- 1Centre for Chemical Biology, Universiti Sains Malaysia, Bayan Lepas, Malaysia
- 2Institute of Tropical Biodiversity and Sustainable Development, Universiti Malaysia Terengganu, Kuala Terengganu, Malaysia
- 3Faculty of Science and Marine Environment, Universiti Malaysia Terengganu, Kuala Terengganu, Malaysia
- 4The Habitat Foundation, George Town, Malaysia
- 5School of Biological Sciences, Universiti Sains Malaysia, Minden, Malaysia
Introduction
Decapoda order (Crustacea and Malacostraca) is the most diverse and species-rich taxonomy group comprising many of the well-known invertebrates such as shrimps, prawns, crayfishes, lobsters, and true crabs (Shen et al., 2013). The true crabs belong to Brachyura, the largest infraorder within Decapoda with approximately 7,200 described species and some of which are economically important (De Grave et al., 2009; Ahyong et al., 2011). Brachyura crabs have a diverse range of forms and adaptations, making them one of the most important groups to study in terms of evolution (Castro et al., 2015). Brachyura encompasses Podotremata, Heterotremata, and Thoracotremata, the latter two of which are collectively referred to as Eubrachyura (de Saint Laurent, 1980; Ahyong et al., 2007). Within Eubrachyura, the family Sesarmidae includes terrestrial, semiterrestrial, or tree-climbing species occurring mainly in the mangroves (Cumberlidge et al., 2005; Serrano-Sánchez et al., 2016). Sesarmid crabs, due to their role in nutrient cycling, have been considered keystone species in the mangrove ecosystem (Smith et al., 1991). Multiple transition routes have been proposed for this family, which comprises members that have colonized land via both marine and freshwater environments (Watson-Zink, 2021).
The genus Geosesarma De Man, 1892, belonging to the Sesarmidae family, is represented by 67 species at present. The genus is distributed in Malaysia, Indonesia, Philippines, Thailand, Papua New Guinea, and the Andamans (Ng et al., 2008; Ng and Wowor, 2019; Shy and Ng, 2019; Naruse and Ng, 2020). Geosesarma species are commonly known as vampire crabs due to the bright yellow eyes of some species (Ng et al., 2015). Geosesarma species are amphibious or terrestrial, and they sometimes live far from water (Hartnoll, 1988). As many of the species display abbreviated or no larval development and low fecundity, Geosesarma species often have restricted distributions and a high degree of endemism (Schubart and Ng, 2014). Geosesarma faustum and Geosesarma penangensis are found on Penang Hill, Malaysia, with G. faustum associated with phytotelms and living at altitudes greater than 700 m, whereas G. penangensis at lower altitudes (Ng, 2017). Geosesarma species are small crabs with carapace widths that rarely exceed 10 mm (Hartnoll, 1988). Their small size, high level of endemism, and cryptic nature account for the scarcity of information on the genus.
The animal mitogenomes are circular double-stranded molecules of 15–18 kb in size and generally contain 37 genes such as 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes, 2 ribosomal RNA (12S rRNA and 16S rRNA) genes, and an A + T-rich control region (Boore, 1999). Crustacean mitogenomes exhibit exceptions to this organization with instances of duplications of tRNA gene and rearrangements of the gene order (Segawa and Aotsuka, 2005; Basso et al., 2017). Due to its smaller effective population size and lack of recombination, mitochondrial DNA (mtDNA) is more strongly influenced by evolutionary processes than nuclear DNA and has thus been used to analyze genetic diversity and adaptive evolution (Shen et al., 2019; Peng et al., 2021). The high level of endemism exhibited by Geosesarma crabs makes them an index for the study of adaptation to the local environment. Nevertheless, genomic and genetic resources are lacking for the genus Geosesarma. Previous studies have assigned Geosesarma to Sesarmidae based on morphology (Ng et al., 2008; Ng, 2017; Ng and Wowor, 2019; Shy and Ng, 2019); nevertheless, no genetic analysis has been performed to show the position of Geosesarma within Sesarmidae. In this study, the complete mitogenomes of G. faustum and G. penangensis were analyzed and compared with other Brachyura mitogenomes. The results illustrate the taxonomic placement of Geosesarma within Brachyura, gene arrangement of Geosesarma, and adaptive evolution in mitochondrial genes related to adaptation to hill habitat.
Materials and Methods
Sample Collection and Mitogenome Sequencing
Adult specimens of G. faustum and G. penangensis were collected from Penang Hill, Malaysia (5.4085° N, 100.2773° E) in April 2021. Voucher specimens of G. faustum and G. penangensis were deposited at the Zoological Reference Museum Collection, Universiti Sains Malaysia. Total DNA was extracted from muscle tissues of three individuals in total for each species using the DNeasy Blood and Tissue Kit (QIAGEN, Germany) according to the instructions of the manufacturer and pooled together. DNA quality was assessed using gel electrophoresis, and DNA concentration was quantified using the dsDNA HS Assay Kit on the Qubit Fluorometer (Thermo Fisher, United States). Sequencing libraries were constructed using the TruSeq PCR-Free DNA Library Kit (Illumina, United States) and sequenced on an Illumina NovaSeq 6000 platform. Paired-end 150-bp sequencing yielded approximately 5.8 Gb data for each species.
