- Department of Human Evolutionary Biology, Nutritional and Microbial Ecology Lab, Harvard University, Cambridge, MA, United States
Diet shapes the gut microbiome, which in turn influences host phenotype. Accordingly, there is much interest in leveraging diet to modulate gut microbial communities and host biology. However, recent approaches have not fully appreciated that hosts and gut microbes experience diet differently. Whether dietary nutrients reach the gut microbiota, which primarily resides in the colon in humans and other hindgut fermenters, depends on nutrient absorption in the small intestine. That gut microbes utilize the fraction of diet that escapes host-driven digestion creates a paradigm where host nutrient status is decoupled from, and often negatively correlated with, gut microbiota nutrient status. Here, we present a framework based on this concept of decoupled nutrient status (DNS), which can be used to understand distinct host and gut microbial phenotypes that are ultimately mediated by the small intestinal digestibility of the diet. We evaluate our framework against existing research employing diets of varying digestibility and demonstrate convergence of host phenotypes and gut microbial signatures across studies. Further, we highlight that gut microbial signatures predicted by DNS manifest most strongly in humans living industrialized lifestyles and in captive animals that habitually consume diets with high host-driven digestibility. We posit that the evolutionary decoupling of nutritional status between hosts and their gut microbiota has likely been especially pronounced in humans due to our intensified pursuit of calorie-rich, easy-to-digest diets. We conclude by proposing future research directions to better capture diet as it appears to gut microbes, a perspective likely to deliver new understanding of diet-microbiome interactions.
Introduction
Effective and efficient extraction of nutrients from the environment is fundamental to organismal fitness. Therefore, evolution has selected for diverse and innovative foraging strategies across species. Diet is likewise essential to the fitness of microbes inhabiting the gastrointestinal (GI) tract, which subsist off the influx of diet-derived nutrients and endogenous host secretions and tissues (Sonnenburg and Sonnenburg, 2014). Further, gut microbial communities and their metabolites affect host fitness through critical interactions with the immune system and energy balance, as demonstrated by impaired immune cell development and low body weights despite high caloric intake in germ-free animals (Bäckhed et al., 2004; Kennedy et al., 2018). Thus, to evaluate the fitness of a host, we must also consider the fitness impacts of gut microbes and their collective ability to adapt to dietary flux.
However, there is inherent competition among gut microbes and between hosts and their gut microbes over the energetic substrates that enter the GI tract. Dietary components that are readily digested by the host in the small intestine do not reach downstream gut microbial communities, which are concentrated in the colon. In contrast, dietary components inaccessible to host absorption in the small intestine are available to the colonic gut microbiota (Wong and Jenkins, 2007). In other words, a nutrient-rich diet from a host perspective is a nutrient-poor diet from a gut microbial perspective, and vice versa. Thus, to better appreciate how diets of variable digestibility impact host-microbial interactions, we must conceptually decouple host nutrient status from microbial nutrient status. Here, we present our framework of decoupled nutrient status (DNS) and evaluate it using prior research, demonstrating convergent effects of diverse diets mediated by their relative availability to the host or gut microbiota. We then propose ways to test our framework centered around quantifying diet as it appears to gut microbes.
Anatomical and physiological basis for DNS
Our DNS framework is rooted in spatial and temporal separations between host digestion and absorption and gut microbial metabolism of dietary substrates dictated by mammalian gut physiology. Certain ruminant hosts that consume fiber-rich diets harbor foregut fermentation chambers that offer microbes primary access to nutrients (“first dibs”) as a way of leveraging their comparatively vast genomic capacities for carbohydrate digestion (Figure 1). Accordingly, ruminants derive the majority of their daily energy needs from the short-chain fatty acids (SCFAs) produced during microbial fermentation of plant polysaccharides and other complex carbohydrates. SCFAs serve as energetic substrates for diverse host tissues, albeit at a lesser rate of return (∼2 kcal/g) compared with the chemical value of the source carbohydrate (∼4 kcal/g) (Cummings and Macfarlane, 1997). SCFAs also function as pleiotropic signaling molecules, affecting host energy intake, energy allocation across tissues, and energy expenditure (Carmody and Bisanz, 2023; Carmody et al., 2024).
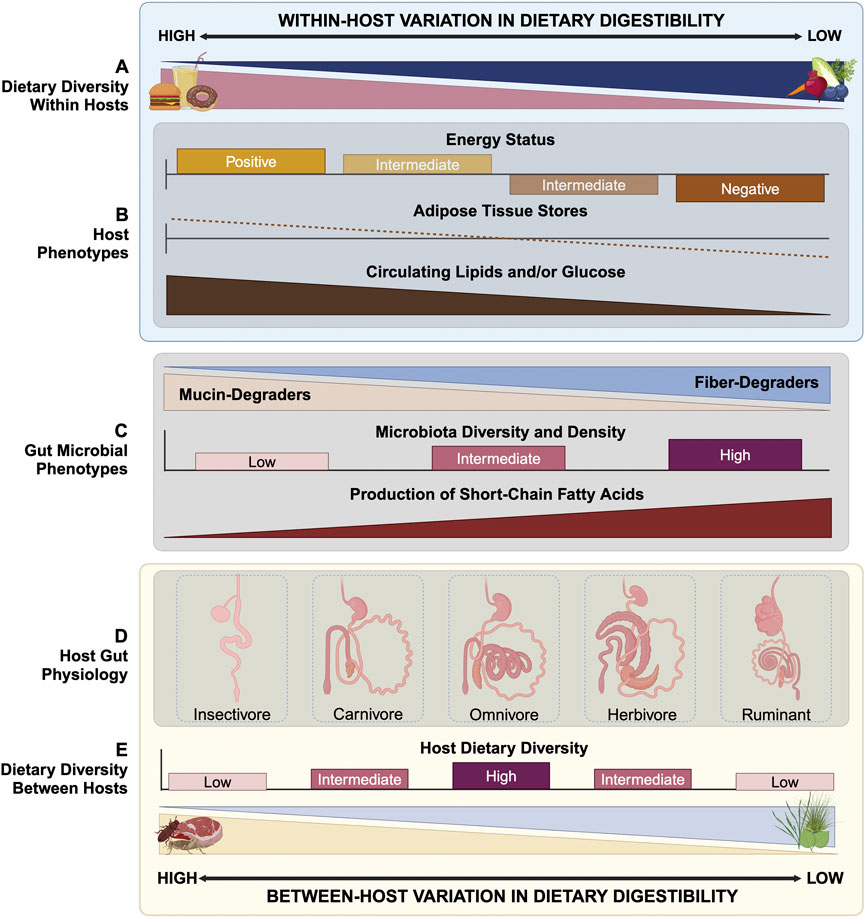
Figure 1. Relationship between host-driven digestibility (HDD) and host and gut microbial phenotypes. (A) Within hindgut fermenting omnivorous hosts, overall dietary digestibility can fluctuate across time, with hosts consuming items with relatively high or low HDD. (B) In general, diets with high HDD will be absorbed primarily by the host, leading to higher levels of circulating glucose and/or lipids for use by host tissues and host energy storage. By contrast, diets with low HDD, including those rich in fiber or resistant starch, lead to relatively low direct energy uptake by the host. (C) High HDD diets reduce diet-derived nutrient availability for gut microbial communities, reducing their density and shifting overall microbial community composition, particularly from fiber-degraders to mucin-degraders. In response, gut microbial production of short-chain fatty acids (SCFAs) decreases, with diverse downstream impacts on host biology. By contrast, low HDD diets provide the gut microbiome with a rich source of nutrients, promoting high gut microbial diversity and density. Likewise, fermentation products rise, including SCFAs, which provide hosts with approximately half the energy of their carbohydrate precursors. (D, E) Gastrointestinal (GI) tract physiology of diverse mammalian species reflects host versus microbial nutrient availability of evolved diets and overall dietary diversity. The digestive tracts of hosts consuming high HDD diets, such as insectivores and carnivores, have relatively elongated small intestines and shortened colons, prioritizing host-driven digestion. Alternatively, hosts consuming low HDD diets, such as herbivores, have enlarged colons with auxiliary chambers for microbial fermentation and, in the case of ruminants, provide microbes primary access to dietary nutrients via foregut fermentation chambers. Omnivores, including humans, have intermediate distributions of small intestinal and colonic tissue, reflecting dietary plasticity. Humans exhibit elongated small intestines and truncated colons compared to closely related primates when adjusted for body size, the result of our increasing commitment to calorie-rich, high HDD diets during human evolution. Digestive tracts in (D) are not depicted at scale relative to each other. Created in BioRender.com.
By contrast, many hosts, including humans, that consume diets largely accessible to their own digestive enzymes secure primary access to ingested nutrients via adaptations that restrict gut microbes to the distal GI tract (Walter and Ley, 2011). For instance, pH gradients prevent most gut microbes (which are acid-sensitive) from inhabiting the upper GI tract. Likewise, bile acids, which are secreted into the duodenum, have canonical functions in dietary fat emulsion and absorption but are also strongly antimicrobial. Bile acids levels remain high until the ileum, where they are either resorbed via enterohepatic circulation or deconjugated by bile-tolerant gut microbes (Chadaideh and Carmody, 2021). Together, pH and bile acid gradients restrict gut microbial biomass proximal to the ileocecal valve, which then increases by roughly three orders of magnitude in the colon (Walter and Ley, 2011). GI sphincters, including the ileocecal valve, provide further physical barriers limiting microbial encroachment into the upper GI tract. These features jointly allow host digestive enzymes to engage with dietary substrates without robust competition from microbial enzymes (Walter and Ley, 2011).
