- 1School of Minerals Processing and Bioengineering, Central South University, Changsha, China
- 2Key Laboratory of Biometallurgy of Ministry of Education, Central South University, Changsha, China
- 3Changsha Tobacco Company of Hunan Province, Changsha, China
- 4Yongzhou Tobacco Company of Hunan Province, Yongzhou, China
- 5Changde Tobacco Company of Hunan Province, Changde, China
- 6Institute of Grassland, Flower and Landscape Ecology, Beijing Academy of Agricultural and Forestry Sciences, Beijing, China
- 7Hunan Tobacco Science Institute, Changsha, China
Virus-borne auxiliary genes have been demonstrated to influence their hosts’ characteristics and flexibility, resulting in an elevated ability to withstand diverse conditions and competition. However, the research on the soil viral community and its influencing factors is not extensive. The effects of bacterium-phage co-evolution in the metal contaminated environment have yet to be fully understood. In our research, numerous viral genes with conserved functional residues/motifs working on detoxifying heavy metals were pinpointed in the virome recovered from 58 contaminated soil metagenomic samples of mineral area, including tellurite resistance genes (e.g., terC, terD), copper resistance genes (e.g., copC, copA), and arsenate resistance genes (e.g., arsC). Moreover, viral proteins involved in key processes of nutrient uptake and metabolism, cellular function, polysaccharides production and biomineralization were also detected, which may also contribute to the response of microbial community to heavy metal stress. Our research showed that viruses have helped their hosts to acquire novel metal-resistance abilities through horizontal gene transfer (HGT) during the adaptation to challenging metal-rich environments. This was also evidenced by the high Codon adaption index (CAI) values across metal-resistance-related genes. This study has advanced our understanding on virus-host interplay within heavy metal contaminated soils.
1 Introduction
Viruses are abundant and pervasive in Earth’s ecosystems and can have a substantial effect on the composition, activity, and performance of microbial communities, offering an untapped potential for applications in health, industry, and agriculture (Jones et al., 2007; Taati et al., 2020). Viruses are the most abundant and diverse creatures on earth, with a population amount of approximately 1030–1032, a density ten times that of bacteria (Hemminga et al., 2010; Rakonjac et al., 2016). Besides, it is estimated that about twenty percent of prokaryotic genomes were obtained via virus-related elements (Brüssow et al., 2004). Viruses are divided into lysogenic and lytic, which respectively modulate microbial population through infection and cell death, guide microbial metabolism using auxiliary metabolic genes, and facilitate inter-host gene transfer (Thompson et al., 2011; Feiner et al., 2015; Puxty et al., 2015; Jin et al., 2019). Viruses persist longer in the environment than other mobile genetic elements (MGEs) like plasmids. Of particular interest is the lysogenic virus, as it is heavily involved in horizontal gene transfer (HGT). The incorporation of novel genes by lysogenic virus may give their host a beneficial phenotype and help them survive in difficult conditions. In return, the host can provide a safe haven for the virus (Gao et al., 2022; López-Leal et al., 2022). A most prominent instance is that the marine viruses encode and transfer many key components of photosystem, a common target of photo-inhibition. The presence of these viruses in the ocean may help prevent photo-inhibition of infected algae, allowing photosynthesis to continue and providing the energy needed for the virus to replicate (Puxty et al., 2015).
Soils can contain up to ten billion viruses per Gram, while geographical location and climatic condition are the major determinants of the spread of viruses in the soil. Recent studies propose that viruses may play a crucial role in both carbon and nutrient cycling in terrestrial environments, similar to their role in marine systems (Ashelford et al., 2003; Williamson et al., 2017; Attai and Brown, 2019; Starr et al., 2019; Hillary et al., 2022). Viruses can also affect the phosphorus cycling of agricultural soils since viral-encoded phosphorus metabolism genes are unexpectedly abundant (Han et al., 2022). Viruses in soil environment were also regarded as an underestimated reservoir for the storage and transmission of antibiotic resistance determinants (Ross and Topp, 2015; Zhang et al., 2022). Even though viruses have a significant role in the microbial evolution and global biogeochemical cycles, our knowledge of soil-borne viruses and the factors influencing their spread is far less than our understanding of marine viruses (Thompson et al., 2011; Breitbart et al., 2018; Jin et al., 2019). Compared with viruses in the marine environment, the types, quantity, and diversity of viruses in soil are more complex. This makes it more difficult to conduct systematic studies on soil viruses.
Environmental stress has a significant impact on the survival rate of microbes and can drastically change the microbial community structure (Harrison et al., 2013). For instance, the unfavorable geochemical conditions of tailing can inhibit biological activities and alter the keystone taxa of the microbial community (Sun et al., 2020). Likewise, multiple environmental factors, including host, pH, water, metal ion concentration, climate and site altitude, are linked to viral community composition (Mojica and Brussaard, 2014; Adriaenssens et al., 2017; Emerson et al., 2018; Trubl et al., 2018). The accumulation of heavy metals in soils among mineral area poses a grave risk to public health and ecological stability due to its prevalence, durability, and toxicity (Ye et al., 2015; Nolos et al., 2022). Soils contaminated with heavy metals in mineral area usually show a geochemical gradient from the source of contamination (Xu et al., 2020). These contaminated soils among mineral area are special ecosystems under intense pressure and are ideal for studying the interaction between viruses, bacteria, and environmental stressors. In order to combat heavy metal toxicity, microbe has developed complex defense systems such as metal chelation and precipitation, molecular pumps that expel toxic metals from cells, enzymatic conversion of metal ions into less dangerous forms, and construct more resilient cell walls and membranes that can serve as a barrier against toxic metals (Navarro et al., 2013). The lysogenic phage-infected bacteria may acquire heavy metal resistance genes in the viral genome when the lysogens rupture, facilitating the transduction of these genes among bacteria and therefore enhancing the viral host’s resistance against heavy metal toxicity (Tang et al., 2019; Tang et al., 2021). Viruses such as temperate phages can establish a long-term partnership with bacterial hosts which allows for efficient reproduction of both the viruses and their hosts (Feiner et al., 2015). This has opened up a potential route for remediating (mineral) environments through massive application of lysogenic viral communities.
The presence of heavy metals in soils among mineral area significantly affects the composition of virome (Huang et al., 2021), yet little is known about the virus-bacterium interactions under heavy metal-induced stress. Many aspects of virus-borne genetic makeup and their influences on the host remain poorly understood: how does environmental stress affect bacteriophage collaboration, particularly in terms of survival under extreme conditions among mineral area? What role does the virus’s genetic makeup play in the virus-host interaction?
To gain a better understanding of viral capacity for further lysogenic phage application, it is important to first figure out how the microbial hosts and viruses cooperate, such as how environmental stress and viral genome compositions influence the host’s survival in mineral area. Understanding virus-host interactions in heavy metal stress scenarios can also improve our assessment of the eco-toxicity of heavy metals and the viability of using microbiome engineering for contaminated soil remediation among mineral area. Against such background, we focus on the putative heavy metal resistance related components encoded by viral genomes recovered from soil metagenome among heavy metal contaminated area (58 Genbank metagenomes) to investigate the adaptation processes of soil-borne microbiome to hazardous environments and the importance of viruses for biogeochemical processes. Further phylogenetic analysis, three-dimensional (3D) modeling of their protein structures along with selection pressure assessment/codon adaption index (CAI) analysis were carried out to investigate the diversity, functional conserved motifs, the strength of natural selection and expression possibility of the interested heavy metal resistance related genes identified in viral sequences. This work could aid us in comprehending the intricacies of bacterium-virus collaboration and perhaps provide a theoretical foundation for the wide application of remediating lysogenic virus in the future.
2 Results
2.1 Overall status
We first queried and retrieved all the items in the Genbank public database (Benson et al., 2018) with the keywords “soil” and “metal” within the “biosample” regions. The out-coming 58 soil metagenomes of microbiome as listed in Supplementary Table S1 at https://doi.org/10.6084/m9.figshare.22226998.v3 was used for downstream analyses. High radionuclide (137Cs, 210Pb, 226Ra, 228Ra, 60Co, 241Am, 238U, 228Th, and 232Th) levels ranging from 30 to 3750 Bq/kg and high toxic metal (Co, Ni, Cu, Zn, As, Cd, Hg, Pb, and Cr) levels ranging from 60 to 2500 mg/kg soil are present in the tested soil samples (Rogiers et al., 2021). We then successfully recovered a total of 785 viral scaffolds from the tested metagenomes, with 2.5% of them has complete circular and 6.6% of them has medium or high quality. The pangenome of the viromes derived from the tested 58 soil metagenomes possessed 27,404 gene families. Pangenome mathematical modeling of the retrieved 58 viromes revealed an ‘‘open’’ pangenome fitted into a power-law regression function [Ps (n) = 2620.57n0.56], while the core genome was fitted into an exponential regression [Fc (n) = 2306.47 e −0.38n] (Figure 1A). These results indicate vast genetic diversity within the soil viromes from heavy metal contaminated soils and that the currently characterized features of the viromes are still far from saturation. Viral families Zierdtviridae, Orlajensenviridae, Mesyanzhinovviridae, Peduoviridae were the most abundant in the tested soil samples from mineral area (Figure 1B). Our results are quite different from previous reports on soil samples from Antarctic, in which viral families Siphoviridae, Myoviridae and Podoviridae were dominant (Adriaenssens et al., 2017). Siphoviridae and Podoviridae are mild viruses, while Myoviridae is a lytic virus in polar marine viruses (Luo et al., 2022). Functional annotation based on clusters of orthologous group (COG) revealed that the accessory gene of tested viromes had a higher proportion of genes classified in COG categories L (replication, recombination, and repair, 24.53%), K (Transcription, 10.03%), and M (Cell wall/membrane/envelope biogenesis, 8.41%) (Figure 1C), which were probably related to the mutualism and co-evolution of virus-host interplay to oligotrophic and metal-laden conditions in mineral area that often cause DNA damages needed rapid response and restoration. Principal coordinate analysis (PCoA) based on the numerical distributions of viral protein families in tested viromes showed that the viromes appeared to be separated by the geographical locations (Figure 1D, analysis of similarity [ANOSIM], R > 0, p < 0.05). Previous research has revealed that the viral communities and functions from nineteen soil samples across China can be grouped into clusters corresponding well with the geographical locations (Han et al., 2022). This suggests that viral community composition and function might be driven by geographic-related factors. Interestingly, virome was reported to more sensitive than total metagenome in reflecting the spatiotemporal patterns of soil samples (Santos-Medellin et al., 2021). We fail to detect any positive selection signal across virome in this study with Posigene software (Sahm et al., 2017). Also, the codon adaption index (CAI) was used to measure the synonymous codon usage bias for the viral gene sequences, comparing it to the reference (host codon usage bias) data set to indicate bias (expression level) (Figure 1E). Results showed that many functional genes encoded by viral sequences possess relatively high CAI values: averagely, the COG categories [U] Intracellular trafficking, secretion, and vesicular transport (0.70) [C] Energy production and conversion (0.67) [G] Carbohydrate transport and metabolism (0.67) have relatively higher predicted CAI values (higher expression level). The viral genes related with microbial metal resistance as annotated against the BacMet database (Pal et al., 2014) also present high CAI values (average 0.68). However, these viral gene families related with metal resistance distribute unevenly among tested contaminated soil samples (Figure 1F).
