- 1Smithsonian Tropical Research Institute, Panama City, Panama
- 2Department of Genetics and Molecular Biology, University of Panama, Panama City, Panama
Biodiversity surveys of aquatic systems often include DNA metabarcoding analyses of environmental samples that are collected through filtration of large volumes of water. The standard practice of sterile collection and filtration in or near the field sites is challenging to implement in remote locations, and filtration of large volumes is a limiting step, especially for water from highly productive systems or with high suspended sediment loads. Recent trials have shown that passive samplers can be effective for aquatic metabarcoding to document metazoan diversity, but that this approach needs to be trialed under a wider variety of conditions and across more diverse taxa. Here we assess the utility of passive sampling for documenting the diversity of bacteria in six tropical aquatic environments (one lake, one reservoir, two mountain streams and two blackwater rivers). We find that passive collectors generally recover significantly higher diversity of Bacteria compared to filtered samples, despite capturing significantly less overall DNA than active water filtering. However, the communities captured by the two methods show significant differences within sites, with only 26% of the Bacteria ASVs recovered by both methods. These differences were largely driven by relative abundances of taxa within Actinobacteriota, Campilobacterota, Desulfobacterota, and Proteobacteria. Our results demonstrate that passive collectors can be a cost-effective solution for monitoring aquatic microbial diversity but that the two methods are not interchangeable. Additional work is necessary to understand the selectivity of both passive collectors and active water filtering for eDNA studies.
Introduction
Microbial communities can be incredibly diverse and molecular methods are revolutionizing the endeavor of describing the biodiversity and understanding the microbial ecology of aquatic systems. Metabarcoding analysis of environmental DNA (eDNA) obtained from filtered water samples has allowed the characterization of diverse and remote systems on a temporal and spatial scale difficult to implement with classical approaches to biodiversity discovery and monitoring (e.g., Sales et al., 2021; Bista et al., 2017; Li et al., 2023). Nevertheless, logistical constraints for sample collection and processing may still limit the temporal and spatial resolution or even prohibit this approach from being applied in very remote or under-resourced regions.
In many cases, the limiting step in the traditional approach is the collection and filtering of water samples. As the number of species detected generally increases with the volume of water filtered, large volumes (often 1 L) are typically vacuum filtered in preference to syringe filtering smaller volumes in the field (Bairoliya et al., 2022; Patin and Goodwin, 2023). For large scale sampling efforts this involves transporting cumbersome (large and heavy) numbers of sample bottles on ice. To process large numbers efficiently, filtration is often set up in parallel, further increasing the challenge of maintaining sterility and avoiding cross contamination of water samples, a particular concern for microbial samples (Alexander et al., 2023). Water with high levels of suspended solids or high concentrations of phytoplankton can rapidly clog filters, which increases processing time and opportunities for contamination as filters are changed (Patin and Goodwin, 2023; Chen et al., 2024). For water with high turbidity this approach may not be feasible at all. Finally, if large sample sizes are required these approaches can involve significant investment in disposable plastics and can produce significant plastic waste (Chen et al., 2022). These processing challenges particularly limit our ability to sample remote locations where access to sterile lab space and power to support hours or days of filtration may not be available.
Recent studies have explored the use of natural eDNA collectors like living sponges which process large volumes of seawater during their feeding and retain residual eDNA in their bodies (Mariani et al., 2019; Turon et al., 2020), or collection of natural biofilms which may also trap eDNA (Rivera et al., 2022). Sampling modules with high surface areas have been manufactured with the aim of trapping eDNA either through towing or static deployments (Alexander et al., 2023; van der Heyde et al., 2023), and small bags of high surface area materials like hemp or active carbon (Bessey et al., 2022) have also been trialed. The simplest and cheapest approach to passive sampling, suspension of glass fiber, cellulose or other kinds of filter membranes in the water for a few hours to days, has also proved effective (Bessey et al., 2021; Chen et al., 2022, 2024; Redden et al., 2023). When tested in laboratory mesocosms or in the field these approaches have been found to detect the local diversity of fish (Bessey et al., 2021; Chen et al., 2024; Zhang et al., 2024) and other metazoans (van der Heyde et al., 2023) as effectively as standard water filtration. This approach has thus far been applied primarily for metabarcoding of eukaryotes (see Discussion), and its efficacy for capturing microbial diversity has yet to be explored in a natural environmental setting. Researchers who have used this passive sampling approach successfully have pointed to the importance of conducting trials across a wider array of environmental conditions to confirm its utility and to document any potential environmental limitations (Bessey et al., 2021; Chen et al., 2024). To this end, we compared bacterial communities characterized with metabarcoding from traditional actively filtered water samples and from passive membrane collectors that were deployed in six tropical aquatic sites reflecting the range of common environmental conditions experienced in lowland tropical environments.
