- College of Science and Engineering, James Cook University, Townsville, QLD, Australia
Passive acoustic monitoring has emerged as a useful technique for monitoring vocal species and contributing to biodiversity monitoring goals. However, finding target sounds for species without pre-existing recognisers still proves challenging. Here, we demonstrate how the embeddings from the large acoustic model BirdNET can be used to quickly and easily find new sound classes outside the original model’s training set. We outline the general workflow, and present three case studies covering a range of ecological use cases that we believe are common requirements in research and management: monitoring invasive species, generating species lists, and detecting threatened species. In all cases, a minimal amount of target class examples and validation effort was required to obtain results applicable to the desired application. The demonstrated success of this method across different datasets and different taxonomic groups suggests a wide applicability of BirdNET embeddings for finding novel sound classes. We anticipate this method will allow easy and rapid detection of sound classes for which no current recognisers exist, contributing to both monitoring and conservation goals.
Introduction
Vertebrate biodiversity is declining worldwide, necessitating tools for effective monitoring to detect declines and evaluate management interventions (Schmeller et al., 2015). Passive acoustic monitoring has recently emerged as a useful technique for monitoring vocal species, and given that many vertebrate taxa readily vocalise, it has the potential to allow monitoring at the large spatial and temporal scales needed, which are typically not possible using traditional survey methods (Sugai et al., 2019; Hoefer et al., 2023). However, while the ease of collecting and storing audio recordings has increased dramatically, automating the detection of individual species from large audio data sets can still prove challenging, hampering efforts to efficiently monitor vertebrate species.
While there has been much recent progress in automated species recognition (Priyadarshani et al., 2018; Brodie et al., 2020; Kahl et al., 2021; Stowell, 2022; Nieto-Mora et al., 2023; Xie et al., 2023), most acoustic studies still employ manual methods of analysis (Sugai et al., 2019) as there are few well developed and tested artificial intelligence classifiers or ‘recognisers’ for most vocal taxa. This is particularly true for vocal taxa that lack extensive call libraries (e.g., frogs and mammals), as well as many rare and threatened species. This leaves researchers interested in utilising passive-acoustic monitoring for many under-studied species with the choice of developing their own recogniser or using existing tools to find their sound class of interest. Recogniser development can require considerable investment and specialist training lacked by many ecologists and management practitioners. Additionally, there is often a lack of sufficient example vocalisations from target species required to build traditional bespoke artificial intelligence recognisers (Priyadarshani et al., 2018). Therefore, tools flexible enough to allow detection of a wide array of taxa, and relatively straightforward to implement, with little user input, are needed to allow acoustic monitoring of many taxa.
There are a number of tools available for users to find sound classes of interest with few or even no examples. One approach is template matching, such as that implemented in the R package monitoR (Katz et al., 2016) or the ARBIMON platform (https://arbimon.rfcx.org/; e.g., Vélez et al., 2024), where at least one example is used to create a template that can be run over an unlabelled audio data set returning a similarity score for each window of time. While this method can be successful in finding the target sound class, templates do not recognise spectral and temporal variations present in many animal vocalisations, often produce too many false-positives, and typically require many example templates to be created and validated to achieve acceptable performance (Teixeira et al., 2022; De Araújo et al., 2023; Linke et al., 2023). Another approach is unsupervised clustering, such as that implemented in the commercial software Kaleidoscope Pro (Wildlife Acoustics, https://www.wildlifeacoustics.com), in which the user does not even need an example vocalisation, instead, similar sounds present in the unlabelled data set are clustered together (Pérez-Granados and Schuchmann, 2020; Rowe et al., 2023). However, this approach may not work well for rare sound classes because there may not be enough detections in the data to form a cluster, potentially requiring additional known examples to be added to achieve sufficient results (e.g., Bobay et al., 2018). Additionally, all output clusters need to be manually searched at least partially, to determine whether the target sound class has been successfully clustered or not. Deep-learning CNNs promise much better performance than these previous methods but typically require lots of training data to train from scratch, although transfer learning is possible with a smaller set of example data (Arora and Haeb-Umbach, 2017; Ghani et al., 2023). Another option with deep-learning models can be to use their embeddings for search, such that a single example may be used to find similar vocalisations in a data set.
Embeddings, in the context of deep-learning acoustic models, are the lower dimensional representations of sound data learned by the model during its training process (Stowell, 2022; Ghani et al., 2023). The embeddings from these models can be used to search for novel sound classes outside the original model’s training set, by measuring the similarity or distance between the embeddings of a target sound class (e.g., a species of interest), and the embeddings of unknown sound classes. Unknown sound classes with a high similarity, or low distance, to the target class are the most likely to be from the target sound class and are worth listening to. While the use of embeddings for similarity search in other applications is common (e.g., semantic search for text - Shen et al., 2014), we are not aware of its use for processing data from passive acoustic monitoring, perhaps due to the relatively recent development of very large acoustic models specific to wildlife sounds.
Large acoustic models such as BirdNET (Kahl et al., 2021) have been trained on thousands of vocal species from around the world (e.g., there are over 6.5k sound classes in BirdNET v2.4) potentially allowing researchers to rapidly analyse long-duration recordings to monitor species richness and study community change (see Pérez-Granados, 2023 for a review). The embeddings from these deep-learning models are likely to be useful for searching for new faunal sounds due to domain similarity. In a similar vein, BirdNET embeddings have successfully been used to distinguish within-species sound classes, such as call types and life stage (McGinn et al., 2023), although in this case the species investigated were already present in the training data of the model.
In this study we present the general workflow, as well as three case studies utilising BirdNET embeddings to detect species from single examples in long-duration audio recordings across different ecological settings. In the first, we map the distribution of an introduced reptile on Christmas Island, in the second, we conduct species inventories of amphibians and vocal mammals, and in the third, we search for a threatened bird species in a continental acoustic observatory. The demonstrated success of this method across different datasets and different taxonomic groups suggests a wide applicability of BirdNET embeddings for finding novel sound classes.