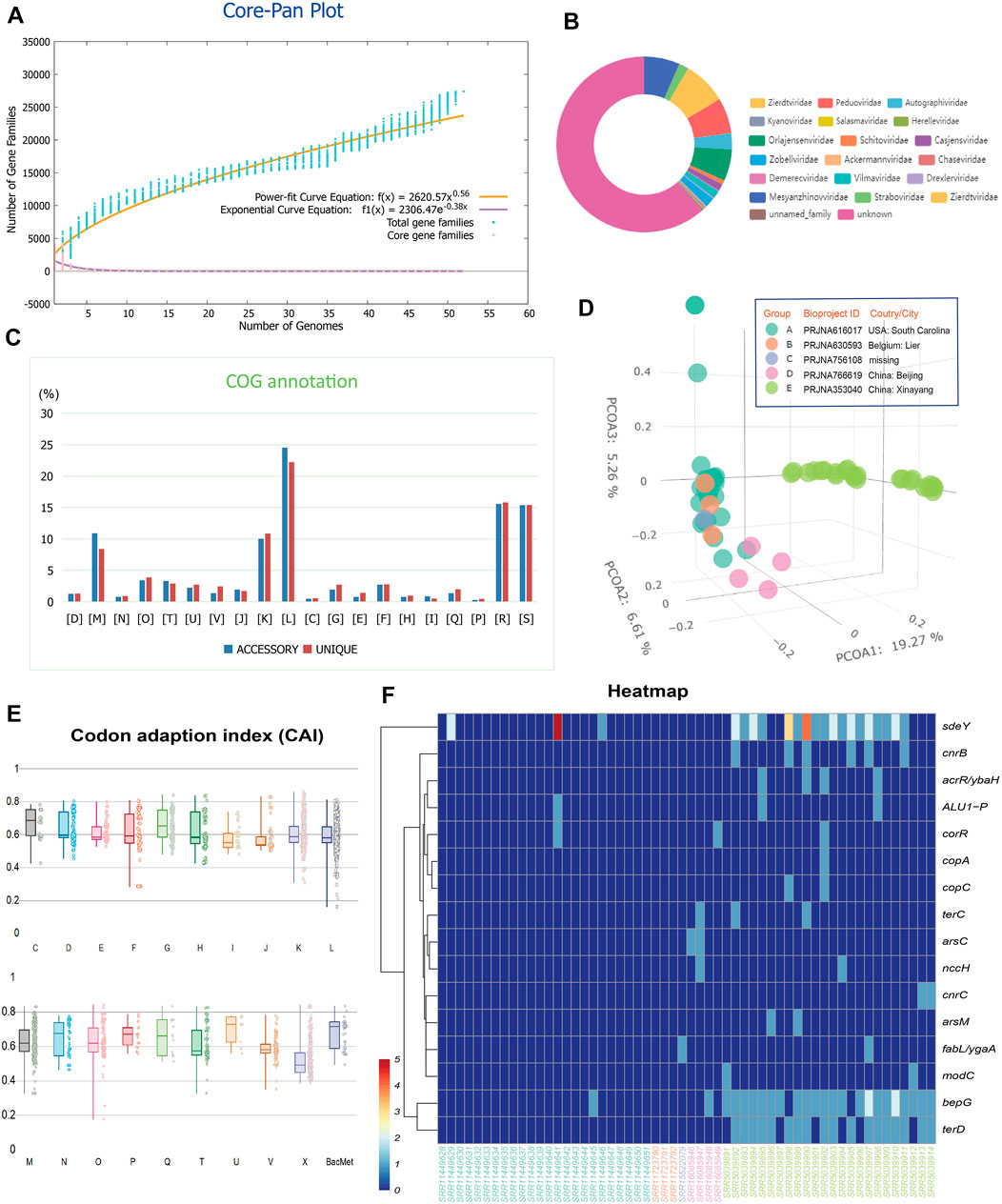
FIGURE 1. (A) Pangenome analyses and mathematical modeling of the size succession of the pangenome and core genome of the 57 viromes. (B) Taxonomic annotation and statistics of the scaffolds from the viromes using PhaGCN. (C) Bar chart showing functional proportions (based on COG categories) of different parts of the pangenome (i.e., accessory, unique). Detailed descriptions of the COG categories: [D] Cell cycle control, cell division, chromosome partitioning; [M] Cell wall/membrane/envelope biogenesis; [N] Cell motility; [O] Post-translational modification, protein turnover, and chaperones; [T] Signal transduction mechanisms; [U] Intracellular trafficking, secretion, and vesicular transport; [V] Defense mechanisms; [J] Translation, ribosomal structure and biogenesis; [K] Transcription; [L] Replication, recombination and repair; [C] Energy production and conversion; [G] Carbohydrate transport and metabolism; [E] Amino acid transport and metabolism; [F] Nucleotide transport and metabolism; [H] Coenzyme transport and metabolism; [I] Lipid transport and metabolism; [Q] Secondary metabolites biosynthesis, transport, and catabolism; [P] Inorganic ion transport and metabolism; [R] General function prediction only; [S] Function unknown. (D) Principal coordinate analysis (PCoA) based on the numerical distributions of viral protein families in tested viromes. (E) Distribution of CAI values measuring the synonymous codon usage bias for the viral gene sequences classified by functional categories. (F) Heatmap showing uneven distribution of viral gene families related with metal resistance among tested samples.
2.2 Metal resistance genes in viral sequences
Previous studies indicated that HGT was a main adaptive mechanism for microbes to cope with heavy-metal-rich environments (Li et al., 2019). Viruses can be regarded as a particular type of HGT vectors to facilitate the spread of resistance among microbial community through transduction (Ross and Topp, 2015; Zhang et al., 2022). By annotation against the BacMet database (Pal et al., 2014), we have detected in the tested viromes multiple gene families related with microbial metal resistance (Figure 1F).
2.2.1 Tellurite resistance
Tellurite is highly toxic due to the similarity to sulfur, which can interfere with the thiol inside cells (Zannoni et al., 2008). Even low concentration (1 μg/mL) of tellurite can severely inhibit the cell growth of E. coli (Chasteen et al., 2009). The ter gene cluster is considered essential for tellurite resistance, allowing for the high tellurite resistance (Peng et al., 2022). Consistently, we have identified abundant tellurite resistance genes (mainly terC and terD) in the viral sequences, which are further verified by bioinformatics approaches (Figure 2).
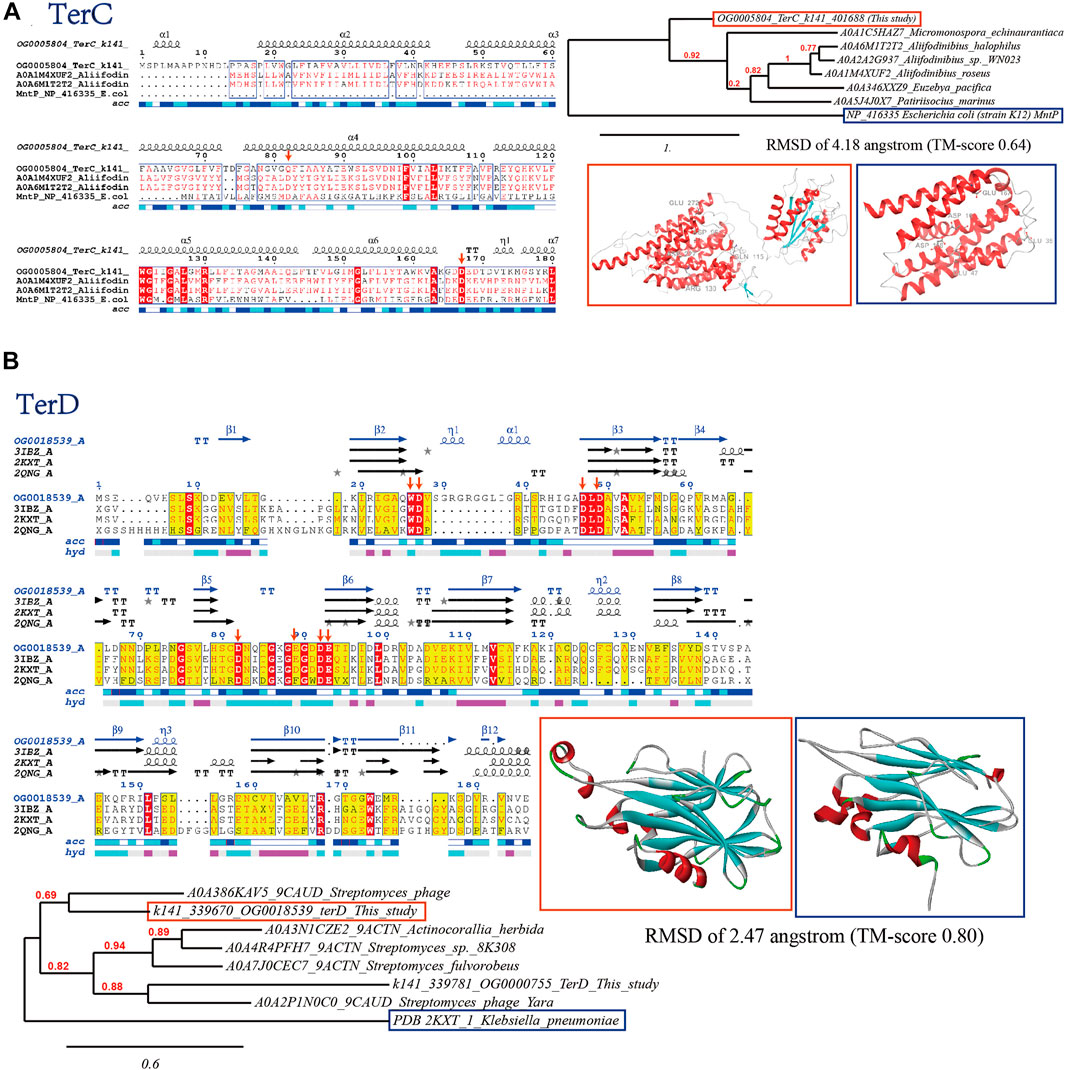
FIGURE 2. Bioinformatics verification of tellurite resistance proteins (A) TerC and (B) TerD through multiple sequence alignment (with verified conserved key residues marked with red arrows), phylogenetic reconstruction and 3D protein model comparison.
