
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
CURRICULUM, INSTRUCTION, AND PEDAGOGY article
Front. Educ., 09 April 2025
Sec. STEM Education
Volume 9 - 2024 | https://doi.org/10.3389/feduc.2024.1380117
This article is part of the Research TopicUsing Case Study and Narrative Pedagogy to Guide Students Through the Process of ScienceView all 5 articles
Microbiome data is increasingly important in health and environmental research. This data can be used to teach students both microbiology content as well as bioinformatics competencies. A narrative case study, Learning from Feces: the Effects of Antibiotics on Gut Microbiomes, was developed where publicly available human gut microbiome data from infants treated with antibiotics is analyzed using the student-accessible, user-friendly DNA Subway Purple line. Students, through this case study, learn about microbiome analysis including how to choose samples and process sequence data through a pipeline and finally interpret and explain the biological relevance of this data. This case study was successfully implemented in Microbiology courses for both majors and non-majors as a multi-step module on microbiomes. This manuscript presents the case development process that includes undergraduate authors, learning objectives, teaching materials and the results of implementation. Reflections on the implementation in two different courses are shared with notes for future users. It is hoped that this case will be effective in student learning and helpful for other instructors, especially those with limited bioinformatics training and the time and financial resources required for a full wet lab.
The human microbiome is most heavily studied because of its role in human health, and disease, and usefulness in diagnosis, prognosis and development of treatments (Reviewed in Gilbert et al., 2018; Cullen et al., 2020). However, notably microbiomes associated with soil and plants are also of great interest for their role in the sustainability and production of crops in the face of climate change (Reviewed in Compant et al., 2019; Dubey et al., 2019). Introducing microbiomes in the classroom is therefore crucial to keeping undergraduates informed of current research methods. Further, microbiomes provide an opportunity to teach both a range of biology content and bioinformatics skills [Reviewed in Muth and Caplan (2020)]. Unfortunately, completing the steps involved in microbiome research requires funding unavailable to some and analysis requires bioinformatics skills that are often beyond those trained in biological disciplines including instructors [Williams et al. (2019), Reviewed in Muth and Caplan (2020)]. Thus, inexpensive resources accessible to instructors inexperienced in microbiome analysis that engage different levels of students are valuable. To address this need and in response to this call for case studies, we developed a microbiome analysis case study “Learning from Feces: The Effects of Antibiotics on the Gut Microbiome,” where students are guided through analysis of publicly available research-level microbiome data from the paper by Yassour et al. (2016) using the web-based DNA Subway purple line (DNA Subway, 2024). This case study was co-authored by two undergraduate students and their instructor who were experienced in using the purple line for microbiome analysis and an instructor new to all aspects of the case study who implemented it in two different class contexts. The goal was to create a case study that was engaging and accessible to students, easy for an instructor unfamiliar with microbiome analysis to implement and that addressed learning objectives grounded in published curricular frameworks relevant to the specific courses. Herein we describe the case study development process, the case, curricular alignment, implementation, learning outcome results and a discussion to help others use the case with their students.
Since microbiome analysis is now a major part of Microbiology research it follows that many instructors have used it to varying degrees in undergraduate research [Reviewed in Muth and Caplan (2020)]. Muth and Caplan (2020) collated 26 articles describing diverse approaches to undergraduate research with microbiomes as well as the possible resulting data science skills and content learning gains. Despite these advantages of microbiome research in the classroom and the desire for of data science skills at the undergraduate level [e.g., BioSkills by Clemmons et al. (2020)], some instructors report poor preparation (Williams et al., 2019). The DNA Learning Center at Cold Spring Harbor Laboratory addresses this gap, through workshops that provide training and support for instructors to involve students in microbiome work. This group uses DNA Subway, a free, publicly available, web-based workspace to analyze different types of sequence data that was developed by the DNA Learning Center for the Cyverse (formerly iPlant Collaborative; DNA Subway, 2024). The purple line provides a student-friendly QIIME 2-based pipeline to analyze microbiome data where students can use their own or imported data to learn as they do analysis (DNA Subway b, 2024). There is published curriculum such as that by Zelaya et al. (2022) where the purple line is used as a tool for analyzing your own or public datasets. We add to available curricular material with this narrative-based case study approach.
While the most authentic route would be to actually have students employ the scientific method, create their own original research questions, conduct wet lab work, and generate their own dataset, high throughput sequencing can be quite expensive for some budgets and time a possible constraint tied to a case study can be an effective, time-saving and cost-saving strategy (Muth and Caplan, 2020; Robertson et al., 2021). In case-based learning, the student drives learning more than the instructor (Allchin, 2013) and this approach has been shown to be better at teaching (Bonney, 2015), even though this may not always be the case (Rhodes et al., 2020). Smith and Murphy (1998) discuss that case studies can be used in varying types of class contexts (e.g., lecture or lab) and assessments (e.g., report, exam) to engage students. In our case study, students learn and do microbiome analysis as they follow along with Dr. Jones, a fictional pediatrician doing her own analysis. Dr. Jones is concerned about the effect of antibiotics on her young patients since treatment is known to instigate resistance in bacteria which results in loss of life when medicines fail to protect individuals from the detrimental effects of infectious agents (Ventola, 2015). Understanding the impact of repeated antibiotic treatments on the gut microbiome, especially at the initial stage of human development outside of the womb, can provide much insight into the onset of chronic illnesses and disorders that plague humans (Yassour et al., 2016). After reading the article by Yassour et al. (2016), Natural history of the infant gut microbiome and impact of antibiotic treatment on bacterial strain diversity and stability, Dr. Jones chooses to learn how to do the analysis herself by practicing with data from the paper. She chooses data from the larger dataset discussed in the article (Yassour et al., 2016) and completes the purple line steps over the course of a week around her clinical duties. The narrative format works well for this multi-step process and to model a scientist’s approach (Allchin, 2013) as students similarly choose or are assigned patient data from the same study and are guided to complete the pipeline on the purple line alongside Dr. Jones.
