
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Comput. Sci., 22 September 2021
Sec. Digital Public Health
Volume 3 - 2021 | https://doi.org/10.3389/fcomp.2021.672867
This article is part of the Research TopicExtracting Insights from Digital Public Health Data using Artificial IntelligenceView all 15 articles
 A. Ziletti1*
A. Ziletti1* C. Berns2
C. Berns2 O. Treichel3
O. Treichel3 T. Weber3
T. Weber3 J. Liang4
J. Liang4 S. Kammerath5
S. Kammerath5 M. Schwaerzler1
M. Schwaerzler1 J. Virayah6
J. Virayah6 D. Ruau1
D. Ruau1 X. Ma7
X. Ma7 A. Mattern4
A. Mattern4Millions of unsolicited medical inquiries are received by pharmaceutical companies every year. It has been hypothesized that these inquiries represent a treasure trove of information, potentially giving insight into matters regarding medicinal products and the associated medical treatments. However, due to the large volume and specialized nature of the inquiries, it is difficult to perform timely, recurrent, and comprehensive analyses. Here, we combine biomedical word embeddings, non-linear dimensionality reduction, and hierarchical clustering to automatically discover key topics in real-world medical inquiries from customers. This approach does not require ontologies nor annotations. The discovered topics are meaningful and medically relevant, as judged by medical information specialists, thus demonstrating that unsolicited medical inquiries are a source of valuable customer insights. Our work paves the way for the machine-learning-driven analysis of medical inquiries in the pharmaceutical industry, which ultimately aims at improving patient care.
Every day pharmaceutical companies receive numerous medical inquiries related to their drugs from patients, healthcare professionals, research institutes, or public authorities from a variety of sources (e.g., websites, e-mail, phone, social media channels, company personnel, telefax). These medical inquiries may relate to drug-drug-interactions, availability of drugs, side effects of pharmaceuticals, clinical trial information, product quality issues, comparison with competitor products, storage conditions, dosing regimen, and the like. On the one hand, a single medical inquiry is simply a question of a given person searching for a specific information related to a medicinal product. On the other hand, a plurality of medical inquiries from different persons may provide useful insight into matters related to medicinal products and associated medical treatments. Examples of these insights could be early detection of product quality or supply chain issues, anticipation of treatment trends and market events, improvement of educational material and standard answers/frequently asked question coverage, potential changes in treatment pattern, or even suggestions on new possible indications to investigate. From a strategic perspective, this information could enable organizations to make better decisions, drive organization results, and more broadly create benefits for the healthcare community.
However, obtaining high-level general insights is a complicated task since pharmaceutical companies receive copious amounts of medical inquiries every year. Machine learning and natural language processing represent a promising route to automatically extract insights from these large amounts of unstructured (and noisy) medical text. Natural language processing and text mining techniques have been widely used in the medical domain (Allahyari et al., 2017; Luque et al., 2019), with emphasis on electronic health records (Sun et al., 2017; Landi et al., 2020; Mascio et al., 2020; Kormilitzin et al., 2021). In particular, deep learning has been successfully applied to medical text, with the overwhelming majority of works in supervised learning, or representation learning (in a supervised or self-supervised setting) to learn specialized word vector representations (i.e. word embeddings) (Alsentzer et al., 2019; Beltagy et al., 2019; Neumann et al., 2019; Weng and Szolovits, 2019; Wu et al., 2019). Conversely, the literature on unsupervised learning for medical text is scarce despite the bulk of real-world medical text being unstructured, without any labels or annotations. Unsupervised learning from unstructured medical text is mainly limited to the development of topic models based on latent Dirichlet allocation (LDA) (Blei et al., 2003). Examples of applications in the medical domain are clinical event identification in brain cancer patients from clinical reports (Arnold and Speier, 2012), modeling diseases (Pivovarov et al., 2015) and predicting clinical order patterns (Chen et al., 2017) from electronic health records, or detecting cases of noncompliance to drug treatment from patient forums (Abdellaoui et al., 2018). Only recently, word embeddings and unsupervised learning techniques have been combined to analyze unstructured medical text to study the concept of diseases (Shah and Luo, 2017), medical product reviews (Karim et al., 2020), or to extract informative sentences for text summarization (Moradi and Samwald, 2019).
In this work, we combine biomedical word embeddings and unsupervised learning to discover topics from real-world medical inquiries received by Bayer™. A real-world corpus of medical inquiries presents numerous challenges. From an inquirer (e.g. healthcare professional or patient) perspective, often the goal is to convey the information requested in as few words as possible to save time. This leads to an extensive use of acronyms and abbreviations, sentences with atypical syntactic structure, occasionally missing verb or subject, or inquiries comprising exclusively a single noun phrase. Moreover, since medical inquiries come from different sources, it is common to find additional (not relevant) information related to the text source; examples are references to internal computer systems, form frames (i.e. textual instructions) alongside with the actual form content, lot numbers, email headers and signatures, city names. The corpus contains a mixture of layman and medical language depending (mostly) on the inquirer being either a patient or a healthcare professional. Style and content of medical inquiries vary quite substantially according to which therapeutic areas (e.g. cardiovascular vs oncology) a given medicinal drug belongs to.
As already mentioned, medical inquiries are short. More specifically, they comprise less than fifteen words in most cases. Standard techniques for topic modelling based on LDA (Blei et al., 2003) do not apply, since the main assumption - each document/text is a distribution over topics - clearly does not hold given that the text is short (Qiang et al., 2019). Approaches based on pseudo-documents (Mehrotra et al., 2013) or using auxiliary information (Phan et al., 2008; Jin et al., 2011) are also not suitable since no meaningful pseudo-document nor auxiliary information are available for medical inquiries. Moreover, these models aim to learn semantics (e.g. meaning of words) directly from the corpus of interest. However, the recent success of pretrained embeddings (Peters et al., 2018; Devlin et al., 2019) shows that it is beneficial to include semantics learned on a general (and thus orders of magnitude larger) corpus, thus providing semantic information difficult to obtain from smaller corpora. This is particularly important for limited data and short text settings. To this end, there has been recently some work aimed at incorporating word embeddings into probabilistic models similar to LDA (Dirichlet multinomial mixture model (Yin and Wang, 2014)) and that - contrary to LDA - satisfies the single topic assumption (i.e. one document/text belong to only one topic) (Nguyen et al., 2015; Li et al., 2016). Even though these models include (some) semantic information in the topic model, it is not evident how to choose the required hyperparameters, for example determining an appropriate threshold when filtering semantically related word pairs (Li et al., 2016). Concurrently to our work, document-level embeddings and hierarchical clustering have been combined to obtain topic vectors from news articles and a question-answer corpus (Angelov, 2020).
