- 1Department of Nephrology, Shengjing Hospital of China Medical University, Shenyang, China
- 2Department of Oncology, Shengjing Hospital of China Medical University, Shenyang, China
N6-methyladensine (m6A) has been identified as the best-characterized and the most abundant mRNA modification in eukaryotes. It can be dynamically regulated, removed, and recognized by its specific cellular components (respectively called “writers,” “erasers,” “readers”) and have become a hot research field in a variety of biological processes and diseases. Currently, the underlying molecular mechanisms of m6A epigenetic modification in diabetes mellitus (DM) and diabetic microvascular complications have not been extensively clarified. In this review, we focus on the effects and possible mechanisms of m6A as possible potential biomarkers and therapeutic targets in the treatment of DM and diabetic microvascular complications.
1 Introduction
Diabetes mellitus (DM) is an international health problem characterized by insulin resistance (IR) and insulin deficit (1). It has been estimated by the International Diabetes Federation that 537 million individuals worldwide are living with diabetes in 2021. By 2045, 784 million people will be affected by diabetes (2). DM can lead to macrovascular and microvascular complications. Macrovascular complications include coronary heart disease, strokes, and peripheral arterial disease. Microvascular complications include diabetic kidney disease (DKD), diabetic retinopathy (DR), and diabetic peripheral neuropathy (DPN) (3).
Methylation is an important modification of nucleic acids and proteins. It can regulate the expression and inhibition of genes and be involved in a variety of diseases, such as DM, cancer, aging, and so on (1, 4–6). RNA epigenetics modification has become a regulatory mechanism to coordinate cell transcriptome and proteome in different physiological processes. Similar to DNA methylation and histone modifications, RNA modifications can be dynamically regulated, removed, and recognized by its specific cellular components (respectively called “writers,” “eraser,” “readers”) and affect RNA splicing, stability, localization, translation, and transcription of mRNAs (7). RNA methylation includes N6-methyladensine (m6A), 5-methylcytosine, N1-methyladenosine, N7-methylguanosine, etc. (8). Among these modifications, m6A has been identified as the best-characterized and the most abundant mRNA modification in eukaryotes (9–11). And m6A methylation has become a hot research field in a variety of biological processes and diseases, such as aging, lipid metabolism, the development of hematopoietic system, central nervous system and reproductive system, obesity, cardiovascular diseases, cancers, renal diseases and et al. (7, 11–16). We find that five reviews on m6A and diabetes have been published (1, 17–20). However, there’re still some gaps that the existing reviews mainly focused on the relationship between m6A modification and DM and did not provide a detailed summary about the advancement of m6A and diabetic microvascular complications. Microvascular injury is very important for the prognosis of DM. In consequence, this review highlights the molecular mechanisms and potential therapeutic targets of m6A and diabetic microvascular complications.
2 m6A
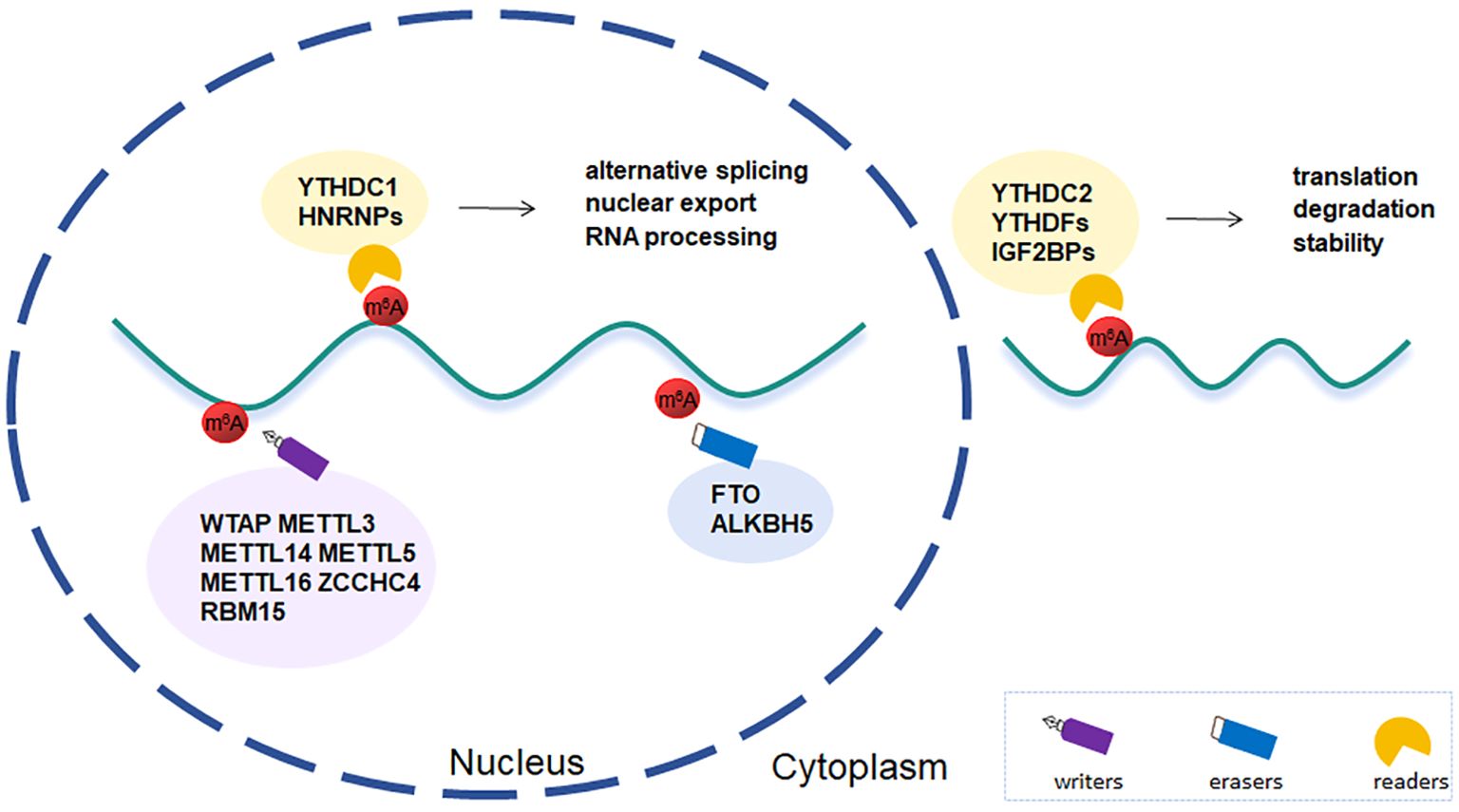
m6A, first discovered in Novikoff hepatic cancer cells (21), is an internal modification and highly clustered in near stop codons and in 3’UTRs of mRNAs (22). With the constant sequence RRACH (where R stands for A or G and H for A, C, or U), it is found in highly conserved sections and is dynamically regulated by particular methyltransferases and demethylases, which interacts to maintain RNA methylation homeostasis (10, 23) (Figure 1).
2.1 Writers
The m6A modification is post-transcriptionally installed by methyltransferase complex(MTC), which composes of METTL3, METTL14, Wilms’ tumor 1-associated protein (WTAP) (24). Either METTL3 or METTL14 alone exhibits fairly weak catalytic activity in vitro. However, the METTL3-METTL14 complex displayed significantly higher activity. Meanwhile, METTL14 can offer an RNA-binding scaffold to enhanced activity of METTL3 methylation (25). METTL3 and METTL14 are the core subunits of MTC and play a key role in different biological processes. In mouse embryonic brains, knockout of METTL3 and METTL14 can prolong the cell cycle of radial glial cells and extend cortical neurogenesis into the postnatal stage in a m6A-dependent manner (26). Besides, the METTL3-METTL14 heterodimer complex is closely related to the most of m6A sites in mRNA. More than 99% of the total m6A was lost in mouse embryonic stem cells upon genomic deletion of METTL3 or CRISPR-mediated silencing of METTL14 (27). WTAP is the third subunit of MTC. Although WTAP has no catalytic activity against RNA targets, it can facilitate the accumulation of METTL3-METTL14 heterodimer complex in nuclear speckles (28). WTAP depletion led to a marked decrease of m6A levels in mRNA and the buildup of both METTL3 and METTL14 in nuclear speckles (29). Besides, WTAP is involved in regulating transcription and alternative splicing of mRNA (29, 30). Therefore, WTAP, as a regulatory subunit, may play a key role in RNA epigenetic modification.
Apart from MTC, other methyltransferases include METTL5, METTL16, Zinc finger CCHC-type containing 4 (ZCCHC4), RNA-binding motif protein 15 (RBM15), and others. The miCLIP analysis confirmed that METTL5 and ZCCHC4 are highly specific methyltransferases which can respectively install 18S rRNA and 28S rRNA. TRMT112 is indispensable to stability and activation of METTL5 in order to achieve metabolic capacity in cells (31). Recent studies demonstrated the crucial function of METTL5-mediated 18S rRNA m6A modification in regulation of tumor development and immune microenvironment (32, 33). It has been identified METTL16 is a conserved eukaryotic methyltransferase. MAT2A transcript encoding SAM synthetase and U6 snRNA are the two methylation targets. METTL16 can bind to mRNA MAT2A 3’UTR hairpins, thereby affecting the splicing and stability of MAT2A pre-mRNA and regulating SAM homeostasis (34). Another methyltransferase, RBM15, belongs to the split ends protein family. The long non-coding RNA X-inactive specific transcript (XIST)-mediated gene silencing requires RBM15 and its paralogue-mediate dadenosine methylation. On the contrary, knockdown of both RBM15 and RBM15b blocked XIST-mediated gene silencing (35). So RBM15 is essential for XIST-mediated X chromosome inactivation. In addition, by interacting with intron-binding splicing factor, SF3B1, RBM15 regulates alternative splicing and megakaryocyte differentiation (36).
2.2 Erasers
m6A demethylases–the fat mass and obesity-associated protein (FTO) and AlkB homolog 5 (ALKBH5), can directly reverse adenosine methylation, so called erasers. Both belong to the AlkB family dioxygenases (37, 38). FTO distributes in the nucleus and cytoplasm and has different substrates. FTO can mediate nuclear m6A and cytoplasmic m6Am and m6A in mRNA, m6A in U6RNA, m6Am in snRNA and m1A in tRNA (39). Early genome-wide related studies have demonstrated the impact of FTO on human obesity and homeostasis (40, 41). Overexpression of FTO in mice led to increased food consumption and obesity whereas inactivation of FTO resulted in significant weight loss and growth retardation (41, 42). Recent studies have shown that FTO is involved in the occurrence and development of various biological processes, such as neuropsychiatric disorders and tumorigenesis and development, etc. (43, 44). Similar to METTL3, ALKBH5 collocates with nuclear speckles and affects mRNA processing, and eventually has an impact on mRNA export and RNA metabolism. ALKBH5 is highly specific for the demethylation of m6A mRNA and no other substrates have been found (38).
2.3 Readers
The reader’s ability to recognize is a determining factor in how the m6A modification affects targeted RNA metabolism. Currently, the most well characterized readers in eukaryotes include YT521-B homology (YTH) domain family, heterogeneous nuclear ribonucleoproteins (HNRNPs), and insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs). These readers alter RNA function by attaching either directly or indirectly to the m6A motifs (45). YTH domain family consists of YTH domain family protein 1-3 (YTHDF1-3) and YTH domain containing protein 1-2 (YTHDC1-2). They process similar and highly conserved YTH domain structure, which is mainly composed of four α-helices and four β-strands (46, 47). YTHDC1 is the only member of the YTH family localizing in the nucleus. The N- or C-terminal sequence of YTHDC1 interacts with hypo- or de-phosphorylated RS domain of SRSF3, which mediates alternative splicing and mRNA export (48–50). YTHDC2 binds U-rich motifs in 3’UTRs of RNAs using a DExD box helicase domain and interacts with the 5’ to 3’ exoribonuclease XRN1, thus promoting translation and degradation of mRNAs (51, 52). There are different views on the effect of YTHDF proteins on m6A mRNA. Prevailing canonical model think YTHDF1 enhances mRNA translation, YTHDF2 promotes mRNA degradation, and YTHDF3 has both functions (53–55). A unified model proposed by Zaccara et al. demonstrates that YTHDF proteins are closely related to the degradation of m6A-RNAs but not the translation (56). Besides, some evidence pointed the function of YTHDF proteins depends on the context in which they are located (57). YTHDF1 can interact with argonaute 2 and contribute to P-body (a membrane-free organelle involved on post-transcriptional regulation of mRNAs) formation, finally promoting the degradation of the target mRNAs (58). The interaction of YTHDF3 with eukaryotic translation initiation factor 2 alpha kinase 2 facilitates translational processes in oxaliplatin-resistant colorectal cancer (59). HNRNPs include HNRNPAB, HNRNPC, HNRNPG, the most abundant protein of which is HNRNPAB. HNRNPs do not directly bind to m6A, but through a mechanism called “m6A switch”. That is, m6A-dependent RNA structural remodeling can regulates RNA-HNRNPs interactions, thus influencing nuclear events such as gene expression, maturation and processing (60–62). Each of HNRNPs processes high- or low-affinity nucleic acid binding sites that can bind a variety of RNA and DNA sequences (63). For instance, HNRNP A2/B1 contains two RNA recognition motif (RRM), RRM1 and RRM2, which can respectively recognize the AGGG motif and UAG motif (60). Besides, it can regulate alternative splicing and promote processing of pri-miRNAs via interacting with DGCR8 protein in a METTL3-dependent manner (64). HNRNPC is involved in the regulation of premRNA splicing and is crucial for the development of tumors (65, 66). HNRNPG contains the extensive low-complexity regions, N-terminal ~300 amino acids and Cterminal ~58 amino acids (62). HNRNPG can bind a purine-rich motif and indirectly recognize the N6-methyl group through a low-complexity region. Besides, Using an RGG region in the low-complexity region, HNRNPG regulates alternative splicing by interacting with phosphorylated C-terminal domain of RNA polymerase II and m6A-modified nascent pre-mRNA (67). IGF2BPs and YTHDF2 impose an opposite role in m6A function. YTHDF2 contributes to RNA degradation (53, 56). whereas IGF2BPs can regulate stability and translation of target RNAs (68). Besides, they recognize different targets and share only a small number of binding sites (68).
