- Department of Biochemistry, School of Medicine, Case Western Reserve University, Cleveland, OH, USA
We have exploited a prandial insulin analog to elucidate the underlying structure and dynamics of insulin as a monomer in solution. A model was provided by insulin lispro (the active component of Humalog®; Eli Lilly and Co.). Whereas NMR-based modeling recapitulated structural relationships of insulin crystals (T-state protomers), dynamic anomalies were revealed by amide-proton exchange kinetics in D2O. Surprisingly, the majority of hydrogen bonds observed in crystal structures are only transiently maintained in solution, including key T-state-specific inter-chain contacts. Long-lived hydrogen bonds (as defined by global exchange kinetics) exist only at a subset of four α-helical sites (two per chain) flanking an internal disulfide bridge (cystine A20–B19); these sites map within the proposed folding nucleus of proinsulin. The anomalous flexibility of insulin otherwise spans its active surface and may facilitate receptor binding. Because conformational fluctuations promote the degradation of pharmaceutical formulations, we envisage that “dynamic re-engineering” of insulin may enable design of ultra-stable formulations for humanitarian use in the developing world.
Introduction
We describe the structure and dynamics of a rapid-acting (prandial) insulin analog based on 1H-NMR spectroscopy. Structure-based design of such analogs has enhanced the safety and efficacy of glycemic control in the treatment of diabetes mellitus (DM; Selden et al., 1986; Tripathi and Srivastava, 2006). The importance of tight control in Type 1 DM has been demonstrated by clinical trials, including the landmark DCCT (The Diabetes Control Group and Complications Trial Research Group, 1993) and its successor, the Epidemiology of Diabetes Interventions and Complications Study (Nathan et al., 2005). Although the safety of protocols to achieve tight control in long-established Type 2 DM may vary based on individual metabolic and clinical features [e.g., transient excursions in blood glucose concentration, duration of disease, and co-morbidities (Saltiel, 2001)], the United Kingdom Prospective Diabetes Study (Nicollerat, 2000) and subgroup analysis in the recent Action to Control Cardiovascular Risk in Diabetes Study (Riddle, 2010; Gerstein et al., 2011) support the appropriateness of moderate glycemic targets (Ismail-Beigi et al., 2011). To achieve these clinical objectives, rapid- and long-acting insulin analogs have been designed based on classical crystal structures (Baker et al., 1988) and general physico-chemical principles (Creighton, 1997).
To obtain a model monomer, our studies employed insulin lispro (Ciszak et al., 1995; Campbell et al., 1996), the active component of Humalog® (Eli Lilly and Co.); its sequence is shown in Figure 1A. Approved by the United States Food and Drug Administration in 1996 (Campbell et al., 1996; Holleman and Hoekstra, 1997) and currently in broad clinical use (Hartman, 2008), this analog (also designated KP-insulin) contains paired substitutions near the C-terminus of the B-chain (ProB28 → Lys and LysB29 → Pro; asterisk in Figure 1A and black spheres in Figure 1B). Interchange of residues B28 and B29, which recapitulates a feature of insulin-like growth factor I, does not perturb the receptor-binding surface of the hormone (Baker et al., 1988), and so the analog retains native potency (Shoelson et al., 1992). Subtle perturbation of the dimer interface (Ciszak et al., 1995) nonetheless leads to accelerated disassembly of the zinc insulin analog hexamer on subcutaneous injection (DeFelippis et al., 2001). Accordingly, the analog is absorbed into the bloodstream more rapidly than is wild-type insulin (Bakaysa et al., 1996). Although intrinsic rates of clearance from the bloodstream are presumably unaffected, enhanced absorption also leads to more rapid exhaustion of the subcutaneous depot and thus more limited duration of action (DeFelippis et al., 2001). The structural basis of such altered pharmacokinetics (PK) and pharmacodynamics (PD) has been investigated by X-ray crystallography (Figure 2; Ciszak et al., 1995). Whereas the overall structure of the variant hexamer closely resembles that of wild-type insulin (Figures 2A,B), superposition of component dimers reveals splaying of the C-terminal segments of the B-chain (box in Figure 2C) with weakening of β-sheet-related hydrogen bonds (dotted lines; Dodson et al., 1993; Ciszak et al., 1995). Such analysis highlighted the subtle contributions of ProB28 and LysB29 to the wild-type dimer interface (Figure 2D). Structure-based design of insulin lispro and related analog insulin aspart (AspB28-insulin, the active component of Novolog®; Novo-Nordisk, Ltd.) represented pioneering examples of protein engineering in molecular pharmacology (Brange et al., 1988, 1992a; DeFelippis et al., 2001).
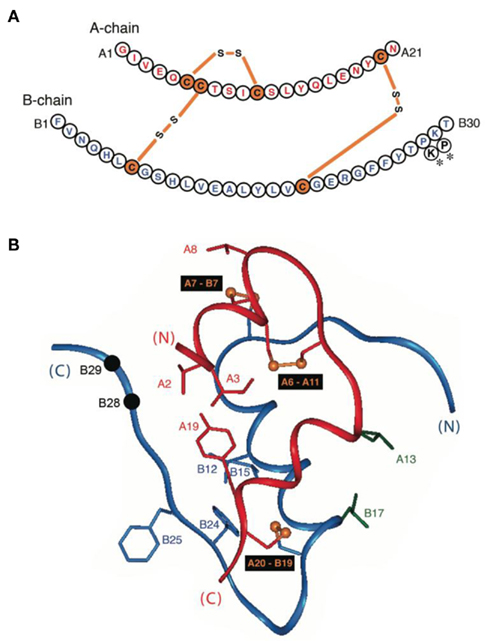
Figure 1. Sequence and structure of insulin. (A) Sequence of human insulin with A-chain at top (red) and B-chain beneath (blue). Cysteines are highlighted in orange with linkages as indicated (bars). Sites of substitution in KP-insulin are indicated (asterisks). (B) Ribbon model of insulin (T-state protomer; Protein Databank accession code 4INS; Baker et al., 1988). The A-chain is shown in red, and the B-chain in blue. Disulfide bridges are indicated by gold spheres (sulfur atoms); pairings are shown in black boxes. Black spheres indicate Cα carbons of residues B28 and B29 (site of sequence interchange in KP-insulin). Selected side chains at the receptor-binding surface are also shown as labeled.
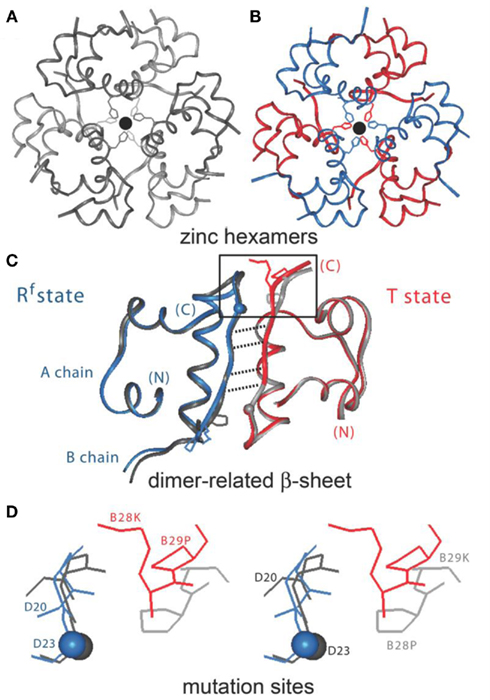
Figure 2. Design of insulin lispro. (A,B) Ribbon models of the wild-type zinc insulin hexamer (A) and KP-insulin (B). The hexamer type is in each case  The A- and B-chains in (A) are shown in light and dark gray, respectively; the A- and B-chains in (B) are shown in red and blue. In each hexamer the overlying positions of the axial zinc ions are shown as central black circles coordinated by the side chains of HisB10 (three per metal-ion binding site). Respective Protein Databank accession codes are 1TRZ (wild-type) and 1LPH (KP-insulin; Ciszak et al., 1995). (C) Structure of component TRf insulin dimer in the wild-type hexamer (light and dark gray) and variant hexamer (red and blue). The structures were aligned according to the main-chain atoms of residues B3–B28 and A2–A21. T- and Rf-protomers are shown at right and left, respectively, in an orientation that highlights the dimer-related β-sheet (residues B24–B28) and its four inter-molecular hydrogen bonds (dotted lines; Baker et al., 1988). Boxed region contains sites of substitution in KP-insulin. (D) Enlargement of boxed region in (C). The KP substitutions are associated with lengthening of the inter-molecular hydrogen bonds and subtle repositioning of residues B28–B30 (Ciszak et al., 1995). Respective GlyB23 Cα atoms in corresponding Rf protomers are indicated by gray (wild-type) and blue (variant) spheres. In the wild-type hexamer electron density (accession code 1TRZ) was not well defined for the Cγ, Cδ, and Nε atoms of LysB29.
The A- and B-chains in (A) are shown in light and dark gray, respectively; the A- and B-chains in (B) are shown in red and blue. In each hexamer the overlying positions of the axial zinc ions are shown as central black circles coordinated by the side chains of HisB10 (three per metal-ion binding site). Respective Protein Databank accession codes are 1TRZ (wild-type) and 1LPH (KP-insulin; Ciszak et al., 1995). (C) Structure of component TRf insulin dimer in the wild-type hexamer (light and dark gray) and variant hexamer (red and blue). The structures were aligned according to the main-chain atoms of residues B3–B28 and A2–A21. T- and Rf-protomers are shown at right and left, respectively, in an orientation that highlights the dimer-related β-sheet (residues B24–B28) and its four inter-molecular hydrogen bonds (dotted lines; Baker et al., 1988). Boxed region contains sites of substitution in KP-insulin. (D) Enlargement of boxed region in (C). The KP substitutions are associated with lengthening of the inter-molecular hydrogen bonds and subtle repositioning of residues B28–B30 (Ciszak et al., 1995). Respective GlyB23 Cα atoms in corresponding Rf protomers are indicated by gray (wild-type) and blue (variant) spheres. In the wild-type hexamer electron density (accession code 1TRZ) was not well defined for the Cγ, Cδ, and Nε atoms of LysB29.
