
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Food. Sci. Technol. , 23 November 2023
Sec. Food Safety and Quality Control
Volume 3 - 2023 | https://doi.org/10.3389/frfst.2023.1239884
This article is part of the Research Topic The Future of Foods View all 9 articles
Introduction: The clean label trend emphasizes the need for natural approaches to combat pathogenic bacteria in food. This study explores the potential of inducing prophages within bacterial genomes as a novel strategy to control pathogenic and spoilage bacterial growth.
Methods: A luminescence-based high-throughput assay was developed to identify natural compounds capable of inducing prophages. Bioactive compounds from four chemical libraries were screened at a final concentration of 10 µM. The assay measured luminescence production in Escherichia coli BR513, a genetically modified strain producing β-galactosidase upon prophage λ induction. Luminescence values were normalized to cell concentration (OD600) and the interquartile mean of each 384-well plate. A cut-off for normalized luminescence values, set at 2.25 standard deviations above the mean, defined positive prophage induction.
Results: Four naturally-derived compounds (osthol, roccellic acid, galanginee, and sclareol) exhibited positive prophage induction, along with previously identified inducers, rosemary, and gallic acid. Dose-response experiments were conducted to determine optimal concentrations for prophage induction. However, the results could not distinguish between prophage-induced cell death and other mechanisms, making it challenging to identify ideal concentrations.
Discussion: The high-throughput luminescent prophage induction assay serves as a valuable tool for the initial screening of natural bioactive compounds that have the potential to enhance food safety and quality by inducing prophages. Further research is required to understand the mechanism of bacterial cell death and to establish optimal concentrations for prophage induction in a food preservation context.
In developed countries, consumers are increasingly demanding that foods only contain natural and easily recognizable ingredients (Asioli et al., 2017). This consumer demand, known as the clean label trend, is largely driven by an increasing awareness of the potential toxicity of certain synthetic food additives (Carocho et al., 2014). As such, consumers tend to choose additive-free foods or foods containing natural additives over products that contain synthetic additives (Carocho et al., 2014). The food industry is thus shifting toward the use of naturally-sourced antimicrobials as replacements for commonly-used synthetic compounds, such as benzoates, sorbates, propionates, nitrites, and parabens (Carocho et al., 2015) that have traditionally been used to control the growth of foodborne bacterial pathogens (Emerton and Choi, 2008; Garcia et al., 2008).
The food industry’s transition to the strict use of naturally-sourced additives should not compromise food safety. Natural compounds from plants, animals, and microorganisms are known to have antimicrobial properties (Carocho et al., 2015). Varying mechanisms for the antimicrobial action of natural compounds have been proposed, which may involve interactions with proteins, enzymes, or membrane function (Fan et al., 2018). Bacteriophages (phages) have also emerged as effective natural antimicrobials (Sillankorva et al., 2012; Quinto et al., 2019) that can be added to foods to eliminate foodborne bacterial pathogens. Virulent phages introduce their DNA into cells of the target bacterial pathogens to produce progeny phage that ultimately lyse the host cells, in what is termed the lytic cycle (Salmond and Fineran, 2015; Moye et al., 2018). A number of commercialized phage products are employed to reduce the presence of bacterial pathogens in foods (Moye et al., 2018).
One limitation of the current phage control approach is the fact that due to the limits of phage host range, only a single bacterial species can be targeted at a single time. This limits applications of phage control in the food industry. For example, the use of phages to control bacterial spoilage is impractical because many different bacterial species are typically involved in the spoilage of a food (Odeyemi et al., 2020). Induction of temperate phages is an alternative phage-based antimicrobial mechanism of action (MOA) that was recently proposed for controlling bacterial pathogens on foods (Cadieux et al., 2018). Temperate phages are viruses of bacteria that infect their hosts by integrating their DNA into the bacterial host chromosome and immediately enter a lysogenic life cycle, in which no harm is inflicted on the host bacterial cell (Feiner et al., 2015). Various environmental stresses trigger the chromosomally-embedded phage (prophage) to enter into the lytic cycle, in which the phage DNA excises from the bacterial DNA and forms progeny that eventually lyse the host cell (Salmond and Fineran, 2015). Activation of the lytic cycle, known as prophage induction, can be initiated by the addition of bioactive compounds, which could be used as an approach for eliminating bacterial pathogens in food (Cadieux et al., 2018), since the majority of bacteria possess prophages (Kang et al., 2017).
It is well known that certain antibiotics induce prophages of foodborne bacterial pathogens (Cone et al., 1976; Raya and H'Bert E, 2009; McDonald et al., 2010); however, fewer natural compounds have been shown to induce prophages. We previously demonstrated that various teas, coffee, gallic acid, rosemary and cranberry juice are prophage inducers of prophage λ (Tompkins et al., 2018). These natural compounds have been described in the literature as having antimicrobial activity against bacterial pathogens (Côté et al., 2011; Nieto et al., 2018), and our results suggest that the MOA of the compounds is through the induction of prophages in the target bacteria.
In this work, a high-throughput luminescent prophage induction assay was modified in order to render it amenable to automated systems so that thousands of known bioactive compounds from chemical libraries could be screened. The high-throughput luminescent prophage induction assay serves as a preliminary screen to identify natural compounds that could be used to induce prophages from foodborne pathogenic and spoilage bacteria, thereby improving the safety and quality of foods using sustainable approaches.
The assay used in this work is based on the use of the genetically engineered E. coli BR513 (ATCC 33312). E. coli BR513 carries a lacZ-prophage λ gene fusion, and cleavage of the lambdoid phage repressor, CI, results in the synthesis of β-galactosidase (Elespuru and Yarmolinsky, 1979). The cleavage of the CI repressor, and thus prophage induction, may be caused by treatments that cause DNA damage. β-galactosidase produced by E. coli BR513 can be measured using luminescent substrates and taken as an indication of prophage λ induction. E. coli BR513 was stored at −80°C in 20% glycerol and cells were revived by streaking a loopful of frozen stock onto tryptic soy agar (TSA). Plates were incubated at 37°C for 18–24 h.
Compounds from the Centre for Microbial Chemical Biology (CMCB, McMaster University, Hamilton, Ontario, Canada) bioactives collection were screened for their ability to induce prophage λ using the luminescent high-throughput prophage induction assay. The CMCB bioactives collection (Table 1) contains 3,747 compounds from four vendor libraries, including the Prestwick Chemical Library, the BIOMOL2865 Natural Products Library, the Lopac1280 (International Version) Library, and the Spectrum Collection, and comprises Food and Drug Administration (FDA)-approved drugs, off-patent drugs, natural products, and other compounds with demonstrated biological activity (Miller et al., 2009; Davenport et al., 2014; Torres et al., 2016).
