
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Food. Sci. Technol. , 05 May 2023
Sec. Food Safety and Quality Control
Volume 3 - 2023 | https://doi.org/10.3389/frfst.2023.1165871
Staphylococcus (S.) aureus can proliferate in a broad range of food and contact surfaces. The ability to grow as a biofilm enhances its resistance to cleaning agents and the chance to persist on food facility contact surfaces and enter the food chain. This presents a risk to the health of food workers and consumers, considering that this pathogen has been associated with a wide variety of local and systemic human infections, as well as with food poisoning caused by the production of enterotoxins. In particular, ready-to-eat (RTE) food, that does not undergo further processing capable of reducing bacterial contamination, may be of particular concern since its consumption poses a direct microbiological risk to consumers. To worsen this scenario, S. aureus harbors several biocide and antimicrobial resistance genes (BRGs and ARGs), which, respectively, reduce the efficacy of sanitizing agents during cleaning procedures and antimicrobial treatments when infections occur. Considering this, several novel methods have recently been investigated to control S. aureus contamination in food and contact surfaces in food facilities in order to overcome the limitations of traditional sanitizing protocols and improve the safety of the produced food products. In this review, we will provide an overview of S. aureus ARGs and BRGs and whole-genome sequence (WGS)-based methods recently implemented for their surveillance. Furthermore, we will describe the presence of antimicrobial-resistant S. aureus in RTE food and food-contact surfaces and present novel natural or chemical compounds, new food-contact materials, and innovative physical methods to control the contamination of this pathogen in the food sector. Finally, we will also discuss if S. aureus complex-related species are emerging as new antimicrobial-resistant pathogens of the food chain.
Staphylococcus aureus is an opportunistic pathogen that causes several diseases such as skin and soft tissue infections (Lacey et al., 2016), food poisoning, and life-threatening complications, such as pneumonia, endocarditis (Grapsa et al., 2021), osteomyelitis, and toxic shock syndrome, due to its large arsenal of exotoxins, including enterotoxins, as well as invasion, immune evasion, and antibiotic resistance mechanisms (Fusco et al., 2011; Chieffi et al., 2020).
Due to its ability to proliferate in a wide range of temperatures, pH levels, and salt concentrations, S. aureus can contaminate a wide variety of food and contact surfaces in the food facility environment. Indeed, it has been reported in meat products (Silva-de-Jesus et al., 2022), fruit juices, ready-to-eat (RTE) food, salad dressings, milk and dairy products (Chieffi et al., 2020; Mekhloufi et al., 2021), seafood, and freshwater fish (Kukułowicz et al., 2021; Rashid et al., 2021). In particular, RTE food that does not undergo further processing capable of reducing bacterial contamination is of particular concern, and their consumption may pose a direct risk to consumer health.
S. aureus can aggregate and form biofilm on food facility surfaces becoming less susceptible to biocides, sanitizers, and antimicrobials in general (Fux et al., 2004) than planktonic cells dispersed in the environment. This increases the persistence of the bacterial cells on such surfaces and the possibility of cross-contamination with other materials and food (Van-Houdt and Michiels, 2010; Vázquez-Sánchez et al., 2014; Bridier et al., 2015; Gutiérrez et al., 2021).
Pathogenic foodborne bacteria can acquire resistance to antimicrobial agents and biocides through horizontal gene transfer from antimicrobial-resistant bacteria, or via adaptive mutation. The growth of S. aureus as a biofilm enhances the possibility of transferring antimicrobial resistance genes (ARGs) and biocide resistance genes (BRGs) to both pathogenic and non-pathogenic bacteria in food products and contact surfaces in food facility environments and, being a reservoir of these genes, presents a serious health risk for consumers (Savage et al., 2013). In addition, the presence of ARGs and BRGs has been demonstrated to be correlated with an increase in pathogenicity (Beceiro et al., 2013; Alenizi, 2014; Thompson and Brown, 2017; Rasmi et al., 2022).
Herein, we provide a summary of ARGs and BRGs in S. aureus and on whole-genome sequence (WGS)-based methods for the surveillance of such resistances. An overview of novel strategies to control S. aureus biofilm and contamination in food and food-contact surfaces is also provided. Moreover, we discuss the possibility of the S. aureus complex-related species emerging as new antimicrobial-resistant pathogens of the food chain.
Glycopeptide antibiotics, such as vancomycin and teicoplanin, bind with high affinity to the dipeptide D-Ala4-D-Ala5 of lipid II that forms complexes with peptidoglycan precursors, inhibiting cell wall synthesis (Loll and Axelsen, 2000). There are two different types of glycopeptide resistance described in S. aureus. Genetic bases of the intermediate vancomycin (glycopeptide)-resistant (VISA/GISA) S. aureus strains, which result in an increased cell wall thickness, involve stepwise mutations in genes encoding molecules mainly implicated in cell wall biosynthesis and its regulation (McGuinness et al., 2017). The high vancomycin resistance mechanism is based, as in other microorganisms, on the presence of van genes, located on mobile elements that encode for enzymes that replace D-ala-D-ala with a low-affinity depsipeptide, D-alanyl-D-lactate (D-ala-D-lac) (Bugg et al., 1991). The first high-level vancomycin-resistant S. aureus (VRSA) strain carrying vanA operon was isolated in 2002 (Chang et al., 2003). This high-level vancomycin-resistant S. aureus isolate harbored a 57.9-kilobase multiresistance conjugative plasmid (pLW1043) from Enterococcus faecalis, within which it was integrated into the Tn1546, encoding the vancomycin resistance gene cluster (Weigel et al., 2003). A genetic study by Bakthavatchalam et al. (2018) in methicillin-resistant S. aureus (MRSA) with reduced teicoplanin susceptibility identified mutations in the tcaA and tcaB genes of the tcaRAB operon as determinants of teicoplanin resistance. The tcaRAB operon was identified by Brandenberger et al. (2000); insertional inactivation of tcaA or deletion of the entire operon increased teicoplanin resistance in S. aureus in a strain-dependent way, and, in the methicillin-resistant strain COL, it was coupled with a remarkable decrease in methicillin resistance.
Tetracycline resistance is determined by two different mechanisms: the action of efflux pumps, encoded by tetK and tetL genes that are plasmid-located, and ribosomal protection by elongation factor-like proteins, encoded by tet(O)/tet(M) genes, typically chromosomally located on conjugative transposons such as Tn916 and Tn1545 (Jensen and Lyon, 2009). TetK is located on the small pT181 plasmid, which has also been found integrated within the SCCmecIII cassette of MRSA strains (Jensen and Lyon, 2009). TetO/M binds to the EF-G binding site on the ribosome, thus dislodging tetracycline from the ribosome (Burdett, 1996; Trieber and Taylor, 2002). Resistance can also be determined by mutations causing the increased expression of chromosomally encoded efflux pumps, such as Tet38 (Truong-Bolduc et al., 2022).
Chloramphenicol acetyltransferase (CAT) is an inducible detoxifying enzyme that inactivates by acetylation of chloramphenicol, which inhibits protein biosynthesis by binding with the peptidyltransferase center at the 50S ribosomal subunit of 70S ribosomes (Schlünzen et al., 2001; Schwarz et al., 2004). cat genes, alone or in combination with streptomycin resistance (Gillespie and Skurray, 1988; Schwarz and Grölz-Krug, 1991), are carried by RC plasmids, of which pC221, pC223, pUB112, and pC194 have been characterized (Horinouchi and Weisblum, 1982; Brückner and Matzura, 1985; Projan et al., 1985; Schwarz et al., 2004; Smith and Thomas, 2004). In addition, the 23S rRNA methyltransferase gene cfr was shown to confer resistance to chloramphenicol, as well as efflux systems, inactivation by phosphotransferases, mutations of the target site, permeability barriers, and the presence of the non-enzymatic inducible chloramphenicol resistance determinant (clmA) carried on the transposon Tn1696 (Bissonnette et al., 1991; Stokes and Hall, 1991).
Aminoglycoside resistance in S. aureus is conferred by the action of aminoglycoside-modifying enzymes (AMEs) (aminoglycoside phosphotransferase, acetyltransferases, and nucleotidyl-transferase) (Ardic et al., 2006). Streptomycin is an aminoglycoside antibiotic that irreversibly binds to the 16S rRNA and S12 protein within the bacterial 30S ribosomal subunit (Demirci et al., 2013). While high-level streptomycin resistance in clinical S. aureus is associated with a chromosomal mutation affecting ribosome affinity (Lacey and Chopra, 1972), low-level resistance is associated with the streptomycin adenylyltransferase-encoding str gene, which is plasmid-located (Projan et al., 1988). The kanamycin resistance plasmid was first isolated and characterized in 1974 by Stiffler et al. (1974). This plasmid carries the Tn4001 transposon, which also mediates resistance to gentamicin and tobramycin (Lyon et al., 1984). Resistance is achieved by the action of a bifunctional protein with aminoglycoside acetyltransferase [AAC(6′)] and aminoglycoside phosphotransferase [APH(2″)] activities encoded by aacA-aphD (Rouch et al., 1987). Additionally, aadD (also referred to as ant(6) or ant(6)Ia) and aphA-3 code for an aminoglycoside adenyltransferase and a phosphotransferase, respectively, involved in neomycin and kanamycin resistance. These genes are, respectively, carried by the Tn5405 transposon and the plasmid pUB110, whose integration was mediated by IS257 (Byrne et al., 1991; Derbise et al., 1996). The transposon Tn5405 also carries aadE and sar4, which code for a streptomycin adenylyltransferase and the streptothricin acetyltransferase, respectively, involved in antibiotic resistance.
Fluoroquinolone resistance in S. aureus is due to mutations in the gyrA, gyrB, and parC genes, causing the synthesis of proteins with reduced susceptibility to this class of antibiotics (Lowry, 2003). Resistance can also be ascribable to the overexpression of fluoroquinolone efflux pumps encoded by norA, norB, norC, sdrM, and other major facilitator superfamilies (MFSs) (Tanaka et al., 2000; Ding et al., 2008).
The target of macrolides (such as erythromycin), common also to lincosamides and streptogramin B, is the V domain of the 23S rRNA of the subunit 50S. Resistance is thus achieved by four types of mechanisms: 1) the action of rRNA methyltransferases (coded by erm genes), which modify an adenine residue of the 23S rRNA, reducing the affinity with the antibiotic (Roberts et al., 1999); 2) antibiotic resistance ATP-binding cassette subfamily F proteins, encoded by msr genes that protect the ribosomal site, displacing the antibiotic from the target (Feßler et al., 2018); 3) MFS membrane transporters, encoded by mef genes, involved in macrolide efflux (Feßler et al., 2018); and 4) enzymatic inactivation by macrolide phosphotransferase or macrolide esterase, encoded by mph(C) or ere(A) and ere(B) genes, respectively (Feßler et al., 2018). Many of these genes can be transferred across strains, species, and even genera since they are located on mobile elements such as plasmids, transposons, or genomic islands (Schwendener et al., 2020).
Beta-lactam antibiotics, such as penicillin and methicillin, impair the synthesis of the bacterial cell wall by targeting enzymes involved in the synthesis of peptidoglycans (transpeptidases, carboxypeptidases, and transglycosylases) (Zeng and Lin, 2013). Several mechanisms are associated with beta-lactam resistance, including the synthesis of additional penicillin-binding proteins (PBP2a), the synthesis of beta-lactamases, and, rarely reported a mutation in the PBP coding genes, which reduce their affinity to antibiotics. S. aureus beta-lactamases are synthesized by the blaZ gene, which is usually located within the blaI–blaR1–blaZ operon in plasmids and transposons, comprising the regulatory gene coding for the DNA binding protein BlaI and the gene coding for the signal transducer BlaR1 (Hao et al., 2012). PBP2a proteins, expressed by MRSA, are low-affinity transpeptidases conferring resistance to almost all ß-lactams, including methicillin, cefoxitin, and oxacillin. They are synthesized by mecA within the mec gene complex (mecA, mecRI, and mecI) located in the staphylococcal chromosomal cassette mec (SCCmec) (Hanssen and Sollid, 2006). SCCmec are transferable genomic islands, classified into main types according to the combination of the ccr chromosomal recombinase gene complex (ccrA, ccrB, and ccrC) and mec genes harbored. To date, 14 major types of SCCmec have been described and sequenced (Mlynarczyk-Bonikowska et al., 2022). SCCmec can additionally harbor genes conferring resistance to other groups of antibiotics, such as macrolides, aminoglycosides, tetracyclines, lincosamides, and streptogramin B (Hiramatsu et al., 2001; Hanssen and Sollid, 2006; Liu et al., 2016). For additional genes involved in specific resistance to less relevant antibiotics from a clinical and epidemiological perspective, please refer to other available exhaustive reviews on this topic (Foster, 2017; Mlynarczyk-Bonikowska et al., 2022).
The presence of antimicrobial-resistant S. aureus is being constantly reported in RTE food of different categories, such as meat-based products, seafood, fruits and vegetables, egg- and milk-derived food, bakery and confectionery products, and mixed RTE food made with a variety of ingredients, as well as from food-contact surfaces in food facility environment (Table 1), the latter representing sources of cross-contamination for food, other utensils or surfaces, and humans, especially employed in the food industry (Di Ciccio et al., 2015; Ho et al., 2015; Plaza-Rodríguez et al., 2019). In particular, considering all the tested isolates and the most frequently investigated antimicrobials in the recently reported studies (Table 1), S. aureus shows relatively high rates of resistance toward beta-lactams [penicillin (80.7%); ampicillin (27.1%); oxacillin (25%) and cefoxitin (31.4%)]; macrolides [azithromycin (49.5%) and erythromycin (33.6%)]; folate pathway inhibitors [trimethoprim/sulfamethoxazole (48.7%)]; tetracyclines [tetracycline (39.5%)]; lincosamides [clindamycin (38.1%)]; and aminoglycosides [gentamicin (21%)], while slightly lower resistance is observed for fluoroquinolones, including ciprofloxacin (12.9%) and levofloxacin (12.3%). Although the detection of ARGs is not systematically addressed, to date, several genes have been identified in RTE food- and contact surface-derived S. aureus (Table 1) that present a risk for the spreading of antimicrobial resistance in both foodborne and human bacterial communities. Studies are mainly focused on the detection of the mecA gene (Table 1) since MRSA, able to resist methicillin, oxacillin, cefoxitin, and almost all the other beta-lactam antibiotics, has emerged as a growing public health issue since the 1990s (Oniciuc et al., 2017). Vancomycin has been selected as the main drug to treat serious infections caused by MRSA (Mahros et al., 2021); thus, the emerging occurrence of vancomycin-resistant S. aureus (VRSA) in food, including RTE food (Mahros et al., 2021; Table 1), is alarming and raises concern related to their overall spread. Therefore, although the rates of vancomycin resistance are relatively quite low (2.2%) in the recently published studies (Table 1), the detection of VRSA deserves significant attention, and systematic surveillance may allow a deeper understanding of the health burden that VRSA, beyond MRSA, might pose to consumers.
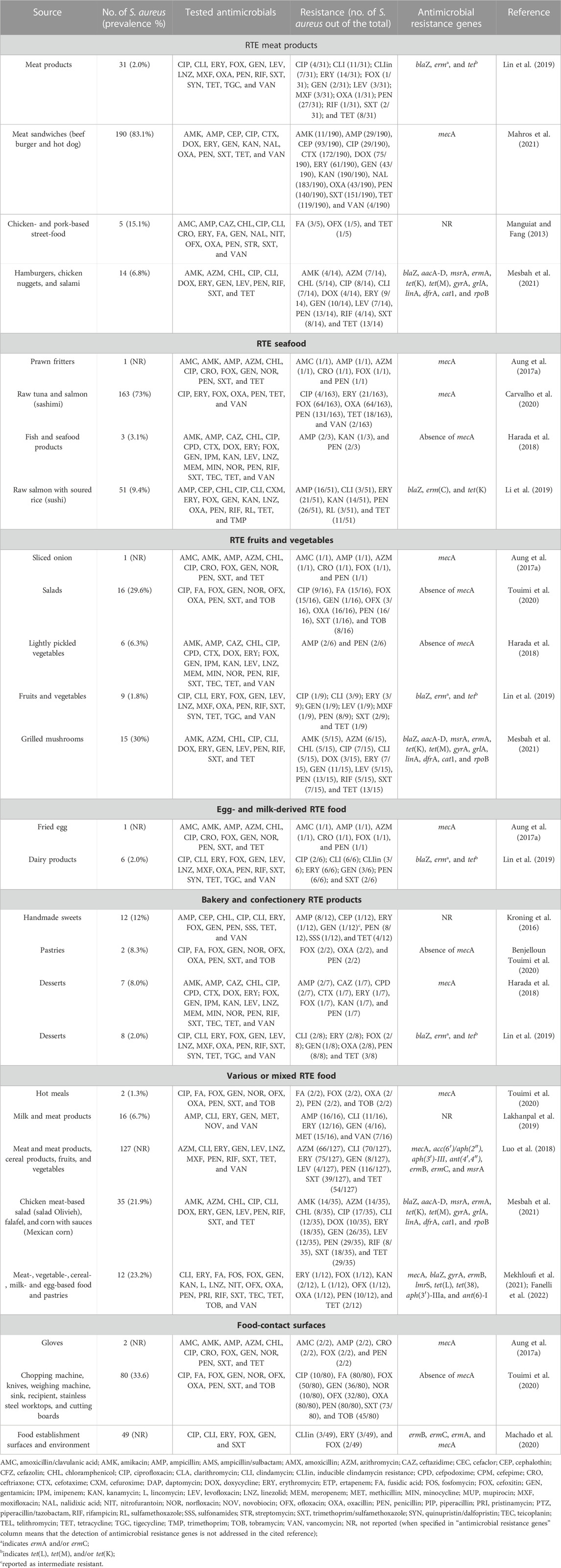
TABLE 1. Recently reported S. aureus in ready-to-eat food and food-contact surfaces, and related antimicrobial resistance.