The hydrophobic TerC protein is likely a membrane protein that transports tellurite, and is the only one of its kind in the ter gene cluster, suggesting its role is essential. The TerC domains have a common core of seven TM helices, characterized by two extra transmembrane helices inserted between helices three and four of the conserved core, and an N-terminal hydrophobic helix which might be either an additional TM segment or part of an intracellular signal peptide (Figure 2A). The similarity between TerC protein and manganese transporter MntP suggests that TerC may be involved in metal transportation. The 3D structure of the viral TerC domain exhibited the classical transmembrane helices with TM-aligned score of 0.64 to Escherichia coli (strain K12) MntP (Zeinert et al., 2018). Both TerC and MntP proteins harbor two conserved acidic and negatively charged Asp residues (Figure 2A, marked with red arrows, in MntP order), Asp16 and Asp118, from the first helix of each of the trihelical units. These residues form a negatively charged pore within the membrane, allowing the metal cation to be expelled whereas mutation of Asp16 or Asp118 would completely abolish MntP activity (Zeinert et al., 2018). Protein TerD also contributes to tellurite resistance in addition to TerC (Turkovicova et al., 2016). Multiple sequence alignment (MSA) analysis showed that the reported metal-binding sites vital for the function of TerD concealing in the inter-strand loops are conserved within viral homologs detected in this study and TerD with structures available in PDB database (Figure 2B, marked with red arrows), which are respectively constituted by Asp12, Asp67 and Asp70 (site 1) and Asp22, Asp24, Asp60, and Glu71 (site 2), with Trp11 acting as a hydrophobic structural bridge between them and facilitate cooperative metal-binding by the two sites (Pan et al., 2011). These results indicated that viral TerD-like proteins may bind calcium through the same pattern. Besides, the predicted 3D structure of the viral TerD domain exhibited the classical ß-sandwich fold and shared RMSD of 2.47 angstrom (TM-score 0.80) to Klebsiella pneumonia TerD (PDB 2KXT) (Pan et al., 2011). Phylogenetic analysis showed that identified viral TerD-like proteins in this study are mostly clustering with homologs related to Streptomyces (the indicated bacterial host).
2.2.2 Copper resistance
We have also detected copper resistance proteins CopC and CopA in our retrieved viral sequences from soil samples contaminated with heavy metals, which are further verified by bioinformatics approaches (Figure 3A). The copper binding proteins (CopC) found in the outer membrane is part of the cop system that control copper availability. Like the bacterial homolog, viral CopC found in current study contains the conserved histidine brace (His-brace), a copper-binding motif that is associated with both oxidative enzymes and proteinaceous copper chaperones. CopC is composed of two domains, an N-terminal domain and a C-terminal domain. The N-terminal domain is composed of two alpha helices and two beta strands, while the C-terminal domain is composed of three alpha helices and two beta strands (Figure 3A). The key residues of CopC are located in the N-terminal domain and are involved in the binding of copper ions (Ipsen et al., 2021). The key residues include histidine, aspartate, and glutamate, which are all conserved in our viral homologs (Figure 3A, marked with red arrows). The functions of these key residues are to bind copper ions and to regulate the activity of the protein. The binding of copper ions is important for the proper functioning of the protein, as it is involved in the transport of copper ions across the outer membrane of E. coli. Additionally, the regulation of the protein’s activity is important for the proper functioning of the protein, as it helps to ensure that the protein is active when needed and inactive when not needed. Histidine is important for the coordination of copper ions, while aspartate and glutamate are involved in the stabilization of the copper-binding site. Additionally, these residues are involved in the regulation of the protein’s activity. As verified by previous site-directed mutation study (Ipsen et al., 2021), the residues His1 and His83 are essential for the extremely high copper affinity of CopC. Mutations in the conserved Glu27 affect the oxidation rate of ascorbate and Apo D83A was unable to bind Cu(II) since Asp83 residue directly coordinates copper through its carboxylic group. The other copper resistance proteins found to be encoded by virus in our study is CopA, which displays domains that are characteristic of P-type ATPases containing conserved functional residues as visualized after structure modeling (Figure 3B). Val 74 of CopA is reported to initiate the long and curved helix, and the N-terminal part provides a short transmembrane helix kinking at the cytosolic membrane interface facilitated by two conserved glycines, Gly129 and Gly130. Conserved residues Met 148, Glu 205 and Asp 337 from the entry are responsible for initial Cu+ coordination. Cu+ was then transferred from Met 148 to Cys 382, stabilized by hydrogen bonds of Leu 151 and Ile 152. Negatively charged residue Glu 189 further stimulated Cu+ release to the extracellular side. Finally, the conserved residue Met 717 was suggested to serve to guide copper to the exit (Gourdon et al., 2011). These verified conserved key residues are marked with red arrows in the multiple sequence alignment graph (Figure 3B).
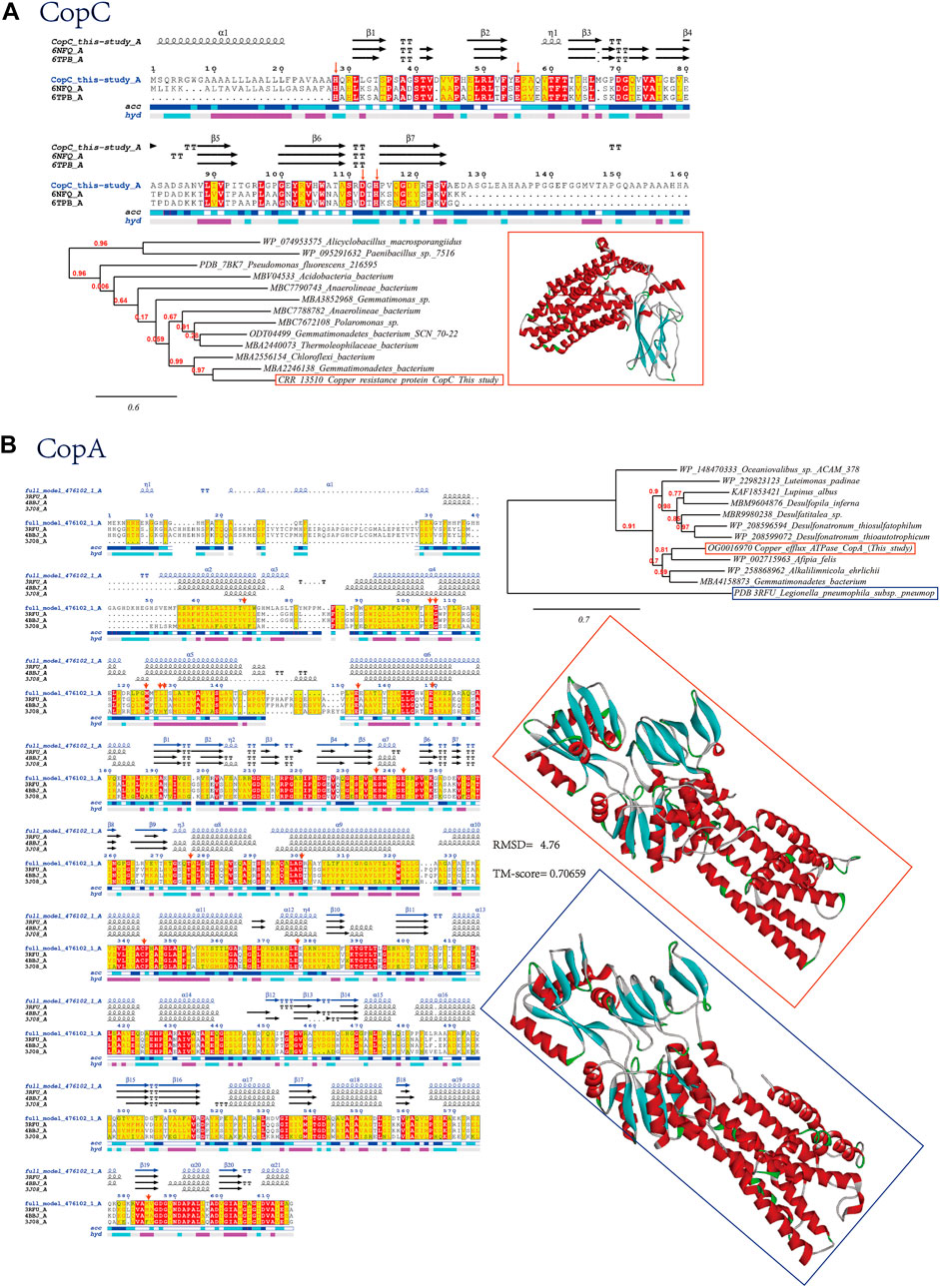
FIGURE 3. Bioinformatics verification of copper resistance proteins (A) CopC and (B) CopA through multiple sequence alignment (with verified conserved key residues marked with red arrows), phylogenetic reconstruction and 3D protein model comparison.
2.2.3 Arsenate resistance
Arsenate reductase (ArsC) catalyzes the reduction of arsenic-containing compounds for detoxification, which is a common contaminant in metal-contaminated soils. The key residue in ArsC enzymes is typically a cysteine residue, which is responsible for binding and reducing the arsenic compound. The initial step of arsenate reduction by ArsC involves the active cysteines Cys8 and Cys80 (the substitutions of Cys8 and Cys80 by serine led to inactive reactions), and the reaction results in the formation of a Cys8-Cys80 disulfide-bridged intermediate (Hu et al., 2015). Consistently, these two Cys residues are conserved in the viral homolog (Figure 4). The predicted structure of viral ArsC is in highly homologous to Synechocystis ArsC (RMSD 1.67, TM-align score 0.85) (Hu et al., 2015): a central ß-sheet is formed by four parallel ß-strands, which are flanked by three a-helices and extended loops. The secondary structural elements are generally similar but differ in local conformations. Besides, S-adenosyl-L-methionine-dependent methyltransferases (ArsM) detected in the virome that contain conserved cysteine residues (see Supplementary Figure S1 at https://doi.org/10.6084/m9.figshare.22226998.v3), might also enhance the ability of virus-bacteria consortium to survive in metal contaminated soils by transferring methyl groups from S-adenosyl-L-methionine to metals like arsenite to reduce the toxicity (Struck et al., 2012).
