- 1CSIRO Environment, Hobart, TAS, Australia
- 2National Collections and Marine Infrastructure, Hobart, TAS, Australia
Rapid development and deployment of marine carbon dioxide removal (mCDR) approaches will be required to prevent the worst consequences of climate change and meet national treaty obligations under the Paris agreement. However, approaches to monitor the efficacy and environmental safety of mCDR are not being developed with the same intensity as the technology. Verification will be required to convince a sceptical public and regulatory community of the overall benefit of mCDR as well as provide the regulatory community a basis for risk assessments that will be required for at scale deployments. In this perspective, we posit that genomics-based approaches can be used to assess the efficacy of carbon sequestration and monitor for the possibility of unintended consequences. By adopting these approaches, it will be feasible to develop the evidence portfolio necessary to underpin assessments of the risks, benefits and trade-offs involved in responsible deployment of mCDR.
1 Introduction
Global surface temperatures are projected to exceed 1.5°C above preindustrial levels (which we may have already temporarily exceeded) (IPCC, 2021). This increase will impact global ecological systems and the human activities (including agriculture), as well as increase the frequency and intensity of fires, floods, droughts and intense storms. Even with greatly accelerated emissions reductions, nearly all future scenarios that limit warming to 1.5–2 degrees contain an “overshoot” period, where the mean global temperature target is exceeded (Smith et al., 2023).
Active atmospheric carbon dioxide removal is needed to avoid the worst consequences of global climate change (Nemet et al., 2018; National Academies of Sciences, Engineering, and Medicine, 2022; Smith et al., 2023). Carbon dioxide removal (CDR) is defined as intentional capture of atmospheric carbon dioxide (CO2) and retention for time scales of decades to millennia (Smith et al., 2023).
Our preference for marine CDR (henceforth mCDR) is explained by our brief summary of some of the ethical arguments. The majority of CDR projects undertaken to date have been afforestation (Smith et al., 2023). There are concerns about the permanence carbon sequestered via afforestation given the increased risk of forests fires, and potential changes to land management practices (Fawzy et al., 2020; Fuss et al., 2018; Deprez et al., 2024). Land-based CDR (including afforestation and the growth of fuel stocks for Bioenergy with Carbon Capture and Storage) will compete for arable land and freshwater resources, with conservation, and agriculture. Anticipated future global shortages of fresh water, in particular, raise potential ethical concerns about land-based CDR (Minx et al., 2018). Any gains made in climate should not unnecessarily jeopardise other UN sustainability goals.1 Other approaches, such as Direct Air Capture or Enhanced Weathering and in situ mineralisation are also being developed, but no one technology alone is considered sufficient to sequester carbon dioxide at a gigaton scale. Therefore, throughout this perspective, we argue for the increased ethical use of mCDR. While we do not advocate for unnecessarily altering the ocean’s biogeochemistry, affecting fisheries productivity, or harming deep sea ecosystems, the consequences of inaction on climate change will be much worse for marine ecosystems than ethical deployment of CDR (Cullen and Boyd, 2008).
In addition to developing the technology needed for mCDR, we will need to establish methodologies to demonstrate that the mCDR approach is effectively sequestering carbon (here and throughout; carbon sequestration refers to removal of carbon dioxide from the atmosphere into a durable form) without producing unintended consequences. Much of the work to develop mCDR is undertaken by the oceanographic community. Current approaches utilised in oceanography are frequently “macro” and rely on technologies such as remote sensing and deployed sensors. Large scale models are frequently used in oceanography because of the ocean’s scale and the cost involved in accessing remote areas (Siegel et al., 2023). Many mCDR approaches, most notably Ocean Iron Fertilisation, are considered to be prohibited by the London Protocol against the dumping of wastes at sea (LCP 1.1 and LCP 4.8) due to concerns about the approaches efficacy and uncertainty about environmental impacts (Dixon et al., 2014). Exceptions to the protocol require extensive impact assessments and biological monitoring. The regulatory community—who will be approving mCDR approaches—is by contrast, accustomed to site-specific evidence-based criteria utilised in environmental risk assessments (e.g., Directive 2011/92/EU of the European Parliament, 2014; Directive 2014/52/EU of the European Parliament, 2014; Australia Environment Protection and Biodiversity Conservation Act (AEPBCA), 2014). This disparity contributes to the regulatory hurdles in mCDR implementation due to uncertainty in degrees of risk associated with mCDR approaches and how those risks could be mitigated. In addition, the public has a mixed perception of mCDR (Nawaz et al., 2023; Smith et al., 2023): they understand the need to urgently act on climate change, but also are reluctant to adopt untested approaches with perceived environmental risks.
Approaches for mCDR have been recently and thoroughly reviewed elsewhere (Nemet et al., 2018; National Academies of Sciences, Engineering, and Medicine, 2022; Smith et al., 2023). In this perspective, we focus on a set of approaches that can be used for measuring efficacy as well as the potential risk of unintended consequences, giving the regulatory community and ultimately, the public at large confidence in assessing the risks and impacts of using mCDR. We argue that using environmental genomics (in this case, metabarcoding and metatranscriptomics, illustrated in Figure 1) on samples collected from mCDR field trials and deployments, in parallel with other domains such analytical chemistry or remote sensing, can be used to achieve these goals. The use of genomics-based tools as a line of evidence for verification of mCDR should be developed in parallel with the sequestration approaches so that the regulatory community gains confidence in them. These approaches will also provide the ability to compare mCDR deployments to the risks and impacts of unmitigated climate change.
