
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Educ., 29 October 2021
Sec. STEM Education
Volume 6 - 2021 | https://doi.org/10.3389/feduc.2021.711535
This article is part of the Research TopicOriginal Strategies for Training and Educational Initiatives in BioinformaticsView all 14 articles
 Ellen G. Dow1*
Ellen G. Dow1* Elisha M. Wood-Charlson1*
Elisha M. Wood-Charlson1* Steven J. Biller2
Steven J. Biller2 Timothy Paustian3
Timothy Paustian3 Aaron Schirmer4
Aaron Schirmer4 Cody S. Sheik5
Cody S. Sheik5 Jason M. Whitham6
Jason M. Whitham6 Rose Krebs7
Rose Krebs7 Carlos C. Goller7,8
Carlos C. Goller7,8 Benjamin Allen9
Benjamin Allen9 Zachary Crockett9
Zachary Crockett9 Adam P. Arkin1,10
Adam P. Arkin1,10Over the past year, biology educators and staff at the U.S. Department of Energy Systems Biology Knowledgebase (KBase) initiated a collaborative effort to develop a curriculum for bioinformatics education. KBase is a free web-based platform where anyone can conduct sophisticated and reproducible bioinformatic analyses via a graphical user interface. Here, we demonstrate the utility of KBase as a platform for bioinformatics education, and present a set of modular, adaptable, and customizable instructional units for teaching concepts in Genomics, Metagenomics, Pangenomics, and Phylogenetics. Each module contains teaching resources, publicly available data, analysis tools, and Markdown capability, enabling instructors to modify the lesson as appropriate for their specific course. We present initial student survey data on the effectiveness of using KBase for teaching bioinformatic concepts, provide an example case study, and detail the utility of the platform from an instructor’s perspective. Even as in-person teaching returns, KBase will continue to work with instructors, supporting the development of new active learning curriculum modules. For anyone utilizing the platform, the growing KBase Educators Organization provides an educators network, accompanied by community-sourced guidelines, instructional templates, and peer support, for instructors wishing to use KBase within a classroom at any educational level–whether virtual or in-person.
Modern biology is becoming more reliant on “big data” to answer a range of questions. The ability to generate or re-analyze large data sets is quickly becoming a mainstay in many areas, from cellular biology to ecosystem ecology. This is especially true for the field of genomics and molecular biology, where large data sets are now commonplace. With the rapid growth and free online availability of biological data derived from DNA sequencing technologies, the need for skilled researchers to analyze these data is growing as well (Kodama et al., 2012; Koboldt et al., 2013). Topics in bioinformatics have therefore become a key feature of biology curriculum development in recent years (Maloney et al., 2010; Machluf et al., 2017). When the 2020 global pandemic drastically increased the need for virtual, learn-from-home coursework, bioinformatics also provided an attractive option for hands-on experience outside of a traditional wet lab. However, bioinformatics and the techniques used to analyze data advance quickly, and it can be difficult to incorporate the most cutting edge resources into the classroom.
One of the core challenges of teaching data analysis courses is that many students may not have access to a computer outside of their school’s computer labs. During the pandemic, many schools did what they could to provide computer access for students at home, but school-loaned laptops are likely unable to support many of the bioinformatics software programs necessary to run analyses locally. For those students with more sophisticated computers at home, instructors still have the challenge of helping them troubleshoot integration of bioinformatics software across a variety of operating systems (e.g., Windows, macOS, Chrome OS, or Unix-based systems) (Cummings and Temple, 2010). In addition, many bioinformatics programs or pipelines are coded in languages such as Java or R, or rely heavily on Perl (Perl Programming Language, RRID:SCR_018313), Python (Python Programming Language, RRID:SCR_008394), Ruby or Julia programming languages that require an understanding of command-line interfaces to operate (Ken Arnold et al., 2000; Wall et al., 2000; Flanagan and Matsumoto, 2008; Van Rossum and Drake, 2009; Bezanson et al., 2017; R Core Team, 2021). These tools and resources advance quickly, with new versions becoming available regularly, requiring updates or deprecation, depending on whether a different tool is adopted by the community as the new standard. Finally, some of the most powerful bioinformatic programs require vast amounts of memory or processing power, and larger data sets often require more computing power than is available on a personal computer. High-performance computing (HPC) clusters are often available at large research centers, which may offer these resources to students, but are relatively uncommon at teaching colleges or high schools. While cloud resources, such as Azure through Microsoft or AWS through Amazon, are more readily available, the monetary costs and time and resources to set up the workflows may still impede adoption by schools and institutions (Raj et al., 2020).
To date, acquiring a depth of knowledge in bioinformatics has largely been contingent on prerequisite computational knowledge about scripting, programming, data typology, and database management, alongside statistics and of course biology (Wilson Sayres et al., 2018). Although fundamental to research development work in bioinformatics, students and faculty are often missing one or more of these skill sets. From the lack of training and access to adequate computing resources for faculty, to the constantly changing software ecosystem, these problems can seem insurmountable for teaching and create significant challenges to the effective instruction of practical data science knowledge (Cummings and Temple, 2010; Williams et al., 2019).
For students and educators looking to improve their command of the command line, the Bioinformatics Virtual Coordination Network provides peer-support and resources (BVCN, https://biovcnet.github.io), or they can turn to training portals like the Global Organisation for Bioinformatics Learning (Corpas et al., 2015). For everyone else, online or graphic user interfaces (GUI) platforms [i.e., KBase, Nephele (Nephele, RRID:SCR_016595], Galaxy (Galaxy, RRID:SCR_006281), PATRIC (Pathosystems Resource Integration Center, RRID:SCR_004154), CLC Genomics (CLC Genomics Workbench, RRID:SCR_011853), or Geneious (Geneious, RRID:SCR_010519) may address some of these challenges by facilitating greater access to bioinformatic tools and data (Afgan et al., 2018; Arkin et al., 2018; Weber et al., 2018; Davis et al., 2019; Kearse et al., 2012; QIAGEN CLC Workbench, 2013). While some of these services are provided for free, several require the purchase of a software license, which can be expensive for faculty and students already struggling with the costs of education. Thus, as the community seeks to bring modern bioinformatic approaches to students in the classroom, the community should leverage resources that address many of the barriers to entry and strive to empower students.
The U.S. Department of Energy (DOE) Systems Biology Knowledgebase (KBase) is a free knowledge creation and discovery environment designed for both biologists and bioinformaticians (Arkin et al., 2018). KBase integrates a variety of data and analysis tools, from DOE and other public services, into an easy-to-use platform that employs scalable computing infrastructure to perform sophisticated systems biology analyses. KBase uses a dynamic GUI based on the Jupyter Notebooks framework (Kluyver et al., 2016) to provide documentable, shareable, and reproducible analysis workflows called Narratives. This integration allows for hosting code and programs outside of a traditional command-line interface. Data brought into KBase are transformed into a unique data model, with specifications for each type of biological data (reads, genomes, metabolic models, etc.), structured around the FAIR–findable, accessible, interoperable, and reusable–data principles (Wilkinson et al., 2016) to facilitate discovery, access, and interoperability throughout the system. Data from different sources or file formats can be used interchangeably and downloaded in standard formats, and data workflows can be directly copied to reproduce the data provenance. All the data, tools, commentary, and code used for analysis are stored as KBase Narratives on the KBase servers, for easy, persistent access with minimal requirements for the user (more information at: https://docs.kbase.us/getting-started/browsers).