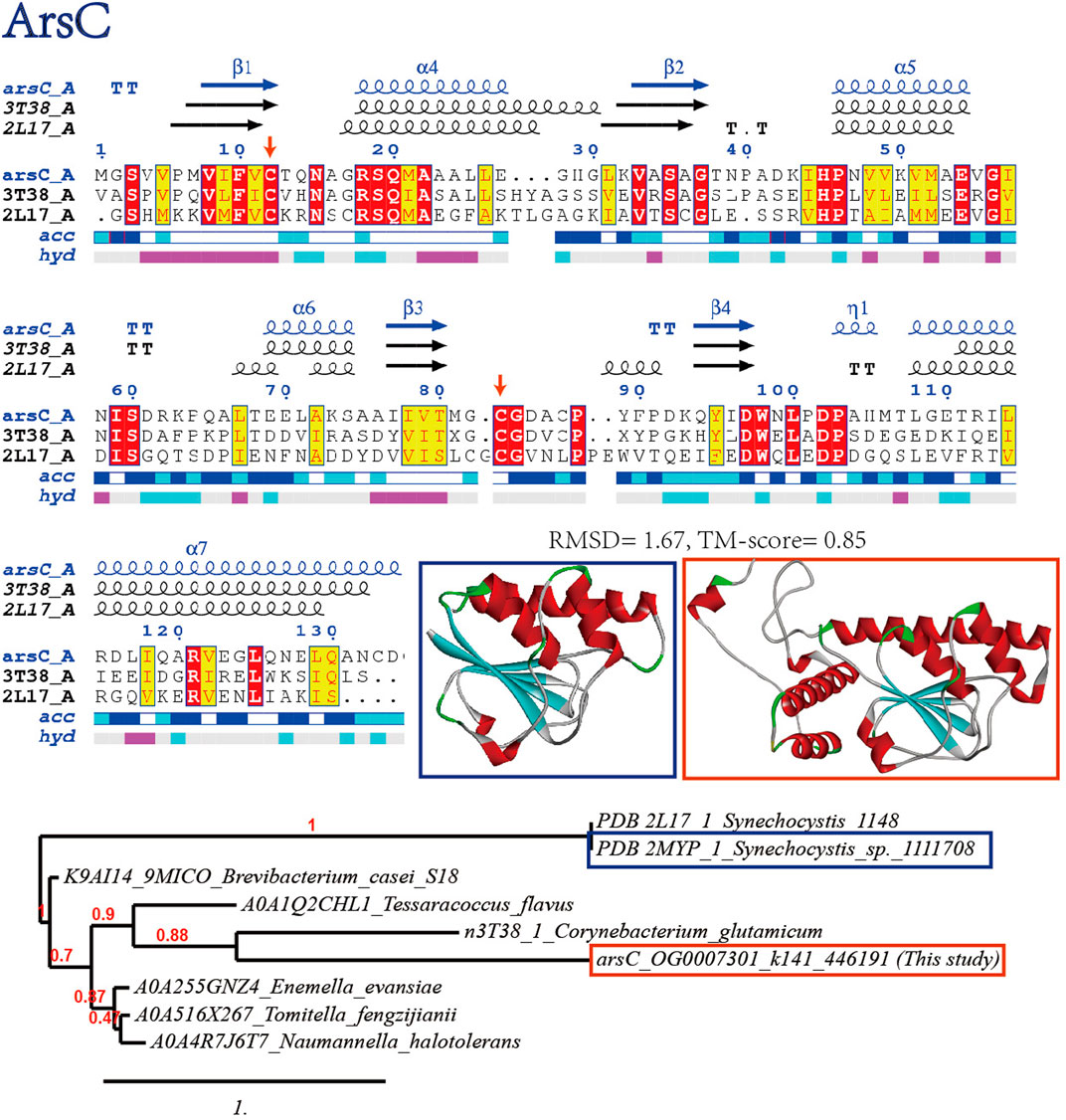
FIGURE 4. Bioinformatics verification of arsenate resistance proteins ArsC through multiple sequence alignment (with verified conserved key residues marked with red arrows), phylogenetic reconstruction and 3D protein model comparison.
2.2.4 Other metal resistance proteins
Metal resistance proteins with wider metal substrate specificity are also found (Figure 1F). Anion transmembrane transporters (ALU1-P) are proteins that facilitate the movement of anions (negatively charged ions) across the cell membrane. By having this activity, the virus can more easily move anions in and out of the cell, which can help it to survive in metal contaminated soils. For example, anion transmembrane transporters can help the virus to take up essential nutrients, such as iron, from the soil, which can help it to survive in metal contaminated soils. Additionally, anion transmembrane transporters can help the virus to expel toxic metals from the cell. Another protein, CutA1 with divalent ion tolerance potential, is a ubiquitous trimeric protein that binds to heavy metals, such as zinc, copper and helps to prevent them from entering the cell and damaging its genetic materials (Tanaka et al., 2004). This helps to increase the virus-bacteria consortium’s ability to survive in metal contaminated soils.
2.3 Viral genes indirectly related with metal resistance
Apart from the viral genes directly involved in metal efflux and metabolism, there are other viral genes from the virome of metal contaminated soils putatively response to heavy metal stress (see Supplementary Table S2 at https://doi.org/10.6084/m9.figshare.22226998.v3), which can be classified into categories as followed: gene regulation, auxiliary metabolism, molecular chaperone, antioxidant and DNA repair.
2.3.1 Gene regulation
Viral genes encoding LysR-type and LuxR family transcriptional regulators, restriction modification, SOS response components, and ROK family proteins that may function in host gene expression regulation are detected in the virome from the metagenomes of metal contaminated soils (see Supplementary Table S2 at https://doi.org/10.6084/m9.figshare.22226998.v3).
LysR-type transcriptional regulator (Lsr) family proteins are involved in the regulation of a variety of metabolic processes, including the utilization of alternative carbon sources, the production of secondary metabolites, and the response to environmental stress. Previous results have suggested that the Lsr proteins help Mycobacterium smegmatis cells cope with stress conditions, including hypoxia and exposure to toxic compounds (Kołodziej et al., 2021). Restriction modification systems (RMS), consisting of restriction endonucleases (REs) and DNA methylases (MTases), regulate virulence, plasmid mobilization, bacterial immunity, DNA repair, transcription, replication, regulation and resistance in bacteria. The SOS response associated peptidase (SRAP) is a type of enzyme that helps to repair the damage and restore the cell’s normal functioning by cleaving specific peptides that are produced during the SOS response. SRAP also acts as a DNA-associated autoproteolytic switch that recruits diverse repair enzymes to efficiently fix crosslinking DNA damage caused by the metals (Kühbacher and Duxin, 2020). The LuxR family proteins are transcriptional regulators that bind to the lux promoter and activate the expression of genes involved in quorum sensing in response to environmental signals. Quorum sensing (QS) is a process of cell-to-cell communication in bacteria that allows them to coordinate their behaviors and expression of a variety of genes in response to changes in their environment (e.g., heavy metal stress), including those genes encoding motility, nodulation, plasmid transfer, bioluminescence, and biofilm formation against environmental turbulence (Chen and Xie, 2011), enhancing the virus-bacteria consortium’s ability to resist surrounding challenges, and thereby increasing viral ability to infect, replicate and spread in different host cells. In metal contaminated soils, the LuxR proteins of QS system might also help the virus-bacteria consortium to survive by regulating the expression of genes involved in metal resistance, such as those encoding metal-chelating proteins, efflux pumps, and other proteins involved in metal detoxification, allowing their bacterial hosts to better tolerate the toxic effects of metals. The ROK family of genes encodes repressor, open reading frame, and kinase proteins, which are important for regulating gene expression and controlling the activity of enzymes. These proteins can help to regulate the expression of genes involved in viral replication and host metabolism. Genes that encode transcriptional regulator CopR, CzcR, and AcrR/ybaH TetR family transcriptional regulators, would help to regulate the expression of genes involved in the metal stress response. These proteins bind to specific DNA sequences and control the expression of genes involved in metabolic pathways in response to toxic compounds.
By controlling the expression of genes, the virus-bacteria consortium can better adapt to the metal contaminated environment and increase its ability to survive. The expression of genes involved in metal resistance can be upregulated to better tolerate the metal-contaminated environment. Additionally, the expression of genes involved in nutrient acquisition can also be upregulated, allowing the virus-bacteria consortium to better utilize the available resources.
2.3.2 Auxiliary metabolism
Auxiliary metabolic genes (AMGs) are genes that are carried by lysogenic viruses and are involved in the metabolism of the host cell. These genes are not essential for the survival of the virus, but they can provide the virus-bacteria consortium with an advantage in the environment, which can be involved in a variety of metabolic pathways, including the synthesis of amino acids, vitamins, and other metabolites. They can also be involved in the degradation of toxic compounds, the production of antibiotics, and the regulation of gene expression. AMGs can be horizontally transferred between different virus-bacteria consortiums, allowing them to adapt to different environments. Consistently, a wide range of AMGs are detected in the virome from the metagenomes of metal contaminated soils (see Supplementary Table S2 at https://doi.org/10.6084/m9.figshare.22226998.v3).
Chitinase class I is an enzyme that breaks down chitin, a polysaccharide found in the cell walls of fungi and other organisms. By breaking down chitin, the host can gain access to nutrients and other resources that would otherwise be unavailable. Overexpressing chitinase has shown to confer resistance to salinity and heavy metals (Song et al., 2011). UDP-galactose 4-epimerase (GalE) is a gene that encodes a protein involved in the synthesis of galactose, an energy source sugar. By encoding from virus the gene for GalE, the viral hosts can utilize galactose more efficiently as an energy source, which can help it to survive in metal contaminated soils. Cellulose 1,4-beta-cellobiosidase is an enzyme that breaks down cellulose, a major component of cell walls. By having this enzyme, the virus-bacteria consortium can more easily break down the cell walls of dead microbial residues. The dTDP-4-dehydrorhamnose 3,5-epimerase helps to convert dTDP-4-dehydrorhamnose into dTDP-4-keto-6-deoxy-D-glucose, which is an essential precursor for the synthesis of the surface polysaccharides and biofilm components. By having this enzyme present in the viral sequences, the virus-bacteria consortium would synthesize the cell barrier more efficiently, which would in turn enhance its ability to survive in metal contaminated soils. Beta (1–6) glucans synthase is an enzyme that helps to synthesize beta-glucans, which are polysaccharides that are found in the cell walls of bacteria, fungi, and algae. Beta-glucans are known to have a variety of beneficial properties, including the strong ability to bind to and neutralize (forming a complex) toxins and metal ions, such as lead, mercury, and cadmium (Ayangbenro and Babalola, 2017). Likewise, alpha-1,4-glucan-protein synthase (UDP-forming) is an enzyme that helps to synthesize alpha-glucan found in protective coatings of many bacteria and fungi against the toxic effects of the metal. By encoding this enzyme in the viral sequences, it could help the virus-bacteria consortium to survive in metal contaminated soils by providing protection from the toxins and stimulating the resilience response. Besides, oxidoreductases such as Fe(II)-dependent oxygenase, tryptophan halogenase, and thioredoxin, which are enzymes that promote oxidative reactions of the hosts, forming cascades of signalization and reprogramming host metabolism to facilitate viral infection (Silva et al., 2017; Wang Q. et al., 2022).