Here, we propose an approach - schematically depicted in Figure 1 - to discover topics from short, unstructured, real-world medical inquiries. Our methodology consists of the following steps: medical inquiries are preprocessed (via lemmatization, stopword removal) and converted to vectors via a biomedical word embedding (scispacy (Neumann et al., 2019)), a dimensionality reduction is then applied to lower the dimensionality of the embedded vectors (via UMAP (McInnes et al., 2018a; McInnes et al., 2018b)), clustering is performed in this lower dimensional space to group together similar inquiries (via HDBSCAN (Campello et al., 2013; Melo et al., 2016; McInnes et al., 2017)). These clusters of similar inquiries are then merged based on semantic similarity: we define these (merged) clusters as topics. Topics are then quantitatively evaluated via two novel quantities: topic semantic compactness and name saliency, introduced in this work. Finally, for visualization purposes, another dimensionality reduction is applied to visualize topics in a topic map. This methodology is used to discovery topics in medical inquiries received by Bayer™ Medical Information regarding the oncology drug regorafenib.
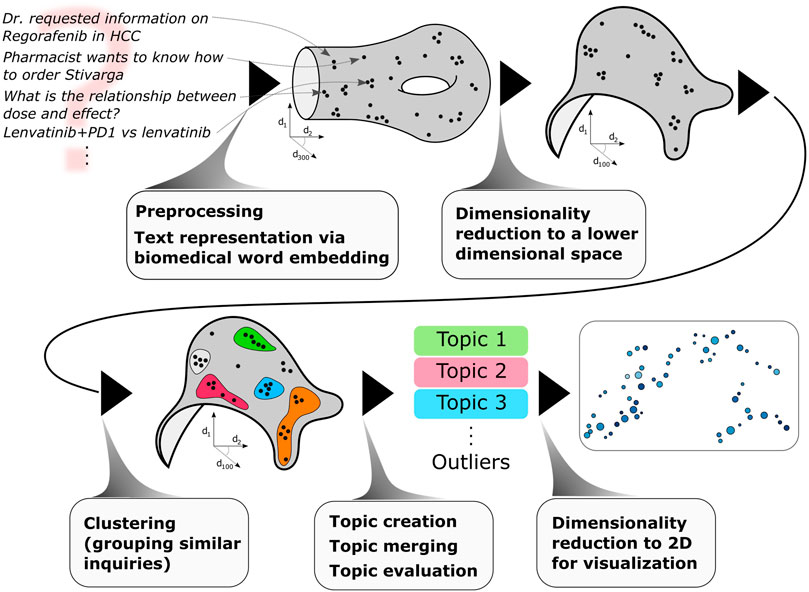
FIGURE 1. From medical inquiries to medical topics via natural language processing and machine learning.
One of the main challenges of topic discovery in short text is sparseness: it is not possible to extract semantic information from word co-occurrences because words rarely appear together since the text is short. In our case, the sparseness problem is exacerbated by two following aspects. First, the amount of data available is limited: most medicinal products receive less than 4,000 medical inquiries yearly. Second, medical inquiries are sent by patients as well as healthcare professionals (e.g. physicians, pharmacists, nurses): this leads to inquiries with widely different writing styles, containing a mixture of common and specialized medical text. The sparsity problem can be tackled by leveraging word embedding models trained on large corpora; these embeddings have been shown to learn semantic similarities directly from data, even for specialized biomedical text (Alsentzer et al., 2019; Beltagy et al., 2019; Lee et al., 2019; Neumann et al., 2019). Specifically, we use the scispaCy word embedding model (Neumann et al., 2019), which was trained on a large corpus containing scientific abstracts from medical literature (PubMed) as well as web pages (OntoNotes 5.0 corpus (Pradhan et al., 2013)). This assorted training corpus enables the model to treat specialized medical terminology and layman terms on the same footing, so that medical topics are discovered regardless of the writing style.
One of the main disadvantages of word vector (word2vec) models - like the (scispaCy) model used in this work - is their inability to handle out-of-vocabulary (oov) words: if a word appearing in the text is not included in the model vocabulary, it is effectively skipped from the analysis (i.e. a vector of all zeros is assigned to it). To tackle this issue, several models have been proposed, initially based on chargram level embeddings (FastText (Bojanowski et al., 2017)), and more recently contextual embeddings based on character (ELMO (Peters et al., 2018)), or byte pair encoding (Sennrich et al., 2016) representations (BERT (Devlin et al., 2019)). Even though other advancements - namely word polysemy handling and the use of attention (Vaswani et al., 2017) - were arguably the decisive factors, improvements in oov word handling also contributed in making ELMO and BERT the de facto gold standard for natural language processing, at least for supervised learning tasks.
Even though the use of contextual word embeddings is generally beneficial and can be readily incorporated in our approach (simply substituting the word representation), we notice that - given the large amount of noise present and the purely unsupervised setting - a word2vec model is actually advantageous for the task of extracting medical topics from real-word medical inquiries. Indeed, using a model with a limited yet comprehensive vocabulary (the scispaCy model used in this work includes 600 k word vectors) constitutes a principled, data-driven, efficient, and effective way to filter relevant information from the noise present in the corpus. This filtering is principled, and data driven because the words (and vectors) included in the model vocabulary are automatically determined in the scispaCy training procedure by optimizing the performance on biomedical text benchmarks (Neumann et al., 2019). This also leads to harmonization of the medical inquiry corpus by eliminating both non-relevant region-specific terms, and noise introduced by machine translation (words or expressions are sometimes not translated but simply copied still in the original language (Knowles et al., 2018)). Clearly, in this context it is of paramount importance to use specialized biomedical embeddings so that the word2vec model has a comprehensive knowledge of medical terms despite its relatively limited vocabulary.