In S. aureus, resistance to commonly used disinfectants is mediated by efflux pumps generally encoded by plasmid-borne genes (Vijayakumar and Sandle, 2019).
Qac genes encode efflux pumps capable of expelling many quaternary ammonium compounds (QACs), such as benzalkonium chloride and cetrimide, from bacterial cells (Wassenaar et al., 2015). The first study on this efflux system was performed by assaying the ethidium bromide efflux (Johnston and Dyke, 1969); the gene involved was identified on a plasmid in 1989 and thus named ebr by Sasatsu et al. (1989).
The qacA gene is located on the psK1 plasmid (Rouch et al., 1990) and codes for the production of a transmembrane efflux protein of the major facilitator superfamily named QacA (Tennent et al., 1989). qacA requires the activity of a transcriptional regulator coded by qacR (Peters et al., 2009). The qacB gene was sequenced from the plasmid psK23 in 1996 (Paulsen et al., 1996) and found to share a high degree of homology with qacA; its sequence differs from that of qacA in only seven nucleotides, resulting in changes in six amino acid positions. Despite this genetic similarity, qacB has a different substrate specificity, recognizing monovalent organic cations due to the presence of an uncharged residue, alanine, instead of aspartic acid as found in QacA (Paulsen et al., 1996). The qacC gene was first sequenced from the plasmid pSK89 (Littlejohn et al., 1991) but then isolated from other S. aureus plasmids and reported with alternative names, such as smr (staphylococcal multidrug resistance) or qacD (Grinius et al., 1992; Grinius and Goldberg, 1994). The smr gene has been detected on large conjugative multiresistance plasmids (Lyon and Skurray, 1987; Evans and Dyke, 1988) and on small non-conjugative plasmids (<3 kb; Emslie et al., 1986; Leelaporn et al., 1994). This transporter functions as a homodimer and does not require any transcriptional regulator. While QacA and QacB proteins are members of the MFS, QacC belongs to the small multidrug resistance (SMR) protein family.
The intact qacE gene was only recently detected in S. aureus, isolated from a clinical setting (Sarwar et al., 2022). QuaE is a four-transmembrane segment SMR protein and has a partially active deletion derivative (Paulsen et al., 1993). The qacJ gene was detected from a newly discovered RC plasmid (pNVH01) and then identified in several equine isolates of S. aureus (Bjorland et al., 2003); the qacH gene was identified on pST94 in food industry staphylococcal isolates by Heir et al. (1999), but not yet reported in S. aureus.
Horizontal gene transfer is considered one of the major factors responsible for the spread of antimicrobial resistance (AMR) in bacterial species (Sun et al., 2019; von Wintersdorff et al., 2016).
ARGs are mainly located in mobile genetic elements (MGEs), which, in Staphylococcus, can be classified into genomic islands, transposons, phages, plasmids, integrative conjugative elements (Sansevere and Robinson, 2017), integrons, and staphylococcal chromosomal cassettes (SCCs) (Lindsay, 2010; Alibayov et al., 2014). Approximately 15%–20% of the S. aureus genome is composed of MGEs, whose diversity confers the genome’s high variability (Chambers and DeLeo, 2009; McCarthy et al., 2014).
Although bacteriophages rarely carry ARGs, they play pivotal roles in the mobility of ARGs in S. aureus (Deghorain and Van Melderen, 2012; Xia and Wolz, 2014). S. aureus pathogenicity islands (SaPIs), which use helper phages for replication and dissemination or remain integrated into the bacterial chromosome (Penadés and Christie, 2015), can harbor ARGs such as aad, ermA, fosfomycin resistance genes, or multidrug exporters (Novick et al., 2010).
Transfer of MGEs mainly occurs during colonization of human or animal hosts, as evidenced by epidemiological studies (Knight et al., 2012; Lindsay et al., 2012; Stanczak-Mrozek et al., 2015), or during biofilm formation (Savage et al., 2013).
ARGs carried on S. aureus MGE and mechanisms of gene transfer were recently reviewed by Haaber et al. (2017).
Plasmid-carrying ARGs can be transferred between distantly related bacteria (Dahlberg et al., 1998), even between Gram-positive and Gram-negative bacteria, and this raises further concern in relation to the spread of resistance (Courvalin, 1994). An example of this was reported by Bes et al. (2021), who recently demonstrated the in vitro conjugative transfer of the plasmid p_8N_qac carrying the qacA gene from S. aureus to E. coli, highlighting how this issue is also associated with the dissemination of resistance to common sanitizers.
The role of biocides in the spread of AMR is controversial and argued. Some AMR and biocide resistance mechanisms may share a common mechanism based on the action of efflux pumps, changes in the permeability, and biofilm formation; MGEs carrying biocide resistance genes can also harbor some ARG, as in the case of qacE (Pal et al., 2015); furthermore, the exposure to biocides can induce the expression of efflux pumps involved in AMR (Paul et al., 2019).
In 2009, the Scientific Committee on Emerging and Newly Identified Health Risks (SCENIHR, 2009) established that the use or misuse of certain active substances in biocidal products in various settings may contribute to increasing the opportunities for co-selection both in humans and in the environment across taxonomic groups and different types of biocides. Indeed, the co-selection potential has been discussed in several papers (Jones and Joshi, 2021) and contexts: in farms (reviewed by Davies and Wales, 2019), in clinical practice (Russell, 2002), and in community environments (Chen et al., 2021).
A focused analysis by Pal et al. (2015) clarifies that the Staphylococcus genus is one of the bacterial groups in which the higher proportion of plasmids hosted (approximately 20%) tended to carry both BRGs and ARGs on the same plasmid. Plasmids with co-selection potential tend to be conjugative and more often carry toxin–antitoxin systems, which have a role in stabilizing plasmids in their hosts by killing daughter cells that do not inherit the plasmids (Gerdes et al., 1986). Results of this study, however, suggest that plasmids provide limited opportunities for biocides and metals to promote the horizontal transfer of antibiotic resistance through co-selection (Gullberg et al., 2014), whereas wide possibilities exist for indirect selection via chromosomal biocide/metal resistance genes.
Some studies demonstrated that the use of biocides can produce selective pressure on S. aureus. Ciusa et al. (2012) demonstrated that the biocide triclosan produces a selective pressure on S. aureus, identifying novel resistance mechanisms with high potential for horizontal gene transfer linked to the presence of a mutated fab gene, coding for the NADH-dependent trans-2-enoyl-acyl carrier protein reductase, or an additional sh-fabI allele, derived from S. haemolyticus.
Hardy et al. (2018) demonstrated that the increased use of antiseptics (chlorhexidine and octenidine) was associated with reduced susceptibility in clinical isolates of S. aureus. Biocide susceptibility did not correlate with the carriage of qac efflux pump genes, but mutations within the NorA or NorB efflux pumps, associated with chlorhexidine export, were suggested to exert an important mechanism of biocide tolerance. Htun et al. (2019) demonstrated a positive association between qacA/B carriage and chlorhexidine/octenidine exposure. Chlorhexidine exposure was associated with reduced chlorhexidine susceptibility (MIC >4 mg/L), and carriage of qacA/B or qacC was associated with reduced chlorhexidine susceptibility. Neither octenidine exposure nor carriage of qacA/B or qacC genes was associated with reduced susceptibility to octenidine; on the contrary, isolates exposed to octenidine were four times less likely to have reduced susceptibility to octenidine than unexposed isolates.
Concerning food production, the report of the Joint FAO and WHO Expert Meeting on foodborne antimicrobial resistance held in Rome (FAO and WHO, 2019) declared that “insufficient evidence is available to identify biocide use in food production as a driver of AMR. However, the identified association between biocide tolerance and resistance to one or more classes of antimicrobials underscores the need for increased awareness and prudent use of these products”.
AMR has several routes to enter the food chain: selective pressure by overuse and misuse of antimicrobials in farms and exposure to biocides or cationic compounds used as disinfectants, antiseptics, preservatives, supplements for livestock, and crop protectants. With the emergence of AMR, food may play an underestimated role as a reservoir and hotspot in the spread of resistance.
Many studies reported the WGS-based prediction of S. aureus AMR in food (Zhang F. et al., 2022; Fanelli et al., 2022; Li et al., 2022; Sri Prabakusuma et al., 2022; Wu et al., 2022). Resistance prediction databases, such as CARD, MEGARes, and AMRFinder, generally rely on BLASTn analysis and require significant computational resources and time (Gordon et al., 2014; Babiker et al., 2019). Depending on the tools used, the accuracy of these methods can vary significantly (Mason et al., 2018) and relies on the quality of the available database (Hendriksen et al., 2019); furthermore, studies on the concordance between phenotypic assessment and genomic prediction provided discordant results (Mason et al., 2018; Babiker et al., 2019). In 2022, Wang et al. (2022) created a novel S. aureus prediction model based on the analysis of antimicrobial-resistant phenotypes of training isolates and k-mer calculation, combined with machine learning algorithms, to predict the resistome of S. aureus.
Nowadays, automated pipelines have also been introduced in clinical environments to detect transmission chains and subtypes and monitor outbreaks of S. aureus, providing fast and cost-effective surveillance of MRSA (Leopold et al., 2014; Dymond et al., 2020; Slott Jensen et al., 2020).
These novel typing methods are based on WGS sequencing data and are defined as core genome multi-locus sequence typing (cgMLST) and target 1,861 S. aureus core loci (https://www.cgmlst.org/ncs/schema/141106/https://www.cgmlst.org/ncs/schema/141106/) (Leopold et al., 2014). cgMLST pipelines have been implemented within many bioinformatics software, such as BioNumerics (bioMérieux SA, Sint-Martens-Latem, Belgium), SeqSphere+ (Ridom, Germany), PHYLOViZ (http://www.phyloviz.net/), Galaxy@Sciensano (http://galaxy.sciensano.be/), and chewBBACA (Silva et al., 2018).
Since the development of the S. aureus cgMLST scheme (Leopold et al., 2014), a few dozen papers have been published on the cgMLST for epidemiological study. The majority of these are associated with CA-MRSA or MSSA outbreaks (Park et al., 2017; Madigan et al., 2018; Cheng et al., 2019; Cho et al., 2020; Slingerland et al., 2020).
Konstantinovski et al. (2021) used this method to monitor the transmission of oxacillin-resistant SA in hospital environments, while Kinnevey et al. (2021) studied the transmission of MRSA in healthcare workers, patients, and environments in non-outbreak settings. Zhu et al. (2021), by performing cgMLST analysis to map the transmission of CA-MRSA in households, revealed that the home environment was an important MRSA reservoir. Slott Jensen (2020) evaluated the transmission of livestock-associated MRSA CC398 in hospitals by calculating the cost of the intervention to stop the transmission and demonstrated the utility of cgMLST for the surveillance of transmission of LA-MRSA in hospitals.
Effelsberg et al. (2019), by analyzing the LA-MRSA CC398 in pigsty fieldworkers, traced the origin of the Western German LA-MRSA CC398 back to the 1990s. This clone diversified into farm-specific genotypes, which stayed relatively consistent over time. Loncaric et al. (2019) investigated the diversity of Australian MRSA from companion animals, highlighting the predominance of the ST398 lineage and the presence of new clones.
Other studies on animals are those carried out by i) Scholtzek et al. (2019), which characterized equine SA isolates, exhibiting reduced oxacillin susceptibility, ii) Leijon et al. (2021) and Ndahetuye et al. (2021) on SA associated with bovine clinical mastitis, iii) Kaiser-Thom et al. (2022) on SA isolated from horses with equine pastern dermatitis, and iv) Ozawa et al. (2022) on MRSA isolated from pigs in Japan.
Only three cgMLST studies have so far been reported on food. Tegegne et al. (2021) characterized 34 livestock-associated MRSA spa Type t899 strains belonging to different sequence types isolated from humans, animals, and a few food items, of which 20 belonged to ST398, 13 to ST9, and one to ST4034; all t899 isolates harbored the mecA gene on a SCCmecIVa (2B) element, except for two, which presented either the SCCmecV element or an undefined cassette. The SNP-based phylogeny analysis was consistent with the core genome multilocus sequence typing (cgMLST) analysis, with S. aureus isolates clustering apart based on STs. Phiri et al. (2022) investigated the prevalence and diversity of SA in the dairy value chain in Zambia, with a focus on raw milk. cgMLST performed on 93 isolates indicated transmission of strains along the dairy chain in Zambia with possible persistence in the chain over time. cgMLST also revealed a very close relatedness between some isolates from milkers, raw milk, or milk buckets, confirming a possible transmission between the milker and milk chain and clearly supporting the hypothesis of a direct or indirect transfer of human-derived S. aureus to cows, raw milk, or the surface of milk-handling equipment. The predominant spa type varied depending on the province, underlining the local characteristics of the traditional Zambian dairy chain. Mikhaylova et al. (2022) evaluated the genomic relatedness of 35 SA isolated from RTE food in Russia. The isolates belonged to 15 different MLST-based types, with the predominant ones belonging to clonal complex 22. The authors, examining the isolates belonging to the same/single strain based on cgMLST analysis, identified the differences in their accessory genomes, marking their dynamics and plasticity. A total of 14 samples (40%) carried at least one enterotoxin gene; additionally, a major portion of the isolates harboring the tsst1 gene were MRSA. cgMLST-based approaches have been demonstrated to provide high-resolution typing and be efficient and cost-effective in the surveillance of SA transmission in different environments, including clinical settings, animals, and food.
In 2022, Lagos et al. (2022) performed a comparative analysis between cgMLS- and SNP-based methods, using two cgMLST schemes and two SNP pipelines. The authors concluded that, independently from the approach used, an estimated genomic variation rate of 2.0–5.8 genetic events per year (without recombination) is a general guideline to be used for surveillance and outbreak investigation at clinical laboratories. The authors also highlighted the importance of selecting a reference genome with a well-defined core genome closely related to the sequences analyzed in order to avoid bias that can influence the subsequent analysis, such as the detection of genetic events.
Eradication of AMR infections is challenged by the existence of asymptomatic colonization (MacKinnon and Allen, 2000; Smith et al., 2004), and time is an essential factor for the success of intervention measures against the spread of AMR, especially in healthcare-associated infections (Bootsma et al., 2006). While WGS data production has become increasingly fast and accurate, limitations occurred when all the data produced had to be analyzed and interpreted. In this scenario, the need to develop user-friendly bioinformatics platforms, which allow the management and interrogation of data for workers that do not have bioinformatics expertise, remains pivotal.
Biofilm is a bacterial community embedded in a self-produced matrix made of extracellular polymeric substances (EPSs). The main constituents of EPSs are polysaccharide intercellular adhesin or poly-N-acetylglucosamine, as well as proteins, lipids, RNA, and extracellular DNA (Forson et al., 2020). Biofilm formation is controlled by the quorum sensing system that confers the ability to sense the bacterial cell density and respond to environmental stimuli through a cell-to-cell communication using small diffusible chemical signaling molecules called autoinducers (Peng et al., 2022). In Staphylococcus, quorum sensing is regulated by the accessory gene regulator (agr) system (Le and Otto, 2015).
