- 1Ottawa Laboratory—Fallowfield, Canadian Food Inspection Agency, Ottawa, ON, Canada
- 2Fertilizer Safety Section, Plant Health and Biosecurity Directorate, Canadian Food Inspection Agency, Ottawa, ON, Canada
Dissemination of microorganisms with antimicrobial resistance genes (ARGs) through the food chain has been recognized as a growing public health concern worldwide. Biosolids, a product of wastewater treatment process, have been used as fertilizers in agriculture globally and have also been considered as a potential source of pathogens and ARGs for horizontal transfer across various environments. This study characterized antibiotic resistomes and microbiota in 24 biosolids samples collected from two Canadian waste water treatment plants in different cities in 2009 and 2019. The ARGs were detected using a qPCR array kit, and microbiota was analyzed using 16S ribosomal RNA gene amplicon sequencing. Furthermore, correlation analysis of ARG abundance and bacterial genera abundance was explored to predict potential hosts of ARGs. Seventy-one of 84 ARGs were detected in at least one or more samples with 12 ARGs being detected in all samples. Antibiotic resistomes did not show a statistically significant distinction between different collection years, sites, or year and site combined in principle coordinate analysis. The microbiota communities were significantly different between samples collected in different years, sites, or year and site combined. In total 34 phyla were detected with 13 genera among the top three phyla were typically related to the human gut microbiota and seven of them showing strong correlation with ARGs related to aminoglycoside and beta-lactam resistance. This study provides valuable baseline information and consistent trend on ARGs and bacterial communities in biosolid fertilizers in Canada, indicating that the biosolid fertilizer could potentially be a source of ARGs in the agricultural soils and may leading to potential contamination of plant-based food chains.
Introduction
Antimicrobial resistance (AMR) is currently a significant public health challenge, with an estimated 700,000 human deaths and trillions of dollars of economic burdens every year in the world (Wozniak, 2018; Ruckert et al., 2020). In Canada, AMR-related infections or diseases contributed to over 14,000 deaths and cost about 1.4 billion in 2018 (CCA, 2019). Humans can acquire antibiotic-resistant bacteria (ARB) from various sources, including infections, food, and water intake (Singer et al., 2016; Bennani et al., 2020; Nappier et al., 2020; Larsson and Flach, 2021). During the last decades, a new concern has arisen regarding the critical role of the food chain played in the transmission of ARB to humans (Greig et al., 2015; Bennani et al., 2020; Panel, et al., 2021), since food can be easily contaminated by ARGs and/or ARB derived from different environmental sources including agriculture (Law et al., 2021; Zielinski et al., 2021). Biosolids, nutrient-rich organic fertilizer, and soil conditioner, obtained from the solid effluents after municipal wastewater treatment, have been considered as an alternative source of nutrients to chemical fertilizer and have been widely and directly applied to land, including forestry and farmland, in almost every province in Canada and throughout the United States (Lu et al., 2012; Egan, 2013; Mason-Renton and Luginaah, 2016; Whitehouse et al., 2022). Canadian agricultural industries spend around $3 billion on fertilizers and supplements, and some of these fertilizers contain biosolids produced by wastewater treatment plants (WWTP) (CFPF, 2015). It was reported (CIELAP, 2009) that approximately 300,000 dry tons of municipal sewage biosolids were generated per year in Ontario, Canada, with about 40% for land application, 40% landfilled and 20% being incinerated. Among the biosolids applied on land, around 50% of 120,000 tons of biosolids were used by farmers and spread on 15,000 ha of land per year in the province of Ontario, Canada before 2008 (Vyhnak, 2008). By 2012, there are over 660,000 metric tons of biosolids are generated per year in Canada (CCME, 2012) in Canada. The economic and environmental costs of landfilling and incineration are leading municipalities to pursue more land application of biosolids as a means of disposal at least in Ontario (CIELAP, 2009).
Wastewater including sewage needs to be adequately treated and environmentally managed to reduce the negative impacts of their application or disposal. It is well known that raw sewage, is mainly produced by human activity, and contains human pathogenic and commensal bacteria. Therefore, biosolids production and usage all over the world is regulated by various rules, acts or directives defined and implemented by different countries to prevent harmful effects on soil, vegetation, animals and people (Iranpour et al., 2004; CCME, 2010). Different WWTPs may go through multiple and different treatment processes for biosolid production, including hygienization process. The common used methods of hygienization include, digestion pretreatments (sonication and thermal hydrolysis) with two sequential biological processes (mesophilic/thermophilic and anaerobic/aerobic digestion), irradiation and oxidation processes by chemical disinfectants, and high pressure/temperature treatment (Al-Gheethi et al., 2018). Different WWTPs may use different processes. For example, in Ontario, Canada, chemical disinfectants and UV irradiation are commonly used for disinfection (Ontario, G.O., 2019). Therefore, biosolids are generally considered safe for agriculture use (Gerba and Pepper, 2009; Egan, 2013; Brooks et al., 2015; Al-Gheethi et al., 2018; Whitehouse et al., 2022). However, it is shown that some pathogenic bacteria could still survive these processes and end up in biosolid-amended soils (Alam et al., 2007; Viau and Peccia, 2009a; Duceppe et al., 2019), some of which, including Listeria monocytogenes (Al-Ghazali and Al-Azawi, 1990) and Staphylococcus aureus (Lewis and Gattie, 2002) contaminate crops and environment. Moreover, many bacteria within the sewage matrix could carry ARGs, and some of these ARGs get concentrated during treatment processes resulting in a higher concentration of ARGs in biosolids (Munir et al., 2011). Consequently, biosolids have also been considered a reservoir of ARGs and many pathogenic bacteria (Viau and Peccia, 2009a; Agga et al., 2015; Yergeau et al., 2016; Al-Gheethi et al., 2018; Law et al., 2021; Jankowski et al., 2022). And it is possible that ARGs, ARB and pathogenic bacteria could enter food chains through biosolid application to crop lands (EFSA BIOHAZ Panel et al., 2021). Meanwhile, there have been no published guidelines or standards regarding the control of ARGs in biosolids. The WWTP treatment procedures are also not designed to remove ARGs (Uluseker et al., 2021). Therefore, there is a need for long-term monitoring of ARGs and identification of their hosts, especially pathogenic bacteria, present in biosolids, such a risk information is essential for the risk assessment and the effective control of the dissemination of ARGs.
This study investigated the distributions of various ARGs and the microbiota in biosolids collected from two WWTPs at different Canadian cities collected in 2009 and 2019, respectively. The main goals of this study were to understand: 1) antibiotic resistomes and bacterial taxa in biosolids produced by two WWTPs in Canada, 2) variations of the ARGs and biosolids microbiota between two WWTPs over 10 years interval; 3) co-occurrence between ARGs and bacterial taxa to explore a likelihood of ARG hosts.
Materials and methods
Study sites and sample collection
Twenty-four solid end products (biosolids) from two municipal wastewater treatment plants (WWTPs) located at city A (WWTP-A) and city B (WWTP-B), within 100 km distance, were collected at 10-year intervals in 2009 and 2019, respectively. The WWPT-A at a more prominent city treats waste-activated sludge by anaerobic digestion and dewatering process with the end product of wet pellets. WWTP-B treats waste-activated sludge in a dewatering/pelletization process involving the belt-filter press and final pelletization process by thermal drier (250–450°C at the entry, and 80–130°C at the exit). Samples were taken in triplicate twice a year with an interval of 5-6 moths per plant (n = 24 in total). 500 g per sample were taken, and 10 g were used for DNA extraction. The biosolids were either wet or dry pellets in irregular shape with approximately diameters between 0.1—0.5 center meter and the moisture contents approximately ranging from 59—73% for WWTP A and 3—11% for WWTP B. The samples were transported on ice and were stored at −20°C until further use.