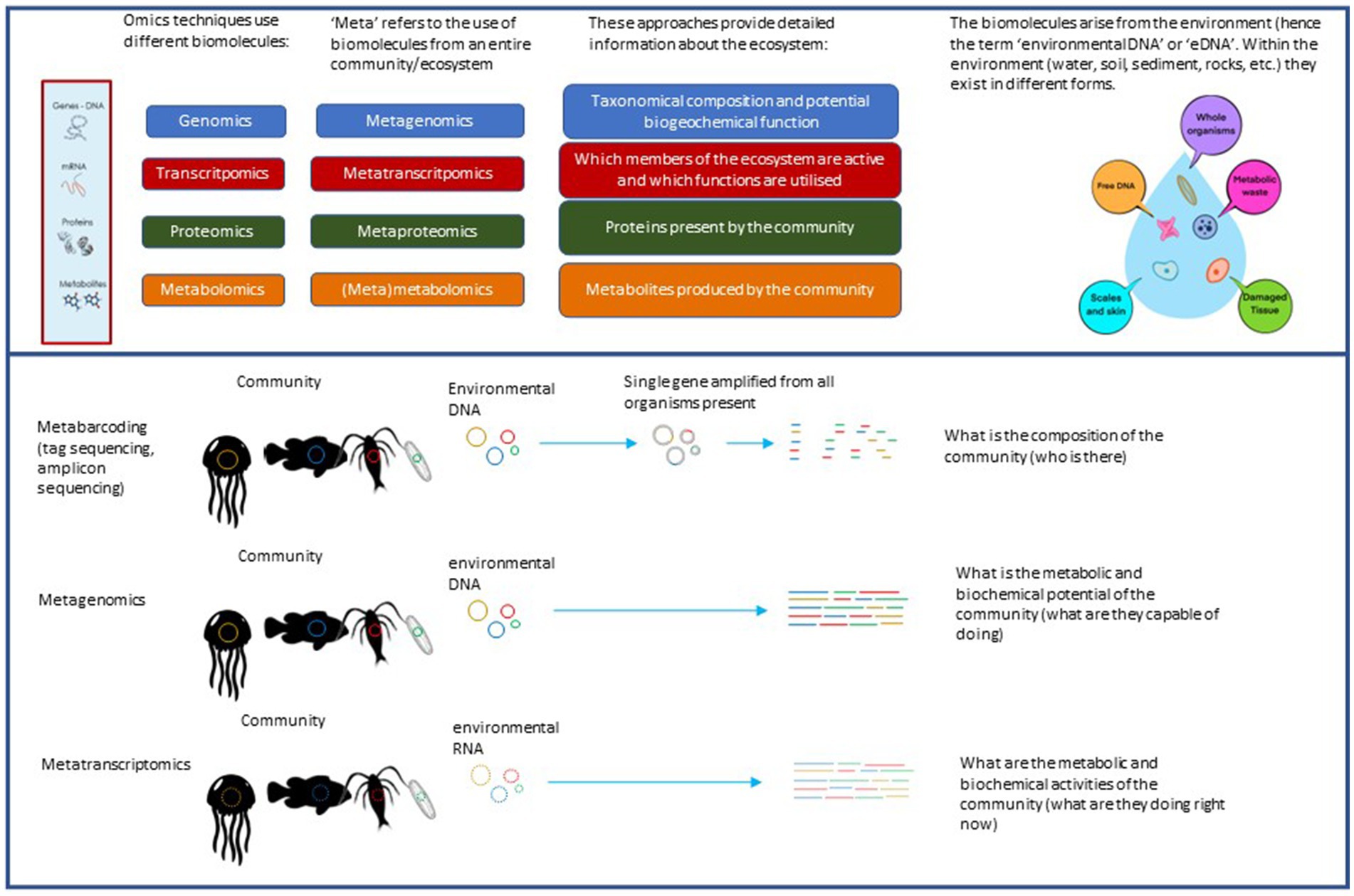
Figure 1. An illustration of the genomics approaches we are advocating for adoption in this perspective. Metabarcoding (sometimes called tag sequencing or amplicon sequencing) uses DNA and PCR to amplify all the different copies of a specific gene from a sample, which is subsequently used to differentiate and identify the various taxa present in the sample and compare the different samples. Metagenomics sequences the entirety of DNA in the sample, providing a holistic view of community function. Metatranscriptomics sequences all the RNA in a sample. Based on the broad assumption that genes are only transcribed to RNA, it is used as an approach to assess the active functions of a community.
2 Verification needs
2.1 Determining the efficacy of sequestration
2.1.1 Iron fertilisation
To demonstrate that iron fertilisation is effective, we will need to show that the growth of iron-limited phytoplankton cells is stimulated, and that these cells sink to depths necessary for carbon sequestration. While most studies have found an increase in phytoplankton growth following iron additions (reviewed in Boyd et al., 2007), the estimated amounts of photosynthetically-fixed carbon that has sunk to the sea floor have been inconsistent (Buesseler et al., 2008; Smetacek et al., 2012; Williamson et al., 2022); in general, fertilisation efforts that stimulate diatom growth have more export than those that stimulated growth of smaller cells (Mari et al., 2017; Guérin et al., 2022).
Recent studies illustrate how genomics-based approaches could be used to verify the growth of target species and export of photosynthetically fixed material. For instance, Guidi et al. (2016) used both genomics and metagenomics to identify plankton associated with particles that sink out of the upper surface waters. Metabarcoding was also used to examine the composition of sinking particles in mesotrophic and oligotrophic oceanic environments (Valencia et al., 2022). These approaches could be used to verify the export of stimulated phytoplankton and the acceleration of the biological pump if iron fertilisation were utilised in mCDR.
A recent study (Hook et al., 2021) used metatranscriptomics to identify phytoplankton responding to ammonia released by aquaculture. The study found changes in community composition, increased photosynthesis and carbon fixation in dinoflagellates, and an increased abundance of transcripts involved in ammonia uptake. A similar metatranscriptomic approach could be used to identify which phytoplankton are responding to additions of iron along a transect away from the site of fertilisation. It could also be used to identify the physiological pathways that underpin the differences in response in to iron (both as altered growth rates and differences in the amounts of carbon exported).
2.1.2 Restoration of blue carbon ecosystems and optimisation of their storage potential
The restoration of high-carbon sequestering coastal habitats (mangrove forests, seagrass meadows, salt marshes) is considered as one of the most environmentally friendly approaches to mCDR, as these approaches have positive environmental side effects, including shoreline protection, creation of habitat, and providing nursery for juvenile fisheries species. However, much of the carbon sequestered in Blue Carbon Ecosystem Restoration is sequestered for 100 years or less (Johannessen, 2022) and the carbon storage capacity can vary significantly spatially and temporally within these ecosystems (Owers et al., 2020; Ricart et al., 2020).