S. aureus biofilm may form on food and food facility equipment, facilitating the persistence of bacterial cells, including antimicrobial-resistant forms, on such surfaces and the possibility that they could cross-contaminate other materials and food. This increases the spreading of S. aureus and the possible consequent onset of S. aureus-related diseases. Moreover, biofilm protects cells against adverse environmental conditions such as nutrient limitation, change in temperature, and dehydration (Idrees et al., 2021); therefore, cells living in biofilm exhibit a greater resistance than their planktonic form (Liu et al., 2019).
Stainless steel, glass, polystyrene, and polyvinyl chloride (PVC) are commonly used materials in food facilities and, although not unequivocally demonstrated (da Silva Meira et al., 2012; Pagedar et al., 2010; Cha et al., 2019; Lee et al., 2015), surfaces with high free energy, such as those made of stainless steel and glass, exhibit greater hydrophilicity, which may allow a better bacterial attachment, which easily leads to biofilm formation (Lee et al., 2015; Cha et al., 2019).
Traditionally, various cleaning agents and biocides such as quaternary ammonium compounds, sodium hypochlorite (NaClO) and other chlorine-based compounds, sodium hydroxide (NaOH), nitric acid (HNO3), anionic acids, and iodophors (Martin et al., 2016) are used in the food sector for surface decontamination. However, beyond the issue related to the onset of the genetic resistance to the utilized biocides (Butucel et al., 2022), their effectiveness is actually hampered by the reduced penetrability of these agents toward biofilm due to its peculiar structure that protects the embedded cells (Idrees et al., 2021).
It has been shown that a commercial chlorine-based sanitizer applied on stainless steel (type 304, no. 4 finish) for 1 min at room temperature, when compared with distilled water able to reduce S. aureus biofilm cells by 0–2 Log colony-forming units (CFU), decreased S. aureus biofilm cells approximately by only 1–3 Log CFU (starting from an S. aureus biofilm cell density of approximately 3–6 Log CFU/10 cm2) (Lee et al., 2015). Martin et al. (2016) assessed the efficacy of a cleaning protocol employing sodium hypochlorite in cheese-producing dairy plants. Although a certain efficacy was observed in removing adhered S. aureus cells from stainless steel and polypropylene surfaces at a temperature of 5°C, the protocol was ineffective in removing the S. aureus cells at the cheese-making temperature of 35°C, which is probably due to the higher number of cells adhering at this higher temperature (1.95–4.98 versus 5.15–6.58 Log CFU/cm2 at 5°C and 35°C, respectively) (Martin et al., 2016), suggesting that traditional cleaning protocols should be revised in order to control S. aureus more effectively. Considering this, researchers are driven to seek novel strategies to address the issue related to S. aureus contamination and its spread in the food sector (Figure 1; Table 2).
Lactic acid bacteria may produce molecules with antibacterial and antibiofilm activities so that cell-free supernatants (CFSs) may be effective against S. aureus. In particular, it has been demonstrated that CFSs of Lactobacillus acidophilus LA5 and Lactobacillus casei 431 possess antibacterial activity, being able to inhibit S. aureus growth, as determined by the agar-well diffusion test, and reduce 2-day-old biofilms grown on polystyrene and glass surfaces by 45%–70%, depending on the CFS concentrations used (40%–100%) (Koohestani et al., 2018).
Specific bacterial molecules have also been evaluated for their activity against S. aureus. Surfactin produced by Bacillus subtilis has been proposed as a potential coating agent for contact surfaces to prevent S. aureus biofilm formation (Liu et al., 2019). In particular, this environmentally friendly low-toxicity biosurfactant inhibited S. aureus growth, showing a minimum-inhibitory concentration (MIC) of 32 μg/mL and a minimum bactericidal concentration (MBC) of 128 μg/mL. Moreover, likely due to its effect on the S. aureus quorum sensing system and the ability to downregulate the icaA and icaD gene expression, impairing the biofilm polysaccharide production, this molecule decreased the formation of S. aureus biofilm by 40 to >80% and decreased the formation of mature biofilm on glass, polystyrene, and stainless-steel surfaces by < 10 to 90%, depending on surfactin concentrations and duration of treatment (Liu et al., 2019).
Strategies employing viruses have also been evaluated against S. aureus. When used on poultry products in combination with 1% thyme essential oil, a bacteriophage of the family Myoviridae (vB_SauM_CP9) able to resist different food processing stress factors (such as pH and temperature changes), showed lytic activity on several multidrug-resistant S. aureus strains resistant to several antibiotics, including methicillin, nalidixic acid, sulfamethoxazole, amoxicillin, doxycycline, cefotaxime, erythromycin, norfloxacin, gentamicin, and ciprofloxacin. Thus, it was recommended for application in the food sector by spraying or via packaging materials (Abdallah et al., 2021). A phage-encoded endolysin (LysCSA13) from an S. aureus-specific bacteriophage (CSA13) showed activity against S. aureus biofilm and planktonic cells, including MRSA, when applied on polystyrene, glass, and stainless-steel surfaces, demonstrating its potential use in food facility environments (Cha et al., 2019).
Recent studies have indicated that various natural compounds not only inhibit biofilm formation but also eradicate mature biofilm produced by several bacterial species, including S. aureus (Mastoor et al., 2022). Essential oils are mixtures of secondary metabolites obtained from plants, which can exert an antimicrobial activity directly when used in food products and in active food packaging, or indirectly when used as sanitizing or antibiofilm agents on food- and other contact surfaces (Pedonese et al., 2017). In particular, it has been found that manuka leaf (Leptospermum scoparium J. R. et G. Forst), thymus (Thymus vulgaris L.), and cinnamon bark (Cinnamomum zeylanicum L.) essential oils may have a better growth inhibition activity against foodborne MRSA and foodborne biofilm-producing S. aureus strains than bergamot (Citrus bergamia Risso) essential oils, likely due to the presence of terpene (carvacrol, leptospermone, p-cymene, thymol, trans-Calamenene, and ß-caryophyllene) and non-terpene ((E)-cinnamaldehyde) derivatives as major compounds, being therefore proposed as food additives, food packaging constituents, and possible surface sanitizers in the food sector (Pedonese et al., 2017).
In addition, organic acids extracted from plants may have good inhibitory activities against foodborne pathogens. Quinic acid, widely spread in plants, has been shown to inhibit biofilm formation, mainly decreasing the initial S. aureus adhesion, as well as reducing the metabolic activity and the viability of the S. aureus biofilm cells (Bai et al., 2019). Moreover, a certain ability to reduce established S. aureus cells adhered to stainless steel has been demonstrated. Those results suggested that quinic acid can be used in food facilities as an antibiofilm agent, especially for preventing surface biofilm formation (Bai et al., 2019).
Among plant extracts, environmentally friendly molecules can be extracted using green technologies in order to reduce the impact of pollution derived from the widespread usage of traditional chemical sanitizers. Moringa oleifera seed oil extracted using pressurized n-propane instead of toxic solvents (n-hexane, ethanol, and DMSO) could be used in the food sector, providing an option for a safe and efficient large-scale producible antibiofilm agent (de Oliveira et al., 2021). Probably due to its high percentage of fatty acids (especially oleic acid, behenic acid, and palmitic acid), Moringa oleifera seed oil extract showed activity against S. aureus biofilm, being able to inhibit its formation or eliminate the formed biofilm, depending on its concentration (0.25, 0.5, and 1%) and contact time (10, 20, 40, and 60 min), on polystyrene or PVC (de Oliveira et al., 2021). In addition, the bulb extract from the plant Eleutherine americana Merr., which is commonly used in Thai cuisine and in folk medicine, has been suggested as a potential natural preservative to be used in food against S. aureus, due to the ability of the ethanol crude extract (0.25 mg/mL–4 mg/mL) to reduce the concentration of foodborne S. aureus by approximately 3–6 log cfu/mL, depending on the strain tested (Ifesan et al., 2009).
Enzymes, derived mainly from animals, fungi, or bacteria, may also represent a promising technology to be used in food facilities. It was found that 1 U/100 μL of a-amylase, cellulase R-10 and DNase I, and 1 milli-Anson unit/100 μL of proteinase K could have a certain inhibitory effect on the formation or exert a certain degradative effect on the formed S. aureus biofilm on polystyrene, while amyloglucosidase showed only an inhibitory effect on biofilm formation (Kim M. J. et al., 2019). However, it should be noted that the work by Kim M. J. et al. (2019) reported a strong strain-dependent effect and, in one case, also a paradox effect occurred since the enzymatic treatment (α-amylase, amyloglucosidase, cellulase, and proteinase K) enhanced the biofilm formation by S. aureus ATCC 12600 (Kim M. J. et al., 2019).
Apart from microbiological agents and natural substances, new chemical compounds and new food-contact materials are exploitable to control S. aureus contamination. Recently, a new derivative belonging to the oxadiazole class, i.e., the synthetic 1,3,4-oxadiazole compound LMM6, previously reported for its fungicidal activity against Candida albicans (Faria et al., 2021), has been described to inhibit both bacterial growth in liquid culture and biofilm formation, as well as reduce the biomass of the already formed biofilm, probably interfering with the exopolysaccharide matrix, leading to biofilm disruption. Therefore, it was proposed as an innovative synthetic molecule to be used also in combination with other sanitizers or methods for S. aureus control (Dante Formagio et al., 2022).
In addition, novel food-contact surfaces may be used in food facilities. In particular, a novel type of stainless steel containing copper (304CuSS) has been shown to have an antibacterial ability against S. aureus when compared to traditional stainless steel, reducing the formation of biofilm even after a 7-day-long exposure to S. aureus (Nan et al., 2015). Such activity is likely due to the presence of saturated Cu-rich precipitates that continuously release Cu2+ ions that bind to EPS and bacterial proteins, thus altering their properties (Nan et al., 2015). Molybdenum disulfide (MoS2) surfaces, although depending on the size and concentration of the constitutive MoS2 particles, were demonstrated to reduce S. aureus retention and biofilm formation, as well as having no cytotoxic activity on eukaryotic cells (renal human cells HK-2), therefore being potentially safe for consumers and exploitable in the food sector (Amin et al., 2020). Additionally, silver (Ag-NPs) and zinc oxide (ZnO-NPs) nanoparticles can be incorporated into a polymer matrix (polyester) and then exploited as an antimicrobial food-contact surface due to the release and migration of biocidal ions (Fontecha-Umaña et al., 2020). Ag-NPs showed better antimicrobial activity against S. aureus at all the tested concentrations (400 ppm–850 ppm), but the usage in combination with ZnO-NPs (400 ppm) resulted in a synergistic action which further increased the antimicrobial efficacy (Fontecha-Umaña et al., 2020). Chemical elements could also be added to other disposable materials. For instance, bacterial growth can occur on paper products, such as wrappings and towels used in the food sector, that, therefore, may represent sources of S. aureus contamination during food packaging, surface cleaning, or hand drying (Wang and Webster, 2013; Wang et al., 2015). Considering this, paper towels coated with selenium (Se) nanoparticles were shown to be able to inhibit the growth of S. aureus by 89% when compared to Se-uncoated paper towels (Wang and Webster, 2013; Wang et al., 2015). The ability to adsorb large amounts of proteins is probably one of the mechanisms by which the Se-coated paper towels inhibited bacterial growth, and they were proposed as a promising selenium-based antibacterial strategy that may prevent bacterial spreading in the food sector (Wang and Webster, 2013; Wang et al., 2015).
Steam has been evaluated for its anti-S. aureus activity. In particular, the so-called superheated steam (SHS) able to reach 150 °C, above the saturation temperature, is more effective than saturated steam (SS) at 100 °C (Kim S. H. et al., 2019). SHS reduced, below the detection limit, multi-strain 5-day-old S. aureus biofilms, grown on stainless steel, polypropylene, and high-density polyethylene, in 10 (stainless steel) to 15 (polypropylene and high-density polyethylene) seconds (Kim S. H. et al., 2019). It resulted in a time-saving and non-polluting technology, as it does not employ any chemical compound, and it has the potential to be used in the food sector.
On the other hand, low-temperature sterilization techniques are gaining increasing attention for being able to preserve materials sensitive to high temperatures. Electric field techniques are being considered and, contrary to ultraviolet light (UV) sterilization, they prevent radiation damage (Qi et al., 2022). Recently, the high-voltage prick electrostatic field (HVPEF), which has a simpler generation system than that of the previously investigated pulsed electric field (PEF) (Khan et al., 2016; Guo et al., 2018; Wang et al., 2018), has been evaluated against S. aureus present on surfaces and on food (Qi et al., 2021; Qi et al., 2022). HVPEF showed activity against S. aureus contamination on cheese, sausage, salmon, and griskin, and the mortality rate increased with the increase of the electric voltage (10–13 kV) and treatment time (15–30 min), reducing the S. aureus contamination up to 98%–99% (Qi et al., 2021). HVPEF was also effective against S. aureus in the biofilm form (Qi et al., 2022). In particular, it reduced 24- and 48-h-old biofilms established on different surfaces, mainly by killing the embedded bacteria. In addition, it was able to inhibit the S. aureus biofilm formation, likely due to its direct effect on the planktonic cells, together with the ability to reduce the release of the key components of EPS and regulate the expression of biofilm formation-related genes (icaA, ebh, cidA, sarA, icaR, and sigB) (Qi et al., 2022).
Electricity could also be employed in another way. It can be used to produce electrolyzed water (EW), which is a low-cost and environmentally friendly product obtained by electrolyzing water containing NaCl or other salts (e.g., NaNO3) (Sun et al., 2012). It has been shown that the EW acidic fraction (i.e., acidic electrolyzed water, AEW) had a bactericidal effect on the S. aureus cells present in biofilm, while the EW basic fraction (i.e., basic electrolyzed water, BEW) had the ability to directly remove biofilm biomass formed on polystyrene (Sun et al., 2012), indicating that AEW and BEW could be applied as a bactericidal and removing agent against S. aureus biofilm (Sun et al., 2012).
High pressure is being explored as a means for S. aureus decontamination. In particular, it has been shown that ultra-high-pressure homogenization (UHPH) may have the ability to decrease the number of S. aureus planktonic cells that may contaminate liquid food, but such technology showed poor effectiveness in reducing the number of S. aureus cells detached from the biofilm that may form within pipes and that may demonstrate a higher resistance to pressure (Zhang et al., 2021).
Overall, many methods have been investigated to prevent or control S. aureus contamination in food and on food-contact surfaces. The use of natural substances draws much attention, along with the possibility of employing new chemical compounds and new materials, as well as applying physical methods in innovative ways. It must be emphasized that these methods could be considered at an early stage of implementation because there is a substantial lack of practical application in the food establishments to verify (i) their in situ effectiveness, (ii) the economic viability for the food company, (iii) the ability to preserve the organoleptic characteristics of the wide variety of food products that can potentially be treated, and (iv) the absence of short- and long-term undesirable effects on the consumers. Nevertheless, the investigation of these new technologies represents a crucial area to be explored in order to address not only the issue related to chemical residues in food and the environmental impact but also to overcome the known mechanisms of genetic resistance that S. aureus, and bacteria in general, may exhibit to traditional biocides. Such novel technologies are paving the way to improve the safety of food products, preventing contamination by S. aureus, including its antimicrobial-resistant forms, and the onset of S. aureus- related diseases.