1H-NMR spectroscopy in principle provides an opportunity to probe the structure and dynamics of insulin as a monomer in solution and so complements crystallographic studies of insulin self-assembly (Hua et al., 1991). Such spectroscopic studies were long hampered by the complex self-association equilibria characteristic of wild-type insulin at micromolar-to-millimolar protein concentrations (Weiss et al., 1991). In the present study we have explored a broad range of conditions to enable rigorous 1H-NMR characterization of insulin lispro in aqueous solution without the need for additional amino-acid substitutions or addition of organic co-solvents (Hua and Weiss, 1991). Whereas the solution structure of monomeric insulin lispro closely resembles a crystallographic T-state protomer (Figures 1B and 2C, right; Baker et al., 1988), analysis of amide-proton exchange in D2O demonstrated novel dynamic features. This technique focuses on the kinetics of exchange between peptide N-1H protons by solvent-derived deuterons as a measure of both global stability and site-specific conformational fluctuations (Wüthrich, 1986; Englander et al., 1996). Remarkably, the majority of main-chain hydrogen-bonds observed in the crystal structure of insulin are only transiently maintained in solution: long-lived hydrogen bonds are restricted to a subset of α-helical residues flanking the A20–B19 disulfide bridge. This pattern of core conformational stability and flanking fluctuations is discussed in relation to mechanisms of receptor binding (De Meyts and Whittaker, 2002; Nakagawa et al., 2005) and proinsulin folding (Steiner, 1967; Qiao et al., 2003; Weiss, 2009; Yang et al., 2010a).
Conformational stability and fluctuations within insulin are pertinent not only to in vivo biosynthesis and signaling, but also to pharmaceutical formulation (Brange et al., 1987). Rates of chemical and physical degradation, key determinants of shelf-life above room temperature, are enhanced by transient partial unfolding processes (Brange and Langkjoer, 1993). Such degradation is accelerated among rapid-acting insulin analogs, including insulin lispro, and poses a particular challenge among indigent patients in the developing world (Hua et al., 2008). Indeed, the same structural perturbations introduced by substitutions at positions B28 and/or B29 to achieve accelerated absorption lead at the same time to compromised pharmaceutical stability. How may this trade-off, seemingly intrinsic to the logic of protein design, be avoided? The dynamic view of insulin provided herein by 1H-NMR analysis promises to provide an opportunity for the “dynamic re-engineering” of favorable pharmacokinetic- and stability properties. We envisage that such second-generation analog formulations would circumvent the cold chain of insulin delivery and thereby facilitate their storage and use to address the emerging global pandemic of DM (Osei, 2003; Lefebvre and Pierson, 2004).
Materials and Methods
Synthesis of Insulin Analogs
Human insulin and insulin lispro were obtained from Eli Lilly Co. (Indianapolis, IN, USA). Synthetic analogs, kindly provided by Prof. P. G. Katsoyannis and coworkers, were prepared as described (Hu et al., 1993). Predicted molecular masses were in each case verified by mass spectrometry.
Circular Dichroism
Circular dichroism (CD) spectra were obtained as described (Hua et al., 2001). Samples were dissolved in 10 mM acetic acid (0.14% v/v) at pH 3.0; the protein concentration was ca. 25 μM. For equilibrium denaturation studies, samples were diluted in the same buffer to 5 μM; guanidine-HCl was employed as denaturant (Hua et al., 2002a, 2006c). Data were obtained at 25 or 37°C and fitted by non-linear least squares to a two-state model (Sosnick et al., 2000).
NMR Spectroscopy
Spectra were obtained at 700 MHz in 10 mM deuterioacetic acid (pH/pD 3.0; direct meter reading) at 25 and 37°C (Hua et al., 1996a); the protein concentration was ca. 0.5 mM. Resonance assignment was obtained by standard methods (Wüthrich, 1986). Sites of hydrogen bonds were inferred from the pattern of amide-proton protection in D2O (Hua and Weiss, 1990). Spectra were also acquired at pH 7.4 and in 20% (v/v) deuterioacetic acid (pH 2.0).
Structure Calculations
Distance-geometry/simulated annealing calculations (DG/SA) were performed using program DG-II (Havel and Snow, 1991); restrained molecular dynamics (RMD) were calculated using X-PLOR (Brunger, 1992). Nuclear overhauser enhancement (NOE)-related and dihedral-angle restraints were used for molecular modeling as described (Hua et al., 1998).
Amide-proton Exchange
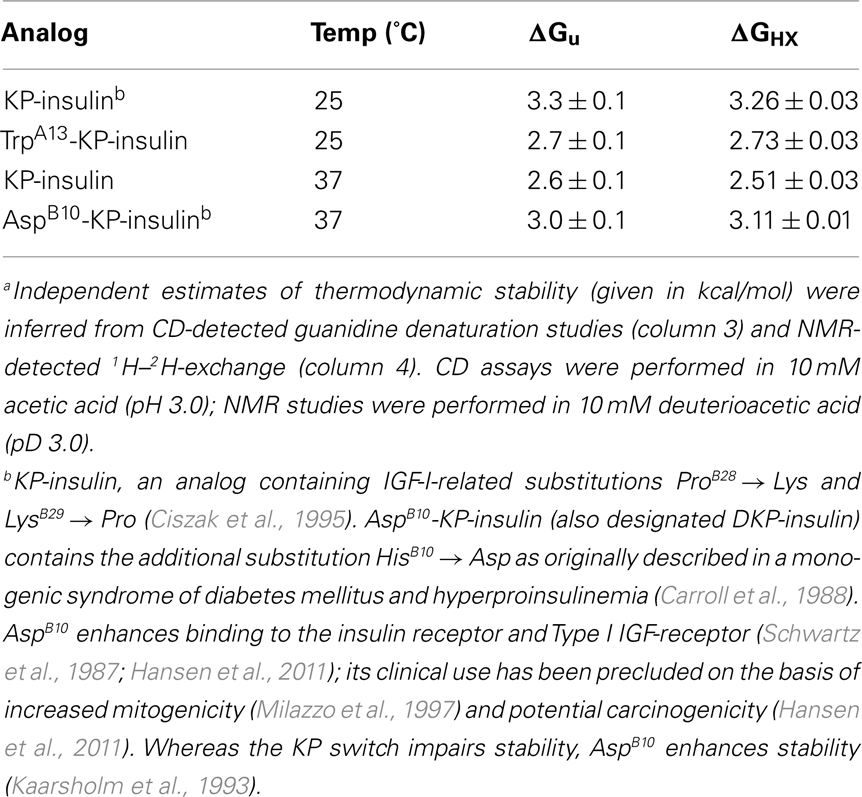
1H–2H-exchange was monitored in 10 mM deuterioacetic acid (pD 3.0) at either 25 or 37°C. The protocol is outlined in schematic form in Figure S1 in Supplementary Material. Intrinsic hydrogen exchange rates (kint) were estimated using the Sphere program (H. Roder, Fox Chase Cancer Center, Philadelphia, PA, USA). Protection factors (PFs) are defined as the ratio of intrinsic exchange rate constants:observed exchange rate constants (kint/kobs; Englander et al., 1996). Sites of global exchange were identified on the basis of comparative studies of protein analogs differing in thermodynamic stability but otherwise similar in structure; at such sites values of ΔGu derived from denaturation studies were in accordance with values of ΔGHX predicted by global PFs according to:
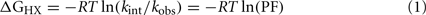
where R is the Rydberg constant, and T is the absolute temperature. Values of kobs and PF are provided in Table S1 in Supplementary Material.
Results
Twenty years ago protein engineering enabled the application of 2D 1H-NMR methods to insulin (Weiss et al., 1991). Simultaneous substitutions were introduced at successive self-association surfaces (such as the classical B28-related dimer contact and B10-related trimer contact; Figures 2A–C) to achieve the millimolar protein concentrations then required for 2D 1H-NMR study (Hua et al., 1996a). Advances in 1H-NMR instrumentation (particularly development of ultra-high sensitivity cryogenic probes) presently enable such study at protein concentrations as low as 0.1–0.5 mM. Aided by this technology, we have found that KP-insulin is tractable in the absence of ancillary substitutions at the trimer interface, providing an opportunity to characterize a prandial insulin in clinical use.
Solution Structure
Representative 1D 1H-NMR spectra of KP-insulin under diverse solution conditions are shown in Figure 3; these include aqueous solution at pH 7.4 (pertinent to conditions in the bloodstream) and at pH 3.0 (optimal for analysis of amide-proton exchange). In each case KP-insulin exhibits patterns of chemical shifts, resonance line widths, and NOEs consistent with an isolated monomer (Hua et al., 1996a). Short- and medium-range NOEs characteristic of secondary structure were in each case consistent with the classical T state (Figure S2 in Supplementary Material). The set of inter-residue NOEs was found to be similar to that previously observed in 2D 1H-NMR studies of AspB10-KP-insulin (also designated DKP-insulin; Hua et al., 1996a). Structural models were calculated by DG/RMD based on 803 distance restraints, 47 dihedral-angular restraints, and 31 hydrogen-bond restraints (Table S2 in Supplementary Material); a total of 881 restraints (ca. 17.3 restraints per residue) were thus employed. As expected, the resulting ensemble of structures (Figure 4B) closely resembles a collection of crystallographic T-state protomers (Figure 4A).