Methods
Study sites
We sampled in three artificial water bodies and three natural waterways in Panama (Supplementary Table S1). Lake Gatun (425 km2), which we sampled at Gamboa in central Panama, was formed in 1913 as part of the Panama Canal. The Big Creek Reservoir (<0.1 km2), on Isla Colon in Bocas del Toro Province, is a small man-made reservoir adjacent to the ocean and provides much of the drinking water to the island. The other four sampling sites are on the mainland of Bocas del Toro (see Clark et al., 2022 for detailed descriptions). Río Nigua and Río Pastores are mountain streams that drain small steep catchments into Bahia Almirante. The catchment of Río Nigua is more highly impacted by human activities than Río Pastores (Clark et al., 2022). The Black River drains the peat swamp forest of the San San Pond Sak Ramsar site, and the Changuinola (Soropta) Canal, a 30 m wide artificial canal constructed in 1903 to transport bananas, connects the Río Changuinola to the Bahia Almirante through the swamp forest. Both have dark, relatively hypoxic and acidic waters typical of blackwater rivers. The tidal influence is evident, as saltwater wedges can extend significantly into these two waterways (Clark et al., 2022).
Field sampling
We accessed each field site either by boat or from docks or piers to ensure samples were taken from the water column without interference from the substrate. For each field site we sampled on 2 days, at least 10 days apart (Supplementary Table S1), between 9 a.m. and 3 p.m. and at a depth between 0.7 and 1.0 m.
All of the equipment used in the field and for filtration (collection bottles, clamps, frame, mesh bags, glass, hoses) was cleaned with Alconox soap, rinsed with deionized (DI) water, soaked for a minimum of 20 min in a 20% bleach solution, and rinsed four times with DI water. The rubber adapters, which were used to adjust the vacuum pump were only washed with DI water. After washing, all glassware and bottles were autoclaved for 20 min at 120°C.
For passive eDNA collection (hereafter passive samples), we used a metal frame with clips to suspend six positively charged 47 mm mixed cellulose filters (Millipore) with a pore size of 0.8 mm into the water (Figure 1). Filters were collected after 24 h of deployment and stored frozen in sterile 5 ml tubes until DNA was extracted. All filters were certified sterile before deployment and were moved with sterilized forceps.

Figure 1. Passive sampling set up: (A) View of the passive collection frame that was deployed for 24 h at each site; (B) Passive collection frame deployed at Río Nigua, September 27, 2022.
For active filtered eDNA collection (hereafter filtered samples), we collected six 1 L bottles of water when we recovered the passive filter membranes at the same depth used for passive sampling at each location. The van Dorn bottle used for sampling was rinsed three times with water from the collection site before making the final collection and the water was transferred to sterile 1 L bottles immediately after collection. In addition, control bottles of autoclaved water were taken to each sampling site (n =7 total) and had their lids removed while the other samples were collected. Water samples were placed on ice immediately and stored at 4°C until filtering, within 24 h of collection. Filtering occurred in the laboratory using a vacuum pump to draw each sample through stacked 47 mm mixed cellulose filters with pore sizes of 0.8 and 0.22 mm (Millipore), which aided in reducing clogging of the lower filter. Filters were stored frozen in dry, sterile 5 ml tubes until DNA was extracted from both filters in a single extraction.
DNA extraction, amplification and sequencing
Total DNA was extracted from all filters using the DNeasy PowerSoil Kit (Qiagen), following the manufacturer's protocols with several adjustments. To maximize DNA recovery, the filters were cut into smaller pieces under sterile conditions in a laminar flow hood prior to adding PowerSoil beads and bead solution (as in Chavarria et al., 2021). Final elution volume was 70 ml in C6 buffer. Two DNA extraction blanks using sterile ddH20 were also performed. DNA concentration was quantified on a NanoDrop (Thermo Fisher).
Library preparation for Illumina sequencing was done using a 2-Stage PCR protocol. Amplifications of the bacterial 16S rRNA V4 region were performed using 1 ml of DNA extract in 35 cycles of 10 ml reactions using KAPA 3G Plant 2X Mastermix (KAPA) and the 515F-806R primer set (Caporaso et al., 2011) with Illumina P5 and P7 adaptors included. A second PCR with six cycles was then performed using 1 ml of PCR1 as template to add on unique barcode indices and Illumina flow cell adaptors. Products were cleaned and normalized using the Just-A-Plate 96 PCR Purification and Normalization Kit (Charm Biotech), pooled to make a sequencing library, and concentrated with KAPAPure magnetic beads. The library was sequenced on a paired end 2 × 250 bp Illumina MiSeq run at the Naos Molecular Laboratory (Smithsonian Tropical Research Institute, Panama).
Analysis
All sequence data analysis was done using R v. 4.41. Primer trimming, sequence quality filtering, and read merging, and taxonomy assignment was performed using the default parameters in cutadapt (Martin, 2011) and DADA2 (Callahan et al., 2016) to produce amplicon sequence variants (ASVs). Taxonomy was assigned using the SILVA 138.1 database (Quast et al., 2013). Downstream analysis was done in phyloseq (McMurdie and Holmes, 2013) and associated packages such as decontam (Davis et al., 2018), vegan (Oksanen et al., 2020) and microbiome (Lahti et al., 2017). Field and extraction blanks and positive and negative controls, as well as all ASVs with <10 counts were eliminated prior to diversity analyses. Finally, samples were rarefied to 5,000 seqs/sample (Supplementary Figures S1a, b) resulting in 94.6% of the bacterial ASVs being retained.
