
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Endocrinol. , 09 January 2025
Sec. Cardiovascular Endocrinology
Volume 15 - 2024 | https://doi.org/10.3389/fendo.2024.1444940
This article is part of the Research Topic Epigenetic Modifications and Cardiovascular Diseases View all articles
Cardiac hypertrophy is an adaptive response to pressure or volume overload such as hypertension and ischemic heart diseases. Sustained cardiac hypertrophy eventually leads to heart failure. The pathophysiological alterations of hypertrophy are complex, involving both cellular and molecular systems. Understanding the molecular events that inhibit or repress cardiac hypertrophy may help identify novel therapeutic strategies. Increasing evidence has indicated that extracellular vesicle (EV)-derived microRNAs (miRNAs) play a significant role in the development and progression of cardiac hypertrophy. In this review, we briefly review recent advancements in EV research, especially on biogenesis, cargoes and its role in cardiac hypertrophy. We then describe the latest findings regarding EV-derived miRNAs, highlighting their functions and regulatory mechanisms in cardiac hypertrophy. Finally, the potential role of EV-derived miRNAs as targets in the diagnosis and treatment of cardiac hypertrophy will be discussed.
Cardiac hypertrophy serves as a compensatory mechanism to address the reduced cardiac output and represents a restricted form of adjustment in response to increased cardiac stress. Pathological cardiac hypertrophy as a well-recognized risk factor for cardiovascular diseases (CVDs) (1, 2), generally accompanies cardiomyocyte enlargement and ventricular remodeling (3, 4). Subsequently, long-term pathological factors contributed to extensive cardiac remodeling, including the development of cardiac fibrosis, hypertrophy, and ultimately heart failure (5). The heart is composed of a diverse combination of cell types, such as cardiomyocytes, fibroblasts, endocardial and epicardial cells, inflammatory cells, and immune cells. These cellular components within the cardiac system communicate extensively to ensure proper cardiac function through direct cellular interaction or paracrine signaling (6). Previous studies have documented the existence of intercellular communication between cardiomyocytes and noncardiomyocytes during the development of cardiac hypertrophy (7–10). This intercellular communication mainly occurs through direct cell-to-cell interaction, specific molecules, or extracellular vesicles (EVs) (11–13).
EVs, which are membranous particles composed of lipid bilayers, are released by various cell types in organisms ranging from bacteria to humans and plants (14–16). Based on their dimensions, EVs have been categorized into three groups comprising exosomes (sizes of 30–150 nm), microvesicles (sizes of 100–1000 nm), and apoptotic bodies (sizes of 50 nm–10 μm) (17, 18). Being the primary classifications, exosomes and microvesicles share the same membrane orientation that is identical to the cell surface, but they vary in terms of size and cargo compositions (RNAs, proteins, lipids, or metabolites) (19). The composition of these functional components may vary based on the cellular source and specific pathophysiological conditions during EV packaging and release (20). EVs have been employed as potential signaling mediators, biomarkers, and potential therapeutic agents for cardiovascular physiology and pathology (21). The abundance of evidence suggests that EVs play a pivotal role in various cardiovascular processes, encompassing both physiological and pathological aspects. These include the regulation of angiogenesis and blood pressure, cardiomyocyte hypertrophy, apoptosis/survival, and cardiac fibrosis (22–26). In the past ten years, EVs have gained increasing recognition for their potential roles as important autocrine and paracrine intercellular communicators. They deliver molecular cargoes from source cells to neighboring cells or even to distant organs and influence the programming of the cardiac microenvironment (27, 28). MicroRNAs (miRNAs) are small noncoding RNAs, which are 22 to 25 nucleotides in length. They can bind to the 3′ untranslated regions of their target messenger RNAs (mRNAs), leading to mRNA degradation or translational repression (29). Direct regulation by miRNAs occurs in over 60% of the protein-coding genes (30). Furthermore, the miRNAs present in cardiovascular exosomes have been shown to regulate target genes within recipient cardiac cells, thereby influencing both physiological and pathophysiological processes (31). In this review, we summarize the current understanding of the roles of EVs in cardiac diseases, with a particular emphasis on pathological cardiac hypertrophy. Specifically, we will attempt to provide an overview of the potential values of EVs-derived miRNAs in the field of pathological cardiac hypertrophy as therapeutic agents and biomarkers.
The generation of exosomes is linked to the endosomal network. Exosomes are generated through the inward budding of the endosome’s limiting membrane into its lumen, as intraluminal vesicles (ILV) (32, 33). Further continued inward budding of the endosomal membrane leads to the formation of the multivesicular body (MVB). Finally, the MVBs merge with the cellular membrane and release the ILVs as exosomes into the extracellular environment. Alternatively, the MVBs can combine with lysosomes inside the cell and release the ILVs for either degradation or recycling within the cellular context. Both ESCRT-dependent and ESCRT-independent mechanisms contribute to the packaging of cargoes into endosomes and the formation of exosomes in multivesicular bodies (MVBs) (34–36). The ESCRT-dependent pathway, consisting of subcomplexes ESCRT-0, ESCRT-I, ESCRT-II, and ESCRT-III, is responsible for the sorting of various nucleic acids and proteins. ESCRT-dependent pathway also has been shown to regulate the inward budding of intraluminal vesicles (ILVs) into the lumen of endosomes and the final forming of MVBs (37, 38). An alternative ESCRT-independent pathway requires the involvement of the cellular Golgi apparatus and the tetraspanin family proteins (CD63, CD81, CD82 and CD9), facilitating the formation of membrane microdomains and sorting cargo to MVBs (39). Although exosome biogenesis is commonly classified as ESCRT-dependent or ESCRT-independent, these pathways may not be entirely distinct. Whereas, the generation of microvesicles occurs within the extracellular space through direct outward budding of the cell’s plasma membrane, which exhibits key characteristics of stepwise vesiculation observed in exosome formation and employs the same machinery (40). Exosomes and microvesicles are considered dynamic carriers of cellular communication owing to their release and uptake by living cells. The formation of apoptotic bodies, on the contrary, is generated from the cell membrane, as a result of cellular disassembly during the apoptosis (41).
EVs transport both membrane-bound and soluble cargoes, which are characterized by distinct compositions of lipids, proteins, and nucleic acids. The lipid composition of their membranes exhibits similarities to raft microdomains, displaying higher levels of cholesterol, sphingomyelin, phosphatidylserine, and ceramide compared to the plasma membrane (42). It is widely recognized that EVs possess a rich assortment of proteins, encompassing those derived from the plasma membrane, cytosol, cytoskeleton, and proteins engaged in vesicle trafficking (43). Exosomal protein markers encompass the ESCRT machinery proteins (Alix, TSG101, and VSP40), Syntenin-1 (44), heat-shock proteins (Hsp20, Hsp60, Hsp70, and Hsp90), transmembrane tetraspanins (CD62, CD63, CD9, and CD81) (45), and blood circulating EVs-derived cytokines (IL-1b, IL-6, TGF-β, and TNF) (46, 47). EV-carried cell-type specific proteins, especially membrane surface proteins, have been very useful for identifying and isolating cell-type specific EVs (48). The cargo nucleic acids possess significant potential as efficient facilitators of inter-tissue communication, despite indications that the levels of RNAs in EVs are generally modest (49). The transfer of miRNAs and mRNAs to target cells through EV-mediated mechanisms has been extensively acknowledged for its functional importance (50, 51). Other types of RNA in EVs are tRNAs, tRNA fragments, fragmented mRNAs, long non-coding RNAs (lncRNAs), and ribosomal RNAs, mitochondrial RNAs. EVs traverse the extracellular fluid, attach to the matrix surrounding cells and intercellular junctions, migrate towards adjacent tissue regions, or gain access to major body fluids to reach distant organs (52, 53). In recent years, EVs have been widely acknowledged for their crucial involvement in intercellular signaling, facilitating the transfer of various biomolecules such as lipids, DNA, mRNA, miRNA, siRNA, and lncRNA to recipient cells. Generally, the transfer of biological signals from EVs to target cells occurs through various mechanisms that rely on the composition of proteins and lipids present on both the surfaces of EVs and target cells. The interaction between EVs and target cells occurs via membrane-bound ligand-receptor pairs, which subsequently initiate intracellular signaling pathways. Target cells can internalize EVs via diverse endocytic pathways, including macropinocytosis, phagocytosis, clathrin-mediated endocytosis, caveolin-dependent endocytosis, and caveolin-independent endocytosis (54).
Pathological cardiac hypertrophy is distinguished by the cardiomyocytes’ enlargement, the existence of interstitial fibrosis, and insufficient blood supply to the heart. These factors collectively contribute to the development of cardiac dilation, impaired systolic and diastolic function, and ultimately heart failure (3, 4). Cardiac hypertrophy is accompanied by disrupted electrophysiology and metabolism during adverse remodeling. While cardiac fibroblasts transform into activated myofibroblasts, exhibiting proliferation and excessive production of extracellular matrix (3, 55).
