
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
METHODS article
Front. Comput. Sci., 09 December 2021
Sec. Computer Vision
Volume 3 - 2021 | https://doi.org/10.3389/fcomp.2021.777837
This article is part of the Research TopicMethods and Tools for Bioimage AnalysisView all 13 articles
Neuronal synapses are highly dynamic communication hubs that mediate chemical neurotransmission via the exocytic fusion and subsequent endocytic recycling of neurotransmitter-containing synaptic vesicles (SVs). Functional imaging tools allow for the direct visualization of synaptic activity by detecting action potentials, pre- or postsynaptic calcium influx, SV exo- and endocytosis, and glutamate release. Fluorescent organic dyes or synapse-targeted genetic molecular reporters, such as calcium, voltage or neurotransmitter sensors and synapto-pHluorins reveal synaptic activity by undergoing rapid changes in their fluorescence intensity upon neuronal activity on timescales of milliseconds to seconds, which typically are recorded by fast and sensitive widefield live cell microscopy. The analysis of the resulting time-lapse movies in the past has been performed by either manually picking individual structures, custom scripts that have not been made widely available to the scientific community, or advanced software toolboxes that are complicated to use. For the precise, unbiased and reproducible measurement of synaptic activity, it is key that the research community has access to bio-image analysis tools that are easy-to-apply and allow the automated detection of fluorescent intensity changes in active synapses. Here we present SynActJ (Synaptic Activity in ImageJ), an easy-to-use fully open-source workflow that enables automated image and data analysis of synaptic activity. The workflow consists of a Fiji plugin performing the automated image analysis of active synapses in time-lapse movies via an interactive seeded watershed segmentation that can be easily adjusted and applied to a dataset in batch mode. The extracted intensity traces of each synaptic bouton are automatically processed, analyzed, and plotted using an R Shiny workflow. We validate the workflow on time-lapse images of stimulated synapses expressing the SV exo-/endocytosis reporter Synaptophysin-pHluorin or a synapse-targeted calcium sensor, Synaptophysin-RGECO. We compare the automatic workflow to manual analysis and compute calcium-influx and SV exo-/endocytosis kinetics and other parameters for synaptic vesicle recycling under different conditions. We predict SynActJ to become an important tool for the analysis of synaptic activity and synapse properties.
Neuronal synapses are important cellular communication hubs that transmit signals between neurons to enable brain function (Figure 1A). The transmission of the signal is mediated by chemical neurotransmitters that are synthesized in the presynaptic neuron and stored in the lumen of synaptic vesicles (SVs). When an action potential reaches the synapse, calcium enters the presynaptic terminal via voltage-gated calcium channels and triggers the exocytic fusion of release-ready SVs with the plasma membrane (Sudhof, 2013). The released neurotransmitter molecules rapidly diffuse through the synaptic cleft to activate postsynaptic receptors of the receiving neuron. Depending on its type, the neurotransmitter can either help (i.e. excitatory transmitters) or hinder (i.e. inhibitory transmitters) the postsynaptic neuron from firing its own action potential. At the presynaptic terminal, shortly after their fusion with the plasma membrane, SVs are recycled via endocytosis and refilled with neurotransmitter to sustain synaptic transmission (Soykan et al., 2016).
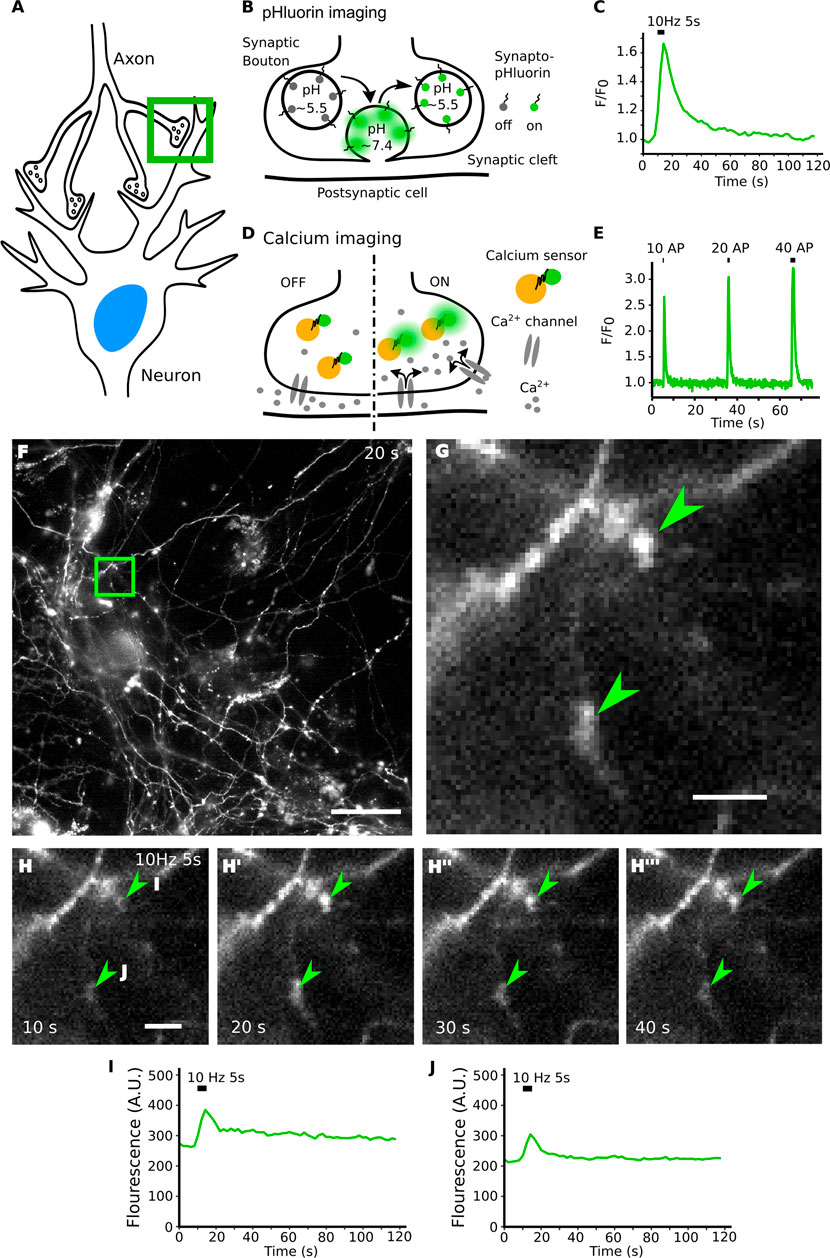
FIGURE 1. Live imaging of synaptic activity. (A) Synaptic signal transmission between neurons depends on the function of synaptic vesicles and on the activity of ion channels. (B) Functional imaging by synapto-pHluorin allows studying synaptic vesicle exocytic fusion and subsequent endocytic recycling and acidification using a pH sensitive GFP genetically fused to the luminal domain of a synaptic vesicle membrane protein. The GFP fluorescence is quenched under physiological steady-state conditions in the acidic vesicle lumen. (B,C) Upon fusion pHluorin is exposed to the higher extracellular pH, becomes fluorescent and is rapidly quenched when the vesicle is endocytosed, and its lumen returns to its normal acidic pH. (D,E) The influx of calcium elicits synaptic vesicle exocytosis and can be detected using calcium sensors. (F) A single frame from a full field of view time-lapse movie of a hippocampal neuronal culture transfected with Synaptophysin-pHluorin. [(G), arrowheads] Acquisition with fast and sensitive widefield live imaging using low light exposure leads to images with many irrelevant and out of focus structures around the objects of interest. (H–H‴) The increase in fluorescence of Synaptophysin-pHluorin after electrical stimulation is small in size and intensity. (I,J) Raw individual traces shown from the top ROI and bottom ROI in panel H. Peak intensity is relatively small over background signal and varies between ROIs. Scale bars: 50 µm (F) and 5 µm (G,H).