Table 1 presents a qualitative comparison of a standard embedding (en core web lg, trained on the Common Crawl) and a specialized biomedical embedding (scispaCy en core sci lg, trained also on PubMed). Specifically, for a given probe word (i.e. leukemia, carcinoma, blood), the words most semantically similar to it - measured by the cosine similarity between word vectors - are retrieved, together with their similarity with the probe word (shown in parenthesis, 1.0 being the highest possible similarity). It is evident that the biomedical embedding returns much more relevant and medically specific terms. For instance, given the probe word leukemia, the standard embedding returns generic terms like cancer, tumor, chemotherapy which are broadly related to oncology, but not necessarily to leukemia. In contrast, the biomedical embedding returns more specialized (and medically relevant) terms like lymphoblastic, myelomonocytic, myelogenous, myeloid, promyelocytic: acute lymphoblastic, chronic myelomonocytic, chronic myelogenous, adult acute myeloid, and acute promyelocytic are all types of leukemia.
We have shown in the previous section that word embeddings provide a natural way to include semantic information (i.e. meaning of individual words) in the modeling. Medical inquiries comprise multiple words, and therefore a semantic representation for each inquiry needs to be computed from the word-level embeddings. We accomplish this by simply averaging the embeddings of the words belonging to the inquiry, thus obtaining one vector for each inquiry. Since these vectors capture semantic information, medical inquiries bearing similar meaning are mapped to nearby vectors in the high-dimensional embedding space. To group similar inquiries, clustering is performed in this embedding space, and for each medicinal product separately.
Before clustering is performed, a non-linear dimensionality reduction is applied to lower the dimensionality of the text representation, similar to Ref. 29. We utilize the UMAP algorithm (McInnes et al., 2018a; McInnes et al., 2018b) because of its firm mathematical foundations from manifold learning and fuzzy topology, ability to meaningfully project to any number of dimensions (not only two or three like t-SNE (van der Maaten and Hinton, 2008)), and computational efficiency. Reducing the dimensionality also considerably improves the clustering computational performance, greatly easing model deployment to production, especially for drugs with more than 5,000 inquiries.
Usually, it is not conducive to define an appropriate number of clusters a priori. A reasonable number of clusters depends on various interdependent factors: number of incoming inquiries, therapeutic area of the medicine, time frame of the analysis, and intrinsic amount of information (i.e. variety of the medical inquiries). For a given medicinal product, typically a handful of frequently asked questions covers a large volume of inquiries, accompanied by numerous low-volume and less cohesive inquiry clusters. These low-volume clusters often contain valuable information, which might not even be known to medical experts: their low volume makes it difficult to detect them via manual inspection. To perform clustering in the embedding space, we use the hierarchical, density-based clustering algorithm HDBSCAN (Campello et al., 2013; Melo et al., 2016; McInnes et al., 2017). As customary in unsupervised learning tasks, one needs to provide some information on the desired granularity, i.e. how fine or coarse the clustering should be. In HDSBCAN, this is accomplished by specifying a single, intuitive hyper-parameter (min cluster size). In our case, the objective is to obtain approximately 100 clusters so that the results can be easily analyzed by medical experts. Thus, the main factor in defining min cluster size is the number of inquiries for a given medicinal drug: the larger the medical inquiry volume, the larger the parameter min cluster size. Note that min cluster size is not a strict controller of cluster size (and thus how many clusters should be formed), but rather a guidance provided to the algorithm regarding the desired clustering granularity. It is also possible to combine different min cluster size for the same dataset, i.e. using a finer granularity for more recent inquiries, thus enabling the discovery of new topics when only few inquiries are received, at a price however of an increase in noise given the low data volume. Moreover, min cluster size is very slowly varying with data (medical inquiry) volume, which facilitate its determination (see Supplementary Material). At the end of this step, for each drug a set of clusters is returned, each containing a collection of medical inquiries. A given medical inquiry is associated to one topic only, in accordance with the single topic assumption.
In order to convey the cluster content to users, a name (or headline) needs to be determined for each cluster. To this end, the top-five most recurring words for each cluster are concatenated, provided that they appear in at least 20% of the inquiries belonging to that cluster; this frequency threshold is set to avoid to include in the topic name words that appear very infrequently but are still in the top-five words. Thus, if a word does not fulfill the frequency requirement, it is not included in the topic name (resulting in topic names with less than five words). By such naming (topic creation), the clusters are represented by a set of words, which summarize their semantic content.
From this list of candidate topics, the vector representation for each word in the topic name is calculated; the topic name vector is then obtained by averaging the word vectors of the words present in the topic name. Topics are merged if their similarity evaluated as cosine similarity between their topic name vectors - is larger than a threshold. Threshold values range between 0.8 and 0.95 depending on the drug considered. This is done to limit the number of topics to be presented to medical experts. We favor this simple method over applying again HDSBCAN because the clustering would have to operate on very few datapoints (∼100 topics). We also notice that HDBSCAN tends to group topics quite aggressively (even with min cluster size = 2), which would result in potentially loosing important information.
After the topics are merged, new topic names are generated according to the procedure outlined above. The final result is a list of topics defined by a given name, each containing a set of similar medical inquiries. The list of discovered topics is then outputted and presented to medical experts.
Since the goal is to extract as much knowledge as possible from incoming medical inquiries, a relatively large number of topics (typically around 100) is returned to medical experts for each medicinal product. To facilitate topic exploration and analysis, topics are visualized on a map that reflects the similarity between topics (Figure 2A): topics close to each other in this map are semantically similar. To obtain this semantic map, first topic vectors are computed by averaging the text representation of all inquiries belonging to a given topic; then, a dimensionality reduction to two dimensions via UMAP is performed.