While many biologists have taken advantage of KBase for academic and industry-related research, there is a growing cohort of educators using KBase in the classroom to demonstrate fundamental content knowledge for bioinformatics. Narratives are well-suited to use in educational settings as they can be shared with other individuals, groups, or made public. Educators can leverage Narrative features to re-run workflows, modify a workflow with new data or tools, extend workflows to incorporate new analyses, and add contextual information ranging from simple text to embedded videos and images, using Markdown or HyperText Markup Language (HTML). The visual display of provenance for data objects allows instructors to see connections between data, which can be used to supplement lectures describing the workflows. For example, due to the immediate relevance of the SARS-CoV-2 pandemic, one instructor created a Narrative to show students how to obtain sequencing data from public repositories, trim the reads, assess quality, and align to a reference genome (Pasqualoni, 2020).
Over the past year, KBase staff have worked with educators to identify how the platform could support their classroom goals. The need for tools and resources that enable virtual, interactive bioinformatics courses was a common request from each instructor. Therefore, as a community, we developed a modular curriculum framework with a variety of resources to empower students (and educators) to grow their bioinformatics content knowledge across a range of instructional levels, including high school, undergraduate, and graduate courses.
The KBase Narrative platform was used to modularize several complete and complex bioinformatic workflows (Dow et al., 2021). Each larger concept was divided into smaller parts to ensure that students could understand the scalability of the content while retaining the ability to answer biological questions. Working groups of instructors and the KBase team developed example teaching workflows by assembling a set of Narratives for a variety of bioinformatics concepts: genome assembly, annotation, and analysis; metagenomics; phylogenetics; and metabolic pathway analysis. The Narratives include a range of the tools available within KBase that are wrapped as applications, referred to as KBase Apps. Each workflow was centered around creating engaging content that met core curriculum concepts using backward design to address learning goals within individual modules that connected to form the complete concept (McTighe and Wiggins, 1998). Alongside FAIR data principles, these Narratives can be used directly as teaching tools, be modified by individual instructors to fit their student population and class focus, or simply serve as inspiration.
The resources developed over the summer were used in a pilot program during Fall 2020 to gather and implement instructor and student feedback, and to develop supporting guidelines that included best practices for educators (https://doi.org/10.25982/1668075). Program resources, including modularized workflows, guiding documentation, the KBase Educators Organization, and KBase User Slack workspace for support and networking were made available in December of 2020 for all educators interested in using KBase and joining the community (see KBase for Educators Webinar: https://youtu.be/K9FxPc_2jzI).
To evaluate the use of KBase in the classroom, a subset of the KBase Educators Working Group developed a post-course student survey (Supplementary Materials). The survey was a student self-assessment that focused on how effective the piloted KBase learning modules were at achieving common learning objectives, the approachability of the KBase platform, and changes in student perceptions and self-confidence in the field of computational biology after completing the modules. The survey used a Likert scale to quantify student responses through self-assessment on awareness of content and growth post-course, confidence around concepts and using tools, and using KBase. The survey study was reviewed and approved by the Human Subjects Committee Institutional Review Board at Lawrence Berkeley National Lab (LBNL) (339NR001-2AP22).
The survey data was used to create frequency distributions of student responses using histograms to gauge student reception for introducing KBase into curriculum and how students measured their own self-efficacy. Likert scale data was converted to numerical equivalents to measure distribution of the data and run Wilcoxon signed rank test with continuity correction between student responses using R v3.1 (R Project for Statistical Computing, RRID:SCR_001905) (R Core Team, 2021).
Members of the KBase Educators Working Group also informally assessed how effective KBase was at addressing the challenges of teaching data science in a virtual classroom mentioned in the introduction, ranging from totally solved (5) to not solved at all (1).
An example of tailoring these resources to a course was provided by the Biotechnology Program (BIT) 477/577 Metagenomics class at North Carolina State University (NC State), which adapted the Metagenomics modules workflow along with additional analyses using Phylogenetics and Metabolic Modeling tools. The goal of these activities was to provide hands-on experience with computational tools to study relationships between microbial communities and ecosystems.
Fall 2020 was the first semester BIT 477/577 students used KBase. Students also had weekly topic modules with graded activities in Nephele (Nephele, RRID:SCR_016595) (Weber et al., 2018) and QIIME2 (QIIME, RRID:SCR_008249) (Caporaso et al., 2010; Bolyen et al., 2019), as well as social annotation and collaborative note-taking assignments with Hypothes.is (Goller et al., 2021), video lectures, interviews of bioinformaticians, creation of podcasts, and a group data analysis project. The course was taught asynchronously online due to the pandemic.
Two KBase Narratives were created for this course, Gold (https://doi.org/10.25982/67335.259/1773074) and Silver (https://doi.org/10.25982/68579.143/1766297), based on frameworks from the KBase Educators Organization and adapted for specific research and course objectives. The Gold Narrative focused on genome assembly and evaluation, through an exploration of available high-throughput metagenomic sequencing data sets, to identify Delftia acidovorans sequences (a bacterium capable of precipitating liquid gold into nanoparticles). In the Silver Narrative, students binned a high-quality metagenome-assembled genome (MAG) from a Silvergrass hybrid soil metagenome, determined its near relatives with a phylogenomic tree, and generated metabolic models to identify the media components required to isolate this potentially important microbiome member. This provided students with the opportunity to learn skills often used by applied scientists leveraging metagenomics. The concepts of MAGs, binning, and metabolic modeling were new to the course. Therefore, a week-long module was designed to introduce key definitions and procedures through videos, a reading assignment, and slides before engaging students in KBase activities. The KBase module addressed the learning objectives listed in Table 1.

TABLE 1. Module Learning Objectives (MO) and instructional materials (resources linked within text).
Prior to the beginning of the course, students were sent information about the study and the approved informed consent form. Thirteen of the 14 students agreed to participate in the study, including five undergraduates and eight graduate students from various programs. The study was approved by the NC State Institutional Review Board (IRB, # 20309). Pre- and post-course quizzes were used to assess the students’ familiarity with various general concepts and ideas in bioinformatics prior to using the Narratives. The results of the pre- and post-course knowledge surveys were analyzed using Fisher’s exact test and t-test to compare averages and variance. An open response section afforded students the opportunity to provide anonymous feedback on aspects of their experience, such as where they had the most difficulty, providing a basis for determining where future improvements to lessons could be most effectively targeted. Quiz questions are included in the Supplementary Data Sheet 1.
The KBase Educators program was initiated to gather information from educators teaching bioinformatics on how KBase could support the community of users with the pandemic shifting classrooms from in-person to online. KBase advertised an open call to instructors to hold a discussion of their needs and speculate around how KBase, as a research platform, could be applied to course curriculum. In late Spring 2020, a kick-off community meeting was held for instructors that expressed the shared need for tools and resources that enable interactive bioinformatic experiences and transferable analytical skills. From the initial conversation, about a dozen educators began the KBase Educators Working Group to develop workflows that connect across basic bioinformatic concepts. Working Group members with classes in the Fall of 2020 participated in the pilot program, which prompted the creation of best practices and supporting guidelines for the community as it grows. Alongside guidelines, KBase implemented infrastructure to support members and promote collaboration and networking within the broader KBase Educator community. Many of the Working Group members co-authored this manuscript.