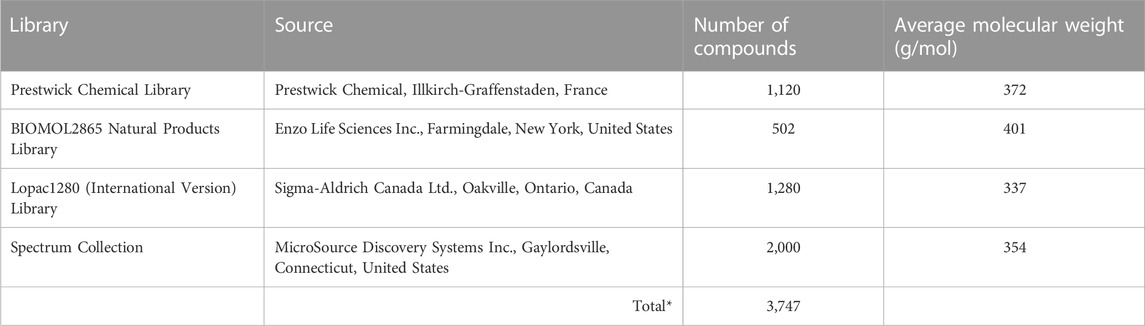
TABLE 1. The Centre for microbial chemical biology bioactives collection consisting of four vendor libraries was used in the high-throughput luminescent prophage induction assay. *The total number does not include duplicate compounds.
The high-throughput luminescent prophage induction assay was adapted for use with an automated robotic system at the CMCB. To begin the assay, duplicate cultures of E. coli BR513 were grown overnight in trypticase soy broth (TSB) supplemented with 0.2 M glucose (TSB 0.2 M glucose) at 37°C with shaking. The next morning, cells were diluted in fresh TSB 0.2 M glucose to an optical density (OD600) of 0.1. The cultures were grown at 37°C with shaking at 250 RPM until the OD600 reached 0.5. A Multidrop Combi Reagent Dispenser (Thermo Fisher Scientific, Waltham, Massachusetts, United States) was used to add 50 μL of culture to the wells of 384-well plates. Next, 0.5 μL of each test compound from the CMCB bioactives library (Table 1), having been dissolved in dimethyl sulfoxide (DMSO), were added to the cultures using a Biomek FXP liquid handler (Beckman Coulter, Indianapolis, Indiana, United States) to achieve a final concentration of 10 μM. Plates were incubated statically at 37°C for 24 h. Positive and negative controls were included in each 384-well plate. Streptonigrin (final concentration of 2 μg/mL) was added to appropriate wells as a positive control and DMSO was used as the negative control.
The OD600 was measured following the 24-h incubation period, followed by measurement of luminescence. For this, 50 μL of the Gal-Screen β-Galactosidase Reporter Gene Assay System (Thermo Fisher Scientific) were added to each well. This assay system includes a lysis buffer and the β-galactosidase substrate, Galacton-Star®, which are used in a single-step reaction that emits light proportional to the amount of the lactose-degrading enzyme. Luminescence readings were measured 5 min after the addition of the Gal-Screen β-Galactosidase Reporter Gene Assay System using an Envision plate reader (Perkin Elmer, Waltham, Massachusetts, United States).
Relative light units (RLUs) generated by the E. coli BR513 cultures incubated with the respective compounds were normalized to the OD600 to account for any effects of cell concentration on β-galactosidase production. Additionally, the OD 600-normalized RLU results were further normalized to the interquartile mean to account for variation between plates. For this, the OD600-normalized RLU from each well were divided by the interquartile mean of the corresponding 384-well plate (excluding controls). The interquartile mean is defined as the mean of the middle 50% of the rank-ordered data (Mangat et al., 2014). All bioactive compounds for which both replicates had a value greater than 2.25 standard deviations from the mean of treated E. coli BR513 samples were considered positive for prophage induction, as described by the following equation:
Where; RLU/OD is the OD600-normalized RLU, μiq is the interquartile mean.
Dose response experiments were performed to investigate the effect of compound concentration on prophage induction capacity and to verify whether naturally-derived compounds with inconclusive results (i.e., compounds for which only one replicate generated a normalized RLU value above the cut-off) were prophage-inducing agents. These compounds included osthol, sclareol, galangine, and roccellic acid. In addition to generating RLU vales above the cutoff in one of the two replicates, these compounds were also included in the dose response experiments because they all have antimicrobial activity (Choudhary et al., 2006; Cushnie and Lam, 2006; Sweidan et al., 2017; Tan et al., 2017), and we hypothesized that the mode of action of these compounds at the appropriate dose could be explained by prophage induction and subsequent cell death. Rosemary and gallic acid were also included in the dose response experiments because they were previously identified as being prophage inducers. Since they are also natural food-grade compounds, they were included in the dose response screen so they could be directly compared to the compounds selected from the high-throughput luminescent prophage induction assay.
The response to varying doses of selected compounds (Table 2) was evaluated by incubating duplicate cultures of E. coli BR513 with 11 concentrations (half-log dilutions of the preceding concentration) of each compound following the methods for the high-throughput luminescent prophage induction as described in Section 2.3. The highest tested concentration of each compound varied between 2 and 100 μg/mL. Streptonigrin, ciprofloxacin, chloramphenicol, and ampicillin (dissolved in DMSO) were also included in the dose response experiments as positive controls since they have been described in the literature as being prophage-inducing antibiotics (Levine and Borthwick, 1963; Goerke et al., 2006; Maiques et al., 2006).