Mitogenome Assembly, Annotation, and Comparative Analyses
Raw reads were assembled by baiting and iterative mapping to the reference mitogenome sequence of Sesarmops sinensis (NC_030196) using MITObim (v1.9.1) (Hahn et al., 2013) and MIRA (v4.0.2). Annotations were performed with the MITOS pipeline (Bernt et al., 2013) using the invertebrate mitochondrial genetic code. The tRNA genes were annotated, and their secondary structures were inferred using the MiTFi module in the MITOS. Secondary structures of the predicted tRNAs were visualized on the Forna web server. Tandem Repeats Finder was used to detect repeat sequences in the control region, and their secondary structures were predicted with Mfold software. The base composition and the relative synonymous codon usage were determined using MEGA X. The skew in the nucleotide composition was calculated according to the formulas as follows: AT-skew = (A–T)/(A + T) and GC-skew = (G–C)/(G + C). Circular mitogenomes representations were drawn using CGView online server. The ratio of non-synonymous to synonymous substitutions (ω = dN/dS) was estimated using the codon-based maximum likelihood (ML) method in PAML (v4.9) (Yang and Nielsen, 2000). The branch-site test was applied with G. faustum or G. penangensis as the foreground branch and the rest of the Sesarmidae as the background branch. A likelihood ratio test was used to compare model A against the null model A. The posterior probability of the positively selected sites was computed by the Bayes Empirical Bayes method.
Phylogenetic Analyses
The phylogenetic analyses were performed using 13 PCG sequences from 76 species covering representative Brachyura families, and Clibanarius infraspinatus was used as the out-group (Supplementary Table 1). The nucleotide sequences of the PCGs were aligned using MAFFT (v7.313) in auto mode and then concatenated. The ambiguous sequences or poorly aligned regions in the alignment were removed using Gblocks (v0.91b) with the default setting. Potential saturation in the nucleic acid sequences was assessed using DAMBE (v7.3.5). None of the tests yielded the index of substitution saturation (Iss) greater than the critical one (Iss < Iss.cSym or Iss.cAsym) except for the third codon position in asymmetrical topology (Iss > Iss.cAsym) (Supplementary Table 2). The best-fitting evolutionary model was determined using ModelFinder (v2.1.1) based on the Akaike Information Criterion. The ML tree was built in IQ-Tree (v1.6.8) (Nguyen et al., 2015) with 1,000 bootstrap replicates and the GTR + F + I + G4 model. The Bayesian inference (BI) was performed with MrBayes (v3.2.7) (Ronquist et al., 2012) using the GTR + I + G model. Markov Chain Monte Carlo was run for 10 million generations with four independent chains. Sampling was performed every 1,000 generations, and the initial 25% of the generations were discarded as burn-in. The convergence of the runs was assessed using Tracer (v1.7.2). The phylogenetic trees were visualized using FigTree (v1.4.4) software1.
Results and Discussion
Genome Structure, Organization, and Composition
In this study, the complete mitochondrial genome sequences of the vampire crabs G. faustum and G. penangensis were determined. The mitogenomes of G. faustum and G. penangensis are 15,880- and 15,955-bp long (Figure 1A), respectively, which fall within the range of other Sesarmidae mitogenomes from 15,611 bp (Parasesarma pictum) to 15,920 bp (Chiromantes neglectum) (Supplementary Table 3). The mitogenomes follow a typical metazoan mtDNA structure comprising 37 genes (13 PCGs, 22 tRNAs, and 2 RNAs) and a major non-coding region known as the control region (Supplementary Table 4). As a result of the compactness of mitogenomes, there are a total of 45- to 71-bp overlap between genes, and the longest overlap occurs between trnL1 and large rRNA (rrnL) (25–55 bp). The mitogenomes also contain 331- to 372-bp intergenic sequences distributed in 20 regions ranging from 1 to 187 bp in size. The whole mitogenomes of Geosesarma are biased toward AT nucleotides (78.44–78.49%) (Supplementary Table 5), showing significant strand asymmetry in a similar manner to other Sesarmidae mitogenomes. The overall AT- and GC-skews for the mitogenomes were also negative, indicating higher occurrences of T than A and C than G. The AT- and GC-skews for all sequenced Sesarmidae mitogenomes were calculated and compared (Supplementary Table 3). The GC-skews for all Sesarmidae are negative, ranging from –0.194 (P. pictum) to –0.232 (Metopaulias depressus), and the AT-skews are weakly negative, ranging from –0.010 (Chiromantes dehaani and C. neglectum) to –0.032 (P. pictum).