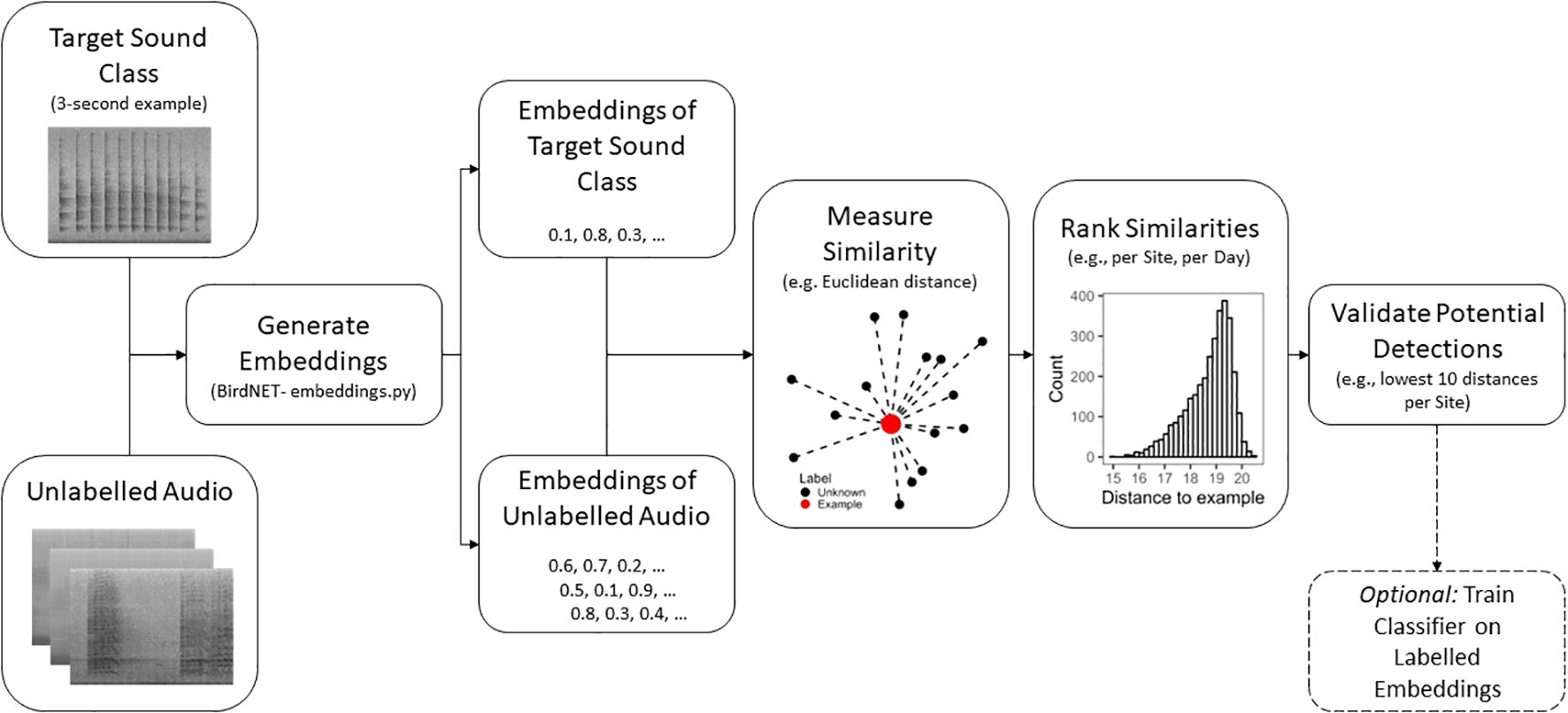
General methodology
The basic workflow required to use BirdNET embeddings to find vocalisations presented here involves four steps: example call(s) selection, embeddings generation, measuring similarity, and validation (Figure 1).
Example call selection
The first step in using this method is to select an example of the target sound class to be searched. This may come from the user’s own recording, or perhaps an online repository of animal sounds, such as xeno-canto (https://xeno-canto.org/). The BirdNET model (Kahl et al., 2021; https://github.com/kahst/BirdNET-Analyzer) runs at a 3-second temporal resolution, so some editing may be required to select an appropriate-length section of audio containing the target sound class. The example call process may require the removal of non-target sound classes present. If available, multiple example calls can be used to generate separate embeddings for search when the target species has different vocalisations.
Embeddings generation
The embeddings of both the target sound class and the unlabelled audio dataset to be searched can be generated using BirdNET’s ‘embeddings.py’ script. This script will generate a text file containing the 1024-length embeddings vector for each 3 seconds of audio; for the example call this will be a single vector, for the audio being searched it will be as many 3- second segments as are contained in the audio recording (examples of such text files for a 2-hour recording and an example call are included in Supplementary Material 1). When generating the embeddings file, outputting a file with the same name as the original recording is useful for ensuring segments identified as being similar to the example call can be traced back to the correct 3-seconds of audio for playback and validation.
Measuring similarity
To determine the similarity between the embeddings vector of our target class and our unlabelled audio, we can calculate the Euclidean distance, this results in a distance value for every 3 seconds of unlabelled audio, such that more similar embeddings have lower distance values. There are other similarity measures available, such as cosine similarity, where a higher value equals greater similarity. We used R (version 4.1.2) to measure Euclidean distance between the embeddings vector of the target vocalisation and each embeddings vector of the unlabelled audio (we provide an example with embeddings files and R script in Supplementary Material 1).
Validation
Once the distances between the embeddings of the target sound class and all the unlabelled audio have been calculated, a validation protocol can then be established based on the needs of the study. Using the names of the original recordings and the start and end values provided in the BirdNET embeddings files it is possible to identify the 3-second segments of audio desired for listening (e.g., those with the lowest distances to the example call). They can then, for example, be programmatically cut out from the original recording and placed into a folder for validation using the researcher’s software of choice (e.g. Audacity, Kaleidoscope Pro, Raven Pro etc.). Simply validating the lowest returned distances may be enough for some studies. For example, if only presence/absence at a location is required, then labelling from the lowest distance for all audio at that location until a detection is made (with some pre-determined stopping point) can be a rapid way to check a large audio data set for the presence of a certain sound class. If presence/absence was desired on a smaller timescale (e.g., daily), then the lowest distance example per day could be checked. There is no golden distance known a-priori for which we can expect to only get true-positive detections of the target class, the distance will vary based on the variation in the target sound class and the similarity of non-target sound classes in the soundscape (Figure 2A). However, a ‘threshold’ distance can be derived from labelling a sample of the unlabelled audio across the range of distances returned, to determine the point at which detections returned have a desired property (e.g., a certain probability of correct prediction; Figure 2B).
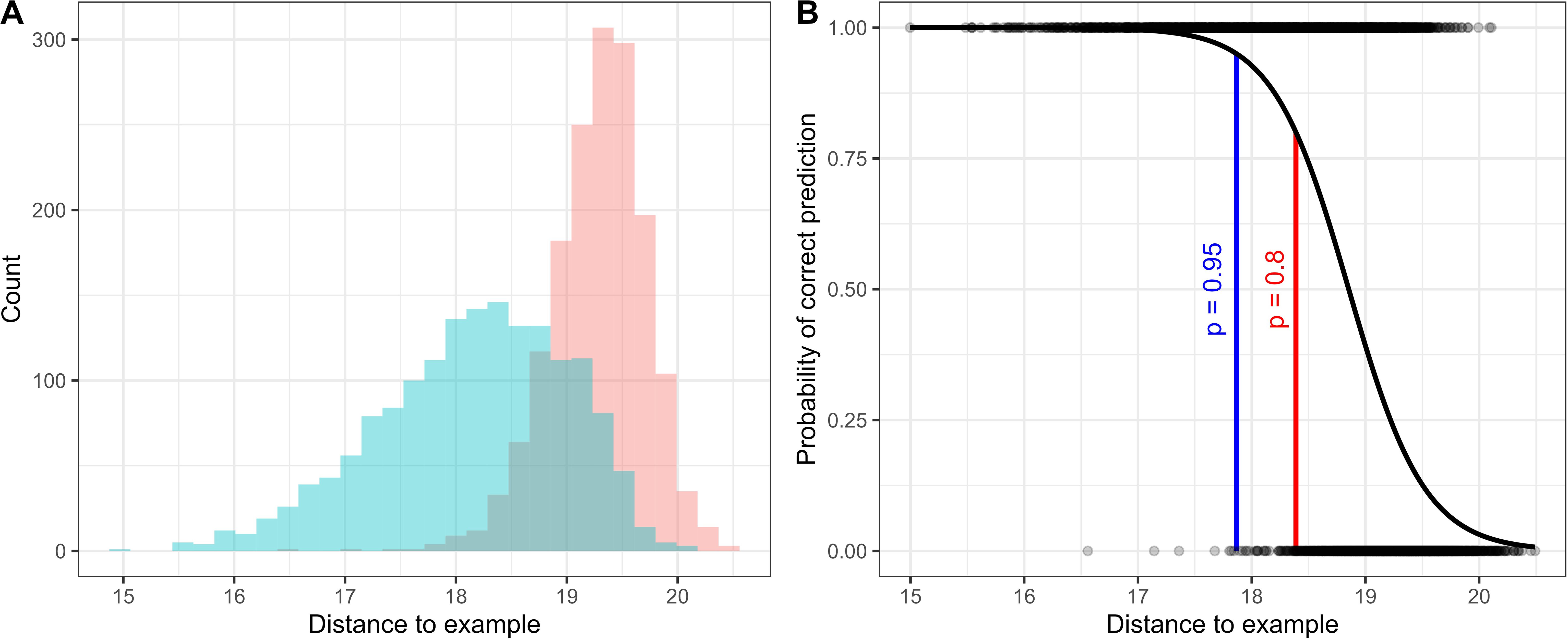
Figure 2. Example plots showing the relationship between distance to example call and the true class. (A) Euclidean distances of unlabelled audio compared to a target sound class coloured by the true label for the audio (target class = blue, non-target class = red). (B) A logistic regression fit to data labelled across a range of distances can be used to determine a threshold distance value for a certain desired property (e.g., 95% or 80% probability of correct prediction).