Chemical labeling and sequencing of m6A is crucial for studying the function of m6A. The chemical inertness of m6A makes it difficult to label directly. The most commonly used high-throughput sequencing technique is methylated RNA immunoprecipitation sequencing (MeRIP-Seq) depending on m6A antibody, which only provides 100-200 nucleotide resolution. Based on MeRIP-seq, several strategies, including miCLIP, PA-m6A-seq and tMeRIP-seq, improve resolution but cannot quantify m6A. And there are some antibody-independent strategies which have the advantage of single-base resolution, such as MAZTER-seq and m6A-REF-seq. In addition, several novel chemical labeling methods for m6A have emerged. m6A-SEAL, a FTO-assisted m6A selective chemical labelling method, can specifically enrich m6A, but cannot achieve single-base resolution and quantify m6A. Compared to m6A-SEAL, NOseq and m6A-label-seq typify single-base resolution feature. On the downside, NOseq Lacks of specificity and sensitivity and cannot distinguish m6A and m6Am, while m6A-label-seq can only be applied to cellular systems and requires the metabolism of Se-allyl-L-selenohomocysteine (69). Current chemical labelling strategies still have much room for improvement. It is essential and urgent to develop a strategy to achieve single-base resolution and specifically enrich m6A independent of antibodies.
3 m6A and DM
Epigenetics of β-cell include DNA methylation, histone modification, chromatin remodeling and accessibility, mRNA and non-coding RNAs (ncRNAs) modification, etc. (70). It can impact β-cell function and adaptation, and be involved in regulating glycometabolism and insulin secretion (71). m6A is the most studied RNA modification and closely related to regulation of islet β-cell function and the progression of DM. Studies have shown that m6A content in RNA was differentially expressed in different tissues. It was reduced in the peripheral blood of type 2 diabetes (T2D) patients compared with healthy controls (72, 73) and elevated in the livers of high fat diet (HFD) mice (74, 75).
3.1 The writers in DM
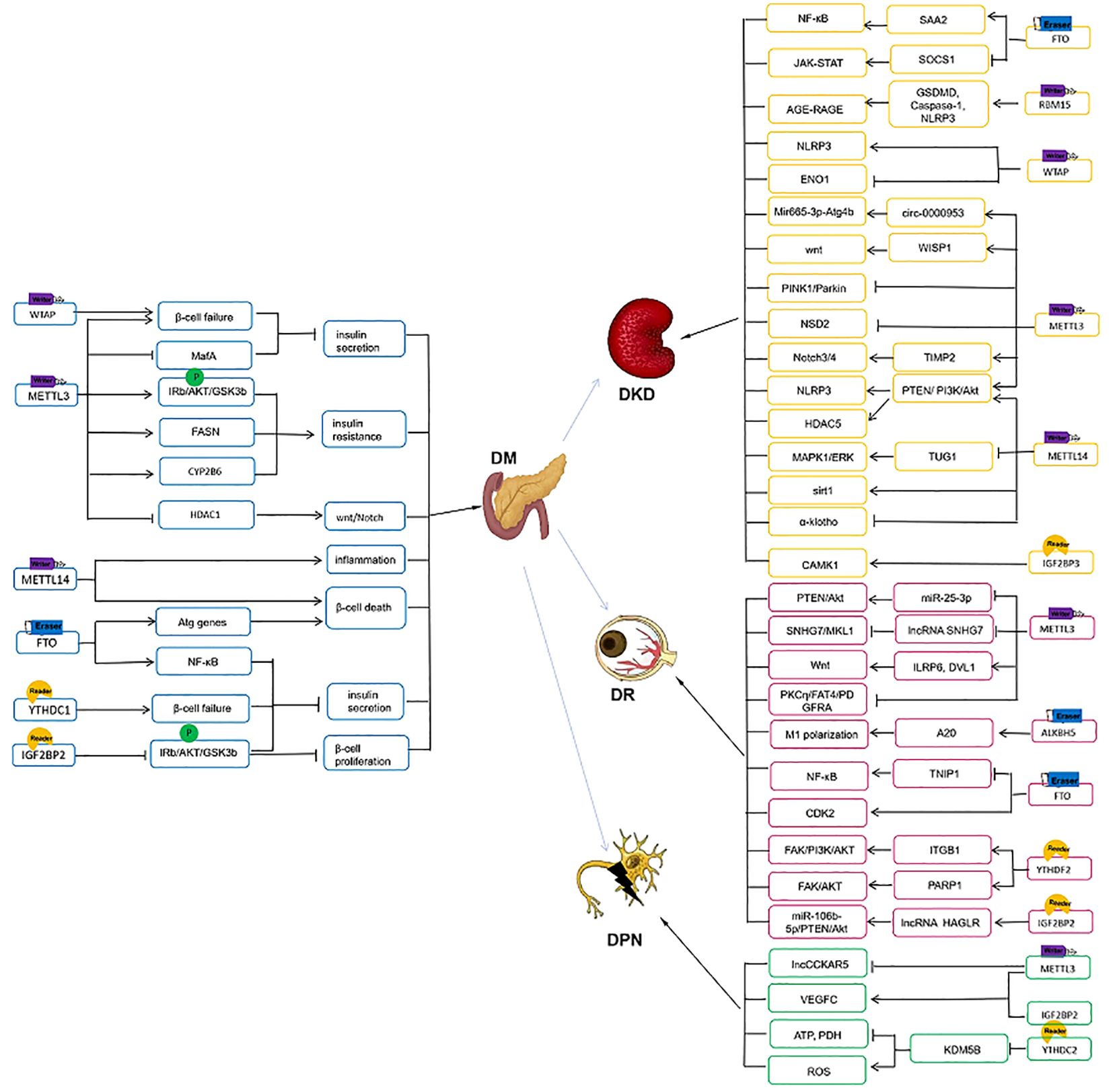
mRNA m6A methylation plays a major role in the pathogenesis of T2D. METTL3 and METTL14 protein levels were downregulated in whole islets from patients with T2D (76, 77). Knowdown of METTL3 and METTL14 in EndoC-βH1 cells inhibited the insulin/IGF1–AKT–PDX1 signaling and led to the cell cycle arrest and impaired insulin secretion in β-cells (76). Methylglyoxal (MG), as a precursor of advanced glycation end products, is significantly increased in patients with newly diagnosed T2D (78). MG-induced downregulation of METTL3 expression promoted decrease in m6A levels in β cells. Besides, METTTL3 plays a protected effect on insulin secretion of β-cell with the evidence that silencing of METTL3 significantly reduced glucose-stimulated insulin secretion (GSIS) through regulating musculoaponeurotic fibrosarcoma oncogene family A (MafA), whereas this process could be reversed by upregulation of METTL3 (79). Li et al. reported similar results that islet β-cell-specific deletion of METTL3 induced β-cell failure, decreased insulin secretion and hyperglycemia (80). Meanwhile, in Pdx1+ pancreatic progenitor cells, absence of METTL3 could inhibit Hdac1 expression and further activate wnt/β-catenin and Notch/Hes1 pathways, leading to hyperglycemia and hypoinsulinemia, along with an atrophic pancreas, reduced islet mass, and abnormal increase in ductal formation (81). Remarkably, METTL3 levels increase significantly in β-cells at the onset of type 1 diabetes but quickly decrease with disease progression. METTL3 silencing enhanced the level of 2′-5′-oligoadenylate synthetase (OAS, an innate immune mediators) by increasing its mRNA stability. Hence m6A methylation regulates the OAS innate immune response as a β-cell protective mechanism. In β-cell METTL14 knockout mouse lines, glucose intolerance, decreased insulin secretion and lower body weight could be observed (82, 83). RNA sequencing showed METTL14 deficiency led to the upregulation of genes related to β cell death and inflammatory response (82). The loss of METTL3 and METTL14 suppressed the expression of critical β cell transcription factors Pdx1, MafA, and Nkx6.1 as well as mature β-cell markers Ucn3 and GLUT2 (77). These studies indicate METTL3 and METTL14 are essential for maintaining β cell function and maturation. WTAP, another m6A writer, has a similar effect on modulating β cell function. WTAP was downregulated in islet β cells of T2D patients due to lipotoxicity and chronic inflammation. WTAP-betaKO mice displayed severe glucose intolerance and reduction in pancreatic insulin content. So WTAP deletion leads to β cell failure and diabetes (84).
METTL3 is also a key factor in regulating IR. In the liver tissues from patients with T2D and HFD mice, the level of m6A and METTL3 was consistently elevated (74, 75). FASN is a metabolism-related protein and its m6A modification is involved with the development of IR and T2D (85). METTL3 deletion in HepG2 cells and primary hepatocytes dramatically reduced the phosphorylation of IRb, AKT, and GSK3b and the expression of FASN, thereby improving glucose homeostasis and insulin sensitivity (74). Another study showed METTL3 overexpression brought about liver metabolic disorders and IR. On the contrary, METTL3 ablation plays a protective role through increasing the stability of key genes involved in hepatic lipid and glucose metabolism (75). IR is also one of the key immunopathogenesis of nonalcoholic fatty liver disease (NAFLD) (86). Li et al. used a NAFLD model to investigate the biological function of METTL3-mediated m6A methylation in IR. The overexpression of METTL3-mediated CYP2B6 suppressed phosphorylation of the insulin receptor substrate, finally leading to hepatic IR (87).
3.2 The erasers in DM
RNA sequencing showed significant associations of variants in FTO and T2D and diabetic nephropathy (DN) (88, 89). Setum FTO level was significantly downregulated in T2D patients and negatively correlated with m6A levels (72, 73, 90). However, The expression of FTO in islets and the interaction between FTO expression and insulin secretion is controversial. Taneera et al. found FTO expression was lower in T2D islets than in non-diabetic islets from cadaver donors. And in glucose-responsive insulin-secreting C-peptide modified human proinsulin (GRINCH) cells and INS-1 cells, silencing of FTO expression led to a reduction in insulin secretion (91, 92). Mechanistically, FTO silencing led to a significant decrease in β-cell functional genes, which compromises pancreatic β-cell function. Meanwhile, the dysregulation of FTO expression leads to impaired mitochondrial function and reduced ATP production, possibly contributing to the pathogenesis of T2D (92). The findings of Fan were at odds with those of Taneera et al., observing that the expression of FTO was high in mouse MIN6 cells. And FTO overexpression significantly inhibited insulin secretion and targeted activating NF-κB pathway via reactive oxygen species (ROS) generation, whereas FTO silence had no effect on insulin secretion (93). This difference may be due to different approach of FTO expression (FTO silence by siRNA, vs. overexpression by lentivirus) and different derivation of in vitro models (GRINCH cells were obtained from a clonal rat, while MIN6 cells were derived from murine). Wu et al. revealed autophagy overload could trigger β cell apoptosis and decrease insulin secretion. In glucolipotoxic stress conditions, enhanced-NR3C1 significantly upregulated FTO expression in β-cells and further diminished m6A modifications on autophagy related genes(Atg12, Atg5, Atg9a, Atg16l2), which induced hyperactive autophagy and β-cell failure (94). And it is observed that (–)-epigallocatechin 3-gallate, the most predominately active catechin in green tea, promoted FTO degradation and prevented the NR3C1 enhancement-induced oxidative stress, thereby exerting a protective effect on glucose tolerance and β-cell function in β-cell-specific NR3C1-overexpressing mice (95). Therefore, targeting FTO provides new insights into the treatment of diabetes.
Although FTO gene has been implicated in the regulation of β-cell function and insulin secretion, the precise mechanism not fully clarified yet. Additional investigations are required to comprehend the regulation of FTO expression and its potential interactions with other transcription factors influencing β-cell survival, metabolism, and function.
3.3 The readers in DM
It has been demonstrated that m6A reader proteins are crucial in regulating β cell activity and glucose metabolism. In pancreatic β cells from T2D patients, Li et al. found a substantial drop in YTHDC1, which is linked to lipotoxicity and chronic inflammation. In β-cell specific YTHDC1 knockout mice, GSIS was reduced and serum glucagon levels were increased dramatically (96). Similarly, another study showed the expression of m6A and YTHDC1 was downregulated in white blood cells from T2D patients. Ablation of YTHDC1 in β-cells of adult mice exhibits a significant decrease in insulin synthesis and secretion, as well as glucose intolerance. On a molecular level, multiple genes correlated with β-cell maturity, such as MafA, Gck and Glut2, were decreased, indicating that β-cell maturity is impacted by YTHDC1 loss (97).