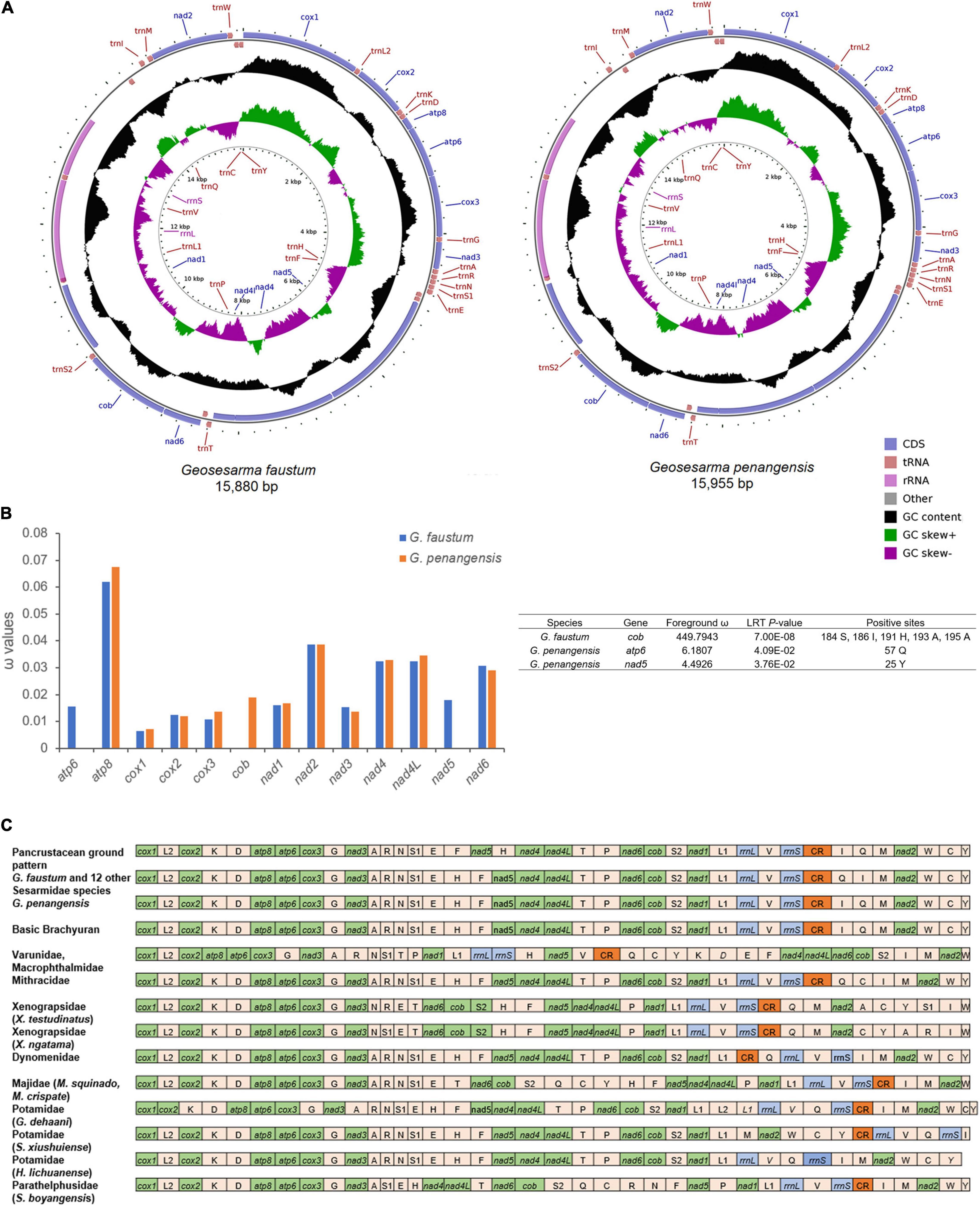
Figure 1. The complete mitochondrial genomes of Geosesarma faustum and Geosesarma penangensis. (A) Circular map of the G. faustum and G. penangensis mitogenomes. (B) Non-synonymous/synonymous substitution (ω) ratios for G. faustum and G. penangensis mitochondrial protein-coding genes (PCGs) and possible sites under positive selection (p-value < 0.05). The ω values for the cob of G. faustum, nad5, and atp6 of G. penangensis not plotted in the chart are shown in the table. Only posterior probability > 0.95 sites is shown. (C) The linear representation of the mitochondrial gene arrangement in Brachyura. Cox1 has been designated the start point for all mitogenomes.
The mitogenomes of G. faustum and G. penangensis have 13 typical PCGs found in brachyuran species consisting of seven NADH dehydrogenases (nad1-6 and nad4L), two ATPases (atp6 and atp8), three cytochrome c oxidases (cox1–3), and one cytochrome b (cob), which account for 11,178–11,181 bp in length. Nine PCGs (i.e., cox1, cox2, atp8, atp6, cox3, nad3, nad6, cob, and nad2) are encoded on the majority strand and the remainings (nad5, nad4, nad4L, and nad1) on the minority strand. While cox1, cox2, atp8, cox3, nad4, and nad4L initiated by the standard ATG codon, atp6, nad3, nad5, nad6, cob, and nad1 (G. penangensis) used ATA, and nad6 (G. faustum) used GTG. Most PCGs, i.e., atp8, atp6, cox3, nad3, nad5, nad4, nad6, nad1, and cox1 (G. penangensis), were terminated by TAA, whereas nad2 used TAG as stop codon, and cox2, cob, and cox1 (G. faustum) terminated by a single T. These incomplete stop codons have been observed in many brachyuran mitogenomes (Wang et al., 2019; Zhang Y. et al., 2020) and are thought to be completed by posttranscriptional polyadenylation (Ojala et al., 1981).