We applied a suite of diagnostic tests to examine how the diversity recovered by the two methods compared. We used a mixed model with site as a random effect to compare the quantity of DNA extracted from the passive and filtered samples. To assess differences in passive vs. active sampling, we first created a Venn diagram to illustrate the number of shared and unique ASVs in each sampling type. We then compared the alpha diversity in each sample type by calculating the Shannon, Inverse Simpson and Chao1 diversity indices and testing for effects of site and sampling method using Kruskal-Wallis and pairwise Wilcox tests with Bonferroni correction. We used principal coordinates analysis (PCoA) based on Bray Curtis distance and PERMANOVA, with subsequent tests for beta dispersion (betadisper followed by a permutation test), to compare how the community composition of the samples varied across sites and between collection methods, in addition to visual inspection of relative abundance plots and Linear Discriminant Analysis Effect Size analysis (LefSe; Mandal et al., 2015), as implemented in MicrobiomeAnalyst using the default parameters.
Results
The concentration of extracted DNA varied with sample collection type with passive DNA collection recovering less DNA than active filtered samples (Table 1). Mixed model analysis with site as a random effect found a significant effect of sampling method (p < 0.001), with active sampling recovering an average of 30.9 ng/μl (SD = 29.0) and passive sampling recovering 4.3 ng/μl (SD = 2.9).
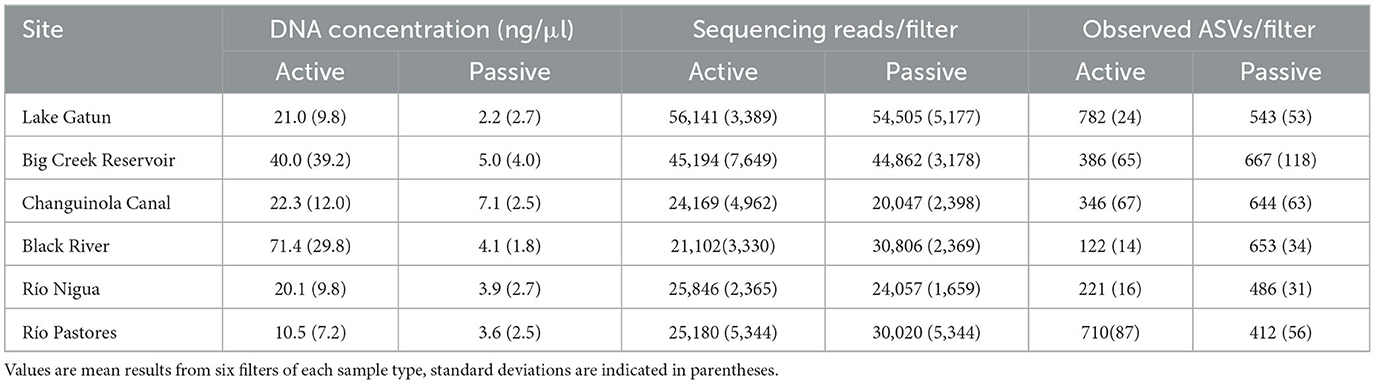
Illumina sequencing produced a total of 4,754,262 high-quality reads that were assigned to 51,393 amplicon sequence variants (ASVs), across 143 water samples and controls (mean 33,480 sequences per sample). After filtering out ASVs with fewer than 10 counts and including only sequences assigned to the Kingdom Bacteria, 14,551 ASVs remained. Rarefaction analysis indicated that sequencing efforts were sufficient to capture the diversity of these communities (Supplementary Figure S1a) but accumulation curves did not asymptote indicating that, for both methods, additional sampling would increase diversity (Supplementary Figure S1b). However, passive sampling detected 46% more ASVs (10,933) than active filtration (7,452) across the entire dataset. Overall, only 26% of the Bacteria ASVs were collected by both active and passive sampling (Table 1, Figure 2A).
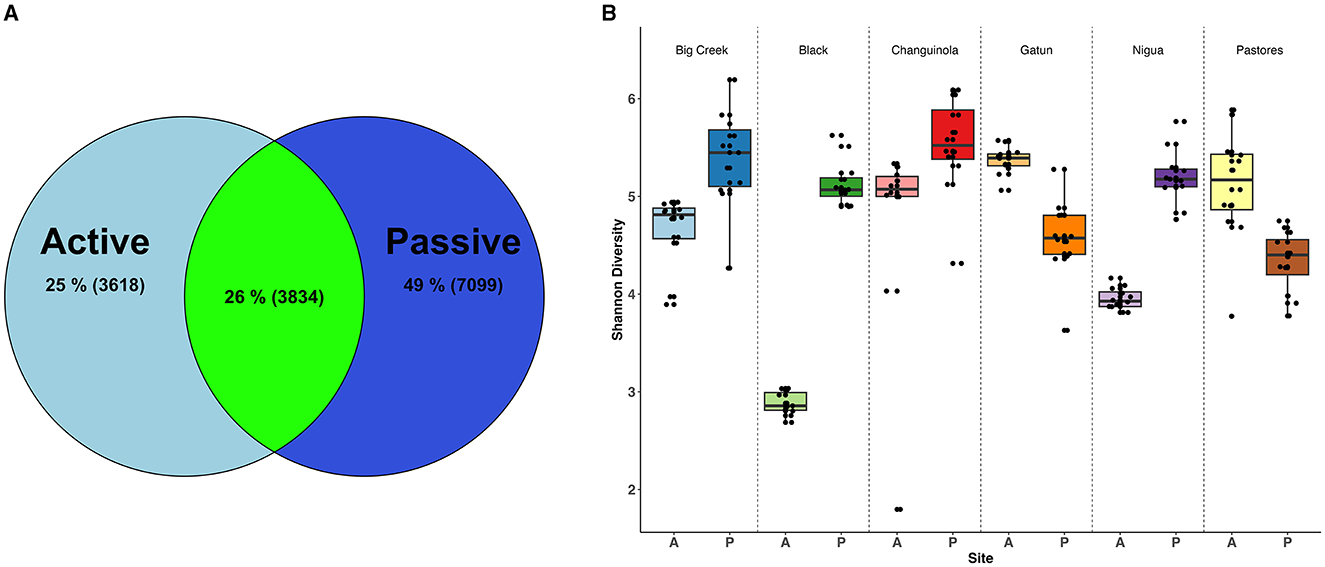
Figure 2. Diversity of bacteria identified with active and passive sampling at six Neotropical waterways: (A) Venn diagram of bacteria ASVs, including the percentage of the total ASV count that is unique to each sampling type (light and dark blue) and shared between sampling types (green). Numbers in parentheses represent ASV counts; (B) Shannon diversity of samples collected with the two methods across sites. A, active sampling; P, passive sampling.
