- 1Institute of Farmland Water Conservancy and Soil-Fertilizer, Xinjiang Academy of Agricultural and Reclamation Science, Shihezi, Xinjiang, China
- 2College of Water Hydraulic and Architectural Engineering, Tarim University, Alar, Xinjiang, China
- 3Western Agricultural Research Center, Chinese Academy of Agricultural Sciences, Changji, Xinjiang, China
- 4Key Laboratory of Northwest Oasis Water-Saving Agriculture, Ministry of Agriculture and Rural Affairs, Shihezi, Xinjiang, China
- 5Xinjiang Production & Construction Corps Key Laboratory of Efficient Utilization of Water and Fertilizer, Shihezi, Xinjiang, China
Introduction: Soil salinization is a critical environmental issue affecting agricultural productivity worldwide, particularly in arid and semi-arid regions. This study focuses on the Xinjiang region of China, specifically the Xiao Haizi and Sha Jingzi irrigation areas, to explore the use of remote sensing technology for surface soil salinity estimation.
Methods: Exhaustive and filter-based feature selection methods were employed by integrating soil salinity data measured on the ground with 32 spectral features derived from Landsat 8 OLI remote sensing images. A 5-fold cross-validation method was used to identify feature combinations that resulted in higher R2 values. Moreover, the inversion accuracy of soil salinization monitoring models built using different feature combinations was compared across five machine learning algorithms: Support Vector Machine (SVM), XGBoost, Decision Tree (DT), Random Forest (RF), and AdaBoost.
Results: The results revealed that: (1) The AdaBoost and DT algorithms demonstrated high efficacy and precision in the prediction of soil salinity, with AdaBoost outperforming other algorithms in the validation set (R2 value of 0.892, MAE of 1.558, RMSE of 2.043), and DT showing the best performance in the training set (R2 value of 0.917, MAE of 0.838, RMSE of 1.182). (2) Feature combination 3, consisting of Salinity Index 5, Salinity Index 1, and Salinity Index 8, not only effectively extracted soil salinity information but also significantly improved the accuracy and efficiency of model estimations, effectively reflecting the actual situation of soil salinization in the irrigation area.
Discussion: This research provides robust methodological support for using remote sensing technology for soil salinity monitoring and management.
1 Introduction
Soil salinization is a global environmental issue that severely impacts human society and natural ecosystems. According to data from the Food and Agriculture Organization of the United Nations, over 1.3 billion hectares of soil globally suffer from salinization, while the area of soil salinization is growing at an annual rate of 2 million hectares, affecting more than 100 countries (Singh, 2018; Hammam and Mohamed, 2020). This problem is particularly pronounced in arid and semi-arid regions, such as parts of Asia, including China, Kazakhstan, and Iran, as well as in certain areas of Africa (Ivushkin et al., 2019; Cackett et al., 2022). In China, soil salinization is widespread, with significant implications for agriculture and environmental conservation. The Xinjiang region, in particular, is heavily affected, with saline-alkali soils covering an area of 36.7 million hectares, over 50% of which is concentrated in this province (Peng et al., 2016; Haj-Amor et al., 2022). Therefore, improving the accuracy of salinization information acquisition and monitoring in real-time is crucial for the sustainable development of agriculture and the protection of ecosystem functions in Xinjiang, China. Currently, soil salinity content (SSC) is the primary method for determining soil salinity levels. However, this method requires traditional soil sampling, processing, and laborious laboratory analyses, making it challenging to meet the requirements for large-scale, long-term SSC monitoring (Zhang et al., 2005; Bannari and Al-Ali, 2020).
Recent studies on remote sensing of soil salinity suggest that this approach may be more effective over large areas than traditional methods. This advantage lies in the ability of remote sensing to monitor the Earth’s surface at different spatial scales and temporal resolutions (Stavi et al., 2021; Paz et al., 2023). Where multispectral data are widely utilized in soil salinity and alkalinity studies, this includes auxiliary information such as vegetation indices, salinity indices, and band reflectance extracted from satellite images, all playing crucial roles in soil salinity monitoring (Wang F. et al., 2017; Stavi et al., 2021; Zhou et al., 2021; Measho et al., 2022). However, models constructed from original bands (such as the red and near-infrared bands) and standard vegetation indices (e.g., EVI and NDVI, etc.) along with salinity indices (SI, SI1) fail to make full use of the multispectral band information, leading to current research staying only at the stage of spectral feature construction. With the increasing number of relevant features, many researchers have begun to explore the impact of high-dimensional feature modeling on the accuracy of soil salinization inversion. They have found that the complexity of feature dimensions could increase model complexity, thereby reducing predictive performance. Selecting appropriate features reduces the dimensionality of the input data, decreases computational load, and also aids in identifying the most suitable variables for environmental monitoring and mapping. Understanding the optimal variables can contribute to designing more efficient remote sensing monitoring programs tailored for specific applications. Various feature selection methods have been proposed (Sun and Du, 2019; Kumar et al., 2020; Esmaeili et al., 2023), with some studies suggesting that combining multiple approaches can yield superior outcomes. For instance, Bajcsy and Groves (2004) integrated several feature selection techniques within their regression model to estimate soil conductivity. Similarly, Thenkabail et al. (2004) utilized multiple methods, including Principal Component Analysis (PCA), λ-λ R2 modeling, and stepwise discriminant analysis, to optimize feature selection for vegetation classification. Chen et al. (2022) calculated 55 environmental features from Landsat and terrain data, employing a hybrid TPE-XGBoost model, and selected 19, 11, 25, and 15 features in four different regions, achieving good performance in predicting soil salinity (R2 > 0.8). Wang et al. (2023) Using the Mixup-LGBM model combined with feature importance evaluation, it was found that among 62 original feature sets in the study area, DEM and human activities had a high impact on soil salinization. Therefore, it is necessary to consider different spectral band combinations, as well as the redundant information generated by various combinations of spectral bands, and use data dimensionality reduction techniques to eliminate redundant data and thus improve the estimation accuracy of the soil salinization monitoring model.
Furthermore, numerous studies (Farifteh et al., 2007; P. Leone et al., 2012; Yu et al., 2016) have successfully constructed soil salinity monitoring models using various independent variables (salinity indices, vegetation indices, original bands) combined with multiple regression methods to achieve good predictive results. Among them, Partial Least Squares Regression (PLSR) is widely used in soil salinity inversion modeling, a robust multivariate regression method particularly suitable for cases where predictor variables exhibit multicollinearity, with many studies reporting successful cases of soil salinity assessment using PLSR (Udelhoven et al., 2003; Zhang et al., 2011; Sawut et al., 2014). However, the relationship between remote sensing images and soil salinity is nonlinear, and these regression methods only focus on the relationship between covariates and soil salinity content, ignoring the fact that the formation of soil salinity is a complex process controlled by multiple factors. Thus, a simple linear summation of various aspects may not reveal the actual situation, whereas nonlinear models can better fit the contributions of numerous factors affecting soil salinity. Machine learning (ML), a branch of artificial intelligence, is particularly suited to dealing with the complex, non-linear relationships between soil salinity and remotely sensed features. Unlike traditional regression methods, which assume linear relationships, ML models can automatically detect and analyze intricate patterns in the data, making them highly effective in dealing with the many factors that influence soil salinity (Chlingaryan et al., 2018). Currently, machine learning techniques combined with various remote sensing images have successfully predicted soil salinity; for example, Wang J. et al. (2021) used random forest combined with remote sensing data to successfully predict soil salinity at multiple depths in the Tarim River Basin in southern Xinjiang, China. Wang J. et al. (2020) used the Cubist model combined with Sentinel-2 MSI to map soil salinity in the Abinur Lake Wetland National Nature Reserve with satisfactory accuracy. However, independent variables often fall short of fully revealing soil salinization patterns. Applying multiple features or feature combinations can improve the accuracy of soil salinization modeling. Notably, no universal feature set is suitable for salinization monitoring across all environments (Chen and Seo, 2023). Therefore, adaptively selecting an optimal subset of features based on local conditions is essential for enhancing salinity prediction models. Feature selection methods can identify representative input variables from numerous salinization factors. Using this refined feature set and various machine learning algorithms may improve the accuracy and timeliness of salinization information retrieval and monitoring at the irrigation district scale.
In this study, five machine learning algorithms, namely Random Forest (RF), Adaptive Boosting (AdaBoost), Support Vector Machine (SVM), Extreme Gradient Boosting (XGBoost), and Decision Tree (DT), were applied to select the spectral index and the combined spectral index that have the highest correlation with soil salinity among a total of 32 variables, including vegetation index, salinity index, and reflectance bands by using an exhaustive combination of features and a cross-validation method. The objective is to estimate the soil salinization in the Xiao Haizi irrigation area using these powerfully explanatory independent and combined variables, improving the accuracy and speed of soil salinity estimation. The study also validated and assessed the accuracy of these five different machine learning algorithms and selected the best-performing model based on the relationship with the indices and accuracy assessments, thus enabling the monitoring and inversion of soil salinization in the Xiao Haizi irrigation area. To validate the model’s adaptability and generalization ability, the model inversion results were also validated in the Sha Jingzi irrigation area (the flowchart of the study is shown in Figure 1). The results of this study will facilitate the acquisition of soil salinization information in irrigation districts and help mobilize local farmers, decision-makers, and environmental managers in this region to address soil salinity issues.

Figure 1. Flowchart of the proposed method for estimating the soil salinity based on machine learning models.
2 Materials and methods
2.1 Study area
The Xiao Haizi irrigation area (78°47′–79°34′E, 39°36′–40°4′N) is situated in Tumxuk City, part of the Third Division of the Xinjiang Production and Construction Corps in northwest China (Figure 2). This region lies at a geographically significant junction, bordered by the Taklamakan Desert to the east, the Pamir Plateau to the west, the Tianshan Mountains to the north, and the Karakoram Mountains to the south. It is home to the largest plain reservoir in northwest China, comprising the Xiao Haizi Reservoir and Yong’anba Reservoir, which collectively have a total storage capacity of 700 million cubic meters. This irrigation area is characterized by a temperate desert climate, marked by extended sunshine durations and pronounced diurnal temperature variations. The average annual temperature is 11.6°C, with a frost-free period of approximately 225 days. Annual rainfall is minimal, ranging from 34 to 39 mm, while evaporation is exceptionally high, reaching 2030–3,318 mm per year. The region includes 73,370 ha of cultivated land and 50,025 ha of ecological land, with rich natural resources such as Populus euphratica forests, Tamarisks, natural grasslands, and various wild plant species, covering an area of 80,040 ha. The topography varies in altitude from 1,024 to 1,075 m, sloping from southwest to northeast. However, the combination of low and flat terrain, arid climatic conditions, and shallow groundwater levels has resulted in severe soil salinization in this region.