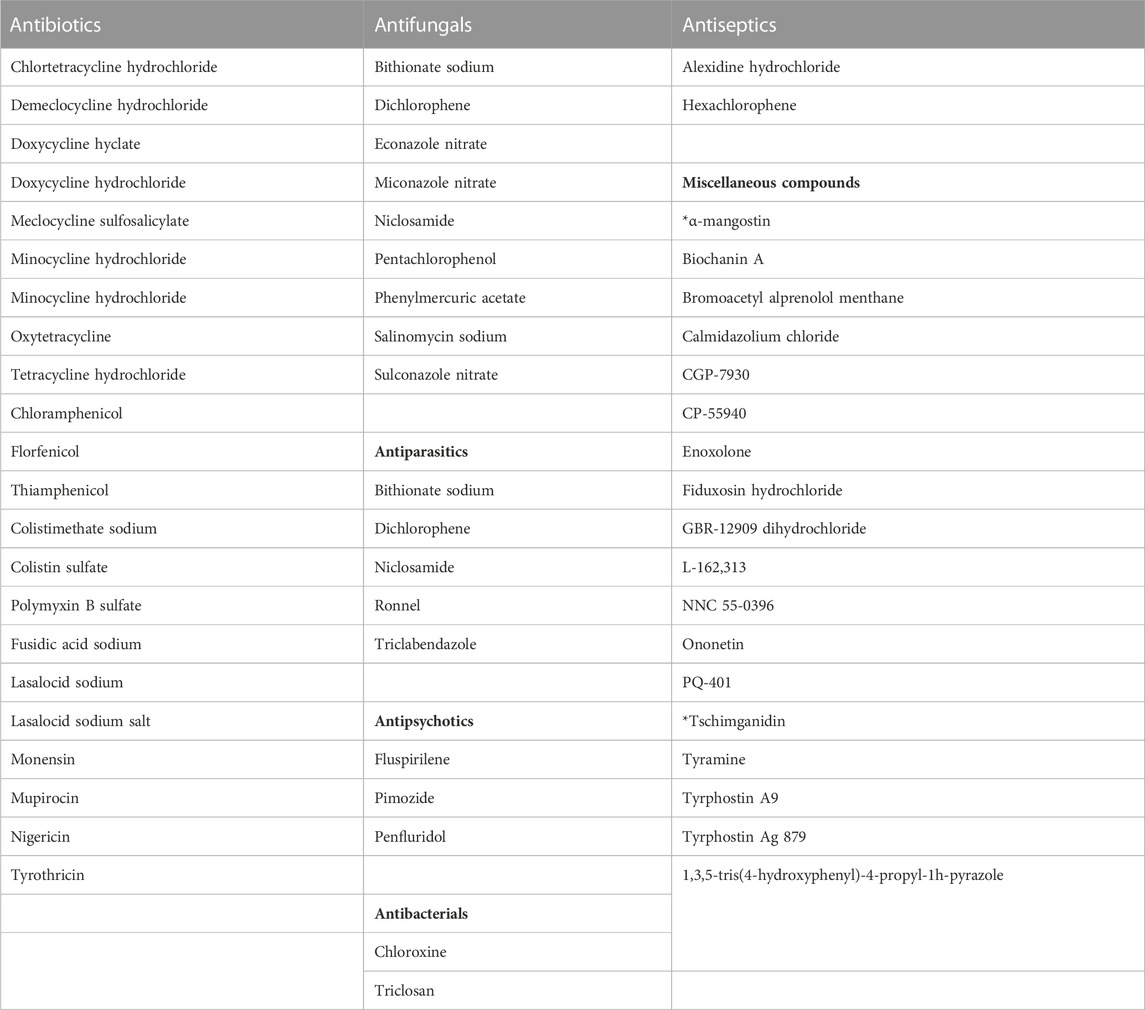
TABLE 2. Prophage λ inducers, based on results from the high-throughput luminescent prophage induction assay. *Naturally-derived compounds.
The entire CMCB bioactives collection, consisting of 3,747 compounds was screened using the high-throughput luminescent prophage induction assay to identify natural compounds that could be used in the induction of prophages from bacterial pathogens and spoilage bacteria that contaminate foods. Compounds were considered positive hits for prophage induction when both replicate signals were greater than the respective cut-offs. The hit cut-offs, calculated as 2.25 standard deviations from the mean, were 2.7 and 2.6 for replicates 1 and 2, respectively. Several structurally-related compounds were identified (Table 2; Figure 1), indicating that certain chemical classes contribute to prophage induction activity. Sixty-one bioactive compounds were identified as potential prophage-inducing agents, including 22 antibiotics, 9 antifungals, 5 antiparasitics, 3 antipsychotics, 2 antibacterials, 2 antiseptics, and 18 miscellaneous compounds, two of which were of natural origin (α-mangostin and tschimganidin). In addition to the positive hits, there were 22 compounds that had inconclusive results (Table 3), which included 5 antibiotics, 2 antifungals, 1 antiparasitic, 3 antibacterials, 1 antiseptic, and 10 miscellaneous compounds. Inconclusive, or non-replicating hits, were those for which only one replicate signal surpassed its cut-off. Six of the non-replicating hits were natural compounds that are derived from plants and lichens, including osthol, roccellic acid, galanginee, sclareol, leoidin, and xanthone (data not shown). The results suggest that similarities in chemical structure amongst the identified compounds may nevertheless indicate which chemical moieties may be responsible for prophage λ induction.
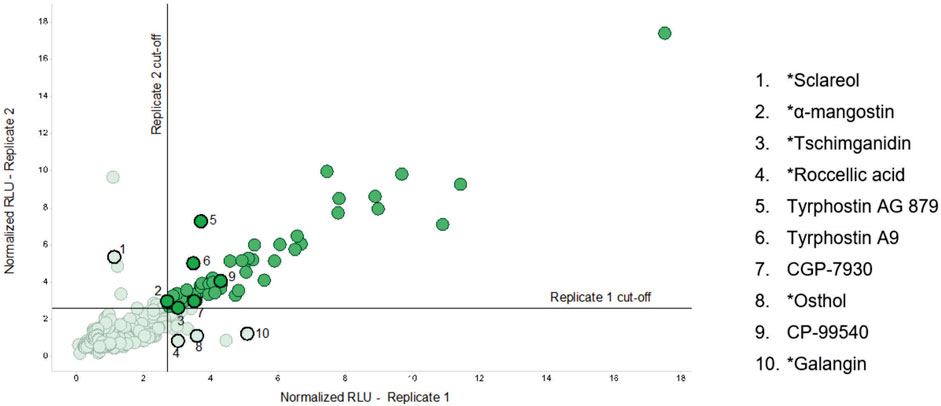
FIGURE 1. Replicate plot showing the normalized RLU signal in the prophage indicator strain Escherichia coli BR513 resulting from the addition of each compound tested in the high-throughput luminescent prophage induction assay. For each compound, two replicates were conducted. The x-axis represents the signal from replicate 1 and the y-axis represents that of replicate 2. RLU values were normalized to OD600 and the interquartile mean, and those in solid green represent positive hits, or compounds that had normalized RLU values greater than 2.25 standard deviations of the mean of each replicate. The numbered points outlined in solid black highlight positive and inconclusive hits that were further tested in the dose response experiments. *Naturally-derived compounds.
