
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Food. Sci. Technol. , 06 April 2022
Sec. Food Safety and Quality Control
Volume 2 - 2022 | https://doi.org/10.3389/frfst.2022.842063
 Raghu Hirikyathanahalli Vishweswaraiah1*
Raghu Hirikyathanahalli Vishweswaraiah1* Shivendra Tenguria2
Shivendra Tenguria2 B. Chandrasekhar1
B. Chandrasekhar1 C. G. Harshitha3
C. G. Harshitha3 Kamal Gandhi3
Kamal Gandhi3 Naresh Kumar1
Naresh Kumar1 Rotimi E. Aluko4*
Rotimi E. Aluko4* Anil Kumar Puniya5*
Anil Kumar Puniya5*Food-borne diseases are on the rise, and these will likely continue as a public health concern into the coming decades. Majority of foodborne outbreaks are linked to infections by emerging foodborne pathogens such as Campylobacter, Salmonella, Listeria monocytogenes, and Escherichia coli O157:H7. Foodborne pathogen identification becomes crucial in such scenarios to control these pathogens, associated outbreaks, and diseases. Pathogen detection systems have evolved as essential food safety tools to combat microbial threats and experts are striving to develop robust, accurate and ergonomic rapid pathogen-detection kits. Lectin, a ubiquitous biomolecule (sugar binding proteins) present in almost all domains of life is a promising alternative to molecular based methods as a bio-recognition molecule in detection of foodborne pathogens for biosensor applications, owing to its multivalency and spatial organization of ligands. Due to their extensive prevalence, lectin-based biosensors have become the most sought-after bio-recognition molecules in biosensor applications because of increased sensitivity and reduced cost when compared to immune-based biosensors. The current paper discusses the claimed benefits of lectin as a superior bio-recognition molecule, as well as its numerous applications in biosensor creation.
⁃ The food safety researchers are working hard to create rapid pathogen detection tests that are reliable, accurate, and easy to use.
⁃ Lectin, a common biomolecule also known as sugar binding proteins, has been proven to be a promising bio-recognition molecule in the detection of foodborne infections.
⁃ Lectins are the most sought-after bio-recognition molecules in biosensor applications due to their increased sensitivity and reduced cost.
⁃ For the detection of food-borne diseases, lectins have been widely used in the fabrication of piezoelectric, optical, and electrochemical biosensors.
Foodborne diseases have become a major global public health concern in the last three decades, with considerable increases in incidence (Ramirez-Hernandez et al., 2020). Consumption of tainted food and water has resulted in 3-5 billion illnesses and almost 1.8 million deaths in young children per year, primarily owing to diarrheal disorders (WHO, 2015). About thirty-one identified food-borne diseases (mostly bacteria and viruses) have been reported in the United States alone (Sachdev et al., 2021). The bulk of disease outbreaks in impoverished nations go unreported and unrecognized as part of the health or financial losses sustained due to a lack of awareness and insufficient medical services. Controlling and managing outbreaks by effective tracing and recalling of contaminated goods become difficult in these circumstances, which exacerbates foodborne diseases. Moreover, developing strategies for identifying the causes linked to contamination, preventing future incidents, and reinforcing food safety rules and platforms is becoming a difficult issue (Vemula et al., 2012).
As a result, it is critical to diagnose food products, particularly ready-to-eat (RTE) foods, for contaminating microorganisms and other chemical contaminants in order to prevent these new foodborne infections and disorders. The farm-to-fork approach to food safety is becoming increasingly popular (Faour-Klingbeil, 2017). Old culture-based methods for detecting bacterial diseases still rely on traditional methods, needing a 5–7-day regimen for successful identification of the causal bacterium (Wu and Zeng, 2017). Though these approaches are recognized as gold standards because they are generally well established and validated, their length and poor speed make them ineffective and outdated in the face of food production and storage deadlines. Due to their increasing sensitivity and speed, genotypic approaches for detecting these pathogenic bacteria have recently surpassed classic microbiological methods. Because of their great accuracy and sensitivity, molecular approaches based on polymerase chain reaction (PCR) and 16S rDNA/amplicon sequencing have surpassed time-consuming conventional procedures (Hameed et al., 2018). However, there is a catch: these approaches necessitate the use of complicated devices and expert employees.
In addition, using biosensors to identify microbiological and chemical pollutants in food is a unique technique and a possible alternative to traditional analytical approaches. In terms of sensitivity, specificity, selectivity, reliability, efficiency, and robustness, biosensors are quick and outstanding. Furthermore, biosensors allow for real-time monitoring of food pollutants and have revolutionized detection procedures, resulting in the creation of quick and specific detection tests that provide precise and immediate results. A biological component (a bio-recognition molecule that identifies and creates a signal), a signal transducer, and a reading device make up a biosensor. For the identification of microbiological or chemical contamination in food items, the interaction and affinity between the bio-recognition molecule and the contaminant is critical. Many of these bio-recognition molecules, such as proteins (lectins, antibodies, enzymes), and nucleic acids, are found in nature. Other than enzymes and antibodies, lectins are carbohydrate-binding proteins. Lectins are found in many aspects of life, from viruses and bacteria to plants and mammals (Raghu and Kumar, 2020). Lectins are used to detect bacterial infections in food because of their capacity to attach to carbohydrate moieties on bacterial cell walls. Their capacity to distinguish cells has aided in the quick detection of pathogens. Therefore, the use of lectins in the surveillance of food-borne bacterial infections is discussed in this article.
Lectins are usually divalent or polyvalent, with two or more carbohydrate binding sites per molecule. Because each reaction site is capable of connecting to two or more carbohydrates on the bacterial cell surface to generate aggregation and precipitation, they have a one-of-a-kind ability to agglutinate cells. Lectins have multivalent carbohydrate-binding sites that are arranged on the surface. The binding of lectins to carbohydrates involves a variety of chemical interactions, including hydrophobic interactions, hydrogen bonds, van der Waals’ interactions, and metal ion coordination (Roy et al., 2016). Hydrogen bonds are mostly formed between carbohydrate hydroxyl groups and lectin protein amine group clusters. Despite the fact that carbohydrates are primarily polar molecules, hydrophobic patches arise as a result of the steric disposition of hydroxyl groups on sugar surfaces, which can interact with lectin hydrophobic areas. Metal ions like Mg2+ and Ca2+ are found at sugar binding sites of lectins (Figure 1), but they rarely contribute directly to carbohydrate binding (Siebert et al., 2002). The importance of cations in supporting the proper placement of amino acid residues in order to improve interactions with sugars cannot be overstated. Furthermore, water bridges have a role in the interaction between ligands and proteins. Water is analogous to molecular “mortar” because of its small size and ability to operate as both a hydrogen donor and acceptor. As a result, water is crucial in lectin recognition (Slominska-Wojewodzka and Sandvig, 2015).