Few novel species are closely phenotypically and genotypically related to S. aureus, being part of the so-called Staphylococcus aureus complex. In 2015, this complex was conceived to include S. aureus and the two novel species Staphylococcus argenteus and Staphylococcus schweitzeri (Tong et al., 2015). Very recently, Staphylococcus singaporensis (Chew et al., 2021) and Staphylococcus roterodami (Schutte et al., 2021) were added to this complex, although, due to phylogenetic analyses, the classification into these two latter species seems questionable (Monecke et al., 2022). However, to date, the Staphylococcus aureus complex comprises S. aureus as well as S. argenteus, S. schweitzeri, S. singaporensis, and S. roterodami. With the exception of S. schweitzeri, the other three novel species (i.e., S. argenteus, S. singaporensis, and S. roterodami) are all associated to human infections (Becker et al., 2019; Chew et al., 2021; Schutte et al., 2021), highlighting that similar to S. aureus, they may act as pathogenic bacteria. Nevertheless, S. schweitzeri was found to have virulence abilities comparable to those of S. aureus, i.e., similar cellular invasion, pro-inflammatory cellular activation, and intracellular cytotoxicity, as well as higher extracellular cytotoxicity (Grossmann et al., 2021). It could also escape from phagolysosomes, coagulate plasma, and form biofilm; therefore Grossmann et al. (2021) warned that it may become an emerging pathogen in the near future. Considering the possibility that the S. aureus complex-related species may act as pathogens, the onset of antimicrobial resistance is a critical issue since it may hinder recovery if a pathological condition occurs. Interestingly, however, S. schweitzeri seems to be antimicrobial-susceptible to all the tested antibiotics (Table 3), while S. roterodami and S. singaporensis strains were found to be resistant or intermediate resistant to some antibiotics (Table 3). It should be noticed that S. roterodami and S. singaporensis were, so far, isolated from humans, fruit bats (Eidolon helvum), and a captive bird (diamond firetail finch, Stagonopleura guttata) in the few studies available to date (Chew et al., 2021; Schutte et al., 2021; Monecke et al., 2022). S. schweitzeri was reported in Africa, isolated from fruit bats (Rousettus aegyptiacus; Eidolon helvum) (Held et al., 2017; Olatimehin et al., 2018), non-human primates (Schaumburg et al., 2015; Tong et al., 2015), and surfaces of fomites (currency note and computer keyboard) (Shittu et al., 2020), while the transmission to humans appeared to be occasional (Schaumburg et al., 2015). Although, to the best of our knowledge, the presence of S. roterodami, S. singaporensis, and S. schweitzeri has not been reported so far in food, most likely due to lack of relevant targeted studies, it should be pointed out that bats and monkeys are food sources in some areas of Africa (Schaumburg et al., 2015; Held et al., 2017). Therefore, bushmeat could actually be a possible source of contamination and spreading of such novel species, deserving investigation in the near future. Conversely, S. argenteus, known for its ability to cause various pathological conditions in humans (e.g., sepsis, joint infection, endocarditis, and lymphadenitis) (Chantratita et al., 2016; Jiang et al., 2018; Ohnishi et al., 2018; Hirai et al., 2022) and animals (e.g., abscess, wound infection, and bovine mastitis) (Indrawattana et al., 2019; Pumipuntu et al., 2019; Meijer et al., 2022), is being reported from food, and it is increasingly recognized as an emerging foodborne pathogen (Shi and Zhang, 2018; Fusco et al., 2020), being able to cause staphylococcal food poisoning due to the production of enterotoxins, as assessed in three outbreaks reported so far in Japan (Suzuki et al., 2017; Wakabayashi et al., 2018). Although the actual prevalence and distribution of S. argenteus in food products and food-related environments is still unclear (Shi and Zhang, 2018), S. argenteus has been recently reported from different food sources, including RTE food, comprising those involved in food poisoning, as well as contact surfaces of kitchen utensils and specimens from food handlers (Table 3). These isolates also demonstrated resistance to several antimicrobials (Table 3), therefore highlighting the possibility that food and contact surfaces may represent sources of S. argenteus and that the problem of antimicrobial resistance should not be overlooked for this emerging pathogenic bacterium. Nevertheless, to the best of our knowledge, the resistance to critical antimicrobials such as penicillinase-stable penicillins (e.g., methicillin, oxacillin, and cefoxitin) and vancomycin has not been reported so far in isolates from food and food-related environments, although methicillin-resistant S. argenteus strains are being found among clinical isolates (Giske et al., 2019; Witteveen et al., 2022), sharing this antimicrobial resistance feature with the related species S. aureus. Foodborne S. argenteus isolates were able to form a biofilm, showing weak-to-strong biofilm production as assessed on polystyrene (Wu et al., 2020), that, therefore, may allow survival in hostile conditions and the persistence of S. argenteus in food and food facility environments. Considering this, strategies to control S. argenteus in the food sector should be contemplated and implemented, and next to methods employing classical biocides, novel methods may represent a thriving sector to be explored as being conducted for S. aureus. The first attempt was made by evaluating a Lactobacillus plantarum-derived bacteriocin (LSB1) that inhibited the growth of S. argenteus planktonic cells and inhibited the biofilm formation up to 80% (Zhang Y. M. et al., 2022), being a candidate to be used as antibacterial agent or natural food preservative for S. argenteus control.
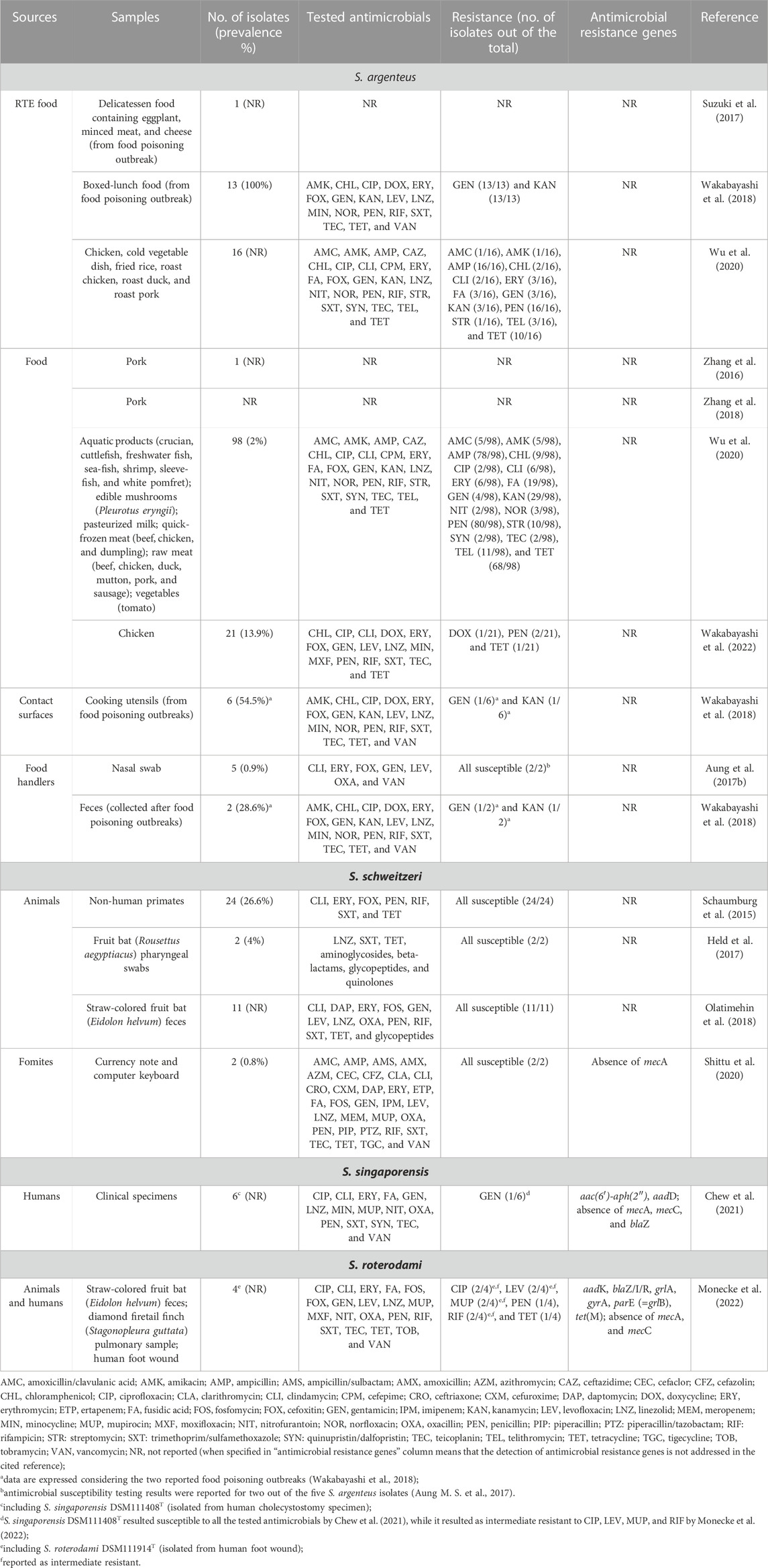
TABLE 3. S. aureus complex-related species (S. argenteus, S. schweitzeri, S. singaporensis, and S. roterodami): sources and reported antimicrobial resistance.
The novel species of the S. aureus complex may therefore represent an emerging issue for public health, also involving the food sector. The constant development and improvement of methods for their accurate identification, such as those based on mass spectrometry (MS) and polymerase chain reaction (PCR) (Zhang et al., 2016; Schuster et al., 2017; Chen et al., 2018), will help define their actual prevalence in food and food-related environments and understand if they represent ecological niches for these microorganisms. Although first genome sequences of strains belonging to these novel species have been deposited in publicly available databases (Chew et al., 2021; Goswami et al., 2021; Grossmann et al., 2021; Monecke et al., 2022), investigations on their antimicrobial resistance genes are lacking (Table 3), deserving future attention. These studies will help understand the antimicrobial resistance potential of these species and evaluate their role in the context of public health and the spreading of antibiotic resistance, allowing us to gain further knowledge on their overall virulence and pathogenic traits.
S. aureus is a pathogenic microorganism that can contaminate both food-contact surfaces and food, including RTE products whose consumption may pose a direct risk to consumer health. Different antimicrobial as well as biocide resistance genes are responsible for i) resistance to antibiotic treatments if infections occur and ii) resistance to sanitizers during cleaning procedures, which facilitates S. aureus persistence on surfaces, especially in the biofilm form. WGS-based analyses, mainly based on the development of resistance prediction pipelines and genome-based typing, represent valuable tools to understand the actual arsenal of resistance genes and allow the epidemiological surveillance of S. aureus, aimed at deeply understanding the public health burden represented by this antimicrobial-resistant pathogenic bacterium. Novel strategies for S. aureus decontamination, employing natural substances, new chemical compounds, or innovative physical methods, represent a new exploration area that may overcome the drawbacks derived from traditional ineffective sanitizing protocols and need to be implemented in the food sector to prevent the spreading of S. aureus, including its antimicrobial-resistant forms, in both food and food-related environment. Novel phenotypically and genotypically related species belonging to the S. aureus complex (S. argenteus, S. schweitzeri, S. singaporensis, and S. roterodami) may represent emerging antimicrobial-resistant foodborne pathogens, whose prevalence in RTE food and role in threatening consumers’ health needs to be defined, requiring targeted studies and the development of methods for their simple, fast, and accurate identification.
VF conceived the work. DC and FF wrote the first draft of the manuscript. VF revised the overall manuscript and is responsible for the overall review quality control. All authors contributed to the manuscript revision, and read and approved the submitted manuscript.
This review was prepared within the project PE13 “One Health Basic and Translational Research Actions Addressing Unmet Needs on Emerging Infectious Diseases” funded by the Italian National Recovery and Resilience Plan (PNRR).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abdallah, K., Tharwat, A., and Gharieb, R. (2021). High efficacy of a characterized lytic bacteriophage in combination with thyme essential oil against multidrug-resistant Staphylococcus aureus in chicken products. Iran. J. Vet. Res. 22 (1), 24–32. doi:10.22099/ijvr.2020.38083.5543
Alenizi, D. A. (2014). Prevalence of Staphylococcus aureus and antibiotic resistance in children with atopic dermatitis in Arar, Saudi Arabia. J. Dermatol. Dermatol. Surg. 18, 22–26. doi:10.1016/j.jssdds.2013.11.001
Alibayov, B., Baba-Moussa, L., Sina, H., Zdeňková, K., and Demnerová, K. (2014). Staphylococcus aureus mobile genetic elements. Mol. Biol. Rep. 41 (8), 5005–5018. doi:10.1007/s11033-014-3367-3
Amin, M., Rowley-Neale, S., Shalamanova, L., Lynch, S., Wilson-Nieuwenhuis, J. T., El Mohtadi, M., et al. (2020). Molybdenum disulfide surfaces to reduce Staphylococcus aureus and Pseudomonas aeruginosa biofilm formation. ACS Appl. Mat. Interfaces 12 (18), 21057–21069. doi:10.1021/acsami.0c02278
Ardic, N., Sareyyupoglu, B., Ozyurt, M., Haznedaroglu, T., and Ilga, U. (2006). Investigation of aminoglycoside modifying enzyme genes in methicillin-resistant staphylococci. Microbiol. Res. 161, 49–54. doi:10.1016/j.micres.2005.05.002
Aung, K. T., Hsu, L. Y., Koh, T. H., Hapuarachchi, H. C., Chau, M. L., Gutiérrez, R. A., et al. (2017). Prevalence of methicillin-resistant Staphylococcus aureus (MRSA) in retail food in Singapore. Antimicrob. Resist. Infect. Control. 6, 94. doi:10.1186/s13756-017-0255-3
Aung, M. S., San, T., Aye, M. M., Mya, S., Maw, W. W., Zan, K. N., et al. (2017). Prevalence and genetic characteristics of Staphylococcus aureus and Staphylococcus argenteus isolates harboring panton-valentine leukocidin, enterotoxins, and TSST-1 genes from food handlers in Myanmar. Toxins 9 (8), 241. doi:10.3390/toxins9080241
Babiker, A., Mustapha, M. M., Pacey, M. P., Shutt, K. A., Ezeonwuka, C. D., Ohm, S. L., et al. (2019). Use of online tools for antimicrobial resistance prediction by whole-genome sequencing in methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant enterococci (VRE). J. Glob. Antimicrob. Resist. 19, 136–143. doi:10.1016/j.jgar.2019.04.006
Bai, J. R., Wu, Y. P., Elena, G., Zhong, K., and Gao, H. (2019). Insight into the effect of quinic acid on biofilm formed by Staphylococcus aureus. RSC Adv. 9 (7), 3938–3945. doi:10.1039/c8ra09136f
Bakthavatchalam, Y. D., Ramaswamy, B., Janakiraman, R., Steve, R. J., and Veeraraghavan, B. (2018). Genomic insights of community-acquired methicillin-resistant Staphylococcus aureus (MRSA) with reduced teicoplanin susceptibility: A case of fatal necrotizing fasciitis. J. Glob. Antimicrob. Resist. 14, 242–245. doi:10.1016/j.jgar.2018.05.006
Beceiro, A., Tomás, M., and Bou, G. (2013). Antimicrobial resistance and virulence: A successful or deleterious association in the bacterial world? Clin. Microbiol. Rev. 26, 185–230. doi:10.1128/CMR.00059-12
Becker, K., Schaumburg, F., Kearns, A., Larsen, A. R., Lindsay, J. A., Skov, R. L., et al. (2019). Implications of identifying the recently defined members of the Staphylococcus aureus complex S. argenteus and S. schweitzeri: A position paper of members of the ESCMID study group for staphylococci and staphylococcal diseases (ESGS). Clin. Microbiol. Infect. 25 (9), 1064–1070. doi:10.1016/j.cmi.2019.02.028
Bes, T. M., Nagano, D. S., Marchi, A. P., Camilo, G., Perdigão-Neto, L. V., Martins, R. R., et al. (2021). Conjugative transfer of plasmid p_8N_qac(MN687830.1) carrying qacA gene from Staphylococcus aureus to Escherichia coli C600: Potential mechanism for spreading chlorhexidine resistance. Rev. Inst. Med. Trop. Sao Paulo. 63, e82. doi:10.1590/S1678-9946202163082
Bissonnette, L., Champetier, S., Buisson, J. P., and Roy, P. H. (1991). Characterization of the nonenzymatic chloramphenicol resistance (cmlA) gene of the In4 integron of Tn1696: Similarity of the product to transmembrane transport proteins. J. Bacteriol. 173 (14), 4493–4502. doi:10.1128/jb.173.14.4493-4502.1991
Bjorland, J., Steinum, T., Sunde, M., Waage, S., and Heir, E. (2003). Novel plasmid-borne gene qacJ mediates resistance to quaternary ammonium compounds in equine Staphylococcus aureus, Staphylococcus simulans, and Staphylococcus intermedius. Antimicrob. Agents Chemother. 47, 3046–3052. doi:10.1128/AAC.47.10.3046-3052.2003
Bootsma, M. C., Diekmann, O., and Bonten, M. J. (2006). Controlling methicillin-resistant Staphylococcus aureus: Quantifying the effects of interventions and rapid diagnostic testing. Proc. Natl. Acad. Sci. U.S.A. 103 (14), 5620–5625. doi:10.1073/pnas.0510077103
Brandenberger, M., Tschierske, M., Giachino, P., Wada, A., and Berger-Bächi, B. (2000). Inactivation of a novel three-cistronic operon tcaR–tcaA–tcaB increases teicoplanin resistance in Staphylococcus aureus. Biochim. Biophys. Acta. 1523, 135–139. doi:10.1016/s0304-4165(00)00133-1
Bridier, A., Sánchez-Vizuete, P., Guilbaud, M., Piard, J. C., Naitali, M., and Briandet, R. (2015). Biofilm-associated persistence of food-borne pathogens. Food Microbiol. 45, 167–178. doi:10.1016/j.fm.2014.04.015