For all metrics tested (Shannon, Inverse Simpson, Chao1), alpha diversity of Bacteria was significantly higher in the passive samples than in the active filtered samples (Shannon: Kruskal-Wallis χ2 = 97.35, df = 11, p < 0.001; Inverse Simpson: χ2 = 103.37, df = 11, p < 0.001; Chao1: χ2 =78.30, df = 11, p < 0.001), except for Gamboa and Pastores where active samples had higher diversity (Pairwise Wilcox test, p < 0.001; Figure 2B).
Passive sampling also detected more phyla than active sampling (55 vs. 47 respectively) and the relative abundance of many taxa differed between active and passive samples (LDA = 4.0, p < 0.05; Figure 3A). Active samples had a higher relative abundance of Actinobacteriota and Campilobacterota compared to passive samples, while the passive samples had higher relative abundances of Proteobacteria, Myxococcota, and Acidobacteriota. Pairwise comparisons of the two sampling methods showed differences in relative abundances of taxa within the phyla Actinobacteriota, Campilobacterota, Desulfobacterota, and Proteobacteria to be driving most of the differences between the communities recovered by the two methods (LDA = 4.0, p < 0.05; Figure 3A).
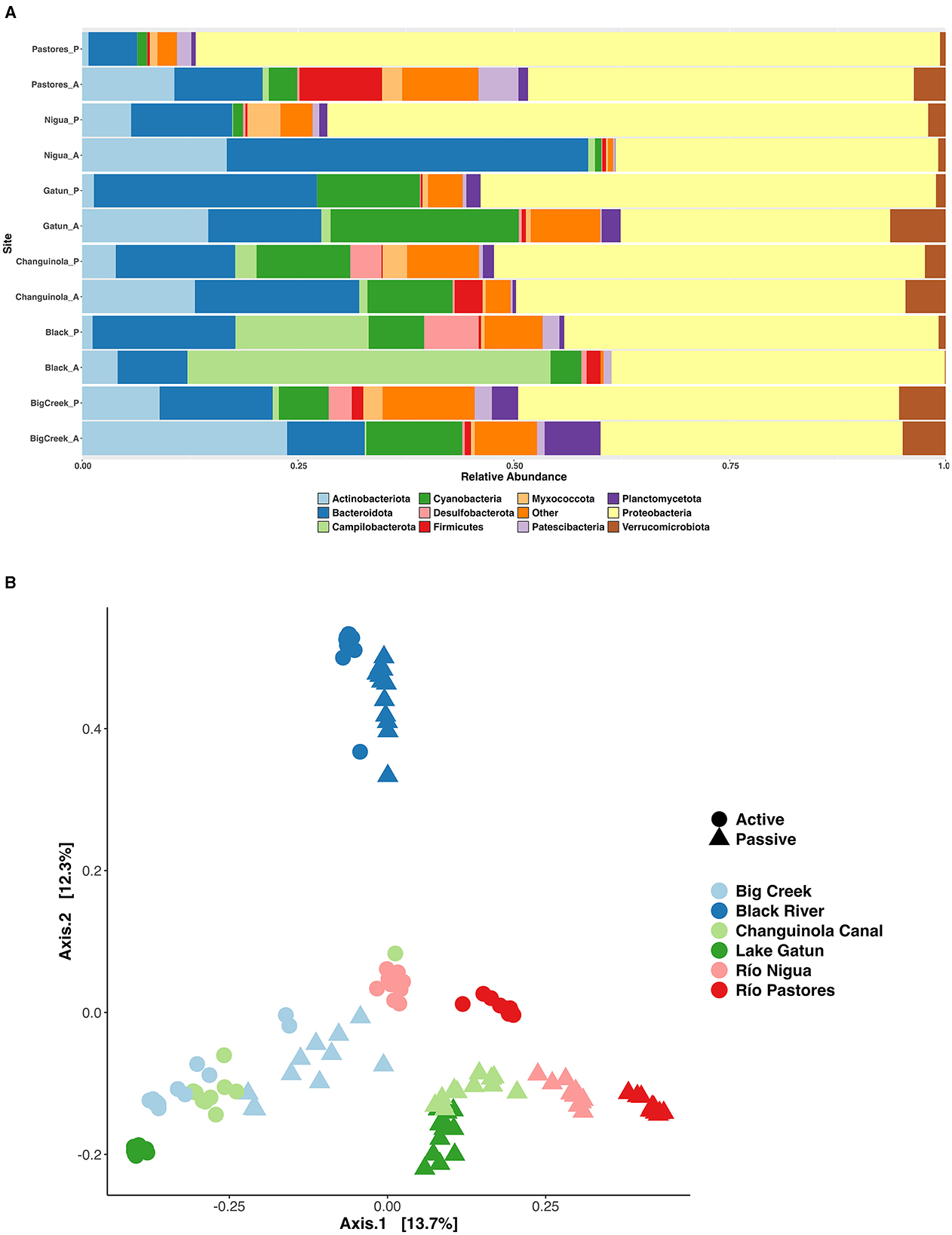
Figure 3. Communities of bacteria identified with active and passive sampling at six Neotropical waterways: (A) Relative abundance of dominant phyla; (B) PCoA ordination of all sites based on Bray Curtis distance. A, active sampling; P, passive sampling.