For carbon sequestration to occur in Blue Carbon ecosystems, organic carbon must be deposited in sediment, without being respired by the bacteria and fungi associated with the rhizosphere. Consequently, the carbon sequestration rates of coastal ecosystems vary depending on site characteristics, like sediment composition, nutrient and oxygen availability (Kida and Fujitake, 2020), hydrology (Reithmaier et al., 2020) and the plant species and composition. These factors affect the soil microbiota which ultimately drives the biogeochemistry determining the fate of sedimentary organic matter (Friesen et al., 2018; Hurtado-McCormick et al., 2022; Trivedi et al., 2013). There is uncertainty about the microbial activity in the rhizosphere, and thus, to what degree carbon sequestration is occurring in many Blue Carbon projects. There is also uncertainty regarding the impact of anthropogenic disturbance on the permanence of CDR in blue carbon ecosystems.
The rhizosphere associated microbial composition and activity can be used as a predictor for remineralisation rates as well as for methane (CH4) and nitrous oxide (N2O) production (offsetting carbon sequestration) (Allard et al., 2020; Reis et al., 2017). Despite this, there has been very limited research linking carbon sequestration (including offsetting Greenhouse gas [GHG] emissions) in coastal ecosystems to the soil microbiota (Allard et al., 2020; Trevathan-Tackett et al., 2019). Genomics based assessments of the sediment microbiota at restoration sites, compared to sites of similar characteristics (climate, hydrology) and their carbon sequestration rates may provide a good proxy for potential success as activity may negatively correlate with the amount of recalcitrant sequestered carbon.
Carbon storage outcomes in coastal ecosystem restoration projects also depend on management choices (de los Santos et al., 2022). Carbon sequestration outcomes could be increased by both managing soil microbiota and reducing environmental disturbances [such as dredging, wetland drainage which increase O2 availability thus microbial activity (Macreadie et al., 2019; Macreadie et al., 2015)] to minimise remineralisation. Management strategies that promote carbon sequestration include amending the soil/sediment with lignin derived phenolic compounds or clay particles (Min et al., 2015; Dunn and Freeman, 2018; Freeman et al., 2012; Freeman et al., 2001). If effective, these low cost, easily applied, and nontoxic amendments could minimise emissions from coastal ecosystems. However, soil microbiota also support plant growth, so actions that promote sequestration by decreasing soil microbial activity may cause detriment to the habitat (Allard et al., 2020; Birnbaum and Trevathan-Tackett, 2022). Genomics based approaches (metatranscriptomics, in particular) could be used to monitor changes in microbial activity to prevent this outcome.
2.2 Unintended negative consequences of mCDR
2.2.1 Nutrient robbing and altered ecosystem dynamics
Much of the hesitancy around deploying mCDR stems from the possibility of unintended consequences to marine ecosystems, in particular changes in primary producers. For instance, ocean fertilisation could result in changes in the phytoplankton size spectrum (Boyd et al., 2007; Chisholm et al., 2001) or the depletion of other nutrients (called nutrient robbing) leading to a decrease in phytoplankton productivity (Boyd et al., 2007). Minerals, such as olivine, used in Ocean Alkalinity Enhancement (OAE) are frequently high in Fe, and may stimulate primary production (Bach et al., 2019), which could also change phytoplankton size classes or causing nutrient depletion.
Hypothetically, increasing the pH of surface waters could also alter primary productivity, as carbon concentrating mechanisms in microalgae are optimised to the pH of surface seawaters. Even small increases above pH 8.5 would decrease the available carbonate ions, and may lead to carbon availability limiting algal growth (Liu et al., 2022). In these scenarios, smaller phytoplankton with efficient carbon uptake replace large algae, such as diatoms, causing reduced carbon export from upper surface waters (Bach et al., 2019). Furthermore, increasing the alkalinity of surface waters could favour the growth of coccolithophores; which are abundant in the comparatively alkaline Black Sea (Bach et al., 2019). Lipid rich diatoms are good food sources for many marine organisms, so their replacement could have trophic consequences.
Seaweed aquaculture causes additional concerns about nutrient robbing, as uptake of growth-limiting nutrients by cultivated algae could reduce future net primary production, and its associated carbon sequestration. This is a critical knowledge gap in understanding the climate change mitigation potential of seaweed aquaculture (Ross et al., 2022).
Metabarcoding could be used to compare the composition of eukaryotic and prokaryotic plankton upstream and downstream of mCDR deployments to determine if the species composition “downstream” are changing in ways that indicate nutrient limitation (due to nutrient robbing).
While, to our knowledge, the metabarcoding approach has not been used to measure unintended consequences of any mCDR projects, it has been routinely used to determine the dynamics of planktonic communities in other studies. Metabarcoding has been used to measure changes in zooplankton populations in a boreal lake following simulated spills of diluted bitumen and to different response technologies (Ankley et al., 2020), and to assess the impacts of nutrients and metals on the composition of planktonic communities in coastal China (Zhang et al., 2023). Other studies have used eDNA surveys to measure seasonal dynamics and the influence of a marine heatwave on biological community composition and species abundance (Berry et al., 2019) and to show changes in the bacterioplankton following a marine heatwave (Brown et al., 2024). Metabarcoding was also used to show that changing concentrations of CO2 has little impact on bacterioplankton composition (Lin et al., 2018). Taken together, the aforementioned studies demonstrate how metabarcoding could be used to measure changes in plankton composition, providing the regulatory community and the public at large evidence for the safety, or alternatively, unintended consequences, of mCDR techniques.
Metatranscriptomics could be used to directly measure changes in resource competition “in patch” and “out of patch” areas anticipated to be impacted by mCDR. Although this approach has not been used (to our knowledge) in mCDR approaches, it has been used to study niche partitioning in the plankton communities (Alexander et al., 2015), adaptive responses in different phytoplankton to variations in Fe availability (Caputi et al., 2019; Kolody et al., 2022). Landscape-scale metatransciptomic profiles were also used to determine the influence of land use patterns on microbial communities in small ponds in Germany (Bizic et al., 2022). Transcriptomic studies have also been used to study nutrient limitation (Harke et al., 2017), circadian rhythms (Hernandez Limon et al., 2020) and co-ordination of nutrient uptake with diazotrophs (Harke et al., 2019) and to study bloom dynamics (Ji et al., 2018).