The KBase teaching Narratives are organized as a series of interconnected workflows, illustrated by the Educators Concept Overview (https://doi.org/10.25982/1668075). This Narrative provides a centralized landing page for the modules and allows educators and students to choose the analysis pipelines they wish to perform. It also contains links to resources and suggestions for teaching fundamental concepts, and is easily extensible so educators can collect and share example data with others in the community or expand existing analyses. The overview Narrative contains a flow diagram that shows the connections between concepts and modules with links to each module (Figure 1). The concepts include broad bioinformatics topics: Genomics, Metagenomics, Phylogenetics, Pangenomes, and Metabolic Modeling (Table 2, with complete overview in Supplementary Table S1). For each concept there are one or more modules to demonstrate the roadmap of analysis within that concept. From there, educators can copy any of the module Narratives and modify the learning concepts and example questions, populate the Narrative with new data related to a topic of interest to the class, or add text to create an explanatory Narrative.
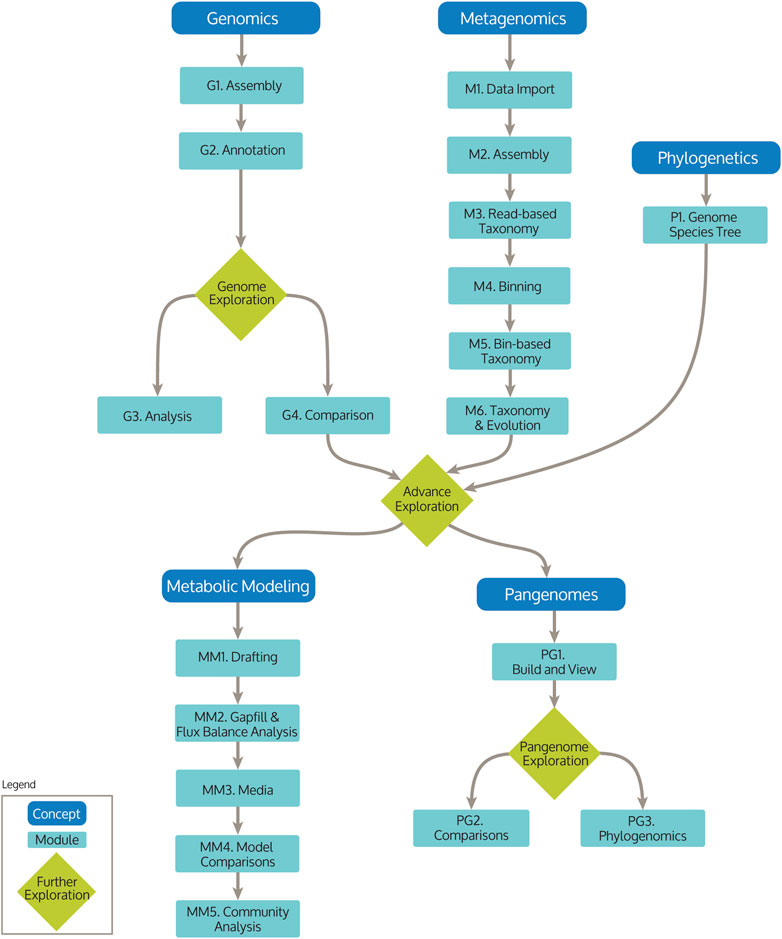
FIGURE 1. KBase Educators Concept Workflows and their modules, including Genomics, Metagenomics, Phylogenetics, Pangenomes, and Metabolic Modeling.

TABLE 2. Example Concept Workflow in KBase using the Narrative on Metagenomics, including module topics and corresponding Learning Objectives.
Narratives are connected to each other through HTML or Markdown hyperlinks (see resources linked in Dow et al., 2021) so that related workflows can be separated into modular components. By breaking complex analysis pipelines into smaller, distinct units, this structure allows students to focus on understanding the inputs, outputs, and rationale behind each step in the process. This also enables educators to more easily pick and choose the aspects most relevant to their particular course learning goals, or to fit analyses into particular timeframes, while ensuring students understand how each step connects to the larger concept and analytical goals.
The primary tutorial Narratives are designed for instructors and contain teaching notes that include explanations and alternatives or suggestions. For each tutorial Narrative there is a paired student version, which maintains the framework of the original Narrative in nascent App workflows without data products or results. Questions are embedded within the Narratives for students to answer as they explore the workflow and data, prompting them to perform the analysis on their own. Narratives can be shared with data and analysis results preloaded, inviting students to focus on interpreting results, or empty of data and analyses, allowing students to explore data and workflow options on their own. Student versions may also provide additional resources, such as links to literature, data sources, and guides to using the tools. To create new student Narratives, educators simply copy an existing Narrative and edit the data and Apps included within the Narrative. Because of this feature, educators can place students at the beginning of a particular workflow or at any point within it, and have the ability to combine workflows for students to follow longer analyses.
Instructors from the Working Group implemented KBase resources in a variety of settings and educational contexts during the 2020–2021 academic year. Contributors represented about a dozen institutions, with six institutions piloting KBase in the classroom in 2020–2021. For example, students at Boca Raton Community High School (Boca Raton, FL), Northeastern Illinois University (Chicago, IL), and graduate students participating in the 2020 Multiscale Microbial Dynamics Modeling course (hosted by the Environmental Molecular Sciences Laboratory (EMSL) and Pacific Northwest National Laboratory (PNNL) Subsurface Biogeochemical Research group) all simultaneously used KBase to reach different learning objectives and educational goals. At Boca Raton Community High School, students used KBase as a “one-stop shop” to explore genomic data. Undergraduate Genomic and Proteomics students at Northeastern Illinois University (Chicago, IL) used KBase as part of a flexible set of semester-long assignments focused on using real-world data to learn about genome assembly, annotation, and comparison. Graduate students and early career researchers participating in the 2020 Multiscale Microbial Dynamics Modeling course (https://www.kbase.us/multiscale-microbial-dynamics-modeling/) used newly implemented Apps, such as DRAM (Shaffer et al., 2020), in KBase for metagenomic data processing and analysis. This range of contexts and approaches highlights the scalability, accessibility, and versatility of the KBase platform.
Several undergraduate courses participated in a student survey to assess student perception of learning with KBase. So far, the survey has received 156 responses (excluding one response that only provided the course name) from students in courses covering Genomics, Metagenomics, Microbiology, and Molecular Biology (Figure 2).