To study these highly dynamic processes that occur on timescales of milliseconds to seconds and to monitor the molecular components that orchestrate them, functional imaging tools are crucial. For instance, the exocytosis and endocytosis of SVs can be tracked using synapto-pHluorins, chimeric proteins comprising a pH sensitive variant of GFP genetically fused to the luminal domain of a SV membrane protein. As the pH of a SV under physiological conditions is acidic (∼pH 5.5) the fluorescence of pHluorin is quenched at steady-state (Miesenböck et al., 1998). Upon arrival of an action potential (e.g. electrical stimulation of a neuron) SVs release their neurotransmitter content by fusing with the plasma membrane, thereby exposing their lumen to the neutral pH of the extracellular medium (∼pH 7.4), upon which pHluorin becomes strongly fluorescent. Subsequent endocytosis of SV membrane proteins from the plasma membrane and rapid acidification of the endocytic vesicle lumen via the vesicular ATPase results in re-quenching of the fluorescence signal elicited from pHluorin molecules (Figures 1B,C) (Miesenböck et al., 1998; Wienisch and Klingauf, 2006; Kavalali and Jorgensen, 2014). Other assays for synaptic function include molecular sensors that detect the pre- or postsynaptic influx of calcium (Figures 1D,E) (Zhao et al., 2011), as well as reporters that monitor changes in membrane voltage or detect the released neurotransmitters (Lin and Schnitzer, 2016). These tools all have in common that they need to detect fast changes in fluorescence signals in small, often diffraction-limited structures to enable the visualization of specific synaptic functions.
The optical detection of synaptic fluorescence is in general performed on live cell or tissue cultures using widefield microscopy to capture as much signal as possible on large, fast and highly sensitive detectors with a diffraction limited resolution of ∼200 nm. In order to reduce bleaching and phototoxicity, the light exposure is kept to a minimum. This produces relatively noisy images with abundant out-of-focus signal and many cellular structures that are irrelevant for the downstream analysis (Figures 1F–H). The resulting time-lapse movies are thus hard to analyze via automated image analysis workflows that only rely on intensity thresholding and detection, often resulting in the analysis being performed manually (Balaji, 2007; Soykan et al., 2017), using custom scripts not easily usable for a wider research community (Chanaday and Kavalali, 2018), or (i.e. for calcium imaging) via advanced software toolboxes that require programming expertise (Kaifosh et al., 2014; Pachitariu et al., 2016; Giovannucci et al., 2019; Robbins et al., 2021) (Supplementary Table S1). To eliminate the need for tedious, time consuming and biased manual image analysis, the key challenge is to develop robust and easy-to-use software tools that do not require in-depth expertise in programming and image analysis. These tools need to be implemented rigorously, using scientific software engineering to enable easy, transparent and fully reproducible analysis of synaptic imaging data. In recent years the research community has recognized this need and more of such tools have become available for different applications. For example, EZcalcium has been developed for the analysis of calcium imaging data (Cantu et al., 2020). However, due to its implementation in MATLAB, EZcalcium and similar tools require expensive software licenses, thereby limiting their accessibility.
Here we present SynActJ an easy-to-use, entirely free and open source Fiji plugin (Schindelin et al., 2012a) combined with an R shiny app (R-Core-Team, 2020; RStudio-Team, 2020; Chang et al., 2021) that allows the fully automatic detection and analysis of synaptic activity in time-lapse movies. The segmentation parameters can be easily and conveniently adjusted and verified over an entire dataset after which the image analysis can be performed in batch. It is fully reproducible by allowing the saving and loading of the segmentation parameters. SynActJ is implemented as a Fiji plugin in Java and can thus be easily installed via a Fiji update site. For the necessary data processing and quality control of the extracted traces a convenient R Shiny app is provided as well as an example script for more detailed analysis and plotting.
The Fiji plugin can be accessed via the Cellular Imaging update site: https://sites.imagej.net/Cellular-Imaging/. The code for the Shiny app can be downloaded from github: https://github.com/schmiedc/SynActJ_Shiny. Documentation for using the plugin and the app can be found here: https://schmiedc.github.io/SynActJ/. A test dataset is made available on Zenodo: https://doi.org/10.5281/zenodo.5644945. The code is distributed free and open-source under the MIT license: https://github.com/schmiedc/SynActJ/blob/master/LICENSE. The source code of the Fiji plugin can be downloaded from github: https://github.com/schmiedc/SynActJ. Contact and support: https://forum.image.sc/u/schmiedc/.
Typically, live cell neuronal cultures are imaged using a widefield system with a large field of view. The tissue is illuminated with as little light as possible, to reduce bleaching and phototoxicity. The data sets, while rich in quantitative data, contain out-of-focus signal and many different biological structures that are irrelevant to the analysis (Figures 1F,G). For synapto-pHluorin imaging, upon applying field stimulation to the live neuron culture with an electric current, the synaptic boutons respond with a sharp elevated fluorescence signal corresponding to the exocytic fusion and subsequent release of neurotransmitter. Following stimulation, the signal rapidly drops back to baseline on a timescale of seconds as the pHluorin tag gets quenched when SVs are endocytosed and acidified (Figures 1H–H‴). These events of briefly elevated signal with relatively small responses over the baseline signal are hard to detect by eye and require that brightness contrast settings are set correctly for perceiving the specific signal responses (Figures 1I,J).
