ2 (F) and mRNA levels in EC samples. * means p value < 0.05 between the two groups.
Furthermore, we performed bioinformatics analysis to reveal the potential functions of KIAA1109, CACNA1C, BSN, AKAP13, CELSR2, and HELZ2 using their co-expressing mRNAs in EC patients. The present study selected the top 500 correlated genes as the potential targets of KIAA1109, CACNA1C, BSN, AKAP13, CELSR2, and HELZ2. Bioinformatics analysis showed KIAA1109 was involved in regulating rRNA processing, translation, transcription, NIK/NF-kappaB signaling, and histone acetylation. The results were shown in Figure 6. CACNA1C was involved in regulating collagen fibril organization, cell-matrix adhesion, cellular response to amino acid stimulus, cell adhesion, and negative regulation of cell proliferation. BSN was involved in regulating epidermis development, cilium movement, smoothened signaling pathway, Wnt signaling pathway, planar cell polarity pathway, and cilium morphogenesis. AKAP13 was involved in regulating translation, mRNA nonsense-mediated decay, rRNA processing, translational initiation, and mRNA splicing via spliceosome. CELSR2 was involved in regulating cell–cell adhesion, keratinocyte differentiation, spliceosomal snRNP assembly, nuclear import, and protein folding. HELZ2 was involved in regulating type I interferon signaling pathway, innate immune response, immune response, inflammatory response, and T cell activation.
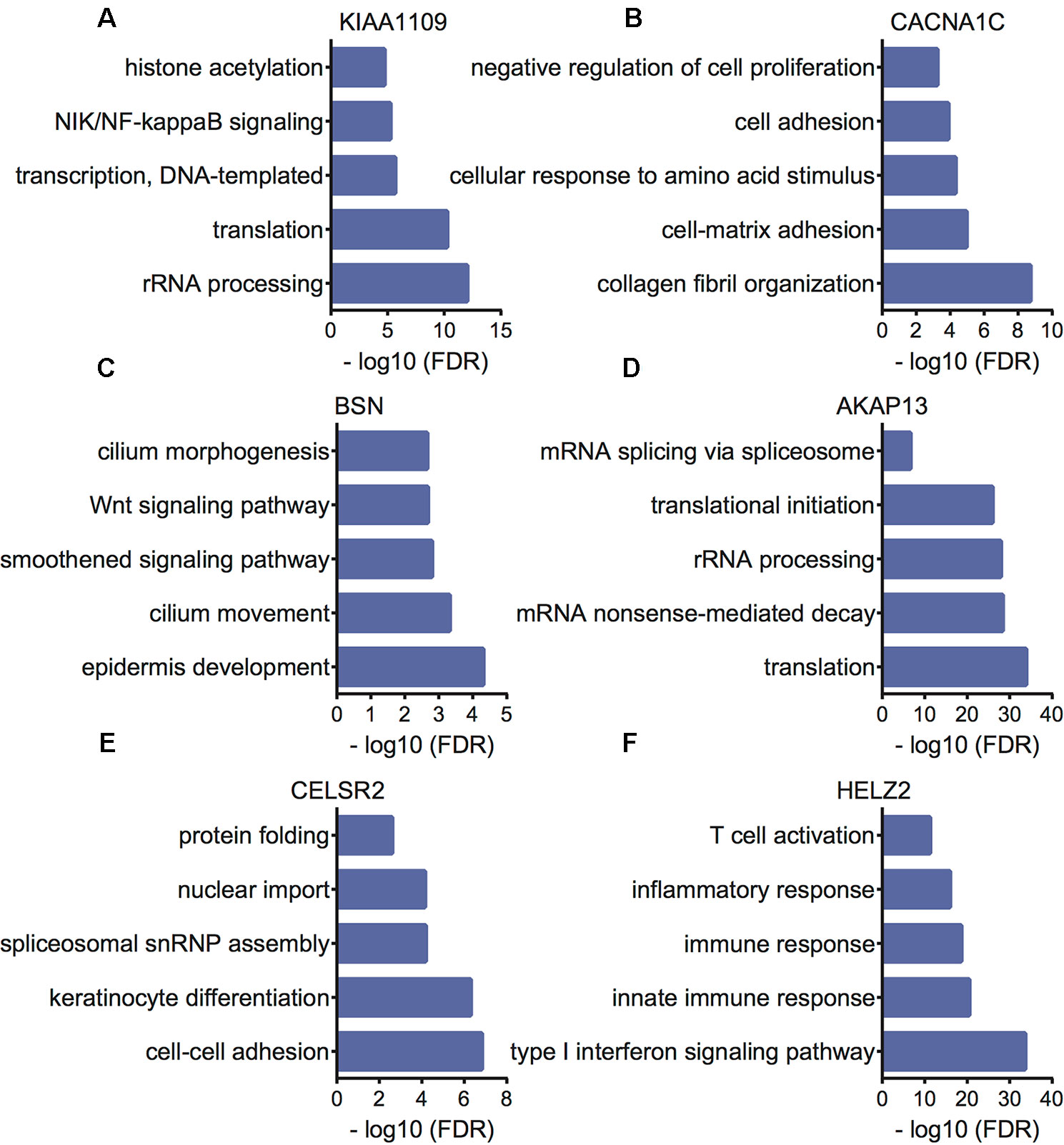
Figure 6 Bioinformatics analysis of KIAA1109 (A), CACNA1C (B), BSN (C), AKAP13 (D), CELSR2 (E) and HELZ2 (F) in EC samples.
Endometrial cancer (EC) is one of the most common gynecologic malignancies. However, the mechanisms underlying EC progression remained unclear. Previous studies had showed the mutations in several genes were related to EC. For example, MUC16 mutations improve EC prognosis through enhancing the infiltration of cytotoxic T lymphocytes. PTEN and PIK3CA mutations played crucial roles in grade 3 EC (Jing and Jing, 2014). The present study screened mutated genes in EC. Our results showed TTN, MUC4, and PIK3CA were the most frequently mutated genes in the EC, which was consistent with previous studies. Moreover, we identified the mutations in 6 genes were associated with the prognosis of EC. The results showed that mutations in KIAA1109, CACNA1C, BSN, AKAP13, and HELZ2 were significantly associated with the longer overall survival time in EC patients. However, mutations in CELSR2 were significantly associated with the shorter overall survival time in EC patients. These results suggested the important roles of these genes in the progression and prognosis of EC.