In addition, for the samples used in the current study, live coliforms, generic E. coli and Salmonella were detected for the biosolids collected in 2009 as described in a separate publication for which the WWTPs were named as WWTP A and C (Yergeau et al., 2016; Duceppe et al., 2019). Similar results for coliforms and generic E. coli have been obtained for the samples used in 2019 with the WWTPs referred as WWTP A and B respectively) (data not shown).
DNA extraction
Biosolids were first ground manually with mortar and pestle to a fine powder. DNA was extracted using the PowerMax® Soil DNA Isolation Kit following the manufacturer’s instructions (QIAGEN, Germany) and quantified using Qubit™ four Fluorimeter (Life Technologies, United States). The DNA extracts were stored at −20 °C and retrieved for ARGs detection using qPCR array and for bacterial host analysis using 16S ribosomal RNA (rRNA) gene sequencing.
ARG detection
Clinically relevant 84 ARGs (Supplementary Table S1) were detected with a commercial ARG qPCR array kit from QIAGEN (Cat# 330261 BAID-1901ZR) using ABI 7500 Fast real-time PCR system (Applied Biosystems, United States), according to the manufacturer’s instruction. Briefly, the 25 μl reaction volume consisted of 2x microbial qPCR master mix (QIAGEN, Germany) and 52 ng DNA for each well. The manufacturer’s instruction recommended minimum of 500 ng/plate or 5.2 ng/well. In order to increase the detection sensitivity, various concentrations of DNA extracts were tested in our preliminary study which showed the increased numbers of AMR genes detected with the increase of the DNA concentrations. 5,000 ng/plates or 5.2 ng/well (or reaction) was used based on the numbers of AMR gene detected and concentrations of the DNA extract obtained from the same DNA extraction. Plates were incubated at 95 °C for 10 min followed by 40 cycles of 95°C for 15 s and 60°C for 2 min. Threshold cycle (Ct) values were validated and normalized using an Excel data analysis template provided by QIAGEN. A Ct value below 34 was considered positive based on the manufacturer’s instruction, indicating an ARG was present in the sample. The heatmap was plotted using the R package pheatmap v1.0.12 (https://github.com/raivokolde/pheatmap). In addition, the binary data of ARG presence and absence were imported into R v4.1.2, and principal coordinates analysis (PCOA) was performed using Vegan v2.5-7 (https://github.com/vegandevs/vegan).
16S rRNA gene amplicon sequencing and data analysis
16S rRNA gene amplicon sequencing was performed as described previously (Laskey et al., 2020). Briefly, the V3–V4 region of the 16S rRNA gene was amplified by PCR. Libraries were prepared and sequenced using a MiSeq system (Illumina, United States). Raw read data was demultiplexed and then analyzed by Qiime2 (Bolyen et al., 2019) using a modified version of the Qiime2 pipeline (AmpliconPipeline, https://github.com/forestdussault/AmpliconPipeline). The biom, tree, and metadata files generated from this pipeline were combined into a phyloseq object using the R package microbiomeMarker v1.0.1 (https://github.com/yiluheihei/microbiome Marker). R packages phyloseq v1.38.0 (McMurdie and Holmes, 2013), ggplots2 v3.3.5 (https://github.com/tidyverse/ggplot2), and vegan v2.5-7 (https://github.com/vegandevs/vegan) packages were used for further analysis and visualization.
Biomarker signature analysis
Biomarker analysis was performed using the linear discriminant analysis (LDA) effect size (LEfSe) method (Segata et al., 2011) using the R package microbiomeMarker v1.0.1 (https://github.com/yiluheihei/microbiomeMarker). A threshold score of LDA score = 4 and Kruskal-Wallis test α value = 0.05 were set as thresholds to detect differences.
Correlation between ARG subtypes and microbial taxa
A correlation matrix and significance levels were generated using the R Hmisc package (https://github.com/harrelfe/Hmisc) to calculate Pearson correlation with significance levels (p-value) between 259 bacterial genera and 71 ARGs presented in 24 samples. A correlation was considered statistically robust if the Pearson’s correlation coefficient (r) was ≥0.8 and the p-value was ≤0.01 (Junker and Schreiber, 2008). The significant and robust correlation visualization was performed using GraphPad Prism 7.0 software (United States).
Statistical analysis
Differences in the alpha diversity (Chao1 and inverse Simpson index) between the groups were analyzed using the Wilcoxon tests in the R package ggpubr v 0.4.0 (https://github.com/kassambara/ggpubr). The difference in Beta-diversity (PCOA plot) between different years, locations, or years and location combined were analyzed using PERMANOVA followed by Betadisper tests (Vegan v 2.5-7). The data should be homoscedastic (p-value was not significant in the Betadisper test) to apply a PERMANOVA. A p-value < 0.05 was considered statistically significant.
Results and discussions
Diversity of antibiotic resistomes in WWTPs
A total of 71 out of 84 targeted ARGs, representing nine different antibiotic classifications, were detected with various abundances (Figure 1A and Supplementary Table S1), which was consistent with a previous study that reported the detection of 70 ARGs (Agga et al., 2015). Moreover, only 12 ARGs (aadA1, blaGES, blaVEB, blaOXA-2, ereB, qnrB-5, qnrB-8, qnrS, ermB, mefA, tetA, and tetB) were detected in all individual samples (Figure 1A and Supplementary Table S1). Five macrolide resistance genes (ermA, ermB, ermC, mefA, and msrA) were detected in most samples. Particularly, ermB has the highest target copy numbers (the lowest average Ct value), a typical characteristic gene detected in urban sewages in North America and Europe (Hendriksen et al., 2019). Numerous studies on the characterization of ARG or ARB in wastewater have been conducted over the past decades using various methods to facilitate the detection of ARGs in biosolids or sludge (Viau and Peccia, 2009a; Novo et al., 2013; Agga et al., 2015; Yergeau et al., 2016; Li et al., 2020; Law et al., 2021; Zielinski et al., 2021; Jankowski et al., 2022). In contrast to these studies, we investigated the changes of the antibiotic resistomes at two different facilities over the 10-year interval and detected a total of 58 and 53 ARGs in samples collected from WWTP-A in 2009 and 2019, respectively. Whereas 66 and 53 ARGs were detected in samples from WWTP-B in 2009 and 2019, respectively. A slight reduction of the total number of ARGs from 69 (in 2009) to 63 (in 2019) was observed in the two facilities combined. Only two genes, vanC and mecA believed to be exclusively found in Gram-positive cocci were detected in the samples collected in 2009 at both WWTPs (Figure 1A and Supplementary Table S1), indicating that Gram-positive bacteria containing these ARGs were probably present in both WWTPs at the early sampling date but not 10 years later or the wastewater treatment efficiencies improved over time and subsequently eliminated the bacteria. Moreover, several ARGs, such as bla BES-1, blaIMP-5, blaCMY-10, and blaFOX, were consistently detected in the samples from WWTP-B, while the bla ACC-1 group was only present in the samples from WWTP-A (Figure 1A and Supplementary Table S1).