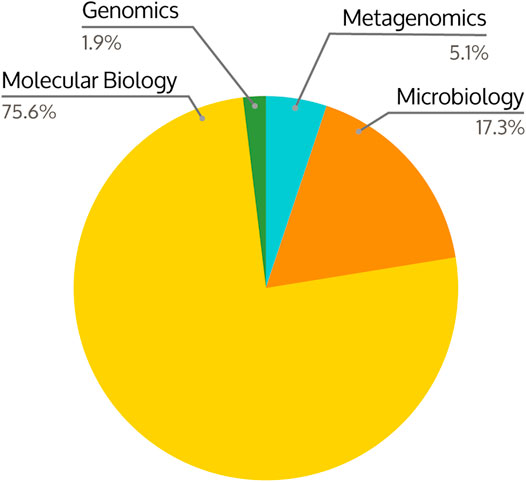
FIGURE 2. Distribution of undergraduate course subjects using KBase within the curriculum across 156 responses. The courses included Genomics, Metagenomics, Microbiology, and Molecular Biology.
Students were asked to assess their own experience and knowledge after the course. Coming into the course, 87% of students reported that the concepts covered were “extremely or moderately new” to them (Figure 3A). After the course, familiarity with bioinformatics concepts and workflows improved for around 96% of responses, with the majority of students responding that they now felt “somewhat or moderately familiar” with the concepts covered (Figure 3B). The change in student perception of course knowledge from pre to post-course was significant (Figures 3A,B; Wilcoxon test p < 0.001). At the end of their courses, approximately 39% of students felt they were able to describe the steps of a bioinformatics workflow, 34% were uncertain, and 27% did not feel that they were able. However, students’ confidence in their ability to use bioinformatics analysis and tools to answer research questions improved in 68% of responses (Figure 3C). Approximately 90% of students described at least some confidence in their ability to continue to use KBase to analyze data and find answers (Figure 4A), with the majority of students agreeing that overall they were able to use KBase effectively (Figure 4B). Student responses describing their change in confidence reflected a similar pattern to their perceived ability to use KBase (Figure 3C, Figure 4B; Wilcoxon test p = 0.649). Overall, approximately 30% of the students reported that KBase was easy to use, 41% had no strong opinion, and 30% had some difficulty using KBase.
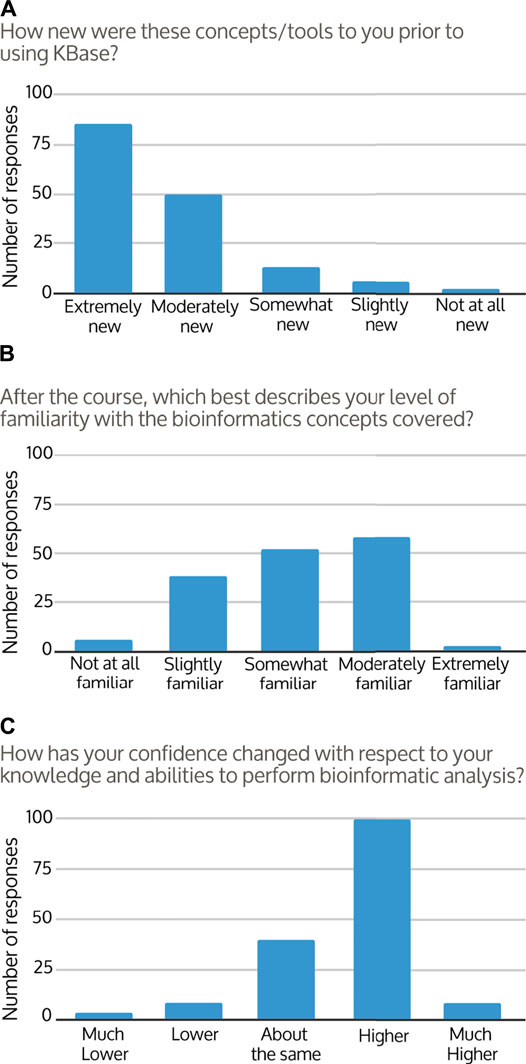
FIGURE 3. Frequency distributions of student responses to survey questions comparing (A) pre-course to (B) post-course knowledge and (C) self-assessed confidence. There were 155–157 responses per question as each question was optional for students to answer.
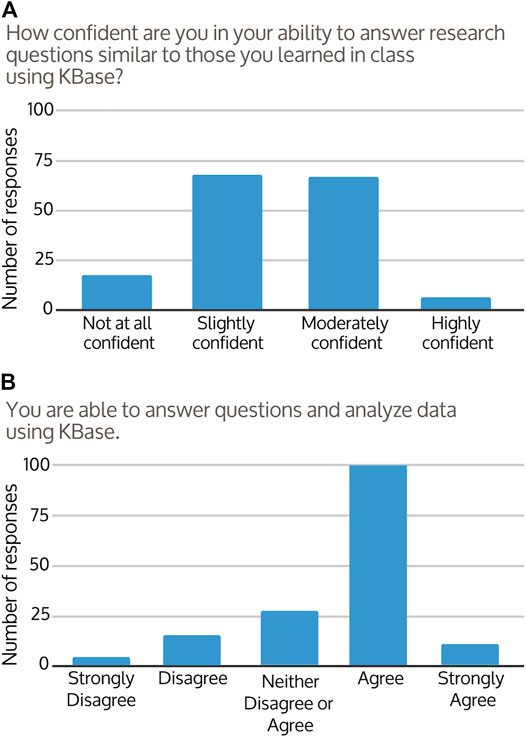
FIGURE 4. Frequency distributions of 156 student responses to their (A) confidence and (B) ability to use KBase post-course.
From an Educator’s perspective (n = 6, working group members), KBase was able to address many of the challenges facing data science in a virtual classroom for undergraduate courses. During the pilot, the platform and available resources were mostly able to address challenges (5 = totally solved, 1 = not solved at all) related to: working across different operating systems (4.3), access to and accessibility of consistent bioinformatic tools and resources (4.8), access to computational resources (4.5), and providing bioinformatics options for students without coding experience (5). Success at granting students access to computers was variable and likely dependent on pre-existing circumstances. Specifically, the education-based co-authors appreciated the ability to easily share Narratives with students; the option to gradually reduce the scaffolding as student knowledge of the platform, data, and analyses increased; and how the platform allowed students to focus on why they were learning bioinformatic concepts, instead of spending time on setup and maintenance of the tools and resources.
At the beginning of the metagenomics course, students (n = 13) had different knowledge gaps related to the instructional concepts. For example, Figure 5A shows students 2, 7, and 11 each had three incorrect answers in the pre-course quiz, but only one of those overlapped. Likewise, students 3 and 8 had five and four incorrect answers, respectively, and only one overlapped. Knowledge gaps were mostly filled by the end of the course based on comparison of pre- and post-course question scores (Figure 5B). The highest number of students that answered correctly on any one question was 11 out of 13 on the pre-test. In contrast, three questions were 100% correctly answered in the post-test. Correct answers increased by as many as six students in the post-test for some questions. On a per question basis, there was a higher frequency of pre/post-quiz questions incorrectly then correctly answered (36) compared to incorrect/incorrect (12) and correct/incorrect (4) answers. A significant imbalance of these ratios (Fisher’s test, p-value = 0.0024) supports student learning during the course. Furthermore, statistical differences in pre and post-course quiz averages and variance (Figure 5C, t-test, p-value = 0.0004, F-test, p-value = 0.0091) indicated significant improvement in students’ understanding of the concepts. While sample size was limited during this pilot study, student responses and feedback were encouraging.
