- 1Department of Entomology, Michigan State University, East Lansing, MI, United States
- 2Ecology, Evolutionary Biology, and Behavior Program, Michigan State University, East Lansing, MI, United States
- 3Department of Biology, Northern Michigan University, Marquette, MI, United States
- 4Department of Biological Sciences, University of Notre Dame, Notre Dame, IN, United States
- 5Department of Osteopathic Medical Specialties, Michigan State University, East Lansing, MI, United States
Salmon decomposition is traditionally viewed through the lens of energy and nutrient subsidies, but not as a potential “microbial subsidy.” Microbial communities residing on and within spawning salmon are directly introduced into streams after host death. This incorporation takes the form of microbes sloughing off and integrating into substrate biofilms, or indirectly, by macroinvertebrates facilitating dispersal via consumption. The objective of this study was to determine the effects of salmon carcass-derived microbial communities on stream biofilms and macroinvertebrates during an experimental salmon carcass addition in a naïve stream (i.e., no evolutionary history of salmon). Microbial communities [epilithic biofilms and within macroinvertebrates (internal)] were sampled at treatment and control sites before (September), during (October), and after (November to following August) a salmon carcass subsidy introduction in 2 successive years (September 2014-August 2016). We found a significant interaction between carcass addition and time on microbial and macroinvertebrate communities. Heptagenia (Heptageniidae: grazer) density was five times higher in the salmon reach compared to the control. In the salmon reach during year one, Stramenopiles (i.e., eukaryotic microbes) decreased in biofilm communities after 2 weeks of decomposition. The internal microbiome of Stegopterna mutata (Simuliidae: collector-filterer) varied between years but was significantly different between reaches over time during year two of the study, with four times greater abundance of melanogenesis functional pathways (function determined in silico) in the control reach. Although unique microbial taxa, introduced to this naïve stream via salmon carrion, persisted in biofilms on benthic substrate and internal to insects during both years, those taxa represented <2% of the relative abundance in microbial communities. These results highlight the importance of allochthonous carrion resources in the microbial ecology of lotic biofilms and macroinvertebrates. Furthermore, this study contributes to previous research into the complex interkingdom interactions in stream communities in response to a novel allochthonous resource.
Introduction
Headwater streams are highly reliant on allochthonous organic matter as an energy base for consumers. Shading from riparian trees restricts the amount of sunlight to most headwater streams thereby limiting autochthonous primary production (Vannote et al., 1980). Therefore, stream trophic networks rely on organic matter decomposition from outside sources. Organic matter decomposition has traditionally been viewed through the lens of carbon and nutrient subsidies (e.g., leaf litter) into the system in ways that alter macrobenthic communities (Polis and Strong, 1996; Hagen et al., 2012; Benbow et al., 2018). Allochthonous organic matter may also act as a “microbial subsidy” source to streams, by transferring novel microbes from one ecosystem to another (Steffan et al., 2017; Figure 1). Due to the high diversity of microbes on Earth, each allochthonous resource has an individual microbial community residing on and within it (Lindström and Langenheder, 2012; Locey and Lennon, 2016; Thompson et al., 2017). These novel microbes are hypothesized to be introduced into streams through the addition and transport of the allochthonous resources from adjacent or upstream habitats (e.g., riparian zones or tributaries) (Ruiz-González et al., 2015), but energy, nutrients, and microbes can also arrive in the form of decomposing heterotrophic biomass, such as carrion (Pechal and Benbow, 2016; Benbow et al., 2018).
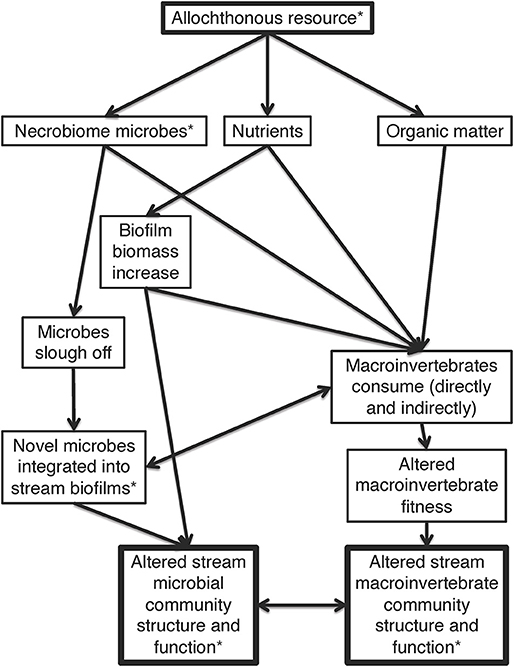
Figure 1. Conceptual framework of allochthonous resources altering microbial and macroinvertebrate communities via nutrients, organic matter, and microbes. Arrows represent directional links of effects. An asterisk represents factors directly measured in this study, while those without asterisks are hypothesized. Hypotheses on the overall importance of each linked agent are not offered, as these may vary over time and space, and the overall figure represents how these components fit into the larger ecosystem context.
Macroinvertebrate consumers may directly ingest and subsequently disperse allochthonous resource microbes throughout a stream (McEwen and Leff, 2001). Insects are vectors of microbes from diverse substrates involved in decomposition, transferring the microbes that come into contact from one environment to another [e.g., insects on food: (Blazar et al., 2011); blow flies (Diptera: Calliphoridae) on agar: (Junqueira et al., 2017)]. For example, in a microcosm experiment, dispersal of marked microbes by mayfly (Baetis sp.), stonefly (Pteronarcys sp.), dragonfly (Aeshnidae) nymphs, and glass shrimp (Palaeomonetes sp.), exhibited considerable variation in the abundance of macroinvertebrate-associated bacteria transferred to other surfaces (Leff et al., 1994). In some trials, there was a large pulse of marked microbes dispersed, and in others, no marked microbes were detected. It remains unknown how these occasional releases of macroinvertebrate-associated bacteria ultimately influence microbial structure on downstream surfaces. Stream macroinvertebrates also act as important consumers of biofilms and can alter microbial community structure by selectively consuming taxa (Mulholland et al., 1991; Feminella and Hawkins, 1995; Rosemond et al., 2000; Lang et al., 2015). Alternatively, microbes may slough off the host resource and become integrated into the water column and benthic biofilm microbial communities (Leff et al., 1998; Crump et al., 2012). These changes to microbial communities alter the functional base of the aquatic food web, which may have far reaching effects throughout the stream network (Hall and Meyer, 1998; Meyer et al., 2007). Therefore, it is important to understand how microbial subsidies associated with allochthonous resources influence stream communities, from riparian leaf litter to carrion generated through mass mortalities, such as annual salmon runs.
Pacific salmon (Oncorhynchus spp.) are an important annual input of allochthonous nutrients and organic matter into streams. Particularly in the Pacific Northwest and Alaska, which have a several thousand year long history of native salmon, and other watersheds in North America where they have been anthropogenically introduced as recreational and economic fisheries (Cederholm et al., 1999; Gende et al., 2002; Moore et al., 2004). Benthic biofilms increase in biomass and have lower nutrient limitation when salmon carrion is present (Wipfli et al., 1998; Johnston et al., 2004; Mitchell and Lamberti, 2005; Rüegg et al., 2011). Similarly, macroinvertebrate community structure, function, and growth rates are influenced by decomposing salmon organic matter (Chaloner and Wipfli, 2002; Chaloner et al., 2002; Lessard and Merritt, 2006), often with contrasting effects within different regions and streams (Bilby et al., 1996; Claeson et al., 2006; Janetski et al., 2009, 2013). Temporal dynamics are important in mediating the influence of carcass additions. For example, peak salmon derived nutrient enrichment (determined by stable isotopes) in grazers occurred 1–2 months after carcass introduction, while it occurred 2–3 months after introduction in predatory macroinvertebrates (Morley et al., 2016). Microbes associated with salmon carcasses were detected in aquatic macroinvertebrates located within Alaskan salmon-bearing streams (Pechal and Benbow, 2016), which demonstrates the potential for salmon carcasses to act as conduits for new microbe introduction into streams. Yet, this potential has not been tested in streams without a historical salmon run, such as those found in the Laurentian Great Lakes watershed, where Pacific salmon have been introduced.
Since first introduced to the Great Lakes region in 1966–1970 to control invasive alewife (Parsons, 1973), chinook (O. tshawytscha) and coho (O. kisutch) salmon have been naturalized to many tributary streams of the watersheds where they now spawn and die. Therefore, salmon carrion has been a non-native resource subsidy to some Michigan streams for only the last 50 years. After spawning and death, the resulting carcasses may structurally and functionally impact the aquatic communities residing in naïve headwater streams (Cederholm et al., 1989). These annual pulses of salmon increase the nutrients and organic matter inputs to recipient streams, which can have far reaching effects on both aquatic and terrestrial ecosystems throughout the food web (Schuldt and Hershey, 1995; Bilby et al., 1996). The salmon resource may be directly used by both stream macroinvertebrates and microbes, as well as indirectly through nutrient and dissolved organic matter subsidy pathways (Collins et al., 2011; Levi and Tank, 2013; Levi et al., 2013). However, salmon do not migrate to all Michigan streams, such as those with dams, providing an opportunity to investigate salmon carrion effects on microbial and macroinvertebrate communities in historically naïve systems through carrion subsidy introduction and monitoring.
The objective of this study was to evaluate allochthonous salmon carcass resource subsidy effects on aquatic macroinvertebrate and microbial communities in a naïve Michigan stream. We postulated these communities would demonstrate short- and long-term responses to introduced salmon carcasses. Specifically, we predicted in carcass-introduced habitats that: (1) macroinvertebrate communities would be initially dominated by shredders and transition to an increase in grazers and collectors; (2) biofilm communities would be dominated by heterotrophic bacteria compared to the control habitats; (3) the internal macroinvertebrate microbiomes would be supplemented with salmon carcass-associated microbes after carcass introduction; and (4) salmon carcasses would introduce microbes to the stream, some of which would persist and become more abundant, while others diminish.