While the process described here is primarily about using the distance values returned directly, an additional step can be added wherein the search results are used to label both positive and negative examples and train a simple classifier using the 1024-length embeddings vectors as predictor features (a brief example is included in Case Study 3).
Case study 1: mapping the distribution of the introduced Asian house gecko (Hemidactylus frenatus) on Christmas Island
Background/motivation
Monitoring invasive species is important for understanding their distribution and spread, particularly for those species that are causing deleterious impacts on native fauna or are being actively managed. Many introduced vertebrate species are vocal, therefore PAM offers a useful tool for monitoring the spread and distribution of these invasives (e.g., Bota et al., 2024). The Asian house gecko (Hemidactylus frenatus) is native to southeast Asia, but has spread globally, after introduction to Australia, the southern United States, and many islands (Rödder et al., 2008). They have been implicated in negative impacts on native geckos in their introduced range (Cole et al., 2005; Hoskin, 2011; Nordberg, 2019). The Asian house gecko was introduced to Christmas Island in 1979, where they may compete with endemic geckos, and have been detected occupying sites previously occupied by native geckos (Cogger, 2006). Asian house geckos have caused declines of geckos on other islands where they have been introduced (Case et al., 1994). Unlike most reptiles, the Asian house gecko has a loud and distinct vocalization that should lend itself well to detection during passive acoustic monitoring. In this case study, we used a recently established network of acoustic recorders to survey for Asian house geckos to understand their distribution around Christmas Island.
Data and methods
In 2023 a network of audio recorders (Frontier Labs, BioAcoustic Recorder-LT™ – 44.1kHz sampling rate), were deployed across Christmas Island, recording continuously for two 1-month periods during May-August. To determine the distribution of the Asian house gecko around Christmas Island, we searched the audio from 76 sites using an example vocalisation from a previous lab study conducted in Townsville, Queensland (Hopkins et al., 2020). BirdNET embeddings for all available recordings collected between 18:00 and 06:00 local time were calculated (mean number of nights: 43.2), as Asian house geckos are most active at night (Hopkins et al., 2020). To determine which sites Asian house geckos were present, as well as obtain an estimate of their occupancy, we validated the lowest distance detection per site per night of recording.
Results
A total of 3286 apparent nightly detections were validated across all sites. Asian house geckoes were detected at 63 out of 76 sites surveyed, with an average of 49 percent of nights having positive detections and 9 sites having detections on every night of recording (Figure 3).
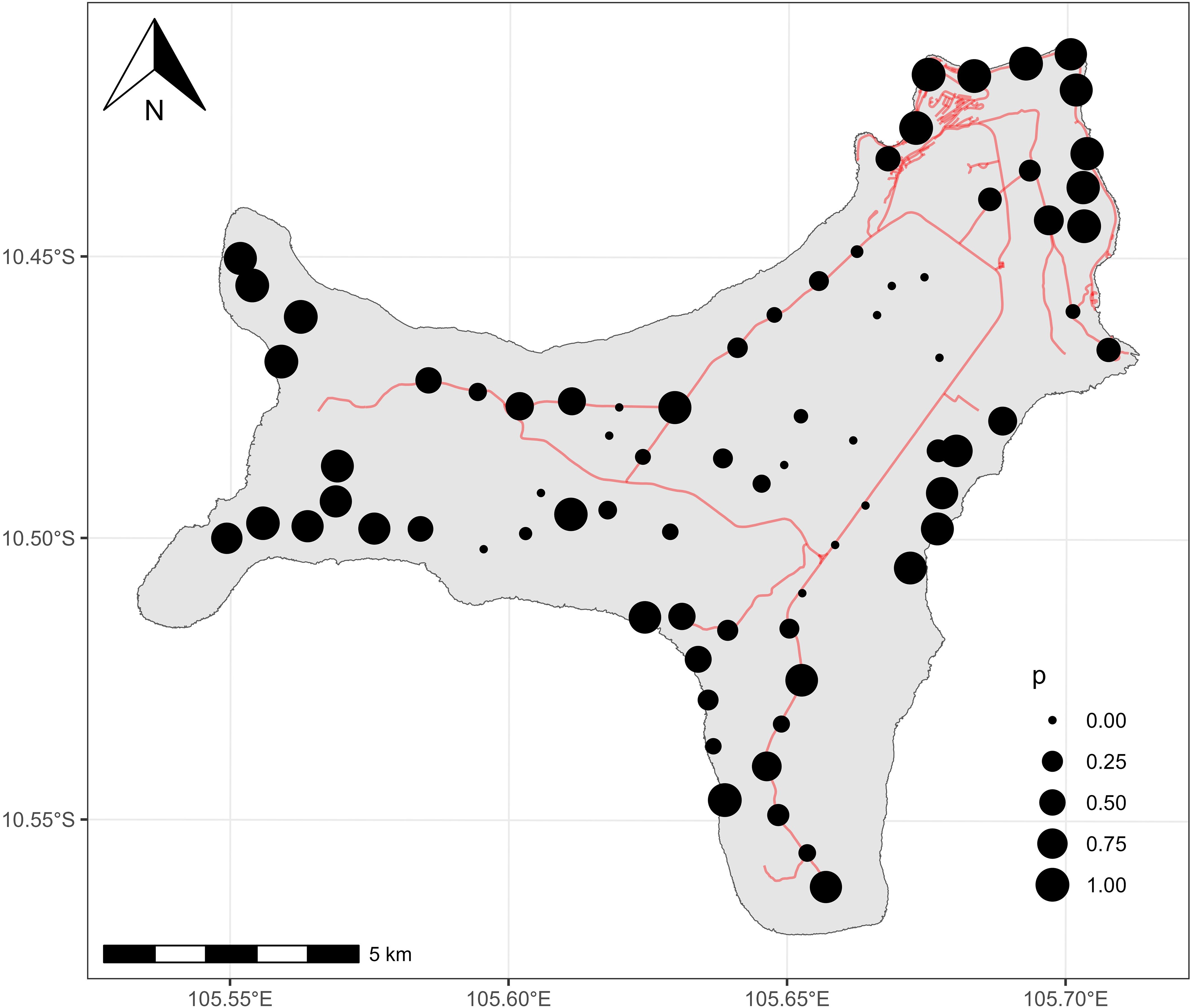
Figure 3. Christmas Island showing the detections of Asian House Geckos at 76 locations. Size of points represents the proportion of surveyed nights (mean 43.2) with positive detections.
Case study 2: generating static and temporal variance estimates around species richness for mammals and amphibians
Background/motivation
The species richness of habitats can be a good indicator of ecosystem health (Carignan and Villard, 2002), and methods that monitor richness though time may be useful for detecting declines and evaluating the success or failure of measures for conservation (Schmeller et al., 2015). However, monitoring species richness using PAM can be quite challenging as it either requires a significant effort to manually listen and label audio for all or most species present, or automated methods that can do the same (Sugai et al., 2019). While acoustic recognition of birds has received a lot of attention from the scientific community, acoustic studies of amphibians and mammals have received less (Hoefer et al., 2023). This is worrying as many amphibian and mammal species are highly threatened in Australia (Woinarski et al., 2015; Luedtke et al., 2023) and having a reliable way to estimate the richness and diversity of these taxa is needed. While the most recent BirdNET versions have been used to detect frog and mammal species elsewhere (Pérez-Granados et al., 2023; Sossover et al., 2024), and also now include Australian bird species ‘out-of-the-box’, it is currently not possible to directly monitor other Australian vocal taxa such as mammals and frogs like this at the moment. Using embeddings from example calls makes this possible. In this case study, we used example calls for a range of amphibian and mammal species to estimate species richness of these taxa at three sites of the Australian Acoustic Observatory (Roe et al., 2021).