The case study was tailored for 2 different courses (majors and non-majors) where antibiotic resistance is covered with related but different goals, reflecting the different types of questions and analysis that can be tackled using this method. Students therefore learn about choosing data that aligns with a research question, processing sequence data through a pipeline, data quality, cleaning data and different types of analysis depending on the research question. Students finally interpret their results and present it either as an infographic or short report depending on the course.
The initial prompt for this case was encouragement of AT while attending a Research Experiences in Microbiome Network (REMNET) workshop to develop a case study for this call. Prioritizing student interest in designing microbiome teaching activities was one of the concepts introduced at the workshop. Student authorship of case studies does occur [e.g., Riley et al. (2021)] and is thought to increase student learning (Escartín et al., 2015). Our reasoning was that two student co-authors who had done microbiome analysis for independent research would be assets in developing material that their peers would find engaging and understandable. Although the two faculty authors worked alongside the students and made final decisions, as detailed in Figure 1, the student authors were extensively involved in the case development process. High-throughput Discovery Science & Inquiry-based Case Studies for Today’s Students (HITS) network workshop materials [Updated materials from 2021 and 2022 HITS workshops orignally described in Robertson et al. (2021) and QUBES b (2024)] were helpful in case study development. The final case materials with minor editing are presented in the Supplementary material. The Learning Management System (LMS; Canvas by Instructure©) version of the introduction to the case study (Supplementary material) included a picture (Science Translational Medicine cover page, volume 8 number 343, https://www.science.org/toc/stm/8/343) which is not a part of the published case study materials. The technical instructions included in the case (Supplementary material) are heavily based on the DNA Subway Manual (DNA Subway b, 2024) as they were essentially supplemented to explain concepts and help guide students. As the four co-authors developed the case together the instructor who was new to microbiome analysis learned the analysis process which partially simulated how we hope this case study is used. Our ultimate goal is to flatten the learning curve by providing sound comprehensive simplified curricular materials such that microbiome analysis is accessible to more instructors and thus their students.
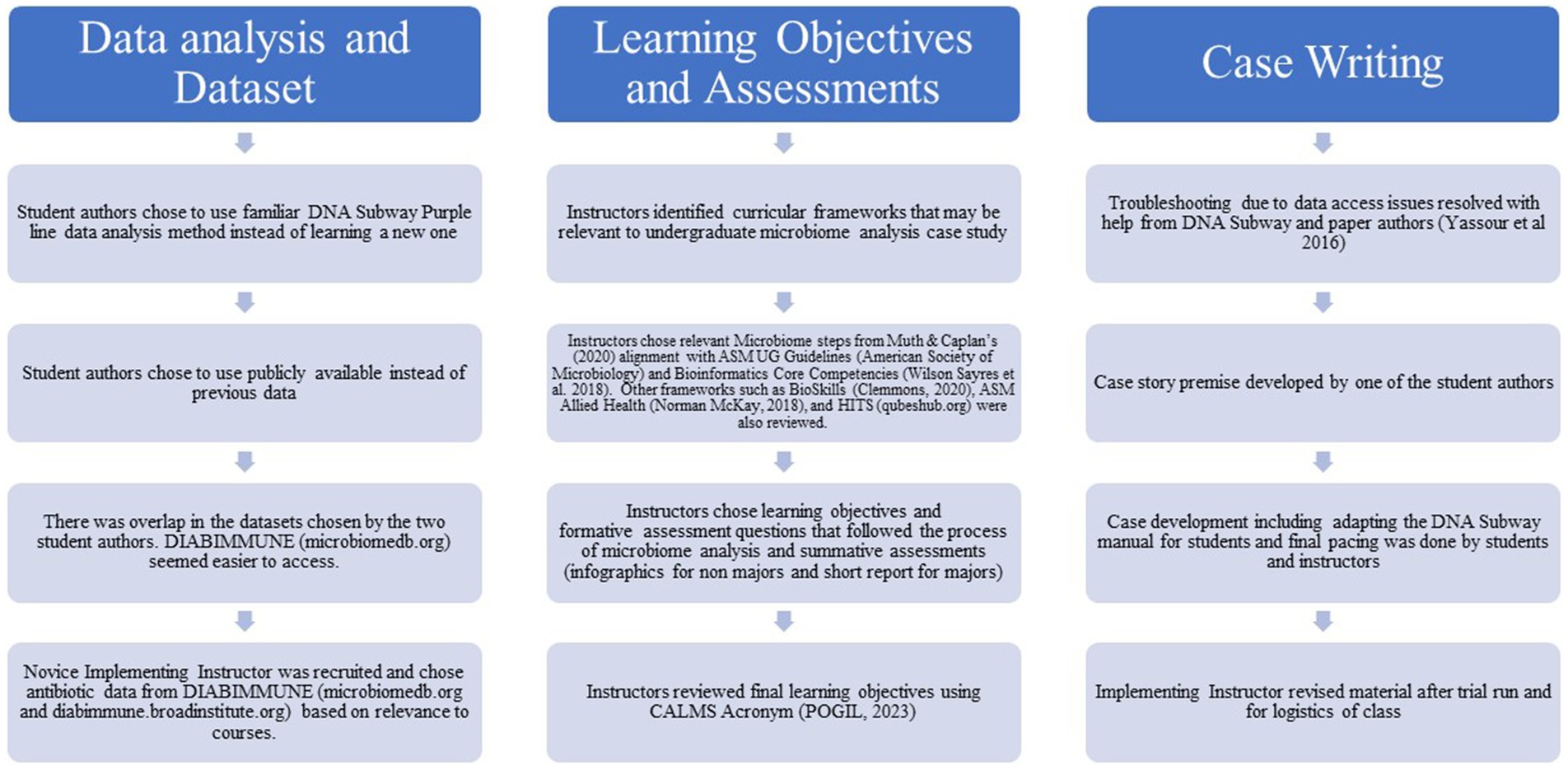
Figure 1. Outline of the case development process. Steps involved in making choices on datasets, learning objectives and assessments and creating the case itself are included and although chronological order is suggested, this was not always the case.