In turn, the colonic gut microbiota specializes in metabolizing the components of diet that routinely escape small intestinal digestion, including oligosaccharides, resistant starches, and non-starch polysaccharides (Wong and Jenkins, 2007; Kennedy and Chang, 2020; Carmody et al., 2019). Gut microbial genomes encode thousands of carbohydrate-active enzymes (CAZymes), allowing them to metabolize complex carbohydrate structures inaccessible to the 17 CAZymes encoded in the human genome (Sonnenburg and Sonnenburg, 2014; Kaoutari et al., 2013).
Diets at opposing ends of host-driven digestibility: evidence from humans and rodents
While dietary substrates vary in their small intestinal digestibility, diets that are comprised primarily of substrates readily digestible by the host can be expected to consistently decouple energetic states for hosts and gut microbes (Figure 1). This decoupling leads to several discordant phenotypic predictions for hosts and their gut microbes (Box 1). Numerous studies have administered diets varying in digestibility and have reported phenotypes consistent with these predictions. On the high end of the digestibility spectrum are “ultra-processed” foods – now dominant in industrialized food systems (Monteiro et al., 2013) – which are comprised of manufactured ingredients extracted from whole foods or synthesized from organic substrates and contain little to no microbiome-sustaining fiber, micronutrients, or phytochemicals (Hall et al., 2019; Dagbasi et al., 2020; Boutari and Mantzoros, 2022; Martínez Leo and Segura Campos, 2020). Nutrients in ultra-processed foods have typically been released from their cellular matrix, expediting their absorption in the small intestine (Monteiro et al., 2013; Dagbasi et al., 2020). The resultant maximization of small intestinal digestibility favors host energy surplus. For example, Hall and colleagues fed macronutrient- and calorie-matched diets composed of unprocessed whole foods versus ultra-processed foods for 2 weeks each to 20 adults in a metabolic ward (Hall et al., 2019). Despite participants reporting comparable levels of hunger and satiety, participants consumed more calories, gained more weight and fat, and had higher post-intervention levels of cholesterol, free fatty acids, and adiponectin when fed the ultra-processed diet (Hall et al., 2019). Inversely, Corbin and colleagues designed a diet that prioritized nutrient availability to the gut microbiome by selecting minimally processed fiber-rich and resistant starch-rich foods with large particle sizes (Corbin et al., 2023). In a crossover study design, they fed 17 participants either their “Microbiome Enhancer Diet” or a macronutrient- and calorie-matched Western diet for 23 days each. The Western diet increased the fraction of energy that was absorbed by the host and host weight gain relative to the Microbiome Enhancer Diet, with the latter increasing gut microbial biomass and both circulating and fecal SCFAs. Additionally, fiber-degrading microbial taxa (e.g., Prevotella and Lachnospira) bloomed on the Microbiome Enhancer Diet whereas mucin-degrading taxa (e.g., Blautia and Bifidobacterium) were more abundant on the Western diet. Thus, enhancement of host-driven digestibility (HDD) led to greater host energy status and favored mucin-degrading taxa whereas diets enhancing microbial metabolism supported denser microbial communities and enriched for fiber-degrading taxa.
Box 1 | Phenotypic Predictions of Decoupled Nutrient Status (DNS).
Our DNS model predicts certain host and microbial responses to dietary digestibility. Specifically, within humans and other hindgut fermenting host species:
• Effects of host energy status and microbiota energy status can be distinguished through the experimental administration of diets that deliver similar calories to the host while altering nutrient delivery to the colon (e.g., comparing refined low-fat and unrefined high-fat diets).
• Diets inducing severe microbial nutrient limitation (e.g., refined high-fat and refined high-sugar diets) will have similar effects on gut microbiota community structure regardless of the macronutrient composition of their digestible fraction.
• Consumption of diets with relatively high host-driven digestibility (HDD) will tend to induce:
o In the host: Positive energy balance with increased energy stores, higher circulating levels of lipids and/or glucose, lower proportion of energy derived from short-chain fatty acids (SCFAs), and potentially increased host-derived secretions that facilitate small intestinal digestion, such as bile acids.
o In the gut microbiome: Decreased community density (absolute abundance), taxonomic diversity (α-diversity), carbohydrate-active enzyme (CAZyme) richness, and SCFA production, with changes in microbial community composition favoring protein- and mucin-degraders, such as Bacteroides and Akkermansia.
• Consumption of diets with relatively low HDD will tend to induce:
o In the host: Negative energy balance with decreased energy stores, lower circulating levels of lipids and/or glucose, and a greater proportion of energy derived from SCFA-driven colonic energy salvage.
o In the gut microbiome: Increased community density, diversity, CAZyme richness, and SCFA production, with changes in microbial community composition favoring fiber-degraders, such as Prevotella.
These broad effects on hosts and their gut microbiomes have been widely reported (Sonnenburg and Sonnenburg, 2014; Monteiro et al., 2013; Hall et al., 2019; Dagbasi et al., 2020; Boutari and Mantzoros, 2022; Martínez Leo and Segura Campos, 2020; David et al., 2014; Sonnenburg et al., 2016; Fragiadakis et al., 2019; Jha et al., 2018; Deehan et al., 2024; Sonnenburg and Bäckhed, 2016; Barber et al., 2020; Cronin et al., 2021; O’Keefe et al., 2015). Myriad studies of laboratory rodents have demonstrated that digestible high-fat/high-sugar diets enrich for mucin-degrading microbes, decrease fiber-degrading microbes and SCFA production, and lead to increased host body weights, adipose stores, and fasting blood glucose and/or lipid levels (Rodriguez et al., 2019; Carmody et al., 2015; Laffin et al., 2019; Jaja-Chimedza et al., 2018; He et al., 2018; Daniel et al., 2014; Shan et al., 2019). Notably, mouse chows high in fat or sugar have been shown to exert similar effects on the gut microbiome relative to standard chows (Shan et al., 2019; Dalby et al., 2017; Morrison et al., 2020), supporting our DNS framework, which predicts that microbial responses to nutrient limitation will be induced by high HDD diets irrespective of their macronutrient composition.
Some rodent studies have directly compared diets with high versus low HDD by modulating macronutrient composition and incorporating refined ingredients that remove nutrients from their cellular-matrix, thus increasing HDD. Dalby and colleagues fed mice refined high-fat (60% kcal) or refined low-fat (10% kcal) diets with 5% fiber or an unrefined chow with 15% fiber for 8 weeks (Dalby et al., 2017). Increases in body mass, fat mass, and fasting blood glucose were specific to the refined, high-fat diet. However, gut microbiome composition of mice fed high-fat and low-fat refined diets converged and were distinct from those of mice fed the unrefined chow, exhibiting reduced SCFA concentrations. Mice fed refined diets had shorter colons and colonic content mass, consistent with reduced nutrient flow to the colonic microbiome. Likewise, Morrison and colleagues fed mice an unrefined chow followed by a refined low-fat diet for 1 week each and then either kept mice on the refined low-fat diet or switched mice to a high-fat refined diet for 4 weeks (Morrison et al., 2020). Again, body weight increased more in mice fed the refined high-fat diet, indicative of host positive energy balance, but the refined low-fat diet restructured the gut microbial community in a manner that remained unchanged in mice subsequently fed the refined high-fat diet. Together, these studies disentangle the effects of host nutrient surplus (induced by refined high-fat diets) from microbial nutrient limitation (induced by both refined high-fat and refined low-fat diets), highlighting microbial nutrient limitation as a key driver of gut microbial community composition as predicted by our DNS model.
Diets modulating host or gut microbial nutrient availability
Several studies have altered colonic nutrient availability more specifically by supplementing diets with substrates known to be indigestible to the host (Li et al., 2024; Xie et al., 2023; Chambers et al., 2015; Liu et al., 2024; Nakajima et al., 2022). Li and colleagues supplemented the diets of 37 overweight adults with 40 g of either indigestible resistant starch or digestible starch for 8 weeks in a crossover study (Li et al., 2024). Participants had significantly lower fat mass, body weight, and waist circumference when supplemented with resistant starch versus digestible starch. Moreover, relative abundances of starch-degrading Ruminococcus bromii as well as Bifidobacterium adolescentis and Bifidobacterium longum increased. Likewise, mice reared for 8 weeks on a high-fat diet supplemented with either resistant starch bound to the SCFA propionate or unbound resistant starch harbored increased Ruminococcus and exhibited higher SCFA production relative to mice fed an unsupplemented high-fat diet (Xie et al., 2023). Weight gain and fat pad mass was highest in mice fed the unsupplemented high-fat diet, intermediate in mice supplemented with resistant starch, and lowest in mice supplemented with propionate-bound resistant starch. Carmody and colleagues interrogated gradients of host versus microbial nutrient availability by feeding mice re-blended diets containing various proportions of high-fat/high-sugar chow versus low-fat/high-fiber chow (Carmody et al., 2015). As the high-fat/high-sugar content increased from 0% to 100%, host adipose tissue stores increased and gut microbial community composition shifted in a dose-response manner.
Beyond increasing dietary fiber, researchers have modulated HDD by supplementing diets with polyphenols, which chelate nutrients and thereby inhibit host-driven digestion, or via bypassing host digestion through the administration of enemas containing buttermilk directly to the colonic environment (Jaja-Chimedza et al., 2018; Liu et al., 2021; Brennan, 2024; Mane et al., 2021). As predicted by DNS, supplementing chows with polyphenols led to decreased body mass and fat mass in mice but increased microbial SCFA production (Jaja-Chimedza et al., 2018; Liu et al., 2021), whereas delivering nutrients directly to the colon in humans via enemas had no impact on host weight or body composition but likewise altered gut microbial composition (Mane et al., 2021).
