
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Ecol. Evol., 26 November 2020
Sec. Phylogenetics, Phylogenomics, and Systematics
Volume 8 - 2020 | https://doi.org/10.3389/fevo.2020.602190
This article is part of the Research TopicBiology, Systematics, Taxonomy, and Evolution of Insect VectorsView all 11 articles
The identification of vector species and their natural infection with arboviruses results in important data for the control of their transmission. However, for the eastern region of Colombia, this information is limited. Therefore, this study morphologically and molecularly identified species of the genus Aedes and the detection of arboviruses (Dengue, Chikungunya, Zika, and Mayaro) in female mosquitoes (individually) present in three municipalities (Saravena, Arauquita, and Tame) by amplifying the genetic material using RT-PCR (reverse transcriptase polymerase chain reaction) in the department of Arauca, eastern Colombia. Inconsistencies between morphological and molecular identification were detected in 13 individuals with Aedes albopictus initially determined as Aedes aegypti based on morphology (n = 13). Molecular identification showed the simultaneous presence of A. aegypti (n = 111) and A. albopictus (n = 58) in the urban municipalities of Saravena and Arauquita. These individuals were naturally infected with Dengue virus type 1 (DENV-1) and Chikungunya virus (CHIKV). The most frequent arbovirus was DENV-1 with an infection rate of 40.7% (11/27) for A. aegypti and 39.7% (23/58) for A. albopictus, which was followed by CHIKV with an infection rate of 1.8% for A. aegypti (2/111) and 6.9% for A. albopictus (4/58). Additionally, a mixed infection of DENV-1 and CHIKV was obtained in 4.5% of A. aegypti (5/111). Zika virus (ZIKV) and Mayaro virus (MAYV) infections were not detected. This study found that barcoding (fragment gene COI) is a successful method for identifying Aedes species. Additionally, we recommend the individual processing of insects as a more accurate strategy for arboviruses detection since the infection rate is obtained and co-infection between DENV-1 and CHIKV is also possible.
Arboviruses (arthropod-borne diseases) have a major impact on public health in Latin America (Shope and Meegan, 1997; Espinal et al., 2019). DENV, ZIKV, and CHIKV have similar epidemiological cycles and result in epidemics approximately every 3 years (Villar et al., 2015). DENV has the highest incidence due to the interaction of different factors such as socioeconomic conditions, arbovirus control plans and the circulation of four serotypes in Latin America where Dengue virus type 1 (DENV-1) has the highest prevalence (Azevedo et al., 2009; de la Cruz et al., 2019; Liu et al., 2020).
Individuals of the Aedes genus are found in areas with high arbovirus rates, which is why it is associated with an infection cycle in urban areas (Beaty et al., 2016). Within the Aedes genus, there are approximately 190 species; however, Aedes aegypti and Aedes albopictus are important for their vector capacity of CHIKV, DENV, ZIKV, and MAYV (Freitas, 2010; Ramasamy et al., 2011; Vega-Rúa et al., 2014; Lounibos and Kramer, 2016). The morphological identification of these two species is complex where individuals are severely damaged, or their external morphological characteristics are altered such as the pattern in the scutum (mesothorax sclerite) (Patsoula et al., 2006; Sumruayphol et al., 2016). Due to these difficulties, different authors mention that the identification of insects based on deoxyribonucleic acid (DNA) can become a more efficient technique when classifying individuals (Rolo, 2020). For this reason, the concept of the DNA barcoding emerges as a new tool for species discrimination. This tool uses a small fragment of DNA which functions as a specific barcode for each species. In insects, this fragment corresponds to a sequence of ∼ 650 base pairs (bp) located at the 5′ end of the cytochrome c oxidase subunit I gene (COI) (Joyce et al., 2018). Therefore, it is important to conduct molecular identification of species using barcoding to accurately identify individuals. The precise identification of A. aegypti and A. albopictus is relevant because this information defines characteristics for these species such as their biology and vector capacity.
The positive detection of arbovirus and the frequency of infection in vectors is reported as the minimum infection rate (MIR), which is defined as the relationship between positive pools and the total number per 1,000 individuals (Gu et al., 2004). However, there are no studies with methodologies that detect natural infection for arboviruses in single insects. In Latin America, A. aegypti has higher MIR values for CHIKV, DENV, ZIKV, and MAYV compared to A. albopictus. Furthermore, no CHIKV and MAYV infection has been detected in the latter. In A. aegypti, DENV presents a higher range of MIR compared to other viruses and ranges from 0.2 to 26 infected individuals per 1,000 individuals. Alternatively, ZIKV has a MIR value ranging from 3 to 17 individuals and CHIKV ranges from 2 to 3 individuals. Finally, for MAYV, the only reported value is 1.75 (Diallo et al., 2014; Guerbois et al., 2016; Huerta et al., 2017; Cevallos et al., 2018; Eiras et al., 2018; Garcia-Rejon et al., 2018). These data show that A. aegypti presents a higher rate of natural infection by arboviruses compared to A. albopictus and this is attributed to the greater presence of this species in urban areas, which indicates that A. aegypti is the main vector of arboviruses and A. albopictus is the secondary vector in Latin America (Black et al., 2002; Paupy et al., 2009).
Focusing on both the frequency of the viral infection and the incriminated vector species in endemic areas is important to propose reliable prevention and control measures. Also, there is a pivotal need to understand infection rates not only on humans but also on the arbovirus vectors to reveal disease transmission dynamics. The eastern departments of Colombia present the highest incidence of DENV, CHIKV, and ZIKV. Additionally, due to its close proximity to Venezuela, attempting to identify MAYV is worthwhile (Lorenz et al., 2019). Therefore, this study morphologically and molecularly identifies Aedes species circulating in three municipalities of the department of Arauca (Colombia) and obtained sequences that were used to assess the precision of the molecular test used to identify A. aegypti and A. albopictus. In addition, the infection by DENV-1, CHIKV, ZIKV, and MAYV was attempted.
The insects were captured in the department of Arauca in the municipalities of Saravena (6° 57′17″ N, 71° 52′36″ W), Arauquita (7° 01′34″ N, 71° 25′38″ W), and Tame (6° 27′30″ N, 71° 44′41″ W) (Figure 1). These sites were chosen due to the high incidence of arboviral cases. The collection was conducted in the urban area of three municipalities; however, in the municipality of Saravena, individuals were also captured in rural areas, because in this municipality there is not a defined urban area, thus in the rural area there are different farms that have potential breeding sites. Two samplings were conducted, one in 2018 and the other in 2019, and each BG-Mosquitaire was installed for three consecutive months during the rainy season (April–May–June). Six traps were installed in the municipality of Saravena, 5 in Tame and 4 in Arauquita. In each trap, only adult females of the genus Aedes were chosen, and subsequently, the pools were formed using only individuals belonging to the same species and according to their morphological identification. The insects or pools were stored in Invitrogen® RNAlater (Salehi and Najafi, 2014) in jars marked with the coordinates and information regarding the house and/or the collection site and transported to the microbiology laboratory of the Universidad del Rosario. Once in the laboratory, they were individually separated in Eppendorf tubes for molecular identification of species, and in total, 169 specimens were analyzed.