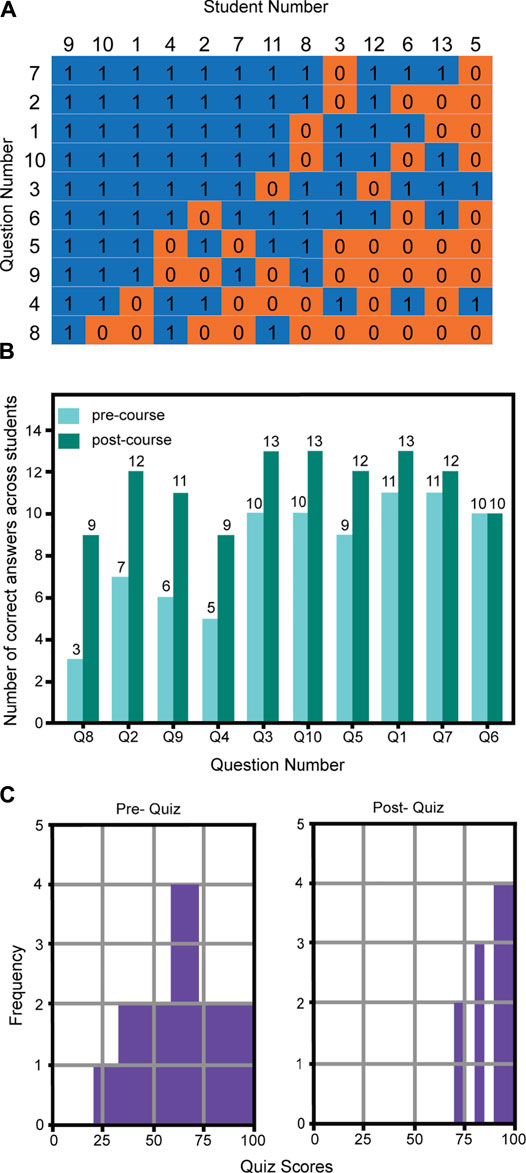
FIGURE 5. Pre- and post-course knowledge survey of 13 students in NC State’s Biotechnology (BIT) 477/577 Fall 2020 course. (A) Pre-quiz scores arranged by students and questions to highlight diversity in pre-knowledge. Responses are sorted to visually show pattern and clustering of correct and incorrect responses. 1 = correct, 0 = incorrect responses, n = 13 students. (B) Correct and incorrect pre- and post-course quiz questions. Questions can be found in the Supplemental Materials. Questions were sorted to show greatest improvement in correct answers. (C) Pre- and post-course quiz grade distributions. Student scores transitioned from nearly normal with a wide variance in the pre-quiz to highly skewed left with low variance in post-quiz.
At the end of the course, students completed their own projects by applying data analysis methods to another data set. Students chose between KBase or QIIME2 (run on a local HPC cluster) to complete their final project. Of the 13 students, seven chose to complete their project through KBase, some actually expressing an aversion to using the command-line and terminal interfaces during office hours. Some students explained that they would have chosen the KBase option for their data analysis project if they had managed their time better. The KBase assignment would take more time to complete because it was more open-ended, and involved analyzing a set of data in a novel way, whereas the QIIME assignment had specific questions to answer and a prepared script that needed minimal additions from the student to work properly.
Open-ended student responses indicated new knowledge and understanding of computational pipelines for metagenomics analysis (Table 3). Overall, testimonials were telling about the impact of KBase. Six out of 13 students used the word “comfort” in describing their familiarity with KBase and other tools taught in the course (Table 3).
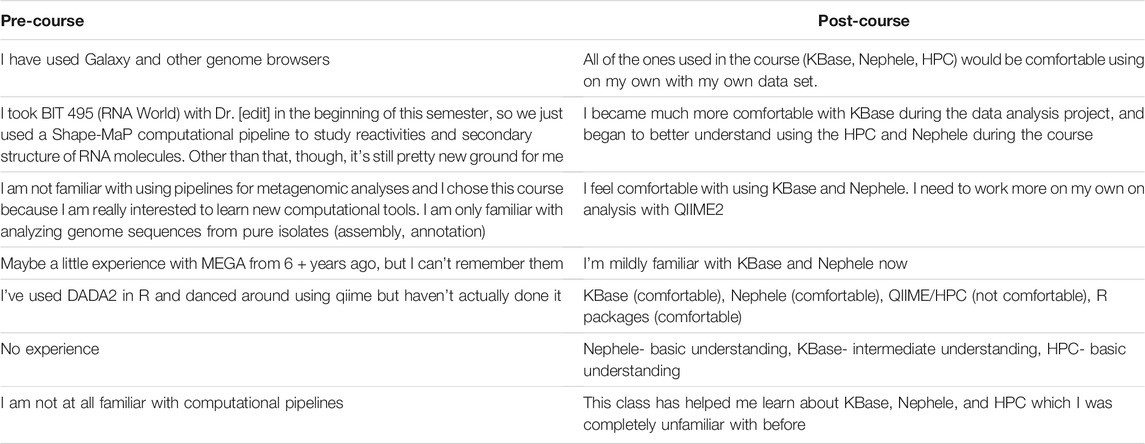
TABLE 3. Select student responses to the question “List and describe your familiarity with computational pipelines for metagenomic analyses.”
To facilitate the use of KBase Narratives, the Working Group developed a set of guidelines and resources associated with the Educators organization. An educator’s “Standard Operating Procedures” document (SOP) includes crowd-sourced suggestions for best practices and detailed logistics for using KBase Narratives in the classroom setting. This guide includes suggestions for organizing classes within KBase, guidelines for registering student accounts, and other key details. Guidance is also provided on estimated time requirements for running different modules with students at different educational levels to help educators plan their initial rollout. These represent “living documents” which can be continually improved with feedback from the community of users.
A number of open resources are provided that educators can adopt directly to their curriculum, reducing the barriers associated with incorporating new material into a class. Examples include links to videos and specific step-by-step instructions, which can be provided to students to support their use of KBase. Guidance within the SOP describes common problems encountered in creating, modifying, and using Narratives including links to the KBase documentation site (http://docs.kbase.us). Some Narratives have associated introductory slides which can be used directly or customized by an instructor to introduce the concepts and tools utilized. Examples of student assessment questions, along with model answers, are also available. Educators interested in sharing additional or modified materials associated with the use of these Narratives in different classroom settings are encouraged to contribute these within the KBase Educators Organization, described below.
The KBase Educators Organization is a community of educators, representing diverse types of educational institutions and levels, who are interested in using KBase to teach bioinformatics concepts (Figure 6). To provide a common location where educational Narratives could be freely shared, the “Organization” framework within KBase was used to create a central homepage for the KBase Educators Organization. This Educators Organization represents a space where conceptual tutorial Narratives, student Narratives, or other resources can be shared in a moderated fashion. The Educators Organization homepage (https://narrative.kbase.us/#/orgs/kbase-educators) has open resources from educator guides to using the Narrative and Markdown language to instructional Narratives that can be accessed by anyone, but contributions to more detailed resources come from members of the community.