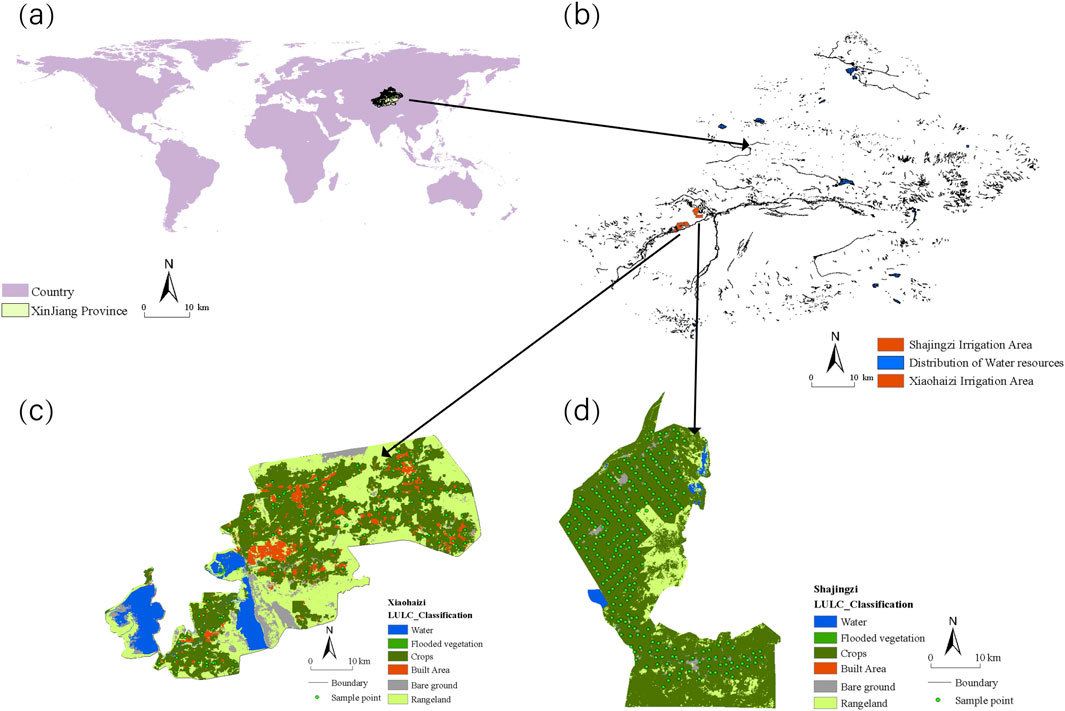
Figure 2. Location of study areas and sample points, (A) Xinjiang Province and (B) Distribution of Water Resources (C) Xiao Haizi LULC_Classification and Sample points (D) Sha Jingzi LULC_Classification and Sample points.
Model inversion validation was conducted in another challenging area, the Sha Jingzi Irrigation District (79°22′–80°16′E, 40°20′–40°26′N), located in the middle and lower reaches of the Aksu River Basin, about 60 km southwest of Aksu City (Figure 2). This region is geographically defined by Aisiman Lake to the east, flood protection barriers to the west, the Southern Xinjiang Railway to the north, and Dahalakule to the south, spanning an area of 99,000 ha. The district has a temperate continental arid climate, with annual precipitation concentrated mainly from June to August and an average rainfall of 62.9 mm. Like Xiao Haizi, evaporation rates are extremely high, averaging 1950 mm per year. The landscape is predominantly composed of forests and farmland, with relatively regular patterns of land parcels.
Both the Xiao Haizi and Sha Jingzi irrigation areas are located in southern Xinjiang, between the Tianshan and Kunlun mountains. Due to its unique geological and climatic conditions, this region is particularly vulnerable to soil salinization, with salt-affected soils accounting for 41.21% of the total arable land, well above the regional average. In addition, intensified land development and land use changes have altered the type, quantity and distribution of salt-affected soils in this area, posing a significant threat to the sustainability of local agriculture. As a result, rapid and accurate measurement of SSC is critical for managing soil salinization.
2.2 Data acquisition and preprocessing
2.2.1 Sample collection and measurement
In the Sha Jingzi and Xiao Haizi irrigation districts, spring brings high evaporation rates, frequent winds, and minimal rainfall, resulting in the lowest soil moisture levels of the year. The accumulation of soil salts on the surface due to capillary action results in a peak in salinization. Furthermore, farmland is typically unplanted and exposed during this season, making spring an optimal period for field sampling. In order to ensure that soil samples are representative and scientifically robust, the sampling design incorporated two key considerations. Firstly, using Google Earth imagery and considering local conditions and land use types, sampling points were systematically spaced every 200 ha to reconcile the point-scale observations with the spatial resolution of remote sensing imagery. Secondly, based on the results of previous inversions, the sampling points were stratified by salinization levels, including non-salinized, lightly, moderately and heavily salinized, as well as saline soil. This ensured a balanced representation across the salinity categories. This integrated approach provides a robust basis for accurate salinization monitoring and analysis. Of these, 115 soil samples from the 0–20 cm layer were collected in the Xiao Haizi irrigation area from April 13 to 25, 2021, and 250 soil samples were collected for soil salinity determination in the Sha Jingzi irrigation area from March 20 to 30, 2023. The five-point sampling method was used to collect 0–20 cm soil layer samples, which were mixed to create a representative composite soil sample. The geographic locations of these samples were recorded using a handheld GPS. Before further laboratory analysis, the samples were air-dried, crushed, and sieved through a 2 mm mesh. Potassium (K⁺) and sodium (Na⁺) ions were measured using an FP640 flame photometer, calcium (Ca2⁺) and magnesium (Mg2⁺) ions were determined with a Z-2000 atomic absorption spectrophotometer, sulfate (SO₄2⁻) ions were measured by indirect titration with EDTA, chloride (Cl⁻) ions were determined by silver nitrate titration. The dual indicator method was used to determine carbonate (CO₃2⁻) and bicarbonate (HCO₃⁻) ions. The detection limits for each ion were as follows: 0.05 mg/kg, 0.02 mg/kg, 0.03 mg/kg, 0.005 mg/kg, 0.07 mg/kg, 0.09 mg/kg, 0.07%, and 0.05%. The total soil salinity was determined by preparing a soil solution with a 1:5 soil-to-water mass ratio, which was then stirred, settled, precipitated, and filtered. Fifty ml of clear leachate was drawn and placed in a glass evaporation dish, which was dried to constant weight at 105°C–110 °C. The leachate was evaporated in a water bath, and small amounts of hydrogen peroxide were slowly added with a pipette while swirling the dish to ensure complete contact with the dried residue, oxidizing all organic matter. The dish was then placed in an oven at 105°C–110 °C for 2 hours, cooled in a desiccator for 30 min, and weighed. The amount of soil water-soluble salt, i.e., soil salinity content (Equation 1), was calculated as follows:
where m1, m2, t, m, and k correspond to glass evaporating dish mass (g), whole salt plus glass evaporating dish mass (g), fractionation times, air-dried soil sample mass (g), and moisture conversion coefficients of air-dried soil samples converted to dried soil samples, respectively.
2.2.2 Acquisition of multispectral images and calculation of feature variables
With its 16-day revisit cycle and 30-meter spatial resolution, the Landsat data provide high-quality geographic data for monitoring soil salinity. To reduce the uncertainty in predicting soil salinity, the acquisition dates of the Landsat images (Xiao haizi: Landsat 8 OLI 24 April 2021, Sha Jingzi: Landsat 8 OLI 29 March 2023) should be close to the sampling dates, and the cloud cover should be less than 10%. Meanwhile, to enhance monitoring accuracy, this paper used the RF classification algorithm under supervised classification to classify the study area into Water, Flooded, Crops, Built Area, Bare ground, and Poplar Forest Areas, with a classification accuracy of 92% and a Kappa coefficient of 0.93. The classification process was completed using Google Earth Engine (GEE), and the planting areas were extracted using GIS.
The smooth surface of saline soils typically exhibits a higher degree of reflectance in the visible and near-infrared spectral regions than non-saline soils. Specifically, within the visible spectrum (0.45–0.68 mm), saline soils reflect a higher proportion of incoming light, thereby providing a clear basis for distinguishing them from other surface features, such as average soils and vegetation. Furthermore, the research conducted by Masoud (2014) illustrates that saline soils subjected to low moisture conditions exhibit remarkably high reflectance within the blue and red spectral bands, a crucial attribute for precisely assessing salinization levels. Alterations in vegetation cover serve as a vital indirect indicator in salinization monitoring, as saline soils frequently demonstrate inadequate vegetation growth. Vegetation indices are an effective means of capturing this condition. Moreover, salinity index and other specifically developed spectral indices facilitate the direct detection of salt characteristics, thereby enabling the precise characterization of soil salinization levels. In order to gather detailed surface salinity information to build soil salinity prediction models in the Xiao Haizi and Sha Jingzi irrigation areas, a total of 32 features were selected from relevant studies. These features, which are highly correlated with salinization, serve as input variables for the salinity monitoring model. The aforementioned features, which included vegetation indices, salinity indices, and band reflectance values, were calculated using GEE and exported locally (Table 1).
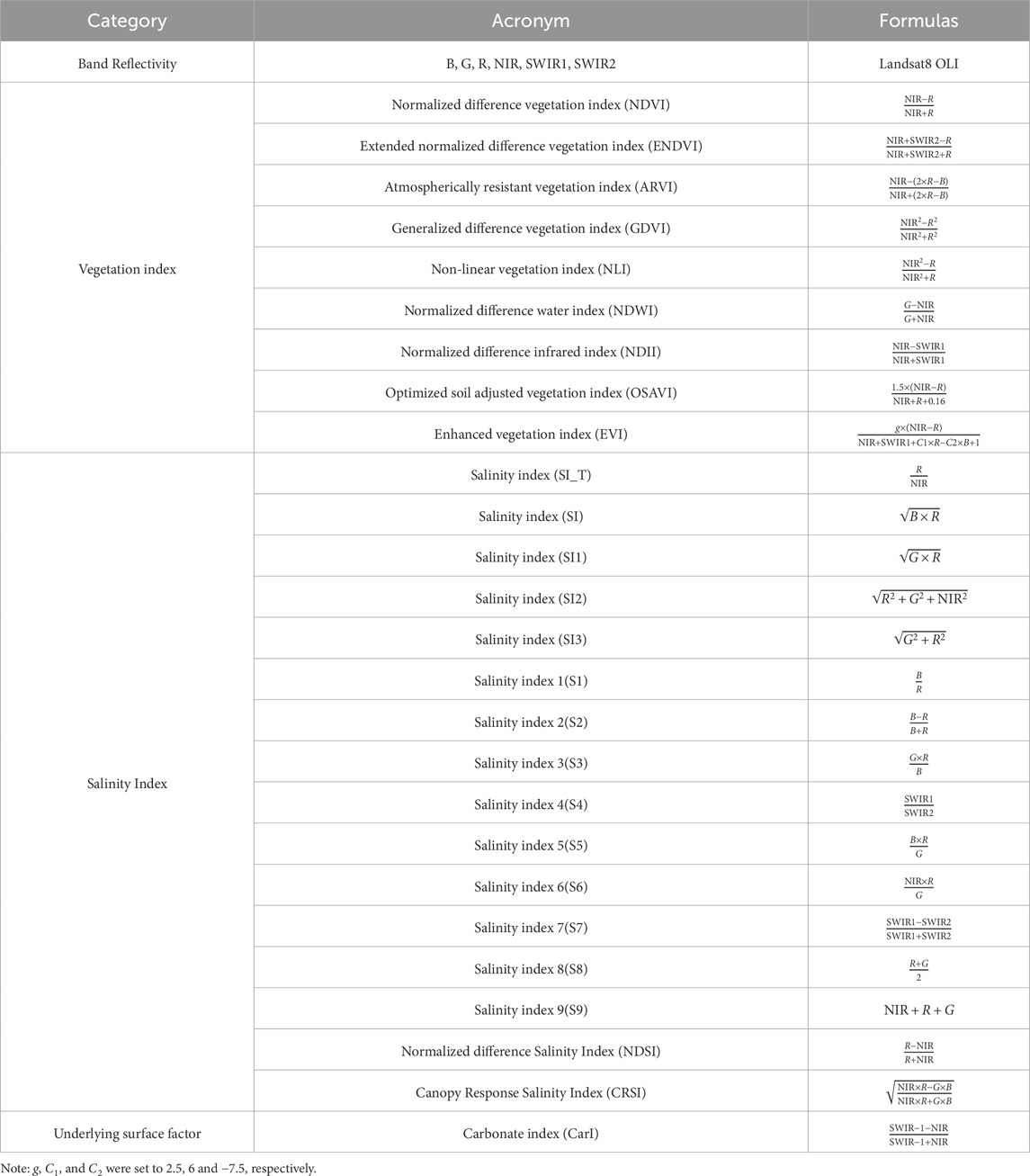
Table 1. Spectral reflectance bands, vegetation indices, and salinity indices were obtained from Landsat 8 OLI satellite data.
