
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
BRIEF RESEARCH REPORT article
Front. Educ., 30 September 2021
Sec. STEM Education
Volume 6 - 2021 | https://doi.org/10.3389/feduc.2021.725591
This article is part of the Research TopicOriginal Strategies for Training and Educational Initiatives in BioinformaticsView all 14 articles
The development and application of bioinformatics has been growing steadily, but its learning and training has been lagging. We have approached this problem through a bi-annual event, called EGB (Escola Gaúcha de Bioinformática), dedicated to undergraduate and graduate students (mainly from biology, biomedicine, chemistry, physics, and computer sciences), as well as professionals, to mingle and be presented to bioinformatics from sequence, structure, and computational standpoints simultaneously. The interactive environment provided by EGB allows for participants mingling, independently from their training background, fostering collaborative learning and experience exchange. Both lecturers and students are encouraged to collaborate and communicate, with no formal acknowledgement of “status differentiation”.
Bioinformatics can be defined as simply as the “application of tools of computation and analysis to the capture and interpretation of biological data” (Bayat, 2002). Nonetheless, such definition is not as clear cut as it seems, with some authors being much more detailed in their description, e.g., “Bioinformatics is conceptualizing biology in terms of macromolecules (in the sense of physical chemistry) and then applying “informatics” techniques (derived from disciplines such as applied maths, computer science, and statistics) to understand and organize the information associated with these molecules, on a large-scale” (Luscombe et al., 2001). More recently, a definition of bioinformatics has been proposed as “an interdisciplinary field that is concerned with the development and application of algorithms that analyze biological data to investigate the structure and function of biological polymers and their relationships to living systems” (Tapprich et al., 2021). Contrasting with its array of definitions, there is no contending that bioinformatics has been steadily growing, both in its use and as a research field, at least since the early 2000s (Hodcroft et al., 2021; Wilson Sayres et al., 2018; Brusic, 2007; Perez-Iratxeta et al., 2007). This lack of agreement reflects the quick paced development of an evolving discipline whose target is not well defined yet. This lack of a settled focus is one of the major problems in teaching Bioinformatics: despite bioinformatics’ half-century history (Gauthier et al., 2019), its learning and teaching seem to be trailing behind its observed growth and use (Hack and Kendall, 2005; Wilson Sayres et al., 2018). This trend is observed especially in undergraduate courses focused in biological and health sciences (Madlung, 2018), but computational science courses also lack intersection with biological applications and implications (Atwood et al., 2019). Thus, while students of life sciences lack formal training in data management and programming, computer science students have little to no contact with biological data and its intrinsic variability. This poses the immediate problem: how to address the shortcomings in bioinformatics teaching and learning when dealing with such diverse set of actor backgrounds?Considering such problem, we herein present the results from a bioinformatics learning model based on: 1) a space for undergraduate and graduate students interactions, among each other and with professionals; 2) integration of participants originated from different backgrounds; 3) exposition of the participants, simultaneously, to bioinformatics from sequence, structure and computational standpoints, through 4) both theoretical lectures and hands-on courses, from introductory and advanced, methodological and applied standpoints. Encompassing three editions so far of the Escola Gaúcha de Bioinformática (EGB), or “Southernmost Brazilian School of Bioinformatics” in a free translation, such model has been applied with variations in Brazil since the 1980s through multiple fields, such as physics, chemistry, molecular simulation and others, and has been able to offer a generational impact on the formation of highly qualified researchers.
Different strategies to consolidate bioinformatics as part of the main curriculum of different courses, especially in the biological sciences, have been proposed through the years. These include the definition of its core competencies (Welch et al., 2014), its defining elements (Tapprich et al., 2021), the inspection of successful teaching cases in the US and United Kingdom (Hack and Kendall, 2005), the need to go beyond traditional classroom short courses (Atwood et al., 2019), and the analysis of a dedicated learning module of bioinformatics for biology students (Madlung, 2018). The frequently mentioned transdisciplinary nature of the field, however, remains observational rather than practical in most of these propositions, with disciplines borrowing techniques from each other, but without forming a single, major area of knowledge. Thus, in order to show explicitly the combination of technical fields that make up this fast-paced field, we decided that, during the EGB, the transdisciplinarity should be presented as a living experience, bringing together participants from the multiple backgrounds that encompass the definition of bioinformatics.