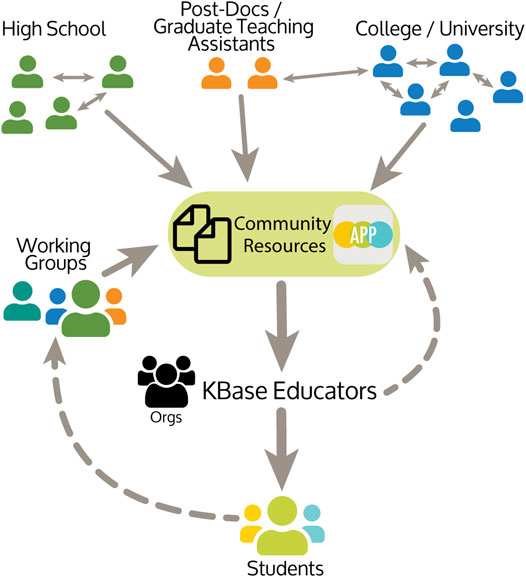
FIGURE 6. KBase Educators Community: The community includes instructors that teach at High School and college levels. Resources are developed by the community, including teaching Narratives using bioinformatics tools wrapped as KBase Apps and supporting documentation. Working groups have developed workflows across common topics. From student and community feedback, resources including teaching Narratives, are improved and added to the KBase Educators Organization.
KBase Organizations can also be created for individual classes. Educators can provide both informative and evaluative Narratives to their own organization, which students can copy, and then add their assigned projects to the organization or share with their instructor. Access to these organizations can be restricted, or not, as any instructor chooses.
The KBase team also supports educators and scientists to create working groups focused on specific topics. For example, the KBase Educators Working Group developed the existing resources (Figure 6). A subset of the Working Group recently developed a workflow for Metabolic Modeling (Supplementary Table S1), and other members have an interest in working on RNAseq. These collaborations are invaluable at providing ideas, exploring questions that arise during the implementation, and novel ways to use KBase Apps and Narratives.
Communication between instructors and KBase staff is a central feature of the KBase Educators Organization. To facilitate these interactions, KBase hosts a Slack workspace open to KBase users which includes a specific channel for education purposes. This Slack workspace is monitored by KBase staff and represents an important avenue for users to obtain technical support with any aspect of the KBase website, Narrative questions, or issues with specific tools. Importantly, it also serves as a venue for educators to seek advice from other educators and to serve as a meeting space for individuals with shared interests, facilitating collaborative development of new teaching modules.
KBase provides a free, streamlined entry point for students (and educators) to explore and perform sophisticated bioinformatics analyses (Dow, et al., 2021). The KBase Narrative interface allows instructors to break down complex real-world data sets and create analyses that are approachable and easily digested by students. From the students’ perspective, their familiarity with the concepts and confidence increased from the start of the courses surveyed. Most students began the course with little to no knowledge of the bioinformatics concepts covered. By the end, students gained confidence and familiarity with bioinformatics workflows overall, and in their ability to do bioinformatics analysis within KBase. By lowering many of the barriers students face when getting started in bioinformatics, it is possible to successfully increase students’ confidence in bioinformatics within a single course offering. Overall, students using KBase appreciated the workflow architecture and visualizations available within the Narrative. They especially appreciated being able to complete an entire sequencing analysis process in a relatively short amount of time. Beyond the ease of using a streamlined GUI (as compared to command line), students were able to perform research using the same tools as world-class researchers in a framework that aligns the learning process with applicable experience to empower students to get excited about their own education. The web-based nature of KBase also reduced the “barriers to entry” for instructors by minimizing the time spent on setup and maintenance of resources.
KBase provides a standardized framework for the integration of tools, resulting in a modular, yet interoperable and easy-to-follow interface that connects analyses and data to results and visualizations. KBase, and the network of community developers that contribute to the platform, continually strive to update software and Apps, incorporate new resources as they are developed, and ensure computational requirements are met for each App. The KBase servers include over 6,500 total cores, which run over 660,000 central processing unit hours per month, and over 400 terabytes of storage (and growing). Students only need a computer that can run a browser and an internet connection to access the resources behind KBase. This democratization of hardware enables a greater diversity of students to explore data science and bioinformatics, to run computationally demanding programs, and to open up much deeper analysis questions than previously possible.
In the case study at NC State, KBase was introduced as a course module component, as well as an option for the students’ final data analysis project. One key difference with using KBase for the course was that the Narratives enabled text and pictures to be incorporated within the analysis workflow. Students were introduced to Narratives through instructor-created videos, and then asked to copy the example Narratives for their own exploration. Embedded challenge questions allowed students to investigate publicly available data and encouraged them to be creative, while still providing scaffolding and structure via tutorial Narratives. Future course adaptations and modifications could expand the assignment by including similar individual or group challenges that require students to develop their own Narrative workflows to demonstrate an appropriate application and sequence of tools.
KBase was a favored tool for case study students. Where tools were mentioned in the post-course open-response questions, students most frequently referenced KBase by name, often first within lists, and articulated their comfort using the platform for bioinformatics analyses. Students indicated more confidence with using KBase over other tools used in the course during office hours. Notably, KBase was chosen for final projects more frequently than the HPC option by students with less coding experience.
The pivot towards confidence and comfort when using data science tools was an encouraging discovery. The perception of coding or using a computer for data analysis outside of basic spreadsheets may elicit feelings of inadequacy in the students (Cline and Prokop, 2019). These deep-seated feelings could translate into the students disregarding the assignment or not fully understanding the greater concepts of the exercise, so it is important to provide an accessible and inclusive framework that focuses on knowledge generation (Wright et al., 2020).
The interactive nature of KBase Narratives reduces the cognitive load placed on students learning bioinformatics, which allows students to focus on learning biology. Therefore, the emphasis shifts to biological concepts and the process of biological data analysis using a GUI, that provides exposure to bioinformatic tools without requiring students to become proficient with coding or command line tools. Students can run high-memory, computationally intensive applications on KBase that would not be possible on their own computers, or even in computer labs at many institutions. Access to institutional HPC resources that can handle applications requiring higher computational power may also be restricted to prevent large numbers of inexperienced users from degrading the performance for more senior researchers.
KBase is also versatile. The Narrative workflow format, where data and Apps can be easily added or removed, allows KBase to be implemented in a variety of educational environments ranging from the high school general biology classroom to advanced graduate courses in bioinformatics. For example, each App has detailed explanations describing its purpose and links to research articles appropriate for advanced courses that need to explore the design and logic behind each analysis. In addition, students can view and analyze publicly available data sets, or they can conduct de novo analyses using novel data sets. In support of novel analyses, KBase is scalable–both in the size or number of data sets, as well as the functional areas available for instruction. This provides faculty members with the ability to incorporate a single platform at multiple levels and in multiple contexts within their curriculum. This flexibility and scalability provides the opportunity for a variety of both formative and summative assessments in the classroom and laboratory. Taken together, feedback from the pilot was extremely positive and demonstrated that KBase can be a powerful educational tool that can be easily applied to a variety of educational contexts.