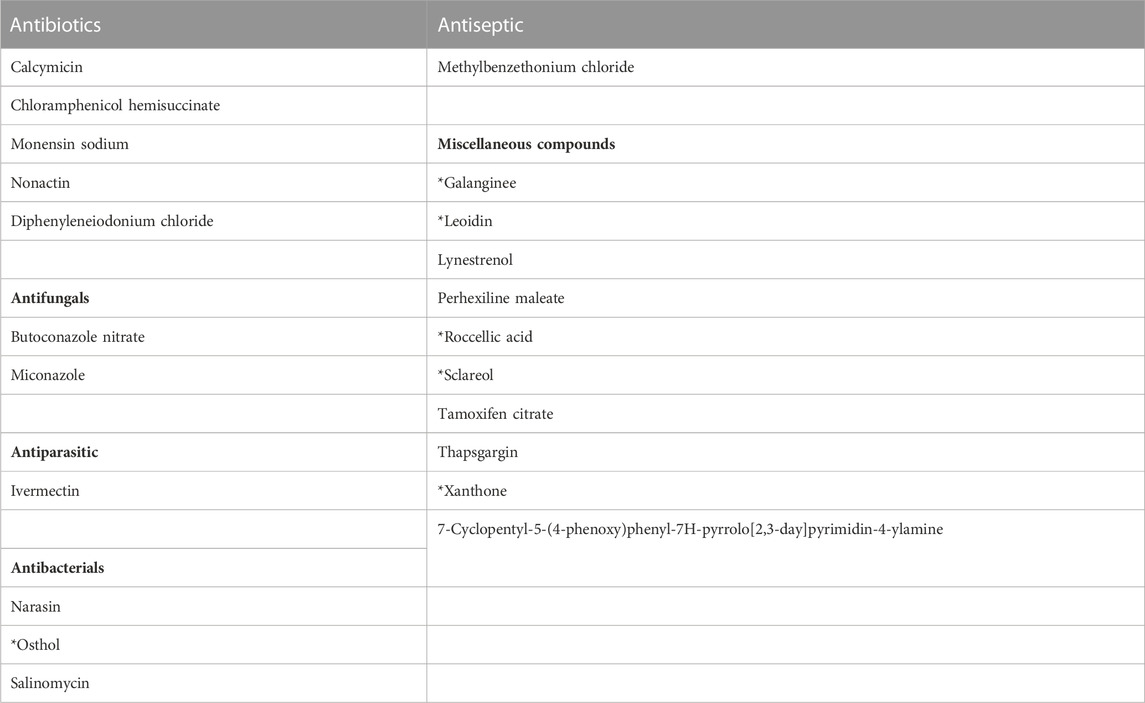
TABLE 3. Compounds identified as inconclusive hits in the high-throughput luminescent prophage induction assay. *Naturally-derived compounds.
A subset of positive hits from the high-throughput assay was selected for evaluation in dose response experiments. The selection included compounds with structural similarities, as well as compounds that were natural in origin, since they could have potential for use in clean label food applications. For example, two tyrphostins and CGP-7930 all contain a di-tert-butyl phenol group and were thus evaluated and compared. Four of the inconclusive hits that were compounds of natural origin (osthol, roccellic acid, galanginee, and sclareol) were also included to confirm whether they were indeed capable of inducing prophage λ. Tschimganidin and α-mangostin were selected from the positive hits since they are naturally-derived compounds, whereas streptonigrin, ciprofloxacin, chloramphenicol and ampicillin were used as positive controls since they have all been described as agents that can induce prophages (Levine and Borthwick, 1963; Goerke et al., 2006; Maiques et al., 2006).
The dose response curves (Figure 2) confirmed the four natural compound inconclusive hits as being prophage λ-inducing agents, since the 10 μM concentration, as used in the initial screen, generated a normalized RLU response greater than the 2.25 standard deviation cut-off for both replicates. For most of the compounds included in the dose response experiments, the OD600-normalized RLU increased and the OD600 decreased as the concentration of bioactive compounds increased. This was the case for streptonigrin, indicating that a higher concentration resulted in higher production of β-Galactosidase due to the induction of prophage λ. The only compound that did not generate a dose response in RLU/OD600 was rosemary, which was included in the dose response experiments because it is a known antimicrobial compound and was previously shown to induce prophage λ. Gallic acid, an organic acid naturally found in teas, was the only other compound that did not cause a significant response in OD600, suggesting that a higher concentration may be required to cause cell death.
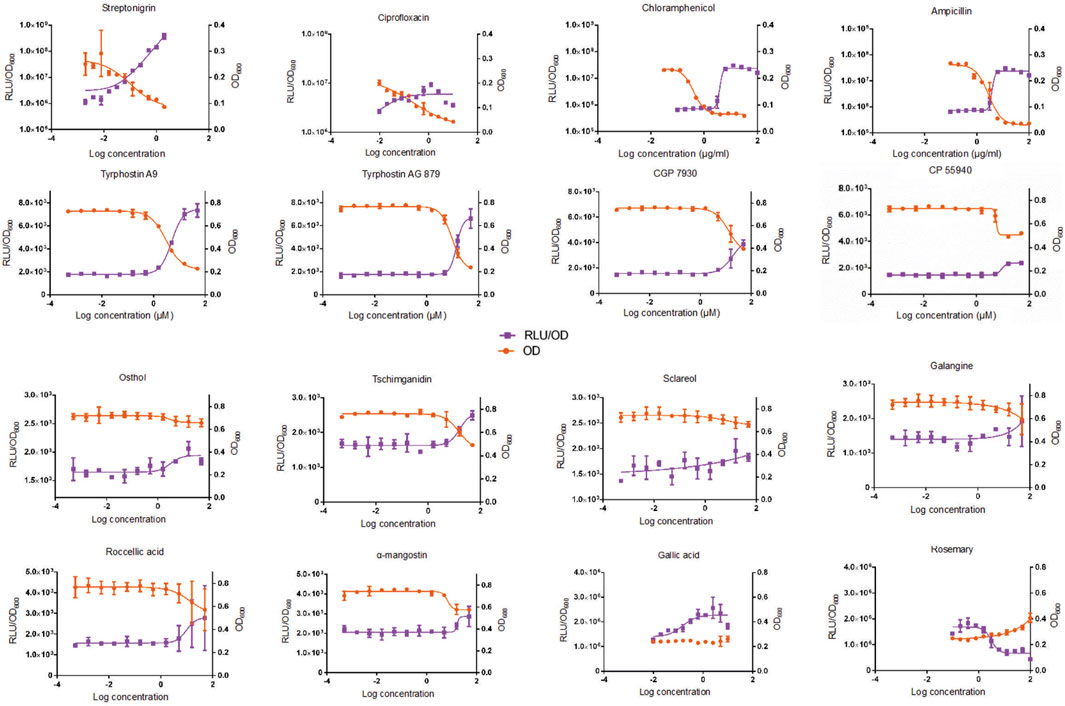
FIGURE 2. The average response in RLU/OD600 and in OD600 in Escherichia coli BR513 after 24-h exposure to varying concentrations of bioactive compounds from the Centre for Microbial Chemical Biology (CMCB, McMaster University, Hamilton, Ontario, Canada) bioactives collection, four antibiotics that were used as positive controls (top row) and two compounds (gallic acid and rosemary) that were previously shown to be prophage inducers. The purple squares represent values that correspond to RLU/OD600 values, while the orange squares represent values that correspond to OD600. Error bars represent standard deviations of the duplicate cultures that were incubated in the dose response experiments. Compounds in the bottom two rows are naturally-derived.
