- 1Department of Chemistry, University of Louisiana at Lafayette, Lafayette, LA, United States
- 2High Performance Computing, Frey Computing Services Center, Louisiana State University, Baton Rouge, LA, United States
Given the exponential growth of biochemical data and deep effect of computational methods on life sciences, there is a need to rethink undergraduate curricula. A project-oriented learning approach based on the Triangular Spatial Relationship (TSR) algorithm has been developed. The TSR-based method was designed for protein 3D structural comparison, motif discovery and probing molecular interactions. The uniqueness of the method benefits students’ learning of big data and computational methods. Specifically, students learn (i) how to search proteins of interest from the PDB archive, (ii) basic supercomputer skills, (iii) how to prepare datasets, (iv) how to perform protein structure and sequence analyses, (v) how to interpret the results, visualize protein structures and make graphs. Five specific strategies have been developed to achieve students’ highest potentials. (i) This lab exercise is designed as a project-oriented learning approach. (ii) The skills-first and concept-second approach is used. (iii) Students choose the proteins based on their interests. (iv) Students are encouraged to learn from each other to promote student–student interactions. (v) Students are required to write a report and/or present their studies. To assess students’ performance, we have developed an assessment rubric that includes (i) demonstration of supercomputer skills in job script preparation, submission and monitoring, (ii) skills in preparation of datasets, (iii) data analytical skills, (iv) project report, (v) presentation, and (vi) integration of the TSR-based method with other computational methods (e.g., molecular 3D structural visualization and protein sequence analysis). This project has been introduced in undergraduate biochemistry research and teaching labs for 4 years. Most students have learned the basic supercomputer skills as well as structure data analysis skills. Students’ feedback is positive and encouraging. It can be further developed as a module for an integrated computational chemistry lecture course.
1 Introduction
Big data, computational (bio)chemistry has not only spurred exponential growth in the life sciences but also has revolutionized the research process. Despite the spotlight on big data and computational science in both basic and applied life science fields, training for undergraduate students remains limited in the undergraduate curriculum at most universities. Protein sequence and structure data (Supplementary Figure 1) are becoming rapidly and increasingly available. As of November 2024, there are more than 227,000 structures freely available in the Protein Data Bank (PDB) (Berman et al., 2000) that promise to accelerate scientific discoveries in all areas of life sciences. Each protein structure solved by experiment is expected to require extensive specialized effort. Nearly every stage of the structure determination process needs to be refined and optimized by specialists in the field of protein structures. To make effective use of these experimental data, there is a growing need for more sensitive and automated comparison, search, and analysis tools for studying protein structures.
The Triangular Spatial Relationship (TSR)-based method was developed for comparing molecular 3D structures and probing molecular interactions (Kondra et al., 2021; Sarkar et al., 2021; Kondra et al., 2022; Sarkar et al., 2022; Sarkar et al., 2023; Milon et al., 2024). The input data for the TSR-based method are experimentally determined 3D structures from the PDB. A triangle using Cα atoms of the amino acids as the vertices is constructed for every combination of three amino acids of a protein structure. A TSR key (an integer) is computed using the length, angle and vertex labels of each triangle using a rule-based label-assignment formula, which ensures the assignment of the same key to identical TSRs across different proteins. Life sciences are going through a dramatic biotechnological revolution, producing huge amounts of data, which is often deposited in public databases. Biological or chemical science curricula for undergraduate students have hardly been altered to reflect this revolution (Rubinstein and Chor, 2014). The TSR-based method along with other computational software packages have been brought into the undergraduate research classes as well as the teaching lab in the past 4–5 years. In review of the first paper on the TSR-based method (Kondra et al., 2021), Dr. Xu designed a 1-credit hour research course offering a project-based learning experience for undergraduate students to use the TSR method. Sophomores, juniors or seniors majoring in biology or chemistry can take this course. Project-based learning introduces real-world problems and captures the students’ curiosity, motivating them to recognize and investigate abstract concepts and principles. This experience cultivates critical thinking skills during the course of their training in order to prepare them for their future careers.
Most biochemistry education research literature has been based on research in the lecture/classroom setting (Lang and Bodner, 2020). In contrast, the focuses of this study are to research students’ ability in the laboratory setting to (i) identify the proteins in which they are interested from the PDB, (ii) formulate their own questions and (iii) answer their own questions after they have learned the skills through hands-on training of analyzing big data using the computational tools. The learning objectives of the course include (i) introducing students to basic supercomputer skills, (ii) preparing students for independent data analysis and visualization of biological and chemical data, (iii) showing students how skills-based computational techniques can lead to biological or chemical discoveries, concomitantly emphasizing the importance of interdisciplinary training as well as integration of experimental and computational approaches in the future, (iv) stimulating student interests, encouraging creative ideas and training them to manage their time in order to accomplish specific and planned tasks. The strategies for achieving the learning objectives can be summarized as follows: (i) This lab exercise or undergraduate research experience is designed as a project-oriented learning approach. Traditionally, instructor-focused lecturing in STEM classrooms is effective. In contrast, the project-oriented learning lays the foundation for students to become the driving force for their own learning. Studies show that active learning benefits students’ education (Nguyen et al., 2021). (ii) The skills-first and concept-second approach is used. Skill learning through hands-on training often facilitates longer knowledge retention longer. (iii) Students are encouraged to develop their interests and take charge of the project through allowing them to choose the proteins based on their interests. (iv) Students are encouraged to learn from each other to promote student–student interactions. (v) Students are required to write a report and/or present their work. This provides students an opportunity for recognizing biological and/or chemical theories by drawing conclusions from their own results. In order for the instructors, whose research and teaching expertise is not strictly computational, to be able to use this method, we intend to provide the necessary technical details. Most details will be included in the Supplementary figures and files. Some of the Supplementary files also follow the styles and standards of the tutorial articles (Fox and Ouellette, 2013) where theoretical context as well as the type of questions and how to answer them are provided. It is expected that this study may be of value to others teaching traditional biochemistry.
