
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Comput. Sci. , 03 March 2022
Sec. Computer Vision
Volume 4 - 2022 | https://doi.org/10.3389/fcomp.2022.805166
This article is part of the Research Topic Methods and Tools for Bioimage Analysis View all 13 articles
The remarkable performance of Convolutional Neural Networks on image segmentation tasks comes at the cost of a large amount of pixelwise annotated images that have to be segmented for training. In contrast, feature-based learning methods, such as the Random Forest, require little training data, but rarely reach the segmentation accuracy of CNNs. This work bridges the two approaches in a transfer learning setting. We show that a CNN can be trained to correct the errors of the Random Forest in the source domain and then be applied to correct such errors in the target domain without retraining, as the domain shift between the Random Forest predictions is much smaller than between the raw data. By leveraging a few brushstrokes as annotations in the target domain, the method can deliver segmentations that are sufficiently accurate to act as pseudo-labels for target-domain CNN training. We demonstrate the performance of the method on several datasets with the challenging tasks of mitochondria, membrane and nuclear segmentation. It yields excellent performance compared to microscopy domain adaptation baselines, especially when a significant domain shift is involved.
Semantic segmentation—partitioning the image into areas of biological (semantic) meaning—is a ubiquitous problem in microscopy image analysis. Compared to natural images, microscopy segmentation problems are particularly well-suited for feature-based (“shallow”) machine learning, as the difference between semantic classes can often be captured in local edge, texture, or intensity descriptors (Belevich et al., 2016; Arganda-Carreras et al., 2017; Berg et al., 2019). While convolutional neural networks (CNNs) have long overtaken feature-based approaches in segmentation accuracy and inference speed, interactive feature-based solutions continue to attract users due to the low requirements to training data volumes, nearly real-time training speeds and general simplicity of the setup, which does not require computational expertise.
CNNs are made up of millions of learnable parameters which have to be configured based on user-provided training examples. With insufficient training data, CNNs are very prone to overfitting, “memorizing” the training data instead of deriving generalizable rules. Strategies to suppress overfitting include data augmentation (Ronneberger et al., 2015), incorporation of prior information (El Jurdi et al., 2021), dropout and sub-network re-initialization (Han et al., 2016; Taha et al., 2021) and, in case a similar task has already been solved on sufficiently similar data, domain adaptation, and transfer learning. In the latter case, the network exploits a large amount of labels in the so called “source” domain to learn good parameter values for the task at hand, which are further adapted for the unlabeled or sparsely labeled “target” domain through unsupervised or weakly supervised learning. For microscopy images, the adaptation is commonly achieved by bringing the distributions of the source and target domain data closer to each other, either by forcing the network to learn domain-invariant features (Long et al., 2015; Roels et al., 2019; Liu et al., 2020) or by using generative networks and cycle consistency constraints (Zhang et al., 2018; Chen et al., 2019; Januszewski and Jain, 2019). Alternatively, the domain shift can be explicitly learned in a part of the network (Rozantsev et al., 2018). In addition to labels in the source domain, pseudo-labels in the target domain are often used for training (Choi et al., 2019; Xing et al., 2019). Pseudo-labels can be computed from the predictions of the source domain network (Choi et al., 2019) or predictions for pixels similar to source domain labels (Bermúdez-Chacón et al., 2019).
In contrast, Random Forest (RF), one of the most popular “shallow” learning classifiers (Breiman, 2001), does not overfit on small amounts of training data and trains so fast that in practice no domain adaptation strategies are applied—the classifier is instead fully retrained with sparse labels in the target domain. However, unlike a CNN, it cannot fully profit from large amounts of training data. The aim of our contribution is to combine the best of both worlds, exploiting fast training of the Random Forest for domain adaptation and excellent performance of CNNs for accurate segmentation with large amounts of training data. We use the densely labeled source domain to train many Random Forests for segmentation and then train a CNN for Random Forest prediction enhancement (see Figure 1). On the target domain, we train a new Random Forest from a few brushstroke labels and simply apply the pre-trained Prediction Enhancer (PE) network to improve the probability maps. The enhanced predictions are substantially more accurate than the Random Forest or a segmentation CNN trained only on the source domain. Furthermore, a new CNN can be trained using enhanced predictions as pseudo-labels, achieving an even better accuracy with no additional annotation cost. Since the Prediction Enhancer is only trained on RF probability maps, it remains agnostic to the appearance of the raw data and can therefore be applied to mitigate even very large domain gaps between source and target datasets, as long as the segmentation task itself remains similar. To illustrate the power of our approach, we demonstrate domain adaptation between different datasets of the same modality, and also from confocal to light sheet microscopy, from electron to confocal microscopy and from fluorescent light microscopy to histology. From the user perspective, domain adaptation is realized in a straightforward, user-friendly setting of training a regular U-Net, without adversarial elements or task re-weighting. Furthermore, a well-trained Prediction Enhancer network can be used without retraining, only requiring training of the Random Forest from the user. Our Prediction Enhancer networks for mitochondria, nuclei, or membrane segmentation tasks are available at the BioImage Model Zoo (https://bioimage.io) and can easily be applied to improve predictions of the Pixel Classification workflow in ilastik or of the Weka Trainable Segmentation plugin in Fiji.
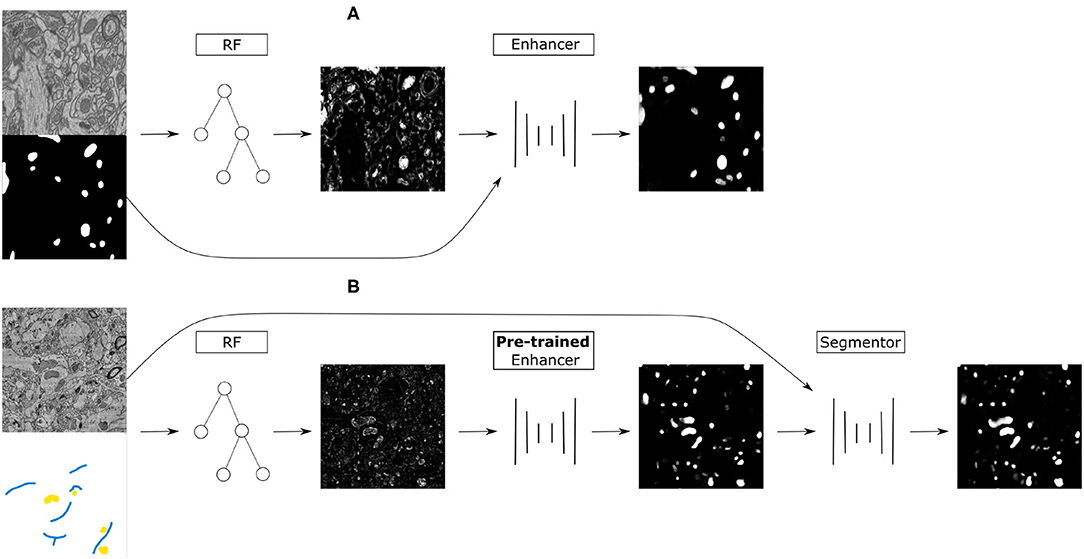
Figure 1. (A) Training on the source dataset: many Random Forests are trained by subsampling patches of raw data and dense groundtruth segmentation. Random Forest predictions are used as inputs and groundtruth segmentation as labels to train the Prediction Enhancer CNN to improve RF segmentations. (B) Domain adaptation to the target dataset: a RF is trained interactively with brushstroke labels. The pre-trained PE is applied to improve the RF predictions. Optionally, PE predictions are used as pseudo-labels to train a segmentation network for even better results with no additional annotations, but using a larger computational budget.