A modified backward design approach (Wiggins and McTighe, 2005) was used since our starting point was our larger aim of using a case study to teach microbiomes using DNA Subway (Figure 1). Muth and Caplan (2020) highlight curricular frameworks applicable to teaching microbiomes: ASM Undergraduate Microbiology Curriculum Guidelines (American Society for Microbiology, 2012) and the Bioinformatics Core Competencies (Wilson Sayres et al., 2018). There are others such as BioSkills (Clemmons et al., 2020), BioCore (Brownell et al., 2014), Coursesource Bioinformatics Learning Framework (Rosenwald et al., 2016), Bioinformatics core competencies (Welch et al., 2014), ASM Allied Health Undergraduate Curriculum Guidelines (Norman-McKay, 2018) and HITS (QUBES, 2024). Some of these were helpful as the instructors determined final learning objectives. The instructors found ASM Allied Health Undergraduate Curriculum Guidelines (Norman-McKay, 2018) and Muth and Caplan’s (2020) stepwise process and alignment of the microbiome research process steps to the ASM Undergraduate Microbiology Curriculum Guidelines (American Society for Microbiology, 2012) and the Bioinformatics Core Competencies (Wilson Sayres et al., 2018) particularly valuable. We determined that most of the steps listed (Muth and Caplan, 2020) could be accomplished in a case study. The only step that could not be accomplished was “DNA/RNA Isolation” however we were still able to include aspects of the “sample selection” and “sequencing” steps due to the large size of the dataset from Yassour et al. (2016) with multiple microbiome samples per patient and having access to the sequence data. Because the goal was a case study where students would complete the steps in microbiome analysis, the learning objectives and formative assessment questions were developed to parallel the process used by the DNA Subway purple line, as indicated in Table 1. Learning objectives are similar to other curriculum (e.g. LO1 “Define a microbiome” is also used by Lentz et al., 2017). The case was broken down into days of the week to align with Dr. Jones as she walked through the steps in her work week. We think this strategy helps to keep steps in order while recognizing the need to run some of the steps overnight since processing times for some steps can be long. Since we knew that a non-majors and a science majors class would be our first implementations, that also informed our choice of learning objectives and that the summative assessment would be an infographic and short report accordingly since these are activities that the instructor had used in these types of classes. Students should have mastered the assumed prerequisite knowledge: (1) Basic understanding of what a microbiome is and (2) Identification of organisms using DNA sequences of biomarker genes, e.g., 16S. The students should then be able to navigate this case as shown in the alignment (Table 1) of the final learning objectives, activities with suggested modality and case study assessment.
In Summer 2023 the Learning from Feces Case Study was designed and subsequently implemented in the Fall semester at Wingate University, a small comprehensive liberal arts institution in Wingate, North Carolina. Implementation occurred over a two-week period in one section each of two courses with face to face modality: Microbe Hunters – Antibiotic Discovery Course (BIO 105) and Microbiology (BIO 320). These courses were taught by the same instructor and are Course Undergraduate Research Experiences with the Tiny Earth (TE) curriculum implemented in the laboratory portion (Tiny Earth, 2024). The TE curriculum (Tiny Earth, 2024) is a student sourcing initiative focused on antibiotic discovery in response to the antibiotic resistance global health crisis; students mine soil samples in search of novel antibiotic producing bacteria. With this focus, the student populations were considered to be well suited for the case study reviewing bacterial populations in infants treated with a multitude of antibiotic therapies from Yassour et al. (2016). The summative assignments, infographic and short report, as typical scientific formats were assessed using standard rubrics that were not original to the instructor and were modified for this implementation (SCTL, 2014; TET, 2014) and so are not included in the case study materials. The student responses were evaluated for this manuscript to focus on the learning objectives.