2.2.2 The potential for toxicity
Although OAE has great potential for carbon sequestration, there is the potential for localised harm to marine organisms through elevated pH or trace metals. The impact of elevated pH on marine organisms has not been well studied (Kitidis et al., 2024). The minerals be used in OAE contain trace metals, which could be toxic or bioaccumulated (National Academies of Sciences, Engineering, and Medicine, 2022; Bach et al., 2019; Ferderer et al., 2022; Kitidis et al., 2024). Recent studies have examined the impacts of OAE on primary production and have found comparatively minor impacts (Ferderer et al., 2022; Gore et al., 2019; Hutchins et al., 2023). Others have found direct mortality of invertebrate macroorganisms (Jones et al., 2024) or altered timings of the Spring bloom (González-Santana et al., 2024). These studies frequently employ environmentally unrealistic exposure durations, however.
Metatranscriptomics studies would identify the major biochemical pathways changing due to OAE in planktonic organisms. Transcriptomic and proteomic-based approaches could also be used to determine whether larger organisms are responding to increases in ocean pH or metal exposure, as has been demonstrated in the ocean acidification literature (e.g., Schwaner et al., 2023; Wong and Hofmann, 2021). For example, a transcriptomic study identified pathways of shell formation, bicarbonate transport, cytoskeleton, immunity, stress and metabolism as conferring resilience to decreased ocean pH in clams (Mercinaria mercinaria) (Schwaner et al., 2023). Similar approaches have been used to study sea urchin larvae responses to increased pCO2 (and as a consequence, decreased pH) (Wong and Hofmann, 2021). Laboratory exposures to OAE materials could be used to identify targets of potential adverse outcomes (Ankley et al., 2010), followed by subsequent field-based monitoring programmes screening potentially impacted organisms for these targets.
2.2.3 Potential for production of other greenhouse gases
For mCDR approaches to be effective, the carbon sequestered must be stored in either sediments or deep waters, not remineralised into CO2, N2O or CH4. If the quantity of photosynthetically produced carbon is sufficient, there will not be sufficient oxygen for remineralisation, and other diagenetic processes will be utilised. As a consequence, there is also concern that the breakdown of sequestered material may lead to the release of N2O and CH4—both of which are more potent greenhouse gases than CO2 (Gnanadesikan and Marinov, 2008; Boyd et al., 2022; Fuhrman and Capone, 1991). In addition, some studies have found that mangroves can be a net source of CO2, CH4 and N2O (Reithmaier et al., 2020) depending on the composition and activity the sediment microbiota (Alongi, 1988; Malerba et al., 2022; Reis et al., 2017; Rosentreter et al., 2018). Credible quantification of carbon sequestration by coastal restoration projects should include consideration of microbial activity (Johannessen, 2022; Mazarrasa et al., 2018; Ricart et al., 2020).
Metabarcoding could be used to determine whether there are increases in bacteria that could produce CH4 and N2O associated with sequestered carbon. Metatranscriptomics studies would also highlight when and where mCDR can lead to increased production of CH4 or N2O, counteracting the effect of CO2 removal. These approaches could also be used to measure the abundance and activity of methanotrophs and N2O reducing bacteria.
3 Discussion
CDR strategies will need to be rapidly developed and implemented if we are to avoid the worse consequences of climate change, as well as to allow many countries and industries to meet their “Net Zero” pledges. To avoid competition with other UN Sustainability Goals by land-based CDR, we argue for the rapid development of mCDR. However, mCDR approaches can only be deployed at scale following a thorough risk assessment of their benefits, risks, and trade-offs, and have not been allowed under the London Protocol because of uncertainty about their efficacy and off-target environmental impacts. Resolving this uncertainty is not possible with the current set of available data and the approaches used to collect it. There is uncertainty about the potential scale of the carbon removal (e.g., whether the approaches are working); “nutrient robbing” (i.e., diverting nutrients away from other ecosystem processes) as well as uncertainty about the unintended impacts to other parts of the ecosystem.
Marine CDR interventions will require close monitoring to demonstrate that unintended consequences are within the risk tolerance for social acceptance, and that they are effective in permanently sequestering carbon. We suggest that the monitoring, reporting, and verification (MRV) approaches be conducted using a multiple line of evidence approach. The sensor, satellite, and other approaches that are being implemented now provide information over a geographic scale and frequently in close to real time, neither of which is currently feasible with the techniques highlighted in this perspective. Nonetheless, we suggest that using modern genomics-based approaches on samples collected from in situ deployments in conjunction with biomarkers and isotopic methods could help resolve much of the uncertainty around the potential unintended consequences of using mCDR, as well as help quantify the degree to which carbon is being sequestered. Although use of genomics based assays will require continuous sampling over a wide spatial footprint, all MRV approaches will have similar requirements, so this condition alone should not be an impediment to adoption of the technique, especially as they may provide the certainty required for social acceptance or palatability. This line of evidence should be developed in parallel with the development of mCDR approaches. Similar approaches are currently being used to monitor the impacts of offshore energy on fish and wildlife species (Sepulveda et al., 2024) and in community based monitoring by the New Zealand EPA.2 Our suggestions are summarised in Table 1. These approaches can provide detail on the microbial processes that would otherwise be unattainable. They can help resolve uncertainty around the potential for unintended consequences of environmental interventions, as well as provide a line of evidence for their efficacy. This additional information will lend us the support needed to act on climate change.