EVs play important roles in intercellular communication among nearby and distant cells, including cardiac cells. They have been implicated in the regulation of various processes such as cardiomyocyte hypertrophy, apoptosis, cardiac fibrosis, and angiogenesis (56–58). EVs can be separated from various cell types present in the heart, including cardiac fibroblasts, cardiomyocytes, endothelial cells, vascular smooth muscle cells, and Mesenchymal stem cells (MSCs) (59–71).
As the predominant cell type in the heart, Cardiomyocytes play crucial roles in various pathological processes, including cardiac hypertrophy, angiogenesis, cardiac fibrosis, autophagy, and apoptosis (72). The release of specific exosomes by cardiomyocytes has been investigated in the context of pathological cardiac hypertrophy (73, 74). Furthermore, the production and release of EVs originating from cardiomyocytes are impacted by various factors. For example, Hsp20 facilitated the initiation of exosome formation and their release from cardiomyocytes through its interaction with Tsg101, a key regulator of exosome biogenesis (75). Besides, cardiomyocytes can selectively modify the contents of their exosomes and enhance the exosomes’ release in response to various stressors such as glucose deficiency, oxygen deprivation, inflammatory conditions, physical trauma, or elevated angiotensin II levels (76–78). Specific proteins, including two members of the heat shock protein family (Hsp20 and Hsp90), can be secreted by cardiomyocytes to facilitate the restoration of cardiac function in the context of pathological cardiac hypertrophy (79–82). One research study demonstrated that diabetic cardiomyocytes can release exosomes with lower levels of Hsp20. Conversely, the upregulation of Hsp20 has been found to mitigate cardiac dysfunction, hypertrophy, apoptosis in cardiomyocytes, fibrosis in the heart tissue, and decreased density of microvessels induced by STZ (75). Additionally, previous research has indicated that Hsp90 plays a pivotal role in modulating ventricular hypertrophy by activating the MAPK pathway, NF-κB pathway, STAT-3 pathway, and stabilizing HIF-1 alpha. Exosomal Hsp90 derived from cardiomyocytes, along with secreted IL-6, participates in the activation of STAT-3 signaling within cardiac fibroblasts, leading to enhanced synthesis and accumulation of collagen during cardiac hypertrophy (81).
On the other hand, exosomes derived from fibroblasts have a crucial function in facilitating communication between cardiomyocytes and fibroblasts throughout the hypertrophic procedure (83–87). The administration of angiotensin II to fibroblasts was found to enhance the release of exosomes, thereby inducing cardiomyocyte hypertrophy in vitro through the activation of Akt and mitogen-activated protein kinases (MAPKs), as well as increased expression of the renin-angiotensin system (RAS) in cardiomyocytes (63). The study mentioned above shows that the fibroblast-derived exosome proteins osteopontin (Spp1) and epidermal growth factor receptor (EGFR) can trigger the PI3K/Akt and MAPK pathways, resulting in the upregulation of RAS in cardiomyocytes.
MiRNAs, as small endogenous oligonucleotides ranging from 21 to 25 nucleotides in length, play a crucial role in the post-transcriptional regulation of genes by binding to or inhibiting target mRNAs (88, 89). MiRNAs genes are initially transcribed by RNA polymerase II as primary miRNAs (pri-miRNA), the long transcripts containing either non-coding or coding hairpins (90). Then, pri-miRNAs are enzymatically processed to form precursor miRNA (pre-miRNA) by the functional Microprocessor complex, comprising RNase III Drosha and DiGeorge syndrome critical region gene 8 (DGCR8) (91). The pre-miRNA is subsequently exported to the cytoplasm by exportin 5 (Exp-5) and is finally processed into a mature miRNA by the RNase III Dicer (91). Non-canonical miRNA biogenesis, including the Drosha-independent and the Dicer-independent pathways, are also observed (92).
It is well known that miRNA-mediated transcriptional and post-transcriptional gene regulation within the cells. Additionally, miRNAs can be released from cells into the circulation in EVs. Recently, Garcia-Martin et al. reported that that miRNAs possess sorting sequences determined their EV secretion or cellular retention (93). However, the mechanisms by which miRNAs are secreted into EVs or retained in cells not well understood and require further investigation.
Altered expression of miRNAs has been linked to cardiovascular disease conditions, such as heart failure and cardiac hypertrophy (94–98). For example, Bernardo et al. reported that inhibition of miR-34a could attenuate cardiac dysfunction in a mouse model with pre-existing pathological hypertrophy (99). In addition, miRNA expression analysis in the human heart found a correlation between downregulated miRNAs in cardiospheres/cardiosphere-derived cells and patient age (100). Apart from these proteins mentioned above aforementioned proteins, specialized miRNAs-containing EVs can be released by cardiomyocytes to facilitate the restoration of cardiac function in cases of pathological hypertrophy (101–103). Furthermore, compelling evidence suggests that in pathological conditions, miRNAs are secreted from various cell types, including fibroblasts, endothelial cells, stem cells, and immune cells (104). These miRNAs are then transferred to cardiomyocytes through EV-mediated trafficking (Figure 1), directly promoting the hypertrophic growth of cardiomyocytes (86, 105). EV-derived miRNAs that have been found to regulate cardiac hypertrophy are shown in Table 1.
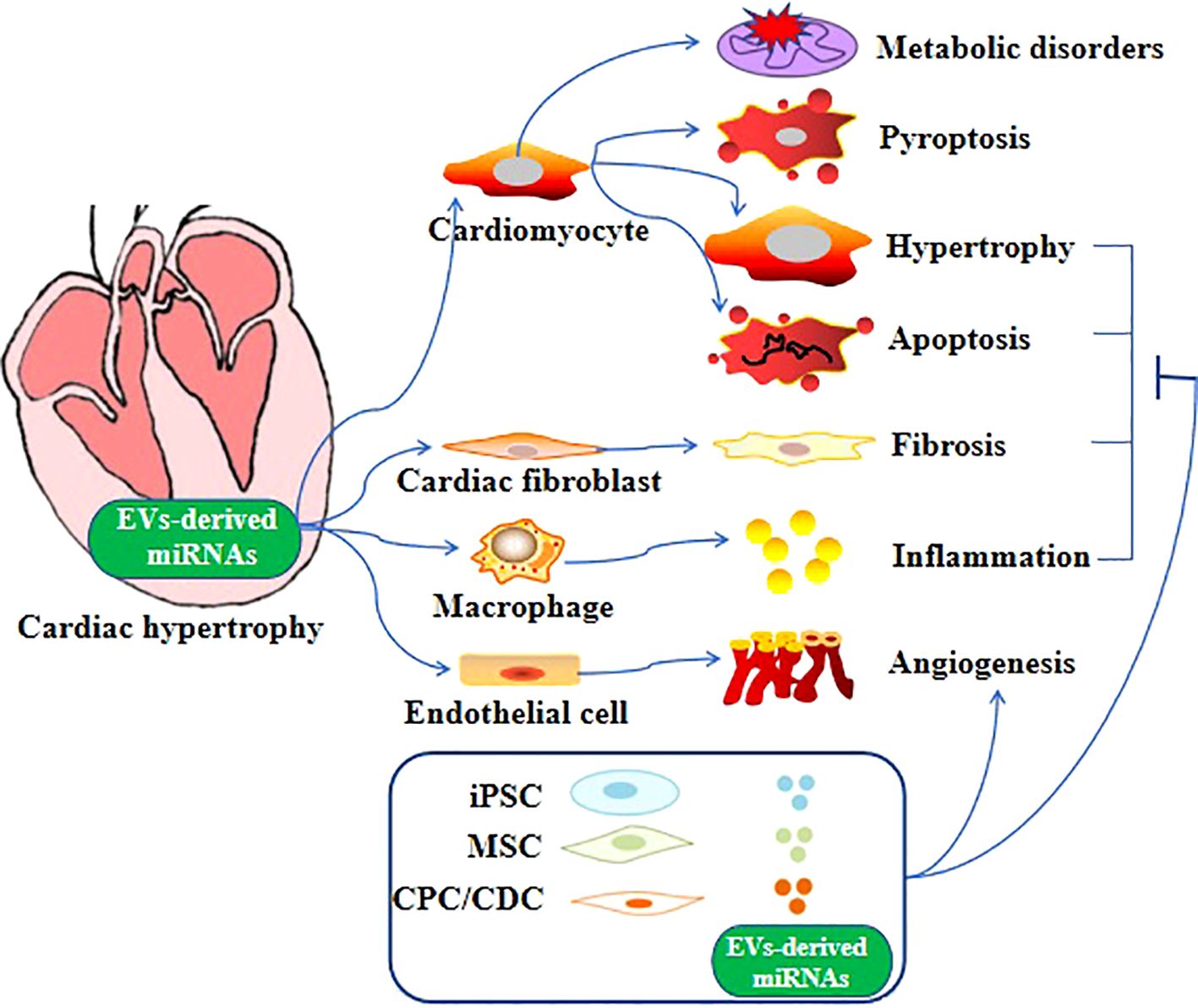
Figure 1. Role of EV-derived miRNAs in cardiac hypertrophy. Different EV-derived miRNAs cause cardiac hypertrophy through different mechanisms and even multiple mechanisms.