KIAA1109, located on the chromosome 4, was reported to be associated with susceptibility to celiac disease. Of note, 2 recent studies indicated KIAA1109 was associated with the prognosis of human cancers. For example, Qing et al. reported mutations in KIAA1109, DNAH5 and KCNH7 were associated with poor survival of Chinese esophageal squamous cell carcinoma patients (Tao et al., 2017). Tindall et al. found genetic variation of KIAA1109 might be associated with prostate cancer susceptibility in men with a family history of the disease (Tindall et al., 2010). CACNA1C gene encodes an alpha-1 subunit of a voltage-dependent calcium channel (Fayi et al., 2016). The mutations in CACNA1C were observed in various types of human diseases, such as ventricular fibrillation, and schizophrenia (Charles et al., 2007). Previous studies showed CACNA1C was down-regulated in multiple human cancers (Fastje et al., 2009), including brain tumors, kidney cancers and lung cancers, suggested its regulatory roles in cancer progression. BSN encoded a scaffolding protein involved in organizing the presynaptic cytoskeleton. BSN has been demonstrated to have chemo-preventive, antiproliferative, antifungal, and anti-carcinogenic activities. In addition, BSN has been reported to induce G1 phase arrest through increase of p21 and p27. In PCa, BSN was involved in regulating cell apoptosis in cancer cells (Xu et al., 2016). The dysregulation and mutation of AKAP13 were found to be associated with the progression of colorectal cancer and breast cancer. Bentin et al. showed AKAP13 is essential for the phosphorylation of ERαS305 (Toaldo et al., 2015), which leads to tamoxifen resistance in breast cancer. HELZ2 encoded b a nuclear transcriptional co-activator for peroxisome proliferator activated receptor alpha (Jakobsson et al., 2010). However, its roles in human cancers remained largely unclear. CELSR2 was found to be dysregulated in breast cancer (Jiang et al., 2018). However, the potential functions of CELSR2 in EC remained unknown.