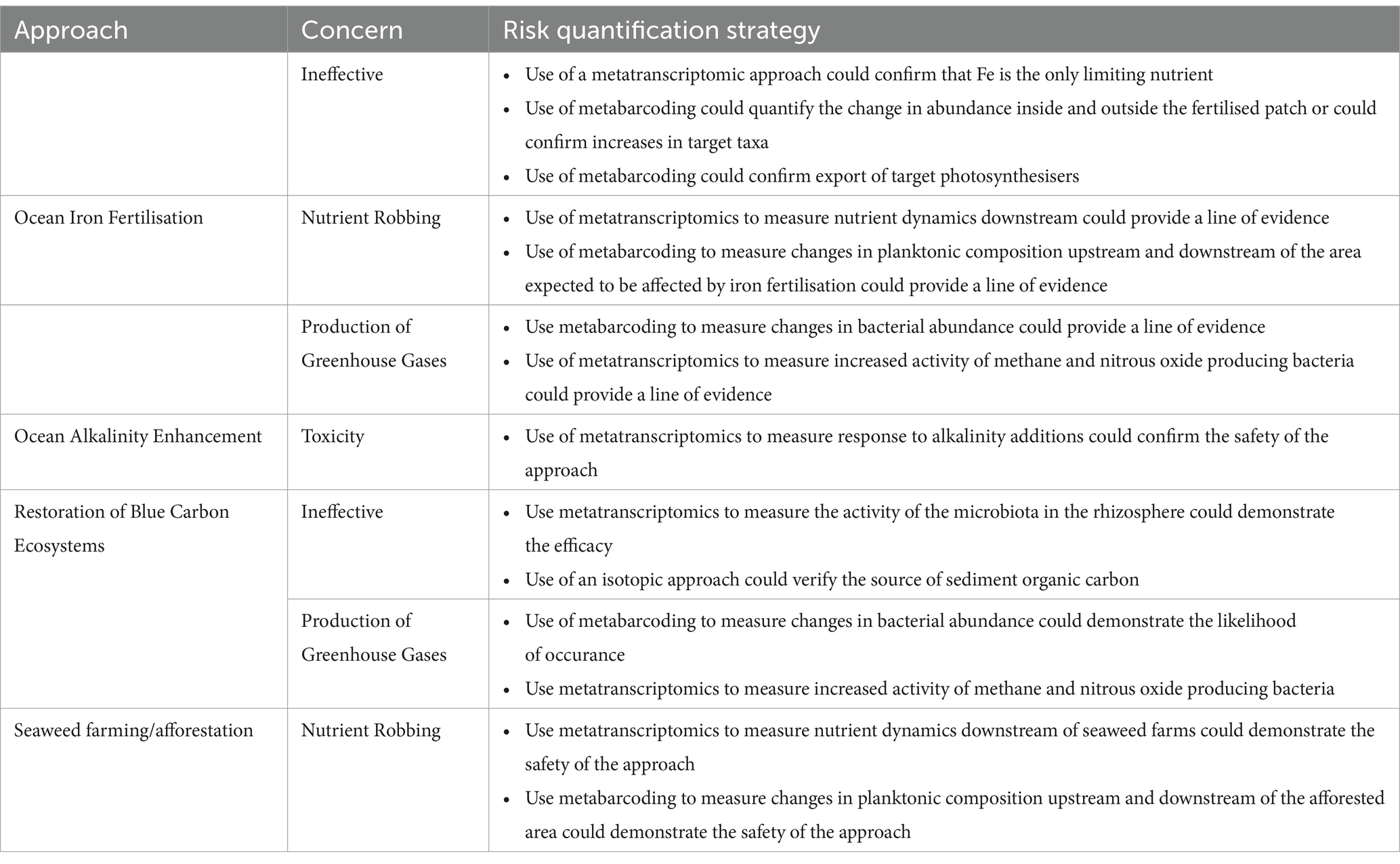
Table 1. Summary of the concerns regarding mCDR approaches, and how the risk could be quantified using a genomics based approach.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
Author contributions
SH: Conceptualization, Funding acquisition, Project administration, Writing – original draft, Writing – review & editing. LB: Writing – original draft, Writing – review & editing. EB: Writing – original draft, Writing – review & editing. AW: Conceptualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. Funding for this work was provided by CSIRO’s Permanent Carbon Locking Future Science Platform.
Acknowledgments
The authors would like to thank Joey Crosswell, Audrey Bester, Yuwan Malakar, Michael Battaglia, and Mat Vanderklift (CSIRO) for helpful comments on a previous version of this manuscript, and Andrew Lenton (CSIRO) for valuable discussion. Moira Hodgkins provided assistance with the illustration.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
References
Alexander, H., Jenkins, B. D., Rynearson, T. A., and Dyhrman, S. T. (2015). Metatranscriptome analyses indicate resource partitioning between diatoms in the field. Proc. Natl. Acad. Sci. USA 112, E2182–E2190. doi: 10.1073/pnas.1421993112
Allard, S. M., Costa, M. T., Bulseco, A. N., Helfer, V., Wilkins, L. G. E., Hassenruck, C., et al. (2020). Introducing the mangrove microbiome initiative: identifying microbial research priorities and approaches to better understand, protect, and rehabilitate mangrove ecosystems. mSystems 5:e00658–20. doi: 10.1128/mSystems.00658-20
Alongi, D. M. (1988). Bacterial productivity and microbial biomass in tropical mangrove sediments. Microb. Ecol. 15, 59–79. doi: 10.1007/BF02012952
Ankley, G. T., Bennett, R. S., Erickson, R. J., Hoff, D. J., Hornung, M. W., Johnson, R. D., et al. (2010). Adverse outcome pathways: a conceptual framework to support ecotoxicology research and risk assessment. Environ. Toxicol. Chem. 29, 730–741. doi: 10.1002/etc.34
Ankley, G. T., Blackwell, B. R., Cavallin, J. E., Doering, J. A., Feifarek, D. J., Jensen, K. M., et al. (2020). Adverse outcome pathway network-based assessment of the interactive effects of an androgen receptor agonist and an aromatase inhibitor on fish endocrine function. Environ. Toxicol. Chem. 39, 913–922. doi: 10.1002/etc.4668
Australia Environment Protection and Biodiversity Conservation Act (AEPBCA). (2014). Australia Environment Protection and Biodiversity Conservation Act 1999. Australia: Federal Register of Legislation.