After the add/drop period there were 5 students enrolled in this non-majors course, BIO 105 (one student subsequently withdrew after the case study was implemented), a lecture/laboratory hybrid tailored for non-science majors who are fulfilling their general education credit. The course met twice a week for 3 h and was primarily focused on the TE curriculum; there were weeks in which students were immersed in laboratory work and others when a lecture/lab hybrid format was employed. Prior to implementing the Learning from Feces case study, students were introduced to the concept of microbiomes and the use of DNA gene sequences as taxonomic biomarkers. When the case study was implemented, 3 partial class sessions were utilized over varying times depending on the progress of the case study steps (see Table 1 – pacing and Supplementary material). At first, to meet LO1 (Table 1) students were required to individually complete the pre-knowledge assessment which was one question “What is a microbiome?.” Once this was completed, they were required to read the case study introduction (Supplementary material) in the Learning Management System (LMS; Canvas by Instructure©). Prior to the class meeting, students were not required to read the research study (Yassour et al., 2016) however, they were provided with a summary and explanation of the primary figures followed by a class discussion and the pre-work in-class assignment was administered. This assignment gave students the opportunity to provide us with consent to use their assignment submissions for case study evaluation, consider the experimental design and create their DNA Subway/Cyverse account.
Three patients were then randomly assigned to each student from the DIABIMMUNE pool of 20 that received 9 or more antibiotic treatments over their first 3 years of life (Yassour et al., 2016). Once patients were assigned the students completed the Monday afternoon instructions of the case study (Supplementary material) where they collected demographic data, identified corresponding 16S gene sequence files in the DNA Subway platform for the first (BEG), midpoint (MID) and final (END) antibiotic treatments of each patient and sorted via demultiplexing. In the next class meeting Tuesday morning and Wednesday steps for data validation and diversity analyses were executed (Supplementary material). Students were instructed to follow and complete Thursday instructions (Supplementary material) to set up core metric analyses (Beta and Alpha diversity) before the next class when data analysis (Supplementary material) was to be conducted. The diversity measurements (Beta and Alpha parameters), operational taxonomic units frequency (OTU) bar charts, and Unweighted Unifracs Emperor plots were evaluated by students, with the instructor’s assistance, and a post-analysis homework was assigned. The final assignment was an Infographic summarizing their findings and providing recommendations for an audience of hypothetical parents of patients and medical professionals. To help students with this assignment infographic examples from the Centers of Disease Control were provided.
There were 14 students enrolled in BIO 320, an elective course for students in the science majors. The laboratory class meetings were twice a week, 1.5 h/meeting and also primarily focused on the TE curriculum, however there is a stand-alone lecture portion in this course. The lecture genetics module was introduced earlier than usual in the semester to help students understand the basis of the research study and microbiomes. To implement the case study, 2.5 laboratory class sessions were used (Table 1) and in the first session students were required to complete the pre-knowledge assessment (as in the non-majors course) and read the introduction in the LMS module. Students were assigned pre-work homework which included all the same questions as administered to the non-majors course, however they were required to read the research article by Yassour et al. and additional questions were included that related to the experimental design of the research study and all panels in Figure 1 of the article (Yassour et al., 2016).
In the next class meeting students were randomly assigned one patient of the same pool of 20 described earlier and directed to conduct the Monday afternoon (Supplementary material) instructions of the case study. All other steps, Tuesday through Friday (Supplementary material), were completed as homework and students uploaded their resulting patient diversity data to a group folder so that the gut microbiomes from all 14 patients together could be analyzed in the classroom for later comparison with the data of individual patients. The post analysis assignment was a short report that focused on describing the findings of the diversity data analysis of the patient at each of the 3 stages described previously, BEG, MID and END, with recommendations for hypothetical physicians.
Wingate University Institutional Research Review Board reviewed and exempted the procedures for data collection and participant recruitment (Protocol #DD080323). All students enrolled in both courses (majors and non-majors) provided consent for their assignment submissions to be included in the assessment of the case study implementation. The execution of the case study was quite the success as students in both courses followed the written instructions easily and there were only a few questions, mostly regarding errors that were not preempted which are included in Teacher Notes (Supplementary material).
Case study learning objectives were assessed using qualitative data from assignment submissions. The participants’ ability to define microbiomes (LO1) was evaluated by comparing the pre-knowledge assessment and the post analysis assignments. The initial responses of the non-major students were either accurate or focused primarily on bacteria as the organisms of a microbiome; this was likely due to the previous discussions in the class prior to case study implementation. The post-analysis responses defined microbiomes using all microbial types (archaea, bacteria, fungi, protists and viruses), referred to the ecosystems and/or environments where they exist, and some students even discussed the possibility of harmful organisms being present. The responses of all the science major students pre-knowledge assessment defined microbiomes as containing all microorganisms in a particular environment; this common thread is likely due to the definition provided in lecture material. In the post-analysis assignments, microbiome definitions were focused on bacterial isolates and intestinal/gut environments. This information was to be included in the background section of the short report and it is assumed that since the data being analyzed was solely of bacterial origin the students focused on this microbial type.