Brückner, R., and Matzura, H. (1985). Regulation of the inducible chloramphenicol acetyltransferase gene of the Staphylococcus aureus plasmid pUB112. EMBO J. 4, 2295–2300. doi:10.1002/j.1460-2075.1985.tb03929.x
Bugg, T. D., Wright, G. D., Dutka-Malen, S., Arthur, M., Courvalin, P., and Walsh, C. T. (1991). Molecular basis for vancomycin resistance in Enterococcus faecium BM4147: Biosynthesis of a depsipeptide peptidoglycan precursor by vancomycin resistance proteins VanH and VanA. Biochemistry 30 (43), 10408–10415. doi:10.1021/bi00107a007
Burdett, V. (1996). Tet(M)-promoted release of tetracycline from ribosomes is GTP dependent. J. Bacteriol. 178, 3246–3251. doi:10.1128/jb.178.11.3246-3251.1996
Butucel, E., Balta, I., Ahmadi, M., Dumitrescu, G., Morariu, F., Pet, I., et al. (2022). Biocides as biomedicines against foodborne pathogenic bacteria. Biomedicines 10 (2), 379. doi:10.3390/biomedicines10020379
Byrne, M. E., Gillespie, M. T., and Skurray, R. A. (1991). 4',4'' adenyltransferase activity on conjugative plasmids isolated from Staphylococcus aureus is encoded on an integrated copy of pUB110. Plasmid 25 (1), 70–75. doi:10.1016/0147-619x(91)90008-k
Carvalho, J. S., Neto, A. F. L., Melo, I. M., Varjão, L. M., Andrade, C. A. D. N., Xavier, D. E., et al. (2020). Occurrence of methicillin-resistant Staphylococcus aureus in ready-to-eat raw fish from Japanese cuisine restaurants in salvador, Brazil. J. Food Prot. 83 (6), 991–995. doi:10.4315/0362-028X.JFP-19-375
Cha, Y., Son, B., and Ryu, S. (2019). Effective removal of staphylococcal biofilms on various food contact surfaces by Staphylococcus aureus phage endolysin LysCSA13. Food Microbiol. 84, 103245. doi:10.1016/j.fm.2019.103245
Chambers, H. F., and DeLeo, F. R. (2009). Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat. Rev. Microbiol. 7, 629–641. doi:10.1038/nrmicro2200
Chang, S., Sievert, D. M., Hageman, J. C., Boulton, M. L., Tenover, F. C., Downes, F. P., et al. (2003). Infection with vancomycin-resistant Staphylococcus aureus containing the vanA resistance gene. N. Engl. J. Med. 348 (14), 1342–1347. doi:10.1056/NEJMoa025025
Chantratita, N., Wikraiphat, C., Tandhavanant, S., Wongsuvan, G., Ariyaprasert, P., Suntornsut, P., et al. (2016). Comparison of community-onset Staphylococcus argenteus and Staphylococcus aureus sepsis in Thailand: A prospective multicentre observational study. Clin. Microbiol. Infect. 22 (5), e11–e19. doi:10.1016/j.cmi.2016.01.008
Chen, B., Han, J., Dai, H., and Jia, P. (2021). Biocide-tolerance and antibiotic-resistance in community environments and risk of direct transfers to humans: Unintended consequences of community-wide surface disinfecting during COVID-19? Environ. Pollut. 283, 117074. doi:10.1016/j.envpol.2021.117074
Chen, S. Y., Lee, H., Teng, S. H., Wang, X. M., Lee, T. F., Huang, Y. C., et al. (2018). Accurate differentiation of novel Staphylococcus argenteus from Staphylococcus aureus using MALDI-TOF MS. Future Microbiol. 13, 997–1006. doi:10.2217/fmb-2018-0015
Cheng, V. C. C., Wong, S. C., Cao, H., Chen, J. H. K., So, S. Y. C., Wong, S. C. Y., et al. (2019). Whole-genome sequencing data-based modeling for the investigation of an outbreak of community-associated methicillin-resistant Staphylococcus aureus in a neonatal intensive care unit in Hong Kong. Eur. J. Clin. Microbiol. Infect. Dis. 38 (3), 563–573. doi:10.1007/s10096-018-03458-y
Chew, K. L., Octavia, S., Lai, D., Lin, R. T. P., and Teo, J. W. P. (2021). Staphylococcus singaporensis sp. nov., a new member of the Staphylococcus aureus complex, isolated from human clinical specimens. Int. J. Syst. Evol. Microbiol. 71 (10). doi:10.1099/ijsem.0.005067
Chieffi, D., Fanelli, F., Cho, G. S., Schubert, J., Blaiotta, G., Franz, C. M. A. P., et al. (2020). Novel insights into the enterotoxigenic potential and genomic background of Staphylococcus aureus isolated from raw milk. Food Microbiol. 90, 103482. doi:10.1016/j.fm.2020.103482
Cho, H. K., Yang, J. N., Cunningham, S. A., Greenwood-Quaintance, K. E., Dalton, M. L., Collura, C. A., et al. (2020). Molecular epidemiology of methicillin-susceptible Staphylococcus aureus in infants in a neonatal intensive care unit. Infect. Control Hosp. Epidemiol. 41 (12), 1402–1408. doi:10.1017/ice.2020.355
Ciusa, M. L., Furi, L., Knight, D., Decorosi, F., Fondi, M., Raggi, C., et al. (2012). A novel resistance mechanism to triclosan that suggests horizontal gene transfer and demonstrates a potential selective pressure for reduced biocide susceptibility in clinical strains of Staphylococcus aureus. Int. J. Antimicrob. Agents. 40 (3), 210–220. doi:10.1016/j.ijantimicag.2012.04.021
Courvalin, P. (1994). Transfer of antibiotic resistance genes between gram-positive and gram-negative bacteria. Antimicrob. Agents Chemother. 38 (7), 1447–1451. doi:10.1128/AAC.38.7.1447
da Silva Meira, Q. G. I., de Medeiros Barbosa, M. B., Athayde, J. P., de Siqueira-Júnior, A. J. A. A., and de Souza., E. L. (2012). Influence of temperature and surface kind on biofilm formation by Staphylococcus aureus from food-contact surfaces and sensitivity to sanitizers. Food Cont. 25 (2), 469–475. doi:10.1016/j.foodcont.2011.11.030
Dahlberg, C., Bergström, M., and Hermansson, M. (1998). In situ detection of high levels of horizontal plasmid transfer in marine bacterial communities. Appl. Environ. Microbiol. 64 (7), 2670–2675. doi:10.1128/AEM.64.7.2670-2675.1998
Dante Formagio, M., de Oliveira Silva, J. V., Fortunato Prohmann, L., Zanetti Campanerut-Sá, P. A., Grenier Capoci, I. R., Seki Kioshima Cotica, É., et al. (2022). New 1,3,4-oxadiazole compound with effective antibacterial and antibiofilm activity against Staphylococcus aureus. Lett. Appl. Microbiol. 75 (4), 957–966. doi:10.1111/lam.13766
Davies, R., and Wales, A. (2019). Antimicrobial resistance on farms: A review including biosecurity and the potential role of disinfectants in resistance selection. Compr. Rev. Food Sci. Food Saf. 18 (3), 753–774. doi:10.1111/1541-4337.12438
de Oliveira, A. M., Anjos Szczerepa, M. M. D., Bronharo Tognim, M. C., Abreu Filho, B. A., Cardozo-Filho, L., Gomes, R. G., et al. (2021). Moringa oleifera seed oil extracted by pressurized n-propane and its effect against Staphylococcus aureus biofilms. Environ. Technol. 44, 1083–1098. doi:10.1080/09593330.2021.1994653
Deghorain, M., and Van Melderen, L. (2012). The staphylococci phages family: An overview. Viruses 4 (12), 3316–3335. doi:10.3390/v4123316
Demirci, H., Murphy, F., Murphy, E., Gregory, S. T., Dahlberg, A. E., and Jogl, G. (2013). A structural basis for streptomycin-induced misreading of the genetic code. Nat. Commun. 4, 1355. doi:10.1038/ncomms2346
Derbise, A., Dyke, K. G., and el Solh, N. (1996). Characterization of a Staphylococcus aureus transposon, Tn5405, located within Tn5404 and carrying the aminoglycoside resistance genes, aphA-3 and aadE. Plasmid 35 (3), 174–188. doi:10.1006/plas.1996.0020
Di Ciccio, P., Vergara, A., Festino, A. R., Paludi, D., Zanardi, E., Ghidini, S., et al. (2015). Biofilm formation by Staphylococcus aureus on food contact surfaces: Relationship with temperature and cell surface hydrophobicity. Food control. 50, 930–936. doi:10.1016/j.foodcont.2014.10.048
Ding, Y., Onodera, Y., Lee, J. C., and Hooper, D. C. (2008). NorB, an efflux pump in Staphylococcus aureus strain MW2, contributes to bacterial fitness in abscesses. J. Bacteriol. 190 (21), 7123–7129. doi:10.1128/JB.00655-08
Dymond, A., Davies, H., Mealing, S., Pollit, V., Coll, F., Brown, N. M., et al. (2020). Genomic surveillance of methicillin-resistant Staphylococcus aureus: A mathematical early modeling study of cost-effectiveness. Clin. Infect. Dis. 70 (8), 1613–1619. doi:10.1093/cid/ciz480
Effelsberg, N., Udarcev, S., Müller, H., Kobusch, I., Linnemann, S., Boelhauve, M., et al. (2019). Genotypic characterization of livestock-associated methicillin-resistant Staphylococcus aureus isolates of clonal complex 398 in pigsty visitors: Transient carriage or persistence? J. Clin. Microbiol. 58 (1), 012766. doi:10.1128/JCM.01276-19
Emslie, K. R., Townsend, D. E., and Grubb, W. B. (1986). Isolation and characterisation of a family of small plasmids encoding resistance to nucleic acid-binding compounds in Staphylococcus aureus. J. Med. Microbiol. 22 (1), 9–15. doi:10.1099/00222615-22-1-9
Evans, J., and Dyke, K. G. (1988). Characterization of the conjugation system associated with the Staphylococcus aureus plasmid pJE1. J. Gen. Microbiol. 134 (1), 1–8. doi:10.1099/00221287-134-1-1
Fanelli, F., Chieffi, D., Cho, G. S., Schubert, J., Mekhloufi, O. A., Bania, J., et al. (2022). First genome-based characterisation and staphylococcal enterotoxin production ability of methicillin-susceptible and methicillin-resistant Staphylococcus aureus strains isolated from ready-to-eat foods in algiers (Algeria). Toxins (Basel). 14 (11), 731. doi:10.3390/toxins14110731
FAO and WHO (2019). “Joint FAO/WHO Expert meeting in collaboration with OIE on foodborne antimicrobial resistance: Role of the environment, crops and biocides – meeting report,” in Microbiological risk assessment series no. 34. Rome.
Faria, D. R., Melo, R. C., Arita, G. S., Sakita, K. M., Rodrigues-Vendramini, F. A. V., Capoci, I. R. G., et al. (2021). Fungicidal activity of a safe 1,3,4-oxadiazole derivative against Candida albicans. Pathogens 10 (3), 314. doi:10.3390/pathogens10030314
Feßler, A. T., Wang, Y., Wu, C., and Schwarz, S. (2018). Mobile macrolide resistance genes in staphylococci. Plasmid 99, 2–10. doi:10.1016/j.plasmid.2018.05.001
Fontecha-Umaña, F., Ríos-Castillo, A. G., Ripolles-Avila, C., and Rodríguez-Jerez, J. J. (2020). Antimicrobial activity and prevention of bacterial biofilm formation of silver and zinc oxide nanoparticle-containing polyester surfaces at various concentrations for use. Foods 9 (4), 442. doi:10.3390/foods9040442
Forson, A. M., van der Mei, H. C., and Sjollema, J. (2020). Impact of solid surface hydrophobicity and micrococcal nuclease production on Staphylococcus aureus Newman biofilms. Sci. Rep. 10 (1), 12093. doi:10.1038/s41598-020-69084-x
Foster, T. J. (2017). Antibiotic resistance in Staphylococcus aureus. Current status and future prospects. FEMS Microbiol. Rev. 41 (3), 430–449. doi:10.1093/femsre/fux007
Fusco, V., Chieffi, D., Fanelli, F., Logrieco, A. F., Cho, G.-S., Kabisch, J., et al. (2020). Microbial quality and safety of milk and milk products in the 21st century. Comp. Rev. Food Sci. Food Saf. 19, 2013–2049. doi:10.1111/1541-4337.12568
Fusco, V., Quero, G. M., Morea, M., Blaiotta, G., and Visconti, A. (2011). Rapid and reliable identification of Staphylococcus aureus harbouring the enterotoxin gene cluster (egc) and quantitative detection in raw milk by real time PCR. Int. J. Food Microbiol. 144, 528–537. doi:10.1016/j.ijfoodmicro.2010.11.016
Fux, C. A., Wilson, S., and Stoodley, P. (2004). Detachment characteristics and oxacillin resistance of Staphylococcus aureus biofilm emboli in an in vitro catheter infection model. J. Bacteriol. 186 (14), 4486–4491. doi:10.1128/JB.186.14.4486-4491.2004
Gerdes, K., Rasmussen, P. B., and Molin, S. (1986). Unique type of plasmid maintenance function: Postsegregational killing of plasmid-free cells. Proc. Natl. Acad. Sci. U.S.A. 83 (10), 3116–3120. doi:10.1073/pnas.83.10.3116
Gillespie, M. T., and Skurray, R. A. (1988). Structural relationships among chloramphenicol resistance plasmids of Staphylococcus aureus. FEMS Microbiol. Lett. 51, 205–210. doi:10.1111/j.1574-6968.1988.tb02998.x
Giske, C. G., Dyrkell, F., Arnellos, D., Vestberg, N., Hermansson Panna, S., Fröding, I., et al. (2019). Transmission events and antimicrobial susceptibilities of methicillin-resistant Staphylococcus argenteus in Stockholm. Clin. Microbiol. Infect. 25 (10), 1289.e5–1289.e8. doi:10.1016/j.cmi.2019.06.003
Gordon, N. C., Price, J. R., Cole, K., Everitt, R., Morgan, M., Finney, J., et al. (2014). Prediction of Staphylococcus aureus antimicrobial resistance by whole-genome sequencing. J. Clin. Microbiol. 52 (4), 1182–1191. doi:10.1128/JCM.03117-13
Goswami, C., Fox, S., Holden, M., Leanord, A., and Evans, T. J. (2021). Genomic analysis of global Staphylococcus argenteus strains reveals distinct lineages with differing virulence and antibiotic resistance gene content. Front. Microbiol. 12, 795173. doi:10.3389/fmicb.2021.795173
Grapsa, J., Blauth, C., Chandrashekhar, Y. S., Prendergast, B., Erb, B., Mack, M., et al. (2021). Staphylococcus aureus infective endocarditis: JACC patient pathways. JACC Case Rep. 4 (1), 1–12. doi:10.1016/j.jaccas.2021.10.002
Grinius, L., Dreguniene, G., Goldberg, E. B., Liao, C. H., and Projan, S. J. (1992). A staphylococcal multidrug resistance gene product is a member of a new protein family. Plasmid 27 (2), 119–129. doi:10.1016/0147-619x(92)90012-y
Grinius, L. L., and Goldberg, E. B. (1994). Bacterial multidrug resistance is due to a single membrane protein which functions as a drug pump. J. Biol. Chem. 269 (47), 29998–30004. doi:10.1016/s0021-9258(18)43980-4
Grossmann, A., Froböse, N. J., Mellmann, A., Alabi, A. S., Schaumburg, F., and Niemann, S. (2021). An in vitro study on Staphylococcus schweitzeri virulence. Sci. Rep. 11 (1), 1157. doi:10.1038/s41598-021-80961-x
Gullberg, E., Albrecht, L. M., Karlsson, C., Sandegren, L., and Andersson, D. I. (2014). Selection of a multidrug resistance plasmid by sublethal levels of antibiotics and heavy metals. mBio 5 (5), 019188. doi:10.1128/mBio.01918-14
Guo, J., Dang, J., Wang, K., Zhang, J., and Fang, J. (2018). Effects of nanosecond pulsed electric fields (nsPEFs) on the human fungal pathogen Candida albicans: An in vitro study. J. Phys. D. Appl. Phys. 51, 185402. doi:10.1088/1361-6463/aab8c8
Gutiérrez, D., Delgado, S., Vázquez-Sánchez, D., Martínez, B., Cabo, M. L., Rodríguez, A., et al. (2021). Incidence of Staphylococcus aureus and analysis of associated bacterial communities on food industry surfaces. Appl. Environ. Microbiol. 78 (24), 8547–8554. doi:10.1128/AEM.02045-12
Haaber, J., Penadés, J. R., and Ingmer, H. (2017). Transfer of antibiotic resistance in Staphylococcus aureus. Trends Microbiol. 25 (11), 893–905. doi:10.1016/j.tim.2017.05.011
Hanssen, A. M., and Sollid, J. U. E. (2006). SCC mec in staphylococci: Genes on the move. FEMS Immunol. Med. Microbiol. 46 (1), 8–20. doi:10.1111/j.1574-695x.2005.00009.x
Hao, H., Dai, M., Wang, Y., Huang, L., and Yuan, Z. (2012). Key genetic elements and regulation systems in methicillin-resistant Staphylococcus aureus. Future Microbiol. 7 (11), 1315–1329. doi:10.2217/fmb.12.107