Our approach combines the advantages of feature-based and end-to-end segmentation methods by training a Prediction Enhancer network to predict one from the other. On the target dataset, retraining can be limited to the feature-based classifier as its predictions—unlike the raw data—do not exhibit a significant domain shift if the same semantic classes are being segmented. In more detail, we propose the following sequence of steps (see also Figure 1):
1. Create training data for the Prediction Enhancer CNN by training multiple Random Forests on random samples of the densely labeled source domain.
2. Train the Prediction Enhancer using the RF predictions as input and the ground-truth segmentation as labels.
3. Train a Random Forest on the target dataset with a few brushstroke labels and use the pre-trained Prediction Enhancer to improve the predictions.
4. Use the improved predictions as pseudo-labels to train a CNN on the target dataset. This step is optional and trades improved quality for the computational cost of training a CNN from scratch.
Note that the Prediction Enhancer only takes the predictions of the Random Forest as input. Neither raw data nor labels of the source dataset are needed to apply it to new data. Our method can therefore be classified as source-free domain adaption, but the additional feature-based learning step allows us to avoid training set estimation or reconstruction, commonly used in other source-free or knowledge distillation-based approaches like Du et al. (2021) and Liu et al. (2021). At the same time, we can fully profit from all advances in the field of pseudo-label rectification (Prabhu et al., 2021; Wu et al., 2021; Zhang et al., 2021; Zhao et al., 2021), applying those to pseudo-labels generated by the PE network.
The Prediction Enhancer is based on the U-Net architecture (Ronneberger et al., 2015). To create training data, we train multiple Random Forests on the dense labels of the source domain, using the same pixel features as in the ilastik pixel classification workflow (Berg et al., 2019). To obtain a diverse set of shallow classifiers we sample patches of various size and train a classifier for each patch based on the raw data and dense labels. Typically, we train 500–1,000 different classifiers. Next, we train the U-Net following the standard approach for semantic segmentation, using Random Forest predictions (but not the raw data) as input and the provided dense labels of the source domain as the groundtruth. To create more variability, we sample from all previously trained classifiers. We use either the binary cross entropy or the Dice score as loss function.
Segmentation of a new dataset only requires training a single Random Forest; its predictions can directly be improved with the pre-trained Prediction Enhancer. Here, we use ilastik pixel classification workflow, which enables training a Random Forest interactively from brushstroke user annotations.
The Prediction Enhancer can improve the segmentation results significantly, as shown in Section 3. However, it relies only on the Random Forest predictions, and can thus not take intensity, texture or other raw image information into account. To make use of such information and further improve segmentation results, we can use the predictions of the Enhancer as pseudo-labels and train a segmentation U-Net on the target dataset. We use either Dice score or binary cross entropy as loss and make the following adjustments to the standard training procedure to enable training from noisy pseudo-labels:
• Use the RF predictions as soft labels in range [0, 1] instead of hard labels in {0, 1}.
• Use a simple label rectification strategy to weight the per-pixel loss based on the prediction confidence (see Section 2.2.1).
• In the final loss, add a consistency term similar to Tarvainen and Valpola (2017) that compares the current predictions to the predictions of the network's exponential moving average (see Section 2.2.2).
Label rectification is a common strategy in self-learning-based domain adaptation methods, where predictions from the source model are used as pseudo-labels on the target domain. Rectification is then used to correct for the label noise. Several strategies have been proposed, for example based on the distance to class prototypes in the feature space (Zhang et al., 2021) or prediction confidence after several rounds of dropout (Wu et al., 2021).
Here, we adopt a simple label rectification strategy based on the prediction confidence to weight the pseudo-labels y:
where k is the class index. The pseudo labels yk correspond to the predictions of the Prediction Enhancer and are continuous in the range [0, 1]. For the case of foreground/background segmentation k ∈ {0, 1} and we define the per-pixel weight for the foreground class as
Here, p1 is the foreground probability map predicted by the segmentation network and η1 a scalar value, defined as the exponentially weighted average computed over the foreground mask S:
Here, X is the set of all pixels in the input image. We set λ = 0.999 in all experiments. The weight ω0 for the background class is computed in the same manner.
For training with pseudo-labels we introduce a consistency term in the loss function, which is based on the “Mean Teacher” training procedure for semi-supervised classification (Tarvainen and Valpola, 2017). The loss term compares the output of the network f with the output of the network g, defined as the exponential moving average (EMA) of f. This method promotes more consistent predictions across training iterations. We make use of this method for training a segmentation network (parameterized by θf) from pseudo-labels. Its EMA, g is parameterized by
where we set the smoothing coefficient α to 0.999 following (Tarvainen and Valpola, 2017).
Given that we are comparing the per pixel predictions of the current network and its EMA, we use the loss function that is also employed for comparing to the pseudo labels: we either use the Dice loss
or the binary cross entropy loss
Here x denotes the input image, pf = f(x), pg = g(x), and N is the number of pixels. The combined loss function is
where R is either Dice or BCE. The term LR compares the output from f with pseudo-labels defined in Equation 1 and LR,c is the consistency term.
We evaluate the proposed domain adaptation method on challenging semantic segmentation problems, including mitochondria segmentation in Electron Microscopy (EM), membrane segmentation in electron, and light microscopy (LM) as well as nucleus segmentation in LM. Table 1 summarizes all datasets used for the experiments. Table A1 lists the data size as well as the train, validation, and test splits for all datasets.