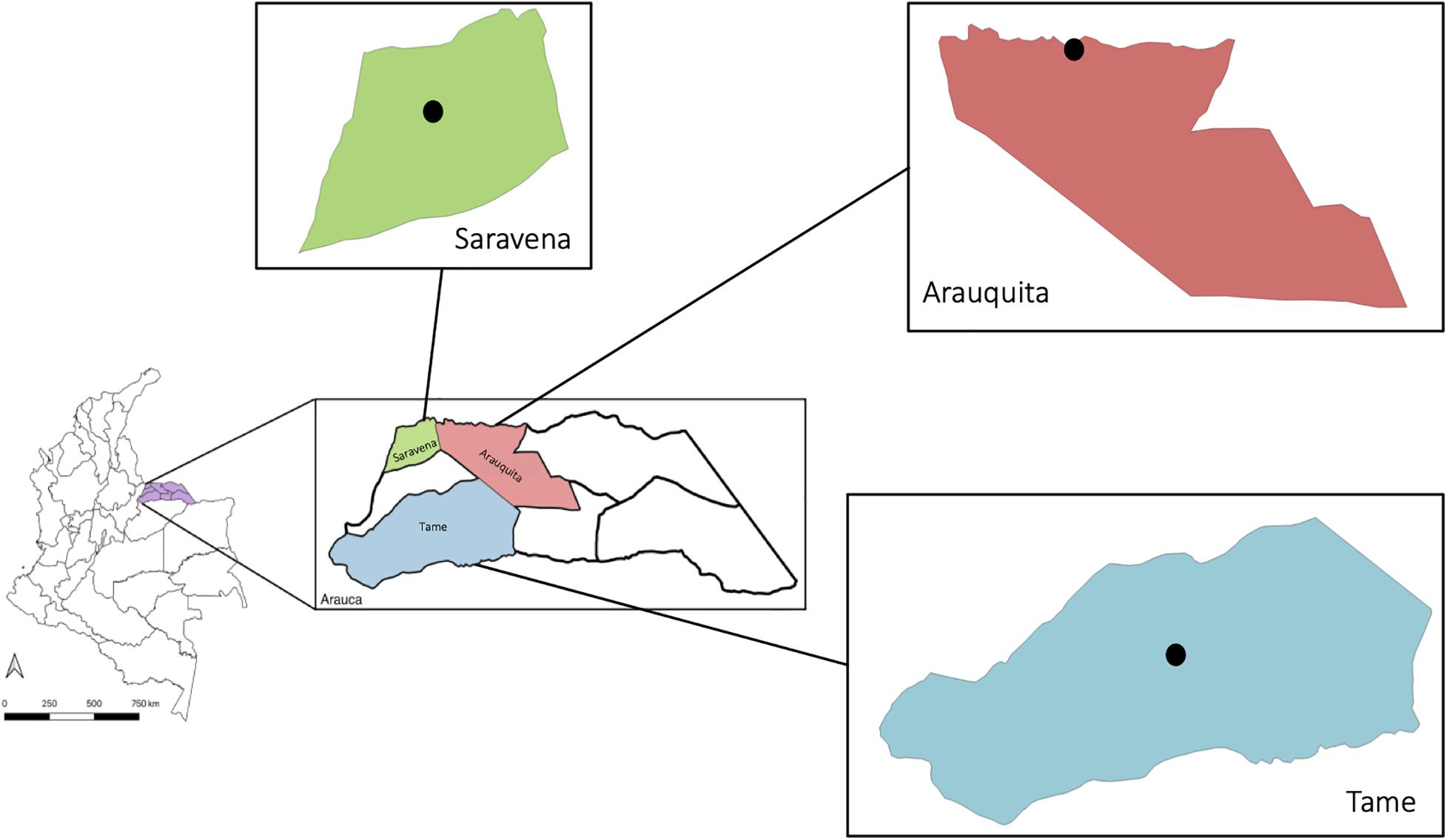
Figure 1. Municipalities where captures were conducted: Saravena (green), Arauquita (pink), and Tame (blue) in the department of Arauca (purple), Colombia. The black dot indicates the urban area where the capture of individuals was made.
The morphological identification of Aedes genus individuals was performed using a stereomicroscope following the taxonomic key proposed by Forattini (1965) to identify insects to the genus level, and the taxonomic review by Cova-Garcia et al. (1966) was used to identify species. Molecular identification of individual Aedes species was conducted by amplifying a fragment of the COI gene (barcoding) from the RNA of the insect. For technical reasons, only the RNA of the insect could be obtained.
RNA extraction was conducted with the Quick-RNATissue/Insect Microprep Kit (Zymo, # R2030) following the manufacturer’s recommended instructions, and the RNA concentration was quantified in a NanoDrop (Thermo ScientificTM). Subsequently, One-Step RT-PCR was conducted from the RNA to generate cDNA (complementary DNA) and amplify the COI gene. PCR reactions were performed in a final volume of 20 μL containing 10 μL qScriptTM One-Step, Low ROXTM (Quantabio, Carlsbad, CA, United States), 10 μM of each primer and 5 μL of RNA. The primers LCO1490 (5′-GGT CAA ATC ATA AAG ATA TTG G-3) and HCO2198 (5′-TAA ACT TCA GGG TGA CCA AAA AAT CA-3′) were also used (Joyce et al., 2018). The thermal profile consisted of the following: one cycle at 50°C for reverse transcriptase activation for 10 min followed by initial denaturation at 95°C for 1 min, 45 cycles of 94°C for 10 s, 60°C for 1 min and final extension at 72°C for 10 min.
The amplification of the gene fragment was conducted using 2.5% agarose gel electrophoresis in TBE 1x buffer and SYBR® Safe (Invitrogen®, Carlsbad, CA, United States) as the intercalating agent and then visualized under UV light to observe a band of ∼650 base pairs (bp). Subsequently, the samples were sequenced by the Sanger method if the band corresponding to the fragment of the COI gene was observed.
Sequence cleaning and alignment was performed in Seqman software (Lasergene) and the nucleotide sequence was compared to the Nucleotide Blast database1; the species were assigned considering the percentage of identity, the coverage value and e-value. The sequences were aligned using the MUSCLE algorithm in the MEGA X v10.1 software (Sohpal et al., 2010). From the alignments, a network of haplotypes was generated in PorpArt v1.7 software using the TCS algorithm (Clement et al., 2002), and haplotype diversity (h) was calculated in DNAsp v6 (Librado and Rozas, 2009). A Maximum Likelihood (ML) tree was built in the software IQtree (Nguyen et al., 2014) using the Jukes–Cantor substitution model, and the support of the nodes was established from 5,000 bootstrap replicates. The external group for the phylogenetic tree was Culex spinosus (KM593059.1).
The detection of each arbovirus (DENV-1/CHKV/ZIKV/MAYV) was conducted on the RNA collected by qRT-PCR One Step using the corresponding primers and Taqman probes (Table 1). Since Endogenous Flaviviral Elements have been identified in the mosquito genome (Houé et al., 2019), the chosen primers have an analytical validation to avoid cross reactions. The qRT-PCR reactions for DENV-1, CHIKV, and ZIKV (per sample) were performed in a final volume of 20 μL containing 10 μL qScriptTM One-Step, Low ROXTM (Quantabio, Carlsbad, CA, United States), 10 μM of each primer, 10 μM of the probe and 5 μL of RNA. The protocol recommended by the WHO (World Health Organization [WHO], 2020) was used for the MAYV detection. The thermal profile used for all arboviruses consisted of the following: one cycle for 50°C for reverse transcriptase activation for 10 min, followed by initial denaturation at 95°C for 1 min, 40 cycles of 95°C for 15 s, 60°C for 1 min and final extension at 72°C for 10 min. The viral RNA from each arbovirus was used as a positive control and was provided by the virology group of the National Institute of Health and the confirmation of positive tests was performed in duplicate. Finally, the infection frequency for each arbovirus species was expressed as the infection rate (infected individual/total individuals).
Descriptive analyses of the frequency of each species and natural infection of each arbovirus were performed based on percentages. In addition, the kappa index was calculated to identify the percentage of agreement between the identification of A. aegypti and A. albopictus species by taxonomic code and molecular marker. A logistic regression, including the intercept, was performed to identify the statistical associations between arbovirus infection as a composite variable (y) and the explanatory variables: vector species, municipality and collection area. The species variable and collection area were transformed into dummies (1/0) using A. albopictus and the rural area as references (0). Preliminarily, a main effects model was run to identify variables potentially associated with the established outcome and a p-value < 0.2 was significant. Subsequently, a step-by-step, second-level interaction model was executed, which included interactions between vector species and the collection area or municipality.
In the two sampling periods, 169 specimens of Aedes were captured with 68% (n = 115) collected in the municipality of Saravena, 15.4% (n = 26) in Arauquita and 16.6% (n = 28) in Tame. Overall, differences were observed in the number of individuals identified as A. aegypti and A. albopictus between molecular and morphological identification methods (Figure 2). A kappa index of 0.764 and agreement of 91.1% (n = 154) were obtained between species identification methods (taxonomic key vs. molecular) (Cook, 2005). Inconsistencies were mainly evident in the identification of 13 individuals (7.7%), where they were initially identified through morphology as A. aegypti while the molecular marker analysis indicated that they were A. albopictus.