Jointly, these studies capture inverse relationships between host nutrient availability and energy status and microbial nutrient availability and metabolism as predicted by our DNS framework (Box 1).
DNS is predicted to vary across host species and be exacerbated in humans
Comparing gut anatomies across mammals of diverse foraging strategies provides a window into dietary digestibility and the extent to which hosts are reliant on microbial metabolism for energy harvest (Muegge et al., 2011; Ley et al., 2008) (Figure 1). Ruminants and non-ruminant herbivores, which consume fibrous plant materials inaccessible to mammalian enzymes, have adopted bifurcated strategies for leveraging the large enzymatic arsenal of the gut microbiota to break down complex carbohydrates. Whereas ruminants have evolved a large microbial fermentation chamber (rumen) upstream of the small intestine that grants microbes “first dibs” on dietary nutrients (McKenzie et al., 2017), non-ruminant herbivores largely restrict gut microbial communities to the distal GI tract but provide expanded chambers for fermentation, including enlarged ceca and elongated, sacculated colons. Although divergent in form, ruminant and non-ruminant herbivore guts share the outcome of providing additional niche space and time for microbial fermentation and nutrient absorption, which helps them optimize energy harvest from diets with low HDD. At the other end of the spectrum, carnivores and insectivores, which consume diets with high HDD, have low stomach pH, elongated small intestines, and reduced ceca and colons – a bauplan prioritizing host absorption of dietary substrates and reducing niche space for gut microbes (Reese et al., 2021). In the middle fall omnivores, which consume diets varying along a spectrum of moderately low to moderately high HDD according to host feeding niche (Hutchinson et al., 2022), and may exhibit substantial day-to-day or season-to-season variability in HDD due to stochasticity in optimal foraging (Remonti et al., 2016; Jensen et al., 2024; Coogan et al., 2014). Accordingly, omnivorous guts exhibit digestive features that are generally intermediate between those of herbivores and carnivores/insectivores, with structures and physiologies that grant the host priority access to nutrients in the small intestine while supporting some degree of microbial fermentation the distal gut. While omnivorous species vary widely in their habitual diets, gut anatomy, and digestive physiology, they share the ecological condition of relatively high variability in HDD, and therefore benefit from adapting to dietary flux on short timescales (Carmody et al., 2019; David et al., 2014).
Derived human gut anatomy and physiology suggest that the decoupling of host and microbial nutrient status may have been exacerbated over human evolution. Humans are unique among primates in that we routinely consume dietary substrates with high HDD and caloric density (e.g., animal foods, nuts, honey), and we additionally process our diet heavily through thermal and physical means (Carmody et al., 2019; Dagbasi et al., 2020; Carmody and Wrangham, 2009; Carmody et al., 2011; Wrangham and Carmody, 2010). Thermal and physical processing of food increases its net caloric value by increasing the susceptibility of nutrients to digestive enzymes and decreasing host digestive effort (Carmody et al., 2011; Groopman et al., 2015; Boback et al., 2007; Barr and Wright, 2010). Humans have adapted anatomically and physiologically to these dietary innovations (Wrangham and Carmody, 2010; Carmody et al., 2016; Aiello and Wheeler, 1995; Zink and Lieberman, 2016). For instance, compared with our australopithecine ancestors and our closest living relatives in Pan, humans have energetically expensive features (e.g., large body size, relatively large brains) yet markedly reduced structures for mastication and digestion (e.g., small oral cavities, small chewing muscles, small GI tracts) (Aiello and Wheeler, 1995). In addition, our GI tracts have been reproportioned to favor small intestinal over colonic tissue, with the small intestine being ∼52% larger (by mass) than expected for a great ape of our body size, but the colon being ∼74% smaller (Bryant et al., 2023). These anatomical changes have effectively committed humans to a calorie-rich and relatively high HDD diet, concomitantly reducing the fraction of dietary nutrients reaching colonic microbes. Habitually high HDD in the human diet has likely exacerbated the decoupling of host and microbial nutrient status, placing humans in a uniquely persistent state of energy surplus relative to our gut microbial communities. Though increasing HDD is hypothesized to have fueled the evolution of modern humans (Wrangham and Carmody, 2010; Aiello and Wheeler, 1995; Zink and Lieberman, 2016; Carmody et al., 2017), an underappreciated consequence is that our evolved dietary niche has effectively “starved” our microbes. We might therefore predict that human physiology has evolved to depend less heavily on the products of microbial fermentation than is the case for many other omnivorous species, and further, that the human gut microbiome has experienced unique selection pressures related to nutrient limitation over the course of our evolution (Carmody et al., 2019; Amato and Carmody, 2023). These hypotheses await empirical testing, but may help to explain why human gut microbiota exhibit less restructuring in response to dietary change compared to the microbiotas of other omnivorous model organisms, such as mice (Carmody and Bisanz, 2023).
Convergent gut microbial phenotypes of diverse species on high HDD diets
Over the past few thousand years, consumption of human-modified diets has expanded beyond humans to diverse host species that humans have domesticated or held in captivity (Frankel et al., 2019). While studies have found variable effects of captivity and domestication on the gut microbiomes of diverse animal species (Reese et al., 2021; Diaz and Reese, 2021; Alberdi et al., 2021), those that have considered dietary change due to captivity or domestication have generally reported convergent results (Table 1). Multiple studies have demonstrated that many animals, especially primates, shift towards “human-like” gut microbiome profiles in captivity, when consuming human-modified diets (Houtz et al., 2021; Clayton et al., 2016). Notably, consistent with predictions of our DNS model, multiple studies have found that the effects of captivity on the gut microbiome are exacerbated among hindgut fermenting species that typically consume low HDD diets in the wild (Frankel et al., 2019; Greene et al., 2019). Moreover, some ruminant species have not replicated such microbial shifts in the colon despite the consumption of human-modified diets (McKenzie et al., 2017; Trevelline and Moeller, 2022), which is consistent with the idea that the ruminant digestive tract allows other gut microbes first access to nutrients.
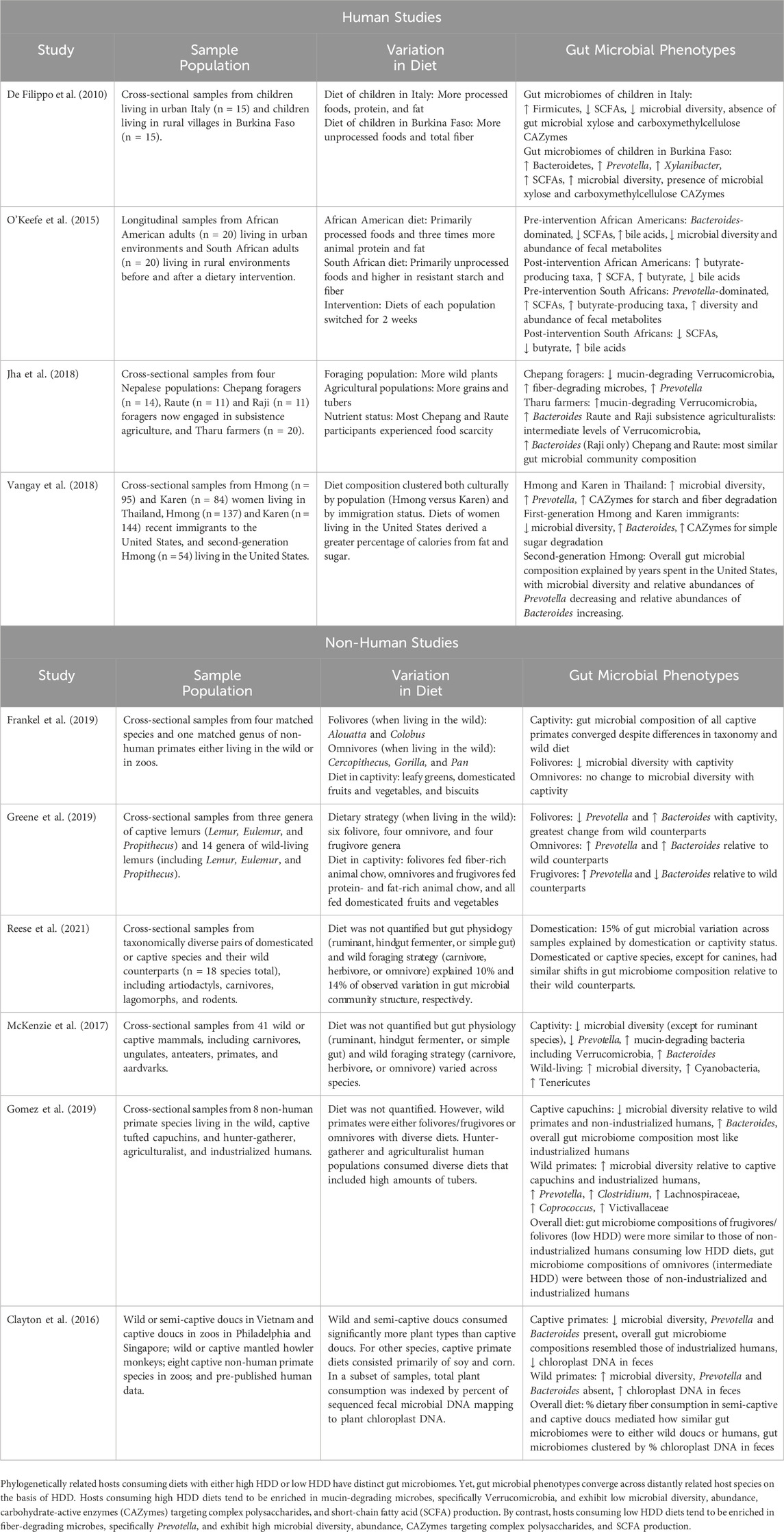
Table 1. Diets with high host-driven digestibility (HDD) mediate convergence of the gut microbiome across host species.