The relative synonymous codon usage for 13 PCGs was analyzed (Supplementary Figure 1). Both the mitogenomes of G. faustum and G. penangensis consist of 3,716 codons (Supplementary Figure 2). In the 13 PCGs, the most frequently utilized amino acids were Leu (12.94%), Ile (10.09–10.15%), and Phe (9.58–9.85%). The usages of both the twofold and fourfold degenerate codons are biased toward codons abundant in A or T. This amino acid bias trend is consistent with the higher representation of nucleotides A and T in the whole mitogenome.
Similar to other Brachyura mtDNAs, G. faustum and G. penangensis mitogenomes contain a complete set of 22 tRNA genes ranging in size from 63 bp (trnF) to 73 bp (trnV). The majority of tRNAs fold into a typical cloverleaf secondary structure, although trnD, trnH, and trnR (G. faustum) have truncated dihydrouridine (DHU) loop compared with normal tRNA (Supplementary Figure 3). The trnS also lacks the thymidine-pseudouridine-cytidine (TΨC) stem and loop, whereas trnC (G. penangensis) has reduced the DHU stem. Aberrant tRNA structures in mitogenomes appear to be a common phenomenon in many species (Li et al., 2020; Wang Z. et al., 2020); previous research on nematodes and metazoans suggests that such features have no effect on tRNA function (Watanabe et al., 2014; Fourdrilis et al., 2018).
The rrnL and small rRNA (rrnS) genes of G. faustum and G. penangensis are 1,364–1,394 bp and 825–844 bp in length, respectively, which both are located between trnL1 and control region, and separated by trnV. The control region located between rrnS and trnQ spans 712–785 bp and has functional importance related to the initiation of replication and transcription of the mitogenome (Clayton, 1992). This region exhibits high A + T content (82.56–83.69%) and contains features such as tandem repeat, microsatellite sequence (TA)n, and stem-loop structure (Supplementary Figure 4).
Selection Analyses
To analyze the selective pressure on G. faustum and G. penangensis compared with other Sesarmidae species, the non-synonymous/synonymous substitution ratios of the mitochondrial PCGs were estimated under the branch-site test (Figure 1B). The analysis detected signals of positive selection in cob of G. faustum, as well as nad5 and atp6 of G. penangensis at seven codon sites. The different positively selected sites detected in the two Geosesarma species could be attributed to differences in the mechanisms used by crabs endemic to different locations. Previous studies have found evidence for positive selection in mitochondrial genes that may play a role in adaptive evolution in organisms inhabiting extreme environments or with requirements for extreme metabolic demands (Li et al., 2018; Sun et al., 2018; Shen et al., 2019). In Chinese snub-nosed monkeys and galliform birds living in high-altitude environments, the selections on mitochondrial genes are driven by the requirements for higher energy demand and oxygen metabolism (Yu et al., 2011; Zhou et al., 2014). In brachyurans, a positive selection of mitochondrial genes has only been reported in a deep-sea representative (Zhang B. et al., 2020). We showed here, for the first time, the occurrence of positive selection detected in a land crab, which suggests a relationship between altered metabolic requirements and high-altitude colonization. Terrestrial brachyurans display morphological, behavioral, and physiological adaptations to sustain prolonged periods from the water source (Lim and Goh, 2021). Among these, osmotic and ionic regulation, waste excretion, aerial respiration, and thermoregulation are pivotal changes (Watson-Zink, 2021). The positive selection of PGCs in the two Geosesarma species showed the importance of adaptation in respiratory physiology for survival at higher altitudes. The differences in positive selection sites and genes between G. faustum and G. penangensis, which occupy different altitude niches, warrant further investigation.