Data and method
BirdNET embeddings were generated for all available audio for three Australian Acoustic Observatory (https://data.acousticobservatory.org/) sites in North Queensland (Chillagoe, 17.2°S, 144.5°E; Fletcherview, 19.88°S, 146.18°E; Spyglass, 19.5°S, 145.7°E), totalling over 120,000 h of audio. Each site has four sensors per site, with two sensors located close to a water source and two sensors located in a relatively dry location (Roe et al., 2021). We generated BirdNET embeddings for 17 mammalian sound classes (59 total example vocalisations), and 30 amphibian sound classes (67 total example vocalisations) that may occur across the distribution of the three sites. Some of the mammalian classes used were not classified to species-level (macropods, pteropodids), due to the similarity of vocalisations of closely related species making species identity difficult to determine with audio alone. To determine overall sensor level richness, the top detection per example used was validated per sensor for the entire available audio data set, leading to the validation of 708 audio files for mammals and 804 audio files for amphibians. To determine temporal trends in mammalian and amphibian richness, the top detection per example was validated per month for one sensor (Fletcherview - Dry B), resulting in 132 mammal validations and 156 amphibian validations.
Results
A total of 11 mammalian and 16 amphibian sound classes were detected across the three sites (Figures 4A, C). At one sensor, validated at a monthly timescale, richness of mammalian sound classes peaked towards the middle of the year (~April-June), while richness of amphibian sound classes peaked at the end and beginning of the year (~December-March; Figures 4B, D).
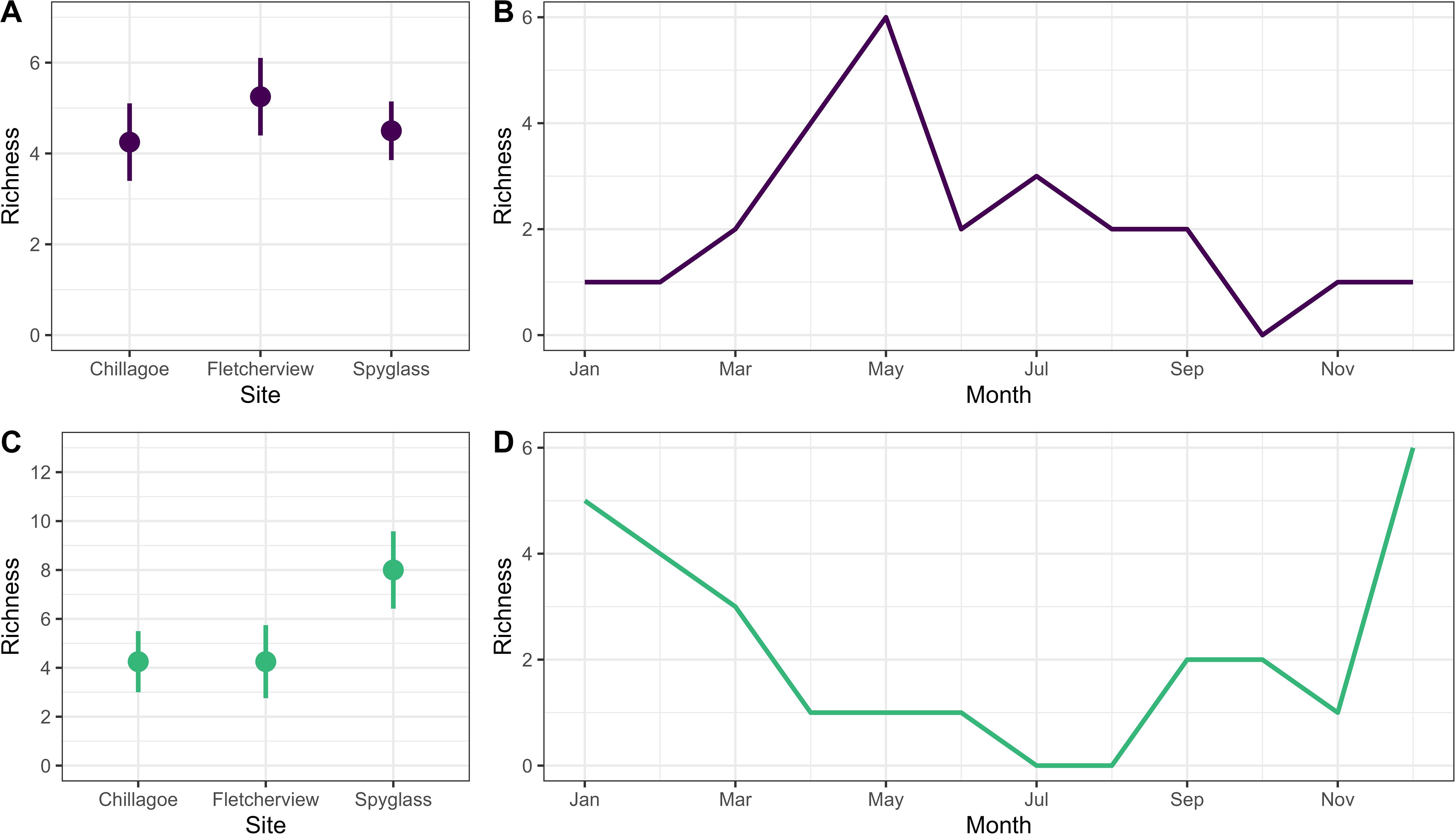
Figure 4. Mean (± SE) richness of mammalian (A) and amphibian (C) sound classes determined using BirdNET embeddings at 12 sensors across 3 Australian Acoustic Observatory sites. Temporal trends in richness of mammalian (B) and amphibian (D) sound classes detected at a single sensor (Fletcherview - Dry B) validated using the top detection per month.
Case study 3: detecting the threatened Blue-winged parrot (Neophema chrysostoma) using an acoustic observatory
Background/motivation
Continuous acoustic monitoring has the potential to be very valuable for monitoring threatened species, as they are often rare and are difficult to detect using infrequent in-person surveys (Klingbeil and Willig, 2015; Gibb et al., 2019; Manzano-Rubio et al., 2022; Allen-Ankins et al., 2024). Australia has one of the highest rates of vertebrate decline worldwide (Ritchie et al., 2013), and long-term passive acoustic monitoring networks such as the Australian Acoustic Observatory have huge potential to detect threatened species and track declines in site occupancy (Schwarzkopf et al., 2023). Unfortunately, many rare and threatened species do not have extensive call libraries available for training custom deep-learning recognisers, necessitating other approaches for detecting their calls in long-duration audio recordings. The Blue-winged parrot (Neophema chrysostoma) is currently listed as Vulnerable on both the IUCN Red List and under Australia’s EPBC Act due to undergoing a population decline over the last three generations (decline of 30–50%) that is primarily attributed to habitat loss (Holdsworth et al., 2021). The species is not currently included in BirdNET as of version 2.4 and therefore cannot be readily detected in audio recordings using existing tools. In this case study, we used BirdNET embeddings to determine Blue-winged parrot site presence across a range of Australian Acoustic Observatory sites within their predicted distribution.