It has been found that EVs can be transferred from tubular epithelial cells to cardiomyocytes and facilitate the development of cardiomyocyte hypertrophy when renal tubular cells are treated with TGF-b1 (106). However, this effect could be eliminated by a miR-21 inhibitor, suggesting that miR-21 derived from EVs can be transmitted to recipient cardiomyocytes and contribute to cardiomyocyte hypertrophy. Through further investigation, Jia and colleagues have substantiated that the presence of miR-21 in EVs can induce cardiomyocyte hypertrophy by targeting SORBS2 and ENH. Moreover, the investigation conducted by Chuppa et al. revealed that inhibition of miR-21 safeguards rats with 5/6 nephrectomy against the progression of left ventricular hypertrophy and deterioration in left ventricular function (107). It has also been found that the pericardial fluid of mice with induced hypertrophy through transverse aortic constriction exhibited an elevated level of subtypes of miR-21 (miR-21-3p, miR-21*) compared to sham-operated mice. Additionally, the inhibition of using miR-21-3p antagomir effectively prevented Ang II-induced cardiac hypertrophy (59). In their study, Bang et al. revealed the selective enrichment of miR-21-3p in fibroblast-derived exosomes and emphasized that factors such as temperature and actin dynamics are crucial for cardiomyocyte uptake of exosomes. Upon internalization into cardiomyocytes, the presence of miR-21-3p derived from exosomes resulted in a significant augmentation of cardiomyocyte cell size (108). Specifically, previous studies identified that exosomal miR-21-3p is transported into cardiomyocytes, where it targets SORBS2 and PDLIM5 (PDZ and LIM domain 5), contributing to cardiac hypertrophy (59, 109). However, many important questions remain elusive. For example, multiple types of pro-fibrotic cells are involved in the development of fibroblast during cardiomyopathy, it is therefore essential to explore the effects of exosomes on such a process of cardiac fibrosis (110). In addition, Yan et al. investigated the functional role of miR-21-3p in cardiac hypertrophy and demonstrated that its overexpression significantly mitigates TAC (transverse aortic constriction)-induced cardiac hypertrophy and Ang II-induced cardiac hypertrophy by suppressing HDAC8 expression to activate the Akt/Gsk3b pathway (111).
Previous studies by Tian et al. demonstrated that there was an increase in the expression levels of miRNA-27a in both infarcted heart tissue and circulating samples in rat models of chronic heart failure (112). They observed a significant upregulation of miR-27a in cultured cardiac fibroblasts following proinflammatory stimulation. miR-27a, as the predominant miRNA contained in cardiac fibroblast-derived EVs, was selectively incorporated into EVs and released into the surrounding environment. These EVs were then taken up by cardiomyocytes, leading to increased oxidative stress and expression of genes associated with hypertrophy in these cells by targeting Nrf2 signaling. In their subsequent investigations, Tian et al. made the discovery that miR-27a* (the passenger strand of miR-27a) functions as an additional star miRNA, which can exacerbate cardiac hypertrophy in a chronic heart failure model induced by myocardial infarction. In the presence of Ang II stimulation, cardiac fibroblasts exhibited an increase in miR-27a* expression (113). Subsequently, miR-27a* was selectively encapsulated within EVs and then released into the extracellular space. miR-27a*-enriched EVs were then internalized by cardiac myocytes, leading to the development of a hypertrophic phenotype through differentially targeting Nrf2/ARE signaling pathway and PDLIM5. It has also been discovered that the upregulation of miR-27a-3p can serve as a diagnostic indicator for cardiac hypertrophy in an in vitro model of cardiac hypertrophy by treating H9c2 cardiomyocytes with Ang II, as well as in an in vivo model achieved through chronic infusion of Ang II into mice. In addition, the upregulation of miR-27a-3p promotes cardiac hypertrophy by targeting NOVA1 (neurooncological ventral antigen 1) (114). Moreover, pericardial adipose tissue derived exosomal miR-27a-3p regulated myocardial hypertrophy and fibrosis via AMPK/PINK1/Parkin signaling (115).
MiR-29a-containing exosomes derived from MSCs exhibited a significant cardioprotective effect by preventing pathological remodeling of the myocardium and maintaining heart function under conditions of pressure overload induced by TAC (116). In addition, cardiomyocyte-derived exosomal miR-29a inhibited the angiogenic ability of cardiac microvascular endothelial cell (CMEC) by targeting vascular endothelial growth factor (VEGFA) (117). Moreover, exosomal-miR-29 was identified as crucial targets from exercised hearts, which can facilitate cardiomyocyte growth and physiologic hypertrophy (118).
MiR-126 plays a key role in modulating cardiac function. Chen et al. reported that low expression level of heart miR-126 was associated with cardiomyocyte hypertrophy, fibrosis, and inflammation. They further found that miR-126 was highly enriched in exosomes derived from endothelial cells. Meanwhile, exosome-enriched miR-126 inhibited cardiomyocyte hypertrophy by targeting vascular cell adhesion protein-1 (VCAM-1) and monocyte chemotactic protein-1 (MCP-1) (119). Moreover, previous study revealed that the expression of miR-126 was significantly decreased in exosomes derived from diabetic Goto-Kakizaki (GK) cardiomyocytes. Of interest, exosomal miR-126 derived from healthy cardiomyocytes can promoted normal angiogenesis (120). Previous study also observed that exosomal miR-126 derived from CD34+ peripheral blood mono-nuclear cells (PBMCs) exhibits pro-angiogenic effects. Moreover, pro-angiogenic effects of CD34+ PBMCs were blocked by anti-miRNA-126 treatment (120).
Previously recognized as a suppressor of innate immune responses, miR-146a exerts its regulatory effects through direct interactions with interleukin-1 receptor-associated kinase 1 (IRAK1) and TNF receptor-associated factor 6 (TRAF6) (121). Its roles in cardiac pathophysiology have been demonstrated to exhibit variability across different models of heart disease (122, 123). In a recent investigation, it was observed that miR-146a contributes to the development of cardiac dysfunction in maladaptive hypertrophy by reducing SUMO1 expression and regulating Ca2+ cycling. MiR-146a is released from transdifferentiated fibroblasts and subsequently transferred into cardiomyocytes through EV-mediated trafficking in the failing heart (86). In addition, miR-146a-enriched exosomes released from endothelial cells or fibroblasts contribute to improve contractile function in cardiomyocytes by targeting Erbb4, Notch1, and Irak1 (124). At the same time, cardiosphere-derived exosomal miR-146a-5p was associated with myocardial repair in pediatric dilated cardiomyopathy (125). Moreover, exosomal miR-146a could also serve as a potential biomarker for future heart failure diagnosis (126).
In the case of the uremic heart, macrophage-derived miR-155–containing exosomes could enhance both pyroptosis and hypertrophy in the uremic cardiomyopathy model (105). Wang et al. found that miR-155 was synthesized and loaded into exosomes in increased infiltration of macrophages in a uremic heart. The fusion of exosomes with the plasma membrane results in the liberation of miR-155 into the cytosol, leading to the inhibition of translation for FoxO3a in cardiomyocytes. Ultimately, exosomes carrying miR-155 from macrophages were found to induce pyroptosis in cardiomyocytes and contribute to the development of uremic cardiomyopathy symptoms such as cardiac hypertrophy and fibrosis. This effect was achieved through direct inhibition of FoxO3a in uremic mice. Furthermore, inhibition of miR-155 using a specific inhibitor or the use of mice lacking miR-155 demonstrated that FoxO3a levels were partially recovered and reduced cardiomyocyte pyroptosis, hypertrophy, and fibrosis in uremic mice. Moreover, previous studies have suggested that exosomes derived from macrophages containing miR-155 act as a paracrine modulator of fibroblast proliferation and inflammation. This modulation results in the inhibition of fibroblast proliferation and an increase in fibroblast inflammation, ultimately leading to compromised cardiac repair following myocardial infarction (127). As it is widely acknowledged, the pathways leading to cardiac hypertrophy are closely linked to the pathogenesis of myocardial infarction. Intriguingly, the activation of the MAPK pathway may be triggered by miR-155-enriched exosomes derived from Ang II-induced hypertrophic cardiomyocytes, thereby inducing inflammation in macrophages (128). Taken together, microRNA-155, derived from EVs, plays a crucial role in facilitating communication between macrophages and fibroblasts or cardiomyocytes during the development of pathological cardiac hypertrophy. Relevant prior studies had shown that the activation of the Jaird2 signaling pathway in cardiomyocytes was facilitated by miR-155, leading to cardiac hypertrophy (129). Inhibition of endogenous Jarid2 partially ameliorated the impact of miR-155 deficiency in isolated cardiomyocytes.