Bach, L. T., Gill, S. J., Rickaby, R. E. M., Gore, S., and Renforth, P. (2019). CO2 removal with enhanced weathering and ocean alkalinity enhancement: potential risks and co-benefits for marine pelagic ecosystems. Front. Clim. 1:7. doi: 10.3389/fclim.2019.00007
Berry, T. E., Saunders, B. J., Coghlan, M. L., Stat, M., Jarman, S., Richardson, A. J., et al. (2019). Marine environmental DNA biomonitoring reveals seasonal patterns in biodiversity and identifies ecosystem responses to anomalous climatic events. PLoS Genet. 15:e1007943. doi: 10.1371/journal.pgen.1007943
Birnbaum, C., and Trevathan-Tackett, S. M. (2022). Aiding coastal wetland restoration via the belowground soil microbiome: an overview. Restor. Ecol. 31:e13824. doi: 10.1111/rec.13824
Bizic, M., Ionescu, D., Karnatak, R., Musseau, C. L., Onandia, G., Berger, S. A., et al. (2022). Land-use type temporarily affects active pond community structure but not gene expression patterns. Mol. Ecol. 31, 1716–1734. doi: 10.1111/mec.16348
Boyd, P. W., Bach, L. T., Hurd, C. L., Paine, E., Raven, J. A., and Tamsitt, V. (2022). Potential negative effects of ocean afforestation on offshore ecosystems. Nat. Ecol. Evol. 6, 675–683. doi: 10.1038/s41559-022-01722-1
Boyd, P. W., Jickells, T., Law, C. S., Blain, S., Boyle, E. A., Buesseler, K. O., et al. (2007). Mesoscale iron enrichment experiments 1993-2005: synthesis and future directions. Science 315, 612–617. doi: 10.1126/science.1131669
Brown, M. V., Ostrowski, M., Messer, L. F., Bramucci, A., van de Kamp, J., Smith, M. C., et al. (2024). A marine heatwave drives significant shifts in pelagic microbiology. Commun. Biol. 7:125. doi: 10.1038/s42003-023-05702-4
Buesseler, K. O., Doney, S. C., Karl, D. M., Boyd, P. W., Caldeira, K., Chai, F., et al. (2008). Environment - ocean iron fertilization - moving forward in a sea of uncertainty. Science 319:162. doi: 10.1126/science.1154305
Caputi, L., Carradec, Q., Eveillard, D., Kirilovsky, A., Pelletier, E., Karlusich, J. J. P., et al. (2019). Community-level responses to iron availability in open ocean plankton ecosystems. Glob. Biogeochem. Cycles 33, 391–419. doi: 10.1029/2018GB006022
Chisholm, S. W., Falkowski, P. G., and Cullen, J. J. (2001). Oceans - dis-crediting ocean fertilization. Science 294, 309–310. doi: 10.1126/science.1065349
Cullen, J. J., and Boyd, P. W. (2008). Predicting and verifying the intended and unintended consequences of large-scale ocean iron fertilization. Mar. Ecol. Prog. Ser. 364, 295–301. doi: 10.3354/meps07551
de los Santos, C. B., Egea, L. G., Martins, M., Santos, R., Masqué, P., Peralta, G., et al. (2022). Sedimentary organic carbon and nitrogen sequestration across a vertical gradient on a temperate wetland seascape including salt marshes, seagrass meadows and Rhizophytic macroalgae beds. Ecosystems 26, 826–842. doi: 10.1007/s10021-022-00801-5
Deprez, A., Leadley, P., Dooley, K., Williamson, P., Cramer, W., Gattuso, J.-P., et al. (2024). Sustainability limits needed for CO2 removal. Science 383, 484–486. doi: 10.1126/science.adj6171
Dixon, T., Garrett, J., and Kleverlaan, E. (2014). Update on the London Protocol – Developments on Transboundary CCS and on Geoengineering. Energy Procedia, 63, 6623–6628.
Dunn, C., and Freeman, C. (2018). The role of molecular weight in the enzyme-inhibiting effect of phenolics: the significance in peatland carbon sequestration. Ecol. Eng. 114, 162–166. doi: 10.1016/j.ecoleng.2017.06.036
Fawzy, S., Osman, A. I., Doran, J., and Rooney, D. W. (2020). Strategies for mitigation of climate change: a review. Environ. Chem. Lett. 18, 2069–2094. doi: 10.1007/s10311-020-01059-w
Ferderer, A., Chase, Z., Kennedy, F., Schulz, K. G., and Bach, L. T. (2022). Assessing the influence of ocean alkalinity enhancement on a coastalphytoplankton community. Biogeosciences 19, 5375–5399. doi: 10.5194/bg-19-5375-2022
Freeman, C., Fenner, N., and Shirsat, A. H. (2012). Peatland geoengineering: an alternative approach to terrestrial carbon sequestration. Philos. Trans. R. Soc. A Math. Phys. Eng. Sci. 370, 4404–4421. doi: 10.1098/rsta.2012.0105
Freeman, C., Ostle, N., and Kang, H. (2001). An enzymic ‘latch’ on a global carbon store. Nature 409:149. doi: 10.1038/35051650
Friesen, S. D., Dunn, C., and Freeman, C. (2018). Decomposition as a regulator of carbon accretion in mangroves: a review. Ecol. Eng. 114, 173–178. doi: 10.1016/j.ecoleng.2017.06.069
Fuhrman, J. A., and Capone, D. G. (1991). Possible biogeochemical consequences of ocean fertilization. Limnol. Oceanogr. 36, 1951–1959. doi: 10.4319/lo.1991.36.8.1951
Fuss, S., Lamb, W. F., Callaghan, M. W., Hilaire, J., Creutzig, F., Amann, T., et al. (2018). Negative emissions—part 2: costs, potentials and side effects. Environ. Res. Lett. 13:063002. doi: 10.1088/1748-9326/aabf9f
Gnanadesikan, A., and Marinov, I. (2008). Export is not enough: nutrient cycling and carbon sequestration. Mar. Ecol. Prog. Ser. 364, 289–294. doi: 10.3354/meps07550
González-Santana, D., Segovia, M., González-Dávila, M., Ramírez, L., González, A. G., Pozzo-Pirotta, L. J., et al. (2024). Ocean alkalinity enhancement using sodium carbonate salts does not lead to measurable changes in Fe dynamics in a mesocosm experiment. Biogeosciences 21, 2705–2715. doi: 10.5194/bg-21-2705-2024
Gore, S., Renforth, P., and Perkins, R. (2019). The potential environmental response to increasing oceanalkalinity for negative emissions. Mitig. Adapt. Strateg. Glob. Chang. 24, 1191–1211. doi: 10.1007/s11027-018-9830-z
Guérin, N., Ciccarella, M., Flamant, E., Frémont, P., Mangenot, S., Istace, B., et al. (2022). Genomic adaptation of the picoeukaryote to iron-poor oceans revealed by a chromosome-scale genome sequence. Commun. Biol. 5:983. doi: 10.1038/s42003-022-03939-z
Guidi, L., Chaffron, S., Bittner, L., Eveillard, D., Larhlimi, A., Roux, S., et al. (2016). Plankton networks driving carbon export in the oligotrophic ocean. Nature 532, 465–470. doi: 10.1038/nature16942