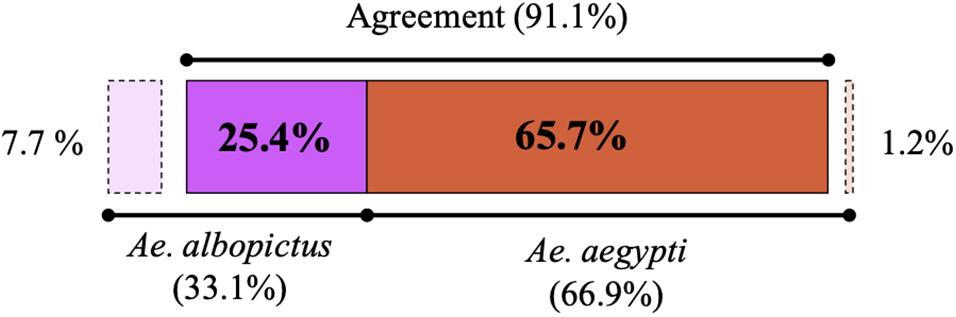
Figure 2. Percentage of A. albopictus (purple) and A. aegypti (orange) individuals identified by morphological and molecular testing. The dotted lines mark the inconsistencies where individuals were initially assigned by morphology to a different species compared to the one detected by molecular methods, and the latter is the technique by which individuals were ultimately assigned for subsequent sequence analysis.
A 338 bp fragment of the COI gene was obtained. In the haplotype network constructed from the sequences of the individuals of the genus Aedes, 19 haplotypes were found (Figure 3). We observed two groups that corresponded to the species A. aegypti (Orange) and A. albopictus (Purple) and are differentiated by 36 bp (Figure 3A). Additionally, the highest number of haplotypes of the two species was found in the municipality of Saravena (Azul) with 8 haplotypes of the species A. aegypti and 9 haplotypes of the species A. albopictus (Figure 3B). The diversity of haplotypes found in Saravena was h = 0.806 and in Arauquita h = 0.498, and only one haplotype was found in Tame. Furthermore, the global diversity of haplotypes for all individuals captured in Arauca was h = 0.639.
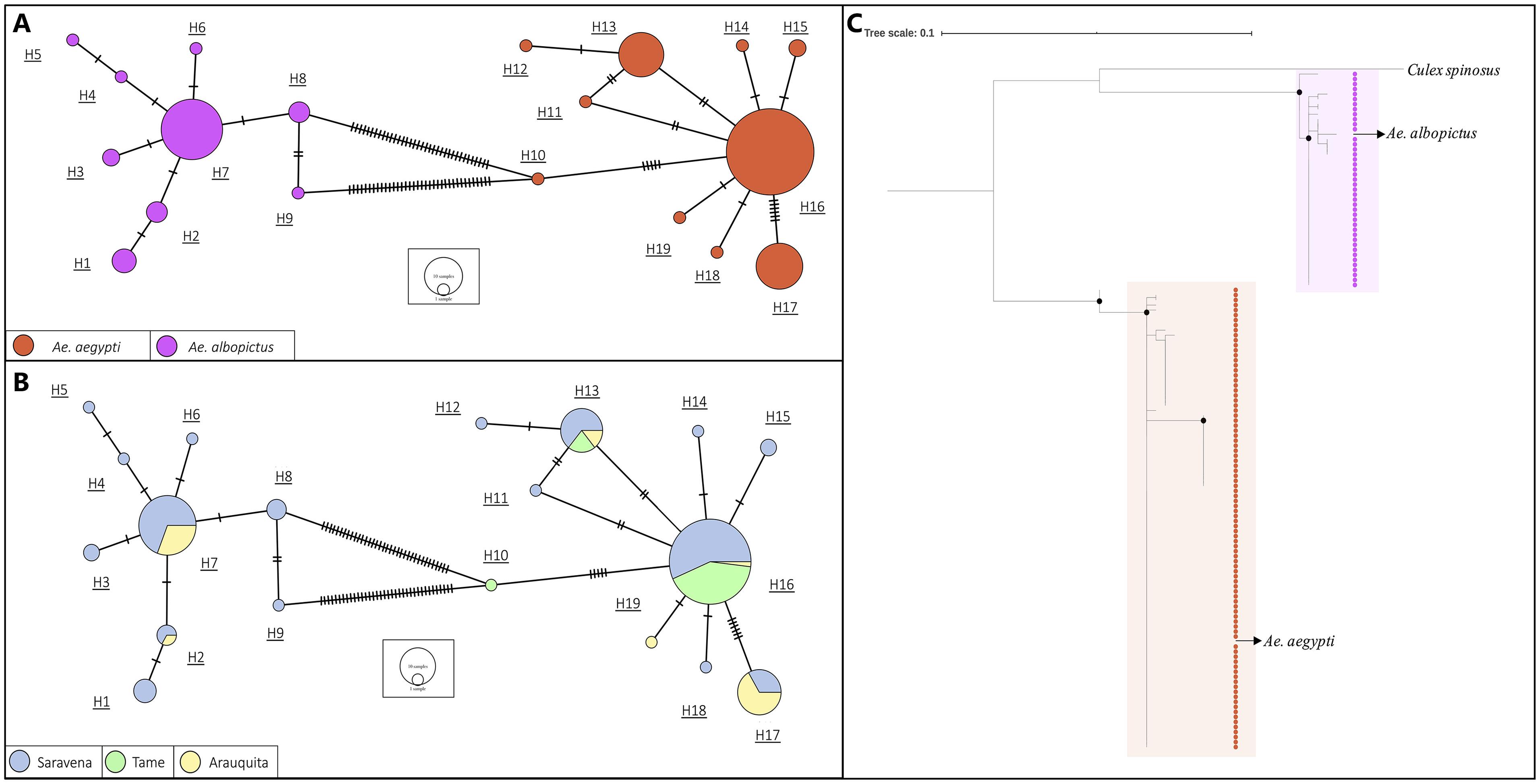
Figure 3. The haplotype network based on sequences of A. aegypti and A. albopictus collected in municipalities of the department of Arauca and colored by (A) species and by (B) collection municipality. The lines in each branch indicate the number of mutational steps between haplotypes, and the size of the circle is proportional to the number of individuals. (C) A maximum likelihood tree based on the JC model of A. aegypti and A. albopictus indivuduals from each place where they were collected. Black dots indicate support bootstrap ≥ 90.
The topology of the phylogenetic tree obtained from the COI sequence alignment revealed two well-supported clusters (bootstrap ≥ 90) that correspond to A. aegypti and A. albopictus (Figure 3C) with a genetic distance of 0.107 between the species. Additionally, the genus Culex (included as an outgroup) formed an independent clade in the species of genus Aedes.
For the results of the geographic distribution of individuals and their natural infection by arboviruses, molecular identification of the species was considered. Table 2 shows the information regarding the frequency of A. aegypti individuals (n = 111) and A. albopictus (n = 58) by area and the municipality of capture. The species A. aegypti was present in the three municipalities and only in urban areas (Figure 4A) and there was a greater frequency of this species in the municipality of Saravena at 64.9% (72/111). However, A. albopictus, which is only present in the municipalities of Saravena and Arauquita in both urban and rural areas (Figure 4A), showed a frequency of 74.1% (43/58) in the municipality of Arauquita. The above results show clear evidence of the presence of these two species in the urban area of the municipality of Saravena and Arauquita.