Infographics were created by 4 non-science major students, they were engaging and visually appealing; however in regards to meeting LO5 (discussing beta and alpha diversity in the gut microbiomes over the course of multiple antibiotic treatments across their 3 patients each) there was great variability in their abilities to do so. The objective of the post-analysis homework was to address these concepts and provide feedback for students that they could use to create infographics that effectively communicated their findings. Consequently, all students were able to discuss what the Faith Phylogenetic Diversity (Faith PD) and Shannon Diversity index values suggested was the impact of antibiotics on the diversity of their developing gut microbiomes. In regard to beta diversity analyses using Unweighted Unifracs Emperor plots, only one student correctly discussed this outcome. Students were able to successfully identify taxonomic groups that were significantly impacted by antibiotic use but struggled to connect the implications of the identified group on patient health and future microbiomes. Only one student understood the concept of potential limitations of the study regarding sample size and randomization (LO6), however none attempted to include this in their infographic (likely a space issue) despite receiving feedback in their post-analysis homework assignment.
There was variability in the short report submissions of the majors course and 13 students completed the assignment. The average score was 81 and 54% of the submissions earned a grade in the range of 79–70%; it seemed their greatest challenge was fitting their findings into a 3-page document. Students either really understood their findings and excelled in the assignment, or had a difficult time, however they all understood the objective of the activity (background, Figure 2). Students could explain alpha diversity results (Shannon diversity and Faith PD; LO5) while identifying taxonomic groups that were impacted over time (Results, Figure 2), however relating this to antibiotic use was somewhat more challenging (Discussion, Figure 2). The results section of the report had the widest variability in scores as shown in Figure 2. The median line of the box and whiskers plot for results is quite high and close to the upper quartile and maximum, and the interquartile range is larger than that of the background or discussion sections. Students who understood their alpha and beta diversity results chose representative figures well (LO7) and were able to communicate what the data was suggesting was the effect of the antibiotics on the developing gut microbiome (LO5). Those students who did not understand the outcomes of their analyses were unable to effectively communicate this information and earned lower scores. The background section had the lowest scores overall and a wider range based on the minimum and maximum quartiles even though its median line was close to the value of that in the discussion box (Figure 2). In comparison to the non-majors group, the science majors were able to more effectively communicate in the discussion section how the use of the data of 1 patient was limiting as in some cases the data trends were completely different from the data provided when all 14 patients were compiled (LO6). The non-majors, however, did not review data analysis output of all of their patients combined.
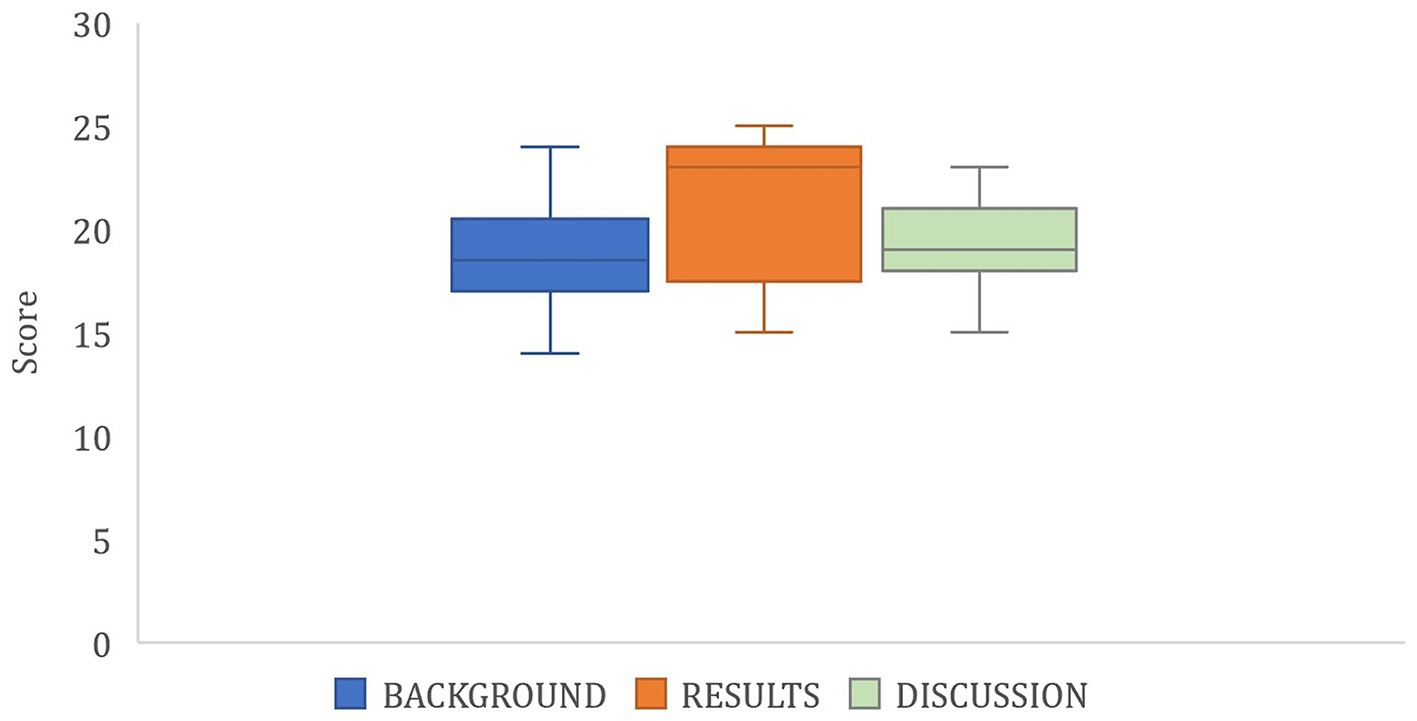
Figure 2. Box plot of scores earned in each section of the majors course (BIO 320) short report assignment, n = 13. Each section, background, results and discussion, were scored out of a possible 25 points.