Gene Rearrangement
Compared to the hypothetical ancestral pancrustacean gene order (Boore et al., 1998), it is evident that the Geosesarma experienced mitochondrial gene rearrangement (Figure 1C). Typically, trnH located between nad4 and nad5 in the pancrustacean ground pattern had been shifted into the trnE and trnF junction in the Geosesarma mitogenomes. This translocation is a relatively common event in the brachyuran mitogenomes (Chen et al., 2019; Wang Q. et al., 2020). In the most common brachyuran gene order, the tRNA gene arrangement between the control region and nad2 is trnI-trnQ-trnM. Nevertheless, G. faustum and all Sesarmidae mitogenomes (except G. penangensis) shared a transposition of trnQ that resulted in the trnQ-trnI-trnM order. The tandem duplication-random loss model is one of the most widely used mechanisms to explain mitochondrial gene arrangement (Moritz and Brown, 1987; Macey et al., 1997). In this case, tandem duplication in the trnF-nad5-trnH region, followed by loss of supernumerary genes, is the most likely process for forming trnH-trnF-nad5 gene order. Meanwhile, the trnQ-trnI-trnM arrangement could also be caused by slipped-strand mispairing followed by gene deletion. Despite belonging to the Geosesarma genus, G. penangensis differs from G. faustum and the rest of the Sesarmidae by showing trnI-trnQ-trnM. This feature raises the possibility of a secondary independent reversal to the plesiomorphic brachyuran gene arrangement. Such reversion is extremely rare; endemism in Geosesarma species may have promoted the evolutionary transition. Brachyura families such as Varunidae, Macropthalmidae, Xenograpsidae, Majidae, Potamidae, and Parathelphusidae also showed signatures of gene arrangement with respect to the ancestral pancrustacean pattern.
Phylogenetic Analyses
The phylogenetic relationships were analyzed based on the nucleotide sequences of 13 PCGs from 76 Brachyura species and one out-group. ML and BI analyses resulted in tree topologies that were largely congruent (Figure 2 and Supplementary Figure 5). In this study, G. faustum and G. penangensisclustered in one branch with high support values, indicating that the two have a sister group relationship. This is the first phylogenomic analysis that placed Geosesarma as the sister group to the [(C. dehaani + C. neglectum) + (Chiromantes haematocheir + S. sinensis)] clade. This result confirmed that Geosesarma belongs to Grapsoidea, Sesarmidae, congruent with the morphological assignments (Ng et al., 2008; Ng, 2017; Ng and Wowor, 2019; Shy and Ng, 2019). All Sesarmidae species cluster into a clade consisting of two sister groups, which was consistent with previous findings (Wang et al., 2019; Li et al., 2020; Lu et al., 2020; Wang Z. et al., 2020). Together, ((((C. dehaani + C. neglectum) + (C. haematocheir + S. sinensis)) + (G. faustum + G. penangensis)) + Chiromantes eulimene) and ((((Parasesarma affine + Perisesarma bidens) + Nanosesarma minutum) + (P. pictum + Parasesarma tripectinis)) + (Clistocoeloma sinense + M. depressus)) groups form the polyphyletic Sesarmidae clade.

Figure 2. Phylogenetic tree inferred from the nucleotide sequences of 13 PCGs of the mitogenomes using the maximum-likelihood analysis. Bootstrap values greater than 50 are indicated at nodes, and sequences generated in this study are indicated in bold.
Sesarmidae species are most closely related to Gecacinidae, constituting part of the Grapsoidea group, which is in accordance with the results of the studies by Chen et al. (2019), Wang et al. (2019), Lu et al. (2020), and Zhang Y. et al. (2020). The mitochondrial phylogenomic analyses including our dataset have shown the divergent phylogenetic status of Grapsidae and Dotilidae (Wang et al., 2019; Li et al., 2020; Wang Z. et al., 2020; Zhang Y. et al., 2020). In the ML tree, Grapsidae and Dotilidae form a sister group and constitute the base of the (((((Sesarmidae) + Gecarcinidae) + Xenograpsidae) + Ocypodidae) + (Varunidae + (Macropthalmidae + Mictyridae))) clade. Nevertheless, the BI tree placed Grapsidae and Dotilidae group at the basal to the ((((Sesarmidae) + Gecarcinidae) + Xenograpsidae) + Ocypodidae) clade. While ML tree recovered Dynomenidae + [Homolidae + (Raninidae)] relationship that was similar to those reported in the studies by Lu et al. (2020) and Wang Z. et al. (2020), BI tree exhibited Homolidae + [Dynomenidae + (Raninidae)] topology. The presence of only one or a small number of representatives for certain families could have resulted in unstable phylogenetic relationships.
In this study, the complete mitogenomes of G. faustum and G. penangensis were characterized and compared with that of other Brachyura. The phylogenetic analyses confirmed that Geosesarma belongs to Grapsoidea, Sesarmidae. Evidence of positive selection was detected in Geosesarma mitochondrial genes, highlighting adaptive evolution in crabs for the colonization of terrestrial hill habitat. Unlike G. faustum and all other Sesarmidae mitogenomes, G. penangensis has a trnI-trnQ-trnM gene arrangement, suggesting a reversal to the plesiomorphic brachyuran pattern. Importantly, our study provides genomic resources that are pivotal to understand the biology of endemic Geosesarma crabs.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI (accession: MZ725940, MZ725941, and SRR15514280-1).
Author Contributions
ACS-C and N-SL conceived and designed the experiments. N-SL analyzed the data and wrote the manuscript. K-KS performed the experiments. AA, K-AS, and AWAZ collected and identified the samples. All authors reviewed the manuscript.