Harada, T., Taguchi, M., Kawahara, R., Kanki, M., and Kawatsu, K. (2018). Prevalence and antimicrobial susceptibility of bacterial pathogens in ready-to-eat foods retailed in osaka prefecture, Japan. J. Food Prot. 81 (9), 1450–1458. doi:10.4315/0362-028X.JFP-18-035
Hardy, K., Sunnucks, K., Gil, H., Shabir, S., Trampari, E., Hawkey, P., et al. (2018). Increased usage of antiseptics is associated with reduced susceptibility in clinical isolates of Staphylococcus aureus. mBio 9 (3), 008944–e918. doi:10.1128/mBio.00894-18
Heir, E., Sundheim, G., and Holck, A. L. (1999). Identification and characterization of quaternary ammonium compound resistant staphylococci from the food industry. Int. J. Food Microbiol. 48 (3), 211–219. doi:10.1016/s0168-1605(99)00044-6
Held, J., Gmeiner, M., Mordmüller, B., Matsiégui, P. B., Schaer, J., Eckerle, I., et al. (2017). Bats are rare reservoirs of Staphylococcus aureus complex in Gabon. Infect. Genet. Evol. 47, 118–120. doi:10.1016/j.meegid.2016.11.022
Hendriksen, R. S., Bortolaia, V., Tate, H., Tyson, G. H., Aarestrup, F. M., and McDermott, P. F. (2019). Using genomics to track global antimicrobial resistance. Front. Publ. Health. 7, 242. doi:10.3389/fpubh.2019.00242
Hirai, J., Suzuki, H., Sakanashi, D., Kuge, Y., Kishino, T., Asai, N., et al. (2022). The first case report of community-acquired infective endocarditis due to sequence type 1223 Staphylococcus argenteus complicated with convexity subarachnoid hemorrhage. Infect. Drug Resist. 15, 4963–4970. doi:10.2147/IDR.S373352
Hiramatsu, K., Cui, L., Kuroda, M., and Ito, T. (2001). The emergence and evolution of methicillin-resistant Staphylococcus aureus. Trends Microbiol. 9 (10), 486–493. doi:10.1016/s0966-842x(01)02175-8
Ho, J., Boost, M. V., and O'Donoghue, M. M. (2015). Tracking sources of Staphylococcus aureus hand contamination in food handlers by spa typing. Am. J. Infect. control 43 (7), 759–761. doi:10.1016/j.ajic.2015.03.022
Horinouchi, S., and Weisblum, B. (1982). Nucleotide sequence and functional map of pC194, a plasmid that specifies inducible chloramphenicol resistance. J. Bacteriol. 150, 815–825. doi:10.1128/jb.150.2.815-825.1982
Htun, H. L., Hon, P. Y., Holden, M. T. G., Ang, B., and Chow, A. (2019). Chlorhexidine and octenidine use, carriage of qac genes, and reduced antiseptic susceptibility in methicillin-resistant Staphylococcus aureus isolates from a healthcare network. Clin. Microbiol. Infect. 25 (9), 1154.e1–1154. doi:10.1016/j.cmi.2018.12.036
Idrees, M., Sawant, S., Karodia, N., and Rahman, A. (2021). Staphylococcus aureus biofilm: Morphology, genetics, pathogenesis and treatment strategies. Int. J. Environ. Res. Public health 18 (14), 7602. doi:10.3390/ijerph18147602
Ifesan, B. O., Hamtasin, C., Mahabusarakam, W., and Voravuthikunchai, S. P. (2009). Inhibitory effect of Eleutherine americana Merr. extract on Staphylococcus aureus isolated from food. J. Food Sci. 74 (1), M31–M36. doi:10.1111/j.1750-3841.2008.01004.x
Indrawattana, N., Pumipuntu, N., Suriyakhun, N., Jangsangthong, A., Kulpeanprasit, S., Chantratita, N., et al. (2019). Staphylococcus argenteus from rabbits in Thailand. MicrobiologyOpen 8 (4), e00665. doi:10.1002/mbo3.665
Jensen, S. O., and Lyon, B. R. (2009). Genetics of antimicrobial resistance in Staphylococcus aureus. Fut. Microbiol. 4 (5), 565–582. doi:10.2217/fmb.09.30
Jiang, B., You, B., Tan, L., Yu, S., Li, H., Bai, G., et al. (2018). Clinical Staphylococcus argenteus develops to small colony variants to promote persistent infection. Front. Microbiol. 9, 1347. doi:10.3389/fmicb.2018.01347
Johnston, L. H., and Dyke, K. G. (1969). Ethidium bromide resistance, a new marker on the staphylococcal penicillinase plasmid. J. Bacteriol. 100 (3), 1413–1414. doi:10.1128/jb.100.3.1413-1414.1969
Jones, I. A., and Joshi, L. T. (2021). Biocide use in the antimicrobial era: A review. Molecules 26 (8), 2276. doi:10.3390/molecules26082276
Kaiser-Thom, S., Gerber, V., Collaud, A., Hurni, J., and Perreten, V. (2022). Prevalence and WGS-based characteristics of Staphylococcus aureus in the nasal mucosa and pastern of horses with equine pastern dermatitis. BMC Vet. Res. 18 (1), 79. doi:10.1186/s12917-021-03053-y
Khan, S. I., Blumrosen, G., Vecchio, D., Golberg, A., McCormack, M. C., Yarmush, M. L., et al. (2016). Eradication of multidrug-resistant pseudomonas biofilm with pulsed electric fields. Biotechnol. Bioeng. 113 (3), 643–650. doi:10.1002/bit.25818
Kim, M. J., Lim, E. S., and Kim, J. S. (2019). Enzymatic inactivation of pathogenic and nonpathogenic bacteria in biofilms in combination with chlorine. J. Food Prot. 82 (4), 605–614. doi:10.4315/0362-028X.JFP-18-244
Kim, S. H., Park, S. H., Kim, S. S., and Kang, D. H. (2019). Inactivation of Staphylococcus aureus biofilms on food contact surfaces by superheated steam treatment. J. Food Prot. 82 (9), 1496–1500. doi:10.4315/0362-028X.JFP-18-572
Kinnevey, P. M., Kearney, A., Shore, A. C., Earls, M. R., Brennan, G., Poovelikunnel, T. T., et al. (2021). Methicillin-resistant Staphylococcus aureus transmission among healthcare workers, patients and the environment in a large acute hospital under non-outbreak conditions investigated using whole-genome sequencing. J. Hosp. Infect. 118, 99–107. doi:10.1016/j.jhin.2021.08.020
Knight, G. M., Budd, E. L., Whitney, L., Thornley, A., Al-Ghusein, H., Planche, T., et al. (2012). Shift in dominant hospital-associated methicillin-resistant Staphylococcus aureus (HA-MRSA) clones over time. J. Antimicrob. Chemother. 67 (10), 2514–2522. doi:10.1093/jac/dks245
Konstantinovski, M. M., Veldkamp, K. E., Lavrijsen, A. P. M., Bosch, T., Kraakman, M. E. M., Nooij, S., et al. (2021). Hospital transmission of borderline oxacillin-resistant Staphylococcus aureus evaluated by whole-genome sequencing. J. Med. Microbiol. 70 (7), 001384. doi:10.1099/jmm.0.001384
Koohestani, M., Moradi, M., Tajik, H., and Badali, A. (2018). Effects of cell-free supernatant of Lactobacillus acidophilus LA5 and Lactobacillus casei 431 against planktonic form and biofilm of Staphylococcus aureus. Vet. Res. Forum. 9 (4), 301–306. doi:10.30466/vrf.2018.33086
Kroning, I. S., Iglesias, M. A., Sehn, C. P., Valente Gandra, T. K., Mata, M. M., and da Silva, W. P. (2016). Staphylococcus aureus isolated from handmade sweets: Biofilm formation, enterotoxigenicity and antimicrobial resistance. Food Microbiol. 58, 105–111. doi:10.1016/j.fm.2016.04.001
Kukułowicz, A., Steinka, I., and Siwek, A. (2021). Presence of antibiotic-resistant Staphylococcus aureus in fish and seafood originating from points of sale in the tri-city area (Poland). J. Food Prot. 84 (11), 1911–1914. doi:10.4315/JFP-21-115
Lacey, K. A., Geoghegan, J. A., and McLoughlin, R. M. (2016). The role of Staphylococcus aureus virulence factors in skin infection and their potential as vaccine antigens. Pathogens 5 (1), 22. doi:10.3390/pathogens5010022
Lacey, R. W., and Chopra, I. (1972). Evidence for mutation to streptomycin resistance in clinical strains of Staphylococcus aureus. J. Gen. Microbiol. 73 (1), 175–180. doi:10.1099/00221287-73-1-175
Lagos, A. C., Sundqvist, M., Dyrkell, F., Stegger, M., Söderquist, B., and Mölling, P. (2022). Evaluation of within-host evolution of methicillin-resistant Staphylococcus aureus (MRSA) by comparing cgMLST and SNP analysis approaches. Sci. Rep. 12 (1), 10541. doi:10.1038/s41598-022-14640-w
Lakhanpal, P., Panda, A. K., Chahota, R., Choudhary, S., and Thakur, S. D. (2019). Incidence and antimicrobial susceptibility of Staphylococcus aureus isolated from ready-to-eat foods of animal origin from tourist destinations of North-western Himalayas, Himachal Pradesh, India. J. Food Sci. Technol. 56 (2), 1078–1083. doi:10.1007/s13197-018-03556-x
Le, K. Y., and Otto, M. (2015). Quorum-sensing regulation in staphylococci-an overview. Front. Microbiol. 6, 1174. doi:10.3389/fmicb.2015.01174
Lee, J. S., Bae, Y. M., Lee, S. Y., and Lee, S. Y. (2015). Biofilm Formation of Staphylococcus aureus on various surfaces and their resistance to chlorine sanitizer. J. Food Sci. 80 (10), M2279–M2286. doi:10.1111/1750-3841.13017
Leelaporn, A., Paulsen, I. T., Tennent, J. M., Littlejohn, T. G., and Skurray, R. A. (1994). Multidrug resistance to antiseptics and disinfectants in coagulase-negative staphylococci. J. Med. Microbiol. 40 (3), 214–220. doi:10.1099/00222615-40-3-214
Leijon, M., Atkins, E., Persson Waller, K., and Artursson, K. (2021). Longitudinal study of Staphylococcus aureus genotypes isolated from bovine clinical mastitis. J. Dairy Sci. 104 (11), 11945–11954. doi:10.3168/jds.2021-20562
Leopold, S. R., Goering, R. V., Witten, A., Harmsen, D., and Mellmann, A. (2014). Bacterial whole-genome sequencing revisited: Portable, scalable, and standardized analysis for typing and detection of virulence and antibiotic resistance genes. J. Clin. Microbiol. 52 (7), 2365–2370. doi:10.1128/JCM.00262-14
Li, H., Stegger, M., Dalsgaard, A., and Leisner, J. J. (2019). Bacterial content and characterization of antibiotic resistant Staphylococcus aureus in Danish sushi products and association with food inspector rankings. Int. J. Food Microbiol. 305, 108244. doi:10.1016/j.ijfoodmicro.2019.108244
Li, X., Zhang, J., Zhang, H., Shi, X., Wang, J., Li, K., et al. (2022). Genomic analysis, antibiotic resistance, and virulence of Staphylococcus aureus from food and food outbreaks: A potential public concern. Int. J. Food Microbiol. 377, 109825. doi:10.1016/j.ijfoodmicro.2022.109825
Lin, Q., Sun, H., Yao, K., Cai, J., Ren, Y., and Chi, Y. (2019). The prevalence, antibiotic resistance and biofilm formation of Staphylococcus aureus in bulk ready-to-eat foods. Biomolecules 9 (10), 524. doi:10.3390/biom9100524
Lindsay, J. A. (2010). Genomic variation and evolution of Staphylococcus aureus. Int. J. Med. Microbiol. 300 (2-3), 98–103. doi:10.1016/j.ijmm.2009.08.013
Lindsay, J. A., Knight, G. M., Budd, E. L., and McCarthy, A. J. (2012). Shuffling of mobile genetic elements (MGEs) in successful healthcare-associated MRSA (HA-MRSA). Mob. Genet. Elem. 2 (5), 239–243. doi:10.4161/mge.22085
Littlejohn, T. G., DiBerardino, D., Messerotti, L. J., Spiers, S. J., and Skurray, R. A. (1991). Structure and evolution of a family of genes encoding antiseptic and disinfectant resistance in Staphylococcus aureus. Gene 101, 59–66. doi:10.1016/0378-1119(91)90224-y
Liu, J., Chen, D., Peters, B. M., Li, L., Li, B., Xu, Z., et al. (2016). Staphylococcal chromosomal cassettes mec (SCCmec): A mobile genetic element in methicillin-resistant Staphylococcus aureus. Microb. Pathog. 101, 56–67. doi:10.1016/j.micpath.2016.10.028
Liu, J., Li, W., Zhu, X., Zhao, H., Lu, Y., Zhang, C., et al. (2019). Surfactin effectively inhibits Staphylococcus aureus adhesion and biofilm formation on surfaces. Appl. Microbiol. Biotechnol. 103 (11), 4565–4574. doi:10.1007/s00253-019-09808-w
Loll, P. J., and Axelsen, P. H. (2000). The structural biology of molecular recognition by vancomycin. Annu. Rev. Biophys. Biomol. Struct. 29, 265–289. doi:10.1146/annurev.biophys.29.1.265
Loncaric, I., Lepuschitz, S., Ruppitsch, W., Trstan, A., Andreadis, T., Bouchlis, N., et al. (2019). Increased genetic diversity of methicillin-resistant Staphylococcus aureus (MRSA) isolated from companion animals. Vet. Microbiol. 235, 118–126. doi:10.1016/j.vetmic.2019.06.013
Lowry, D. (2003). Antimicrobial resistance: The example of Staphylococcus aureus. J. Clin. Investig. 111, 1265–1273. doi:10.1172/JCI18535
Luo, K., Shao, F., Kamara, K. N., Chen, S., Zhang, R., Duan, G., et al. (2018). Molecular characteristics of antimicrobial resistance and virulence determinants of Staphylococcus aureus isolates derived from clinical infection and food. Clin. Lab. Anal. 32 (7), e22456. doi:10.1002/jcla.22456
Lyon, B. R., May, J. W., and Skurray, R. A. (1984). Tn4001: A gentamicin and kanamycin resistance transposon in Staphylococcus aureus. Mol. Gen. Genet. 193 (3), 554–556. doi:10.1007/BF00382099
Lyon, B. R., and Skurray, R. (1987). Antimicrobial resistance of Staphylococcus aureus: Genetic basis. Microbiol. Rev. 51 (1), 88–134. doi:10.1128/mr.51.1.88-134.1987
Machado, V., Pardo, L., Cuello, D., Giudice, G., Luna, P. C., Varela, G., et al. (2020). Presence of genes encoding enterotoxins in Staphylococcus aureus isolates recovered from food, food establishment surfaces and cases of foodborne diseases. Rev. Inst. Med. Trop. Sao Paulo. 62, e5. doi:10.1590/S1678-9946202062005
MacKinnon, M. M., and Allen, K. D. (2000). Long-term MRSA carriage in hospital patients. J. Hosp. Infect. 46 (3), 216–221. doi:10.1053/jhin.2000.0807
Madigan, T., Cunningham, S. A., Patel, R., Greenwood-Quaintance, K. E., Barth, J. E., Sampathkumar, P., et al. (2018). Whole-genome sequencing for methicillin-resistant Staphylococcus aureus (MRSA) outbreak investigation in a neonatal intensive care unit. Infect. Control Hosp. Epidemiol. 39 (12), 1412–1418. doi:10.1017/ice.2018.239
Mahros, M. A., Abd-Elghany, S. M., and Sallam, K. I. (2021). Multidrug-methicillin-and vancomycin-resistant Staphylococcus aureus isolated from ready-to-eat meat sandwiches: An ongoing food and public health concern. Int. J. Food Microbiol. 346, 109165. doi:10.1016/j.ijfoodmicro.2021.109165
Manguiat, L. S., and Fang, T. J. (2013). Microbiological quality of chicken- and pork-based street-vended foods from Taichung, Taiwan, and Laguna, Philippines. Food Microbiol. 36 (1), 57–62. doi:10.1016/j.fm.2013.04.005
Martin, J. G., de Oliveira E Silva, G., da Fonseca, C. R., Morales, C. B., Souza Pamplona Silva, C., Miquelluti, D. L., et al. (2016). Efficiency of a cleaning protocol for the removal of enterotoxigenic Staphylococcus aureus strains in dairy plants. Int. J. Food Microbiol. 238, 295–301. doi:10.1016/j.ijfoodmicro.2016.09.018
Mason, A., Foster, D., Bradley, P., Golubchik, T., Doumith, M., Gordon, N. C., et al. (2018). Accuracy of different bioinformatics methods in detecting antibiotic resistance and virulence factors from Staphylococcus aureus whole-genome sequences. J. Clin. Microbiol. 56 (9), e01815-17–e01817. doi:10.1128/JCM.01815-17
Mastoor, S., Nazim, F., Rizwan-Ul-Hasan, S., Ahmed, K., Khan, S., Ali, S. N., et al. (2022). Analysis of the antimicrobial and anti-biofilm activity of natural compounds and their analogues against Staphylococcus aureus isolates. Molecules 27 (20), 6874. doi:10.3390/molecules27206874
McCarthy, A. J., Loeffler, A., Witney, A. A., Gould, K. A., Lloyd, D. H., and Lindsay, J. A. (2014). Extensive horizontal gene transfer during Staphylococcus aureus co-colonization in vivo. Genome Biol. Evol. 6, 2697–2708. doi:10.1093/gbe/evu214
McGuinness, W. A., Malachowa, N., and DeLeo, F. R. (2017). Vancomycin resistance in Staphylococcus aureus. Yale J. Biol. Med. 90 (2), 269–281.