Data and method
Using existing information about the range and distribution of the Blue-winged parrot (Australia - Species of National Environmental Significance Distributions (public grids)) ten Australian Acoustic Observatory sites were selected that provided the best chance of detection (Figure 5). An example vocalisation for the species was obtained from xeno-canto and the distance between its BirdNET embeddings and the embeddings of all available audio for the chosen sites was calculated (total >360,000 hours of audio). To determine the site presence of Blue-winged parrots, the top 100 lowest-distance detections were validated per site. Additionally, to improve our ability to acoustically survey for this species, we fit and tested a simple classifier (linear support vector machine) to the labelled embeddings using a 70:30 train:test split, and report on precision and recall of this method compared to just using the distance to the example call.
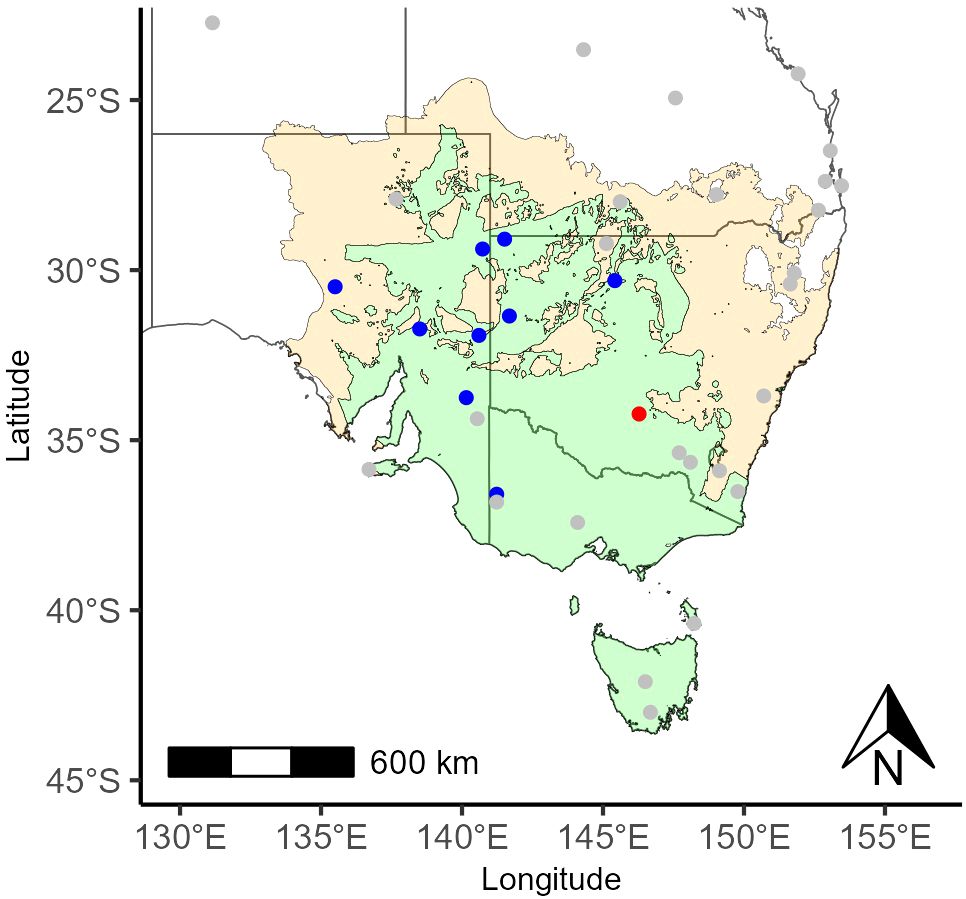
Figure 5. Southeast Australia with Australian Acoustic Observatory locations overlayed on the predicted distribution of Blue-winged parrot. Tan represents areas where ‘Species or species habitat may occur’, and green represents areas where ‘Species or species habitat likely to occur’. Blue points represent successful detection at a site, red points represent unsuccessful detection at a site, and grey points represent unsearched sites.
Results
Using the embeddings for a single example call, Blue-winged parrots were successfully detected at nine out of ten Australian Acoustic Observatory sites searched (Figure 5).
True-positive detections of Blue-winged parrot were typically a lower distance to our target call embeddings than false-positive detections (Figure 6A). Indeed, at eight out of the nine sites where they were detected, it was only necessary to look at the top detection to confirm site presence. However, there is a reasonably sharp drop in precision that begins at moderate levels of recall (~40%, Figure 6C). A simple classifier trained using the BirdNET embeddings as features dramatically improved the ability to distinguish between true- and false-positives, as evidenced by the strong separation of model confidence scores between the two classes (Figure 6B) and precision remaining high at high recall (Figure 6C).
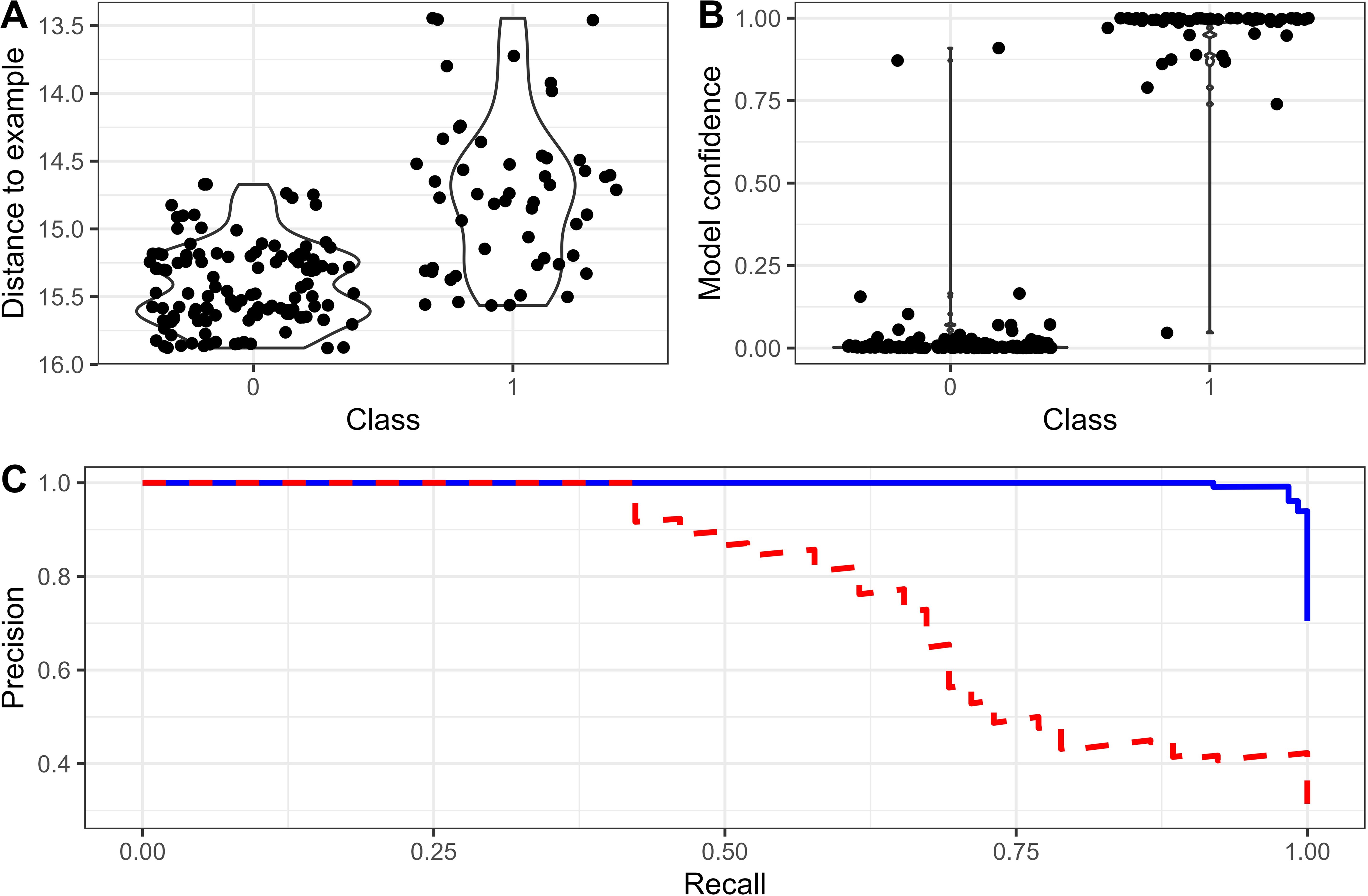
Figure 6. Comparison of performance of using embeddings distance and trained classifier on labelled embeddings for Blue-winged parrot. (A) Relationship between distance and true-positive/false-positive label, (B) relationship between classifier confidence score and true-positive/false-positive label, and (C) precision-recall curves comparing distance measures only (red), and classifier (blue). All data is from 30% test split of the full labelled data.