Some of the datasets we use represent image stacks and could be processed as 3D volumes with different levels of anisotropy. We choose to process them as independent 2D images instead to enable a wider set of source/target domain pairs. If not noted otherwise, training from pseudo-labels is performed using the consistency loss term and label rectification (Equation 7). We use a 2D U-Net architecture (Ronneberger et al., 2015) with 64 features in the initial layer, four downsampling/upsampling levels and double the number of features per level for all networks. The network and training code is based on the PyTorch implementation from Wolny et al. (2020). For all training runs we use the Adam optimizer with initial learning rate of 0.0002, weight decay of 0.00001. Furthermore, we decrease the learning rate by a factor of 0.2 if the validation metric is not improving for a dataset dependent number of iterations. We use binary cross entropy as a loss function for the mitochondria (Section 3.2) and nucleus (Section 3.4) segmentation and dice loss for the membrane segmentation (Section 3.3).
We first perform mitochondria segmentation in EM. We train the Prediction Enhancer on the EPFL dataset (the only FIB/SEM dataset in the collection) and then perform source-free domain adaptation on the VNC, MitoEM-R, MitoEM-H, and Kasthuri datasets. For domain adaption, the Random Forest for initial target prediction is trained interactively in ilastik using a separate train split. The RF predictions are then improved by the PE and the improved predictions are used as pseudo-labels for a U-Net trained from scratch (Pseudo-label Net). We compare to direct predictions of a U-Net trained for Mitochondria segmentation on the source domain EPFL (Source Net) and to the Y-Net (Roels et al., 2019), a different method for domain adaptation, which is unsupervised on the target domain, but not source-free. We also indicate the performance of a U-Net trained on the target dataset as an estimate of the upper bound of the achievable performance (a separate train split is used).
Table 2 summarizes the resulting F1 scores (higher is better) for the source dataset and all target datasets. The Enhancer improves the Random Forest predictions significantly on all target datasets and the CNN trained from pseudo-labels further improves the results. The pseudo-label CNN always performs better than the source network or the Y-Net, which fails completely for the Kasthuri dataset where the domain gap is particularly large. Figure 2 shows an example of the improvements from RF to PE and PE to Pseudo-label Net.
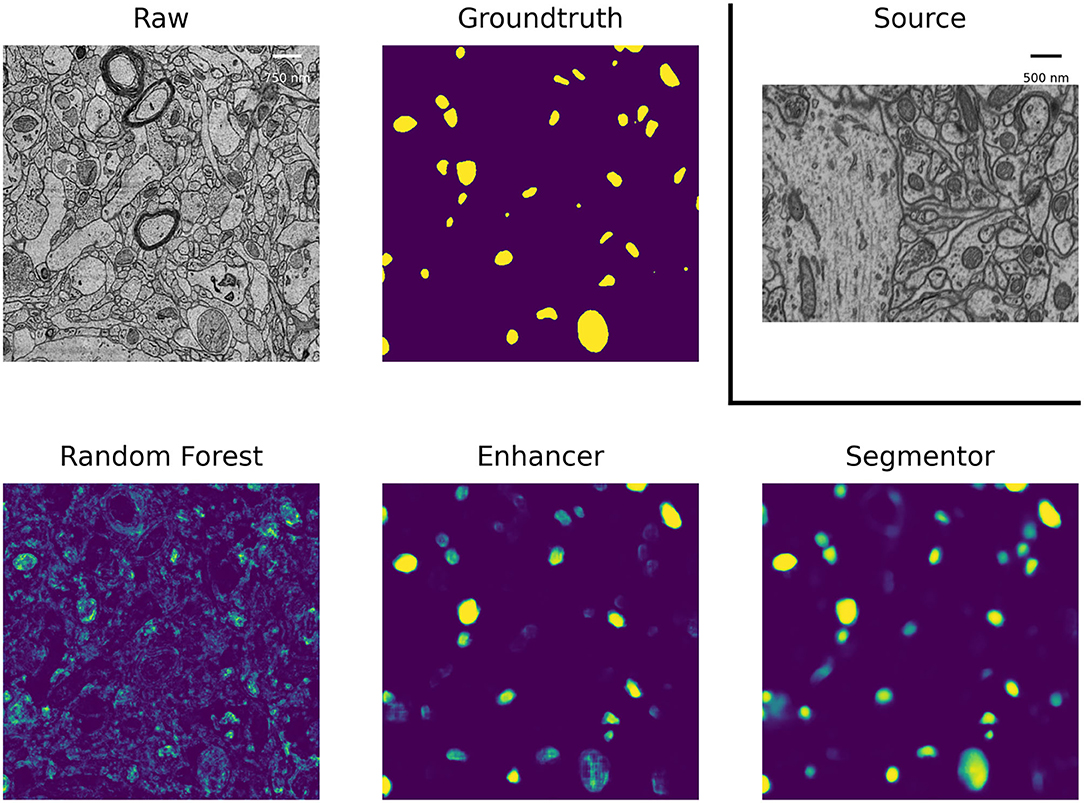
Figure 2. Mitochondria predictions of the Random Forest trained in ilastik, Prediction Enhancer, and Pseudo-label CNN (“Segmentor”) as well as the groundtruth segmentation, on the MitoEM-H dataset. The Enhancer was pre-trained on the EPFL dataset; EPFL raw data shown under Source.
For the mitochondria segmentation task we also check if training the PE on multiple source datasets improves results. Table 3 shows that this is indeed the case, especially for the Kasthuri dataset.
We perform membrane segmentation both in EM and LM data. Obtaining a (semantic) membrane segmentation is often the first step in methods for instance segmentation of neurons or cells as direct prediction of an instance segmentation with a CNN is highly non-trivial due to the label invariance problem. As a consequence we are interested in the quality of the final instance segmentation, not the intermediate boundary segmentation, in these experiments and set up a up a Multicut based post-processing procedure similar to Beier et al. (2017) to obtain instances from the boundary predictions. We then evaluate the instance segmentation using the Variation of Information (Meilă, 2003). Direct evaluation of the boundary predictions via the F1-score is often not indicative of the quality of the resulting instance segmentation due to the large influence of relatively small prediction errors, such as holes (Arganda-Carreras et al., 2015). For the Variation of Information lower values correspond to a better segmentation.
In EM we perform boundary segmentation of neural tissue using the VNC dataset as source and three different datasets from the CREMI challenge (cremi.org) as target. Table 4 shows that the PE significantly improves the RF predictions for all three target datasets. The network trained on pseudo-labels can further improve results, especially for CREMI B and C, which pose a more challenging segmentation problem due to more irregular and elongated neurites compared to CREMI A. Both PE and Pseudo-label Net perform significantly better than a segmentation network trained on the source dataset. The segmentation results of a segmentation network trained on a separate split of the target dataset are shown to indicate an upper bound of the segmentation performance. Figure 3 shows the improvement brought by the PE and the Pseudo-label Net on an image from CREMI C.