2.3 Feature selection
It should be noted that not all features contribute equally to the prediction of soil salinity in the context of modelling soil salinization monitoring and prediction. The application of feature selection techniques enables the identification of the most valuable feature subset, the optimization of model performance, the reduction of dimensionality and the lowering of computational costs. This process reduces redundancy and noise and mitigates the ‘curse of dimensionality’, enhancing the model’s generalization and interpretability. This study introduces an innovative approach to feature selection by employing a multi-stage combination of filter-based selection and exhaustive search methods, ensuring scientific rigor, thoroughness and efficiency in the feature selection process.
The filter-based feature selection method employs statistical correlations to identify essential features prior to model training. This is achieved by analyzing the relationships between features and the target variable. Given the multitude of factors that influence soil salinity, including soil properties, climate conditions, and topographical features, the filter-based method effectively excludes variables that are not directly related to salinity. This reduces the feature space and enhances the efficiency and stability of the modelling process. In this study, Pearson correlation coefficients were selected as the primary measure of feature relevance. The absolute value of the Pearson coefficient ranges from 0 to 1, with values closer to 1 indicating a stronger linear correlation with soil salinity content. By calculating the Pearson correlation between each feature and SSC, we reduced the initial 32 features to 16, which showed a significant correlation with SSC (|p| > 0.1). This process excluded non-significant features, enhancing the effectiveness and computational efficiency of subsequent modelling.
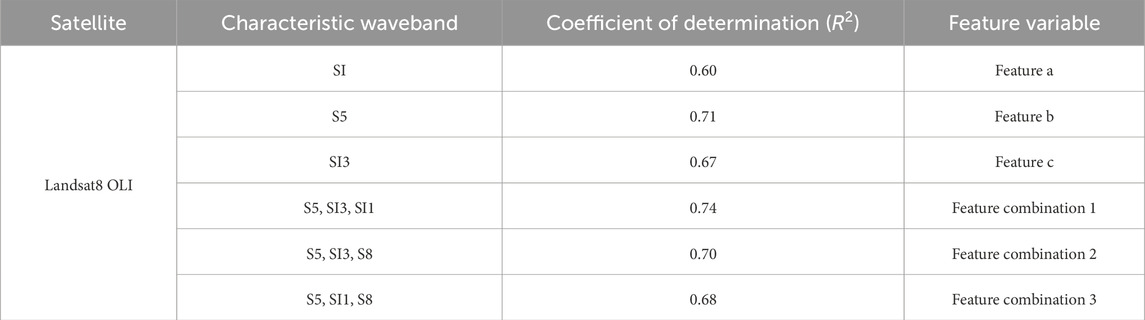
The 16 features selected through filter-based methods will serve as the original dataset, which will then be divided into training and testing sets at a ratio of 5:1. Subsequently, five-fold cross-validation and exhaustive search are applied to the training set to determine the optimal model parameters. The fundamental concept of cross-validation entails the further partitioning the training set into five subsets, with four subsets employed iteratively for training and one for validation. An exhaustive search is conducted for each training subset to test various model parameter combinations. The performance of each combination is evaluated against the validation data to identify the optimal parameters. As a brute-force approach, an exhaustive search systematically assesses all possible combinations, making it suitable for achieving precision on small-scale features or parameter sets. Ultimately, only feature sets with an average R2 more significant than 0.6 are retained for each study area (see Table 2).
2.4 Construction of soil salinity inversion model
Five machine learning algorithms were employed to address the nonlinear relationships and multivariate characteristics inherent in soil salinization modelling: SVM, XGBoost, DT, RF, and AdaBoost. The selection of each algorithm was based on its specific advantages, with parameters fine-tuned to enhance robustness and efficiency.
SVM was selected for its nonlinear mapping capability in high-dimensional spaces, rendering it an appropriate choice for complex multidimensional data. The RBF kernel was employed, with a C value 10 set to balance margin maximization, and error minimization, and gamma set to 5 to control the scope of nonlinear transformations, enhancing fine-grained feature detection. However, SVM can be computationally intensive for large datasets and is sensitive to hyperparameter selection (such as C and gamma), which may lead to overfitting. Cross-validation was applied to optimize these parameters to mitigate this risk, thereby improving generalization.
XGBoost, as a gradient boosting framework, leverages efficient residual learning and refined loss function optimization, maintaining high predictive accuracy and computational efficiency for large, complex datasets (Mantena et al., 2023). The key parameters were optimized through 5-fold cross-validation, with grounds set at 100, max_depth at 4, and eta at 0.2. This approach was employed to capture complex patterns while avoiding overfitting. However, it should be noted that XGBoost is sensitive to data noise, particularly in small or unbalanced datasets. This can result in a tendency for overfitting. To counteract this, training iterations and model complexity were carefully controlled, enhancing robustness.
DT was selected for its interpretability and clear visual structure, which reveals the layered relationships between input features and the response variable, making it suitable for identifying key drivers. Although DT effectively models hierarchical associations, a single tree is susceptible to sample fluctuations, leading to high variance and limited generalization. To improve model robustness, we employed ensemble strategies, specifically RF and AdaBoost.
RF utilizes Bagging to construct multiple decision trees, reducing variance through random sampling and feature selection, thereby enhancing generalization (Wang N. et al., 2020). In this study, we set 100 trees and randomly selected nine features at each split to maintain feature diversity while limiting node size to control complexity. However, RF may be influenced by irrelevant features in high-dimensional data, increasing computational load. We adjusted feature selection and parameter settings to ensure efficiency and precision.
AdaBoost, employing Boosting, incrementally adjusts error weights to aggregate weak classifiers (DT in this study), making it better suited for capturing data details and outliers (Haq et al., 2023). The maximum tree depth was set to 15 to identify complex patterns, while the random state was fixed to enhance repeatability and consistency. However, AdaBoost is sensitive to noise and may overfit when outliers are present. In order to mitigate this risk, the maximum depth and the number of weak classifiers were controlled.
2.5 Model accuracy evaluation parameters
It is crucial to assess the discrepancies between the predicted and actual values to evaluate the model’s accuracy and performance. In order to ascertain the most appropriate model for the prediction of soil salinity, this study employs three principal metrics: The metrics employed are R2, RMSE, and MAE. The R2 statistic measures the overall fit of the model, with a high R2 value indicating an effective capture of the data trends. However, R2 does not provide information regarding the specific magnitude of errors; therefore, its exclusive use may obscure essential details regarding the nature of these errors. The RMSE is a statistical measure that evaluates the magnitude of prediction errors. It is susceptible to outliers, data points that deviate significantly from the rest of the data set. By amplifying more significant errors, RMSE highlights the model’s performance in cases with substantial deviations. MAE quantifies the average absolute difference between predicted and actual values, indicating the model’s overall error magnitude. Unlike RMSE, MAE is less influenced by outliers, offering a balanced view of average prediction errors. The combination of these three metrics addresses the limitations of any single measure, creating a more thorough model evaluation. This balanced approach ensures the chosen soil salinity inversion model is accurate and reliable.
3 Results and analysis
3.1 Characteristics of salt-based ion distribution in soils of Xiao Haizi irrigation district
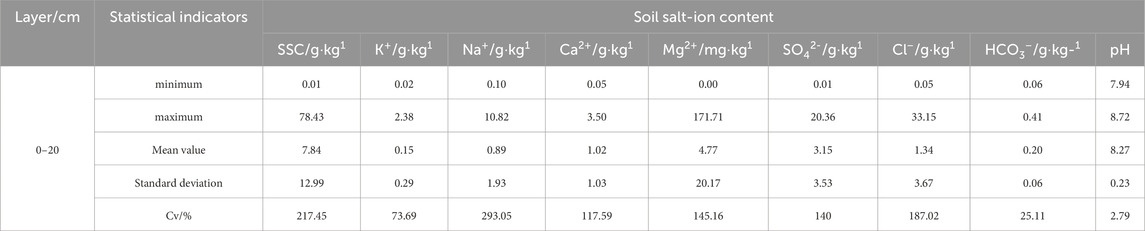
This article utilizes descriptive statistical analysis to investigate the variability characteristics of salt ions in the soil of the Xiao Haizi irrigation district. Table 3 shows that in the 0–20 cm soil layer of the study area, cations including K⁺, Na2⁺, Ca⁺, and Mg2⁺ are present. Among them, Na⁺ has the highest content with a mean value of 1.93 g·kg⁻1, followed by Ca⁺ with a mean value of 1.03 g·kg⁻1. The mean values for K⁺ and Mg2⁺ are 0.29 g·kg⁻1 and 20.17 mg·kg⁻1, respectively. The main anions are SO₄2⁻ and Cl⁻, with mean ion contents of 3.53 g·kg⁻1 and 1.34 g·kg⁻1, respectively. The content of CO₃2⁻ ions is almost zero. The ion content of salt indicates that sodium ions and sulfate ions are essential components of the local soil, and the salinization type of the study area belongs to the chloride sulfate type, with a mean total salt content of 7.84 g·kg⁻1. Additionally, there is a significant difference between the maximum and minimum values of salt ion content, reflecting the region’s uneven spatial distribution of salt ions. Moreover, the study area’s total salt content and the variation coefficients of salt ions (K⁺, Na⁺, Ca2⁺, Mg2⁺, SO₄2⁻, Cl⁻) exceed 100%, indicating substantial variability. The variation coefficient of HCO₃⁻ is between 10% and 100%, indicating moderate variability. Salt ions can affect the pH value of soil. Analysis of soil pH in the Xiao Haizi irrigation district reveals that the pH values of cultivated land soil range from 7.94 to 8.72, with a coefficient of variation of 2.79%. This indicates that the soil in the study area is mainly alkaline, with a small portion strongly alkaline. The variability of soil pH is low, indicating that the soil acidity and alkalinity distribution in cultivated land in the irrigation district is relatively consistent, with no significant spatial heterogeneity.
3.2 Results of spatial interpolation of soil salinity in irrigation district
SSC is a commonly used index to assess the degree of soil salinization (Taghizadeh-Mehrjardi et al., 2021). However, each region has classification standards (Nabiollahi et al., 2021). This study, based on the standards published by the Xinjiang Agricultural and Rural Department (Wu et al., 2018; Yu et al., 2018; Chi et al., 2019; Peng et al., 2019; Gharaibeh et al., 2021), soil salinity values were divided into five categories: non-salinized (SSC < 3 g/kg), slightly saline (3 g/kg < SSC < 6 g/kg), moderately saline (6 g/kg < SSC < 10 g/kg), intensely saline (10 g/kg < SSC < 20 g/kg), and highly saline (SSC > 20 g/kg).