Overall, the students in both non-majors and science majors courses were able to comprehend the concept of the impact of antibiotic use on developing microbiomes, while some learning objectives were met at varying levels of success. Additionally, at the end of the semester it was clear that they reflected on this experience in their TE data analysis communications; TE is the antibiotic discovery student sourcing initiative that was also being run in parallel on both courses. For both sets of students, recognizing that antibiotic use had a significant impact on the human microbiome influenced their understanding of how the global crisis of antibiotic resistance developed and had a greater impression on their newly developed opinions of antibiotic use. This case also provides an opportunity for students to go from raw data to conclusions, use actual patient data and to gain a more authentic perspective on the scientific process.
One consideration is that while students were able to use alpha diversity indices to explain the data trends they were not as skilled in using the Emperor and OTU (taxonomic) diversity plots to describe the changes occurring within the microbiome at the end of the case study. In retrospect, this is likely a reflection of the skill set of the instructor and for science major students the limitations of using the dataset of only one patient. In the non-majors course, students used the data of 3 patients and were able to compare their data and draw more effective conclusions, the Emperor plots were more informative and the changes in the OTU plots were visibly more clear. This type of limit to calculations was also encountered by Zelaya et al. (2022) using a different microbiome analysis platform indicating that this is not uncommon. For the science majors course, the instructor did compile the data output for all 14 patients, but they were only able to show the Shannon Diversity and Faith PD data as the Emperor and OTU plots would require them running their own DNA Subway analyses of the randomly chosen patients. At the point in the implementation when this was realized, there was not enough time to do this. In addition, for students the identification of changes in individual groups was a bit more difficult to discern because the instructor and students were learning how to navigate DNA Subway together. While the instructor did conduct trials in the months leading up to the case study implementation, they were still a novice user of the platform and was learning tricks to manipulate the data from the more tech-savvy students in their classroom.
It is important to note that this case study is relatively easy to follow and implement. The implementing instructor had a very limited skill set and while well-versed in the subject matter of microbiology had never used a case study to teach concepts in their courses, taught a bioinformatics module or course, or used DNA Subway for any purpose prior to developing and implementing this case study. The recommendations in the Teaching Notes (Supplementary material) are written from their perspective with the intention of assisting other instructors with limited skill sets prepare and effectively execute the case study in their classrooms. In summary, it is imperative that instructors familiarize themselves with the 16S DNA sequence data and the steps of the DNA Subway analysis (Supplementary material) prior to implementing the case study. When this case study is used again the instructor will of course be more experienced with the DNA Subway platform, however it is recommended that novice users similarly run the analysis themselves ahead of time not only so they are familiar but also to provide a comparison dataset to walk through the data analysis process with students. This will reduce student anxiety in analyzing the abundance of data that is produced and help the instructor more effectively realize the learning objectives. Two formats were used in this implementation and while the 3 patient model allowed for comparison and clearly provided a more robust data set, depending on the number of students in a course, time available to conduct the case study and the expertise of the instructor, this may not always be possible. It is recommended that instructors tailor the learning objectives and output to be analyzed based on their course parameters and experience. When the students’ data analysis resulted in unexpected results, we returned to the demographic information of the patient(s) and looked at the number of antibiotic treatments, antibiotics used at each stage (in some cases it was the same) and the changes in the taxonomy charts to explain a lack of diversity values or inconclusive results. Regarding your student population you will want to confirm whether there are students under 18 in your course and how you will acquire parental permission for them to use DNA Subway. Pacing steps are outlined (Table 1) based on the experience with the two courses (majors and non-majors). Further guidance is included in the Teaching Notes (Supplementary material) including descriptions of unusual examples and additional suggestions to help others with implementation.
This case can be used as a standalone introduction to microbiome analysis, even with non-majors as modeled here, provided students have been exposed to basic microbiology concepts. Instructors who may want other case studies on microbiomes for students can use the National Center for Case Study Teaching in Science Collection (2024) which also has resources on case study teaching (https://www.nsta.org/case-studies). There is also published curriculum that skips the pipeline and permits simple analyses (Lentz et al., 2017) that AT has used with students as an introduction before using the purple line. Zelaya et al. (2022) have curriculum using the purple line and another analysis platform with students. These instructions were written using the expertly written DNA Subway Manual (DNA Subway b, 2024) with supplements to assist students and instructors navigate the steps of the analysis and is another great resource. Finally, the review by Muth and Caplan (2020) cites many examples of microbiomes in education beyond the analysis, and cost-saving approaches which others may find helpful.
In summary, our hope is that in this case study and its supporting materials, we have streamlined microbiome analysis so that it can be used with minimum instructor preparation and training. We further hope that the narrative case study approach with the heavy influence of our student authors will help engage students as they learn by doing, and see the value of this type of analysis, as demonstrated in their own reflections, on real research-level data.
De-identified student data is available on request. The rubrics used for analysis are however not original to the authors and so cannot be similarly distributed. The sources of the dataset used in the case study have been cited in the manuscript and Supplementary material.