The quick development of Bioinformatics is also often uneven spatially, with some regions developing faster than others, and often each group specializing in a very limited scope of the field of temporary interest for their research, neglecting other possibilities that might ‒ if considered ‒ improve their capacities. For example, as a research field, bioinformatics has been concentrated in the Southeast Region of Brazil, especially in the states of São Paulo and Minas Gerais (Bicudo, 2016). Even with government initiatives to spread research facilities all over the country (from 2007 onwards), such as BioMicro nucleation effort (CAPES, 2008) and a major Computational Biology financial laid call (CAPES, 2013), other regions remained underrepresented (Bicudo, 2016). This also results in numerous groups working in specialized subfields, with little knowledge of other areas, which further hampers development of usefully complete Bioinformatics curricula. By taking advantage of the budding bioinformatics groups in the South Region of Brazil, particularly those in the southernmost state of Rio Grande do Sul, and connecting them with professional from all over Brazil and Latin America, EGB presents itself as a networking environment for the supervised development of abilities required to navigate the field. It is noteworthy that AB3C (Brazilian Association for Bioinformatics and Computational Biology) via its annual X-meeting event, provides training and networking opportunities. Likewise, the Brazilian ISCB Regional Student Group has been connecting bioinformatics students regionally. Nonetheless, these initiatives target subjects already working with bioinformatics, not specifically beginner, non-experienced users. The EGB works both as a scientific event (such as a symposium) and as a teaching-learning environment (like a “school”). This model has been successfully applied to more specific study areas in Brazil, for instance in the Brazilian School of Electronic Structure (since 1987) (dos Santos et al., 2003; EBEE, 2018) and in the School for Molecular Modeling in Biological Systems (since 2002) (EMMSB, 2021). Participation can include research paper submission and poster presentation, but these are not mandatory activities. The integration of all participants in an experience-sharing environment with provocative ideas is the main goal of EGB. A regular day at the “school” would include three to four lectures in the morning, followed by a lunch break, and flash courses in the afternoon, with both morning and afternoon periods having coffee break intermissions. These intermissions are particularly relevant for allowing participants to mingle in a friendly, casual environment. The school duration is proposed to be of 5 days (Monday to Friday), taking place during the Brazilian college winter break (in July). From the practical standpoint, the school requires a large auditorium for lectures, open halls for intermission activities and small individual computer labs for flash courses. Participants have the option to validate their participation as a credit in partner undergraduate and graduate programs. Thus far, three editions of EGB were organized (in 2015, 2017, and 2019), taking place in the Informatics Institute of the Federal University of Rio Grande do Sul, in Porto Alegre, Brazil. Supporting lab spaces were also provided by the Center of Biotechnology and the Institute of Biosciences at the same university. Financial management and enrollment assistance was provided the Brazilian Genetics Society (SBG).
Conceptually, the transdisciplinary characteristic of bioinformatics is approached by a simultaneous exposition of the students to lectures from introductory to advanced graduate levels, and from methodological to applied standpoints. A particular care is taken to include, in the same morning, lecturers from multiple fields in bioinformatics, as those working with sequence- or structure-based approaches, from biological to computer science backgrounds. Consequently, for example, some undergraduate biology student will be familiarized with coding while a computer science graduate student will be presented to protein biochemistry and molecular biology. During the intermissions, a more experienced student could help a newcomer to a particular area of bioinformatics to better understand previously discussed concepts, while the lecturer could both deepen some methodological details or explain in an even more basic level some aspect for participants. Finally, in the afternoon, the aspects from bioinformatics discussed in the morning lectures will be sedimented in hands-on courses on computer labs, also from the basic to more advanced levels. While it may be particularly challenging for some students to have a first contact to advanced coding, protein structure modeling or genome annotation using this approach, it is simultaneously a highly contextualized introduction to such field supported by the student familiarity with some of the other fields discussed during the event. In addition, these interactions have the benefit of encouraging and fostering inter-group cooperation.