The spatial characteristics of soil properties make them particularly amenable to geostatistical analysis, thereby rendering Geostatitics a valuable tool for the study of soil distribution patterns and spatial variability. Geostatistics elucidates the spatial distribution patterns of soil properties and establishes a correlation between these patterns and ecological processes, thereby facilitating a more comprehensive comprehension of soil distribution dynamics. It is a widely used tool in salt accumulation research, particularly for interpolating salt buildup from soil sample analysis. Kriging, a term encompassing generalized least-squares regression methods, is an effective approach, providing linear, unbiased estimates while accounting for clustering by weighting nearby sample points. SK represents the most basic form of Kriging and is designed to extend the technique to multivariate data sets, including auxiliary information, to enhance predictive performance. SK is particularly well-suited to situations where only a limited number of auxiliary variables are available and only cover some sample points. Given the markedly elevated and depressed soil salinity values observed in the samples, utilizing the mean as a predictor could potentially introduce bias. Consequently, this study employs the median soil salinity values for the Sha Jingzi and Xiao Haizi irrigation districts as the anticipated values within the SK model, thereby enhancing the representation of the data’s central tendency, with median values of 3.79 g/kg and 3.44 g/kg, respectively. This approach allows for the direct visualization of soil salinization distribution patterns by comparing the spatial interpolation results derived from ground sampling monitoring data with the inversion results of five machine learning models. This allows for an assessment of the accuracy of each model, thereby further enhancing our understanding of soil salinization dynamics across the study areas.
The results of the interpolation analysis indicate that the soil salinity in the Xiao Haizi irrigation district (Figure 3) displays notable banded and patchy distribution patterns. The highest salinity levels are concentrated in the northern and eastern regions, represented by prominent red and orange areas, indicating marked spatial heterogeneity. The elevated salinity in these areas suggests severe soil degradation, significantly impacting crop growth. In contrast, non-saline regions, characterized by lower salinity levels and relatively favorable soil conditions, are primarily located in the south and southwest. Moderate salinity regions are scattered, mainly in the north and east, where larger patches of moderate salinization create an observable clustering effect.

Figure 3. Spatial distribution of soil salinity in the 0–20 cm soil layer of Xiao Haizi Irrigation Area.
By contrast, the Sha Jingzi irrigation district (Figure 4) exhibits a distinct spatial configuration, characterized by elevated salinity levels in a non-uniform distribution, predominantly concentrated in the central and southern regions. This observation suggests the existence of localized instances of severe salinization. The non-saline areas, representing better soil conditions, are primarily located along the eastern edge and northwest corner. The low-salinity areas, predominantly indicated by the color green, are concentrated in the northern and central regions, reflecting a minor salt accumulation. The moderately saline areas, indicated by yellow, are concentrated in the central part, where soil salinity is relatively high. The areas of high salt accumulation are indicated by red and orange in the southern and central parts, underscoring an urgent need for remediation, such as desalinization projects, salt-tolerant crops, or land-use adjustments to mitigate soil salinization risks.

Figure 4. Spatial distribution of soil salinity in the 0–20 cm soil layer of Sha Jingzi Irrigation Area.
3.3 Model accuracy evaluation analysis
This study selected 32 variables, including vegetation indices, salinity indices, and reflectance bands. Six key input variables with strong explanatory power were identified using a multi-stage combination of filter-based feature selection and exhaustive search methods (see Section 2.3). These include three individual variables (Feature a, Feature b, Feature c) and three combined variables (Combination 1, Combination 2, Combination 3). These variables were then applied as inputs for five machine learning algorithms (RF, AdaBoost, SVM, XGBoost, DT) to predict soil salinity in the Xiao Haizi irrigation district.
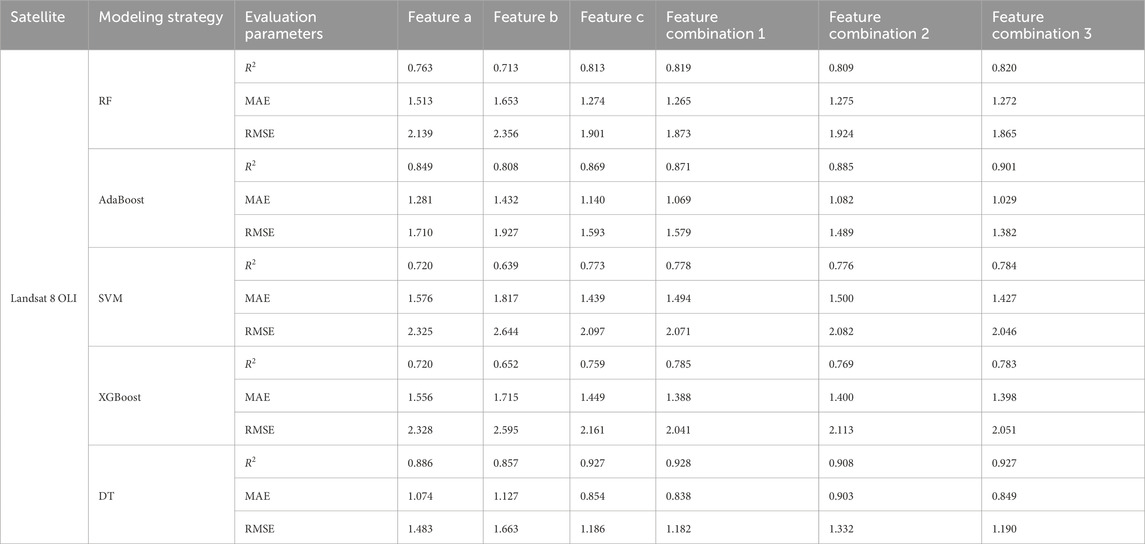
The training results (Table 4) indicate that the DT model outperformed the other algorithms, achieving R2 values above 0.857 for all six input variables. For Features c, Combinations 1, 2, and 3, R2 values exceeded 0.908, with corresponding MAE and RMSE values of 0.854, 0.838, 0.903, 0.849 for MAE, and 1.186, 1.182, 1.332, 1.19 for RMSE, respectively. Among these, Combination 1 yielded the highest R2 (0.917) along with the lowest MAE (0.838) and RMSE (1.182), suggesting that the DT model with Combination 1 was the most effective for extracting soil salinity information. The training accuracy ranking across models was DT > AdaBoost > RF > SVM > XGBoost.
Validation results (Figures 5–9) revealed that the AdaBoost algorithm outperformed other models, achieving R2 values above 0.7 for all six input variables. For Combinations 1, 2, and 3, R2 values were above 0.870, with MAE values below 1.667 and RMSE values below 2.170. Notably, Combination 3 achieved an R2 of 0.892, an MAE of 1.558, and an RMSE of 2.043. These results demonstrate that AdaBoost, combined with Feature Combination 3, has a strong potential for accurate soil salinity extraction with acceptable accuracy. The validation accuracy ranking was AdaBoost > DT > RF > SVM > XGBoost.
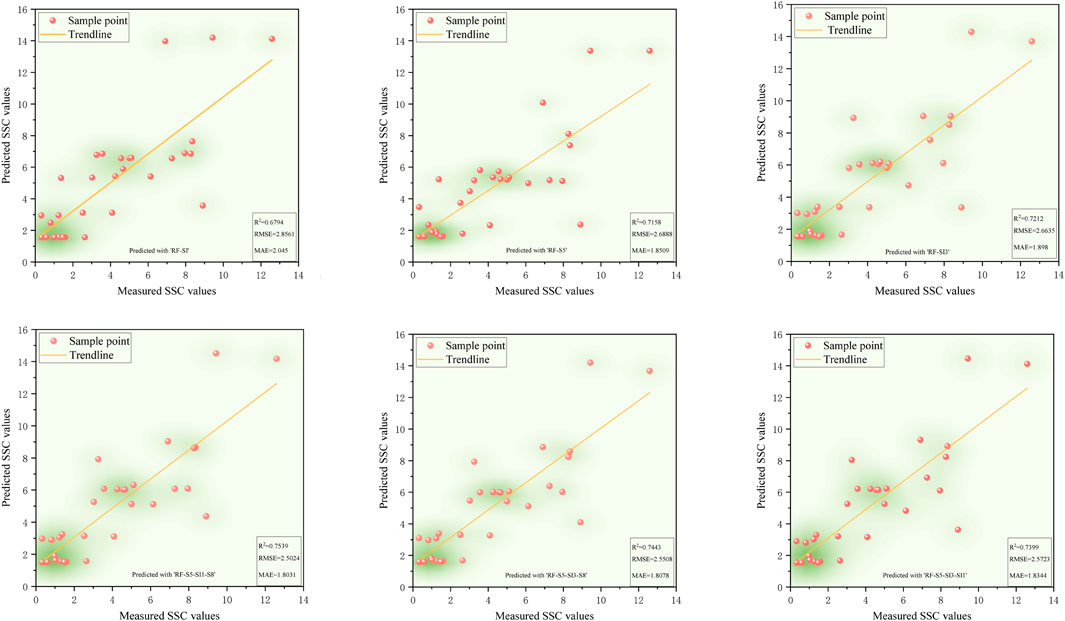
Figure 5. The scatter plot of measured soil salinity values versus predicted values in the RF model.
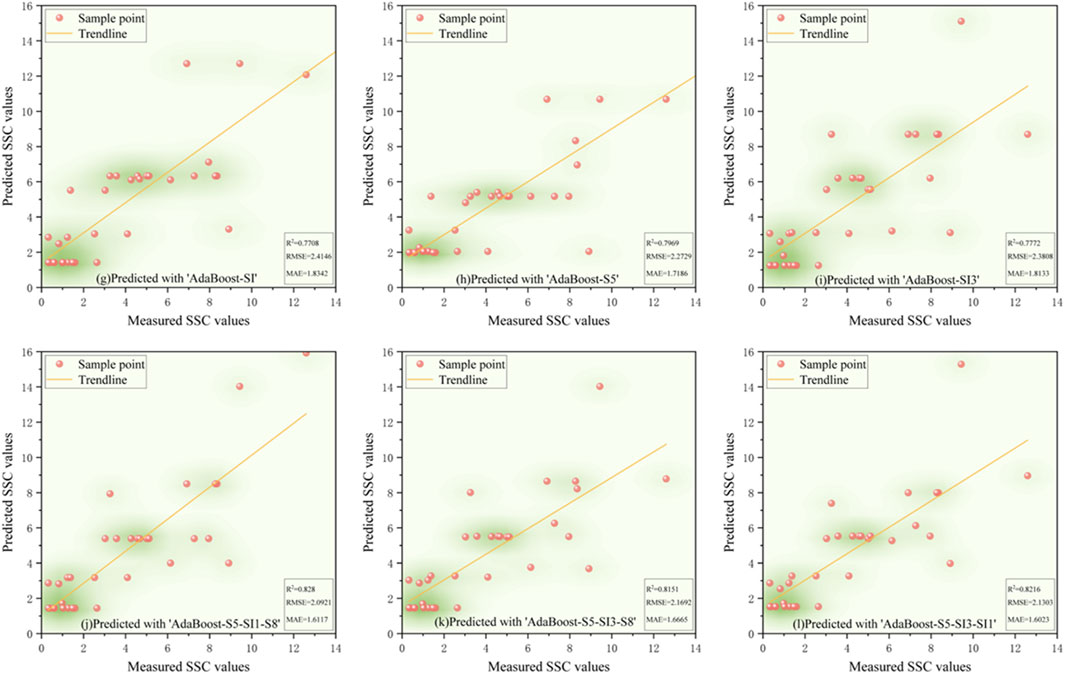
Figure 6. The scatter plot of measured soil salinity values versus predicted values in the AdaBoost model.