SynActJ achieves accurate automatic segmentation of active synaptic boutons (Figures 2A,B). It solves the segmentation problem by carving out the portion of the image signal that increases around the time of stimulation (Figure 2C). This is achieved by dividing the maximum intensity projection of 10 frames post-stimulation by the maximum intensity projection of the frames before the stimulation. This allows the program to selectively detect responding boutons within the image while removing any non-relevant signal from the remainder of the field of view (Figures 2E,F). In order to improve detection of weakly responding synapses an ImageJ implementation of the Laplacian-of-Gaussian (LoG) filter (Sage et al., 2005) is applied to enhance blob-like structures of a defined size range followed by a maximum detection to spot individual events within the diffraction limited resolution of ∼200 nm (Figure 2G). To create an accurate segmentation of the bouton area, an intensity threshold is applied. The detection and segmentation masks are then used in a marker-controlled watershed using the MorpholibJ Fiji plugin (Legland et al., 2016). This limits the segmentation to objects that have passed the detection threshold and enables the separation of touching detected structures (Figure 2H). The segmentation can be further refined by performing a size and object circularity filter. To quantitatively measure the background the maximum intensity projection of the movie is filtered with a large Gaussian blur and an intensity threshold is used to segment the broad signal of the tissue. The resulting mask of the field of view is finally inverted to create a segmentation of the background.
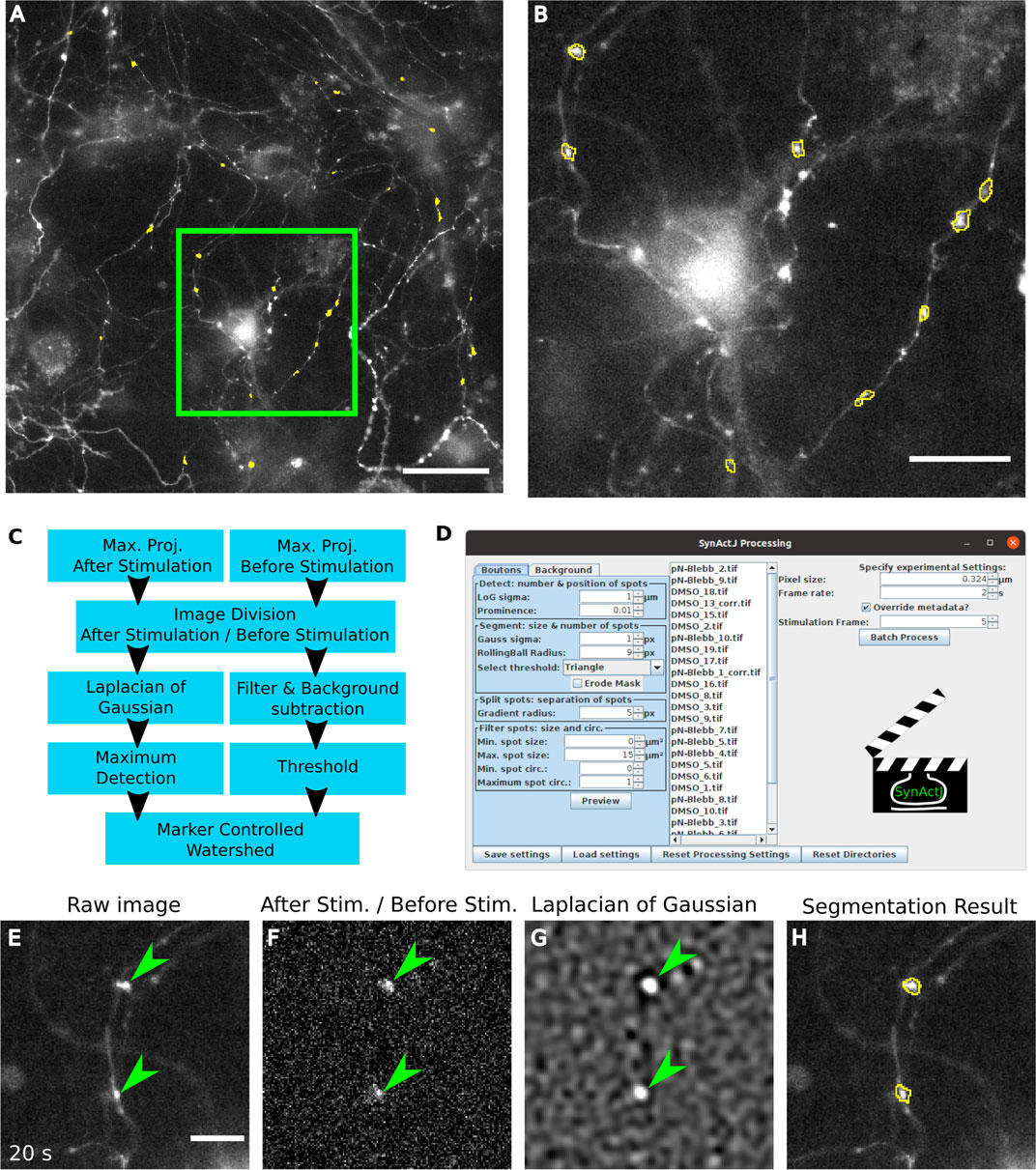
FIGURE 2. Fast, automatic and easy-to-use image analysis of synaptic activity using SynActJ. (A) Hippocampal neuronal cultures transfected with Synaptophysin-pHluorin. [(A,B), yellow outlines] Structures showing a signal response upon stimulation, corresponding to active synaptic boutons are segmented with SynActJ. (C) Segmentation is achieved by detecting blob-like structures that display an increase in signal after the stimulation frame and by separating touching structures using a marker-controlled watershed. (D) The segmentation parameters can be adjusted and verified on the entire dataset using a GUI that allows testing each segmentation parameter easily, after which the workflow can be executed in batch. [(E), arrowheads] Crop of a raw image showing Synaptophysin-pHluorin with peak fluorescence after electrical stimulation. [(F), arrowheads] The increase in signal around the time point of stimulation is enhanced by dividing the maximum projection of time points after the stimulation by the time points before the stimulation. [(G), arrowheads] Blob-like structures of a specific size are enhanced using a LoG filter]. [(H), yellow outlines] Active boutons are segmented using an intensity threshold and touching objects split with a seeded watershed. Scale bars: 50 µm (A), 20 µm (B) and 10 µm (E–H).
SynActJ provides a graphical user interface which enables the user to interactively adjust the parameters of the workflow via a preview (Figure 2D). Importantly the user has access to the entire dataset and, thus, is able to quickly verify and adjust the parameters on any of the movies the analysis is applied to. SynActJ ensures full reproducibility of the workflow by allowing the user to conveniently save, load and access all the processing settings. Once suitable segmentation parameters have been defined by the user, the entire image analysis can be executed on the dataset in batch, e.g. different movies of different stimulations or treatments (Figure 2D). The segmentation exactly outlines the area of the signal response after stimulation (Figures 2A,B,H) and the raw average signal is measured over time in this area. Since the segmentation is outlining the bouton, it is vital that any drift is corrected prior to segmentation using plug-ins such as Correct 3D drift (Parslow et al., 2014).