Many studies have likewise documented differences among human gut microbiomes along gradients of industrialization (Fragiadakis et al., 2019; Jha et al., 2018; O’Keefe et al., 2015; Smits et al., 2017; Moraïs et al., 2024; De Filippo et al., 2010; Rosas-Plaza et al., 2022). While many aspects of ecology differ between industrialized and non-industrialized populations, the gut microbiomes of diverse human populations generally reflect whether diet is comprised of primarily high HDD or low HDD substrates (Table 1). Low HDD diets, which are more common among non-industrialized populations, broadly increase gut microbial α-diversity, fiber-degrading capacity, and SCFA production, all indicators of increased colonic fermentation and gut health in industrialized settings (Sonnenburg and Sonnenburg, 2019a; Sonnenburg and Sonnenburg, 2019b). By contrast, high HDD diets, which are more common among industrialized populations, reduce microbial α-diversity and shift microbial metabolism towards protein- and mucin-degradation, reflecting the increased proportion of host-derived endogenous secretions versus residual dietary substrates suitable for fermentation in the colon (Sonnenburg and Sonnenburg, 2019a; Sonnenburg and Sonnenburg, 2019b). Comparisons of the industrialized and non-industrialized populations sampled to date have found these patterns, with the guts of non-industrialized populations consuming fiber-rich (i.e., low HDD) diets enriched in carbohydrate-degrading taxa like Prevotella, whereas the guts of industrialized populations consuming less fibrous and ultra-processed (i.e., high HDD) diets are enriched in protein- and mucin-degrading taxa like Bacteroidaceae and Verrucomicrobia (Fragiadakis et al., 2019; Jha et al., 2018; O’Keefe et al., 2015; Smits et al., 2017; Rosas-Plaza et al., 2022; Wu et al., 2011). Similarly, industrialized populations have higher ratios of mucin-degrading CAZymes to plant-degrading CAZymes and lower ratios of plant-degrading CAZymes to animal-degrading CAZymes compared to Hadza hunter-gatherers (Smits et al., 2017).
Importantly, our DNS framework predicts that high-latitude hunter-gatherers and pastoralists - non-industrialized populations who habitually consume high HDD diets rich in fat and protein - will have gut microbiomes that more closely resemble those of industrialized populations than is the case for other non-industrialized groups. Though few such studies exist, present evidence suggests that this is the case. Like the gut microbiomes of industrialized populations, the gut microbiomes of pastoralists have relatively low taxonomic richness and diversity (Rosas-Plaza et al., 2022; Rubel et al., 2020; Liu et al., 2016). One meta-analysis found that relative to agriculturalists, agropastoralists, and tropical hunter-gatherers, pastoralists have more gut microbial taxa in common with urbanites (Rosas-Plaza et al., 2022). Further, the Mboro Fulani pastoralists in Cameroon had significantly higher levels of Bacteroides and lower levels of Prevotella compared to Cameroon hunter-gatherer and subsistence agriculturalist populations (Rubel et al., 2020), and Mongolian pastoralists harbored microbiomes enriched in amino acid- and lipid-degrading genes (Liu et al., 2016).
Collectively, while these findings suggest that industrialization-mediated trends in the gut microbiome across human populations are mediated by HDD, few studies have directly quantified diet. For example, a recent analysis reported that cellulose-degrading Ruminococcus bacteria are more prevalent in non-industrialized populations, but notably some industrialized populations measured had comparable prevalence (Moraïs et al., 2024). Without data on dietary intake of cellulose, the factors driving differences in prevalence across populations remain uncertain. Human diets are highly variable across populations, and may be highly variable within populations across time, making it challenging to model diet-microbiome interactions in human populations based on subsistence style-based expectations or even rigorously documented long-term averages (Lieberman et al., 2023).
Direct tests of the DNS framework
Probing host and microbial responses to dietary digestibility will require carefully crafted diets that manipulate HDD as well as the development of accurate and economical methods to index diet as it appears to the colonic gut microbiome. Nutrition research is ripe with tools to modulate HDD, including processing dietary substrates thermally or non-thermally (Carmody et al., 2011; Groopman et al., 2015; Zink and Lieberman, 2016; Evenepoel et al., 1998; Evenepoel et al., 1999), exchanging simple versus complex carbohydrates (Dalby et al., 2017; Morrison et al., 2020), substituting simple starch for resistant starch (DeMartino and Cockburn, 2020; Maier et al., 2017; Haralampu, 2000), supplementing diets with dietary compounds (such as polyphenols) that inhibit HDD (Jaja-Chimedza et al., 2018; Liu et al., 2021), and bypassing small intestinal digestion through colonic instillation of dietary compounds (Mane et al., 2021). Additionally, some pharmaceuticals can modulate gut transit time, affecting the duration that nutrients are available to host or microbial enzymes without altering diet composition (Cummings and Macfarlane, 1997; El Oufir et al., 1996). Thus, our DNS model can begin to be tested directly by performing dietary interventions using diets that reflect gradients of HDD, with downstream measurement of host and microbial responses. Critically, capturing how dietary substrates are transformed by host-driven digestion and the microbiota of the small intestine, which differs significantly from the colonic microbiota (Kohl et al., 2018; Anders et al., 2021; Shalon et al., 2023), will identify which dietary components are available to the colonic gut microbiome.
Prior studies have attempted to isolate HDD by collecting samples from cannulated animals (Sutton and Oldham, 1977; Ipharraguerre et al., 2007), human ileostomates (Wolever et al., 1986; Jenkins et al., 1987), or through detection of isotopically labeled nutrients in circulation (Hui et al., 2017; Klein and Klein, 1985). However, such studies have limited ability to generalize to healthy hosts and are unable to examine hosts in conjunction with their colonic microbes. Two emerging methodological approaches may facilitate the indexing of diet under physiological conditions and could potentially be combined to disentangle host and gut microbial nutrient availability: swallowable capsules that permit non-invasive sampling of effluent along the GI tract (Shalon et al., 2023; Folz et al., 2023) and diet-targeted DNA metabarcoding (Reese et al., 2019a; Petrone et al., 2023; Schneider et al., 2021).
Recently developed swallowable sample collection bladders coated in pH-sensitive capsules can be used to collect effluent along the GI tract (Shalon et al., 2023; Folz et al., 2023). Capsules have been designed to dissolve at a specific pH between 5.5 and 7.5, corresponding to conditions in the duodenum, jejunum, ileum, and/or ascending colon. Once dissolved, the collapsed collection bladder within the intact capsule is exposed to the GI lumen and expands to draw in ∼400 μL of effluent prior to closing via a one-way valve. Collection bladders containing the sample are then retrieved from stool. Swallowing multiple capsules with different pH targets thereby enables non-invasive sampling along the GI tract. A human trial demonstrated the utility of these capsules for probing the gut microbiome and metabolome along the GI tract (Shalon et al., 2023; Folz et al., 2023), finding host and microbial metabolic shifts predicted by decades of prior research. However, to our knowledge these capsules have not previously been employed in studies specifically designed to track the fate of diet-derived substrates in the lumen.
One possible approach for indexing dietary residues in effluent is DNA metabarcoding, which enables the tracking of DNA from plant and animal foods as they traverse the gut by amplifying and sequencing specific marker genes. Though multiple plant and animal marker genes exist, the P6 loop of the plant chloroplast trnL gene (10–143 base pairs) and the V5 region of the animal mitochondrial 12S gene (56–132 base pairs) are economical because their variable regions are short enough for high-throughput Illumina sequencing (Schneider et al., 2021; Taberlet et al., 2007). DNA metabarcoding of plant and animal genes has been widely used in nutritional and behavioral ecology to identify the dietary niches of animals inhabiting an ecosystem (Ficetola and Taberlet, 2023; de Sousa et al., 2019; Evans et al., 2016; Liu et al., 2020). More recently, diet-derived DNA has been amplified from human stomach (Schneider et al., 2021) and fecal (Petrone et al., 2023; Reese et al., 2019b) contents to provide an objective measure of dietary substrates consumed. Despite the ability of human subjects to relay information about their diet, objective measures of food intake are nevertheless powerful because humans often have poor dietary recall (Shim et al., 2014).
DNA metabarcoding has important limitations, including overrepresentation of dietary components with high chloroplast densities such as leaves (Nichols et al., 2018; Ma and Li, 2015), unspecific mapping to the plant or animal part consumed [which can vary widely in digestibility (Ogawa et al., 2018)], and limited taxonomic granularity as related species often share identical DNA sequences within common marker genes of interest (Petrone et al., 2023; Reese et al., 2019b). However, if critical future research validates the basic principle that residual DNA in effluent is inversely correlated with nutrient uptake, DNA metabarcoding could represent a powerful new tool for discriminating the components of diet available to hosts versus colonic microbes. Notably, human subjects in prior studies of DNA metabarcoding reported consuming coffee (Reese et al., 2019b) and fruit juices or sugar cane (Petrone et al., 2023), but no DNA from these substrates with high HDD was recovered in the feces whereas DNA was recovered from nearly all plant-derived foods with relatively low HDD, demonstrating the promise of applying DNA metabarcoding to interrogate HDD.
