- Department of Neurobiology and Behavior, University of California, Irvine, Irvine, CA, United States
Neuroscience courses can be enriched by including an evolutionary perspective. To that end, this essay identifies several concepts critical to understanding nervous system evolution and offers numerous examples that can be used to illustrate those concepts. One critical concept is that the distribution of features among today’s species can be used to reconstruct a feature’s evolutionary history, which then makes it possible to distinguish cases of homology from convergent evolution. Another key insight is that evolution did not simply add new features to old nervous systems, leaving the old features unchanged. Instead, both new and old features have changed, and they generally did so along divergent trajectories in different lineages, not in a linear sequence. Some changes in nervous system organization can be linked to selective pressures (i.e, adaptation), especially if they occurred convergently in different lineages. However, nervous system evolution has also been subject to various constraints, which is why many neural features are, in a sense, suboptimal. An overarching theme is that evolution has brought forth tremendous diversity across all levels of the nervous system and at all levels of organization, from molecules to neural circuits and behavior. This diversity provides excellent research opportunities, but it can also complicate the extrapolation of research findings across species.
1. Introduction
Evolution has been identified by the American Association for the Advancement of Science as a core concept to emphasize in undergraduate biology education (Cary and Branchaw, 2017), and many biologists are fond of citing Dobzhansky’s dictum that “Nothing in biology makes sense except in the light of evolution” (Dobzhansky, 1973). A longer quote from that same article elucidates: “Seen in the light of evolution, biology is, perhaps, intellectually the most satisfying and inspiring science. Without that light it becomes a pile of sundry facts – some of them interesting or curious but making no meaningful picture as a whole” (p. 129).
Evolution has been recognized as a core concept also in neuroscience (Chen et al., 2023). Nonetheless, most neuroscience courses ignore or neglect evolution, perhaps because both psychology and medicine – two cornerstones of modern neuroscience – have historically been slow to incorporate evolutionary thought. This has begun to change (Gosling and Graybeal, 2007; Zampieri, 2009), but many who teach neuroscience remain unfamiliar with central aspects of nervous system evolution. To improve this situation, the present paper identifies several concepts that lie at the heart of nervous system evolution, as well as many concrete examples. Given the time and content constraints most neuroscience courses face, it is recommended that instructors pick whatever examples their students are most likely to find compelling and use them to illustrate at least a subset of the key concepts. Some overarching aims are to make students aware that nervous systems are incredibly diverse, that understanding evolution requires “tree thinking” (rather than thinking of evolution producing ladders), that both homology and independent (convergent) evolution are prominent features of nervous system evolution, and that adaptation through natural selection acts within the context of numerous constraints.
2. Key concepts (and misconceptions) in the study of nervous system evolution
The following concepts are a subset of the evolutionary principles needed to illuminate biology in general (see Cary and Branchaw, 2017), and they are relevant to the evolution of many different biological features, not solely neural traits. However, the listed concepts are most applicable to the kinds of content likely to be covered in a neuroscience course.
2.1. Concept #1: homologous traits have unique evolutionary trajectories
Most courses on evolution emphasize the origin and diversification of species, including gene flow through populations and the history of life in general. In contrast, evolutionary neuroscience is more concerned with the evolution of specific neuronal features, aka traits or characters (Wagner, 2000). These two perspectives are different sides of the same coin, but anyone interested in the evolution of biological traits must grapple with the question of how to study trait evolution. For that, the concept of homology is key.
Although debates about the definition of homology have a long history (Brigandt, 2002, 2003), evolutionary biologists generally agree that homologous traits are those that share a common evolutionary origin (Ereshefsky, 2012). That is, a trait is homologous in two or more species if it can be traced back to the same (corresponding) trait in the presumptive common ancestor of the species (see Striedter and Northcutt, 1991). This definition implies that homologous traits have a unique origin, a distinct evolutionary history, and may sometimes be lost (Ghiselin, 2005). Importantly, homologous traits may change in structure, function, or both. Often those changes are relatively minor, making it easy to identify the homologs in a variety of descendent species. However, it is possible for evolutionary changes to be so extensive that the homologies become cryptic.
How can evolutionary biologists reconstruct which traits emerged in which ancestral species and how they might have changed in the descendent lineages? This question is especially acute in evolutionary neurobiology since most neural features do not fossilize. The answer involves mapping the distribution of the trait onto a well-established phylogenetic tree of the relevant species (i.e., which species have the trait, and which do not) and then determining which underlying history is most likely to have given rise to the observed distribution (Figure 1; Northcutt, 1984; Cunningham et al., 1998). For example, the fact that all placental mammals, but no other vertebrates, have a corpus callosum (interconnecting the two cerebral hemispheres) strongly suggests that this trait first appeared in the last common ancestor of all placental mammals and was then retained, varying mainly in size (Aboitiz and Montiel, 2003). More complex is the phylogenetic history of retinal foveas, defined as a portion of the retina densely packed with photoreceptors and containing a pit in the overlying retinal layers (see Table 1 #1–2; going forward, items in that table will be cited as T#1, T#2, etc.). Among mammals, such foveas are found only in primates; however, some birds also have retinal foveas (Bringmann, 2019). The most likely interpretation of this phylogenetic distribution is that retinal foveas evolved early during primate evolution (Wu et al., 2017), and a second time in birds. Given this scenario, the foveas of birds and primates are not homologous to one another, since the last common ancestor of birds and primates almost certainly lacked a fovea.
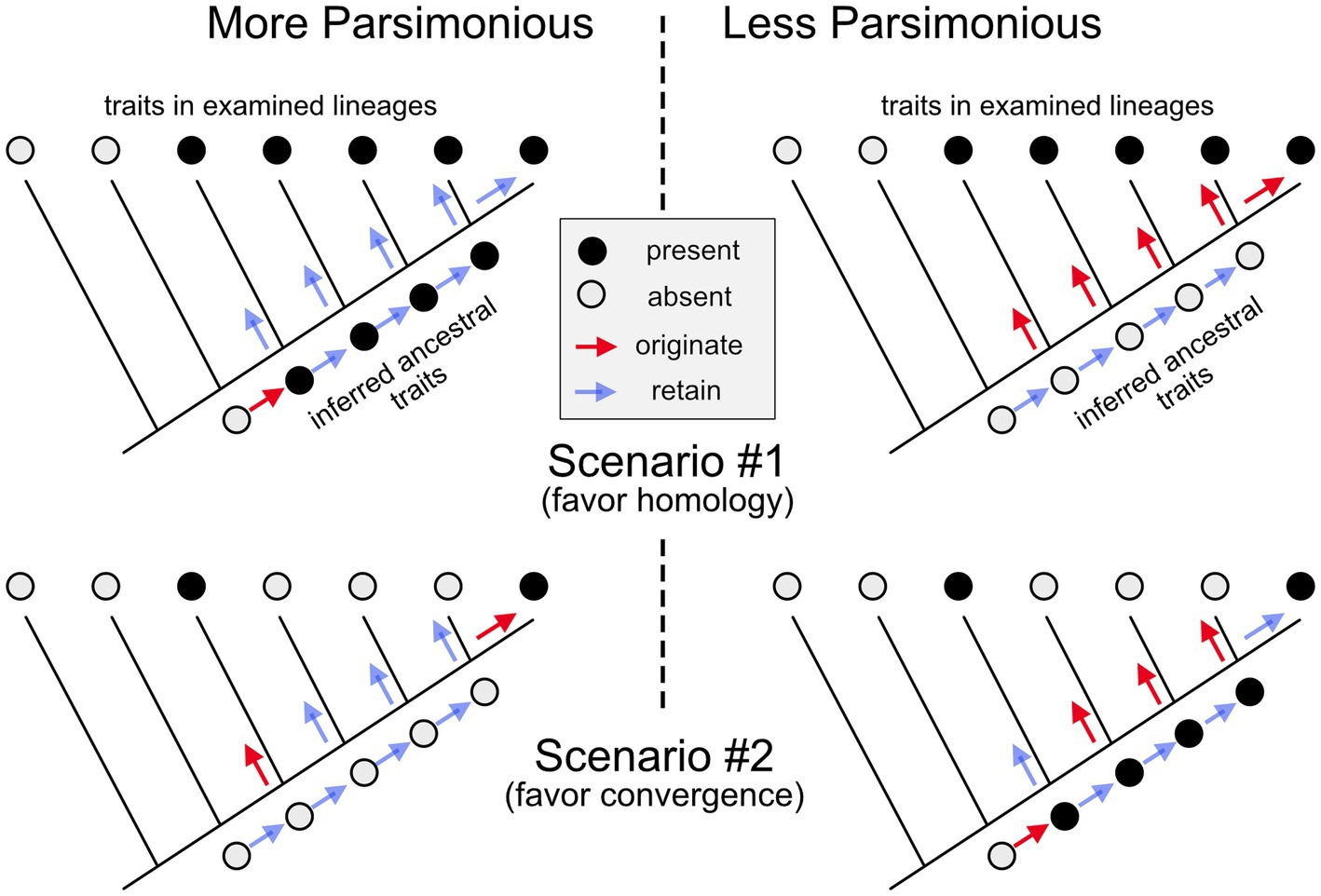
Figure 1. Inferring homology or convergence. In scenario #1, a trait (black circles) is present in 5 of the examined species (or larger lineages) but absent in the two most basal branches of the illustrated phylogenetic “tree.” Given this phylogenetic distribution, it requires fewer evolutionary changes to suppose that the trait originated in the last common ancestor of the species that exhibit it, and that it was then retained (unchanged) in the subsequent lineages (left side), than it is to conclude that it emerged independently in all the examined lineages (right side). Therefore, it is most parsimonious to conclude that the trait is homologous among the examined lineages. In scenario #2, the trait is found only in two lineages that are relatively distant relatives of one another. Given this phylogenetic distribution, it is more parsimonious to conclude that the trait in this second scenario originated twice independently and is, therefore, the result of convergent (or parallel) evolution.
One common misconception related to Concept #1 is that traits can be partially homologous (e.g., 60%). This is often said of genes, but the percentage in those instances refers only to the degree of similarity and not to the homology, which is all-or-nothing. Also confusing is that morphological structures and genes are sometimes said to be “functionally homologous,” even if the structures themselves are not homologous (or of uncertain homology). In most of these cases, the functions are merely attributes of the structures, rather than independent traits that can be subjected to a homology analysis. In general, it is better to refer to similar functions subserved by non-homologous structures as instances of convergent evolution; another widely used term for independently derived functional similarities is “analogy.”
2.2. Concept #2: lineages form nested sets, and so do their traits
As Darwin argued lucidly, species do not just transform from one into another; they often branch, forming a family tree (aka phylogenetic tree). Because each node within this tree defines a lineage that includes all subsequent branches, the lineages collectively form a hierarchically nested set of species, genera, families, orders, classes, and so forth. In parallel with this taxonomic hierarchy, homologous traits also form nested sets. In molecular evolution, these are represented by gene trees (i.e., gene family trees; Huang et al., 2018).
In comparative anatomy, lineage-typical sets of features are called Baupläne (plural of Bauplan, a German word meaning “construction plan”). To illustrate, biologists may contrast the Bauplan for vertebrates with that for insects. One may also hypothesize about the Bauplan of the last common ancestor of insects and vertebrates, or of the Bauplan for specific orders within those larger lineages. It is important to realize, however, that each Bauplan is an abstraction, not an actual organism (or part thereof). Missing from each Bauplan are the innumerable details that vary between the species in the specified lineage.
One common misconception related to concept #2 is that a single species can adequately represent a larger lineage, which is implied when someone talks about laboratory mice as if they were THE rodent. Rather that talking about THE rodent brain, for example, it is better to talk about rodent brains in general or, ideally, to specify which rodents are being discussed. A related fallacy is to assume that modern species can be arranged in a linear phylogenetic series, such as fish-frog-lizard-mouse-monkey-human. This kind of “phylogenetic scale” is incompatible with Darwin’s view of species forming a family tree and not consistent with our modern understanding of how new species and lineages form. To illustrate the point, ask your students to sort their extended family into a linear series; different criteria would lead to different arrangements, none of which are an accurate representation of the underlying genealogy. Another instructive classroom activity is to ask students to create an “orderly arrangement” of the individual vertebrate brain drawings included on three “posters” published as part of a three-volume text on vertebrate brain evolution by Nieuwenhuys et al. (1998; unfortunately, this book is expensive and not always easy to obtain from libraries; T#3).
2.3. Concept #3: evolution does not merely add new parts to old systems
Soon after Darwin wrote his Origin of Species, it became popular to believe that each organism during its embryonic development goes through a series of stages that recapitulate the species’ evolutionary history, that ontogeny recapitulates phylogeny. This idea has turned out to be wrong (Gould, 1977). Mammals do look fish-like at some early stages of development, but that is because vertebrate embryos in general go through an early “phylotypic period” (T#4; Richardson, 1995) when they are far more similar to one another than they are as adults. After (and even before) this phylotypic period, development diverges, often dramatically. As a general rule, novel adult forms are created not by adding new stages to the end of ancestral ontogenies, but by modifying development at various developmental stages such that different adult forms result. In the words of Walter Garstang, “ontogeny does not recapitulate phylogeny, it creates it” (T#5; Garstang, 1922, p. 82). This realization is at the core of evo-devo biology, which has blossomed in recent years.
Closely related is the fact that evolution does not merely add novel bits to an ancestral set. For instance, bird wings were not produced by the phylogenetic addition of wings to ancestral bodies with four legs. Instead, avian wings resulted from evolutionary changes in forelimb development. This may be obvious, but many educated people still believe that early mammals added the hippocampus and amygdala (and other components of the limbic system) to an ancestral “reptilian core,” and that later mammals then added the neocortex. This triune brain theory (MacLean, 1990) has been amazingly persistent in popular imagination, perhaps because it appears to provide a mechanistic basis for the ancient observation that humans often struggle to suppress their baser, animalistic desires. However, the core tenets of this theory are simply incorrect (T#6; Cesario et al., 2020).