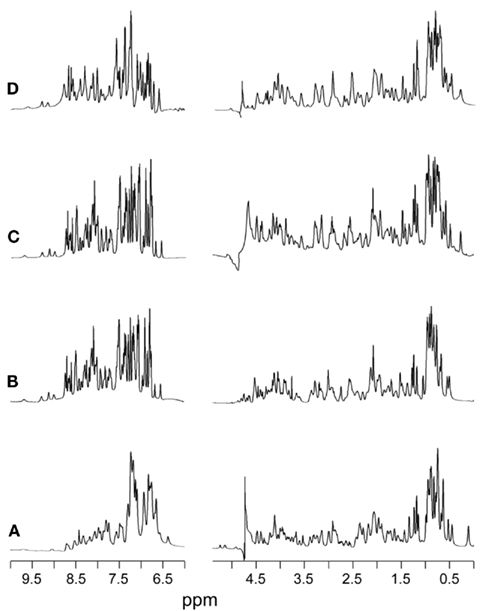
Figure 3. 1H-NMR spectra of KP-insulin at 700 MHz. Spectra were obtained under a variety of solution conditions: (A) aqueous solution at pH 7.4 and 32°C, (B) 20% (v/v) deuterioacetic acid (pH 2.0) at 32°C, (C) 10 mM deuterioacetic acid (pH 3.0) at 32°C, and (D) 10 mM deuterioacetic acid (pH 3.0) at 25°C. In each panel the amide-aromatic spectrum in H2O is shown at left, and the aliphatic spectrum in D2O is shown at right.
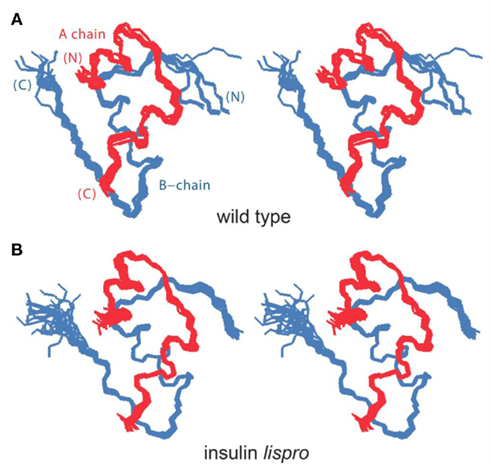
Figure 4. NMR-derived structure of KP-insulin. (A) Reference collection of crystallographic T-state protomers (stereo pair). The structures (wild-type accession codes 1MSO, 4INS, 1APH, 1BPH, 1CPH, 1DPH, 1TRZ, 1TYL, 1TYM, 2INS, 1ZNI, 1LPH, 1G7A; KP-insulin accession code 2JMN) were aligned according to the main-chains within α-helices (residues A2–A7, A13–A19, and B9–B19). The A-chains are shown in red and B-chains in blue. (B) DG/RMD-derived ensemble of structures of KP-insulin as a monomer in solution. The color code and alignment scheme are as in (A).
Distance-geometry/RMD ensembles of KP-insulin exhibit peptide segments of varying precision and so are evocative of dynamic variability. The C-terminal residues of the B-chain (residues B25–B30) are less well defined in solution than among crystal structures, presumably due to stabilizing effects of self-assembly (Baker et al., 1988). Conversely, positioning of the N-terminal arm of the B-chain in solution (residues B1–B4) is more precisely defined than would be expected based on the diversity of crystallographic conformations (Figures 4A,B). The latter difference may reflect the DG/RMD protocol (which enforced all restraints simultaneously and so may underestimate conformational fluctuations), or conversely, variable positioning of the arm in crystals may be enjoined by lattice contacts (Baker et al., 1988). Because DG/RMD ensembles do not directly provide dynamic information, we sought an independent probe of protein stability and dynamics. 1H-NMR analysis of amide-proton exchange in D2O in principle provides such a probe (Wüthrich and Wagner, 1979; Englander et al., 1996).
Amide-Proton Exchange
The amide proton (HN) within each peptide-bond unit is labile, i.e., able to exchange with a solvent-derived 1H or 2H (Englander et al., 1996). The rate of such exchange is typically measured under EX2 conditions wherein the observed rate constant (kobs) is the product of a pH-dependent intrinsic rate constant (kint) and the site-specific opening rate constant (kop; Bai et al., 1993). The extent to which kobs differs from kint enables calculation of individual PFs, defined as kint/kobs (Roder et al., 1985). PF values depend on local structural environment and so define a residue-specific probe. In small globular proteins non-negligible PFs are ordinarily restricted to sites of hydrogen bonding. Protection within KP-insulin might therefore be expected at HN sites within α-helices (A3–A8, A15–A19, and B12–B19, excluding N-terminal residues), within β-turns (B7–B10 and B20–B23), or participating in inter-chain hydrogen bonds; dimer-specific protection would not be expected within the isolated B-chain β-strand (B24–B28; Figure 2C). The extent of protection at a given site depends on rates of structural fluctuations leading to hydrogen-bond breakage and exposure to solvent. Two limiting cases are of interest: (i) local stability is sufficiently marked that exchange requires global unfolding and (ii) local fluctuations are sufficiently marked that protection is negligible even in the presence of a non-random mean structure.
1H–2H-exchange in KP-insulin is illustrated in Figure 5 (1D spectra) and Figure 6 (2D spectra). The studies were conducted at pD 3.0 and 25°C to optimize the quality of the 1H-NMR spectrum in a pH regime between the extremes of acid and base catalysis. The majority of amide protons were found to exchange within 5 min, and so their resonances were not observed in the initial spectrum. Rates of exchange were thus measurable only for a minority of amide resonances. These reside within or adjoining α-helices (Figure 5B) and at inter-chain hydrogen bonds A11 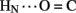 B4 and B6
B4 and B6 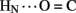 A6 (vertical dashed lines in Figure 5A). Resonance overlap among amide resonances in α-helices was resolved in successive TOCSY spectra (Figure 6). A broad range of PFs was observed (Figure 7A; Table S1 in Supplementary Material), suggesting uncorrelated and inequivalent segmental fluctuations. Whereas the central B-chain α-helix and C-terminal A-chain α-helix contain sites of marked protection, exchange was rapid within the A1–A8 α-helix and the two B-chain β-turns. A summary of sites of observable protection is provided in Figure 7B in relation to hydrogen bonds as defined in crystal structures and as inferred from DG/RMD models.
A6 (vertical dashed lines in Figure 5A). Resonance overlap among amide resonances in α-helices was resolved in successive TOCSY spectra (Figure 6). A broad range of PFs was observed (Figure 7A; Table S1 in Supplementary Material), suggesting uncorrelated and inequivalent segmental fluctuations. Whereas the central B-chain α-helix and C-terminal A-chain α-helix contain sites of marked protection, exchange was rapid within the A1–A8 α-helix and the two B-chain β-turns. A summary of sites of observable protection is provided in Figure 7B in relation to hydrogen bonds as defined in crystal structures and as inferred from DG/RMD models.
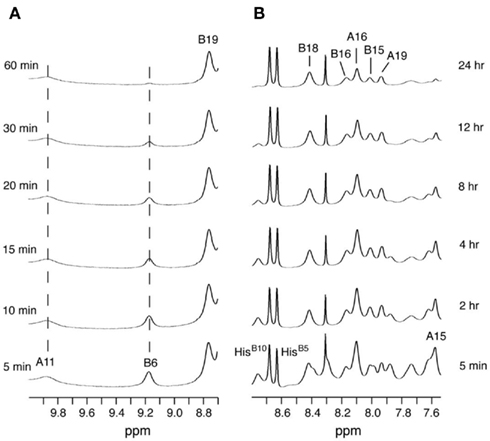
Figure 5. One-dimensional 1H-NMR studies of amide-proton exchange in D2O (pH 3.0 and 25°C). (A) Spectra of downfield amide region as observed after successive times in D2O. Vertical dashed lines indicate the amide protons of CysA11 (9.86 ppm; left) and LeuB6 (9.17 ppm; center), which are engaged in T-state-specific inter-chain hydrogen bonds (see text). (B) Spectra of major amide region at corresponding times; selected assignments are as indicated at top. The two sharp downfield resonances between 8.6 and 8.7 ppm belong to the Hε ring resonances of HisB5 and HisB10; the sharp resonance at 8.3 ppm is an impurity.
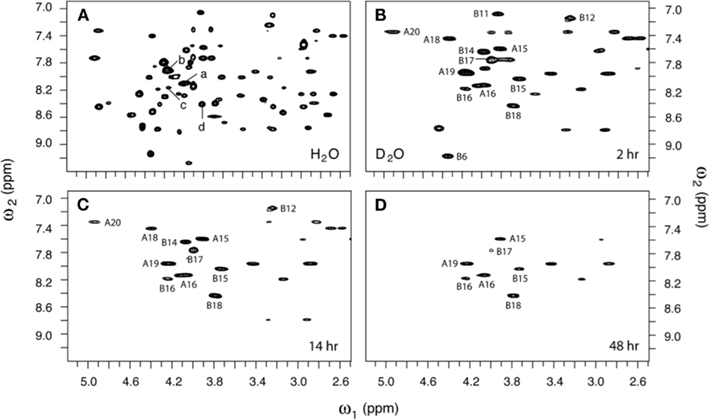
Figure 6. Two-dimensional 1H-NMR studies of amide-proton exchange in D2O (pH 3.0 and 25°C). (A) Baseline TOCSY spectrum in H2O. Selected HN–Hα cross-peak assignments are as follows: (a) LeuA16, (b) TyrA19, (c) TyrB16, and (d) ValB18. (B–D) TOCSY spectra as acquired at successive times after dissolution in D2O: (B) 2 h, (C) 14 h, and (D) 48 h. Assignments are as indicated.