Harke, M. J., Frischkorn, K. R., Haley, S. T., Aylward, F. O., Zehr, J. P., and Dyhrman, S. T. (2019). Periodic and coordinated gene expression between a diazotroph and its diatom host. ISME J. 13, 118–131. doi: 10.1038/s41396-018-0262-2
Harke, M. J., Juhl, A. R., Haley, S. T., Alexander, H., and Dyhrman, S. T. (2017). Conserved transcriptional responses to nutrient stress in bloom-forming algae. Front. Microbiol. 8:1279. doi: 10.3389/fmicb.2017.01279
Hernandez Limon, M. D., Hennon, G. M. M., Harke, M. J., Frischkorn, K. R., Haley, S. T., and Dyhrman, S. T. (2020). Transcriptional patterns of Emiliania huxleyi in the North Pacific subtropical gyre reveal the daily rhythms of its metabolic potential. Environ. Microbiol. 22, 381–396. doi: 10.1111/1462-2920.14855
Hook, S. E., White, C., and Ross, D. J. (2021). A metatranscriptomic analysis of changing dynamics in the plankton communities adjacent to aquaculture leases in southern Tasmania, Australia. Mar. Genom. 59:100858. doi: 10.1016/j.margen.2021.100858
Hurtado-McCormick, V., Trevathan-Tackett, S. M., Bowen, J. L., Connolly, R., Duarte, C. M., and Macreadie, P. I. (2022). Pathways for understanding blue carbon microbiomes with amplicon sequencing. Microorganisms 10:2121. doi: 10.3390/microorganisms10112121
Hutchins, D. A., Fu, F.-X., Yang, S.-C., John, S. G., Romaniello, S. J., Andrews, M. G., et al. (2023). Responses of globally important phytoplankton species to olivine dissolution products and implications for carbon dioxide removal via ocean alkalinity enhancement. Biogeosciences 20, 4669–4682. doi: 10.5194/bg-20-4669-2023
IPCC (2021). “Summary for policymakers” in Climate change 2021: the physical science basis. contribution of working group i to the sixth assessment report of the intergovernmental panel on climate change. eds. V. Masson-Delmotte, P. Zhai, A. Pirani, S. L. Connors, C. Péan, and S. Berger, et al. (UK and New York, NY, USA: Cambridge University Press). 3–33.
Ji, N. J., Lin, L. X., Li, L., Yu, L. Y., Zhang, Y. Q., Luo, H., et al. (2018). Metatranscriptome analysis reveals environmental and diel regulation of a Heterosigma akashiwo (raphidophyceae) bloom. Environ. Microbiol. 20, 1078–1094. doi: 10.1111/1462-2920.14045
Johannessen, S. C. (2022). How can blue carbon burial in seagrass meadows increase long-term, net sequestration of carbon? A critical review. Environ. Res. Lett. 17:093004. doi: 10.1088/1748-9326/ac8ab4
Jones, K., Hemery, L., Ward, N., Regier, P., Ringham, M., and Eisaman, M. (2024). Biological response of eelgrass epifauna, Taylor’s sea hare (Phyllaplysia taylori) and eelgrass isopod (Idotea resecata), to elevated ocean alkalinity. EGUsphere. doi: 10.5194/egusphere-2024-972
Kida, M., and Fujitake, N. (2020). Organic carbon stabilization mechanisms in mangrove soils: a review. Forests 11:981. doi: 10.3390/f11090981
Kitidis, V., Rackley, S. A., Burt, W. J., Rau, G. H., Fawcett, S., Taylor, M., et al. (2024). Magnesium hydroxide addition reduces aqueous carbon dioxide in wastewater discharged to the ocean. Commun. Earth Environ. 5:354. doi: 10.1038/s43247-024-01506-4
Kolody, B. C., Smith, S. R., Allen, L. Z., McCrow, J. P., Moustafa, A., Shi, D., et al. (2022). Nitrogen and iron availability drive metabolic remodeling and natural selection of diverse phytoplankton during experimental upwelling. Msystems 7:e0072922. doi: 10.1128/msystems.00729-22
Lin, X., Huang, R., Li, Y., Li, F., Wu, Y., Hutchins, D. A., et al. (2018). Interactive network configuration maintains bacterioplankton community structure under elevated CO2 in a eutrophic coastal mesocosm experiment. Biogeosciences 15, 551–565. doi: 10.5194/bg-15-551-2018
Liu, J., Yin, W., Zhang, X., Xie, X., Dong, G., Lu, Y., et al. (2022). RNA-seq analysis reveals genes related to photosynthetic carbon partitioning and lipid production in Phaeodactylum tricornutum under alkaline conditions. Front. Microbiol. 13:969639. doi: 10.3389/fmicb.2022.969639
Macreadie, P. I., Atwood, T. B., Seymour, J. R., Fontes, M. L. S., Sanderman, J., Nielsen, D. A., et al. (2019). Vulnerability of seagrass blue carbon to microbial attack following exposure to warming and oxygen. Sci. Total Environ. 686, 264–275. doi: 10.1016/j.scitotenv.2019.05.462
Macreadie, P. I., Trevathan-Tackett, S. M., Skilbeck, C. G., Sanderman, J., Curlevski, N., Jacobsen, G., et al. (2015). Losses and recovery of organic carbon from a seagrass ecosystem following disturbance. Proc. R. Soc. B Biol. Sci. 282:20151537. doi: 10.1098/rspb.2015.1537
Malerba, M. E., Friess, D. A., Peacock, M., Grinham, A., Taillardat, P., Rosentreter, J. A., et al. (2022). Methane and nitrous oxide emissions complicate the climate benefits of teal and blue carbon wetlands. One Earth 5, 1336–1341. doi: 10.1016/j.oneear.2022.11.003
Mari, X., Passow, U., Migon, C., Burd, A. B., and Legendre, L. (2017). Transparent exopolymer particles: effects on carbon cycling in the ocean. Prog. Oceanogr. 151, 13–37. doi: 10.1016/j.pocean.2016.11.002
Mazarrasa, I., Samper-Villarreal, J., Serrano, O., Lavery, P. S., Lovelock, C. E., Marba, N., et al. (2018). Habitat characteristics provide insights of carbon storage in seagrass meadows. Mar. Pollut. Bull. 134, 106–117. doi: 10.1016/j.marpolbul.2018.01.059
Min, K., Freeman, C., Kang, H., and Choi, S.-U. (2015). The regulation by phenolic compounds of soil organic matter dynamics under a changing environment. Biomed. Res. Int. 2015:825098, 1–11. doi: 10.1155/2015/825098
Minx, J. C., Lamb, W. F., Callaghan, M. W., Fuss, S., Hilaire, J., Creutzig, F., et al. (2018). Negative emissions—part 1: research landscape and synthesis. Environ. Res. Lett. 13:063001. doi: 10.1088/1748-9326/aabf9b
National Academies of Sciences, Engineering, and Medicine (2022). A research strategy for ocean-based carbon dioxide removal and sequestration. Washington, DC: The National Academies Press.