A cluster of single nucleotide polymorphisms in the second intron of IGF2BP2 found by genome-wide association studies (GWAS) are the susceptibility gene regions of T2D and closely associated with development of T2D/glucose metabolism (98–102). As demonstrated by Regué et al. IGF2BP2 is strongly expressed in pancreatic β cells, which stimulates insulin production through the upregulation of the AKT-GSK3b-PDX1 pathway (103). PDX1 is a critical transcription regulator for the development and maturation of β cell (104). IMP2 deficiency led to a decrease in Pdx1, in turn affecting β-cell proliferation and function (103). Taken together, IGF2BP2 is a human T2D-associated gene. Targeting IGF2BP2 is a promising avenue to improve β-cell function and the development of T2D (Figure 2).
ncRNAs are crucial regulatory RNA, including microRNAs (miRNAs), long noncoding RNAs (lncRNAs) and circular RNAs (circRNAs) and exert a potential role in the occurrence and development of DM and its complications (105, 106). A variety of ncRNAs regulate pancreatic β cell survival and insulin secretion (105). There are few studies associated to the effect of m6A on ncRNAs in diabetes. A study showed LncRNA XIST was upregulated in the peripheral blood of gestational DM patients and HG-cultured HTR8/SVneo cells, and METTL14 facilitated proliferation and migration and inhibited cell apoptosis and cell cycle arrest by impeding XIST expression (107).
Taken together, these data indicate that m6A and its downstream pathways are important regulatory mechanisms in the occurrence and development of diabetes. There are still unresolved issues in this field. In the current investigations, hyperglycemia and hyperlipemia are the most widely used stimulation conditions in experimental model. The majority of research focus on how one enzyme contributes to the pathophysiology of DM. However, DM is a complex and heterogeneous disorder that can be caused by several different factors, such as autoimmune, genetics, environment, lifestyle, etc. (108). There may exist differences in m6A modification network under the single condition and the complex pathogenesis of DM. Exploring the pathogenesis of m6A in DM under different backgrounds is conducive to elucidate the pathophysiological mechanisms of different diabetes subtypes, so as to provide precise and individualized management strategies for patients in the future.
4 m6A and microvascular complications of DM
4.1 m6A and DKD
DKD is associated with an immune cell-mediated inflammatory response. One study has proved that m6A-modified lncRNA could mediated the expression and inflammatory response of macrophages in patients with DN (109). DN has multiple morphological changes, including thickened glomerular basement membrane, mesangial expansion, podocyte injury, tubulointerstitial fibrosis, epithelial-to-mesenchymal transition (EMT), etc. (110). EMT is thought to be a key factor in renal fibrosis (111). Some researchers have confirmed m6A epigenetics has an important impact on the development of DN through a variety of mechanisms.
4.1.1 The writers in DKD
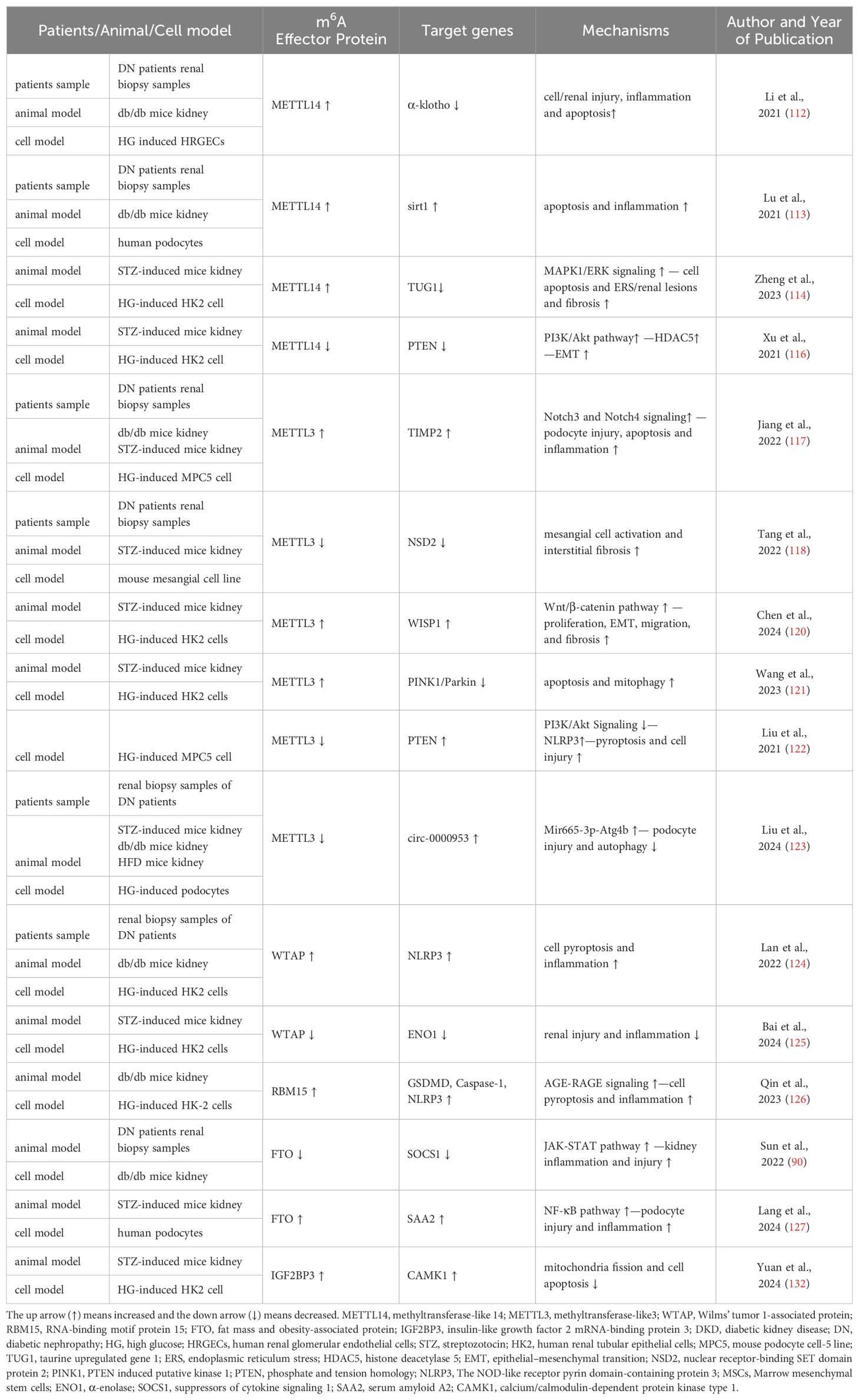
METTL3/METTL4/WTAP complex, as writers of m6A, acted as a regulator in the pathogenesis of DKD. METTL14 expression and m6A RNA levels were upregulated in DKD model. METTL14 could promote cell apoptosis and inflammation and aggravated renal injury of DN through three mechanisms (112–114). First, Overexpression of METTL14 increased inflammatory factors levels and apoptosis in human renal glomerular endothelial cells via downregulating m6A modification of α−klotho (112). Second, METTL14-mediated RNA m6A modification inhibited autophagy and increased apoptosis and inflammation in podocytes and db/db mice through promoting Sirt1 mRNA m6A modification and degradation (113). Third, endoplasmic reticulum stress (ERS) can lead to cell apoptosis and is a vital pathogenic mediator of DN (115). METTL14 regulated the m6A modification of TUG1 and activated the MAPK1/ERK signaling, which aggravated high glucose (HG)-induced renal tubular epithelial cell apoptosis and ERS (114). According to another study, FTO, METTL3 and METTL14 mRNA were shown to be considerably lower in HK2 cells treated with HG as opposed to normal glucose, whereas only METTL14 overexpression could inhibit the expression of EMT-related proteins, such as TGF-β1 and α-SMA, as well as HDAC5 by regulating Akt pathway (116). Similar to METTL14, METTL3 were also involved in the pathogenesis of DN through several pathways. First, upregulation of METTL3 promoted podocytes apoptosis and inflammation factors levels. And TIMP2-mediated Notch signaling pathway was the downstream target of METTL3 in DN (117). Second, nuclear receptor-binding SET domain protein 2 (NSD2), a SET histone methyltransferase family member, was down-regulated in T2D and promoted the proliferation of pancreatic β cell lines and the release of insulin (118). In DN, METTL3 promotes NSD2 expression to lessen mesangial cell activation and interstitial fibrosis under the HG treatment (119). Third, METTL3 silencing could suppress the proliferation, EMT, migration, and fibrosis of HG-treated HK2 cells through mediate m6A modification of WISP1 mRNA, thus alleviating renal injury of DN (120). Fourth, METTL3 could induce apoptosis and mitophagy of renal tubular epithelial cells through modulating the PINK1/Parkin signaling pathway in an YTHDF2-dependent manner, whereas METTL3 knockdown inhibited the progression of DKD (121). Besides, Liu et al. has showed a renoprotective effect of the total flavones of Abelmoschus manihot (TFA) on DN. Mechanistically, TFA could ameliorate pyroptosis and podocytes injury in HG circumstances by downregulating METTL3-dependent m6A modification and activating NLRP3-inflammasome and PTEN/PI3K/Akt signaling (122). Another study revealed that silencing of METTL3 suppressed the degradation of circ-0000953 in HG-stimulated podocytes. And the overexpression of circ-0,000,953 ameliorated podocyte injury and autophagy disorder by targeting Mir665-3p-Atg4b (123). It has been proved WTAP is highly expressed in DN patients and in HG−induced HK−2 cells. WTAP promoted cell pyroptosis and inflammation by targeting NLRP3 in DN. Otherwise, WTAP silencing could inhibit DN progression (124). Bai et al. confirmed marrow mesenchymal stem cells (MSCs) administration could alleviate HG-induced HK-2 cells injury and renal injury in DN mice. Mechanistically, MSCs could repress WTAP expression via inactivating Smad2/3 and thus alleviate the development of DN (125). So targeting m6A through the writer is a prospective therapy strategy for DN.
RBM15 is also a member of the m6A methyltransferases. Qin et al. proved the expression of METTL16 and RBM15 was elevated in the model group of DN mice. In HG-induced HK-2 cells, cell viability was suppressed and the expression of inflammatory factors and pyroptosis-related proteins were elevated, which could be reversed by RBM15 silence. AGE-RAGE signaling pathway activated by RBM15 participated in the pathogenesis of DN (126).
4.1.2 The erasers in DKD
The biological role of FTO-mediated m6A modification in DKD are controversary. serum FTO level was decreased in DKD patients (72, 90), whereas the expression of FTO was increased in high glucose-induced podocytes, and FTO upregulation enhanced serum amyloid A2 mRNA stability by regulating the NF-κB pathway, thus participating in podocyte injury and the progression of DKD (127). Another study showed FTO have a protective effect on DKD pathogenesis with evidence that FTO overexpression significantly attenuates kidney injuries and inflammation of DKD via inhibiting SOCS1/JAK/STAT axis (90).
4.1.3 The readers in DKD
Podocyte is an important component of GBM. Podocyte loss and foot process effacement contribute to the development of DKD (128). Insulin-like growth factor-2 is identified to be produced by the glomerular podocyte and is important for maintaining podocyte survival and glomerular function (129). Previous study showed that calcium/calmodulin-dependent protein kinases (CAMK), belongs to CAMKs Ser/Thr protein kinase family, play an important role in maintaining mitochondrial homeostasis and regulating inflammation and oxidative response (130, 131). IGF2BP3 promoted the stability of CAMK1 mRNA by m6A modification and further alleviated DN progression via inhibiting mitochondria fission and cell apoptosis (132). Lin et al. found circUBXN7 was significantly upregulated in DKD plasma. And upregulated circUBXN7 enhanced the binding of IGF2BP2 and SP1 mRNA, which promoted macrophage infiltration, tubular EMT and fibrosis and accelerated the progression of DKD (133) (Figure 2, Table 1).
Current researches have showed that m6A methylation is involved in the pathogenesis of DKD through regulating cell injury, inflammation, apoptosis, EMT, interstitial fibrosis and etc., which holds promising implications for its diagnosis and treatment. We found that the same effector protein was differentially expressed in DKD (See Table 1) This may be caused by the heterogeneity of different cell or animal models and stimulation conditions, etc. The targeting of m6A methylation and effector protein is a promising regulatory mechanism, which will facilitate the advancement of future therapies for DKD, delay the progression of DKD to end-stage renal disease and enhance the overall prognosis of DKD.
4.2 m6A and DR
DR, a major ocular complication of diabetes, is one of the main causes of visual loss and blindness and accounts for about 30% to 40% of all diabetes cases (134).