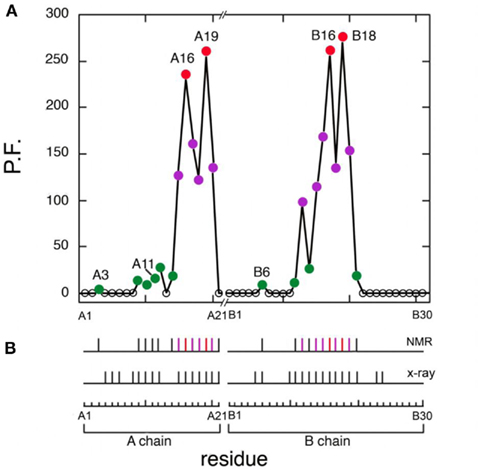
Figure 7. Summary of protection factors (PF) and survey of hydrogen bonds. (A) Plot of residue-specific PFs in KP-insulin (vertical axis) versus sequence (horizontal axis). Color code: (red circles) sites of global exchange, (purple) subglobal exchange, and (green) local fluctuations. (B) Summary of amide hydrogen bonds (vertical segments) as inferred from NMR studies (top) and as predicted by crystal structures (bottom). The color code is as in (A). Sites of global exchange in the A- and B-chains cluster near cystine A20–B19.
Dynamic Classification
Protection factors may be interpreted in relation to protein stability (at sites of global exchange) or protein dynamics (at sites of subglobal or local exchange; Roder et al., 1985). Global refers to an overall equilibrium between native and unfolded structures; such exchange implies that local or segmental conformational fluctuations either do not involve the breakage of hydrogen bonds or do not enable their exposure to solvent. Subglobal pertains to cooperative segmental fluctuations whereas local refers to uncorrelated local motions leading to hydrogen-bond breakage and exposure. The thermodynamic stability of a protein imposes an upper bound on observable PFs; intrinsic (random-coil) exchange rates impose a lower bound.
To distinguish between these modes of exchange, the global thermodynamic stability of KP-insulin was probed by chemical denaturation (Figure S3 in Supplementary Material). Cooperative unfolding at successive concentrations of guanidine-HCl was observed by CD at a helix-sensitive wavelength (222 nm). Application of a two-state model (N ⇔ U) enabled an estimate of the free energy of unfolding (ΔGu) to be obtained as extrapolated to zero denaturant concentration (Hua et al., 2002a). The resulting estimate (3.3 ± 0.1 kcal/mol; see Table 1) implies a PF ceiling between 250 and 300. Because the 1H-NMR studies were conducted in D2O, comparative CD-detected studies in H2O and D2O were undertaken (each in 10 mM acetic acid at pH/pD 3.0); the results suggested that solvent isotope effects on stability are negligible (Figure S4 in Supplementary Material).
Four sites of global exchange were identified as depicted in Figure 8A. Their identification was based on PFs at or near the stability ceiling as imposed by ΔGu: LeuA16 and TyrA19 (the C-terminal segment of the second A-chain α-helix) and TyrB16 and ValB18 (the C-terminal segment of the central B-chain α-helix). Global stability as estimated based on the average of the PFs at these sites (ΔGHX 3.26 ± 0.03 kcal/mol) is in accordance with the results of chemical denaturation (ΔGu 3.3 ± 0.1 kcal/mol; Table 1). Similar consistency was observed at 37°C wherein KP-insulin exhibits both reduced stability to chemical denaturation (ΔGu 2.6 ± 0.1 kcal/mol) and proportionately accelerated global exchange (ΔGHX 2.51 ± 0.03 kcal/mol; Table 1). These sites of global exchange kinetics flank internal cystine A20–B19; their nascent folding is proposed to stabilize the folding nucleus of proinsulin (see Discussion; Hua et al., 2006a). To verify their global status, comparative amide-proton exchange studies were undertaken of analogs of KP-insulin of lower or greater thermodynamic stability (TrpA13-KP-insulin and AspB10-KP-insulin; Table 1); surface substitutions were chosen to avoid perturbations within the hydrophobic core. Whereas substitution HisB10 → Asp provides a more favorable C-cap to the B-chain α-helix (Weiss et al., 1990), substitution LeuA13 → Trp would be expected to introduce a destabilizing “reverse hydrophobic effect” (Chu et al., 1992) and potentially perturb the long-range packing of PheB1. These modifications were each associated with commensurate changes in ΔGHX and ΔGu in accordance with the global exchange model. Representative 1H-NMR data are respectively provided in Figures S5 and S6 in Supplementary Material.
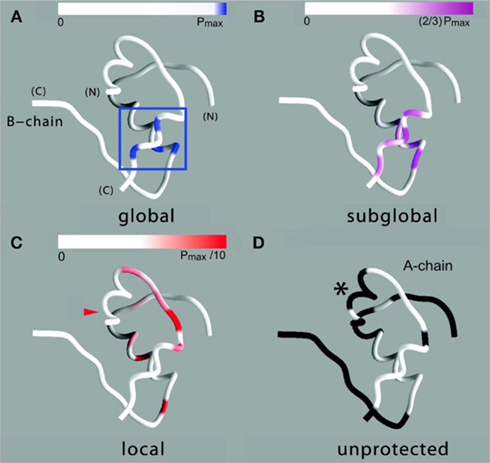
Figure 8. Structural map of amide-proton exchange. (A) Sites of global exchange are shown in relation to a ribbon model of the T state: residues A16, A19, B16, and B18 cluster near cystine A20–B19 (box). (B) Corresponding ribbon model showing sites of subglobal exchange: residues A15, A17, A18, A20, B12, B14, B15, B17, and B19. (C) Sites of local exchange: residues A3, A9–A12, A14, B6, B11, B13, and B20. (D) Sites for which HN resonances were not observed in D2O, presumably due to rapid exchange (i.e., within 5 min). Color bars at top in (A–C) indicate range of PFs shown relative to the maximum value set by global stability; normalized protection factors are given in Table S1 in Supplementary Material. Asterisk in (D) highlights lack of protection within the A1–A8 α-helix; ValA3 exhibits slight protection [arrowhead in (C)].
Sites of subglobal exchange were identified as depicted in Figure 8B. Such sites (heuristically defined by PF values between 100 and 200; Table S1 in Supplementary Material) adjoin the global sites in the A-chain α-helix (residues A15, A17, A18, and A20) and B-chain α-helix (residues B12, B14, B15, B17, and B19). Sites of local exchange (3 < PFs < 30) were likewise identified as depicted in Figure 8C. These sites were observed in the A-chain at residues A3, A9–A11, A13, and A14; and in the B-chain at B6, B11, B13, and B20. Structural correlates are as follows. (i) Protection is negligible or absent within the A1–A8 α-helix; only ValA3 HN was observed in D2O (PFA3 3.6 ± 0.2). Such rapid exchange stands in contrast to retention of canonical helix-related NOEs in this segment (see above; Figure S2 in Supplementary Material). (ii) β-Turn probes LeuB11 (hydrogen bonded to the carbonyl oxygen of GlyB8) and GlyB20 (hydrogen bonded to the carbonyl oxygen of B23) exhibit PFs between 10 and 20 (PFB11 11.4 ± 0.4 and PFB20 19.0 ± 0.6). (iii) Inter-chain hydrogen bonds (A11 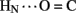 B4 and B6
B4 and B6 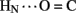 A6) are likewise transient (PFA11 14.5 ± 0.5 and PFB6 4.3 ± 0.3). No PFs were observed with values between 30 and 100, providing a clear empirical demarcation between segments exhibiting subglobal fluctuations and sites of local exchange.
A6) are likewise transient (PFA11 14.5 ± 0.5 and PFB6 4.3 ± 0.3). No PFs were observed with values between 30 and 100, providing a clear empirical demarcation between segments exhibiting subglobal fluctuations and sites of local exchange.
No protection was observed at the remaining sites (Figure 8D), the majority of which are uninvolved in hydrogen bonding in crystal structures (Baker et al., 1988). Conspicuous by their absence, however, is lack of observable HN protection at six sites of hydrogen-bonds characteristic of the classical 2-Zn (T6) insulin hexamer. These map both within the A-chain (A20 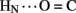 A17) and B-chain (B22
A17) and B-chain (B22 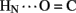 B19 and B23
B19 and B23 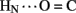 B20). Accelerated 1H–2H-exchange within the central B-chain β-turn correlates with enhanced crystallographic thermal B-factors at residues B20–B23 (Baker et al., 1988). Anomalous lack of protection was also observed at sites of inter-chain hydrogen bonds (A20
B20). Accelerated 1H–2H-exchange within the central B-chain β-turn correlates with enhanced crystallographic thermal B-factors at residues B20–B23 (Baker et al., 1988). Anomalous lack of protection was also observed at sites of inter-chain hydrogen bonds (A20 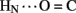 B23, B4
B23, B4 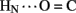 A11, and B25
A11, and B25 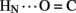 A19). Because the latter two inter-chain hydrogen bonds are broken in the crystallographic TR transition (Baker et al., 1988), such allosteric reorganization may exploit intrinsic sites of flexibility in an insulin protomer as originally envisaged by Hodgkin and colleagues (Chothia et al., 1983).
A19). Because the latter two inter-chain hydrogen bonds are broken in the crystallographic TR transition (Baker et al., 1988), such allosteric reorganization may exploit intrinsic sites of flexibility in an insulin protomer as originally envisaged by Hodgkin and colleagues (Chothia et al., 1983).