Meijer, E. F. J., van Renssen, A., Maat, I., van der Graaf-van Bloois, L., Duim, B., and Broens, E. M. (2022). Canine Staphylococcus argenteus: Case report from The Netherlands. Pathogens 11 (2), 153. doi:10.3390/pathogens11020153
Mekhloufi, O. A., Chieffi, D., Hammoudi, A., Bensefia, S. A., Fanelli, F., and Fusco, V. (2021). Prevalence, enterotoxigenic potential and antimicrobial resistance of Staphylococcus aureus and methicillin-resistant Staphylococcus aureus (MRSA) isolated from Algerian ready to eat foods. Toxins (Basel). 13 (12), 835. doi:10.3390/toxins13120835
Mesbah, A., Mashak, Z., and Abdolmaleki, Z. (2021). A survey of prevalence and phenotypic and genotypic assessment of antibiotic resistance in Staphylococcus aureus bacteria isolated from ready-to-eat food samples collected from Tehran Province, Iran. Iran. Trop. Med. Health 49 (1), 81. doi:10.1186/s41182-021-00366-4
Mikhaylova, Y., Shelenkov, A., Chernyshkov, A., Tyumentseva, M., Saenko, S., Egorova, A., et al. (2022). Whole-genome analysis of Staphylococcus aureus isolates from ready-to-eat food in Russia. Foods 11 (17), 2574. doi:10.3390/foods11172574
Mlynarczyk-Bonikowska, B., Kowalewski, C., Krolak-Ulinska, A., and Marusza, W. (2022). Molecular mechanisms of drug resistance in Staphylococcus aureus. Int. J. Mol. Sci. 23 (15), 8088. doi:10.3390/ijms23158088
Monecke, S., Schaumburg, F., Shittu, A. O., Schwarz, S., Mühldorfer, K., Brandt, C., et al. (2022). Description of staphylococcal strains from straw-coloured fruit bat (Eidolon helvum) and diamond firetail (Stagonopleura guttata) and a review of their phylogenetic relationships to other staphylococci. Front. Cell. Infect. Microbiol. 12, 878137. doi:10.3389/fcimb.2022.878137
Nan, L., Yang, K., and Ren, G. (2015). Anti-biofilm formation of a novel stainless steel against Staphylococcus aureus. Mat. Sci. Eng. C Mat. Biol. Appl. 51, 356–361. doi:10.1016/j.msec.2015.03.012
Ndahetuye, J. B., Leijon, M., Båge, R., Artursson, K., and Persson, Y. (2021). Genetic characterization of Staphylococcus aureus from subclinical mastitis cases in dairy cows in Rwanda. Front. Vet. Sci. 8, 751229. doi:10.3389/fvets.2021.751229
Novick, R. P., Christie, G. E., and Penadés, J. R. (2010). The phage-related chromosomal islands of Gram-positive bacteria. Nat. Rev. Microbiol. 8 (8), 541–551. doi:10.1038/nrmicro2393
Ohnishi, T., Shinjoh, M., Ohara, H., Kawai, T., Kamimaki, I., Mizushima, R., et al. (2018). Purulent lymphadenitis caused by Staphylococcus argenteus, representing the first Japanese case of Staphylococcus argenteus (multilocus sequence type 2250) infection in a 12-year-old boy. J. Infect. Chemother. 24 (11), 925–927. doi:10.1016/j.jiac.2018.03.018
Olatimehin, A., Shittu, A. O., Onwugamba, F. C., Mellmann, A., Becker, K., and Schaumburg, F. (2018). Staphylococcus aureus complex in the straw-colored fruit bat (Eidolon helvum) in Nigeria. Front. Microbiol. 9, 162. doi:10.3389/fmicb.2018.00162
Oniciuc, E.-A., Nicolau, A. I., Hernández, M., and Rodríguez-Lázaro, D. (2017). Presence of methicillin-resistant Staphylococcus aureus in the food chain. Trends Food Sci. Technol. 61, 49–59. doi:10.1016/j.tifs.2016.12.002
Ozawa, M., Furuya, Y., Akama, R., Harada, S., Matsuda, M., Abo, H., et al. (2022). Molecular epidemiology of methicillin-resistant Staphylococcus aureus isolated from pigs in Japan. Vet. Microbiol. 273, 109523. doi:10.1016/j.vetmic.2022.109523
Pagedar, A., Singh, J., and Batish, V. K. (2010). Surface hydrophobicity, nutritional contents affect Staphylococcus aureus biofilms and temperature influences its survival in preformed biofilms. J. Basic Microbiol. 50, S98–S106. doi:10.1002/jobm.201000034
Pal, C., Bengtsson-Palme, J., Kristiansson, E., and Larsson, D. G. (2015). Co-occurrence of resistance genes to antibiotics, biocides and metals reveals novel insights into their co-selection potential. BMC Genomics 16, 964. doi:10.1186/s12864-015-2153-5
Park, K. H., Greenwood-Quaintance, K. E., Uhl, J. R., Cunningham, S. A., Chia, N., Jeraldo, P. R., et al. (2017). Molecular epidemiology of Staphylococcus aureus bacteremia in a single large Minnesota medical center in 2015 as assessed using MLST, core genome MLST and spa typing. PLoS One 12 (6), e0179003. doi:10.1371/journal.pone.0179003
Paul, D., Chakraborty, R., and Mandal, S. M. (2019). Biocides and health-care agents are more than just antibiotics: Inducing cross to co-resistance in microbes. Ecotoxicol. Environ. Saf. 174, 601–610. doi:10.1016/j.ecoenv.2019.02.083
Paulsen, I. T., Brown, M. H., Littlejohn, T. G., Mitchell, B. A., and Skurray, R. A. (1996). Multidrug resistance proteins QacA and QacB from Staphylococcus aureus: Membrane topology and identification of residues involved in substrate specificity. Proc. Natl. Acad. Sci. U.S.A. 93, 3630–3635. doi:10.1073/pnas.93.8.3630
Paulsen, I. T., Littlejohn, T. G., Rådström, P., Sundström, L., Sköld, O., Swedberg, G., et al. (1993). The 3' conserved segment of integrons contains a gene associated with multidrug resistance to antiseptics and disinfectants. Antimicrob. Agents Chemother. 37 (4), 761–768. doi:10.1128/AAC.37.4.761
Pedonese, F., Fratini, F., Pistelli, L., Porta, F. M., Ciccio, P. D., Fischetti, R., et al. (2017). Antimicrobial activity of four essential oils against pigmenting Pseudomonas fluorescens and biofilmproducing Staphylococcus aureus of dairy origin. Ital. J. Food Saf. 6 (4), 6939. doi:10.4081/ijfs.2017.6939
Penadés, J. R., and Christie, G. E. (2015). The phage-inducible chromosomal islands: A family of highly evolved molecular parasites. Annu. Rev. Virol. 2 (1), 181–201. doi:10.1146/annurev-virology-031413-085446
Peng, Q., Tang, X., Dong, W., Sun, N., and Yuan, W. (2022). A review of biofilm formation of Staphylococcus aureus and its regulation mechanism. Antibiot. (Basel) 12 (1), 12. doi:10.3390/antibiotics12010012
Peters, K. M., Sharbeen, G., Theis, T., Skurray, R. A., and Brown, M. H. (2009). Biochemical characterization of the multidrug regulator QacR distinguishes residues that are crucial to multidrug binding and induction of qacA transcription. Biochemistry 48 (41), 9794–9800. doi:10.1021/bi901102h
Phiri, B. S. J., Hang'ombe, B. M., Mulenga, E., Mubanga, M., Maurischat, S., Wichmann-Schauer, H., et al. (2022). Prevalence and diversity of Staphylococcus aureus in the Zambian dairy value chain: A public health concern. Int. J. Food Microbiol. 375, 109737. doi:10.1016/j.ijfoodmicro.2022.109737
Plaza-Rodríguez, C., Kaesbohrer, A., and Tenhagen, B. A. (2019). Probabilistic model for the estimation of the consumer exposure to methicillin-resistant Staphylococcus aureus due to cross-contamination and recontamination. MicrobiologyOpen 8 (11), e900. doi:10.1002/mbo3.900
Projan, S. J., Kornblum, J., Moghazeh, S. L., Edelman, I., Gennaro, M. L., and Novick, R. P. (1985). Comparative sequence and functional analysis of pT181 and pC221, cognate plasmid replicons from Staphylococcus aureus. Mol. Gen. Genet. 199, 452–464. doi:10.1007/BF00330758
Projan, S. J., Moghazeh, S., and Novick, R. P. (1988). Nucleotide sequence of pS194, a streptomycin-resistance plasmid from Staphylococcus aureus. Nucleic Acids Res. 16 (5), 2179–2187. doi:10.1093/nar/16.5.2179
Pumipuntu, N., Tunyong, W., Chantratita, N., Diraphat, P., Pumirat, P., Sookrung, N., et al. (2019). Staphylococcus spp. associated with subclinical bovine mastitis in central and northeast provinces of Thailand. PeerJ 7, e6587. doi:10.7717/peerj.6587
Qi, M., Liu, Q., Liu, Y., Yan, H., Zhang, Y., and Yuan, Y. (2022). Staphylococcus aureus biofilm inhibition by high voltage prick electrostatic field (HVPEF) and the mechanism investigation. Int. J. Food Microbiol. 362, 109499. doi:10.1016/j.ijfoodmicro.2021.109499
Qi, M., Zhao, R., Liu, Q., Yan, H., Zhang, Y., Wang, S., et al. (2021). Antibacterial activity and mechanism of high voltage electrostatic field (HVEF) against Staphylococcus aureus in medium plates and food systems. Food Cont. 120, 107566. doi:10.1016/j.foodcont.2020.107566
Rashid, N., Shafee, M., Iqbal, S., Samad, A., Khan, S. A., Hasni, M. S., et al. (2021). Enterotoxigenic methicillin resistant Staphylococcus aureus contamination in salted fish from Gwadar Balochistan. Braz. J. Biol. 83, e247701. doi:10.1590/1519-6984.247701
Rasmi, A. H., Ahmed, E. F., Darwish, A. M. A., and Gad, G. F. M. (2022). Virulence genes distributed among Staphylococcus aureus causing wound infections and their correlation to antibiotic resistance. BMC Infect. Dis. 22, 652. doi:10.1186/s12879-022-07624-8
Roberts, M. C., Sutcliffe, J., Courvalin, P., Jensen, L. B., Rood, J., and Seppala, H. (1999). Nomenclature for macrolide and macrolide-lincosamide-streptogramin B resistance determinants. Antimicrob. Agents Chemother. 43 (12), 2823–2830. doi:10.1128/AAC.43.12.2823
Rouch, D. A., Byrne, M. E., Kong, Y. C., and Skurray, R. A. (1987). The aacA-aphD gentamicin and kanamycin resistance determinant of Tn4001 from Staphylococcus aureus: Expression and nucleotide sequence analysis. J. Gen. Microbiol. 133 (11), 3039–3052. doi:10.1099/00221287-133-11-3039
Rouch, D. A., Cram, D. S., DiBerardino, D., Littlejohn, T. G., and Skurray, R. A. (1990). Efflux-mediated antiseptic resistance gene qacA from Staphylococcus aureus: Common ancestry with tetracycline- and sugar-transport proteins. Mol. Microbiol. 4 (12), 2051–2062. doi:10.1111/j.1365-2958.1990.tb00565.x
Russell, A. D. (2002). Introduction of biocides into clinical practice and the impact on antibiotic-resistant bacteria. J. Appl. Microbiol. 31, 121S–135S. doi:10.1046/j.1365-2672.92.5s1.12.x
Sansevere, E. A., and Robinson, D. A. (2017). Staphylococci on ICE: Overlooked agents of horizontal gene transfer. Mob. Genet. Elem. 7 (4), 1–10. doi:10.1080/2159256X.2017.1368433
Sarwar, S., Saleem, S., Shahzad, F., and Jahan, S. (2022). Identifying and elucidating the resistance of Staphylococcus aureus isolated from hospital environment to conventional disinfectants. Am. J. Infect. Control. S0196-6553 (22), 178–183. doi:10.1016/j.ajic.2022.05.018
Sasatsu, M., Shima, K., Shibata, Y., and Kono, M. (1989). Nucleotide sequence of a gene that encodes resistance to ethidium bromide from a transferable plasmid in Staphylococcus aureus. Nucleic Acids Res. 17, 10103. doi:10.1093/nar/17.23.10103
Savage, V. J., Chopra, I., and O'Neill, A. J. (2013). Staphylococcus aureus biofilms promote horizontal transfer of antibiotic resistance. Antimicrob. Agents Chemother. 57 (4), 1968–1970. doi:10.1128/AAC.02008-12
SCENIHR (Scientific Committee on Emerging and Newly Identified Health Risks) (2009). Assessment of the antibiotic resistance effects of biocides. Brussels: European Commission, 1–87.