Phosphoadenosine phosphosulfate reductase (PAPSR) is an enzyme that helps to reduce sulfate to sulfide, which is an important source of energy for many viruses. The key residues of the PAPSR are cysteine residues, which act as cofactors to reduce PAPS to sulfite and adenosine monophosphate (AMP). These cysteine residues form a network of hydrogen bonds that facilitate the transfer of electrons (Yu et al., 2008). By having PAPSR, the virus-bacteria consortium can use sulfate as an energy source and use sulfide to chelate metal ions, such as cadmium, mercury, or lead due to the formation of strong bonds between metals and sulfuric complexes (Escobar-Ledesma et al., 2020), which can help them survive in metal contaminated soils from mineral area. Glutamine synthetase is an enzyme that helps to convert ammonia into glutamine, which is an important amino acid for the synthesis of proteins. By having this enzyme, the virus-bacteria consortium can use the ammonia present in the metal contaminated soils to replace damaged proteins and survive. Additionally, glutathione synthesized from glutamine can also help to detoxify toxic elements in the environment. Glutamine amidotransferase (class-II) is enzyme involved in the metabolism of nitrogen-containing compounds and the synthesis of amino acids. Lactoylglutathione lyase helps to break down lactoylglutathione, a compound that is toxic to many organisms. The lactoylglutathione lyase family is reported to be involved in tolerance to salinity and heavy metals (Heidel et al., 2016). These enzymes can help to enhance the ability of viruses to survive in metal contaminated soils by providing the necessary amino acids for its survival. Alkaline phosphatase is an enzyme that helps to break down phosphates for many metabolic processes. Additionally, alkaline phosphatase can help to neutralize the acidic environment of metal contaminated soils, which can also help the virus to survive. The pstS gene that viruses carry is known to increase phosphorus uptake in times of phosphorus deficiency (Nilsson et al., 2022). Viruses can use these enzymes to help their host access and uptake more nutrients. Meanwhile, the production of glycoproteins and cell membrane glycogens would form complexes to immobilize heavy metals and therefore reduce their toxicity. These extended metabolic potentials can help the virus to survive in metal contaminated soils, where the availability of nutrients is limited.
2.3.3 Molecular chaperone and antioxidant
Heat/cold shock proteins and protective redox proteins providing protection from the damaging effects of the environment are also encoded by genes from the virome. Hsp20/alpha crystallin family proteins, small heat shock proteins (sHSPs) are a type of heat shock proteins that help protect cells from stress and damage caused by environmental factors such as extreme temperatures, heavy metals, oxidative stress, and other toxins. By encoding these proteins, the virus can better survive in metal contaminated soils by protecting itself from the damaging effects of the metals. The Cold-shock DNA-binding domain is a protein domain that helps viruses to survive under cold temperatures. The proteins may also help the virus to better survive in extreme environments, such as temperature perturbation. Glutaredoxin-like proteins, such as YruB-family, are involved in the regulation of redox reactions, which are essential for the survival of viruses in metal contaminated soils. These proteins can help to reduce the oxidative stress caused by the metals, allowing the virus to survive in the contaminated environment. Additionally, glutaredoxin-like proteins can also help to scavenge reactive oxygen species (ROS) and reactive nitrogen species (RNS), which can further enhance the virus’ ability to survive in the metal contaminated soils. Superoxide dismutase (SOD) is an enzyme that helps to protect cells from oxidative damage of proteins and genetic materials caused by reactive oxygen species (ROS). By providing protection from ROS, SOD can help viral hosts to survive in metal contaminated soils.
2.3.4 DNA repair
DNA repair proteins of various functions are also detected in the virome from the metagenomes of metal contaminated soils (see Supplementary Table S2 at https://doi.org/10.6084/m9.figshare.22226998.v3). By repairing damages of DNA caused by metal-induced oxidative stress, the virus-bacteria consortium can better survive the metal contaminated soils by being able to replicate and reproduce stably. This helps the virus to survive in harsh environments by more efficiently repairing any damage to its genetic material caused by metal contamination.
Endonuclease resolves Holliday junction intermediates during homologous genetic recombination and DNA repair. DNA ligase then repairs the broken strands of DNA. These enzymes can also help the microbial consortium to more quickly replicate and spread, as it can help to speed up the process of homologous recombination, helping the virus to better adapt to its environment by allowing it to more quickly evolve and acquire new genetic material. DNA excision and crossover junction endodeoxyribonuclease activity helps to enhance the ability of virus-bacteria consortium to survive by allowing them to repair the genetic material: break down the damaged DNA strands and replace them with new, healthy strands. DNA mismatch endonuclease Vsr helps to repair mismatched DNA bases in the genome. This enzyme helps to ensure that the genome is accurately replicated and that any mutations that occur are corrected. DNA and RNA non-specific endonucleases are enzymes that can cleave DNA and RNA molecules at soundings. These enzymes can help to survive by allowing them to break down and degrade the DNA and RNA molecules of other organisms in the soil, thus providing them with a source of nucleotides and nutrients. Uracil DNA glycosylases (UNG) may function in the repair of viral DNA during periods of latency in non-dividing cells (Sire et al., 2008). The helicase nuclease enzyme would also help to enhance the ability of virus-bacteria consortium to survive by allowing them to quickly and efficiently repair any damage to their genetic material caused by metal contamination. A helicase nuclease prepares dsDNA breaks (DSB) for recombinational DNA repair and then binds to DSBs and unwinds DNA via a highly rapid and processive ATP-dependent bidirectional helicase activity. The enzyme would also help to facilitate homologous recombination.
3 Discussion
Viral sequences integrated into the genomes of cellular organisms and passed down through generations were previously considered as a specific category of “junk DNA” [“What is that junk DNA doing in the genomes?” was listed as one of the most advanced scientific questions by the Editorial of Science (2005)]. However, ongoing studies have unraveled the putative roles of these co-evolution products as drivers of genetic innovation (Moniruzzaman et al., 2020). In our study, we have discovered various virus-borne genes putatively contributing to the metal resistance of the microbial community in the metagenomes recovered from heavy metal contaminated soil samples. Further selection pressure and CAI analysis showed that all metal resistance related genes that virus encoded exhibited a dN/dS ratio of less than the cutoff value 1 and relatively high CAI values. Most non-synonymous mutations have adverse effects on functionally important genes and are often removed by purifying (negative) selection (dN/dS ratio <1). We fail to detect any positive selection signal across virome in this study with Posigene software (Sahm et al., 2017), suggesting that purifying (negative) selection might be dominant and acts on functionally viral genes retrieved from metal-contaminated soil samples since even minor changes in the gene sequences might significantly affect the functionality of crucial proteins. Besides, our results showed that around 53.2% of metal resistance genes were predicted by the CAI to be highly expressed (Figure 1E).
Consistent with our results, many studies has reported that viruses can contribute to the stress resistance of microbial communities by transferring genetic material that encodes for stress (e.g., antibiotics and heavy metal) resistance genes (Huang et al., 2021). This can occur through horizontal gene transfer (HGT), where a virus carrying a stress resistance gene infects a host cell and incorporates the gene to the host’s genome, and thereby increase the stress resistance by providing the host cells with the ability to resist the toxic effects of environmental stressors (e.g., antibiotics or metals). Additionally, viruses can also act as vectors for the spread of resistance genes among microbial communities, allowing for the spread of resistance genes to other microbial populations. It has been shown that the interaction between viruses and bacteria was enhanced along with the increase of chromium contamination (Huang et al., 2021). Besides, the abundance of lysogenic components also increased with increasing chromium contamination levels among samples. The authors proposed that lysogenic phages may be the host’s detoxification resort, since the abundance of genes related to inorganic ion transport and reductase increased significantly in the conditions with high heavy metal pollution (Huang et al., 2021). Likewise, it was found in previous research that when self-transduced into the soil microbial genome, prophages (lysogenic phage in lysogenic cycling) produced by arsenic-resistant microorganisms can assist the host in coping with arsenic stress and the abundance of arsC and arsM in microcosm spiked with active lysogenic phage carrying arsC and arsM were significantly higher (by 122-fold and 575-fold) than control (no arsenic added), indicating that viruses are involved in the HGT of arsenic biotransformation genes and subsequent metal resistance gene replication in microorganisms. Moreover, the soil arsenic level was positively correlated with metal resistance gene propagation rate (Tang et al., 2019; Tang et al., 2021). Similar promotion effects on the virus-mediated transfer of stress resistance genes were observed in samples treated with heavy metals such as Zn, Cr, and Cu (Yang et al., 2021), nano-metal oxide particles (e.g., ZnO, TiO2, Al2O3) (Han et al., 2020; Otinov et al., 2020; Ding et al., 2021; Li and Zhang, 2022), or organic chemicals (Sun et al., 2018). The mechanisms of transduction induced by nanometal-particle may be that the reactive oxygen radicals (ROS) induced by metals has caused phospholipid peroxidation and increased the bacterial membrane permeability (Han et al., 2020; Xiao et al., 2021). This might explain the tripartite interaction among the viruses, their host microbe and heavy metals, in which nano-metal particles have aided viral infection by changing the physical properties of microbial cell barrier, while viruses in the other hand have endorsed metal resistance and prolong the life span of their hosts for better reproduction. Consistently, our results have shown that tellurium resistance proteins (e.g., TerD was present in 32.1% tested samples, Figure 1F) was among the most often detected metal resistance genes that viruses carried, while the causes of the enrichment remains unknown. Previous study has also found that terC and terD located on mobile genetic elements are closely related with bacteriophage infection and cellular response in addition to tellurium resistance (Whelan et al., 1997). Besides, experiment study has confirmed that recombinant plasmids carrying terCD-like genes can endow microbes with high tellurite resistance (Peng et al., 2022). Moreover, terC and terD genes are identified with versatile functions beyond tellurium resis tance, which might have explained their enrichment in virus-related sequences. For example, the terC homologue identified in the thylakoid membrane of Arabidopsis thaliana plays a important role in the formation of chloroplast, and terD homologue in S. coelicolor was suggested to take part in the differentiation, cell growth, and spores forming (Kwon and Cho, 2008; Sanssouci et al., 2011; Schneider et al., 2014).
Superoxide dismutase (SOD) and Glutaredoxin (Grx) with antioxidant activity are key players in regulating cellular oxidative stress, which might be crucial in preventing damage caused by metal-induced oxidative stress to the cellular machineries. These proteins are also encoded by viral sequences from the metagenomes of metal contaminated soils in the current study (see Supplementary Table S2 at https://doi.org/10.6084/m9.figshare.22226998.v3). SOD was biochemically characterized in Megavirus to reduce the cellular oxidative stress after the viral infection (Moniruzzaman et al., 2020). Likewise, it was demonstrated in previous studies that viral genes conferring to bacterial survival benefits (e.g., DNA repair proteins), or enhancement of fitness (e.g., carbohydrate degradation) were also enriched upon antibiotic stress (Becattini et al., 2016), indicating that viruses might be a key reservoir for environmental adaptation genes to be retrieved by bacterial hosts in times of need (Moniruzzaman et al., 2020). Additionally, we have discovered in the viral sequences genes encoding DNA repair proteins (e.g., DNA mismatch endonuclease Vsr), molecular chaperone (e.g., DnaJ, CspA), carbohydrate metabolism and cell wall remodeling (e.g., RfbC) and gene regulation proteins (e.g., luxR family) (see Supplementary Table S2 at https://doi.org/10.6084/m9.figshare.22226998.v3), which may also contribute to the response of microbial community to heavy metal stress. This is consistent with the consensus that microbial heavy metal stress response involves not only specialized metal transporters, but also various cellular processes associated with systems-level maintenance. For instance, viruses might regulate host cell density by encoding QS components (Wang Y. et al., 2022). Viral QS peptide Rapφ-Phrφsimilar to the bacterial homolog is widely used in phages and can be horizontal gene transferred (Bernard et al., 2021). It has also been reported that soil nutrient cycling can be promoted by stimulating enzyme activities of ß-glucosidase activity and alkaline phosphatase in metal contaminated soil to meet C, N, and P requirements for microbial activity (Duan et al., 2021). Soil viruses have an important role in depleting P from the soil ecosystem and the concentration of Pi in soil is correlated with progeny produced by viruses (Han et al., 2022).