In this study, a high-throughput luminescent prophage induction assay was developed to identify compounds capable of inducing prophages as a way to inhibit the growth and survival of pathogenic and spoilage bacteria that contaminate foods. The use of automated equipment improved the efficiency of the assay, and allowed for rapid screening of thousands of bioactive compounds. The compounds that resulted in the highest normalized luminescent signals were considered positive hits for prophage induction. Data analysis involved normalization to the interquartile mean to reduce variation between plates, as systematic error is inherent in high-throughput screening (HTS) data (Mangat et al., 2014). Luminescence was measured using the Gal-Screen β-Galactosidase Reporter Gene Assay System (Thermo Fisher Scientific) because this system was specifically designed for HTS applications. The dose response experiments were conducted to confirm the prophage inducing capacity of inconclusive hits, as well as two natural compounds (gallic acid and rosemary) that were selected based on previous results.
Most of the compounds identified by the assay are not suitable for use in foods, however some natural compounds were positive hits. The most abundant class of compounds identified by the assay were antibiotics, but antiseptics, antifungals, antiparasitics, antipsychotics, antibacterials, antiseptics and miscellaneous drugs were also identified as prophage inducers. Although not all the bioactive compounds identified in the screen may be used in foods, the results are still useful because they identify which types of compounds and/or which chemical moieties may possess prophage inducing characteristics. The notion that chemical structure may be correlated with the ability to induce prophages is supported by the fact that three compounds identified as positive hits in the current screen, tyrphostin A9, tyrphostin AG 879, and CGP-7930, contain a common chemical moiety: a di-tert-butyl phenol group. In total, twenty tyrphostin analogues were screened in the assay, but the only two tyrphostins (A9 and AG879) identified as positive hits contain di-tert-butyl phenol groups. Although these positive hits are not natural compounds, the fact that they were positive hits suggest that naturally-derived compounds with similar chemical structure may be useful as suitable food additives for the control of the growth of bacterial pathogens by means of prophage induction. For example, 2,4-di-tert-butylphenol (DTBP) is an antioxidant that may be extracted from seeds, fruits and sweet potato and can be produced as a fermentation product of certain lactic acid bacteria (Choi, et al., 2013; Varsha et al., 2015). It has been shown to exhibit antioxidant and antifungal activity, thus demonstrating its potential as a natural food additive. (Varsha et al., 2015). Contrasting these results, other compounds that contain a di-tert-butyl phenol group were not observed to induce prophages. The widely-used synthetic phenolic antioxidant, butylated hydroxytoluene (BHT), also contains a di-tert-butyl phenol group. BHT is used as a preservative in foods and in food packaging and acts as an antioxidant as well as an antimicrobial (Ayaz et al., 1980), but its antimicrobial activity has not yet been linked to prophage induction. In this study, we did not observe prophage induction during incubation with BHT. As an antioxidant BHT likely binds free radicals, thereby reducing DNA damage, which is a major cause of prophage induction (Nanda et al., 2014).
The synthetic cannabinoid, CP-55940, was also identified as a positive hit in the high-throughput luminescent prophage induction assay. CP-55940 was developed by Pfizer, Inc. in the 1970s but was never marketed. CP-55940 shares similar structural features and mimics the euphoric effects of the naturally occurring cannabinoid, tetrahydrocannabinol (THC) (Debruyne and Le Boisselier, 2015). Since the two compounds exhibit similar toxicological effects and are structurally similar, the naturally-occurring cannabinoids, THC and cannabidiol (CBD), could be screened for their ability to induce prophages. Cannabis became legal in Canada with the assent of the Cannabis Act in June 2018 (Cannabis Act, 2018). Cannabis edibles became legal in 2019, and a number of foods and drinks that have been infused with cannabis may be legally sold. Therefore, naturally-occurring cannabinoids that are structurally similar to CP-55940 may serve as antimicrobials to reduce the presence of bacterial foodborne pathogens in cannabis food products.
Two natural compounds that originate from plants within the Umbelliferae or Apiaceae family were identified as positive hits. A terpenoid, tschimganidin was identified in the assay, and a coumarin derivative, osthol, was an inconclusive hit that was subsequently confirmed as a prophage-inducing agent in the dose response experiment. Tschimganidin and osthol may be extracted from the roots of Ferula tschimganica and Ferula campestris, respectively (Kadyrov et al., 1972; Basile et al., 2009). Both compounds are also found in other umbellifers, but have a common source: root resin of Ferula persica (Kerminov, 1992). The plant may therefore represent an important source for the extraction of effective natural prophage-inducing compounds.
Tan and others (2017) elucidated the structures of osthol and nine other prenylated coumarins by spectroscopic methods and direct comparison with reference compounds, and showed that the root extract and its prenylated coumarins exhibited antimicrobial activity against clinically relevant bacteria including the foodborne pathogens Bacillus subtilis and Klebsiella pneumoniae at concentrations between 5 and 125 μg/mL. Interestingly, green teas contain coumarins (Yang et al., 2009), and our group identified various green tea as potent inducers of prophage λ (Tompkins et al., 2018). We previously identified gallic acid as a strong natural prophage inducer. However, in this study, gallic acid was not identified as a positive hit. The reason for the discrepancy is likely due to the fact that different concentrations of gallic acid were used in the different studies. For example, the 10 μM concentration used in the current study is equivalent to 1.7 μg/mL, while the concentration used in our earlier work was 10 μg/mL. These results suggest that the prophage induction activity of gallic acid may be dose-dependent, and gallic acid was thus included in the dose response experiment, in which an increase in RLU/OD was observed with increasing concentration. Increased concentration of gallic acid did not however, significantly affect the OD600. Higher concentrations should be tested to confirm whether gallic acid is able to cause cell death. Gallic acid is a component of green tea, meaning that green teas may represent an important group of food-grade ingredients that should be the focus of future prophage induction studies.
The fact that gallic acid is a known antimicrobial (Borges et al., 2013) with DNA-damaging properties (Hossain et al., 2013) elucidates, at least partially, its effectiveness as a prophage inducer, as DNA damage is the first step in the predominant mechanism of prophage induction. DNA damage, whether caused by exposure to UV irradiation or treatment with a specific compound, triggers the SOS response in bacterial cells, thus initiating the production of the RecA protein, in turn causing autoproteolytic cleavage of prophage repressors that are responsible for the lysogenic state (Nanda et al., 2014). The cleavage of phage repressors thus allows the prophage to excise from the bacterial host chromosome and enter the lytic cycle (Sassanfar and Roberts, 1990; Campoy et al., 2006).
Mitomycin C, streptonigrin and norfloxacin are antibiotics that are known to activate the recA-dependent pathway and prophage induction (Levine and Borthwick, 1963; Raya and H'Bert E, 2009; McDonald et al., 2010). These compounds were also included in the current screen as part of the BIOMOL 2865 Natural Products Library and the Prestwick Chemical Library. However, in this work they were not identified as positive hits as the normalized RLUs for streptonigrin, mitomycin C and norfloxacin did not surpass the required cut-offs to be considered a positive result. Earlier, we confirmed each antibiotic as being a prophage-inducing compound (Tompkins et al., 2018). The concentration used in the earlier work was 2 μg/mL for each antibiotic, while in the current screen, the concentrations used were 5.1 μg/mL, 3.6 μg/mL, and 4.4 μg/mL for streptonigrin, mitomycin C and norfloxacin, respectively. Since the concentrations of the three antibiotics used in the current screen were higher than the previous work, it was expected that all three compounds would be identified as prophage inducing compounds. False negative results may be attributed to the age of the chemical stocks used in the bioactives library, as freezing and thawing, as well as hydration of compounds stored in DMSO over time may diminish compound concentration, solubility, and potency (Kozikowski et al., 2003). This is supported by the fact that the streptonigrin used as a positive control and in the dose response experiment was prepared as a fresh stock and resulted in a positive hit for prophage induction.