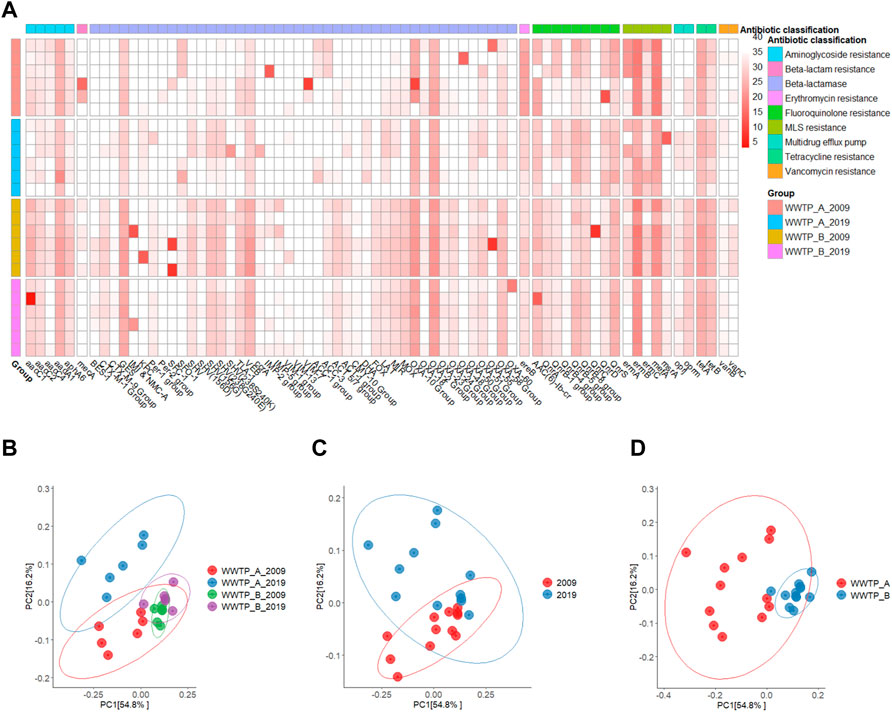
FIGURE 1. Diversity of antibiotic resistomes in biosolids samples collected from WWTP-A and -B in 2009 and 2019, respectively. (A) Heatmap analysis of qPCR Ct values of 71 ARGs representing nine different antibiotic classifications detected at 24 biosolids samples. Lighter colors indicate low abundance or absence (high Ct value), while darker colors indicate high abundance (low Ct value). Ct value below 34 was evaluated as positive. Principal coordinates analysis (PCoA) plot of antibiotic resistome between site and year (B), year (C), and site (D) based on Bray-Curtis dissimilarity. The percentage of variation explained is noted on the axis labels.
β-diversity analysis was used in this study to estimate the overall similarity in the antibiotic resistomes between the samples collected from two different sites on two occasions 10 years apart. There was no clear distinction between the antibiotic resistomes clustered for the grouped samples based on the year and site combined (Figure 1B), years (Figure 1C), or sites alone (Figure 1D), indicating that the antibiotic resistomes are similar to each compared group. Additionally, antibiotic resistome in samples collected from WWTP-A in both 2019 and 2009 have a longer distance to the centroid, indicating antibiotic resistome in samples collected from WWTP-A are more variable than WWTP-B (Supplementary Figure 1S). Compared to antibiotic resistomes of WWTPs in the world (Hendriksen et al., 2019), both WWTP-A and -B show very similar ARG profiling to that of WWTPs in Canada and United States, which further confirm diversity of antibiotic resistomes is consistent in the close geographical regions.
In recent years, effective actions, including a drastic reduction in antibiotics consumption, reduction in the uncontrolled use in humans and animals, and the ban of the sub-clinical dosage growth promoter) usage of antibiotics for animal and crop productions to promote growth have been in place to reduce the spread of ARGs globally (WHO, 2015; Merlin, 2020) and in Canada (McCubbin et al., 2021). It seems that the measures to reduce the use of AMRs in animals were effective in reducing ARGs in these animals (Dorado-Garcia et al., 2016; McDonnell et al., 2017; Bennani et al., 2020). However, it is not clear whether this translated into ARG reduction in humans. In this study, a slight reduction of the total number of ARGs was observed, but overall, there were no apparent shifts of ARGs in biosolids over 10 years in two WWTPs in Canada. ARGs or related genetic elements have existed in the environment for a very long time, even before the antibiotic era. These genes can be transferred from the environment to human-related activities or the opposite direction, but it is not easy to get rid of them from our world. Eventually, their concentrations on earth will increase if more clinically important ARGs or ARBs show up due to the misuse in non-clinical settings. Our results showed no significant increase in the total number of ARGs, and there were no apparent shifts of ARGs in biosolids over 10 years in two WWTPs in Canada, suggesting a reduction or slowing down or stabilization of the speed of ARGs dissemination from human sources into the environment (Viau and Peccia, 2009b). More investigations are needed to evaluate the potential sources that contribute to the circulation and accumulation of ARGs in biosolids, and the potentially transfer to food crops.
Characterization of bacterial communities in WWTPs
V3-V4 hypervariable regions of the 16S rRNA gene amplicons sequencing were used to characterize bacteria communities in WWTPs at different sites and years. Overall, compared to the WWTP-A, higher microbial richness (Chao1 index) was observed in the samples collected from WWTP-B in 2009 and 2019, indicating more diverse bacterial species present in the biosolids samples collected from WWTP-B (Figure 2A), compared to that of WWTP-A.
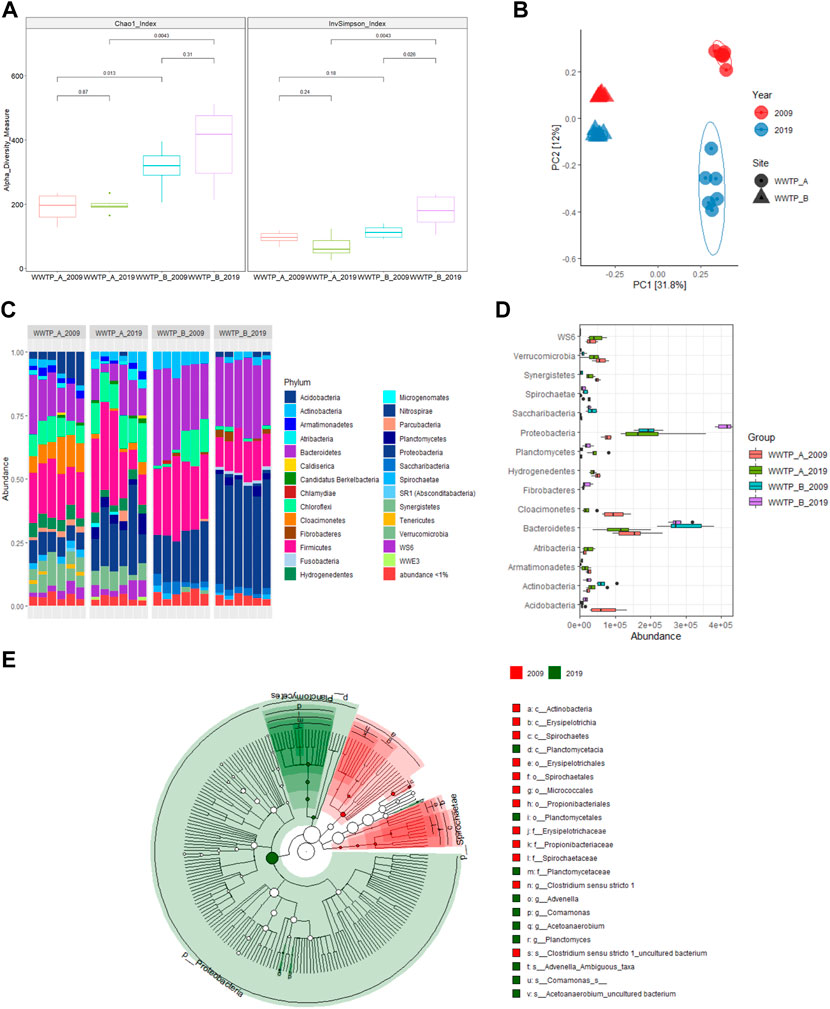
FIGURE 2. Characterization of bacterial communities in biosolids samples collected from WWTP-A and B in 2009 and 2019, respectively. (A) Alpha-diversity assessed by richness (Chao1) and diversity (inverse Simpson) observed in different sites and years. Kruskal-Wallis tests were used to calculate the p-value between two comparisons. Boxes represent the interquartile range, lines indicate medians, and whiskers indicate the range. (B) Principal coordinates analysis of biosolids microbiota between different sites and years based on Bray-Curtis dissimilarity. The percentage of variation explained is noted on the axis labels. (C) Relative abundance of the bacterial phyla in each sample. (D) LEfSe analysis of significantly different abundant phyla of bacteria between different sites and years. (E) The cladogram generated by LEfSe represents statistically significant taxa differences between 2009 and 2019. Each successive circle is one step lower phylogenetically [phylum (p), class(c), order (o), family (f), and species(s)]. Regions in red indicate taxa enriched in 2009, while regions in green indicate taxa enriched in 2019. Each circle’s diameter is proportional to the taxon’s abundance.