Additionally, in previous studies a strong correlation between the relative abundance of metal resistance genes and lysogenic phages was identified, which is most likely due to the fact that most metal resistance genes are harbored by lysogenic phages (Huang et al., 2021), which is also reflected by our results that up to 11.4% detected viruses are lysogenic. This indicates a mutualism between phage and hosts in the context of lysogenic integration and rising metal pollution in soils. Bacteria lysogenized by viruses offer a steady environment for the housing viruses, in exchange for benefit from auxiliary genes provided by the viruses. Additionally, metal resistance genes found in the virome indicate viruses contributing to biogeochemical processes in polluted sites. This might stir up interest in virus-based strategies (such as virome transplantation) for microbiome control. Moreover, many of these mentioned virus-borne proteins have been experimentally proven to be highly expressible and industrially valuable (Schvarcz and Steward, 2018; Ha et al., 2021; Al-Shayeb et al., 2022; de Oliveira et al., 2022), which is also reflected by the results of our CAI analysis (Figure 1E). Lastly, it should be emphasized that experimental confirmations of the functioning and expression of metal-resistance-related genes located on soil-borne viral sequences recovered in the current study are required for a better understanding and future application of these genes.
4 Materials and methods
We first queried and retrieved all the items in the Genbank public database (Benson et al., 2018) with the keywords “soil” and “metal” within the “biosample” regions. We then chosen and downloaded from the Genbank public database the out-coming 58 soil metagenomes of microbiome as listed in Supplementary Table S1 at https://doi.org/10.6084/m9.figshare.22226998.v3 for downstream analyses. Metagenome reads were assembled with MEGAHIT version 1.1.1 with default k-mer options (Li et al., 2015). A combination of VIBRANT (Kieft et al., 2020) and DeepVirFinder (Ren et al., 2020) were applied for viral scaffold recovery and analyses under default parameters. CheckV v.1.0 was applied for quality assessment (Nayfach et al., 2021) and PhaGCN was applied for virus taxonomic assignment (Shang et al., 2021), which was followed by protein sequence clustering and analysis through software BPGA v.1.0 (Chaudhari et al., 2016) by default procedures, as well as functional annotation against the EggNOG database version 5.0 (Huerta-Cepas et al., 2019). Codon adaption index (CAI) was used as a numerical estimator of gene expression level (Hiraoka et al., 2009; Zhou et al., 2013), and correspondingly, the webserver CAIcal (Puigbò et al., 2008) (http://genomes.urv.es/CAIcal/calc.php) was applied to calculate respective CAI viral genes, as conducted previously (Li et al., 2019). We used the PosiGene pipeline (Sahm et al., 2017) for genome-wide detection of positively selected genes. Origin Pro 2020 software (OriginLab, Northampton, MA, USA) was used for data analysis and figure creation. The phylogeny of different kinds of abiotic resistance gene was constructed based on gene-translating protein sequences using PhyML program (Guindon et al., 2010) with the Maximum Likelihood (ML) method and 1,000 bootstrap replicates (with bootstrap values in red showing on respective branches) and visualized with iTOL (Letunic and Bork, 2021). Sequences were aligned with MUSCLE (Edgar, 2004) and trimmed with Gblocks (Talavera and Castresana, 2007) prior to tree construction. AlphaFold2 (Jumper et al., 2021) and RoseTTAFold (Baek et al., 2021) were applied to generate the full-length 3D structure of proteins encoded by virus in this study.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author contributions
HZ, JH, TL, and HY conceived and designed the research. WZ, QX, YZ, WK, and JZ analyzed the data. HZ and JH wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
The authors declare that this study received funding from Science and Technology of Hunan Branch of China National Tobacco Corporation. The funder was not involved in the study design, collection, analysis, interpretation of data, the writing of this article, or the decision to submit it for publication.
Conflict of interest
Authors WZ, XX, and YZ are employed by Tobacco Company of Hunan Province.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fenvs.2023.1182673/full#supplementary-material
References
Adriaenssens, E. M., Kramer, R., Van Goethem, M. W., Makhalanyane, T. P., Hogg, I., and Cowan, D. A. (2017). Environmental drivers of viral community composition in Antarctic soils identified by viromics. Microbiome 5, 83. doi:10.1186/s40168-017-0301-7
Al-Shayeb, B., Skopintsev, P., Soczek, K. M., Stahl, E. C., Li, Z., Groover, E., et al. (2022). Diverse virus-encoded CRISPR-Cas systems include streamlined genome editors. Cell 185, 4574–4586.e16. doi:10.1016/j.cell.2022.10.020
Ashelford, K. E., Day, M. J., and Fry, J. C. (2003). Elevated abundance of bacteriophage infecting bacteria in soil. Appl. Environ. Microbiol. 69, 285–289. doi:10.1128/AEM.69.1.285-289.2003
Attai, H., and Brown, P. J. (2019). Isolation and characterization T4-and T7-like phages that infect the bacterial plant pathogen Agrobacterium tumefaciens. Viruses 11 (6), 528–541. doi:10.3389/fmicb.2018.01861
Ayangbenro, A. S., and Babalola, O. O. (2017). A new strategy for heavy metal polluted environments: A review of microbial biosorbents. Int. J. Environ. Res. Public Health 14, 94. doi:10.3390/ijerph14010094
Baek, M., DiMaio, F., Anishchenko, I., Dauparas, J., Ovchinnikov, S., Lee, G. R., et al. (2021). Accurate prediction of protein structures and interactions using a three-track neural network. Science 373, 871–876. doi:10.1126/science.abj8754
Becattini, S., Taur, Y., and Pamer, E. G. (2016). Antibiotic-Induced changes in the intestinal microbiota and disease. Trends Mol. Med. 22, 458–478. doi:10.1016/j.molmed.2016.04.003
Benson, D. A., Cavanaugh, M., Clark, K., Karsch-Mizrachi, I., Ostell, J., Pruitt, K. D., et al. (2018). GenBank. Nucleic Acids Res. 46, D41–D47. doi:10.1093/nar/gkx1094
Bernard, C., Li, Y., Lopez, P., and Bapteste, E. (2021). Beyond arbitrium: Identification of a second communication system in Bacillus phage phi3T that may regulate host defense mechanisms. Isme J. 15, 545–549. doi:10.1038/s41396-020-00795-9
Breitbart, M., Bonnain, C., Malki, K., and Sawaya, N. A. (2018). Phage puppet masters of the marine microbial realm. Nat. Microbiol. 3, 754–766. doi:10.1038/s41564-018-0166-y
Brüssow, H., Canchaya, C., and Hardt, W. D. (2004). Phages and the evolution of bacterial pathogens: From genomic rearrangements to lysogenic conversion. Microbiol. Mol. Biol. Rev. 68, 560–602. doi:10.1128/MMBR.68.3.560-602.2004
Chasteen, T. G., Fuentes, D. E., Tantaleán, J. C., and Vásquez, C. C. (2009). Tellurite: History, oxidative stress, and molecular mechanisms of resistance. FEMS Microbiol. Rev. 33, 820–832. doi:10.1111/j.1574-6976.2009.00177.x
Chaudhari, N. M., Gupta, V. K., and Dutta, C. (2016). BPGA-an ultra-fast pan-genome analysis pipeline. Sci. Rep. 6, 24373. doi:10.1038/srep24373
Chen, J., and Xie, J. (2011). Role and regulation of bacterial LuxR-like regulators. J. Cell Biochem. 112, 2694–2702. doi:10.1002/jcb.23219
de Oliveira, E. G., Carvalho, J., Botelho, B. B., Da, C. F. C., Henriques, L. R., de Azevedo, B. L., et al. (2022). Giant viruses as a source of novel enzymes for biotechnological application. Pathogens 11, 1453. doi:10.3390/pathogens11121453
Ding, C., Jin, M., Ma, J., Chen, Z., Shen, Z., Yang, D., et al. (2021). Nano-Al(2)O(3) can mediate transduction-like transformation of antibiotic resistance genes in water. J. Hazard Mater 405, 124224. doi:10.1016/j.jhazmat.2020.124224
Duan, C., Mei, Y., Wang, Q., Wang, Y., Li, Q., Hong, M., et al. (2021). Rhizobium inoculation enhances the resistance of alfalfa and microbial characteristics in Copper-Contaminated soil. Front. Microbiol. 12, 781831. doi:10.3389/fmicb.2021.781831
Edgar, R. C. (2004). Muscle: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi:10.1093/nar/gkh340
Emerson, J. B., Roux, S., Brum, J. R., Bolduc, B., Woodcroft, B. J., Jang, H. B., et al. (2018). Host-linked soil viral ecology along a permafrost thaw gradient. Nat. Microbiol. 3, 870–880. doi:10.1038/s41564-018-0190-y
Escobar-Ledesma, F. R., Aragón-Tobar, C. F., Espinoza-Montero, P. J., and de la Torre-Chauvin, E. (2020). Increased recovery of gold thiosulfate alkaline solutions by adding thiol groups in the porous structure of activated carbon. Molecules 25, 2902. doi:10.3390/molecules25122902
Feiner, R., Argov, T., Rabinovich, L., Sigal, N., Borovok, I., and Herskovits, A. A. (2015). A new perspective on lysogeny: Prophages as active regulatory switches of bacteria. Nat. Rev. Microbiol. 13, 641–650. doi:10.1038/nrmicro3527
Gao, C., Liang, Y., Jiang, Y., Paez-Espino, D., Han, M., Gu, C., et al. (2022). Virioplankton assemblages from challenger deep, the deepest place in the oceans. IScience 25, 104680. doi:10.1016/j.isci.2022.104680
Gourdon, P., Liu, X., Skjørringe, T., Morth, J. P., Møller, L. B., Pedersen, B. P., et al. (2011). Crystal structure of a copper-transporting PIB-type ATPase. Nature 475, 59–64. doi:10.1038/nature10191