A previous study conducted by Evgeniya et al. discovered that the administration of sacubitril/valsartan in human iCM (human-induced pluripotent stem cell-derived cardiomyocytes) led to a reduction in the expression of exosomal miR-181a. This decrease was found to contribute to the mitigation of myocardial fibrosis and hypertrophy (103). The downregulation of exosomal miR-181a was also observed by Evgeniya et al. in a rodent model of chronic myocardial infarction following sacubitril/valsartan treatment. Furthermore, the downregulation of exosomal miR-181a attenuates myocardial fibrosis and pathological hypertrophy, thereby restoring the chronic heart failure model in Sprague-Dawley rats. In Evgeniya et al. studies, the subsequent investigations utilized the chronic rodent myocardial injury model to explore the functionality of exosomal miR-181a. The validation was performed by employing miR-181a antagomir, which demonstrated favorable effects of exosomal miR-181a on cardiac function, volumes, and morphology. Other former relevant studies have demonstrated that miR-181a derived from cardiomyocytes possesses the potential to alleviate pathological hypertrophy in the heart by modulating multiple targets. For example, miR-181a as the newly discovered controller of cardiac remodeling, played a role in the regulation of autophagy, the p53-p21 pathway, PTEN/PI3K/AKT signaling, and the aldosterone-mineralocorticoid receptor (AldoMR) pathway in myocardial hypertrophy (130, 131).
As a response to pathological hypertrophy, miR-217 was released from cardiomyocytes. The exosomes derived from cardiomyocytes, which contain miR-217, have been observed to enhance fibroblast proliferation and potentially contribute to cardiac fibrosis through the modulation of PTEN (132). Moreover, Nie et al. observed a significant upregulation of miR-217 in the cardiac tissues of patients with chronic heart failure. In vivo, overexpression of miR-217 exacerbated pressure overload-induced cardiac hypertrophy and dysfunction by repressing PTEN expression (101).
Fang et al. demonstrated that activation of PPARγ signaling in adipocytes enhanced the expression and secretion of miR-200a by employing cocultures comprising adipocytes and cardiomyocytes. Exosomal delivery of miR-200a from adipocytes to cardiomyocytes played a role in the development of cardiomyocyte hypertrophy by reducing TSC1 levels and subsequently activating mTOR (133). In addition, the expression of various miRNAs, particularly miR-22, a crucial controller of cardiac hypertrophy and remodeling, could be influenced by exosomes released from induced pluripotent stem cells derived from cardiac fibroblasts (CF-iPSCs) (134). Exosomal miR-133a released from ischemic cardiomyocytes or cardiac progenitor cells (CPCs) exerts inhibitory effects on hypertrophy (132, 135). Wu et al. showed that serum exosomal miR-92b-5p is a potential biomarker for the diagnosis of acute heart failure caused by dilated cardiomyopathy (136). In addition, another study has revealed that miR-27b participated in cardiac hypertrophy via the TGF-β1-PPARγ pathway (137). There is evidence to suggest that exosomal miR-212/-132 family may also induce cardiac hypertrophy (138, 139). Moreover, exosomal miR-1, miR-222, and miR-208a released from skeletal muscle were also involved in the regulation of cardiac hypertrophy (138). Exercise altered circulating exosomal miRNAs expression, including miR-486-5p, miR-215-5p, miR-941, and miR-151b. These exosomal miRNAs modulated cardiac hypertrophy through IGF-1 signaling (140). MiR-199a induces cardiac hypertrophy through mammalian target of rapamycin (mTOR) signaling pathway (132). Exosomal miRNA-148a from CDCs improves TAC-induced myocardial hypertrophy via down-regulation of GP130, leading to the inhibition of STAT3/ERK1/2/AKT signaling pathway (141). MiR-4731 overexpression promoted cardiac hypertrophy by targeting sirtuin 2 (SIRT2) (142). Exosomal miR 331-5p is a critical regulator of hypertrophy and fibrosis (143).
Considerable advancements have been made in the past decade regarding our understanding of the biology associated with miRNAs derived from EVs and their pivotal role, particularly as proficient mediators of inter-tissue communication, in the physiology and pathological progression of cardiac hypertrophy (144–146). In this review, we focus on elucidating the biological functions and pivotal roles of extracellular EVs, encompassing their biogenesis, molecular cargoes, and facilitation of intercellular communication within the cardiovascular system. Furthermore, miRNAs derived from EVs exhibit promising potential in regulating pathological cardiac hypertrophy. These miRNAs-enriching EVs are involved in various functional behaviors of cardiac cells and intercellular communication, playing a critical role in the pathophysiological progression of cardiac hypertrophy.
The investigation into the biology of miRNAs derived from EVs holds significant potential for future therapeutic interventions aimed at cardioprotection, despite the limited research on EVs harboring miRNAs in cardiac hypertrophy. Further and in-depth research on the mechanism and role of miRNAs derived from EVs can contribute to the identification of novel interacting molecules and signal transduction pathways involved in cardiac hypertrophy, thereby offering innovative ideas and methodologies for its diagnosis and treatment. In the future, EV-derived miRNAs could potentially function as clinical indicators for cardiac hypertrophy owing to their distinctive inclusiveness. To establish EV-derived miRNAs as diagnostic or prognostic biomarkers in pathological cardiac hypertrophy, several technical challenges persist in this research field and more efforts are needed to conduct large randomized clinical trials.
HH: Formal Analysis, Validation, Writing – original draft. XW: Funding acquisition, Visualization, Writing – original draft. HY: Formal Analysis, Funding acquisition, Writing – review & editing. ZW: Conceptualization, Funding acquisition, Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. The present study was supported by the National Natural Science Foundation of China (grant nos., 82170296, 82060084 and 82260092), the Natural Science Foundation of Inner Mongolia Autonomous Region of China (grant no., 2024LHMS08020).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. van Berlo JH, Maillet M, Molkentin JD. Signaling effectors underlying pathologic growth and remodeling of the heart. J Clin Invest. (2013) 123:37–45. doi: 10.1172/JCI62839
2. Vega RB, Konhilas JP, Kelly DP, Leinwand LA. Molecular mechanisms underlying cardiac adaptation to exercise. Cell Metab. (2017) 25:1012–26. doi: 10.1016/j.cmet.2017.04.025
3. Nakamura M, Sadoshima J. Mechanisms of physiological and pathological cardiac hypertrophy. Nat Rev Cardiol. (2018) 15:387–407. doi: 10.1038/s41569-018-0007-y
4. Waldenström A, Ronquist G. Role of exosomes in myocardial remodeling. Circ Res. (2014) 114:315–24. doi: 10.1161/CIRCRESAHA.114.300584
5. Zhang P, Su J, Mende U. Cross talk between cardiac myocytes and fibroblasts: from multiscale investigative approaches to mechanisms and functional consequences. Am J Physiol Heart Circ Physiol. (2012) 303:H1385–96. doi: 10.1152/ajpheart.01167.2011
6. Talman V, Kivelä R. Cardiomyocyte-endothelial cell interactions in cardiac remodeling and regeneration. Front Cardiovasc Med. (2018) 5:101. doi: 10.3389/fcvm.2018.00101
7. Coles B, Fielding CA, Rose-John S, Scheller J, Jones SA, O’Donnell VB. Classic interleukin-6 receptor signaling and interleukin-6 trans-signaling differentially control angiotensin II-dependent hypertension, cardiac signal transducer and activator of transcription-3 activation, and vascular hypertrophy. vivo. Am J Pathol. (2007) 171:315–25. doi: 10.2353/ajpath.2007.061078