One problem with the triune brain theory is that reptiles clearly possess hippocampus and amygdala homologs and, most likely, a neocortex homolog as well (Medina et al., 2022). It’s just that these structures diverged substantially from their mammalian counterparts (Figure 2; Striedter and Northcutt, 2020). This is not to say that evolution never produces anything new (Wagner, 2015). Neurons, for example, had a distinct phylogenetic origin (see below). It is also true that evolutionary neurobiologists continue to debate when in evolution a neocortex homolog originated (see Section 3.8). In any case, given that all mammals possess a neocortex, this trait is very unlikely to have originated late in mammalian phylogeny, as the triune brain theory supposed. Moreover, even when a novel brain region (or other neural trait) appears, the other traits need not remain unchanged. Very few neural traits are truly invariant across long evolutionary distances.
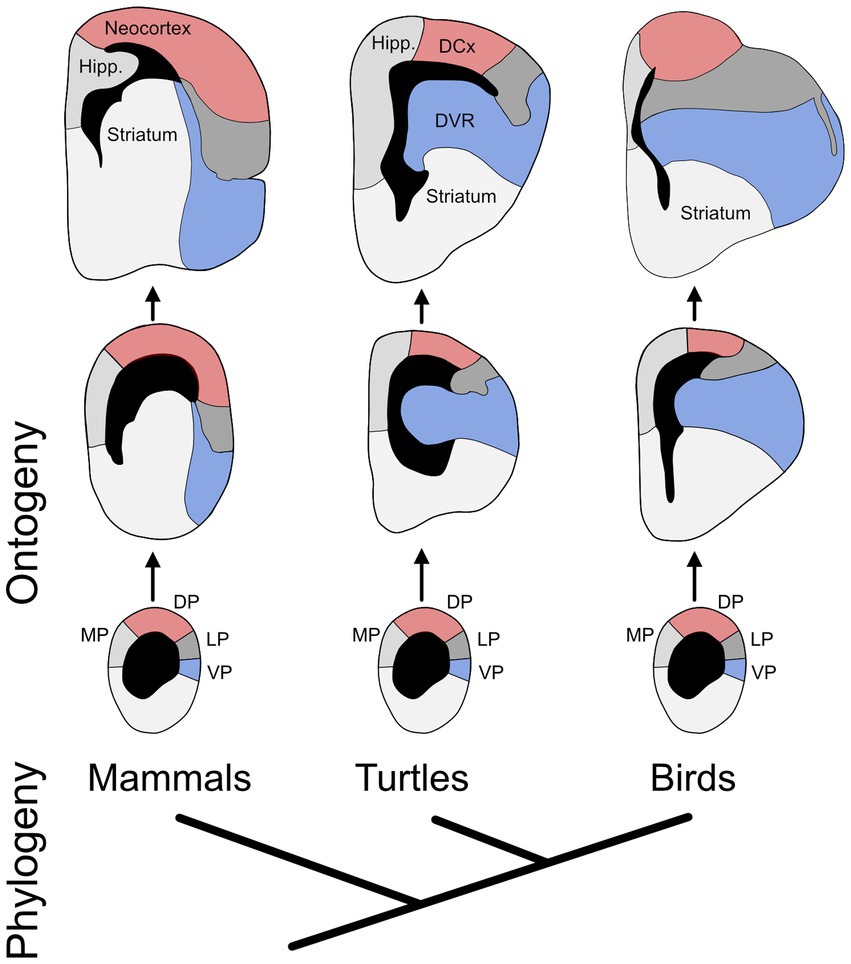
Figure 2. The evolution of developmental divergence. The phylogenetic relationship of birds, turtles, and mammals is shown at the bottom. The rest of the diagram shows the developmental (ontogenetic) trajectories of the telencephalon in those three lineages (oldest stages at the top). The individual diagrams represent cross sections through one hemisphere, showing the cerebral ventricles in black. Highlighted in red are the mammalian neocortex and its most likely homologs in turtles and birds, as well as their developmental precursors; collectively, they are called the dorsal pallium (DP). Marked in blue are the ventral pallium (VP) and its adult derivatives in the three lineages. The main point of the figure is that, during development, the VP enlarges more in birds and turtles than in mammals (it also bulges into the ventricle), while the DP enlarges more in mammals. In other words, the ontogenetic trajectories of these two pallial divisions diverge phylogenetically. Additional abbreviations: DCx – dorsal cortex; DVR – dorsal ventricular ridge; Hipp. – hippocampus; LP – lateral pallium; MP – medial pallium. Adapted from Puelles et al. (2017) and Striedter (1997).
2.4. Concept #4: biological traits are shaped by natural selection, but constrained
Most species-typical traits are thought to be (or have been) adaptive, in the sense that they were shaped by natural selection (Gregory, 2009). Some adaptive traits are optimal in the engineering sense of the term, or nearly so. Vertebrate rod photoreceptors, for instance, can signal reliably when they are struck by a single photon (T#7; Morshedian and Fain, 2017); it is theoretically impossible for rods to be any more sensitive to light. However, rods become essentially useless in bright light, which is where cone photoreceptors function effectively. Thus, evolution did not optimize vertebrate photoreceptors for all kinds of lighting conditions. Instead, it has produced a compromise: retinas containing rods as well as cones (i.e., duplex retinas).
Among the most important constraints on neurobiological evolution are energetic costs. In humans, for example, the brain consumes about 20–25% of the body’s resting energy budget (Leonard et al., 2007). Therefore, the benefits of evolutionary increases in brain size must be balanced against the metabolic costs of building and maintaining a large brain. Large-brained species have generally solved this problem by increasing parental care, eating more nutritious food, and reducing the size of other metabolically expensive organs, such as the gut (Isler and van Schaik, 2009; Tsuboi et al., 2015). Within the lineage leading to modern humans, neonatal brain size has also been constrained by the need for the baby’s head to pass through the mother’s birth canal, which in turn is constrained by factors related to bipedal locomotion (T#8; Haeusler et al., 2021).
Engineers face analogous constraints, but they can generally build their devices from scratch, whereas evolution can only modify the ancestral system. Because evolutionary history has generally constrained a trait’s evolutionary path, that history is causally relevant (Cisek, 2019). For example, the eye of a giant squid is well adapted to detecting predatory whales over great distances (Nilsson et al., 2012), but it cannot resolve small details as well as vertebrate eyes can. The squid cannot solve this problem by evolving a vertebrate-type retina, because cephalopods and vertebrates long ago embarked on divergent trajectories, each imposing different constraints (Gregory, 2008). Jumping from one path to the other after millions of years of evolutionary divergence is effectively impossible. Put differently, the Baupläne mentioned under Concept #2 are not mere abstractions; they causally constrain phylogeny (Wagner, 2014).
It is technically difficult to demonstrate that a specific trait is adaptive, because this requires showing that the trait is both hereditary and improves an organism’s chances of survival and reproduction (i.e., fitness). However, good circumstantial evidence for adaptation exists when functionally similar features evolved convergently under similar ecological conditions. For example, in most mammals the sodium channel Nav1.7 is involved in sensing pain, including the sensing of (painful!) tissue acidity. However, naked mole rats and various hibernating mammals have independently evolved similar changes in the amino acid sequence of Nav1.7 that make it acid-insensitive (T#9; Liu et al., 2014b). As a result, these animals can spend long, painless stretches of time underground, where CO2 levels are likely to be high and cause tissue acidity. Collectively, these data make a strong circumstantial case for adaptation.
2.5. Concept #5: convergent evolution happens at all levels of biological organization
Detailed similarity between features often suggests homology but, as noted in the previous section, it is possible for such similarities to arise by evolutionary convergence (or parallelism, if they arose independently from homologous ancestral features). When convergent features exhibit a very high degree of similarity, it is tempting to conclude that they must be homologous, because it is difficult to imagine that such high similarity could have arisen by chance. The puzzle is resolved by recognizing that, although evolution depends on random mutations, its long-term outcomes are usually non-random. If different lineages experience very similar selective pressures, then similar outcomes are likely to arise, especially if there is only a limited number of ways for organisms to address those selective pressures (Losos, 2011; Blount et al., 2018).
Biologists used to believe that convergent evolution is relatively rare at the molecular level, mainly because they assumed that selection acts only on phenotypes, while gene and protein evolution is governed almost entirely by chance (see Castoe et al., 2010). However, numerous examples of convergent molecular evolution have now been revealed (Zakon, 2002). One excellent example is related to the independent evolution of echolocation in several lineages of bats and in toothed whales (e.g., dolphins). These echolocating species have undergone parallel evolutionary changes in the amino acid sequence of a protein called Prestin that is expressed in the outer hair cells of mammalian cochleas and facilitates the kind of high frequency hearing important for echolocation (Figure 3; T#10; Liu et al., 2014a).
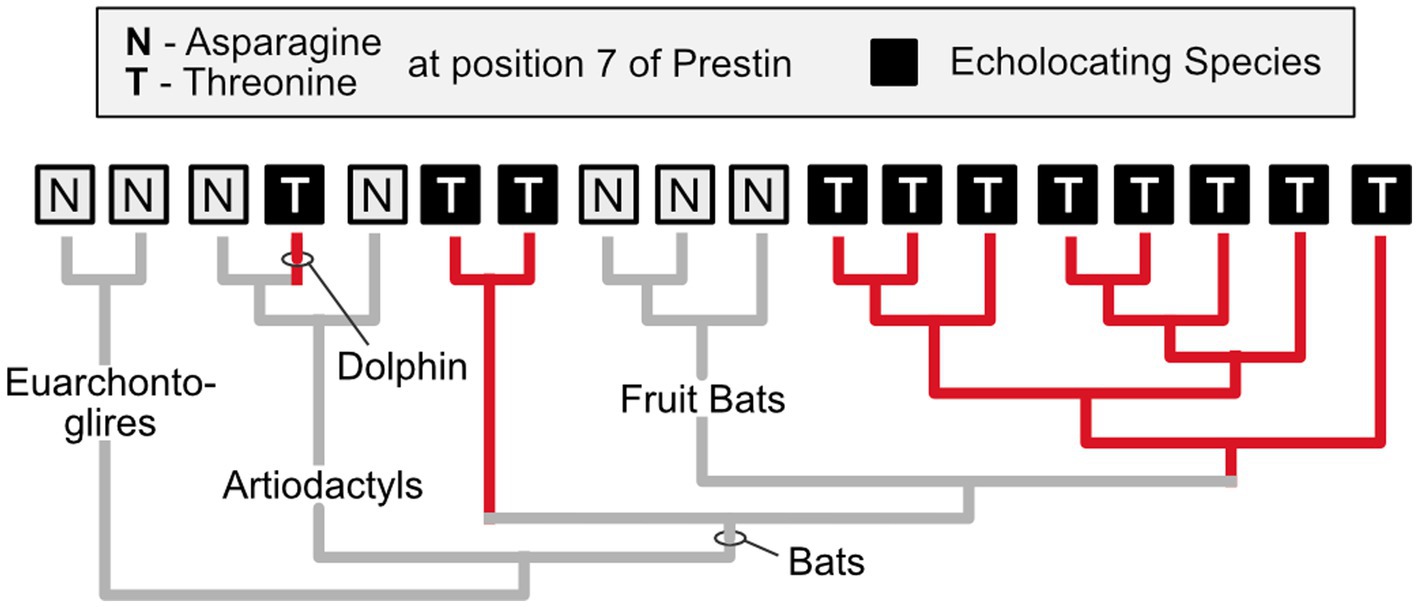
Figure 3. Independent evolution of identical amino acid substitutions. Prestin, a protein that is found in the outer hair cells of mammals, exhibits an identical amino acid substitution (from asparagine to threonine at position #7) in echolocating bats (excluding the non-echolocating fruit bats) and dolphins, which also engage in echolocation. Given this phylogenetic distribution, it is parsimonious to infer that this substitution arose independently in at least two of the echolocating lineages (it may have been lost in the fruit bats). It is a compelling case of parallel evolution at the molecular level (parallel because in all instances the inferred ancestral condition is asparagine at position 7). Based on Li et al. (2010).
2.6. Concept #6: trait divergence provides research opportunities but also complicates cross-species extrapolation
Some species are exceptionally convenient for experimental studies (Krebs, 1975). Indeed, the study of model organisms like E. coli, Drosophila, and mice has yielded innumerable insights that have been broadly applicable. However, not all attempts to extrapolate findings species have fared so well (Striedter, 2022). For example, neuroscientists have long tried to model traumatic brain injury in non-humans, but very few of the treatments that had been effective in animal models have passed clinical trials (Bragge et al., 2016). Part of the problem is that many of the animal studies were conducted on rodents, which have smooth cortices that do not respond to injury in the same manner as the complexly folded cortices of humans (Kochanek et al., 2018). To avoid this problem, scientists have turned to non-human primates, but working with them raises complicated ethical issues.
A more general problem with the use of supposedly simple “model species” to learn about the human condition is the assumption that our more complex system consists of an essentially unchanged simple system, on top of which evolution has layered additional complexities (Bargmann, 2000). As noted in relation to the triune brain theory (Section 2.3), this is not how evolution works. Instead, the simple systems we study today have usually diverged significantly from the homologous systems in our own species (Striedter, 2019). Although biologists often talk about genes or other traits as being “broadly conserved,” this does not mean that those traits have remained unchanged; it simply means that their homologs can be recognized in a broad array of species. It is somewhat misleading, therefore, to talk of “human disease genes” in fruit flies, sea anemones, or yeast (T#11; Rubin et al., 2000; Maxwell et al., 2014). These species may have homologs of genes that have been linked to human diseases, but the DNA sequences and functions of these genes have almost always diverged. For example, nematodes have a homolog of the human presenilin gene, which has been linked to Alzheimer’s disease, but this gene in nematodes is linked to egg-laying (Levitan et al., 1996).