Discussion
The zinc insulin hexamer exhibits representative structural features of globular proteins in general, including canonical elements of secondary structure, well-organized hydrophobic core, ligand-binding sites, and capacity for allosteric reorganization (Chothia et al., 1983). A model critical to the development and application of X-ray crystallographic techniques (Baker et al., 1988), the hexamer provides a stable storage form of the hormone both in the secretory granules of pancreatic β-cells (Steiner, 1967; Dodson and Steiner, 1998) and in pharmaceutical formulations (Brange et al., 1987). Structure-based mutagenesis of the zinc insulin hexamer (Brange et al., 1988) enabled development of rapid-acting insulin analog formulations for prandial injection and use in pumps (Zinman et al., 1997). Although originally described as “monomeric” (Brange et al., 1988), such analogs are formulated as zinc insulin hexamers (Humalog® and Novolog®; DeFelippis et al., 2001) or as an equilibrium distribution of zinc-free oligomers (Apidra®; Garg et al., 2005). The overall goal of such protein engineering is enhanced glycemic control with reduced risk of major hypoglycemic events per decrement in hemoglobin A1c (Melki et al., 1998; Renner et al., 1999; Hartemann-Heurtier et al., 2003).
The present study has exploited a rapid-acting analog in current clinical use as a biophysical model of the insulin monomer. Insulin lispro contains paired substitutions near the C-terminus of the B-chain [ProB28 → Lys and LysB29 → Pro; (Bakaysa et al., 1996)]. These changes are peripheral to the receptor-binding surface (Shoelson et al., 1992) and so are compatible with native hypoglycemic potency (Campbell et al., 1996). Although dimerization is markedly impaired in the absence of zinc ions, zinc-dependent hexamer assembly (critical to the stability of its pharmaceutical formulation) is maintained on binding of phenolic ligands (Ciszak et al., 1995). The structure and dynamics of the zinc-free monomer are of broad interest in relation to receptor-binding in vivo (Dodson and Steiner, 1998) and mechanisms of degradation in vitro (Brange and Langkjoer, 1993). Structural relationships within and between A- and B-chains are also pertinent to the folding of proinsulin (as a nascent monomer in the endoplasmic reticulum; Dodson and Steiner, 1998) and its aggregation-coupled misfolding in a newly recognized monogenic diabetes syndrome (Stoy et al., 2007; Weiss, 2009).
The solution structure of insulin lispro closely resembles the crystallographic T state as observed within structures of zinc insulin dimers and hexamers (Baker et al., 1988). This result was expected in light of previous NMR studies of related and unrelated engineered insulin monomers under a variety of solution conditions by several laboratories (for review, see Baker et al., 1988; De Meyts and Whittaker, 2002). Characterization of insulin lispro as a native-like monomer at pH 3.0 and in the absence of an organic co-solvent nonetheless provided an opportunity to obtain a quantitative assessment of amide-proton exchange in D2O. Use of an acidic pH facilitates observation of 1H–2H-exchange in flexible systems due to reduction in baseline rates of exchange from the random-coil state; between pH 3.0 and 4.0 the simultaneous efficiency of acid-catalyzed and base-catalyzed exchange was thus minimized (Roder et al., 1985; Wüthrich, 1986; Englander et al., 1996). In addition, the absence of an organic co-solvent permitted application of tabulated random-coil exchange rate constants (Englander et al., 1996) and hence calculation of site-specific PFs in the native state (Bai et al., 1993; Englander et al., 1996). Strikingly, at the majority of potential sites protection could not be observed, due either to absence of hydrogen bonding or local conformational fluctuations. Evidence for the latter is provided by the conspicuous absence of observable protection at sites otherwise structurally compatible with hydrogen-bond formation in the majority of DG/RMD models (e.g., A20 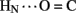 A17 and A21
A17 and A21 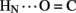 B23; see Results). Because insulin is more stable under acidic conditions than at neutral pH (due to protonation of HisB5; Bryant et al., 1993), the present results may underestimate the extent of conformational fluctuations in the bloodstream.
B23; see Results). Because insulin is more stable under acidic conditions than at neutral pH (due to protonation of HisB5; Bryant et al., 1993), the present results may underestimate the extent of conformational fluctuations in the bloodstream.
From the present analysis three structural classes of hydrogen bonds may be defined: (i) those within stable α-helices, (ii) those within β-turns, or (iii) those between the A- and B-chains. Surprisingly, the majority of such hydrogen bonds are labile in solution (i.e., exhibit PFs smaller than would be predicted based on the global thermodynamic stability of the protein) irrespective of structural class. Global exchange kinetics was observed only at four sites (two per chain) flanking internal cystine A20–B19 (Figure 8A). Successive classes of exchange kinetics provide a dynamic map (Figures 8B–D). The lability of the majority of hydrogen bonds in monomeric insulin stands in contrast to the greater PFs observed in pioneering studies of bovine pancreatic trypsin inhibitor (Wagner and Wüthrich, 1986; Roder, 1989). Previous studies have suggested that hydrogen bonds within insulin are stabilized on self-assembly (Hua et al., 1993) in accordance with high-resolution crystal structures (Baker et al., 1988).
Sites of global exchange kinetics in KP-insulin coincide with the proposed folding nucleus of proinsulin (Hua et al., 2006b). Although the latter precursor comprises a single polypeptide chain, the success of insulin chain combination (Katsoyannis, 1966) implies that chemical information required for folding is contained within the sequences of the A- and B domains (Tang and Tsou, 1990; Wang and Tsou, 1991). Studies of the disulfide-coupled folding of proinsulin and related polypeptides – foreshortened single-chain analogs (mini-proinsulin; Huang et al., 2001) and insulin-like growth factors – are notable for populated one- and two-disulfide intermediates (Hober et al., 1992; Miller et al., 1993; Qiao et al., 2001). Such intermediates are thought to acquire structure stepwise on successive disulfide pairing (for reviews see Jia et al., 2003; Weiss, 2009; Hua, 2010).
Spectroscopic studies of equilibrium peptide models support a structural pathway of disulfide pairing. In this process a key role is played by preferred initial formation of cystine A20–B19. In the native state this internal bridge connects the C-terminal α-helix of the A domain to the central α-helix of the B domain, connecting sites of global exchange kinetics as defined in the present study (Hua et al., 1995, 2002a,b). Although studies of proinsulin itself have been limited by aggregation (imposing a requirement for pH conditions >9), the disulfide pathways of mini-proinsulin and IGF-I are well characterized near neutral pH (Hober et al., 1992, 1994; Miller et al., 1993; Qiao et al., 2001). In such studies cystine A20–B19 is the only one-disulfide species to accumulate (Milner et al., 1999). We therefore envisage that the dynamic “core” of KP-insulin in its native state reflects its role in folding (Hua et al., 1996b, 2001). Impaired folding of the A20–B19-associated folding nucleus may account for the proteotoxicity and β-cell dysfunction caused by GlyB23 → Val and related B-domain mutations in patients with permanent neonatal-onset neonatal DM (Weiss, 2009).
Sites of rapid, local, and subglobal exchange are prominent within the N-terminal segments of the A- and B-chains. Only limited protection was observed, for example, at the sites of key T-state-specific inter-chain hydrogen bonds (A11 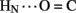 B4 and B6
B4 and B6 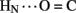 A6); no protection was observed corresponding to B4
A6); no protection was observed corresponding to B4 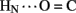 A11. Breakage of such contacts (a prominent feature of the TR transition among zinc hexamers; Chothia et al., 1983) has been proposed to accompany binding of insulin to the insulin receptor (Baker et al., 1988; Nakagawa et al., 2005). Although co-crystal structures of the hormone-receptor complex have not been determined, evidence for such induced fit in the B-chain has been obtained through characterization of substitutions of GlyB8. Formation of the A11
A11. Breakage of such contacts (a prominent feature of the TR transition among zinc hexamers; Chothia et al., 1983) has been proposed to accompany binding of insulin to the insulin receptor (Baker et al., 1988; Nakagawa et al., 2005). Although co-crystal structures of the hormone-receptor complex have not been determined, evidence for such induced fit in the B-chain has been obtained through characterization of substitutions of GlyB8. Formation of the A11 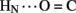 B4 and B6
B4 and B6 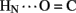 A6 hydrogen bonds is coupled to maintenance of the adjoining T-state-specific β-turn (residues B7–B10). Remarkably, stabilization (or destabilization) of this turn by respective D- (or L) amino-acid substitutions at GlyB8 is associated with reciprocal effects on activity. On the one hand, because in the T state GlyB8 exhibits a negative ɸ dihedral angle, characteristic of D-amino acids, diverse D-substitutions at B8 are compatible with native-like structure but low activity (Nakagawa et al., 2005; Hua et al., 2006c). On the other hand, L-amino-acid substitutions, consistent with the positive B8 ɸ angle of GlyB8 in the R state, can exhibit high activities despite markedly impaired stability and low yields of chain combination or yeast expression (Kristensen et al., 1997; Guo et al., 2005; Hua et al., 2006c). Accordingly, we have proposed that GlyB8 functions an “ambidextrous switch” between folding-competent (T-like) and active (R-like) conformations. In this model nascent T-like local structure in the proximal arm (including inter-domain hydrogen bonding and contacts by the imidazole ring of HisB5; Hua et al., 2006c) enhance the efficiency and fidelity of disulfide pairing. The present 1H–2H-exchange data highlight the flexibility of T-like structure in the B-chain and hence its amenability for structural reorganization on receptor binding.