The target audience for EGB was conceived to be wide, from undergraduate to graduate students and professionals. Hence, since its first edition in 2015, similar amounts of undergraduate and graduate students have participated in the event (Figure 1), simultaneously to a steady increase in the interest for the school, at an approximate 25% rate per edition, ranging from 174 participants in 2015 to 251 participants in 2019. We have observed a slight increase in graduate students as event editions progressed (Figures 1A,C). The same tendency was observed for the interest in focused flash courses (Figure 1A). More data is required to interpret these trends, but we hypothesize that graduate courses (especially those in the biological areas) are being insufficient in providing their students with the bioinformatics training they need or aspire. Since it is not directly enforced, the varied audience emerges as reflection of the public interest in the subject. Admissions are processed in first-come, first-served basis, with the only selection criteria in place being the preference for new participants (in lieu of returning ones). So far, no exclusion has occurred in the admissions process.
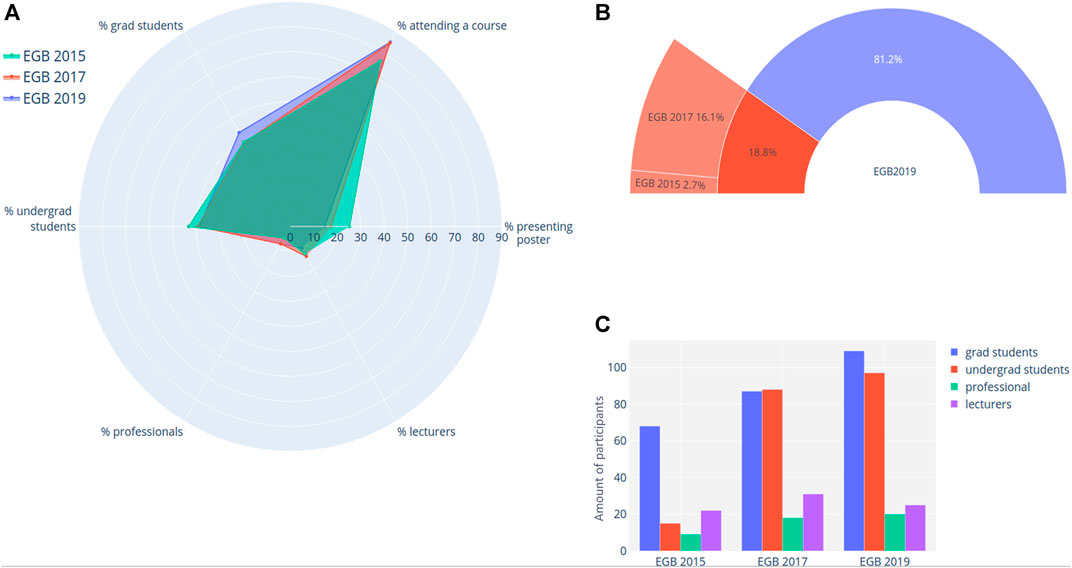
FIGURE 1. EGB participant demographics. (A) Radar plot highlighting shifts in participant profiles; (B) ratio of participants returning from previous EGB editions; (C) Total participant amount by year, indicating growth in participation at each edition of the event.
Around 90% of the participants were Brazilians, with three quarters of them being from the Rio Grande do Sul State. These participants listed traditional University cities as their origin, with a concentration in the capital city of Porto Alegre and its surroundings. Non-Brazilian participants were Latin-American, coming from Argentina, Bolivia, Chile, Colombia, Mexico, Peru, and Uruguay (Figure 2). Direct involvement of international participants remains dependent on additional funding for travel expenses, something that is yet to be secured. The origin institutions of the participants (universities, research centers, etc) were also listed, forming a total of 42 different affiliations. At the end of each event, all participants were invited to provide anonymous evaluations on multiple aspects of the school. This qualitative data has been used to adjust and improve subsequent editions of EGB. These observations are reported in Section 3. Participant demographic questionnaire and obtained results are presented in the Supplementary Material. The full questionnaire was not applied in the first edition of the event.