A few hands-on computational experiences on concepts such as 3D molecular structures, stereochemistry and data analyses have achieved positive impacts on students’ learning (Esselman and Hill, 2016; Winfield et al., 2019; Esselman and Hill, 2019; Wright et al., 2019; Rodríguez-Becerra et al., 2020). Computational approaches are not the mere use of software tools, but the integration of computational algorithms and mathematical formulas to experimental design. If only use of software package, students may employ computational tools as “black boxes” without a deep understanding of the computational concepts and underlying assumptions (Rubinstein and Chor, 2014; May, 2004). Section 2 focuses on the use of software packages while Section 3 is to achieve in-depth understanding of 3D structural relationships. In fact, employers are looking for well-rounded graduates who not only understand the practical aspects of computational biochemistry, but also understand the concepts (Holien et al., 2023).
2 The specific computational skills that students have learned
Students in the undergraduate research course first go through a hands-on tutorial using their own laptops following along with the instructor going through the steps of protein 3D structure analysis for a case that has been published. Students then, either individually or in groups, choose their own protein structure to investigate. By doing this they develop the following computational skills.
2.1 Students have learned how to search proteins of interest from the PDB archive
The search can be carried out through key words or an input of a protein amino acid sequence.1 Students can learn this step quickly without difficulty.
2.2 Students have learned how to connect their own computer to supercomputers for job submission and for file transfer between their own computers and supercomputers
The example we present is for a windows computer. Students need to download two software packages: WinSCP and Putty.2 WinSCP allows students to connect their own computers to the supercomputers and to transfer files between the local computers and supercomputers. Putty allows students to submit a job to the supercomputers and monitor their jobs. Most college students use Mac Book computers in school. For a Mac Book user, we have also prepared detailed instructions.
2.3 Students have learned how to prepare datasets, submit and monitor jobs
Biochemistry has transformed from insular entities into interdisciplinary sciences, which in turn demand cross-disciplinary training for future work force in modern sciences. Despite its importance, computational (bio)chemistry still has a somewhat limited presence in undergraduate curricula (Qin, 2009; Lehtola and Karttunen, 2022). The TSR-based software package for this study can be downloaded from GitHub.3 The detailed instructions for downloading this software package can be found in Supplementary File 1. The package contains six Python codes. The first code (Code #1) is to download the input structural files from the PDB. The second code (Code #2) is to generate key and triplet files for each of the downloaded PDB files using the key generation mathematic formula (Kondra et al., 2021). The third code (Kondra et al., 2021) (Code #3) is to perform pairwise structural comparisons using Generalized Jaccard coefficient method and hierarchical clustering. The fourth code (Code #4) is to show clustering results. The fifth code (Code #5) is to calculate the common substructures through computing the numbers of commonly shared TSR keys. The sixth code (Code #6) is to calculate the unique substructures exclusively belonging to a certain (sub)type of proteins. We have prepared detailed instructions on the input files, output files and procedures of running each code (Supplementary File 2).
2.4 Students have learned how to perform protein sequence analyses
Genomics including structural biology are big data science that has reshaped all disciplines of life sciences, facilitating an explosion of sequence data that can be generated for nearly any organisms (Koboldt et al., 2013). Conservative estimates put the volume genomic data doubling every 7 months, with zetabase-levels (1021) of sequences and associated heterogeneous metadata generated for 1.2 million species by the year 2025 (Stephens et al., 2015). The achievements of ‘big data science’ require integration of computer skills across several fields (Schatz, 2012). In fact, computer skills have become ubiquitous in many areas of biological and chemical research (Gallagher et al., 2011). In this study, students have learned how to perform multiple protein alignment using SnapGene. They have also learned how to conduct sequence alignment and phylogenetic analyses using MEGA7 (Kumar et al., 2016).
2.5 Students have learned how to visualize protein structures
One of the most important skills for a biochemist is to be able to visualize a protein, find a ligand binding site, and show 3D interactions with the coordinated amino acid residues (Abdinejad et al., 2021). It is important to educate students how to benefit from the up-to-date scientific data and software (Pine and Paina, 2020). Students have learned hierarchical cluster analysis (Ackerman and Ben-David, 2016) and reduced protein 3D structure dimensions using the Multidimensional Scaling (MDS) method (Kruskal and Wish, 1978). Students have prepared structural images using the Visual Molecular Dynamics (VMD) package (Humphrey et al., 1996).
2.6 Students have learned how to make graphs
Students have learned how to make different types of graphs using OriginPro and have learned how to make Venn diagram using Venn Diagram Plotter.4
3 Tutorial: a case study using caspases as an example for showing the capacities and applications of the TSR-based algorithm in undergraduate education
One student group chose to investigate caspases for their project and we use that student-led study as an example for demonstrating what students can learn from using the TSR-based computational software package and other computational tools.
3.1 Preparation of the caspase dataset
The name caspase is an abbreviation of cysteine-dependent, aspartate-specific peptidase, because caspases have a dominant specificity for protein substrates (P4P3P2P1) that contain a highly conserved aspartate at the P1 position (the cleavage site) (Thornberry, 1997; Nicholson, 1999; Chéreau et al., 2003) and a preferable glutamate at the P3 position (Nicholson, 1999). Functionally, the caspases are major regulators of apoptotic cell death pathways, proliferation and inflammation, which play vital roles in the life and death of animal cells (McLuskey and Mottram, 2015). Caspases were classified as either inflammatory (caspases 1, 4 and 5) or apoptotic caspases, with the latter being further organized into initiator (caspases 2, 8, 9 and 10) and effector (caspases 3, 6 and 7) caspases (Pop and Salvesen, 2009). The students initially prepared a small caspase dataset and expanded the small dataset to a large caspase dataset. The students hierarchically arranged the small dataset based on their functional classification (Supplementary Figure 2).
3.2 Illustration of caspase structural relationship using a hierarchical cluster analysis
Hierarchical cluster analysis is a popular machine learning method for big data research aiming to establish a hierarchy of clusters (Petushkova et al., 2014). The result shows one large cluster for caspases 4, 6, 7, 8 and 9, and three small clusters for caspases 1, 2 and 3. Caspases 3, 6 and 7 are effectors. Caspases 6 and 7 are grouped together while caspases 3 are separated from caspases 6 and 7. A similar situation is observed for initiators (caspases 2, 8 and 9). Inflammation caspases 1 and 4 are not grouped together (Supplementary Figure 2A). The MDS analysis of caspase structures agrees with the hierarchical cluster analysis (Supplementary Figure 2B). The structural clusters of the caspases do not perfectly agree with their functional classifications (Supplementary Figures 2A, 3). One of the reasons is due to the difference in size of the caspases. Caspases 4, 6, 7, 8 and 9 are larger than caspases 1, 2 and 3 (Supplementary Figure 2A). Their corresponding amino acid sequence analysis using MEGA does not perfectly match with their functional classifications (Supplementary Figures 3, 4) as well as structural clusters (Supplementary Figures 2A, 4). Students were asked to suggest how to improve the method for addressing the mismatches between structures and functions as well as sequences and functions. However, students were not required to implement their hypotheses.