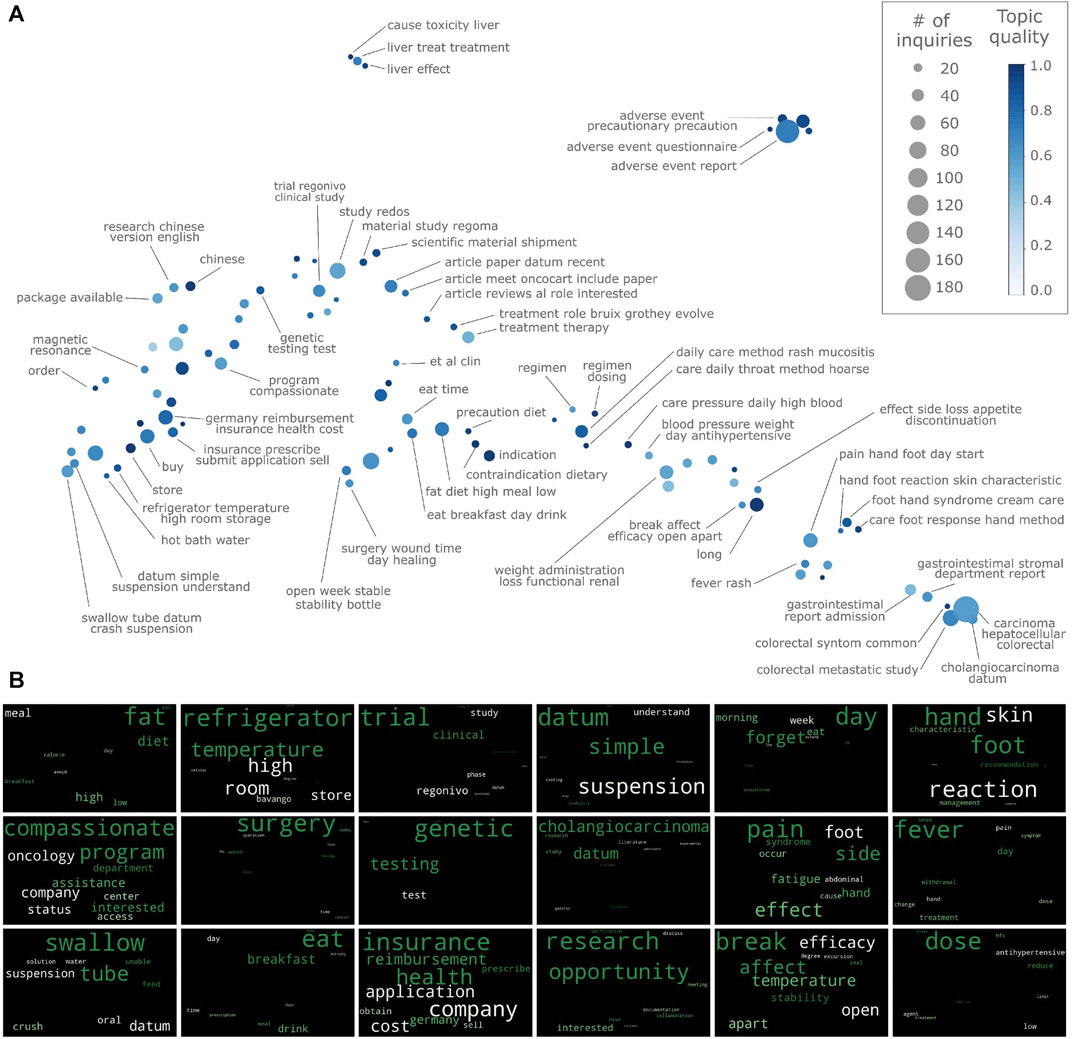
FIGURE 2. Topic discovery for medical inquiries on regorafenib. (A) Topic semantic map. Axis are the non-linear dimensionality components and thus are not shown. (B) Wordclouds for selected topics.
Once topics are discovered, it is desirable to provide medical experts with information regarding the quality of a given topic.
The most popular topic evaluation metrics for topic modelling on long text are UCI (Newman et al., 2010) and UMass (Mimno et al., 2011). However, both UCI and UMass metrics are not good indicators for quality of topics in short text topic modelling due to the sparseness problem (Quan et al., 2015). In Ref. 44, a purity measure is introduced to evaluate short text topic modelling; however, it requires pairs of short and long documents (e.g. abstract and corresponding full text article), and thus it is not applicable here because there is no long document associated to a given medical inquiry. Indeed, evaluation of short text topic modelling is an open research problem (Qiang et al., 2019). An additional challenge is the absence of labels. Performing annotations would require substantial manual effort by specialized medical professionals and would be of limited use because one of the main goals is to discover previously unknown topics as new inquiries are received. The absence of labels precludes the use of the metrics based on purity and normalized mutual information proposed in Ref. Rosenberg and Hirschberg (2007), Huang et al. (2013), Yin and Wang (2014), Aletras et al. (2013). bring forward the valuable idea of using distributional semantic to evaluate topic coherence, exploiting the semantic similarity learned by word2vec models. Topic coherence is assessed by calculating the similarity among the top n-words of a given topic: semantically similar top n-words lead to higher topic coherence. If this might be in general desirable, in the case of discovering medical topics it is actually detrimental: interesting (and potentially previously unknown) topics are often characterized by top n-words which are not semantically similar. For example, a medical topic having as top 2-words rivaroxaban (an anticoagulant medication) and gluten is clearly relevant from a medical topic discovery standpoint. However, rivaroxaban and gluten are not semantically similar, and thus the metric proposed in Ref. 47 would consider this as a low coherence (and thus low quality) topic, in stark contrast with human expert judgment. Analogous considerations apply to the indirect confirmation measures in Roeder et al. (2015): words emerging in novel topics would have rarely appeared before in a shared context. For this reason, we introduce a new measure of topic compactness which takes into account the semantics of the inquiries, and does not require any labeled data. Specifically, we compute the similarity of all inquiries belonging to a given topic with each other (excluding self-similarity), sum the elements of the resulting similarity matrix, and divide by the total number of elements in this matrix. The topic semantic compactness γα of topic α reads
where |Cα| is the cardinality of topic α (how many inquiries are in topic α), qi (and qj) is the word vector representing inquiry i (j), and S is a function quantifying the semantic similarity between inquiry qi and qj, taking values between 0 and 1 (S = 1 when qi and qj are identical, and S = 0 being the lowest possible similarity). Given the chosen normalization factor (i.e. the denominator in Eq. 1), 0 ≤ γα ≤ 1 and thus γα can be directly used as (a proxy for) topic quality score. The topic compactness maximum (γα = 1) is attained if and only if every sentence (after preprocessing) contains exactly the same words. It is important to point out that γα automatically takes semantics into account: different but semantically similar medical inquiries would still have high similarity score, and thus would lead (as desired) to a high topic semantic compactness, despite these inquiries using different words to express similar content. Contrary to Ref. 47, the topic semantic compactness γα introduced in Eq. 1 does not artificially penalize novel topics just because they associate semantically different words appearing in the same inquiry. To come back to the previous example, if numerous inquiries in a discovered topic contain the words rivaroxaban and gluten, the topic semantic compactness would be high (as desired), regardless from the fact that the top 2-words are not semantically similar since the similarity is evaluated at the inquiry level (by S (qi,qj) in Eq. 1).