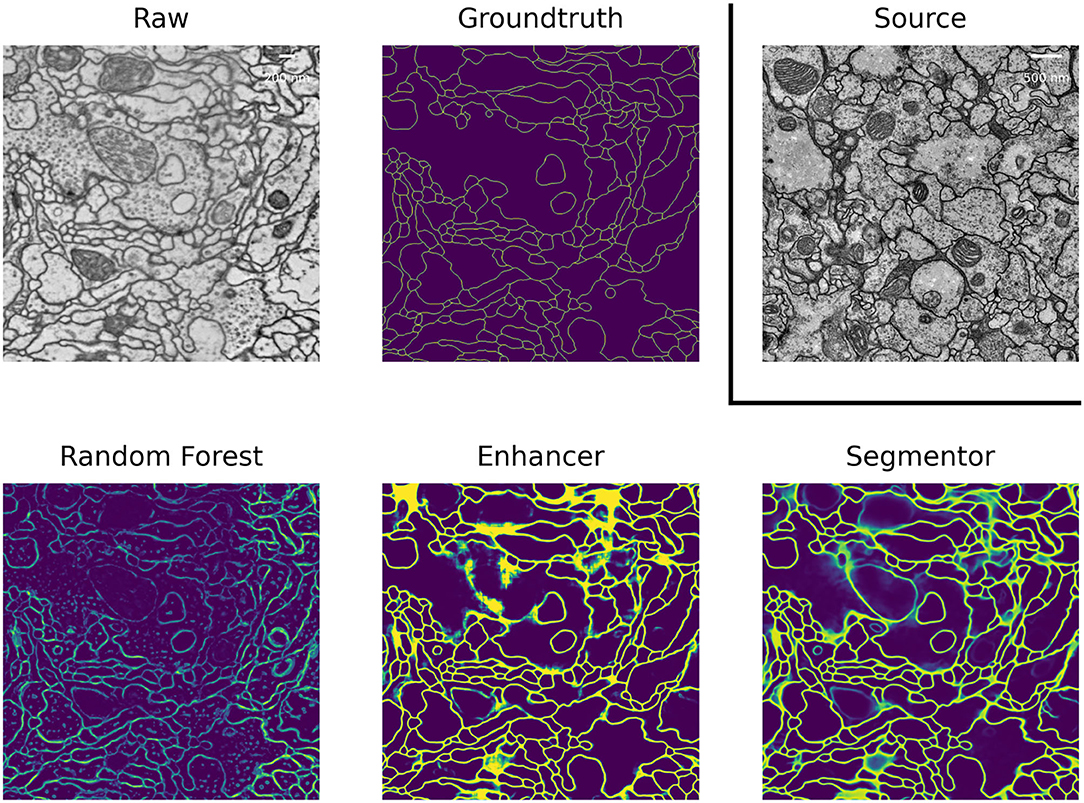
Figure 3. Boundary predictions of the Random Forest trained in ilastik, Prediction Enhancer, and Pseudo-label Net, as well as the groundtruth segmentation, on the CREMI C dataset. The Enhancer was pre-trained on VNC, VNC raw data shown under Source.
In LM we perform boundary segmentation of cells in a confocal microscopy image stack of Arabidopsis thaliana ovule tissue. We use a light-sheet microscopy image stack of Arabidopsis root tissue as source data. Note that we downsample the Ovules dataset by a factor of 2 to match the resolution of the Root dataset (see Table 1B). The results are shown in the “Root (LM)” column in Table 5. The PE significantly improves the RF results and pseudo-label training improves them even further. In this case the quality of the pseudo-label net almost reaches the target network. Note that the overall quality of results reported here is inferior compared to the results reported in Wolny et al. (2020). This can be explained by the fact that all models only receive 2D input, whereas the state-of-the-art uses 3D models.
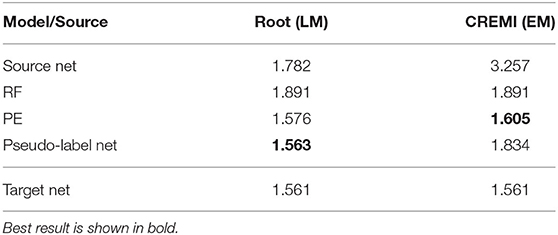
Table 5. LM-Boundaries and cross modality experiments: Variation of Information after applying graph partitioning (Multicut) to the boundary predictions.
We also experiment with a much larger domain shift and apply a PE that was trained on the EM dataset CREMI A as source. The results are shown in the “CREMI (EM)” column in Table 5. As expected, transfer of the source network fails, because it was trained on a completely different domain. However, the PE successfully improves RF predictions. The fact that the PE only receives the RF predictions as input enables successful transfer in this case; while the image data distribution is very different in source and target domain, RF probability maps look sufficiently similar. Furthermore, the resolution of the two domains differs by almost three orders of magnitude. However, the size of the structures in pixels is fairly similar, enabling successful domain adaptation. Figure 4 shows RF, PE and Pseudo-label Net predictions next to the source and target domain data. In this case, training with pseudo-labels does not improve the result, probably because the predictions get smoothed significantly compared to the PE, as can be seen in the figure.