Discussion
Here we have demonstrated how the embeddings from a deep-learning model such as BirdNET can be effectively used to find a range of new sound classes from multiple taxonomic groups. This method presents an opportunity for rapid detection of new sound classes with minimal user input and validation required, and without the need to develop complex, bespoke recognisers. BirdNET is free to use, and requires no deep-learning expertise to run, potentially allowing ecologists and conservation practitioners to monitor acoustically for any vocal species, either present in the model (Kahl et al., 2021), or for which they can obtain a single example call. The three case-studies we have selected demonstrate three different possible use cases for automated sound recognition, which we suspect are common requirements in research and management.
Understanding the distribution and spread of invasive species is critical to management and control, and ecoacoustic recordings are extremely useful for detecting vocal invasive species (Ribeiro et al., 2022; Bota et al., 2024). Here we used recordings made for a different purpose (detecting rare birds and mammals) to detect invasive house geckos (Hemidactylus frenatus) on Christmas Island, a location characterised by several species of endangered native geckos (Cyrtodactylus sadleri and Lepidodactylus listeri; Andrew et al., 2018), using recorders placed in a wide range of locations recording 24/7 to maximise coverage. We used nightly detections collected over a 2-month period to determine site occupancy, but because it was comparatively easy to validate calls nightly, we could also obtain a measure of the relative activity of geckos at a site. Asian house geckos likely do not move long distances (Frenkel, 2006), so high daily activity at a site likely indicates that they are present there, not just passing through. Indeed, vocal activity rates of various taxa have been shown to correlate well with abundance (Nelson and Graves, 2004; Pérez-Granados et al., 2019). Comparing levels of activity at different sites could, for example, be used to select sites for control or to quantify the impact of control measures once implemented. While the Asian house gecko was found all over Christmas Island owing to the long-time since their introduction, this method could be used in other locations to identify areas of incursion. Additionally, acoustic recognisers paired with on-device detection (e.g., with a BirdNET-Pi) could provide real-time data for early detection and removal of invasive species before they can establish. We suspect this method will prove useful for other projects monitoring invasive species activity and movement.
Identifying species and describing community composition at a site are critical aspects of monitoring (e.g., Pollock et al., 2002). Manual surveys typically record species presence, and a wide range of global databases record species detections for mapping and monitoring (Moussy et al., 2022). Thus, we included here an example in which passive acoustic monitoring can be used for species identification and estimation of site richness. Our method provided the variation in richness estimates of species among four sites at each of three locations in Queensland, Australia, and allowed the user to examine this variation over an extended time (1-1.5 years). Conducting these species richness estimates long-term may be useful for identifying changes to community composition or detecting the loss of species from a site. Capturing large spatial and fine temporal variation is a strength of acoustic monitoring (e.g., Gibb et al., 2019), which we demonstrate here can be accessed in a fairly straightforward manner requiring little validation. Of course, a thorough understanding of the relationship between the species richness that can be obtained using acoustic methods and the actual on-ground species richness is needed to ensure detected patterns are reliable (Hoefer et al., 2023).
Another common use of passive acoustic monitoring is to detect threatened species, and to determine their site occupancy and activity (Sugai et al., 2019). Here we demonstrate the use of our method to detect the calls of a vulnerable bird species, the Blue-winged Parrot, (Neophema chrysostoma) (Radford and Bennett, 2005). The ability of our method to increase detection of species for which there are few recorded calls, which is the case for many endangered species is a great strength of this approach. Once more calls have been detected and verified, those calls, in addition to the similar but incorrect calls detected, can be combined, as we did here, to create and train a recogniser. This approach could be reiterated, if necessary, to increase the precision and recall of the initial recogniser to desired levels. Whereas with standard complex-neural-network style recognisers, improving a recogniser may take a great deal of specialist programming ability, and many iterations (e.g., Eichinski et al., 2022) it may be shortened and simplified for finding rare species in large datasets using the proposed approach.
One time-consuming aspect of acoustic monitoring is the effort of detecting calls. Manually processing raw audio, until recently one of the most common means of detecting calls (Sugai et al., 2019), can be quick when focused on a single species with distinct calls that can be detected visually on a spectrogram, but can take at least as long as the audio itself, or significantly longer when targeting many species with diverse vocalisations (e.g., 120 seconds per 30-second sample, Linke and Deretic, 2020). Validating detections from more automated methods is faster, but still needs to occur (e.g., Campos-Cerqueira and Aide, 2016). The approach outlined here allows the user to determine the amount of validation required, depending on the question, but, in each case, comparatively little validation effort was needed to obtain the results presented. The detection of Asian house geckos (Case study 1) at 76 sites only took 3.5 hours, including procuring information at a nightly resolution to estimate relative prevalence. The enumeration of mammalian and amphibian species richness (27 species detected; Case study 2) at 12 sensors only took 3.5 hours, with validation of monthly detections at a single sensor taking an additional 1.5 hours. The validation of apparent detections of the vulnerable Blue-winged parrot (Case study 3) at 10 sensors only took 10 hours, and this included extra validation beyond establishing site presence to create a dataset for training a recogniser. Each project can predetermine the information they actually require and validate detections specific to the question. The finer the granularity of calling information that is required the more validation effort that will be needed to obtain it. For large datasets it may be necessary to determine the relationship between the distance to the example call and the precision of the results using a subset of detections, such that a threshold of a suitably high precision can be applied without validating the entire dataset. The strong focus on achieving both high precision and recall in machine learning applications (e.g., Juba and Le, 2019) may not be required for every biological application. High precision and adequate recall may be enough to ensure a call is detected daily or weekly to answer specific questions, say on site occupancy or species presence.
The main advantage of using a large pre-trained acoustic model such as BirdNET to search for a target vocalisation is the ability to use very few examples, even a single example, and to rapidly detect many instances of those vocalisations. As we have shown, the distance between the embeddings of the target class and the audio being searched may alone be enough for many applications (Case studies 1 & 2), however it is also a useful method for obtaining both examples of the target class and non-target class for rapidly training a recogniser using the embeddings as features (Case study 3). The non-target class examples that have relatively low distances to the target class for which you are searching, are likely to be very useful for training a recogniser, as they probably share some similar features with the target class, hence the similarity of the BirdNET embeddings.
As the embeddings are dependent on the pre-trained model (i.e., BirdNET), rather than the sound classes for which you are searching, they can be computed for an entire audio dataset once, and then stored, so that new examples, such as different species or different call types can be found readily. This differs from other approaches, such as unsupervised clustering with Kaleidoscope Pro, which may require species-specific settings for ideal clustering, thus requiring reanalysis for new tasks. It is also more efficient than using call templates, such as those implemented in monitoR, which requires new templates to be run over the entire audio set each time a new variant is attempted, a much more time-consuming process than measuring the Euclidean distances between a new target class and precomputed embeddings.