That said, research on model species has been enormously influential. An excellent example is the work of Hodgkin and Huxley on squid giant axons (Raman and Ferster, 2022). These axons are ideally suited for studying the mechanisms underlying action potentials, because they are large enough to insert an electrode into the axon’s interior, and because they continue to function as an isolated, heavily manipulated preparation. By measuring the flows of sodium and potassium ions across the membrane of these axons during individual action potentials, Hodgkin and Huxley (1952) were able to reconstruct the fundamental features of squid action potentials, which have turned out to be broadly conserved. It is worth noting, however, that action potentials in mammals and some arthropods are metabolically more efficient, because they have reduced the temporal overlap between sodium influx and potassium efflux, two processes that can cancel each other out in terms of their effect on the axon’s membrane potential (T#12; Sengupta et al., 2010). Although this demonstrates that the squid’s action potentials are suboptimal, they are (most of the time) sufficient for the squid, which use their giant axons to generate escape behaviors.
3. Evolutionary sketches well suited for a neuroscience course
An influential paper in 1979 accused evolutionary biologists of telling too many “just-so stories” that exaggerated the role of adaptation in shaping biological traits (Gould and Lewontin, 1979). This critique of the “adaptationist paradigm” rightfully led to more emphasis on Baupläne and other constraints, and it made biologists construct more careful arguments about natural selection’s influence. As a result, biologists have now produced a broad array of well-documented sketches about the evolutionary history of diverse traits, including many aspects of nervous system evolution. I here offer a sampling of such sketches that can be incorporated into a general neuroscience course to make the key concepts concrete. Many sketches illustrate multiple key concepts, and each key concept can be introduced through multiple examples. Sketch selection and concept mapping are, therefore, left to individual instructors. Cited works with potentially helpful illustrations are listed in Table 1 (T#1, T#2, etc.).
3.1. Ancient origins of neurons, synapses, and action potentials
All multicellular animals, except sponges and placozoans, have neurons. Although the phylogeny of the major animal lineages remains somewhat uncertain, neurons almost certainly appeared more than 500 million years ago, either in the last common ancestor of all multicellular animals (with sponges and placozoans later losing them), or independently in ctenophores (comb jellies) and the last common ancestor of all other multicellular animals (T#13; Moroz et al., 2015; Burkhardt and Sprecher, 2017). Supporting the latter hypothesis is the observation that ctenophore neurons are very different from those of other animals. Most relevant to an introductory neurobiology course is that ctenophore neurons form a syncitial nerve net in which neurons connect to one another via direct cellular anastomoses, rather than synapses (T#14–15; Burkhardt et al., 2023). Thus, the ctenophore nerve net is consistent with Golgi’s reticular theory, rather than Cajal’s neuron doctrine. The ctenophore nerve net does connect to sensory and motor cells via synapse-like structures that are thought to release glutamate or glycine, as well as various ctenophore-specific peptides (Sachkova et al., 2021), but ctenophore neurons do not express acetylcholine, dopamine, serotonin, norepinephrine, or histamine.
Many molecular components of synapses originated long before neurons (T#15–16). Specifically, a substantial fraction of all the molecules typically associated with synapse formation and function have direct homologs (orthologs) in unicellular organisms such as yeast and choanoflagellates, as well as sponges (Ryan and Grant, 2009). This discovery implies that the evolution of chemical synapses involved “deploying existing genes and modules in new cellular contexts” (Colgren and Burkhardt, 2022, 787). The evolutionary process of creating novelties by combining old parts in new ways is what Francois Jacob called “tinkering” (Jacob, 1977). Compared to the more ancient mode of paracrine peptide secretion, chemical synapses with narrow synaptic clefts were surely adaptive, because they allowed for temporally more precise and metabolically efficient neural signaling, especially in larger animals (T#17; Moroz and Romanova, 2021).
Many of the ion channels underlying neuronal signaling also have ancient origins (T#18–19). Voltage-gate calcium and potassium channels evolved prior to the emergence of multicellular animals, whereas voltage-gated sodium channels came later, with the origin of bilaterally symmetrical animals (Nishino and Okamura, 2018). Sodium channels are not required for the generation of action potentials, which can be recorded even in unicellular organisms and plants (Brunet and Arendt, 2016). However, the use of voltage-gated sodium channels was probably adaptive for early bilaterian animals because sodium ions were so abundant in their extracellular environment that, in conjunction with the pre-existing sodium-potassium pump (Sáez et al., 2009), it became relatively easy for these animals to generate a steep transmembrane concentration gradient for sodium. This gradient, in turn, steepened the rising phase of action potentials and, thus, increased their temporal precision and responsiveness without the potentially troublesome effects of increasing intracellular calcium. Later in phylogeny, voltage-gated sodium channels diversified significantly, mainly by gene duplication and divergence (Zakon et al., 2011; Zakon, 2013). Curiously, voltage-gated sodium channels were lost in nematodes (T#20; Hobert, 2018).
3.2. An arsenal of natural neurotoxins
Venoms and neurotoxins have evolved in many different lineages. Their most common function is to paralyze prey, but some species, including pufferfish and toxic amphibians, use it for defense (Itoi et al., 2014). The modes of action for these toxins are diverse. The tetrodotoxin found in pufferfish and several species of newts blocks voltage-gated sodium channels; the botulinum toxin (Botox) produced by the bacterium Clostridium botulinum inhibits acetylcholine release at vertebrate neuromuscular junctions; snakes of the genus Bangarus produce α-bungarotoxin, which blocks nicotinic acetylcholine receptors; and black mamba venom includes a toxin that selectively blocks L-type calcium channels. In most lineages, venoms evolved early and then became increasingly complex, mainly by means of gene duplication and the co-option of additional compounds (Kordiš and Gubenšek, 2000; Fry et al., 2006). A single predatory cone snail species, for instance, may produce 200 different peptides that interfere with various ion channels and receptors (Olivera et al., 1999).
An intriguing aspect of toxin evolution is the evolution of toxin resistance (T#21–22). For instance, rough-skinned newts obtain tetrodotoxin through their diet, but evolved resistance to it (Hanifin and Gilly, 2015). Garter snakes like to eat these newts and can do so, because the snakes likewise evolved tetrodotoxin resistance. Not to be outdone, the fittest newts accumulated ever higher levels of tetrodotoxin, leading to an evolutionary arms race between the two species (Brodie et al., 2005). This explains why rough-skinned newts contain what at first appears to be an excess of toxin, enough to kill several humans: this amount is what is needed to deter the snakes! A similar evolutionary drama involves scorpions whose venom causes intense pain by activating a type of voltage-gated sodium channel that triggers action potentials in pain-sensing axons (Nav1.7). Grasshopper mice, which prey on these scorpions, evolved resistance to their venom by modifying a closely related sodium channel (Nav1.8) so that it is blocked by the scorpion venom, thereby preventing transmission of pain signals to the brain (Rowe et al., 2013).
The astounding diversity of natural toxins has provided valuable opportunities for translational research. For example, the development of angiotensin-converting enzyme (ACE) inhibitors to lower blood pressure in humans (by promoting vasodilation) was prompted by the study of pit viper venom, which incapacitates prey by collapsing their blood pressure (Péterfi et al., 2019). More recently, researchers have isolated a pain-inhibiting peptide from predatory centipedes and are attempting to develop this compound into a potent analgesic for humans (T#23; Wang et al., 2017). Natural neurotoxins can also function as insecticides. For example, chrysanthemum flowers contain toxins called pyrethrins, which bind to the voltage-gated sodium channels of insects and keep them in their open position (Field et al., 2017). Humans use pyrethrins to kill lice, ticks, mosquitoes, and diverse other insects. Importantly, pyrethrins are not nearly as toxic to humans, because they are an order of magnitude less able to bind mammalian voltage-gated sodium channels; this species difference, in turn, results from a point mutation that altered a single amino acid (Soderlund and Lee, 2001).
3.3. Sensory receptor diversity
Like natural toxins, sensory receptors are incredibly diverse. Many animal lineages have gained new types of sensors, creating new possibilities for intraspecific communication and locating resources. Other sensor types were lost, most likely because they were no longer advantageous and eventually succumbed to relaxed selection (Lahti et al., 2009). Because of this diversity, the perceptual worlds of different species may be radically different, even if those species live in outwardly similar environments (Caves et al., 2019).
A good example of sensor loss is the evolutionary demise of the lateral line systems, which fishes use to sense water pressure and flow, as well as weak electrical signals (T#24; Dijkgraaf, 1963). These systems became essentially useless in terrestrial vertebrates and were consequently lost. Another good example involves whales, which lost most of their olfactory and taste sensors (T#25–26; Feng et al., 2014; Kishida et al., 2015). Whales also lost the cone photoreceptors that sense wavelengths other than blue (T#27; Meredith et al., 2013), presumably because whales commonly swim at depths where blue light predominates (Dungan et al., 2016). Similarly interesting is the loss of sweet taste receptors in many carnivores, including cats and many birds (Jiang et al., 2012). In addition, most birds modified one of their molecular pain sensors so that it is no longer sensitive to capsaicin, the active ingredient in spicy peppers (Jordt and Julius, 2002). Clever entrepreneurs have taken advantage of this species difference to create bird seed that is dusted with capsaicin (Curtis et al., 2000): most birds do not care, but squirrels quickly learn to stay away.
Some lineages have re-gained long-lost sensory abilities. Electroreception, for example, was lost in early ray-finned fishes (for reasons that remain unclear) but then re-evolved in two groups of teleost fishes (Baker and Modrell, 2018) and duck-billed platypus (Pettigrew, 1999). Similarly, hummingbirds and some songbirds independently re-acquired the ability to taste sweets, which helps them evaluate the sugar content of nectar and fruit; however, they did so by modifying the ancestral umami taste receptor, rather than bringing back the lost sweet receptor (T#28; Toda et al., 2021). An important point here is that the re-evolution of lost senses does not, in general, re-create the ancestral condition. Instead, evolution finds a different route to achieving the same ends; it is a good biological of what philosophers call multiple realizability.
Not surprisingly, evolution also brought forth some totally new sensor types. Particularly interesting is the evolution of infrared (i.e., radiant heat) sensors in fire beetles, which use them to locate recent burn areas (yet avoid actual fires; T#29; Hinz et al., 2018; Bell, 2023), and in pit vipers, which use them to hunt rodents in darkness (T#30; Newman and Hartline, 1982). The details of these novel sensor types differ between the lineages (Gracheva et al., 2010), as one would expect, given their independent evolution.
Especially diverse are vertebrate odorant receptors (T#31; Niimura, 2012). Given that the set of odors an animal can smell depends largely on the mix of odorant receptors (OR) its olfactory sensory neurons express, it is interesting that almost half of the olfactory receptor genes in humans have become pseudogenes, leaving a functional set of <350 OR genes, compared to >900 in mice (Godfrey et al., 2004). It is tempting to conclude from this information that mice have a generally better sense of smell than humans, but humans can smell some odorants nearly as well as mice (Laska, 2017). Therefore, it is better to interpret the larger repertoire of OR genes in mice as allowing these animals to sense and discriminate a wider variety of odorants. This hypothesis has not been tested directly, but many species can detect odorants to which humans are insensitive. For example, storm petrels and some other pelagic seabirds can find schools of fish in the open ocean by following the scent of dimethyl sulfide (Nevitt, 2008); human noses would not pick up this clue.
3.4. Vertebrate retinas
Vertebrate photoreceptors have undergone numerous evolutionary changes (T#32–35; Lamb, 2013; Kelber and Jacobs, 2016). Most non-mammalian vertebrates express four different opsin genes, and lampreys express five of them (Hart et al., 2008; Collin et al., 2009; Davies et al., 2012), making it quite likely that the earliest vertebrates already possessed good color vision. Early mammals later lost two of the ancestral vertebrate opsin genes. The best explanation for this gene loss is that early mammals were predominantly active during the night, when it is adaptive to sacrifice color vision in favor of visual sensitivity (Heesy and Hall, 2010). However, early mammals did not become completely color-blind. They all possess at least two cone opsins that are sensitive to short and long wavelengths, respectively. Roughly speaking, they are red-green colorblind (T#36).
When the ancestors of Old World monkeys and apes later became more diurnal, they duplicated the long-wavelength opsin gene and shifted the sensitivity of one duplicate (paralog) toward shorter wavelengths (T#37; Jacobs, 2008; Carvalho et al., 2017). Thus, these primates re-evolved the ability to discriminate red from green, which probably helped them identify ripe fruit and young leaves, which are often red. Some New World primates, as well as some marsupials, likewise re-evolved trichromatic vision, but they did so independently, using different genetic mechanisms (Arrese et al., 2002; Carvalho et al., 2017). Another fascinating twist on color vision is that birds shifted the sensitivity of their short wavelength-sensitive opsin away from the ultraviolet (UV) range toward violet, and then back toward UV in select lineages (T#38; Ödeen et al., 2012; Cronin and Bok, 2016).
The photoreceptors in vertebrate retinas are positioned in such a way that the incoming light must pass through the overlying retinal layers to reach the rod and cone outer segments, where the opsins are embedded. This arrangement is suboptimal because some light will be scattered by the retinal tissue before it can reach the outer segments, and because it forces axons leaving the retina to pass through the photoreceptor layer, creating a blind spot. Moreover, it need not be this way, as cephalopods have a retina in which the light sensing components are placed closest to the incoming light (Hanke and Kelber, 2020). Some authors have argued that the vertebrate retina’s “inverted” arrangement stems from an ancient developmental constraint (T#39; Cisek and Hayden, 2021). This is probably correct, but the vertebrate arrangement is really not so bad. For one thing, it made it easier for vertebrates to evolve lenses that are far enough away from the photoreceptors to allow for focused pattern vision (Baden and Nilsson, 2022). It also allowed the light-absorbing pigment epithelium, which helps recycle key elements of the phototransduction cascade, to be placed right next to the outer segments without obstructing the light path (Striedter, 2015). In addition, vertebrates evolved glial light guides that minimize light scattering by channeling light toward the back of the retina (T#40; Franze et al., 2007).