8. Meléndez GC, McLarty JL, Levick SP, Du Y, Janicki JS, Brower GL. Interleukin 6 mediates myocardial fibrosis, concentric hypertrophy, and diastolic dysfunction in rats. Hypertension. (2010) 56:225–31. doi: 10.1161/HYPERTENSIONAHA.109.148635
9. Ma F, Li Y, Jia L, Han Y, Cheng J, Li H, et al. Macrophage-stimulated cardiac fibroblast production of IL-6 is essential for TGF β/Smad activation and cardiac fibrosis induced by angiotensin II. PloS One. (2012) 7:e35144. doi: 10.1371/journal.pone.0035144
10. Kamo T, Akazawa H, Komuro I. Cardiac nonmyocytes in the hub of cardiac hypertrophy. Circ Res. (2015) 117:89–98. doi: 10.1161/CIRCRESAHA.117.305349
11. Poe AJ, Knowlton AA. Exosomes as agents of change in the cardiovascular system. J Mol Cell Cardiol. (2017) 111:40–50. doi: 10.1016/j.yjmcc.2017.08.002
12. Chen G, Xu C, Gillette TG, Huang T, Huang P, Li Q, et al. Cardiomyocyte-derived small extracellular vesicles can signal eNOS activation in cardiac microvascular endothelial cells to protect against Ischemia/Reperfusion injury. Theranostics. (2020) 10:11754–74. doi: 10.7150/thno.43163
13. Chen P, Wang L, Fan X, Ning X, Yu B, Ou C, et al. Targeted delivery of extracellular vesicles in heart injury. Theranostics. (2021) 11:2263–77. doi: 10.7150/thno.51571
14. Deatherage BL, Cookson BT. Membrane vesicle release in bacteria, eukaryotes, and archaea: a conserved yet underappreciated aspect of microbial life. Infect Immun. (2012) 80:1948–57. doi: 10.1128/IAI.06014-11
15. Robinson DG, Ding Y, Jiang L. Unconventional protein secretion in plants: a critical assessment. Protoplasma. (2016) 253:31–43. doi: 10.1007/s00709-015-0887-1
16. O’Brien K, Breyne K, Ughetto S, Laurent LC, Breakefield XO. RNA delivery by extracellular vesicles in mammalian cells and its applications. Nat Rev Mol Cell Biol. (2020) 21:585–606. doi: 10.1038/s41580-020-0251-y
17. Chen Q, Wang ZY, Chen LY, Hu HY. Roles of high mobility group box 1 in cardiovascular calcification. Cell Physiol Biochem. (2017) 42:427–40. doi: 10.1159/000477591
18. de Abreu RC, Fernandes H, da Costa Martins PA, Sahoo S, Emanueli C, Ferreira L. Native and bioengineered extracellular vesicles for cardiovascular therapeutics. Nat Rev Cardiol. (2020) 17:685–97. doi: 10.1038/s41569-020-0389-5
19. Zaborowski MP, Balaj L, Breakefield XO, Lai CP. Extracellular vesicles: Composition, biological relevance, and methods of study. Bioscience. (2015) 65:783–97. doi: 10.1093/biosci/biv084
20. de Boer C, Calder B, Blackhurst D, Marais D, Blackburn J, Steinmaurer M, et al. Analysis of the regenerative capacity of human serum exosomes after a simple multistep separation from lipoproteins. J Tissue Eng Regener Med. (2021) 15:63–77. doi: 10.1002/term.3155
21. Zhang Y, Liu Y, Liu H, Tang WH. Exosomes: biogenesis, biologic function and clinical potential. Cell Biosci. (2019) 9:19. doi: 10.1186/s13578-019-0282-2
22. de Couto G, Jaghatspanyan E, DeBerge M, Liu W, Luther K, Wang Y, et al. Mechanism of enhanced MerTK-dependent macrophage efferocytosis by extracellular vesicles. Arterioscler Thromb Vasc Biol. (2019) 39:2082–96. doi: 10.1161/ATVBAHA.119.313115
23. Eguchi S, Takefuji M, Sakaguchi T, Ishihama S, Mori Y, Tsuda T, et al. Cardiomyocytes capture stem cell-derived, anti-apoptotic microRNA-214 via clathrin-mediated endocytosis in acute myocardial infarction. J Biol Chem. (2019) 294:11665–74. doi: 10.1074/jbc.RA119.007537
24. Dekker M, Waissi F, van Bennekom J, Silvis MJM, Timmerman N, Bank IEM, et al. Plasma extracellular vesicle proteins are associated with stress-induced myocardial ischemia in women presenting with chest pain. Sci Rep. (2020) 10:12257. doi: 10.1038/s41598-020-69297-0
25. De Vita A, Liverani C, Molinaro R, Martinez JO, Hartman KA, Spadazzi C, et al. Lysyl oxidase engineered lipid nanovesicles for the treatment of triple negative breast cancer. Sci Rep. (2021) 11:5107. doi: 10.1038/s41598-021-84492-3
26. Düsing P, Zietzer A, Goody PR, Hosen MR, Kurts C, Nickenig G, et al. Vascular pathologies in chronic kidney disease: pathophysiological mechanisms and novel therapeutic approaches. J Mol Med (Berl). (2021) 99:335–48. doi: 10.1007/s00109-021-02037-7
27. Sluijter JP, Verhage V, Deddens JC, van den Akker F, Doevendans PA. Microvesicles and exosomes for intracardiac communication. Cardiovasc Res. (2014) 102:302–11. doi: 10.1093/cvr/cvu022
28. Poe AJ, Knowlton AA. Exosomes and cardiovascular cell-cell communication. Essays Biochem. (2018) 62:193–204. doi: 10.1042/EBC20170081
29. Yates LA, Norbury CJ, Gilbert RJ. The long and short of microRNA. Cell. (2013) 153:516–9. doi: 10.1016/j.cell.2013.04.003
30. Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. (2009) 19:92–105. doi: 10.1101/gr.082701.108
31. Bollini S, Smits AM, Balbi C, Lazzarini E, Ameri P. Triggering endogenous cardiac repair and regeneration via extracellular vesicle-mediated communication. Front Physiol. (2018) 9:1497. doi: 10.3389/fphys.2018.01497
32. Klumperman J, Raposo G. The complex ultrastructure of the endolysosomal system. Cold Spring Harb Perspect Biol. (2014) 6:a016857. doi: 10.1101/cshperspect.a016857
33. Frydrychowicz M, Kolecka-Bednarczyk A, Madejczyk M, Yasar S, Dworacki G. Exosomes - structure, biogenesis and biological role in non-small-cell lung cancer. Scand J Immunol. (2015) 81:2–10. doi: 10.1111/sji.12247
34. Hurley JH. ESCRT complexes and the biogenesis of multivesicular bodies. Curr Opin Cell Biol. (2008) 20:4–11. doi: 10.1016/j.ceb.2007.12.002
36. Jakhar R, Crasta K. Exosomes as emerging pro-tumorigenic mediators of the senescence-associated secretory phenotype. Int J Mol Sci. (2019) 20:2547. doi: 10.3390/ijms20102547
37. Kowal J, Tkach M, Théry C. Biogenesis and secretion of exosomes. Curr Opin Cell Biol. (2014) 29:116–25. doi: 10.1016/j.ceb.2014.05.004
38. Villarroya-Beltri C, Baixauli F, Gutiérrez-Vázquez C, Sánchez-Madrid F, Mittelbrunn M. Sorting it out: regulation of exosome loading. Semin Cancer Biol. (2014) 28:3–13. doi: 10.1016/j.semcancer.2014.04.009
39. van Niel G, Bergam P, Di Cicco A, Hurbain I, Lo Cicero A, Dingli F, et al. Apolipoprotein E regulates amyloid formation within endosomes of pigment cells. Cell Rep. (2015) 13:43–51. doi: 10.1016/j.celrep.2015.08.057
40. Colombo M, Raposo G, Théry C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol. (2014) 30:255–89. doi: 10.1146/annurev-cellbio-101512-122326
41. Lee Y, El Andaloussi S, Wood MJ. Exosomes and microvesicles: extracellular vesicles for genetic information transfer and gene therapy. Hum Mol Genet. (2012) 21:R125–34. doi: 10.1093/hmg/dds317
42. Skotland T, Sandvig K, Llorente A. Lipids in exosomes: Current knowledge and the way forward. Prog Lipid Res. (2017) 66:30–41. doi: 10.1016/j.plipres.2017.03.001
43. Yáñez-Mó M, Siljander PR, Andreu Z, Zavec AB, Borràs FE, Buzas EI, et al. Biological properties of extracellular vesicles and their physiological functions. J Extracell Vesicles. (2015) 4:27066. doi: 10.3402/jev.v4.27066
44. Kugeratski FG, Hodge K, Lilla S, McAndrews KM, Zhou X, Hwang RF, et al. Quantitative proteomics identifies the core proteome of exosomes with syntenin-1 as the highest abundant protein and a putative universal biomarker. Nat Cell Biol. (2021) 23:631–41. doi: 10.1038/s41556-021-00693-y
45. Raimondo F, Morosi L, Chinello C, Magni F, Pitto M. Advances in membranous vesicle and exosome proteomics improving biological understanding and biomarker discovery. Proteomics. (2011) 11:709–20. doi: 10.1002/pmic.201000422
46. Zhang HG, Liu C, Su K, Yu S, Zhang L, Zhang S, et al. A membrane form of TNF-alpha presented by exosomes delays T cell activation-induced cell death. J Immunol. (2006) 176:7385–93. doi: 10.4049/jimmunol.176.12.7385