The topic name is one of the main information shown to the users to summarize the semantic content of a discovered medical topic. It is therefore of interest to quantify how representative the name is for a given medical topic. This is tackled by answering the following question: how similar is the name with the inquiries grouped in the topic it represents? To this end, we calculate the name saliency τα for medical topic α by calculating the similarity of the word vector representing the topic name with the word vectors representing the inquiries in the topic, sum these similarity values, and divide by the total number of inquiries in the topic. This reads
where |Cα| is the cardinality of topic α (how many inquiries are in topic α), tα is the word vector representing the name of topic α, and qi is the vector representing inquiry i. This returns a score (0 ≤ τα ≤ 1) which quantifies how representative (salient) the name is for the topic it represents. As in the case of the topic semantic compactness, the name saliency τα takes natively semantics (e.g. synonyms) into account via S (tα, qi) in Eq. 2. In both Eqs. 1,2, the cosine similarity is used as similarity measure.
As a real-world example of topic discovery, we present the results for medical inquiries on the oncology drug regorafenib (Bekaii-Saab et al., 2019). Regorafenib is an oral multikinase inhibitor which inhibits various signal pathways responsible for tumor growth.
In this work, all unsolicited medical inquiries received by Bayer™ worldwide in the time frame July 2019-June 2020 are considered (3,880 medical inquiries, see Figure 3). All non-English inquiries are translated to English using machine translation. These inquiries are then pre-processed: acronyms and abbreviations are resolved; non-informative phrases, words or patterns are removed; text is tokenized and lemmatized. Additional details are provided in Supplementary Material. Then, the topic discovery algorithm introduced above is applied with min cluster size = 6 and the UMAP dimensionality reduction to 100 components.
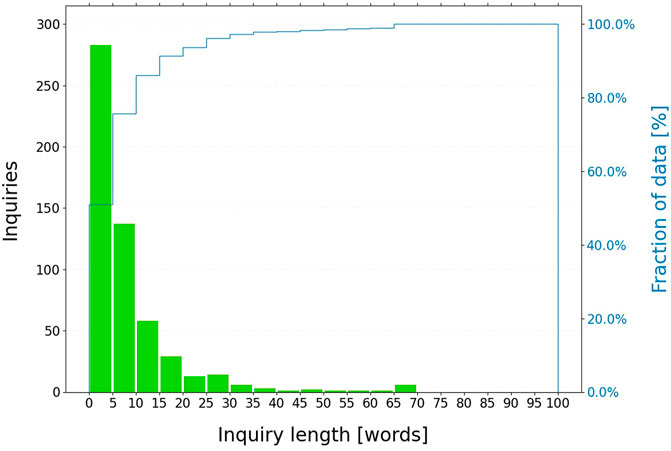
FIGURE 3. Length distribution of medical inquiries on regorafenib. The dataset contains 3,880 medical inquiries; most of the inquiries (∼90%) contains less than fifteen words after preprocessing.
The semantic map with the discovered topics is shown in Figure 2A. These topics span a relatively large variety of themes, ranging from interactions with food and adverse drug reactions to purchase costs and literature requests. The topics are judged as meaningful and medically relevant by medical information specialists, based on their expert knowledge of the medicinal product.
Topics are also specific: the unsupervised learning approach allows information to emerge directly from the data, without recurring to predefined lists of keywords or classes, as required when using ontologies or supervised learning. An example of a very specialized topic for inquiries on scientific literature is treatment role bruix grothey evolve: 12 requests related to the review article on the treatment of advanced cancer with regorafenib published on February 2020 (Grothey et al., 2020). Other examples are the five topics fat diet high meal low, eat breakfast day drink, precaution diet, eat time, contraindication diet. Even though all these topics relate to nutrition, they are addressing different aspects. It is quite advantageous that they are identified as distinct since medical recommendations will likely differ across these five topics.
Thanks to the inclusion of semantics via word embedding, the algorithm is able to group together inquiries having similar meaning, even though the actual words in them are distinct. For instance, the topic pain side effect foot fatigue comprises 21 inquiries on medical issues (which may or may not be related with the medicine), in which the following words appear: pain (seven times), side effect (six times) nausea (three times), fatigue (five times), dysphonia (two times). The algorithm is able to cluster these inquiries together because similar inquiries are mapped close to each other in the high dimensional semantic space where clustering is performed. This is corroborated by the relatively high similarity score between the terms appearing in these inquiries (pain-nausea: 0.66, nausea-fatigue:0.61, pain-fatigue:0.71, dysphonia-pain:0.55, dysphonia-fatigue:0.49), scores much higher than zero, zero being the score expected for unrelated terms(cf. pain-day:0.05, nausea-sun:0.08). Conversely, if there is a moderate number of inquiries on a specific medical matter, the algorithm is generally able to detect that signal, as in the case of mucositis and hoarse in the two topics daily care method rash mucositis, and daily care method rash hoarse.
As shown in Figure 2A, the automatically generated topic names provide a reasonably good insight into the semantic content of their respective topics. However, one needs to be mindful that the topic might - and usually will - contain additional information of relevance. To convey this information in a simple yet effective way to the users, wordclouds are generated for each topic; examples of wordcloud are shown in Figure 2B. For example, in the wordcloud of topic compassionate program (Figure 2B, 1st column-2nd row), concepts not included in the topic name (e.g. assistance, interested, access, status) appear, thus giving further insight into the topic content. In some cases, even the wordcloud might not convey the topic meaning: users will then resort to manually inspect the inquiries belonging to the topic. For instance, the content of topic chinese is not clear, neither from the topic name nor from the wordcloud; however, inspection of the actual inquiries quickly reveals that they refer to the interaction between Chinese medicine and regorafenib (the word medicine does not appear since it is a stopword). Other examples are al et clin and long, which group together requests for scientific articles and product durability, respectively. Topic quality provides a useful guidance when exploring topics. If topic quality is close to one, medical inquiries in that topic are all very similar, and the topic name is expected to summarize the topic content well. Conversely, topics with low quality will contain inquiries that might differ quite substantially yet are similar enough to be clustered together by the algorithm. In these cases, manual inspection of the underlying medical inquiries may be a good strategy. From Figure 2A, it appears that smaller topics tend to have higher topic scores, although no clear trend emerges.