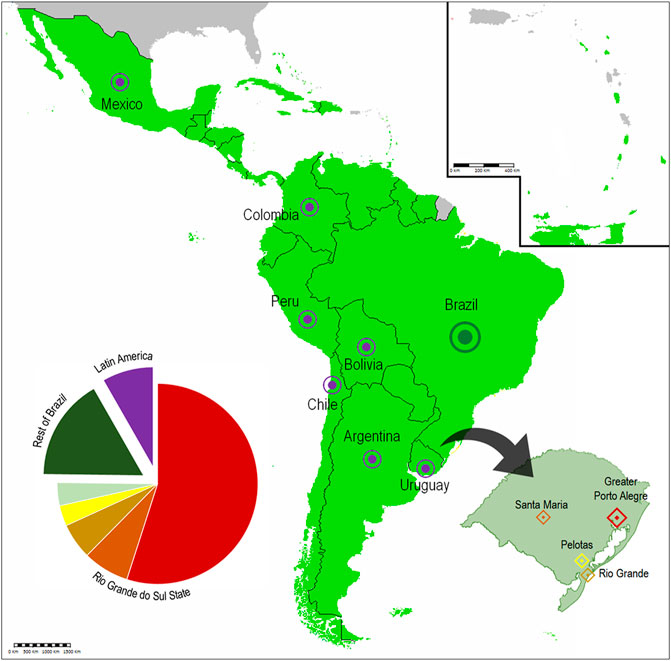
FIGURE 2. EGB participant coverage. The pie chart is color-coded as depicted in the map and reflects relative proportion of participants from each location. The State of Rio Grande do Sul is highlighted, pinpointing University cities that most contributed participants to the event. (Incorporates “Map for Community of Latin American and Caribbean States”, licensed under the Creative Commons Attribution-Share Alike 4.0 International, by Roblespepe).
Following the general concept of “mixing and matching”, the lectures were planned to expand the participants’ perspectives on the field. Accordingly, during each morning talks from specialists were given to the students covering multiple bioinformatics aspects, under three main standpoints: sequence or structure-based methods, biological applications and questions, and computational approaches to solve them. The main goal here is that the students get familiarized with multiple perspectives of bioinformatics, which are reinforced during the event. Thus, a lecture cycle could begin with observations on ways bioinformatics assisted zoologists to solve how tigers got their stripes (based, for example, on genomic data), followed by drug discovery against a specific pharmacological target (supported by docking, for instance), and ending with examples of computer clusters being used for crop enhancement via synthetic biology. Themes covered by lectures included (to cite a few) agricultural enhancements via genetic manipulation, big data in health and ecology, chemoinformatics, evolution, forensic applications of bioinformatics, massive parallel processing for bioapplications, molecular docking, multiscale molecular modeling and simulation, protein engineering, and vaccinology. Themes were changed at each event, and while one theme was given in a basic level in one edition, in other edition it could be offered in an intermediate level. Such approach accommodates students from multiple backgrounds and stimulate the return of participants for a new addition of the event with, accordingly, a continuous learning on the field. A similar approach was used for the flash courses, that include mandatorily some level of hands-on activity, from physical modeling of protein structures to viral genome assembly. The main themes covered by flash courses included introductory biochemistry, genome assembly and annotation, molecular dynamics, Python programming, structural biology, systems biology, and transcriptome data processing. All flash courses have allotted time at their initial session for familiarization with computer use and (when needed) with nongraphic interfaces. The ratio of 1 assistant for every five flash course participants is preconized. One day of the event was reserved to lectures from researchers in industry, emphasizing commercial applications and opportunities for the students. Also, these entrepreneurs reserved part of the day to mix with the participants. Course instructors and lecturers comprised Brazilian and Latin American experts from various fields pertaining to bioinformatics. These included professors, researchers, product managers, and industry representatives. Each lecture had an expected length of 50 min, followed by around 10 minutes of questioning, while flash courses lasted around 15 hours in total (3 h s/day), with participants opting for one course to follow along the 5 days. The lectures are of general attendance, while participants are invited to choose one flash course to follow through. Lecturers were selected based on suggestions from the Organizing and Scientific Committees and approved upon members’ voting. Training assistants for flash courses were selected among volunteers with a track record of work with the computational tools. Lectures were given mostly in Portuguese and Spanish.