Ultimately, the combination of effluent collection along the GI tract and methods of quantifying diet residues in effluent could help answer key unknowns in nutrition and microbiome research, including: a) how the nutrient milieu changes along the GI tract; b) the extent to which dietary substrates are digested in the small intestine, and therefore their caloric value to the host; c) whether dietary residues entering the colon are a stronger predictor of diet-microbiome interactions than diet as quantified on the plate; and d) the ecological and evolutionary pressures on microbes driven by evolutionary changes in host-driven digestion, including anatomical changes in the GI tract across species but also population-variable genetic adaptations such as lactase persistence (Tishkoff et al., 2007) or salivary amylase production (Perry et al., 2007) in humans.
Outlook
Small intestinal digestibility mediates nutrient availability to humans and other hindgut fermenting hosts versus their colonic gut microbiota. Critically, in these host taxa, host and gut microbial experiences of nutrient status are decoupled from each other. Few studies examining intraspecific variation in gut microbial communities have directly quantified host diet, and virtually all that have quantified diet have indexed it as it appeared on the plate. Indexing the residues of diet that reach gut microbial communities in the small intestine and colon is necessary to accurately capture gut microbial responses to diet, diet-microbe interactions, and downstream implications for host physiology. For instance, identifying which dietary substrates reach gut microbes will expedite our ability to pinpoint the dietary sources of bioactive microbial metabolites (Rowland et al., 2018; Noerman et al., 2020; Canfora et al., 2019) and dietary levers for rationally manipulating the gut microbiota (Reese and Carmody, 2019). Importantly, dietary components digested and absorbed by the host can simultaneously be inferred, providing a new lens into human nutrition and novel insights into digestive physiology across individuals with predicted variation in digestive capabilities, including those with genetic differences in digestive enzymes (Schmidt et al., 2020; Poole et al., 2019; Hjorth et al., 2020) and digestive diseases (Bitzer et al., 2016; Balestrieri et al., 2020; Malterre, 2009). More broadly, establishing the small intestinal digestibility of nutrients affords greater accuracy in calculating the metabolizable energy content of the diet, which at present can only be estimated using coarse-grained, outdated multipliers (Carmody et al., 2024). Finally, our DNS framework provides a rationale and blueprint for controlled tests that probe how gut microbial communities respond to reductions in diet-derived nutrients entering the colon, as we posit happened over human evolution. Such testing could potentially identify host-microbiome interactions that are uniquely human. Together, separating host from microbial responses to diet and reevaluating them through the lens of DNS will promote a richer and more nuanced understanding of host biology, nutrition, and host-microbe interactions, as well as explicate physiological responses to HDD diets in humans, a pattern now reaching extremes in industrialized contexts.
Author contributions
EV: Conceptualization, Data curation, Funding acquisition, Writing–original draft, Writing–review and editing. RC: Conceptualization, Funding acquisition, Supervision, Writing–original draft, Writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by an NSF Graduate Research Fellowship to EV and grants from the William F. Milton Fund and Harvard Dean’s Competitive Fund for Promising Scholarship to RC.
Acknowledgments
We thank Amar Sarkar, Cary Allen-Blevins, Laura Schell, Neil Ibata, and other members of the Nutritional and Microbial Ecology Lab for conceptual discussions and comments on the manuscript. We thank Yi Jia Liow for graphic design support and Neil Ibata for pointing us to research on pastoralist populations.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
DNS, Decoupled Nutrient Status; GI, Gastrointestinal; HDD, Host-Driven Digestibility; SCFAs, Short-Chain Fatty Acids; CAZymes, Carbohydrate-Active Enzymes.
References
Aiello, L. C., and Wheeler, P. (1995). The expensive-tissue hypothesis: the brain and the digestive system in human and primate evolution. Curr. Anthropol. 36 (2), 199–221. doi:10.1086/204350
Alberdi, A., Martin Bideguren, G., and Aizpurua, O. (2021). Diversity and compositional changes in the gut microbiota of wild and captive vertebrates: a meta-analysis. Sci. Rep. 11 (1), 22660. doi:10.1038/s41598-021-02015-6
Amato, K. R., and Carmody, R. N. (2023). Gut microbial intersections with human ecology and evolution. Annu. Rev. Anthropol. 52 (1), 295–311. doi:10.1146/annurev-anthro-052721-085122
Anders, J. L., Moustafa, M. A. M., Mohamed, W. M. A., Hayakawa, T., Nakao, R., and Koizumi, I. (2021). Comparing the gut microbiome along the gastrointestinal tract of three sympatric species of wild rodents. Sci. Rep. 11 (1), 19929. doi:10.1038/s41598-021-99379-6
Bäckhed, F., Ding, H., Wang, T., Hooper, L. V., Koh, G. Y., Nagy, A., et al. (2004). The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl. Acad. Sci. U. S. A. 101 (44), 15718–15723. doi:10.1073/pnas.0407076101
Balestrieri, P., Ribolsi, M., Guarino, M. P. L., Emerenziani, S., Altomare, A., and Cicala, M. (2020). Nutritional aspects in inflammatory bowel diseases. Nutrients 12 (2), 372. doi:10.3390/nu12020372
Barber, T. M., Kabisch, S., Pfeiffer, A. F. H., and Weickert, M. O. (2020). The health benefits of dietary fibre. Nutrients 12 (10), 3209. doi:10.3390/nu12103209
Barr, S. B., and Wright, J. C. (2010). Postprandial energy expenditure in whole-food and processed-food meals: implications for daily energy expenditure. Food Nutr. Res. 54, 5144. doi:10.3402/fnr.v54i0.5144
Bitzer, Z. T., Elias, R. J., Vijay-Kumar, M., and Lambert, J. D. (2016). (-)-Epigallocatechin-3-gallate decreases colonic inflammation and permeability in a mouse model of colitis, but reduces macronutrient digestion and exacerbates weight loss. Mol. Nutr. Food Res. 60 (10), 2267–2274. doi:10.1002/mnfr.201501042
Boback, S. M., Cox, C. L., Ott, B. D., Carmody, R., Wrangham, R. W., and Secor, S. M. (2007). Cooking and grinding reduces the cost of meat digestion. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 148 (3), 651–656. doi:10.1016/j.cbpa.2007.08.014
Boutari, C., and Mantzoros, C. S. (2022). A 2022 update on the epidemiology of obesity and a call to action: as its twin COVID-19 pandemic appears to be receding, the obesity and dysmetabolism pandemic continues to rage on. Metabolism 133 (155217), 155217. doi:10.1016/j.metabol.2022.155217
Brennan, C. S. (2024). The role of valorised plant proteins and phenolic compounds on the digestibility of foods: a short review of recent trends and future opportunities in addressing sustainability issues. Front. Food Sci. Technol. 4. doi:10.3389/frfst.2024.1354391
Bryant, K. L., Hansen, C., and Hecht, E. E. (2023). Fermentation technology as a driver of human brain expansion. Commun. Biol. 6 (1), 1190. doi:10.1038/s42003-023-05517-3
Canfora, E. E., Meex, R. C. R., Venema, K., and Blaak, E. E. (2019). Gut microbial metabolites in obesity, NAFLD and T2DM. Nat. Rev. Endocrinol. 15 (5), 261–273. doi:10.1038/s41574-019-0156-z
Carmody, R. N. (2017). “9. Evolution of the human dietary niche: quest for high quality,” in Chimpanzees and human evolution. Editors M. N. Muller, R. W. Wrangham, and D. R. Pilbeam (Cambridge, MA and London, England: Harvard University Press), 311–338.