The resulting measurements are saved in the specified output folder and can then be processed easily using the R Shiny app provided (Figure 3A). The app allows rapid processing of the individual traces extracted by the image analysis and provides overview plots for quick and efficient quality control of the data. For each movie the number of ROIs, area of segmented structures and background are plotted (Figure 3B). Furthermore, for each movie the raw traces, raw background signals, as well as the average raw traces and background values are plotted and provided as a data table (Figure 3C). The raw values can be further filtered and processed in custom data analysis workflows.
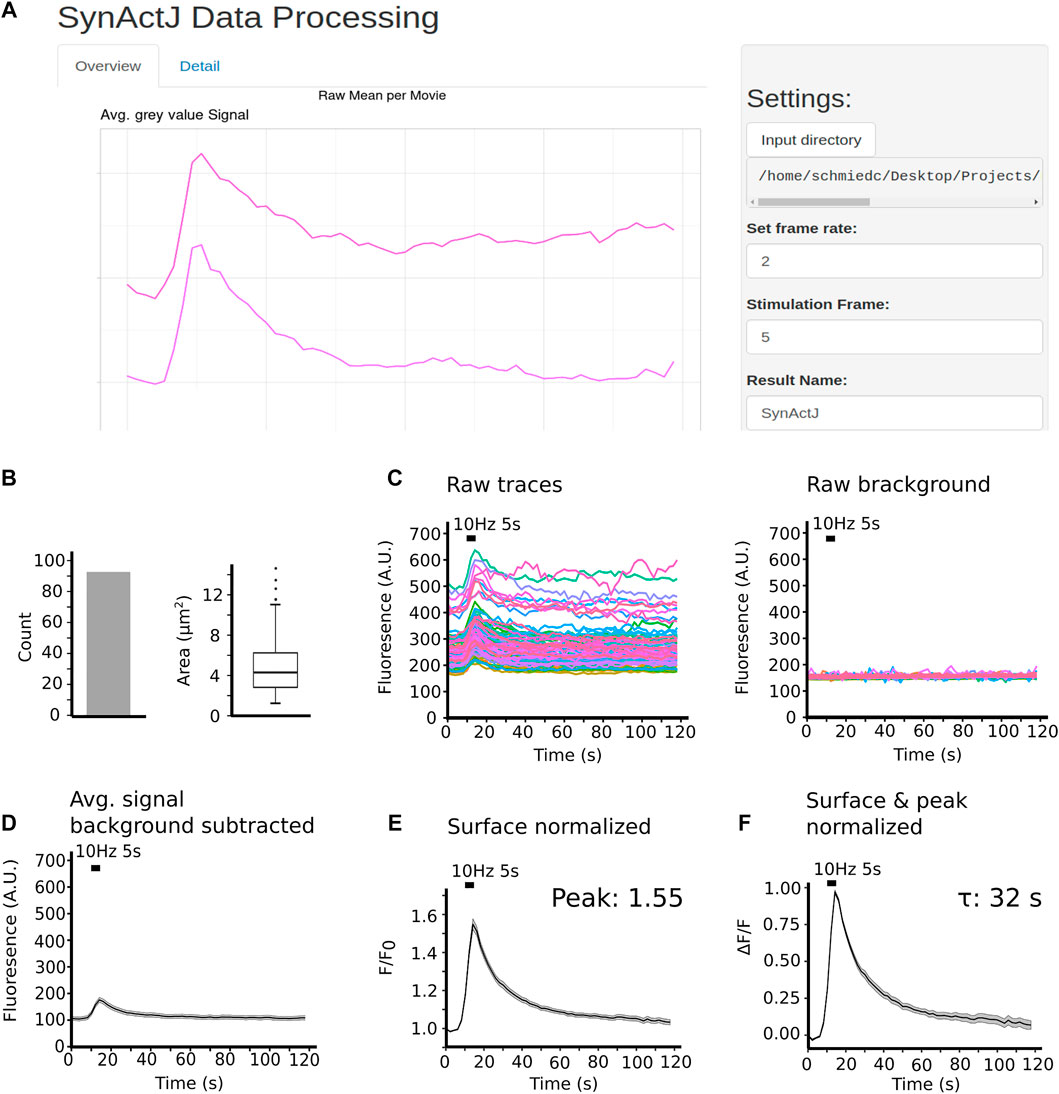
FIGURE 3. SynActJ enables easy and rapid data processing and analysis via an R Shiny app. (A) The shiny app allows users to process and plot the result of the image analysis using a GUI. (B) Vital parameters for quality control such as number of ROIs and mean area of ROIs per movie are plotted per time-lapse. (C) Raw traces of the signal as well as the background are extracted and plotted over time. (D) Per movie the average signal is computed, and the background value is subtracted. The average and background subtracted signal traces are surface (E) as well as peak normalized (F).
The Shiny App also performs basic as well as advanced data processing on each individual movie. The average background value is subtracted from the average signal (Figure 3D). To compare the differences in peak responses to the stimulation, the background-corrected signal is surface-normalized by dividing the signal by the average value of the trace in the frames prior to stimulation (Figure 3E). To compare how fast the signal returns to baseline a further normalization to the peak of the trace is performed (Figure 3F). The result of this data processing is again provided as a data table accessible for further custom statistical analysis and plotting. For instance in order to characterize and quantitatively compare the rate of endocytosis one can fit an exponential decay function to the peak-normalized data and compute the time constant τ (tau: time it took the signal to return to 1/e of the peak) (Figure 3F).
To benchmark our workflow we applied SynActJ to an existing dataset that has previously been analyzed manually (Soykan et al., 2017). In this dataset, cultured hippocampal neurons expressing Synaptophysin-pHluorin were treated with DMSO or para-nitro-Blebbistatin (Kepiro et al., 2014), a specific inhibitor for Myosin-II, and stimulated with 50 APs (10 Hz, 5 s) to determine whether Myosin-II activity is involved in SV exo-/endocytosis. In line with the previously proposed role for Myosin-II at synapses (Chandrasekar et al., 2013; Flores et al., 2014; Yue and Xu 2014), para-nitro-Blebbistatin treatment led to reduced Synaptophysin-pHluorin exocytosis after stimulation, as well as delayed endocytosis. Our automated workflow was able to segment more boutons compared to the previously performed manual analysis, which was limited to about 20 ROIs for responding boutons and five background ROIs (Figure 4A). For further validation we asked how many of the manually selected ROIs also contain an automatically detected ROI. We found that 64% of the manually selected ROIs overlapped with the automatically detected ones (Figure 4B). We further determined which objects the automatic analysis missed in the manual dataset and found that many of those structures were large objects such as clusters of boutons or large uniform, potentially out of focus objects (Figure 4C). Overall the automatic segmentations of SynActJ seemed to be more restrictive in terms of object size and shape compared to the manual segmentation. To remove false positive automatic detections that by chance showed an increase around the stimulation frame, we removed traces in which the response was below two standard deviations of the variation of the background. Further, we removed traces with peaks much later than the stimulation frame. This filter operations removed 312 traces from the original set, leaving 6,623 traces from the automatic segmentation that conform to the necessary response upon stimulation.