While we have used BirdNET here to demonstrate this approach, it is applicable to other large acoustic models (e.g. Google Perch; Ghani et al., 2023). As new deep-learning models come out (potentially with better embeddings that have improved search performance), the same approach can be used. We hope this method will allow easy and rapid detection of sound classes for which no current recognisers exist, contributing to monitoring and conservation goals.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
The animal study was approved by James Cook University Animal Ethics Committee. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
SA-A: Conceptualization, Data curation, Formal analysis, Methodology, Validation, Visualization, Writing – original draft, Writing – review & editing. SH: Data curation, Methodology, Validation, Writing – original draft, Writing – review & editing. JB: Validation, Writing – original draft, Writing – review & editing. SB: Validation, Writing – original draft, Writing – review & editing. LS: Conceptualization, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The authors would like to acknowledge the Anglo American Foundation, the National Environmental Science Program (NESP) and Parks Australia for supporting this research.
Acknowledgments
We acknowledge the traditional custodians of the lands where we conducted this research and pay our respects to elders past and present. We recognize their continued connection to land, water, and culture and acknowledge them as Australia’s first scientists. We express our gratitude to J. Grant for assistance with validating detections of the Blue-winged parrot. Audio recordings on Christmas Island were collected under approval by James Cook University Animal Ethics Committee (A2907).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2024.1409407/full#supplementary-material
References
Allen-Ankins S., Laguna J. M., Schwarzkopf L. (2024). Towards automated detection of the endangered southern black-throated finch (poephila cincta cincta). Wildlife Res. 51. doi: 10.1071/WR23151
Andrew P., Cogger H., Driscoll D., Flakus S., Harlow P., Maple D., et al. (2018). Somewhat saved: A captive breeding programme for two endemic Christmas Island lizard species, now extinct in the wild. Oryx 52, 171–174. doi: 10.1017/S0030605316001071
Arora P., Haeb-Umbach R. (2017). A study on transfer learning for acoustic event detection in a real life scenario (IEEE), 1–6.
Bobay L. R., Taillie P. J., Moorman C. E. (2018). Use of autonomous recording units increased detection of a secretive marsh bird. J. Field Ornithology 89, 384–392. doi: 10.1111/jofo.2018.89.issue-4
Bota G., Manzano-Rubio R., Fanlo H., Franch N., Brotons L., Villero D., et al. (2024). Passive acoustic monitoring and automated detection of the american bullfrog. Biol. invasions 26, 1269–1279. doi: 10.1007/s10530-023-03244-8
Brodie S., Allen-Ankins S., Towsey M., Roe P., Schwarzkopf L. (2020). Automated species identification of frog choruses in environmental recordings using acoustic indices. Ecol. Indic. 119, 106852. doi: 10.1016/j.ecolind.2020.106852
Campos-Cerqueira M., Aide T. M. (2016). Improving distribution data of threatened species by combining acoustic monitoring and occupancy modelling. Methods Ecol. Evol. 7, 1340–1348. doi: 10.1111/mee3.2016.7.issue-11
Carignan V., Villard M.-A. (2002). Selecting indicator species to monitor ecological integrity: A review. Environ. Monit. Assess. 78, 45–61. doi: 10.1023/A:1016136723584
Case T. J., Bolger D. T., Petren K. (1994). Invasions and competitive displacement among house geckos in the tropical pacific. Ecology 75, 464–477. doi: 10.2307/1939550
Cogger H. G. (2006). National recovery plan for lister’s gecko lepidodactylus listeri and the Christmas Island blind snake typhlops exocoeti (Canberra: Department of the Environment of Heritage).
Cole N. C., Jones C. G., Harris S. (2005). The need for enemy-free space: The impact of an invasive gecko on island endemics. Biol. Conserv. 125, 467–474. doi: 10.1016/j.biocon.2005.04.017
De Araújo C., Zurano J., Torres I., Simões C., Rosa G., Aguiar A., et al. (2023). The sound of hope: Searching for critically endangered species using acoustic template matching. Bioacoustics 32, 708–723. doi: 10.1080/09524622.2023.2268579
Eichinski P., Alexander C., Roe P., Parsons S., Fuller S. (2022). A convolutional neural network bird species recognizer built from little data by iteratively training, detecting, and labeling. Front. Ecol. Evol. 133. doi: 10.3389/fevo.2022.810330
Frenkel C. (2006). Hemidactylus frenatus (squamata: Gekkonidae): Call frequency, movement and condition of tail in Costa Rica. Rev. Biol. Trop. 54, 1125–1130. doi: 10.15517/rbt.v54i4.14086
Ghani B., Denton T., Kahl S., Klinck H. (2023). Global birdsong embeddings enable superior transfer learning for bioacoustic classification. Sci. Rep. 13, 22876. doi: 10.1038/s41598-023-49989-z
Gibb R., Browning E., Glover-Kapfer P., Jones K. E. (2019). Emerging opportunities and challenges for passive acoustics in ecological assessment and monitoring. Methods Ecol. Evol. 10, 169–185. doi: 10.1111/mee3.2019.10.issue-2
Hoefer S., McKnight D. T., Allen-Ankins S., Nordberg E. J., Schwarzkopf L. (2023). Passive acoustic monitoring in terrestrial vertebrates: A review. Bioacoustics 32, 506–531. doi: 10.1080/09524622.2023.2209052
Holdsworth M., Newman M., Menkhorst P., Morley C., Cooper R., Starks J., et al. (2021). “Blue-winged parrot neophema chrysostoma,” in The action plan for Australian birds 2020 (Melbourne: CSIRO Publishing), 447–450.