Carmody, R. N., and Bisanz, J. E. (2023). Roles of the gut microbiome in weight management. Nat. Rev. Microbiol. 21, 535–550. doi:10.1038/s41579-023-00888-0
Carmody, R. N., Bisanz, J. E., Bowen, B. P., Maurice, C. F., Lyalina, S., Louie, K. B., et al. (2019). Cooking shapes the structure and function of the gut microbiome. Nat. Microbiol. 4 (12), 2052–2063. doi:10.1038/s41564-019-0569-4
Carmody, R. N., Dannemann, M., Briggs, A. W., Nickel, B., Groopman, E. E., Wrangham, R. W., et al. (2016). Genetic evidence of human adaptation to a cooked diet. Genome Biol. Evol. 8 (4), 1091–1103. doi:10.1093/gbe/evw059
Carmody, R. N., Gerber, G. K., Luevano, J. M., Gatti, D. M., Somes, L., Svenson, K. L., et al. (2015). Diet dominates host genotype in shaping the murine gut microbiota. Cell Host Microbe 17 (1), 72–84. doi:10.1016/j.chom.2014.11.010
Carmody, R. N., Varady, K., and Turnbaugh, P. J. (2024). Digesting the complex metabolic effects of diet on the host and microbiome. Cell 187 (15), 3857–3876. doi:10.1016/j.cell.2024.06.032
Carmody, R. N., Weintraub, G. S., and Wrangham, R. W. (2011). Energetic consequences of thermal and nonthermal food processing. Proc. Natl. Acad. Sci. U. S. A. 108 (48), 19199–19203. doi:10.1073/pnas.1112128108
Carmody, R. N., and Wrangham, R. W. (2009). The energetic significance of cooking. J. Hum. Evol. 57 (4), 379–391. doi:10.1016/j.jhevol.2009.02.011
Chadaideh, K. S., and Carmody, R. N. (2021). Host-microbial interactions in the metabolism of different dietary fats. Cell Metab. 33 (5), 857–872. doi:10.1016/j.cmet.2021.04.011
Chambers, E. S., Viardot, A., Psichas, A., Morrison, D. J., Murphy, K. G., Zac-Varghese, S. E. K., et al. (2015). Effects of targeted delivery of propionate to the human colon on appetite regulation, body weight maintenance and adiposity in overweight adults. Gut 64 (11), 1744–1754. doi:10.1136/gutjnl-2014-307913
Clayton, J. B., Vangay, P., Huang, H., Ward, T., Hillmann, B. M., Al-Ghalith, G. A., et al. (2016). Captivity humanizes the primate microbiome. Proc. Natl. Acad. Sci. U. S. A. 113 (37), 10376–10381. doi:10.1073/pnas.1521835113
Coogan, S. C. P., Raubenheimer, D., Stenhouse, G. B., and Nielsen, S. E. (2014). Macronutrient optimization and seasonal diet mixing in a large omnivore, the grizzly bear: a geometric analysis. PLoS One 9 (5), e97968. doi:10.1371/journal.pone.0097968
Corbin, K. D., Carnero, E. A., Dirks, B., Igudesman, D., Yi, F., Marcus, A., et al. (2023). Host-diet-gut microbiome interactions influence human energy balance: a randomized clinical trial. Nat. Commun. 14 (1), 3161. doi:10.1038/s41467-023-38778-x
Cronin, P., Joyce, S. A., O’Toole, P. W., and O’Connor, E. M. (2021). Dietary fibre modulates the gut Microbiota. Nutrients 13 (5), 1655. doi:10.3390/nu13051655
Cummings, J. H., and Macfarlane, G. T. (1997). Role of intestinal bacteria in nutrient metabolism. Clin. Nutr. 16 (1), 357–365. doi:10.1177/0148607197021006357
Dagbasi, A., Lett, A. M., Murphy, K., and Frost, G. (2020). Understanding the interplay between food structure, intestinal bacterial fermentation and appetite control. Proc. Nutr. Soc. 79 (4), 514–530. doi:10.1017/S0029665120006941
Dalby, M. J., Ross, A. W., Walker, A. W., and Morgan, P. J. (2017). Dietary uncoupling of gut microbiota and energy harvesting from obesity and glucose tolerance in mice. Cell Rep. 21 (6), 1521–1533. doi:10.1016/j.celrep.2017.10.056
Daniel, H., Gholami, A. M., Berry, D., Desmarchelier, C., Hahne, H., Loh, G., et al. (2014). High-fat diet alters gut microbiota physiology in mice. ISME J. 8 (2), 295–308. doi:10.1038/ismej.2013.155
David, L. A., Maurice, C. F., Carmody, R. N., Gootenberg, D. B., Button, J. E., Wolfe, B. E., et al. (2014). Diet rapidly and reproducibly alters the human gut microbiome. Nature 505 (7484), 559–563. doi:10.1038/nature12820
Deehan, E. C., Mocanu, V., and Madsen, K. L. (2024). Effects of dietary fibre on metabolic health and obesity. Nat. Rev. Gastroenterol. Hepatol. 21 (5), 301–318. doi:10.1038/s41575-023-00891-z
De Filippo, C., Cavalieri, D., Di Paola, M., Ramazzotti, M., Poullet, J. B., Massart, S., et al. (2010). Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl. Acad. Sci. U. S. A. 107 (33), 14691–14696. doi:10.1073/pnas.1005963107
DeMartino, P., and Cockburn, D. W. (2020). Resistant starch: impact on the gut microbiome and health. Curr. Opin. Biotechnol. 61, 66–71. doi:10.1016/j.copbio.2019.10.008
de Sousa, L. L., Silva, S. M., and Xavier, R. (2019). DNA metabarcoding in diet studies: unveiling ecological aspects in aquatic and terrestrial ecosystems. Environ. DNA 1 (3), 199–214. doi:10.1002/edn3.27
Diaz, J., and Reese, A. T. (2021). Possibilities and limits for using the gut microbiome to improve captive animal health. Anim. Microbiome 3 (1), 89. doi:10.1186/s42523-021-00155-8
El Oufir, L., Flourie, B., Bruley des Varannes, S., Barry, J. L., Cloarec, D., Bornet, F., et al. (1996). Relations between transit time, fermentation products, and hydrogen consuming flora in healthy humans. Gut 38 (6), 870–877. doi:10.1136/gut.38.6.870
Evans, D. M., Kitson, J. J. N., Lunt, D. H., Straw, N. A., and Pocock, M. J. O. (2016). Merging DNA metabarcoding and ecological network analysis to understand and build resilient terrestrial ecosystems. Funct. Ecol. 30 (12), 1904–1916. doi:10.1111/1365-2435.12659
Evenepoel, P., Claus, D., Geypens, B., Hiele, M., Geboes, K., Rutgeerts, P., et al. (1999). Amount and fate of egg protein escaping assimilation in the small intestine of humans. Am. J. Physiol. 277 (5), G935–G943. doi:10.1152/ajpgi.1999.277.5.G935
Evenepoel, P., Geypens, B., Luypaerts, A., Hiele, M., Ghoos, Y., and Rutgeerts, P. (1998). Digestibility of cooked and raw egg protein in humans as assessed by stable isotope techniques. J. Nutr. 128 (10), 1716–1722. doi:10.1093/jn/128.10.1716
Ficetola, G. F., and Taberlet, P. (2023). Towards exhaustive community ecology via DNA metabarcoding. Mol. Ecol. 32 (23), 6320–6329. doi:10.1111/mec.16881
Folz, J., Culver, R. N., Morales, J. M., Grembi, J., Triadafilopoulos, G., Relman, D. A., et al. (2023). Human metabolome variation along the upper intestinal tract. Nat. Metab. 5 (5), 777–788. doi:10.1038/s42255-023-00777-z
Fragiadakis, G. K., Smits, S. A., Sonnenburg, E. D., Van Treuren, W., Reid, G., Knight, R., et al. (2019). Links between environment, diet, and the hunter-gatherer microbiome. Gut Microbes 10 (2), 216–227. doi:10.1080/19490976.2018.1494103
Frankel, J. S., Mallott, E. K., Hopper, L. M., Ross, S. R., and Amato, K. R. (2019). The effect of captivity on the primate gut microbiome varies with host dietary niche. Am. J. Primatol. 81 (12), e23061. doi:10.1002/ajp.23061
Gomez, A., Sharma, A. K., Mallott, E. K., Petrzelkova, K. J., Jost Robinson, C. A., Yeoman, C. J., et al. (2019). Plasticity in the human gut microbiome defies evolutionary constraints. mSphere 4 (4). doi:10.1128/msphere.00271-19
Greene, L. K., Bornbusch, S. L., McKenney, E. A., Harris, R. L., Gorvetzian, S. R., Yoder, A. D., et al. (2019). The importance of scale in comparative microbiome research: new insights from the gut and glands of captive and wild lemurs. Am. J. Primatol. 81 (10–11), e22974. doi:10.1002/ajp.22974
Groopman, E. E., Carmody, R. N., and Wrangham, R. W. (2015). Cooking increases net energy gain from a lipid-rich food. Am. J. Phys. Anthropol. 156 (1), 11–18. doi:10.1002/ajpa.22622
Hall, K. D., Ayuketah, A., Brychta, R., Cai, H., Cassimatis, T., Chen, K. Y., et al. (2019). Ultra-processed diets cause excess calorie intake and weight gain: an inpatient randomized controlled trial of ad libitum food intake. Cell Metab. 30 (1), 226–277.e3. doi:10.1016/j.cmet.2019.05.020
Haralampu, S. G. (2000). Resistant starch—a review of the physical properties and biological impact of RS3. Carbohydr. Polym. 41 (3), 285–292. doi:10.1016/s0144-8617(99)00147-2
He, C., Cheng, D., Peng, C., Li, Y., Zhu, Y., and Lu, N. (2018). High-fat diet induces dysbiosis of gastric Microbiota prior to gut Microbiota in association with metabolic disorders in mice. Front. Microbiol. 9, 639. doi:10.3389/fmicb.2018.00639
Hjorth, M. F., Christensen, L., Larsen, T. M., Roager, H. M., Krych, L., Kot, W., et al. (2020). Pretreatment Prevotella-to-Bacteroides ratio and salivary amylase gene copy number as prognostic markers for dietary weight loss. Am. J. Clin. Nutr. 111 (5), 1079–1086. doi:10.1093/ajcn/nqaa007
Houtz, J. L., Sanders, J. G., Denice, A., and Moeller, A. H. (2021). Predictable and host-species specific humanization of the gut microbiota in captive primates. Mol. Ecol. 30 (15), 3677–3687. doi:10.1111/mec.15994
Hui, S., Ghergurovich, J. M., Morscher, R. J., Jang, C., Teng, X., Lu, W., et al. (2017). Glucose feeds the TCA cycle via circulating lactate. Nature 551 (7678), 115–118. doi:10.1038/nature24057
Hutchinson, M. C., Dobson, A. P., and Pringle, R. M. (2022). Dietary abundance distributions: dominance and diversity in vertebrate diets. Ecol. Lett. 25 (4), 992–1008. doi:10.1111/ele.13948
Ipharraguerre, I. R., Reynal, S. M., Liñeiro, M., Broderick, G. A., and Clark, J. H. (2007). A comparison of sampling sites, digesta and microbial markers, and microbial references for assessing the postruminal supply of nutrients in dairy cows. J. Dairy Sci. 90 (4), 1904–1919. doi:10.3168/jds.2006-159
Jaja-Chimedza, A., Zhang, L., Wolff, K., Graf, B. L., Kuhn, P., Moskal, K., et al. (2018). A dietary isothiocyanate-enriched moringa (Moringa oleifera) seed extract improves glucose tolerance in a high-fat-diet mouse model and modulates the gut microbiome. J. Funct. Foods 47, 376–385. doi:10.1016/j.jff.2018.05.056
Jenkins, D. J., Cuff, D., Wolever, T. M., Knowland, D., Thompson, L., Cohen, Z., et al. (1987). Digestibility of carbohydrate foods in an ileostomate: relationship to dietary fiber, in vitro digestibility, and glycemic response. Am. J. Gastroenterol. 82 (8), 709–717.