Guindon, S., Dufayard, J. F., Lefort, V., Anisimova, M., Hordijk, W., and Gascuel, O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321. doi:10.1093/sysbio/syq010
Ha, A. D., Moniruzzaman, M., and Aylward, F. O. (2021). High transcriptional activity and diverse functional repertoires of hundreds of giant viruses in a coastal marine system. MSystems 6, e0029321. doi:10.1128/mSystems.00293-21
Han, L. L., Yu, D. T., Bi, L., Du, S., Silveira, C., Cobián, G. A., et al. (2022). Distribution of soil viruses across China and their potential role in phosphorous metabolism. Environ. Microbiome 17, 6. doi:10.1186/s40793-022-00401-9
Han, X., Lv, P., Wang, L., Long, F., Ma, X., Liu, C., et al. (2020). Impact of nano-TiO2 on horizontal transfer of resistance genes mediated by filamentous phage transduction. Environ. Science-Nano 7, 1214–1224. doi:10.1039/c9en01279f
Harrison, J. P., Gheeraert, N., Tsigelnitskiy, D., and Cockell, C. S. (2013). The limits for life under multiple extremes. Trends Microbiol. 21, 204–212. doi:10.1016/j.tim.2013.01.006
Heidel, A. J., Kiefer, C., Coupland, G., and Rose, L. E. (2016). Pinpointing genes underlying annual/perennial transitions with comparative genomics. BMC Genomics 17, 921. doi:10.1186/s12864-016-3274-1
Hemminga, M. A., Vos, W. L., Nazarov, P. V., Koehorst, R. B., Wolfs, C. J., and Spruijt, R. B., et al. (2010). Viruses: Incredible nanomachines. New advances with filamentous phages. Eur. Biophys. J. 39, 541–550. doi:10.1007/s00249-009-0523-0
Hillary, L. S., Adriaenssens, E. M., Jones, D. L., and McDonald, J. E. (2022). RNA-viromics reveals diverse communities of soil RNA viruses with the potential to affect grassland ecosystems across multiple trophic levels. ISME Commun 2, 34. doi:10.1038/s43705-022-00110-x
Hiraoka, Y., Kawamata, K., Haraguchi, T., and Chikashige, Y. (2009). Codon usage bias is correlated with gene expression levels in the fission yeast Schizosaccharomyces pombe. Genes Cells 14, 499–509. doi:10.1111/j.1365-2443.2009.01284.x
Hu, C., Yu, C., Liu, Y., Hou, X., Liu, X., Hu, Y., et al. (2015). A hybrid mechanism for the synechocystis arsenate reductase revealed by structural snapshots during arsenate reduction. J. Biol. Chem. 290, 22262–22273. doi:10.1074/jbc.M115.659896
Huang, D., Yu, P., Ye, M., Schwarz, C., Jiang, X., and Alvarez, P. (2021). Enhanced mutualistic symbiosis between soil phages and bacteria with elevated chromium-induced environmental stress. Microbiome 9, 150. doi:10.1186/s40168-021-01074-1
Huerta-Cepas, J., Szklarczyk, D., Heller, D., Hernández-Plaza, A., Forslund, S. K., Cook, H.j, et al. (2019). EggNOG 5.0: A hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 47, D309–D314. doi:10.1093/nar/gky1085
Ipsen, J. Ø., Hernández-Rollán, C., Muderspach, S. J., Brander, S., Bertelsen, A. B., Jensen, P. E., et al. (2021). Copper binding and reactivity at the histidine brace motif: Insights from mutational analysis of the Pseudomonas fluorescens copper chaperone CopC. FEBS Lett. 595, 1708–1720. doi:10.1002/1873-3468.14092
Jin, M., Guo, X., Zhang, R., Qu, W., Gao, B., and Zeng, R. (2019). Diversities and potential biogeochemical impacts of mangrove soil viruses. Microbiome 7, 58. doi:10.1186/s40168-019-0675-9
Jones, J. B., Jackson, L. E., Balogh, B., Obradovic, A., Iriarte, F. B., and Momol, M. T. (2007). Bacteriophages for plant disease control. Annu. Rev. Phytopathol. 45, 245–262. doi:10.1146/annurev.phyto.45.062806.094411
Jumper, J., Evans, R., Pritzel, A., Green, T., Figurnov, M., Ronneberger, O., et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589. doi:10.1038/s41586-021-03819-2
Kieft, K., Zhou, Z., and Anantharaman, K. (2020). Vibrant: Automated recovery, annotation and curation of microbial viruses, and evaluation of viral community function from genomic sequences. Microbiome 8, 90. doi:10.1186/s40168-020-00867-0
Kołodziej, M., Łebkowski, T., Płociński, P., Hołówka, J., Paściak, M., Wojtaś, B., et al. (2021). Lsr2 and its novel paralogue mediate the adjustment of mycobacterium smegmatis to unfavorable environmental conditions. MSphere 6, 002900–e321. doi:10.1128/mSphere.00290-21
Kühbacher, U., and Duxin, J. P. (2020). How to fix DNA-protein crosslinks. DNA Repair (Amst) 94, 102924. doi:10.1016/j.dnarep.2020.102924
Kwon, K. C., and Cho, M. H. (2008). Deletion of the chloroplast-localized AtTerC gene product in Arabidopsis thaliana leads to loss of the thylakoid membrane and to seedling lethality. Plant J. 55, 428–442. doi:10.1111/j.1365-313X.2008.03523.x
Letunic, I., and Bork, P. (2021). Interactive Tree of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296. doi:10.1093/nar/gkab301
Li, D., Liu, C. M., Luo, R., Sadakane, K., and Lam, T. W. (2015). Megahit: An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31, 1674–1676. doi:10.1093/bioinformatics/btv033
Li, L., Liu, Z., Meng, D., Liu, X., Li, X., Zhang, M., et al. (2019). Comparative genomic analysis reveals the distribution, organization, and evolution of metal resistance genes in the genus acidithiobacillus. Appl. Environ. Microbiol. 85, 021533–e2218. doi:10.1128/AEM.02153-18
Li, W., and Zhang, G. (2022). Detection and various environmental factors of antibiotic resistance gene horizontal transfer. Environ. Res. 212, 113267. doi:10.1016/j.envres.2022.113267
López-Leal, G., Camelo-Valera, L. C., Hurtado-Ramírez, J. M., Verleyen, J., Castillo-Ramírez, S., and Reyes-Muñoz, A. (2022). Mining of thousands of prokaryotic genomes reveals high abundance of prophages with a strictly narrow host range. MSystems 7, e0032622. doi:10.1128/msystems.00326-22
Luo, X. Q., Wang, P., Li, J. L., Ahmad, M., Duan, L., Yin, L. Z., et al. (2022). Viral community-wide auxiliary metabolic genes differ by lifestyles, habitats, and hosts. Microbiome 10, 190. doi:10.1186/s40168-022-01384-y
Mojica, K. D., and Brussaard, C. P. (2014). Factors affecting virus dynamics and microbial host-virus interactions in marine environments. FEMS Microbiol. Ecol. 89, 495–515. doi:10.1111/1574-6941.12343
Moniruzzaman, M., Martinez-Gutierrez, C. A., Weinheimer, A. R., and Aylward, F. O. (2020). Dynamic genome evolution and complex virocell metabolism of globally-distributed giant viruses. Nat. Commun. 11, 1710. doi:10.1038/s41467-020-15507-2
Navarro, C. A., von Bernath, D., and Jerez, C. A. (2013). Heavy metal resistance strategies of acidophilic bacteria and their acquisition: Importance for biomining and bioremediation. Biol. Res. 46, 363–371. doi:10.4067/S0716-97602013000400008
Nayfach, S., Camargo, A. P., Schulz, F., Eloe-Fadrosh, E., Roux, S., and Kyrpides, N. C. (2021). CheckV assesses the quality and completeness of metagenome-assembled viral genomes. Nat. Biotechnol. 39, 578–585. doi:10.1038/s41587-020-00774-7
Nilsson, E., Li, K., Hoetzinger, M., and Holmfeldt, K. (2022). Nutrient driven transcriptional changes during phage infection in an aquatic Gammaproteobacterium. Environ. Microbiol. 24, 2270–2281. doi:10.1111/1462-2920.15904
Nolos, R. C., Agarin, C., Domino, M., Bonifacio, P. B., Chan, E. B., Mascareñas, D. R., et al. (2022). Health risks due to metal concentrations in soil and vegetables from the six municipalities of the island province in the Philippines. Int. J. Environ. Res. Public Health 19, 1587. doi:10.3390/ijerph19031587
Otinov, G. D., Lokteva, A. V., Petrova, A. D., Zinchenko, I. V., Isaeva, M. V., Kovtunov, E. A., et al. (2020). Positive and negative effects of metal oxide nanoparticles on antibiotic resistance genes transfer. Antibiot. (Basel) 9, 742. doi:10.3390/antibiotics9110742
Pal, C., Bengtssonpalme, J., Rensing, C., Kristiansson, E., and Larsson, D. G. J. (2014). BacMet: Antibacterial biocide and metal resistance genes database. Nucleic Acids Res. 42, 737–743. doi:10.1093/nar/gkt1252
Pan, Y. R., Lou, Y. C., Seven, A. B., Rizo, J., and Chen, C. (2011). NMR structure and calcium-binding properties of the tellurite resistance protein TerD from Klebsiella pneumoniae. J. Mol. Biol. 405, 1188–1201. doi:10.1016/j.jmb.2010.11.041
Peng, W., Wang, Y., Fu, Y., Deng, Z., Lin, S., and Liang, R. (2022). Characterization of the Tellurite-Resistance properties and identification of the core function genes for tellurite resistance in pseudomonas citronellolis SJTE-3. Microorganisms 10, 95. doi:10.3390/microorganisms10010095
Puigbò, P., Bravo, I. G., and Garcia-Vallve, S. (2008). CAIcal: A combined set of tools to assess codon usage adaptation. Biol. Direct 3, 38. doi:10.1186/1745-6150-3-38
Puxty, R. J., Millard, A. D., Evans, D. J., and Scanlan, D. J. (2015). Shedding new light on viral photosynthesis. Photosynth Res. 126, 71–97. doi:10.1007/s11120-014-0057-x
Rakonjac, J., Das, B., and Derda, R. (2016). Editorial: Filamentous Bacteriophage in Bio/Nano/Technology, Bacterial Pathogenesis and Ecology. Front. Microbiol. 7, 2109. doi:10.3389/fmicb.2016.02109
Ren, J., Song, K., Deng, C., Ahlgren, N. A., Fuhrman, J. A., Li, Y., et al. (2020). Identifying viruses from metagenomic data using deep learning. Quant. Biol. 8, 64–77. doi:10.1007/s40484-019-0187-4
Rogiers, T., Claesen, J., Van Gompel, A., Vanhoudt, N., Mysara, M., Williamson, A., et al. (2021). Soil microbial community structure and functionality changes in response to long-term metal and radionuclide pollution. Environ. Microbiol. 23, 1670–1683. doi:10.1111/1462-2920.15394
