- 1Department of Food Science and Technology, Institute of Food Science, University of Natural Resources and Life Sciences, Vienna, Austria
- 2Department of Food Science and Biotechnology, Faculty of Agro-Industry, RUA—Royal University of Agriculture, Phnom Penh, Cambodia
The development of indigenous starter cultures for producing fermented foods that meet the expectations of Cambodians is necessary to preserve the country’s food supply. In this study, the safety of 46 lactic acid bacteria strains based on the phenotypic antibiotic susceptibility to clinically relevant antibiotics was assessed. The antibiotic susceptibility of 39 lactobacilli and seven pediococci isolated from Cambodian fermented foods to 16 antibiotics was studied according to ISO 10932/IDF 233. The results were interpreted based on the minimal inhibition concentrations obtained, using differently defined breakpoints and concentration distributions as well as data from the scientific literature. Applying only breakpoints, the results demonstrated two Lactiplantibacillus pentosus, three Companilactobacillus futsaii, three Levilactobacillus namurensis and seven Pediococcus pentosaceus strains with acquired resistance. However, considering further information, one Companilactobacillus futsaii, one Limosilactobacillus fermentum and respectively three Lactiplantibacillus pentosus and Levilactobacillus namurensis strains would possess an acquired resistance. The genetic background for the absence of transmissible antibiotic resistances in lactic acid bacteria strains intended for food application must be confirmed by molecular methods for potential starter cultures.
1 Introduction
In Cambodia, spontaneous fermentation or back-slopping is commonly used to produce fermented foods for preservation and storage as well as maintenance of the food supply (Ly D. et al., 2018). However, the growth of the involved and uncharacterized microbiota, both safe and unsafe microorganisms is completely uncontrolled and unpredictable, resulting in less uniform sensory food characteristics and compositions (Radulović et al., 2011). With rapid urbanization and increasing exports, the demand for fermented foods of good quality is increasing in Cambodia. To make safe, acceptable and consistent products with a long shelf life, controlled and accelerated fermentation processes by the application of starter cultures are needed (Holzapfel, 2002; Ammor and Mayo, 2007; Corsetti et al., 2012). Research and optimization in the development of starter cultures that produce the unique taste, aroma and texture of Cambodian foods is important to meet consumers’ expectations, preserve the variability of fermented foods, and increase competitiveness in local and international markets (Ly et al., 2019). Until now, however, only few studies have been performed on the role of microbiota in Cambodian food fermentation (Ly S. et al., 2018).
Lactic acid bacteria (LAB) produce lactic acid, acetic acid, ethanol, aroma compounds, exopolysaccharides, bacteriocins and enzymes. Resultingly, they improve the texture, increase the shelf-life and microbial safety, and contribute to a pleasant sensory profile of the final products (Leroy et al., 2006). To assure high metabolic performance as well as microbial dominance against possible undesirable microorganisms, starter cultures are added in large quantities to the initial food product (106–107 bacterial cells per gram) (Ramesh, 2007). Some LAB species belong to those mentioned on the qualified presumption of safety (QPS) list of the European Food Safety Authority (EFSA) (Adams, 1995; EFSA, 2018a). Strains of these species that fall within a QPS group do not require a full safety assessment. Regardless of the QPS status, the presence of transferable antibiotic resistance (ABR) must be tested strain by strain (Laulund et al., 2017). The absence of phenotypic ABR in strains intended for use as starters is of course preferred, but strains with phenotypic resistance may still be suitable if the transferability of the observed ABR can be excluded by analyzing the underlying genetic mechanism of the ABR in combination with additional tests (Laulund et al., 2017). Silent, pseudo-, partial or unexpressed genes may cause false positive results, leading to discordance between phenotypic and genotypic susceptibility data. Thus, uninvestigated resistance genes and mutations; novel, unknown or unusual resistance determinants; and non-specific resistance mechanisms such as multidrug resistance transporters or genes associated with oxidative stress can also lead to inconsistencies (Campedelli et al., 2019; El Issaoui et al., 2021; Flórez et al., 2021; Stefańska et al., 2021). Another challenge in resistance determination is the unavailability of standardized cutoff values for most LAB and commonly used antibiotics (ABs) in susceptibility testing (de Castilho et al., 2019; Das et al., 2020). These standardized cutoffs should also account for intrinsic resistance (IR) in cases of inherent structural and functional features (Campedelli et al., 2019; El Issaoui et al., 2021; Stefańska et al., 2021). Since it can affect the decision to consider a bacterium susceptible or resistant, the correct determination of minimal inhibition concentration (MIC) cutoff values is important (de Castilho et al., 2019; Colautti et al., 2022). Due to the great complexity of lactobacilli, comprising most food-fermenting LAB, it has been shown that susceptibility to ABs is species dependent and varies highly among species (Anisimova and Yarullina, 2019; Stefańska et al., 2021). Thus, establishing species-specific cutoffs for lactobacilli is recommended (Ma et al., 2021; Stefańska et al., 2021).
This complexity and extreme heterogeneity of lactobacilli is also the reason for a recent significant change in the taxonomy of this genus, which consisted of 261 species at the time of change (Zheng et al., 2020). Based on a polyphasic approach, Zheng et al. (2020) reclassified the former genus Lactobacillus into 25 genera, including the genus Paralactobacillus and 23 new ones. The amended genus Lactobacillus itself currently consists of species that have been assigned to the Lactobacillus delbrueckii group (Zheng et al., 2020). The updated nomenclature of the genera lactobacilli proposed by Zheng et al. is given in the parentheses, namely Lb. acidipiscis (Ligilactobacillus acidipiscis), Lb. fermentum (Limosilactobacillus fermentum), Lb. pentosus (Lactiplantibacillus pentosus), Lb. plantarum (Lactiplantibacillus plantarum), Lb. namurensis (Levilactobacillus namurensis), Lb. zymae (Levilactobacillus zymae), Lb. futsaii (Companilactobacillus futsaii). From now on, the updated nomenclature of this study is used in reference to microbiological cutoff values from EFSA guidance.
The autochthonous microbial population of Cambodian fermented foods, a valuable source of starter cultures, was examined for the presence of LAB (Ly et al., 2020). In order to exploit strains as potential starter cultures, the assessment of 46 LAB strains must be performed to guarantee safety and avoid harmful questions. Therefore, the aim of the present study is to critically assess the safety of lactobacilli and pediococci based on the phenotypic antibiotic susceptibility to clinically relevant ABs and those used in agriculture and aquaculture.