Neither samples collected from the two WWTPs showed a significant difference for each site between 2009 and 2019 based on the Chao1 index (Figure 2A). Similar to the Chao1 index findings, microbial diversity (Inverse Simpson index) was similar in the samples collected from WWTP-A within 10 years. However, for the WWTP-B, samples collected in 2019 had higher microbial diversity than that of 2009, indicating that the microbiota community is more heterogeneous in the 2019 samples than those in 2009. The samples collected in 2019 from WWTP-B also showed different microbial diversity compared to the samples collected from WWTP-A in 2019 (Figure 2A). The two WWTPs used different treatment procedures resulting in different moisture contents as explained above, and different bacteria counts, both higher for samples from WWTP-A (Yergeau et al., 2016; Materials and methods—Study site and sample collect). These results indicate that the different WWTP procedures could be one of the factors contributing to the differences of microbiota and the subsequent differences of ARBs from the samples of these two facilities.
Using Bray-Curtis distances, the PCoA of bacterial microbiota divided the samples into four separate clusters based on years and sites (Figure 2B). The bacterial microbiota compositions in the samples collected from WWTP-A and -B differed in 2009 (Figure 2B). After 10 years, there were significant parallel shifts in the microbiota composition in both facilities, especially samples collected from WWTP-A (Figure 2B). Additionally, a permutational multivariate ANOVA (PERMANOVA) exhibits a significant difference in the microbiota composition in biosolid samples collected from different sites and years, respectively (Table 1).
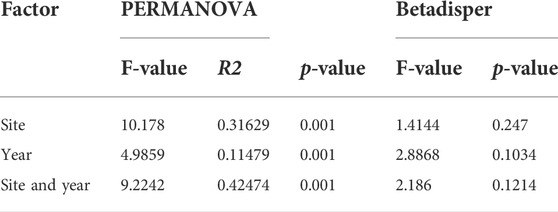
TABLE 1. PERMANOVA and Betadisper results based on Bray-Curtis dissimilarities for the bacterial community in biosolids samples collected from different sites and years.
A total of 34 phyla were observed across all biosolid samples, with most sequences classified to one of three phyla: Proteobacteria, Firmicutes, and Bacteroidetes (Figure 2C), which were also the most frequently identified in a variety of samples collected from different WWTPs (Nascimento et al., 2018; de Celis et al., 2020; Niestepski et al., 2020). Within these dominant phyla, taxa typically associated with human gut microbiota (e.g., Aeromonas, Acidovorax, Dechloromonas, Acinetobacter, Arcobacter, Flavobacterium, Bacteroides, Bacillus, Christensensellaceae, Clostridium, Eubacterium, Erysipelotrichaceae, and Ruminococcaceae) existed in our samples, which is in agreement with previous publications, showing the detection of human gut microbiota in the urban sewage (Newton et al., 2015; Su et al., 2017). The prevalence of human gut microbiota in biosolid samples is not surprising as most human wastes enter WWTPs directly. Additionally, bacteria associated with the environment, such as water and soil (e.g., Zoogloea, Devosia, Ferruginibacter), were also found in our biosolids samples. These results suggest that microbial communities in municipal biosolids are predominantly composed of human and environmental-associated bacteria.
Characterizing microbial communities between different years and sites is key to identifying unique microbial contributions. Predominant taxa differentially associated with each group were determined by LEfSe. The results showed that the bacterial community in samples from WWTP-A mainly belonged to the phyla Cloacimonetes, Verrucomicrobia, and Acidobacteria for 2009 and the phyla WS6, Planctomycetes, and Atribacteria for 2019 (Figure 2D). In contrast, biosolids samples collected from WWTP-B were better characterized by the presence of Bacteroidetes, Actinobacteria, and Saccharibacteria for 2009 and Proteobacteria and Fibrobacteres for 2019 (Figure 2D). Further LDA effect size-based cladogram showed Proteobacteria and Planctomycetes phyla, especially Planctomycetacia class, enriched in biosolid sample from 2019, while bacterial species enriched in biosolid samples collected from 2009 were taxa in the Spirochaetae phylum and Actinobacteria class (Figure 2E). The bacterial communities inhabiting biosolid collected from WWTPs can vary between different treatment facilities over time, due to the differences in treatment procedures between various facilities and the sources of WWTP influents (Yergeau et al., 2016; de Celis et al., 2020; Wolters et al., 2022). However, interestingly, given the fact that there is no significant differences in antibiotic resistome between these two facilities (Figure 1D), observation of significantly different microbiota compositions between these two facilities indicated potential ARG transfer between different bacterial species or existence of large amount of cell-free ARGs in biosolids.
It is well known that a particular microbial community in biosolids-derived fertilizers is thought to play an essential role in enabling and facilitating particular functions to enrich nutrition in the soil (Hu et al., 2019; Pan et al., 2021). In our data, a significant increase in Proteobacteria and Planctomycetes phyla populations was found in 2019 at both facilities. The members of Planctomycetes and Proteobacteria phyla are capable of several metabolisms and biosynthesis, such as nitrogen-fixation and ammonia oxidation (Delmont et al., 2018; Rivas-Marin et al., 2019; Stultiens et al., 2020), and provide an advantage to biosolids as fertilizers to enrich soil microbial community. Future studies predicting the functions of these populations or communities can potentially improve the efficiency of WWTP processes.
Many studies have already demonstrated the presence of a broad range of human bacterial pathogens in wastewater and biosolids samples such as Escherichia coli, Mycobacterium tuberculosis, Pseudomonas aeruginosa, Salmonella enterica, Shigella flexneri, Staphylococcus aureus, Yersinia enterocolitica, etc. (Law et al., 2021; Nguyen et al., 2021; Young et al., 2021; Bai et al., 2022; Jankowski et al., 2022). In our study, due to the limitations of the partial regions of 16S rRNA amplicon sequencing, it was impossible to achieve microbiota’s taxonomic resolution at the species level. However, the microbial community in these biosolids samples consists of several genera, such as Bacteroides, Clostridium, Escherichia, Mycobacterium, which contain some human pathogens. Further studies, such as the full-length 16S rRNA gene amplicon sequences and analysis using Nanopore sequencing platform or bacterial culture, needed to be done to assess the potential environmental risks.