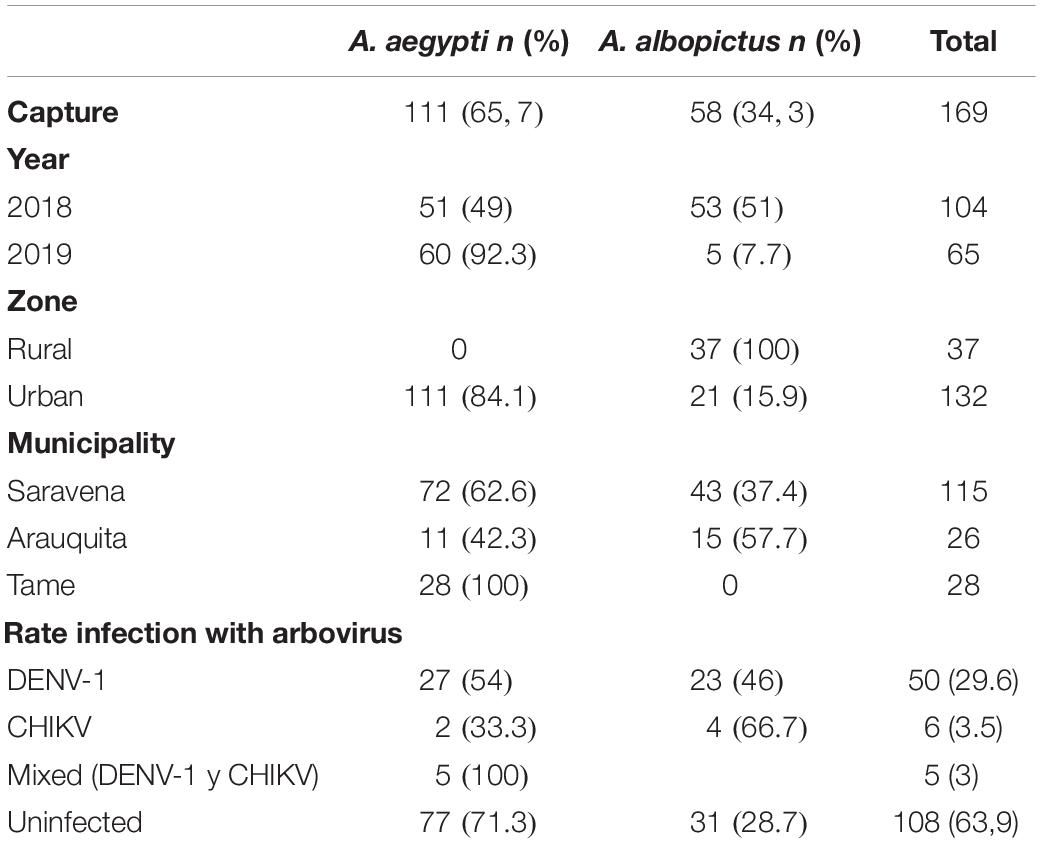
Table 2. Number and percentage of A. aegypti and A. albopictus captured per year, area, municipality and infected with arboviruses.
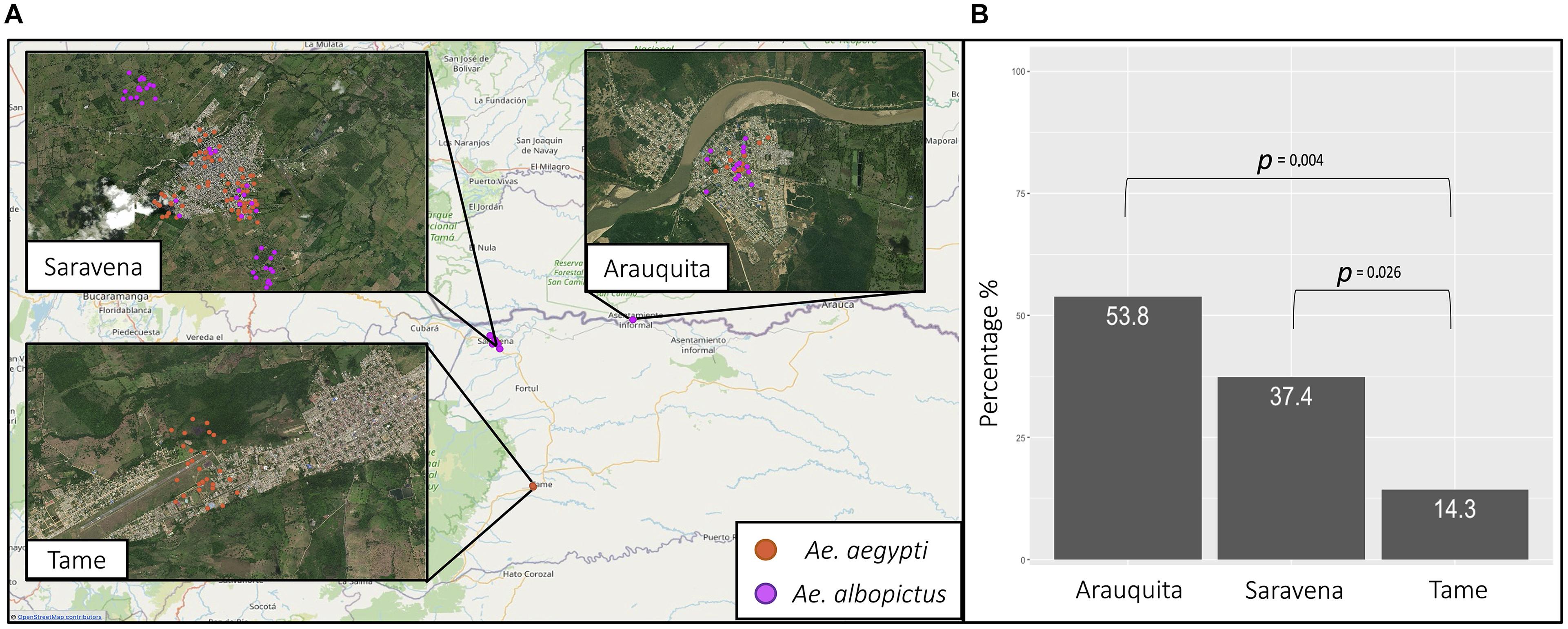
Figure 4. (A) The geographic location of A. aegypti (orange) and A. albopictus (purple) collected from the municipalities of Saravena, Arauquita, and Tame. (B) The percentage of individuals infected by the municipalities of Arauquita, Saravena, and Tame.
Of the 169 individuals collected from the genus Aedes, the overall rate of arboviruses (DENV-1 and CHIKV) infection was 36.1% (61/169). The arbovirus with the highest frequency was DENV-1, which was found in 82% (50/61) of infected individuals, while 9.8% (6/61) were infected by CHIKV and none of the individuals were infected with ZIKV or MAYV. In addition, a 4.5% frequency (5/111) of mixed infection by arboviruses DENV-1 and CHIKV was found in A. aegypti. Table 2 shows the arbovirus infection rates in each species, and there was a higher infection rate observed in individuals of the species A. albopictus. In Saravena was found 18 individuals of A. aegypti and 17 of A. albopictus infected by DENV-1, while for CHIKV one individual of A. aegypti and two individuals of A. albopictus were found. Only 5 individuals of A. aegypti were found coinfected by DENV-1 and CHIKV. In Arauquita were found 5 individuals of A. aegypti and 6 individuals of A. albopictus infected by DENV-1, while for CHIKV one individual of A. aegypti and two individuals of A. albopictus were found in Tame, only 4 individuals of A. aegypti were found infected for DENV-1. The logistic regression results showed that the municipality of Arauquita is 7 times more likely to find infected individuals (OR = 7; 95% CI = 1.890–25.932; p = 0.004) compared to Tame. However, the municipality of Saravena was 3.6 more likely to find infected individuals compared to Tame (OR = 3.6; 95% CI = 1.165–11.025; p = 0.026) (Figure 4B) in the Supplementary Table 1 are shown all the parameters of the models.
The A. aegypti and A. albopictus classification of individuals is mainly determined by morphological identification (Kraemer et al., 2015), and few reports use molecular identification (Zamora-Delgado et al., 2015; Atencia et al., 2018). Although, morphological identification is the most widely used classification technique, it has different limitations. During the transport of individuals, they are susceptible to damage and modifications in their external parts, which can generate confusion during the classification of individuals (Sumruayphol et al., 2016). Taking this into account, it is necessary to use complementary tools that allow a correct identification of individuals. Among these tools it is recommended to use molecular techniques such as DNA barcoding (Cook et al., 2005; Anoopkumar et al., 2019). In this study was described the precision of taxonomic keys and DNA barcoding to classify individuals of the genus Aedes.
Regarding the analysis of the COI sequences, a global diversity of haplotypes value of 0.639 was evident for species A. aegypti and A. albopictus. This value is similar to that reported by Caldera et al. (2019) for individuals captured in Colombia and Joyce et al. (2018) for El Salvador; these values are relatively high. This high haplotype diversity may be due to adaptation processes for different environmental conditions and ecosystems where the insect is exposed due to its cosmopolitan distribution. Accordingly, the high number and diversity of haplotypes present in Saravena (h = 0.806, Figure 3B) may be due to evolutionary pressures that generate genetic changes in these dipteran species such as the use of insecticides. Between the two species, 36 mutations (Figure 3A) and a genetic distance of 0.107 were found. According to Hoyos-Lopez et al. (2015), this value is found for inter-species ranges (0.022–0.2565) that were previously reported in individuals of the Culicidae family. The tree topology (Figure 3C) is consistent with these results since it is evident that the individuals constitute two clusters grouped by species. Considering the groups formed by species in the haplotype network (Figure 3A) and the clustering by species in the phylogenetic tree (Figure 3C), it is possible to determine the precision of molecular tests to identify A. aegypti and A. albopictus.