2 Materials and Methods
2.1 Bacterial Strains and Growth Conditions
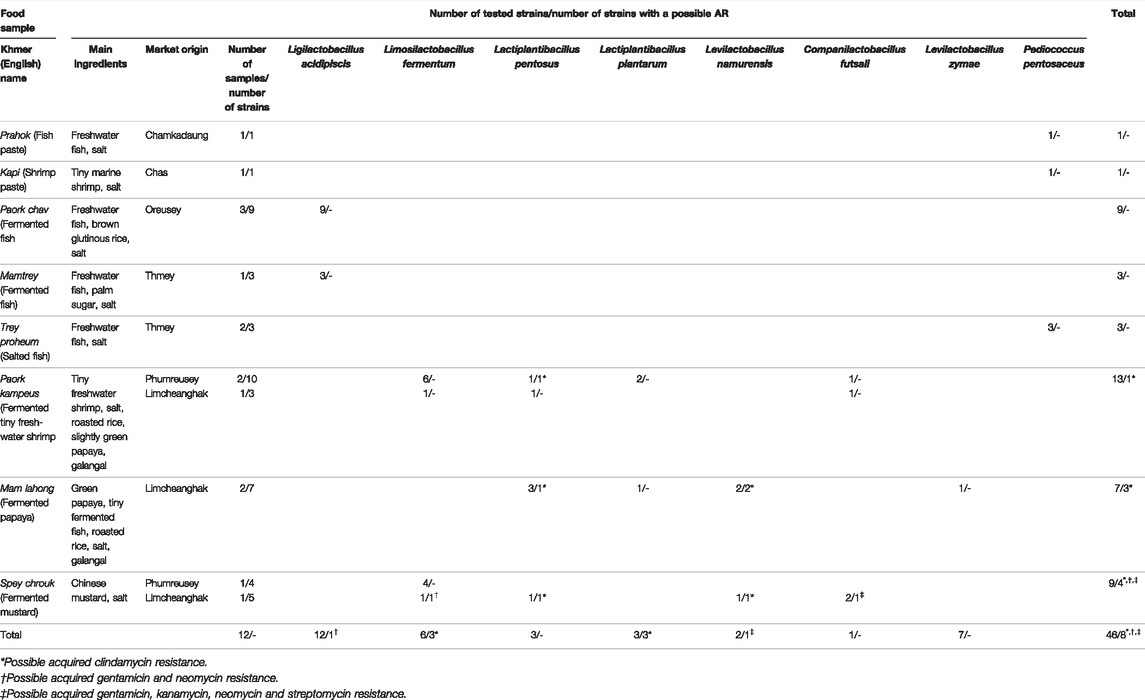
A total of 46 strains of lactobacilli and pediococci were isolated and identified in our previous report (Ly et al., 2019). These strains are members of the species Limosilactobacillus fermentum (n = 12), Ligilactobacillus acidipiscis (n = 12), Lactiplantibacillus pentosus (n = 6), Lactiplantibacillus plantarum (n = 3), Levilactobacillus namurensis (n = 3), Companilactobacillus futsaii (n = 2), Levilactobacillus zymae (n = 1), and Pd. pentosaceus (n = 7). These strains were isolated from eight fermented fishery products, two fermented vegetables and five fermented mixed products made from fish or shrimp and vegetables (Table 1) and identified at species level by partial 16S rDNA sequencing and matrix-assisted laser desorption/ionization–time of flight mass spectrometry (MALDI-TOF MS) by a Bruker MALDI Biotyper (Bruker Daltonics, Bremen, Germany). Subsequently, they were typed at strain level by (GTG)5-PCR (Ly et al., 2019). Except for one Lactobacillus strain, the results of both identification methods were consistent. Since MALDI-TOF MS with a Bruker Biotyper proved to be superior in identifying Lactobacillus species, species names determined by MALDI-TOF MS were applied during further investigation of these strains (Ly et al., 2019). In addition to these strains, the reference strains Lacticaseibacillus paracasei ATCC 334 and Lactiplantibacillus plantarum ATCC 14917 were included as quality-control strains for antimicrobial susceptibility testing according to the ISO 10932/IDF 233 standard (ISO/IDF, 2010).
Strains were maintained in DeMan Rogosa Sharpe (MRS) broth (Merck, Darmstadt, Germany) containing 20% glycerol at −80°C. The bacteria were resuscitated on MRS agar (Merck, Darmstadt, Germany) and incubated for 48 h at 28°C (Lactiplantibacillus plantarum, Levilactobacillus namurensis and Levilactobacillus zymae) or 37°C (Limosilactobacillus fermentum, Ligilactobacillus acidipiscis, Companilactobacillus futsaii and Pediococcus pentosaceus). All incubations were performed in an anaerobic cabinet (80% N2, 10% CO2 and 10% H2; Scholzen, Necker, Sankt Gallen, Switzerland).
2.2 Antimicrobial Susceptibility Testing and Minimal Inhibition Concentration Determination
Minimum inhibitory concentrations (µg ml−1) were determined in a LAB-susceptibility test medium [LSM, 90% of Iso-Sensitest (IST) broth (Oxoid, Hampshire, UK) and 10% MRS broth (Merck, Darmstadt, Germany)] according to the ISO 10932/IDF 233 standard (ISO/IDF, 2010).
Antibiotics representing the classes of aminoglycosides [gentamicin (GEN; 0.5–256 µg ml−1), kanamycin (KAN; 2–1,024 µg ml−1), streptomycin (STR; 0.5–256 µg ml−1), and neomycin (NEO; 0.5–256 µg ml−1)], β-lactams [ampicillin (AMP; 0.032–16 µg ml−1)], macrolides [erythromycin (ERY; 0.016–8 µg ml−1)], fluoroquinolones [ciprofloxacin (CIP; 0.25–128 µg ml−1)], lincosamides [clindamycin (CLI; 0.032–16 µg ml−1)], tetracyclines [tetracycline (TET; 0.125–64 µg ml−1)], phenicols [chloramphenicol (CHL; 0.125–64 µg ml−1)], glycopeptides [vancomycin (VAN; 0.25–128 µg ml−1)], oxazolidinones [linezolid (LNZ; 0.032–16 µg ml−1)], folate pathway inhibitors [trimethoprim (TMP; 0.125–64 µg ml−1), and sulfamethoxazole (SMX; 2–1,024 µg ml−1)], ansamycins [rifampicin (RIF; 0.125–64 µg ml−1)], and nitrofurans [nitrofurantoin (NIT; 0.5–256 µg ml−1)] were tested. With the exception of LNZ (Pfizer, New York, NY, United States), all ABs originated from Sigma-Aldrich (Saint Louis, MO, United States). For the production of the test plates, AB stock solutions were prepared in 20-fold concentration. GEN, KAN, NEO, STR, CLI, LNZ, TET, AMP, and VAN were dissolved in sterile distilled water. To dissolve CHL, ERY, RIF, CIP, TMP, NIT, and SMX, 95% ethanol (CHL, ERY), 100% methanol (RIF), 0.05 mol L−1 HCl (TMP), 0.1 mol L−1 HCl (CIP), 0.1 mol L−1 sodium phosphate buffer (NIT), or 2.5 mol L−1 NaOH and hot water (SMX) were required in volumes as low as possible. The remaining volume was filled with sterile distilled water. Each AB stock solution was then diluted with sterile distilled water to yield AB solutions at concentrations twice the intended concentration in the final microtiter plate. Likewise, a double-strength LSM medium was used for preparing the inoculum to get a final single-strength medium in the microtiter plate after inoculation. Fifty microliters of the double-strength AB solutions were dispensed in each well of the microtiter plates, and these plates were used immediately or frozen until use.
Inocula of the strains were prepared by suspending colonies from MRS agar plates incubated for 24–48 h into 5 ml of a 0.85% (w/v) NaCl solution to obtain a turbidity of McFarland standard 1, equivalent to an optical density of 0.16–0.2 at 625 nm by spectrophotometer (Tecan, Zürich Switzerland). Subsequently, adjusted inocula were diluted 1:500 in a double-strength LSM medium, and the microdilution plates were inoculated with 50 µl portions of the diluted inoculum. After incubating the plates under anaerobic conditions at 28°C or at 37°C for 48 h, MIC values were read as the lowest concentration of an AB agent at which visible growth was totally inhibited. For only TMP and SMX, MIC values corresponded to the concentration at which more than 80% of growth was reduced compared to the positive control. For simplification, TMP and SMX MICs were determined using a spectrophotometer (Tecan, Zürich, Switzerland).
All these analyses were performed twice. The accuracy and performance of susceptibility testing for lactobacilli and pediococci were monitored by parallel use of the quality-control strains Lactiplantibacillus plantarum ATCC 14917 (28°C) and Lacticaseibacillus paracasei ATCC 334 (37°C).
2.3 Susceptibility Interpretation
2.3.1 Description of Minimal Inhibition Concentration Distributions
A first assessment of the obtained MIC profiles for the classification of individual strains into susceptible or resistant categories was conducted by visual inspection (Stock and Wiedemann, 2001). For this purpose, the numbers of strains of a species with the same MIC value were plotted against the respective MIC values obtained for a specific AB, as shown in Supplementary Table S1 of the supplementary material. The assessment was based on the formation of uni-, bi- or multimodal distributions, whereby a unimodal distribution with one peak describes either a uniformly susceptible or intrinsic-resistant wild-type (WT) population. IR is caused by genetic errors that accumulate in existing genes of bacterial cells during the evolutionary process and are transferred to progeny cells via vertical gene transfer (Founou et al., 2016). A bimodal distribution with two distinct peaks enables the characterization of a population with low MICs, which normally corresponds to the susceptible WT population, and outliers or a small subpopulation with higher MICs, which represent non-wild-type (NWT) strains with acquired resistance (AR). In contrast to IR, AR involves the exchange of new resistance genes within and between bacterial species by horizontal gene transfer (Founou et al., 2016). Several existing AR mechanisms can lead to a multimodal distribution with three or more peaks (Sirot et al., 1996). Based on these MIC distributions, it could be decided whether a strain belongs to the susceptible WT population or not. However, the visual inspection of MIC distributions only works properly if MIC distributions are formed by several (>10) strains under test (Klare et al., 2007), which was the case with the species Ligilactobacillus acidipiscis and Limosilactobacillus fermentum.