Co-occurrence patterns between ARGs and bacterial communities
To identify potential ARG hosts, correlation analysis of 71 ARGs abundance and 259 bacterial genera abundance was evaluated based on strong (r > 0.8) and significant (p < 0.01) correlations (Figure 3). Co-occurrence patterns of ARGs and microbial genera in the biosolids could indicate possible host information of ARGs if the ARGs and microbial taxa display strong and significant positive correlations (Nguyen et al., 2021). Exploration of connections between ARGs and bacterial genera showed that 17 out of 20 genera were from the top three phyla: Firmicutes, Bacteroidetes, and Proteobacteria, and most of ARGs belonged to the beta-lactam resistance group (Figure 3). Interestingly, seven genera, including Ruminococcaceae NK4A214 group, Ruminococcaceae UCG 013, Fluviicola, Bacteroides, Fibrobacter, Aeromonas, and Dechloromonas, associated with human gut microbiota, contain ARGs related to aminoglycoside and beta-lactam resistance. In addition, members of the Ruminococcaceae NK4A214 genus are possible hosts for multiple genes responsible for the resistance of β-lactamase-resistant enzymes (Figure 3). Previous studies have revealed the presence of ARB, including specific pathogens in WWTPs (Nguyen et al., 2021). Similar to our results, Proteobacteria have been shown to contain the highest number of ARB, followed by Firmicutes and Bacteroidetes (Liu et al., 2019). In the Firmicutes phylum, Ruminococcaceae NK4A214 and Ruminococcaceae UCG 013 are members of the Ruminococcaceae family, a significant butyrate producer and essential bacteria involved in intestinal health (Vital et al., 2017). However, the Ruminococcaceae family in WWTP might be the host of qnrS, blaTEM, blaSHV (Narciso-da-Rocha et al., 2018). Moreover, members of Aeromonas, Bacteroides, and Mycobacterium genera containing human pathogens have been reported to carry several ARGs (Barancheshme and Munir, 2017; Hultman et al., 2018; Stalder et al., 2019). Our results also revealed typical environmental bacteria species belonging to Ferruginibacter, Rhodobacter, and Arenimonas phyla might carry genes that resist multiple types of beta-lactam antibiotics (Figure 3). Further Hi-C (a high throughput sequencing technique which captures in DNA-DNA proximity) and shotgun metagenomics approaches (Stalder et al., 2019) will be used to confirm the correlated hosts in the near future.
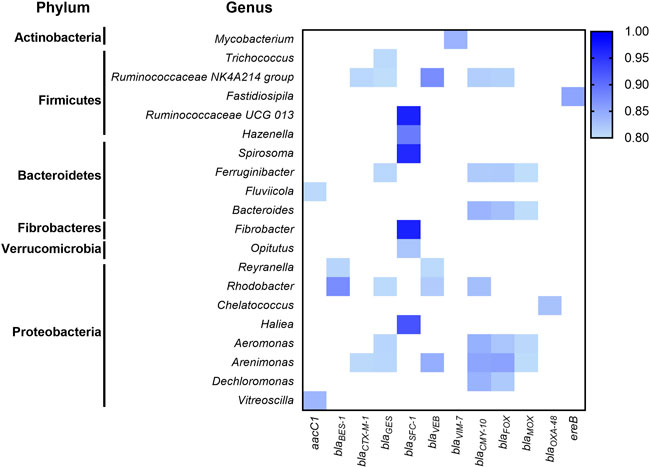
FIGURE 3. Co-occurrence patterns between ARGs and taxa (genus level) analyzed by Pearson’s correlations. The blue gradient color represents a robust (r ≥ 0.8) and significant (p ≤ 0.01) correlation - the darker the color, the higher correlation between the ARG and taxa. Only robust and significant correlations between ARG and taxa are presented here.
Several studies have shown the presence of live bacteria and ARBs in biosolids, indicating that treatments in WWTPs cannot completely inactivate all the bacteria (Alam et al., 2007; Viau and Peccia, 2009b; Yergeau et al., 2016). For the samples used in the current study, live coliforms, generic E. coli and Salmonella were detected for the biosolid samples collected in 2009 as described in separate publications for which the WWTPs were named as WWTP A and C (Yergeau et al., 2016; Duceppe et al., 2019). Similar results for coliform and generic E. coli have been obtained for the samples used in 2019 with the WWTPs referred as WWTP-A and -B respectively) (data not shown). Live ARBs have an opportunity to interact with soil microbiota and spread ARGs into the soil bacterial community via the horizontal gene transfer (Woegerbauer et al., 2020). With the annual application of biosolid fertilizers, the ARGs from biosolids may persist (Mays et al., 2021) or accumulate in the soil (Xie et al., 2016; Zhuang et al., 2021), which may continue to disseminate ARGs to humans potentially through various potential ways including food chains (EFSA BIOHAZ Panel et al., 2021). Because AMR is an increasingly severe threat to public health globally, concurrent actions from all the countries in the world using a One Health approach are necessary (Prestinaci et al., 2015; Murray et al., 2022). Currently, most developed countries have established surveillance programs for ARGs or ARBs. Still, these programs only investigate limited and common ARBs, such as pathogenic E.coli and Salmonella (PHAC, 2020), and do not cover the majority of AMR hosts. Our study has detected many environmental and gut commensal bacteria potential carrying ARGs, pressing the need to conduct comprehensive investigation or detection of ARBs, including pathogenic and non-pathogenic ARBs in the monitoring programs to assess the risks to the food chain. Compared to stable antibiotic resistome in these biosolid samples (Figure 1), significant changes of biosolid microbiota composition between the two WWTPs were observed in our study (Figure 2), which may explain why only a few possible host information of ARGs were identified using the correlation approach (Figure 3). Further studies are required to explore the linkage between ARGs and possible hosts using a combination of proximity ligation data with shotgun sequencing data, as explained above (Stalder et al., 2019) and further more by isolation.
Conclusion
Municipal biosolids are collection point showing the comprehensive picture of a given community. This study conducted a retrospective study in an interval of 10 years. This novel approach was to investigate the changes of AMR during the 10-year period, which is essential in understanding the impact of the measures used to control the misuse of antibiotics in humans, animals and crops in recent years. Although retrospective studies on AMR using isolated pathogenic bacteria have been conducted extensively, it is very rare to use biosolid samples for retrospective studies. There is a slight reduction of ARGs in both WWTPs in 10 years. However there is no significant change of AMRs indicating that overall, current AMR control measure may start to work although not obvious yet for whole human population. This study showed that the bacterial antibiotic resistomes are stable in two WWTPs at different cities over 10 years, while substantial variations in bacterial communities were observed in biosolids collected from different facilities and years. This may indicate that the microbiota carrying the ARGs are present consistently in biosolids. The results from co-occurrence analysis may provide the potential correlation between certain ARGs and bacteria taxa belonging to human gut microbiota and environmental bacteria, which may subsequently indicate the potential bacterial hosts and sources for certain AMRs in biosolids. Such results may enhance the studies to further confirm and characterize the hosts. The data in this study provide dynamic background for ARGs and ARB in biosolid fertilizers from Canadian WWTPs, which can shed light on the need for the future research on and risk analysis of the utilization of biosolid fertilizers in agriculture, including ARGs and ARBs dissemination into food chains. In addition, the results in this study showed that the real-time PCR array is a useful tool for rapid routine surveillance purpose.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession numbers can be found below: BioProject database, accession number PRJNA817560. https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA817560.
Author contributions
MK and HH designed the experiments. HH, EM, and DO arranged resources and biosolids samples. SN and IF conducted DNA extraction and qPCR experiments. MK, SN, and HH analyzed qPCR data. MK and AH performed 16S rRNA gene amplicon sequencing and data analysis. MK and HH prepared the draft of the manuscript, and all authors contributed to the preparation of the manuscript article and approved the submitted version.