According to the kappa index obtained in this study between the identification methods (k = 0.7643) (Cook, 2005), a deficiency in identification through morphology can occur (Figure 2). This could be an indicator of an overestimate of A. aegypti by traditional morphology-based techniques. The main characteristic that permits differentiation between these two species is the pattern in the scutum (mesothorax sclerite). These patterns are described as susceptible to blurring during insect storage processing (Patsoula et al., 2006). Therefore, there may be a risk of incorrect morphological identification and any error in this process has a negative effect on studies that make conclusions on the biology and vector capacity of each of these insect species. In this scenario, molecular identification may become more accurate (Chan et al., 2014). However, molecular techniques are too expensive for routine use and especially in areas with high vector infestation. Therefore, we propose to use this complementary tool mainly in three situations; (i) generate a baseline on the proportions of individuals per species in areas where this information is not available, (ii) as a quality control of morphological identification, choosing a random percentage of individuals to be confirmed by DNA barcoding and (iii) when individuals are damaged and morphological identification cannot be performed.
Studies focusing on the distribution of A. aegypti and A. albopictus conclude that their presence is due to the interaction of different biotic and abiotic factors (Lozano-Fuentes et al., 2012; Liang et al., 2019). This interaction is shown in the models that show an association between land use and the presence of A. aegypti and A. albopictus and the prevalence of arboviruses. The main aspects at risk are human settlements, horticulture and livestock (Cheong et al., 2014), and this is mainly because these activities generate micro-habitats that favor the reproduction of the vector (Sarfraz et al., 2012). In accordance with the above, there is a coexistence of A. aegypti and A. albopictus in urban areas of the Saravena and Arauquita municipalities (Figure 4A). Countries, such as Brazil and Panama, show evidence of this observation (Li et al., 2014; Whiteman et al., 2019). Therefore, the coexistence of these two species in the same area generates a public health risk since they are two vectors contributing to the transmission of arboviruses in the same urban area. Future studies should consider evaluating the transmission dynamics of arboviruses in this region by examining the plausible biological competition of the two species.
However, in the municipality of Arauquita the species that is found in higher frequency is A. albopictus (Table 2). This may be due to the different land uses in each municipality. For example, in Arauquita, there is a high availability of natural water due to the presence of the Arauca river in the northern part of the municipality. The above benefits the reproduction of A. albopictus since this species has preferences for these bodies of water compared to A. aegypti (Claeys et al., 2016). In this study we do not report data about immature collections to identify their most common breeding sites, however, this phenomenon has been previously reported in Vietnam and Brazil (Higa et al., 2010; Barbosa et al., 2020), and in Brazil, a spatial analysis was conducted that determined an association between the presence of vectors, depending on the type of water body (Lozano-Fuentes et al., 2012; Liang et al., 2019). Considering the aforementioned, it is possible that other factors are interacting in the municipalities that influence the geographical distribution of these species and subsequently the transmission dynamics of the arboviruses.
The logistic regression results show that in the municipality of Arauquita there is a higher risk of finding individuals infected with arboviruses (Figure 4B). In accordance with our results, different models (Barmak et al., 2016; Massaro et al., 2019; Zhu et al., 2019) show that human mobility is an important factor for the increase in the incidence of DENV even over short distances, i.e., between cities. Considering the aforementioned, the municipality of Arauquita is in a border area with Venezuela, and therefore, there is a high movement of humans that may maintain the virus in these areas. In accordance with the aforementioned, Lizarazo et al. (2019) showed that DENV-1 strains isolated in Venezuela were closely related to previously published sequences of this serotype in Colombia. This corroborates the influence that the movement of people between these two countries has on the prevalence of infection in insects.
DENV-1 was the arbovirus with the highest frequency in the population of infected individuals at 82% (50/61) and the overall infection rate in A. aegypti and A. albopictus was 29.6% (50/169) (Table 2). The DENV-1 serotype has also been detected more frequently than other arboviruses in Indonesia, Mexico, Cuba, Brazil, and Colombia (Velandia-Romero et al., 2017; Eiras et al., 2018; Garcia-Rejon et al., 2018; Gutiérrez-Bugallo et al., 2018; Rahayu et al., 2019). The high frequency of this arbovirus may be because the transmission in the urban cycle of DENV is provided by the level of viremia of the hosts (humans) since individuals at the beginning of infection have high rates of viremia (Duong et al., 2015; Martínez-Vega et al., 2015). This agrees with the results obtained in this study, given that DENV-1 is the most prevalent arbovirus in the department of Arauca (Boletín Epidemiológico Semanal (BES) Semana Epidemiológica 22, 24 al 30 de mayo, 2020). Furthermore, there is a need to conduct complementary entomo-virological surveillance to actively search for human cases and to have a complete overview of the urban transmission of arboviruses. Nevertheless, future studies should consider the detection of the other 3 DENV serotypes in vectors to complement the understanding of transmission dynamics in this endemic region and due to its proximity to Venezuelan states that are highly endemic for DENV-4 (Grillet et al., 2019).
The results in this study show a higher infection rate by DENV-1 and CHIKV in A. albopictus (Table 2) compared to A. aegypti. This does not agree with the values previously reported since the MIR values in A. aegypti are higher (Guerbois et al., 2016; Huerta et al., 2017; Garcia-Rejon et al., 2018). However, it was experimentally shown that A. albopictus is a vector more susceptible to infection than A. aegypti because the arbovirus dissemination rates were higher in nearly all strains of A. albopictus tested (Turell et al., 1992). The dissemination rate is associated with the CHIKV genotype, where A. albopictus is more susceptible to infection of the Asian genotype and this genotype is reported more frequently than the African genotype in Colombia (Camacho Burgos, 2019). The lack of natural ZIKV infection is due to the low number of cases reported in humans in the department of Arauca over the last 2 years (Boletín Epidemiológico Semanal (BES) Semana Epidemiológica 22, 24 al 30 de mayo, 2020). This confirms that this arbovirus is not circulating in the urban cycle of the three sampled municipalities.
However, the absence of individuals infected by MAYV may be because the insects processed in this study are part of the urban cycle and no studies demonstrate the efficiency of viral transmission in this cycle. However, the study by Pereira et al. (2020) shows that under laboratory conditions, A. aegypti and A. albopictus have the ability to become infected and the potential to transmit MAYV genotype D. Likewise, it is possible to infect mosquitoes with the virus from the saliva of individuals who initially tested negative. This indicates that for the infection to occur, a low viral load is necessary, which cannot be detected by PCR, and this is reflected in the MIR values of 0.4 (95% CI = 0.0–1.4) that are reported for this species (Maia et al., 2019). This is why different studies propose viral enrichment techniques that allow the viral sequences to increase, which is corroborated by next generation sequencing (NGS) (Hall et al., 2014). Therefore, it is important that future studies implement viral enrichment techniques that allow virus detection even at low concentrations.
Natural coinfection between DENV-2 and CHIKV has been previously reported in A. albopictus, where an association with simultaneous circulation of these two arboviruses during a dengue outbreak in Central Africa is possible (Caron et al., 2012). In this study, a mixed DENV-1 and CHIKV coinfection rate of 4.5% (5/111) was found in A. aegypti. This shows a simultaneous circulation of these two arboviruses within one specimen, which can lead to coinfection in humans, where an increase in coinfection cases would lead to rapid genetic evolution of the virus (Caron et al., 2012). There are two methods to estimate the infection rate from detection by pools: MIR (Minimum Infection Rate) and MLE (Maximum Likelihood Estimate). However, to produce a correct estimate, it is necessary to obtain a sampling ≥ 1,000 individuals and sizes of small pools. Processing individually allows us to naturally observe mixed infections in vectors including the option to conduct molecular discrimination of Aedes species (Walter et al., 1980; Gu et al., 2004). Therefore, the importance of conducting surveillance of arboviruses in the individual vector in areas where there is no information on natural infection in the vectors is important. This generates a baseline and avoids erroneous data regarding natural arbovirus infection in each of the vectors. In places where such a baseline exists and there are large samplings, the processing of insects by pools is possible. Future studies should compare the most reliable method for expressing infection rates in arboviral diseases in terms of sensitivity and cost-effectiveness.