2.3.2 Classification of Results by Different Breakpoints
Microbiological breakpoints termed as microbiological cutoffs were defined by the Panel on Additives and Products or Substances Used in Animal Feed (FEEDAP) of the EFSA to distinguish between susceptible lactobacilli or pediococci with MICs equal or lower than the established cutoff (susceptible: x µg ml−1 ≤ cutoff) and resistant lactobacilli or pediococci with MICs higher than the cutoff (resistant: x μg ml−1 > cutoff) (EFSA, 2018b). The EFSA specifies these values for the Pediococcus genus; Lactobacillus acidophilus group; and species Lb. casei/Lb. paracasei, Lb. plantarum/Lb. pentosus, Lb. reuteri and Lb. rhamnosus (EFSA, 2018b). For other species of the genus Lactobacillus, the EFSA refers to the fermentation categories, defining cutoffs for obligate heterofermentative, obligate homofermentative (OHO), and facultative heterofermentative lactobacilli (Campedelli et al., 2019). The last fermentation category includes the homofermentative species Lb. salivarius (Ligilactobacillus salivarius) (EFSA, 2018b), the type species of the genus Ligilactobacillus. Microbiological cutoffs exist for the ABs CHL, ERY, GEN, KAN, STR, CLI, TET, and AMP. VAN cutoffs are only required for the Lb. acidophilus group and other OHO lactobacilli such as Companilactobacillus futsaii. Except for KAN, equivalent microbiological breakpoints, which are referred to as epidemiological cutoff values (ECOFFs), were set by Klare et al. (2007) for these ABs and additionally for LNZ regarding the species Limosilactobacillus fermentum, Lactiplantibacillus plantarum and Pediococcus pentosaceus. ECOFFs enable differentiation between WT strains lacking AR, thus those susceptible or intrinsic resistant, and NWT strains containing AR. Moreover, Rozman et al. (2020) adopted microbiological cutoff values for lactobacilli and ABs not covered by the EFSA recommendation from the scientific literature.
For some ABs relevant to the treatment of infections caused by gram-positive anaerobes (e.g., AMP, VAN, CLI, and CHL), the European Committee on Antimicrobial Susceptibility Testing (EUCAST; https://www.eucast.org/) indicated clinical breakpoints (EUCAST, 2020a). These are best established for clinically important microorganisms and not for lactobacilli, which are rarely associated with a clinical infection (Stefańska et al., 2021). Different information such as the pharmacokinetic and pharmacodynamic of the AB, concentration-dependent toxicity and microbiological breakpoint are used for setting clinical breakpoints (Turnidge and Paterson, 2007). This means that clinical breakpoints and microbiological breakpoints do not always match (e.g., VAN). As a result, there is also a difference in terminology, and susceptible strains describe those for which there is a high likelihood of therapeutic success. Accordingly, a resistant strain is associated with a high likelihood of failure. Nevertheless, clinical breakpoints were used within this study, especially in situations where microbiological breakpoints have not been defined.
The classification into susceptible WT or resistant NWT strains can only be done reliably for AB– species combinations for which such breakpoints have been defined.
2.3.3 Classification Based on Wild-Type Populations on the European Committee on Antimicrobial Susceptibility Testing Website
In the absence of breakpoints for AB–species combinations, the EUCAST MIC distribution website (EUCAST, 2020b) was accessed and the name of the AB entered. If no distributions of the relevant species or genera were found, those of the related Enterococcus species were chosen, and the specified ECOFFs were used. However, interpretation should be done with caution (EUCAST, 2020c).
2.4 Classification of Strains With Possible AR
Strains from different fermented fishery and vegetable products were classified as resistant or susceptible to NIT, considering the description of MIC distributions and the ECOFFs on the EUCAST MIC distribution website for related Enterococcus species. AR to all other ABs was determined based on MIC distributions and available breakpoints.
3 Results
3.1 Antimicrobial Susceptibility Testing and Minimal Inhibition Concentration Determination
MIC results obtained by testing the antimicrobial susceptibility of 46 LAB (39 lactobacilli and seven pediococci) strains to 16 ABs are presented in Supplementary Table S1 of the supplementary material.
3.2 Susceptibility Interpretation
3.2.1 Description of Minimal Inhibition Concentration Distributions
Depending on the species and AB, the phenotypic susceptibility profiles of the tested LAB strains varied considerably (Supplementary Table S1). A unimodal MIC distribution at the low-end concentration range is representative for a susceptible WT population, which was observed for all investigated species and ERY (0.125–1 μg ml−1) as well as RIF (≤0.125–2 µg ml−1; Supplementary Table S1). This type of distribution was also detected for CHL and CIP but at higher concentration ranges (2–8 µg ml−1 and 2–64 µg ml−1).
Similarly, unimodal MIC distributions were found for LNZ (1–8 µg ml−1), TET (lactobacilli: 2–16 µg ml−1; pediococci: 32–64 µg ml−1) and AMP (0.125–4 µg ml−1), but respectively one species (LNZ: Ligilactobacillus acidipiscis, 0.125–0.25 µg ml−1, 2 µg ml−1; TET: Levilactobacillus namurensis, 4 µg ml−1, 16 µg ml−1; AMP: Lactiplantibacillus pentosus, 1 µg ml−1, 4–8 µg ml−1) showed a bimodal distribution. In contrast, unimodal MIC distributions were at the high-end concentration range of VAN (≥128 µg ml−1). The only species that displayed a bimodal distribution for this AB was Ligilactobacillus acidipiscis (2 µg ml−1, ≥128 µg ml−1).
MIC values from the low to medium or even high concentration range were determined for the aminoglycoside ABs GEN (≤0.5–8 µg ml−1), KAN (≤2–256 µg ml−1), NEO (≤0.5–16 µg ml−1) and STR (≤0.5–64 µg ml−1). Next to unimodal distributions, also bimodal MIC distributions were verified for GEN and the species Limosilactobacillus fermentum (≤0.5–1 µg ml−1, 4 µg ml−1) and Companilactobacillus futsaii (≤0.5 µg ml−1, 4 µg ml−1), KAN and Ligilactobacillus acidipiscis (≤2 µg ml−1, 8–16 µg ml−1) and Companilactobacillus futsaii (32 µg ml−1, 256 µg ml−1), NEO and Limosilactobacillus fermentum (≤0.5–1 µg ml−1, 8 µg ml−1), Ligilactobacillus acidipiscis (≤0.5 µg ml−1, 2–4 µg ml−1) and Companilactobacillus futsaii (2 µg ml−1, 16 µg ml−1) as well as STR and Ligilactobacillus acidipiscis (≤0.5 µg ml−1, 4–16 µg ml−1) and Companilactobacillus futsaii (8 µg ml−1, 32 µg ml−1). Bimodal distributions were also observed for CLI and Lactiplantibacillus plantarum (1 µg ml−1, 4 µg ml−1), Lactiplantibacillus pentosus (≤0.032 µg ml−1, 4–8 µg ml−1) and Ligilactobacillus acidipiscis (peaks: ≤ 0.032 µg ml−1, 0.12 µg ml−1), while the CLI MIC distributions of all other species were unimodal (≤0.032–16 µg ml−1).
Multimodal distributions were described for some species and the ABs NIT, SMX and TMP. Thus, low (4 µg ml−1), moderately high (32 µg ml−1), and high (128 µg ml−1) MICs were obtained for NIT and the species Lactiplantibacillus pentosus. In addition, a bimodal distribution was found for the tested Ligilactobacillus acidipiscis species (4 µg ml−1, 64–128 µg ml−1). Generally, the NIT MICs were spread over a wide range (1–128 µg ml−1), while those for SMX were at a higher concentration range or even higher than the highest concentration investigated (64– > 1,024 µg ml−1). A multimodal MIC distribution with three peaks at 64 µg ml−1, 256 µg ml−1 and 1,024 µg ml−1 was determined for Limosilactobacillus fermentum and TMP. The species Lactiplantibacillus pentosus (64–128 µg ml−1, 512 µg ml−1) and Lb. acidpiscis (peaks: 256 µg ml−1, 1,024 µg ml−1) displayed bimodal distributions. The TMP MICs were distributed over the entire range tested (≤0.125–64 µg ml−1). Multimodal and bimodal distributions were detected for Ligilactobacillus acidipiscis (0.5–1 µg ml−1, 4–8 µg ml−1, 64 µg ml−1) and Companilactobacillus futsaii (peaks: 0.5 µg ml−1, 2 µg ml−1), respectively. For these three ABs, the distributions of all species not mentioned were unimodal (NIT: 1–64 µg ml−1; SMX; 128– >1,024 µg ml−1; TMP: lactobacilli: ≤ 0.125–2 µg ml−1, pediococci: 8–32 µg ml−1).