Schaumburg, F., Pauly, M., Anoh, E., Mossoun, A., Wiersma, L., Schubert, G., et al. (2015). Staphylococcus aureus complex from animals and humans in three remote African regions. Clin. Microbiol. Infect. 21 (4), 345.e1–e8. doi:10.1016/j.cmi.2014.12.001
Schlünzen, F., Zarivach, R., Harms, J., Bashan, A., Tocilj, A., Albrecht, R., et al. (2001). Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature 413 (6858), 814–821. doi:10.1038/35101544
Scholtzek, A. D., Hanke, D., Walther, B., Eichhorn, I., Stöckle, S. D., Klein, K. S., et al. (2019). Molecular characterization of equine Staphylococcus aureus isolates exhibiting reduced oxacillin susceptibility. Toxins (Basel) 11 (9), 535. doi:10.3390/toxins11090535
Schuster, D., Rickmeyer, J., Gajdiss, M., Thye, T., Lorenzen, S., Reif, M., et al. (2017). Differentiation of Staphylococcus argenteus (formerly: Staphylococcus aureus clonal complex 75) by mass spectrometry from S. aureus using the first strain isolated from a wild african great ape. Int. J. Med. Microbiol. 307 (1), 57–63. doi:10.1016/j.ijmm.2016.11.003
Schutte, A. H. J., Strepis, N., Zandijk, W. H. A., Bexkens, M. L., Bode, L. G. M., and Klaassen, C. H. W. (2021). Characterization of Staphylococcus roterodami sp. nov., a new species within the Staphylococcus aureus complex isolated from a human foot infection. Int. J. Syst. Evol. Microbiol. 71 (9). doi:10.1099/ijsem.0.004996,
Schwarz, S., and Grölz-Krug, S. (1991). A chloramphenicol-streptomycin resistance plasmid from a clinical strain of Staphylococcus sciuri and its structural relationships to other staphylococcal resistance plasmids. FEMS Microbiol. Lett. 66, 319–322. doi:10.1016/0378-1097(91)90281-e
Schwarz, S., Kehrenberg, C., Doublet, B., and Cloeckaert, A. (2004). Molecular basis of bacterial resistance to chloramphenicol and florfenicol. FEMS Microbiol. Rev. 28 (5), 519–542. doi:10.1016/j.femsre.2004.04.001
Schwendener, S., Donà, V., and Perreten, V. (2020). The novel macrolide resistance genes mef(D), msr(F), and msr(H) are present on resistance islands in Macrococcus canis, Macrococcus caseolyticus, and Staphylococcus aureus. Antimicrob. Agents Chemother. 64 (5), 00160–e220. doi:10.1128/AAC.00160-20
Shi, X., Zhang, D. F., Li, X., Wang, M., and Qin, R. (2018). Staphylococcus argenteus: An emerging foodborne pathogen? Curr. Opin. Food Sci. 20, 76–85. doi:10.1016/j.canlet.2018.06.015
Shittu, A. O., Mellmann, A., and Schaumburg, F. (2020). Molecular characterization of Staphylococcus aureus complex from fomites in Nigeria. Infect. Genet. Evol. 85, 104504. doi:10.1016/j.meegid.2020.104504
Silva, M., Machado, M. P., Silva, D. N., Rossi, M., Moran-Gilad, J., Santos, S., et al. (2018). chewBBACA: A complete suite for gene-by-gene schema creation and strain identification. Microb. Genom 4 (3), e000166. doi:10.1099/mgen.0.000166
Silva-de-Jesus, A. C., Ferrari, R. G., Panzenhagen, P., and Conte-Junior, C. A. (2022). Staphylococcus aureus biofilm: The role in disseminating antimicrobial resistance over the meat chain. Microbiol. Read. 168 (10). doi:10.1099/mic.0.001245
Slingerland, B. C. G. C., Vos, M. C., Bras, W., Kornelisse, R. F., De Coninck, D., van Belkum, A., et al. (2020). Whole-genome sequencing to explore nosocomial transmission and virulence in neonatal methicillin-susceptible Staphylococcus aureus bacteremia. Antimicrob. Resist. Infect. Control. 9 (1), 39. doi:10.1186/s13756-020-0699-8
Slott Jensen, M. L., Nielsine Skov, M., Pries Kristiansen, H., Toft, A., Lundgaard, H., Gumpert, H., et al. (2020). Core genome multi-locus sequence typing as an essential tool in a high-cost livestock-associated meticillin-resistant Staphylococcus aureus CC398 hospital outbreak. J. Hosp. Infect. 104 (4), 574–581. doi:10.1016/j.jhin.2019.12.009
Smith, D. L., Dushoff, J., Perencevich, E. N., Harris, A. D., and Levin, S. A. (2004). Persistent colonization and the spread of antibiotic resistance in nosocomial pathogens: Resistance is a regional problem. Proc. Natl. Acad. Sci. U.S.A. 101 (10), 3709–3714. doi:10.1073/pnas.0400456101
Smith, M. C., and Thomas, C. D. (2004). An accessory protein is required for relaxosome formation by small staphylococcal plasmids. J. Bacteriol. 186 (11), 3363–3373. doi:10.1128/JB.186.11.3363-3373.2004
Sri Prabakusuma, A., Zhu, J., Shi, Y., Ma, Q., Zhao, Q., Yang, Z., et al. (2022). Prevalence and antimicrobial resistance profiling of Staphylococcus aureus isolated from traditional cheese in Yunnan, China. 3 Biotech. 12 (1), 1. doi:10.1007/s13205-021-03072-4
Stanczak-Mrozek, K. I., Manne, A., Knight, G. M., Gould, K., Witney, A. A., and Lindsay, J. A. (2015). Within-host diversity of MRSA antimicrobial resistances. J. Antimicrob. Chemother. 70 (8), 2191–2198. doi:10.1093/jac/dkv119
Stiffler, P. W., Sweeney, H. M., Schneider, M., and Cohen, S. (1974). Isolation and characterization of a kanamycin resistance plasmid from Staphylococcus aureus. Antimicrob. Agents Chemother. 6 (4), 516–520. doi:10.1128/AAC.6.4.516
Stokes, H. W., and Hall, R. M. (1991). Sequence analysis of the inducible chloramphenicol resistance determinant in the Tn1696 integron suggests regulation by translational attenuation. Plasmid 26, 10–19. doi:10.1016/0147-619x(91)90032-r
Sun, D., Jeannot, K., Xiao, Y., and Knapp, C. W. (2019). Editorial: Horizontal gene transfer mediated bacterial antibiotic resistance. Front. Microbiol. 10, 1933. doi:10.3389/fmicb.2019.01933
Sun, J. L., Zhang, S. K., Chen, J. Y., and Han, B. Z. (2012). Efficacy of acidic and basic electrolyzed water in eradicating Staphylococcus aureus biofilm. Can. J. Microbiol. 58 (4), 448–454. doi:10.1139/w2012-005
Suzuki, Y., Kubota, H., Ono, H. K., Kobayashi, M., Murauchi, K., Kato, R., et al. (2017). Food poisoning outbreak in Tokyo, Japan caused by Staphylococcus argenteus. Int. J. Food Microbiol. 262, 31–37. doi:10.1016/j.ijfoodmicro.2017.09.005
Tanaka, M., Wang, T., Onodera, Y., Uchida, Y., and Sato, K. (2000). Mechanism of quinolone resistance in Staphylococcus aureus. J. Infect. Chemother. 6 (3), 131–139. doi:10.1007/s101560070010
Tegegne, H. A., Koláčková, I., Florianová, M., Wattiau, P., Gelbíčová, T., Boland, C., et al. (2021). Genomic insights into methicillin-resistant Staphylococcus aureus spa type t899 isolates belonging to different sequence types. Appl. Environ. Microbiol. 87 (6), 01994–e2020. doi:10.1128/AEM.01994-20
Tennent, J. M., Lyon, B. R., Midgley, M., Jones, I. G., Purewal, A. S., and Skurray, R. A. (1989). Physical and biochemical characterization of the qacA gene encoding antiseptic and disinfectant resistance in Staphylococcus aureus. J. Gen. Microbiol. 135 (1), 1–10. doi:10.1099/00221287-135-1-1
Thompson, T. A., and Brown, P. D. (2017). Association between the agr locus and the presence of virulence genes and pathogenesis in Staphylococcus aureus using a Caenorhabditis elegans model. Int. J. Infect. Dis. 54, 72–76. doi:10.1016/j.ijid.2016.11.411
Tong, S. Y. C., Schaumburg, F., Ellington, M. J., Corander, J., Pichon, B., Leendertz, F., et al. (2015). Novel staphylococcal species that form part of a Staphylococcus aureus-related complex: The non-pigmented Staphylococcus argenteus sp. nov. And the non-human primate-associated Staphylococcus schweitzeri sp. nov. Int. J. Syst. Evol. Microbiol. 65, 15–22. doi:10.1099/ijs.0.062752-0
Touimi, G. B., Bennani, L., Berrada, S., Moussa, B., and Bennani, B. (2020). Prevalence and antibiotic resistance profiles of Staphylococcus sp. isolated from food, food contact surfaces and food handlers in a Moroccan hospital kitchen. Lett. Appl. Microbiol. 70 (4), 241–251. doi:10.1111/lam.13278
Trieber, C. A., and Taylor, D. E. (2002). Mutations in the 16S ribosomal RNA genes of Helicobacter pylori mediate resistance to tetracycline. J. Bacteriol. 184, 2131–2140. doi:10.1128/JB.184.8.2131-2140.2002
Truong-Bolduc, Q. C., Wang, Y., and Hooper, D. C. (2022). Role of Staphylococcus aureus Tet38 in transport of tetracycline and its regulation in a salt stress environment. J. Bacteriol. 204 (7), e0014222. doi:10.1128/jb.00142-22
Van Houdt, R., and Michiels, C. W. (2010). Biofilm formation and the food industry, a focus on the bacterial outer surface. J. Appl. Microbiol. 109 (4), 1117–1131. doi:10.1111/j.1365-2672.2010.04756.x
Vázquez-Sánchez, D., Cabo, M. L., Ibusquiza, P. S., and Rodríguez-Herrera, J. J. (2014). Biofilm-forming ability and resistance to industrial disinfectants of Staphylococcus aureus isolated from fishery products. Food control. 39, 8–16. doi:10.1016/j.foodcont.2013.09.029
Vijayakumar, R., and Sandle, T. (2019). A review on biocide reduced susceptibility due to plasmid-borne antiseptic-resistant genes-special notes on pharmaceutical environmental isolates. J. Appl. Microbiol. 126 (4), 1011–1022. doi:10.1111/jam.14118
von Wintersdorff, C. J., Penders, J., van Niekerk, J. M., Mills, N. D., Majumder, S., van Alphen, L. B., et al. (2016). Dissemination of antimicrobial resistance in microbial ecosystems through horizontal gene transfer. Front. Microbiol. 7, 173. doi:10.3389/fmicb.2016.00173
Wakabayashi, Y., Takemoto, K., Iwasaki, S., Yajima, T., Kido, A., Yamauchi, A., et al. (2022). Isolation and characterization of Staphylococcus argenteus strains from retail foods and slaughterhouses in Japan. Int. J. Food Microbiol. 363, 109503. doi:10.1016/j.ijfoodmicro.2021.109503
Wakabayashi, Y., Umeda, K., Yonogi, S., Nakamura, H., Yamamoto, K., Kumeda, Y., et al. (2018). Staphylococcal food poisoning caused by Staphylococcus argenteus harboring staphylococcal enterotoxin genes. Int. J. Food Microbiol. 265, 23–29. doi:10.1016/j.ijfoodmicro.2017.10.022
Wang, Q., Larese-Casanova, P., and Webster, T. J. (2015). Inhibition of various gram-positive and gram-negative bacteria growth on selenium nanoparticle coated paper towels. Int. J. Nanomedicine. 10, 2885–2894. doi:10.2147/IJN.S78466
Wang, Q., Li, Y., Sun, D. W., and Zhu, Z. (2018). Enhancing food processing by pulsed and high voltage electric fields: Principles and applications. Crit. Rev. Food Sci. Nutr. 58 (13), 2285–2298. doi:10.1080/10408398.2018.1434609
Wang, Q., and Webster, T. J. (2013). Short communication: Inhibiting biofilm formation on paper towels through the use of selenium nanoparticles coatings. Int. J. Nanomedicine. 8, 407–411. doi:10.2147/IJN.S38777
Wang, S., Zhao, C., Yin, Y., Chen, F., Chen, H., and Wang, H. (2022). A practical approach for predicting antimicrobial phenotype resistance in Staphylococcus aureus through machine learning analysis of genome data. Front. Microbiol. 13, 841289. doi:10.3389/fmicb.2022.841289
Wassenaar, T. M., Ussery, D., Nielsen, L. N., and Ingmer, H. (2015). Review and phylogenetic analysis of qac genes that reduce susceptibility to quaternary ammonium compounds in Staphylococcus species. Eur. J. Microbiol. Immunol. Bp. 5 (1), 44–61. doi:10.1556/EUJMI-D-14-00038
Weigel, L. M., Clewell, D. B., Gill, S. R., Clark, N. C., McDougal, L. K., Flannagan, S. E., et al. (2003). Genetic analysis of a high-level vancomycin-resistant isolate of Staphylococcus aureus. Science 302 (5650), 1569–1571. doi:10.1126/science.1090956
Witteveen, S., Hendrickx, A. P. A., de Haan, A., Notermans, D. W., Landman, F., van Santen-Verheuvel, M. G., et al. (2022). Genetic characteristics of methicillin-resistant Staphylococcus argenteus isolates collected in the Dutch national MRSA surveillance from 2008 to 2021. Microbiol. Spectr. 10 (5), e0103522. doi:10.1128/spectrum.01035-22
Wu, S., Huang, J., Zhang, F., Dai, J., Pang, R., Zhang, J., et al. (2020). Staphylococcus argenteus isolated from retail foods in China: Incidence, antibiotic resistance, biofilm formation and toxin gene profile. Food Microbiol. 91, 103531. doi:10.1016/j.fm.2020.103531
Wu, S., Huang, J., Zhang, F., Zhang, J., Yang, R., Pang, R., et al. (2022). Emergence of extensive multidrug-resistant Staphylococcus aureus carrying novel Sa-MRRlsa(E) in retail food. J. Glob. Antimicrob. Resist. 30, 205–213. doi:10.1016/j.jgar.2022.06.011
Xia, G., and Wolz, C. (2014). Phages of Staphylococcus aureus and their impact on host evolution. Infect. Genet. Evol. 21, 593–601. doi:10.1016/j.meegid.2013.04.022
Zeng, X., and Lin, J. (2013). Beta-lactamase induction and cell wall metabolism in Gram-negative bacteria. Front. Microbiol. 4, 128. doi:10.3389/fmicb.2013.00128
Zhang, D. F., Xu, X., Song, Q., Bai, Y., Zhang, Y., Song, M., et al. (2016). Identification of Staphylococcus argenteus in Eastern China based on a nonribosomal peptide synthetase (NRPS) gene. Future Microbiol. 11, 1113–1121. doi:10.2217/fmb-2016-0017
Zhang, D. F., Yang, X. Y., Zhang, J., Qin, X., Huang, X., Cui, Y., et al. (2018). Identification and characterization of two novel superantigens among Staphylococcus aureus complex. Int. J. Med. Microbiol. 308 (4), 438–446. doi:10.1016/j.ijmm.2018.03.002
Zhang, F., Wu, S., Lei, T., Wu, Q., Zhang, J., Huang, J., et al. (2022). Presence and characterization of methicillin-resistant Staphylococcus aureus co-carrying the multidrug resistance genes cfr and lsa(E) in retail food in Chin. Int. J. Food Microbiol. 363, 109512. doi:10.1016/j.ijfoodmicro.2021.109512a
Zhang, L., Zhu, C., Chen, X., Xu, X., and Wang, H. (2021). Resistance of detached-cells of biofilm formed by Staphylococcus aureus to ultra high pressure homogenization. Food Res. Int. 139, 109954. doi:10.1016/j.foodres.2020.109954
Zhang, Y. M., Jiang, Y. H., Li, H. W., Li, X. Z., and Zhang, Q. L. (2022). Purification and characterization of Lactobacillus plantarum-derived bacteriocin with activity against Staphylococcus argenteus planktonic cells and biofilm. J. Food Sci. 87 (6), 2718–2731. doi:10.1111/1750-3841.16148
Keywords: Staphylococcus aureus, ready-to-eat food, food-contact surfaces, co-selection, cgWGS, antimicrobial resistance, biocide resistance, biofilm
Citation: Chieffi D, Fanelli F and Fusco V (2023) Antimicrobial and biocide resistance in Staphylococcus aureus: genomic features, decontamination strategies, and the role of S. aureus complex-related species, with a focus on ready-to-eat food and food-contact surfaces. Front. Food. Sci. Technol. 3:1165871. doi: 10.3389/frfst.2023.1165871
Received: 14 February 2023; Accepted: 14 April 2023;
Published: 05 May 2023.
Edited by:
Frederick Tawi Tabit, University of South Africa, South AfricaReviewed by:
Ahmad Cheikhyoussef, University of Namibia, NamibiaCopyright © 2023 Chieffi, Fanelli and Fusco. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Francesca Fanelli, ZnJhbmNlc2NhLmZhbmVsbGlAaXNwYS5jbnIuaXQ=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.