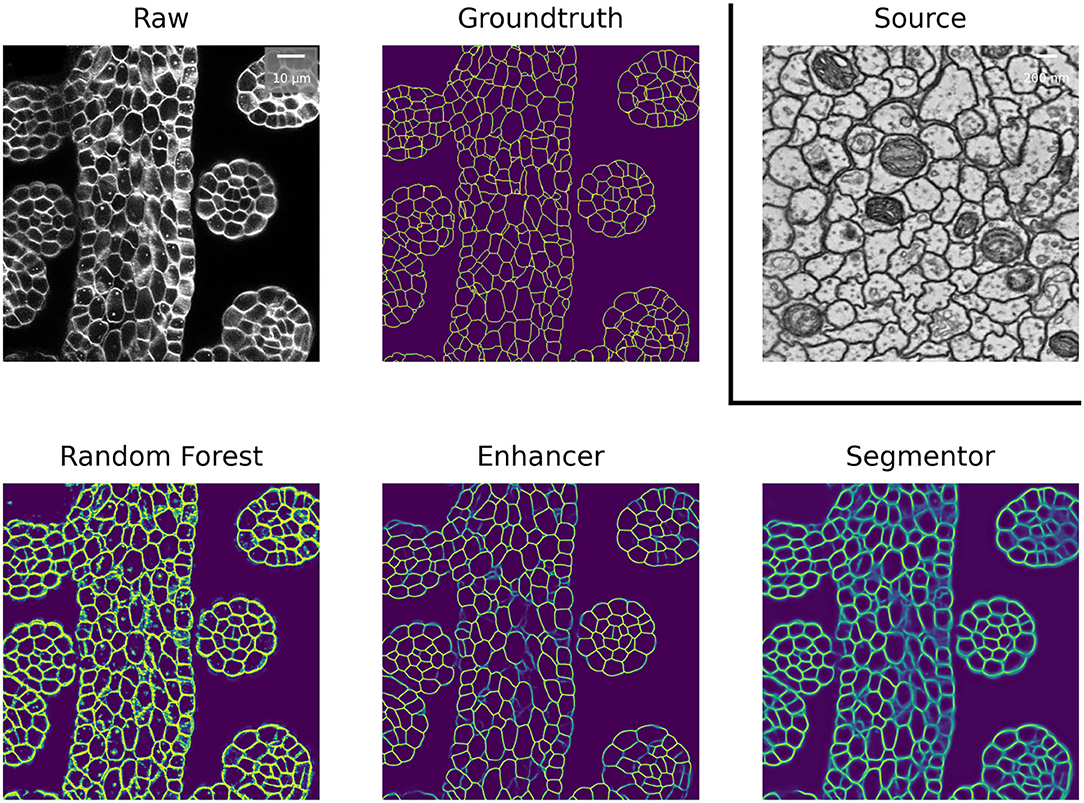
Figure 4. Boundary predictions of the Random Forest trained in ilastik, Prediction Enhancer, and Pseudo-label Net, as well as groundtruth segmentation, on the Ovules dataset. The Enhancer was pre-trained on CREMI A, CREMI A raw data shown under Source.
As another example of cross-modality adaptation, we perform nucleus segmentation between fluorescence microscopy images from Caicedo et al. (2019) (DSB-FL) and histopathology images of the human kidney from Kumar et al. (2019) (Monuseg). Table 6 shows the results for using Monuseg as source and DSB-FL as target (column “DSB-FL”) and vice versa (column “Monuseg”). The Enhancer and pseudo-label training offer a modest improvement for the transfer from Monuseg to DSB-FL. For the transfer in the opposite direction the Enhancer yields inferior results compared to ilastik predictions and consequently also inferior results for pseudo-label training. This observation can be explained by the fact that the images in the DSB-FL dataset were acquired with different microscopy modalities and resolutions, resulting in significantly different nuclei sizes across the dataset. In contrast, the size of nuclei in the Monuseg dataset is uniform and closest to the smallest nuclei in DSB-FL. We identify this behavior as a limitation of our method and further investigate the results in Table 9.
In the following, we perform ablation studies to determine the impact of some of our design choices on the overall performance of the method.
First, we investigate if the consistency loss (CL, Equation 6) and label rectification (LR, Equation 1) improve the accuracy obtained after pseudo-label training. We perform pseudo-label training for mitochondria segmentation on the VNC and MitoEM-R datasets using the PE trained on VNC to generate the pseudo-labels. We perform the training without any modification of the loss, adding only CL, adding only LR and adding both CL and LR. The results in Table 7 show that both CL and LR improve performance on their own. Combining them leads to an additional small improvement on VNC and to a slight decrease in quality on MitoEM-R.
Using the same experiment setup, we also investigate whether using the PE enhancer for generating the pseudo-labels is actually beneficial compared to using the RF trained on target or using the source network. Table 8 shows that using the PE for pseudo-label generation significantly improves over the two other approaches. We have also studied the influence of the size of the Random Forests used for training the PE, but found that it did not have a significant influence on PE performance. See Table A2 for details.
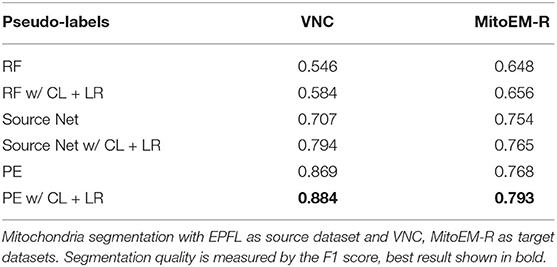
Table 8. Results of pseudo-label network training using RF, Source Network, and PE for label generation.
The high number of layers, their interconnections and especially skip-connections between them allow the U-net to implicitly learn a strong shape prior for the objects of interest. This effect is exacerbated in our Prediction Enhancer network as it by design does not observe the raw pixel properties and has to exploit shape cues even more than a regular segmentation U-net. While this effect is clearly advantageous for same-task transfer learning, it can lead to catastrophic network hallucinations if very differently shaped objects of interest need to be segmented in the target domain. To illustrate this point, we show the transfer of a PE learned for mitochondria on the EPFL dataset to predict boundaries on the VNC dataset and vice versa in Figure 5. The PE amplifies/hallucinates the structures it was trained on while suppressing all other signal in the prediction.
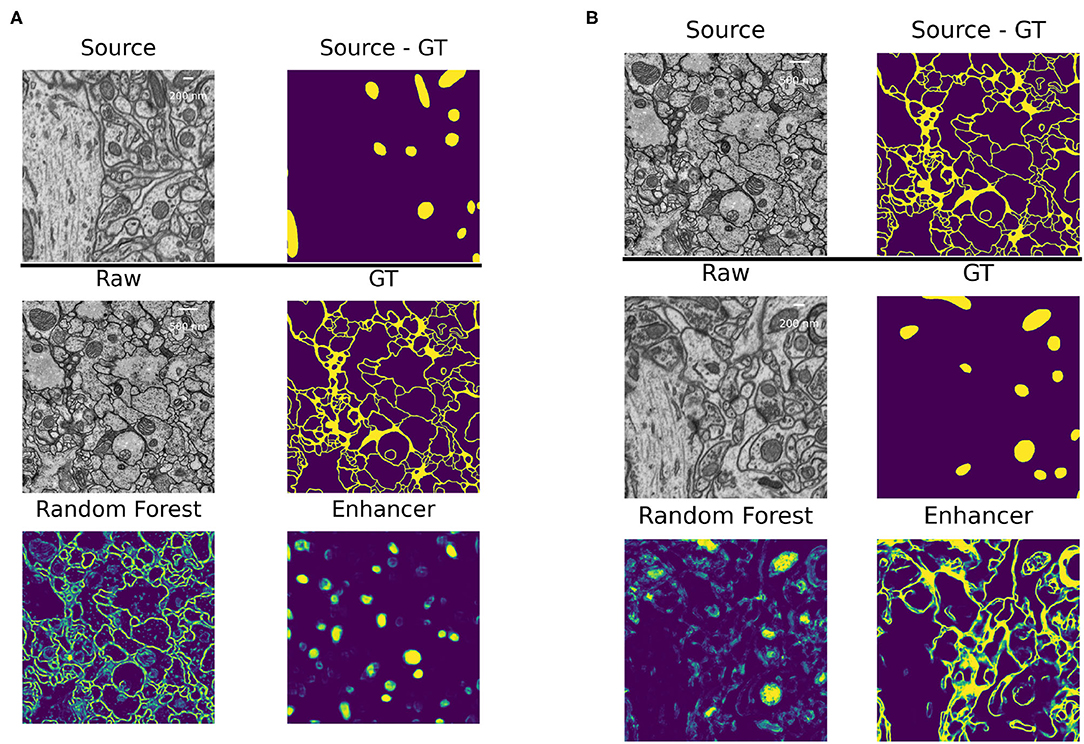
Figure 5. Failure case: different segmentation tasks in source and target datasets. (A) Domain adaptation of a PE trained for mitochondria segmentation on the EPFL dataset to boundary prediction task on the VNC dataset. (B) Domain adaptation of a PE trained for boundary prediction on the VNC dataset to mitochondria segmentation task on the EPFL dataset.