Materials and Methods
Experimental Design
This study was conducted in Hunt Creek on the property of the Hunt Creek Fisheries Research Station near Lewiston, Michigan, USA (44.86, −84.16). Hunt Creek is a groundwater fed second-order stream in the Thunder Bay River watershed and has never received an annual salmon run (Grossman et al., 2012). Several barriers to upstream movement of fish preclude colonization of Hunt Creek by salmon, which were reasonably abundant in Lake Huron before a major decline in the early to mid-2000s (Cwalinski et al., 2006). A Before-After-Control-Intervention field study design was implemented for this study (Stewart-Oaten et al., 1986). Chinook and Coho (n = 120; 50/50 species split) salmon carcasses were introduced into the same salmon “treatment” reach in October 2014 and October 2015 (the typical timing of Michigan salmon runs; Gerig et al., 2018) using loading rates (~1 kg m−2 of stream) approximate to that of a typical salmon run in a Lake Michigan tributary (Janetski et al., 2012; Gerig, 2017). Michigan Department of Natural Resources hatcheries were the source of the salmon carcasses, and salmon died of natural causes. For around 1 year, carcasses were frozen to prevent inadvertent disease introduction to waters that do not have migratory fish runs. Carcasses were then brought to ambient temperature before being staked with rebar in reach habitats, including pools, undercut banks, and debris jams, as has been performed in similar salmon carrion studies (Tiegs et al., 2011). A control reach lacking salmon carcass introduction was located 600 m upstream of the salmon reach (Figure S1). Both control and salmon reaches were 90 m long. The average width and depth of the control reach were 0.18 and 3.21 m, and the average width and depth of the treatment reach were 0.19 and 3.61 m, respectively.
Field Sample Collections
Prior to salmon carcass introduction, epinecrotic microbial communities of each carcass were aseptically sampled with sterile and DNA-free cotton swabs using the methods of Pechal and Benbow (2016). Swab samples were individually stored in 200 μL of molecular grade ethanol (>96%) at −20°C. Internal salmon carcass microbial samples were not sampled, so as to not influence the decomposition process by physically altering the salmon carcasses. Microbial and macroinvertebrate communities were sampled at three sub-reaches within the treatment and control reaches: once before (September), once during (October), and four times after (March through August) carcass introduction each year. Sterilized hexagonal unglazed ceramic tiles (29.25 cm2) were deployed in the stream to characterize epilithic microbial communities (Lang et al., 2016). Six tiles were secured to a brick using a silicone adhesive; five bricks were placed along a transect perpendicular to stream flow in the center of each sub-reach. Bricks were introduced into the stream 2 weeks prior to the first sample collection to establish baseline communities in both reaches. During each collection, the bricks were removed from the stream, a tile was collected and placed in a sterile 188 mL WhirlPak bag (Nasco, Fort Atkinson, WI, USA), kept on ice during transport, and stored at −20°C until DNA extraction. Bricks with the remaining tiles were returned to the same location within the stream. After all tiles were collected, the biofilms were scraped from tiles in the laboratory using autoclaved sterile and decontaminated razor blades into a 2 mL microcentrifuge tube for immediate DNA processing. We did not quantify the amount of microbial growth on any of the collected samples, due to the small amount of growth. During each sampling event, water chemistry parameters of dissolved oxygen (mg/L), pH, conductivity (mS/cm), and temperature (°C) were determined using a YSI 6-Series multiparameter water quality 6600 V2-4 sonde (Table S1).
Macroinvertebrates were sampled using a modified Hess sampler (Merritt et al., 2008). At each sub-reach, three riffle habitats were sampled for 30 s each and combined into a single composite for that location (total area = 0.3 m2). Individual specimens that represented dominant taxa over a variety of feeding groups were hand-picked from the composite Hess sampler collection to ensure adequate sample sizes from representative groups to obtain internal microbial communities; samples were immediately stored in molecular grade ethanol for subsequent internal microbial community analysis. The remainder of the composite Hess sample was stored in 70% ethanol and hand-sorted in the laboratory. Macroinvertebrates were identified to the lowest taxonomic level (genus), except for those used for internal microbiome analyses, which were identified to species (Merritt et al., 2008; Bright, 2016). Functional feeding group was also determined using Merritt et al. (2008) (Table S2).Three species were used for internal microbiome analysis due to their abundance and to represent different functional feeding groups: Heptagenia flavescens (Walsh) (Ephemeroptera: Heptageniidae; grazer), Baetis brunneicolor McDunnough (Ephemeroptera: Baetidae; collector-gatherer), and Stegopterna mutata (Malloch) (Diptera: Simuliidae; collector-filterer).
DNA Processing and Targeted 16S rRNA Gene Amplicon Sequencing
For insects, three identified individuals were pooled into one sample and surface sterilized using a 10% bleach rinse followed by three sterile deionized water rinses (Ridley et al., 2012). The insects were air-dried and ground in a 1.7 mL tube using a sterile pestle. DNA extraction was performed with the Qiagen PowerSoil DNA extraction kit® (Qiagen, Inc., Valencia, CA, USA) using a modified manufacturer's protocol: 20 mg mL−1 of lysozyme was added during the lysis step and the final DNA was eluted in 50 μL of C6. DNA quantification was performed using the Quanti-iT dsDNA HS Assay kit and a Qubit 2.0 (Grand Island, NY, USA); a concentration of 0.1 ng μL−1 was used as a minimum threshold for subsequent sequencing procedures. All DNA preparations were stored at −20°C.
Illumina MiSeq 16S library construction (2 × 250 bp paired-end reads) and sequencing was performed at the MSU Genomics Core using a modified version of the Illumina MiSeq protocol (Caporaso et al., 2011a). The variable region 4 of the 16S rRNA gene was amplified with region-specific primers, 515F/806R (5′-GTGCCAGCMGCCGCGG-3′, 5′-TACNVGGGTATCTAATCC-3′) (Claesson et al., 2010; Caporaso et al., 2011b, 2012). The resulting 16S rRNA amplicon sequencing data were assembled, quality-filtered, and demultiplexed using QIIME2 version 19.1 (Kuczynski et al., 2012). Default settings were used, unless specified in the following methods. DADA2 was used to discard chimeric reads and other sequencing artifacts (Callahan et al., 2016). Taxonomy was assigned using a Naïve Bayes classifier trained using the 16S rRNA region, primer set, read length, and Greengenes 99% reference set version 13.8 (DeSantis et al., 2006; McDonald et al., 2012; Werner et al., 2012), including taxonomy for chloroplasts from eukaryotic microbes. Singletons were removed and samples rarefied to 2,500 sequences, which was the highest sequencing depth that included all biofilm samples (Figure S2). Relative abundance was determined by the number of reads in the rarefied dataset. Five samples (four carcass and one internal H. flavescens) were excluded due to insufficient sequence reads as a result of extraction or sequencing errors. Sequence files and metadata for all samples used in this study have been deposited in the NCBI SRA under number PRJNA526072.
Carrion-introduced operational taxonomic units (OTUs) in year one were determined by identifying those OTUs detected on salmon carrion prior to deposition in the stream, but not found in samples from either the control reach anytime during year one or the salmon reach before salmon were introduced (September). For year two, carrion-introduced OTUs were those OTUs not detected in year two in the control reach (background OTUs for year two) or during all of year one (both control and salmon reaches and carcasses—background OTUs resulting from any OTUs introduced in year one). The reasoning for excluding year one carcass-associated OTUs from year two carcass introduced, unique OTUs was to evaluate the integration of OTUs that the carrion introduces into biofilms and insects, rather than carrion associated OTUs themselves. Therefore, our strategy was to investigate the microbes completely naïve to the stream biofilms and internal insects during each year's salmon carcass introduction. These targeted sets of unique carrion introduced OTUs (year one and year two) were evaluated for presence in the downstream biofilms and internal insect microbiomes after carcass introduction. In addition, year one unique carrion introduced OTUs were evaluated for presence in year two carrion, to determine what OTUs not found in biofilms or internal insects were introduced both years.
Functional composition of the microbiome was predicted in silico using Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) on the 16S rRNA amplicon sequences (Langille et al., 2013), using default settings in the online Galaxy version (http://galaxy.morganlangille.com/). PICRUSt analysis requires closed-reference OTU picking using the Greengenes database, thus clustering was conducted on 97% similarity OTUs using VSEARCH in QIIME2 (Rognes et al., 2016). OTUs were normalized by copy number, and predicted functional categories were assigned using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database to predict KEGG orthologs, which were then collapsed at level 3 into hierarchical KEGG pathways by function (Kanehisa and Goto, 2000).
Statistical Analyses
Mean ± standard error (MSE) was calculated for each individual taxon. Estimates of α-diversity in microbial communities [observed OTUs, Chao 1 richness, Shannon H', and Faith's phylogenetic diversity (PD) indices] were calculated in QIIME2 based on OTU sequence read matrices (Caporaso et al., 2010). Statistical analyses were performed using R version 1.1.442 (R Core Team, 2018). Estimates of α-diversity in macroinvertebrate communities (genus richness, Simpson's diversity index) were calculated using the vegan 2.5-2 library diversity function in R (Oksanen et al., 2019). Differences in α-diversity metrics for each year's carcass epinecrotic microbial community prior to deposition were tested using unpaired, two-tailed t-tests, while differences in α-diversity metrics in each reach over time were tested separately using repeated measures ANOVA. Residuals vs. fit and normal probability plots were examined to determine if the assumptions of each statistical test were met. When assumptions were not met a Poisson distribution (count data) or log10 transformation (non-count data) was used. Confidence intervals (95%) were determined for coefficient estimates for log10-transformed data. Population densities of the three genera used for internal microbiome testing were examined using the same methods as α-diversity assessments.