47. Clayton A, Mitchell JP, Court J, Mason MD, Tabi Z. Human tumor-derived exosomes selectively impair lymphocyte responses to interleukin-2. Cancer Res. (2007) 67:7458–66. doi: 10.1158/0008-5472.CAN-06-3456
48. Loyer X, Zlatanova I, Devue C, Yin M, Howangyin KY, Klaihmon P, et al. Intra-cardiac release of extracellular vesicles shapes inflammation following myocardial infarction. Circ Res. (2018) 123:100–6. doi: 10.1161/CIRCRESAHA.117.311326
49. Chevillet JR, Kang Q, Ruf IK, Briggs HA, Vojtech LN, Hughes SM, et al. Quantitative and stoichiometric analysis of the microRNA content of exosomes. Proc Natl Acad Sci U.S.A. (2014) 111:14888–93. doi: 10.1073/pnas.1408301111
50. Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. (2007) 9:654–9. doi: 10.1038/ncb1596
51. Skog J, Würdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. (2008) 10:1470–6. doi: 10.1038/ncb1800
52. Mulcahy LA, Pink RC, Carter DR. Routes and mechanisms of extracellular vesicle uptake. J Extracell Vesicles. (2014) 3. doi: 10.3402/jev.v3.24641
53. Meldolesi J. Exosomes and ectosomes in intercellular communication. Curr Biol. (2018) 28:R435–44. doi: 10.1016/j.cub.2018.01.059
54. Bang C, Thum T. Exosomes: new players in cell-cell communication. Int J Biochem Cell Biol. (2012) 44:2060–4. doi: 10.1016/j.biocel.2012.08.007
55. Takeda N, Manabe I. Cellular interplay between cardiomyocytes and nonmyocytes in cardiac remodeling. Int J Inflam. (2011) 2011:535241. doi: 10.4061/2011/535241
56. Emanueli C, Shearn AI, Angelini GD, Sahoo S. Exosomes and exosomal miRNAs in cardiovascular protection and repair. Vascul Pharmacol. (2015) 71:24–30. doi: 10.1016/j.vph.2015.02.008
57. Deddens JC, Vrijsen KR, Colijn JM, Oerlemans MI, Metz CH, van der Vlist EJ, et al. Circulating extracellular vesicles contain miRNAs and are released as early biomarkers for cardiac injury. J Cardiovasc Transl Res. (2016) 9:291–301. doi: 10.1007/s12265-016-9705-1
58. Firoozi S, Pahlavan S, Ghanian MH, Rabbani S, Barekat M, Nazari A, et al. Mesenchymal stem cell-derived extracellular vesicles alone or in conjunction with a SDKP-conjugated self-assembling peptide improve a rat model of myocardial infarction. Biochem Biophys Res Commun. (2020) 524:903–9. doi: 10.1016/j.bbrc.2020.02.009
59. Bang C, Batkai S, Dangwal S, Gupta SK, Foinquinos A, Holzmann A, et al. Cardiac fibroblast-derived microRNA passenger strand-enriched exosomes mediate cardiomyocyte hypertrophy. J Clin Invest. (2014) 124:2136–46. doi: 10.1172/JCI70577
60. Barile L, Lionetti V, Cervio E, Matteucci M, Gherghiceanu M, Popescu LM, et al. Extracellular vesicles from human cardiac progenitor cells inhibit cardiomyocyte apoptosis and improve cardiac function after myocardial infarction. Cardiovasc Res. (2014) 103:530–41. doi: 10.1093/cvr/cvu167
61. Fertig ET, Gherghiceanu M, Popescu LM. Extracellular vesicles release by cardiac telocytes: electron microscopy and electron tomography. J Cell Mol Med. (2014) 18:1938–43. doi: 10.1111/jcmm.12436
62. Khan M, Nickoloff E, Abramova T, Johnson J, Verma SK, Krishnamurthy P, et al. Embryonic stem cell-derived exosomes promote endogenous repair mechanisms and enhance cardiac function following myocardial infarction. Circ Res. (2015) 117:52–64. doi: 10.1161/CIRCRESAHA.117.305990
63. Lyu L, Wang H, Li B, Qin Q, Qi L, Nagarkatti M, et al. A critical role of cardiac fibroblast-derived exosomes in activating renin angiotensin system in cardiomyocytes. J Mol Cell Cardiol. (2015) 89:268–79. doi: 10.1016/j.yjmcc.2015.10.022
64. Kervadec A, Bellamy V, El Harane N, Arakélian L, Vanneaux V, Cacciapuoti I, et al. Cardiovascular progenitor-derived extracellular vesicles recapitulate the beneficial effects of their parent cells in the treatment of chronic heart failure. J Heart Lung Transplant. (2016) 35:795–807. doi: 10.1016/j.healun.2016.01.013
65. Xiao J, Pan Y, Li XH, Yang XY, Feng YL, Tan HH, et al. Cardiac progenitor cell-derived exosomes prevent cardiomyocytes apoptosis through exosomal miR-21 by targeting PDCD4. Cell Death Dis. (2016) 7:e2277. doi: 10.1038/cddis.2016.181
66. Agarwal U, George A, Bhutani S, Ghosh-Choudhary S, Maxwell JT, Brown ME, et al. Experimental, systems, and computational approaches to understanding the microRNA-mediated reparative potential of cardiac progenitor cell-derived exosomes from pediatric patients. Circ Res. (2017) 120:701–12. doi: 10.1161/CIRCRESAHA.116.309935
67. Gallet R, Dawkins J, Valle J, Simsolo E, de Couto G, Middleton R, et al. Exosomes secreted by cardiosphere-derived cells reduce scarring, attenuate adverse remodelling, and improve function in acute and chronic porcine myocardial infarction. Eur Heart J. (2017) 38:201–11. doi: 10.1093/eurheartj/ehw240
68. Barile L, Cervio E, Lionetti V, Milano G, Ciullo A, Biemmi V, et al. Cardioprotection by cardiac progenitor cell-secreted exosomes: role of pregnancy-associated plasma protein-A. Cardiovasc Res. (2018) 114:992–1005. doi: 10.1093/cvr/cvy055
69. Gao L, Gregorich ZR, Zhu W, Mattapally S, Oduk Y, Lou X, et al. Large cardiac muscle patches engineered from human induced-pluripotent stem cell-derived cardiac cells improve recovery from myocardial infarction in swine. Circulation. (2018) 137:1712–30. doi: 10.1161/CIRCULATIONAHA.117.030785
70. Namazi H, Mohit E, Namazi I, Rajabi S, Samadian A, Hajizadeh-Saffar E, et al. Exosomes secreted by hypoxic cardiosphere-derived cells enhance tube formation and increase pro-angiogenic miRNA. J Cell Biochem. (2018) 119:4150–60. doi: 10.1002/jcb.26621
71. Zhu J, Lu K, Zhang N, Zhao Y, Ma Q, Shen J, et al. Myocardial reparative functions of exosomes from mesenchymal stem cells are enhanced by hypoxia treatment of the cells via transferring microRNA-210 in an nSMase2-dependent way. Artif Cells Nanomed Biotechnol. (2018) 46:1659–70. doi: 10.1080/21691401.2017.1388249
72. Stastna M, Chimenti I, Marbán E, Van Eyk JE. Identification and functionality of proteomes secreted by rat cardiac stem cells and neonatal cardiomyocytes. Proteomics. (2010) 10:245–53. doi: 10.1002/pmic.200900515
73. Garcia NA, Moncayo-Arlandi J, Sepulveda P, Diez-Juan A. Cardiomyocyte exosomes regulate glycolytic flux in endothelium by direct transfer of GLUT transporters and glycolytic enzymes. Cardiovasc Res. (2016) 109:397–408. doi: 10.1093/cvr/cvv260
74. Yang J, Yu X, Xue F, Li Y, Liu W, Zhang S. Exosomes derived from cardiomyocytes promote cardiac fibrosis via myocyte-fibroblast cross-talk. Am J Transl Res. (2018) 10:4350–66.
75. Wang X, Gu H, Huang W, Peng J, Li Y, Yang L, et al. Hsp20-mediated activation of exosome biogenesis in cardiomyocytes improves cardiac function and angiogenesis in diabetic mice. Diabetes. (2016) 65:3111–28. doi: 10.2337/db15-1563
76. Gupta S, Knowlton AA. HSP60 trafficking in adult cardiac myocytes: role of the exosomal pathway. Am J Physiol Heart Circ Physiol. (2007) 292:H3052–6. doi: 10.1152/ajpheart.01355.2006
77. Yu X, Deng L, Wang D, Li N, Chen X, Cheng X, et al. Mechanism of TNF-α autocrine effects in hypoxic cardiomyocytes: initiated by hypoxia inducible factor 1α, presented by exosomes. J Mol Cell Cardiol. (2012) 53:848–57. doi: 10.1016/j.yjmcc.2012.10.002
78. Gennebäck N, Hellman U, Malm L, Larsson G, Ronquist G, Waldenström A, et al. Growth factor stimulation of cardiomyocytes induces changes in the transcriptional contents of secreted exosomes. J Extracell Vesicles. (2013) 2. doi: 10.3402/jev.v2i0.20167
79. Lee KH, Jang Y, Chung JH. Heat shock protein 90 regulates IκB kinase complex and NF-κB activation in angiotensin II-induced cardiac cell hypertrophy. Exp Mol Med. (2010) 42:703–11. doi: 10.3858/emm.2010.42.10.069