Antibodies are commonly utilized in the construction of biosensors to detect food safety risks such as microorganisms due to their strong and unique interactions. However, there are certain drawbacks to employing antibodies as biosensors for microbial detection in meals, such as the considerable time it takes to separate and refine antibodies. There is also the challenge of regenerating these biosensors, which makes it tough to stabilize the antibodies as part of the lengthy procedure. As a result, using lectins to solve these challenges and detect microbial infections in food is a good idea because lectins are more stable than monoclonal antibodies and there are hundreds of lectins accessible commercially (Masarova et al., 2004).
Because of their high affinity for sugar fractions assisted by polyvalent interfaces originating from the three-dimensional interaction of carbohydrate molecules, lectins make excellent bio-recognition molecules. The unique binding of lectins to glycoprotein terminal sugar moieties on cell surfaces has been thoroughly explained (Bertozzi and Kiessling, 2001). Table 1 lists lectins utilized as bio-recognition molecules, as well as their carbohydrate specificity. Concanavalin A (ConA) is one of the most extensively used lectins in the creation of various sensors for ligand identification using fluorescent markers and fluorometer detection.
Lectins have multivalent and non-covalent interactions with glycoproteins, which are important in distinguishing bacterial pathogens from contaminated food. This feature of lectins has been exploited to isolate gut bacteria by conjugating them with magnetic beads (Gorakshakar and Ghosh, 2016). Lectins have been immobilized on magnetic beads by adsorption, covalent immobilization on tosylactivated beads or biotinylated lectins were coupled to magnetic particles with immobilized streptavidin. Magnetic particles can be separated later using a free competing sugar (Safarik and Safarikova, 1999). A battery of pathogenic and commensal bacteria was tested using lectins extracted from plants such as Canavalia ensiformis, Datura stramonium, Vicia faba, Solanum tuberosum, Helix aspersa, Cytisus scoparius, Erythrina cristagall, Bauhnia purpurea, Limulus polyphemus, Caragana arborescens. Lycopersicon Con A lectin, for example, has a higher ability to bind with E. coli and S. aureus at rates of 0–98% and 50–100%, respectively (Cheung et al., 2015; Malik et al., 2016). Hyun et al. (2020) devised a method for selectively eliminating harmful bacteria by using bacterial lectin-targeting glycoconjugates containing an epitope or a photosensitizer to enhance antibody-dependent cellular cytotoxicity (ADCC) or photodynamic treatment (PDT). Furthermore, Yang et al. (2019a) employed wheat germ agglutinin lectin (WGA) as a bio-recognition agent for S. aureus enrichment and found that even at 100–1000 CFU/ml of S. aureus, WGA demonstrated high enrichment efficiency. Later, this unique enrichment was applied in a quantitative and selective detection technique to detect S. aureus with a detection limit of 3.5 102 CFU/ml using double stranded DNA (dsDNA) stabilized gold nanoparticles (AuNPs). To improve the efficacy of magnetic nanoparticles in investigating the scope of bacterial pathogen detection based on bio-recognition, a lectin magnetic separation (LMS) technique was developed for S. aureus (Yang et al., 2019b) and the main aim was to improve the efficiency of magnetic nanoparticles and to expand the scope of bacterial recognition. In conventional method, i.e., using Baird–Parker (B-P) method, S. aureus can be detected at limit of 3 × 10⁰ CFU·mL⁻1 within 15 h; the polymerase chain reaction (PCR) method can be finished within 4 h, with the limit of detection (LOD) of 3 × 102 CFU·mL⁻1. The LOD of HRP-pig IgG-based colorimetric method is 3 × 10⁵ CFU·mL⁻1, and the method only lasts for 2 h. If combined with specific detection methods, it meets different needs for rapid detection of S. aureus.
Listeria monocytogenes is categorized into four serological groups and 13 serovars based on somatic and flagellar antigen components (Lopez Valladares, 2019). These are Serogroup 1 (1/2a and 1/2b serovars), serogroup 2 (serovar 1/2c), serogroup 3 (serovars 3a, 3b, and 3c), and serogroup 4 (serovars 4a, 4 ab, 4b, 4c, 4) (Li et al., 2020). Nonetheless, epidemiologic studies benefit little from serotyping because only three serovars, 1/2a, 1/2b, and 4b account for over 90% of all clinical isolates (Braga et al., 2017). Changes in glycosidic substituents coupled to a polyribitol phosphate (Rbop) backbone cause the most variation in wall teichoic acids (WTA) (Brown et al., 2013). WTAs in L. monocytogenes are made up of repetitive Rbop units with variations in carbohydrate molecules such as galactose, rhamnose, glucose, and N-acetyl glucosamine, which vary a lot according to the strain and serovar. On the C-2 and C-4 sites of the thiol group, serovars 12 and 3 have rhamnose sugars and GlcNAc. Serovars 4, 5, and 6 have a more complex WTA assembly in the centre of the GlcNAc chain, which is fused as a chunk of the polyribitol chain and may contain Glc and/or Gal substituents. Listeria WTA does not have any D-alanine substituents (Bielmann et al., 2015). Through a GlcNAc moiety, wheat germ agglutinin (WGA) binds to WTAs (Murakami et al., 2014). Flannery (2019) used an agglutination assay and a florescence-based assay to investigate cell wall diversity of Listeria spp., Canavalia ensiformis, Griffonia simpliciforlia lectin I, Helix pomatia agglutinin, Ficinus cummunis agglutinin, and Ricinus communis agglutinin in clinical, food, and environmental isolates.