Hopkins J. M., Higgie M., Hoskin C. J. (2020). Calling behaviour in the invasive asian house gecko (hemidactylus frenatus) and implications for early detection. Wildlife Res. 48(2), 152–162. doi: 10.1071/WR20003
Hoskin C. J. (2011). The invasion and potential impact of the asian house gecko (hemidactylus frenatus) in Australia. Austral Ecol. 36, 240–251. doi: 10.1111/j.1442-9993.2010.02143.x
Juba B., Le H. S. (2019). Precision-recall versus accuracy and the role of large data sets. 33, 4039–4048. doi: 10.1609/aaai.v33i01.33014039
Kahl S., Wood C. M., Eibl M., Klinck H. (2021). Birdnet: A deep learning solution for avian diversity monitoring. Ecol. Inf. 61, 101236. doi: 10.1016/j.ecoinf.2021.101236
Katz J., Hafner S. D., Donovan T. (2016). Tools for automated acoustic monitoring within the r package monitor. Bioacoustics 25, 197–210. doi: 10.1080/09524622.2016.1138415
Klingbeil B. T., Willig M. R. (2015). Bird biodiversity assessments in temperate forest: The value of point count versus acoustic monitoring protocols. PeerJ 3, e973. doi: 10.7717/peerj.973
Linke S., Deretic J. A. (2020). Ecoacoustics can detect ecosystem responses to environmental water allocations. Freshw. Biol. 65, 133–141. doi: 10.1111/fwb.13249
Linke S., Teixeira D., Turlington K. (2023). Evaluating and optimising performance of multi-species call recognisers for ecoacoustic restoration monitoring. Ecol. Evol. 13, e10309. doi: 10.1002/ece3.10309
Luedtke J. A., Chanson J., Neam K., Hobin L., Maciel A. O., Catenazzi A., et al. (2023). Ongoing declines for the world’s amphibians in the face of emerging threats. Nature 622, 308–314. doi: 10.1038/s41586-023-06578-4
Manzano-Rubio R., Bota G., Brotons L., Soto-Largo E., Pérez-Granados C. (2022). Low-cost open-source recorders and ready-to-use machine learning approaches provide effective monitoring of threatened species. Ecol. Inf. 72, 101910. doi: 10.1016/j.ecoinf.2022.101910
McGinn K., Kahl S., Peery M. Z., Klinck H., Wood C. M. (2023). Feature embeddings from the birdnet algorithm provide insights into avian ecology. Ecol. Inf. 74, 101995. doi: 10.1016/j.ecoinf.2023.101995
Moussy C., Burfield I. J., Stephenson P., Newton A. F., Butchart S. H., Sutherland W. J., et al. (2022). A quantitative global review of species population monitoring. Conserv. Biol. 36, e13721. doi: 10.1111/cobi.13721
Nelson G. L., Graves B. M. (2004). Anuran population monitoring: Comparison of the north american amphibian monitoring program’s calling index with mark-recapture estimates for rana clamitans. J. Herpetology 38, 355–359. doi: 10.1670/22-04A
Nieto-Mora D., Rodríguez-Buritica S., Rodríguez-Marín P., Martínez-Vargaz J., Isaza-Narváez C. (2023). Systematic review of machine learning methods applied to ecoacoustics and soundscape monitoring. Heliyon. 9(10), e20275. doi: 10.1016/j.heliyon.2023.e20275
Nordberg E. J. (2019). Potential impacts of intraguild predation by invasive asian house geckos. Austral Ecol. 44, 1487–1489. doi: 10.1111/aec.12826
Pérez-Granados C. (2023). Birdnet: Applications, performance, pitfalls and future opportunities. Ibis 165, 1068–1075. doi: 10.1111/ibi.v165.3
Pérez-Granados C., Bota G., Giralt D., Barrero A., Gómez-Catasús J., Bustillo-De La Rosa D., et al. (2019). Vocal activity rate index: A useful method to infer terrestrial bird abundance with acoustic monitoring. Ibis 161, 901–907. doi: 10.1111/ibi.v161.4
Pérez-Granados C., Feldman M. J., Mazerolle M. J. (2023). Combining two user-friendly machine learning tools increases species detection from acoustic recordings. Can. J. Zoology 102, 403–409. doi: 10.1139/cjz-2023-0154
Pérez-Granados C., Schuchmann K.-L. (2020). Monitoring the annual vocal activity of two enigmatic nocturnal neotropical birds: The common potoo (nyctibius griseus) and the great potoo (nyctibius grandis). J. Ornithology 161, 1129–1141. doi: 10.1007/s10336-020-01795-4
Pollock K. H., Nichols J. D., Simons T. R., Farnsworth G. L., Bailey L. L., Sauer J. R. (2002). Large scale wildlife monitoring studies: Statistical methods for design and analysis. Environmetrics: Off. J. Int. Environmetrics Soc. 13, 105–119. doi: 10.1002/env.v13:2
Priyadarshani N., Marsland S., Castro I. (2018). Automated birdsong recognition in complex acoustic environments: A review. J. Avian Biol. 49, jav–01447. doi: 10.1111/jav.2018.v49.i5
Radford J. Q., Bennett A. F. (2005). Terrestrial avifauna of the gippsland plain and strzelecki ranges, victoria, Australia: Insights from atlas data. Wildlife Res. 32, 531–555. doi: 10.1071/WR04012
Ribeiro J., José W., Harmon K., Leite G. A., de Melo T. N., LeBien J., et al. (2022). Passive acoustic monitoring as a tool to investigate the spatial distribution of invasive alien species. Remote Sens. 14, 4565. doi: 10.3390/rs14184565
Ritchie E. G., Bradshaw C. J., Dickman C. R., Hobbs R., Johnson C. N., Johnston E. L., et al. (2013). Continental-scale governance and the hastening of loss of Australia’s biodiversity. Conserv. Biol. 27, 1133–1135. doi: 10.1111/cobi.2013.27.issue-6
Rödder D., Solé M., Böhme W. (2008). Predicting the potential distributions of two alien invasive housegeckos (gekkonidae: Hemidactylus frenatus, hemidactylus mabouia). North-Western J. Zoology 4, 236–246.
Roe P., Eichinski P., Fuller R. A., McDonald P. G., Schwarzkopf L., Towsey M., et al. (2021). The Australian acoustic observatory. Methods Ecol. Evol. 12, 1802–1808. doi: 10.1111/2041-210X.13660
Rowe K. M., Selwood K. E., Bryant D., Baker-Gabb D. (2023). Acoustic surveys improve landscape-scale detection of a critically endangered Australian bird, the plains-wanderer (Pedionomus torquatus). Wildlife Research. 51 (1). doi: 10.1071/WR22187
Schmeller D. S., Julliard R., Bellingham P. J., Böhm M., Brummitt N., Chiarucci A., et al. (2015). Towards a global terrestrial species monitoring program. J. Nat. Conserv. 25, 51–57. doi: 10.1016/j.jnc.2015.03.003
Schwarzkopf L., Roe P., Mcdonald P. G., Watson D. M., Fuller R. A., Allen-Ankins S. (2023). Can an acoustic observatory contribute to the conservation of threatened species? Austral Ecology. 48 (7), 1230–1237. doi: 10.1111/aec.13398
Shen Y., He X., Gao J., Deng L., Mesnil G. (2014). Learning semantic representations using convolutional neural networks for web search. 373–374. doi: 10.1145/2567948.2577348
Sossover D., Burrows K., Kahl S., Wood C. M. (2024). Using the birdnet algorithm to identify wolves, coyotes, and potentially their interactions in a large audio dataset. Mammal Res. 69, 159–165. doi: 10.1007/s13364-023-00725-y
Stowell D. (2022). Computational bioacoustics with deep learning: A review and roadmap. PeerJ 10, e13152. doi: 10.7717/peerj.13152
Sugai L. S. M., Silva T. S. F., Ribeiro J. W. Jr., Llusia D. (2019). Terrestrial passive acoustic monitoring: Review and perspectives. BioScience 69, 15–25. doi: 10.1093/biosci/biy147
Teixeira D., Linke S., Hill R., Maron M., van Rensburg B. J. (2022). Fledge or fail: Nest monitoring of endangered black-cockatoos using bioacoustics and open-source call recognition. Ecol. Inf. 69, 101656. doi: 10.1016/j.ecoinf.2022.101656
Vélez J., McShea W., Pukazhenthi B., Stevenson P., Fieberg J. (2024). Implications of the scale of detection for inferring co-occurrence patterns from paired camera traps and acoustic recorders. Conserv. Biol. 38, e14218. doi: 10.1111/cobi.14218
Woinarski J. C., Burbidge A. A., Harrison P. L. (2015). Ongoing unraveling of a continental fauna: Decline and extinction of Australian mammals since european settlement. Proc. Natl. Acad. Sci. 112, 4531–4540. doi: 10.1073/pnas.1417301112
Keywords: acoustic recognition, bioacoustics, biodiversity, conservation, deep learning, passive acoustic monitoring
Citation: Allen-Ankins S, Hoefer S, Bartholomew J, Brodie S and Schwarzkopf L (2025) The use of BirdNET embeddings as a fast solution to find novel sound classes in audio recordings. Front. Ecol. Evol. 12:1409407. doi: 10.3389/fevo.2024.1409407
Received: 30 March 2024; Accepted: 08 November 2024;
Published: 16 January 2025.
Edited by:
Almo Farina, University of Urbino Carlo Bo, ItalyReviewed by:
Stefan Kahl, Cornell University, United StatesCristian Pérez-Granados, University of Alicante, Spain
Copyright © 2025 Allen-Ankins, Hoefer, Bartholomew, Brodie and Schwarzkopf. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Slade Allen-Ankins, c2xhZGUuYWxsZW5hbmtpbnNAamN1LmVkdS5hdQ==
 Slade Allen-Ankins
Slade Allen-Ankins Sebastian Hoefer
Sebastian Hoefer Sheryn Brodie
Sheryn Brodie Lin Schwarzkopf
Lin Schwarzkopf