As stated earlier, the caspase dataset was hierarchically arranged (Figure 1A). The root, family, subfamily and leaf node of the caspase hierarchical organization are shown in Figure 1B. At the root of the caspase dataset, the numbers of distinct and total TSR keys were calculated by without and with counting the key occurrence frequencies, respectively (Figure 1C). The distinct TSR keys represent unique smallest substructures (triangles) of the caspase dataset and the total TSR keys represent all the smallest substructures. A triangle constituted by three Cα atoms is considered as the smallest substructure in this study. At the family level, the numbers of specific and Common TSR keys were calculated. The specific TSR keys represent the substructures that are exclusively found in the family of either inflammation caspases or initiators (Figure 1D). The Common TSR keys represent the substructures shared by inflammation caspases and initiators (Figure 1E). Similar to the family level, the numbers of specific (Figure 1F) and Common (Figure 1G) TSR keys were calculated for the caspase subfamily level. At the leaf node level, the numbers of specific TSR keys representing unique substructures were calculated for each type of caspases (Figure 1H). The distinct and total common TSR keys represent the substructures that can be found in every structure at the level of family, subfamily or leaf node (Figure 1I). The numbers of common TSR keys decreases or remain the same from the root to the family level and then to the subfamily level (Figure 1I). Both common and Common TSR keys represent common substructures. However, the common TSR keys represent common substructures that can be found in every structure while the Common TSR keys represent common substructures that are shared by two or more families (apoptotic and inflammation), subfamilies (initiator, effector and inflammation) or types (caspases 1, 2, 3, 4, 6, 7, 8, and 9) and may not be present in every structure of each (sub)family or each type.
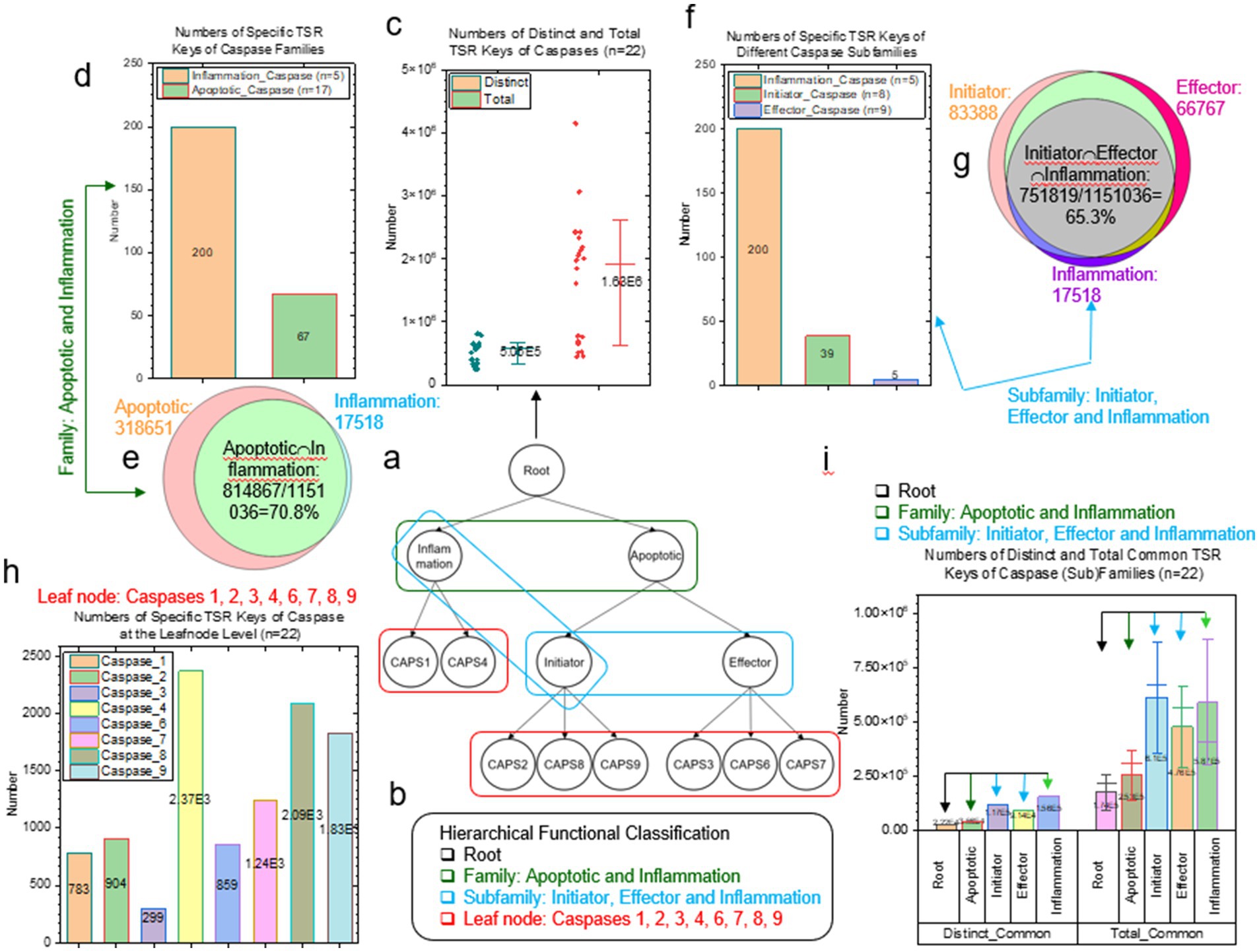
Figure 1. Different types of TSR keys were used to interpret the hierarchical structural relationships of the caspases. (A) The small caspase dataset was hierarchically arranged. The different colors illustrate the levels of the hierarchical arrangement. Black, green, blue and red colors represent the root, the level 1, the level 2 and the leaf node; (B) The functional classifications of the caspases at the levels of the root, family, subfamily and leaf node; (C) Distinct and total TSR keys of the dataset (the root) were calculated and are present. The number of the caspases in the dataset, the average values and SDs are labeled; (D) The specific TSR keys of the caspases at the family level were calculated and are present. The average values and the numbers of apoptotic and inflammatory caspase families are labeled; (E) The Venn diagram shows the numbers of the TSR keys exclusively belonging to each of the groups of apoptotic and inflammatory caspases and the intersection between the two groups. The percentage of the intersection is shown; (F) The specific TSR keys of the caspases at the subfamily level were calculated and are present. The average values and the numbers of inflammatory, initiator and effector caspases are labeled; (G) The Venn diagram shows the numbers of the TSR keys exclusively belonging to each of the groups of inflammatory, initiator and effector caspases and the intersection among the three groups. The percentage of such the intersection is shown; (H) The specific TSR keys of each type of caspases were calculated and are present. The caspases and the average values are labeled; (I) Distinct and total common TSR keys were calculated for the root, families and subfamilies of the caspases. The average values and SDs are labeled. The color arrows represent the levels of the hierarchical arrangement.