The qualitative evaluation by the participants of the event was highly positive, with courses and lectures being considered “excellent” or “good” by more than 90% of the participants. Likewise, the scientific program and the venue were deemed excellent, and about three quarters of the participants expressed interest in participating in future editions of EGB. This interest is confirmed by the amount of returning participants at each new edition (Figure 1B). From the open-ended questionnaire, some aspects were brought up and have helped shape a more inclusive program in bioinformatics. Such aspects encompass the requirement for more physically accessible auditoria for mobility challenged individuals, the option for sign language interpreters and commitment to gender-balanced lineup of lecturers and instructors. Considering the need for training in bioinformatics and the observed success of the EGB strategy, impacting directly around 200 people per edition, plans are underway for expanding its geographical coverage. Initial arrangements are in progress for providing simultaneous instances of the event in different regions/countries, with live webcasting of lectures and local offerings of flash courses. Nonetheless, with the lengthening of the COVID-19 pandemic and its ongoing effects (Zoumpourlis et al., 2020), these plans were halted. Considering the social mingling that has been considered the highlight of EGB, by providing ways of connecting people with different backgrounds with a common interest in bioinformatics, and the additional need for supervised activities in the flash courses, the total conversion of the event to distance activities was deemed inadequate by the organizing committee. Despite still lacking proper assessment, information volunteered by former participants indicate that the interaction opportunities provided by EGB have fostered collaborations among researchers and students from different institutions, allowing for multiple groups joining forces for topical research projects. Such collaborations were also stimulated with the bridging of academia and different economic sectors, with students familiarizing themselves with the entrepreneurial environment. Undergrad students were also able to socialize with graduate students, sparkling interest in pursuing further academic career pathways. Still, the enrolled students came majorly from life sciences backgrounds, with computer science students representing a minor portion of the attending public. Even if the social component of the event may not apply to all participants, the knowledge gathered can supplement the current curricula by providing up-do-date lectures and high-level discussions. We hope these strategies may aid other endeavors for consolidating bioinformatics as a perennial knowledge in students and professionals alike.
In conclusion, the opportunity for interaction between people from biological and computational science backgrounds in a casual environment emerges as the main advantage of the model for bioinformatics training, teaching, and learning presented in the EGB school events.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
All authors listed have made a substantial, direct, and intellectual contribution to the work, and approved it for publication.
Previous editions of EGB were funded by grants from FAPERGS (570-2551/15-6 and 17/2551-0000373-0), CAPES (23038.001974/2015-73, 88881.123406/2016-01, and 88881.290880/2018-01) and Asociación de Universidades Grupo Montevideo (AUGM). Support was also provided by the Universidade Federal do Rio Grande do Sul (UFRGS) via its Institute of Informatics, Institute of Biosciences, and Center of Biotechnology.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors are extremely grateful to all students who worked in the organization of previous editions of EGB, and to all lecturers and course tutors for accepting the challenge. The authors acknowledge Diego Bonatto for his involvement in the first edition of the event, and the Brazilian Genetics Society (SBG) for providing instrumental support for the second and third editions of the event.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/feduc.2021.725591/full#supplementary-material
Attwood, T. K., Blackford, S., Brazas, M. D., Davies, A., and Schneider, M. V. (2019). A Global Perspective on Evolving Bioinformatics and Data Science Training Needs. Brief Bioinform 20, 398–404. doi:10.1093/bib/bbx100
Bayat, A. (2002). Science, Medicine, and the Future: Bioinformatics. BMJ 324, 1018–1022. doi:10.1136/bmj.324.7344.1018
Bicudo, E. (2016). Genomics Politics through Space and Time: The Case of Bioinformatics in Brazil. Public health genomics 19, 81–92. doi:10.1159/000443472
CAPES (2008). Doutorados em bioinformática e microeletrônica (BioMicro), Available at: https://www.gov.br/capes/pt-br/acesso-a-informacao/acoes-e-programas/bolsas/programas-estrategicos/outras-informacoes/programas-encerrados-estrategicos/biomicro (Accessed June 7, 2021).