80. Eschricht S, Jarr KU, Kuhn C, Lehmann L, Kreusser M, Katus HA, et al. Heat-shock-protein 90 protects from downregulation of HIF-1α in calcineurin-induced myocardial hypertrophy. J Mol Cell Cardiol. (2015) 85:117–26. doi: 10.1016/j.yjmcc.2015.05.018
81. Datta R, Bansal T, Rana S, Datta K, Datta Chaudhuri R, Chawla-Sarkar M, et al. Myocyte-derived Hsp90 modulates collagen upregulation via biphasic activation of STAT-3 in fibroblasts during cardiac hypertrophy. Mol Cell Biol. (2017) 37:e00611–16. doi: 10.1128/MCB.00611-16
82. Tamura S, Marunouchi T, Tanonaka K. Heat-shock protein 90 modulates cardiac ventricular hypertrophy via activation of MAPK pathway. J Mol Cell Cardiol. (2019) 127:134–42. doi: 10.1016/j.yjmcc.2018.12.010
83. Kakimoto Y, Ito S, Abiru H, Kotani H, Ozeki M, Tamaki K, et al. Sorbin and SH3 domain-containing protein 2 is released from infarcted heart in the very early phase: proteomic analysis of cardiac tissues from patients. J Am Heart Assoc. (2013) 2:e000565. doi: 10.1161/JAHA.113.000565
84. Duygu B, Da Costa Martins PA. miR-21: a star player in cardiac hypertrophy. Cardiovasc Res. (2015) 105:235–7. doi: 10.1093/cvr/cvv026
85. Roncarati R, Viviani Anselmi C, Losi MA, Papa L, Cavarretta E, Da Costa Martins P, et al. Circulating miR-29a, among other up-regulated microRNAs, is the only biomarker for both hypertrophy and fibrosis in patients with hypertrophic cardiomyopathy. J Am Coll Cardiol. (2014) 63:920–7. doi: 10.1016/j.jacc.2013.09.041
86. Oh JG, Watanabe S, Lee A, Gorski PA, Lee P, Jeong D, et al. miR-146a suppresses SUMO1 expression and induces cardiac dysfunction in maladaptive hypertrophy. Circ Res. (2018) 123:673–85. doi: 10.1161/CIRCRESAHA.118.312751
87. Cao X, Zhang Z, Wang Y, Shan W, Wang R, Mao S, et al. MiR-27a-3p/hoxa10 axis regulates angiotensin II-induced cardiomyocyte hypertrophy by targeting Kv4.3 expression. Front Pharmacol. (2021) 12:680349. doi: 10.3389/fphar.2021.680349
88. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. (2004) 116:281–97. doi: 10.1016/s0092-8674(04)00045-5
89. Pu M, Chen J, Tao Z, Miao L, Qi X, Wang Y, et al. Regulatory network of miRNA on its target: coordination between transcriptional and post-transcriptional regulation of gene expression. Cell Mol Life Sci. (2019) 76:441–51. doi: 10.1007/s00018-018-2940-7
90. Bhaskaran M, Mohan M. MicroRNAs: history, biogenesis, and their evolving role in animal development and disease. Vet Pathol. (2014) 51:759–74. doi: 10.1177/0300985813502820
91. Shang R, Lee S, Senavirathne G, Lai EC. microRNAs in action: biogenesis, function and regulation. Nat Rev Genet. (2023) 24:816–33. doi: 10.1038/s41576-023-00611-y
92. Yang JS, Lai EC. Alternative miRNA biogenesis pathways and the interpretation of core miRNA pathway mutants. Mol Cell. (2011) 43:892–903. doi: 10.1016/j.molcel.2011.07.024
93. Garcia-Martin R, Wang G, Brandão BB, Zanotto TM, Shah S, Kumar Patel S, et al. MicroRNA sequence codes for small extracellular vesicle release and cellular retention. Nature. (2022) 601:446–51. doi: 10.1038/s41586-021-04234-3
94. Thum T, Galuppo P, Wolf C, Fiedler J, Kneitz S, van Laake LW, et al. MicroRNAs in the human heart: a clue to fetal gene reprogramming in heart failure. Circulation. (2007) 116:258–67. doi: 10.1161/CIRCULATIONAHA.107.687947
95. Thum T, Gross C, Fiedler J, Fischer T, Kissler S, Bussen M, et al. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature. (2008) 456:980–4. doi: 10.1038/nature07511
96. Matkovich SJ, Van Booven DJ, Youker KA, Torre-Amione G, Diwan A, Eschenbacher WH, et al. Reciprocal regulation of myocardial microRNAs and messenger RNA in human cardiomyopathy and reversal of the microRNA signature by biomechanical support. Circulation. (2009) 119:1263–71. doi: 10.1161/CIRCULATIONAHA.108.813576
97. Tijsen AJ, Creemers EE, Moerland PD, de Windt LJ, van der Wal AC, Kok WE, et al. MiR423-5p as a circulating biomarker for heart failure. Circ Res. (2010) 106:1035–9. doi: 10.1161/CIRCRESAHA.110.218297
98. Ganesan J, Ramanujam D, Sassi Y, Ahles A, Jentzsch C, Werfel S, et al. MiR-378 controls cardiac hypertrophy by combined repression of mitogen-activated protein kinase pathway factors. Circulation. (2013) 127:2097–106. doi: 10.1161/CIRCULATIONAHA.112.000882
99. Bernardo BC, Gao XM, Tham YK, Kiriazis H, Winbanks CE, Ooi JY, et al. Silencing of miR-34a attenuates cardiac dysfunction in a setting of moderate, but not severe, hypertrophic cardiomyopathy. PloS One. (2014) 9:e90337. doi: 10.1371/journal.pone.0090337
100. Iannolo G, Sciuto MR, Cuscino N, Carcione C, Coronnello C, Chinnici CM, et al. miRNA expression analysis in the human heart: Undifferentiated progenitors vs. bioptic tissues-Implications for proliferation and ageing. J Cell Mol Med. (2021) 25:8687–700. doi: 10.1111/jcmm.16824
101. Nie X, Fan J, Li H, Yin Z, Zhao Y, Dai B, et al. miR-217 promotes cardiac hypertrophy and dysfunction by targeting PTEN. Mol Ther Nucleic Acids. (2018) 12:254–66. doi: 10.1016/j.omtn.2018.05.013
102. Yuan J, Liu H, Gao W, Zhang L, Ye Y, Yuan L, et al. MicroRNA-378 suppresses myocardial fibrosis through a paracrine mechanism at the early stage of cardiac hypertrophy following mechanical stress. Theranostics. (2018) 8:2565–82. doi: 10.7150/thno.22878
103. Vaskova E, Ikeda G, Tada Y, Wahlquist C, Mercola M, Yang PC. Sacubitril/valsartan improves cardiac function and decreases myocardial fibrosis via downregulation of exosomal miR-181a in a rodent chronic myocardial infarction model. J Am Heart Assoc. (2020) 9:e015640. doi: 10.1161/JAHA.119.015640
104. Gao R, Li X. Extracellular vesicles and pathological cardiac hypertrophy. Adv Exp Med Biol. (2023) 1418:17–31. doi: 10.1007/978-981-99-1443-2_2
105. Wang B, Wang ZM, Ji JL, Gan W, Zhang A, Shi HJ, et al. Macrophage-derived exosomal mir-155 regulating cardiomyocyte pyroptosis and hypertrophy in uremic cardiomyopathy. JACC Basic Transl Sci. (2020) 5:148–66. doi: 10.1016/j.jacbts.2019.10.011
106. Di J, Yang M, Zhou H, Li M, Zhao J. MicroRNA-21-containing microvesicles from tubular epithelial cells promote cardiomyocyte hypertrophy. Ren Fail. (2021) 43:391–400. doi: 10.1080/0886022X.2021.1891098
107. Chuppa S, Liang M, Liu P, Liu Y, Casati MC, Cowley AW, et al. MicroRNA-21 regulates peroxisome proliferator-activated receptor alpha, a molecular mechanism of cardiac pathology in Cardiorenal Syndrome Type 4. Kidney Int. (2018) 93:375–89. doi: 10.1016/j.kint.2017.05.014
108. Wu C, Arora P. MicroRNA passenger strand: orchestral symphony of paracrine signaling. Circ Cardiovasc Genet. (2014) 7:567–8. doi: 10.1161/CIRCGENETICS.114.000805
109. Zhang X, Sun S, Ren G, Liu W, Chen H. Advances in intercellular communication mediated by exosomal ncRNAs in cardiovascular disease. Int J Mol Sci. (2023) 24:16197. doi: 10.3390/ijms242216197
110. Ailawadi S, Wang X, Gu H, Fan GC. Pathologic function and therapeutic potential of exosomes in cardiovascular disease. Biochim Biophys Acta. (2015) 1852:1–11. doi: 10.1016/j.bbadis.2014.10.008
111. Yan M, Chen C, Gong W, Yin Z, Zhou L, Chaugai S, et al. miR-21-3p regulates cardiac hypertrophic response by targeting histone deacetylase-8. Cardiovasc Res. (2015) 105:340–52. doi: 10.1093/cvr/cvu254
112. Tian C, Gao L, Zimmerman MC, Zucker IH. Myocardial infarction-induced microRNA-enriched exosomes contribute to cardiac Nrf2 dysregulation in chronic heart failure. Am J Physiol Heart Circ Physiol. (2018) 314:H928–39. doi: 10.1152/ajpheart.00602.2017
113. Tian C, Hu G, Gao L, Hackfort BT, Zucker IH. Extracellular vesicular MicroRNA-27a* contributes to cardiac hypertrophy in chronic heart failure. J Mol Cell Cardiol. (2020) 143:120–31. doi: 10.1016/j.yjmcc.2020.04.032