Variation in microbial and macroinvertebrate community composition was visualized using non-metric multidimensional scaling (NMDS) and statistically evaluated for β-diversity metrics with PERMANOVA, a phylogeny based matrix (weighted UniFrac) for microbial communities and Bray-Curtis dissimilarity matrix for macroinvertebrate communities (standard for macroinvertebrate community analysis) using 99,999 permutations in the vegan 2.5-2 library “adonis” function in R (Anderson, 2001). For those samples where stream reach was statistically significant, we identified taxa (genus level for macroinvertebrates and family level for microbes) found in the salmon reach over time via indicator species analysis (ISA) with Indicator Value (IndVal) Index and its significance using 99,999 permutations in the “signassoc” function in the R package indicspecies, with p-value adjustments for multiple comparisons using the Sidak method (Dufrêne and Legendre, 1997; Cáceres and Legendre, 2009; Cáceres et al., 2010). All other statistical tests were considered significant at α = 0.05. Due to statistically significant differences in salmon carrion conditions during each year of the study (see section Results, Tables S3–S5), separate analyses were conducted for each year for all response variables.
Results
Macroinvertebrate Community Composition
A total of 13,730 aquatic macroinvertebrates were identified comprising 49 taxa, with Chironomidae the most relative abundant and a mean of 17% (±2%) (Table S6). No covariate (salmon treatment, time, or their interaction) was found to significantly influence macroinvertebrate density or richness in either year of the study (p > 0.05). Although no covariate influenced diversity during year one, in year two macroinvertebrate diversity increased by 0.0015 (± 0.0005) each day (p < 0.01), and there was a significant time × treatment interaction (p = 0.04). Only time had significant effects on macroinvertebrate community structure during both years (PERMANOVA: p < 0.01, Table 1). In year two, Brachycentrus (collector-filterer) density (individuals per 0.3 m2) was lower in the salmon reach relative to the control reach [S = 0 (±0), C = 6 (±3), ISA: p = 0.03, Figure S3]. Heptagenia population density significantly increased in year one and was also significantly higher in the salmon reach during both years [S = 14 (±4), C = 3 (±1), ANOVA: p < 0.02, Figure S3]. Baetis density significantly increased each year, with a significant time × treatment interaction due to a higher abundance in the control reach 9 months after salmon introduction during both years [S = 41 (±14), C = 191 (±41), p < 0.01, Figure S3]. We found a significant time × treatment interaction influencing Stegopterna density in year one, due to decreased abundance in the salmon reach 9–11 months after salmon introduction [S = 19 (±6), C = 109 (±44), p < 0.01]. Stegopterna density also significantly increased over time during year two (p < 0.01).
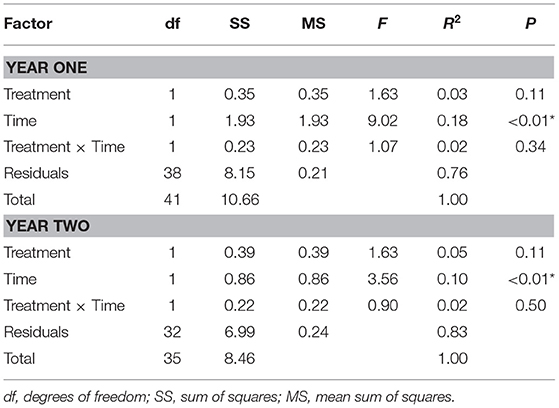
Table 1. PERMANOVA results testing benthic macroinvertebrate community structure based on Bray-Curtis distances for each year of the study with significant results (p < 0.05) indicated by an asterisk.
Salmon Carcass Epinecrotic Community Composition
A total of 11,219 microbial OTUs representing 51 phyla were identified in the carcass microbial communities. Moraxellaceae (γ-Proteobacteria) had the highest relative abundance [16% (±4%)]. While the diversity metrics Faith's PD and Chao 1 were not significantly different for each year of introduction (t-test: p > 0.1), the epinecrotic microbial communities were different between years, both taxonomically (OTU level) and functionally (PERMANOVA: p < 0.01, Table S3). Twenty microbial families were indicators of the year in the epinecrotic communities (ISA: p < 0.05, Table S4). Ruminocaccaceae, Geobaceracae, Succinivibrionaceae, Spirochaetaceae, an unknown family in Bacteroidales, and an unknown family in YS2 were the most significant indicator families (p < 0.01) and were all only found in year one carcasses. The most abundant indicator family, Sphingomonadaceae, had 2.7 times higher relative abundance in year one carcasses than year two carcasses [Y1 = 17% (±3%), Y2 = 6% (±1%)]. Functionally, 135 KEGG orthologs were indicators of salmon carcass introduction year (ISA: p < 0.05, Table S5). The most significant KEGG orthologs were caffeine metabolism, ether lipid metabolism, ethyl benzene degradation, isoflavonoid biosynthesis, mineral absorption and proteasome, all of which were greatest in year two carcass microbial communities (p < 0.01). The most abundant salmon indicator KEGG ortholog was DNA repair and recombination protein, which was 1.4 times higher in year two carcasses [Y1 = 41,587 (±2,865), Y2 = 57,353 (±3,244), ISA: p = 0.02]. Melanogenesis, a salmon indicator KEGG ortholog, was 3 times higher in year two [Y1 = 8 (±2), Y2 = 25 (±4), ISA: p < 0.01].
Biofilm Community Composition
A total of 11,051 and 9,434 OTUs represented epilithic biofilm communities in year one and year two, respectively, from 72 total samples (36 per year). The most abundant family was an unnamed family in the order Stramenopiles, representing 17% (±2%) of the community. Faith's PD significantly decreased over time during both years (year one: 2–3%, year two: 1–2%, ANOVA: p < 0.01), while Chao 1 richness increased over year one (0.6–0.8%, p < 0.01) and decreased in year two (0.4–0.6%, p < 0.01), but salmon treatment did not have an effect on Faith's PD or Chao 1 richness (p > 0.05). Treatment, time, and a treatment × time interaction influenced community composition during year one (PERMANOVA, p < 0.05, Table 2A, Figure S4), but only time was significant in year two.
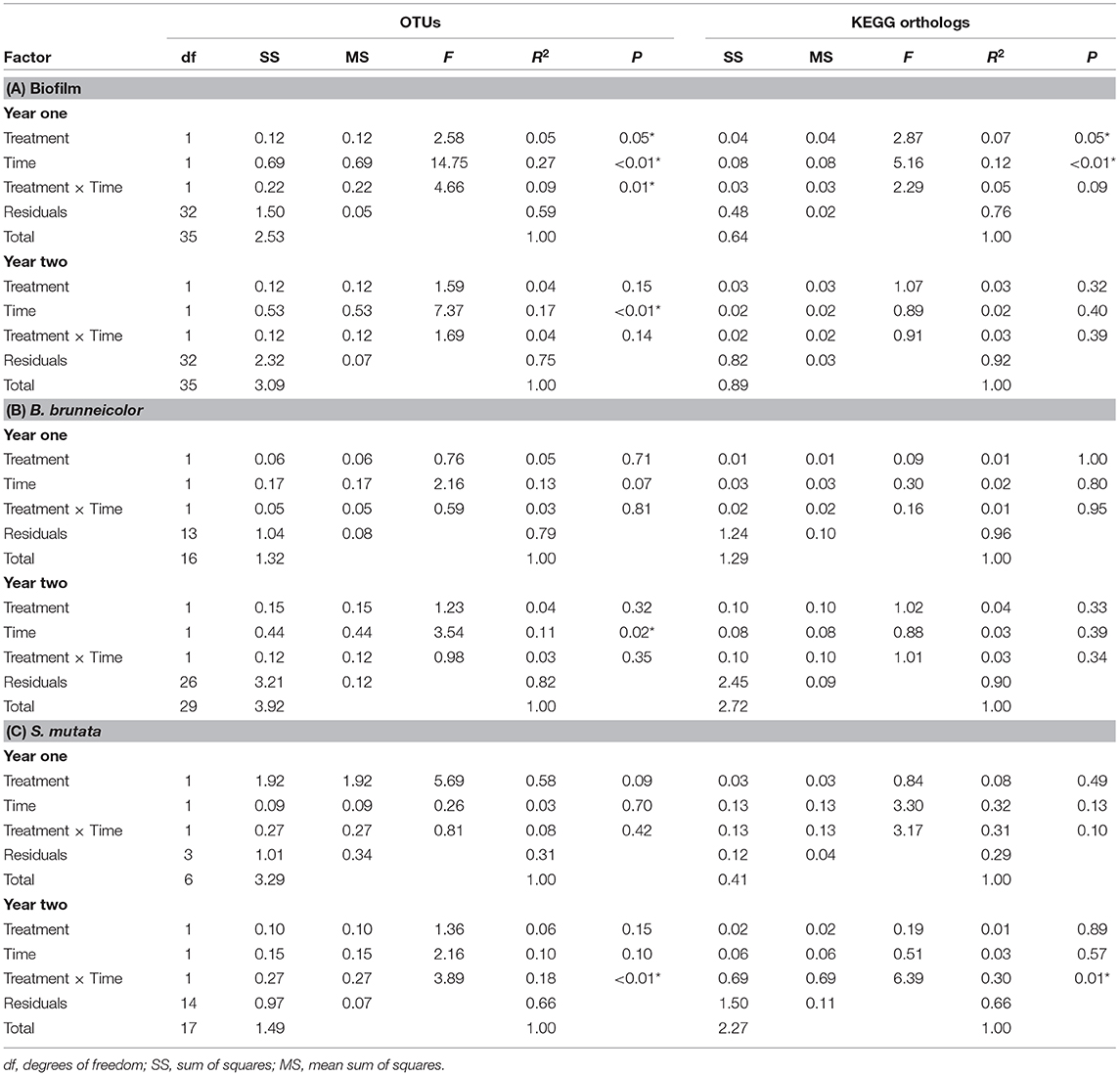
Table 2. PERMANOVA results testing microbial community structure based on the weighted phylogenetic distance (UniFrac) matrix for β-diversity and Jaccard distance matrix for KEGG orthologs among the microbial communities from the 2 years' biofilms (A), internal B. brunneicolor (B), and internal S. mutata (C), with significant results (p < 0.05) indicated by an asterisk.