Principal Coordinates analysis (PCoA) using Bray Curtis distance showed clear differences along axis 1 (Figure 3B), with variation in the communities across sites (PERMANOVA, Pseudo-F = 30.15, R2 = 0.409, p < 0.001), between those captured by active and passive sampling within sites (PERMANOVA, Pseudo-F = 32.28, R2 = 0.088, p < 0.001) and, in some cases, the sampling date (Supplementary Figure S2). PCoA analysis using a distance metric weighted for phylogenetic relationships of community members (i.e. weighted Unifrac, not shown) yielded similar results. The Black River had the most unique communities across sites (Figures 3A, B), possibly related to the low pH, high levels of Total Dissolved Solids, and low levels of dissolved Oxygen found at this site (Supplementary Table S1).
Discussion
Use of passive filters for eDNA collection in aquatic systems is a potentially low-cost and efficient way to sample remote locations where access to equipment is limited. Our results show that the application of the simplest type of passive sampler, the suspended membrane filter, can effectively sample microbial communities across a variety of environmental conditions typical of aquatic systems in the tropics. Previous work has demonstrated that passive sampling approaches can effectively detect the eDNA of marine and freshwater fishes (Bessey et al., 2021; Chen et al., 2022, 2024; Zhang et al., 2024) and invertebrates (Alexander et al., 2023). Similar to other studies with metazoans (Alexander et al., 2023; van der Heyde et al., 2023), we found that passive samplers recovered less total DNA than active filtration. However, in contrast to comparisons of metazoan diversity which have found similar levels of diversity or species richness in both passive and active sampling methods (Rivera et al., 2022; van der Heyde et al., 2023; Bessey et al., 2022), the microbial diversity that we recovered was generally higher from passive samples than from active filtering.
Effective sampling of eDNA may require different conditions for different types of organisms. Both passive and active sampling appears to work well for fish and benthic metazoans, because both methods are capturing DNA from large organisms that is floating free, such as in mucus, small particles of tissue or feces in the water column. In low diversity systems, like the coastal waters studied by Zhang et al. (2024), the two methods have demonstrated very high (~90%) overlap between the fish species sampled, and similarly good performance has been reported in mesocosms for amphibian DNA (Chen et al., 2022). However, previous work in high-diversity natural systems often reports only partial overlap in the fish taxa recovered (e.g., Bessey et al., 2021; Chen et al., 2024). This may be due to insufficient sampling to recover the entire community. The distribution of eDNA is not homogeneous in any environment and can change quickly depending on local conditions (Barnes et al., 2014; Joseph et al., 2022). Physiochemical stratification, such as salinity and temperature gradients, and turbidity have been shown to affect eDNA movement and longevity in the water column (Canals et al., 2021; Jeunen et al., 2020; Kumar et al., 2021). In our study, while we did not collect sufficient samples with either method to sample the communities completely, as evidenced by the ASV accumulation curves (Supplementary Figure S1a), the differences in the composition of the communities between methods are striking and are unlikely to result from insufficient sampling.
For single celled eukaryotes and prokaryotes, it is plausible that passive and active sampling may sample different communities. The water column is full of organic material, both living and dead, from a variety of organisms. Therefore, it is entirely possible that active filtration could increase the likelihood of sampling an exclusively planktonic community with lower prokaryotic diversity but higher DNA concentrations, due to other organisms or extracellular tissues being captured. Indeed, our results show high variability in the concentration of DNA obtained across our actively filtered samples, suggesting that some replicates captured more organic material than others. In addition, diverse microbial communities quickly colonize surfaces in aquatic systems, and these taxa may not be present in the adjacent waters at high densities (Zhang et al., 2019). The biofilms produced by benthic fouling communities can incorporate extracellular DNA (Das et al., 2013; Decho and Gutierrez, 2017; Hancock, 2001) and have been shown, for example, to include DNA for the local fish fauna (Rivera et al., 2022). As such, passive collection may better capture not only microbial communities from the water column, but also the fouling community as well as some fragments from the planktonic community, resulting in lower DNA yield but higher overall diversity.