Mammalian retinas also contain a small percentage of retinal ganglion cells (RGCs) that are depolarized by light even when rods and cones are deleted experimentally (Berson et al., 2002). These intrinsically photosensitive RGCs convey information about ambient light levels to the brain, which then uses it to control day-night cycles and pupillary constriction (T#41; Gooley et al., 2003). These RGCs express melanopsin, which is more closely related to the rhabdomeric opsins mediating light sensitivity in the main eyes of invertebrates than to the ciliary opsins in vertebrate rods and cones (Bellingham and Foster, 2002). This observation has led to the hypothesis that photosensitive RGCs are homologous to the rhabdomeric photoreceptors of invertebrates (Arendt, 2003). Since ciliary photoreceptors (expressing ciliary opsins) coexist with rhabdomeric photoreceptors not only in vertebrates but also in invertebrates (Arendt et al., 2004; Bellingham et al., 2006), both cell types have probably coexisted in animals since their earliest days (T#42). Later invertebrates utilized mainly the rhabdomeric type, whereas vertebrates emphasized ciliary photoreceptors (Davies et al., 2010; Do, 2019). The functional factors underlying this divergent trajectory remain uncertain. However, rhabdomeric photoreceptors in early invertebrates probably functioned over a greater range of wavelengths and light intensities than their ciliary counterparts; vertebrate rods and cones collectively achieved similar sensitivity, while being energetically more efficient (Fain et al., 2010).
3.5. Hindbrain and spinal cord
Much of what has been learned about the spinal cord in non-mammalian vertebrates, notably lampreys (T#43; Grillner et al., 1995), applies also to mammals. In particular, it is now clear that vertebrate spinal cords contain groups of neurons that collectively create the rhythms underlying locomotion, and that the activity of these central pattern generators (CPGs) is modulated by both sensory inputs and axons descending from the brain (Dietz, 2003).
Despite this broad conservation, spinal cords vary between species. Significant changes accompanied the transition from swimming to walking, as leg muscle control requires more complex patterns of motor activity than swimming by lateral undulation. Birds further modified their spinal cords so that their legs during walking move in a left–right alternating rhythm, whereas the wings move synchronously on the two sides (T#44; Haimson et al., 2021). Birds also modified part of their lumbar spinal cord into something that looks like a second vestibular apparatus that, apparently, helps birds maintain their balance as they walk (Necker, 2006; Stanchak et al., 2020). Less is known about how humans modified their spinal cord. However, spinal motor neurons receive more direct descending neocortical input in primates than in other mammals (T#45), and primate spinal cords are more dependent on this input for organized activity (Lemon and Griffiths, 2005). Indeed, functional recovery after spinal cord injury is more difficult for humans than for mice or rats. In this context, it is interesting that the spinal cords of mice exhibit several highly unusual features that likely favor axon regeneration and recovery (Inman and Steward, 2003; Sroga et al., 2003). Many teleost fishes and some amphibians (but not lizards!) go even further: they can regenerate entire sections of their spinal cord (Simpson, 1968; Sîrbulescu et al., 2009).
Much like the spinal cord, the hindbrain is broadly conserved but not invariant. Most of the sensory and motor nuclei associated with the cranial nerves are found in topologically equivalent locations in the hindbrain of all vertebrates (T#46; Nieuwenhuys, 2011). A major exception is that the sensory nuclei associated with the mechanosensory lateral line system were lost in terrestrial vertebrates, together with the lateral line receptors and nerves. Other hindbrain nuclei emerged de novo during vertebrate phylogeny. For instance, novel electrosensory lateral line lobes evolved in the hindbrain of weakly electric fishes (Berman and Maler, 1999; Meek et al., 1999), and cochlear nuclei emerged with the independent evolution of high-frequency hearing in frogs, reptiles, and mammals (T#47; Manley, 2017). The lineage leading to turtles and birds evolved an additional auditory hindbrain region called nucleus laminaris, whose role in sound localization has been extensively studied in birds and involves an elegant delay-line mechanism (Carr and Konishi, 1990). Curiously, mammals localize sounds by means of a significantly different hindbrain mechanism (Grothe and Pecka, 2014). Also interesting is that the dorsal cochlear nucleus, a mammalian innovation, lost its laminar organization in the lineage leading to humans (T#48; Moore, 1980).
Some hindbrain nuclei are broadly conserved but dramatically enlarged in a subset of species. For example, goldfishes evolved an enormously hypertrophied vagal sensory nucleus, called the vagal lobe (T#49). It processes information from a specialized taste organ in the roof of the mouth that allows these animals to identify and retain tasty morsels in mouthfuls of mud (Finger, 2008). This vagal lobe features multiple cellular laminae, whereas its homolog in other fishes does not. In an African teleost named Heterotis niliticus, the vagal lobe is even more hypertrophied and assumes a most peculiar spiral shape (Braford, 1986). Among the hindbrain’s motor nuclei, the trigeminal motor nucleus is unusually large in lampreys, presumably because it controls their rasping sucker mouth. Even more enlarged are the motor nuclei that control a Torpedo ray’s electric organ, which is composed of modified pectoral fin muscles and innervated by cranial nerves VII, IX, and X (T#46; Nieuwenhuys, 2011). Incidentally, it was the Torpedo ray’s electric organ from which acetylcholine receptors were first isolated (Miyazawa et al., 2003).
3.6. Cerebellum and midbrain
Jawless vertebrates do not possess a proper cerebellum (Wicht, 1996), which means that the cerebellum is not part of the vertebrate brain Bauplan; it is part of the jawed vertebrate brain plan. The cerebellum probably began as a relatively small structure in the dorsal part of the most anterior hindbrain (T#50; Hibi and Shimizu, 2012) and then expanded independently in cartilaginous fishes, birds, and mammals (Iwaniuk et al., 2006; Yopak et al., 2016). As it expanded, the cerebellar cortex became increasingly folded, presumably because of some (still unspecified) developmental constraint. The most dramatically enlarged cerebellum is found in the weakly electric mormyrid teleosts, which cover their entire brain with a sheet of cerebellar cortex that is folded back on itself and almost crystalline in the precision of its internal circuitry (T#51; Meek and Nieuwenhuys, 1991; Zhang et al., 2011). This “gigantocerebellum” probably plays a major role in electrolocation, which is the use of a self-generated electric field to sense external objects.
Perhaps the most fascinating aspect of cerebellar evolution is that its internal circuitry has been remarkably stable, with only minor modifications (T#50; Hashimoto and Hibi, 2012; Hibi et al., 2017). This circuit-level conservation strongly suggests that the cerebellum performs an ancient, invaluable computational function (Dean et al., 2010; Porrill et al., 2013). Despite this elemental conservation, the cerebellum’s connections with other brain regions are numerous and phylogenetically variable, suggesting that the cerebellum’s conserved computational function can be usefully deployed in a variety of biological contexts. Most likely, the cerebellum originally functioned mainly in the control of balance and locomotion (Striedter and Northcutt, 2020) but then acquired some additional behavioral functions, notably a role in complex cognition (Schmahmann, 1991). In support of this hypothesis, the lateral portion of the mammalian cerebellum evolved a set of multi-synaptic loops that connect it with the neocortex, including parts of prefrontal cortex (Strick et al., 2009). This lateral cerebellum became selectively enlarged in humans and great apes (MacLeod et al., 2003; Balsters et al., 2010), and independently in several other mammalian lineages (T#52; Smaers et al., 2018), presumably because doing so provided significant cognitive benefits.
Neuroscience courses tend to give short shrift to the midbrain, perhaps because it is quite small in primates, relative to the neocortex (Schiller, 2010). However, the dorsal midbrain is large in many non-mammals. This is true especially of the optic tectum, which is homologous to the mammalian superior colliculus. In most fishes, reptiles, and birds, the optic tectum is not only large but also contains numerous cellular laminae. Functionally, the optic tectum is involved in orienting and stimulus-tracking behaviors, as demonstrated elegantly in larval zebrafish using activity-sensing calcium indicators (T#53–54; Muto et al., 2013; Förster et al., 2020). It probably retains these functions in mammals, but in primates the retinal input to the superior colliculus is much reduced, while retinal projections to the thalamus increased (Perry and Cowey, 1984). This evolutionary shift in connectivity suggests that some of the ancestral functions of the optic tectum have been subsumed by the neocortex in primates (Knudsen, 2020). Unfortunately, this functional neocorticalization hypothesis has not been tested rigorously. What is clear is that the superior colliculus in primates retains a relatively ancient role in directing visual spatial attention (Shipp, 2004; Knudsen and Schwarz, 2016).
A major component of the mammalian ventral midbrain is a group of dopaminergic neurons that project to the telencephalon and have been studied extensively for their role in reward signaling and reinforcement learning. Similar dopaminergic neurons are found in most vertebrates, but their rostrocaudal position varies considerably, rendering one-to-one homologies uncertain (T#55; Yamamoto and Vernier, 2011). Beyond the vertebrates, the homologies become even murkier. Dopamine and dopamine receptors clearly have ancient origins (Yamamoto et al., 2015), and in most animal phyla dopamine modulates behavior in ways that maximize feeding efficiency (Barron et al., 2010; Solvi et al., 2016). These data suggest that dopamine’s role in regulating foraging is broadly conserved across all animals but gained additional functions in later lineages (Hills et al., 2015). Unfortunately, this subsequent history is difficult to unravel, because dopamine is released from many different cell types (including neurons in the retina and olfactory bulb) and has many functions, some of which remain poorly understood (Miller, 2020).
A simpler evolutionary perspective to take on the midbrain dopaminergic system is to stress how this system can be hijacked by addictive drugs. Most drugs of abuse trigger dopamine release either directly or indirectly (Schultz, 2011). This dopamine release makes the user seek and take the drug again in the future, thus reinforcing drug taking in the same way as dopamine release reinforces feeding and sex (Wise, 2006; Beny-Shefer et al., 2017). However, drugs of abuse do not increase evolutionary fitness as food and sex tend to do. Why, then, have humans not evolved resistance to these drugs? The likely answer is that drugs have become much more potent and widely available in recent years, far faster than the rate of human evolutionary change (Nesse and Berridge, 1997). From this perspective, drug addiction results from an evolutionary mismatch between our current environment and the environment that shaped most human traits (Perry, 2021).
3.7. Forebrain evo-devo biology
The forebrain of vertebrates includes the diencephalon and telencephalon, and each of these contains additional subdivisions. Because some of these subdivisions vary substantially in size, complexity, and connectivity, it can be difficult to identify their homologs across distantly related species. A major step toward solving this problem was the realization that the species differences in forebrain morphology are much smaller in embryos than in adults (see Section 2.3). Moreover, embryonic vertebrate brains are divisible into relatively discrete patches of precursor cells that express different combinations of transcription factors, which helps researchers homologize them across species (Figure 2; T#56; Rubenstein et al., 1994). Based on these comparative molecular data, researchers have identified a broadly conserved Bauplan for embryonic vertebrate brain forebrains (T#57; Puelles, 2001; Puelles et al., 2013).
A major remaining challenge is to construct a fate map that specifies which embryonic precursor zones develop into which sets of adult brain regions (Puelles, 2013). Such fate mapping experiments are difficult (Puelles et al., 2016; Bloch et al., 2020), but the task is simplified by the fact that some genes maintain their expression for significant stretches of developmental time (Medina et al., 2011). Furthermore, cells derived from the various embryonic zones tend not to migrate far or intermingle much as they assume their final, adult position. A major exception is that most of the inhibitory interneurons in the dorsal telencephalon (called the pallium) migrate to their adult position from the ventral portion of the embryonic telencephalon (i.e., the subpallium), and do so in all vertebrates examined so far (T#58; Moreno et al., 2008; Hansen et al., 2013). Given all these observations, comparative neurobiologists now recognize a Bauplan for adult vertebrate brains that includes most of the major forebrain divisions, including thalamus, striatum, hippocampus, and amygdala.
Another serious challenge is that the telencephalon of teleost fishes (roughly half of all vertebrate species!) does not develop by the balloon-like evagination that occurs in other vertebrates. Instead, the telencephalon of teleosts everts (T#59; Striedter and Northcutt, 2006). As part of this eversion, the dorsomedial aspects of the left and right telencephalon move laterally apart as development proceeds, rather than staying next to one another (as they do during a typical evagination). Consequently, the dorsolateral portion of the telencephalon in teleosts corresponds topologically to the dorsomedial portion of the telencephalon in other vertebrates (T#60; Nieuwenhuys, 2009). Since the mammalian hippocampus develops from the dorsomedial pallium, researchers have proposed that the hippocampus homolog in teleosts should be located dorsolaterally, and experimental data on spatial memory have generally supported this hypothesis (T#61; Broglio et al., 2015). Homologies for other regions of the teleostean forebrain have been more difficult to demonstrate (Striedter and Northcutt, 2021).
3.8. Neocortex origin and expansion
The neocortex is sometimes said to be the crowning achievement of the mammalian lineage and, indeed, humans born without a neocortex are dramatically impaired (though they retain a primitive form of consciousness; Merker, 2007). But is the neocortex “new” with mammals (as implied by the triune brain theory), or did it have a more ancient evolutionary origin? If we define neocortex as the kind of 6-layered structure we see in modern mammals, then the neocortex is indeed limited to mammals. However, this is really a question about neocortex homologs, and homologs may change in any aspect of structure or function, thus defeating any simple definition based on “essential” characteristics (Wilson et al., 2007). So instead of a strict anatomical definition, reconstructing neocortex evolution requires a broad comparative analysis that allows for extensive, multidimensional variation across species.
Such an analysis reveals that invertebrates do not possess a neocortex homolog, even though some invertebrate lineages have independently evolved large and complex integrative brain regions (T#62–64; Strausfeld et al., 2020; Harzsch and Krieger, 2021). Among the vertebrates, putative homologs of mammalian neocortex have been proposed for most lineages, including lampreys (Suryanarayana et al., 2020), but several of the proposed homologies remain controversial (Striedter and Northcutt, 2021). By far the least controversial neocortex homolog outside of mammals is the dorsal cortex of reptiles, which develops from the dorsal pallium, contains three distinct layers, and receives visual input from the thalamus (T# 65; Fournier et al., 2018). Moreover, individual neurons in the anterior dorsal cortex of turtles cluster with human neocortical neurons in a comprehensive comparative analysis of transcription factor expression (Tosches et al., 2018). Thus, the available data indicate that the last common ancestor of mammals and reptiles already possessed a neocortex homolog. Whether the neocortex evolved even earlier remains debatable (Woych et al., 2022).