4.2.1 The writers in DR
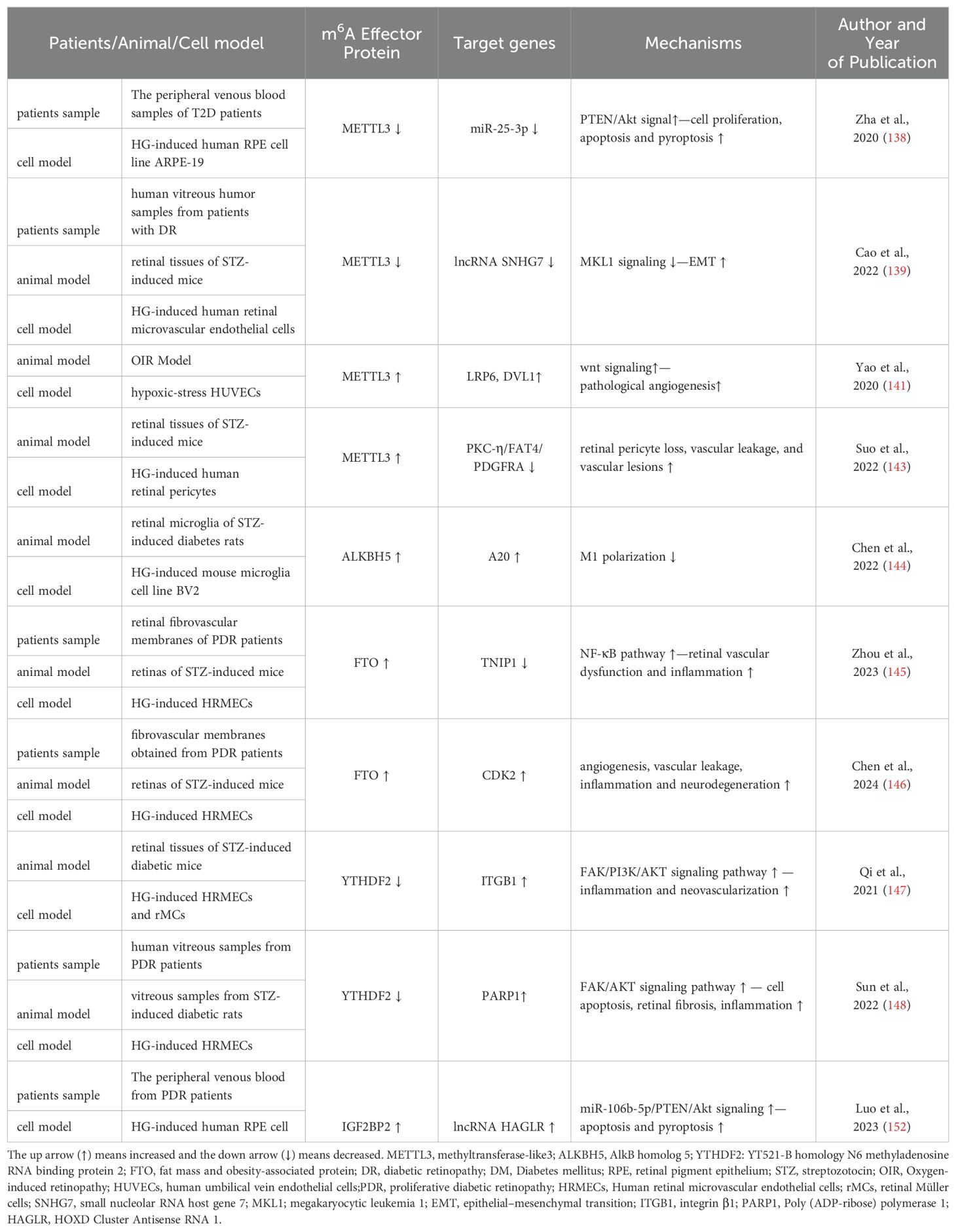
Endothelial dysfunction and EMT are the prominent factors in the pathogenesis of DR (135, 136). Retinal pigment epithelium (RPE) cells are essential for the development and maintenance of adjacent photoreceptors in the vertebrate retina and frequently utilized in vitro cellular models in DR research (137). METTL3 expression levels were lowered in RPE cells treated with HG in a time-dependent manner. Mechanistically, overexpression of METTL3 in RPE cells attenuated HG-induced cell proliferation, apoptosis, and pyroptosis by regulating miR-25-3p/PTEN/Akt signal pathway (138). Similarly, another study revealed METTL3 expression was downregulated in DR patients, mice and human retinal microvascular endothelial cells. METTL3 overexpression could suppress EMT-related molecules levels via the SNHG7/MKL1 signaling pathway (139). Therefore, METTL3 play a protective role on endothelial dysfunction and EMT.
Oxidative stress is a key event that contributes to DR pathogenesis (140). Under the hypoxic-stress condition, m6A methylation and METTL3 in endothelial cells and mouse retinas were upregulated, which contributed to the progression of pathological angiogenesis by regulating wnt signaling activation (a significant increase in LRP6 and DVL1 levels) in a YTHDF1-dependent manner. Conversely, METTL3 silencing suppresses pathological angiogenesis (141). Adequate pericyte attachment was critical for maintenance of blood-retinal barrier (BRB) integrity and maturation. Pericyte dropout impaired BRB, eventually leading to blindness, which is involved in DR pathogenesis and accelerates DR progression (142). Suo et al. reported that m6A modification level and METTL3 were increased in retinal pericyte dysfunction under HG condition and retinal vessels of diabetic mice. Pericyte-specific METTL3 deletion resulted in a high expression of PKC-η, FAT4, and PDGFRA via YTHDF2-dependent pathway, which could minimize pericyte apoptosis via impacting their proliferation, viability, and differentiation, and alleviate retinal vascular leakage (143).
4.2.2 The erasers in DR
Retinal microglia’s M1 polarization was enhanced while M2 polarization was suppressed by HG. A20, anti-inflammatory molecule, was negatively correlated with M1 polarization. Besides, inhibiting ALKBH5 in microglia led to higher m6A modification level, which decreased A20 expression and further enhanced M1 polarization of retinal microglia of DR. Therefore, targeting A20 is a promising therapeutic means for DR (144). FTO is regarded as an essential epitranscriptomic regulator in diabetes-induced vascular endothelial dysfunction. Zhou et al. identified high glucose could induce retinal vascular leakage and enhance inflammation cytokine (IL-1β, IL-18) secretion and apoptosis of human retinal microvascular endothelial cells (HRMECs). FTO silencing could alleviate diabetes-related retinal vascular dysfunction and inflammation both in vivo and in vitro by inhibiting NF-κB pathway (145). Besides, Chen et al. found in neural retinas collected from STZ mice FTO overexpression contributed to DR phenotypes, including angiogenesis, vascular leakage, inflammation and neurodegeneration by enhancing CDK2 mRNA stability in an YTHDF2-dependent manner (146).
4.2.3 The readers in DR
YTHDF2, m6A reader, plays a significant role in the progress of DR. Qi et al. reported the expression of YTHDF2 was significantly decreased in the retinal tissues of STZ-induced mice and HG-treated HRMECs and retinal Müller cells (rMCs). YTHDF2 silencing enhanced expression of pro-inflammatory factors in rMCs and induced proliferation, migration and invasion in HRMECs (147). Besides, high glucose promotes poly (ADP-ribose) polymerase (PARP) expression, which participates in HRMECs apoptosis and mediates retinal fibrosis and inflammation. YTHDF2- mediated m6A modification epigenetically may regulate stabilization of m6A methylated PARP1 transcripts and activate FAK/AKTsignaling pathway in the pathogenesis of DR (148). Previous study showed the activation of PI3K/AKT pathway led to RPE cells damage and was involved to DR progression (149). YTHDF2 promoted instability of integrin β1 mRNA, which further suppressed FAK/PI3K/AKT pathway and alleviated the progression of DR (147). Dysregulation of autophagy and pyroptosis in RPE cells was a significant pathological mechanism of DR (138, 150). It was reported CircFAT1 bound to YTHDF2 to promote autophagy and suppress pyroptosis of HG-induced RPE cells, thereby alleviating DR progression (151) Another study showed IGF2BP2 may positively regulate lncRNA HOXD Cluster Antisense RNA 1 (HAGLR) via a m6A‐dependent manner. Knockdown of HAGLR inhibited HG‐induced HRPE cells apoptosis and pyroptosis via targeting miR‐106b‐5p/PTEN/Akt signaling, thereby alleviating DR pathology (152) (Figure 2, Table 2).
The above findings reveal m6A RNA modification influences various factors associated with early DR pathogenesis like inflammation, oxidative stress, and neurogenesis, suggesting m6A may play a crucial role in metabolic memory of DR. Thus far, only a small number of pathways related to the pathogenesis of DR have been identified. Therefore, to ascertain the underlying regulatory mechanisms of m6A methylation in DR, more research is necessary.
4.3 m6A and DPN
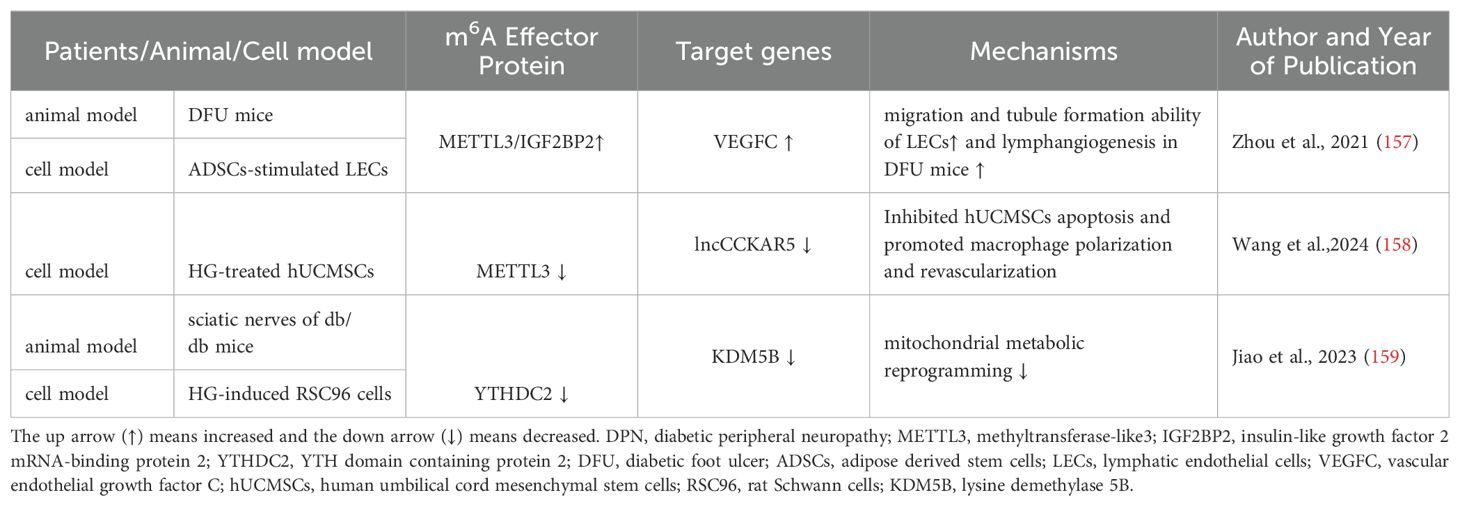
DPN is among the most common long-term complications of diabetes and is at higher risk of all-cause and cardiovascular mortality. Mild symptoms encompass numbness and tingling. Even in some patients it can cause diabetic foot ulcers (DFU), disabling neuropathic pain and lower-limb amputation (153). DPN was characterized by the increase of oxidative stress, mitochondrial damage, and neuron apoptosis (154). Adipose derived stem cells (ADSCs) play a vital role in wound repair by secreting some natural growth factors (155) and activating the PI3K/Akt signaling pathway (156). Zhou et al. discovered a novel link between ADSCs and wound repair with evidence that ADSCs promoted the expression of vascular endothelial growth factor C and lymphangiogenesis marker, LYVE-1, via METTL3/IGF2BP2-m6A pathway in DFU mice (157). Wang et al. ‘s study revealed that knocking down METTL3 substantially reduced the abundance of lncCCKAR5, which further inhibited human umbilical cord mesenchymal stem cells apoptosis and promoted macrophage polarization and revascularization under the conditions of HG stimulation. So m6A-modification of lncCCKAR-5 is a potential therapeutic target of diabetic wound healing (158). Another study showed HG-treatment resulted in a decrease in ATP as well as PDH activity and an increase in ROS in RSC96 cells, which were reversed by YTHDC2 overexpression. It means YTHDC2 overexpression improved mitochondrial metabolic reprogramming in DPN (159) (Figure 2, Table 3).
Overall, the evidence so far suggests that m6A RNA modification process is emerging as a novel mechanism in DPN, but there are still few relevant studies. Therefore, further probing the molecular mechanism of m6A in DPN is of great significance for elucidating the pathogenesis and discovering new therapeutic strategies for DPN.