Funding
This work was funded by the Canadian Food Inspection Agency (CFIA).
Acknowledgments
We thank Beverley Phipps-Todd and Chris Grenier at Ottawa laboratory—Fallowfield, CFIA for technical support, and staff at Fertilizer Safety Section, CFIA, for arranging biosolid samples.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/frfst.2022.894671/full#supplementary-material
References
Agga, G. E., Arthur, T. M., Durso, L. M., Harhay, D. M., and Schmidt, J. W. (2015). Antimicrobial-resistant bacterial populations and antimicrobial resistance genes obtained from environments impacted by livestock and municipal waste. PLoS One 10 (7), e0132586. doi:10.1371/journal.pone.0132586
Al-Ghazali, M. R., and Al-Azawi, S. K. (1990). Listeria monocytogenes contamination of crops grown on soil treated with sewage sludge cake. J. Appl. Bacteriol. 69 (5), 642–647. doi:10.1111/j.1365-2672.1990.tb01557.x
Al-Gheethi, A. A., Efaq, A. N., Bala, J. D., Norli, I., Abdel-Monem, M. O., and Ab. Kadir, M. O. (2018). Removal of pathogenic bacteria from sewage-treated effluent and biosolids for agricultural purposes. Appl. Water Sci. 8 (2), 74. doi:10.1007/s13201-018-0698-6
Alam, M. Z., Muyibi, S. A., and Jamal, P. (2007). Biological treatment of sewage treatment plant sludge by pure bacterial culture with optimum process conditions in a stirred tank bioreactor. J. Environ. Sci. Health. A Tox. Hazard. Subst. Environ. Eng. 42 (11), 1671–1679. doi:10.1080/10934520701518232
Bai, H., He, L. Y., Wu, D. L., Gao, F. Z., Zhang, M., Zou, H. Y., et al. (2022). Spread of airborne antibiotic resistance from animal farms to the environment: Dispersal pattern and exposure risk. Environ. Int. 158, 106927. doi:10.1016/j.envint.2021.106927
Barancheshme, F., and Munir, M. (2017). Strategies to combat antibiotic resistance in the wastewater treatment plants. Front. Microbiol. 8, 2603. doi:10.3389/fmicb.2017.02603
Bennani, H., Mateus, A., Mays, N., Eastmure, E., Stark, K. D. C., and Hasler, B. (2020). Overview of evidence of antimicrobial use and antimicrobial resistance in the food chain. Antibiot. (Basel) 9 (2), E49. doi:10.3390/antibiotics9020049
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al-Ghalith, G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37 (8), 852–857. doi:10.1038/s41587-019-0209-9
Brooks, J. P., Gerba, C. P., and Pepper, I. L. (2015). “Chapter 26 - land application of organic residuals: Municipal biosolids and animal manures,” in Environmental microbiology. Editors I. L. Pepper, C. P. Gerba, and T. J. Gentry. Third Edition (San Diego: Academic Press), 607–621.
CCA (2019). When antibiotics fail- the expert panel on the potential socio-economic impacts of antimicrobial resistance in Canada. Ottawa, Canada: Council of Canadian Academies. Available at: https://cca-reports.ca/wp-content/uploads/2018/10/When-Antibiotics-Fail-1.pdf.
CCME (2010). A review of the current Canadian legislative framework for wastewater biosolids [online]. Available: https://silo.tips/download/a-review-of-the-current-canadian-legislative-framework-for-wastewater-biosolids (Accessed May 27, 2022).
CCME (2012). Canada-wide approach for the management of wastewater biosolids [Online]. Available: https://ccme.ca/en/res/biosolids_cw_approach_e.pdf (Accessed July 14, 2022).
CFPF (2015). Working group report on communications. Available at: https://www.ohchr.org/en/hr-bodies/hrc/complaint-procedure/wg-situations.
CIELAP (2009). CIELAP (Canadian institute for environmental Law and policy) brief on biosolids management in Ontario june 2009 [online]. Available: https://www.cielap.org/pdf/Brief_Biosolids.pdf (Accessed July 14, 2022).
de Celis, M., Belda, I., Ortiz-Alvarez, R., Arregui, L., Marquina, D., Serrano, S., et al. (2020). Tuning up microbiome analysis to monitor WWTPs' biological reactors functioning. Sci. Rep. 10 (1), 4079. doi:10.1038/s41598-020-61092-1
Delmont, T. O., Quince, C., Shaiber, A., Esen, O. C., Lee, S. T., Rappe, M. S., et al. (2018). Author Correction: Nitrogen-fixing populations of Planctomycetes and Proteobacteria are abundant in surface ocean metagenomes. Nat. Microbiol. 3 (8), 963. doi:10.1038/s41564-018-0209-4
Dorado-Garcia, A., Mevius, D. J., Jacobs, J. J., Van Geijlswijk, I. M., Mouton, J. W., Wagenaar, J. A., et al. (2016). Quantitative assessment of antimicrobial resistance in livestock during the course of a nationwide antimicrobial use reduction in The Netherlands. J. Antimicrob. Chemother. 71 (12), 3607–3619. doi:10.1093/jac/dkw308
Duceppe, M. O., Phipps-Todd, B., Carrillo, C., and Huang, H. (2019). Draft genome sequences of eight Canadian Citrobacter braakii and Citrobacter freundii strains. Microbiol. Resour. Announc. 8 (27), e00273-19. doi:10.1128/MRA.00273-19
Egan, M. (2013). Biosolids management strategies: An evaluation of energy production as an alternative to land application. Environ. Sci. Pollut. Res. Int. 20 (7), 4299–4310. doi:10.1007/s11356-013-1621-1
Gerba, C. P., and Pepper, I. L. (2009). Wastewater treatment and biosolids reuse. Environ. Microbiol., 503–530. doi:10.1016/B978-0-12-370519-8.00024-9
Greig, J., Rajic, A., Young, I., Mascarenhas, M., Waddell, L., and LeJeune, J. (2015). A scoping review of the role of wildlife in the transmission of bacterial pathogens and antimicrobial resistance to the food Chain. Zoonoses Public Health 62 (4), 269–284. doi:10.1111/zph.12147
Panel, EFSA BIOHAZ, Koutsoumanis, K., Allende, A., Alvarez-Ordonez, A., Bolton, D., Bover-Cid, S., et al. (2021). Role played by the environment in the emergence and spread of antimicrobial resistance (AMR) through the food chain. EFSA J. 19 (6), e06651. doi:10.2903/j.efsa.2021.6651
Hendriksen, R. S., Munk, P., Njage, P., van Bunnik, B., McNally, L., Lukjancenko, O., et al. (2019). Global monitoring of antimicrobial resistance based on metagenomics analyses of urban sewage. Nat. Commun. 10 (1), 1124. doi:10.1038/s41467-019-08853-3
Hu, Y., Pang, S., Yang, J., Zhao, X., and Cao, J. (2019). Changes in soil microbial community structure following amendment of biosolids for seven years. Environ. Pollut. Bioavailab. 31 (1), 24–31. doi:10.1080/26395940.2019.1569478
Hultman, J., Tamminen, M., Parnanen, K., Cairns, J., Karkman, A., and Virta, M. (2018). Host range of antibiotic resistance genes in wastewater treatment plant influent and effluent. FEMS Microbiol. Ecol. 94 (4), fiy038. doi:10.1093/femsec/fiy038
Iranpour, R., Cox, H. H. J., Kearney, R. J., Clark, J. H., Pincince, A. B., and Daigger, G. T. (2004). Regulations for biosolids land application in US and European Union. J. Residuals Sci. Tech. 1 (4), 209–222.