Early mammals had small bodies and small brains (Rowe et al., 2011). Body and brain size later increased in several mammalian lineages, with brain size sometimes increasing more than predicted from changes in body size. The fitness consequences of increasing body and brain size have been debated extensively and probably differ between lineages. However, increased brain size in multiple vertebrate lineages correlates positively with social complexity and/or diet (T#66; Emery et al., 2007; Shultz and Dunbar, 2007; Gonzalez-Voyer et al., 2009). Importantly, evolutionary increases in overall brain size are consistently associated with changes in brain region proportions (T#67; Finlay and Darlington, 1995). Particularly interesting is that the neocortex scales with the highest slope against total brain size, such that it occupies about 20% of the brain in a tiny shrew but 80% in humans. Also important is that the neocortex expanded much more in surface area than thickness, and that its tangential expansion correlates with increased neocortical folding. In short, as brains grew larger, the neocortex came to dominate the brain and became increasingly folded. The causes underlying these coordinated changes in brain morphology are not yet fully understood but certainly involve some fundamental rules of brain and neocortex development (Striedter et al., 2015).
A comparative analysis suggests that the small neocortex of early mammals contained only about 20 distinct cortical areas, including the primary sensory cortices, as well as prefrontal, cingulate, retrosplenial, and parahippocampal cortices (Kaas, 2019). As the neocortex expanded, additional cortical areas appeared, including a distinct motor cortex. Within the primate lineage, new areas emerged primarily in the expanded frontal, parietal, and temporal regions (T#68–69; Sherwood et al., 2012; Buckner and Krienen, 2013). Functionally, having more cortical areas probably allowed individual areas to become more specialized for specific functions, thereby overcoming the constraints imposed by multifunctionality (Kaas, 1989, 2019). In addition, the added areas probably allowed for additional kinds of perceptual representation and memory (Murray et al., 2016).
3.9. Prefrontal cortices and complex cognition
Early primates did not merely expand their prefrontal cortex (Passingham and Smaers, 2014). They also added new prefrontal areas, and the lineage containing Old world monkeys and humans later added several more (Wise, 2017). Collectively, these additional areas probably augmented the ability to make and achieve complex plans. It came as a surprise, therefore, when researchers discovered that birds are capable of similarly complex cognition and possess a telencephalic brain region, called caudolateral nidopallium (NCL), that exhibits many of the morphological and functional features of prefrontal cortex (Güntürkün, 2005). For example, this region receives dopaminergic innervation, integrates a variety of sensory inputs, and exhibits patterns of neural activity linked to working memory. Lesions of this area also cause behavioral deficits resembling those observed after prefrontal cortex lesions in mammals.
Despite these similarities, the avian NCL is not homologous to the mammalian prefrontal cortex. For one thing, lizards and turtles, which are phylogenetically intermediate between birds and mammals (T#70; Brusatte et al., 2015), do not possess an NCL. For another, the NCL does not develop from the embryonic dorsal pallium, which gives rise to mammalian neocortex; instead, it develops from the ventral pallium, which gives rise to parts of the mammalian amygdala and claustrum (Figure 2; T#71; Puelles et al., 2017; Puelles, 2021). This difference in developmental origin is supported by comparative analyses of single-cell gene expression data (Colquitt et al., 2021; Tosches, 2021). For both of these reasons, comparative neurobiologists now view the similarities between NCL and prefrontal cortex as the result of convergent evolution. It is an excellent example of how convergent evolution can produce astounding similarities from very different developmental and phylogenetic starting points.
4. Conclusion
One simple, overarching aim in teaching students about the evolutionary aspects of neurobiology is to make them aware of how incredibly diverse nervous systems are. It is tempting, especially in introductory courses, to teach about “the brain,” but even when the focus is on human brains, our knowledge often comes from the study of non-human brains. Much of this knowledge can be extrapolated to humans, but human brains have had a distinctive phylogenetic history and are, therefore, divergent in numerous respects (Preuss, 2016). It is important, for example, to appreciate that humans are unusually susceptible to Alzheimer’s disease (Walker and Jucker, 2017). Even apes rarely exhibit its symptoms, and among rodents only the South American degu naturally falls victim to this terrible disease (Hurley et al., 2018).
Nature’s great diversity can also be appreciated for its own sake, even if it is not to our medical benefit. As Darwin wrote at the end of his Origin of Species (Darwin, 1859):
“It is interesting to contemplate a tangled bank, clothed with many plants of many kinds, with birds singing on the bushes, with various insects flitting about, and with worms crawling through the damp earth, and to reflect that these elaborately constructed forms, so different from each other, and dependent upon each other in so complex a manner, have all been produced by laws acting around us…”
As neurobiologists, we may marvel at the myriad differences between the nervous systems of the animals that share our planet. Making sense of this variety requires evolutionary theory.
Author contributions
GS: Writing – original draft.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Acknowledgments
I thank the scholars who proclaimed that evolution is a concept at the core of both biology and, more specifically, neuroscience. Having worked for most of my career to make evolutionary neurobiology more accessible to a broad audience, and hence more relevant, I greatly appreciate the opportunity to make the present contribution. I know that much of the information presented here is rather dense and will require some additional reading before it can be incorporated into a neuroscience course; still, I hope that it remains accessible enough to pique the reader’s interest. Space constraints did not permit more elaboration, and narrowing the range of content seemed counterproductive, given the vast range of neurobiological phenomena evolution has shaped.
Conflict of interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Aboitiz, F., and Montiel, J. (2003). One hundred million years of interhemispheric communication: the history of the corpus callosum. Braz. J. Med. Biol. Res. 36, 409–420. doi: 10.1590/S0100-879X2003000400002
Arendt, D. (2003). Evolution of eyes and photoreceptor cell types. Int. J. Dev. Biol. 47, 563–571. doi: 10.1387/ijdb.14756332
Arendt, D., Tessmar-Raible, K., Snyman, H., Dorresteijn, A. W., and Wittbrodt, J. (2004). Ciliary photoreceptors with a vertebrate-type opsin in an invertebrate brain. Science 306, 869–871. doi: 10.1126/science.1099955
Arrese, C., Hart, N., Thomas, N., Beazley, L., and Shand, J. (2002). Trichromacy in Australian marsupials. Curr. Biol. 12, 657–660. doi: 10.1016/S0960-9822(02)00772-8
Baden, T., and Nilsson, D.-E. (2022). Is our retina really upside down? Curr. Biol. 32, R300–R303. doi: 10.1016/j.cub.2022.02.065
Baker, C. V. H., and Modrell, M. S. (2018). Insights into electroreceptor development and evolution from molecular comparisons with hair cells. Integr. Comp. Biol. 58, 329–340. doi: 10.1093/icb/icy037
Balsters, J. H., Cussans, E., Diedrichsen, J., Phillips, K. A., Preuss, T. M., Rilling, J. K., et al. (2010). Evolution of the cerebellar cortex: the selective expansion of prefrontal-projecting cerebellar lobules. NeuroImage 49, 2045–2052. doi: 10.1016/j.neuroimage.2009.10.045
Barron, A. B., Søvik, E., and Cornish, J. L. (2010). The roles of dopamine and related compounds in reward-seeking behavior across animal phyla. Front. Behav. Neurosci. 4:163. doi: 10.3389/fnbeh.2010.00163
Bell, A. J. (2023). Like moths to a flame: a review of what we know about pyrophilic insects. For. Ecol. Manag. 528:120629. doi: 10.1016/j.foreco.2022.120629
Bellingham, J., Chaurasia, S. S., Melyan, Z., Liu, C., Cameron, M. A., Tarttelin, E. E., et al. (2006). Evolution of melanopsin photoreceptors: discovery and characterization of a new melanopsin in nonmammalian vertebrates. PLoS Biol. 4:e254. doi: 10.1371/journal.pbio.0040254
Bellingham, J., and Foster, R. G. (2002). Opsins and mammalian photoentrainment. Cell Tissue Res. 309, 57–71. doi: 10.1007/s00441-002-0573-4
Beny-Shefer, Y., Zilkha, N., Lavi-Avnon, Y., Bezalel, N., Rogachev, I., Brandis, A., et al. (2017). Nucleus accumbens dopamine signaling regulates sexual preference for females in male mice. Cell Rep. 21, 3079–3088. doi: 10.1016/j.celrep.2017.11.062
Berman, N. J., and Maler, L. (1999). Neural architecture of the electrosensory lateral line lobe: adaptations for coincidence detection, a sensory searchlight and frequency-dependent adaptive filtering. J. Exp. Biol. 202, 1243–1253. doi: 10.1242/jeb.202.10.1243
Berson, D. M., Dunn, F. A., and Takao, M. (2002). Phototransduction by retinal ganglion cells that set the circadian clock. Science 295, 1070–1073. doi: 10.1126/science.1067262
Bloch, S., Hagio, H., Thomas, M., Heuzé, A., Hermel, J.-M., Lasserre, E., et al. (2020). Non-thalamic origin of zebrafish sensory nuclei implies convergent evolution of visual pathways in amniotes and teleosts. elife 9:e54945. doi: 10.7554/eLife.54945
Blount, Z. D., Lenski, R. E., and Losos, J. B. (2018). Contingency and determinism in evolution: replaying life’s tape. Science 362, 1–10. doi: 10.1126/science.aam5979
Braford, M. R. (1986). De gustibus non Est disputandem: a spiral center for taste in the brain of the teleost fish, Heterotis niloticus. Science 232, 489–491. doi: 10.1126/science.232.4749.489
Bragge, P., Synnot, A., Maas, A. I., Menon, D. K., Cooper, D. J., Rosenfeld, J. V., et al. (2016). A state-of-the-science overview of randomized controlled trials evaluating acute management of moderate-to-severe traumatic brain injury. J. Neurotrauma 33, 1461–1478. doi: 10.1089/neu.2015.4233
Brigandt, I. (2002). Homology and the origin of correspondence. Biol. Philos. 17, 389–407. doi: 10.1023/A:1020196124917
Brigandt, I. (2003). Homology in comparative, molecular, and evolutionary developmental biology: the radiation of a concept. J. Exp. Zoolog. B Mol. Dev. Evol. 299, 9–17. doi: 10.1002/jez.b.36
Bringmann, A. (2019). Structure and function of the bird fovea. Anat. Histol. Embryol. 48, 177–200. doi: 10.1111/ahe.12432
Brodie, E. D., Feldman, C. R., Hanifin, C. T., Motychak, J. E., Mulcahy, D. G., Williams, B. L., et al. (2005). Parallel arms races between garter snakes and newts involving tetrodotoxin as the phenotypic interface of coevolution. J. Chem. Ecol. 31, 343–356. doi: 10.1007/s10886-005-1345-x
Broglio, C., Martín-Monzón, I., Ocaña, F. M., Gómez, A., Durán, E., Salas, C., et al. (2015). Hippocampal pallium and map-like memories through vertebrate evolution. J. Behav. Brain Sci. 5, 109–120. doi: 10.4236/jbbs.2015.53011
Brunet, T., and Arendt, D. (2016). From damage response to action potentials: early evolution of neural and contractile modules in stem eukaryotes. Philos. Trans. R. Soc. B 371:20150043. doi: 10.1098/rstb.2015.0043
Brusatte, S. L., O’Connor, J. K., and Jarvis, E. D. (2015). The origin and diversification of birds. Curr. Biol. 25, R888–R898. doi: 10.1016/j.cub.2015.08.003
Buckner, R. L., and Krienen, F. M. (2013). The evolution of distributed association networks in the human brain. Trends Cogn. Sci. 17, 648–665. doi: 10.1016/j.tics.2013.09.017
Burkhardt, P., Colgren, J., Medhus, A., Digel, L., Naumann, B., Soto-Angel, J. J., et al. (2023). Syncytial nerve net in a ctenophore adds insights on the evolution of nervous systems. Science 380, 293–297. doi: 10.1126/science.ade5645
Burkhardt, P., and Sprecher, S. G. (2017). Evolutionary origin of synapses and neurons – bridging the gap. BioEssays 39:1700024. doi: 10.1002/bies.201700024
Carr, C. E., and Konishi, M. (1990). A circuit for detection of interaural time differences in the brain stem of the barn owl. J. Neurosci. 10, 3227–3246. doi: 10.1523/JNEUROSCI.10-10-03227.1990
Carvalho, L. S., Pessoa, D. M. A., Mountford, J. K., Davies, W. I. L., and Hunt, D. M. (2017). The genetic and evolutionary drives behind primate color vision. Front. Ecol. Evol. 5:34. doi: 10.3389/fevo.2017.00034
Cary, T., and Branchaw, J. (2017). Conceptual elements: a detailed framework to support and assess student learning of biology core concepts. CBE—Life Sci. Educ. 16:ar24. doi: 10.1187/cbe.16-10-0300
Castoe, T. A., de Koning, A. P. J., and Pollock, D. D. (2010). Adaptive molecular convergence. Commun. Integr. Biol. 3, 67–69. doi: 10.4161/cib.3.1.10174
Caves, E. M., Nowicki, S., and Johnsen, S. (2019). Von Uexküll revisited: addressing human biases in the study of animal perception. Integr. Comp. Biol. 59, 1451–1462. doi: 10.1093/icb/icz073