Funding
This study was supported by the Universiti Sains Malaysia under the 10 Q1/Q2 Initiatives grant (311.PCCB.411954).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2021.778570/full#supplementary-material
Footnotes
References
Ahyong, S. T., Lai, J. C., Sharkey, D., Colgan, D. J., and Ng, P. K. (2007). Phylogenetics of the brachyuran crabs (Crustacea: Decapoda): the status of Podotremata based on small subunit nuclear ribosomal RNA. Mol. Phylogenet. Evol. 45, 576–586. doi: 10.1016/j.ympev.2007.03.022
Ahyong, S. T., Lowry, J. K., Alonso, M., Bamber, R. N., Boxshall, G. A., and Castro, P. (2011). “Subphylum Crustacea Brünnich, 1772,” in Animal Biodiversity: An Outline of Higher-Level Classification and Survey of Taxonomic Richness, ed. Z.-Q. Zhang (Auckland: Magnolia Press), 165–191. doi: 10.11646/zootaxa.3148.1.33
Basso, A., Babbucci, M., Pauletto, M., Riginella, E., Patarnello, T., and Negrisolo, E. (2017). The highly rearranged mitochondrial genomes of the crabs Maja crispata and Maja squinado (Majidae) and gene order evolution in Brachyura. Sci. Rep. 7:4096. doi: 10.1038/s41598-017-04168-9
Bernt, M., Donath, A., Jühling, F., Externbrink, F., Florentz, C., Fritzsch, G., et al. (2013). MITOS: improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 69, 313–319. doi: 10.1016/j.ympev.2012.08.023
Boore, J. L. (1999). Animal mitochondrial genomes. Nucleic Acids Res. 27, 1767–1780. doi: 10.1093/nar/27.8.1767
Boore, J. L., Lavrov, D. V., and Brown, W. M. (1998). Gene translocation links insects and crustaceans. Nature 392, 667–668. doi: 10.1038/33577
Castro, P., Davie, P. J. F., Guinot, D., Schram, F. R., and Von Vaupe Klei, J. C. (2015). The Crustacea, Complementary to the Volumes Translated From the French of the Traité de Zoologie. Leiden: Brill, 1–1221.
Chen, J., Xing, Y., Yao, W., Xu, X., Zhang, C., Zhang, Z., et al. (2019). Phylomitogenomics reconfirm the phylogenetic position of the genus Metaplax inferred from the two grapsid crabs (Decapoda: Brachyura: Grapsoidea). PLoS One 14:e0210763. doi: 10.1371/journal.pone.0210763.g003
Clayton, D. A. (1992). Transcription and replication of animal mitochondrial DNAs. Int. Rev. Cytol. 141, 217–232. doi: 10.1016/s0074-7696(08)62067-7
Cumberlidge, N., Fenolio, D. B., Walvoord, M. E., and Stout, J. (2005). Tree-climbing crabs (Potamonautidae and Sesarmidae) from phytotelmic microhabitats in rainforest canopy in Madagascar. J. Crustacean Biol. 25, 302–308. doi: 10.1651/c-2532
De Grave, S., Dean, P. N., Shane, A., Tin-Yam, C., Keith, C. A., and Peter, D. C. (2009). A classification of living and fossil genera of decapod crustaceans. Raffles Bull. Zool. 21, 1–109. doi: 10.5479/si.00963801.78-2851.1
De Man, J. G. (1892). “Decapoden des Indischen archipels,” in Zoologische Ergebnisse Einer Reise in Niederlandisch OstIndien, Vol. 2, ed. M. Weber, (Leyden: Leyden Museum), 265–527, pls. 15–29.
de Saint Laurent, M. (1980). Sur la classification et la phylogénie des crustacés décapodes brachyoures. I. Podotremata Guinot, 1977, et Eubrachyura sect. nov. CR Hebd. Séances Acad. Sci.(Paris) Sen. D 290, 1265–1268.
Fourdrilis, S., De Frias Martins, A. M., and Backeljau, T. (2018). Relation between mitochondrial DNA hyperdiversity, mutation rate and mitochondrial genome evolution in Melarhaphe neritoides (Gastropoda: Littorinidae) and other Caenogastropoda. Sci. Rep. 8:17964. doi: 10.1038/s41598-018-36428-7
Hahn, C., Bachmann, L., and Chevreux, B. (2013). Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads–a baiting and iterative mapping approach. Nucleic Acids Res. 41, e129–e129. doi: 10.1093/nar/gkt371
Hartnoll, R. G. (1988). “Evolution, systematics, and geographical distribution,” in Biology of the Land Crab, eds W. W. Burggren and B. R. Mcmahon (New York, NY: Cambridge University Press), 6–54. doi: 10.1017/cbo9780511753428.003
Li, X.-D., Jiang, G.-F., Yan, L.-Y., Li, R., Mu, Y., and Deng, W.-A. (2018). Positive selection drove the adaptation of mitochondrial genes to the demands of flight and high-altitude environments in grasshoppers. Front. Genet. 9:605. doi: 10.3389/fgene.2018.00605
Li, Y.-T., Xin, Z.-Z., Tang, Y.-Y., Yang, T.-T., Tang, B.-P., Sun, Y., et al. (2020). Comparative mitochondrial genome analyses of sesarmid and other brachyuran crabs reveal gene rearrangements and phylogeny. Front. Genet. 11:536640. doi: 10.3389/fgene.2020.536640
Lim, S. S. L., and Goh, S. J. A. (2021). Ecomorphological adaptations of second maxilliped-setation: insights from three species of fiddler crabs from Panama. Zool. Stud. 60:48.