114. Li D, Shen M, Deng X, Bai Y. MicroRNA miR-27a-3p accelerates cardiac hypertrophy by targeting neuro-oncological ventral antigen 1. Bioengineered. (2022) 13:8982–93. doi: 10.1080/21655979.2022.2054150
115. Chen Z, Zhang M, Xu Q, Lu P, Liu M, Yin R, et al. Huangqi-Danshen decoction improves heart failure by regulating pericardial adipose tissue derived extracellular vesicular miR-27a-3p to activate AMPKα2 mediated mitophagy. Phytomedicine. (2024) 135:156187. doi: 10.1016/j.phymed.2024.156187
116. Chen F, Li X, Zhao J, Geng J, Xie J, Xu B. Bone marrow mesenchymal stem cell-derived exosomes attenuate cardiac hypertrophy and fibrosis in pressure overload induced remodeling. In Vitro Cell Dev Biol Anim. (2020) 56:567–76. doi: 10.1007/s11626-020-00481-2
117. Li G, Qiu Z, Li C, Zhao R, Zhang Y, Shen C, et al. Exosomal miR-29a in cardiomyocytes induced by angiotensin II regulates cardiac microvascular endothelial cell proliferation, migration and angiogenesis by targeting VEGFA. Curr Gene Ther. (2022) 22:331–41. doi: 10.2174/1566523222666220303102951
118. Wang H, Xie Y, Guan L, Elkin K, Xiao J. Targets identified from exercised heart: killing multiple birds with one stone. NPJ Regener Med. (2021) 6:23. doi: 10.1038/s41536-021-00128-0
119. Chen J, Cui C, Yang X, Xu J, Venkat P, Zacharek A, et al. MiR-126 affects brain-heart interaction after cerebral ischemic stroke. Transl Stroke Res. (2017) 8:374–85. doi: 10.1007/s12975-017-0520-z
120. Salem ESB, Fan GC. Pathological effects of exosomes in mediating diabetic cardiomyopathy. Adv Exp Med Biol. (2017) 998:113–38. doi: 10.1007/978-981-10-4397-0_8
121. Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U.S.A. (2006) 103:12481–6. doi: 10.1073/pnas.0605298103
122. Gao M, Wang X, Zhang X, Ha T, Ma H, Liu L, et al. Attenuation of cardiac dysfunction in polymicrobial sepsis by microRNA-146a is mediated via targeting of IRAK1 and TRAF6 expression. J Immunol. (2015) 195:672–82. doi: 10.4049/jimmunol.1403155
123. Feng B, Chen S, Gordon AD, Chakrabarti S. miR-146a mediates inflammatory changes and fibrosis in the heart in diabetes. J Mol Cell Cardiol. (2017) 105:70–6. doi: 10.1016/j.yjmcc.2017.03.002
124. Aliotta JM, Pereira M, Wen S, Dooner MS, Del Tatto M, Papa E, et al. Exosomes induce and reverse monocrotaline-induced pulmonary hypertension in mice. Cardiovasc Res. (2016) 110:319–30. doi: 10.1093/cvr/cvw054
125. Hirai K, Ousaka D, Fukushima Y, Kondo M, Eitoku T, Shigemitsu Y, et al. Cardiosphere-derived exosomal microRNAs for myocardial repair in pediatric dilated cardiomyopathy. Sci Transl Med. (2020) 12:eabb3336. doi: 10.1126/scitranslmed.abb3336
126. Halkein J, Tabruyn SP, Ricke-Hoch M, Haghikia A, Nguyen NQ, Scherr M, et al. MicroRNA-146a is a therapeutic target and biomarker for peripartum cardiomyopathy. J Clin Invest. (2013) 123:2143–54. doi: 10.1172/JCI64365
127. Wang C, Zhang C, Liu L, A X, Chen B, Li Y, Du J, et al. Macrophage-derived mir-155-containing exosomes suppress fibroblast proliferation and promote fibroblast inflammation during cardiac injury. Mol Ther. (2017) 25:192–204. doi: 10.1016/j.ymthe.2016.09.001
128. Yu H, Qin L, Peng Y, Bai W, Wang Z. Exosomes derived from hypertrophic cardiomyocytes induce inflammation in macrophages via miR-155 mediated MAPK pathway. Front Immunol. (2021) 11:606045. doi: 10.3389/fimmu.2020.606045
129. Seok HY, Chen J, Kataoka M, Huang ZP, Ding J, Yan J, et al. Loss of microRNA-155 protects the heart from pathological cardiac hypertrophy. Circ Res. (2014) 114:1585–95. doi: 10.1161/CIRCRESAHA.114.303784
130. Li AL, Lv JB, Gao L. MiR-181a mediates Ang II-induced myocardial hypertrophy by mediating autophagy. Eur Rev Med Pharmacol Sci. (2017) 21:5462–70. doi: 10.26355/eurrev_201712_13936
131. Garg A, Foinquinos A, Jung M, Janssen-Peters H, Biss S, Bauersachs J, et al. MiRNA-181a is a novel regulator of aldosterone-mineralocorticoid receptor-mediated cardiac remodelling. Eur J Heart Fail. (2020) 22:1366–77. doi: 10.1002/ejhf.1813
132. Xue R, Tan W, Wu Y, Dong B, Xie Z, Huang P, et al. Role of Exosomal miRNAs in heart failure. Front Cardiovasc Med. (2020) 7:592412. doi: 10.3389/fcvm.2020.592412
133. Fang X, Stroud MJ, Ouyang K, Fang L, Zhang J, Dalton ND, et al. Adipocyte-specific loss of PPARγ attenuates cardiac hypertrophy. JCI Insight. (2016) 1:e89908. doi: 10.1172/jci.insight.89908
134. Kurtzwald-Josefson E, Zeevi-Levin N, Rubchevsky V, Bechar Erdman N, Schwartz Rohaker O, Nahum O, et al. Cardiac fibroblast-induced pluripotent stem cell-derived exosomes as a potential therapeutic mean for heart failure. Int J Mol Sci. (2020) 21:7215. doi: 10.3390/ijms21197215
135. Izarra A, Moscoso I, Levent E, Cañón S, Cerrada I, Díez-Juan A, et al. miR-133a enhances the protective capacity of cardiac progenitors cells after myocardial infarction. Stem Cell Rep. (2014) 3:1029–42. doi: 10.1016/j.stemcr.2014.10.010
136. Wu T, Chen Y, Du Y, Tao J, Zhou Z, Yang Z. Serum exosomal MiR-92b-5p as a potential biomarker for acute heart failure caused by dilated cardiomyopathy. Cell Physiol Biochem. (2018) 46:1939–50. doi: 10.1159/000489383
137. Xu MY, Ye ZS, Song XT, Huang RC. Differences in the cargos and functions of exosomes derived from six cardiac cell types: a systematic review. Stem Cell Res Ther. (2019) 10:194. doi: 10.1186/s13287-019-1297-7
138. Wadley GD, Lamon S, Alexander SE, McMullen JR, Bernardo BC. Noncoding RNAs regulating cardiac muscle mass. J Appl Physiol (1985). (2019) 127:633–44. doi: 10.1152/japplphysiol.00904.2018
139. Sygitowicz G, Sitkiewicz D. Involvement of circRNAs in the development of heart failure. Int J Mol Sci. (2022) 23:14129. doi: 10.3390/ijms232214129
140. Nair VD, Ge Y, Li S, Pincas H, Jain N, Seenarine N, et al. Sedentary and trained older men have distinct circulating exosomal microRNA profiles at baseline and in response to acute exercise. Front Physiol. (2020) 11:605. doi: 10.3389/fphys.2020.00605
141. Mao L, Li YD, Chen RL, Li G, Zhou XX, Song F, et al. Heart-targeting exosomes from human cardiosphere-derived cells improve the therapeutic effect on cardiac hypertrophy. J Nanobiotechnology. (2022) 20:435. doi: 10.1186/s12951-022-01630-3
142. Zuo H, Li L, Wang X, Chen S, Liao Z, Wei S, et al. A novel circ_0018553 protects against angiotensin-induced cardiac hypertrophy in cardiomyocytes by modulating the miR-4731/SIRT2 signaling pathway. Hypertens Res. (2023) 46:421–36. doi: 10.1038/s41440-022-01111-y
143. Mallaredy V, Roy R, Cheng Z, Thej C, Benedict C, Truongcao M, et al. Tipifarnib reduces extracellular vesicles and protects from heart failure. Circ Res. (2024) 135:280–97. doi: 10.1161/CIRCRESAHA.123.324110
144. Henning RJ. Cardiovascular exosomes and microRNAs in cardiovascular physiology and pathophysiology. J Cardiovasc Transl Res. (2021) 14:195–212. doi: 10.1007/s12265-020-10040-5
145. Fang J, Zhang Y, Chen D, Zheng Y, Jiang J. Exosomes and exosomal cargos: A promising world for ventricular remodeling following myocardial infarction. Int J Nanomedicine. (2022) 17:4699–719. doi: 10.2147/IJN.S377479
Keywords: cardiac hypertrophy, extracellular vesicle, microRNA, inflammation, regulatory mechanism
Citation: Hu H, Wang X, Yu H and Wang Z (2025) Extracellular vesicular microRNAs and cardiac hypertrophy. Front. Endocrinol. 15:1444940. doi: 10.3389/fendo.2024.1444940
Received: 12 June 2024; Accepted: 23 December 2024;
Published: 09 January 2025.
Edited by:
Indira Pokkunuri, University of Houston–Downtown, United StatesReviewed by:
Gioacchin Iannolo, Mediterranean Institute for Transplantation and Highly Specialized Therapies (ISMETT), ItalyCopyright © 2025 Hu, Wang, Yu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hui Yu, aHVpeXUyMDA4QGhvdG1haWwuY29t; Zhanli Wang, d2FuZy56aGFubGlAaG90bWFpbC5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.