CAPES (2013). Edital Biologia Computacional–CAPES Nº051/2013, Available at: https://www.gov.br/capes/pt-br/centrais-de-conteudo/edital-051-2013-biologiacomputacional-pdf (Accessed June 7, 2021).
Dos Santos, H. F., Coura, P. Z., Dantas, S. O., and Barone, P. M. V. B. (2003). Escola Brasileira de Estrutura Eletrônica. Juiz de Fora: Editora Livraria da Física, 324.
EBEE (2018). XVI Escola Brasileira de Estrutura Eletrônica, Available at: https://sites.google.com/site/xviebee/home (Accessed June 7, 2021).
EMMSB (2021). 10a Escola de Modelagem Molecular em Sistemas Biológicos, Available at: http://www.emmsb.lncc.br/(Accessed June 7, 2021).
Gauthier, J., Vincent, A. T., Charette, S. J., and Derome, N. (2019). A Brief History of Bioinformatics. Brief Bioinform1996 20, 1981–1996. doi:10.1093/bib/bby063
Hack, C., and Kendall, G. (2005). Bioinformatics: Current Practice and Future Challenges for Life Science Education. Biochem. Mol. Biol. Educ. 33, 82–85. doi:10.1002/bmb.2005.494033022424
Hodcroft, E. B., De Maio, N., Lanfear, R., MacCannell, D. R., Minh, B. Q., Schmidt, H. A., et al. (2021). Want to Track Pandemic Variants Faster? Fix the Bioinformatics Bottleneck. Nature 591, 30–33. doi:10.1038/d41586-021-00525-x
Luscombe, N. M., Greenbaum, D., and Gerstein, M. (2001). What Is Bioinformatics? A Proposed Definition and Overview of the Field. Methods Inf. Med. 40, 346–358. doi:10.1055/s-0038-1634431
Madlung, A. (2018). Assessing an Effective Undergraduate Module Teaching Applied Bioinformatics to Biology Students. Plos Comput. Biol. 14, e1005872. doi:10.1371/journal.pcbi.1005872
Perez-Iratxeta, C., Andrade-Navarro, M. A., and Wren, J. D. (2007). Evolving Research Trends in Bioinformatics. Brief Bioinform 8, 88–95. doi:10.1093/bib/bbl035
Tapprich, W. E., Reichart, L., Simon, D. M., Duncan, G., McClung, W., Grandgenett, N., et al. (2021). An Instructional Definition and Assessment Rubric for Bioinformatics Instruction. Biochem. Mol. Biol. Educ. 49, 38–45. doi:10.1002/bmb.21361
Welch, L., Lewitter, F., Schwartz, R., Brooksbank, C., Radivojac, P., Gaeta, B., et al. (2014). Bioinformatics Curriculum Guidelines: Toward a Definition of Core Competencies. Plos Comput. Biol. 10, e1003496. doi:10.1371/journal.pcbi.1003496
Wilson Sayres, M. A., Hauser, C., Sierk, M., Robic, S., Rosenwald, A. G., Smith, T. M., et al. (2018). Bioinformatics Core Competencies for Undergraduate Life Sciences Education. PloS one 13, e0196878. doi:10.1371/journal.pone.0196878
Keywords: bioinformatics, Brazil, education, Latin America, outreach, South America
Citation: Dorn M, Ligabue-Braun R and Verli H (2021) Transdisciplinary Approach for Bioinformatics Education in Southern Brazil. Front. Educ. 6:725591. doi: 10.3389/feduc.2021.725591
Received: 12 July 2021; Accepted: 20 September 2021;
Published: 30 September 2021.
Edited by:
Chi-Cheng Chang, National Taiwan Normal University, TaiwanReviewed by:
Jan Egger, Graz University of Technology, AustriaCopyright © 2021 Dorn, Ligabue-Braun and Verli. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rodrigo Ligabue-Braun, cm9kcmlnb2xiQHVmY3NwYS5lZHUuYnI=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.