3.3 Search of a specific substructure against protein structure datasets
Basic Local Alignment Search Tool (BLAST) is a commonly used search program where searching of similar sequences can be accomplished for an input nucleic acid or polypeptide sequence (Altschul et al., 1990; Altschul et al., 1997). However, the counterpart of BLAST 1D sequence search for an input 3D substructure has not been developed. The unique representation of a protein 3D structure by TSR keys allows for BLAST 3D structure searches. Caspase inhibitors have been designed and synthesized as a potential therapeutic agent for the treatment of various diseases (Dhani et al., 2021). A specific caspase 3 inhibitor (Becker et al., 2004) was chosen as an example of 3D substructure search. Elevated levels of caspase 3 activity were also described in brains of Alzheimer’s disease patients (D’Amelio et al., 2012) and its inhibition was able to restore synaptic transmission and memory deficiency in Alzheimer’s disease transgenic mice (D'Amelio et al., 2011). The five representative residues that are close to the caspase 3 inhibitor (0ZZ) are identified (Figure 2A). Those five residues are conserved in the selected different types of caspases (Supplementary Figure 5). There are 10 distinct TSR keys corresponding to these five residues. Such 10 keys represent the specific substructure for part of the caspase 3 inhibitor binding site. The result by searching 12 datasets demonstrates that these 10 TSR keys are exclusively belonging to caspase 3 with the inhibitor 0ZZ (PDB: 1RHQ) (Figure 2B).
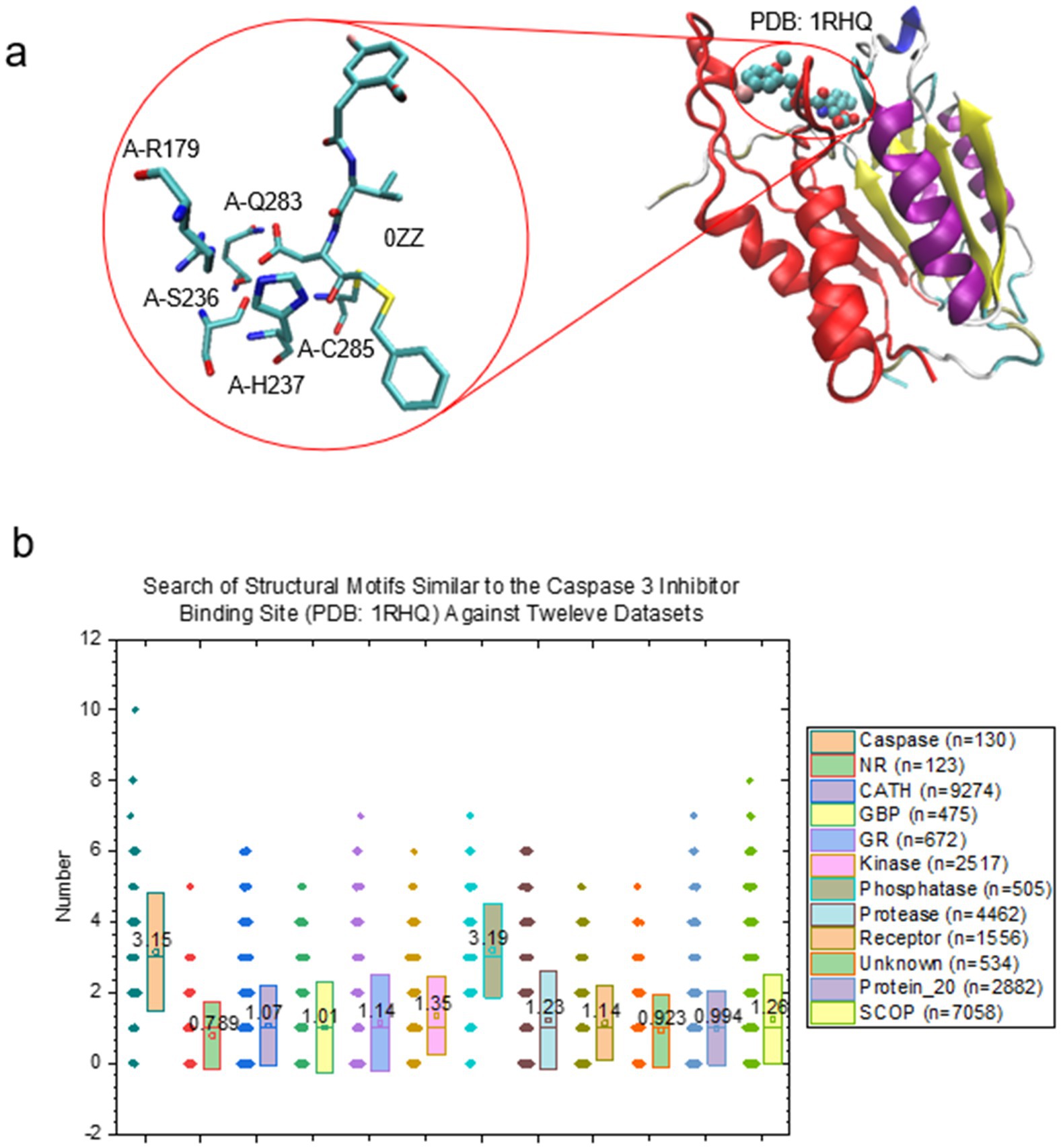
Figure 2. The capacity of key search function using the caspase 3 inhibitor binding site as an example. (A) The substructure of caspase 3 inhibitor 0ZZ. The five residues that closely interact with 0ZZ are labeled. The PDB is 1RHQ; (B) The key search result against the 12 protein structure datasets ranging from 123 to 9,274 structures. NR, neurotransmitter receptors; GBP, glutamate binding protein; GR, glutamate receptor.