This study reports on the natural CHIKV infection of A. aegypti and A. albopictus in Colombia, and a higher infection rate was found in the latter species. This shows the susceptibility of this species to infection by arboviruses. In addition, DENV-1 was the arbovirus with the highest frequency in the collected individuals, which may be associated with different factors such as the transovarian transmission of the vector and human infected with high viremias that can maintain constant transmission of the arbovirus. Expression of the natural infection data using infection rates to obtain a value per individual on arbovirus infection is proposed which increased the infection rate and the finding of viral co-infections. Lastly, we demonstrated the coexistence of A. aegypti and A. albopictus in urban areas of Saravena and Arauquita. This coexistence can be explained by the different land uses in the municipalities and the availability of water bodies that allow correct reproduction and development of the vectors. Our findings could become valuable to improve national control programs for arbovirosis. However, it is important to highlight some limitations of the study, among them the lack of detection of the DENV-2 – 4 serotypes as well as the measurement of other meteorological variables that would allow us to carry out a more robust analysis. Likewise, a longer sampling time that could allow us a greater capture of individuals and a more general view of the behavior of the infection of arboviruses in A. aegypti and A. albopictus. Lastly, the non-sequencing of the positive samples for DENV1 and CHIKV and their comparison with sequences available on GenBank. In the future, studies on this region must be also conducted to depict the molecular epidemiology of this arboviral agents for high resolution epidemiological surveillance.
The datasets generated for this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
YA and AC designed the protocol and carried out the capture of the individuals. DM, CH, and JR performed the processing of the samples in the laboratory. MM carried out the statistical and phylogenetic analysis of the results obtained. DM, CH, MM, and JR wrote the manuscript. All authors read and approved the final version of the manuscript.
We would like to thank the Dirección de Investigación e Innovación from Universidad del Rosario for funding this study and providing the editing English service of ENAGO and cover the publication fees.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2020.602190/full#supplementary-material
Supplementary Table 1 | Logistic regression. The parameters of Main effects model and Second level interaction model.
Anoopkumar, A. N., Puthur, S., Rebello, S., and Aneesh, E. M. (2019). Molecular characterization of Aedes, Culex, Anopheles, and Armigeres vector mosquitoes inferred by mitochondrial cytochrome oxidase I gene sequence analysis. Biologia 74, 1125–1138. doi: 10.2478/s11756-019-00231-0
Atencia, M. C., Pérez, M. D. J., Caldera, S. M., Jaramillo, M. C., and Bejarano, E. E. (2018). Genetic variability of Aedes aegypti in the department of sucre, colombia, by analysis of the nucleotide sequence of the mitochondrial ND4 gene. Biomédica 38, 267–276. doi: 10.7705/biomedica.v38i0.3728
Azevedo, R. S., Silva, E. V., Carvalho, V. L., Rodrigues, S. G., Neto, J. P. N., Monteiro, H. A., et al. (2009). Mayaro fever virus, Brazilian amazon. Emerg. Infect. Dis. 15:1830. doi: 10.3201/eid1511.090461
Barbosa, R. M. R., Melo-Santos, M. A. V. D., Silveira, Jr, J. C., Silva-Filha, M. H. N. L., Souza, W. V., Oliveira, C. M. F. D., et al. (2020). Infestation of an endemic arbovirus area by sympatric populations of Aedes aegypti and Aedes albopictus in Brazil. Mem. Instit. Oswaldo Cruz 115: e190437. doi: 10.1590/0074-02760190437
Barmak, D. H., Dorso, C. O., and Otero, M. (2016). Modelling dengue epidemic spreading with human mobility. Phys. A Statist. Mech. Appl. 447, 129–140. doi: 10.1016/j.physa.2015.12.015
Beaty, B. J., Black, W. C., Eisen, L., Flores, A. E., García-Rejón, J. E., Loroño-Pino, M., and Saavedra-Rodriguez, K. (2016). “The intensifying storm: Domestication of Aedes aegypti, urbanization of arboviruses, and emerging insecticide resistance,” in Proceedings of the Global Health Im-pacts of Vector-Borne Diseases: Workshop Summary (Washington, DC: National Academies Press).
Black, W. C. IV., Bennett, K. E., Gorrochótegui-Escalante, N., Barillas-Mury, C. V., Fernández-Salas, I., de Lourdes Muñoz, M., et al. (2002). Flavivirus susceptibility in Aedes aegypti. Arch. Med. Res. 33, 379–388. doi: 10.1016/S0188-4409(02)00373-9
Boletín Epidemiológico Semanal (BES) Semana Epidemiológica 22, 24 al 30 de mayo (2020). Obtenido de. Available at : https://www.ins.gov.co/buscador-eventos/BoletinEpidemiologico/2020_Bolet%C3%ADn_epidemiológico_semana%2022.pdf (accessed July 20, 2020).
Caldera, S. M., Jaramillo, M. C., Cochero, S., Pérez Doria, A., and Bejarano, E. E. (2019). Diferencias genéticas entre poblaciones de Aedes aegypti de municipios del norte de Colombia, con baja y alta incidencia de dengue. Revista 33, doi: 10.7705/biomedica.v33i0.1573
Camacho Burgos, E. Y. (2019). Caracterización filogenética de los virus dengue, Chikungunya y Zika circulantes en el departamento de Sucre durante el periodo 2013–2015, Colombia: Universidad de Sucre.
Caron, M., Paupy, C., Grard, G., Becquart, P., Mombo, I., Nso, B. B. B., et al. (2012). Recent introduction and rapid dissemination of Chikungunya virus and Dengue virus serotype 2 associated with human and mosquito coinfections in Gabon, central Africa. Clin. Infect. Dis. 55, e45-e53. doi: 10.1093/cid/cis530
Cevallos, V., Ponce, P., Waggoner, J. J., Pinsky, B. A., Coloma, J., Quiroga, C., et al. (2018). Zika and Chikungunya virus detection in naturally infected Aedes aegypti in Ecuador. Acta Trop. 177, 74–80. doi: 10.1016/j.actatropica.2017.09.029
Chan, A., Chiang, L. P., Hapuarachchi, H. C., Tan, C. H., Pang, S. C., Lee, R., et al. (2014). DNA barcoding: complementing morphological identification of mosquito species in Singapore. Paras. Vect. 7, 1–12. doi: 10.1186/s13071-014-0569-4
Cheong, Y. L., Leitão, P. J., and Lakes, T. (2014). Assessment of land use factors associated with dengue cases in Malaysia using boosted regression trees. Spatial Spatio Temp. Epidemiol. 10, 75–84. doi: 10.1016/j.sste.2014.05.002
Claeys, C., Robles, C., Bertaudiere-Montes, V., Deschamps-Cottin, M., Megnifo, H. T., Pelagie-Moutenda, R., et al. (2016). Socio-ecological factors contributing to the exposure of human populations to mosquito bites that transmit dengue fever, chikungunya and zika viruses: a comparison between mainland France and the French Antilles. Environ. Risques Santé 15, 318–325.
Clement, M., Snell, Q., Walker, P., Posada, D., and Crandall, K. (2002). TCS: estimating gene genealogies. Parall. Distrib. Process. Symp. Int. 2:0184. doi: 10.1109/IPDPS.2002.1016585
Cook, S., Diallo, M., Sall, A. A., Cooper, A., and Holmes, E. C. (2005). Mitochondrial markers for molecular identification of Aedes mosquitoes (Diptera: Culicidae) involved in transmission of arboviral disease in West Africa. J. Med. Entomol. 42, 19–28. doi: 10.1603/0022-2585(2005)042[0019:mmfmio]2.0.co;2
Cova-Garcia, P., Sutil, E., and Rausseo, J. A. (1966). Mosquitos (Culicinos) de Venezuela: Tomo I and Tomo II. Caracas: Ministerio de Sanidad y Asistencia Social.
de la Cruz, C. H., Martinez, S. L. A., Failoc-Rojas, V. E., and Aguilar-Gamboa, F. R. (2019). Momento de considerar otras arbovirosis luego del virus mayaro. Rev. Cubana Med. Gen. Integ. 35.