Besides the hallucinations observed in the case of very different shapes of objects in source and target, the size distribution of objects also matters. In Section 3.4, we have investigated transfer between nuclei imaged in histopathology and fluorescence microscopy and observed that the Enhancer yields inferior results for the transfer from histopathology to fluorescence. This can possibly be explained by the fact that the fluorescence dataset contains images of different modalities and resolution, in which the nuclei appear in different sizes. In some of the images the nuclei are small and have a similar average size as in the histopathology dataset, in another one they are of medium size and in yet another of much larger size. We have split the fluorescence dataset into these three modalities (“Small,” “Medium,” “Large”) and list the corresponding results in Table 9. The quality of the Enhancer and pseudo-label network predictions drops dramatically for large nuclei sizes, bringing us to hypothesize that such a significant difference in object size constitutes a domain shift our method cannot easily address, even if the underlying problem is so simple it can almost be solved by the Random Forest alone.
A further potential limitation for our method are systematic differences between the error characteristics of the shallow classifiers used for training on the source dataset and the Random Forest used during inference on the target dataset. We set up a synthetic experiment to investigate this case and train the Enhancer using a mixture of the Random Forest predictions and ground-truth labels as input. Table 10 shows the results for mitochondria prediction using EPFL as source and VNC as target (cf. Section 3.2) as well as for membrane predictions using VNC as source and CremiB as target (cf. Section 3.3). For both experiments we present the Enhancer network with a weighted linear combination of the smoothed groundtruth and the Random Forest predictions during training and tune the weight coefficient between 0 and 100%. For reference we also report the performance of the ilastik Random Forest that is being “enhanced” on the target dataset. We observe that the prediction quality of the Enhancer is significantly better when trained with a large contribution the Random Forest predictions or from pure Random Forest predictions. We conclude that systematic differences in the errors on source and target, especially if the error rate is significantly lower on source, negatively affect the accuracy of our method.
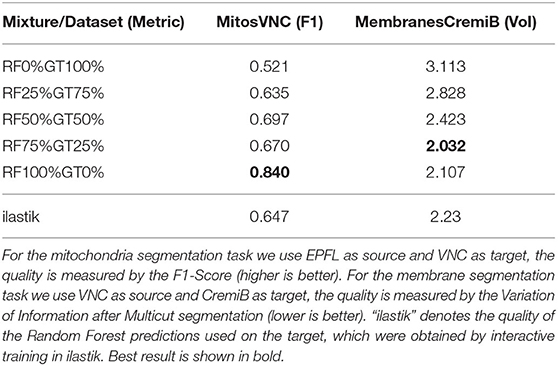
Table 10. The quality of Enhancer predictions on the target data when trained on a mixture of random-forest and ground-truth label on source.
We have introduced a simple, source-free, weakly supervised approach to transfer learning in microscopy which can overcome significant domain gaps and does not require adversarial training. In our setup, the feature-based classifier which is trained from sparse annotations on the target domain acts as an implicit domain adapter for the Prediction Enhancer network. The combination of the feature-based classifier and the prediction enhancer substantially outperforms the segmentation CNN trained on the source domain, with further improvement brought by an additional training step where the Enhancer predictions on the target dataset serve as pseudo-labels. Since the Enhancer network never sees the raw data as input, our method can perform transfer learning between domains of drastically different appearance, e.g., between light and electron microscopy images. By design, this kind of domain gap cannot be handled by unsupervised domain adaptation methods which rely on network feature or raw data alignment. Furthermore, even for small domain gaps and in presence of label rectification strategies, pseudo-labels produced by the Prediction Enhancer lead to much better segmentation CNNs than pseudo-labels of the source network. We expect these results to improve even further with the more advanced label rectification approaches which are now actively introduced in the field.
The major limitation of our approach is the dependency on the quality of the feature-based classifier predictions. We expect that in practice users will train it interactively on the target domain, which already produces better results than “bulk” training: in our mitochondria segmentation experiments, also shown in Table 2, there was commonly a 1.5- to 2-fold improvement in F1-score between interactive ilastik training in the target domain and RF training in a script without seeing the data. In general, the performance of the Prediction Enhancer will lag behind the performance of a segmentation network trained directly on the raw data with dense groundtruth labels except for very easy problems that can be solved by the RF to 100% accuracy. In a way, the Random Forest acts as a lossy compression algorithm for the raw data, which reduces the discriminative power for the Enhancer. However, the pseudo-label training step can again compensate for the “compression” as it allows to train another network on the raw data of the target domain, with pseudo-labels for potentially very large amounts of unlabeled data.
We have also investigated further limitations of our method and found that it is only applicable if the shape and size distribution of objects in the source and target datasets are sufficiently similar. If this is not the case, the accuracy of our method will drop and, in case of dramatic differences between objects of interest, such as membranes vs. mitochondria, it may even hallucinate structures of similar shape as found in the source data. Furthermore, our method relies on the fact that the data distribution of the Random Forest predictions is closer than the raw data distribution between source and target dataset. Given that we always use the same convolutional filter banks for feature computation, the Random Forests on source and target share the same inductive bias and this assumption will most of the time hold up when segmenting the same semantic class (with similar shape and size distributions). However, in some cases systematic differences between Random Forest predictions on source and target may still exist, for example if the source data has much higher signal-to-noise ratio and thus presents an easier segmentation problem. In this case the segmentation accuracy of our method will suffer despite close shape and size distribution.
For simplicity, and also to sample as many source/target pairs with full groundtruth as possible, we have only demonstrated results on 2D data, in a binary foreground/background classification setting. Extension to 3D is straightforward and would not require any changes in our method other than accounting for potentially different z resolution between source and target datasets. Extension to multi-class segmentation would only need a simple update to the pseudo-label training loss.