Jensen, A. J., Muthersbaugh, M., Ruth, C. R., Butfiloski, J. W., Cantrell, J., Adams, J., et al. (2024). Resource pulses shape seasonal and individual variation in the diet of an omnivorous carnivore. Ecol. Evol. 14 (7), e11632. doi:10.1002/ece3.11632
Jha, A. R., Davenport, E. R., Gautam, Y., Bhandari, D., Tandukar, S., Ng, K. M., et al. (2018). Gut microbiome transition across a lifestyle gradient in Himalaya. PLoS Biol. 16 (11), e2005396. doi:10.1371/journal.pbio.2005396
Kaoutari, A. E., Armougom, F., Gordon, J. I., Raoult, D., and Henrissat, B. (2013). The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nat. Rev. Microbiol. 11 (7), 497–504. doi:10.1038/nrmicro3050
Kennedy, E. A., King, K. Y., and Baldridge, M. T. (2018). Mouse Microbiota models: comparing germ-free mice and antibiotics treatment as tools for modifying gut bacteria. Front. Physiol. 9, 1534. doi:10.3389/fphys.2018.01534
Kennedy, M. S., and Chang, E. B. (2020). “The microbiome: composition and locations,” in Progress in molecular biology and translational science (Elsevier), 1–42. (Progress in molecular biology and translational science).
Klein, P. D., and Klein, E. R. (1985). Applications of stable isotopes to pediatric nutrition and gastroenterology: measurement of nutrient absorption and digestion using 13C. J. Pediatr. Gastroenterol. Nutr. 4 (1), 9–19. doi:10.1097/00005176-198502000-00004
Kohl, K. D., Dearing, M. D., and Bordenstein, S. R. (2018). Microbial communities exhibit host species distinguishability and phylosymbiosis along the length of the gastrointestinal tract. Mol. Ecol. 27 (8), 1874–1883. doi:10.1111/mec.14460
Laffin, M., Fedorak, R., Zalasky, A., Park, H., Gill, A., Agrawal, A., et al. (2019). A high-sugar diet rapidly enhances susceptibility to colitis via depletion of luminal short-chain fatty acids in mice. Sci. Rep. 9 (1), 12294. doi:10.1038/s41598-019-48749-2
Ley, R. E., Hamady, M., Lozupone, C., Turnbaugh, P. J., Ramey, R. R., Bircher, J. S., et al. (2008). Evolution of mammals and their gut microbes. Science 320 (5883), 1647–1651. doi:10.1126/science.1155725
Li, H., Zhang, L., Li, J., Wu, Q., Qian, L., He, J., et al. (2024). Resistant starch intake facilitates weight loss in humans by reshaping the gut microbiota. Nat. Metab. 26, 578–597. doi:10.1038/s42255-024-00988-y
Lieberman, D. E., Worthington, S., Schell, L. D., Parkent, C. M., Devinsky, O., and Carmody, R. N. (2023). Comparing measured dietary variation within and between tropical hunter-gatherer groups to the Paleo Diet. Am. J. Clin. Nutr. 118 (3), 549–560. doi:10.1016/j.ajcnut.2023.06.013
Liu, J., Hao, W., He, Z., Kwek, E., Zhu, H., Ma, N., et al. (2021). Blueberry and cranberry anthocyanin extracts reduce bodyweight and modulate gut microbiota in C57BL/6 J mice fed with a high-fat diet. Eur. J. Nutr. 60 (5), 2735–2746. doi:10.1007/s00394-020-02446-3
Liu, M., Clarke, L. J., Baker, S. C., Jordan, G. J., and Burridge, C. P. (2020). A practical guide to DNA metabarcoding for entomological ecologists. Ecol. Entomol. 45 (3), 373–385. doi:10.1111/een.12831
Liu, W., Zhang, J., Wu, C., Cai, S., Huang, W., Chen, J., et al. (2016). Unique features of ethnic Mongolian gut microbiome revealed by metagenomic analysis. Sci. Rep. 6 (1), 34826. doi:10.1038/srep34826
Liu, Z., Dai, J., Liu, R., Shen, Z., Huang, A., Huang, Y., et al. (2024). Complex insoluble dietary fiber alleviates obesity and liver steatosis, and modulates the gut microbiota in C57BL/6J mice fed a high-fat diet. J. Sci. Food Agric. 104 (9), 5462–5473. doi:10.1002/jsfa.13380
Ma, J., and Li, X.-Q. (2015). Organellar genome copy number variation and integrity during moderate maturation of roots and leaves of maize seedlings. Curr. Genet. 61 (4), 591–600. doi:10.1007/s00294-015-0482-1
Maier, T. V., Lucio, M., Lee, L. H., VerBerkmoes, N. C., Brislawn, C. J., Bernhardt, J., et al. (2017). Impact of dietary resistant starch on the human gut microbiome, metaproteome, and metabolome. MBio 8 (5). doi:10.1128/mbio.01343-17
Malterre, T. (2009). Digestive and nutritional considerations in celiac disease: could supplementation help? Altern. Med. Rev. 14 (3), 247–257.
Mane, S., Dixit, K. K., Lathwal, N., Dhotre, D., Kadus, P., Shouche, Y. S., et al. (2021). Rectal administration of buttermilk processed with medicinal plants alters gut microbiome in obese individuals. J. Diabetes Metab. Disord. 20 (2), 1415–1427. doi:10.1007/s40200-021-00879-z
Martínez Leo, E. E., and Segura Campos, M. R. (2020). Effect of ultra-processed diet on gut microbiota and thus its role in neurodegenerative diseases. Nutrition 71 (110609), 110609. doi:10.1016/j.nut.2019.110609
McKenzie, V. J., Song, S. J., Delsuc, F., Prest, T. L., Oliverio, A. M., Korpita, T. M., et al. (2017). The effects of captivity on the mammalian gut microbiome. Integr. Comp. Biol. 57 (4), 690–704. doi:10.1093/icb/icx090
Monteiro, C. A., Moubarac, J.-C., Cannon, G., Ng, S. W., and Popkin, B. (2013). Ultra-processed products are becoming dominant in the global food system. Obes. Rev. 14 (S2), 21–28. doi:10.1111/obr.12107
Moraïs, S., Winkler, S., Zorea, A., Levin, L., Nagies, F. S. P., Kapust, N., et al. (2024). Cryptic diversity of cellulose-degrading gut bacteria in industrialized humans. Science 383 (6688), eadj9223. doi:10.1126/science.adj9223
Morrison, K. E., Jašarević, E., Howard, C. D., and Bale, T. L. (2020). It’s the fiber, not the fat: significant effects of dietary challenge on the gut microbiome. Microbiome 8 (1), 15. doi:10.1186/s40168-020-0791-6
Muegge, B. D., Kuczynski, J., Knights, D., Clemente, J. C., González, A., Fontana, L., et al. (2011). Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science 332 (6032), 970–974. doi:10.1126/science.1198719
Nakajima, H., Nakanishi, N., Miyoshi, T., Okamura, T., Hashimoto, Y., Senmaru, T., et al. (2022). Inulin reduces visceral adipose tissue mass and improves glucose tolerance through altering gut metabolites. Nutr. Metab. (Lond) 19 (1), 50. doi:10.1186/s12986-022-00685-1
Nichols, R. V., Vollmers, C., Newsom, L. A., Wang, Y., Heintzman, P. D., Leighton, M., et al. (2018). Minimizing polymerase biases in metabarcoding. Mol. Ecol. Resour. 18 (5), 927–939. doi:10.1111/1755-0998.12895
Noerman, S., Kolehmainen, M., and Hanhineva, K. (2020). Profiling of endogenous and gut microbial metabolites to indicate metabotype-specific dietary responses: a systematic review. Adv. Nutr. 11 (5), 1237–1254. doi:10.1093/advances/nmaa031
Ogawa, Y., Donlao, N., Thuengtung, S., Tian, J., Cai, Y., Reginio, F. C., et al. (2018). Impact of food structure and cell matrix on digestibility of plant-based food. Curr. Opin. Food Sci. 19, 36–41. doi:10.1016/j.cofs.2018.01.003
O’Keefe, S. J. D., Li, J. V., Lahti, L., Ou, J., Carbonero, F., Mohammed, K., et al. (2015). Fat, fibre and cancer risk in African Americans and rural Africans. Nat. Commun. 6 (1), 6342. doi:10.1038/ncomms7342
Perry, G. H., Dominy, N. J., Claw, K. G., Lee, A. S., Fiegler, H., Redon, R., et al. (2007). Diet and the evolution of human amylase gene copy number variation. Nat. Genet. 39 (10), 1256–1260. doi:10.1038/ng2123
Petrone, B. L., Aqeel, A., Jiang, S., Durand, H. K., Dallow, E. P., McCann, J. R., et al. (2023). Diversity of plant DNA in stool is linked to dietary quality, age, and household income. Proc. Natl. Acad. Sci. 120 (27), e2304441120. doi:10.1073/pnas.2304441120
Poole, A. C., Goodrich, J. K., Youngblut, N. D., Luque, G. G., Ruaud, A., Sutter, J. L., et al. (2019). Human salivary amylase gene copy number impacts oral and gut microbiomes. Cell Host Microbe 25 (4), 553–564. doi:10.1016/j.chom.2019.03.001