Diallo, D., Sall, A. A., Diagne, C. T., Faye, O., Faye, O., Ba, Y., et al. (2014). Zika virus emergence in mosquitoes in southeastern Senegal, 2011. PLoS One 9:e109442. doi: 10.1371/journal.pone.0109442
Duong, V., Lambrechts, L., Paul, R. E., Ly, S., Lay, R. S., Long, K. C., et al. (2015). Asymptomatic humans transmit dengue virus to mosquitoes. Proc. Natl. Acad. Sci. U.S.A. 112, 14688–14693. doi: 10.1073/pnas.1508114112
Eiras, A. E., Pires, S. F., Staunton, K. M., Paixão, K. S., Resende, M. C., Silva, H. A., et al. (2018). A high-risk Zika and dengue transmission hub: virus detections in mosquitoes at a Brazilian university campus. Paras. Vect. 11:359. doi: 10.1186/s13071-018-2883-8
Espinal, M. A., Andrus, J. K., Jauregui, B., Hull Waterman, S., Morens, D. M., Santos, J. I., et al. (2019). Arbovirosis emergentes y reemergentes transmitidas por Aedes en la Región de las Américas: implicaciones en materia de políticas de salud. Rev. Panam. Salud. Publ. 43.
Faye, O., Diallo, D., Diallo, M., Weidmann, M., and Sall, A. A. (2013). Quantitative real-time PCR detection of Zika virus and evaluation with field-caught mosquitoes. Virol. J. 10:311. doi: 10.1186/1743-422x-10-311
Forattini, O. P. (1965). Entomologia médica: culicini: culex, Aedes e Psorophora, Vol. 2. São Paulo: Faculdade de Higiene e Saúde Pública, Depto. de Parasitología.
Freitas, R. M. D. (2010). A review on the ecological determinants of Aedes aegypti (Diptera: Culicidae) vectorial capacity. Oecol. Austr. 14:726–736 doi: 10.4257/oeco.2010.1403.08
Garcia-Rejon, J. E., Ulloa-Garcia, A., Cigarroa-Toledo, N., Pech-May, A., Machain-Williams, C., Cetina-Trejo, R. C., et al. (2018). Study of Aedes aegypti population with emphasis on the gonotrophic cycle length and identification of arboviruses: implications for vector management in cemeteries. Rev. Instit. Med. Trop. São Paul. 60:e44 doi: 10.1590/s1678-9946201860044
Grillet, M. E., Hernández-Villena, J. V., Llewellyn, M. S., Paniz-Mondolfi, A. E., Tami, A., Vincenti-Gonzalez, M. F., et al. (2019). Venezuela’s humanitarian crisis, resurgence of vector-borne diseases, and implications for spillover in the region. Lancet Infect. Dis. 19, e149-e161. doi: 10.1016/S1473-3099(18)30757-6
Gu, W., Lampman, R., and Novak, R. J. (2004). Assessment of arbovirus vector infection rates using variable size pooling. Med. Vet. Entomol. 18, 200-204 doi: 10.1111/j.0269-283X.2004.00482.x
Guerbois, M., Fernandez-Salas, I., Azar, S. R., Danis-Lozano, R., Alpuche-Aranda, C. M., Leal, G., et al. (2016). Outbreak of Zika virus infection, Chiapas State, Mexico, 2015, and first confirmed transmission by Aedes aegypti mosquitoes in the Americas. J. Infect. Dis. 214, 1349–1356. doi: 10.1093/infdis/jiw302
Gutiérrez-Bugallo, G., Rodríguez-Roche, R., Díaz, G., Pérez, M., Mendizábal, M. E., Peraza, I., et al. (2018). Spatio-temporal distribution of vertically transmitted dengue viruses by Aedes aegypti (Diptera: Culicidae) from Arroyo Naranjo, Havana, Cuba. Trop. Med. Int. Health 23, 1342–1349. doi: 10.1111/tmi.13162
Hall, R. J., Wang, J., Todd, A. K., Bissielo, A. B., Yen, S., Strydom, H., et al. (2014). Evaluation of rapid and simple techniques for the enrichment of viruses prior to metagenomic virus discovery. J. Virol. Methods 195, 194–204. doi: 10.1016/j.jviromet.2013.08.035
Higa, Y., Yen, N. T., Kawada, H., Son, T. H., Hoa, N. T., and Takagi, M. (2010). Geographic distribution of Aedes aegypti and Aedes albopictus collected from used tires in Vietnam. J. Am. Mosq. Control Assoc. 26, 1–9. doi: 10.2987/09-5945.1
Houé, V., Bonizzoni, M., and Failloux, A. B. (2019). Endogenous non-retroviral elements in genomes of Aedes mosquitoes and vector competence. Emerg. Microb. Infect. 8, 542–555. doi: 10.1080/22221751.2019.1599302
Hoyos-Lopez, R., Roman Pardo, S., CASTAÑO, J. C., and GALLEGO-GÓMEZ, J. C. (2015). DNA barcode for typing of immature mosquitoes from Armenia and Circasia (Quindío, Colombia). Rev. Colomb. Entomol. 41, 218-227
Huerta, H., González-Roldán, J. F., Sánchez-Tejeda, G., Correa-Morales, F., Romero-Contreras, F. E., Cárdenas-Flores, R., et al. (2017). Detection of Zika virus in Aedes mosquitoes from Mexico. Trans. Royal Soc. Trop. Med. Hygiene 111, 328–331. doi: 10.1093/trstmh/trx056
Joyce, A. L., Torres, M. M., Torres, R., and Moreno, M. (2018). Genetic variability of the Aedes aegypti (Diptera: Culicidae) mosquito in El Salvador, vector of dengue, yellow fever, chikungunya and Zika. Paras. Vect. 11:637. doi: 10.1186/s13071-018-3226-5
Kraemer, M. U., Sinka, M. E., Duda, K. A., Mylne, A. Q., Shearer, F. M., Barker, C. M., et al. (2015). The global distribution of the arbovirus vectors Aedes aegypti and Ae. albopictus. eLife 4:e08347. doi: 10.7554/eLife.08347
Lanciotti, R. S., Kosoy, O. L., Laven, J. J., Panella, A. J., Velez, J. O., Lambert, A. J., et al. (2007). Chikungunya virus in US travelers returning from India, 2006. Emerg. Infect. Dis. 13, 764–767. doi: 10.3201/eid1305.070015
Li, Y., Kamara, F., Zhou, G., Puthiyakunnon, S., Li, C., Liu, Y., et al. (2014). Urbanization increases Aedes albopictus larval habitats and accelerates mosquito development and survivorship. PLoS Negl. Trop. Dis. 8:e3301. doi: 10.1371/journal.pntd.0003301
Liang, L., Jia, P., Tan, X., Chen, J., and Chen, X. (2019). Potential effects of heat waves on the population dynamics of the dengue mosquito Aedes albopictus. PLoS Neglect. Trop. Dis. 13:e0007528. doi: 10.1371/journal.pntd.0007528
Librado, P., and Rozas, J. (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452. doi: 10.1093/bioinformatics/btp187
Liu, Y., Lillepold, K., Semenza, J. C., Tozan, Y., Quam, M. B., and Rocklöv, J. (2020). Reviewing estimates of the basic reproduction number for dengue, Zika and chikungunya across global climate zones. Environ. Res. 182:109114. doi: 10.1016/j.envres.2020.109114
Lizarazo, E., Vincenti-Gonzalez, M., Grillet, M. E., Bethencourt, S., Diaz, O., Ojeda, N., et al. (2019). Spatial dynamics of Chikungunya virus, Venezuela, 2014. Emerg. Infect. Dis. 25:672. doi: 10.3201/eid2504.172121
Long, K. C., Ziegler, S. A., Thangamani, S., Hausser, N. L., Kochel, T. J., Higgs, S., et al. (2011). Experimental transmission of Mayaro virus by Aedes aegypti. Am. J. Trop. Med. Hyg. 85, 750–757. doi: 10.4269/ajtmh.2011.11-0359
Lorenz, C., Ribeiro, A. F., and Chiaravalloti-Neto, F. (2019). Mayaro virus distribution in South America. Acta tropica 198:105093. doi: 10.1016/j.actatropica.2019.105093
Lounibos, L. P., and Kramer, L. D. (2016). Invasiveness of Aedes aegypti and Aedes albopictus and vectorial capacity for chikungunya virus. J. Infect. Dis. 214(suppl_5), S453-S458. doi: 10.1093/infdis/jiw285