Ross, J., and Topp, E. (2015). Abundance of antibiotic resistance genes in bacteriophage following soil fertilization with dairy manure or municipal biosolids, and evidence for potential transduction. Appl. Environ. Microbiol. 81, 7905–7913. doi:10.1128/AEM.02363-15
Sahm, A., Bens, M., Platzer, M., and Szafranski, K. (2017). PosiGene: Automated and easy-to-use pipeline for genome-wide detection of positively selected genes. Nucleic Acids Res. 45, e100. doi:10.1093/nar/gkx179
Sanssouci, E., Lerat, S., Grondin, G., Shareck, F., and Beaulieu, C. (2011). Tdd8: A TerD domain-encoding gene involved in Streptomyces coelicolor differentiation. Ant. Van Leeuwenhoek 100, 385–398. doi:10.1007/s10482-011-9593-y
Santos-Medellin, C., Zinke, L. A., Ter Horst, A. M., Gelardi, D. L., Parikh, S. J., and Emerson, J. B. (2021). Viromes outperform total metagenomes in revealing the spatiotemporal patterns of agricultural soil viral communities. Isme J. 15, 1956–1970. doi:10.1038/s41396-021-00897-y
Schneider, A., Steinberger, I., Strissel, H., Kunz, H. H., Manavski, N., Meurer, J., et al. (2014). The Arabidopsis Tellurite resistance C protein together with ALB3 is involved in photosystem II protein synthesis. Plant J. 78, 344–356. doi:10.1111/tpj.12474
Schvarcz, C. R., and Steward, G. F. (2018). A giant virus infecting green algae encodes key fermentation genes. Virology 518, 423–433. doi:10.1016/j.virol.2018.03.010
Shang, J., Jiang, J., and Sun, Y. (2021). Bacteriophage classification for assembled contigs using graph convolutional network. Bioinformatics 37, i25–i33. doi:10.1093/bioinformatics/btab293
Silva, B., Coutinho, F. H., Gregoracci, G. B., Leomil, L., de Oliveira, L. S., Fróes, A., et al. (2017). Virioplankton assemblage structure in the lower river and ocean continuum of the amazon. MSphere 2, 003666–e417. doi:10.1128/mSphere.00366-17
Sire, J., Quérat, G., Esnault, C., and Priet, S. (2008). Uracil within DNA: An actor of antiviral immunity. Retrovirology 5, 45. doi:10.1186/1742-4690-5-45
Song, Y., Zhang, C., Ge, W., Zhang, Y., Burlingame, A. L., and Guo, Y. (2011). Identification of NaCl stress-responsive apoplastic proteins in rice shoot stems by 2D-DIGE. J. Proteomics 74, 1045–1067. doi:10.1016/j.jprot.2011.03.009
Starr, E. P., Nuccio, E. E., Pett-Ridge, J., Banfield, J. F., and Firestone, M. K. (2019). Metatranscriptomic reconstruction reveals RNA viruses with the potential to shape carbon cycling in soil. Proc. Natl. Acad. Sci. USA 116, 25900–25908. doi:10.1073/pnas.1908291116
Struck, A. W., Thompson, M. L., Wong, L. S., and Micklefield, J. (2012). S-adenosyl-methionine-dependent methyltransferases: Highly versatile enzymes in biocatalysis, biosynthesis and other biotechnological applications. Chembiochem 13, 2642–2655. doi:10.1002/cbic.201200556
Sun, M., Ye, M., Jiao, W., Feng, Y., Yu, P., Liu, M., et al. (2018). Changes in tetracycline partitioning and bacteria/phage-comediated ARGs in microplastic-contaminated greenhouse soil facilitated by sophorolipid. J. Hazard Mater 345, 131–139. doi:10.1016/j.jhazmat.2017.11.036
Sun, X., Xu, R., Dong, Y., Li, F., Tao, W., Kong, T., et al. (2020). Investigation of the ecological roles of putative keystone taxa during tailing revegetation. Environ. Sci. Technol. 54, 11258–11270. doi:10.1021/acs.est.0c03031
Taati, M. M., Amirmozafari, N., Shariati, A., Hallajzadeh, M., Mirkalantari, S., Khoshbayan, A., et al. (2020). How phages overcome the challenges of drug resistant bacteria in clinical infections. Infect. Drug Resist 13, 45–61. doi:10.2147/IDR.S234353
Talavera, G., and Castresana, J. (2007). Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 56, 564–577. doi:10.1080/10635150701472164
Tanaka, Y., Tsumoto, K., Nakanishi, T., Yasutake, Y., Sakai, N., Yao, M., et al. (2004). Structural implications for heavy metal-induced reversible assembly and aggregation of a protein: The case of Pyrococcus horikoshii CutA. FEBS Lett. 556, 167–174. doi:10.1016/s0014-5793(03)01402-9
Tang, X., Yu, P., Tang, L., Zhou, M., Fan, C., Lu, Y., et al. (2019). Bacteriophages from arsenic-resistant bacteria transduced resistance genes, which changed arsenic speciation and increased soil toxicity. Environ. Sci. Technol. Lett. 6, 675–680. doi:10.1021/acs.estlett.9b00600
Tang, X., Zhou, M., Fan, C., Zeng, G., Lu, Y., Dong, H., et al. (2021). The arsenic chemical species proportion and viral arsenic biotransformation genes composition affects lysogenic phage treatment under arsenic stress. Sci. Total Environ. 780, 146628. doi:10.1016/j.scitotenv.2021.146628
Thompson, L. R., Zeng, Q., Kelly, L., Huang, K. H., Singer, A. U., Stubbe, J., et al. (2011). Phage auxiliary metabolic genes and the redirection of cyanobacterial host carbon metabolism. Proc. Natl. Acad. Sci. U. S. A. 108, E757–E764. doi:10.1073/pnas.1102164108
Trubl, G., Jang, H. B., Roux, S., Emerson, J. B., Solonenko, N., Vik, D. R., et al. (2018). Soil viruses are underexplored players in ecosystem carbon processing. MSystems 3, 000766–e118. doi:10.1128/mSystems.00076-18
Turkovicova, L., Smidak, R., Jung, G., Turna, J., Lubec, G., and Aradska, J. (2016). Proteomic analysis of the TerC interactome: Novel links to tellurite resistance and pathogenicity. J. Proteomics 136, 167–173. doi:10.1016/j.jprot.2016.01.003
Wang, Q., Cai, L., Zhang, R., Wei, S., Li, F., Liu, Y., et al. (2022a). A unique set of auxiliary metabolic genes found in an isolated cyanophage sheds new light on marine Phage-Host interactions. Microbiol. Spectr. 10, e0236722. doi:10.1128/spectrum.02367-22
Wang, Y., Dai, J., Wang, X., Wang, Y., and Tang, F. (2022b). Mechanisms of interactions between bacteria and bacteriophage mediate by quorum sensing systems. Appl. Microbiol. Biotechnol. 106, 2299–2310. doi:10.1007/s00253-022-11866-6
Whelan, K. F., Sherburne, R. K., and Taylor, D. E. (1997). Characterization of a region of the IncHI2 plasmid R478 which protects Escherichia coli from toxic effects specified by components of the tellurite, phage, and colicin resistance cluster. J. Bacteriol. 179, 63–71. doi:10.1128/jb.179.1.63-71.1997
Williamson, K. E., Fuhrmann, J. J., Wommack, K. E., and Radosevich, M. (2017). Viruses in soil ecosystems: An unknown quantity within an unexplored territory. Annu. Rev. Virol. 4, 201–219. doi:10.1146/annurev-virology-101416-041639
Xiao, X., Ma, X., Han, X., Wu, L., Liu, C., and Yu, H. (2021). TiO2 photoexcitation promoted horizontal transfer of resistance genes mediated by phage transduction. Sci. Total Environ. 760, 144040. doi:10.1016/j.scitotenv.2020.144040
Xu, R., Li, B., Xiao, E., Young, L. Y., Sun, X., Kong, T., et al. (2020). Uncovering microbial responses to sharp geochemical gradients in a terrace contaminated by acid mine drainage. Environ. Pollut. 261, 114226. doi:10.1016/j.envpol.2020.114226
Yang, Y., Xing, S., Chen, Y., Wu, R., Wu, Y., Wang, Y., et al. (2021). Profiles of bacteria/phage-comediated ARGs in pig farm wastewater treatment plants in China: Association with mobile genetic elements, bacterial communities and environmental factors. J. Hazard Mater 404, 124149. doi:10.1016/j.jhazmat.2020.124149
Ye, X., Xiao, W., Zhang, Y., Zhao, S., Wang, G., Zhang, Q., et al. (2015). Assessment of heavy metal pollution in vegetables and relationships with soil heavy metal distribution in Zhejiang province, China. Environ. Monit. Assess. 187, 378. doi:10.1007/s10661-015-4604-5
Yu, Z., Lemongello, D., Segel, I. H., and Fisher, A. J. (2008). Crystal structure of Saccharomyces cerevisiae 3′-Phosphoadenosine-5′-phosphosulfate reductase complexed with adenosine 3′,5′-bisphosphate. Biochemistry 47, 12777–12786. doi:10.1021/bi801118f
Zannoni, D., Borsetti, F., Harrison, J. J., and Turner, R. J. (2008). The bacterial response to the chalcogen metalloids Se and Te. Adv. Microb. Physiol. 53, 1–72. doi:10.1016/S0065-2911(07)53001-8
Zeinert, R., Martinez, E., Schmitz, J., Senn, K., Usman, B., Anantharaman, V., et al. (2018). Structure-function analysis of manganese exporter proteins across bacteria. J. Biol. Chem. 293, 5715–5730. doi:10.1074/jbc.M117.790717
Zhang, Y., Guo, Y., Qiu, T., Gao, M., and Wang, X. (2022). Bacteriophages: Underestimated vehicles of antibiotic resistance genes in the soil. Front. Microbiol. 13, 936267. doi:10.3389/fmicb.2022.936267
Keywords: heavy metal resistance, virome, contaminated soil, detoxification, auxiliary metabolic gene, metagenome
Citation: Zhang H, Huang J, Zeng W, Xiao Q, Zhu Y, Kong W, Zou J, Liu T and Yin H (2023) Dissecting the metal resistance genes contributed by virome from mining-affected metal contaminated soils. Front. Environ. Sci. 11:1182673. doi: 10.3389/fenvs.2023.1182673
Received: 09 March 2023; Accepted: 17 April 2023;
Published: 23 May 2023.
Edited by:
Ling Xia, Wuhan University of Technology, ChinaReviewed by:
Zhendong Yang, Chengdu University, ChinaXiao Yunhua, Hunan Agricultural University, China
Copyright © 2023 Zhang, Huang, Zeng, Xiao, Zhu, Kong, Zou, Liu and Yin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tianbo Liu, 173298972@qq.com; Huaqun Yin, yinhuaqun_cs@sina.com
†These authors have contributed equally to this work
 Hetian Zhang
Hetian Zhang