Figure 7. The scatter plot of measured soil salinity values versus predicted values in the SVM model.
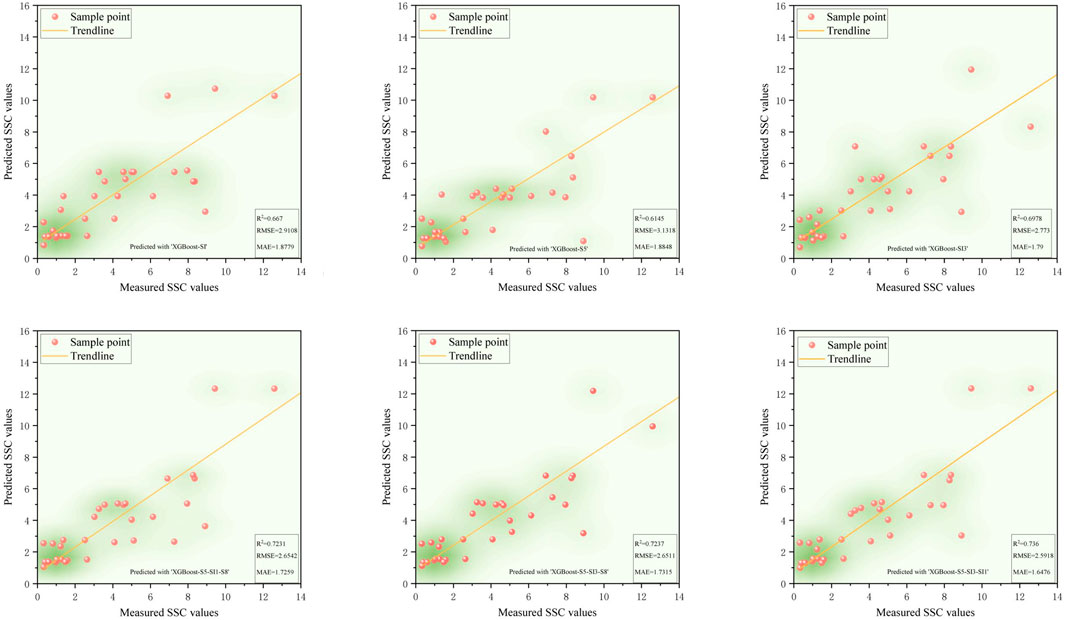
Figure 8. The scatter plot of measured soil salinity values versus predicted values in the XGBoost model.
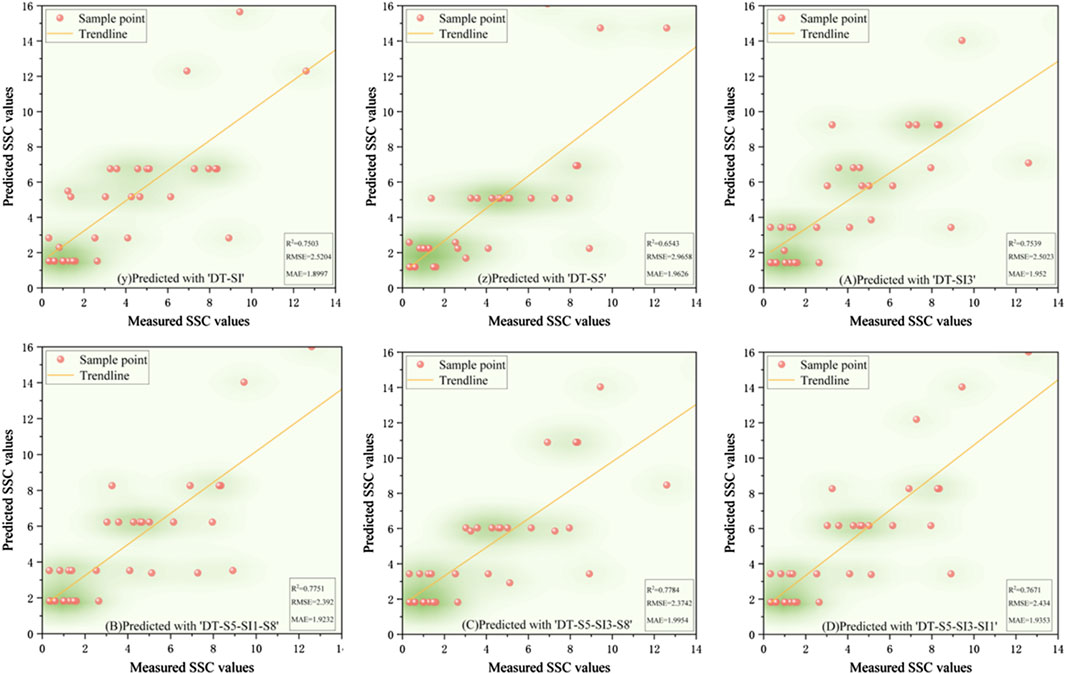
Figure 9. The scatter plot of measured soil salinity values versus predicted values in the DT model.
In summary, both AdaBoost and DT models demonstrated high learning capacity and accuracy for soil salinity inversion. AdaBoost performed superior in the validation, and DT performed best on the training set. Elnaggar and Noller (2009) similar results in salinity mapping with decision trees (DTs) were observed, attributing it to DT’s ability to incorporate diverse predictive factors during modeling. However, DTs are prone to overfitting, meaning they perform well on training data but poorly on untrained data. In contrast, AdaBoost mitigates noise by weighting, combining multiple weak learners into a robust predictor, and enhancing stability on new samples like the validation set. Compared to single models like decision trees or SVM, AdaBoost has a greater tolerance for complex data, effectively reducing overfitting in the validation set. AdaBoost’s strong classification performance has led to applications in ensemble learning, including image recognition, fruit biochemical parameter estimation, and complex change prediction models.
3.4 Spatial mapping results of salt distribution in the Xiao Haizi and Sha Jingzi irrigation districts
The appropriate machine learning algorithm enhances the accuracy and robustness of capturing soil salinization patterns. It effectively reduces model bias across diverse regional conditions, supporting efficient analysis and decision-making in salinization monitoring. As shown in Section 3.3, the AdaBoost model demonstrated excellent fit and low error on training and validation sets, indicating a strong ability to learn and represent soil salinity data. Each feature variable may offer unique spatial information or capture distinct salinity distribution characteristics. Thus, mapping with all selected features (see Table 2) provides a comprehensive view of soil salinization distribution across the study area. As shown in Figure 10, non-saline areas are widely distributed in the western and southeastern Xiao Haizi irrigation districts. At the same time, slightly saline soils are primarily found in the eastern and central regions, typically forming a blue transitional band along the edges of non-saline areas. This pattern is particularly pronounced in the model built with a salinity index of S5, indicating that the S5 index, constructed from blue, red, and green bands, effectively identifies slightly saline soils. Moderately saline soils are scattered across the central and southeastern regions, often adjacent to slightly saline areas. In contrast, intensely saline soils are primarily distributed in the eastern, northern, and parts of the central region. Extremely saline soils are concentrated in specific areas in the north and east, typically within or along the edges of intensely saline zones. Notably, models built with single features show limited ability to identify these two soil types. Figure 11 shows the distribution of soil salinity in the validation area, the Sha Jingzi irrigation district. Non-saline areas are extensively distributed across all models, predominantly in the west and south of the region. Slightly saline soils are more concentrated in the east, north, and parts of the central region. Moderately saline soils cluster in the central and southeastern areas, while intensely and extremely saline soils are sparse. Highly saline soils occur in small clusters in the eastern and northern central regions, while extremely saline soils occur sporadically in the north and east. Models with multiple feature combinations show higher accuracy in identifying high salinity areas, suggesting that multi-feature combinations may improve model prediction accuracy.
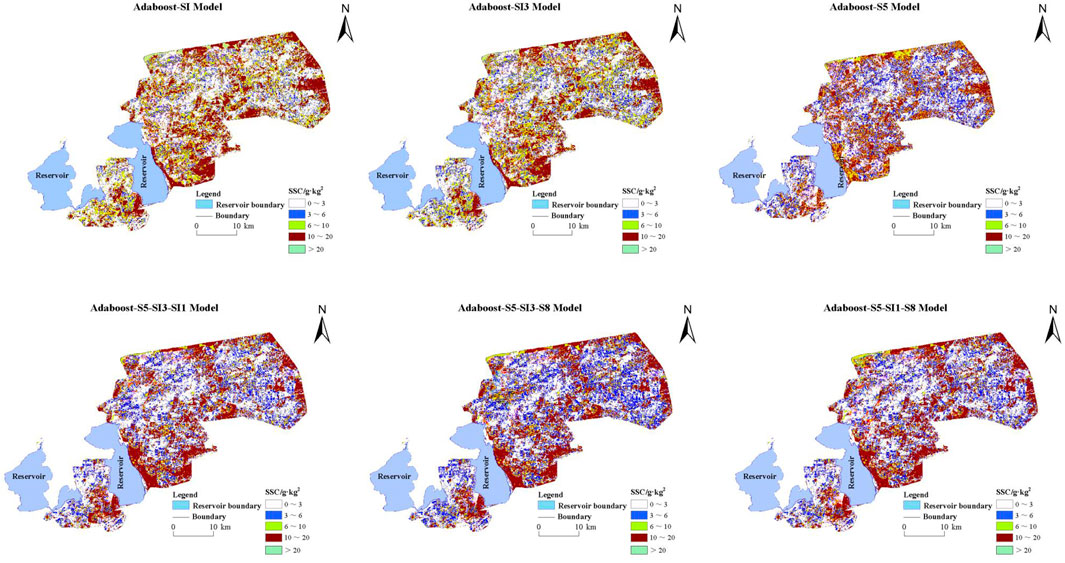
Figure 10. Inversion of soil salinity distribution in Xiao Haizi irrigation area by different models.
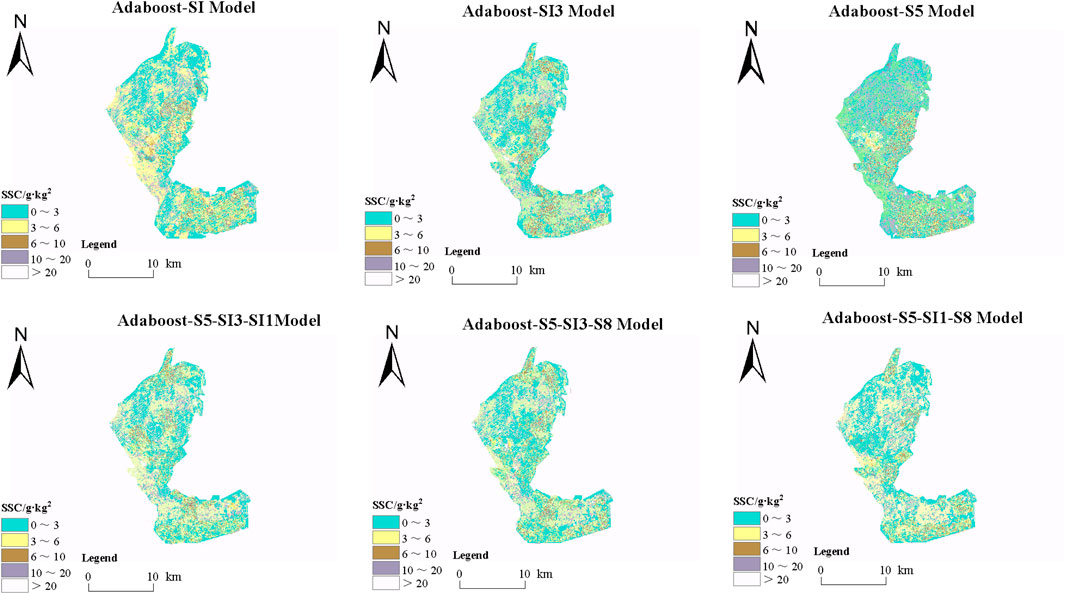
Figure 11. Inversion of soil salinity distribution in Sha Jingzi irrigation area by different models.