Cesario, J., Johnson, D. J., and Eisthen, H. L. (2020). Your brain is not an onion with a tiny reptile inside. Curr. Dir. Psychol. Sci. 29, 255–260. doi: 10.1177/0963721420917687
Chen, A., Phillips, K. A., Schaefer, J. E., and Sonner, P. M. (2023). Community-derived core concepts for neuroscience higher education. CBE Life Sci. Educ. 22:ar18. doi: 10.1187/cbe.22-02-0018
Cisek, P. (2019). Resynthesizing behavior through phylogenetic refinement. Atten. Percept. Psychophys. 81, 2265–2287. doi: 10.3758/s13414-019-01760-1
Cisek, P., and Hayden, B. Y. (2021). Neuroscience needs evolution. Philos. Trans. R. Soc. B Biol. Sci. 377:20200518. doi: 10.1098/rstb.2020.0518
Colgren, J., and Burkhardt, P. (2022). The premetazoan ancestry of the synaptic toolkit and appearance of first neurons. Essays Biochem. 66, 781–795. doi: 10.1042/EBC20220042
Collin, S. P., Davies, W. L., Hart, N. S., and Hunt, D. M. (2009). The evolution of early vertebrate photoreceptors. Phil. Transact. Roy. Soc. B. 364, 2925–2940. doi: 10.1016/0042-6989(84)90098-1
Colquitt, B. M., Merullo, D. P., Konopka, G., Roberts, T. F., and Brainard, M. S. (2021). Cellular transcriptomics reveals evolutionary identities of songbird vocal circuits. Science 371, 1–11. doi: 10.1126/science.abd9704
Cronin, T. W., and Bok, M. J. (2016). Photoreception and vision in the ultraviolet. J. Exp. Biol. 219, 2790–2801. doi: 10.1242/jeb.128769
Cunningham, C. W., Omland, K. E., and Oakley, T. H. (1998). Reconstructing ancestral character states: a critical reappraisal. Trends Ecol. Evol. 13, 361–366. doi: 10.1016/S0169-5347(98)01382-2
Curtis, P. D., Rowland, E. D., Curtis, G. B., and Dunn, J. A. (2000). “Capsaicin-treated seed as a squirrel deterrent at birdfeeders” in Wildlife damage management conferences – Proceedings (U Nebraska Lincoln) Available at: https://digitalcommons.unl.edu/icwdm_wdmconfproc/18?utm_source=digitalcommons.unl.edu%2Ficwdm_wdmconfproc%2F18&utm_medium=PDF&utm_campaign=PDFCoverPages
Davies, W. I. L., Collin, S. P., and Hunt, D. M. (2012). Molecular ecology and adaptation of visual photopigments in craniates. Mol. Ecol. 21, 3121–3158. doi: 10.1111/j.1365-294X.2012.05617.x
Davies, W. L., Hankins, M. W., and Foster, R. G. (2010). Vertebrate ancient opsin and melanopsin: divergent irradiance detectors. Photochem. Photobiol. Sci. 9, 1444–1457. doi: 10.1039/C0PP00203H
Dean, P., Porrill, J., Ekerot, C.-F., and Jörntell, H. (2010). The cerebellar microcircuit as an adaptive filter: experimental and computational evidence. Nat. Rev. Neurosci. 11, 30–43. doi: 10.1038/nrn2756
Dietz, V. (2003). Spinal cord pattern generators for locomotion. Clin. Neurophysiol. 114, 1379–1389. doi: 10.1016/S1388-2457(03)00120-2
Dijkgraaf, S. (1963). The functioning and significance of the lateral-line organs. Biol. Rev. 38, 51–105. doi: 10.1111/j.1469-185X.1963.tb00654.x
Do, M. T. H. (2019). Melanopsin and the intrinsically photosensitive retinal ganglion cells: biophysics to behavior. Neuron 104, 205–226. doi: 10.1016/j.neuron.2019.07.016
Dobzhansky, T. (1973). Nothing in biology makes sense except in the light of evolution. Am. Biol. Teach. 35, 125–129. doi: 10.2307/4444260
Dungan, S. Z., Kosyakov, A., and Chang, B. S. W. (2016). Spectral tuning of killer whale (Orcinus orca) rhodopsin: evidence for positive selection and functional adaptation in a cetacean visual pigment. Mol. Biol. Evol. 33, 323–336. doi: 10.1093/molbev/msv217
Emery, N. J., Seed, A. M., von Bayern, A. M. P., and Clayton, N. S. (2007). Cognitive adaptations of social bonding in birds. Philos. Trans. R. Soc. B Biol. Sci. 362, 489–505. doi: 10.1098/rstb.2006.1991
Fain, G. L., Hardie, R., and Laughlin, S. B. (2010). Phototransduction and the evolution of photoreceptors. Curr. Biol. 20, R114–R124. doi: 10.1016/j.cub.2009.12.006
Farris, S. M. (2008). Evolutionary convergence of higher brain centers spanning the protostome-deuterostome boundary. Brain Behav. Evol. 72, 106–122. doi: 10.1159/000151471
Feng, P., Zheng, J., Rossiter, S. J., Wang, D., and Zhao, H. (2014). Massive losses of taste receptor genes in toothed and baleen whales. Genome Biol. Evol. 6, 1254–1265. doi: 10.1093/gbe/evu095
Field, L. M., Emyr Davies, T. G., O’Reilly, A. O., Williamson, M. S., and Wallace, B. A. (2017). Voltage-gated sodium channels as targets for pyrethroid insecticides. Eur. Biophys. J. 46, 675–679. doi: 10.1007/s00249-016-1195-1
Finger, T. E. (2008). Sorting food from stones: the vagal taste system in Goldfish, Carassius auratus. J. Comp. Physiol. A 194, 135–143. doi: 10.1007/s00359-007-0276-0
Finlay, B. L., and Darlington, R. B. (1995). Linked regularities in the development and evolution of mammalian brains. Science 268, 1578–1584. doi: 10.1126/science.7777856
Förster, D., Helmbrecht, T. O., Mearns, D. S., Jordan, L., Mokayes, N., and Baier, H. (2020). Retinotectal circuitry of larval zebrafish is adapted to detection and pursuit of prey. elife 9:e58596. doi: 10.7554/eLife.58596
Fournier, J., Müller, C. M., Schneider, I., and Laurent, G. (2018). Spatial information in a non-retinotopic visual cortex. Neuron 97, 164–180.e7. doi: 10.1016/j.neuron.2017.11.017
Franze, K., Grosche, J., Skatchkov, S. N., Schinkinger, S., Foja, C., Schild, D., et al. (2007). Müller cells are living optical fibers in the vertebrate retina. Proc. Natl. Acad. Sci. U. S. A. 104, 8287–8292. doi: 10.1073/pnas.0611180104
Fry, B. G., Vidal, N., Norman, J. A., Vonk, F. J., Scheib, H., Ramjan, S. F. R., et al. (2006). Early evolution of the venom system in lizards and snakes. Nat. Cell Biol. 439, 584–588. doi: 10.1038/nature04328
Garstang, W. (1922). The theory of recapitulation: a critical re-statement of the biogenetic law. J. Linn. Soc. Lond. Zool. 35, 81–101. doi: 10.1111/j.1096-3642.1922.tb00464.x
Ghiselin, M. T. (2005). Homology as a relation of correspondence between parts of individuals. Theory Biosci. 124, 91–103. doi: 10.1007/BF02814478
Godfrey, P. A., Malnic, B., and Buck, L. B. (2004). The mouse olfactory receptor gene family. Proc. Natl. Acad. Sci. U. S. A. 101, 2156–2161. doi: 10.1073/pnas.0308051100
Gonzalez-Voyer, A., Winberg, S., and Kolm, N. (2009). Social fishes and single mothers: brain evolution in African cichlids. Proc Roy Soc B 276, 161–167. doi: 10.1098/rspb.2008.0979
Gooley, J. J., Lu, J., Fischer, D., and Saper, C. B. (2003). A broad role for melanopsin in nonvisual photoreception. J. Neurosci. 23, 7093–7106. doi: 10.1523/JNEUROSCI.23-18-07093.2003
Gosling, S. D., and Graybeal, A. (2007). Tree thinking: a new paradigm for integrating comparative data in psychology. J. Gen. Psychol. 134, 259–277. doi: 10.3200/GENP.134.2.259-278
Gould, S. J., and Lewontin, R. C. (1979). The spandrels of san Marco and the Panglossian paradigm: a critique of the adaptationist programme. Proc. R. Soc. Lond. Ser. B 205, 581–598. doi: 10.1098/rspb.1979.0086
Gracheva, E. O., Ingolia, N. T., Kelly, Y. M., Cordero-Morales, J. F., Hollopeter, G., Chesler, A. T., et al. (2010). Molecular basis of infrared detection by snakes. Nature 464, 1006–1011. doi: 10.1038/nature08943
Gregory, T. R. (2008). The evolution of complex organs. Evol. Educ. Outreach 1, 358–389. doi: 10.1007/s12052-008-0076-1
Gregory, T. R. (2009). Understanding natural selection: essential concepts and common misconceptions. Evol. Educ. Outreach 2, 156–175. doi: 10.1007/s12052-009-0128-1
Grillner, S., Deliagina, T., el Manira, A., Hill, R. H., Orlovsky, G. N., Wallén, P., et al. (1995). Neural networks that co-ordinate locomotion and body orientation in lamprey. Trends Neurosci. 18, 270–279. doi: 10.1016/0166-2236(95)80008-P
Grothe, B., and Pecka, M. (2014). The natural history of sound localization in mammals--a story of neuronal inhibition. Front. Neural Circuits 8:116. doi: 10.3389/fncir.2014.00116
Güntürkün, O. (2005). The avian ‘prefrontal cortex’ and cognition. Curr. Opin. Neurobiol. 15, 686–693. doi: 10.1016/j.conb.2005.10.003
Haeusler, M., Grunstra, N. D. S., Martin, R. D., Krenn, V. A., Fornai, C., and Webb, N. M. (2021). The obstetrical dilemma hypothesis: there’s life in the old dog yet. Biol. Rev. Camb. Philos. Soc. 96, 2031–2057. doi: 10.1111/brv.12744
Haimson, B., Meir, O., Sudakevitz-Merzbach, R., Elberg, G., Friedrich, S., Lovell, P. V., et al. (2021). Natural loss of function of ephrin-B3 shapes spinal flight circuitry in birds. Sci. Adv. 7:eabg5968. doi: 10.1126/sciadv.abg5968
Hanifin, C. T., and Gilly, W. F. (2015). Evolutionary history of a complex adaptation: tetrodotoxin resistance in salamanders. Evolution 69, 232–244. doi: 10.1111/evo.12552
Hanke, F. D., and Kelber, A. (2020). The eye of the common octopus (Octopus vulgaris). Front. Physiol. 10:1637. doi: 10.3389/fphys.2019.01637
Hansen, D. V., Lui, J. H., Flandin, P., Yoshikawa, K., Rubenstein, J. L., Alvarez-Buylla, A., et al. (2013). Non-epithelial stem cells and cortical interneuron production in the human ganglionic eminences. Nat. Neurosci. 16, 1576–1587. doi: 10.1038/nn.3541
Hart, N. S., Bailes, H. J., Vorobyev, M., Marshall, N. J., and Collin, S. P. (2008). Visual ecology of the Australian lungfish (Neoceratodus forsteri). BMC Ecol. 8, 21–14. doi: 10.1186/1472-6785-8-21
Harzsch, S., and Krieger, J. (2021). Genealogical relationships of mushroom bodies, hemiellipsoid bodies, and their afferent pathways in the brains of Pancrustacea: recent progress and open questions. Arthropod Struct. Dev. 65:101100. doi: 10.1016/j.asd.2021.101100
Hashimoto, M., and Hibi, M. (2012). Development and evolution of cerebellar neural circuits. Dev. Growth Amp. Differ. 54, 373–389. doi: 10.1111/j.1440-169X.2012.01348.x
Heesy, C. P., and Hall, M. I. (2010). The nocturnal bottleneck and the evolution of mammalian vision. Brain Behav. Evol. 75, 195–203. doi: 10.1159/000314278
Hibi, M., Matsuda, K., Takeuchi, M., Shimizu, T., and Murakami, Y. (2017). Evolutionary mechanisms that generate morphology and neural-circuit diversity of the cerebellum. Dev. Growth Diff. 59, 228–243. doi: 10.1111/dgd.12349
Hibi, M., and Shimizu, T. (2012). Development of the cerebellum and cerebellar neural circuits. Dev. Neurobiol. 72, 282–301. doi: 10.1002/dneu.20875
Hills, T. T., Todd, P. M., Lazer, D., Redish, A. D., and Couzin, I. D., Cognitive Search Research Group (2015). Exploration versus exploitation in space, mind, and society. Trends Cogn. Sci. 19, 46–54. doi: 10.1016/j.tics.2014.10.004
Hinz, M., Klein, A., Schmitz, A., and Schmitz, H. (2018). The impact of infrared radiation in flight control in the Australian “firebeetle” Merimna atrata. PLoS One 13:e0192865. doi: 10.1371/journal.pone.0192865
Hobert, O. (2018). “The neuronal genome of Caenorhabditis elegans,” in WormBook: the online review of C. elegans biology [internet] (WormBook). Available at: https://www.ncbi.nlm.nih.gov/books/NBK154158/ (Accessed July 19, 2023).
Hodgkin, A. L., and Huxley, A. F. (1952). A quantitative description of membrane current and its application to conduction and excitation in nerve. J. Physiol. 117, 500–544. doi: 10.1113/jphysiol.1952.sp004764
Huang, X., Albou, L.-P., Mushayahama, T., Muruganujan, A., Tang, H., and Thomas, P. D. (2018). Ancestral genomes: a resource for reconstructed ancestral genes and genomes across the tree of life. Nucleic Acids Res. 47, D271–D279. doi: 10.1093/nar/gky1009
Hurley, M. J., Deacon, R. M. J., Beyer, K., Ioannou, E., Ibáñez, A., Teeling, J. L., et al. (2018). The long-lived Octodon degus as a rodent drug discovery model for Alzheimer’s and other age-related diseases. Pharmacol. Ther. 188, 36–44. doi: 10.1016/j.pharmthera.2018.03.001
Inman, D. M., and Steward, O. (2003). Physical size does not determine the unique histopathological response seen in the injured mouse spinal cord. J. Neurotrauma 20, 33–42. doi: 10.1089/08977150360517164
Isler, K., and van Schaik, C. P. (2009). The expensive brain: a framework for explaining evolutionary changes in brain size. J. Hum. Evol. 57, 392–400. doi: 10.1016/j.jhevol.2009.04.009
Itoi, S., Yoshikawa, S., Asahina, K., Suzuki, M., Ishizuka, K., Takimoto, N., et al. (2014). Larval pufferfish protected by maternal tetrodotoxin. Toxicon 78, 35–40. doi: 10.1016/j.toxicon.2013.11.003
Iwaniuk, A. N., Hurd, P. L., and Wylie, D. R. W. (2006). Comparative morphology of the avian cerebellum: I. Degree of foliation. Brain. Behav. Evol. 68, 45–62. doi: 10.1159/000093530
Jacobs, G. H. (2008). Primate color vision: a comparative perspective. Vis. Neurosci. 25, 619–633. doi: 10.1017/S0952523808080760
Jiang, P., Josue, J., Li, X., Glaser, D., Li, W., Brand, J., et al. (2012). Major taste loss in carnivorous mammals. Proc. Natl. Acad. Sci. U. S. A. 109, 4956–4961. doi: 10.1073/pnas.1118360109
Jordt, S.-E., and Julius, D. (2002). Molecular basis for species-specific sensitivity to “hot” chili peppers. Cells 108, 421–430. doi: 10.1016/s0092-8674(02)00637-2
Kaas, J. H. (1989). Why does the brain have so many visual areas? J. Cogn. Neurosci. 1, 121–135. doi: 10.1162/jocn.1989.1.2.121
Kaas, J. H. (2019). “Chapter 3 – the origin and evolution of neocortex: from early mammals to modern humans” in Progress in brain research evolution of the human brain: from matter to mind. ed. M. A. Hofman (Amsterdam: Elsevier), 61–81.