Dose-response experiments are used to determine the response of an organism, to a stimulus or stressor, following exposure for a set exposure time (Peleg, 2021). In this study, “dose” is defined as the quantity of a compound that results in prophage induction. Determining the dose response, and developing dose–response models, is typically used to assess the concentrations of drugs required to inhibit or kill microorganisms. Dose–response data can be visualized by dose–response curves, which relate the magnitude of a dose (stimulus or stressor) to the response of a biological system. The applied dose is usually plotted on the X-axis and the biological response is plotted on the Y-axis (Hamilton et al., 1977; Altshuler, 1981).
In this study, the dose response experiment demonstrated that prophage inducing compounds inhibited growth of the indicator strain, E. coli BR513, as previously reported (Elespuru and Yarmolinsky 1979), but could not distinguish between the causes of cell death. Since RLU values were normalized to OD600, the responses in normalized RLU could have been caused by cell death as a result of other direct mechanisms. Although certain compounds at sublethal concentrations induce prophages and lyse the host cell from the inside, additional mechanisms could also cause cell death when a given compound is used at higher concentrations. For example, chloramphenicol, which was identified as a positive hit, is known to generally inhibit protein synthesis (Shinagawa et al., 1977), suggesting it could inhibit the production of repressor proteins that keep prophages in their lysogenic state, thus causing induction of the lytic cycle. This may be the case when used at sublethal levels, but at higher concentrations, chloramphenicol kills bacterial cells by inhibition of protein synthesis required for cell wall formation, which is an additional pathway to cell death that does not involve phages (Schwarz et al., 2016.
In order to determine whether differing concentrations of a respective compound kill bacterial cells through prophage induction or another mechanism, the dose response experiment will need to be modified by including a negative control strain that does not carry prophages. In this scenario, if the compound kills bacterial cells through prophage induction, we would expect that the OD of the prophage negative control strain would not decrease, as there would be no prophages to induce and subsequently lyse the cell. Future experiments aimed at further studying the prophage inducing activities of the natural compounds we identified will include a prophage negative E. coli strain.
Overall, the naturally-derived compounds that were included in the dose response experiments did not exhibit as marked of a response as the synthetic compounds, which parallels the results from the initial screen. Sclareol, roccellic acid, osthol, and galangine were all inconclusive hits in the high-throughput luminescent prophage induction assay, thus their bacterial killing capacity was expected to be less potent than the synthetically-derived compounds included in the screen. While these compounds were confirmed to be positive for bacterial inhibition in the dose response experiments, they should be considered as hits of lower intensity, since their normalized RLU values were closer to the positive cut-off values (data not shown). Since these plant-derived and lichen-derived (roccellic acid) bioactive compounds generally demonstrated the expected profile and are known to exhibit antimicrobial properties (Cushnie et al., 2003; Hayet et al., 2007; Sweidan et al., 2017; Tan et al., 2017), they should be considered for use in future prophage induction screens.
The dose response curves for osthol, α-mangostin, and CP-55940 reached a plateau in both RLU/OD600 and OD600 signals, suggesting that these compounds elicited a maximum response, or that higher concentrations would have no significant effect on inhibition of cell growth or ability to induce prophages. In contrast, the dose response for rosemary demonstrated the opposite effect: as the normalized RLU/OD600 values decreased the OD600 values increased. This is due to the fact that the signals were likely skewed due to the colour of the rosemary extract powder used for its preparation. The OD600 and the RLU/OD600 results in the dose response experiments for rosemary should thus not be considered accurate, as an increased concentration of the pigmented solution could have caused an increase in OD600 as well as a decrease in the luminescent signal.
In conclusion, several libraries of bioactive compounds were screened for capacity to induce prophage λ, as indicated by the production of β-galactosidase in the indicator organism E. coli BR513. This work identified several compounds that could be used to induce prophages in foodborne bacterial pathogens and spoilage bacteria. The naturally-derived prophage-inducing compounds identified in this work, as well as other naturally-derived compounds with chemical structures similar to that of the synthetically-derived positive hits should be further tested for their ability to induce prophages, as a sustainable approach to improvement of food safety and quality.
The raw data supporting the conclusion of this article will be made available by the authors, without undue reservation.
ET conducted experiments, analyzed data, and developed the initial manuscript draft. BC provided advice on experimental design, and analyzed data. LG provided conceptualization of the study, experimental resources, funding acquisition, project administration, parts of initial manuscript draft, significant review and editing. All authors contributed to the article and approved the submitted version.
These experiments were funded by grants provided to LG by the Genome Canada, by the Génome Québec provincial genome center, and by the National Sciences and Engineering Council of Canada Discovery Grants Program (grant number RGPIN-2014-0574).
We express our gratitude to Tracey Campbell at the Centre for Microbial Chemical Biology (CMCB, McMaster University, Hamilton, Ontario, Canada) for conducting the high-throughput screen and for helpful comments regarding experimental design and data analysis.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Altshuler, B. (1981). Modeling of dose-response relationships. Environ. Health Perspect. 42, 23–27. doi:10.1289/ehp.814223
Asioli, D., Aschemann-Witzel, J., Caputo, V., Vecchio, R., Annunziata, A., Næs, T., et al. (2017). Making sense of the “clean label” trends: A review of consumer food choice behavior and discussion of industry implications. Food Res. Int. 99, 58–71. doi:10.1016/j.foodres.2017.07.022
Ayaz, M., Luedecke, L. O., and Branen, A. L. (1980). Antimicrobial effect of butylated hydroxyanisole and butylated hydroxytoluene on Staphylococcus aureus. J. Food Prot. 43 (1), 4–6. doi:10.4315/0362-028x-43.1.4
Basile, A., Sorbo, S., Spadaro, V., Bruno, M., Maggio, A., Faraone, N., et al. (2009). Antimicrobial and antioxidant activities of coumarins from the roots of Ferulago campestris (Apiaceae). Molecules 14 (3), 939–952. doi:10.3390/molecules14030939
Borges, A., Ferreira, C., Saavedra, M. J., and Simões, M. (2013). Antibacterial activity and mode of action of ferulic and gallic acids against pathogenic bacteria. Microb. Drug Resist. 19 (4), 256–265. doi:10.1089/mdr.2012.0244
Cadieux, B., Colavecchio, A., Jeukens, J., Freschi, L., Emond-Rheault, J.-G., Kukavica-Ibrulj, I., et al. (2018). Prophage induction reduces shiga toxin producing Escherichia coli (stec) and Salmonella enterica on tomatoes and spinach: A model study. Food control. 89, 250–259. doi:10.1016/j.foodcont.2018.02.001
Campoy, S., Hervàs, A., Busquets, N., Erill, I., Teixidó, L., and Barbé, J. (2006). Induction of the SOS response by bacteriophage lytic development in Salmonella enterica. Virology 351 (2), 360–367. doi:10.1016/j.virol.2006.04.001
Cannabis Act, (2018). Cannabis act (S.C. 2018, c. 16). Available from: https://laws-lois.justice.gc.ca/eng/annualstatutes/.