Sun et al. (2017) isolated and separated enteric bacteria using lectin-labeled magnetic beads. They utilised sixteen different types of lectins and found that magnetic beads labelled with Con A produced the most agglutinations Figure 2. From river water and laboratory cultures, this approach could extract pure and culturable cells. The findings are equivalent to those obtained using antibody-labeled beads. The findings are encouraging, implying that lectins could be employed as an alternate cell discriminator. Sun et al. (2017) separated coliform from fresh water and effluent from a water treatment plant using lectin-labeled magnetic beads. Direct magnetic capture was shown to be less reliable than indirect magnetic capture. Harito et al. (2017) employed the lectin magnetic separation (LMS) approach to isolate Toxoplasma gondii oocysts from water sample concentrates, with microscopy or molecular methods used for identification. Castillo-Torres et al. (2019) used lectin-functionalized and aptamer-functionalized microdiscs to demonstrate selective separation of generic coliforms and E. coli cells, respectively. They also discovered a direct association between bacterial concentration and low signal in control samples in the form of fluorescence signals, which are directly correlated to bacterial concentrations and visible in control samples (negative samples). Using Con A–linked magnetic beads to detect Hepatitis A Virus (HAV), Ko et al. (2018) discovered that it had a significantly higher binding affinity for HAV than reverse transcriptase PCR, with a detection concentration of 10−4 of the virus stock. Magnetic beads containing fragment crystalizable-mannose-binding lectin (FcMBL) were used to capture and magnetically remove bacteria from purified cultures of 12 S. aureus strains, as well as from 8 articular fluid samples and 4 synovial tissue samples collected from patients with osteoarthritis or periprosthetic infections at 85% efficiency (Bicart-See et al., 2016). Tateno et al. (2018) employed lectin to bind malignant cells based on cell aggregation, which resulted in cell death, according to a US patent.
Hendrickson et al. (2019) used a ligand–bioreceptor interaction to develop a flow cytometer analysis of the bacterial pathogens E. coli and S. aureus. Plant lectins with fluorescent labels were used as natural receptors that could precisely bind with bacteria’s cell wall polysaccharides. Additionally, epifluorescence microscopy was employed to validate and observe lectin–carbohydrate interactions. The binding specificity of plant lectins to E. coli and S. aureus cells was investigated, and wheat germ agglutinin was chosen as a receptor since it provided high-affinity interactions. Bacterial pathogens can be detected up to 106 cells/mL in 5 min using this approach. Lectin reagents are a promising method for controlling a wide spectrum of bacteria due to their accessibility and universality.
The inclusion of 4 tryptophan, 12 cysteine, and glycine residues in the amino acid sequence of wheat germ agglutinin (WGA) may explain its natural florescent ability (Bogoeva et al., 2004). WGA has a characteristic tryptophan emission at 348 nm in its fluorescence spectra. The results of the fluorescence study led to the conclusion that fluorescence and phosphorescence are caused by at least two of the three tryptophan residues in WGA (Liener, 2012). With increased amounts of log cfu/ml of L. monocytogenes cells, the binding constant of WGA was increased. The affinity of carbohydrate binding domains (CBDs) to their carbohydrate ligands is equivalent to affinity-matured antibodies to bacterial cell surface antigens (Bi and Zimmer, 2020). Another study by Loessner et al. (2002) found that at increasing carbohydrate concentrations, CBDs have a higher association constant with their carbohydrate ligands. Another study found that E. coli O157:H7 that binds to lectins had a higher association constant (Ka) (Wang et al., 2013). In the presence of particular sugar and L. monocytogenes, Raghu (2018) detected maximum fluorescence in the spectra using 290 nm as excitation and 350 nm as emission wavelength with increasing WGA concentration. Therefore, the principle of WGA fluorescence could also be a technique for developing fluorometric tools for detecting L. monocytogenes.
Using platforms such as electrical oscillation (Bhalla et al., 2016), piezoelectric crystal, and micro-calorimetrics, several modules of lectin-based biosensors have been constructed (Vereshchagina et al., 2015). Table 2 shows an overview of biosensors created for the detection of infections employing lectins. Biosensors are seen as a promising alternative to the traditional procedures that have been used in the past. The biorecognition element must be fixed on the sensor surface to provide high selectivity. Lectins are a type of biorecognition element that is more stable and covers greater surface area than other biomolecules like antibodies (Sondhi et al., 2020). The sections that follow will go over the many types of transducers that can be utilized to detect bacterial infections using lectins.
Piezoelectric biosensors are analytical devices that work on the idea of analyte and ligand molecule affinity, which causes a change in mass/weight on vibrating quartz crystals, changing the frequency of oscillation. The change in oscillation frequency is a measure of the analyte concentration in the samples (Skladal, 2016), and Figure 3 is a graphical depiction of a piezoelectric lectin-based sensor. A weight indicator made from a vibrating quartz crystal is extremely sensitive and accurate. Mass changes can be detected utilizing a crystal-controlled oscillator and the piezoelectric biosensor’s resonance frequency (Petr, 2015). Serra et al. (2008) suggested a piezoelectric biosensor for the detection and quantification of bacteria using an electrochemical quartz crystal microbalance (EQCM). Con A and E. coli were used to test the efficacy of lectin-based quartz crystal microbalance. Shen et al. (2007) employed a non-labeled quartz crystal microbalance (QCM) biosensor to detect bacterial infections using lectins as bio-receptors. The bio-recognition of O-antigen using a carbohydrate non-labeled mass sensor packed with lectin could be a promising technique with a reduced detection limit and maximum selectivity for high molecular weight bacterial targets. With the use of Con A lectin as the bio-recognition molecule, Shen et al. (2007) developed a QCM biosensor. It did, however, reveal a low detection limit for E. coli (limited to a few hundred bacterial cells). These proteins were employed by Serra et al. (2008) to make biosensors for detecting bacterial infections. Using an EQCM, gold-plated quartz crystals were immobilized with lectins. The Con A was used to test the efficiency of the lectin immobilization approach and the biosensor performance for E. coli detection. Adsorption of polarized and non-polarized (0.200 V) gold-empaneled quartz crystals through avidin–biotin binding was used to ensure attachment of Con A. By measuring the change in frequency with E. coli concentration 1 h after bacterial binding, a linear calibration plot in the range of 5.0 × 106 to 2.0 × 107 cfu/mL was obtained, with the limit of detection of this developed sensor at around 1.0 × 104 cfu/mL.
Using four different carbohydrate binding lectins, namely WGA, MAL, LCA, and Con A, a QCM biosensor was applied to distinguish seven Campylobacter jejuni strains (Yakovleva et al., 2011). The frequency change in the Con A lectin was higher at 34 Hz (Wang et al., 2013). The use of a carbohydrate epitope in conjunction with a lectin-bacterial O-antigen interaction in the development of sensitive and specific non-labeled biosensors for the detection of bacterial pathogens has been demonstrated using this patented technology for “Microorganism Detection and Analysis Using Carbohydrate and Lectin Recognition” (Zeng and Shen, 2016). Many other exo- or endogenous toxins produced by or present in bacterial cells may detect the carbohydrate ligand and generate increased cross-reactivity, leading to false positive and negative results.