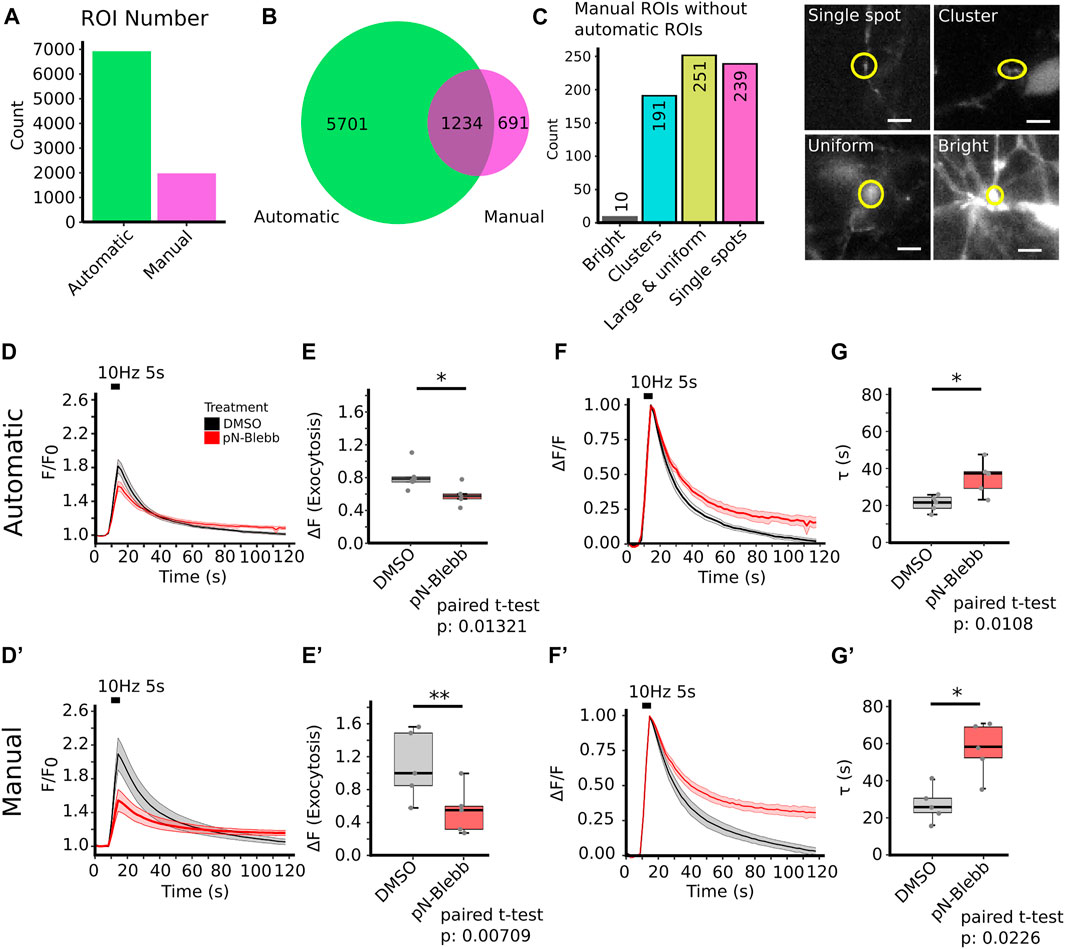
FIGURE 4. SynActJ allows the rapid analysis of synaptic activity using synapto-pHluorin imaging and reproduces the results of a comparable manual analysis. (A) The automatic image segmentation allows extracting many more ROIs from the image data compared to a manual segmentation. (B) 64% of the manual ROIs have been detected in the automated analysis. (C) Many of the manual ROIs without automatic ROIs are clusters of boutons or relatively large, potential out of focus objects, aside from single boutons (false negative automatic segmentation), example images of these cases are provided. [(D,E) compare to (D′,E′)] The automatic image analysis is able to reproduce the results from a manually performed image analysis (Soykan et al., 2017) showing significant decrease in peak signal increase after stimulation. (F,G) compared to (F′,G′) A significant delay of SV endocytosis and reacidification can be detected in the automatic and manual analysis. Data are presented as mean ± standard error of the mean (SEM) in all panels; n. s not significant; *p < 0.05; **p < 0.01. N = 5 independent experiments; paired t-test. Scale bar corresponds to 5 µm (C).
The automatic analysis was able to reproduce the results of the manual analysis: Surface-normalized plots revealed a decreased exocytic response to stimulation (Figures 4D,D′) while peak-normalized signals displayed a delay in the return to baseline (Figures 4F,F′). The automatic analysis also showed a reduced peak signal (Figures 4E,E′) and a significant increase in the time constant for returning to baseline intensity upon para-nitro-Blebbistatin application (Figures 4G,G′). Interestingly, automatic analysis detected a lower peak signal than analysis by manual assessment. This likely is attributed to the fact our automated analysis workflow is superior with respect to the detection of weakly responding synaptic events, thereby increasing the accuracy of the analysis and reducing bias towards larger and strong responding structures (Figure 4C). This is particularly overt for the DMSO control, for which manual detection overall used much fewer active boutons (Figures 4E,E′). Finally, the variability in the extracted parameters is greatly reduced with the standard deviation dropping by a factor of 1.5–2.4 (Figures 4E,G; Tables 1, 2), further illustrating the increased precision of the automated approach.
To test if our plugin can be applied to other modes of functional imaging of synaptic activity, we further validated our approach on calcium imaging data. Calcium imaging suffers from similar challenges as those seen for Synapto-pHluorins such as relatively noisy and background-rich live images with fast, yet comparably small fluorescence signal changes over baseline (Figure 5A–C). We recorded time-lapse movies of cultured hippocampal neurons expressing the presynaptic calcium sensor Synaptophysin-RGECO and stimulated successively with 10, 20 and 40 APs (all at 40 Hz and 0.25, 0.5 and 1 s, respectively). Similar to the data from Synapto-pHluorin imaging experiments, we were able to rapidly produce accurate segmentations of active boutons in which calcium influx was revealed by RGECO (Figures 5A,B). Automated image analysis by SynActJ successfully extracted traces thereby enabling the detection of calcium spikes with overall results near-identical to those produced by manual analysis (Figures 5D,E), yet in only a fraction of the time.