Jankowski, P., Gan, J., Le, T., McKennitt, M., Garcia, A., Yanac, K., et al. (2022). Metagenomic community composition and resistome analysis in a full-scale cold climate wastewater treatment plant. Environ. Microbiome 17 (1), 3. doi:10.1186/s40793-022-00398-1
Junker, B. H., and Schreiber, F. (2008). Analysis of biological networks. Hoboken, NJ: John Wiley & Sons.
Larsson, D. G. J., and Flach, C. F. (2021). Antibiotic resistance in the environment. Nat. Rev. Microbiol. 20, 257–269. doi:10.1038/s41579-021-00649-x
Laskey, A., Ottenbrite, M., Devenish, J., Kang, M., Savic, M., Nadin-Davis, S., et al. (2020). Mobility of beta-lactam resistance under bacterial Co-infection and ampicillin treatment in a mouse model. Front. Microbiol. 11, 1591. doi:10.3389/fmicb.2020.01591
Law, A., Solano, O., Brown, C. J., Hunter, S. S., Fagnan, M., Top, E. M., et al. (2021). Biosolids as a source of antibiotic resistance plasmids for commensal and pathogenic bacteria. Front. Microbiol. 12, 606409. doi:10.3389/fmicb.2021.606409
Lewis, D. L., and Gattie, D. K. (2002). Pathogen risks from applying sewage sludge to land. Environ. Sci. Technol. 36 (13), 286A–293A. doi:10.1021/es0223426
Li, W., Su, H., Cao, Y., Wang, L., Hu, X., Xu, W., et al. (2020). Antibiotic resistance genes and bacterial community dynamics in the seawater environment of Dapeng Cove, South China. Sci. Total Environ. 723, 138027. doi:10.1016/j.scitotenv.2020.138027
Liu, Z., Klumper, U., Liu, Y., Yang, Y., Wei, Q., Lin, J. G., et al. (2019). Metagenomic and metatranscriptomic analyses reveal activity and hosts of antibiotic resistance genes in activated sludge. Environ. Int. 129, 208–220. doi:10.1016/j.envint.2019.05.036
Lu, Q., He, Z. L., and Stoffella, P. J. (2012). Land application of biosolids in the USA: A review. Appl. Environ. Soil Sci. 2012, 1–11. doi:10.1155/2012/201462
Mason-Renton, S., and Luginaah, I. (2016). Interfering with therapeutic tranquility: Debates surrounding biosolid waste processing in rural Ontario. Health Place 41, 42–49. doi:10.1016/j.healthplace.2016.07.004
Mays, C., Garza, G. L., Waite-Cusic, J., Radniecki, T. S., and Navab-Daneshmand, T. (2021). Impact of biosolids amendment and wastewater effluent irrigation on enteric antibiotic-resistant bacteria—a greenhouse study. Water Res. X 13, 100119. doi:10.1016/j.wroa.2021.100119
McCubbin, K. D., Anholt, R. M., de Jong, E., Ida, J. A., Nobrega, D. B., Kastelic, J. P., et al. (2021). Knowledge gaps in the understanding of antimicrobial resistance in Canada. Front. Public Health 9, 726484. doi:10.3389/fpubh.2021.726484
McDonnell, L., Armstrong, D., Ashworth, M., Dregan, A., Malik, U., and White, P. (2017). National disparities in the relationship between antimicrobial resistance and antimicrobial consumption in Europe: An observational study in 29 countries. J. Antimicrob. Chemother. 72 (11), 3199–3204. doi:10.1093/jac/dkx248
McMurdie, P. J., and Holmes, S. (2013). phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8 (4), e61217. doi:10.1371/journal.pone.0061217
Merlin, C. (2020). Reducing the consumption of antibiotics: Would that Be enough to slow down the dissemination of resistances in the downstream environment? Front. Microbiol. 11, 33. doi:10.3389/fmicb.2020.00033
Munir, M., Wong, K., and Xagoraraki, I. (2011). Release of antibiotic resistant bacteria and genes in the effluent and biosolids of five wastewater utilities in Michigan. Water Res. 45 (2), 681–693. doi:10.1016/j.watres.2010.08.033
Murray, C. J. L., Ikuta, K. S., Sharara, F., Swetschinski, L., Robles Aguilar, G., Gray, A., et al. (2022). Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 399, 629–655. doi:10.1016/S0140-6736(21)02724-0
Nappier, S. P., Liguori, K., Ichida, A. M., Stewart, J. R., and Jones, K. R. (2020). Antibiotic resistance in recreational waters: State of the science. Int. J. Environ. Res. Public Health 17 (21), E8034. doi:10.3390/ijerph17218034
Narciso-da-Rocha, C., Rocha, J., Vaz-Moreira, I., Lira, F., Tamames, J., Henriques, I., et al. (2018). Bacterial lineages putatively associated with the dissemination of antibiotic resistance genes in a full-scale urban wastewater treatment plant. Environ. Int. 118, 179–188. doi:10.1016/j.envint.2018.05.040
Nascimento, A. L., Souza, A. J., Andrade, P. A. M., Andreote, F. D., Coscione, A. R., Oliveira, F. C., et al. (2018). Sewage sludge microbial structures and relations to their sources, treatments, and chemical attributes. Front. Microbiol. 9, 1462. doi:10.3389/fmicb.2018.01462
Newton, R. J., McLellan, S. L., Dila, D. K., Vineis, J. H., Morrison, H. G., Eren, A. M., et al. (2015). Sewage reflects the microbiomes of human populations. mBio 6 (2), e02574. doi:10.1128/mBio.02574-14
Nguyen, A. Q., Vu, H. P., Nguyen, L. N., Wang, Q., Djordjevic, S. P., Donner, E., et al. (2021). Monitoring antibiotic resistance genes in wastewater treatment: Current strategies and future challenges. Sci. Total Environ. 783, 146964. doi:10.1016/j.scitotenv.2021.146964
Niestepski, S., Harnisz, M., Ciesielski, S., Korzeniewska, E., and Osinska, A. (2020). Environmental fate of Bacteroidetes, with particular emphasis on Bacteroides fragilis group bacteria and their specific antibiotic resistance genes, in activated sludge wastewater treatment plants. J. Hazard. Mat. 394, 122544. doi:10.1016/j.jhazmat.2020.122544
Novo, A., Andre, S., Viana, P., Nunes, O. C., and Manaia, C. M. (2013). Antibiotic resistance, antimicrobial residues and bacterial community composition in urban wastewater. Water Res. 47 (5), 1875–1887. doi:10.1016/j.watres.2013.01.010
Ontario, G.O. (2019). Removal of pathogenic bacteria from sewage-treated effluent and biosolids for agricultural purposes [Online]. Available: https://www.ontario.ca/document/design-guidelines-sewage-works/disinfection-and-supplement-treatment-processes (Accessed February 9, 2022).
Pan, M., Yau, P. C., Lee, K. C., Zhang, H., Lee, V., Lai, C. Y., et al. (2021). Nutrient accumulation and environmental risks of biosolids and different fertilizers on horticultural plants. Water Air Soil Pollut. 232 (12), 480. doi:10.1007/s11270-021-05424-5
PHAC (2020). Canadian antimicrobial resistance surveillance system report. Ottawa, Canada: Public Health Agency of Canada. Available at: https://www.canada.ca/content/dam/hc-sc/documents/services/drugs-health-products/canadian-antimicrobial-resistance-surveillance-system-2020-report/CARSS-2020-report-2020-eng.pdf (Accessed February 1, 2022).