3.2.2 Classification of Results by Different Breakpoints
Microbiological breakpoints and even clinical breakpoints of various organizations were used to distinguish between susceptible WT and resistant NWT lactobacilli and pediococci (Supplementary Table S1). While several of these different breakpoints are available for the same AB–species combinations, others have none, e.g., NIT and SMX. As the breakpoints of these organizations are not always concordant, the highest was used for classification (Table 2).
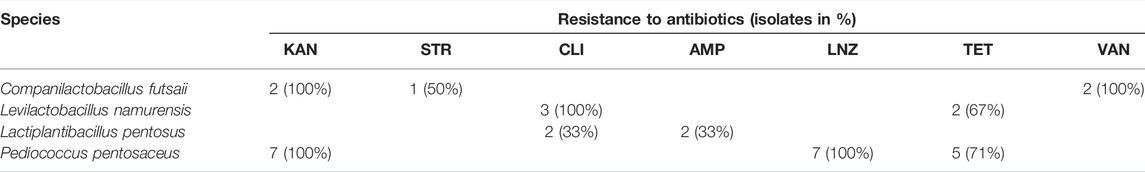
TABLE 2. Classification of LAB isolates as resistant to tested antibiotics based on available breakpoints.
Using all available breakpoints, no resistances were detected for all strains and the ABs CHL, ERY and GEN, as well as all lactobacilli tested and NEO, CIP, RIF and TMP. No corresponding breakpoints were found for pediococci and the last four ABs. If the same as for lactobacilli are applied, all seven Pediococcus pentosaceus strains would also have been susceptible. The affiliation of these seven strains to the WT populations regarding these four ABs is reinforced by the appearance of unimodal MIC distributions.
3.2.3 Classification Based on Wild-Type Populations of the European Committee on Antimicrobial Susceptibility Testing Website
Fairly high ECOFFs were set for NIT and the specified Enterococcus spp. on the EUCAST website (e.g., 32 µg ml−1: E. faecalis; 256 µg ml−1: E. faecium). With an ECOFF of 32 µg ml−1 for lactobacilli and pediococci, all strains would be susceptible to NIT except one Lactiplantibacillus pentosus (16.7%) and 10 Ligilactobacillus acidipiscis (83.3%) strains, whereas all strains tested would be susceptible when applying an ECOFF of 256 µg ml−1 (EUCAST, 2020d). No suitable ECOFF was found for SMX. However, since all strains had MICs between 64 and ≥ 1,024 µg ml−1, it could be assumed that all lactobacilli and pediococci tested are intrinsically resistant to SMX (Supplementary Table S1).
3.3 Identification of Strains With Possible AR
Taking the MIC distributions and breakpoints into account, one Companilactobacillus futsaii strain appears to possess an AR against all aminoglycoside ABs examined in this study, while one Limosilactobacillus fermentum strain may be resistant to GEN and NEO. Three Levilactobacillus namurensis and three Lactiplantibacillus pentosus strains could be resistant to CLI.
Respectively, one strain of each of these species (32a, 32b, 32c, 32e) was isolated from the same sample. This was a spey chrouk (fermented mustard) sample made from Chinese mustard purchased at the Limcheanghak market (Table 1). One Levilactobacillus namurensis (36d) and one Lactiplantibacillus pentosus (36f) strain were also isolated from the same sample, which was a mam lahong (fermented green papaya) bought at the same market. In addition, another CLI-resistant Levilactobacillus namurensis (37b) strain was isolated from mam lahong from the Limcheanghak market. The ingredients of mam lahong are green papaya, tiny slightly fermented fish, salt, roasted rice and galangal. Another possibly CLI-resistant Lactiplantibacillus pentosus strain (45e) was detected in paork kampeus (fermented tiny freshwater shrimp), which contains shrimp, roasted rice, mostly ripe papaya, galangal and salt. This sample, however, comes from another market, Phumreusey (Table 1).
4 Discussion
Lactobacilli are usually susceptible to cell-wall-targeting ß-lactams (AMP) and inhibitors of protein synthesis like phenicols (CHL), macrolides (ERY), lincosamides (CLI), and tetracyclines (TET) (Anisimova and Yarullina, 2019). Except for CLI, mainly unimodal distributions of MICs below the corresponding breakpoint were found for these ABs. Therefore, most lactobacilli in this study were also susceptible to AMP, CHL, ERY and TET. The sole bimodal MIC distributions in the case of the AB–species combinations AMP– Lactiplantibacillus pentosus and TET–Levilactobacillus namurensis resulted from rare strains possessing MICs outside the unimodal MIC distribution. This could be due to the small number of strains tested per species, which should be at least 10 in order to obtain acceptable MIC ranges for the definition of WT populations (Klare et al., 2007). According to Klare et al. (2007), such “outsiders” can be ignored if their MICs are ± one dilution step away from the values of the other strains, as this is the normal standard deviation for MIC determinations. This was the case for both AB–species combinations. It is therefore questionable whether the two TET-resistant Levilactobacillus namurensis or two AMP-resistant Lactiplantibacillus pentosus strains are indeed resistant. The MICs of these strains were only one dilution step higher than the respective breakpoint. Furthermore, the WT distribution of the AMP–Lactiplantibacillus pentosus combination is split by the breakpoint, which shouldn’t be (EUCAST, 2019). However, a splitting of WT populations through breakpoints cannot be completely avoided. Due to differences in the applied susceptibility testing method as well as intra- and inter-laboratory variations when using the same method, there will never be a perfect correlation between phenotypic data and breakpoints (Turnidge, 2016). For example, although there is a standard for non-enterococcal LAB for determining MICs to ABs that uses the widely applied microdilution broth method and the specially defined LSM medium (ISO/IDF, 2010), the MRS medium is still used (Colautti et al., 2022), leading to inconsistent MIC determination in LAB (Álvarez-Cisneros and Ponce-Alquicira, 2018). Bimodal MIC distributions were also observed for CLI and Lactiplantibacillus plantarum, Lactiplantibacillus pentosus and Ligilactobacillus acidipiscis. In the case of Lactiplantibacillus plantarum and Ligilactobacillus acidipiscis, nearby outsiders may in turn be responsible for the observed profile. However, several dilution steps lie between the respective three Lactiplantibacillus pentosus strains with low and higher MICs. In addition to all three Levilactobacillus namurensis strains, two of the three Lactiplantibacillus pentosus strains with higher MICs were also CLI resistant according to the corresponding breakpoint. This suggests an AR.