Lozano-Fuentes, S., Hayden, M. H., Welsh-Rodriguez, C., Ochoa-Martinez, C., Tapia-Santos, B., Kobylinski, K. C., et al. (2012). The dengue virus mosquito vector Aedes aegypti at high elevation in Mexico. Am. J. Trop. Med. Hyg. 87, 902-909. doi: 10.4269/ajtmh.2012.12-0244
Maia, L. M. S., Bezerra, M. C. F., Costa, M. C. S., Souza, E. M., Oliveira, M. E. B., Ribeiro, A. L. M., et al. (2019). Natural vertical infection by dengue virus serotype 4, Zika virus and Mayaro virus in Aedes (Stegomyia) aegypti and Aedes (Stegomyia) albopictus. Med. Vet. Entomol. 33, 437–442. doi: 10.1111/mve.12369
Martínez-Vega, R. A., Danis-Lozano, R., Díaz-Quijano, F. A., Velasco-Hernández, J., Santos-Luna, R., Román-Pérez, S., et al. (2015). Peridomestic infection as a determining factor of dengue transmission. PLoS Neglect. Trop. Dis. 9:e0004296. doi: 10.1371/journal.pntd.0004296
Massaro, E., Kondor, D., and Ratti, C. (2019). Assessing the interplay between human mobility and mosquito borne diseases in urban environments. Sci. Rep. 9, 1–13 doi: 10.1038/s41598-019-53127-z
Nguyen, LT, Schmidt, HA, von Haeseler, A, Minh, BQ. (2014). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Patsoula, E., Samanidou-Voyadjoglou, A., Spanakos, G., Kremastinou, J., Nasioulas, G., and Vakalis, N. C. (2006). Molecular and morphological characterization of Aedes albopictus in northwestern Greece and differentiation from Aedes cretinus and Aedes aegypti. J. Med. Entomol. 43, 40–54. doi: 10.1603/0022-2585(2006)043[0040:mamcoa]2.0.co;2
Paupy, C., Delatte, H., Bagny, L., Corbel, V., and Fontenille, D. (2009). Aedes albopictus, an arbovirus vector: from the darkness to the light. Microbes Infect. 11, 1177–1185. doi: 10.1016/j.micinf.2009.05.005
Pereira, T. N., Carvalho, F. D., De Mendonça, S. F., Rocha, M. N., and Moreira, L. A. (2020). Vector competence of Aedes aegypti, Aedes albopictus, and Culex quinquefasciatus mosquitoes for Mayaro virus. PLoS Negl. Trop. Dis. 14:e0007518. doi: 10.1371/journal.pntd.0007518
Rahayu, A., Saraswati, U., Supriyati, E., Kumalawati, D. A., Hermantara, R., Rovik, A., et al. (2019). Prevalence and distribution of dengue virus in Aedes aegypti in yogyakarta city before deployment of Wolbachia infected Aedes aegypti. Int. J. Environ. Res. Public Health 16:1742. doi: 10.3390/ijerph16101742
Ramasamy, R., Surendran, S. N., Jude, P. J., Dharshini, S., and Vinobaba, M. (2011). Larval development of Aedes aegypti and Aedes albopictus in peri-urban brackish water and its implications for transmission of arboviral diseases. PLoS Negl. Trop. Dis. 5:e1369. doi: 10.1371/journal.pntd.0001369
Rolo, E. S. A. (2020). DNA Barcoding and Forensic Entomology: A Molecular Approach for Diptera Species’ Identification. Doctoral dissertation, University of Cape Town.
Salehi, Z, Najafi, M. (2014). RNA preservation and stabilizaton. Biochem. Physiol. Open Access. 3, 1–4.
Santiago, G. A., Vergne, E., Quiles, Y., Cosme, J., Vazquez, J., Medina, J. F., et al. (2013). Analytical and clinical performance of the CDC real time RT-PCR assay for detection and typing of dengue virus. PLoS Negl. Trop. Dis. 7:e2311. doi: 10.1371/journal.pntd.0002311
Sarfraz, M. S., Tripathi, N. K., Tipdecho, T., Thongbu, T., Kerdthong, P., and Souris, M. (2012). Analyzing the spatio-temporal relationship between dengue vector larval density and land-use using factor analysis and spatial ring mapping. BMC Publ. Health 12:853. doi: 10.1186/1471-2458-12-853
Shope, R. E., and Meegan, J. M. (1997). Arboviruses in Viral Infections of Humans. Boston, MA: Springer, 151–183. doi: 10.1007/978-1-4899-0036-4_6
Sohpal, V. K., Dey, A., and Singh, A. (2010). MEGA biocentric software for sequence and phylogenetic analysis: a review. Int. J. Bioinform. Res. Appl. 6, 230–240. doi: 10.1504/ijbra.2010.034072
Sumruayphol, S., Apiwathnasorn, C., Ruangsittichai, J., Sriwichai, P., Attrapadung, S., Samung, Y., et al. (2016). DNA barcoding and wing morphometrics to distinguish three Aedes vectors in Thailand. Acta Trop. 159, 1–10. doi: 10.1016/j.actatropica.2016.03.010
Turell, M. J., Beaman, J. R., and Tammariello, R. F. (1992). Susceptibility of selected strains of Aedes aegypti and Aedes albopictus (Diptera: Culicidae) to chikungunya virus. J. Med. Entomol. 29, 49–53. doi: 10.1093/jmedent/29.1.49
Vega-Rúa, A., Zouache, K., Girod, R., Failloux, A. B., and Lourenço-de-Oliveira, R. (2014). High level of vector competence of Aedes aegypti and Aedes albopictus from ten American countries as a crucial factor in the spread of Chikungunya virus. J. Virol. 88, 6294–6306. doi: 10.1128/jvi.00370-14
Velandia-Romero, M. L., Olano, V. A., Coronel-Ruiz, C., Cabezas, L., Calderón-Peláez, M. A., Castellanos, J. E., et al. (2017). Detección del virus del dengue en larvas y pupas de Aedes aegypti recolectadas en áreas rurales del municipio de Anapoima, Cundinamarca, Colombia. Biomédica 37, 193–200. doi: 10.7705/biomedica.v37i0.3584
Villar, L. A., Rojas, D. P., Besada-Lombana, S., and Sarti, E. (2015). Epidemiological trends of dengue disease in Colombia (2000-2011): a systematic review. PLoS Negl. Trop. Dis. 9: e0003499. doi: 10.1371/journal.pntd.0003499
Walter, SD, Hildreth, SWY and Beaty, BJ. (1980). Estimación de las tasas de infección en poblaciones de organismos utilizando grupos de tamaño variable. Am. J. Epidemiol. 112, 124–128. doi: 10.1093/oxfordjournals.aje.a112961
Whiteman, A., Gomez, C., Rovira, J., Chen, G., McMillan, W. O., and Loaiza, J. (2019). Aedes mosquito infestation in socioeconomically contrasting neighborhoods of panama city. EcoHealth 16, 210–221. doi: 10.1007/s10393-019-01417-3
World Health Organization [WHO] (2020). Real-Time RT-PCR (TaqManTM) protocol – Mayaro virus (MAYV). Geneve: WHO.
Zamora-Delgado, J., Castaño, J. C., and Hoyos-López, R. (2015). DNA barcode sequences used to identify Aedes (Stegomyia) albopictus (Diptera: Culicidae) in La Tebaida (Quindío, Colombia). Rev. Colomb. Entomol. 41, 212-217
Keywords: arbovirus, dengue, Zika, Chikungunya, Mayaro, Aedes
Citation: Martínez D, Hernández C, Muñoz M, Armesto Y, Cuervo A and Ramírez JD (2020) Identification of Aedes (Diptera: Culicidae) Species and Arboviruses Circulating in Arauca, Eastern Colombia. Front. Ecol. Evol. 8:602190. doi: 10.3389/fevo.2020.602190
Received: 02 September 2020; Accepted: 04 November 2020;
Published: 26 November 2020.
Edited by:
Jader Oliveira, Sao Paulo State University, BrazilReviewed by:
Jinbao Gu, Southern Medical University, ChinaCopyright © 2020 Martínez, Hernández, Muñoz, Armesto, Cuervo and Ramírez. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Juan David Ramírez, anVhbmQucmFtaXJlekB1cm9zYXJpby5lZHUuY28=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.