Fifteen families were significant representatives of salmon reach biofilm communities in year one (p < 0.05, Table S7). Alteromonadaceae was 3.5 times more abundant in the salmon reach [ISA: p < 0.01, Figure 2]. Only two indicator families represented >10% of the community composition: Saprospiraceae and the abovementioned unnamed family in the order Stramenopiles. Saprospiraceae was 17 times lower in abundance in the salmon reach [0.7% (±0.2%)] compared to the control reach [12% (±3%)] 2 weeks after salmon introduction in year one, while an unnamed family in the order Stramenopiles exhibited the same pattern during both years, having 30 and 2.5 times lower abundance in the salmon reach in year one [S = 2% (±2%), C = 61% (±9%)] and year two [S = 25% (±13%), C = 60% (±4%)], respectively. Three indicator families identified from biofilms after carcass introduction were also indicator families of the salmon carcass microbial communities: Sphingomonadaceae [S = 1.8% (±0.4%), C = 0.9% (±0.1%)], Geobacteraceae [S = 0.12% (±0.04%), C = 0.04% (±0.02%)], and Xanthomonadaceae [S = 1.0% (±0.2%), C = 0.6% (±0.1%)], all of which had higher mean relative abundance in the salmon reach.
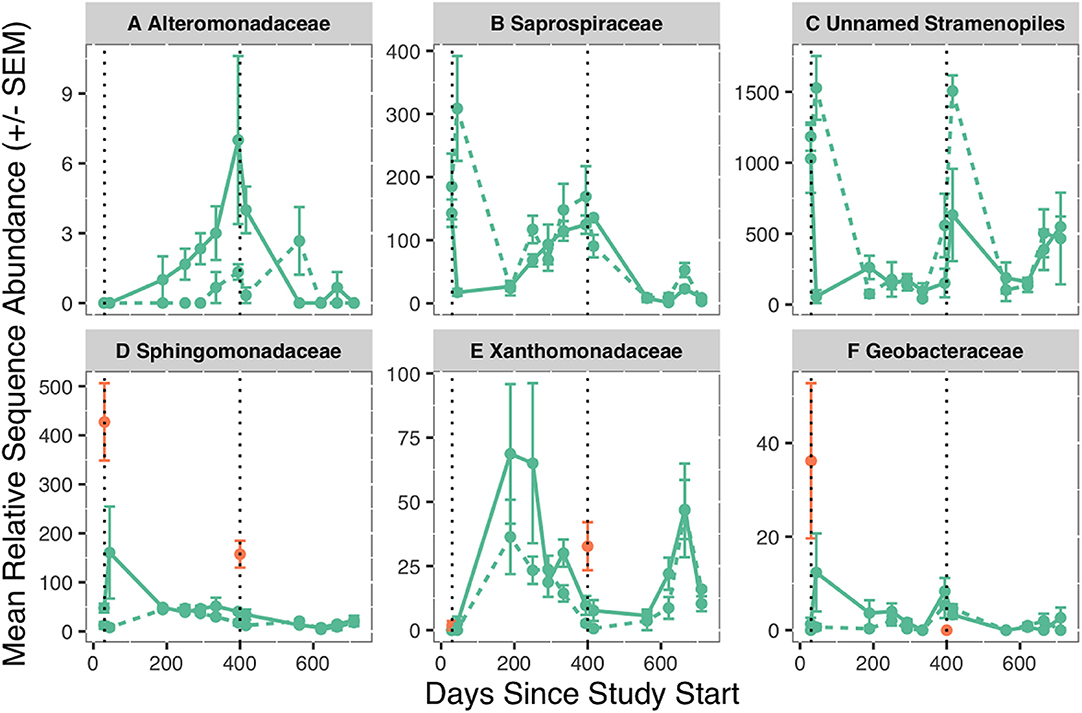
Figure 2. Mean relative sequence abundance (±SEM) of Alteromonadaceae (A), Saprospiraceae (B), Unnamed Stramenopiles family (C), Sphingomonadaceae (D), Xanthomonadaceae (E), and Geobacteraceae (F) in biofilms (green) over time. Sphingomonadaceae, Xanthomonadaceae, and Geobacteraceae that were part of the carrion associated communities are shown in orange, as these families were also indicator taxa for year of salmon carrion addition within the salmon carrion microbial communities. The dashed line is the control reach, and the solid line is the salmon reach. Black, dotted vertical lines represent the day of salmon introduction. Relative sequence abundance is the number of reads in the rarefied (2,500 reads) dataset.
In year one, treatment and time (but not interaction) significantly influenced the composition of KEGG orthologs (PERMANOVA, p < 0.05, Table 2A, Figure S4), yet there were no significant effects in year two. In year one biofilms, 113 indicator KEGG orthologs (p < 0.05, Table S8) were identified, with the most significant carcass KEGG ortholog indicator being fluorobenzoate degradation [S = 1,760 (±36), C = 1,580 (±42), ISA: p < 0.01], and the most abundant was the two-component system [S = 61,151 (±148), C = 55,800 (±154), ISA: p = 0.04], both of which were higher in the control reach. A total of 41 KEGG orthologs indicated salmon treatment biofilm communities, as well as year of carcass introduction. Of those shared indicator KEGG orthologs with higher abundance in the salmon reach, phosphotransferase system was the most abundant [S = 1,805 (±106), C = 1,411 (±163), ISA: p < 0.01]. Another one of those shared KEGG orthologs was melanogenesis, which was 1.7 times higher in abundance in salmon reach biofilms [S = 17 (±3), C = 10 (±2), ISA: p < 0.01].
Aquatic Insect Internal Microbial Community Composition
In the mayfly B. brunneicolor, 1,898 and 2,269 OTUs were detected in year one and year two, respectively (47 total samples with 3 individuals each). In year one, an unnamed family in Mollicutes was the most relatively abundant family [13% (±4%)], while in year two the most abundant was Pseudomonadaceae [23% (±4%)]. Neither time nor treatment significantly influenced Chao 1 richness (ANOVA: p > 0.05), yet Faith's PD decreased over time in year one (2.2–9.3%, p < 0.01) and year two (0.4–1.7%, p < 0.01). In year two, we also observed 99% lower Faith's PD in the salmon reach (31–100%), and a significant time × treatment interaction (p < 0.05).
Six families were indicators of internal microbial communities of B. brunneicolor in the salmon reach (p < 0.05, Table S9). Of these six, the most significant and abundant was the aforementioned unnamed family in Mollicutes, which was five times greater in the control reach [S = 5% (±2%), C = 25% (±7%), ISA: p < 0.01]. An unknown family in Rhizobiales (α-Proteobacteria) was an indicator of microbial communities in both internal B. brunneicolor from the salmon reach and from salmon carcass communities of year one, with 3.5 times higher abundance in the salmon reach [S = 7% (±2%), C = 2% (±1%), ISA: p < 0.01]. Only time significantly influenced the microbial community composition of the B. brunneicolor internal microbiome during year two (PERMANOVA: p = 0.02, Table 2B), while neither time nor treatment had significant effects in year one or influenced the KEGG orthologs detected in biofilms in either year (p > 0.05).
In the black fly S. mutata, a total of 449 and 1224 OTUs were detected in year one and year two, respectively (23 total samples with 3 individuals each), with Firmicutes being the predominant phylum [32% (±5%)]. Although no significant factors influenced Faith's PD in year one, in year two, mean diversity was 72 (±33) times higher in internal S. mutata in the salmon reach compared to the control reach (ANOVA: p = 0.05). Chao 1 richness was not influenced by time or treatment during either year (p > 0.05).
Treatment, time, nor their interaction significantly affected the microbial composition or functional KEGG ortholog community composition of internal S. mutata in year one (PERMANOVA: p > 0.05). In year two, the treatment × time interaction significantly influenced both the internal microbial community structure and function (p < 0.02, Table 2C). An unnamed family in Streptophyta was an indicator family of treatment in year two, with four times greater relative abundance in the control reach internal S. mutata [S = 1.1% (±0.4%), C = 4% (±1%), ISA: p < 0.03]. Melanogenesis was the only indicator KEGG ortholog for S. mutata internal communities from the salmon reach in year two, with four times greater relative abundance in the control reach [S = 21 (±4), C = 61 (±16), ISA: p = 0.05], which was also an indicator KEGG ortholog in salmon carcasses and biofilms (Figure S5).
The internal microbiome of the mayfly H. flavescens could not be compared between treatment and control reaches due to low-yield microbial DNA and a low sample size (n = 8).
Introduced Salmon Carcass Microbes
Of the total 686 salmon carcass-derived OTUs introduced in year one, 645, representing an average relative abundance of 63 ± 3%, were not found in biofilm or internal insect samples in non-salmon reaches (unique) (Figure S6). During year two, 1,786 [51% (±6%)] of a total of 2,196 were OTUs associated with introduced carcasses and not found in biofilm or internal insect samples anytime during year one, or in non-salmon reaches. Of unique OTUs introduced via carrion in the first year, 31 were detected in treatment biofilms (Table S10), of which 21 were only found in year one biofilms, six were only found in year two biofilms, and four were found in both years (Figure S6). Of the unique OTUs introduced via carrion in year two, 25 were detected in year two treatment biofilms. However, all unique OTUs introduced via salmon carrion and found in biofilms represented <2% of biofilm communities, except for a pulse 2 weeks after carcass introduction in year one when they increased to 5% (±1%) (Figures 3A and 3D). Three year one, salmon carrion unique OTUs found in biofilms were found in the upstream control reach in year two.