A6 hydrogen bonds is coupled to maintenance of the adjoining T-state-specific β-turn (residues B7–B10). Remarkably, stabilization (or destabilization) of this turn by respective D- (or L) amino-acid substitutions at GlyB8 is associated with reciprocal effects on activity. On the one hand, because in the T state GlyB8 exhibits a negative ɸ dihedral angle, characteristic of D-amino acids, diverse D-substitutions at B8 are compatible with native-like structure but low activity (Nakagawa et al., 2005; Hua et al., 2006c). On the other hand, L-amino-acid substitutions, consistent with the positive B8 ɸ angle of GlyB8 in the R state, can exhibit high activities despite markedly impaired stability and low yields of chain combination or yeast expression (Kristensen et al., 1997; Guo et al., 2005; Hua et al., 2006c). Accordingly, we have proposed that GlyB8 functions an “ambidextrous switch” between folding-competent (T-like) and active (R-like) conformations. In this model nascent T-like local structure in the proximal arm (including inter-domain hydrogen bonding and contacts by the imidazole ring of HisB5; Hua et al., 2006c) enhance the efficiency and fidelity of disulfide pairing. The present 1H–2H-exchange data highlight the flexibility of T-like structure in the B-chain and hence its amenability for structural reorganization on receptor binding.
The flexibility of insulin may also contribute to its susceptibility to degradation. Whereas zinc hexamer assembly permits pharmaceutical formulation of adequate shelf life, degradation at elevated temperatures complicates the transport, storage, and use of insulin in the developing world. Brange and colleagues have characterized two modes of degradation, chemical processes leading to changes in covalent structure (such as formation of iso-aspartic acid or disulfide-liked polymers; Brange et al., 1992b) and physical processes leading to aggregation-coupled denaturation and fibrillation (Brange et al., 1997). Conformational fluctuations enhance rates of both chemical and physical degradation. The transient nature of the majority of hydrogen bonds in an insulin monomer is thus likely to play a central role in such mechanisms. In this regard the anomalous dynamic properties of the A2–A8 α-helix (as manifest by rapid amide-proton exchange and attenuated 13Cα/β chemical shifts despite maintenance of helix-related NOE patterns) represent an “Achilles’ Heel” (Yang et al., 2010b). We speculate the molecular repair of this nascent helix may permit design of second-generation insulin analogs that are refractory to thermal degradation and so of humanitarian value in the developing world. One step in this direction has been taken based on solid-state NMR studies of insulin fibrils and analysis of β-branched side chains (Yang et al., 2010b). An alternative route toward stabilization of this segment may be provided by its tethering to the B-chain through the engineering of active single-chain analogs (Hua et al., 2008; Rajpal et al., 2009).
Following submission of this article, a complementary analysis of hydrogen bonds in insulin was presented (Antolíková et al., 2011). Remarkably, this and the present studies have put forward a similar perspective based on unrelated biophysical methodologies. Employing isothermal microcalorimetry, Brzozowski, Jirácek, and their respective colleagues exploited N-methylated insulin analogs to distinguish between the roles of inter- and intra-molecular hydrogen bonds at the dimer interface. Of particular interest, formation (or disruption) of the intra-molecular B25 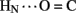 A19 hydrogen bond (unprotected in insulin lispro) was observed to correlate with the relative competency of the B-chain to form inter-molecular β-sheet-related hydrogen bonds (Antolíková et al., 2011)). Such coupling, supported by crystallographic analyses of variant zinc hexamers, highlights the non-local nature of a hydrogen-bond network and its potential functional relevance. The particular N-methylated insulin analogs studied by Antolíková et al. 2011 exemplify the power of chemical modification to dissect specific dynamic features of a globular protein.
A19 hydrogen bond (unprotected in insulin lispro) was observed to correlate with the relative competency of the B-chain to form inter-molecular β-sheet-related hydrogen bonds (Antolíková et al., 2011)). Such coupling, supported by crystallographic analyses of variant zinc hexamers, highlights the non-local nature of a hydrogen-bond network and its potential functional relevance. The particular N-methylated insulin analogs studied by Antolíková et al. 2011 exemplify the power of chemical modification to dissect specific dynamic features of a globular protein.
Conclusion
In the present study we have presented the solution structure of a prandial insulin analog as a monomer in solution. Dynamic characterization by amide-proton exchange has provided surprising insights into the lability of hydrogen bonds within and between the insulin chains. Although these hydrogen bonds are prominent features of classical crystal structures (Baker et al., 1988) and contribute to self-assembly (Dodson et al., 1993), their breakage is likely to contribute to the biological activity of the hormone as well as its degradation in pharmaceutical formulations. To our knowledge, this report represents the first quantitative analysis of 1H–2H PFs in an insulin analog. We envisage that a deeper understanding of the dynamics of insulin will enable design of second-generation analogs with application to the developing world.
Conflict of Interest Statement
Dr. Weiss holds stock in and is Chief Scientific Officer of Thermalin Diabetes, L.L.C.; he has also been a consultant to Merck, Inc. and the DEKA Research and Development Corp. The authors otherwise declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Prof. P. G. Katsoyannis and members of his laboratory at the Mt. Sinai School of Medicine (Y. C. Chu, S. Q. Hu, R. Y. Wang, and S. Wang) for synthesis of insulin analogs; S. H. Nakagawa and L. Whittaker for insulin receptor-binding assays; N. F. Phillips for assistance with biochemical procedures; Eli and Lilly and Co. for human insulin; Novo-Nordisk for radio-labeled insulin; J. Lee, D. N. Jones, K. Hallenga, and Y. Yang for NMR facility management and advice during this long-term research program; P. Arvan, G. G. Dodson, E. Dodson, B. H. Frank, and D. F. Steiner for helpful discussions; and M. Karplus and K. Wüthrich for encouragement in the early stages of this work. This work was supported in part by grants from the National Institutes of Health to Michael A. Weiss (DK040949 and DK079233) and represents a contribution from the Cleveland Center for Membrane and Structural Biology.
References
Antolíková, E., Žáková, L., Turkenburg, J. P., Watson, C. J., HanŽlová, I., Šanda, M., Cooper, A., Kraus, T., Brzozowski, A. M., and Jiráček, J. (2011). Non-equivalent role of inter- and intra-molecular hydrogen bonds in the insulin dimer interface. J. Biol. Chem. [Epub ahead of print].
Bai, Y., Milne, J. S., Mayne, L., and Englander, S. W. (1993). Primary structure effects on peptide group hydrogen exchange. Proteins 17, 75–86.
Bakaysa, D. L., Radziuk, J., Havel, H. A., Brader, M. L., Li, S., Dodd, S. W., Beals, J. M., Pekar, A. H., and Brems, D. N. (1996). Physicochemical basis for the rapid time-action of LysB28ProB29-insulin: dissociation of a protein-ligand complex. Protein Sci. 5, 2521–2531.
Baker, E. N., Blundell, T. L., Cutfield, J. F., Cutfield, S. M., Dodson, E. J., Dodson, G. G., Hodgkin, D. M., Hubbard, R. E., Isaacs, N. W., and Reynolds, C. D. (1988). The structure of 2Zn pig insulin crystals at 1.5 Å resolution. Philos. Trans. R. Soc. Lond. B Biol. Sci. 319, 369–456.
Brange, J., Andersen, L., Laursen, E. D., Meyn, G., and Rasmussen, E. (1997). Toward understanding insulin fibrillation. J. Pharm. Sci. 86, 517–525.
Brange, J., Hansen, J. F., Langkjaer, L., Markussen, J., Ribel, U., and Sorensen, A. R. (1992a). Insulin analogues with improved absorption characteristics. Horm. Metab. Res. Suppl. 26, 125–130.
Brange, J., Havelund, S., and Hougaard, P. (1992b). Chemical stability of insulin. 2. Formation of higher molecular weight transformation products during storage of pharmaceutical preparations. Pharm. Res. 9, 727–734.
Brange, J., and Langkjoer, L. (1993). Insulin structure and stability. Pharm. Biotechnol. 5, 315–350.
Brange, J., Ribel, U., Hansen, J. F., Dodson, G., Hansen, M. T., Havelund, S., Melberg, S. G., Norris, F., Norris, K., and Snel, L. (1988). Monomeric insulins obtained by protein engineering and their medical implications. Nature 333, 679–682.
Brange, J., Skelbaek-Pedersen, B., Langkjaer, L., Damgaard, U., Ege, H., Havelund, S., Heding, L. G., Jorgensen, H., Lykkeberg, J., Markussen, M., Pingel, M., and Rasmussen, E. (1987). Galenics of Insulin: The Physico-Chemical and Pharmaceutical Aspects of Insulin and Insulin Preparations. Berlin: Springer-Verlag.
Brunger, A. T. (1992). X-PLOR (Version 3.1) a System for X-Ray Crystallography and NMR. New Haven, CT: Yale University Press.
Bryant, C., Spencer, D. B., Miller, A., Bakaysa, D. L., McCune, K. S., Maple, S. R., Pekar, A. H., and Brems, D. N. (1993). Acid stabilization of insulin. Biochemistry 32, 8075–8082.
Campbell, R. K., Campbell, L. K., and White, J. R. (1996). Insulin lispro: its role in the treatment of diabetes mellitus. Ann. Pharmacother. 30, 1263–1271.
Carroll, R. J., Hammer, R. E., Chan, S. J., Swift, H. H., Rubenstein, A. H., and Steiner, D. F. (1988). A mutant human proinsulin is secreted from islets of Langerhans in increased amounts via an unregulated pathway. Proc. Natl. Acad. Sci. U.S.A. 85, 8943–8947.
Chothia, C., Lesk, A. M., Dodson, G. G., and Hodgkin, D. C. (1983). Transmission of conformational change in insulin. Nature 302, 500–505.