4 Educational outcomes
To assess the students’ performances, we have developed a rubric (Supplementary File 3) that includes six criteria: (i) basic supercomputer skills, (ii) skills in preparation of datasets, (iii) data analytical skills, (iv) students’ report, (v) students’ presentation and (vi) the ability to integrate two or more different computational software packages. Students are encouraged to achieve discoveries, to gain teaching and communication experiences by training other students or performing group studies. However, these are not used to score their performances. The outcomes of the project-oriented learning were summarized in Figure 3. From the spring of 2021 to the fall of 2023, 44 undergraduate students selected the proteins based on their interests and studied protein structures using the TSR-based method. After completing undergraduate research class (CHEM 362) for one semester, all 44 students can independently prepare protein datasets by searching for proteins of interest from the PDB, correctly and independently submit and monitor jobs, transfer files between local computers and supercomputers, and independently perform cluster analysis, interpret clustering results analyze results and draw the conclusions based on the data and results (Figure 3). All the students gave a 10-min presentation at the end. Students have created a learning environment where students help to train students. Besides the TSR-based method for protein structure analysis,5 protein sequence analysis software, e.g., MEGA, SnapGene, and visualization software package, e.g., VMD, have been introduced to the students. 15–25% of the students can independently perform 3D visualization (~25%), sequence analysis (~20%), or both (~15%) (Figure 3). The detailed assessment rubric basic supercomputer skills, preparation of dataset structural analysis, report, presentation, structural visualization and sequence analyses were developed (Supplementary File 3). Students’ feedback for this exercise can be found in Supplementary File 4.
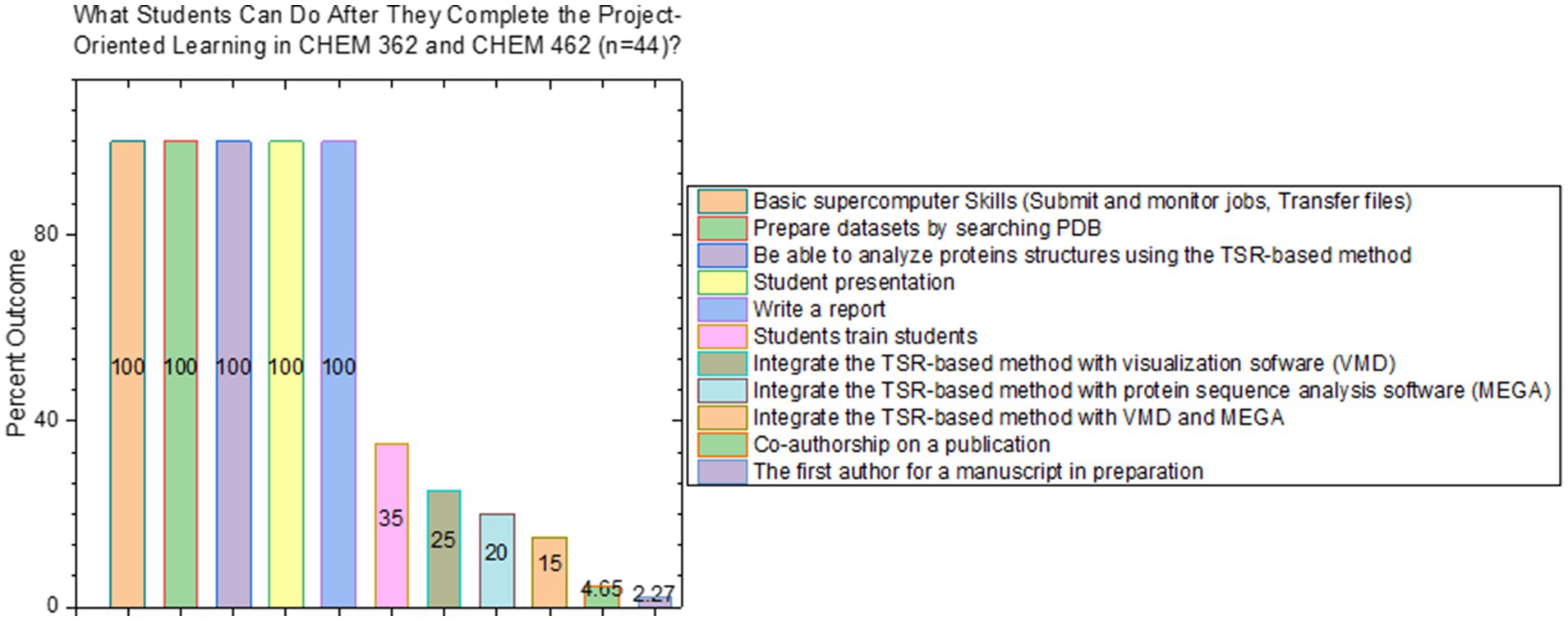
Figure 3. Summary of the outcomes when the project-oriented learning strategy has been introduced in the undergraduate research classes. The x axis shows the categories that students were assessed and the y axis the percentages of the students who have achieved “satisfaction.” “satisfaction” means that students can independently accomplish a task without help or with a minimal help from the instructor.