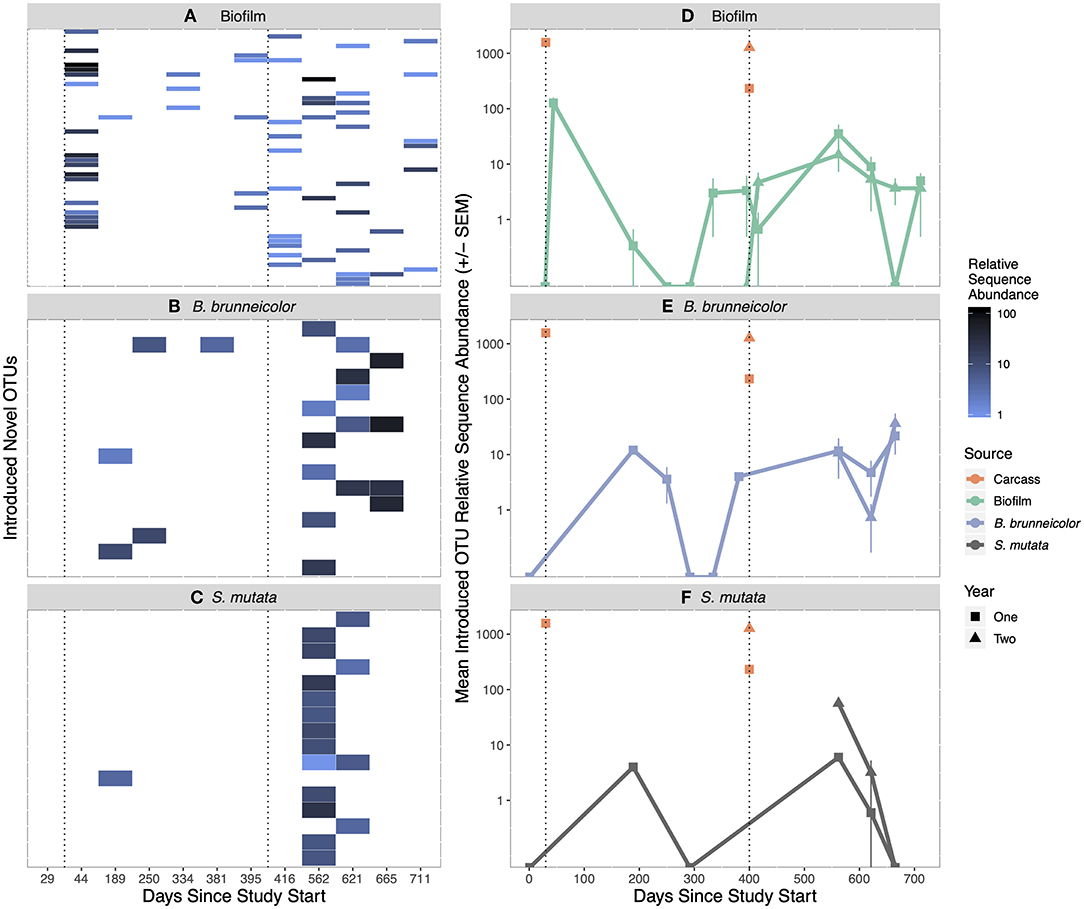
Figure 3. Heatmap of unique salmon carrion introduced OTUs found in treatment biofilm (A), internal B. brunneicolor (C), and internal S. mutata (E) after carcass introduction. Each y-axis row on the heatmap represents one unique salmon carrion introduced OTU. White color in heatmaps represents zero observations. Mean introduced OTU abundance (±SEM) in salmon carcasses and biofilms (B), internal B. brunneicolor (D), and internal S. mutata (F), over time. The orange carcass points represent the unique OTUs introduced to the stream for that year. The carcasses added in year 2 contained OTU's previously introduced in year 1 (orange square), as well as new OTU's not previously found in biofilm or insect samples (orange triangle). The sample (biofilm, internal B. brunneicolor, or internal S. mutata) points (in green, purple or gray, respectively) represent the amount of that year's salmon introduced OTUs found in biofilm samples. Black, dotted vertical lines represent the day of salmon introduction. Relative sequence abundance is the number of reads in the rarefied (2,500 reads) dataset.
Of the OTUs introduced via carrion in year one, nine were found in the internal B. brunneicolor communities collected in the salmon reach: three, five, and one OTU(s) were detected in year one, year two, and both years, respectively (Figure S7, Table S10). Four of these OTUs were also detected in treatment biofilms (Figures 3A and 3B). Of those unique OTUs introduced via carrion in year two, eight persisted in year two treatment B. brunneicolor internal communities, none of which were found in treatment biofilms or S. mutata. However, these carrion-introduced unique OTUs represented <1% of the relative abundance of internal B. brunneicolor communities (Figures 3B and 3E); three year one introduced OTUs found in internal B. brunneicolor were also found in the upstream control reach in year two.
Three OTUs introduced by carrion in year one and not found in biofilm or internal insect samples in non-salmon reaches were detected in internal S. mutata collected in the salmon reach: one in year one and two in year two (Figure S7, Table S10). None of these OTUs were found in biofilms or B. brunneicolor. Of the unique salmon carrion OTUs introduced in year two, 14 were detected in year two treatment internal communities of S. mutata. Three of those fourteen were also detected in treatment biofilms in year two. These introduced OTUs from both years represented a small proportion (<1%) of internal S. mutata communities (Figures 3C and 3F). In year two, four year one introduced OTUs found in internal S. mutata were also found in the upstream control reach.
Discussion
Macroinvertebrate Community Composition
Specific metrics of α-diversity and population density of macroinvertebrate communities were altered by salmon carrion additions, which were influenced by the population dynamics of four taxa: Brachycentrus, Baetis, Stegopterna, and Heptagenia. Higher Brachycentridae abundance has been detected 2–4 weeks after salmon carcass introduction in Idaho (Kohler et al., 2008), and Brachycentrus has been observed feeding on salmon carcasses in Alaska (Kline et al., 1997). Yet, in our study, Brachycentrus density was lower in the salmon reach, never representing more than 1% of the community, which did not support the hypothesis that collectors would increase in the salmon treatment reach. In Michigan, Brachycentrus populations can be drastically reduced by a microsporidium parasite (Kohler and Hoiland, 2001), which has an unknown life cycle. It is possible that salmon carrion introduces microsporidium spores, and a local outbreak could have lowered Brachycentrus population in the salmon reach, although there is no direct historical evidence of this parasite in Hunt Creek (Wills et al., 2006), and the presence of this parasite or its spores were not directly measured in this study. Additional salmon carcass introduction studies in other Great Lakes streams with Brachycentrus are needed to determine whether these observed changes were due to natural environmental shifts that occurred upstream of the salmon reach or a treatment effect. Changes in the phenology of this insect may be more important in structuring the population than the availability of resources. Great Lakes region salmon research shows that salmon carrion has a much smaller impact on stream biota than it does in its native range in the Pacific Northwest and Alaska (Janetski et al., 2013), leaving phenological population changes to have a greater impact.
Although the Brachycentrus population was consistently higher in the control reach, Baetis and Stegopterna populations were higher only during a short time peak in the control reach compared to the salmon reach. Both taxa belong to the collector functional feeding group, and were found to increase in density or have no significant response to salmon carrion subsidies in Alaska (Wipfli et al., 1998, 1999; Minakawa and Gara, 1999; Chaloner et al., 2002, 2004; Lessard et al., 2009). In the few studies that show lower collector densities in salmon-bearing streams, this was attributed to benthic disturbance by live salmon spawning behavior (Honea and Gara, 2009; Collins et al., 2011), which was not a factor in this study, as we introduced salmon carcasses directly to a naïve stream. Earlier insect emergence in streams that experience annual salmon runs could be attributed to an insect evolutionary response to salmon spawning disturbance (Moore and Schindler, 2010). Alternatively, the salmon nutrient subsidy may also lead to earlier emergence because of increased production and faster growth rate in insects. The short 50 years of evolutionary history of salmon in Great Lakes streams may preclude such responses in taxa such as Baetis and Stegopterna.
Although Brachycentridae, Baetis, and Stegopterna populations had higher mean relative abundance in the control reach, Heptagenia were higher in the salmon reach. Mayfly grazers, such as heptageniids, have been found to consume periphyton containing salmon-derived nitrogen (Schuldt and Hershey, 1995). Therefore, a salmon nutrient subsidy may have had positive effects on the Heptagenia population, supporting the hypothesis that grazer macroinvertebrates would increase in abundance in the salmon treatment reach.
Microbial Community Structure
The microbial communities residing in benthic biofilms were altered by carcass introduction over time for both years of the study, but this impact differed in each year. The introduced carcasses supported different microbial communities between the 2 years, which may contribute to this variation. Specifically, melanogenesis, a pathway responsible for pigment production, was an indicator KEGG ortholog in salmon carrion microbial communities, as well as in biofilms and internal S. mutata, but with contrasting effects. Melanin pigment in microbes is associated with virulence in pathogens and protection against environmental stressors (Nosanchuk and Casadevall, 2003). Each year's salmon carcasses were raised in different environments, causing more melanogenesis in year two salmon epinecrotic microbial communities. Then, the melanogenesis performing microbes became integrated into salmon treatment biofilms in year two, so there was increased melanogenesis in salmon treatment biofilms compared to control sites. This functional pathway existed in the stream prior to salmon carrion introduction, but salmon could have enriched the OTUs already present in biofilms, leading to higher abundance in that treatment reach. In contrast, the internal microbial communities within S. mutata had elevated melanogenesis in the control reach. This elevation may be due to an environmental change in the treatment reach due to salmon introduction, such as increased dissolved organic carbon (Schuldt and Hershey, 1995; Collins et al., 2011), which may decrease the abundance of microbes that perform melanogenesis. It should also be noted that KEGG orthologs are predicted via in silico analysis of the microbial community datasets, and further studies directly measuring microbial functions are needed.
Another shift in biofilm composition involved an unnamed family in Stramenopiles in year one, which was lower in the salmon reach compared to the control reach 2 weeks after carcass introduction. Functionally, Stramenopiles are a dominant group of primary producers (Burliga and Kociolek, 2016). We would expect Stramenopiles to be more abundant after leaf fall, due to increases in light with less canopy cover (Sumner and Fisher, 1979). Leaf fall occurred at the same time that salmon carrion was introduced into the stream. Primary production only marginally increases due to nutrients released by salmon carrion in Great Lakes streams (Schuldt and Hershey, 1995; Hershey and Wold, 1999); however, this production can be altered by stream conditions, such as light availability, habitat structure, and organic material (Cederholm et al., 1999). Nutrient addition in a Tennessee stream increased primary production in the autumn with increased light availability, but this effect was significantly lower in the presence of grazers (Rosemond et al., 2000). We detected an increase in grazer Heptagenia in the salmon reach, and thus these grazer communities could have influenced the biofilm response and limited autotrophic microbes despite nutrient inputs from salmon carrion.