In future work, we envision integration of our approach with other pseudo-label training strategies. Furthermore, as pseudo-label training can largely be configured without target domain knowledge, we expect our method to be a prime candidate for user-facing tools which already include interactive feature-based classifier training.
The original contributions presented in the study are included in the article/supplementary materials, further inquiries can be directed to the corresponding author/s.
AK, AM, AW, and CP have conceptualized the method. AM has implemented the method and run the experiments under the supervision of AK, AW, and CP. AM and CP have drafted the manuscript. AK, AW, and CP have written the final manuscript. All authors contributed to the article and approved the submitted version.
AW was funded by DFG FOR2581 for this work.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank the EMBL IT Services for their support.
Arganda-Carreras, I., Kaynig, V., Rueden, C., Eliceiri, K. W., Schindelin, J., Cardona, A., et al. (2017). Trainable Weka segmentation: a machine learning tool for microscopy pixel classification. Bioinformatics 33, 2424–2426. doi: 10.1093/bioinformatics/btx180
Arganda-Carreras, I., Turaga, S. C., Berger, D. R., Cireşan, D., Giusti, A., Gambardella, L. M., et al. (2015). Crowdsourcing the creation of image segmentation algorithms for connectomics. Front. Neuroanat. 9:142. doi: 10.3389/fnana.2015.00142
Beier, T., Pape, C., Rahaman, N., Prange, T., Berg, S., Bock, D. D., et al. (2017). Multicut brings automated neurite segmentation closer to human performance. Nat. Methods 14, 101–102. doi: 10.1038/nmeth.4151
Belevich, I., Joensuu, M., Kumar, D., Vihinen, H., and Jokitalo, E. (2016). Microscopy image browser: a platform for segmentation and analysis of multidimensional datasets. PLoS Biol. 14:e1002340. doi: 10.1371/journal.pbio.1002340
Berg, S., Kutra, D., Kroeger, T., Straehle, C. N., Kausler, B. X., Haubold, C., et al. (2019). Ilastik: interactive machine learning for (bio) image analysis. Nat. Methods 16, 1226–1232. doi: 10.1038/s41592-019-0582-9
Bermúdez-Chacón, R., Altingövde, O., Becker, C., Salzmann, M., and Fua, P. (2019). Visual correspondences for unsupervised domain adaptation on electron microscopy images. IEEE Trans. Med. Imaging 39, 1256–1267. doi: 10.1109/TMI.2019.2946462
Caicedo, J. C., Goodman, A., Karhohs, K. W., Cimini, B. A., Ackerman, J., Haghighi, M., et al. (2019). Nucleus segmentation across imaging experiments: the 2018 data science bowl. Nat. Methods 16, 1247–1253. doi: 10.1038/s41592-019-0612-7
Chen, C., Dou, Q., Chen, H., Qin, J., and Heng, P.-A. (2019). “Synergistic image and feature adaptation: towards cross-modality domain adaptation for medical image segmentation,” in Proceedings of the AAAI Conference on Artificial Intelligence, Vol. 33 (Honolulu, HI), 865–872. doi: 10.1609/aaai.v33i01.3301865
Choi, J., Jeong, M., Kim, T., and Kim, C. (2019). Pseudo-labeling curriculum for unsupervised domain adaptation. arXiv preprint arXiv:1908.00262.
Du, Y., Yang, H., Chen, M., Jiang, J., Luo, H., and Wang, C. (2021). Generation, augmentation, and alignment: a pseudo-source domain based method for source-free domain adaptation. arXiv preprint arXiv:2109.04015.
El Jurdi, R., Petitjean, C., Honeine, P., Cheplygina, V., and Abdallah, F. (2021). High-level prior-based loss functions for medical image segmentation: a survey. Comput. Vis. Image Understand. 210:103248. doi: 10.1016/j.cviu.2021.103248
Gerhard, S., Funke, J., Martel, J., Cardona, A., and Fetter, R. (2013). Segmented anisotropic sstem dataset of neural tissue. figshare. Dataset. doi: 10.6084/m9.figshare.856713.v1
Han, S., Pool, J., Narang, S., Mao, H., Tang, S., Elsen, E., et al. (2016). DSD: regularizing deep neural networks with dense-sparse-dense training flow. arXiv preprint arXiv:1607.04381.
Januszewski, M., and Jain, V. (2019). Segmentation-enhanced cyclegan. bioRxiv 2019:548081. doi: 10.1101/548081
Kasthuri, N., Hayworth, K. J., Berger, D. R., Schalek, R. L., Conchello, J. A., Knowles-Barley, S., et al. (2015). Saturated reconstruction of a volume of neocortex. Cell 162, 648–661. doi: 10.1016/j.cell.2015.06.054
Kumar, N., Verma, R., Anand, D., Zhou, Y., Onder, O. F., Tsougenis, E., et al. (2019). A multi-organ nucleus segmentation challenge. IEEE Trans. Med. Imaging 39, 1380–1391. doi: 10.1109/TMI.2019.2947628
Liu, D., Zhang, D., Song, Y., Zhang, F., O'Donnell, L., Huang, H., et al. (2020). Pdam: a panoptic-level feature alignment framework for unsupervised domain adaptive instance segmentation in microscopy images. IEEE Trans. Med. Imaging 40, 154–165. doi: 10.1109/TMI.2020.3023466
Liu, Y., Zhang, W., and Wang, J. (2021). “Source-free domain adaptation for semantic segmentation,” in Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (Virtual), 1215–1224. doi: 10.1109/CVPR46437.2021.00127
Long, M., Cao, Y., Wang, J., and Jordan, M. (2015). “Learning transferable features with deep adaptation networks,” in International Conference on Machine Learning (Lille), 97-105.
Lucchi, A., Li, Y., and Fua, P. (2013). “Learning for structured prediction using approximate subgradient descent with working sets,” in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (Portland, OR), 1987–1994. doi: 10.1109/CVPR.2013.259
Meilă, M. (2003). “Comparing clusterings by the variation of information,” in Learning Theory and Kernel Machines (Washington, DC: Springer), 173–187. doi: 10.1007/978-3-540-45167-9_14
Prabhu, V., Khare, S., Kartik, D., and Hoffman, J. (2021). S4t: Source-free domain adaptation for semantic segmentation via self-supervised selective self-training. arXiv preprint arXiv:2107.10140.