5 Discussion
The traditional way of teaching strategy is that teachers give a lecture, students take notes and are tested on the information. Research has highlighted the importance of practical skills (White et al., 2013). A specific project-oriented learning strategy has been developed for undergraduate biochemistry research or teaching labs. The results from a previous study showed that project-based learning, compared with the traditional teaching model, significantly improved students’ learning outcomes (Zhang and Ma, 2023). The studies also demonstrated that the course-based undergraduate research experiences improve students’ hands-on abilities (He et al., 2024). As stated earlier, the skills first and concepts second approach is used. First, students will learn computational skills. Second, they will prepare their own protein datasets based on their interests. Third, they will analyze the proteins of interest using the computational skills they have learned. Finally, the basic principles for the computational method will be introduced to the students after they have generated and analyzed their own data. Students will have an opportunity to prepare as many protein datasets as possible for broadening their knowledge as well as an opportunity to deepen the study toward a mechanistic understanding of a subject. On the teachers’ side, instructors will have an opportunity to cultivate students’ interest in science and teach creative thinking skills. This project-oriented learning module complements the traditional teaching approach. When it was used in a research lab, both presentations and formal reports are required. When it was used in a teaching lab, only formal reports were required. Students can be roughly classified into two groups: the primary objective of one group is to pass the course and the primary objective of the other group is to learn research-oriented skills and research thinking. Specific strategies have been developed to motivate both groups to achieve their highest potentials. The data structure of the TSR-based method is a vector of integers that are well-suited for incorporating an artificial intelligence component in the undergraduate curriculum. This project could also be integrated with a computational chemistry lecture course. The limitation of this lab exercise is that an effective study of ≥50 protein structures requires teachers and students to have access to supercomputer clusters. If <50 protein structures, especially <20 protein structures, will be studied, the updated version of the TSR-based method can be executed on personal computers. In summary, the course objectives, knowledge delivery strategies and evaluation plans are illustrated in Figure 4. A lab exercise procedure has been developed for the users of Windows and MacBook computers to use the TSR-based method (Supplementary File 2). Additional educational outcomes can be found in Supplementary File 5. We observed a higher class-attendance and a higher performance in Biochemistry I (CHEM 317) and II (CHEM 417) lecture courses for the students who conducted this lab exercise than the students who did not do this lab exercise. We conclude that the students who have performed better in biochemistry lecture courses want to gain undergraduate research experiences (Supplementary File 5).
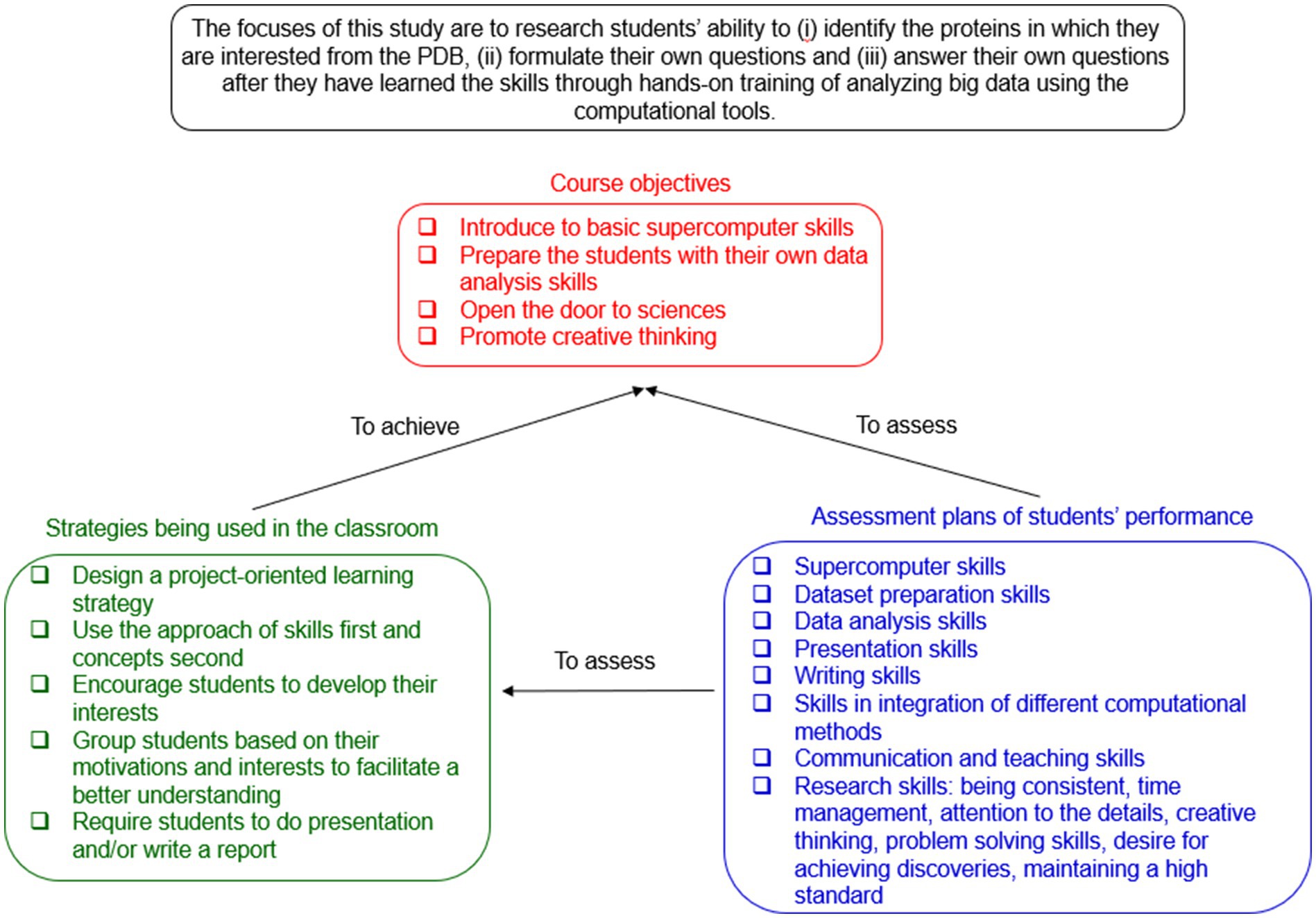
Figure 4. What were researched, the course objectives, knowledge delivery strategies and evaluation plans are summarized. The relationships between objective, strategy and evaluation are illustrated.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
CR: Data curation, Formal analysis, Investigation, Methodology, Validation, Visualization, Writing – original draft, Writing – review & editing. SM: Data curation, Formal analysis, Investigation, Methodology, Validation, Visualization, Writing – original draft, Writing – review & editing. JG: Data curation, Formal analysis, Investigation, Methodology, Validation, Visualization, Writing – original draft, Writing – review & editing, Conceptualization. SD: Data curation, Formal analysis, Investigation, Methodology, Validation, Visualization, Writing – original draft, Writing – review & editing. TM: Methodology, Visualization, Writing – original draft, Writing – review & editing, Resources, Software. FC: Software, Writing – original draft, Writing – review & editing. WX: Writing – original draft, Writing – review & editing, Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Visualization.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study is supported by NIH NIGMS (1R15GM144944-01).