5 Clinical implications of m6A modification
The exploration of molecular mechanism is for clinical application. Developing therapies that targeting m6A modification or related enzymes has been the focus of many research teams and some m6A inhibitors have been discovered (160, 161). Several inhibitors targeting m6A have been reported in metabolic diseases. Dac51, entacaponea and meclofenamic acid are the inhibitors of FTO. Dac51 could protect against excessive autophagy activation and reverse β-cell dysfunction (94). Entacapone could decrease fasting blood glucose and improve glucose tolerance in high-fat diet-induced obese mouse model (162). Meclofenamic acid has been shown to alleviate ROS accumulation and cell apoptosis (163). METTL3-specific inhibitor STM2457 has a significant inhibitory effect on renal fibrosis (164). In addition, some natural compounds have shown potential therapeutic effects via targeting m6A methylation. Epigallocatechin gallate is the most biologically active and abundant catechin in green tea and curcumin is a natural phenolic compound, which both act to inhibit lipogenesis. Mechanistically, epigallocatechin gallate can suppress the protein stability of FTO while curcumin can decrease the expression of ALKHB5 (95, 165, 166). Another study reported that intake of betaine inhibits hepatic fat accumulation and regulates mitochondrial activity by targeting FTO, thereby improving fatty liver disease and metabolic syndrome (167). TFA, a compound that is extracted from abelmoschus manihot, has been identified to ameliorate pyroptosis and podocytes injury in DKD by targeting METTL3-dependent m6A modification (122). The inhibition of dysregulated m6A effector proteins is a possible new treatment approach, but no m6A inhibitors have entered clinical trials to yet. Researchers still need to devote more efforts to finding new methods and drugs that can be put into clinical use as soon as possible.
Although current studies reveals the abnormal expression of m6A effector proteins in peripheral blood mononuclear cells or biopsy specimen of DM and its microvascular complications, limited researches have not been able to confirm whether effector proteins are specifically expressed only in a particular disease or at a certain stage in the process of diseases. Therefore, they cannot be employed as a specific biomarker for early diagnosis in DM and its microvascular complications.
6 Conclusion and perspectives
m6A is a dynamic and reversible epigenetic modification. A growing number of studies have revealed m6A is involved in the occurrence and development of various metabolic diseases, such as obesity, cardiovascular diseases, diabetes, NAFLD and et al., and provides valuable insights into the etiology, pathogenic mechanism and treatment (168). This review summarizes the underlying molecular mechanisms between m6A in DM and its microvascular complications, but many mechanisms remain to be elucidated. Current research exists some limitations. First, in most studies, animal and cell models are used for in vivo and in vitro studies, while patients sample is rarely used. Second, some researchers have discovered the association of genetic variants with DM using GWAS, but have not explored the mechanism in depth. Third, dysfunction of m6A effector proteins have been identified in diabetic microvascular complications, however upstream regulators remain unclear. Fourth, sample size is relatively small. Of note, DKD, DR, DPN are all microvascular complications of diabetes. There are differences in the regulation of RNA methylation in different organs due to heterogeneity in terms of tissue distribution, origin, phenotype and microenvironment, which also increases the difficulty in the study between m6A methylation and microvascular complications of diabetes. We put forward several future research directions. First, with the advancement of gene sequencing technology and the reduction of cost, more metabolism-related genomics will be discovered so as to further explore the biological function of m6A and the mechanism in diabetes and its complications. Second, multiple enzymes together regulate and maintain m6A RNA methylation. Future studies should focus on the interaction between multiple enzymes under multiple incentive conditions and the mutual interplays of m6A and other RNA modifications in DM and diabetic microvascular complications. Third, exploring m6A modification as a specific biomarker that can predict the development of diabetes, the risk of complications, and the response to treatment will be conducive to more precise disease management and intervention. Fourth, due to the heterogeneity of diabetes, it is essential to investigate individual differences of m6A in patients with diabetes and develop individualized diagnosis and treatment strategies. Fifth, develop drugs that target specific pathologic pathways based on in-depth understanding of m6A and DM and its microvascular complications, for instance β cell protection and regeneration, podocyte repair and etc. In conclusion, discovering underlying mechanisms of m6A methylation in DM and its microvascular complications and more upstream regulators and downstream targets of m6A are beneficial for providing more personalized, effective and safe treatment strategies for diabetes patients.
Author contributions
YW: Conceptualization, Writing – original draft, Data curation, Investigation, Formal analysis. JZ: Conceptualization, Writing – original draft, Data curation, Investigation, Formal analysis. HZ: Funding acquisition, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This paper was supported by National Nature Science Foundation, grant number 82170740, Applied Basic Research Program of Liaoning Province, grant number 2022JH2/101300048, Liao Ning Revitalization Talents Program, grant number XLYC2002081, Pandeng Scholar of Liaoning Province, grant number 2013222, Outstanding Scientific Fund of Shengjing Hospital, grant number 202206.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Glossary
m6A: N6-methyladenosine
DM: diabetes mellitus
IR: insulin resistance
DKD: diabetic kidney disease
DR: diabetic retinopathy
DPN: diabetic peripheral neuropathy
MTC: methyltransferase complex
WTAP: Wilms' tumor 1-associated protein
ZCCHC4: Zinc finger CCHC-type containing 4
RBM15: RNA-binding motif protein 15
XIST: X-inactive specific transcript
FTO: fat mass and obesity-associated protein
ALKBH5: AlkB homolog 5
YTH: YT521-B homology
HNRNPs: heterogeneous nuclear ribonucleoproteins
IGF2BPs: insulin-like growth factor 2 mRNA-binding proteins
YTHDF: YTH domain family protein
YTHDC: YTH domain containing protein
RRM: RNA recognition motif
MeRIP-Seq: methylated RNA immunoprecipitation sequencing
ncRNA: non-coding RNAs
T2D: type 2 diabetes
HFD: high fat diet
MG: methylglyoxal
GSIS: glucose-stimulated insulin secretion
MafA: musculoaponeurotic fibrosarcoma oncogene family A
OAS: 2′-5′-oligoadenylate synthetase
NAFLD: nonalcoholic fatty liver disease
DN: diabetic nephropathy
GRINCH: glucose-responsive insulin-secreting C-peptide modified human proinsulin
ROS: reactive oxygen species
EMT: epithelial-to-mesenchymal transition
GWAS: genome-wide association studies
miRNAs: microRNAs
lncRNAs: long noncoding RNAs
circRNAs: circular RNAs
EMT: epithelial-to-mesenchymal transition
ERS: endoplasmic reticulum stress
HG: high glucose
NSD2: nuclear receptor-binding SET domain protein 2
TFA: total flavones of Abelmoschus manihot
MSCs: mesenchymal stem cells
CAMK: calcium/calmodulin-dependent protein kinases
RPE: Retinal pigment epithelium
BRB: blood-retinal barrier
HRMECs: human retinal microvascular endothelial cells
rMCs: retinal Müller cells
PARP: poly (ADP-ribose) polymerase
HAGLR: HOXD Cluster Antisense RNA 1
DFU: diabetic foot ulcers
ADSCs: adipose derived stem cells.
References
1. Zhang W, Zhang S, Dong C, Guo S, Jia W, Jiang Y, et al. A bibliometric analysis of rna methylation in diabetes mellitus and its complications from 2002 to 2022. Front Endocrinol. (2022) 13:997034. doi: 10.3389/fendo.2022.997034
2. Kidney Disease: Improving Global Outcomes (KDIGO) Diabetes Work Group. Kdigo 2022 clinical practice guideline for diabetes management in chronic kidney disease. Kidney Int. (2022) 102:S1–S127. doi: 10.1016/j.kint.2022.06.008
3. Tampi RR. Diabetes, cognition, and mortality. Am J Geriatr Psychiatry. (2023) 31:583–5. doi: 10.1016/j.jagp.2023.05.003
4. Suarez R, Chapela SP, Alvarez-Cordova L, Bautista-Valarezo E, Sarmiento-Andrade Y, Verde L, et al. Epigenetics in obesity and diabetes mellitus: new insights. Nutrients. (2023) 15:811. doi: 10.3390/nu15040811
5. Sun L, Zhang H, Gao P. Metabolic reprogramming and epigenetic modifications on the path to cancer. Protein Cell. (2022) 13:877–919. doi: 10.1007/s13238-021-00846-7
6. Wang K, Liu H, Hu Q, Wang L, Liu J, Zheng Z, et al. Epigenetic regulation of aging: implications for interventions of aging and diseases. Signal Transduct Target Ther. (2022) 7:374. doi: 10.1038/s41392-022-01211-8
7. Luan J, Kopp JB, Zhou H. N6-methyladenine rna methylation epigenetic modification and kidney diseases. Kidney Int Rep. (2023) 8:36–50. doi: 10.1016/j.ekir.2022.10.009
8. Zhao LY, Song J, Liu Y, Song CX, Yi C. Mapping the epigenetic modifications of DNA and rna. Protein Cell. (2020) 11:792–808. doi: 10.1007/s13238-020-00733-7
9. Shi H, Wei J, He C. Where, when, and how: context-dependent functions of rna methylation writers, readers, and erasers. Mol Cell. (2019) 74:640–50. doi: 10.1016/j.molcel.2019.04.025
10. Liu N, Pan T. N6-methyladenosine-encoded epitranscriptomics. Nat Struct Mol Biol. (2016) 23:98–102. doi: 10.1038/nsmb.3162
11. Fan Y, Lv X, Chen Z, Peng Y, Zhang M. M6a methylation: critical roles in aging and neurological diseases. Front Mol Neurosci. (2023) 16:1102147. doi: 10.3389/fnmol.2023.1102147
12. Wang Y, Wang Y, Gu J, Su T, Gu X, Feng Y. The role of rna M6a methylation in lipid metabolism. Front Endocrinol (Lausanne). (2022) 13:866116. doi: 10.3389/fendo.2022.866116
13. Jiang X, Liu B, Nie Z, Duan L, Xiong Q, et al. Jin Z. role m6A modification Biol functions diseases Signal Transduct Target Ther. (2021) 6:74. doi: 10.1038/s41392-020-00450-x
14. Xu Z, Lv B, Qin Y, Zhang B. Emerging roles and mechanism of M6a methylation in cardiometabolic diseases. Cells. (2022) 11:1101. doi: 10.3390/cells11071101
15. Li L, Xu N, Liu J, Chen Z, Liu X, Wang J. M6a methylation in cardiovascular diseases: from mechanisms to therapeutic potential. Front Genet. (2022) 13:908976. doi: 10.3389/fgene.2022.908976
16. Ma S, Chen C, Ji X, Liu J, Zhou Q, Wang G, et al. The interplay between M6a rna methylation and noncoding rna in cancer. J Hematol Oncol. (2019) 12:121. doi: 10.1186/s13045-019-0805-7
17. Benak D, Benakova S, Plecita-Hlavata L, Hlavackova M. The role of M6a and M6am rna modifications in the pathogenesis of diabetes mellitus. Front Endocrinol. (2023) 14:1223583. doi: 10.3389/fendo.2023.1223583
18. Ren Y, Li Z, Li J, Liang R, Wang Z, Bai Y, et al. M6a mrna methylation: biological features, mechanisms, and therapeutic potentials in type 2 diabetes mellitus. Obes Rev. (2023) 24:e13639. doi: 10.1111/obr.13639