Liedberg was the first to propose the SPR sensor for use in biosensors (Prabowo et al., 2018). It has now been thoroughly investigated and is gradually evolving into an inherent label-free device for facilitating chemical interactions between the target analyte and ligand molecules. When a photon of light strikes a metal surface (gold or silver), it interacts with and excites the metal’s electrons, causing them to move as a single electrical unit called a Plasmon, which oscillates parallel to the metal surface (Mitchell and Wu, 2010). Surface Plasmon is a phenomenon in which a Plasmon oscillates and generates an electric field that extends about 300 nm out from the metal surface (Homola, 2003; Hoa et al., 2007). It is based on the fact that at a specific incidence angle and wavelength, electrons in a metal layer become excited and absorb light. Surface Plasmons (SPs) are electron density waves that develop at the interface of a metal (such as gold) and a dielectric (such as glass). Changes in the refractive index caused by an analyte binding to a prism’s metal-coated surface characterize these waves (Mullett et al., 2000; Homola, 2003). A resonance occurs at a specific angle where the energy shift from the photon to the Plasmon is well matched, resulting in significantly increased energy shift and a commensurate decrease in reflectance (Mitchell and Wu, 2010).
If mass is attached to the surface within this 300 nm region, it perturbs the Plasmon and alters the resonance angle for a photon of fixed wavelength. If the metal surface’s plane is fixed, the resonance wavelength can be determined by scanning a range of photon incidence angles and determining the resonance angle (Zeng et al., 2017). An SPR biosensor operates on the same concept as a surface Plasmon resonance sensor, measuring minute changes in refractive index that correspond to binding interactions on the sensor surface (Homola, 2008). In most cases, SPR biosensors can achieve resolutions of 1 resonance unit (RU), or 1 × 10−6 refractive index units (Prabowo et al., 2018). SPR biosensors can detect binding interactions between molecules as small as 2 kDa, but smaller molecules produce insufficient changes in bound mass, which cannot be monitored directly (Perumal and Hashim, 2014).
Only a few reports on how lectins are employed as a bio-receptor for bacterial detection are available to date. Gram-negative bacteria, such as E. coli, can be distinguished by their chemically distinct surface LPS, which can be detected by specific lectins; hence, bio-receptors assist ConA attach to E. coli W1485. Masarova et al. (2004) created a SPR detection system for endotoxins or entire bacterial cells by covalently attaching eight distinct lectins to CM 5 chip surfaces (CM5). These eight lectins were found to be unique. Based on lectin-endotoxin (lipopolysaccharide) interaction, these eight lectins were able to distinguish between the microorganisms. Wang et al. (2013) employed SPR to create a novel type of biosensor for detecting E. coli O157:H7, which utilized lectin as a bio-recognition molecule. Using amine coupling chemistry, lectin was immobilized on the gold chip (CM5) surfaces to capture this bacterial pathogen in the buffer solution. WGA, ConA, UEA, PNA, and MAL were evaluated for the best lectin, one that could efficiently bind to this bacterial pathogen. Among the lectins that were screened, WGA lectin has a wide linear range of 3 × 105 to 3 × 108 cfu/mL with a lower detection limit of 3 × 103 cfu/mL, demonstrating good selectivity by utilizing SPR. SPR has advantages over QCM and electrochemical measurements, according to Raghu and Kumar (2020). This is because biomolecular interactions can be tracked in real time, allowing kinetic parameters to be determined (KD and KA). The affinity constant KA, in particular, aids in performance analysis of the bacteria and carbohydrate binding protein in terms of specificity and utility. In contrast, there are few publications on lectin based SPR sensors. Wang et al. (2013) and Raghu and Kumar (2020) developed a novel SPR biosensor using lectin as the bio-receptor for fast detection of E. coli O157:H7 and L. monocytogenes. Figure 4 depicts the sensor configuration for the detection of L. monocytogenes. Lectins were immobilised on the CM5 chip with the help of self-assembly monolayers to capture E. coli O157:H7 in the buffer solution (SAM). In another investigation, Leonard et al. (2004) used a Biacore SPR 3000 biosensor with a detection limit of 1105 L. monocytogenes cells/ml within 30 min to detect L. monocytogenes with a polyclonal antibody.
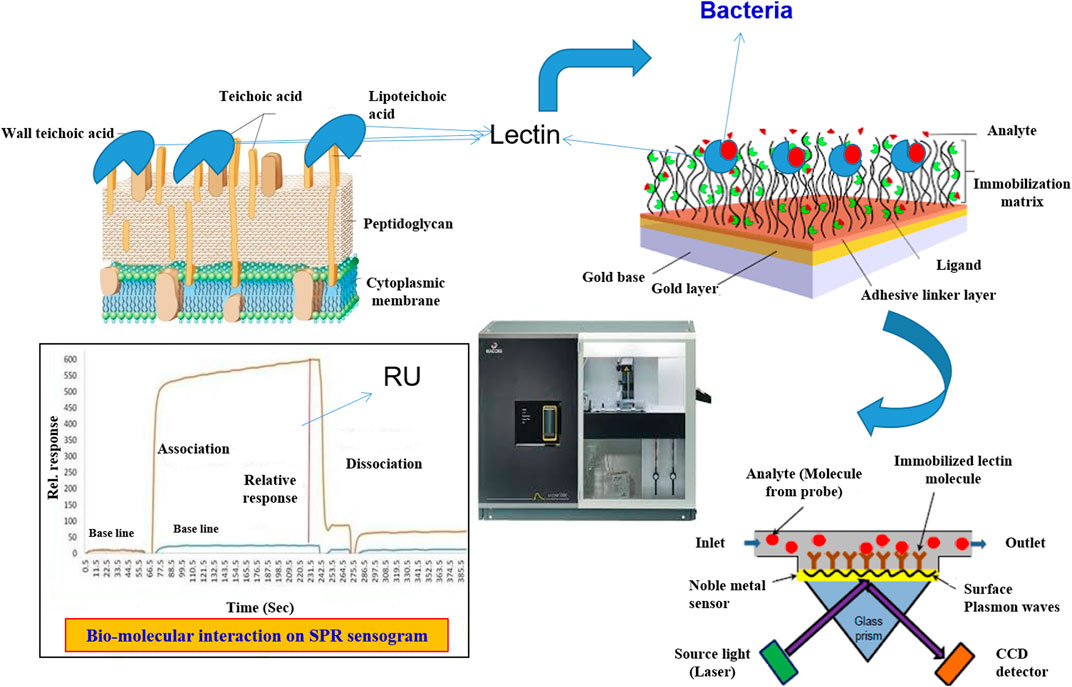
FIGURE 4. Lectin based detection of Bacterial pathogens using Surface Plasmon resonance-based sensor.