Acknowledgments
Most of this research was conducted with high-performance computational resources provided by the Louisiana Optical Network Infrastructure (http://www.loni.org). Here we want to appreciate the LONI support team, especially Jianxiong Li, and Oleg Starovoytov. WX acknowledges Dr. Kathleen D. Knierim (Physical Chemist) for fruitful discussion on this project.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/feduc.2024.1455173/full#supplementary-material
Footnotes
1. ^https://www.rcsb.org/docs/search-and-browse/advanced-search/search-examples
2. ^https://winscp.net/downloads.php
3. ^https://github.com/dbxmcf/wu_sizegap.git
4. ^https://pnnl-comp-mass-spec.github.io/Venn-Diagram-Plotter/
5. ^The source code is available for academic users on GitHub https://github.com/WuXu26/Protein-3D-TSR.
References
Abdinejad, M., Talaie, B., Qorbani, H. S., and Dalili, S. (2021). Student perceptions using augmented reality and 3D visualization technologies in chemistry education. J. Sci. Educ. Technol. 30, 87–96. doi: 10.1007/s10956-020-09880-2
Ackerman, M., and Ben-David, S. (2016). A characterization of linkage-based hierarchical clustering. J. Mach. Learn. Res. 17, 8182–8198. Available at: https://dl.acm.org/doi/abs/10.5555/2946645.3053512
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. doi: 10.1093/nar/25.17.3389
Becker, J. W., Rotonda, J., Soisson, S. M., Aspiotis, R., Bayly, C., Francoeur, S., et al. (2004). Reducing the peptidyl features of caspase-3 inhibitors: a structural analysis. J. Med. Chem. 47, 2466–2474. doi: 10.1021/jm0305523
Berman, H. M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T. N., Weissig, H., et al. (2000). The protein data bank. Nucleic Acids Res. 28, 235–242. doi: 10.1093/nar/28.1.235
Chéreau, D., Kodandapani, L., Tomaselli, K. J., Spada, A. P., and Wu, J. C. (2003). Structural and functional analysis of caspase active sites. Biochemistry 42, 4151–4160. doi: 10.1021/bi020593l
D’Amelio, M., Sheng, M., and Cecconi, F. (2012). Caspase-3 in the central nervous system: beyond apoptosis. Trends Neurosci. 35, 700–709. doi: 10.1016/j.tins.2012.06.004
D'Amelio, M., Cavallucci, V., Middei, S., Marchetti, C., Pacioni, S., Ferri, A., et al. (2011). Caspase-3 triggers early synaptic dysfunction in a mouse model of Alzheimer's disease. Nat. Neurosci. 14, 69–76. doi: 10.1038/nn.2709
Dhani, S., Zhao, Y., and Zhivotovsky, B. (2021). A long way to go: caspase inhibitors in clinical use. Cell Death Dis. 12:949. doi: 10.1038/s41419-021-04240-3
Esselman, B. J., and Hill, N. J. (2016). Integration of computational chemistry into the undergraduate organic chemistry laboratory curriculum. J. Chem. Educ. 93, 932–936. doi: 10.1021/acs.jchemed.5b00815
Esselman, B. J., and Hill, N. J. (2019). Integrating computational chemistry into an organic chemistry laboratory curriculum using WebMO, using computational methods to teach chemical principles NW, Washington, DC: American Chemical Society, 139–162.
Fox, J. A., and Ouellette, B. F. (2013). Education in computational biology today and tomorrow. PLoS Comput. Biol. 9:e1003391. doi: 10.1371/journal.pcbi.1003391
Gallagher, S. R., Coon, W., Donley, K., Scott, A., and Goldberg, D. S. (2011). A first attempt to bring computational biology into advanced high school biology classrooms. PLoS Comput. Biol. 7:e1002244. doi: 10.1371/journal.pcbi.1002244
He, Y., Li, S., Chen, Z., Liu, B., and Luo, X. (2024). Knowledge-map analysis of undergraduate biochemistry teaching research: a bibliometric study from 2012 to 2021. J. Chem. Educ. 101, 307–318.
Holien, J. K., Coff, L., Guy, A. J., and Boer, J. C. (2023). Drug discovery in real life: an online learning activity for bioinformatics students. J. Chem. Educ. 100, 1053–1057. doi: 10.1021/acs.jchemed.2c00993
Humphrey, W., Dalke, A., and Schulten, K. (1996). VMD: visual molecular dynamics. J. Mol. Graph. 14, 33–38. doi: 10.1016/0263-7855(96)00018-5
Koboldt, D. C., Steinberg, K. M., Larson, D. E., Wilson, R. K., and Mardis, E. R. (2013). The next-generation sequencing revolution and its impact on genomics. Cell 155, 27–38. doi: 10.1016/j.cell.2013.09.006
Kondra, S., Chen, F., Chen, Y., Chen, Y., Collette, C. J., and Xu, W. (2022). A study of a hierarchical structure of proteins and ligand binding sites of receptors using the triangular spatial relationship-based structure comparison method and development of a size-filtering feature designed for comparing different sizes of protein structures. Proteins 90, 239–257. doi: 10.1002/prot.26215
Kondra, S., Sarkar, T., Raghavan, V., and Xu, W. (2021). Development of a TSR-based method for protein 3-D structural comparison with its applications to protein classification and motif discovery. Front. Chem. 8:602291. doi: 10.3389/fchem.2020.602291
Kruskal, J. B., and Wish, M. (1978). Multidimensional scaling. Thousand Oaks, California: SAGE Publications, Inc.