In contrast to Stramenopiles, the Sphingomonadaceae (α-Proteobacteria) were over twice as abundant in the salmon reach, suggesting that heterotrophic bacteria respond positively to salmon carrion subsidies. Some of this increase in heterotrophic microbes can be attributed to rare OTUs introduced via salmon carrion, but most are likely due to an increase in organic matter. Benthic biofilms in streams with higher dissolved organic carbon often have higher Proteobacteria relative abundance (Gao et al., 2005). Proteobacteria also was found to be in high relative abundance in the internal microbiome of a predator mayfly in salmon-bearing streams (Pechal and Benbow, 2016); we found a similar trend with an unknown family in Rhizobiales (α-Proteobacteria), which had 3.5 times higher abundance in the internal microbiome of B. brunneicolor in the salmon reach of Hunt Creek.
Temporal Dynamics
Early research into salmon carcass decomposition in streams have indicated that periphyton first use salmon nutrients followed by primary consumers (Juday et al., 1932; Mathisen et al., 1988). Salmon-derived nutrients were found to peak in insects directly feeding on carcasses at 2 weeks after introduction, while biofilms and insects that were indirectly affected by salmon carrion had a peak in salmon derived nutrients at 2 months after introduction (Claeson et al., 2006). We found that biofilms responded 2 weeks after introduction, integrating a small amount (<2%) of unique salmon introduced OTUs into these epilithic communities, suggesting a more direct uptake path. Additionally, we were only able to sample a small amount of the total introduced microbial diversity via salmon in our carcass surface swabs, because we were not able to monitor the introduction of microbes from the gastrointestinal (GI) tract. We speculate the GI microbes may also contribute to novel taxa found in biofilms and internal insects that were not detected in this small relative abundance. The macroinvertebrate communities shifted several months after introduction, suggesting indirect and lagged carcass resource use. In our study, the small number of microbes unique to carrion and integrated into biofilms are subsequently integrated into consumers, as nutrients would be integrated up the food chain. An alternative explanation is that naïve Hunt Creek does not contain the necrophilous invertebrates of a typical Pacific salmon stream, and future research should focus on the direct and indirect pathways of introduced microbes.
It should be noted that the number of reads in a sequencing dataset do not necessarily directly translate to abundance in the environment, but rather serves as a proxy. Additionally, the detection of OTUs in biofilms does not indicate living microbes, but that the DNA of those microbes was present. Residual DNA from the salmon may slough off and be retained in biofilms, without the bacteria reproducing and functioning in the environment. Further studies, using active, transcribed forms of DNA are necessary to mechanistically determine whether these unique OTUs play an environmentally significant role. Past studies have shown that rare microbial taxa may play vital roles in maintaining biodiversity and having functional roles (Shade et al., 2014; Lynch and Neufeld, 2015; Jousset et al., 2017). For example, rare taxa can provide a “seed bank” that may increase in abundance when there is a local extinction of more abundant taxa or immigrate to another habitat where it can outcompete other resident microorganisms. Therefore, despite the low abundances of carrion-introduced OTUs in our system, they may play a more disproportionate role in biodiversity and ecosystem functioning that future studies should investigate.
Conclusions
In this study, we contribute to knowledge on ecology of salmon carrion decomposition by investigating the microbial fauna of a naïve stream following a salmon carcass addition. These data provide evidence that salmon introduce microbial taxa to recipient streams, and a small amount become incorporated into the ecosystem. Further, these taxa may elicit a cascading effect that influences stream producer and consumer communities through direct and indirect pathways. Salmon migration may ignite complex interkingdom interactions in stream communities, necessitating additional field and laboratory studies on allochthonous sources of microbes and their potential importance and mechanisms to ecosystem function. Therefore, the functional roles of these salmon-associated microbial taxa represent a frontier for ecological research.
Data Availability Statement
The datasets generated for this study can be found in the NCBI SRA under number PRJNA526072 [https://www.ncbi.nlm.nih.gov/bioproject/PRJNA526072].
Author Contributions
CL designed the microbial studies, collected the macroinvertebrate and microbial data in the field, analyzed the macroinvertebrate and microbial data, and wrote the paper. JP contributed to the study design, assisted in the field, and edited the paper. BG designed the overall experiment, assisted in the field, and edited the paper. DC assisted with experimental design and edited the paper. GL contributed to experimental design and edited the paper. MB contributed to study design, assisted in the field, and contributed to writing and editing the paper.
Funding
The Great Lakes Fishery Trust (Grant No. 2012.1244) provided funding for the salmon carcass addition. The Hutson Memorial Endowment Fund, Gordon E. Guyer Endowed Fellowship in Aquatic Entomology, and Merritt Endowed Fellowship in Entomology also provided funding for this research. Michigan State University College of Agriculture and Natural Resources, AgBioResearch and the College of Osteopathic Medicine provided additional funding for this project. This material is based upon CL's work supported by the National Science Foundation Graduate Research Fellowship Program under Grant No. DGE-1424871. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Sydney Manning, McKinley Brewer, Ryan Walquist, Caleb Armstrong, and Daniel Hulbert for assistance with fieldwork and sample collection. Gratitude is also expressed for Courtney Weatherbee, Joseph Receveur, Dustin Phelps, and Brianna Timmons for assistance with sample processing in the laboratory. For his assistance with salmon carrion acquisition and stream site access, we thank William Wellenkamp and the Michigan Department of Natural Resources.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2019.00505/full#supplementary-material
References
Anderson, M. J. (2001). A new method for non-parametric multivariate analysis of variance. Austral Ecol. 26, 32–46. doi: 10.1046/j.1442-9993.2001.01070.x
Benbow, M. E., Barton, P. S., Ulyshen, M. D., Beasley, J. C., DeVault, T. L., Strickland, M. S., et al. (2018). Necrobiome framework for bridging decomposition ecology of autotrophically and heterotrophically derived organic matter. Ecol. Monogr. 89:e01331. doi: 10.1002/ecm.1331
Bilby, R. E., Fransen, B. R., and Bisson, P. A. (1996). Incorporation of nitrogen and carbon from spawning coho salmon into the trophic system of small streams: evidence from stable isotopes. Can. J. Fish. Aquat. Sci. 53, 164–173. doi: 10.1139/f95-159
Blazar, J., Allard, M., and Lienau, E. K. (2011). Insects as vectors of foodborne pathogenic bacteria. Terr. Arthropod Rev. 4, 5–16. doi: 10.1163/187498311X543989
Bright, E. (2016). Aquatic Insects of Michigan. Available online at: http://www.aquaticinsects.org/
Burliga, A. L., and Kociolek, J. P. (2016). “Diatoms (Bacillariophyta) in rivers,” in River Algae, ed O. Necchi Jr. (Cham: Springer International Publishing), 93–128. doi: 10.1007/978-3-319-31984-1_5
Cáceres, M. D., and Legendre, P. (2009). Associations between species and groups of sites: indices and statistical inference. Ecology 90, 3566–3574. doi: 10.1890/08-1823.1
Cáceres, M. D., Legendre, P., and Moretti, M. (2010). Improving indicator species analysis by combining groups of sites. Oikos 119, 1674–1684. doi: 10.1111/j.1600-0706.2010.18334.x
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P. (2016). DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Caporaso, J. G., Knight, R., and Kelley, S. T. (2011a). Host-associated and free-living phage communities differ profoundly in phylogenetic composition. PLoS ONE 6:e16900. doi: 10.1371/journal.pone.0016900
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Huntley, J., Fierer, N., et al. (2012). Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6, 1621–1624. doi: 10.1038/ismej.2012.8
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Lozupone, C. A., Turnbaugh, P. J., et al. (2011b). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. U.S.A. 108 (Suppl. 1):4516–4522. doi: 10.1073/pnas.1000080107
Cederholm, C. J., Houston, D. B., Cole, D. L., and Scarlett, W. J. (1989). Fate of coho salmon (Oncorhynchus kisutch) carcasses in spawning streams. Can. J. Fish. Aquat. Sci. 46, 1347–1355. doi: 10.1139/f89-173
Cederholm, C. J., Kunze, M. D., Murota, T., and Sibatani, A. (1999). Pacific salmon carcasses: essential contributions of nutrients and energy for aquatic and terrestrial ecosystems. Fisheries 24, 6–15. doi: 10.1577/1548-8446(1999)024<0006:PSC>2.0.CO;2
Chaloner, D. T., Lamberti, G. A., Merritt, R. W., Mitchell, N. L., Ostrom, P. H., and Wipfli, M. S. (2004). Variation in responses to spawning Pacific salmon among three south-eastern Alaska streams. Freshw. Biol. 49, 587–599. doi: 10.1111/j.1365-2427.2004.01213.x
Chaloner, D. T., and Wipfli, M. S. (2002). Influence of decomposing Pacific salmon carcasses on macroinvertebrate growth and standing stock in southeastern Alaska streams. J. N. Am. Benthol. Soc. 21, 430–442. doi: 10.2307/1468480
Chaloner, D. T., Wipfli, M. S., and Caouette, J. P. (2002). Mass loss and macroinvertebrate colonisation of Pacific salmon carcasses in south-eastern Alaskan streams. Freshw. Biol. 47, 263–273. doi: 10.1046/j.1365-2427.2002.00804.x
Claeson, S. M., Li, J. L., Compton, J. E., and Bisson, P. A. (2006). Response of nutrients, biofilm, and benthic insects to salmon carcass addition. Can. J. Fish Aquat. Sci. 63, 1230–1241. doi: 10.1139/f06-029
Claesson, M. J., Wang, Q., O'Sullivan, O., Greene-Diniz, R., Cole, J. R., Ross, R. P., et al. (2010). Comparison of two next-generation sequencing technologies for resolving highly complex microbiota composition using tandem variable 16S rRNA gene regions. Nucleic Acids Res. 38:e200. doi: 10.1093/nar/gkq873
Collins, S. F., Moerke, A. H., Chaloner, D. T., Janetski, D. J., and Lamberti, G. A. (2011). Response of dissolved nutrients and periphyton to spawning Pacific salmon in three northern Michigan streams. Freshw. Sci. 30, 831–839. doi: 10.1899/10-164.1
Crump, B. C., Amaral-Zettler, L. A., and Kling, G. W. (2012). Microbial diversity in arctic freshwaters is structured by inoculation of microbes from soils. ISME J. 6, 1629–1639. doi: 10.1038/ismej.2012.9
Cwalinski, T. A., Godby, N. A. Jr., and Nuhfer, A. (2006). Thunder Bay River Assessment. Ann Arbor, MI: Michigan Department of Natural Resources.