Kelber, A., and Jacobs, G. H. (2016). “Evolution of color vision,” in Human color vision, eds. J. Kremers Barras, R. C., and Marshall, M. J. Cham (Springer), 317–354.
Kishida, T., Thewissen, J., Hayakawa, T., Imai, H., and Agata, K. (2015). Aquatic adaptation and the evolution of smell and taste in whales. Zool. Lett. 1:9. doi: 10.1186/s40851-014-0002-z
Knudsen, E. I. (2020). Evolution of neural processing for visual perception in vertebrates. J. Comp. Neurol. 528, 2888–2901. doi: 10.1002/cne.24871
Knudsen, E. I., and Schwarz, J. S. (2016). “The optic tectum: a structure evolved for stimulus selection” in Evolution of nervous systems. eds. J. H. Kaas and G. F. Striedter, vol. 1. 2nd ed (Cham: Elsevier), 387–408.
Kochanek, P. M., Dixon, C. E., Mondello, S., Wang, K. K. K., Lafrenaye, A., Bramlett, H. M., et al. (2018). Multi-center pre-clinical consortia to enhance translation of therapies and biomarkers for traumatic brain injury: operation brain trauma therapy and beyond. Front. Neurol. 9:640. doi: 10.3389/fneur.2018.00640
Kordiš, D., and Gubenšek, F. (2000). Adaptive evolution of animal toxin multigene families. Gene 261, 43–52. doi: 10.1016/S0378-1119(00)00490-X
Krebs, H. A. (1975). The august Krogh principle: “for many problems there is an animal on which it can be most conveniently studied”. J. Exp. Zool. 194, 221–226. doi: 10.1002/jez.1401940115
Lahti, D. C., Johnson, N. A., Ajie, B. C., Otto, S. P., Hendry, A. P., Blumstein, D. T., et al. (2009). Relaxed selection in the wild. Trends Ecol. Evol. 24, 487–496. doi: 10.1016/j.tree.2009.03.010
Lamb, T. D. (2013). Evolution of phototransduction, vertebrate photoreceptors and retina. Prog. Retin. Eye Res. 36, 52–119. doi: 10.1016/j.preteyeres.2013.06.001
Laska, M. (2017). “Human and animal olfactory capabilities compared” in Springer Handbook of Odor. ed. A. Buettner (Cham: Springer), 675–689.
Lemon, R. N., and Griffiths, J. (2005). Comparing the function of the corticospinal system in different species: organizational differences for motor specialization? Muscle Nerve 32, 261–279. doi: 10.1002/mus.20333
Leonard, W. R., Snodgrass, J. J., and Robertson, M. L. (2007). Effects of brain evolution on human nutrition and metabolism. Annu. Rev. Nutr. 27, 311–327. doi: 10.1146/annurev.nutr.27.061406.093659
Levitan, D., Doyle, T. G., Brousseau, D., Lee, M. K., Thinakaran, G., Slunt, H. H., et al. (1996). Assessment of normal and mutant human presenilin function in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U. S. A. 93, 14940–14944. doi: 10.1073/pnas.93.25.14940
Li, Y., Liu, Z., Shi, P., and Zhang, J. (2010). The hearing gene Prestin unites echolocating bats and whales. Curr. Biol. 20, R55–R56. doi: 10.1016/j.cub.2009.11.042
Liu, Z., Qi, F.-Y., Zhou, X., Ren, H.-Q., and Shi, P. (2014a). Parallel sites implicate functional convergence of the hearing gene prestin among echolocating mammals. Mol. Biol. Evol. 31, 2415–2424. doi: 10.1093/molbev/msu194
Liu, Z., Wang, W., Zhang, T.-Z., Li, G.-H., He, K., Huang, J.-F., et al. (2014b). Repeated functional convergent effects of NaV1.7 on acid insensitivity in hibernating mammals. Proc. R. Soc. B 281:20132950. doi: 10.1098/rspb.2013.2950
Losos, J. B. (2011). Convergence, adaptation, and constraint. Evolution 65, 1827–1840. doi: 10.1111/j.1558-5646.2011.01289.x
MacLeod, C. E., Zilles, K., Schleicher, A., Rilling, J. K., and Gibson, K. R. (2003). Expansion of the neocerebellum in Hominoidea. J. Hum. Evol. 44, 401–429. doi: 10.1016/S0047-2484(03)00028-9
Manley, G. A. (2017). Comparative auditory neuroscience: understanding the evolution and function of ears. J. Assoc. Res. Otolaryngol. 18, 1–24. doi: 10.1007/s10162-016-0579-3
Maxwell, E. K., Schnitzler, C. E., Havlak, P., Putnam, N. H., Nguyen, A.-D., Moreland, R. T., et al. (2014). Evolutionary profiling reveals the heterogeneous origins of classes of human disease genes: implications for modeling disease genetics in animals. BMC Evol. Biol. 14:212. doi: 10.1186/s12862-014-0212-1
Medina, L., Abellán, A., and Desfilis, E. (2022). Evolving views on the pallium. Brain Behav. Evol. 96, 181–199. doi: 10.1159/000519260
Medina, L., Bupesh, M., and Abellán, A. (2011). Contribution of genoarchitecture to understanding forebrain evolution and development, with particular emphasis on the amygdala. Brain Behav. Evol. 78, 216–236. doi: 10.1159/000330056
Meek, J., Grant, K., and Bell, C. (1999). Structural organization of the mormyrid electrosensory lateral line lobe. J. Exp. Biol. 202, 1291–1300. doi: 10.1242/jeb.202.10.1291
Meek, J., and Nieuwenhuys, R. (1991). Palisade pattern of mormyrid Purkinje cells: a correlated light and electron microscopic study. J. Comp. Neurol. 306, 156–192. doi: 10.1002/cne.903060111
Meredith, R. W., Gatesy, J., Emerling, C. A., York, V. M., and Springer, M. S. (2013). Rod monochromacy and the coevolution of cetacean retinal opsins. PLoS Genet. 9:e1003432. doi: 10.1371/journal.pgen.1003432
Merker, B. (2007). Consciousness without a cerebral cortex: a challenge for neuroscience and medicine. Behav. Brain Sci. 30, 63–81-discussion 81-134. doi: 10.1017/S0140525X07000891
Miller, M. W. (2020). Dopamine as a multifunctional neurotransmitter in gastropod molluscs: an evolutionary hypothesis. Biol. Bull. 239, 189–208. doi: 10.1086/711293
Miyazawa, A., Fujiyoshi, Y., and Unwin, N. (2003). Structure and gating mechanism of the acetylcholine receptor pore. Nature 423, 949–955. doi: 10.1038/nature01748
Moore, J. K. (1980). The primate cochlear nuclei: loss of lamination as a phylogenetic process. J. Comp. Neurol. 193, 609–629. doi: 10.1002/cne.901930303
Moreno, N., González, A., and Rétaux, S. (2008). Evidences for tangential migrations in Xenopus telencephalon: developmental patterns and cell tracking experiments. Dev. Neurobiol. 68, 504–520. doi: 10.1002/dneu.20603
Moroz, L. L., Kocot, K. M., Citarella, M. R., Dosung, S., Norekian, T. P., Povolotskaya, I. S., et al. (2015). The ctenophore genome and the evolutionary origins of neural systems. Nat. Cell Biol. 510, 109–114. doi: 10.1038/nature13400
Moroz, L. L., and Romanova, D. Y. (2021). Selective advantages of synapses in evolution. Front. Cell Dev. Biol. 9:726563. doi: 10.3389/fcell.2021.726563
Morshedian, A., and Fain, G. L. (2017). The evolution of rod photoreceptors. Philos. Trans. R. Soc. B Biol. Sci. 372:20160074. doi: 10.1098/rstb.2016.0074
Murray, E., Wise, S., and Graham, K. (2016). The evolution of memory systems. Oxford Oxford University Press
Muto, A., Ohkura, M., Abe, G., Nakai, J., and Kawakami, K. (2013). Real-time visualization of neuronal activity during perception. Curr. Biol. 23, 307–311. doi: 10.1016/j.cub.2012.12.040
Necker, R. (2006). Specializations in the lumbosacral vertebral canal and spinal cord of birds: evidence of a function as a sense organ which is involved in the control of walking. J. Comp. Physiol. A 192, 439–448. doi: 10.1007/s00359-006-0105-x
Nesse, R. M., and Berridge, K. C. (1997). Psychoactive drug use in evolutionary perspective. Science 278, 63–66. doi: 10.1126/science.278.5335.63
Nevitt, G. A. (2008). Sensory ecology on the high seas: the odor world of the procellariiform seabirds. J. Exp. Biol. 211, 1706–1713. doi: 10.1242/jeb.015412
Newman, E. A., and Hartline, P. H. (1982). The infrared “vision” of snakes. Sci. Am. 246, 116–127. doi: 10.1038/scientificamerican0382-116
Nieuwenhuys, R. (2009). The forebrain of actinopterygians revisited. Brain Behav. Evol. 73, 229–252. doi: 10.1159/000225622
Nieuwenhuys, R. (2011). The structural, functional, and molecular organization of the brainstem. Front. Neuroanat. 5, 1–17. doi: 10.3389/fnana.2011.00033
Nieuwenhuys, R., ten Donkelaar, H.J., and Nicholson, C. (1998). The central nervous system of vertebrates. Springer, Berlin.
Niimura, Y. (2012). Olfactory receptor multigene family in vertebrates: from the viewpoint of evolutionary genomics. Curr. Genomics 13, 103–114. doi: 10.2174/138920212799860706
Nilsson, D.-E., Warrant, E. J., Johnsen, S., Hanlon, R., and Shashar, N. (2012). A unique advantage for giant eyes in giant squid. Curr. Biol. 22, 683–688. doi: 10.1016/j.cub.2012.02.031
Nishino, A., and Okamura, Y. (2018). Evolutionary history of voltage-gated sodium channels. Handb. Exp. Pharmacol. 246, 3–32. doi: 10.1007/164_2017_70
Northcutt, R. G. (1984). Evolution of the vertebrate central nervous system: patterns and processes. Am. Zool. 24, 701–716. doi: 10.1093/icb/24.3.701
Ödeen, A., Pruett-Jones, S., Driskell, A. C., Armenta, J. K., and Håstad, O. (2012). Multiple shifts between violet and ultraviolet vision in a family of passerine birds with associated changes in plumage coloration. Proc. R. Soc. B Biol. Sci. 279, 1269–1276. doi: 10.1098/rspb.2011.1777
Olivera, B. M., Walker, C., Cartier, G. E., Hooper, D., Santos, A. D., Schoenfeld, R., et al. (1999). Speciation of cone snails and interspecific hyperdivergence of their venom peptides: potential evolutionary significance of introns. Ann. N. Y. Acad. Sci. 870, 223–237. doi: 10.1111/j.1749-6632.1999.tb08883.x
Passingham, R. E., and Smaers, J. B. (2014). Is the prefrontal cortex especially enlarged in the human brain allometric relations and remapping factors. Brain Behav. Evol. 84, 156–166. doi: 10.1159/000365183
Perry, V. H., and Cowey, A. (1984). Retinal ganglion cells that project to the superior colliculus and pretectum in the macaque monkey. Neuroscience 12, 1125–1137. doi: 10.1016/0306-4522(84)90007-1
Péterfi, O., Boda, F., Szabó, Z., Ferencz, E., and Bába, L. (2019). Hypotensive snake venom components – a mini-review. Molecules 24:2778. doi: 10.3390/molecules24152778
Pettigrew, J. D. (1999). Electroreception in monotremes. J. Exp. Biol. 202, 1447–1454. doi: 10.1242/jeb.202.10.1447
Porrill, J., Dean, P., and Anderson, S. R. (2013). Adaptive filters and internal models: multilevel description of cerebellar function. Neural Netw. 47, 134–149. doi: 10.1016/j.neunet.2012.12.005
Preuss, T. M. (2016). “The human brain: evolution and distinctive features” in On human nature. eds. M. Tibayrenc and F. J. Ayala (Amsterdam: Elsevier Inc), 125–149.
Puelles, L. (2001). Brain segmentation and forebrain development in amniotes. Brain Res. Bull. 55, 695–710. doi: 10.1016/S0361-9230(01)00588-3
Puelles, L. (2013). “Plan of the developing vertebrate nervous system: relating embryology to the adult nervous system (prosomere model, overview of brain organization)” in Comprehensive developmental neuroscience: patterning and cell type specification in the developing CNS and PNS. eds. J. L. R. Rubenstein and P. Goldman-Rakic (Oxford: Elsevier), 187–209.