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lang, F. K., and Bodner, G. M. (2020). A review of biochemistry education research. J. Chem. Educ. 97, 2091–2103. doi: 10.1021/acs.jchemed.9b01175
Lehtola, S., and Karttunen, A. J. (2022). Free and open source software for computational chemistry education. WIREs Comput. Mol. Sci. 12:e1610. doi: 10.1002/wcms.1610
May, R. M. (2004). Uses and abuses of mathematics in biology. Science 303, 790–793. doi: 10.1126/science.1094442
McLuskey, K., and Mottram, J. C. (2015). Comparative structural analysis of the caspase family with other clan CD cysteine peptidases. Biochem. J. 466, 219–232. doi: 10.1042/BJ20141324
Milon, T. I., Wang, Y., Fontenot, R. L., Khajouie, P., Villinger, F., Raghavan, V., et al. (2024). Development of a novel representation of drug 3D structures and enhancement of the TSR-based method for probing drug and target interactions. Comput. Biol. Chem. 112:108117. doi: 10.1016/j.compbiolchem.2024.108117
Nguyen, K. A., Borrego, M., Finelli, C. J., DeMonbrun, M., Crockett, C., Tharayil, S., et al. (2021). Instructor strategies to aid implementation of active learning: a systematic literature review. Int. J. STEM Educ. 8:9. doi: 10.1186/s40594-021-00270-7
Nicholson, D. W. (1999). Caspase structure, proteolytic substrates, and function during apoptotic cell death. Cell Death Differ. 6, 1028–1042. doi: 10.1038/sj.cdd.4400598
Petushkova, N. A., Pyatnitskiy, M. A., Rudenko, V. A., Larina, O. V., Trifonova, O. P., Kisrieva, J. S., et al. (2014). Applying of hierarchical clustering to analysis of protein patterns in the human cancer-associated liver. PLoS One 9:e103950. doi: 10.1371/journal.pone.0103950
Pine, P., and Paina, L. I. (2020). Computational methods in chemistry and biochemistry education: visualization of proteins. Comput. Sci. Eng. 22, 45–49. doi: 10.1109/MCSE.2019.2962118
Pop, C., and Salvesen, G. S. (2009). Human caspases: activation, specificity, and regulation. J. Biol. Chem. 284, 21777–21781. doi: 10.1074/jbc.R800084200
Qin, H. (2009). Teaching computational thinking through bioinformatics to biology students. SIGCSE Bull. 41, 188–191. doi: 10.1145/1539024.1508932
Rodríguez-Becerra, J., Cáceres-Jensen, L., Díaz, T., Druker, S., Bahamonde Padilla, V., Pernaa, J., et al. (2020). Developing technological pedagogical science knowledge through educational computational chemistry: a case study of pre-service chemistry teachers’ perceptions. Chem. Educ. Res. Pract. 21, 638–654. doi: 10.1039/C9RP00273A
Rubinstein, A., and Chor, B. (2014). Computational thinking in life science education. PLoS Comput. Biol. 10:e1003897. doi: 10.1371/journal.pcbi.1003897
Sarkar, T., Chen, Y., Wang, Y., Chen, Y., Chen, F., Reaux, C. R., et al. (2023). Introducing mirror-image discrimination capability to the TSR-based method for capturing stereo geometry and understanding hierarchical structure relationships of protein receptor family. Comput. Biol. Chem. 103:107824. doi: 10.1016/j.compbiolchem.2023.107824
Sarkar, T., Raghavan, V. V., Chen, F., Riley, A., Zhou, S., and Xu, W. (2021). Exploring the effectiveness of the TSR-based protein 3-D structural comparison method for protein clustering, and structural motif identification and discovery of protein kinases, hydrolase, and SARS-CoV-2’s protein via the application of amino acid grouping. Comput. Biol. Chem. 92:107479. Available at: https://www.sciencedirect.com/science/article/abs/pii/S1476927121000463
Sarkar, T., Reaux, C. R., Li, J., Raghavan, V. V., and Xu, W. (2022). The specific applications of the TSR-based method in identifying Zn2+ binding sites of proteases and ACE/ACE2. Data Brief 45:108629. doi: 10.1016/j.dib.2022.108629
Schatz, M. C. (2012). Computational thinking in the era of big data biology. Genome Biol. 13:177. doi: 10.1186/gb-2012-13-11-177
Stephens, Z. D., Lee, S. Y., Faghri, F., Campbell, R. H., Zhai, C., Efron, M. J., et al. (2015). Big data: astronomical or genomical? PLoS Biol. 13:e1002195. doi: 10.1371/journal.pbio.1002195
Thornberry, N. A. (1997). The caspase family of cysteine proteases. Br. Med. Bull. 53, 478–490. doi: 10.1093/oxfordjournals.bmb.a011625
White, H. B., Benore, M. A., Sumter, T. F., Caldwell, B. D., and Bell, E. (2013). What skills should students of undergraduate biochemistry and molecular biology programs have upon graduation? Biochem. Mol. Biol. Educ. 41, 297–301. doi: 10.1002/bmb.20729
Winfield, L. L., McCormack, K., and Shaw, T. (2019). Using iSpartan to support a student-centered activity on alkane conformations. J. Chem. Educ. 96, 89–92. doi: 10.1021/acs.jchemed.8b00145
Wright, A. M., Schwartz, R. S., Oaks, J. R., Newman, C. E., and Flanagan, S. P. (2019). The why, when, and how of computing in biology classrooms. F1000Res. 8:1854. doi: 10.12688/f1000research.20873.1
Keywords: project-oriented learning, biochemistry lab, protein 3D structure, TSR-based method, big data analysis
Citation: Reaux CR, Meche SA, Grider JM, Dhanabal S, Milon TI, Chen F and Xu W (2025) Design of a TSR-based project learning strategy for biochemistry undergraduate teaching and research labs: a case study. Front. Educ. 9:1455173. doi: 10.3389/feduc.2024.1455173
Edited by:
Xiang Hu, Renmin University of China, ChinaReviewed by:
Ellis Bell, University of San Diego, United StatesLuo Xudong, Hubei University of Medicine, China
Copyright © 2025 Reaux, Meche, Grider, Dhanabal, Milon, Chen and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wu Xu, d3h4Njk0MUBsb3Vpc2lhbmEuZWR1
 Camille R. Reaux
Camille R. Reaux Shelby A. Meche
Shelby A. Meche Jordan M. Grider
Jordan M. Grider Soundharya Dhanabal1
Soundharya Dhanabal1 Feng Chen
Feng Chen Wu Xu
Wu Xu