The selection of pore size is also an important consideration when filtering water for eDNA, particularly when active filtration, which forces the water through the filter, is used. Filter pore sizes of 0.22 μm should retain nearly all bacteria but many eDNA studies use single filters of 0.45 or 0.80 μm (Bessey et al., 2022; Majaneva et al., 2018; Schreiber et al., 2023) as they are targeting larger organisms such as fish. We used membrane filters with a pore size of 0.8 μm as passive samplers as they are similar to those shown to develop a covering of adhered particles and biofilm after short deployments (Bessey et al., 2022). However, when processing our active samples, we used stacked filters with an 0.8 μm filter above to capture larger particles and prevent clogging of the 0.22 μm filter underneath which was intended to capture the free-living bacteria (Chavarria et al., 2021; Urvoy et al., 2022). Overall, we found that only 26% of the ASVs assigned to the Kingdom Bacteria were found in both actively filtered and passively collected samples and the passive samples captured more ASVs and had higher bacterial diversity at four of our six sites. Further, our PCoA ordinations (Figure 3B, Supplementary Figure S2) demonstrate that the two sampling types consistently captured different communities. This suggests that fewer bacteria were captured by the filtration process with active sampling, despite the inclusion of two filters of differing pore sizes in our DNA extractions.
In addition to the physical ability of active and passive sampling to capture different communities, the temporal scale of sampling may be expected to lead to higher diversity measures from passive sampling. Our passive samplers were deployed for 24 h, and therefore each collected a time-integrated sample. Diurnal changes in aquatic microbial community composition have been previously documented (Weber and Apprill, 2020; Becker et al., 2020; Shahraki et al., 2021). If such cycles occur in the aquatic systems studied here, the passive samples would have captured a signature of this cycle in addition to the daytime diversity recorded by the filtered samples. In addition to integrating across diurnal variation, the passive samples could also integrate across changes in community composition reflecting tidal cycles in the two tidally influenced sites (Black River and Changuinola Canal; Clark et al., 2022), as well as any changes in flow rate or water chemistry reflecting runoff from rain events in the catchments that may have occurred during the deployments. It is possible that such events could contribute to the large differences between the actively and passively sampled communities at all of our sites. Our observation that species richness is higher in passive samples is consistent with this temporal integration.
In conclusion, we found that passive sampling by suspension of membrane filters was an effective way to sample microbial diversity in tropical aquatic systems spanning a range of environmental conditions. Although active filtering recovered more DNA and overall communities were not similar, with only ~25% of ASVs recovered using both methods, the levels of diversity in passive samples were higher than from active filtration in most of our sites. Passive filtering appears to be an excellent candidate for new projects focused on documenting and monitoring microbial communities in aquatic systems with eDNA, as it eliminates the time and effort needed for water filtration and reduces associated costs and risks of sample contamination associated with filtering. However, the distinct communities captured by the two methods demonstrate that they are not inter-changeable and transitioning ongoing monitoring projects to passive sampling may not be possible. It is also evident that multiple sampling methods may be needed to adequately describe prokaryote diversity in aquatic systems. Moving forward, optimization of the technique through streamlining of sample collection and laboratory workflows, such as pooling of filters for DNA extraction, should be prioritized. With further optimization and standardization, passive filtration could become an important resource for exploration, monitoring and management of aquatic environments.
Data availability statement
The dataset presented in this study can be found in the NCBI SRA archive at: https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA1163416.
Author contributions
KS: Conceptualization, Data curation, Formal analysis, Methodology, Project administration, Resources, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. JD: Formal analysis, Funding acquisition, Investigation, Methodology, Writing – review & editing. MV: Investigation, Supervision, Writing – review & editing. RC: Conceptualization, Formal analysis, Project administration, Resources, Supervision, Validation, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by a fellowship from SENACYT and was completed in partial fulfillment of the requirements for a Masters degree from the University of Panama.
Acknowledgments