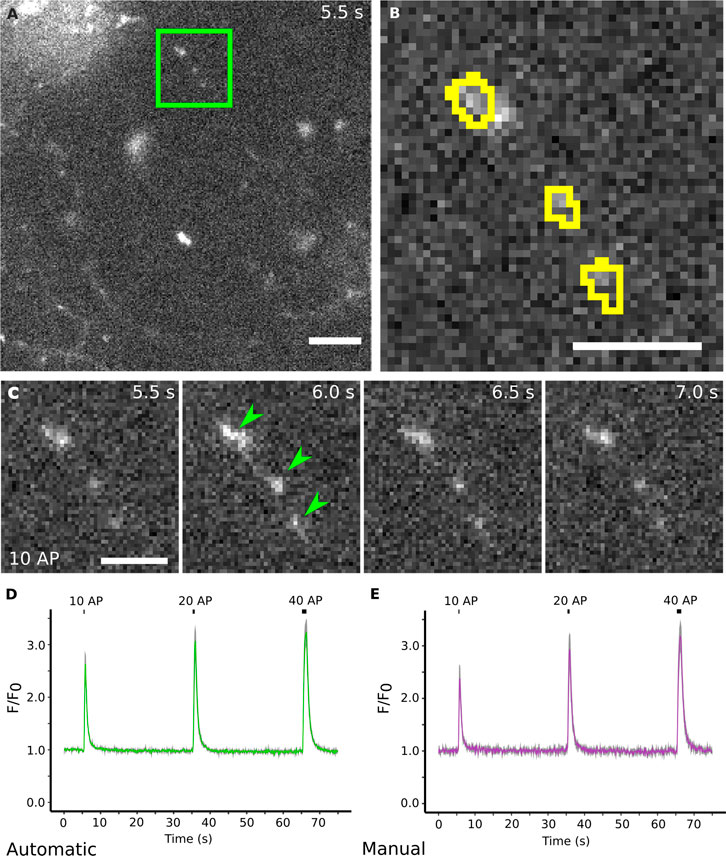
FIGURE 5. SynActJ can readily be applied to other imaging modalities for synaptic activity such as calcium imaging. (A) Cultured mouse neurons transfected with Synaptophysin-RGECO. [(B), yellow outline] Active boutons in calcium imaging data can be successfully segmented]. (C) Electric field stimulation of the neurons leads to an increase in fluorescence in synaptic boutons that rapidly drops down to baseline. (D) The data analysis allows the visualization of the increase in signal that corresponds to the influx of calcium during/post-stimulation, reproducing the results from manual segmentation (E). Scale bars: 10 µm (A) and 5 µm (B,C). Data presented as mean ± SEM. N = 4 images.
Biological image data is notoriously difficult to robustly analyze automatically. Not only do the biological and experimental variability hamper computational analysis but also noise and background introduced by hard limitations in the acquisition of biological data. Thus, it is often easier for highly trained scientists to perform these image analysis tasks by hand. For analyzing synaptic activity another key challenge are the brief changes in fluorescent intensity, limited to many small structures throughout a large field of view with heterogeneous signal. This renders manual detection also biased towards strong and large structures that are easier to detect. The difficulty to manually detecting active boutons effectively also limits the number and completeness of detected events that can be analyzed (Soykan et al., 2017). Furthermore, manual selection of responding synaptic terminals using circular ROIs does not allow for precisely outlining the boutons, which is advantageous if there is a slight sample drift or tissue movement. However, this leads to the inclusion of some background area in the final quantification making the measurement less accurate.
The automated analysis of SynActJ efficiently overcomes these challenges. As an easy-to-use workflow it allows the robust and transparent application of well-tested classical image analysis on complex time-lapse movies. It is delivered as an easily installed Fiji plugin for the image analysis as well as a Shiny app that are centered on easy-to-use GUIs. The segmentation is interactive and can be fine-tuned using particle size and shape filters. Basic as well as advanced data analysis is provided via an R Shiny app. SynActJ thereby replaces the tedious manual analysis, which may be prone to user-dependent bias, increases as demonstrated the overall accuracy and precision of the analysis, and enables a fully reproducible data analysis by allowing a standardized ROI selection. Together with its compatibility with a wide variety of optical sensors, we believe SynActJ will become an important toolset for the analysis of synaptic activity and function.
Hippocampal neuronal cultures were prepared by dissection of hippocampi from postnatal mice at p1-3 and grown in MEM medium (ThermoFisher) supplemented with 5% FCS, 0.5 mM l-glutamine and 2% B-27-supplement. For limiting glial proliferation 2 µM AraC was added at 2 days in vitro (DIV). The hippocampal neurons were transfected at DIV 7-9 with 6 µg plasmid DNA carrying synaptophysin fused to pHluorin (gift from L. Lagnado, Cambride, United Kingdom) or SypHy-RGECO (Jackson and Burrone, 2016) (Addgene Plasmid #84078), using a Calcium Phosphate transfection kit (Promega).
Synapto-pHluorin assay and calcium imaging was performed at DIV13-16 by applying electrical field stimulation to the neuronal culture in a RC-47FSLP stimulation chamber (Warner Instrument) and imaging at 37°C in equilibrated and osmolarity-adjusted basic buffer [170 mM NaCl, 3.5 mM KCl, 0.4 mM KH2PO4, 20 mM N-Tris (hydroxyl-methyl)-methyl-2-aminoethane-sulphonic acid (TES), 5 mM NaHCO3, 5 mM glucose, 1.2 mM Na2SO4, 1.2 mM MgCl2, 1.3 mM CaCl2, 10 mM CNQX and 50 mM AP-5, pH 7.4] using an inverted epifluorescence microscopy (Nikon Eclipse Ti) and 40Χ/NA 1.3 oil-immersion objective. For detecting synapto-pHluorin signal an eGFP filter set F36-526, and a sCMOS camera (Neo, Andor) were used. Image acquisition was performed at 0.5 frames per second with 100 ms exposure. For calcium imaging, a 580 nm LED (pE4000, CoolLED), backilluminated-sCMOS camera (Prime95B, Photometrics), a Penta DAPI/FITC/Cy3/Cy5/Cy7 Penta LED HC Filter Set (AHF 66-615) and 600/52 emission filter (Semrock) were used. Image acquisition was perfomed at 10 frames per second with 20 ms exposure.