The studies involving humans were approved by the Wingate University Institutional Research Review Board who reviewed and exempted the procedures for data collection and participant recruitment (Protocol #DD080323). The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation was not required from the participants or the participants’ legal guardians/next of kin because informed consent was obtained electronically.
AT: Conceptualization, Methodology, Supervision, Writing – original draft, Writing – review & editing, Project administration. DD: Formal analysis, Investigation, Writing – original draft, Writing – review & editing, Methodology. JF: Conceptualization, Methodology, Writing – original draft, Writing – review & editing. SK: Conceptualization, Methodology, Writing – original draft, Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. Publication costs are from the grant Implementing DNA Barcoding for Course-Based Undergraduate Research Experiences NSF IUSE #1821657.
The authors thank Dr. Bruch Nash, Dr. Ray Enke and others from the DNA Barcoding/Metabarcoding Progam for instructor training (AT) and support for teaching undergraduates; Daniel Jacobs, IT Manager, DNA Learning Center from Cold Spring Harbor laboratories for help with troubleshooting and for placing the data in the public section of Cyverse; Tommi Vatanen and Ramnik Xavier and other authors of the article whose data the case study is based on, for supporting our use of the data for this purpose and their quick helpful responses; Davida Smythe from the REMNET Summer 2023 Workshop for encouragement to do a case study for this call; High-throughput Discovery Science and Inquiry Based Case Studies for Today’s Students (HITS) Network Fellow program for training AT in case study writing and big data; Student participants for agreeing to participate. Andy Yang for early help with troubleshooting data format issues. We acknowledge the use of Cyverse (This material is based upon work supported by the National Science Foundation under Award Numbers DBI-0735191, DBI-1265383, and DBI-1743442. www.cyverse.org/); Dr. Don Bouchard for advice on editing.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/feduc.2024.1380117/full#supplementary-material
Allchin, D. (2013). Problem- and case-based learning in science: an introduction to distinctions, values, and outcomes. CBE Life Sci. Educ. 12, 364–372. doi: 10.1187/cbe.12-11-0190
American Society for Microbiology (2012). Recommended curriculum guidelines for undergraduate microbiology education. Washington DC: American Society for Microbiology. Available at: https://asm.org/Guideline/ASM-Curriculum-Guidelines-for-Undergraduate-Microb (Accessed June, 2023).
Bonney, K. M. (2015). Case study teaching method improves student performance and perceptions of learning gains. J Microbiol Biol Educ. 16, 21–28. doi: 10.1128/jmbe.v16i1.846
Brownell, S. E., Freeman, S., Wenderoth, M. P., and Crowe, A. J. (2014). BioCore guide: A tool for interpreting the core concepts of vision and change for biology majors. CBE Life Sci. Educ. 13, 200–11. doi: 10.1187/cbe.13-12-0233
Clemmons, A. W., Timbrook, J., Herron, J. C., and Crowe, A. J. (2020). BioSkills guide: development and National Validation of a tool for interpreting the vision and change Core competencies. CBE Life Sci. Educ. 19:ar53. doi: 10.1187/cbe.19-11-0259
Compant, S., Samad, A., Faist, H., and Sessitsch, A. (2019). A review on the plant microbiome: ecology, functions, and emerging trends in microbial application. J. Adv. Res. 19, 29–37. doi: 10.1016/j.jare.2019.03.004
Cullen, C. M., Aneja, K. K., Beyhan, S., Cho, C. E., Woloszynek, S., Convertino, M., et al. (2020). Emerging priorities for microbiome research. Front. Microbiol. 11:136. doi: 10.3389/fmicb.2020.00136
DIABIMMUNE (2024). Data Set DIABIMMUNE (https://microbiomedb.org/mbio/app). (Accessed January 31, 2024).
DIABIMMUNE (2024). Data Set DIABIMMUNE (https://diabimmune.broadinstitute.org/diabimmune/). (Accessed July, 2024)
DNA Subway (2024). Available at: https://DNASubway.cyverse.org (Accessed January 31, 2024).
DNA Subway b (2024). Manual Purple Line. https://learning.cyverse.org/dna_subway_guide/#walkthrough-of-dna-subway-purple-line-beta-testing-documentation (Accessed January 31, 2024).
Dubey, A., Malla, M. A., Khan, F., Chowdhary, K., Yadav, S., Kumar, A., et al. (2019). Soil microbiome: a key player for conservation of soil health under changing climate. Biodivers. Conserv. 28, 2405–2429. doi: 10.1007/s10531-019-01760-5
Escartín, J., Saldaña, O., Martín-Peña, J., Varela-Rey, A., Jiménez, Y., Vidal, T., et al. (2015). The impact of writing case studies: benefits for students’ success and well-being. Procedia. Soc. Behav. Sci. 196, 47–51. doi: 10.1016/j.sbspro.2015.07.009
Gilbert, J. A., Blaser, M. J., Caporaso, J. G., Jansson, J. K., Lynch, S. V., and Knight, R. (2018). Current understanding of the human microbiome. Nat. Med. 24, 392–400. doi: 10.1038/nm.4517
Lentz, T. B., Ott, L. E., Robertson, S. D., Windsor, S. C., Kelley, J. B., Wollenberg, M. S., et al. (2017). ‘Unique down to our microbes—assessment of an inquiry-based metagenomics activity’, journal of Microbiology & Biology. Education 18. doi: 10.1128/jmbe.v18i2.1284
Muth, T. R., and Caplan, A. J. (2020). Microbiomes for all. Front. Microbiol. 11:3472. doi: 10.3389/fmicb.2020.593472
National Center for Case Study Teaching in Science Collection (2024). Available at: https://www.nsta.org/case-studies (Accessed January 31, 2024).