Lu, X., Gong, L., Zhang, Y., Chen, J., Liu, L., Jiang, L., et al. (2020). The complete mitochondrial genome of Calappa bilineata: the first representative from the family Calappidae and its phylogenetic position within Brachyura. Genomics 112, 2516–2523. doi: 10.1016/j.ygeno.2020.02.003
Macey, J. R., Larson, A., Ananjeva, N. B., Fang, Z., and Papenfuss, T. J. (1997). Two novel gene orders and the role of light-strand replication in rearrangement of the vertebrate mitochondrial genome. Mol. Biol. Evol. 14, 91–104. doi: 10.1093/oxfordjournals.molbev.a025706
Moritz, C., and Brown, W. M. (1987). Tandem duplications in animal mitochondrial DNAs: variation in incidence and gene content among lizards. Proc. Natl. Acad. Sci. U.S.A. 84, 7183–7187. doi: 10.1073/pnas.84.20.7183
Naruse, T., and Ng, P. K. L. (2020). Revision of the sesarmid crab genera Labuanium Serène and Soh, 1970, Scandarma Schubart, Liu and Cuesta, 2003 and Namlacium Serène and Soh, 1970 (Crustacea: Decapoda: Brachyura: Sesarmidae), with descriptions of four new genera and two new species. J. Nat. Hist. 54, 445–532. doi: 10.1080/00222933.2020.1763491
Ng, P. K. L. (2017). On the identities of the highland vampire crabs, Geosesarma foxi (Kemp, 1918) and G. serenei Ng, 1986, with description of a new phytotelmic species from Penang, Peninsular Malaysia (Crustacea: Decapoda: Brachyura: Sesarmidae). Raffles Bull. Zool. 65, 226–242.
Ng, P. K. L., Guinot, D., and Davie, P. J. F. (2008). Systema Brachyurorum: part I. An annotated checklist of extant brachyuran crabs of the world. Raffles Bull. Zool. 17, 1–286.
Ng, P. K. L., Schubart, C. D., and Lukhaup, C. (2015). New species of “vampire crabs” (Geosesarma De Man, 1892) from central Java, Indonesia, and the identity of Sesarma (Geosesarma) nodulifera De Man, 1892 (Crustacea, Brachyura, Thoracotremata, Sesarmidae). Raffles Bull. Zool. 63, 3–13.
Ng, P. K. L., and Wowor, D. (2019). The vampire crabs of Java, with descriptions of five new species from Mount Halimun Salak National Park, West Java, Indonesia (Crustacea: Brachyura: Sesarmidae: Geosesarma). Raffles Bull. Zool. 67, 217–246.
Nguyen, L. T., Schmidt, H. A., Von Haeseler, A., and Minh, B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Ojala, D., Montoya, J., and Attardi, G. (1981). tRNA punctuation model of RNA processing in human mitochondria. Nature 290, 470–474. doi: 10.1038/290470a0
Peng, M., Zhu, W., Yang, C., Yao, J., Chen, H., Jiang, W., et al. (2021). Genetic diversity of mitochondrial D-LOOP sequences in the spotted scat (Scatophagus argus) from different geographical populations along the northern coast of the South China Sea. J. Appl. Ichthyol. 37, 73–82. doi: 10.1111/jai.14121
Ronquist, F., Teslenko, M., Van Der Mark, P., Ayres, D. L., Darling, A., Höhna, S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Schubart, C. D., and Ng, P. K. L. (2014). Two new species of land-dwelling crabs of the genus Geosesarma De Man, 1892 (Crustacea: Brachyura: Thoracotremata: Sesarmidae) from Bintan Island, Indonesia. Raffles Bull. Zool. 62, 615–619.