DeSantis, T. Z., Hugenholtz, P., Larsen, N., Rojas, M., Brodie, E. L., Keller, K., et al. (2006). Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72, 5069–5072. doi: 10.1128/AEM.03006-05
Dufrêne, M., and Legendre, P. (1997). Species assemblages and indicator species:the need for a flexible asymmetrical approach. Ecol. Monogr. 67, 345–366. doi: 10.1890/0012-9615(1997)067[0345:SAAIST]2.0.CO;2
Feminella, J. W., and Hawkins, C. P. (1995). Interactions between stream herbivores and periphyton: a quantitative analysis of past experiments. J. N. Am. Benthol. Soc. 14, 465–509. doi: 10.2307/1467536
Gao, X., Olapade, O. A., and Leff, L. G. (2005). Comparison of benthic bacterial community composition in nine streams. Aquat. Microb. Ecol. 40, 51–60. doi: 10.3354/ame040051
Gende, S. M., Edwards, R. T., Willson, M. F., and Wipfli, M. S. (2002). Pacific salmon in aquatic and terrestrial ecosystems. BioScience 52, 917–928. doi: 10.1641/0006-3568(2002)052[0917:PSIAAT]2.0.CO;2
Gerig, B. S. (2017). Controls of Contaminant Biotransport by Pacific Salmon to Great Lakes Tributaries. Available online at: https://search.proquest.com/docview/2171772692/fulltextPDF/128E53B2306D4B07PQ/1?accountid=12598 (accessed April 9, 2019).
Gerig, B. S., Chaloner, D. T., Janetski, D. J., Moerke, A. H., Rediske, R. R., O'Keefe, J. P., et al. (2018). Environmental context and contaminant biotransport by Pacific salmon interact to mediate the bioaccumulation of contaminants by stream-resident fish. J. Appl. Ecol. 55, 1846–1859. doi: 10.1111/1365-2664.13123
Grossman, G. D., Nuhfer, A., Zorn, T., Sundin, G., and Alexander, G. (2012). Population regulation of brook trout (Salvelinus fontinalis) in Hunt Creek, Michigan: a 50-year study. Freshw. Biol. 57, 1434–1448. doi: 10.1111/j.1365-2427.2012.02806.x
Hagen, E. M., McCluney, K. E., Wyant, K. A., Soykan, C. U., Keller, A. C., Luttermoser, K. C., et al. (2012). A meta-analysis of the effects of detritus on primary producers and consumers in marine, freshwater, and terrestrial ecosystems. Oikos 121, 1507–1515. doi: 10.1111/j.1600-0706.2011.19666.x
Hall, R. O., and Meyer, J. L. (1998). The trophic significance of bacteria in a detritus-based stream food web. Ecology 79, 1995–2012. doi: 10.1890/0012-9658(1998)079[1995:TTSOBI]2.0.CO;2
Hershey, A., and Wold, A. (1999). Effects of salmon carcass decomposition on biofilm growth and wood decomposition. Can. J. Fish. Aquat. Sci. 56, 767–773. doi: 10.1139/f99-030
Honea, J. M., and Gara, R. I. (2009). Macroinvertebrate community dynamics: strong negative response to salmon redd construction and weak response to salmon-derived nutrient uptake. J. N. Am. Benthol. Soc. 28, 207–219. doi: 10.1899/08-030.1
Janetski, D. J., Chaloner, D. T., Moerke, A. H., Levi, P. S., and Lamberti, G. A. (2013). Novel environmental conditions alter subsidy and engineering effects by introduced Pacific salmon. Can. J. Fish. Aquat. Sci. 71, 502–513. doi: 10.1139/cjfas-2013-0292
Janetski, D. J., Chaloner, D. T., Moerke, A. H., Rediske, R. R., O'Keefe, J. P., and Lamberti, G. A. (2012). Resident fishes display elevated organic pollutants in salmon spawning streams of the Great Lakes. Environ. Sci. Technol. 46, 8035–8043. doi: 10.1021/es301864k
Janetski, D. J., Chaloner, D. T., Tiegs, S. D., and Lamberti, G. A. (2009). Pacific salmon effects on stream ecosystems: a quantitative synthesis. Oecologia 159, 583–595. doi: 10.1007/s00442-008-1249-x
Johnston, N. T., MacIsaac, E. A., Tschaplinski, P. J., and Hall, K. J. (2004). Effects of the abundance of spawning sockeye salmon (Oncorhynchus nerka) on nutrients and algal biomass in forested streams. Can. J. Fish. Aquat. Sci. Ott. 61, 384–403. doi: 10.1139/f03-172
Jousset, A., Bienhold, C., Chatzinotas, A., Gallien, L., Gobet, A., Kurm, V., et al. (2017). Where less may be more: how the rare biosphere pulls ecosystems strings. ISME J. 11, 853–862. doi: 10.1038/ismej.2016.174
Juday, C., Rich, W. H., and Kemmerer, G. I. (1932). Limnological studies of Karluk Lake Alaska. Bull. Bur. Fish. 47, 407–436.
Junqueira, A. C. M., Ratan, A., Acerbi, E., Drautz-Moses, D. I., Premkrishnan, B. N. V., Costea, P. I., et al. (2017). The microbiomes of blowflies and houseflies as bacterial transmission reservoirs. Sci. Rep. 7:16324. doi: 10.1038/s41598-017-16353-x
Kanehisa, M., and Goto, S. (2000). KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 28, 27–30. doi: 10.1093/nar/28.1.27
Kline, T. C., Goering, J. J., and Piorkowski, R. J. (1997). “The effect of salmon carcasses on Alaskan freshwaters,” in Freshwaters of Alaska: Ecological Syntheses Ecological Studies, eds A. M. Milner and M. W. Oswood (New York, NY: Springer), 179–204. doi: 10.1007/978-1-4612-0677-4_7
Kohler, A. E., Rugenski, A., and Taki, D. (2008). Stream food web response to a salmon carcass analogue addition in two central Idaho, U.S.A. streams. Freshw. Biol. 53, 446–460. doi: 10.1111/j.1365-2427.2007.01909.x
Kohler, S. L., and Hoiland, W. K. (2001). Population regulation in an aquatic insect: the role of disease. Ecology 82, 2294–2305. doi: 10.1890/0012-9658(2001)082[2294:PRIAAI]2.0.CO;2
Kuczynski, J., Stombaugh, J., Walters, W. A., González, A., Caporaso, J. G., and Knight, R. (2012). Using QIIME to analyze 16S rRNA gene sequences from microbial communities. Curr. Protoc. Microbiol. 27, Unit 10.17. doi: 10.1002/9780471729259.mc01e05s27
Lang, J. M., Erb, R., Pechal, J. L., Wallace, J. R., McEwan, R. W., and Benbow, M. E. (2016). Microbial biofilm community variation in flowing habitats: potential utility as bioindicators of postmortem submersion intervals. Microorganisms 4:1. doi: 10.3390/microorganisms4010001
Lang, J. M., McEwan, R. W., and Benbow, M. E. (2015). Abiotic autumnal organic matter deposition and grazing disturbance effects on epilithic biofilm succession. FEMS Microbiol. Ecol. 91:fiv060. doi: 10.1093/femsec/fiv060
Langille, M. G. I., Zaneveld, J., Caporaso, J. G., McDonald, D., Knights, D., Reyes, J. A., et al. (2013). Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 31, 814–821. doi: 10.1038/nbt.2676
Leff, L. G., McArthur, J. V., Meyer, J. L., and Shimkets, L. J. (1994). Effect of macroinvertebrates on detachment of bacteria from biofilms in stream microcosms. J. North Am. Benthol. Soc. 13, 74–79. doi: 10.2307/1467267
Leff, L. G., McArthur, J. V., and Shimkets, L. J. (1998). Persistence and dissemination of introduced bacteria in freshwater microcosms. Microb. Ecol. 36, 202–211. doi: 10.1007/s002489900107
Lessard, J. A. L., Merritt, R. W., and Berg, M. B. (2009). Investigating the effect of marine-derived nutrients from spawning salmon on macroinvertebrate secondary production in southeast Alaskan streams. Freshw. Sci. 28, 683–693. doi: 10.1899/08-141.1
Lessard, J. L., and Merritt, R. W. (2006). Influence of marine-derived nutrients from spawning salmon on aquatic insect communities in southeast Alaskan streams. Oikos 113, 334–343. doi: 10.1111/j.2006.0030-1299.14512.x
Levi, P. S., and Tank, J. L. (2013). Nonnative Pacific salmon alter hot spots of sediment nitrification in Great Lakes tributaries. J. Geophys. Res. Biogeosci. 118, 436–444. doi: 10.1002/jgrg.20044
Levi, P. S., Tank, J. L., Rüegg, J., Janetski, D. J., Tiegs, S. D., Chaloner, D. T., et al. (2013). Whole-stream metabolism responds to spawning Pacific salmon in their native and introduced ranges. Ecosystems 16, 269–283. doi: 10.1007/s10021-012-9613-4
Lindström, E. S., and Langenheder, S. (2012). Local and regional factors influencing bacterial community assembly. Environ. Microbiol. Rep. 4, 1–9. doi: 10.1111/j.1758-2229.2011.00257.x
Locey, K. J., and Lennon, J. T. (2016). Scaling laws predict global microbial diversity. Proc. Natl. Acad. Sci. U. S. A. 113, 5970–5975. doi: 10.1073/pnas.1521291113
Lynch, M., and Neufeld, J. (2015). Ecology and exploration of the rare biosphere. Nat. Rev. Microbiol. 13, 217–229. doi: 10.1038/nrmicro3400
Mathisen, O. A., Parker, P. L., Goering, J. J., Kline, T. C., Poe, P. H., and Scalan, R. S. (1988). Recycling of marine elements transported into freshwater systems by anadromous salmon. SIL Proc. 1922-2010 23, 2249–2258. doi: 10.1080/03680770.1987.11899884
McDonald, D., Price, M. N., Goodrich, J., Nawrocki, E. P., DeSantis, T. Z., Probst, A., et al. (2012). An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 6, 610–618. doi: 10.1038/ismej.2011.139
McEwen, H., and Leff, L. G. (2001). Colonization of stream macroinvertebrates by bacteria. Arch. Für Hydrobiol. 151, 51–65. doi: 10.1127/archiv-hydrobiol/151/2001/51
Merritt, R. W., Cummins, K. W., and Berg, M. B., (eds.). (2008). An Introduction to the Aquatic Insects of North America, 4th Edn. Dubuque, IA: Kendall Hunt Publishing.