19. Zhang H, Gu Y, Gang Q, Huang J, Xiao Q, Ha X. N6-methyladenosine rna modification: an emerging molecule in type 2 diabetes metabolism. Front Endocrinol. (2023) 14:1166756. doi: 10.3389/fendo.2023.1166756
20. Li Y-L, Zhang Y, Chen N, Yan Y-X. The role of M6a modification in type 2 diabetes: A systematic review and integrative analysis. Gene. (2024) 898:148130. doi: 10.1016/j.gene.2024.148130
21. Desrosiers R, Friderici K, Rottman F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc Natl Acad Sci USA. (1974) 71:3971–5. doi: 10.1073/pnas.71.10.3971
22. Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mrna methylation reveals enrichment in 3’ Utrs and near stop codons. Cell. (2012) 149:1635–46. doi: 10.1016/j.cell.2012.05.003
23. Zhang W, Qian Y, Jia G. The detection and functions of rna modification M(6)a based on M(6)a writers and erasers. J Biol Chem. (2021) 297:100973. doi: 10.1016/j.jbc.2021.100973
24. Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, et al. A mettl3-mettl14 complex mediates mammalian nuclear rna N6-adenosine methylation. Nat Chem Biol. (2014) 10:93–5. doi: 10.1038/nchembio.1432
25. Wang X, Feng J, Xue Y, Guan Z, Zhang D, Liu Z, et al. Structural basis of N(6)-adenosine methylation by the mettl3-mettl14 complex. Nature. (2016) 534:575–8. doi: 10.1038/nature18298
26. Yoon KJ, Ringeling FR, Vissers C, Jacob F, Pokrass M, Jimenez-Cyrus D, et al. Temporal control of mammalian cortical neurogenesis by M(6)a methylation. Cell. (2017) 171:877–89 e17. doi: 10.1016/j.cell.2017.09.003
27. Geula S, Moshitch-Moshkovitz S, Dominissini D, Mansour AA, Kol N, Salmon-Divon M, et al. Stem cells. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science. (2015) 347:1002–6. doi: 10.1126/science.1261417
28. Sorci M, Ianniello Z, Cruciani S, Larivera S, Ginistrelli LC, Capuano E, et al. Mettl3 regulates wtap protein homeostasis. Cell Death Dis. (2018) 9:796. doi: 10.1038/s41419-018-0843-z
29. Ping X-L, Sun B-F, Wang L, Xiao W, Yang X, Wang W-J, et al. Mammalian wtap is a regulatory subunit of the rna N6-methyladenosine methyltransferase. Cell Res. (2014) 24:177–89. doi: 10.1038/cr.2014.3
30. Xie W, Wei L, Guo J, Guo H, Song X, Sheng X. Physiological functions of wilms’ Tumor 1-associating protein and its role in tumourigenesis. J Cell Biochem. (2019) 120:10884–92. doi: 10.1002/jcb.28402
31. van Tran N, Ernst FGM, Hawley BR, Zorbas C, Ulryck N, Hackert P, et al. The human 18s rrna M6a methyltransferase mettl5 is stabilized by trmt112. Nucleic Acids Res. (2019) 47:7719–33. doi: 10.1093/nar/gkz619
32. Wei Z, Chen Y, Zeng Z, Peng Y, Li L, Hu N, et al. The novel M6a writer mettl5 as prognostic biomarker probably associating with the regulation of immune microenvironment in kidney cancer. Heliyon. (2022) 8:e12078. doi: 10.1016/j.heliyon.2022.e12078
33. Dai Z, Zhu W, Hou Y, Zhang X, Ren X, Lei K, et al. Mettl5-mediated 18s rrna M(6)a modification promotes oncogenic mrna translation and intrahepatic cholangiocarcinoma progression. Mol Ther. (2023) 31:3225–42. doi: 10.1016/j.ymthe.2023.09.014
34. Ruszkowska A. Mettl16, methyltransferase-like protein 16: current insights into structure and function. Int J Mol Sci. (2021) 22:2176. doi: 10.3390/ijms22042176
35. Patil DP, Chen CK, Pickering BF, Chow A, Jackson C, Guttman M, et al. M(6)a rna methylation promotes xist-mediated transcriptional repression. Nature. (2016) 537:369–73. doi: 10.1038/nature19342
36. Zhang L, Tran N-T, Su H, Wang R, Lu Y, Tang H, et al. Cross-talk between prmt1-mediated methylation and ubiquitylation on rbm15 controls rna splicing. eLife. (2015) 4:e07938. doi: 10.7554/eLife.07938
37. Bartosovic M, Molares HC, Gregorova P, Hrossova D, Kudla G, Vanacova S. N6-methyladenosine demethylase fto targets pre-mrnas and regulates alternative splicing and 3’-end processing. Nucleic Acids Res. (2017) 45:11356–70. doi: 10.1093/nar/gkx778
38. Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, et al. Alkbh5 is a mammalian rna demethylase that impacts rna metabolism and mouse fertility. Mol Cell. (2013) 49:18–29. doi: 10.1016/j.molcel.2012.10.015
39. Wei J, Liu F, Lu Z, Fei Q, Ai Y, He PC, et al. Differential M6a, M6am, and M1a demethylation mediated by fto in the cell nucleus and cytoplasm. Mol Cell. (2018) 71:973–85.e5. doi: 10.1016/j.molcel.2018.08.011
40. Scuteri A, Sanna S, Chen WM, Uda M, Albai G, Strait J, et al. Genome-wide association scan shows genetic variants in the fto gene are associated with obesity-related traits. PloS Genet. (2007) 3:e115. doi: 10.1371/journal.pgen.0030115
41. Church C, Moir L, McMurray F, Girard C, Banks GT, Teboul L, et al. Overexpression of fto leads to increased food intake and results in obesity. Nat Genet. (2010) 42:1086–92. doi: 10.1038/ng.713
42. Fischer J, Koch L, Emmerling C, Vierkotten J, Peters T, Bruning JC, et al. Inactivation of the fto gene protects from obesity. Nature. (2009) 458:894–8. doi: 10.1038/nature07848
43. Chang R, Huang Z, Zhao S, Zou J, Li Y, Tan S. Emerging roles of fto in neuropsychiatric disorders. BioMed Res Int. (2022) 2022:2677312. doi: 10.1155/2022/2677312
44. Azzam SK, Alsafar H, Sajini AA. Fto M6a demethylase in obesity and cancer: implications and underlying molecular mechanisms. Int J Mol Sci. (2022) 23:3800. doi: 10.3390/ijms23073800
45. Zhao Y, Shi Y, Shen H, Xie W. M(6)a-binding proteins: the emerging crucial performers in epigenetics. J Hematol Oncol. (2020) 13:35. doi: 10.1186/s13045-020-00872-8
46. Wang JY, Lu AQ. The biological function of M6a reader ythdf2 and its role in human disease. Cancer Cell Int. (2021) 21:109. doi: 10.1186/s12935-021-01807-0
47. Ma C, Liao S, Zhu Z. Crystal structure of human ythdc2 yth domain. Biochem Biophys Res Commun. (2019) 518:678–84. doi: 10.1016/j.bbrc.2019.08.107
48. Roundtree IA, Luo GZ, Zhang Z, Wang X, Zhou T, Cui Y, et al. Ythdc1 mediates nuclear export of N(6)-methyladenosine methylated mrnas. Elife. (2017) 6:e31311. doi: 10.7554/eLife.31311
49. Widagdo J, Anggono V, Wong JJL. The multifaceted effects of ythdc1-mediated nuclear M6a recognition. Trends Genet. (2022) 38:325–32. doi: 10.1016/j.tig.2021.11.005
50. Tatsuno T, Ishigaki Y. Multiple phosphorylations of sr protein srsf3 and its binding to M6a reader ythdc1 in human cells. Cells. (2022) 11:1461. doi: 10.3390/cells11091461
51. Larivera S, Meister G. Domain confusion 2: M6a-independent role of ythdc2. Mol Cell. (2022) 82:1608–9. doi: 10.1016/j.molcel.2022.04.012
52. Li L, Krasnykov K, Homolka D, Gos P, Mendel M, Fish RJ, et al. The xrn1-regulated rna helicase activity of ythdc2 ensures mouse fertility independently of M(6)a recognition. Mol Cell. (2022) 82:1678–90 e12. doi: 10.1016/j.molcel.2022.02.034
53. Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, et al. N(6)-methyladenosine modulates messenger rna translation efficiency. Cell. (2015) 161:1388–99. doi: 10.1016/j.cell.2015.05.014
54. Du H, Zhao Y, He J, Zhang Y, Xi H, Liu M, et al. Ythdf2 destabilizes M(6)a-containing rna through direct recruitment of the ccr4-not deadenylase complex. Nat Commun. (2016) 7:12626. doi: 10.1038/ncomms12626
55. Shi H, Wang X, Lu Z, Zhao BS, Ma H, Hsu PJ, et al. Ythdf3 facilitates translation and decay of N6-methyladenosine-modified rna. Cell Res. (2017) 27:315–28. doi: 10.1038/cr.2017.15
56. Zaccara S, Jaffrey SR. A unified model for the function of ythdf proteins in regulating M(6)a-modified mrna. Cell. (2020) 181:1582–95 e18. doi: 10.1016/j.cell.2020.05.012
57. Sikorski V, Selberg S, Lalowski M, Karelson M, Kankuri E. The structure and function of ythdf epitranscriptomic M(6)a readers. Trends Pharmacol Sci. (2023) 44:335–53. doi: 10.1016/j.tips.2023.03.004
58. Li J, Chen K, Dong X, Xu Y, Sun Q, Wang H, et al. Ythdf1 promotes mrna degradation via ythdf1-ago2 interaction and phase separation. Cell Prolif. (2022) 55:e13157. doi: 10.1111/cpr.13157
59. Zhao Y, Zhao H, Zhang D, Quan Q, Ge Y, Li L, et al. Ythdf3 facilitates eif2ak2 and eif3a recruitment on mrnas to regulate translational processes in oxaliplatin-resistant colorectal cancer. ACS Chem Biol. (2022) 17:1778–88. doi: 10.1021/acschembio.2c00131
60. Wu B, Su S, Patil DP, Liu H, Gan J, Jaffrey SR, et al. Molecular basis for the specific and multivariant recognitions of rna substrates by human hnrnp A2/B1. Nat Commun. (2018) 9:420. doi: 10.1038/s41467-017-02770-z
61. Liu N, Dai Q, Zheng G, He C, Parisien M, Pan T. N(6)-methyladenosine-dependent rna structural switches regulate rna-protein interactions. Nature. (2015) 518:560–4. doi: 10.1038/nature14234
62. Liu N, Zhou KI, Parisien M, Dai Q, Diatchenko L, Pan T. N 6-methyladenosine alters rna structure to regulate binding of a low-complexity protein. Nucleic Acids Res. (2017) 45:6051–63. doi: 10.1093/nar/gkx141
63. He Y, Smith R. Nuclear functions of heterogeneous nuclear ribonucleoproteins a/B. Cell Mol Life Sci. (2009) 66:1239–56. doi: 10.1007/s00018-008-8532-1
64. Alarcon CR, Goodarzi H, Lee H, Liu X, Tavazoie S, Tavazoie SF. Hnrnpa2b1 is a mediator of M(6)a-dependent nuclear rna processing events. Cell. (2015) 162:1299–308. doi: 10.1016/j.cell.2015.08.011
65. He Q, Yang C, Xiang Z, Huang G, Wu H, Chen T, et al. Linc00924-induced fatty acid metabolic reprogramming facilitates gastric cancer peritoneal metastasis via hnrnpc-regulated alternative splicing of mnk2. Cell Death Dis. (2022) 13:987. doi: 10.1038/s41419-022-05436-x
66. Huang XT, Li JH, Zhu XX, Huang CS, Gao ZX, Xu QC, et al. Hnrnpc impedes M(6)a-dependent anti-metastatic alternative splicing events in pancreatic ductal adenocarcinoma. Cancer Lett. (2021) 518:196–206. doi: 10.1016/j.canlet.2021.07.016
67. Zhou KI, Shi H, Lyu R, Wylder AC, Matuszek Z, Pan JN, et al. Regulation of co-transcriptional pre-mrna splicing by M(6)a through the low-complexity protein hnrnpg. Mol Cell. (2019) 76:70–81 e9. doi: 10.1016/j.molcel.2019.07.005
68. Huang H, Weng H, Sun W, Qin X, Shi H, Wu H, et al. Recognition of rna N(6)-methyladenosine by igf2bp proteins enhances mrna stability and translation. Nat Cell Biol. (2018) 20:285–95. doi: 10.1038/s41556-018-0045-z
69. Wang Y, Yang W, Zhou X. Chemical labelling for M6a detection: opportunities and challenges. Fundam Res. (2022) 2:56–8. doi: 10.1016/j.fmre.2021.11.034
70. Kim H, Kulkarni RN. Epigenetics in beta-cell adaptation and type 2 diabetes. Curr Opin Pharmacol. (2020) 55:125–31. doi: 10.1016/j.coph.2020.10.008
71. De Jesus DF, Kulkarni RN. Epigenetic modifiers of islet function and mass. Trends Endocrinol Metab. (2014) 25:628–36. doi: 10.1016/j.tem.2014.08.006
72. Shen F, Huang W, Huang JT, Xiong J, Yang Y, Wu K, et al. Decreased N(6)-methyladenosine in peripheral blood rna from diabetic patients is associated with fto expression rather than alkbh5. J Clin Endocrinol Metab. (2015) 100:E148–54. doi: 10.1210/jc.2014-1893
73. Yang Y, Shen F, Huang W, Qin S, Huang JT, Sergi C, et al. Glucose is involved in the dynamic regulation of M6a in patients with type 2 diabetes. J Clin Endocrinol Metab. (2019) 104:665–73. doi: 10.1210/jc.2018-00619
74. Xie W, Ma LL, Xu YQ, Wang BH, Li SM. Mettl3 inhibits hepatic insulin sensitivity via N6-methyladenosine modification of fasn mrna and promoting fatty acid metabolism. Biochem Biophys Res Commun. (2019) 518:120–6. doi: 10.1016/j.bbrc.2019.08.018