Carocho, M., Barreiro, M. F., Morales, P., and Ferreira, I. C. F. R. (2014). Adding molecules to food, pros and cons: A review on synthetic and natural food additives. Compr. Rev. Food Sci. Food Saf. 13 (4), 377–399. doi:10.1111/1541-4337.12065
Carocho, M., Morales, P., and Ferreira, I. C. F. R. (2015). Natural food additives: quo vadis? Trends Food Sci. Technol. 45 (2), 284–295. doi:10.1016/j.tifs.2015.06.007
Choi, S. J., Kim, J. K., Kim, H. K., Harris, K., Kim, C. J., Park, G. G., et al. (2013). 2,4-Di- Tert-butylphenol from sweet potato protects against oxidative stress in PC12 cells and in mice. J. Med. food 16 (11), 977–983. doi:10.1089/jmf.2012.2739
Choudhary, M. I., Siddiqui, Z. A., Hussain, S., and Atta-ur-Rahman, (2006). Structure elucidation and antibacterial activity of new fungal metabolites of sclareol. Chem. Biodivers. 3 (1), 54–61. doi:10.1002/cbdv.200690007
Cone, R., Hasan, S. K., Lown, J. W., and Morgan, A. R. (1976). The mechanism of the degradation of DNA by streptonigrin. Can. J. Biochem. 54 (3), 219–223. doi:10.1139/o76-034
Côté, J., Caillet, S., Doyon, G., Dussault, D., Sylvain, J. F., and Lacroix, M. (2011). Antimicrobial effect of cranberry juice and extracts. Food control. 22 (8), 1413–1418. doi:10.1016/j.foodcont.2011.02.024
Cushnie, T. P., Hamilton, V. E., and Lamb, A. J. (2003). Assessment of the antibacterial activity of selected flavonoids and consideration of discrepancies between previous reports. Microbiol. Res. 158 (4), 281–289. doi:10.1078/0944-5013-00206
Cushnie, T. P., and Lamb, A. J. (2006). Assessment of the antibacterial activity of galangin against 4-quinolone resistant strains of Staphylococcus aureus. Phytomedicine 13 (3), 187–191. doi:10.1016/j.phymed.2004.07.003
Davenport, J., Balch, M., Galam, L., Girgis, A., Hall, J., Blagg, B. S., et al. (2014). High-throughput screen of natural product libraries for hsp90 inhibitors. Biol. (Basel) 3 (1), 101–138. doi:10.3390/biology3010101
Debruyne, D., and Le Boisselier, R. (2015). Emerging drugs of abuse: current perspectives on synthetic cannabinoids. Subst. abuse rehabilitation 6, 113–129. doi:10.2147/SAR.S73586
Elespuru, R. K., and Yarmolinsky, M. B. (1979). A colorimetric assay of lysogenic induction designed for screening potential carcinogenic and carcinostatic agents. Environ. Mutagen 1 (1), 65–78. doi:10.1002/em.2860010113
Fan, X., Ngo, H., and Wu, C. (2018). “Natural and bio-based antimicrobials: A review,” in Natural and bio-based antimicrobials for food applications (Washington, United States: American Chemical Society), 1–24.
Feiner, R., Argov, T., Rabinovich, L., Sigal, N., Borovok, I., and Herskovits, A. A. (2015). A new perspective on lysogeny: prophages as active regulatory switches of bacteria. Nat. Rev. Microbiol. 13 (10), 641–650. doi:10.1038/nrmicro3527
Garcia, P., Martinez, B., Obeso, J. M., and Rodriguez, A. (2008). Bacteriophages and their application in food safety. Lett. Appl. Microbiol. 47 (6), 479–485. doi:10.1111/j.1472-765X.2008.02458.x
Goerke, C., Köller, J., and Wolz, C. (2006). Ciprofloxacin and trimethoprim cause phage induction and virulence modulation in Staphylococcus aureus. Antimicrob. agents Chemother. 50 (1), 171–177. doi:10.1128/AAC.50.1.171-177.2006
Hamilton, M. A., Russo, R. C., and Thurston, R. V. (1977). Trimmed Spearman–Karber method for estimating median lethal concentrations in toxicity bioassays. Environ. Sci. Technol. 11 (7), 714–719. doi:10.1021/es60130a004
Hayet, E., Fatma, B., Souhir, I., Waheb, F. A., Abderaouf, K., Mahjoub, A., et al. (2007). Antibacterial and cytotoxic activity of the acetone extract of the flowers of Salvia sclarea and some natural products. Pak J. Pharm. Sci. 20 (2), 146–148.
Hossain, M. Z., Gilbert, S. F., Patel, K., Ghosh, S., Bhunia, A. K., and Kern, S. E. (2013). Biological clues to potent DNA-damaging activities in food and flavoring. Food Chem. Toxicol. 55, 557–567. doi:10.1016/j.fct.2013.01.058
Kadyrov, A. S., Khasanov, T. K., Saidkhodzhaev, A. I., and Nikonov, G. K. (1972). New phenolic compounds of the roots of Ferula tschimganica. Chem. Nat. Compd. 8 (6), 796–797. doi:10.1007/BF00564621
Kang, H. S., McNair, K., Cuevas, D., Bailey, B., Segall, A., and Edwards, R. A. (2017). Prophage genomics reveals patterns in phage genome organization and replication. bioRxiv. doi:10.1101/114819
Kerminov, Y. B., Abyshev, A. Z., Serkervov, S. V., Isaev, D. I., and Bairamov, P. B. (1992). Phenol derivatives from the roots of Ferula persica. Chem. Nat. Compd. 28 (5), 506.