In the present study, we performed co-expression analysis to reveal the potential roles of these mutated genes in EC. The results showed KIAA1109 was involved in regulating NIK/NF-kappaB signaling. Of note, NF-kappaB signaling had been demonstrated to be a key regulator in cancers. Suppressing of NF-kappaB signaling could inhibit cell growth and invasion in multiple cancers. For example, NF-κB suppresses apoptosis and promotes the proliferation of bladder cancer cells. A recent study showed liposomal curcumin targeting EC through the NF-κB Pathway. Bioinformatics analysis revealed CACNA1C played important roles in regulation of EC metastasis and proliferation. BSN was found to regulate Wnt signaling pathway. Mounting evidence has confirmed the activation of Wnt/β-catenin signaling was associated with multiple cancers, including EC. AKAP13 was predicted as a RNA processing regulator. CELSR2 was involved in regulating cell–cell adhesion, keratinocyte differentiation, spliceosomal snRNP assembly, nuclear import, and protein folding. HELZ2 was involved in regulating type I interferon signaling pathway, innate immune response, immune response, inflammatory response, and T cell activation. These results suggested these mutated genes played important roles in EC tumorigenesis and progression.
Despite that bioinformatics analyses were conducted to predict the potential functions of these mutated genes in EC, several limitations still existed in this study. First, the mutated sites of these genes should be further validated in EC clinical samples using Sanger sequencing. Second, the molecular function of these key mutated genes in EC remained unclear. Therefore, gain or loss of function assays should be further conducted to investigate their important roles in EC.
In conclusion, we screened mutated genes in EC and found that the mutations in KIAA1109, CACNA1C, BSN, AKAP13, CELSR2, and HELZ2 correlated with the overall survival time in patients with EC. Bioinformatics analysis showed KIAA1109 was involved in regulating NIK/NF-kappaB signaling, CACNA1C was found to regulate cell migration and proliferation, BSN was found to regulate Wnt signaling pathway, CELSR2 was involved in regulating cell-cell adhesion, nuclear import, and protein folding, and HELZ2 was found to regulate multiple immune related biological processes. The findings provided a novel therapeutic strategy in patients with EC.
All datasets analysed in this study can be found in the Genomic Data Commons Data Portal (https://portal.gdc.cancer.gov/). We screened the gene mutations in EC. We have downloaded these data from the database and the top 500 mutated genes in EC were listed in Supplementary Table 1.
JZ designed experiments; ZQ and YJ analyzed the data. All authors wrote and approved the manuscript.
This work is supported by the Natural Science Foundation of Liaoning Province, China (Grant no. 20170540570).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2019.00909/full#supplementary-material
Supplementary Table 1 | The top 500 mutated genes in EC.
Attarha, S., Mints, M., Andersson, S., Souchelnytskyi, S., (2011). Endometrial cancer and application of proteomics. Exp. Oncol. 33, 174–177.
Charles, A., Pollevick, G. D., Cordeiro, J. M., Oscar, C., Sanguinetti, M. C., Yoshiyasu, A., et al. (2007). Loss-of-function mutations in the cardiac calcium channel underlie a new clinical entity characterized by ST-segment elevation, short QT intervals, and sudden cardiac death. Circ. 115, 442–449. doi: 10.1161/CIRCULATIONAHA.106.668392
Dennis, G., Jr., Sherman, B. T., Hosack, D. A., Yang, J., Gao, W., Lane, C. H., et al. (2003). DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 4 (9) R60.