All tested lactobacilli were susceptible to LNZ and RIF, which corresponds to the literature (Sirichoat et al., 2020). Even the MIC values obtained for LNZ and RIF in this study fall within the range of the four or five most frequently received LNZ (1–8 µg ml−1) or RIF (0.12–2 µg ml−1) MICs of Campedelli et al. (2019). This may be due to the use of the same method for antimicrobial susceptibility testing with the microdilution broth method, LSM medium, ISO 10932/IDF 233, and 48 h incubation, which may largely avoid variations (Turnidge, 2016). The applied LSM medium supports the growth of some Lactobacillus spp. only weakly or not at all (Mayrhofer et al., 2014; Campedelli et al., 2019). As the MICs of weakly growing strains might be falsely low, the outcomes of these strains are even more confusing than those of strains that fail to grow (Mayrhofer et al., 2014). The bi- and multimodal MIC distributions of the Ligilactobacillus acidipiscis species for more than half of the ABs tested (KAN, NEO, STR, CLI, LNZ, VAN, NIT, SMX, TMP) could be attributed to the poor growth of this species. The subpopulations of this species with higher MIC values, which are mostly in the same concentration ranges as those of other Lactobacillus strains classified as susceptible, could be formed by better growing Ligilactobacillus acidipiscis strains. In particular, the VAN-resistant population with three susceptible outsiders is confusing because the type-species of Ligilactobacillus genus is Ligilactobacillus salivarius, which is normally intrinsically resistant to VAN (Zhang et al., 2018). In addition to its weak growth, incorrect species identification could be another reason for the bi- and multimodal MIC distributions of the Ligilactobacillus acidipiscis species. Proper species identification is a prerequisite for appropriate data evaluation (Laulund et al., 2017). Only representatives of the Lb. acidophilus group, which belong to the OHO lactobacilli, are described as susceptible to VAN (Hamilton and Shah, 1998; Adimpong et al., 2012; Abriouel et al., 2015). However, the three allegedly VAN-susceptible strains were assigned to the species Ligilactobacillus acidipiscis by both methods applied with a high level of reliability (Ly et al., 2019). This supports the assumption of bi- and multimodal distributions due to weakly growing strains. Since the two Companilactobacillus futsaii strains are OHO lactobacilli, it is assumed that they are also VAN susceptible (EFSA, 2018b). However, both strains had MICs > 128 µg ml−1, greatly exceeding the microbiological VAN cutoff value of EFSA for OHO Lactobacillus (2 µg ml−1) (EFSA, 2018b). Relevant susceptibility data for other OHO species of the phylogenetic Lb. alimentarius group (Companilactobacillus genus), which contains the Companilactobacillus futsaii species (Pot et al., 2014), are rare, but the related species Lb. crustorum (Companilactobacillus crustorum) and Lb. farciminis (Companilactobacillus farciminis) were also described as VAN resistant (Scheirlinck et al., 2007; Sandes et al., 2017). Moreover, Zhang et al. (2018) predicted VAN resistance for the Lb. alimentarius group (Companilactobacillus genus) members by investigating the sequences of the active site of the D-alanyl-D-alanine dipeptide ligase, which determines VAN resistance or susceptibility.
Resistance to aminoglycoside ABs is also considered to be intrinsic in Lactobacillus. The reduced susceptibility of lactobacilli to aminoglycosides originates from membrane impermeability due to the lack of cytochrome-mediated drug transport (Elkins and Mullis, 2004; Wolupeck et al., 2017). Campedelli et al. (2019) determined a higher resistance toward KAN and STR than to GEN and NEO. Within this study, the MIC values of GEN and NEO also tended towards the low end of the concentration range examined, while those of STR and KAN were distributed almost over the entire concentration range. The higher susceptibility to GEN is most likely linked to the superior ability of this AB to cross the membrane (Campedelli et al., 2019). Most bimodal distributions for these ABs may again be explained by the small number of strains tested per species and the poor growth of Ligilactobacillus acidipiscis strains. Interestingly, one Limosilactobacillus fermentum strain had higher MIC values for GEN and NEO than the other strains of the same species. Nevertheless, this strain was not classified as resistant because its MICs were lower than the corresponding breakpoints. In contrast, both Companilactobacillus futsaii strains should be KAN resistant based on the breakpoint. However, only the second Companilactobacillus futsaii strain could in fact possess AR because it had a higher KAN MIC in addition to GEN and NEO and was also resistant to STR. AR to aminoglycosides is explained by impaired uptake, efflux, target modification or enzymatic inactivation. Of these, enzymatic inactivation is generally associated with gram-positive bacteria, especially LAB (Aslangul et al., 2006; Jaimee and Halami, 2016).
Unimodal MIC distributions in the concentration range of 2–32 µg ml−1 were determined for CIP. The higher MICs of lactobacilli for quinolones such as CIP are considered to arise from IR due to cell-wall impermeability or efflux mechanisms (Anisimova and Yarullina, 2019). Broad bimodal or multimodal profiles were observed for NIT. The basic action mechanism of nitrofuran ABs such as NIT is unclear (Vardanyan and Hruby, 2016). Multiple mechanisms like the inhibition of many microbial enzyme systems or damage to DNA, RNA and proteins are described (Blass, 2015; Vardanyan and Hruby, 2016). These diverse mechanisms could explain the obtained profiles, since accumulations of different numbers of adequate resistance mechanisms could lead to a broad MIC distribution (Phillips, 1998). Therefore, further studies are required to generally elucidate the mechanisms of this AB and the associated resistances. Next to NIT, MIC profiles that were difficult to evaluate were also observed for TMP and SMX. This may be due to the presence of antagonists (TMP: thymidine; SMX: p-aminobenzoic acid) in the LSM medium (Klare et al., 2007) and reading the MIC at a concentration with a growth inhibition ≥80% (ISO/IDF, 2010). Both ABs act by targeting successive steps in the folic-acid metabolism necessary for DNA synthesis (Minato et al., 2018). Lactobacilli have been described to be intrinsically resistant to both ABs (Rojo-Bezares et al., 2006; Danielsen et al., 2007; Abriouel et al., 2015). TMP resistance is supposed to be due to the presence of a TMP-insensitive target enzyme or the lack of a metabolic pathway of folic-acid synthesis (Guo et al., 2017; Wolupeck et al., 2017; Rozos et al., 2018). Because of a wide MIC range (0.125–64 µg ml−1), which was also obtained by Guo et al. (2017) (0.25–64 µg ml−1), resistance to TMP was not easy to recognize. Contrarily, all SMX MIC values were at the higher concentration range (64–>1,024 µg ml−1). Resistance to this AB is probably due to the cell-wall structure and membrane impermeability, sometimes complemented by efflux mechanisms (Rojo-Bezares et al., 2006).
Most species of Pediococcus are described to be intrinsically resistant to aminoglycoside ABs (GEN, KAN, STR, NEO), VAN, TET, CIP and SMX with or without TMP (Rojo-Bezares et al., 2006; Danielsen et al., 2007; Hummel et al., 2007; Franz et al., 2014; Singla et al., 2018; Zommiti et al., 2018). In contrast, pediococci are mainly found to be susceptible to AMP, CHL and ERY (Singla et al., 2018). Compared to lactobacilli, however, susceptibility data are rare, meaning fewer breakpoints have been defined. Frequently, these are only given at genus level and may even be unsuitable (Ludin et al., 2018), leading to a lack of correlation between phenotypic and genotypic analyses of ABR in pediococci (Akpınar Kankaya and Tuncer, 2020). According to these breakpoints, all seven Pediococcus strains would have AR to KAN and five strains to TET, regardless of their supposed IR. As with the AMP–Lactiplantibacillus pentosus combination, the MIC distribution of the seven Pd. pentosaceus strains tested, which was revealed as unimodal, mostly over two dilutions, was split by the indicated TET breakpoint. Based on the LNZ breakpoint, all strains would also possess AR to this AB. Although another medium and incubation time were used, comparable MIC ranges (4–8 µg ml−1) were obtained by Danielsen et al. (2007) (2–4 µg ml−1) and Klare et al. (2007) (0.5–2 µg ml−1) for this AB. It is therefore questionable whether all these strains have indeed AR to KAN, LNZ and TET. All seven Pediococcus pentosaceus strains seem to be susceptible to CHL, ERY, CLI and AMP as well as IR to GEN, STR and VAN. For ABs for which no categorization of these strains was possible due to missing breakpoints (NEO, CIP, RIF, SMX, TMP), susceptibilities similar to those of the tested lactobacilli and those mentioned in the literature were found. As with the tested lactobacilli, it was not possible to classify the Pediococcus pentosaceus strains for NIT.
Already Laulund et al. (2017) reported a lack of knowledge about normal ABR levels for individual species, which can make a proper assessment of these species difficult. Furthermore, according to EFSA, the microbiological cutoff values, which were last updated in 2018, are only a pragmatic response, that is, reviewed regularly and adjusted if necessary (EFSA, 2012). Thus, many of the species examined in the studies of Campedelli et al. (2019) and Ma et al. (2021) had MICs above the EFSA-recommended cutoffs, suggesting that these should be reviewed with consideration for the genetic basis of resistance (Campedelli et al., 2019) to better distinguish between susceptible strains and those with AR (Ma et al., 2021). The current study also indicates that with the exception of the Lb. acidophilus group, a re-evaluation would be needed for the microbiological VAN cutoffs of OHO lactobacilli as well as for the microbiological KAN, LNZ and TET cutoffs of pediococci. Compared to VAN resistance in lactobacilli, however, further investigations are necessary to determine the underlying genetic mechanism of KAN and TET resistance in Pediococcus pentosaceus as well as exclude AR for LNZ in this species.