Reese, A. T., and Carmody, R. N. (2019). Thinking outside the cereal box: noncarbohydrate routes for dietary manipulation of the gut Microbiota. Appl. Environ. Microbiol. 85 (10). doi:10.1128/aem.02246-18
Reese, A. T., Chadaideh, K. S., Diggins, C. E., Schell, L. D., Beckel, M., Callahan, P., et al. (2021). Effects of domestication on the gut microbiota parallel those of human industrialization. Elife 10, e60197. doi:10.7554/elife.60197
Reese, A. T., Kartzinel, T. R., Petrone, B. L., Turnbaugh, P. J., Pringle, R. M., and David, L. A. (2019a). Using DNA metabarcoding to evaluate the plant component of human diets: a proof of concept. mSystems 4 (5), doi:10.1128/mSystems.00458-19
Reese, A. T., Kartzinel, T. R., Petrone, B. L., Turnbaugh, P. J., Pringle, R. M., and David, L. A. (2019b). Using DNA metabarcoding to evaluate the plant component of human diets: a proof of concept. mSystems 4 (5), doi:10.1128/msystems.00458-19
Remonti, L., Balestrieri, A., Raubenheimer, D., and Saino, N. (2016). Functional implications of omnivory for dietary nutrient balance. Oikos 125 (9), 1233–1240. doi:10.1111/oik.02801
Rodriguez, D. M., Benninghoff, A. D., Aardema, N. D. J., Phatak, S., and Hintze, K. J. (2019). Basal diet determined long-term composition of the gut microbiome and mouse phenotype to a greater extent than fecal microbiome transfer from lean or obese human donors. Nutrients 11 (7), 1630. doi:10.3390/nu11071630
Rosas-Plaza, S., Hernández-Terán, A., Navarro-Díaz, M., Escalante, A. E., Morales-Espinosa, R., and Cerritos, R. (2022). Human gut microbiome across different lifestyles: from Hunter-gatherers to urban populations. Front. Microbiol. 13, 843170. doi:10.3389/fmicb.2022.843170
Rowland, I., Gibson, G., Heinken, A., Scott, K., Swann, J., Thiele, I., et al. (2018). Gut microbiota functions: metabolism of nutrients and other food components. Eur. J. Nutr. 57 (1), 1–24. doi:10.1007/s00394-017-1445-8
Rubel, M. A., Abbas, A., Taylor, L. J., Connell, A., Tanes, C., Bittinger, K., et al. (2020). Lifestyle and the presence of helminths is associated with gut microbiome composition in Cameroonians. Genome Biol. 21 (1), 122. doi:10.1186/s13059-020-02020-4
Schmidt, V., Enav, H., Spector, T. D., Youngblut, N. D., and Ley, R. E. (2020). Strain-level analysis of Bifidobacterium spp. from gut microbiomes of adults with differing lactase persistence genotypes. mSystems 5 (5), doi:10.1128/msystems.00911-20
Schneider, J., Mas-Carrió, E., Jan, C., Miquel, C., Taberlet, P., Michaud, K., et al. (2021). Comprehensive coverage of human last meal components revealed by a forensic DNA metabarcoding approach. Sci. Rep. 11 (1), 8876. doi:10.1038/s41598-021-88418-x
Shalon, D., Culver, R. N., Grembi, J. A., Folz, J., Treit, P. V., Shi, H., et al. (2023). Profiling the human intestinal environment under physiological conditions. Nature 617 (7961), 581–591. doi:10.1038/s41586-023-05989-7
Shan, K., Qu, H., Zhou, K., Wang, L., Zhu, C., Chen, H., et al. (2019). Distinct gut Microbiota induced by different fat-to-sugar-ratio high-energy diets share similar pro-obesity genetic and metabolite profiles in prediabetic mice. mSystems 4 (5), doi:10.1128/msystems.00219-19
Shim, J.-S., Oh, K., and Kim, H. C. (2014). Dietary assessment methods in epidemiologic studies. Epidemiol. Health 36, e2014009. doi:10.4178/epih/e2014009
Smits, S. A., Leach, J., Sonnenburg, E. D., Gonzalez, C. G., Lichtman, J. S., Reid, G., et al. (2017). Seasonal cycling in the gut microbiome of the Hadza hunter-gatherers of Tanzania. Science 357 (6353), 802–806. doi:10.1126/science.aan4834
Sonnenburg, E. D., Smits, S. A., Tikhonov, M., Higginbottom, S. K., Wingreen, N. S., and Sonnenburg, J. L. (2016). Diet-induced extinctions in the gut microbiota compound over generations. Nature 529 (7585), 212–215. doi:10.1038/nature16504
Sonnenburg, E. D., and Sonnenburg, J. L. (2014). Starving our microbial self: the deleterious consequences of a diet deficient in Microbiota-accessible carbohydrates. Cell Metab. 20 (5), 779–786. doi:10.1016/j.cmet.2014.07.003
Sonnenburg, E. D., and Sonnenburg, J. L. (2019a). The ancestral and industrialized gut microbiota and implications for human health. Nat. Rev. Microbiol. 17 (6), 383–390. doi:10.1038/s41579-019-0191-8
Sonnenburg, J. L., and Bäckhed, F. (2016). Diet–microbiota interactions as moderators of human metabolism. Nature 535 (7610), 56–64. doi:10.1038/nature18846
Sonnenburg, J. L., and Sonnenburg, E. D. (2019b). Vulnerability of the industrialized microbiota. Science 366 (6464), eaaw9255. doi:10.1126/science.aaw9255
Sutton, J. D., and Oldham, J. D. (1977). Feed evaluation by measurement of sites of digestion in cannulated ruminants. Proc. Nutr. Soc. 36 (2), 203–209. doi:10.1079/pns19770034
Taberlet, P., Coissac, E., Pompanon, F., Gielly, L., Miquel, C., Valentini, A., et al. (2007). Power and limitations of the chloroplast trnL (UAA) intron for plant DNA barcoding. Nucleic Acids Res. 35 (3), e14. doi:10.1093/nar/gkl938
Tishkoff, S. A., Reed, F. A., Ranciaro, A., Voight, B. F., Babbitt, C. C., Silverman, J. S., et al. (2007). Convergent adaptation of human lactase persistence in Africa and Europe. Nat. Genet. 39 (1), 31–40. doi:10.1038/ng1946
Trevelline, B. K., and Moeller, A. H. (2022). Robustness of mammalian gut Microbiota to humanization in captivity. Front. Ecol. Evol. 9, 785089. doi:10.3389/fevo.2021.785089
Vangay, P., Johnson, A. J., Ward, T. L., Al-Ghalith, G. A., Shields-Cutler, R. R., Hillmann, B. M., et al. (2018). US immigration westernizes the human gut microbiome. Cell 175 (4), 962–972. doi:10.1016/j.cell.2018.10.029
Walter, J., and Ley, R. (2011). The human gut microbiome: ecology and recent evolutionary changes. Annu. Rev. Microbiol. 65, 411–429. doi:10.1146/annurev-micro-090110-102830
Wolever, T. M., Cohen, Z., Thompson, L. U., Thorne, M. J., Jenkins, M. J., Prokipchuk, E. J., et al. (1986). Ileal loss of available carbohydrate in man: comparison of a breath hydrogen method with direct measurement using a human ileostomy model. Am. J. Gastroenterol. 81 (2), 115–122.
Wong, J. M. W., and Jenkins, D. J. A. (2007). Carbohydrate digestibility and metabolic effects. J. Nutr. 137 (11), 2539S-2546S–2546S. doi:10.1093/jn/137.11.2539S
Wrangham, R., and Carmody, R. (2010). Human adaptation to the control of fire. Evol. Anthropol. 19 (5), 187–199. doi:10.1002/evan.20275
Wu, G. D., Chen, J., Hoffmann, C., Bittinger, K., Chen, Y.-Y., Keilbaugh, S. A., et al. (2011). Linking long-term dietary patterns with gut microbial enterotypes. Science 334 (6052), 105–108. doi:10.1126/science.1208344
Xie, Z., Yao, M., Castro-Mejía, J. L., Ma, M., Zhu, Y., Fu, X., et al. (2023). Propionylated high-amylose maize starch alleviates obesity by modulating gut microbiota in high-fat diet-fed mice. J. Funct. Foods 102 (105447), 105447. doi:10.1016/j.jff.2023.105447
Keywords: microbiome, digestion, gut physiology, microbial diversity, host-microbe coevolution
Citation: Venable EM and Carmody RN (2024) Decoupled Nutrient Status: a framework to disentangle host from microbial responses to diets that vary in digestibility. Front. Food. Sci. Technol. 4:1469470. doi: 10.3389/frfst.2024.1469470
Received: 23 July 2024; Accepted: 27 September 2024;
Published: 05 November 2024.
Edited by:
Silvani Verruck, Federal University of Santa Catarina, BrazilReviewed by:
Nicoletta Righini, Universidad de Guadalajara, MexicoCopyright © 2024 Venable and Carmody. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Emily M. Venable, ZW1pbHl2ZW5hYmxlQGcuaGFydmFyZC5lZHU=; Rachel N. Carmody, Y2FybW9keUBmYXMuaGFydmFyZC5lZHU=
 Emily M. Venable
Emily M. Venable Rachel N. Carmody
Rachel N. Carmody