Fastje, C. D., Le, K., Sun, N. N., Wong, S. S., Sheppard, P. R., Witte, M. L. (2009). Prenatal exposure of mice to tungstate is associated with decreased transcriptome-expression of the putative tumor suppressor gene, DMBT1: implications for childhood leukemia. Land Contam. Reclam. 17, 169–178. doi: 10.2462/09670513.931
Fayi, N., Xiaoli, W., Panpan, Z., Hao, Y., Wenhua, Z., Yaling, Z., et al. (2016). Genetic analysis of SNPs in CACNA1C and ANK3 gene with schizophrenia: a comprehensive meta-analysis. Am. J. Med. Genet. B Neuropsychiatr. Genet. 168, 637–648. doi: 10.1002/ajmg.b.32348
Gottwald, L., Pluta, P., Piekarski, J., Spych, M., Hendzel, K., Topczewskatylinska, K., et al. (2010). Long-term survival of endometrioid endometrial cancer patients. Arch. Med. Sci. 6, 937–944. doi: 10.5114/aoms.2010.19305
Jakobsson, L., Franco, C. A., Bentley, K., Collins, R. T., Ponsioen, B., Aspalter, I. M., et al. (2010). Endothelial cells dynamically compete for the tip cell position during angiogenic sprouting. Nat. Cell Biol. 12, 943–953. doi: 10.1038/ncb2103
Jeske, Y. W., Ali, S., Byron, S. A., Gao, F., Mannel, R. S., Ghebre, R. G., et al. (2017). FGFR2 mutations are associated with poor outcomes in endometrioid endometrial cancer: an NRG oncology/gynecologic oncology group study. Gynecol. Oncol. 145, 366–373. doi: 10.1016/j.ygyno.2017.02.031
Jiang, L., Zhang, X., Xiang, C., Geradts, J., Wei, Q., Liang, Y., et al. (2018). Differential cellular localization of CELSR2 and ING4 and correlations with hormone receptor status in breast cancer. Histol. Histopathol. 33, 11979. doi: 10.14670/HH-11-979
Jianjiong, G., Bülent Arman, A., Ugur, D., Gideon, D., Benjamin, G., Onur, S. S., et al. (2013). Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6, pl1–pl1. doi: 10.1126/scisignal.2004088
Jing, H., Jing, S. (2014). MUC16 mutations improve patients’ prognosis by enhancing the infiltration and antitumor immunity of cytotoxic T lymphocytes in the endometrial cancer microenvironment. Oncoimmunology.
Marshall, A. D., Bailey, C. G., Champ, K., Vellozzi, M., O’Young, P., Metierre, C., et al. (2017). CTCF genetic alterations in endometrial carcinoma are pro-tumorigenic. Oncogene 36, 4100–4110. doi: 10.1038/onc.2017.25
Mcconechy, M. K., Jiarui, D., Cheang, M. C. U., Wiegand, K. C., Janine, S., Tone, A. A., et al. (2012). Use of mutation profiles to refine the classification of endometrial carcinomas. J. Pathol. 228, 20–30. doi: 10.1002/path.4056
Tao, Q., Zhu, S., Suo, C., Zhang, L., Zheng, Y., Shi, L. (2017). Somatic mutations in ZFHX4 gene are associated with poor overall survival of Chinese esophageal squamous cell carcinoma patients. Sci. Rep. 7, 4951. doi: 10.1038/s41598-017-04221-7
Tindall, E. A., Hoang, H. N., Southey, M. C., English, D. R., Hopper, J. L., Giles, G. G., et al. (2010). The 4q27 locus and prostate cancer risk. BMC Cancer 10, 69. doi: 10.1186/1471-2407-10-69
Toaldo, C. B., Alexi, X., Beelen, K., Kok, M., Hauptmann, M., Jansen, M., et al. (2015). Protein Kinase A-induced tamoxifen resistance is mediated by anchoring protein AKAP13. BMC Cancer 15, 1–12. doi: 10.1186/s12885-015-1591-4
Keywords: endometrial cancer, bioinformatics analyses, mutation, overall survival time, biomarkers
Citation: Qiao Z, Jiang Y, Wang L, Wang L, Jiang J and Zhang J (2019) Mutations in KIAA1109, CACNA1C, BSN, AKAP13, CELSR2, and HELZ2 Are Associated With the Prognosis in Endometrial Cancer. Front. Genet. 10:909. doi: 10.3389/fgene.2019.00909
Received: 12 May 2019; Accepted: 28 August 2019;
Published: 07 November 2019.
Edited by:
Rui Henrique, Portuguese Oncology Institute, PortugalReviewed by:
Noritaka Yamaguchi, Chiba University, JapanCopyright © 2019 Qiao, Jiang, Wang, Wang, Jiang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jingru Zhang, eWk4NTMxNzg3MEAxNjMuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.