4 Discussion
4.1 The soil salinity spatial distribution characteristics and influencing factors in the Xiao Haizi irrigation district
This study focuses on the Xiao Haizi irrigation district, analyzing the spatial distribution characteristics of soil salinity and its influencing factors using Geostatistics technology. The results indicate an overall trend of lower salinity in the southwest and higher salinity in the northeast of the study area, which is related to factors such as topography, groundwater, and climate. Specifically, the study area is located in the middle and lower reaches of the Yarkand River and Kashgar River basins, with a terrain characterized by a southwest high to northeast low trend (Jiang et al., 2022). The rivers flow west to east, with the southern part near the two major reservoirs. As the altitude decreases, soil salinization becomes more severe, with higher soil salinity content observed in the northeast and lower in the southwest.
The groundwater depth gradually increases from south to north in the study area, ranging from less than 1 m near the watershed to 36 m in the south and exceeding 10 m in the northernmost part. Cong et al. (2023) also found that the surface water mineralization of Xiao Haizi Reservoir and Yong’anba Reservoir ranges from 0.56 to 0.74 g·L⁻1, classified as fresh water. In comparison, the average groundwater mineralization is 4.15 g·L⁻1, falling within the range of salty water. The increase in ground-water mineralization leads to increased salt content in the soil, with higher soil salinity observed in areas with shallower groundwater and lower soil salinity in areas with deeper groundwater.
The study area has a warm, temperate, extremely arid climate, with an average annual precipitation of only 34.1–38.8 mm and an average yearly evaporation of 2030.8–3318.26 mm, far exceeding precipitation. This results in severe water shortage in the soil, and intense evaporation promotes the migration of water in the soil to the surface layer, leading to the dissolution and accumulation of salts in the surface soil layer and higher soil salinity content in cultivated land within the irrigation district (Yu et al., 2018). Moreover, the presence of shallow groundwater facilitates the transport of salts to the surface, while evaporation intensifies the concentration of salts at the topsoil. The increase in shallow groundwater and the high rate of evaporation contribute to the accumulation of salts in the surface soil in the study area.
Furthermore, this study found that the soil’s total salt and various salt ions exhibit highly uneven spatial distribution, showing solid or moderate spatial heterogeneity. This may be related to the distribution of artificial water canals, as irrigation by artificial canals can alter the water-salt balance of the soil and affect the distribution of soil salinity. Additionally, Cong et al. (2023) measured the concentration of significant surface water components in the Yarkand River basin and found that the highest average values of significant cations in reservoir water were K⁺ and Na⁺, followed by Ca2⁺, and the lowest content was Mg2⁺. The average concentrations of the main anions ranged from high to low and were as follows: SO₄2⁻, Cl⁻, HCO₃⁻, which is consistent with the pattern of salt ion content in the soil observed in this study.
4.2 The key role of multi-variable combinations in enhancing the accuracy of soil salinization inversion
Soil salinization is the result of a combination of natural and human factors. The use of auxiliary information extracted from remote sensing images, such as vegetation indices, salinity indices, and reflectance bands, improves the accuracy of soil salinity monitoring. Furthermore, terrain factors, such as elevation, slope, and surface roughness, which affect groundwater flow and salt migration, can be extracted from digital elevation models (DEMs) to support the spatial analysis of soil salinity.
The integration of multiple influencing factors has been demonstrated to markedly enhance the accuracy of soil salinity prediction. To illustrate, Xu et al. (2020) selected 25, 16, and 24 variables for quantitative salinity assessment in the Wei-Ku Oasis, Qitai Oasis, and Sangeng River Basin, respectively. Their findings revealed that specific variables exerted significant impacts on salinization across regions. In the Wei-Ku Oasis, surface temperature was identified as a key factor, while soil texture, irrigation methods, and livestock carrying capacity were identified as necessary in the Qitai Oasis and Sangeng Basin. Similarly, another study employed 46 variables in machine learning models to estimate soil salinity in the Eshtehard Salt River, demonstrating that the integration of multi-source remote sensing and field data markedly enhances prediction accuracy (Zarei et al., 2021).
The findings of this study are in alignment with those of the aforementioned study. A comparison of multi-feature combinations (Combinations 1, 2, 3) with single-feature variables (Feature a, Feature b, Feature c) demonstrated that multi-feature combinations markedly enhanced model fit and predictive robustness. During the construction of the model and the subsequent spatial mapping (see Table 5; Figures 5–11), the multi-feature combinations achieved an average R2 of 0.836 on the test set, which is notably higher than the 0.783 achieved by single features. The MAE was reduced to 1.214 (from 1.393 with single features), and the RMSE was decreased to 1.746 (from 2.007). Similarly, the multi-feature combinations demonstrated superior performance on the validation set, with an average R2 of 0.758 (compared to 0.712 for single features), MAE reduced to 2.013 (from 2.572), and RMSE to 1.9006 (from 2.222). Furthermore, models constructed with single features (e.g., Adaboost-SI, Adaboost-SI3, Adaboost-S5) exhibited high sensitivity and consistent performance in identifying non-saline lands. However, they demonstrated limited capacity to accurately identify intensely saline and highly saline soils. This indicates that integrating spectral features with a stronger correlation to soil salinity can markedly enhance prediction precision. In particular, the Adaboost-S5-SI1-S8 model, constructed with a combination of S5, SI1, and S8, precisely delineated soil salinity distribution in the Xiao Haizi irrigation district, facilitating real-time salinity monitoring and providing invaluable insights for regional management and improvement initiatives.
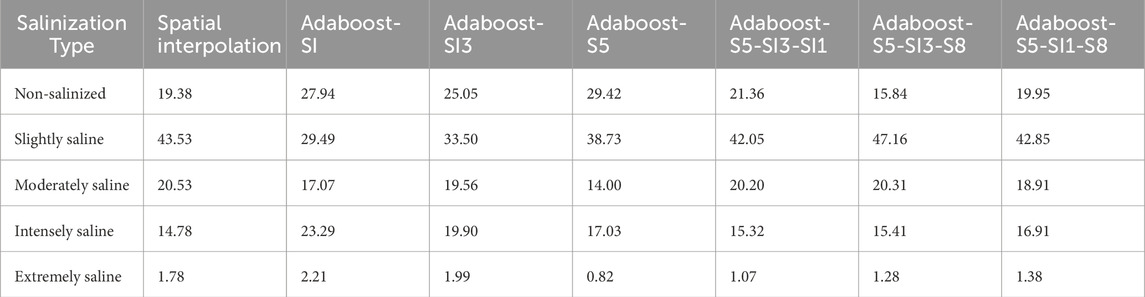
Table 5. Area proportions of different salinization types under various models in the Xiao Haizi irrigation district (%).
Although vegetation indices are commonly employed in the monitoring of soil salinity (Zhang et al., 2011; Allbed et al., 2014; Ramos et al., 2020), this study did not utilize salinity-related vegetation indices and reflectance bands as final features. This may be attributed to the sampling period, as the accuracy of satellite image classification is contingent upon seasonal timing. The optimal monitoring period is typically during the dry season (March to April), whereas high vegetation cover during the wet or hot season can impede the detection of salinity signals. (Khan et al., 2005; Peng et al., 2019; Stavi et al., 2021). During the feature selection process in this study, salinity indices were retained, thereby validating their efficacy under bare soil conditions. Previous studies have demonstrated that the integration of field-measured and spectral data enhances the accuracy of soil salinity monitoring (Gorji et al., 2017).
Future research will further explore how higher-dimensional multi-variable combinations impact model accuracy, particularly addressing potential data redundancy issues among variables such as sensor data types, soil moisture, vegetation types, and climate factors (Sahbeni et al., 2023). This approach aims to improve the temporal and spatial adaptability of soil salinity monitoring.
4.3 Effectiveness of AdaBoost in soil salinization monitoring
In addition to the utilization of multivariate combinations to enhance the precision of soil salinity monitoring, advancements in computer science have yielded novel methodologies for soil salinity monitoring. The advent of ensemble learning methodologies has effectively addressed the issue of the limited generalization capacity of individual learners.
In this study, two ensemble learning models, AdaBoost and RF, exhibited superior estimation results, exceeding those of the single learner SVM. Previous scholars have successfully applied RF to soil salinity monitoring research, achieving higher accuracy than SVR and MLR (Wang F. et al., 2021; Suleymanov et al., 2023). This can be attributed to the capacity of ensemble learning to integrate base learners with disparate hypothesis spaces, thereby expanding the overall model’s hypothesis space and enhancing its resilience to unknown data distributions. Nevertheless, despite RF’s satisfactory performance in this study, AdaBoost demonstrated superior outcomes. This discrepancy may be attributed to RF’s constraints in managing data beyond the training set, resulting in diminished accuracy when validating samples with specific noise (Wang et al., 2019; Shi et al., 2021). In contrast, AdaBoost exhibited superior generalization and stability as a boosting ensemble learning algorithm. By employing an iterative training process involving a series of weak learners and adjusting the weights of high-error samples, AdaBoost effectively shifts the focus in each iteration to poorly performing samples. (Zhao et al., 2023). This mechanism provides robust noise resistance and enhances its capacity to model intricate data. Ultimately, AdaBoost attains final estimates through a weighted average of each weak learner’s predictions. Despite a constrained training set in this experiment, AdaBoost demonstrated remarkable adaptability and noise resistance, attaining the most accurate estimation performance. These findings further substantiate the potential and advantages of AdaBoost in soil salinization monitoring (Jiang et al., 2024b).
4.4 Study significance and limitations in soil salinization monitoring and management
This study employed a multi-stage strategy combining filter-based feature selection and exhaustive search in order to identify the most representative features from a set of candidate variables. The integration of these features into machine learning models resulted in a notable enhancement in the accuracy of soil salinization estimation. Similarly, Wang N. et al. (2020) demonstrated the effectiveness of integrating remote sensing data, landscape characteristics, and machine learning models for soil salinity measurement and monitoring in arid and semi-arid regions, highlighting the importance of feature selection and data integration for improving model performance.