Meyer, J. L., Strayer, D. L., Wallace, J. B., Eggert, S. L., Helfman, G. S., and Leonard, N. E. (2007). The contribution of headwater streams to biodiversity in river networks. J. Am. Water Resour. Assoc. 43, 86–103. doi: 10.1111/j.1752-1688.2007.00008.x
Minakawa, N., and Gara, R. I. (1999). Ecological effects of a chum salmon (Oncorhynchus keta) spawning run in a small stream of the Pacific Northwest. J. Freshw. Ecol. 14, 327–335. doi: 10.1080/02705060.1999.9663687
Mitchell, N. L., and Lamberti, G. A. (2005). Responses in dissolved nutrients and epilithon abundance to spawning salmon in southeast Alaska streams. Limnol. Oceanogr. 50, 217–227. doi: 10.4319/lo.2005.50.1.0217
Moore, J. C., Berlow, E. L., Coleman, D. C., Ruiter, P. C., de Dong, Q., Hastings, A., et al. (2004). Detritus, trophic dynamics and biodiversity. Ecol. Lett. 7, 584–600. doi: 10.1111/j.1461-0248.2004.00606.x
Moore, J. W., and Schindler, D. E. (2010). Spawning salmon and the phenology of emergence in stream insects. Proc. Biol. Sci. 277, 1695–1703. doi: 10.1098/rspb.2009.2342
Morley, S. A., Coe, H. J., Duda, J. J., Dunphy, L. S., McHenry, M. L., Beckman, B. R., et al. (2016). Seasonal variation exceeds effects of salmon carcass additions on benthic food webs in the Elwha River. Ecosphere 7:e01422. doi: 10.1002/ecs2.1422
Mulholland, P. J., Steinman, A. D., Palumbo, A. V., Elwood, J. W., and Kirschtel, D. B. (1991). Role of nutrient cycling and herbivory in regulating periphyton communities in laboratory streams. Ecology 72, 966–982. doi: 10.2307/1940597
Nosanchuk, J. D., and Casadevall, A. (2003). The contribution of melanin to microbial pathogenesis. Cell. Microbiol. 5, 203–223. doi: 10.1046/j.1462-5814.2003.00268.x
Oksanen, J., Blanchet, F. G., Friendly, M., Kindt, R., Legendre, P., McGlinn, D., et al. (2019). vegan: Community Ecology Package. Available online at: https://CRAN.R-project.org/package=vegan
Parsons, J. W. (1973). History of Salmon in the Great Lakes, 1850–1970. U.S. Bureau of Sport Fisheries and Wildlife; for sale by the Superintendent of Documents, U.S. Govt. Print. Office.
Pechal, J. L., and Benbow, M. E. (2016). Microbial ecology of the salmon necrobiome: evidence salmon carrion decomposition influences aquatic and terrestrial insect microbiomes. Environ. Microbiol. 18, 1511–1522. doi: 10.1111/1462-2920.13187
Polis, G. A., and Strong, D. R. (1996). Food web complexity and community dynamics. Am. Nat. 147, 813–846. doi: 10.1086/285880
R Core Team (2018). R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. Available online at: www.R-project.org
Ridley, E. V., Wong, A. C.-N., Westmiller, S., and Douglas, A. E. (2012). Impact of the resident microbiota on the nutritional phenotype of drosophila melanogaster. PLoS ONE 7:e36765. doi: 10.1371/journal.pone.0036765
Rognes, T., Flouri, T., Nichols, B., Quince, C., and Mahé, F. (2016). VSEARCH: a versatile open source tool for metagenomics. PeerJ 4:e2584. doi: 10.7717/peerj.2584
Rosemond, A. D., Mulholland, P. J., and Brawley, S. H. (2000). Seasonally shifting limitation of stream periphyton: response of algal populations and assemblage biomass and productivity to variation in light, nutrients, and herbivores. Can. J. Fish. Aquat. Sci. 57, 66–75. doi: 10.1139/f99-181
Rüegg, J., Tiegs, S. D., Chaloner, D. T., Levi, P. S., Tank, J. L., and Lamberti, G. A. (2011). Salmon subsidies alleviate nutrient limitation of benthic biofilms in southeast Alaska streams. Can. J. Fish. Aquat. Sci. 68, 277–287. doi: 10.1139/F10-145
Ruiz-González, C., Niño-García, J. P., and del Giorgio P. A. (2015). Terrestrial origin of bacterial communities in complex boreal freshwater networks. Ecol. Lett. 18, 1198–1206. doi: 10.1111/ele.12499
Schuldt, J. A., and Hershey, A. E. (1995). Effect of salmon carcass decomposition on Lake Superior tributary streams. J. N. Am. Benthol. Soc. 14, 259–268. doi: 10.2307/1467778
Shade, A., Jones, S. E., Caporaso, J. G., Handelsman, J., Knight, R., Fierer, N., et al. (2014). Conditionally rare taxa disproportionately contribute to temporal changes in microbial diversity. mBio 5:e01371-14. doi: 10.1128/mBio.01371-14
Steffan, S. A., Chikaraishi, Y., Dharampal, P. S., Pauli, J. N., Guédot, C., and Ohkouchi, N. (2017). Unpacking brown food-webs: animal trophic identity reflects rampant microbivory. Ecol. Evol. 7, 3532–3541. doi: 10.1002/ece3.2951
Stewart-Oaten, A., Murdoch, W. W., and Parker, K. R. (1986). Environmental impact assessment: “Pseudoreplication” in time? Ecology 67, 929–940. doi: 10.2307/1939815
Sumner, W. T., and Fisher, S. G. (1979). Periphyton production in Fort River, Massachusetts. Freshw. Biol. 9, 205–212. doi: 10.1111/j.1365-2427.1979.tb01504.x
Thompson, L. R., Sanders, J. G., McDonald, D., Amir, A., Ladau, J., Locey, K. J., et al. (2017). A communal catalogue reveals Earth's multiscale microbial diversity. Nature 551, 457–463. doi: 10.1038/nature24621
Tiegs, S. D., Levi, P. S., Rüegg, J., Chaloner, D. T., Tank, J. L., and Lamberti, G. A. (2011). Ecological effects of live salmon exceed those of carcasses during an annual spawning migration. Ecosystems 14, 598–614. doi: 10.1007/s10021-011-9431-0
Vannote, R. L., Minshall, G. W., Cummins, K. W., Sedell, J. R., and Cushing, C. E. (1980). The river continuum concept. Can. J. Fish. Aquat. Sci. 37, 130–137. doi: 10.1139/f80-017
Werner, J. J., Koren, O., Hugenholtz, P., DeSantis, T. Z., Walters, W. A., Caporaso, J. G., et al. (2012). Impact of training sets on classification of high-throughput bacterial 16S rRNA gene surveys. ISME J. 6, 94–103. doi: 10.1038/ismej.2011.82
Wills, T. C., Baker, E. A., Nuhfer, A. J., and Zorn, T. G. (2006). Response of the benthic macroinvertebrate community in a northern Michigan stream to reduced summer streamflows. River Res. Appl. 22, 819–836. doi: 10.1002/rra.938
Wipfli, M. S., Hudson, J., and Caouette, J. (1998). Influence of salmon carcasses on stream productivity: response of biofilm and benthic macroinvertebrates in southeastern Alaska, U.S.A. Can. J. Fish. Aquat. Sci. 55, 1503–1511. doi: 10.1139/f98-031
Keywords: salmon carcass, decomposition, allochthonous resources, community ecology, insect-microbe interactions
Citation: Larson CE, Pechal JL, Gerig BS, Chaloner DT, Lamberti GA and Benbow ME (2020) Microbial Community Response to a Novel Salmon Resource Subsidy. Front. Ecol. Evol. 7:505. doi: 10.3389/fevo.2019.00505
Received: 05 May 2019; Accepted: 12 December 2019;
Published: 10 January 2020.
Edited by:
Robert Ptacnik, Wasser Cluster Lunz, AustriaReviewed by:
Sofia Duarte, University of Minho, PortugalSarah Wheeler Keenan, South Dakota School of Mines and Technology, United States
Copyright © 2020 Larson, Pechal, Gerig, Chaloner, Lamberti and Benbow. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Courtney E. Larson, Y291cnRuZXllbGFyc29uMTI2QGdtYWlsLmNvbQ==; M. Eric Benbow, YmVuYm93QG1zdS5lZHU=
 Courtney E. Larson
Courtney E. Larson Jennifer L. Pechal
Jennifer L. Pechal Brandon S. Gerig
Brandon S. Gerig Dominic T. Chaloner4
Dominic T. Chaloner4 Gary A. Lamberti
Gary A. Lamberti M. Eric Benbow
M. Eric Benbow