Chu, Y. C., Zong, L., Burke, G. T., and Katsoyannis, P. G. (1992). The A14 position of insulin tolerates considerable structural alterations with modest effects on the biological behavior of the hormone. J. Protein Chem. 11, 571–577.
Ciszak, E., Beals, J. M., Frank, B. H., Baker, J. C., Carter, N. D., and Smith, G. D. (1995). Role of C-terminal B-chain residues in insulin assembly: the structure of hexameric LysB28ProB29-human insulin. Structure 3, 615–622.
Creighton, T. E. (1997). Protein Structure: A Practical Approach. Oxford: IRL Press at Oxford University Press.
De Meyts, P., and Whittaker, J. (2002). Structural biology of insulin and IGF1 receptors: implications for drug design. Nat. Rev. Drug Discov. 1, 769–783.
DeFelippis, M. R., Chance, R. E., and Frank, B. H. (2001). Insulin self-association and the relationship to pharmacokinetics and pharmacodynamics. Crit. Rev. Ther. Drug Carrier Syst. 18, 201–264.
Dodson, G., and Steiner, D. (1998). The role of assembly in insulin’s biosynthesis. Curr. Opin. Struct. Biol. 8, 189–194.
Dodson, G. G., Dodson, E. J., Turkenburg, J. P., and Bing, X. (1993). Molecular recognition in insulin assembly. Biochem. Soc. Trans. 21, 609–614.
Englander, S. W., Sosnick, T. R., Englander, J. J., and Mayne, L. (1996). Mechanisms and uses of hydrogen exchange. Curr. Opin. Struct. Biol. 6, 18–23.
Garg, S. K., Ellis, S. L., and Ulrich, H. (2005). Insulin glulisine: a new rapid-acting insulin analogue for the treatment of diabetes. Expert Opin. Pharmacother. 6, 643–651.
Gerstein, H. C., Miller, M. E., Genuth, S., Ismail-Beigi, F., Buse, J. B., Goff, D. C. Jr., Probstfield, J. L., Cushman, W. C., Ginsberg, H. N., Bigger, J. T., Grimm, R. H. Jr., Byington, R. P., Rosenberg, Y. D., and Friedewald, W. T. (2011). Long-term effects of intensive glucose lowering on cardiovascular outcomes. N. Engl. J. Med. 364, 818–828.
Guo, Z. Y., Zhang, Z., Jia, X. Y., Tang, Y. H., and Feng, Y. M. (2005). Mutational analysis of the absolutely conserved B8Gly: consequence on foldability and activity of insulin. Acta Biochim. Biophys. Sin. (Shanghai) 10, 673–679.
Hansen, B. F., Kurtzhals, P., Jensen, A. B., Dejgaard, A., and Russell-Jones, D. (2011). Insulin X10 revisited: a super-mitogenic insulin analogue. Diabetologia 54, 2226–2231.
Hartemann-Heurtier, A., Sachon, C., Masseboeuf, N., Corset, E., and Grimaldi, A. (2003). Functional intensified insulin therapy with short-acting insulin analog: effects on HbA1c and frequency of severe hypoglycemia. An observational cohort study. Diabetes Metab. 29, 53–57.
Hartman, I. (2008). Insulin analogs: impact on treatment, success, satisfaction, quality of life and adherence. Clin. Med. Res. 6, 54–67.
Havel, T. F., and Snow, M. E. (1991). A new method for building protein conformations from sequence alignments with homologues of known structure. J. Mol. Biol. 217, 1–7.
Hober, S., Forsberg, G., Palm, G., Hartmanis, M., and Nilsson, B. (1992). Disulfide exchange folding of insulin-like growth factor I. Biochemistry 31, 1749–1756.
Hober, S., Hansson, A., Uhlen, M., and Nilsson, B. (1994). Folding of insulin-like growth factor I is thermodynamically controlled by insulin-like growth factor binding protein. Biochemistry 33, 6758–6761.
Hu, S. Q., Burke, G. T., Schwartz, G. P., Ferderigos, N., Ross, J. B., and Katsoyannis, P. G. (1993). Steric requirements at position B12 for high biological activity in insulin. Biochemistry 32, 2631–2635.
Hua, Q. X., Chu, Y. C., Jia, W., Phillips, N. B., Wang, R. Y., Katsoyannis, P. G., and Weiss, M. A. (2002a). Mechanism of insulin chain combination. Asymmetric roles of A-chain alpha-helices in disulfide pairing. J. Biol. Chem. 277, 43443–43453.
Hua, Q. X., Jia, W., Frank, B. H., Phillips, N. B., and Weiss, M. A. (2002b). A protein caught in a kinetic trap: structures and stabilities of insulin disulfide isomers. Biochemistry 41, 14700–14715.
Hua, Q. X., Gozani, S. N., Chance, R. E., Hoffmann, J. A., Frank, B. H., and Weiss, M. A. (1995). Structure of a protein in a kinetic trap. Nat. Struct. Biol. 2, 129–138.
Hua, Q. X., Hu, S. Q., Frank, B. H., Jia, W., Chu, Y. C., Wang, S. H., Burke, G. T., Katsoyannis, P. G., and Weiss, M. A. (1996a). Mapping the functional surface of insulin by design: structure and function of a novel A-chain analogue. J. Mol. Biol. 264, 390–403.
Hua, Q. X., Narhi, L., Jia, W., Arakawa, T., Rosenfeld, R., Hawkins, N., Miller, J. A., and Weiss, M. A. (1996b). Native and non-native structure in a protein-folding intermediate: spectroscopic studies of partially reduced IGF-I and an engineered alanine model. J. Mol. Biol. 259, 297–313.
Hua, Q. X., Hu, S. Q., Jia, W., Chu, Y. C., Burke, G. T., Wang, S. H., Wang, R. Y., Katsoyannis, P. G., and Weiss, M. A. (1998). Mini-proinsulin and mini-IGF-I: homologous protein sequences encoding non-homologous structures. J. Mol. Biol. 277, 103–118.
Hua, Q. X., Jia, W., Frank, B. H., and Weiss, M. A. (1993). Comparison of the dynamics of an engineered insulin monomer and dimer by acid-quenched amide proton exchange. Non-local stabilization of interchain hydrogen bonds by dimerization. J. Mol. Biol. 230, 387–394.
Hua, Q. X., Liu, M., Hu, S. Q., Jia, W., Arvan, P., and Weiss, M. A. (2006a). A conserved histidine in insulin is required for the foldability of human proinsulin. Structure and function of an AlaB5 analog. J. Biol. Chem. 281, 24889–24899.
Hua, Q. X., Mayer, J., Jia, W., Zhang, J., and Weiss, M. A. (2006b). The folding nucleus of the insulin superfamily: a flexible peptide model foreshadows the native state. J. Biol. Chem. 281, 28131–28142.
Hua, Q. X., Nakagawa, S. H., Hu, S. Q., Jia, W., Wang, S., and Weiss, M. A. (2006c). Toward the active conformation of insulin. Stereospecific modulation of a structural switch in the B chain. J. Biol. Chem. 281, 24900–24909.
Hua, Q. X., Nakagawa, S. H., Jia, W., Hu, S. Q., Chu, Y. C., Katsoyannis, P. G., and Weiss, M. A. (2001). Hierarchical protein folding: asymmetric unfolding of an insulin analogue lacking the A7-B7 interchain disulfide bridge. Biochemistry 40, 12299–12311.
Hua, Q. X., Nakagawa, S. H., Jia, W., Huang, K., Phillips, N. B., Hu, S. Q., and Weiss, M. A. (2008). Design of an active ultrastable single-chain insulin analog: synthesis, structure, and therapeutic implications. J. Biol. Chem. 283, 14703–14716.
Hua, Q. X., Shoelson, S. E., Kochoyan, M., and Weiss, M. A. (1991). Receptor binding redefined by a structural switch in a mutant human insulin. Nature 354, 238–241.
Hua, Q. X., and Weiss, M. A. (1990). Toward the solution structure of human insulin: sequential 2D 1H NMR assignment of a des-pentapeptide analogue and comparison with crystal structure. Biochemistry 29, 10545–10555.
Hua, Q. X., and Weiss, M. A. (1991). Comparative 2D NMR studies of human insulin and des-pentapeptide insulin: sequential resonance assignment and implications for protein dynamics and receptor recognition. Biochemistry 30, 5505–5515.
Huang, Y., Liang, Z., and Feng, Y. (2001). The relationship between the connecting peptide of recombined single chain insulin and its biological function. Sci. China C Life Sci. 44, 593–600.
Ismail-Beigi, F., Moghissi, E., Tiktin, M., Hirsch, I. B., Inzucchi, S. E., and Genuth, S. (2011). Individualizing glycemic targets in type 2 diabetes mellitus: implications of recent clinical trials. Ann. Intern. Med. 154, 554–559.
Jia, X. Y., Guo, Z. Y., Wang, Y., Xu, Y., Duan, S. S., and Feng, Y. M. (2003). Peptide models of four possible insulin folding intermediates with two disulfides. Protein Sci. 12, 2412–2419.
Kaarsholm, N. C., Norris, K., Jorgensen, R. J., Mikkelsen, J., Ludvigsen, S., Olsen, O. H., Sorensen, A. R., and Havelund, S. (1993). Engineering stability of the insulin monomer fold with application to structure-activity relationships. Biochemistry 32, 10773–10778.
Kristensen, C., Kjeldsen, T., Wiberg, F. C., Schaffer, L., Hach, M., Havelund, S., Bass, J., Steiner, D. F., and Andersen, A. S. (1997). Alanine scanning mutagenesis of insulin. J. Biol. Chem. 272, 12978–12983.