In the comparative analysis of machine learning models, the AdaBoost algorithm outperformed DT, RF, SVM, and XGBoost, demonstrating superior accuracy and adaptability under varying conditions. A practical application in the Sha Jingzi irrigation district validated the models’ performance. All six models effectively captured the spatial distribution of soil salinity, with the AdaBoost-S5-SI1-S8 model achieving the highest accuracy. Notably, the estimated areas of intensely and extremely saline soils by the models were 5.09%–12.2% and 2.65%–8.34% higher (Table 6), respectively, than those derived from spatial interpolation, highlighting the limitations of sparse sampling in interpolation-based methods (Wang Y. et al., 2017). For non-saline, slightly, and moderately saline soils, the models’ estimates closely aligned with the interpolation results.

Table 6. Area proportions of different salinization types under various models in the Sha Jingzi irrigation district (%).
The findings provide a robust framework for precise soil salinization monitoring, facilitating agricultural managers in promptly implementing measures, such as crop rotation and targeted irrigation, to effectively prevent or mitigate soil salinization (Hussain et al., 2020). This study also underscores the potential of machine learning techniques to enhance the precision and reliability of salinity monitoring, particularly in arid and semi-arid regions (Jiang et al., 2024a).
However, this study focused solely on soil salinization monitoring under bare soil conditions, limiting the comprehensive utilization of the potential offered by multisource and multidimensional remote sensing data. Future research should incorporate more diverse features to better reflect the mechanisms of salinization, such as data from various sensors (including SAR), vegetation types, climatic factors, soil types, soil moisture, site-specific management practices, and multi-year spectral indices (Pôças et al., 2020). Additionally, the exploration of more advanced machine learning algorithms (e.g., deep learning) and the optimization of model parameters could further enhance prediction accuracy and model generalizability (Xiao et al., 2023).
5 Conclusion
This study employs the integration of remote sensing technology and machine learning algorithms to achieve the rapid and accurate inversion of soil salinity in the Xiao Haizi irrigation district. It examines the spatial distribution patterns and critical influencing factors, thereby providing a scientific basis for the monitoring and management of soil salinization. A multi-stage strategy of filter-based feature selection and exhaustive search was employed to identify six highly explanatory input variables. The selected variables demonstrated efficacy in soil salinity prediction, with an R2 value exceeding 0.614. The optimal model attained an R2 of 0.828, markedly enhancing the precision and efficiency of the inversion process. The principal conclusions are as follows:
1. The combination of multiple variables led to a notable enhancement in the accuracy and generalizability of soil salinity estimates, particularly in heterogeneous environments, thereby demonstrating a high degree of adaptability.
2. Among the five machine learning algorithms that were the subject of comparison, the AdaBoost model demonstrated the highest learning capacity and predictive accuracy, indicating that it has strong potential for application.
3. The validation results demonstrate that the model constructed with Feature Combination 3 (S5, SI1, and S8) exhibited an exceptional capacity for extracting soil salinity information in the Sha Jingzi irrigation district. The inversion results were found to be in close alignment with the outcomes of spatial interpolation, thereby confirming the accuracy and reliability of the model.
This study proposes a data-driven framework for soil salinity monitoring based on multi-variable combinations. This framework provides a valuable reference for soil salinity inversion in similar regions, meeting the high precision and efficiency demands of salinization monitoring and showing promising potential for broader applications.
Data availability statement
The data analyzed in this study is subject to the following licenses/restrictions: Data sharing is available upon request. Requests to access these datasets should be directed to anVuYm94aWUxMjNAZ21haWwuY29t.
Author contributions
JX: Data curation, Formal Analysis, Investigation, Methodology, Software, Writing–original draft. CS: Data curation, Methodology, Writing–original draft. YL: Conceptualization, Data curation, Writing–original draft. QW: Conceptualization, Data curation, Writing–original draft. ZZ: Conceptualization, Data curation, Formal Analysis, Writing–original draft. SH: Formal Analysis, Funding acquisition, Project administration, Visualization, Writing–review and editing. XW: Formal Analysis, Project administration, Supervision, Visualization, Writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research was supported by the National Key Research and Development Program (2021YFD1900805) and the Bingtuan Science and Technology Program (2021AB009).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Allbed, A., Kumar, L., and Aldakheel, Y. Y. (2014). Assessing soil salinity using soil salinity and vegetation indices derived from IKONOS high-spatial resolution imageries: applications in a date palm dominated region. Geoderma 230, 1–8. doi:10.1016/j.geoderma.2014.03.025
Bajcsy, P., and Groves, P. (2004). Methodology for hyperspectral band selection. Photogrammetric Eng. and Remote Sens. 70 (7), 793–802. doi:10.14358/pers.70.7.793
Bannari, A., and Al-Ali, Z. M. (2020). Assessing climate change impact on soil salinity dynamics between 1987–2017 in arid landscape using Landsat TM, ETM+ and OLI data. Remote Sens. 12 (17), 2794. doi:10.3390/rs12172794
Cackett, L., Cannistraci, C. V., Meier, S., Ferrandi, P., Pěnčík, A., Gehring, C., et al. (2022). Salt-specific gene expression reveals elevated auxin levels in Arabidopsis thaliana plants grown under saline conditions. Front. plant Sci. 13, 804716. doi:10.3389/fpls.2022.804716
Chen, B., Zheng, H., Luo, G., Chen, C., Bao, A., Liu, T., et al. (2022). Adaptive estimation of multi-regional soil salinization using extreme gradient boosting with Bayesian TPE optimization. Int. J. Remote Sens. 43 (3), 778–811. doi:10.1080/01431161.2021.2009589
Chen, C., and Seo, H. (2023). Prediction of rock mass class ahead of TBM excavation face by ML and DL algorithms with Bayesian TPE optimization and SHAP feature analysis. Acta Geotech. 18 (7), 3825–3848. doi:10.1007/s11440-022-01779-z
Chi, Y., Sun, J., Liu, W., Wang, J., and Zhao, M. (2019). Mapping coastal wetland soil salinity in different seasons using an improved comprehensive land surface factor system. Ecol. Indic. 107, 105517. doi:10.1016/j.ecolind.2019.105517
Chlingaryan, A., Sukkarieh, S., and Whelan, B. (2018). Machine learning approaches for crop yield prediction and nitrogen status estimation in precision agriculture: a review. Comput. Electron. Agric. 151, 61–69. doi:10.1016/j.compag.2018.05.012
Cong, S., Lihan, C., Yifei, Z., Shuai, H., and Haixia, X. (2023). Soil salinization characteristics of cultivated land in Xiaohaizi irrigation area of Xinjiang. Arid. Land Geogr. 46 (8), 1314–1323. doi:10.12118/j.issn.1000–6060.2023.008
Elnaggar, A. A., and Noller, J. S. (2009). Application of remote-sensing data and decision-tree analysis to mapping salt-affected soils over large areas. Remote Sens. 2 (1), 151–165. doi:10.3390/rs2010151
Esmaeili, M., Abbasi-Moghadam, D., Sharifi, A., Tariq, A., and Li, Q. (2023). Hyperspectral image band selection based on CNN embedded GA (CNNeGA). IEEE J. Sel. Top. Appl. Earth Observations Remote Sens. 16, 1927–1950. doi:10.1109/jstars.2023.3242310
Farifteh, J., Van der Meer, F., Atzberger, C., and Carranza, E. (2007). Quantitative analysis of salt-affected soil reflectance spectra: a comparison of two adaptive methods (PLSR and ANN). Remote Sens. Environ. 110 (1), 59–78. doi:10.1016/j.rse.2007.02.005
Gharaibeh, M. A., Albalasmeh, A. A., Pratt, C., and El Hanandeh, A. (2021). Estimation of exchangeable sodium percentage from sodium adsorption ratio of salt-affected soils using traditional and dilution extracts, saturation percentage, electrical conductivity, and generalized regression neural networks. Catena 205, 105466. doi:10.1016/j.catena.2021.105466
Gorji, T., Sertel, E., and Tanik, A. (2017). Monitoring soil salinity via remote sensing technology under data scarce conditions: a case study from Turkey. Ecol. Indic. 74, 384–391. doi:10.1016/j.ecolind.2016.11.043
Haj-Amor, Z., Araya, T., Kim, D.-G., Bouri, S., Lee, J., Ghiloufi, W., et al. (2022). Soil salinity and its associated effects on soil microorganisms, greenhouse gas emissions, crop yield, biodiversity and desertification: a review. Sci. Total Environ. 843, 156946. doi:10.1016/j.scitotenv.2022.156946
Hammam, A., and Mohamed, E. (2020). Mapping soil salinity in the East Nile Delta using several methodological approaches of salinity assessment. Egypt. J. Remote Sens. Space Sci. 23 (2), 125–131. doi:10.1016/j.ejrs.2018.11.002
Haq, Y. u., Shahbaz, M., Asif, H. S., Al-Laith, A., and Alsabban, W. H. (2023). Spatial mapping of soil salinity using machine learning and remote sensing in Kot Addu, Pakistan. Sustainability 15 (17), 12943. doi:10.3390/su151712943
Hussain, M. I., Farooq, M., Muscolo, A., and Rehman, A. (2020). Crop diversification and saline water irrigation as potential strategies to save freshwater resources and reclamation of marginal soils—a review. Environ. Sci. Pollut. Res. 27 (23), 28695–28729. doi:10.1007/s11356-020-09111-6
Ivushkin, K., Bartholomeus, H., Bregt, A. K., Pulatov, A., Kempen, B., and De Sousa, L. (2019). Global mapping of soil salinity change. Remote Sens. Environ. 231, 111260. doi:10.1016/j.rse.2019.111260
Jiang, X., Ma, Y., Li, G., Huang, W., Zhao, H., Cao, G., et al. (2022). Spatial distribution characteristics of soil salt ions in Tumushuke City, Xinjiang. Sustainability 14 (24), 16486. doi:10.3390/su142416486
Jiang, Z., Ding, J., Li, Z., and Liu, J. (2024a). Present knowledge and future challenges in remote sensing for soil salinization monitoring: a review of bibliometric analysis. Int. J. Remote Sens., 1–26. doi:10.1080/01431161.2024.2412804
Jiang, Z., Hao, Z., Ding, J., Miao, Z., Zhang, Y., Alimu, A., et al. (2024b). Weighted variable optimization-based method for estimating soil salinity using multi-source remote sensing data: a case study in the Weiku Oasis, Xinjiang, China. Remote Sens. 16 (17), 3145. doi:10.3390/rs16173145