Although the LAB investigated in this study were isolated from eight fermented fish products, two fermented vegetables and five fermented mixed products made from fish or shrimp and vegetables (Table 1), all possibly resistant lactobacilli originated from fermented vegetables or mixed products. Interestingly, the aminoglycoside AB STR is one of the primary ABs used in vegetable farming. GEN is also used in crops but is not as common (Williams-Nguyen et al., 2016). Environmental pollution could also be the reason for the emergence of lactobacilli with AR (Founou et al., 2016). Since no guidelines address animal waste disposal and irrigation, untreated or insufficiently treated AB-rich wastewater is common in developing countries (Nadimpalli et al., 2018). The susceptibility of LAB strains isolated from fermented fishery products can be explained by the use of freshwater fish or shrimp as ingredients. Captured freshwater fish make up the largest part of Cambodia’s fish supply (Baran and Gallego, 2015). Compared to aquaculture, which is in its infant stage in this country (Lang, 2015), the AB selection pressure in freshwater would be much lower. However, it is difficult to verify these possible explanations for AR in lactobacilli based on general information, as surveillance of AB use and ABR in non-humans has been relatively neglected in Cambodia and relevant data are lacking (Om and McLaws, 2016; Zellweger et al., 2017; Reed et al., 2019).
5 Conclusion
The results demonstrate that two Lactiplantibacillus pentosus, three Companilactobacillus futsaii, three Levilactobacillus namurensis and seven Pediococcus pentosaceus strains possess AR characteristics when applying only breakpoint assessment. Other results were found with one Companilactobacillus futsaii, one Limosilactobacillus fermentum, three Lactiplantibacillus pentosus and three Levilactobacillus namurensis strains that would possess AR characteristics if taking into account further available scientific knowledge. In particular, it must be considered that microbiological cutoff values published by EFSA are only a pragmatic response. They are under review regularly and modified when necessary. Since phenotypic MIC testing alone is only useful as a preliminary screening, a subsequent molecular analysis is usually required to identify the possible presence and nature of genes associated with phenotypic resistance and to differentiate between IR and AR. Therefore, detailed analysis by whole-genome sequencing is the next step in determining the suitability of these LAB strains as potential autochthonous starter cultures for the production of fermented foods that meet the dietary habits of Cambodians.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
Conceptualization, DL, KD, and SM; Investigation, DL; Resources, DL and KD; Writing—Original Draft Preparation, DL and SM; Writing—Review and Editing, DL, SM, and KD; Supervision, KD and SM. All authors agree to be accountable for the content of the work.
Funding
This study was financed by the European Commission for the Erasmus Mundus Action 2 under the ALFABET project with the reference number 552071 and partially funded by the Schlumberger Foundation Faculty for the Future Program in support of the first author conducting a PhD.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors thank the EQ-BOKU VIBT GmbH and the BOKU Core Facility Food & Bio Processing for providing the MALDI-TOF mass spectrometer (Bruker MALDI Biotyper) for the identification of bacterial isolates.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/frfst.2022.892319/full#supplementary-material
References
Abriouel, H., Casado Muñoz, M. d. C., Lavilla Lerma, L., Pérez Montoro, B., Bockelmann, W., Pichner, R., et al. (2015). New Insights in Antibiotic Resistance of Lactobacillus Species from Fermented Foods. Food Res. Int. 78, 465–481. doi:10.1016/j.foodres.2015.09.016
Adams, M. (1995). On the Safety of Lactic Acid Bacteria from Food. Int. J. Food Microbiol. 27, 263–264. doi:10.1016/0168-1605(95)00067-t
Adimpong, D. B., Nielsen, D. S., Sorensen, K. I., Derkx, P. M., and Jespersen, L. (2012). Genotypic Characterization and Safety Assessment of Lactic Acid Bacteria from Indigenous African Fermented Food Products. BMC Microbiol. 12, 1–12. doi:10.1186/1471-2180-12-75
Akpınar Kankaya, D., and Tuncer, Y. (2020). Antibiotic Resistance in Vancomycin‐resistant Lactic Acid Bacteria (VRLAB) Isolated from Foods of Animal Origin. J. Food Process. Preserv. 44, 1–14. doi:10.1111/jfpp.14468
Álvarez-Cisneros, Y. M., and Ponce-Alquicira, E. (2018). “Antibiotic Resistance in Lactic Acid Bacteria,” in Antimicrobial Resistance - A Global Threat. Editor Y. Kumar (London, UK: IntechOpen).
Ammor, M. S., and Mayo, B. (2007). Selection Criteria for Lactic Acid Bacteria to Be Used as Functional Starter Cultures in Dry Sausage Production: An Update. Meat Sci. 76, 138–146. doi:10.1016/j.meatsci.2006.10.022
Anisimova, E. A., and Yarullina, D. R. (2019). Antibiotic Resistance of Lactobacillus Strains. Curr. Microbiol. 76, 1407–1416. doi:10.1007/s00284-019-01769-7
Aslangul, E., Massias, L., Meulemans, A., Chau, F., Andremont, A., Courvalin, P., et al. (2006). Acquired Gentamicin Resistance by Permeability Impairment in Enterococcus faecalis. Antimicrob. Agents Chemother. 50, 3615–3621. doi:10.1128/AAC.00390-06
Baran, E., and Gallego, G. (2015). Cambodia's Fisheries: a Decade of Changes and Evolution. Catch Cult., 28–31. https://digitalarchive.worldfishcenter.org/bitstream/handle/20.500.12348/460/3961_WF-3961.pdf?sequence1=
Blass, B. (2015). “Case Studies in Drug Discovery,” in Basic Principles of Drug Discovery and Development. Editor B. E. Blass (CA, USA: Academic Press), 499–530. doi:10.1016/b978-0-12-411508-8.00013-x
Campedelli, I., Mathur, H., Salvetti, E., Clarke, S., Rea, M. C., Torriani, S., et al. (2019). Genus-wide Assessment of Antibiotic Resistance in Lactobacillus Spp. Appl. Environ. Microbiol. 85, 1–21. doi:10.1128/AEM.01738-18
Colautti, A., Arnoldi, M., Comi, G., and Iacumin, L. (2022). Antibiotic Resistance and Virulence Factors in Lactobacilli: Something to Carefully Consider. Food Microbiol. 103, 103934. doi:10.1016/j.fm.2021.103934
Corsetti, A., Perpetuini, G., Schirone, M., Tofalo, R., and Suzzi, G. (2012). Application of Starter Cultures to Table Olive Fermentation: An Overview on the Experimental Studies. Front. Microbiol. 3, 1–6. doi:10.3389/fmicb.2012.00248
Danielsen, M., Simpson, P. J., O'Connor, E. B., Ross, R. P., and Stanton, C. (2007). Susceptibility of Pediococcus spp. to antimicrobial agents. J. Appl. Microbiol. 102, 384–389. doi:10.1111/j.1365-2672.2006.03097.x
Das, D. J., Shankar, A., Johnson, J. B., and Thomas, S. (2020). Critical insights into antibiotic resistance transferability in probiotic Lactobacillus. Nutrition 69, 110567. doi:10.1016/j.nut.2019.110567
de Castilho, N. P. A., Nero, L. A., and Todorov, S. D. (2019). Molecular screening of beneficial and safety determinants from bacteriocinogenic lactic acid bacteria isolated from Brazilian artisanal calabresa. Lett. Appl. Microbiol. 69, 204–211. doi:10.1111/lam.13194
EFSA (2012). Guidance on the assessment of bacterial susceptibility to antimicrobials of human and veterinary importance. EFSA J. 10, 2740. doi:10.2903/j.efsa.2012.2740
EFSA (2018b). Guidance on the characterisation of microorganisms used as feed additives or as production organisms. EFSA J. 16, 5206. doi:10.2903/j.efsa.2018.5206
EFSA (2018a). Update of the list of QPS‐recommended biological agents intentionally added to food or feed as notified to EFSA 8: suitability of taxonomic units notified to EFSA until March 2018. EFSA J. 16, 5315. doi:10.2903/j.efsa.2018.5315
El Issaoui, K., Khay, E. O., Abrini, J., Zinebi, S., Amajoud, N., Senhaji, N. S., et al. (2021). Molecular identification and antibiotic resistance of bacteriocinogenic lactic acid bacteria isolated from table olives. Arch. Microbiol. 203, 597–607. doi:10.1007/s00203-020-02053-0
Elkins, C. A., and Mullis, L. B. (2004). Bile-mediated aminoglycoside sensitivity in Lactobacillus species likely results from increased membrane permeability attributable to cholic acid. Appl. Environ. Microbiol. 70, 7200–7209. doi:10.1128/AEM.70.12.7200-7209.2004
EUCAST (2020c). Antimicrobial susceptibility tests on groups of organisms or agents for which there are no EUCAST breakpoints. Available at: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/General_documents/Organisms_and_agents_without_breakpoints_20160626.pdf (Accessed November 30, 2020).