Lefebvre, P., and Pierson, A. (2004). The global challenge of diabetes. World Hosp. Health Serv. 40, 37–40, 42.
Melki, V., Renard, E., Lassmann-Vague, V., Boivin, S., Guerci, B., Hanaire-Broutin, H., Bringer, J., Belicar, P., Jeandidier, N., Meyer, L., Blin, P., Augendre-Ferrante, B., and Tauber, J. P. (1998). Improvement of HbA1c and blood glucose stability in IDDM patients treated with lispro insulin analog in external pumps. Diabetes Care 21, 977–982.
Milazzo, G., Sciacca, L., Papa, V., Goldfine, I. D., and Vigneri, R. (1997). ASPB10 insulin induction of increased mitogenic responses and phenotypic changes in human breast epithelial. Mol. Carcinog. 18, 19–25.
Miller, J. A., Narhi, L. O., Hua, Q. X., Rosenfeld, R., Arakawa, T., Rohde, M., Prestrelski, S., Lauren, S., Stoney, K. S., Tsai, L., and Weiss, M. A. (1993). Oxidative refolding of insulin-like growth factor 1 yields two products of similar thermodynamic stability: a bifurcating protein-folding pathway. Biochemistry 32, 5203–5213.
Milner, S. J., Carver, J. A., Ballard, F. J., and Francis, G. L. (1999). Probing the disulfide folding pathway of insulin-like growth factor-I. Biotechnol. Bioeng. 62, 693–703.
Nakagawa, S. H., Zhao, M., Hua, Q. X., Hu, S. Q., Wan, Z. L., Jia, W., and Weiss, M. A. (2005). Chiral mutagenesis of insulin. Foldability and function are inversely regulated by a stereospecific switch in the B chain. Biochemistry 44, 4984–4999.
Nathan, D. M., Cleary, P. A., Backlund, J. Y., Genuth, S. M., Lachin, J. M., Orchard, T. J., Raskin, P., Zinman, B., DCCT/EDIC Study Research Group. (2005). Intensive diabetes treatment and cardiovascular disease in patients with type 1 diabetes. N. Engl. J. Med. 353, 2643–2653.
Nicollerat, J. A. (2000). Implications of the United Kingdom Prospective Diabetes Study (UKPDS) results on patient management. Diabetes Educ. 26(Suppl.), 8–10.
Osei, K. (2003). Global epidemic of type 2 diabetes: implications for developing countries. Ethn. Dis. 13, S102–S106.
Qiao, Z. S., Guo, Z. Y., and Feng, Y. M. (2001). Putative disulfide-forming pathway of porcine insulin precursor during its refolding in vitro. Biochemistry 40, 2662–2668.
Qiao, Z. S., Min, C. Y., Hua, Q. X., Weiss, M. A., and Feng, Y. M. (2003). In vitro refolding of human proinsulin. Kinetic intermediates, putative disulfide-forming pathway, folding initiation site, and protential role of C-peptide in folding process. J. Biol. Chem. 278, 17800–17809.
Rajpal, G., Liu, M., Zhang, Y., and Arvan, P. (2009). Single-chain insulins as receptor agonists. Mol. Endocrinol. 23, 679–688.
Renner, R., Pfutzner, A., Trautmann, M., Harzer, O., Sauter, K., and Landgraf, R. (1999). Use of insulin lispro in continuous subcutaneous insulin infusion treatment. Results of a multicenter trial. German Humalog-CSII Study Group. Diabetes Care 22, 784–788.
Riddle, M. C. (2010). Effects of intensive glucose lowering in the management of patients with type 2 diabetes mellitus in the Action to Control Cardiovascular Risk in Diabetes (ACCORD) trial. Circulation 122, 844–846.
Roder, H. (1989). Structural characterization of protein folding intermediates by proton magnetic resonance and hydrogen exchange. Meth. Enzymol. 176, 446–473.
Roder, H., Wagner, G., and Wüthrich, K. (1985). Amide proton exchange in proteins by EX1 kinetics: studies of the basic pancreatic trypsin inhibitor at variable p2H and temperature. Biochemistry 24, 7396–7407.
Saltiel, A. R. (2001). New perspectives into the molecular pathogenesis and treatment of type-2 diabetes. Cell 104, 517–529.
Schwartz, G. P., Burke, G. T., and Katsoyannis, P. G. (1987). A superactive insulin: [B10-aspartic acid]insulin(human). Proc. Natl. Acad. Sci. U.S.A. 84, 6408–6411.
Selden, R. F., Skokiewicz, M. J., Howie, K. B., Russell, P. S., and Goodman, H. M. (1986). Regulation of human insulin gene expression in transgenic mice. Nature 321, 525–528.
Shoelson, S. E., Lu, Z. X., Parlautan, L., Lynch, C. S., and Weiss, M. A. (1992). Mutations at the dimer, hexamer, and receptor-binding, surfaces of insulin independently affect insulin-insulin and insulin-receptor interactions. Biochemistry 31, 1757–1767.
Sosnick, T. R., Fang, X., and Shelton, V. M. (2000). Application of circular dichroism to study RNA folding transitions. Meth. Enzymol. 317, 393–409.
Steiner, D. F. (1967). Evidence for a precursor in the biosynthesis of insulin. Trans. N. Y. Acad. Sci. 30, 60–68.
Stoy, J., Edghill, E. L., Flanagan, S. E., Ye, H., Paz, V. P., Pluzhnikov, A., Below, J. E., Hayes, M. G., Cox, N. J., Lipkind, G. M., Lipton, R. B., Greeley, S. A., Patch, A. M., Ellard, S., Steiner, D. F., Hattersley, A. T., Philipson, L. H., and Bell, G. I. (2007). Insulin gene mutations as a cause of permanent neonatal diabetes. Proc. Natl. Acad. Sci. U.S.A. 104, 15040–15044.
Tang, J. G., and Tsou, C. L. (1990). The insulin A and B chains contain structural information for the formation of the native molecule. Studies with protein disulphide-isomerase. Biochem. J. 268, 429–435.
The Diabetes Control Group and Complications Trial Research Group. (1993). The effect of intensive treatment of diabetes on the development and progression of long-term complications in insulin-dependent diabetes mellitus. N. Engl. J. Med. 329, 977–986.
Tripathi, B. K., and Srivastava, A. K. (2006). Diabetes mellitus: complications and therapeutics. Med. Sci. Monit. 12, RA130–RA147.
Wagner, G., and Wüthrich, K. (1986). Observation of internal motility of proteins by nuclear magnetic resonance in solution. Meth. Enzymol. 131, 307–326.
Wang, C. C., and Tsou, C. L. (1991). The insulin A and B chains contain sufficient structural information to form the native molecule. Trends Biochem. Sci. 16, 279–281.
Weiss, M. A. (2009). Proinsulin and the genetics of diabetes mellitus. J. Biol. Chem. 284, 19159–19163.
Weiss, M. A., Frank, B. H., Khait, I., Pekar, A., Heiney, R., Shoelson, S. E., and Neuringer, L. J. (1990). NMR and photo-CIDNP studies of human proinsulin and prohormone processing intermediates with application to endopeptidase recognition. Biochemistry 29, 8389–8401.
Weiss, M. A., Hua, Q. X., Lynch, C. S., Frank, B. H., and Shoelson, S. E. (1991). Heteronuclear 2D NMR studies of an engineered insulin monomer: assignment and characterization of the receptor-binding surface by selective 2H and 13C labeling with application to protein design. Biochemistry 30, 7373–7389.
Wüthrich, K., and Wagner, G. (1979). Nuclear magnetic resonance of labile protons in the basic pancreatic trypsin inhibitor. J. Mol. Biol. 130, 1–18.
Yang, Y., Hua, Q. X., Liu, J., Shimizu, E. H., Choquette, M. H., Mackin, R. B., and Weiss, M. A. (2010a). Solution structure of proinsulin: connecting domain flexibility and prohormone processing. J. Biol. Chem. 285, 7847–7851.
Yang, Y., Petkova, A., Huang, K., Xu, B., Hua, Q. X., Ye, I. J., Chu, Y. C., Hu, S. Q., Phillips, N. B., Whittaker, J., Ismail-Beigi, F., Mackin, R. B., Katsoyannis, P. G., Tycko, R., and Weiss, M. A. (2010b). An Achilles’ Heel in an amyloidogenic protein and its repair. Insulin dynamics, misfolding, and therapeutic design. J. Biol. Chem. 285, 10806–10821.
Keywords: diabetes mellitus, NMR spectroscopy, hydrogen bond, amide-proton exchange, protein structure, protein dynamics, protein engineering, protein therapeutics
Citation: Hua Q-X, Jia W and Weiss MA (2011) Conformational dynamics of insulin. Front. Endocrin. 2:48. doi: 10.3389/fendo.2011.00048
Received: 17 August 2011; Paper pending published: 04 September 2011;
Accepted: 18 September 2011; Published online: 18 October 2011.
Edited by:
Kenneth Siddle, University of Cambridge, UKReviewed by:
Andrzej Marek Brzozowski, University of York, UKRamasamy Paulmurugan, Stanford University, USA
Copyright: © 2011 Hua, Jia and Weiss. This is an open-access article subject to a non-exclusive license between the authors and Frontiers Media SA, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and other Frontiers conditions are complied with.
*Correspondence: Michael A. Weiss, Department of Biochemistry, School of Medicine, Case Western Reserve University, 10900 Euclid Avenue – Wood W436, Cleveland, OH 44106-4935, USA. e-mail:bWljaGFlbC53ZWlzc0BjYXNlLmVkdQ==
 Qing-Xin Hua
Qing-Xin Hua