An electrochemical sensor is a device that turns biological events into a signal that can be measured analytically, such as the amount of a certain analyte in a sample or composition, harmful contaminants, or bacterial infections (Thevenot et al., 2001). A distinguishing feature of an electrochemical biosensor is the electrochemical technique used. By using lectins as a bio-recognition molecule, Ertl and Mikkelsen (2001) produced an electrochemical biosensor array, and transduction is assessed by taking the respiratory cycle activity, in which the native respiratory chain of the microbe is interrupted by exogenous oxidants. They discovered a link between agglutination outcomes and the electrochemical assessment of lectin cell binding. To determine how each organism reacts to lectin, chronocoulometric measures were created. Bacillus cereus, S. aureus, Proteus vulgaris, E. coli, Enterobacter aerogenes, and Saccharomyces cerevisiae can all be distinguished using the established method. Ertl et al. (2003) also created an electrochemical biosensor with a screen-printed array whose selectivity was based on lectin affinity for cell-surface lipopolysaccharides. These arrays were used in conjunction with factor analysis to quickly identify four E. coli subspecies (E. coli B, E. coli Neotype, E. coli JM105, and E. coli HB101) in under 40 min (Velusamy et al., 2010). Sa et al. (2020) used Con A and WGA to construct an impedimetric biosensor for detecting pathogenic Candida species. The sensor can distinguish between Candida species with a detection sensitivity of 102–106 CFU/ml. Lopez-Tellez et al. (2020) established a sensitive electrochemical detection approach for Salmonella spp. employing Hechtia argentea lectin immobilized on a screen-printed gold electrode that exhibited selectivity for D-mannose, which is found on the lipopolysaccharide (LPS) layer of Salmonella spp. With increased concentrations of Salmonella spp., the interaction of bacteria with immobilized lectin on a biosensor surface increased the impedance, achieving a detection range of 15–2.57 × 107 CFU/mL, with a 5 CFU/ml limit of detection.
Lectin biosensors have a limited ability to work in a multiplexed fashion, which is their principal drawback. Only SPR imaging (SPRi) and, to a lesser extent, micro-cantilever-based assays allow for highly parallel analysis. More research on electrochemical approaches that work in an array arrangement is required. It is expected that lectin biosensors will be better suited for diagnostic purposes than for preliminary assays of a large number of samples. There is also the additional problem of lectin biosensors and their use in the examination of real complex materials (Belicky et al., 2016). However, controlled immobilization of lectin for the construction of lectin biosensors is more difficult due to the larger complexity of a protein molecule.
Carbohydrate-protein interactions have been identified as critical for a variety of biological activities, including extracellular and intracellular signaling, as well as cell-to-cell recognition. Lectins are useful tools for studying glycoconjugates and understanding the mechanics of numerous biological processes because of their carbohydrate binding selectivity. Lectins are useful tools in glycobiology and are the greatest model system for studying protein-carbohydrate interactions. Lectins have been linked to the creation of lectin-based high-throughput molecular tools and have established themselves as a major glycan decoding tool in glycobiology. However, there is still a lot of room for lectins to be used in numerous biotechnological and medicinal applications. Furthermore, the identification and characterization of novel lectins, as well as in-depth research on existing lectins, will have a significant impact on future research.
The right to food and the right to safe food are two sides of the same coin; one without the other is incomplete. Food security and delivering safe food to consumers are becoming increasingly important to all food supply chain stakeholders. This has turned the focus to developing accurate, precise, and cost-effective quick food pathogen detection systems with an ergonomic design. The search for an effective biosensor has led to the discovery of a number of bio-recognition molecules with excellent specificity and sensitivity. Antibodies performed admirably; however, they have intrinsic limits in terms of synthesis. Sugar binding proteins, such as lectin, have emerged as potential alternatives to antibodies due to their high specificity, abundance, and stability. It is a positive development that lectins can act as bio-receptors in a number of biosensor systems. As a result, lectin research has received a lot of attention, and scientists all over the world are attempting to fully exploit this new line of biosensor technology. These lectins have found widespread application in clinical settings, generating hopes for their potential use in rapid pathogen detection systems in disease diagnostics and food safety. Lectins have been linked to the creation of lectin-based high-throughput molecular tools and have established themselves as a major glycan decoding tool in glycobiology. Furthermore, the identification and characterization of novel lectins, as well as in-depth research on existing lectins, will have a significant impact on future research. Despite some progress, more effort is required to fully realize the potential of lectins as biosensors.
RV and ST wrote the introduction, lectins, types of lectins, biological significance of lectins, bio-recognition molecules, application of lectins in detection of pathogens, lectin-based biosensors, and lateral flow assay. KG, BC, and CH drafted the introduction, conclusion, and references. NK, AP, and RA provided direction and critically revised and edited the manuscript. The final version was approved by all authors.