Khan, N. M., Rastoskuev, V. V., Sato, Y., and Shiozawa, S. (2005). Assessment of hydrosaline land degradation by using a simple approach of remote sensing indicators. Agric. Water Manag. 77 (1-3), 96–109. doi:10.1016/j.agwat.2004.09.038
Kumar, B., Dikshit, O., Gupta, A., and Singh, M. K. (2020). Feature extraction for hyperspectral image classification: a review. Int. J. Remote Sens. 41 (16), 6248–6287. doi:10.1080/01431161.2020.1736732
Mantena, S., Mahammood, V., and Rao, K. N. (2023). Prediction of soil salinity in the Upputeru river estuary catchment, India, using machine learning techniques. Environ. Monit. Assess. 195 (8), 1006. doi:10.1007/s10661-023-11613-y
Masoud, A. A. (2014). Predicting salt abundance in slightly saline soils from Landsat ETM+ imagery using Spectral Mixture Analysis and soil spectrometry. Geoderma 217, 45–56. doi:10.1016/j.geoderma.2013.10.027
Measho, S., Li, F., Pellikka, P., Tian, C., Hirwa, H., Xu, N., et al. (2022). Soil salinity variations and associated implications for agriculture and land resources development using remote sensing datasets in central Asia. Remote Sens. 14 (10), 2501. doi:10.3390/rs14102501
Nabiollahi, K., Taghizadeh-Mehrjardi, R., Shahabi, A., Heung, B., Amirian-Chakan, A., Davari, M., et al. (2021). Assessing agricultural salt-affected land using digital soil mapping and hybridized random forests. Geoderma 385, 114858. doi:10.1016/j.geoderma.2020.114858
Paz, A. M., Amezketa, E., Canfora, L., Castanheira, N., Falsone, G., Gonçalves, M. C., et al. (2023). Salt-affected soils: field-scale strategies for prevention, mitigation, and adaptation to salt accumulation. Italian J. Agron. 18 (2166), 2166. doi:10.4081/ija.2023.2166
Peng, J., Biswas, A., Jiang, Q., Zhao, R., Hu, J., Hu, B., et al. (2019). Estimating soil salinity from remote sensing and terrain data in southern Xinjiang Province, China. Geoderma 337, 1309–1319. doi:10.1016/j.geoderma.2018.08.006
Peng, J., Ji, W., Ma, Z., Li, S., Chen, S., Zhou, L., et al. (2016). Predicting total dissolved salts and soluble ion concentrations in agricultural soils using portable visible near-infrared and mid-infrared spectrometers. Biosyst. Eng. 152, 94–103. doi:10.1016/j.biosystemseng.2016.04.015
P Leone, A., A Viscarra-Rossel, R., Amenta, P., and Buondonno, A. (2012). Prediction of soil properties with PLSR and vis-NIR spectroscopy: application to mediterranean soils from Southern Italy. Curr. Anal. Chem. 8 (2), 283–299. doi:10.2174/157341112800392571
Pôças, I., Calera, A., Campos, I., and Cunha, M. (2020). Remote sensing for estimating and mapping single and basal crop coefficientes: a review on spectral vegetation indices approaches. Agric. Water Manag. 233, 106081. doi:10.1016/j.agwat.2020.106081
Ramos, T. B., Castanheira, N., Oliveira, A. R., Paz, A. M., Darouich, H., Simionesei, L., et al. (2020). Soil salinity assessment using vegetation indices derived from Sentinel-2 multispectral data. application to Lezíria Grande, Portugal. Agric. Water Manag. 241, 106387. doi:10.1016/j.agwat.2020.106387
Sahbeni, G., Ngabire, M., Musyimi, P. K., and Székely, B. (2023). Challenges and opportunities in remote sensing for soil salinization mapping and monitoring: a review. Remote Sens. 15 (10), 2540. doi:10.3390/rs15102540
Sawut, M., Ghulam, A., Tiyip, T., Zhang, Y.-j., Ding, J.-l., Zhang, F., et al. (2014). Estimating soil sand content using thermal infrared spectra in arid lands. Int. J. Appl. Earth Observation Geoinformation 33, 203–210. doi:10.1016/j.jag.2014.05.010
Shi, H., Hellwich, O., Luo, G., Chen, C., He, H., Ochege, F. U., et al. (2021). A global meta-analysis of soil salinity prediction integrating satellite remote sensing, soil sampling, and machine learning. IEEE Trans. Geoscience Remote Sens. 60, 1–15. doi:10.1109/tgrs.2021.3109819
Singh, A. (2018). Alternative management options for irrigation-induced salinization and waterlogging under different climatic conditions. Ecol. Indic. 90, 184–192. doi:10.1016/j.ecolind.2018.03.014
Stavi, I., Thevs, N., and Priori, S. (2021). Soil salinity and sodicity in drylands: a review of causes, effects, monitoring, and restoration measures. Front. Environ. Sci. 9, 712831. doi:10.3389/fenvs.2021.712831
Suleymanov, A., Gabbasova, I., Komissarov, M., Suleymanov, R., Garipov, T., Tuktarova, I., et al. (2023). Random forest modeling of soil properties in saline semi-arid areas. Agriculture 13 (5), 976. doi:10.3390/agriculture13050976
Sun, W., and Du, Q. (2019). Hyperspectral band selection: a review. IEEE Geoscience Remote Sens. Mag. 7 (2), 118–139. doi:10.1109/mgrs.2019.2911100
Taghizadeh-Mehrjardi, R., Hamzehpour, N., Hassanzadeh, M., Heung, B., Goydaragh, M. G., Schmidt, K., et al. (2021). Enhancing the accuracy of machine learning models using the super learner technique in digital soil mapping. Geoderma 399, 115108. doi:10.1016/j.geoderma.2021.115108
Thenkabail, P. S., Enclona, E. A., Ashton, M. S., and Van Der Meer, B. (2004). Accuracy assessments of hyperspectral waveband performance for vegetation analysis applications. Remote Sens. Environ. 91 (3-4), 354–376. doi:10.1016/j.rse.2004.03.013
Udelhoven, T., Emmerling, C., and Jarmer, T. (2003). Quantitative analysis of soil chemical properties with diffuse reflectance spectrometry and partial least-square regression: a feasibility study. Plant soil 251, 319–329. doi:10.1023/a:1023008322682
Wang, F., Ding, J., Wei, Y., Zhou, Q., Yang, X., and Wang, Q. (2017a). Sensitivity analysis of soil salinity and vegetation indices to detect soil salinity variation by using Landsat series images: applications in different oases in Xinjiang, China. Acta Ecol. Sin. 37, 5007–5022. doi:10.5846/stxb201605090890
Wang, F., Yang, S., Wei, Y., Shi, Q., and Ding, J. (2021a). Characterizing soil salinity at multiple depth using electromagnetic induction and remote sensing data with random forests: a case study in Tarim River Basin of southern Xinjiang, China. Sci. Total Environ. 754, 142030. doi:10.1016/j.scitotenv.2020.142030
Wang, J., Ding, J., Yu, D., Teng, D., He, B., Chen, X., et al. (2020a). Machine learning-based detection of soil salinity in an arid desert region, Northwest China: a comparison between Landsat-8 OLI and Sentinel-2 MSI. Sci. Total Environ. 707, 136092. doi:10.1016/j.scitotenv.2019.136092
Wang, J., Peng, J., Li, H., Yin, C., Liu, W., Wang, T., et al. (2021b). Soil salinity mapping using machine learning algorithms with the Sentinel-2 MSI in arid areas, China. Remote Sens. 13 (2), 305. doi:10.3390/rs13020305
Wang, L., Hu, P., Zheng, H., Liu, Y., Cao, X., Hellwich, O., et al. (2023). Integrative modeling of heterogeneous soil salinity using sparse ground samples and remote sensing images. Geoderma 430, 116321. doi:10.1016/j.geoderma.2022.116321
Wang, N., Xue, J., Peng, J., Biswas, A., He, Y., and Shi, Z. (2020b). Integrating remote sensing and landscape characteristics to estimate soil salinity using machine learning methods: a case study from Southern Xinjiang, China. Remote Sens. 12 (24), 4118. doi:10.3390/rs12244118
Wang, S., Chen, Y., Wang, M., and Li, J. (2019). Performance comparison of machine learning algorithms for estimating the soil salinity of salt-affected soil using field spectral data. Remote Sens. 11 (22), 2605. doi:10.3390/rs11222605
Wang, Y., Akeju, O. V., and Zhao, T. (2017b). Interpolation of spatially varying but sparsely measured geo-data: a comparative study. Eng. Geol. 231, 200–217. doi:10.1016/j.enggeo.2017.10.019
Wu, W., Zucca, C., Muhaimeed, A. S., Al-Shafie, W. M., Fadhil Al-Quraishi, A. M., Nangia, V., et al. (2018). Soil salinity prediction and mapping by machine learning regression in C entral M esopotamia, I raq. Land Degrad. and Dev. 29 (11), 4005–4014. doi:10.1002/ldr.3148
Xiao, C., Ji, Q., Chen, J., Zhang, F., Li, Y., Fan, J., et al. (2023). Prediction of soil salinity parameters using machine learning models in an arid region of northwest China. Comput. Electron. Agric. 204, 107512. doi:10.1016/j.compag.2022.107512
Xu, H., Chen, C., Zheng, H., Luo, G., Yang, L., Wang, W., et al. (2020). AGA-SVR-based selection of feature subsets and optimization of parameter in regional soil salinization monitoring. Int. J. remote Sens. 41 (12), 4470–4495. doi:10.1080/01431161.2020.1718239
Yu, H., Liu, M., Du, B., Wang, Z., Hu, L., and Zhang, B. (2018). Mapping soil salinity/sodicity by using Landsat OLI imagery and PLSR algorithm over semiarid West Jilin Province, China. Sensors 18 (4), 1048. doi:10.3390/s18041048
Yu, X., Liu, Q., Wang, Y., Liu, X., and Liu, X. (2016). Evaluation of MLSR and PLSR for estimating soil element contents using visible/near-infrared spectroscopy in apple orchards on the Jiaodong peninsula. Catena 137, 340–349. doi:10.1016/j.catena.2015.09.024
Zarei, A., Hasanlou, M., and Mahdianpari, M. (2021). A comparison of machine learning models for soil salinity estimation using multi-spectral earth observation data. ISPRS Ann. photogrammetry, remote Sens. spatial Inf. Sci. 3, 257–263. doi:10.5194/isprs-annals-v-3-2021-257-2021
Zhang, H., Schroder, J., Pittman, J., Wang, J., and Payton, M. (2005). Soil salinity using saturated paste and 1: 1 soil to water extracts. Soil Sci. Soc. Am. J. 69 (4), 1146–1151. doi:10.2136/sssaj2004.0267
Zhang, T.-T., Zeng, S.-L., Gao, Y., Ouyang, Z.-T., Li, B., Fang, C.-M., et al. (2011). Using hyperspectral vegetation indices as a proxy to monitor soil salinity. Ecol. Indic. 11 (6), 1552–1562. doi:10.1016/j.ecolind.2011.03.025
Zhao, J., Mehmood, A., Dong, Q., and Li, D. (2023). Rapid assessment of chilled chicken spoilage based on hyperspectral imaging technology and AdaBoost-RT. Food Anal. Methods 16 (9), 1504–1511. doi:10.1007/s12161-023-02501-9
Keywords: remote sensing, Landsat 8, agricultural sustainability, soil salinity, machine learning, feature selection
Citation: Xie J, Shi C, Liu Y, Wang Q, Zhong Z, He S and Wang X (2025) Soil salinization prediction through feature selection and machine learning at the irrigation district scale. Front. Earth Sci. 12:1488504. doi: 10.3389/feart.2024.1488504
Received: 30 August 2024; Accepted: 12 December 2024;
Published: 06 January 2025.
Edited by:
Hamdi A. Zurqani, University of Arkansas at Monticello, United StatesReviewed by:
Abdulsalam Albukhari, Omar Al-Mukhtar University, LibyaSegun Adedapo, University of Georgia, United States
I. Putu Sugiana, Bogor Agricultural University, Indonesia
Murad Milad Aburas, Omar Al-Mukhtar University, Libya
Copyright © 2025 Xie, Shi, Liu, Wang, Zhong, He and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xingpeng Wang, MTM5OTkwNjgzNTRAMTYzLmNvbQ==; Shuai He, eGpzaHpoc0AxNjMuY29t
 Junbo Xie
Junbo Xie Cong Shi
Cong Shi Yang Liu1,2
Yang Liu1,2 Shuai He
Shuai He Xingpeng Wang
Xingpeng Wang