Puelles, L. (2021). Current status of the hypothesis of a claustro-insular homolog in sauropsids. Brain Behav. Evol. 96, 212–241. doi: 10.1159/000520742
Puelles, L., Harrison, M., Paxinos, G., and Watson, C. (2013). A developmental ontology for the mammalian brain based on the prosomeric model. Trends Neurosci. 36, 570–578. doi: 10.1016/j.tins.2013.06.004
Puelles, L., Medina, L., Borello, U., Legaz, I., Teissier, A., Pierani, A., et al. (2016). Radial derivatives of the mouse ventral pallium traced with Dbx1-LacZ reporters. J. Chem. Neuroanat. 75, 2–19. doi: 10.1016/j.jchemneu.2015.10.011
Puelles, L., Sandoval, J. E., Ayad, A., del Corral, R., Alonso, A., Ferran, J. L., et al. (2017). “The pallium in reptiles and birds in the light of the updated tetrapartite pallium model” in Evolution of nervous systems. eds. J. H. Kaas and G. F. Striedter, vol. 1. 2nd ed (Oxford: Elsevier), 519–555.
Raman, I. M., and Ferster, D. L. (2022). The annotated Hodgkin and Huxley: a reader’s guide. Princeton, NJ Princeton University Press.
Richardson, M. K. (1995). Heterochrony and the phylotypic period. Dev. Biol. 172, 412–421. doi: 10.1006/dbio.1995.8041
Rowe, T. B., Macrini, T. E., and Luo, Z.-X. (2011). Fossil evidence on origin of the mammalian brain. Science 332, 955–957. doi: 10.1126/science.1203117
Rowe, A. H., Xiao, Y., Rowe, M. P., Cummins, T. R., and Zakon, H. H. (2013). Voltage-gated sodium channel in grasshopper mice defends against bark scorpion toxin. Science 342, 441–446. doi: 10.1126/science.1236451
Rubenstein, J. L., Martinez, S., Shimamura, K., and Puelles, L. (1994). The embryonic vertebrate forebrain: the prosomeric model. Science 266, 578–580. doi: 10.1126/science.7939711
Rubin, G. M., Yandell, M. D., Wortman, J. R., Gabor, G. L., Miklos,, Nelson, C. R., et al. (2000). Comparative genomics of the eukaryotes. Science 287, 2204–2215. doi: 10.1126/science.287.5461.2204
Ryan, T. J., and Grant, S. G. N. (2009). The origin and evolution of synapses. Nat. Rev. Neurosci. 10, 701–712. doi: 10.1038/nrn2717
Sachkova, M. Y., Nordmann, E.-L., Soto-Àngel, J. J., Meeda, Y., Górski, B., Naumann, B., et al. (2021). Neuropeptide repertoire and 3D anatomy of the ctenophore nervous system. Curr. Biol. 31, 5274–5285.e6. doi: 10.1016/j.cub.2021.09.005
Sáez, A. G., Lozano, E., and Zaldívar-Riverón, A. (2009). Evolutionary history of Na,K-ATPases and their osmoregulatory role. Genetica 136, 479–490. doi: 10.1007/s10709-009-9356-0
Schiller, P. H. (2010). “The superior colliculus and visual function” in Handbook of physiology – the nervous system, Suppl. 3. eds. J. Brookhart, V. Mountastle, I. Darian-Smith, and S. Geiger (New York, NY: Wiley Online), 457–505.
Schmahmann, J. D. (1991). An emerging concept. The cerebellar contribution to higher function. Arch. Neurol. 48, 1178–1187. doi: 10.1001/archneur.1991.00530230086029
Schultz, W. (2011). Potential vulnerabilities of neuronal reward, risk, and decision mechanisms to addictive drugs. Neuron 69, 603–617. doi: 10.1016/j.neuron.2011.02.014
Sengupta, B., Stemmler, M., Laughlin, S. B., and Niven, J. E. (2010). Action potential energy efficiency varies among neuron types in vertebrates and invertebrates. PLoS Comput. Biol. 6:e1000840. doi: 10.1371/journal.pcbi.1000840
Sherwood, C. C., Bauernfeind, A. L., Bianchi, S., Raghanti, M. A., and Hof, P. R. (2012). Human brain evolution writ large and small. Prog. Brain Res. 195, 237–254. doi: 10.1016/B978-0-444-53860-4.00011-8
Shipp, S. (2004). The brain circuitry of attention. Trends Cogn. Sci. 8, 223–230. doi: 10.1016/j.tics.2004.03.004
Shultz, S., and Dunbar, R. I. M. (2007). The evolution of the social brain: anthropoid primates contrast with other vertebrates. Proc. R. Soc. B Biol. Sci. 274, 2429–2436. doi: 10.1098/rspb.2007.0693
Simpson, S. B. Jr. (1968). Morphology of the regenerated spinal cord in the lizard, Anolis carolinensis. J. Comp. Neurol. 134, 193–209. doi: 10.1002/cne.901340207
Sîrbulescu, R. F., Ilieş, I., and Zupanc, G. K. H. (2009). Structural and functional regeneration after spinal cord injury in the weakly electric teleost fish, Apteronotus leptorhynchus. J. Comp. Physiol. A 195, 699–714. doi: 10.1007/s00359-009-0445-4
Smaers, J. B., Turner, A. H., Gómez-Robles, A., and Sherwood, C. C. (2018). A cerebellar substrate for cognition evolved multiple times independently in mammals. elife 7:393. doi: 10.7554/eLife.35696
Soderlund, D. M., and Lee, S. H. (2001). Point mutations in Homology Domain II modify the sensitivity of rat Nav1.8 sodium channels to the pyrethroid insecticide cismethrin. Neurotoxicology 22, 755–765. doi: 10.1016/S0161-813X(01)00065-1
Solvi, C., Baciadonna, L., and Chittka, L. (2016). Unexpected rewards induce dopamine-dependent positive emotion–like state changes in bumblebees. Science 353, 1529–1531. doi: 10.1126/science.aaf4454
Sroga, J. M., Jones, T. B., Kigerl, K. A., McGaughy, V. M., and Popovich, P. G. (2003). Rats and mice exhibit distinct inflammatory reactions after spinal cord injury. J. Comp. Neurol. 462, 223–240. doi: 10.1002/cne.10736
Stanchak, K. E., French, C., Perkel, D. J., and Brunton, B. W. (2020). The balance hypothesis for the avian lumbosacral organ and an exploration of its morphological variation. Integr. Org. Biol. 2, 1–15. doi: 10.1093/iob/obaa024
Strausfeld, N. J., Sinakevitch, I., Brown, S. M., and Farris, S. M. (2009). Ground plan of the insect mushroom body: functional and evolutionary implications. J. Comp. Neurol. 513, 265–291. doi: 10.1002/cne.21948
Strausfeld, N. J., Wolff, G. H., and Sayre, M. E. (2020). Mushroom body evolution demonstrates homology and divergence across Pancrustacea. elife 9:e52411. doi: 10.7554/eLife.52411
Strick, P. L., Dum, R. P., and Fiez, J. A. (2009). Cerebellum and nonmotor function. Annu. Rev. Neurosci. 32, 413–434. doi: 10.1146/annurev.neuro.31.060407.125606
Striedter, G. F. (1997). The telencephalon of tetrapods in evolution. Brain Behav. Evol. 49, 179–194. doi: 10.1159/000112991
Striedter, G. F. (2015). Neurobiology: a functional approach. New York, NY: Oxford University Press.
Striedter, G. F. (2019). Variation across species and levels: implications for model species research. Brain Behav. Evol. 93, 1–13. doi: 10.1159/000499664
Striedter, G. F. (2022). Model systems in biology: history, philosophy, and practical concerns. Cambridge, MA MIT Press.
Striedter, G. F., and Northcutt, R. G. (1991). Biological hierarchies and the concept of homology. Brain Behav. Evol. 38, 177–189. doi: 10.1159/000114387
Striedter, G. F., and Northcutt, R. G. (2006). Head size constrains forebrain development and evolution in ray-finned fishes. Evol. Amp Dev. 8, 215–222. doi: 10.1111/j.1525-142X.2006.00091.x
Striedter, G. F., and Northcutt, R. G. (2020). Brains through time: a natural history of vertebrates. New York, NY: Oxford University Press.
Striedter, G. F., and Northcutt, R. G. (2021). The independent evolution of dorsal pallia in multiple vertebrate lineages. Brain Behav. Evol. 96, 200–211. doi: 10.1159/000516563
Striedter, G. F., Srinivasan, S., and Monuki, E. S. (2015). Cortical folding: when, where, how, and why? Annu. Rev. Neurosci. 38, 291–307. doi: 10.1146/annurev-neuro-071714-034128
Suryanarayana, S. M., Pérez-Fernández, J., Robertson, B., and Grillner, S. (2020). The evolutionary origin of visual and somatosensory representation in the vertebrate pallium. Nat. Ecol. Evol. 4, 639–651. doi: 10.1038/s41559-020-1137-2
Toda, Y., Ko, M.-C., Liang, Q., Miller, E. T., Rico-Guevara, A., Nakagita, T., et al. (2021). Early origin of sweet perception in the songbird radiation. Science 373, 226–231. doi: 10.1126/science.abf6505
Tosches, M. A. (2021). Different origins for similar brain circuits. Science 371, 676–677. doi: 10.1126/science.abf9551
Tosches, M. A., Yamawaki, T. M., Naumann, R. K., Jacobi, A. A., Tushev, G., and Laurent, G. (2018). Evolution of pallium, hippocampus, and cortical cell types revealed by single-cell transcriptomics in reptiles. Science 360, 881–888. doi: 10.1126/science.aar4237
Tsuboi, M., Husby, A., Kotrschal, A., Hayward, A., Buechel, S. D., Zidar, J., et al. (2015). Comparative support for the expensive tissue hypothesis: big brains are correlated with smaller gut and greater parental investment in Lake Tanganyika cichlids. Evolution 69, 190–200. doi: 10.1111/evo.12556
Wagner, G. P. (2014). Homology, genes, and evolutionary innovation. Princeton, NJ Princeton University Press.
Wagner, G. P. (2015). Evolutionary innovations and novelties: let us get down to business! Zool. Anz. 256, 75–81. doi: 10.1016/j.jcz.2015.04.006
Walker, L. C., and Jucker, M. (2017). The exceptional vulnerability of humans to Alzheimer’s disease. Trends Mol. Med. 23, 534–545. doi: 10.1016/j.molmed.2017.04.001
Wang, Y., Li, X., Yang, M., Wu, C., Zou, Z., Tang, J., et al. (2017). Centipede venom peptide SsmTX-I with two intramolecular disulfide bonds shows analgesic activities in animal models. J. Pept. Sci. Off. Publ. Eur. Pept. Soc. 23, 384–391. doi: 10.1002/psc.2988
Wicht, H. (1996). The brains of lampreys and hagfishes: characteristics, characters, and comparisons. Brain Behav. Evol. 48, 248–261. doi: 10.1159/000113204
Wilson, R. A., Barker, M. J., and Brigandt, I., University of Arkansas Press (2007). When traditional essentialism fails: biological natural kinds. Philos. Top. 35, 189–215. doi: 10.5840/philtopics2007351/29
Wise, R. A. (2006). Role of brain dopamine in food reward and reinforcement. Philos. Trans. R. Soc. B Biol. Sci. 361, 1149–1158. doi: 10.1098/rstb.2006.1854
Wise, S. P. (2017). “The evolution of the prefrontal cortex in early primates and anthropoids” in Evolution of nervous systems. eds. J. H. Kaas and L. Krubitzer, vol. 3. 2nd edn. (Oxford, UK: Elsevier), 387–422.
Woych, J., Ortega Gurrola, A., Deryckere, A., Jaeger, E. C. B., Gumnit, E., Merello, G., et al. (2022). Cell type profiling in salamanders identifies innovations in vertebrate forebrain evolution. Science 377:eabp9186. doi: 10.1126/science.abp9186
Wu, Y., Wang, H., Wang, H., and Hadly, E. A. (2017). Rethinking the origin of primates by reconstructing their diel activity patterns using genetics and morphology. Sci. Rep. 7:11837. doi: 10.1038/s41598-017-12090-3
Yamamoto, K., Fontaine, R., Pasqualini, C., and Vernier, P. (2015). Classification of dopamine receptor genes in vertebrates: nine subtypes in Osteichthyes. Brain Behav. Evol. 86, 164–175. doi: 10.1159/000441550
Yamamoto, K., and Vernier, P. (2011). The evolution of dopamine systems in chordates. Front. Neuroanat. 5, 1–21. doi: 10.3389/fnana.2011.00021
Yopak, K. E., Galinsky, V. L., Berquist, R. M., and Frank, L. R. (2016). Quantitative classification of cerebellar foliation in cartilaginous fishes (class: Chondrichthyes) using three-dimensional shape analysis and its implications for evolutionary biology. Brain Behav. Evol. 87, 252–264. doi: 10.1159/000446904
Zakon, H. H. (2002). Convergent evolution on the molecular level. Brain Behav. Evol. 59, 250–261. doi: 10.1159/000063562
Zakon, H. H. (2013). “Adaptive evolution of voltage-gated sodium channels: the first 800 million years” in In the light of evolution: Volume VI: Brain and behavior. eds. G. F. Striedter, J. C. Avise, and F. J. Ayala (USA: National Academies Press)
Zakon, H. H., Jost, M. C., and Lu, Y. (2011). Expansion of voltage-dependent Na+ channel gene family in early tetrapods coincided with the emergence of terrestriality and increased brain complexity. Mol. Biol. Evol. 28, 1415–1424. doi: 10.1093/molbev/msq325
Zampieri, F. (2009). Medicine, evolution, and natural selection: an historical overview. Q. Rev. Biol. 84, 333–355. doi: 10.1086/648122
Keywords: neurobiology, core concepts, evolutionary biology, phylogeny, development, molecular, homology, neurotoxin
Citation: Striedter GF (2023) Incorporating evolution into neuroscience teaching. Front. Educ. 8:1278279. doi: 10.3389/feduc.2023.1278279
Edited by:
Kimberley A Phillips, Trinity University, United StatesReviewed by:
Kara Erin Yopak, University of North Carolina Wilmington, United StatesMuhammad A. Spocter, Des Moines University Osteopathic Medical Center, United States
Bruce A. Carlson, Washington University in St. Louis, United States
Copyright © 2023 Striedter. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Georg F. Striedter, Z2Vvcmcuc3RyaWVkdGVyQGdtYWlsLmNvbQ==
 Georg F. Striedter
Georg F. Striedter