Roels, J., Hennies, J., Saeys, Y., Philips, W., and Kreshuk, A. (2019). “Domain adaptive segmentation in volume electron microscopy imaging,” in 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019) (Venice), 1519–1522. doi: 10.1109/ISBI.2019.8759383
Ronneberger, O., Fischer, P., and Brox, T. (2015). “U-net: convolutional networks for biomedical image segmentation,” in International Conference on Medical Image Computing and Computer-Assisted Intervention (Munich: Springer), 234–241. doi: 10.1007/978-3-319-24574-4_28
Rozantsev, A., Salzmann, M., and Fua, P. (2018). Beyond sharing weights for deep domain adaptation. IEEE Trans. Pattern Anal. Mach. Intell. 41, 801–814. doi: 10.1109/TPAMI.2018.2814042
Taha, A., Shrivastava, A., and Davis, L. (2021). “Knowledge evolution in neural networks,” in Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) (Virtual). doi: 10.1109/CVPR46437.2021.01265
Tarvainen, A., and Valpola, H. (2017). “Mean teachers are better role models: weight-averaged consistency targets improve semi-supervised deep learning results,” in Proceedings of the 31st International Conference on Neural Information Processing Systems, NIPS'17 (Red Hook, NY; Long Beach, CA: Curran Associates Inc.), 1195–1204.
Wei, D., Lin, Z., Franco-Barranco, D., Wendt, N., Liu, X., Yin, W., et al. (2020). “Mitoem dataset: large-scale 3d mitochondria instance segmentation from EM images,” in International Conference on Medical Image Computing and Computer-Assisted Intervention (Lima: Springer), 66–76. doi: 10.1007/978-3-030-59722-1_7
Wolny, A., Cerrone, L., Vijayan, A., Tofanelli, R., Barro, A. V., Louveaux, M., et al. (2020). Accurate and versatile 3D segmentation of plant tissues at cellular resolution. Elife 9:e57613. doi: 10.7554/eLife.57613
Wu, S., Chen, C., Xiong, Z., Chen, X., and Sun, X. (2021). “Uncertainty-aware label rectification for domain adaptive mitochondria segmentation,” in International Conference on Medical Image Computing and Computer-Assisted Intervention (Springer) (Virtual), 191–200. doi: 10.1007/978-3-030-87199-4_18
Xing, F., Bennett, T., and Ghosh, D. (2019). “Adversarial domain adaptation and pseudo-labeling for cross-modality microscopy image quantification,” in International Conference on Medical Image Computing and Computer-Assisted Intervention (Shenzhen: Springer), 740–749. doi: 10.1007/978-3-030-32239-7_82
Zhang, P., Zhang, B., Zhang, T., Chen, D., Wang, Y., and Wen, F. (2021). “Prototypical pseudo label denoising and target structure learning for domain adaptive semantic segmentation,” in Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (Virtual), 12414–12424. doi: 10.1109/CVPR46437.2021.01223
Zhang, Y., Miao, S., Mansi, T., and Liao, R. (2018). “Task driven generative modeling for unsupervised domain adaptation: application to x-ray image segmentation,” in International Conference on Medical Image Computing and Computer-Assisted Intervention (Granada: Springer), 599–607. doi: 10.1007/978-3-030-00934-2_67
Zhao, Y., Zhong, Z., Luo, Z., Lee, G. H., and Sebe, N. (2021). Source-free open compound domain adaptation in semantic segmentation. arXiv preprint arXiv:2106.03422.
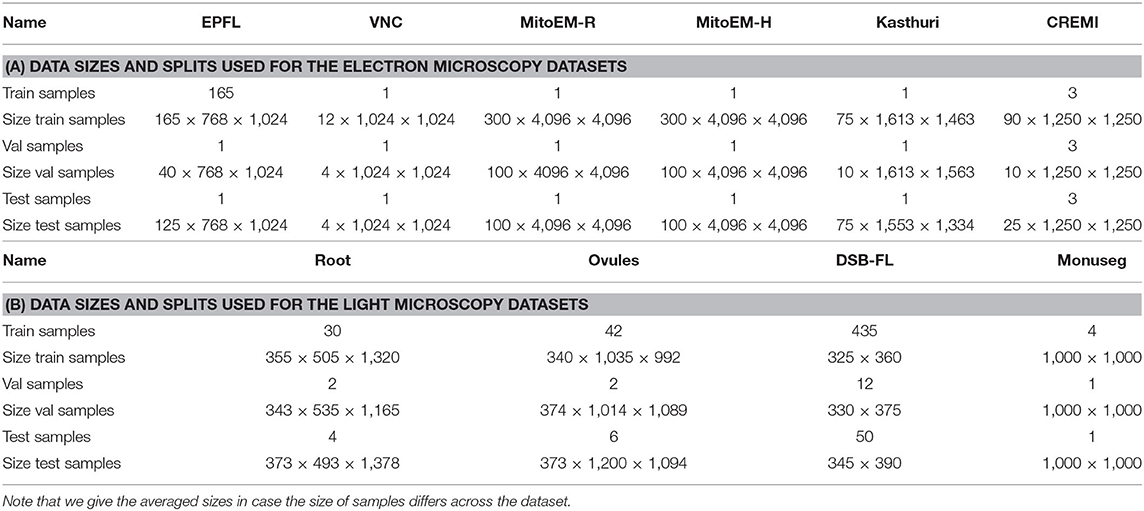
Table A1. The number of samples used for train, validation and test splits as well as the size of one of the sample in pixels.
Here, we study the influence of the number of trees per Random Forest on the Enhancer. We train the Enhancer from RF predictions where each Forest contains 50, 100, 150 or a number of trees drawn randomly from the range 50 to 150. Table A2 shows the results for the same data as used in Section 3.2 where we have used 100 trees per RF. Note that the results do not directly correspond to any of the results in Table 2 where we have used further refined target RFs. Here, we observe that the quality of the enhancer is not systematically influenced by the number of trees.
Keywords: microscopy segmentation, domain adaptation, deep learning, transfer learning, biomedical segmentation
Citation: Matskevych A, Wolny A, Pape C and Kreshuk A (2022) From Shallow to Deep: Exploiting Feature-Based Classifiers for Domain Adaptation in Semantic Segmentation. Front. Comput. Sci. 4:805166. doi: 10.3389/fcomp.2022.805166
Received: 29 October 2021; Accepted: 02 February 2022;
Published: 03 March 2022.
Edited by:
Florian Jug, Human Technopole, ItalyReviewed by:
Dagmar Kainmueller, Max Delbrück Center for Molecular Medicine, Helmholtz Association of German Research Centers (HZ), GermanyCopyright © 2022 Matskevych, Wolny, Pape and Kreshuk. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Constantin Pape, Y29uc3RhbnRpbi5wYXBlQGVtYmwuZGU=; Anna Kreshuk, YW5uYS5rcmVzaHVrQGVtYmwuZGU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.