75. Li Y, Zhang Q, Cui G, Zhao F, Tian X, Sun BF, et al. M(6)a regulates liver metabolic disorders and hepatogenous diabetes. Genomics Proteomics Bioinf. (2020) 18:371–83. doi: 10.1016/j.gpb.2020.06.003
76. De Jesus DF, Zhang Z, Kahraman S, Brown NK, Chen M, Hu J, et al. M(6)a mrna methylation regulates human beta-cell biology in physiological states and in type 2 diabetes. Nat Metab. (2019) 1:765–74. doi: 10.1038/s42255-019-0089-9
77. Wang Y, Sun J, Lin Z, Zhang W, Wang S, Wang W, et al. M(6)a mrna methylation controls functional maturation in neonatal murine beta-cells. Diabetes. (2020) 69:1708–22. doi: 10.2337/db19-0906
78. Kong X, Ma MZ, Huang K, Qin L, Zhang HM, Yang Z, et al. Increased plasma levels of the methylglyoxal in patients with newly diagnosed type 2 diabetes 2. J Diabetes. (2014) 6:535–40. doi: 10.1111/1753-0407.12160
79. Cheng Y, Yao XM, Zhou SM, Sun Y, Meng XJ, Wang Y, et al. The M(6)a methyltransferase mettl3 ameliorates methylglyoxal-induced impairment of insulin secretion in pancreatic beta cells by regulating mafa expression. Front Endocrinol (Lausanne). (2022) 13:910868. doi: 10.3389/fendo.2022.910868
80. Li X, Jiang Y, Sun X, Wu Y, Chen Z. Mettl3 is required for maintaining B-cell function. Metabolism. (2021) 116:154702. doi: 10.1016/j.metabol.2021.154702
81. Sun J, Wang Y, Fu H, Kang F Song J, Xu M, Ning G, et al. METTL3-mediated m6A methylation controls pancreatic bipotent progenitor fate and islet formation. Diabetes. (2024) 73:237–49. doi: 10.2337/db23-0360/738457/db230360
82. Liu J, Luo G, Sun J, Men L, Ye H, He C, et al. Mettl14 is essential for beta-cell survival and insulin secretion. Biochim Biophys Acta Mol Basis Dis. (2019) 1865:2138–48. doi: 10.1016/j.bbadis.2019.04.011
83. Men L, Sun J, Luo G, Ren D. Acute deletion of mettl14 in beta-cells of adult mice results in glucose intolerance. Endocrinology. (2019) 160:2388–94. doi: 10.1210/en.2019-00350
84. Li X, Yang Y, Li Z, Wang Y, Qiao J, Chen Z. Deficiency of wtap in islet beta cells results in beta cell failure and diabetes in mice. Diabetologia. (2023) 66:1084–96. doi: 10.1007/s00125-023-05900-z
85. Li YL, Li L, Liu YH, Hu LK, Yan YX. Identification of metabolism-related proteins as biomarkers of insulin resistance and potential mechanisms of M(6)a modification. Nutrients. (2023) 15:1839. doi: 10.3390/nu15081839
86. Tanase DM, Gosav EM, Costea CF, Ciocoiu M, Lacatusu CM, Maranduca MA, et al. The intricate relationship between type 2 diabetes mellitus (T2dm), insulin resistance (Ir), and nonalcoholic fatty liver disease (Nafld). J Diabetes Res. (2020) 2020:3920196. doi: 10.1155/2020/3920196
87. Li Y, Zhang D, Gao Y, Wang P, Wang Z, Zhang B, et al. Mettl3 exacerbates insulin resistance in hepatocytes by regulating M(6)a modification of cytochrome P450 2b6. Nutr Metab (Lond). (2023) 20:40. doi: 10.1186/s12986-023-00762-z
88. Montesanto A, Bonfigli AR, Crocco P, Garagnani P, De Luca M, Boemi M, et al. Genes associated with Type 2 Diabetes and vascular complications. Aging (Albany NY). (2018) 10:178–96. doi: 10.18632/aging.101375
89. Chen G, Xu Y, Lin Y, Lai X, Yao J, Huang B, et al. Association study of genetic variants of 17 diabetes-related genes/loci and cardiovascular risk and diabetic nephropathy in the chinese she population. J Diabetes. (2013) 5:136–45. doi: 10.1111/1753-0407.12025
90. Sun Q, Geng H, Zhao M, Li Y, Chen X, Sha Q, et al. Fto-mediated M(6) a modification of socs1 mrna promotes the progression of diabetic kidney disease. Clin Transl Med. (2022) 12:e942. doi: 10.1002/ctm2.942
91. Taneera J, Prasad RB, Dhaiban S, Mohammed AK, Haataja L, Arvan P, et al. Silencing of the fto gene inhibits insulin secretion: an in vitro study using grinch cells. Mol Cell Endocrinol. (2018) 472:10–7. doi: 10.1016/j.mce.2018.06.003
92. Taneera J, Khalique A, Abdrabh S, Mohammed AK, Bouzid A, El-Huneidi W, et al. Fat mass and obesity-associated (Fto) gene is essential for insulin secretion and B-cell function: in vitro studies using ins-1 cells and human pancreatic islets. Life Sci. (2024) 339:122421. doi: 10.1016/j.lfs.2024.122421
93. Fan HQ, He W, Xu KF, Wang ZX, Xu XY, Chen H. Fto inhibits insulin secretion and promotes nf-kappab activation through positively regulating ros production in pancreatic beta cells. PloS One. (2015) 10:e0127705. doi: 10.1371/journal.pone.0127705
94. Wu T, Shao Y, Li X, Wu T, Yu L, Liang J, et al. Nr3c1/glucocorticoid receptor activation promotes pancreatic B-cell autophagy overload in response to glucolipotoxicity. Autophagy. (2023) 19:2538–57. doi: 10.1080/15548627.2023.2200625
95. Shao Y, Zhang Y, Zou S, Wang J, Li X, Qin M, et al. (-)-Epigallocatechin 3-Gallate Protects Pancreatic B-Cell against Excessive Autophagy-Induced Injury through Promoting Fto Degradation. Autophagy. (2024), 1–18. doi: 10.1080/15548627.2024.2370751
96. Li X, Yang Y, Chen Z. Downregulation of the M(6)a reader protein ythdc1 leads to islet beta-cell failure and diabetes. Metabolism. (2023) 138:155339. doi: 10.1016/j.metabol.2022.155339
97. Yang K, Sun J, Zhang Z, Xiao M, Ren D, Liu SM. Reduction of mrna M(6)a associates with glucose metabolism via ythdc1 in human and mice. Diabetes Res Clin Pract. (2023) 198:110607. doi: 10.1016/j.diabres.2023.110607
98. Gu T, Horova E, Mollsten A, Seman NA, Falhammar H, Prazny M, et al. Igf2bp2 and igf2 genetic effects in diabetes and diabetic nephropathy. J Diabetes Complications. (2012) 26:393–8. doi: 10.1016/j.jdiacomp.2012.05.012
99. Han X, Luo Y, Ren Q, Zhang X, Wang F, Sun X, et al. Implication of genetic variants near SLC30A8 HHEX CDKAL1 CDKN2AB IGF2BP2 FTO TCF2 KCNQ1 and WFS1 in type 2 diabetes in a Chinese population. BMC Med Genet. (2010) 11:81. doi: 10.1186/1471-2350-11-81
100. Sanghera DK, Ortega L, Han S, Singh J, Ralhan SK, Wander GS, et al. Impact of nine common type 2 diabetes risk polymorphisms in asian Indian sikhs: pparg2 (Pro12ala), igf2bp2, tcf7l2 and fto variants confer a significant risk. BMC Med Genet. (2008) 9:59. doi: 10.1186/1471-2350-9-59
101. van Hoek M, Langendonk JG, de Rooij SR, Sijbrands EJ, Roseboom TJ. Genetic variant in the igf2bp2 gene may interact with fetal malnutrition to affect glucose metabolism. Diabetes. (2009) 58:1440–4. doi: 10.2337/db08-1173
102. Groenewoud MJ, Dekker JM, Fritsche A, Reiling E, Nijpels G, Heine RJ, et al. Variants of cdkal1 and igf2bp2 affect first-phase insulin secretion during hyperglycaemic clamps. Diabetologia. (2008) 51:1659–63. doi: 10.1007/s00125-008-1083-z
103. Regué L, Zhao L, Ji F, Wang H, Avruch J, Dai N. Rna M6a reader imp2/igf2bp2 promotes pancreatic B-cell proliferation and insulin secretion by enhancing pdx1 expression. Mol Metab. (2021) 48:101209. doi: 10.1016/j.molmet.2021.101209
104. Zhu Y, Liu Q, Zhou Z, Ikeda Y. Pdx1, neurogenin-3, and mafa: critical transcription regulators for beta cell development and regeneration. Stem Cell Res Ther. (2017) 8:240. doi: 10.1186/s13287-017-0694-z
105. Tian Y, Xu J, Du X, Fu X. The interplay between noncoding rnas and insulin in diabetes. Cancer Lett. (2018) 419:53–63. doi: 10.1016/j.canlet.2018.01.038
106. Xu Y-X, Pu S-D, Li X, Yu Z-W, Zhang Y-T, Tong X-W, et al. Exosomal ncrnas: novel therapeutic target and biomarker for diabetic complications. Pharmacol Res. (2022) 178:106135. doi: 10.1016/j.phrs.2022.106135
107. Li Y, Liu Y, Yao X, Wang H, Shi Z, He M. Mettl14-mediated lncrna xist silencing alleviates gdm progression by facilitating trophoblast cell proliferation and migration via the mir-497-5p/foxo1 axis. J Biochem Mol Toxicol. (2023) 38:e23621. doi: 10.1002/jbt.23621
108. Leslie RD, Ma RCW, Franks PW, Nadeau KJ, Pearson ER, Redondo MJ. Understanding diabetes heterogeneity: key steps towards precision medicine in diabetes. Lancet Diabetes Endocrinol. (2023) 11:848–60. doi: 10.1016/s2213-8587(23)00159-6
109. Li C, Su F, Liang Z, Zhang L, Liu F, Fan W, et al. Macrophage M1 regulatory diabetic nephropathy is mediated by M6a methylation modification of lncrna expression. Mol Immunol. (2022) 144:16–25. doi: 10.1016/j.molimm.2022.02.008
110. Gupta S, Dominguez M, Golestaneh L. Diabetic kidney disease. Med Clinics North America. (2023) 107:689–705. doi: 10.1016/j.mcna.2023.03.004
111. Lovisa S, LeBleu VS, Tampe B, Sugimoto H, Vadnagara K, Carstens JL, et al. Epithelial-to-mesenchymal transition induces cell cycle arrest and parenchymal damage in renal fibrosis. Nat Med. (2015) 21:998–1009. doi: 10.1038/nm.3902
112. Li M, Deng L, Xu G. Mettl14 promotes glomerular endothelial cell injury and diabetic nephropathy via M6a modification of alpha-klotho. Mol Med. (2021) 27:106. doi: 10.1186/s10020-021-00365-5
113. Lu Z, Liu H, Song N, Liang Y, Zhu J, Chen J, et al. Mettl14 aggravates podocyte injury and glomerulopathy progression through N(6)-methyladenosine-dependent downregulating of sirt1. Cell Death Dis. (2021) 12:881. doi: 10.1038/s41419-021-04156-y
114. Zheng Y, Zhang Z, Zheng D, Yi P, Wang S. Mettl14 promotes the development of diabetic kidney disease by regulating M(6)a modification of tug1. Acta Diabetol. (2023) 60:1567–80. doi: 10.1007/s00592-023-02145-5
115. Taniguchi M, Yoshida H. Endoplasmic reticulum stress in kidney function and disease. Curr Opin Nephrol Hypertens. (2015) 24:345–50. doi: 10.1097/MNH.0000000000000141
116. Xu Z, Jia K, Wang H, Gao F, Zhao S, Li F, et al. Mettl14-regulated pi3k/akt signaling pathway via pten affects hdac5-mediated epithelial-mesenchymal transition of renal tubular cells in diabetic kidney disease. Cell Death Dis. (2021) 12:32. doi: 10.1038/s41419-020-03312-0
117. Jiang L, Liu X, Hu X, Gao L, Zeng H, Wang X, et al. Mettl3-mediated M(6)a modification of timp2 mrna promotes podocyte injury in diabetic nephropathy. Mol Ther. (2022) 30:1721–40. doi: 10.1016/j.ymthe.2022.01.002
118. Shi S, Zhao L, Zheng L. Nsd2 is downregulated in T2dm and promotes beta cell proliferation and insulin secretion through the transcriptionally regulation of pdx1. Mol Med Rep. (2018) 18:3513–20. doi: 10.3892/mmr.2018.9338
119. Tang W, Zhao Y, Zhang H, Peng Y, Rui Z. Mettl3 enhances nsd2 mrna stability to reduce renal impairment and interstitial fibrosis in mice with diabetic nephropathy. BMC Nephrol. (2022) 23:124. doi: 10.1186/s12882-022-02753-3
120. Chen Y, Li P, Lin M, Jiang Y, Tan G, Huang L, et al. Silencing of METTL3 prevents the proliferation, migration, epithelial-mesenchymal transition, and renal fibrosis of high glucose-induced HK2 cells by mediating WISP1 in m6A-dependent manner. Aging (Albany NY). (2024) 16:1237–48. doi: 10.18632/aging.205401
121. Wang F, Bai J, Zhang X, Wang D, Zhang X, Xue J, et al. Mettl3/ythdf2 M6a axis mediates the progression of diabetic nephropathy through epigenetically suppressing pink1 and mitophagy. J Diabetes Invest. (2023) 15:288–99. doi: 10.1111/jdi.14113
122. Liu B-H, Tu Y, Ni G-X, Yan J, Yue L, Li Z-L, et al. Total flavones of abelmoschus manihot ameliorates podocyte pyroptosis and injury in high glucose conditions by targeting mettl3-dependent M6a modification-mediated nlrp3-inflammasome activation and pten/pi3k/akt signaling. Front Pharmacol. (2021) 12:667644. doi: 10.3389/fphar.2021.667644