The KBase Educators program is an on-going effort supported by the KBase team. All biological sciences instructors, with any experience level in bioinformatics, are welcome to join. Joining is easy and includes: 1) access to the KBase Educators Organization, which has a plethora of resources and pre-built teaching Narratives, and 2) an invite to the KBase Users Slack workspace, which has a dedicated channel for educators to converse, ask questions of the community, or get asynchronous support from KBase staff. To join, create a KBase user account at www.kbase.us, then submit a “request to join” to the KBase Educators Org (https://narrative.kbase.us/#org/kbase-educators). Send a follow up email to ZW5nYWdlQGtiYXNlLnVz to introduce yourself. Please include what institution you are affiliated with and what courses you are hoping to teach using KBase. The KBase team will approve your join request and send you an invite to the Slack group.
One of the major benefits of joining the KBase Educators program is the community that has rallied around virtual learning using a GUI. The existing resources are free for use under a Creative Commons by 4.0 license, with attribution by citing this manuscript and (Arkin et al., 2018). As your instructional needs evolve and diversify, the teaching Narratives can be easily adapted and expanded. All work in KBase is backed by free large-scale computing resources and access to the KBase Help Desk (https://kbase-jira.atlassian.net/). KBase also supports the FAIR data principles (Wilkinson et al., 2016) by providing a mechanism to publish reproducible research. Static Narratives are snapshots of the current Narrative analysis that are made visible to anyone on the internet (including search engines) without a KBase account. If your class is working on a publication, it is also possible to request a DOI for a static Narrative(s) for inclusion in the manuscript’s data availability statement and reference section.
Finally, for those of you that are feeling a bit more adventurous and want to also teach your students how to code, the Bioinformatics Virtual Coordination Network (BVCN, https://biovcnet.github.io) is another great pandemic-inspired program led by Dr. Ben Tully (Tully et al., 2021).
The datasets analyzed for the teaching portions of this study can be found through KBase and the publication Narrative [https://doi.org/10.25982/90997.49/1783189]. The study records and raw datasets comprising student survey data are not available to persons outside of the study team under stipulations of the survey study review and approval (LBNL HSC 339NR001-2AP22; NC State IRB, # 20309).
The studies involving human participants were reviewed and approved by the Human Subjects Committee Institutional Review Board at Lawrence Berkeley National Lab (LBNL) (339NR001-2AP22) for the student survey and the NC State Institutional Review Board (IRB, # 20309) for the case study. The participants provided their written informed consent to participate in this study.
ED and EWC conceptualized the project. ED supervised the project. ED, EWC, SB, CG, AS, and CS contributed to developing the student survey. ED contributed to the methodology and performed the statistical analysis for the student survey. RK, JW, and CG contributed to the case study methodology and wrote the case study sections. JW performed statistical analysis for the case study. ED, EWC, BA, ZC, SB, JW, CG, TP, and CS wrote the first draft of the manuscript. ED, EWC, SB, CG, AS, CS, and AA contributed to the manuscript revisions. All authors read and approved the final manuscript.
KBase and its related work is supported as part of the Genomic Sciences Program DOE Systems Biology Knowledgebase (KBase) funded by the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research under Award Numbers DE-AC02-05CH11231, DE-AC02-06CH11357, DE-AC05-00OR22725, and DE-AC02-98CH10886.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We recognize fellow Educator Working Group members Jon Benskin, Jennifer Chase, Kathleen Morrow, and Mike Shaffer for developing these resources and using them within their courses. We thank fellow Educator Org members for taking part in initial discussions and lending their insights, especially Bill Andreopolos, Valerie de Crecy, Adam Deutschbauer, JP Dundore-Arias, Sharon Greenblum, Igor Grigoriev, Ana Juarez-Vazquez, Mark Martin, Victoria Orphan, Ben Tully, and Chris Vaglio. We thank members of the KBase team (https://www.kbase.us/team/) for their involvement and support, especially Dylan Chivian, Paramvir Dehal, Meghan Drake, Janaka Edirisinghe, José Faria, Chris Henry, Sean Jungbluth, Miriam Land, Erik Pearson, Bill Riehl, Boris Sadkhin, and Pamela Weisenhorn. Thank you to the Pacific Northwest National Lab and Environmental Molecular Sciences Laboratory User Facility for using KBase during the 2020 Multiscale Microbial Dynamics Modeling course, led by Tim Scheibe and Nancy Hess.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/feduc.2021.711535/full#supplementary-material
Afgan, E., Baker, D., Batut, B., van den Beek, M., Bouvier, D., Cech, M., et al. (2018). The Galaxy Platform for Accessible, Reproducible and Collaborative Biomedical Analyses: 2018 Update. Nucleic Acids Res. 46, W537–W544. doi:10.1093/nar/gky379
Arkin, A. P., Cottingham, R. W., Henry, C. S., Harris, N. L., Stevens, R. L., Maslov, S., et al. (2018). KBase: The United States Department of Energy Systems Biology Knowledgebase. Nat. Biotechnol. 36, 566–569. doi:10.1038/nbt.4163
Arnold, Ken., Gosling, J., and Holmes, D. (2000). The Java Programming Language. 75 Arlington Street, Suite 300 Boston, MA United States: Addison-Wesley Longman Publishing Company, Inc.
Bezanson, J., Edelman, A., Karpinski, S., and Shah, V. B. (2017). Julia: A Fresh Approach to Numerical Computing. SIAM Rev. 59, 65–98. doi:10.1137/141000671
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al-Ghalith, G. A., et al. (2019). Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat. Biotechnol. 37, 852–857. doi:10.1038/s41587-019-0209-9
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME Allows Analysis of High-Throughput Community Sequencing Data. Nat. Methods 7, 335–336. doi:10.1038/nmeth.f.303
Cline, S. G., and Prokop, J. W. (2019). Framework, Barriers, and Proposed Solutions for Engaging Students in Bioinformatics Research. Red Hook, NY: CSREA Press, 6.
Corpas, M., Jimenez, R. C., Bongcam-Rudloff, E., Budd, A., Brazas, M. D., Fernandes, P. L., et al. (2015). The GOBLET Training portal: a Global Repository of Bioinformatics Training Materials, Courses and Trainers. Bioinformatics 31, 140–142. doi:10.1093/bioinformatics/btu601
Cummings, M. P., and Temple, G. G. (2010). Broader Incorporation of Bioinformatics in Education: Opportunities and Challenges. Brief. Bioinform. 11, 537–543. doi:10.1093/bib/bbq058
Davis, J. J., Wattam, A. R., Aziz, R. K., Brettin, T., Butler, R., Butler, R. M., et al. (2019). The PATRIC Bioinformatics Resource Center: Expanding Data and Analysis Capabilities. Nucleic Acids Res.. doi:10.1093/nar/gkz943
Dow, E., Wood-Charlson, E., Biller, S., Paustian, T., Schimer, A., Sheik, C., et al. (2021). Data from: Bioinformatic Teaching Resources – for Educators, by Educators – Using KBase, a Free, User-Friendly, Open Source Platform. KBase. doi:10.25982/90997.49/17831890
Goller, C. C., Vandegrift, M., Cross, W., and Smyth, D. S. (2021). Sharing Notes Is Encouraged: Annotating and Cocreating with Hypothes.Is and Google Docs †. J. Microbiol. Biol. Educ. 22. doi:10.1128/jmbe.v22i1.2135
KBase for Educators Webinar (2020). YouTube. Available at: https://youtu.be/K9FxPc_2jzI (Accessed May 18, 2021).