EUCAST (2020d). Antimicrobial wild type distributions of microorganisms. Available at: https://mic.eucast.org/search/?search%5Bmethod%5D=mic&search%5Bantibiotic%5D=138&search%5Bspecies%5D=-1&search%5Bdisk_content%5D=-1&search%5Blimit%5D=50 (Accessed November 30th, 2020).
EUCAST (2020a). Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 10.0; 2020.
EUCAST (2020b). MIC and zone distributions and ECOFFs. Available at: https://www.eucast.org/mic_distributions_and_ecoffs/(Accessed: November 30, 2020).
EUCAST (2019). MIC distributions and epidemiological cut-off value (ECOFF) setting. EUCAST Sop. 10 (1).
Florez, A. B., Vazquez, L., Rodriguez, J., and Mayo, B. (2021). Directed recovery and molecular characterization of antibiotic resistance plasmids from cheese bacteria. Int. J. Mol. Sci. 22, 7801. doi:10.3390/ijms22157801
Founou, L. L., Founou, R. C., and Essack, S. Y. (2016). Antibiotic resistance in the food chain: A developing country-perspective. Front. Microbiol. 7, 1–19. doi:10.3389/fmicb.2016.01881
Franz, C. M. A. P., Endo, A., Abriouel, H., Reenen, C. A. V., Gálvez, A., and Dicks, L. M. T. (2014). “The genus Pediococcus,” in Lactic Acid Bacteria: Biodiversity and Taxonomy. Editors W. H. Holzapfel, and B. J. B. Wood (Chennai, India: Wiley Blackwell), 359–376. doi:10.1002/9781118655252.ch21
Guo, H., Pan, L., Li, L., Lu, J., Kwok, L., Menghe, B., et al. (2017). Characterization of antibiotic resistance genes from Lactobacillus isolated from traditional dairy products. J. Food Sci. 82, 724–730. doi:10.1111/1750-3841.13645
Hamilton, J. M., and Shah, S. (1998). Vancomycin susceptibility as an aid to the identification of lactobacilli. Lett. Appl. Microbiol. 26, 153–154. doi:10.1046/j.1472-765X.1998.00297.x
Holzapfel, W. H. (2002). Appropriate starter culture technologies for small-scale fermentation in developing countries. Int. J. Food Microbiol. 75, 197–212. doi:10.1016/s0168-1605(01)00707-3
Hummel, A. S., Hertel, C., Holzapfel, W. H., and Franz, C. M. (2007). Antibiotic resistances of starter and probiotic strains of lactic acid bacteria. Appl. Environ. Microbiol. 73, 730–739. doi:10.1128/aem.02105-06
ISO/IDF (2010). Milk and Milk Products — Determination of the Minimal Inhibitory Concentration (MIC) of Antibiotics Applicable to Bifidobacteria and Non-enterococcal Lactic Acid Bacteria (LAB). 1st ed. Geneva, Switzerland: ISO and IDF. ISO 10932/IDF 233.
Jaimee, G., and Halami, P. M. (2016). Emerging resistance to aminoglycosides in lactic acid bacteria of food origin-an impending menace. Appl. Microbiol. Biotechnol. 100, 1137–1151. doi:10.1007/s00253-015-7184-y
Klare, I., Konstabel, C., Werner, G., Huys, G., Vankerckhoven, V., Kahlmeter, G., et al. (2007). Antimicrobial susceptibilities of Lactobacillus, Pediococcus and Lactococcus human isolates and cultures intended for probiotic or nutritional use. J. Antimicrob. Chemother. 59, 900–912. doi:10.1093/jac/dkm035
Lang, O. (2015). “Current status of sustainable aquaculture in Cambodia,” in Resource Enhancement and Sustainable Aquaculture Practices in Southeast Asia: Challenges in Responsible Production of Aquatic Species: Proceedings of the International Workshop on Resource Enhancement and Sustainable Aquaculture Practices in Southeast Asia 2014 (RESA). Editors M. R. R. Romana-Eguia, F. D. Parado-Estepa, N. D. Salayo, and M. J. H. Lebata-Ramos (Tigbauan, Iloilo, Philippines: Aquaculture Dept., Southeast Asian Fisheries Development Center), 27–40.
Laulund, S., Wind, A., Derkx, P. M. F., and Zuliani, V. (2017). Regulatory and safety requirements for food cultures. Microorganisms 5. doi:10.3390/microorganisms5020028
Leroy, F., Verluyten, J., and De Vuyst, L. (2006). Functional meat starter cultures for improved sausage fermentation. Int. J. Food Microbiol. 106, 270–285. doi:10.1016/j.ijfoodmicro.2005.06.027
Ludin, P., Roetschi, A., Wuthrich, D., Bruggmann, R., Berthoud, H., and Shani, N. (2018). Update on tetracycline susceptibility of Pediococcus acidilactici based on strains isolated from Swiss cheese and whey. J. Food Prot. 81, 1582–1589. doi:10.4315/0362-028x.jfp-18-160
Ly, D., Mayrhofer, S., Agung Yogeswara, I. B., Nguyen, T.-H., and Domig, K. J. (2019). Identification, classification and screening for γ-amino-butyric acid production in lactic acid bacteria from Cambodian fermented foods. Biomolecules 9, 768. doi:10.3390/biom9120768
Ly, D., Mayrhofer, S., and Domig, K. J. (2018a). Significance of traditional fermented foods in the lower Mekong subregion: A focus on lactic acid bacteria. Food Biosci. 26, 113–125. doi:10.1016/j.fbio.2018.10.004
Ly, D., Mayrhofer, S., Schmidt, J. M., Zitz, U., and Domig, K. J. (2020). Biogenic amine contents and microbial characteristics of Cambodian fermented foods. Foods 9, 198. doi:10.3390/foods9020198
Ly, S., Mith, H., Tarayre, C., Taminiau, B., Daube, G., Fauconnier, M. L., et al. (2018b). Impact of microbial composition of Cambodian traditional dried starters (dombea) on flavor compounds of rice wine: Combining amplicon sequencing with HP-SPME-GCMS. Front. Microbiol. 9, 1–15. doi:10.3389/fmicb.2018.00894
Ma, Q., Pei, Z., Fang, Z., Wang, H., Zhu, J., Lee, Y. K., et al. (2021). Evaluation of Tetracycline Resistance and Determination of the Tentative Microbiological Cutoff Values in Lactic Acid Bacterial Species. Microorganisms 9, 2128. doi:10.3390/microorganisms9102128
Mayrhofer, S., Zitz, U., Birru, F. H., Gollan, D., Golos, A. K., Kneifel, W., et al. (2014). Comparison of the CLSI guideline and ISO/IDF standard for antimicrobial susceptibility testing of Lactobacilli. Microb. Drug Resist 20, 591–603. doi:10.1089/mdr.2013.0189
Minato, Y., Dawadi, S., Kordus, S. L., Sivanandam, A., Aldrich, C. C., and Baughn, A. D. (2018). Mutual potentiation drives synergy between trimethoprim and sulfamethoxazole. Nat. Commun. 9, 1003. doi:10.1038/s41467-018-03447-x
Nadimpalli, M., Delarocque-Astagneau, E., Love, D. C., Price, L. B., Huynh, B. T., Collard, J. M., et al. (2018). Combating global antibiotic resistance: Emerging one health concerns in lower- and middle-income countries. Clin. Infect. Dis. 66, 963–969. doi:10.1093/cid/cix879
Om, C., and McLaws, M. L. (2016). Antibiotics: practice and opinions of Cambodian commercial farmers, animal feed retailers and veterinarians. Antimicrob. Resist Infect. Control 5, 1–8. doi:10.1186/s13756-016-0147-y
Phillips, I. (1998). The 1997 Garrod Lecture. The subtleties of antibiotic resistance. J. Antimicrob. Chemother. 42, 5–12. doi:10.1093/jac/42.1.5
Pot, B., Felis, G. E., Bruyne, K. D., Tsakalidou, E., Papadimitriou, K., Leisner, J., et al. (2014). “The genus Lactobacillus,” in Lactic Acid Bacteria: Biodiversity and Taxonomy. Editors W. H. Holzapfel, and B. J. B. Wood (Chennai, India: Wiley Blackwell), 249–353. doi:10.1002/9781118655252.ch19
Radulović, Z., Miočinović, J., Pudja, P., Barać, M., Miloradović, Z., Paunović, D., et al. (2011). The application of autochthonous lactic acid bacteria in white brined cheese production. Mljekarstvo 61, 15–25. https://hrcak.srce.hr/file/97625
Ramesh, K. V. (2007). Food Microbiology Paperback – 1 January 2007, Tamil Nadu 600005, India. Chennai: MJP Publishers.