This publication of this review paper was fully supported by ICAR-National Dairy Research Institute, Karnal, Haryana, India.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Belický, Š., Katrlík, J., and Tkáč, J. (2016). Glycan and Lectin Biosensors. Essays Biochem. 60 (1), 37–47. doi:10.1042/ebc20150005
Bertozzi, C. R., and Kiessling, a. L. L. (2001). Chemical Glycobiology. Science 291 (5512), 2357–2364. doi:10.1126/science.1059820
Bhalla, N., Jolly, P., Formisano, N., and Estrela, P. (2016). Introduction to Biosensors. Essays Biochem. 60 (1), 1–8. doi:10.1042/EBC20150001
Bi, Y., and Zimmer, J. (2020). Structure and Ligand-Binding Properties of the O Antigen ABC Transporter Carbohydrate-Binding Domain. Structure 28 (2), 252–258. doi:10.1016/j.str.2019.11.020
Bicart-See, A., Rottman, M., Cartwright, M., Seiler, B., Gamini, N., Rodas, M., et al. (2016). Rapid Isolation of Staphylococcus aureus Pathogens from Infected Clinical Samples Using Magnetic Beads Coated with Fc-Mannose Binding Lectin. PLoS ONE 11 (6), e0156287. doi:10.1371/journal.pone.0156287
Bielmann, R., Habann, M., Eugster, M. R., Lurz, R., Calendar, R., Klumpp, J., et al. (2015). Receptor Binding Proteins of Listeria Monocytogenes Bacteriophages A118 and P35 Recognize Serovar-specific Teichoic Acids. Virology 477, 110–118. doi:10.1016/j.virol.2014.12.035
Bogoeva, V. P., Radeva, M. A., Atanasova, L. Y., Stoitsova, S. R., and Boteva, R. N. (2004). Fluorescence Analysis of Hormone Binding Activities of Wheat Germ Agglutinin. Biochim. Biophys. Acta (Bba) - Proteins Proteomics 1698 (2), 213–218. doi:10.1016/j.bbapap.2003.12.002
Braga, V., Vázquez, S., Vico, V., Pastorino, V., Mota, M. I., Legnani, M., Schelotto, F., Lancibidad, G., and Varela, G. (2017). Prevalence and Serotype Distribution of Listeria Monocytogenes Isolated from Foods in Montevideo-Uruguay. Braz. J. Microbiol. 48, 689–694. doi:10.1016/j.bjm.2017.01.010
Brown, S., Santa Maria, J. P., and Walker, S. (2013). Wall Teichoic Acids of Gram-Positive Bacteria. Annu. Rev. Microbiol. 67, 313–336. doi:10.1146/annurev-micro-092412-155620
Castillo-Torres, K. Y., McLamore, E. S., and Arnold, D. P. (2019). A High-Throughput Microfluidic Magnetic Separation (ΜFMS) Platform for Water Quality Monitoring. Micromachines 11 (1), 16. doi:10.3390/mi11010016
Cheung, R. C. F., Wong, J. H., Pan, W., Chan, Y. S., Yin, C., Dan, X., et al. (2015). Marine Lectins and Their Medicinal Applications. Appl. Microbiol. Biotechnol. 99 (9), 3755–3773. doi:10.1007/s00253-015-6518-0
Ertl, P., and Mikkelsen, S. R. (2001). Electrochemical Biosensor Array for the Identification of Microorganisms Based on Lectin−Lipopolysaccharide Recognition. Anal. Chem. 73 (17), 4241–4248. doi:10.1021/ac010324l
Ertl, P., Wagner, M., Corton, E., and Mikkelsen, S. R. (2003). Rapid Identification of Viable Escherichia coli Subspecies with an Electrochemical Screen-Printed Biosensor Array. Biosens. Bioelectron. 18 (7), 907–916. doi:10.1016/s0956-5663(02)00206-3
Faour-Klingbeil, D. (2017). The Microbiological Safety of Fresh Produce in Lebanon-A Holistic “Farm-to-fork Chain” Approach to Evaluate Food Safety, Compliance Levels and Underlying Risk Factors. Doctoral dissertation, University of Plymouth.
Flannery, A. (2019). A Study of the Presentation and Biological Effects of Biofilm Component Poly-N-Acetylglucosamine from S. aureus and A. Baumannii Using Novel Multidisciplinary Platforms and Methods. Doctoral dissertation, NUI Galway.
Gorakshakar, A., and Ghosh, K. (2016). Use of Lectins in Immunohematology. Asian J. Transfus. Sci. 10 (1), 12–21. doi:10.4103/0973-6247.172180
Hameed, S., Xie, L., and Ying, Y. (2018). Conventional and Emerging Detection Techniques for Pathogenic Bacteria in Food Science: A Review. Trends Food Sci. Tech. 81, 61–73. doi:10.1016/j.tifs.2018.05.020
Harito, J. B., Campbell, A. T., Tysnes, K. R., Dubey, J. P., and Robertson, L. J. (2017). Lectin-magnetic Separation (LMS) for Isolation of Toxoplasma Gondii Oocysts from Concentrated Water Samples Prior to Detection by Microscopy or qPCR. Water Res. 114, 228–236. doi:10.1016/j.watres.2017.02.044
Hendrickson, O. D., Nikitushkin, V. D., Zherdev, A. V., and Dzantiev, B. B. (2019). Lectin-based Detection of Escherichia coli and Staphylococcus aureus by Flow Cytometry. Arch. Microbiol. 201, 313–324. doi:10.1007/s00203-018-1613-0
Hoa, X. D., Kirk, A. G., and Tabrizian, M. (2007). Towards Integrated and Sensitive Surface Plasmon Resonance Biosensors: a Review of Recent Progress. Biosens. Bioelectron. 23 (2), 151–160. doi:10.1016/j.bios.2007.07.001
Homola, J. (2003). Present and Future of Surface Plasmon Resonance Biosensors. Anal. Bioanal. Chem. 377, 528–539. doi:10.1007/s00216-003-2101-0
Homola, J. (2008). Surface Plasmon Resonance Sensors for Detection of Chemical and Biological Species. Chem. Rev. 108 (2), 462–493. doi:10.1021/cr068107d
Hyun, J. Y., Lee, C.-H., Lee, H., Jang, W.-D., and Shin, I. (2020). Bacterial Lectin-Targeting Glycoconjugates for Selective Elimination of Pathogenic Bacteria. ACS Macro Lett. 9 (10), 1429–1432. doi:10.1021/acsmacrolett.0c00454
I. Liener (Editor) (2012). The Lectins: Properties, Functions, and Applications in Biology and Medicine (Elsevier).
Ko, S. M., Cho, S. Y., Oh, M. J., Kwon, J., Vaidya, B., and Kim, D. (2018). Application of Concanavalin A–Linked.
Leonard, P., Hearty, S., Quinn, J., and O’Kennedy, R. (2004). A Generic Approach for the Detection of Whole Listeria Monocytogenes Cells in Contaminated Samples Using Surface Plasmon Resonance. Biosens. Bioelectron. 19 (10), 1331–1335. doi:10.1016/j.bios.2003.11.009
Li, F., Ye, Q., Chen, M., Shang, Y., Zhang, J., Ding, Y., and Wu, Q. (2020). Real-time PCR Identification of L. Monocytogenes Serotype 4c Using Primers for Novel Target Genes Obtained by Comparative Genomic Analysis. LWT 2020, 110774.
Li, Z., Fu, Y., Fang, W., and Li, Y. (2015). Electrochemical Impedance Immunosensor Based on Self-Assembled Monolayers for Rapid Detection of Escherichia coli O157:H7 with Signal Amplification Using Lectin. Sensors 15, 19212–19224. doi:10.3390/s150819212
Loessner, M. J., Kramer, K., Ebel, F., and Scherer, S. (2002). C-terminal Domains of Listeria Monocytogenes Bacteriophage Murein Hydrolases Determine Specific Recognition and High-Affinity Binding to Bacterial Cell wall Carbohydrates. Mol. Microb. 44 (2), 335–349. doi:10.1046/j.1365-2958.2002.02889.x
Lopez Valladares, G. (2019). Human Listeriosis: Grouping of Human L. Monocytogenes Isolates with PFGE and AscI Restriction Enzyme.