Segawa, R. D., and Aotsuka, T. (2005). The mitochondrial genome of the Japanese freshwater crab, Geothelphusa dehaani (Crustacea: Brachyura): evidence for its evolution via gene duplication. Gene 355, 28–39. doi: 10.1016/j.gene.2005.05.020
Serrano-Sánchez, M. D. L., Guerao, G., Centeno-García, E., and Vega, F. J. (2016). Crabs (Brachyura: Grapsoidea: Sesarmidae) as inclusions in lower miocene amber from Chiapas, Mexico. Boletín de la Soc. Geol. Mex. 68, 37–43. doi: 10.18268/bsgm2016v68n1a6
Shen, H., Braband, A., and Scholtz, G. (2013). Mitogenomic analysis of decapod crustacean phylogeny corroborates traditional views on their relationships. Mol. Phylogenet. Evol. 66, 776–789. doi: 10.1016/j.ympev.2012.11.002
Shen, X., Pu, Z., Chen, X., Murphy, R. W., and Shen, Y. (2019). Convergent evolution of mitochondrial genes in deep-sea fishes. Front. Genet. 10:925. doi: 10.3389/fgene.2019.00925
Shy, J.-Y., and Ng, P. K. L. (2019). Geosesarma mirum, a new species of semi-terrestrial sesarmid crab (Crustacea, Decapoda, Brachyura) from central Taiwan. ZooKeys 858, 1–10. doi: 10.3897/zookeys.858.35198
Smith, T. J., Boto, K. G., Frusher, S. D., and Giddins, R. L. (1991). Keystone species and mangrove forest dynamics: the influence of burrowing by crabs on soil nutrient status and forest productivity. Estuar. Coast. Shelf Sci. 33, 419–432. doi: 10.1016/0272-7714(91)90081-l
Sun, S. E., Hui, M., Wang, M., and Sha, Z. (2018). The complete mitochondrial genome of the alvinocaridid shrimp Shinkaicaris leurokolos (Decapoda, Caridea): insight into the mitochondrial genetic basis of deep-sea hydrothermal vent adaptation in the shrimp. Comp. Biochem. Physiol. Part D Genomics Proteomics 25, 42–52. doi: 10.1016/j.cbd.2017.11.002
Wang, Q., Tang, D., Guo, H., Wang, J., Xu, X., and Wang, Z. (2020). Comparative mitochondrial genomic analysis of Macrophthalmus pacificus and insights into the phylogeny of the Ocypodoidea & Grapsoidea. Genomics 112, 82–91. doi: 10.1016/j.ygeno.2019.12.012
Wang, Z., Shi, X., Guo, H., Tang, D., Bai, Y., and Wang, Z. (2020). Characterization of the complete mitochondrial genome of Uca lacteus and comparison with other Brachyuran crabs. Genomics 112, 10–19. doi: 10.1016/j.ygeno.2019.06.004
Wang, Z., Shi, X., Tao, Y., Wu, Q., Bai, Y., Guo, H., et al. (2019). The complete mitochondrial genome of Parasesarma pictum (Brachyura: Grapsoidea: Sesarmidae) and comparison with other Brachyuran crabs. Genomics 111, 799–807. doi: 10.1016/j.ygeno.2018.05.002
Watanabe, Y., Suematsu, T., and Ohtsuki, T. (2014). Losing the stem-loop structure from metazoan mitochondrial tRNAs and co-evolution of interacting factors. Front. Genet. 5:109. doi: 10.3389/fgene.2014.00109
Watson-Zink, V. M. (2021). Making the grade: physiological adaptations to terrestrial environments in decapod crabs. Arthropod Struct. Dev. 64:101089. doi: 10.1016/j.asd.2021.101089
Yang, Z., and Nielsen, R. (2000). Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol. Biol. Evol. 17, 32–43. doi: 10.1093/oxfordjournals.molbev.a026236
Yu, L., Wang, X., Ting, N., and Zhang, Y. (2011). Mitogenomic analysis of Chinese snub-nosed monkeys: evidence of positive selection in NADH dehydrogenase genes in high-altitude adaptation. Mitochondrion 11, 497–503. doi: 10.1016/j.mito.2011.01.004
Zhang, B., Wu, Y., Wang, X., Jiang, W., Yin, J., and Lin, Q. (2020). Comparative analysis of mitochondrial genome of a deep-sea crab Chaceon granulates reveals positive selection and novel genetic features. J. Oceanol. Limnol. 38, 427–437. doi: 10.1007/s00343-019-8364-x
Zhang, Y., Gong, L., Lu, X., Jiang, L., Liu, B., Liu, L., et al. (2020). Gene rearrangements in the mitochondrial genome of Chiromantes eulimene (Brachyura: Sesarmidae) and phylogenetic implications for Brachyura. Int. J. Biol. Macromol. 162, 704–714.
Keywords: mitochondrial DNA, Sesarmidae, Geosesarma, phylogenetic, gene arrangement
Citation: Lau N-S, Sam K-K, Ahmad AB, Siti K-A, Ahmad Zafir AW and Shu-Chien AC (2021) Gene Arrangement and Adaptive Evolution in the Mitochondrial Genomes of Terrestrial Sesarmid Crabs Geosesarma faustum and Geosesarma penangensis. Front. Ecol. Evol. 9:778570. doi: 10.3389/fevo.2021.778570
Received: 17 September 2021; Accepted: 04 November 2021;
Published: 30 November 2021.
Edited by:
Cristina Grande, Autonomous University of Madrid, SpainReviewed by:
Benny K. K. Chan, Academia Sinica, TaiwanMarta Álvarez-Presas, University of Bristol, United Kingdom
Copyright © 2021 Lau, Sam, Ahmad, Siti, Ahmad Zafir and Shu-Chien. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alexander Chong Shu-Chien, alex@usm.my
 Nyok-Sean Lau
Nyok-Sean Lau