All samples were collected under permit ARG-040-2021 from the Panamanian Ministry of the Environment (MiAmbiente). We thank the Autoridad del Canal de Panamá (ACP) and MiAmbiente for granting permission for our research in Panama, Viviana Bravo, Ximena Boza, and Carolina Cesar for help sampling in Bocas del Toro, and Tania Romero, Milton Sandoval and members of the Collin Lab for logistical support in Panama City and providing feedback.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/ffwsc.2024.1460713/full#supplementary-material
References
Alexander, J. B., Marnane, M. J., McDonald, J. I., Lukehurst, S. S., Elsdon, T. S., Simpson, T., et al. (2023). Comparing environmental DNA collection methods for sampling community composition on marine infrastructure. Estuar. Coast. Shelf Sci. 283:108283. doi: 10.1016/j.ecss.2023.108283
Bairoliya, S., Koh Zhi Xiang, J., and Cao, B. (2022). Extracellular DNA in environmental samples: occurrence, extraction, quantification, and impact on microbial biodiversity assessment. Appl. Environ. Microbiol. 88:e01845-21. doi: 10.1128/aem.01845-21
Barnes, M. A., Turner, C. R., Jerde, C. L., Renshaw, M. A., Chadderton, W. L., Lodge, D. M., et al. (2014). Environmental conditions influence eDNA persistence in aquatic systems. Environ. Sci. Technol. 48, 1819–1827. doi: 10.1021/es404734p
Becker, C. C., Weber, L., Suca, J. J., Llopiz, J. K., Mooney, T. A., Apprill, A., et al. (2020). Microbial and nutrient dynamics in mangrove, reef, and seagrass waters over tidal and diurnal time scales. Aquat. Microb. Ecol. 85, 101–119. doi: 10.3354/ame01944
Bessey, C., Gao, Y., Truong, Y. B., Miller, H., Jarman, S. N., Berry, O., et al. (2022). Comparison of materials for rapid passive collection of environmental DNA. Mol. Ecol. Resour. 22, 2559–2572. doi: 10.1111/1755-0998.13640
Bessey, C., Jarman, S. N., Simpson, T., Miller, H., Stewart, T., Keesing, J. K., et al. (2021). Passive eDNA collection enhances aquatic biodiversity analysis. Commun. Biol. 4:236. doi: 10.1038/s42003-021-01760-8
Bista, I., Carvalho, G. R., Walsh, K., Seymour, M., Hajibabaei, M., Lallias, D., et al. (2017). Annual time-series analysis of aqueous eDNA reveals ecologically relevant dynamics of lake ecosystem biodiversity. Nat. Commun. 8:14087. doi: 10.1038/ncomms14087
Callahan, B., McMurdie, P., Rosen, M., Han, A. W., Johnson, A. J., and Holmes, S. P. (2016). DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Canals, O., Mendibil, I., Santos, M., Irigoien, X., and Rodríguez-Ezpeleta, N. (2021). Vertical stratification of environmental DNA in the open ocean captures ecological patterns and behavior of deep-sea fishes. Limnol. Oceanogr. Lett. 6, 339–347. doi: 10.1002/lol2.10213
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Lozupone, C. A., Turnbaugh, P. J., et al. (2011). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. USA. 108, 4516–4522. doi: 10.1073/pnas.1000080107
Chavarria, K. A., Saltonstall, K., Vinda, J., Batista, J., Lindmark, M., Stallard, R. F., et al. (2021). Land use influences stream bacterial communities in lowland tropical watersheds. Sci. Rep. 11:21752. doi: 10.1038/s41598-021-01193-7
Chen, X., Kong, Y., Zhang, S., Zhao, J., Li, S., Yao, M., et al. (2022). Comparative evaluation of common materials as passive samplers of environmental DNA. Environ. Sci. Technol. 56, 10798–10807. doi: 10.1021/acs.est.2c02506
Chen, X., Li, S., Zhao, J., and Yao, M. (2024). Passive eDNA sampling facilitates biodiversity monitoring and rare species detection. Environ. Int. 187:108706. doi: 10.1016/j.envint.2024.108706
Clark, K. E., Bravo, V. D., Giddings, S. N., Davis, K. A., Pawlak, G., Torres, M. A., et al. (2022). Land use and land cover shape river water quality at a continental Caribbean land-ocean interface. Front. Water 4:737920. doi: 10.3389/frwa.2022.737920
Das, T., Sehar, S., and Manefield, M. (2013). The roles of extracellular DNA in the structural integrity of extracellular polymeric substance and bacterial biofilm development. Environ. Microbiol. Rep. 5, 778–786. doi: 10.1111/1758-2229.12085
Davis, N. M., Proctor, D. M., Holmes, S. P., Relman, D. A., and Callahan, B. J. (2018). Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome 6:226. doi: 10.1186/s40168-018-0605-2
Decho, A. W., and Gutierrez, T. (2017). Microbial extracellular polymeric substances (EPSs) in ocean systems. Front. Microbiol. 8:922. doi: 10.3389/fmicb.2017.00922
Hancock, R. E. W. (2001). A brief on bacterial biofilms. Nat. Genet. 29:360. doi: 10.1038/ng1201-360
Jeunen, G. J., Lamare, M. D., Knapp, M., Spencer, H. G., Taylor, H. R., Stat, M., et al. (2020). Water stratification in the marine biome restricts vertical environmental DNA (eDNA) signal dispersal. Environ. DNA 2, 99–111. doi: 10.1002/edn3.49
Joseph, C., Faiq, M. E., Li, Z., and Chen, G. (2022). Persistence and degradation dynamics of eDNA affected by environmental factors in aquatic ecosystems. Hydrobiologia 849, 4119–4133. doi: 10.1007/s10750-022-04959-w
Kumar, G., Farrell, E., Reaume, A. M., Eble, J. A., and Gaither, M. R. (2021). One size does not fit all: tuning eDNA protocols for high- and low-turbidity water sampling. Environ. DNA 4, 167–180. doi: 10.1002/edn3.235
Lahti, L., Shetty, S., Blake, T., and Salojarvi, J. (2017). Tools for microbiome analysis in R. Version 1.26. Availble at: http://microbiome.github.com/microbiome (accessed June 8, 2024).