Manual quantitative analysis of responding boutons was performed in ImageJ (NIH) (Schneider et al., 2012b) by selecting 20 circular ROIs that exhibit remarkable fluorescence increase at the time point of stimulation and five similar-sized circular ROIs selected from the background areas that display uniform fluorescence values throughout the time-lapse image. Examples of these manual regions of interest are provided here: https://doi.org/10.5281/zenodo.5644945. The mean intensity for each ROI is measured using the built-in tools in ImageJ and raw intensity values were calculated by subtracting the average intensity of background ROIs from responding ROIs. Normalized traces were generated for each image by dividing the raw intensity values to the mean intensity before stimulation (for surface normalization) and the maximum intensity (for peak normalization). For the analysis of synapto-pHluorin images, Prism 5 (Graphpad) software were used to determine the post stimulation time constants by fitting the peak normalized values to a mono-exponential decay curve:
Constraints: y0 = 1 and zero offset.
Movies affected by drift were processed using the Correct 3D drift (Parslow et al., 2014) plugin using multi time scale computation for detecting slow drifts. Since the tissue presented many edge features, the edge enhancement was turned on. Sub pixel drift correction was turned off to avoid interpolation. Only pixels with gray values larger than 130 were considered. The maximum shift was set to 10 pixel (px) in x and y.
The image analysis plugin was developed as ImageJ1 plugin (Schneider et al., 2012b). To create comparable detections over different image magnification the sigma of LoG is input in micrometre. For the analysis of the synapto-pHluorin time-lapse movies a LoG of 1 µm was used. For the maximum detection a prominence of 0.01 (A.U.) was selected. To segment the area a Gaussian filter with a sigma of 1 px, rolling ball background subtraction of 9 px and an automatic intensity threshold using the Triangle algorithm was used. No erosion was applied to the segmentation masks. For splitting detections that are touching a gradient radius with 5 px radius was applied. The final ROIs were filtered for objects larger than 15 μm2, no circularity filter was applied. For segmenting the background a Gaussian filter with a sigma of 4 px and an intensity threshold using the MinError algorithm was applied to the maximum projected movie. An example of the raw synapto-pHluorin movies as well as the image analysis parameters and results is provided here: https://doi.org/10.5281/zenodo.5644945.
For the calcium imaging data a LoG sigma of 1 µm and a prominence of 0.001 A.U. For segmenting the area of the boutons a Gaussian filter with sigma of 2 px and a rolling ball background subtraction with radius 50 px was applied. The Triangle algorithm was used for intensity thresholding without applying an erosion to the binary mask. For the seeded watershed a Gradient radius of 10 px was applied. No circularity and size filter was applied. For the background segmentation a Gaussian filter with a sigma of 4 px and the MinError thresholding was applied. The stimulation frame was set to the first stimulation at frame 55.
For the synapto-pHluorin datasets the extracted traces per ROI were collected using the SynActJ Shiny app. The detection traces were filtered for false positive detections particularly traces that showed a noisy profile without specific response to the stimulation or traces with linear increase over the movie. These would correspond to false detections in the background that by chance had a sufficiently large intensity difference around the stimulation, bright objects that move slightly in their position over the time-lapse as well as structures in the tissue that increase in intensity without responding to stimulation. For this we computed for each trace an average before the stimulation (6–8 s) as well as at the peak of the intensity just after the stimulation (14–16 s). Traces were removed that had a smaller increase in intensity than 2x the standard deviation of the variation of the background signal. Further, traces were rejected that peaked 26 s after the start of the movie. From the mean traces per movie the mean of the background was subtracted. The background corrected mean over time was then divided by the mean intensity of the frames before stimulation (surface normalization). To determine the exocytic response the peak over the baseline was measured. The surface normalized traces were divided by the peak to perform a peak normalization. Examples for the results of the data processing are provided in the example dataset (https://doi.org/10.5281/zenodo.5644945). For determining the stimulation time constant the same method as the manual image analysis was performed.
For the calcium imaging dataset the extracted traces were collected with the SynActJ Shiny app, no filtering for false positive detections was applied for the further analysis. The mean of the detection traces were subtracted by the mean of the background traces per movie. The corrected mean traces per movie were surface normalized and the means over the different movies plotted as line plot with mean ± SEM.
For testing how many manual ROIs also contain an automatic ROI, a binary mask image was first produced for the automatic segmentations. The manual ROIs were then overlaid onto this binary mask image. Since the manual ROIs do not outline the objects of interest precisely nor they were generated on drift corrected movies, any overlap was counted as manual ROI with an automatic ROI. Manual ROIs with overlap to multiple automatic ROIs was counted only once. Manual ROIs without overlap with an automatic ROI were then further visually analyzed to estimate false negatives. The objects outlined by the manual ROIs were grouped into the following classes: 1. Single bouton. 2. Large structure of cluster of boutons. 3. Very large and uniform, potentially out of focus object. 4. Large and bright structures.
All data are presented as mean ± SEM and were obtained from multiple independent experiments (e.g. independent mouse cultures), with total sample numbers provided in the figure legends as N independent experiments. No statistical method was used to pre-determine sample size as sample sizes were not chosen based on pre-specified effect size. Instead, multiple independent experiments were carried out using several sample replicates as detailed in the figure legends. The statistical significance between two groups for all normally distributed data was evaluated with a two-tailed paired Student’s t-test.
The raw data supporting the conclusion of this article will be made available by the authors, without undue reservation.
The animal study was reviewed and approved by the Tierversuchskommission Landesamt für Gesundheit und Soziales (LAGeSo) Berlin.
TS, CS, SB, VH, and ML contributed to conception and design of the project. CS designed and implemented the software. TS and SB performed experiments. SB and TS tested the software. CS, TS, and SB wrote sections of the manuscript. CS and TS performed the data and statistical analysis. All authors contributed to manuscript revision, read, and approved the submitted version.
Supported by grants from the Deutsche Forschungsgemeinschaft (SFB958/A01 to V.H. and HA2686/20-1 to V.H.).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank Delia Löwe (FMP-Berlin) for expert technical assistance. Furthermore, we thank Robert Haase (DFG Cluster of Excellence “Physics of Life”/CSBD Dresden) for feedback to the manuscript and valuable material for writing ImageJ plugins.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcomp.2021.777837/full#supplementary-material
Balaji, J. (2007). Time Series Analyzer. https://imagej.nih.gov/ij/plugins/time-series.html.
Cantu, D. A., Wang, B., Gongwer, M. W., He, C. X., Goel, A., Suresh, A., et al. (2020). EZcalcium: Open-Source Toolbox for Analysis of Calcium Imaging Data. Front. Neural Circuits 14, 25–29. doi:10.3389/fncir.2020.00025
Chanaday, N. L., and Kavalali, E. T. (2018). Optical Detection of Three Modes of Endocytosis at Hippocampal Synapses. eLife 7, 1–24. doi:10.7554/eLife.36097
Chandrasekar, I., Huettner, J. E., Turney, S. G., and Bridgman, P. C. (2013). Myosin II Regulates Activity Dependent Compensatory Endocytosis at Central Synapses. J. Neurosci. 33 (41), 16131–16145. doi:10.1523/jneurosci.2229-13.2013
Chang, W., Cheng, J., Allaire, J. J., Sievert, C., Schloerke, B., Xie, Y., et al. (2021). Shiny: Web Application Framework for R.