Finally, in addition of having similar inquiries within topics, the model captures semantic similarities between topics. This is apparent from Figure 2A: similar topics tend to be close to each other in the semantic map. Even though this feature does not influence the topic discovered, from a user perspective it provides a clear advantage when exploring topics (e.g. compared to reading them from as a simple list).
This study introduces an unsupervised machine learning approach to automatically discover topics from medical inquiries. After the initial (one-time) effort for preprocessing (e.g. abbreviation definition, stopword refinement) and hyper-parameters determination, the algorithm runs without requiring any human intervention, discovering key topics as medical inquiries are received. Topics can be discovered even if only a small number of inquiries is present, and are generally specific, thus enabling targeted, informed decisions by medical experts. Being completely unsupervised, the algorithm can discover topics that were neither known nor expected in advance, topics which often are the most valuable. This is in stark contrast with ontology or supervised based approaches, where topics need to be defined a priori (as collections of keywords or classes), and incoming text can be associated only to these predefined lists of topics, thus hindering the discovery of a priori unknown topics. The machine learning approach introduced here does not use ontologies (which are costly and hard to build, validate, maintain, and difficult to apply when layman and specialized medical terms are combined), and instead it incorporates domain knowledge via specialized biomedical word embeddings. This allows to readily apply the topic discovery algorithm to different drugs, without the burden of having to develop specialized ontologies for each product or therapeutic area. Indeed, the algorithm is periodically analyzing hundreds of thousands of medical inquiries for sixteen Bayer™ medicinal products, encompassing cardiology, oncology, gynecology, hematology, and ophthalmology.
Our approach has several limitations. First, it can happen that a small fraction of inquiries associated to a given topic are actually extraneous to it, especially for semantically broad topics. This is because - due to the noise present in this real-world dataset - the soft clustering HDBSCAN algorithm must be applied with a low probability threshold for cluster assignment to avoid the majority of inquiries being considered as outliers (see Supplementary Material). Second, even though the topic names are generally quite informative, a medical expert needs to read the actual inquiries to fully grasp the topic meaning, especially if a decision will be made on the grounds of the discovered topics. This is however not burdensome because inspection is limited to the inquiries associated to a given topic (and not all inquiries). Last, some discovered topics are judged by medical experts based on their expert knowledge - so similar that they could have been merged in a single topic, but are considered distinct by the algorithm. In these cases, manual topic grouping might be required to determine the top topics by inquiry volumes. Still, these similar topics very often appear close to each other in the topic map.
Despite these limitations, this study demonstrates that medical inquiries contain useful information, and that machine learning can extract this information in an automatic way, discovering topics that are judged by medical information specialists as meaningful and valuable. The hope is that this will stimulate mining of medical inquiries, and more generally the use of natural language processing and unsupervised learning in the medical industry. Interesting future directions are the inclusion of a priori expert knowledge (e.g. a list of expected topics) while at the same time maintaining the ability to discover new and previously unknown topics, and grouping topics in meta-topics though a clustering hierarchy.
The datasets presented in this article are not readily available because the data is proprietary to Bayer AG. Requests to access the datasets should be directed to angelo.ziletti@bayer.com.
AZ. led and thereby ideated and implemented the topic discovery algorithm, and is the main author the manuscript. MS, CB, DR. provided valuable suggestions on the topic discovery algorithm. CB, OT, and TW. designed and implemented the software architecture and data engineering pipeline for the algorithm deployment. TW, JV, JL, SK, XM, AM, DR, and MS. provided the in-house resources for the study, supervised the overall project, and provided domain knowledge expertise. All authors revised and commented on the manuscript.
Funding for the study was provided by Bayer AG.
Financial support for the research was provided by Bayer AG. AZ, OT, TW, JL, SK, MS, JV, DR, and AM work for Bayer AG. CB works for Areto Consulting Gmbh. The authors report a patent application on Topic Modelling of Short Medical Inquiries submitted on April 21st, 2020 (application number EP20170513.4).
The authors declare that this study received funding from Bayer AG. The funder was involved in the study design, model development, data analysis, and the writing of this article.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
AZ. thanks Robin Williams and Nikki Hayward from Bayer™ Medical Information for providing expert insightful and in-depth feedback on the results of topic discovery.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcomp.2021.672867/full#supplementary-material
Abdellaoui, R., Foulquié, P., Texier, N., Faviez, C., Burgun, A., and Schück, S. (2018). Detection of Cases of Noncompliance to Drug Treatment in Patient Forum Posts: Topic Model Approach. J. Med. Internet Res. 20, e85. doi:10.2196/jmir.9222
Aletras, N., and Stevenson, M. (2013). Evaluating Topic Coherence Using Distributional Semantics.,” in IWCS. Editors K. Erk, and A. Koller. (The Association for Computer Linguistics), 13–22.
Allahyari, M., Pouriyeh, S. A., Assefi, M., Safaei, S., Trippe, E. D., Gutierrez, J. B., et al. (2017). A Brief Survey of Text Mining: Classification, Clustering and Extraction Techniques. arXiv [Epub ahead of print]. CoRR.
Alsentzer, E., Murphy, J., Boag, W., Weng, W.-H., Jindi, D., Naumann, T., and McDermott, M. (2019). “Publicly Available Clinical BERT Embeddings,” in Proceedings of the 2nd Clinical Natural Language Processing Workshop, Minneapolis, Minnesota, USA: Association for Computational Linguistics, 72–78.
Arnold, C., and Speier, W. (2012). “A Topic Model of Clinical Reports,” in Proceedings of the 35th International ACM SIGIR Conference on Research and Development in Information Retrieval, New York, NY, USA: SIGIR ’12Association for Computing Machinery), 1031–1032. doi:10.1145/2348283.2348454
Bekaii-Saab, T., Ou, F., Ahn, D., Boland, P., Ciombor, K., Heying, E., et al. (2019). Regorafenib Dose-Optimisation in Patients with Refractory Metastatic Colorectal Cancer (Redos): a Randomised, Multicentre, Open-Label, Phase 2 Study. Lancet Oncol. 20, 1070–1082. doi:10.1016/S1470-2045(19)30272-4
Beltagy, I., Lo, K., and Cohan, A. (2019). Scibert: A Pretrained Language Model for Scientific Text,” in EMNLP/IJCNLP (1). Editors K. Inui, J. Jiang, V. Ng, and X. Wan. (Hong Kong, China: Association for Computational Linguistics), 3613–3618.
Blei, D. M., Ng, A. Y., and Jordan, M. I. (2003). Latent Dirichlet Allocation. J. Machine Learn. Res. 3, 993–1022.