Reed, T. A. N., Krang, S., Miliya, T., Townell, N., Letchford, J., Bun, S., et al. Cambodia Technical Working Group on Antimicrobial, R. (2019). Antimicrobial resistance in Cambodia: a review. Int. J. Infect. Dis. 85, 98–107. doi:10.1016/j.ijid.2019.05.036
Rojo-Bezares, B., Saenz, Y., Poeta, P., Zarazaga, M., Ruiz-Larrea, F., and Torres, C. (2006). Assessment of antibiotic susceptibility within lactic acid bacteria strains isolated from wine. Int. J. Food Microbiol. 111, 234–240. doi:10.1016/j.ijfoodmicro.2006.06.007
Rozman, V., Mohar Lorbeg, P., Accetto, T., and Bogovic Matijasic, B. (2020). Characterization of antimicrobial resistance in lactobacilli and bifidobacteria used as probiotics or starter cultures based on integration of phenotypic and In Silico data. Int. J. Food Microbiol. 314, 108388. doi:10.1016/j.ijfoodmicro.2019.108388
Rozos, G., Voidarou, C., Stavropoulou, E., Skoufos, I., Tzora, A., Alexopoulos, A., et al. (2018). Biodiversity and microbial resistance of lactobacilli isolated from the traditional Greek cheese Kopanisti. Front. Microbiol. 9, 517. doi:10.3389/fmicb.2018.00517
Sandes, S., Alvim, L., Silva, B., Acurcio, L., Santos, C., Campos, M., et al. (2017). Selection of new lactic acid bacteria strains bearing probiotic features from mucosal microbiota of healthy calves: Looking for immunobiotics through In Vitro and In Vivo approaches for immunoprophylaxis applications. Microbiol. Res. 200, 1–13. doi:10.1016/j.micres.2017.03.008
Scheirlinck, I., Van der Meulen, R., Van Schoor, A., Huys, G., Vandamme, P., De Vuyst, L., et al. (2007). Lactobacillus crustorum sp. nov., isolated from two traditional Belgian wheat sourdoughs. Int. J. Syst. Evol. Microbiol. 57, 1461–1467. doi:10.1099/ijs.0.64836-0
Singla, V., Mandal, S., Sharma, P., Anand, S., and Tomar, S. K. (2018). Antibiotic susceptibility profile of Pediococcus spp. from diverse sources. 3 Biotech, 8, 489. doi:10.1007/s13205-018-1514-6
Sirichoat, A., Florez, A. B., Vazquez, L., Buppasiri, P., Panya, M., Lulitanond, V., et al. (2020). Antibiotic susceptibility profiles of lactic acid bacteria from the human vagina and genetic basis of acquired resistances. Int. J. Mol. Sci. 21. doi:10.3390/ijms21072594
Sirot, J., Courvalin, P., and Soussy, C. J. (1996). Definition and determination of In Vitro antibiotic susceptibility breakpoints for bacteria. Clin. Microbiol. Infect. 2 (Suppl. 1), S5–S10. doi:10.1111/j.1469-0691.1996.tb00870.x
Stefańska, I., Kwiecien, E., Jozwiak-Piasecka, K., Garbowska, M., Binek, M., and Rzewuska, M. (2021). Antimicrobial Susceptibility of Lactic Acid Bacteria Strains of Potential Use as Feed Additives-The Basic Safety and Usefulness Criterion. Front. Vet. Sci. 8, 687071. doi:10.3389/fvets.2021.687071
Stock, I., and Wiedemann, B. (2001). Natural antibiotic susceptibilities of Edwardsiella tarda, E. ictaluri, and E. hoshinae. Antimicrob. Agents Chemother. 45, 2245–2255. doi:10.1128/AAC.45.8.2245-2255.2001
Turnidge, J. (2016). “MIC wild type distributions and ECOFFs and their role in setting clinical breakpoints,” in ESCMID/ASM conference on drug development to meet the challenge of antimicrobial resistance, Vienna, Austria.
Turnidge, J., and Paterson, D. L. (2007). Setting and Revising Antibacterial Susceptibility Breakpoints. Clin. Microbiol. Rev. 20, 391–408. doi:10.1128/CMR.00047-06
Vardanyan, R., and Hruby, V. (2016). “Antibacterial drugs,” in Synthesis of Best-Seller Drugs. Editors R. Vardanyan, and V. Hruby (CA, USA: Elsevier), 645–667. doi:10.1016/b978-0-12-411492-0.00031-6
Williams-Nguyen, J., Sallach, J. B., Bartelt-Hunt, S., Boxall, A. B., Durso, L. M., McLain, J. E., et al. (2016). Antibiotics and antibiotic resistance in agroecosystems: State of the science. J. Environ. Qual. 45, 394–406. doi:10.2134/jeq2015.07.0336
Wolupeck, H. L., Morete, C. A., DallaSanta, O. R., Luciano, F. B., Madeira, H. M. F., and de Macedo, R. E. F. (2017). Methods for the evaluation of antibiotic resistance in Lactobacillus isolated from fermented sausages. Ciência Rural. 47, 1–7. doi:10.1590/0103-8478cr20160966
Zellweger, R. M., Carrique-Mas, J., Limmathurotsakul, D., Day, N. P. J., Thwaites, G. E., and Baker, S. (2017). Southeast Asia Antimicrobial Resistance, NA current perspective on antimicrobial resistance in Southeast Asia. J. Antimicrob. Chemother. 72, 2963–2972. doi:10.1093/jac/dkx260
Zhang, S., Oh, J. H., Alexander, L. M., Ozcam, M., and van Pijkeren, J. P. (2018). D-alanyl-d-alanine ligase as a broad-host-range counterselection marker in vancomycin-resistant lactic acid bacteria. J. Bacteriol. 200, e00607–17. doi:10.1128/JB.00607-17
Zheng, J., Wittouck, S., Salvetti, E., Franz, C., Harris, H. M. B., Mattarelli, P., et al. (2020). A taxonomic note on the genus Lactobacillus: Description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 70, 2782–2858. doi:10.1099/ijsem.0.004107
Zommiti, M., Bouffartigues, E., Maillot, O., Barreau, M., Szunerits, S., Sebei, K., et al. (2018). In Vitro assessment of the probiotic properties and bacteriocinogenic potential of Pediococcus pentosaceus MZF16 isolated from artisanal Tunisian meat "Dried Ossban. Front. Microbiol. 9, 2607. doi:10.3389/fmicb.2018.02607
Keywords: Cambodia, fermented fish, fermented vegetables, starter cultures, lactic acid bacteria, transferable antibiotic resistance, safety assessment, phenotypic susceptibility data
Citation: Ly D, Mayrhofer S and Domig KJ (2022) An Evaluation of the Phenotypic Antibiotic Susceptibility of Potential Lactic Acid Bacteria Starter Cultures Isolated From Cambodian Fermented Foods. Front. Food. Sci. Technol. 2:892319. doi: 10.3389/frfst.2022.892319
Received: 08 March 2022; Accepted: 20 June 2022;
Published: 22 July 2022.
Edited by:
Antonello Santini, University of Naples Federico II, ItalyReviewed by:
Frederick Tawi Tabit, University of South Africa, South AfricaJames Owusu-Kwarteng, University of Energy and Natural Resources, Ghana
Copyright © 2022 Ly, Mayrhofer and Domig. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sigrid Mayrhofer, c2lncmlkLm1heXJob2ZlckBib2t1LmFjLmF0
 Dalin Ly
Dalin Ly Sigrid Mayrhofer
Sigrid Mayrhofer Konrad J. Domig
Konrad J. Domig