Prestinaci, F., Pezzotti, P., and Pantosti, A. (2015). Antimicrobial resistance: A global multifaceted phenomenon. Pathog. Glob. Health 109 (7), 309–318. doi:10.1179/2047773215Y.0000000030
Rivas-Marin, E., Stettner, S., Gottshall, E. Y., Santana-Molina, C., Helling, M., Basile, F., et al. (2019). Essentiality of sterol synthesis genes in the planctomycete bacterium Gemmata obscuriglobus. Nat. Commun. 10 (1), 2916. doi:10.1038/s41467-019-10983-7
Ruckert, A., Fafard, P., Hindmarch, S., Morris, A., Packer, C., Patrick, D., et al. (2020). Governing antimicrobial resistance: A narrative review of global governance mechanisms. J. Public Health Policy 41 (4), 515–528. doi:10.1057/s41271-020-00248-9
Segata, N., Izard, J., Waldron, L., Gevers, D., Miropolsky, L., Garrett, W. S., et al. (2011). Metagenomic biomarker discovery and explanation. Genome Biol. 12 (6), R60. doi:10.1186/gb-2011-12-6-r60
Singer, A. C., Shaw, H., Rhodes, V., and Hart, A. (2016). Review of antimicrobial resistance in the environment and its relevance to environmental regulators. Front. Microbiol. 7, 1728. doi:10.3389/fmicb.2016.01728
Stalder, T., Press, M. O., Sullivan, S., Liachko, I., and Top, E. M. (2019). Linking the resistome and plasmidome to the microbiome. ISME J. 13 (10), 2437–2446. doi:10.1038/s41396-019-0446-4
Stultiens, K., van Kessel, M., Frank, J., Fischer, P., Pelzer, C., van Alen, T. A., et al. (2020). Diversity, enrichment, and genomic potential of anaerobic methane- and ammonium-oxidizing microorganisms from a brewery wastewater treatment plant. Appl. Microbiol. Biotechnol. 104 (16), 7201–7212. doi:10.1007/s00253-020-10748-z
Su, J. Q., An, X. L., Li, B., Chen, Q. L., Gillings, M. R., Chen, H., et al. (2017). Metagenomics of urban sewage identifies an extensively shared antibiotic resistome in China. Microbiome 5 (1), 84. doi:10.1186/s40168-017-0298-y
Uluseker, C., Kaster, K. M., Thorsen, K., Basiry, D., Shobana, S., Jain, M., et al. (2021). A review on occurrence and spread of antibiotic resistance in wastewaters and in wastewater treatment plants: Mechanisms and perspectives. Front. Microbiol. 12, 717809. doi:10.3389/fmicb.2021.717809
Viau, E., and Peccia, J. (2009a). Evaluation of the enterococci indicator in biosolids using culture-based and quantitative PCR assays. Water Res. 43 (19), 4878–4887. doi:10.1016/j.watres.2009.09.016
Viau, E., and Peccia, J. (2009b). Survey of wastewater indicators and human pathogen genomes in biosolids produced by class a and class B stabilization treatments. Appl. Environ. Microbiol. 75 (1), 164–174. doi:10.1128/AEM.01331-08
Vital, M., Karch, A., and Pieper, D. H. (2017). Colonic butyrate-producing communities in humans: An overview using omics data. mSystems 2 (6), e00130-17. doi:10.1128/mSystems.00130-17
Vyhnak, C. (2008). Biosolids a 'disaster waiting to happen' [Online]. Toronto Star. Available: https://www.thestar.com/news/gta/2008/07/13/biosolids_a_disaster_waiting_to_happen.html (Accessed February 9, 2022).
Whitehouse, S., Tsigaris, P., Wood, J., and Fraser, L. H. (2022). Biosolids in western Canada: A case study on public risk perception and factors influencing public attitudes. Environ. Manage. 69 (1), 179–195. doi:10.1007/s00267-021-01540-4
WHO (2015). Global action plan on antimicrobial resistance [online]. Available: http://www.emro.who.int/health-topics/drug-resistance/global-action-plan.html (Accessed October 14, 2019).
Woegerbauer, M., Bellanger, X., and Merlin, C. (2020). Cell-Free DNA: An underestimated source of antibiotic resistance gene dissemination at the interface between human activities and downstream environments in the context of wastewater reuse. Front. Microbiol. 11, 671. doi:10.3389/fmicb.2020.00671
Wolters, B., Hauschild, K., Blau, K., Mulder, I., Heyde, B. J., Sorensen, S. J., et al. (2022). Biosolids for safe land application: Does wastewater treatment plant size matters when considering antibiotics, pollutants, microbiome, mobile genetic elements and associated resistance genes? Environ. Microbiol. 24 (3), 1573–1589. doi:10.1111/1462-2920.15938
Wozniak, T. M. (2018). Letter to the editor in response to estimating the burden of antimicrobial resistance: A systematic literature review. Antimicrob. Resist. Infect. Control 7, 91. doi:10.1186/s13756-018-0379-0
Xie, W. Y., McGrath, S. P., Su, J. Q., Hirsch, P. R., Clark, I. M., and Shen, Q. (2016). Long-term impact of field applications of sewage sludge on soil antibiotic resistome. Environ. Sci. Technol. 50 (23), 12602–12611. doi:10.1021/acs.est.6b02138
Yergeau, E., Masson, L., Elias, M., Xiang, S., Madey, E., Huang, H., et al. (2016). Comparison of methods to identify pathogens and associated virulence functional genes in biosolids from two different wastewater treatment facilities in Canada. PLoS One 11 (4), e0153554. doi:10.1371/journal.pone.0153554
Young, B. C., Wu, C. H., Charlesworth, J., Earle, S., Price, J. R., Gordon, N. C., et al. (2021). Antimicrobial resistance determinants are associated with Staphylococcus aureus bacteraemia and adaptation to the healthcare environment: A bacterial genome-wide association study. Microb. Genom. 7 (11), 000700. doi:10.1099/mgen.0.000700
Zhuang, M., Achmon, Y., Cao, Y., Liang, X., Chen, L., Wang, H., et al. (2021). Distribution of antibiotic resistance genes in the environment. Environ. Pollut. 285, 117402. doi:10.1016/j.envpol.2021.117402
Keywords: biosolids, antimicrobial resistance (AMR), antimicrobial resistance genes (ARGs), antibiotic-resistant bacteria (ARB), wastewater treatment plants (WWTPs), food safety
Citation: Kang M, Naushad S, Hartke A, Firth I, Madey E, Ogunremi D and Huang H (2022) Antibiotic resistomes and microbial communities in biosolid fertilizers collected from two Canadian wastewater treatment plants in a 10-years interval-potential risks to food chains?. Front. Food. Sci. Technol. 2:894671. doi: 10.3389/frfst.2022.894671
Received: 12 March 2022; Accepted: 25 July 2022;
Published: 13 September 2022.
Edited by:
Fernando Perez Rodriguez, University of Cordoba, SpainReviewed by:
Adriana Osińska, Norwegian University of Life Sciences, NorwayHaley Sanderson, International Vaccine Centre (VIDO-InterVac), Canada
Copyright © 2022 Kang, Naushad, Hartke, Firth, Madey, Ogunremi and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongsheng Huang, aG9uZ3NoZW5nLmh1YW5nQGluc3BlY3Rpb24uZ2MuY2E=
 Mingsong Kang
Mingsong Kang Sohail Naushad
Sohail Naushad Allison Hartke1
Allison Hartke1 Hongsheng Huang
Hongsheng Huang