Bojanowski, P., Grave, E., Joulin, A., and Mikolov, T. (2017). Enriching Word Vectors with Subword Information. Tacl 5, 135–146. doi:10.1162/tacl_a_00051
Campello, R. J. G. B., Moulavi, D., and Sander, J. (2013). Density-based Clustering Based on Hierarchical Density Estimates. Lect. Notes In Comput. Sci. 7819, 160–172. doi:10.1007/978-3-642-37456-2_14
Chen, J. H., Goldstein, M. K., Asch, S. M., Mackey, L., and Altman, R. B. (2017). Predicting Inpatient Clinical Order Patterns with Probabilistic Topic Models vs Conventional Order Sets. JAMIA 24 (3), 472–480. doi:10.1093/jamia/ocw136
Devlin, J., Chang, M.-W., Lee, K., and Toutanova, K. (2019). {Bert}: Pre-training of Deep Bidirectional Transformers for Language Understanding. Proceedings of the 2019 Conference of the North {A}merican Chapter of the Association for Computational Linguistics: Human Language Technologies (Minneapolis, Minnesota: Association for Computational Linguistics), 4171–4186.
Grothey, A., Blay, J.-Y., Pavlakis, N., Yoshino, T., and Bruix, J. (2020). Evolving Role of Regorafenib for the Treatment of Advanced Cancers. Cancer Treat. Rev. 86, 101993. doi:10.1016/j.ctrv.2020.101993
Huang, R., Yu, G., Wang, Z., Zhang, J., and Shi, L. (2013). Dirichlet Process Mixture Model for Document Clustering with Feature Partition. IEEE Trans. Knowl. Data Eng. 25 (8), 1748–1759. doi:10.1109/tkde.2012.27
Jin, O., Liu, N. N., Zhao, K., Yu, Y., and Yang, Q. (2011). “Transferring Topical Knowledge from Auxiliary Long Texts for Short Text Clustering,” in Proceedings of the 20th ACM International Conference on Information and Knowledge Management, New York, NY, USA: CIKM ’11Association for Computing Machinery, 775–784. doi:10.1145/2063576.2063689
Karim, M. R., Beyan, O., Zappa, A., Costa, I. G., Rebholz-Schuhmann, D., Cochez, M., et al. (2020). Deep Learning-Based Clustering Approaches for Bioinformatics. Brief. Bioinform. 22, 393–415. doi:10.1093/bib/bbz170
Knowles, R., and Koehn, P. (2018). “Context and Copying in Neural Machine Translation.,” in EMNLP. Editors E. Riloff, D. Chiang, J. Hockenmaier, and J. Tsujii. (Brussels, Belgium: Association for Computational Linguistics) 3034–3041. doi:10.18653/v1/d18-1339
Kormilitzin, A., Vaci, N., Liu, Q., and Nevado-Holgado, A. (2021). Med7: a Transferable Clinical Natural Language Processing Model for Electronic Health Records. Artif. Intell. Med. 118, 102086. doi:10.1016/j.artmed.2021.102086
Landi, I., Glicksberg, B. S., Lee, H.-C., Cherng, S., Landi, G., Danieletto, M., et al. (2020). Deep Representation Learning of Electronic Health Records to Unlock Patient Stratification at Scale. NPJ Digit. Med. 3, 96. doi:10.1038/s41746-020-0301-z
Lee, J., Yoon, W., Kim, S., Kim, D., Kim, S., So, C. H., et al. (2019). BioBERT: a Pre-trained Biomedical Language Representation Model for Biomedical Text Mining. Bioinformatics 36, 1234–1240. doi:10.1093/bioinformatics/btz682
Li, H., Wang, H., Zhang, Z., Sun, A., and Ma, Z. (2016). “Topic Modeling for Short Texts with Auxiliary Word Embeddings,” in Proceedings of the 39th International ACM SIGIR Conference on Research and Development in Information Retrieval, New York, NY, USA: SIGIR ’16Association for Computing Machinery, 165–174. doi:10.1145/2911451.2911499
Luque, C., Luna, J. M., Luque, M., and Ventura, S. (2019). An Advanced Review on Text Mining in Medicine. WIREs Data Mining Knowl. Discov. 9 (3), e1302. doi:10.1002/widm.1302
Mascio, A, Kraljevic, Z, Bean, D, Dobson, R. J. B., Stewart, R., Bendayan, R., et al. (2020). “Comparative Analysis of Text Classification Approaches in Electronic Health Records,” in BioNLP. Editors D. Demner-Fushman, K. B. Cohen, S. Ananiadou, and J. Tsujii, (Association for Computational Linguistics), 86–94. doi:10.18653/v1/2020.bionlp-1.9
McInnes, L., Healy, J., and Astels, S. (2017). Hdbscan: Hierarchical Density Based Clustering. Joss 2 (11), 205. doi:10.21105/joss.00205
McInnes, L., Healy, J., and Melville, J. (2018). Umap: Uniform Manifold Approximation and Projection for Dimension Reduction. cite arxiv:1802.03426Comment: Reference implementation available at http://github.com/lmcinnes/umap (Accessed September 9, 2021).
McInnes, L., Healy, J., Saul, N., and Grossberger, L. (2018). Umap: Uniform Manifold Approximation and Projection. Joss 3 (29), 861. doi:10.21105/joss.00861
Mehrotra, R., Sanner, S., Buntine, W., and Xie, L. (2013). “Improving Lda Topic Models for Microblogs via Tweet Pooling and Automatic Labeling,” in Proceedings of the 36th International ACM SIGIR Conference on Research and Development in Information Retrieval, New York, NY, USA: SIGIR ’13Association for Computing Machinery, 889–892. doi:10.1145/2484028.2484166
Melo, D., Toledo, S., Mourão, F., Sachetto, R., Andrade, G., Ferreira, R., et al. (2016). Hierarchical Density-Based Clustering Based on GPU Accelerated Data Indexing Strategy, Proced. Comput. Sci. 80, 951–961. doi:10.1016/j.procs.2016.05.389
Mimno, D. M., Wallach, H. M., Talley, E. M., Leenders, M., and McCallum, A. (2011). “Optimizing Semantic Coherence in Topic Models,” in EMNLP (Edinburgh, United Kingdom: ACL), 262–272.