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious Basic: An Integrated and Extendable Desktop Software Platform for the Organization and Analysis of Sequence Data. Bioinformatics 28, 1647–1649. doi:10.1093/bioinformatics/bts199
Kluyver, T., Ragan-Kelley, B., Pérez, F., Bussonnier, M., Frederic, J., Hamrick, J., et al. (2016). Jupyter Notebooks—A Publishing Format for Reproducible Computational Workflows. In Positioning and Power in Academic Publishing: Players, Agents and Agendas Amsterdam: IOS Press, 87–90.
Koboldt, D. C., Steinberg, K. M., Larson, D. E., Wilson, R. K., and Mardis, E. R. (2013). The Next-Generation Sequencing Revolution and its Impact on Genomics. Cell 155, 27–38. doi:10.1016/j.cell.2013.09.006
Kodama, Y., Shumway, M., and Leinonen, R.on behalf of the International Nucleotide Sequence Database Collaboration (2012). The Sequence Read Archive: Explosive Growth of Sequencing Data. Nucleic Acids Res. 40, D54–D56. doi:10.1093/nar/gkr854
Kluyver, T., Ragan-Kelley, B., Pérez, F., Bussonnier, M., Frederic, J., Hamrick, J., et al. (2016). “Jupyter Notebooks–A Publishing Format for Reproducible Computational Workflows,” in Positioning and Power in Academic Publishing: Players, Agents and Agendas (IOS Press), 87–90.
Lim, Y. W., Haynes, M., Furlan, M., Robertson, C. E., Harris, J. K., and Rohwer, F. (2014). Purifying the Impure: Sequencing Metagenomes and Metatranscriptomes from Complex Animal-Associated Samples. J. Vis. Exp. 52117. doi:10.3791/52117
Machluf, Y., Gelbart, H., Ben-Dor, S., and Yarden, A. (2017). Making Authentic Science Accessible-The Benefits and Challenges of Integrating Bioinformatics into a High-School Science Curriculum. Brief. Bioinform. 18, 145–159. doi:10.1093/bib/bbv113
Maloney, M., Parker, J., LeBlanc, M., Woodard, C. T., Glackin, M., and Hanrahan, M. (2010). Bioinformatics and the Undergraduate Curriculum Essay. CBE Life Sci. Educ. 9, 172–174. doi:10.1187/cbe.10-03-0038
McTighe, J., and Wiggins, G. (1998). “Backward Design,” in Understanding by Design. Alexandria, VA: Association for Supervision and Curriculum Development, 13–34.
QIAGEN CLC Workbench (2013). QIAGEN Bioinformatics Is Now QIAGEN Digital Insights. Available at: https://digitalinsights.qiagen.com/.
R Core Team (2021). R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing. Available at: https://www.R-project.org.
Raj, R. K., Romanowski, C. J., Impagliazzo, J., Aly, S. G., Becker, B. A., Chen, J., et al. (2020). High Performance Computing Education. In Proceedings of the Working Group Reports on Innovation and Technology in Computer Science Education New York, NY: Trondheim Norway: ACM, 51–74. doi:10.1145/3437800.3439203
Shaffer, M., Borton, M. A., McGivern, B. B., Zayed, A. A., La Rosa, S. L., Solden, L. M., et al. (2020). DRAM for Distilling Microbial Metabolism to Automate the Curation of Microbiome Function. Nucleic Acids Res. 48, 8883–8900. doi:10.1093/nar/gkaa621
Tully, B. J., Buongiorno, J., Cohen, A. B., Cram, J. A., Garber, A. I., Hu, S. K., et al. (2021). The Bioinformatics Virtual Coordination Network: An open-source and interactive learning environment. Front. Educ. doi:10.3389/feduc.2021.711618
Van Rossum, G., and Drake, F. L. (2009). Python 3 Reference Manual. 100 Enterprise Way. CA: Suite A200 Scotts ValleyCreateSpace.
Wall, L., Christiansen, T., and Orwant, J. (2000). The Perl Programming Language. Sebastopol, CA: O’Reilly Media, Inc.
Weber, N., Liou, D., Dommer, J., MacMenamin, P., Quiñones, M., Misner, I., et al. (2018). Nephele: a Cloud Platform for Simplified, Standardized and Reproducible Microbiome Data Analysis. Bioinformatics 34, 1411–1413. doi:10.1093/bioinformatics/btx617
Wilkinson, M. D., Dumontier, M., Aalbersberg, I. J., Appleton, G., Axton, M., Baak, A., et al. (2016). The FAIR Guiding Principles for Scientific Data Management and Stewardship. Sci. Data 3, 160018. doi:10.1038/sdata.2016.18
Williams, J. J., Drew, J. C., Galindo-Gonzalez, S., Robic, S., Dinsdale, E., Morgan, W. R., et al. (2019). Barriers to Integration of Bioinformatics into Undergraduate Life Sciences Education: A National Study of US Life Sciences Faculty Uncover Significant Barriers to Integrating Bioinformatics into Undergraduate Instruction. PLOS ONE 14, e0224288. doi:10.1371/journal.pone.0224288
Wilson Sayres, M. A., Hauser, C., Sierk, M., Robic, S., Rosenwald, A. G., Smith, T. M., et al. (2018). Bioinformatics Core Competencies for Undergraduate Life Sciences Education. PLOS ONE 13, e0196878. doi:10.1371/journal.pone.0196878
Wright, A. M., Schwartz, R. S., Oaks, J. R., Newman, C. E., and Flanagan, S. P. (2020). The Why, when, and How of Computing in Biology Classrooms, version 2; peer review: 2 approved. F1000Res 8, 1854. doi:10.12688/f1000research.20873.2
Perl Programming Language, RRID:SCR_018313
Python Programming Language, RRID:SCR_008394
Nephele, RRID:SCR_016595
Galaxy RRID:SCR_006281
PATRIC – Pathosystems Resource Integration Center, RRID:SCR_004154
CLC Genomics, Workbench, RRID:SCR_011853
Geneious, RRID:SCR_010519
R Project for Statistical Computing, RRID:SCR_001905
QIIME2–QIIME, RRID:SCR_008249
Keywords: computational biology, bioinformatics, data science, STEM education and learning, undergraduate education
Citation: Dow EG, Wood-Charlson EM, Biller SJ, Paustian T, Schirmer A, Sheik CS, Whitham JM, Krebs R, Goller CC, Allen B, Crockett Z and Arkin AP (2021) Bioinformatic Teaching Resources – For Educators, by Educators – Using KBase, a Free, User-Friendly, Open Source Platform. Front. Educ. 6:711535. doi: 10.3389/feduc.2021.711535
Received: 18 May 2021; Accepted: 02 September 2021;
Published: 29 October 2021.
Edited by:
Hugo Verli, Federal University of Rio Grande do Sul, BrazilReviewed by:
Dhananjaya Pratap Singh, National Bureau of Agriculturally Important Microorganisms (ICAR), IndiaCopyright © 2021 Dow, Wood-Charlson, Biller, Paustian, Schirmer, Sheik, Whitham, Krebs, Goller, Allen, Crockett and Arkin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ellen G. Dow, ZWdkb3dAbGJsLmdvdg==; Elisha M. Wood-Charlson, ZWxpc2hhd2NAbGJsLmdvdg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.