Kozikowski, B. A., Burt, T. M., Tirey, D. A., Williams, L. E., Kuzmak, B. R., Stanton, D. T., et al. (2003). The effect of freeze/thaw cycles on the stability of compounds in DMSO. J. Biomol. Screen. 8 (2), 210–215. doi:10.1177/1087057103252618
Levine, M., and Borthwick, M. (1963). The action of streptonigrin on bacterial DNA metabolism and on induction of phage production in lysogenic bacteria. Virology 21 (4), 568–574. doi:10.1016/0042-6822(63)90228-9
Maiques, E., Úbeda, C., Campoy, S., Salvador, N., Lasa, Í., Novick, R. P., et al. (2006). β-lactam antibiotics induce the SOS response and horizontal transfer of virulence factors in Staphylococcus aureus. J. Bacteriol. 188 (7), 2726–2729. doi:10.1128/jb.188.7.2726-2729.2006
Mangat, C. S., Bharat, A., Gehrke, S. S., and Brown, E. D. (2014). Rank ordering plate data facilitates data visualization and normalization in high-throughput screening. J. Biomol. Screen 19 (9), 1314–1320. doi:10.1177/1087057114534298
McDonald, J. E., Smith, D. L., Fogg, P. C. M., McCarthy, A. J., and Allison, H. E. (2010). High-throughput method for rapid induction of prophages from lysogens and its application in the study of Shiga toxin-encoding Escherichia coli strains. Appl. Environ. Microbiol. 76 (7), 2360–2365. doi:10.1128/aem.02923-09
Miller, C. H., Nisa, S., Dempsey, S., Jack, C., and O'Toole, R. (2009). Modifying culture conditions in chemical library screening identifies alternative inhibitors of mycobacteria. Antimicrob. Agents Chemother. 53 (12), 5279–5283. doi:10.1128/aac.00803-09
Moye, Z. D., Woolston, J., and Sulakvelidze, A. (2018). Bacteriophage applications for food production and processing. Viruses 10 (4), 205. doi:10.3390/v10040205
Nanda, A. M., Heyer, A., Krämer, C., Grünberger, A., Kohlheyer, D., and Frunzke, J. (2014). Analysis of SOS-induced spontaneous prophage induction in Corynebacterium glutamicum at the single-cell level. J. Bacteriol. 196 (1), 180–188. doi:10.1128/JB.01018-13
Nieto, G., Ros, G., and Castillo, J. (2018). Antioxidant and antimicrobial properties of rosemary (Rosmarinus officinalis, L.): A review. Med. (Basel, Switz. 5 (3), 98. doi:10.3390/medicines5030098
Odeyemi, O. A., Alegbeleye, O. O., Strateva, M., and Stratev, D. (2020). Understanding spoilage microbial community and spoilage mechanisms in foods of animal origin. Compr. Rev. Food Sci. Food Saf. 19 (2), 311–331. doi:10.1111/1541-4337.12526
Peleg, M. (2021). Microbial dose-response curves and disinfection efficacy models revisited. Food Eng. Rev. 13 (2), 305–321. doi:10.1007/s12393-020-09249-6
Quinto, E. J., Caro, I., Villalobos-Delgado, L. H., Mateo, J., De-Mateo-Silleras, B., and Redondo-Del-Río, M. P. (2019). Food safety through natural antimicrobials. Antibiot. (Basel) 8 (4), 208. doi:10.3390/antibiotics8040208
Raya, R. R., and H'Bert E, M. (2009). Isolation of phage via induction of lysogens. Methods Mol. Biol. 501, 23–32. doi:10.1007/978-1-60327-164-6_3
Salmond, G. P., and Fineran, P. C. (2015). A century of the phage: past, present and future. Nat. Rev. Microbiol. 13 (12), 777–786. doi:10.1038/nrmicro3564
Sassanfar, M., and Roberts, J. W. (1990). Nature of the SOS-inducing signal in Escherichia coli: the involvement of dna replication. J. Mol. Biol. 212 (1), 79–96. doi:10.1016/0022-2836(90)90306-7
Schwarz, S., Shen, J., Kadlec, K., Wang, Y., Brenner Michael, G., Fessler, A. T., et al. (2016). Lincosamides, streptogramins, phenicols, and pleuromutilins: mode of action and mechanisms of resistance. Cold Spring Harb. Perspect. Med. 6 (11), a027037. doi:10.1101/cshperspect.a027037
Shinagawa, H., Mizuuchi, K., and Emmerson, P. T. (1977). Induction of prophage lambda by γ-rays, mitomycin C and tif; Repressor cleavage studied by immunoprecipitation. Mol. General Genet. MGG 155 (1), 87–91. doi:10.1007/BF00268564
Sillankorva, S. M., Oliveira, H., and Azeredo, J. (2012). Bacteriophages and their role in food safety. Int. J. Microbiol. 2012, 863945. PMID: 23316235; PMCID: PMC3536431. doi:10.1155/2012/863945
Sweidan, A., Chollet-Krugler, M., Sauvager, A., van de Weghe, P., Chokr, A., Bonnaure-Mallet, M., et al. (2017). Antibacterial activities of natural lichen compounds against Streptococcus gordonii and Porphyromonas gingivalis. Fitoterapia 121, 164–169. doi:10.1016/j.fitote.2017.07.011
Tan, N., Yazici-Tutunis, S., Bilgin, M., Tan, E., and Miski, M. (2017). Antibacterial activities of pyrenylated coumarins from the roots of Prangos hulusii. Molecules 22 (7), 1098. doi:10.3390/molecules22071098
Tompkins, E., Cadieux, B., and Goodridge, L. (2018). Development and evaluation of a novel assay to identify prophage inducers as a new class of antimicrobials in foods. International association for food protection (IAFP) annual meeting, Salt Lake City, abstract T10-09. J. Food Prot. 81, 71. Supplement A.
Torres, N. S., Abercrombie, J. J., Srinivasan, A., Lopez-Ribot, J. L., Ramasubramanian, A. K., and Leung, K. P. (2016). Screening a commercial library of pharmacologically active small molecules against Staphylococcus aureus biofilms. Antimicrob. Agents Chemother. 60 (10), 5663–5672. doi:10.1128/aac.00377-16
Varsha, K. K., Devendra, L., Shilpa, G., Priya, S., Pandey, A., and Nampoothiri, K. M. (2015). 2,4-Di-tert-butyl phenol as the antifungal, antioxidant bioactive purified from a newly isolated Lactococcus sp. Int. J. Food Microbiol. 211, 44–50. doi:10.1016/j.ijfoodmicro.2015.06.025
Keywords: clean label trend, sustainable approaches to food safety, natural compounds, prophage inducers, food safety
Citation: Tompkins E, Cadieux B, Amitrano M and Goodridge L (2023) High-throughput screening of natural compounds for prophage induction in controlling pathogenic bacteria in food. Front. Food. Sci. Technol. 3:1239884. doi: 10.3389/frfst.2023.1239884
Received: 14 June 2023; Accepted: 18 September 2023;
Published: 23 November 2023.
Edited by:
Noemi Elisabet Zaritzky, National University of La Plata, ArgentinaReviewed by:
Sri Charan Bindu Bavisetty, King Mongkut’s Institute of Technology Ladkrabang, ThailandCopyright © 2023 Tompkins, Cadieux, Amitrano and Goodridge. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lawrence Goodridge, Z29vZHJpZGxAdW9ndWVscGguY2E=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.