Lopez-Tellez, J., Sanchez-Ortega, I., Hornung-Leoni, C. T., Santos, E. M., Miranda, J. M., and Rodriguez, J. A. (2020). Impedimetric Biosensor Based on a Hechtia Argentea Lectin for the Detection of Salmonella Spp. Chemosensors 8 (4), 115. doi:10.3390/chemosensors8040115
Malik, S., Petrova, M. I., Imholz, N. C., Verhoeven, T. L., Noppen, S., Van Damme, E. J., Liekens, S., Balzarini, J., Schols, D., Vanderleyden, J., and Lebeer, S. (2016). High Mannose-specific Lectin Msl Mediates Key Interactions of the Vaginal Lactobacillus Plantarum Isolate CMPG5300. Sci. Rep. 6 (1), 37339–37416. doi:10.1038/srep37339
Masarova, J., Dey, E. S., and Danielsson, B. (2004). Optical Lectin Based Biosensor as Tool for Bacteria Identification. Pol. J. Microbiol. 53 Suppl, 23–27.
Mitchell, J. S., and Wu, Y. (2010). Surface Plasmon Resonance Biosensors for Highly Sensitive Detection of Small Biomolecules. Biosensors, 151–168.
Mullett, W. M., Lai, E. P. C., and Yeung, J. M. (2000). Surface Plasmon Resonance-Based Immunoassays. Methods 22 (1), 77–91. doi:10.1006/meth.2000.1039
Murakami, Y., Hasegawa, Y., Nagano, K., and Yoshimura, F. (2014). Characterization of Wheat Germ Agglutinin Lectin-Reactive Glycosylated OmpA-like Proteins Derived from Porphyromonas Gingivalis. Infect. Immun. 82 (11), 4563–4571. doi:10.1128/iai.02069-14
Mutlu, S., Çökeliler, D., Shard, A., Goktas, H., Ozansoy, B., and Mutlu, M. (2008). Preparation and Characterization of Ethylenediamine and Cysteamine Plasma Polymerized Films on Piezoelectric Quartz crystal Surfaces for a Biosensorfilms on Piezoelectric Quartz crystal Surfaces for a Biosensor. Thin Solid Films 516, 1249–1255. doi:10.1016/j.tsf.2007.06.074
Nand, A., Singh, V., Wang, P., Na, J., and Zhu, J. (2014). Glycoprotein Profiling of Stem Cells Using Lectin Microarray Based on Surface Plasmon Resonance Imaging. Anal. Biochem. 465, 114–120. doi:10.1016/j.ab.2014.07.028
Perumal, V., and Hashim, U. (2014). Advances in Biosensors: Principle, Architecture and Applications. Jab 12 (1), 1–15. doi:10.1016/j.jab.2013.02.001
Prabowo, B., Purwidyantri, A., and Liu, K.-C. (2018). Surface Plasmon Resonance Optical Sensor: A Review on Light Source Technology. Biosensors 8 (3), 80. doi:10.3390/bios8030080
Raghu, H. V., and Kumar, N. (2020). Rapid Detection of Listeria Monocytogenes in Milk by Surface Plasmon Resonance Using Wheat Germ Agglutinin. Food Anal. Methods 13, 982–991. doi:10.1007/s12161-020-01717-3
Raghu, H. V. (2018). Surface Plasmon Resonance Based Detection of Listeria Monocytogenes on Biochip. Karnal): Doctoral dissertation, ICAR-NDRI.
Ramirez‐Hernandez, A., Galagarza, O. A., Álvarez Rodriguez, M. V., Pachari Vera, E., Valdez Ortiz, M. D. C., Deering, A. J., et al. (2020). Food Safety in Peru: A Review of Fresh Produce Production and Challenges in the Public Health System. Compr. Rev. Food Sci. Food Saf. 19 (6), 3323–3342.
Rao Vemula, S., Naveen Kumar, R., and Polasa, K. (20122012). Foodborne Diseases in India - a Review. Br. Food J. 114 (5), 661–680. doi:10.1108/00070701211229954
Roy, R., Murphy, P., and Gabius, H.-J. (2016). Multivalent Carbohydrate-Lectin Interactions: How Synthetic Chemistry Enables Insights into Nanometric Recognition. Molecules 21 (5), 629. doi:10.3390/molecules21050629
Sá, S. R., Silva Junior, A. G., Lima-Neto, R. G., Andrade, C. A. S., and Oliveira, M. D. L. (2020). Lectin-based Impedimetric Biosensor for Differentiation of Pathogenic candida Species. Talanta 220, 121375. doi:10.1016/j.talanta.2020.121375
Sachdev, D., Joshi, A., Taneja, N. K., and Pasricha, R. (2021). Emerging Silver Nanomaterials for Smart Food Packaging in Combating Food-Borne Pathogens. Silver Nanomater. Agri-Food Appl. 2021, 147–185. doi:10.1016/b978-0-12-823528-7.00009-3
Safarík, I., and Safaríková, M. (1999). Use of Magnetic Techniques for the Isolation of Cells. J. Chromatogr. B Biomed. Sci. Appl. 722 (1-2), 33–53.
Safina, G., Vanlier, M., and Danielsson, B. (2008). Flow-injection Assay of the Pathogenic Bacteria Using Lectin-Based Quartz crystal Microbalance Biosensor. Talanta 77 (2), 468–472. doi:10.1016/j.talanta.2008.03.033
Serra, B., Gamella, M., Reviejo, A. J., and Pingarrón, J. M. (2008). Lectin-modified Piezoelectric Biosensors for Bacteria Recognition and Quantification. Anal. Bioanal. Chem. 391 (5), 1853–1860. doi:10.1007/s00216-008-2141-6
Shen, Z., Huang, M., Xiao, C., Zhang, Y., Zeng, X., and Wang, P. G. (2007). Nonlabeled Quartz Crystal Microbalance Biosensor for Bacterial Detection Using Carbohydrate and Lectin Recognitions. Anal. Chem. 79 (6), 2312–2319. doi:10.1021/ac061986j
Siebert, H.-C., Lü, S.-Y., Frank, M., Kramer, J., Wechselberger, R., Joosten, J., André, S., Rittenhouse-Olson, K., Roy, R., von der Lieth, C.-W., Kaptein, R., Vliegenthart, J. F. G., Heck, A. J. R., and Gabius, H.-J. (2002). Analysis of Protein−Carbohydrate Interaction at the Lower Size Limit of the Protein Part (15-Mer Peptide) by NMR Spectroscopy, Electrospray Ionization Mass Spectrometry, and Molecular Modeling. Biochemistry 41 (30), 9707–9717. doi:10.1021/bi025891x
Skládal, P. (2016). Piezoelectric Biosensors. Trac Trends Anal. Chem. 79, 127–133. doi:10.1016/j.trac.2015.12.009
Slominska-Wojewodzka, M., and Sandvig, K. (2015). The Role of Lectin-Carbohydrate Interactions in the Regulation of ER-Associated Protein Degradation. Molecules 20 (6), 9816–9846.