Li, F., Qin, S., Wang, Z., Zhang, Y., and Yang, Z. (2023). Environmental DNA metabarcoding reveals the impact of different land use on multitrophic biodiversity in riverine systems. Sci. Total Environ. 855:158958. doi: 10.1016/j.scitotenv.2022.158958
Majaneva, M., Diserud, O. H., Eagle, S. H. C., Boström, E., Hajibabaei, M., and Ekrem, T. (2018). Environmental DNA filtration techniques affect recovered biodiversity. Sci. Rep. 8:4682. doi: 10.1038/s41598-018-23052-8
Mandal, S., Van Treuren, W., White, R. A., Eggesbø, M., Knight, R., Peddada, S. D., et al. (2015). Analysis of composition of microbiomes: a novel method for studying microbial composition. Microb. Ecol. Health Dis. 26:27663. doi: 10.3402/mehd.v26.27663
Mariani, S., Baillie, C., Colosimo, G., and Riesgo, A. (2019). Sponges as natural environmental DNA samplers. Curr. Biol. 29, R401–R402. doi: 10.1016/j.cub.2019.04.031
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12. doi: 10.14806/ej.17.1.200
McMurdie, P. J., and Holmes, S. (2013). phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 8:E61217. doi: 10.1371/journal.pone.0061217
Oksanen, J., Simpson, G., Blanchet, F., Kindt, R., Legendre, P., Minchin, P. R., et al. (2020). vegan Community Ecology Package v. 2.6-6.1. Available at: https://github.com/vegandevs/vegan; https://vegandevs.github.io/vegan/
Patin, N. V., and Goodwin, K. D. (2023). Capturing marine microbiomes and environmental DNA: a field sampling guide. Front. Microbiol. 13:1026596. doi: 10.3389/fmicb.2022.1026596
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Redden, D. J., Stanhope, T., Anderson, L. E., Campbell, J., Krkošek, W. H., Gagnon, G. A., et al. (2023). An innovative passive sampling approach for the detection of cyanobacterial gene targets in freshwater sources. Sci. Total Environ. 892:164593. doi: 10.1016/j.scitotenv.2023.164593
Rivera, S. F., Rimet, F., Vasselon, V., Vautier, M., Domaizon, I., Bouchez, A., et al. (2022). Fish eDNA metabarcoding from aquatic biofilm samples: methodological aspects. Mol. Ecol. Resour. 22, 1440–1453. doi: 10.1111/1755-0998.13568
Sales, N. G., Wangensteen, O. S., Carvalho, D. C., Deiner, K., Præbel, K., Coscia, I., et al. (2021). Space-time dynamics in monitoring neotropical fish communities using eDNA metabarcoding. Sci. Total Environ. 754:142096. doi: 10.1016/j.scitotenv.2020.142096
Schreiber, L., Castellanos-Galindo, G. A., Robertson, D. R., Torchin, M., Chavarria, K., Laakmann, S., et al. (2023). Environmental DNA (eDNA) reveals potential for interoceanic fish invasions across the Panama Canal. Ecol. Evol. 13:e9675. doi: 10.1002/ece3.9675
Shahraki, A. H., Chaganti, S. R., and Heath, D. D. (2021). Diel dynamics of freshwater bacterial communities at beaches in Lake Erie and Lake St. Clair, Windsor, Ontario. Microb. Ecol. 81, 1–13. doi: 10.1007/s00248-020-01539-0
Turon, M., Angulo-Preckler, C., Antich, A., Præbel, K., and Wangensteen, O. S. (2020). More than expected from old sponge samples: a natural sampler DNA metabarcoding assessment of marine fish diversity in Nha Trang Bay (Vietnam). Front. Mar. Sci. 7:605148. doi: 10.3389/fmars.2020.605148
Urvoy, M., Gourmelon, M., Serghine, J., Rabiller, E., L'Helguen, S., and Labry, C. (2022). Free-living and particle-attached bacterial community composition, assembly processes and determinants across spatiotemporal scales in a macrotidal temperate estuary. Sci. Rep. 12:13897. doi: 10.1038/s41598-022-18274-w
van der Heyde, M., Alexander, J., Nevill, P., Austin, A. D., Stevens, N., Jones, M., et al. (2023). Rapid detection of subterranean fauna from passive sampling of groundwater eDNA. Environ. DNA 5, 1706–1719. doi: 10.1002/edn3.491
Weber, L., and Apprill, A. (2020). Diel, daily, and spatial variation of coral reef seawater microbial communities. PLoS ONE 15:e0229442. doi: 10.1371/journal.pone.0229442
Zhang, L., Zhou, W., Jiao, M., Xie, T., Xie, M., Li, H., et al. (2024). Use of passive sampling in environmental DNA metabarcoding technology: monitoring of fish diversity in the Jiangmen coastal waters. Sci. Total Environ. 908:168298. doi: 10.1016/j.scitotenv.2023.168298
Keywords: bacteria, eDNA, metabarcoding, Panama, Bocas del Toro, water filtration
Citation: Saltonstall K, Delgado J, Vargas M and Collin R (2024) Are passive collectors effective samplers of microbes in natural aquatic systems? Front. Freshw. Sci. 2:1460713. doi: 10.3389/ffwsc.2024.1460713
Received: 06 July 2024; Accepted: 10 October 2024;
Published: 24 October 2024.
Edited by:
Javier Atalah, University of Alicante, SpainReviewed by:
Lena Andrea Schallenberg, Cawthron Institute, New ZealandGeorgia Thomson-Laing, Cawthron Institute, New Zealand
Copyright © 2024 Saltonstall, Delgado, Vargas and Collin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kristin Saltonstall, U2FsdG9uc3RhbGxLQHNpLmVkdQ==
†These authors share first authorship
‡ORCID: Kristin Saltonstall orcid.org/0000-0002-1811-4087
Jacqueline Delgado orcid.org/0000-0001-5740-5189
Marta Vargas orcid.org/0009-0009-4286-8987
Rachel Collin orcid.org/0000-0001-5103-4460
 Kristin Saltonstall
Kristin Saltonstall Jacqueline Delgado1,2†‡
Jacqueline Delgado1,2†‡ Rachel Collin
Rachel Collin