Flores, J. A., Balseiro-Gomez, S., Cabeza, J. M., Acosta, J., Ramirez-Ponce, P., and Ales, E. (2014). A New Role for Myosin II in Vesicle Fission. PLoS One 9 (6), e100757. doi:10.1371/journal.pone.0100757
Giovannucci, A., Friedrich, J., Gunn, P., Kalfon, J., Brown, B. L., Koay, S. A., et al. (2019). CaImAn an Open Source Tool for Scalable Calcium Imaging Data Analysis. eLife 8, 1–45. doi:10.7554/eLife.38173
Jackson, R. E., and Burrone, J. (2016). Visualizing Presynaptic Calcium Dynamics and Vesicle Fusion with a Single Genetically Encoded Reporter at Individual Synapses. Front. Synaptic Neurosci. 8, 21. doi:10.3389/fnsyn.2016.00021
Kaifosh, P., Zaremba, J. D., Danielson, N. B., and Losonczy, A. (2014). SIMA: Python Software for Analysis of Dynamic Fluorescence Imaging Data. Front. Neuroinform 8, 80. doi:10.3389/fninf.2014.00080
Kavalali, E. T., and Jorgensen, E. M. (2014). Visualizing Presynaptic Function. Nat. Neurosci. 17, 10–16. doi:10.1038/nn.3578
Képiró, M., Várkuti, B. H., Végner, L., Vörös, G., Hegyi, G., Varga, M., et al. (2014). para-Nitroblebbistatin, the Non-Cytotoxic and Photostable Myosin II Inhibitor. Angew. Chem. Int. Ed. 53 (31), 8211–8215. doi:10.1002/anie.201403540
Legland, D., Arganda-Carreras, I., and Andrey, P. (2016). MorphoLibJ: Integrated Library and Plugins for Mathematical Morphology with ImageJ. Bioinformatics 32, 3532–3534. doi:10.1093/bioinformatics/btw413
Lin, M. Z., and Schnitzer, M. J. (2016). Genetically Encoded Indicators of Neuronal Activity. Nat. Neurosci. 19, 1142–1153. doi:10.1038/nn.4359
Miesenböck, G., De Angelis, D. A., and Rothman, J. E. (1998). Visualizing Secretion and Synaptic Transmission with pH-Sensitive green Fluorescent Proteins. Nature 394, 192–195. doi:10.1038/28190
Pachitariu, M., Stringer, C., Dipoppa, M., Schröder, S., Rossi, L. F., Dalgleish, H., et al. (2016). Suite2p: Beyond 10,000 Neurons with Standard Two-Photon Microscopy. bioRxiv, 061507. doi:10.1101/061507
Parslow, A., Cardona, A., and Bryson-Richardson, R. J. (2014). Sample Drift Correction Following 4D Confocal Time-Lapse Imaging. J. Vis. Exp. 12 (86), 51086. doi:10.3791/51086
R-Core-Team (2020). R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing. https://www.R-project.org/.
Robbins, M., Christensen, C. N., Kaminski, C. F., and Zlatic, M. (2021). Calcium Imaging Analysis - How Far Have We Come? F1000Res 10, 258. doi:10.12688/f1000research.51755.2
RStudio-Team (2020). RStudio: Integrated Development Environment for R. Boston, MA: PBC. http://www.rstudio.com/.
Sage, D., Neumann, F. R., Hediger, F., Gasser, S. M., and Unser, M. (2005). Automatic Tracking of Individual Fluorescence Particles: Application to the Study of Chromosome Dynamics. IEEE Trans. Image Process. 14, 1372–1383. doi:10.1109/tip.2005.852787
Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V., Longair, M., Pietzsch, T., et al. (2012a). Fiji: An Open-Source Platform for Biological-Image Analysis. Nat. Methods 9, 676–682. doi:10.1038/nmeth.2019
Schneider, C. A., Rasband, W. S., and Eliceiri, K. W. (2012b). NIH Image to ImageJ: 25 Years of Image Analysis. Nat. Methods 9, 671–675. doi:10.1038/nmeth.2089
Soykan, T., Maritzen, T., and Haucke, V. (2016). Modes and Mechanisms of Synaptic Vesicle Recycling. Curr. Opin. Neurobiol. 39, 17–23. Elsevier Ltd. doi:10.1016/j.conb.2016.03.005
Soykan, T., Kaempf, N., Sakaba, T., Vollweiter, D., Goerdeler, F., Puchkov, D., et al. (2017). Synaptic Vesicle Endocytosis Occurs on Multiple Timescales and Is Mediated by Formin-Dependent Actin Assembly. Neuron 93, 854–866. Elsevier Inc.e854. doi:10.1016/j.neuron.2017.02.011
Südhof, T. C. (2013). Neurotransmitter Release: The Last Millisecond in the Life of a Synaptic Vesicle. Neuron 80 (3), 675–690. doi:10.1016/j.neuron.2013.10.022
Wienisch, M., and Klingauf, J. (2006). Vesicular Proteins Exocytosed and Subsequently Retrieved by Compensatory Endocytosis Are Nonidentical. Nat. Neurosci. 9 (8), 1019–1027. doi:10.1038/nn1739
Yue, H.-Y., and Xu, J. (2014). Myosin Light Chain Kinase Accelerates Vesicle Endocytosis at the Calyx of Held Synapse. J. Neurosci. 34 (1), 295–304. doi:10.1523/jneurosci.3744-13.2014
Keywords: pHluorin imaging, calcium imaging, synapse, synaptic activity, ImageJ, Fiji, bioimage analysis, computer vision
Citation: Schmied C, Soykan T, Bolz S, Haucke V and Lehmann M (2021) SynActJ: Easy-to-Use Automated Analysis of Synaptic Activity. Front. Comput. Sci. 3:777837. doi: 10.3389/fcomp.2021.777837
Received: 15 September 2021; Accepted: 19 November 2021;
Published: 09 December 2021.
Edited by:
Florian Levet, Institut Interdisciplinaire de Neurosciences (IINS), FranceReviewed by:
Natali Lujan Chanaday Ricagni, Vanderbilt University, United StatesCopyright © 2021 Schmied, Soykan, Bolz, Haucke and Lehmann. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Christopher Schmied, Y2hyaXN0b3BoZXIuc2NobWllZEBmaHQub3Jn; Volker Haucke, SGF1Y2tlQGZtcC1iZXJsaW4uZGU=; Martin Lehmann, TUxlaG1hbm5AZm1wLWJlcmxpbi5kZQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.