123. Liu X, Jiang L, Zeng H, Gao L, Guo S, Chen C, et al. Circ-0000953 deficiency exacerbates podocyte injury and autophagy disorder by targeting mir665-3p-atg4b in diabetic nephropathy. Autophagy. (2023) 20:1072–97. doi: 10.1080/15548627.2023.2286128
124. Lan J, Xu B, Shi X, Pan Q, Tao Q. Wtap-mediated N(6)-methyladenosine modification of nlrp3 mrna in kidney injury of diabetic nephropathy. Cell Mol Biol Lett. (2022) 27:51. doi: 10.1186/s11658-022-00350-8
125. Bai Y, Huang L, Fan Y, Li Y. Marrow mesenchymal stem cell mediates diabetic nephropathy progression via modulation of smad2/3/wtap/M6a/eno1 axis. FASEB J. (2024) 38:e23729. doi: 10.1096/fj.202301773R
126. Qin Y, Wu S, Zhang F, Zhou X, You C, Tan F. N6-methyladenosine methylation regulator rbm15 promotes the progression of diabetic nephropathy by regulating cell proliferation, inflammation, oxidative stress, and pyroptosis through activating the age-rage pathway. Environ Toxicol. (2023) 38:2772–82. doi: 10.1002/tox.23917
127. Lang Y, Wang Q, Sheng Q, Lu S, Yang M, Kong Z, et al. Fto-mediated M6a modification of serum amyloid A2 mrna promotes podocyte injury and inflammation by activating the nf-Kb signaling pathway. FASEB J. (2024) 38:e23409. doi: 10.1096/fj.202301419RR
128. Jiang A, Song A, Zhang C. Modes of podocyte death in diabetic kidney disease: an update. J Nephrol. (2022) 35:1571–84. doi: 10.1007/s40620-022-01269-1
129. Hale LJ, Welsh GI, Perks CM, Hurcombe JA, Moore S, Hers I, et al. Insulin-like growth factor-ii is produced by, signals to and is an important survival factor for the mature podocyte in man and mouse. J Pathol. (2013) 230:95–106. doi: 10.1002/path.4165
130. Yong L, Yu Y, Li B, Ge H, Zhen Q, Mao Y, et al. Calcium/calmodulin-dependent protein kinase iv promotes imiquimod-induced psoriatic inflammation via macrophages and keratinocytes in mice. Nat Commun. (2022) 13:4255. doi: 10.1038/s41467-022-31935-8
131. Yang J, Sun M, Cheng R, Tan H, Liu C, Chen R, et al. Pitavastatin activates mitophagy to protect epc proliferation through a calcium-dependent camk1-pink1 pathway in atherosclerotic mice. Commun Biol. (2022) 5:124. doi: 10.1038/s42003-022-03081-w
132. Yuan D, Li H, Dai W, Zhou X, Zhou W, He L. Igf2bp3-stabilized camk1 regulates the mitochondrial dynamics of renal tubule to alleviate diabetic nephropathy. Biochim Biophys Acta (BBA) - Mol Basis Dis. (2024) 1870:167022. doi: 10.1016/j.bbadis.2024.167022
133. Lin Z, Lv D, Liao X, Peng R, Liu H, Wu T, et al. Circubxn7 promotes macrophage infiltration and renal fibrosis associated with the igf2bp2-dependent sp1 mrna stability in diabetic kidney disease. Front Immunol. (2023) 14:1226962. doi: 10.3389/fimmu.2023.1226962
134. Tan TE, Wong TY. Diabetic retinopathy: looking forward to 2030. Front Endocrinol (Lausanne). (2022) 13:1077669. doi: 10.3389/fendo.2022.1077669
135. Cao Y, Feng B, Chen S, Chu Y, Chakrabarti S. Mechanisms of endothelial to mesenchymal transition in the retina in diabetes. Invest Ophthalmol Vis Sci. (2014) 55:7321–31. doi: 10.1167/iovs.14-15167
136. Shi Y, Vanhoutte PM. Macro- and microvascular endothelial dysfunction in diabetes. J Diabetes. (2017) 9:434–49. doi: 10.1111/1753-0407.12521
137. Dunn KC, Aotaki-Keen AE, Putkey FR, Hjelmeland LM. ARPE-19, A human retinal pigment epithelial cell line with differentiated properties. Exp Eye Res. (1996) 6:155–69. doi: 10.1006/exer.1996.0020
138. Zha X, Xi X, Fan X, Ma M, Zhang Y, Yang Y. Overexpression of METTL3 attenuates high-glucose induced RPE cell pyroptosis by regulating miR-25-3p PTEN Akt signaling cascade through DGCR8. Aging (Albany NY). (2020) 12:8137–50. doi: 10.18632/aging.103130
139. Cao X, Song Y, Huang L-L, Tian Y-J, Wang X-L, Hua L-Y. Ma transferase mettl3 regulates endothelial-mesenchymal transition in diabetic retinopathy via lncrna snhg7/khsrp/mkl1 axis. Genomics. (2022) 114:110498. doi: 10.1016/j.ygeno.2022.110498
140. Kumari N, Karmakar A, Ahamad Khan MM, Ganesan SK. The potential role of M6a rna methylation in diabetic retinopathy. Exp Eye Res. (2021) 208:108616. doi: 10.1016/j.exer.2021.108616
141. Yao MD, Jiang Q, Ma Y, Liu C, Zhu CY, Sun YN, et al. Role of mettl3-dependent N(6)-methyladenosine mrna modification in the promotion of angiogenesis. Mol Ther. (2020) 28:2191–202. doi: 10.1016/j.ymthe.2020.07.022
142. Park DY, Lee J, Kim J, Kim K, Hong S, Han S, et al. Plastic roles of pericytes in the blood-retinal barrier. Nat Commun. (2017) 8:15296. doi: 10.1038/ncomms15296
143. Suo L, Liu C, Zhang QY, Yao MD, Ma Y, Yao J, et al. Mettl3-mediated N(6)-methyladenosine modification governs pericyte dysfunction during diabetes-induced retinal vascular complication. Theranostics. (2022) 12:277–89. doi: 10.7150/thno.63441
144. Chen T, Zhu W, Wang C, Dong X, Yu F, Su Y, et al. Alkbh5-mediated M(6)a modification of A20 regulates microglia polarization in diabetic retinopathy. Front Immunol. (2022) 13:813979. doi: 10.3389/fimmu.2022.813979
145. Zhou C, She X, Gu C, Hu Y, Ma M, Qiu Q, et al. Fto fuels diabetes-induced vascular endothelial dysfunction associated with inflammation by erasing M6a methylation of tnip1. J Clin Invest. (2023) 133:e160517. doi: 10.1172/jci160517
146. Chen X, Wang Y, Wang J-N, Zhang Y-C, Zhang Y-R, Sun R-X, et al. Lactylation-driven fto targets cdk2 to aggravate microvascular anomalies in diabetic retinopathy. EMBO Mol Med. (2024) 16:294–318. doi: 10.1038/s44321-024-00025-1
147. Qi Y, Yao R, Zhang W, Cui Q. Kat1 triggers ythdf2-mediated itgb1 mrna instability to alleviate the progression of diabetic retinopathy. Pharmacol Res. (2021) 170:105713. doi: 10.1016/j.phrs.2021.105713
148. Sun J, Liu G, Chen R, Zhou J, Chen T, Cheng Y, et al. PARP1 is upregulated by hyperglycemia via N6-methyladenosine modification and promotes diabetic retinopathy. Discovery Med. (2022) 34:115–29.
149. Yang Z, Hu H, Zou Y, Luo W, Xie L, You Z. Mir-7 reduces high glucose induced-damage via hoxb3 and pi3k/akt/mtor signaling pathways in retinal pigment epithelial cells. Curr Mol Med. (2020) 20:372–8. doi: 10.2174/1566524019666191023151137
150. Rosa MD, Distefano G, Gagliano C, Rusciano D, Malaguarnera L. Autophagy in diabetic retinopathy. Curr Neuropharmacol. (2016) 14:810–25. doi: 10.2174/1570159x14666160321122900
151. Huang C, Qi P, Cui H, Lu Q, Gao X. Circfat1 regulates retinal pigment epithelial cell pyroptosis and autophagy via mediating M6a reader protein ythdf2 expression in diabetic retinopathy. Exp Eye Res. (2022) 222:109152. doi: 10.1016/j.exer.2022.109152
152. Luo R, Li L, Han Q, Fu J, Xiao F. HAGLR, stabilized by M6a modification, triggers pten-akt signaling cascade-mediated rpe cell pyroptosis via sponging mir-106b-5p. J Biochem Mol Toxicol. (2023) 38:e23596. doi: 10.1002/jbt.23596
153. Elafros MA, Andersen H, Bennett DL, Savelieff MG, Viswanathan V, Callaghan BC, et al. Towards prevention of diabetic peripheral neuropathy: clinical presentation, pathogenesis, and new treatments. Lancet Neurol. (2022) 21:922–36. doi: 10.1016/S1474-4422(22)00188-0
154. Lin Y, Wei Y, Wei Y, Yu H, Zhang W, Li C, et al. Dexmedetomidine alleviates oxidative stress and mitochondrial dysfunction in diabetic peripheral neuropathy via the microrna-34a/sirt2/S1pr1 axis. Int Immunopharmacol. (2023) 117:109910. doi: 10.1016/j.intimp.2023.109910
155. Badhe RV, Nipate SS. Low-intensity current (Lic) stimulation of subcutaneous adipose derived stem cells (Adscs) - a missing link in the course of lic based wound healing. Med Hypotheses. (2019) 125:79–83. doi: 10.1016/j.mehy.2019.02.039
156. Zhang W, Bai X, Zhao B, Li Y, Zhang Y, Li Z, et al. Cell-free therapy based on adipose tissue stem cell-derived exosomes promotes wound healing via the pi3k/akt signaling pathway. Exp Cell Res. (2018) 370:333–42. doi: 10.1016/j.yexcr.2018.06.035
157. Zhou J, Wei T, He Z. Adscs enhance vegfr3-mediated lymphangiogenesis via mettl3-mediated vegf-C M(6)a modification to improve wound healing of diabetic foot ulcers. Mol Med. (2021) 27:146. doi: 10.1186/s10020-021-00406-z
158. Wang J, Wang X, Chen F, Ning Q, Liu Y, Zhu Y, et al. N6-methyladenosine modification of lnccckar-5 regulates autophagy in human umbilical cord mesenchymal stem cells by destabilizing lmna and inhibits diabetic wound healing. J Invest Dermatol. (2024) 144:1148–60.e15. doi: 10.1016/j.jid.2023.11.023
159. Jiao Y, Wang S, Wang X, Yin L, Zhang YH, Li YZ, et al. The M(6)a reader ythdc2 promotes sirt3 expression by reducing the stabilization of kdm5b to improve mitochondrial metabolic reprogramming in diabetic peripheral neuropathy. Acta Diabetol. (2023) 60:387–99. doi: 10.1007/s00592-022-01990-0
160. Deng X, Qing Y, Horne D, Huang H, Chen J. The roles and implications of rna M6a modification in cancer. Nat Rev Clin Oncol. (2023) 20:507–26. doi: 10.1038/s41571-023-00774-x
161. Qiu L, Jing Q, Li Y, Han J. Rna modification: mechanisms and therapeutic targets. Mol Biomedicine. (2023) 4:25. doi: 10.1186/s43556-023-00139-x
162. Peng S, Xiao W, Ju D, Sun B, Hou N, Liu Q, et al. Identification of entacapone as a chemical inhibitor of FTO mediating metabolic regulation through FOXO1. Sci Transl Med. (2019) 11:eaau7116. doi: 10.1126/scitranslmed.aau7116
163. Li H, Song Y, He Z, Chen X, Wu X, Li X, et al. Meclofenamic acid reduces reactive oxygen species accumulation and apoptosis, inhibits excessive autophagy, and protects hair cell-like hei-oc1 cells from cisplatin-induced damage. Front Cell Neurosci. (2018) 12:139. doi: 10.3389/fncel.2018.00139
164. Jung HR, Lee J, Hong S-P, Shin N, Cho A, Shin D-J, et al. Targeting the M6a rna methyltransferase mettl3 attenuates the development of kidney fibrosis. Exp Mol Med. (2024) 56:355–69. doi: 10.1038/s12276-024-01159-5
165. Wu R, Yao Y, Jiang Q, Cai M, Liu Q, Wang Y, et al. Epigallocatechin gallate targets fto and inhibits adipogenesis in an mrna M6a-ythdf2-dependent manner. Int J Obes. (2018) 42:1378–88. doi: 10.1038/s41366-018-0082-5
166. Chen Y, Wu R, Chen W, Liu Y, Liao X, Zeng B, et al. Curcumin prevents obesity by targeting traf4-induced ubiquitylation in M6a-dependent manner. EMBO Rep. (2021) 22:e52146. doi: 10.15252/embr.202052146
167. Zhang L, Qi Y, Aluo Z, Liu S, Zhang Z, Zhou L. Betaine increases mitochondrial content and improves hepatic lipid metabolism. Food Funct. (2019) 10:216–23. doi: 10.1039/c8fo02004c
Keywords: N6-methyladensine, writer, eraser, reader, DM, microvascular complications
Citation: Wang Y, Zou J and Zhou H (2024) N6-methyladenine RNA methylation epigenetic modification and diabetic microvascular complications. Front. Endocrinol. 15:1462146. doi: 10.3389/fendo.2024.1462146
Received: 09 July 2024; Accepted: 20 August 2024;
Published: 04 September 2024.
Edited by:
Yanshan Dai, Bristol Myers Squibb, United StatesReviewed by:
Shengze Yao, Massachusetts General Hospital and Harvard Medical School, United StatesSensen Lin, University of California, San Francisco, United States
Copyright © 2024 Wang, Zou and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hua Zhou, aHVhemhvdV9jbXVAMTYzLmNvbQ==
†These authors have contributed equally to this work and share first authorship
 Yuanyuan Wang
Yuanyuan Wang Jiayun Zou2†
Jiayun Zou2† Hua Zhou
Hua Zhou