Moradi, M., and Samwald, M. (2019). Clustering of Deep Contextualized Representations for Summarization of Biomedical Texts. arXiv [Epub ahead of print]. abs/1908.02286.
Neumann, M., King, , Beltagy, L., and Ammar, W. (2019). “Scispacy: Fast and Robust Models for Biomedical Natural Language Processing,” in BioNLP@ACL. Editors (D. Demner-Fushman, K. B. Cohen, S. Ananiadou, and J. Tsujii. (Florence, Italy: Association for Computational Linguistics), 319–327. doi:10.18653/v1/w19-5034
Newman, D., Lau, J. H., Grieser, K., and Baldwin, T. (2010). “Automatic Evaluation of Topic Coherence,” “ in Human Language Technologies: The 2010 Annual Conference of the North American Chapter of the Association for Computational Linguistics, HLT ’10, Stroudsburg, PA, USA: Association for Computational Linguistics), 100–108.
Nguyen, D. Q., Billingsley, R., Du, L., and Johnson, M. (2015). Improving Topic Models with Latent Feature Word Representations. Tacl 3, 299–313. doi:10.1162/tacl_a_00140
Peters, , Neumann, M., Iyyer, M., Gardner, M., Clark, C., Lee, K., et al. (2018). “Deep Contextualized Word Representations,” in NAACL-HLT. Editors M. A. Walker, H. Ji, and A. Stent. (New Orleans, LA, United States: Association for Computational Linguistics), 2227–2237. doi:10.18653/v1/n18-1202
Phan, X.-H., Nguyen, L.-M., and Horiguchi, S. (2008). “Learning to Classify Short and Sparse Text and Web with Hidden Topics from Large-Scale Data Collections,” in Proceedings of the 17th International Conference on World Wide Web, New York, NY, USA: WWW ’08Association for Computing Machinery), 91–100.
Pivovarov, R., Perotte, A. J., Grave, E., Angiolillo, J., Wiggins, C. H., and Elhadad, N. (2015). Learning Probabilistic Phenotypes from Heterogeneous Ehr Data. J. Biomed. Inform. 58, 156–165. doi:10.1016/j.jbi.2015.10.001
Pradhan, S., Moschitti, A., Xue, N., Ng, H. T., Bjorkelund, A., Uryupina, O., Zhang, Y., and Zhong, Z. (2013). “Towards Robust Linguistic Analysis Using¨ OntoNotes,” in Proceedings of the Seventeenth Conference on Computational Natural Language Learning, Sofia, Bulgaria (Association for Computational Linguistics), 143–152.
Qiang, J., Qian, Z., Li, Y., Yuan, Y., and Wu, X. (2019). Short Text Topic Modeling Techniques, Applications, and Performance: A Survey. arXiv [Epub ahead of print]. abs/1904.07695.
Quan, X., Kit, C., Ge, Y., and Pan, S. J. (2015). Short and Sparse Text Topic Modeling via Self-Aggregation.,” in IJCAI. Editors Q. Yang, and M. J. Wooldridge, (Buenos Aires, Argentina: AAAI Press), 2270–2276.
Roeder, M., Both, A., and Hinneburg, A. (2015). Exploring the Space of Topic Coherence Measures.,” in WSDM. Editors X. Cheng, H. Li, E. Gabrilovich, and J. Tang. (New York, NY, United States: ACM) 399–408.
Rosenberg, A., and Hirschberg, J. (2007). “V-measure: A Conditional Entropy-Based External Cluster Evaluation Measure,” in Proceedings of the 2007 Joint Conference on Empirical Methods in Natural Language Processing and Computational Natural Language Learning (Prague, Czech Republic: EMNLP-CoNLL), 410–420.
Sennrich, R., Haddow, B., and Birch, A. (2016). “Neural Machine Translation of Rare Words with Subword Units,” in ACL (1) (Berlin, Germany: The Association for Computer Linguistics). doi:10.18653/v1/p16-1162
Shah, S., and Luo, X. (2017). Exploring Diseases Based Biomedical Document Clustering and Visualization Using Self-Organizing Maps. 2017 IEEE 19th International Conference on e-Health Networking, Applications and Services (Healthcom), 1–6. doi:10.1109/HealthCom.2017.8210791
Sun, W., Cai, Z., Liu, F., Fang, S., and Wang, G. (2017). “A Survey of Data Mining Technology on Electronic Medical Records,” in 2017 IEEE 19th International Conference on e-Health Networking, Applications and Services (Healthcom), 1–6.
van der Maaten, L., and Hinton, G. (2008). Visualizing Data Using T-SNE. J. Machine Learn. Res. 9, 2579–2605.
Vaswani, A., Shazeer, N., Parmar, N., Uszkoreit, J., Jones, L., Gomez, A. N., et al. (2017). Attention Is All You Need,” in Advances in Neural Information Processing Systems. Editors I. Guyon, U. V. Luxburg, S. Bengio, H. Wallach, R. Fergus, S. Vishwanathanet al. (Long Beach, CA, United States: Curran Associates, Inc.), 5998–6008.
Weng, W.-H., and Szolovits, P. (2019). Representation Learning for Electronic Health Records. arXiv [Epub ahead of print]. abs/1909.09248.
Wu, S., Roberts, K., Datta, S., Du, J., Ji, Z., Si, Y., et al. (2019). Deep Learning in Clinical Natural Language Processing: a Methodical Review. J. Am. Med. Inform. Assoc. 27, 457–470. doi:10.1093/jamia/ocz200
Keywords: natural language processing, machine learning, medical inquiries, clustering, medical information, topic discovery
Citation: Ziletti A, Berns C, Treichel O, Weber T, Liang J, Kammerath S, Schwaerzler M, Virayah J, Ruau D, Ma X and Mattern A (2021) Discovering Key Topics From Short, Real-World Medical Inquiries via Natural Language Processing. Front. Comput. Sci. 3:672867. doi: 10.3389/fcomp.2021.672867
Received: 26 February 2021; Accepted: 06 September 2021;
Published: 22 September 2021.
Edited by:
Steven Fernandes, Creighton University, United StatesReviewed by:
Angelo D'Ambrosio, Freiburg University Medical Center, GermanyCopyright © 2021 Ziletti, Berns, Treichel, Weber, Liang, Kammerath, Schwaerzler, Virayah, Ruau, Ma and Mattern. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: A. Ziletti, YW5nZWxvLnppbGV0dGlAYmF5ZXIuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.