Sondhi, P., Maruf, M. H. U., and Stine, K. J. (2020). Nanomaterials for Biosensing Lipopolysaccharide. Biosensors (Basel) 10 (1), 2. doi:10.3390/bios10010002
Sun, X., Feng, Z., Zhi, S., Lei, C., Zhang, D., and Zhou, Y. (2017). An Integrated Microfluidic System Using a Micro-fluxgate and Micro Spiral Coil for Magnetic Microbeads Trapping and Detecting. Sci. Rep. 7 (1), 12967–12968. doi:10.1038/s41598-017-13389-x
Tateno, H., Hirabayashi, J., Asashima, M., Yuzuru, I. T. O. U., Onuma, Y., Oda, T., et al. (2018). Method for Detecting Cancer Cells, Reagent for Introducing Substance into Cancer Cells, and Composition for Treating Cancer. U.S. Patent Appl. 15/759, 288.
Thévenot, D. R., Toth, K., Durst, R. A., and Wilson, G. S. (2001). Electrochemical Biosensors: Recommended Definitions and Classification. Biosens. Bioelectron. 16 (1-2), 121–131. doi:10.1016/s0956-5663(01)00115-4
Velusamy, V., Arshak, K., Korostynska, O., Oliwa, K., and Adley, C. (2010). An Overview of Foodborne Pathogen Detection: In the Perspective of Biosensors. Biotechnol. Adv. 28 (2), 232–254. doi:10.1016/j.biotechadv.2009.12.004
Vereshchagina, E., Tiggelaar, R. M., Sanders, R. G. P., Wolters, R. A. M., and Gardeniers, J. G. E. (2015). Low Power Micro-calorimetric Sensors for Analysis of Gaseous Samples. Sensors Actuators B: Chem. 206, 772–787. doi:10.1016/j.snb.2014.08.077
Wang, Y., Ye, Z., Si, C., and Ying, Y. (2013). Monitoring of Escherichia coli O157:H7 in Food Samples Using Lectin Based Surface Plasmon Resonance Biosensor. Food Chem. 136 (3-4), 1303–1308. doi:10.1016/j.foodchem.2012.09.069
WHO (2015). WHO Estimates of the Global burden of Foodborne diseasesFoodborne Diseases burden Epidemiology Reference Group 2007–2015. Geneva: World Health Organization 2015.
Wu, W., and Zeng, L. (2017). Current and Emerging Innovations for Detection of Food-Borne Salmonella. Curr. Top. Salmonella Salmonellosis 83. doi:10.5772/67264
Yaghoubi, M., Rahimi, F., Negahdari, B., Rezayan, A. H., and Shafiekhani, A. (2020). A Lectin-Coupled Porous Silicon-Based Biosensor: Label-free Optical Detection of Bacteria in a Real-Time Mode. Sci. Rep. 10, 16017. doi:10.1038/s41598-020-72457-x
Yakovleva, M. E., Moran, A. P., Safina, G. R., Wadström, T., and Danielsson, B. (2011). Lectin Typing of Campylobacter Jejuni Using a Novel Quartz crystal Microbalance Technique. Anal. Chim. Acta 694 (1-2), 1–5. doi:10.1016/j.aca.2011.03.014
Yang, G., Huang, M., Wang, Y., Chen, G., Zhao, Y., and Xu, H. (2019a). Streptavidin-exposed Magnetic Nanoparticles for Lectin Magnetic Separation (LMS) of Staphylococcus aureus Prior to Three Quantification Strategies. Microchim. Acta 186 (12), 813. doi:10.1007/s00604-019-3978-4
Yang, G., Meng, X., Wang, Y., Yan, M., Aguilar, Z. P., and Xu, H. (2019b). 2-Step Lectin-Magnetic Separation (LMS) Strategy Combined with AuNPs-Based Colorimetric System for S. aureus Detection in Blood. Sensors Actuators B: Chem. 279, 87–94. doi:10.1016/j.snb.2018.09.112
Yang, J., Ye, H., Zhao, F., and Zeng, B. (2016). A Novel CuxO Nanoparticles@ZIF-8 Composite Derived from Core-Shell Metal-Organic Frameworks for Highly Selective Electrochemical Sensing of Hydrogen Peroxide. ACS Appl. Mater. Inter. 8 (31), 20407–20414. doi:10.1021/acsami.6b06436
Yazgan, I., Noah, N. M., Toure, O., Zhang, S., and Sadik, O. A. (2014). Biosensor for Selective Detection of E. coli in Spinach Using the strong Affinity of Derivatized Mannose with Fimbrial Lectin. Biosens. Bioelectron. 61, 266–273. doi:10.1016/j.bios.2014.05.008
Zeng, X., and Shen, Z. (2016). Microorganism Detection and Analysis Using Carbohydrate and Lectin Recognition. US patent 9366672.
Keywords: food safety, biosensor, lectin, SPR, food borne pathogens
Citation: Vishweswaraiah RH, Tenguria S, Chandrasekhar B, Harshitha CG, Gandhi K, Kumar N, Aluko RE and Puniya AK (2022) Monitoring of Microbial Safety of Foods Using Lectins: A Review. Front. Food. Sci. Technol. 2:842063. doi: 10.3389/frfst.2022.842063
Received: 24 December 2021; Accepted: 28 February 2022;
Published: 06 April 2022.
Edited by:
Fernando Perez Rodriguez, University of Cordoba, SpainReviewed by:
Jinsong Feng, Zhejiang University, ChinaCopyright © 2022 Vishweswaraiah, Tenguria, Chandrasekhar, Harshitha, Gandhi, Kumar, Aluko and Puniya. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Raghu Hirikyathanahalli Vishweswaraiah, UmFnaHUuVkBpY2FyLmdvdi5pbg==, NHJ2c3kuZG1uZHJpQGdtYWlsLmNvbQ==, Rotimi E. Aluko, cm90aW1pLmFsdWtvQHVtYW5pdG9iYS5jYQ==, Anil Kumar Puniya, YWtwdW5peWFAZ21haWwuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.