Norman-McKay, L. (2018). Microbiology in nursing and allied health (MINAH) undergraduate curriculum guidelines: a call to retain microbiology lecture and laboratory courses in nursing and allied health programs. J. Microbiol. Biol. Educ. 19. doi: 10.1128/jmbe.v19i1.1524
Process Oriented Guided Inquiry Learning (POGIL) (2023). Summer Workshop Materials Writing Content Learning Objectives for POGIL Activities. Available at: https://pogil.org/authoring-materials/writing-guidelines
QUBES (2024). HITS Learning Outcomes and Skills for HT Case Studies Document. Available at: https://qubeshub.org/community/groups/hits/ht_case_studies (Accessed January 31, 2024).
QUBES b (2024). High-throughput Discovery Science & Inquiry-based Case Studies for Today’s Students (HITS) network qubeshub.org/community/groups/hits (Accessed July, 2024).
Rhodes, A., Wilson, A., and Rozell, T. (2020). Value of case-based learning within stem courses: is it the method or is it the student? CBE Life Sci. Educ. 19:44. doi: 10.1187/cbe.19-10-0200
Riley, K. J., Vardar-Ulu, D., Pollock, E., and Dutta, S. (2021). Students authoring molecular case studies as a partial course-based undergraduate research experience (cure) for lab instruction. Biochem. Mol. Biol. Educ. 49, 853–855. doi: 10.1002/bmb.21578
Robertson, S. D., Bixler, A., Eslinger, M. R., Gaudier-Diaz, M. M., Kleinschmit, A. J., Marsteller, P., et al. (2021). HITS: harnessing a collaborative training network to create case studies that integrate high-throughput, complex datasets into curricula. Front. Educ. (Lausanne) 6:711512. doi: 10.3389/feduc.2021.711512
Rosenwald, A. G., Pauley, M. A., Welch, L., Elgin, S. C., Wright, R., and Blum, J. (2016). The CourseSource Bioinformatics Learning Framework. CBE Life Sci. Educ. 15:le2. doi: 10.1187/cbe.15-10-0217
SCTL. (2014). Assignment Design. Available at: https://stearnscenter.gmu.edu/knowledge-center/course-and-curriculum-redesign/assignment-design/ (Accessed November 2014).
Smith, R. A., and Murphy, S. K. (1998). Using case studies to increase Learning & Interest in biology. Am. Biol. Teach. 60, 265–268. doi: 10.2307/4450469
TET. (2014). Assessment by Oral Presentation. Available at: https://serc.carleton.edu/9137 (Accessed November 2014).
Tiny Earth (2024). Available at: www.tinyearth.wisc.edu (Accessed January 31, 2024).
Ventola, C. L. (2015). The antibiotic resistance crisis: part 1: causes and threats. P & T 40, 277–283
Welch, L., Lewitter, F., Schwartz, R., Brooksbank, C., Radivojac, P., Gaeta, B., et al. (2014). Bioinformatics curriculum guidelines: toward a definition of core competencies. PLoS Comput Biol. 10:e1003496. doi: 10.1371/journal.pcbi.1003496
Wiggins, G. P., and McTighe, J. (2005). Understanding by design. Expanded 2nd edn. Alexandria, VA: Association for Supervision and Curriculum Development.
Williams, J. J., Drew, J. C., Galindo-Gonzalez, S., Robic, S., Dinsdale, E., Morgan, W. R., et al. (2019). Barriers to integration of bioinformatics into undergraduate life sciences education: a National Study of us life sciences faculty uncover significant barriers to integrating bioinformatics into undergraduate instruction. PLoS One 14:e0224288. doi: 10.1371/journal.pone.0224288
Wilson Sayres, M. A., Hauser, C., Sierk, M., Robic, S., Rosenwald, A. G., Smith, T. M., et al. (2018). Bioinformatics Core competencies for undergraduate life sciences education. PLoS One 13:e0196878. doi: 10.1371/journal.pone.0196878
Yassour, M., Vatanen, T., Siljander, H., Hämäläinen, A. M., Härkönen, T., Ryhänen, S. J., et al. (2016). Natural history of the infant gut microbiome and impact of antibiotic treatment on bacterial strain diversity and stability. Sci. Transl. Med. 8, 343ra81–343ra381. doi: 10.1126/scitranslmed.aad0917
Keywords: microbiome, case study teaching, antibiotic resistance, bioinformatics, DNA Subway
Citation: Thomas A, Davis D, Faue J and Kicker S (2025) Learning from feces: a case study teaching microbiome analysis. Front. Educ. 9:1380117. doi: 10.3389/feduc.2024.1380117
Received: 01 February 2024; Accepted: 23 May 2024;
Published: 09 April 2025.
Edited by:
Alex L. Deal, Wake Forest University, United StatesReviewed by:
Maurice H. T. Ling, Temasek Polytechnic, SingaporeCopyright © 2025 Thomas, Davis, Faue and Kicker. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Aeisha Thomas, dGhvbWFzYUBjcm93bi5lZHU=
†These authors have contributed equally to this work and share senior authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.