Nawaz, S., Peterson St-Laurent, G., and Satterfield, T. (2023). Public evaluations of four approaches to ocean-based carbon dioxide removal. Climate Policy, 23:379–394.
Nemet, G. F., Callaghan, M. W., Creutzig, F., Fuss, S., Hartmann, J., Hilaire, J., et al. (2018). Negative emissions—part 3: innovation and upscaling. Environ. Res. Lett. 13:063003. doi: 10.1088/1748-9326/aabff4
Owers, C. J., Rogers, K., Mazumder, D., and Woodroffe, C. D. (2020). Temperate coastal wetland near-surface carbon storage: spatial patterns and variability. Estuar. Coast. Shelf Sci. 235:106584. doi: 10.1016/j.ecss.2020.106584
Reis, C. R. G., Nardoto, G. B., and Oliveira, R. S. (2017). Global overview on nitrogen dynamics in mangroves and consequences of increasing nitrogen availability for these systems. Plant Soil 410, 1–19. doi: 10.1007/s11104-016-3123-7
Reithmaier, G. M. S., Ho, D. T., Johnston, S. G., and Maher, D. T. (2020). Mangroves as a source of greenhouse gases to the atmosphere and alkalinity and dissolved carbon to the coastal ocean: a case study from the Everglades National Park, Florida. J. Geophys. Res.-Biogeo. 125:e2020JG005812. doi: 10.1029/2020JG005812
Ricart, A. M., York, P. H., Bryant, C. V., Rasheed, M. A., Ierodiaconou, D., and Macreadie, P. I. (2020). High variability of blue carbon storage in seagrass meadows at the estuary scale. Sci. Rep. 10:5865. doi: 10.1038/s41598-020-62639-y
Rosentreter, J. A., Maher, D. T., Erler, D. V., Murray, R. H., and Eyre, B. D. (2018). Methane emissions partially offset “blue carbon” burial in mangroves. Sci. Adv. 4:eaao4985. doi: 10.1126/sciadv.aao4985
Ross, F., Tarbuck, P., and Macreadie, P. I. (2022). Seaweed afforestation at large-scales exclusively for carbon sequestration: critical assessment of risks, viability and the state of knowledge. Front. Mar. Sci. 9:1015612. doi: 10.3389/fmars.2022.1015612
Schwaner, C., Farhat, S., Barbosa, M., Boutet, I., Tanguy, A., Espinosa, E. P., et al. (2023). Molecular features associated with resilience to ocean acidification in the northern quahog. Mar. Biotechnol. 25, 83–99. doi: 10.1007/s10126-022-10183-3
Sepulveda, A., Morrison, C., Hunter, M., and Khalil, M. (2024). Realizing the potential of eDNA biodiversity monitoring tools in the marine environment with application to offshore renewable energy. Fact Sheet USGS Numbered Ser. :fs20243019. doi: 10.3133/fs20243019
Siegel, D. A., DeVries, T., Cetinić, I., and Bisson, K. M. (2023). Quantifying the ocean's biological pump and its carbon cycle impacts on global scales 15, 329–356. doi: 10.1146/annurev-marine-040722-115226
Smetacek, V., Klaas, C., Strass, V. H., Assmy, P., Montresor, M., Cisewski, B., et al. (2012). Deep carbon export from a Southern Ocean iron-fertilized diatom bloom. Nature 487, 313–319. doi: 10.1038/nature11229
Smith, S.M., Geden, O., Nemet, G., Gidden, M., Lamb, W. F., Powis, C., et al. (2023). The State of Carbon Dioxide Removal – 1st Edition. Available at: https://www.stateofcdr.org/ (Accessed July 3, 2024).
Trevathan-Tackett, S. M., Sherman, C. D. H., Huggett, M. J., Campbell, A. H., Laverock, B., Hurtado-McCormick, V., et al. (2019). A horizon scan of priorities for coastal marine microbiome research. Nat. Ecol. Evol. 3, 1509–1520. doi: 10.1038/s41559-019-0999-7
Trivedi, P., Anderson, I. C., and Singh, B. K. (2013). Microbial modulators of soil carbon storage: integrating genomic and metabolic knowledge for global prediction. Trends Microbiol. 21, 641–651. doi: 10.1016/j.tim.2013.09.005
Valencia, B., Stukel, M. R., Allen, A. E., McCrow, J. P., Rabines, A., and Landry, M. R. (2022). Microbial communities associated with sinking particles across an environmental gradient from coastal upwelling to the oligotrophic ocean. Deep-Sea Res. I Oceanogr. Res. Pap. 179:103668. doi: 10.1016/j.dsr.2021.103668
Williamson, P., Boyd, P. W., Harrison, D. P., Reynard, N., and Mashayek, A. (2022). “Feasibility of using biologically-based processes in the open ocean and coastal seas for atmospheric CO2 removal” in Chapter 10 in Greenhouse gas removal technologies. eds. M. Bui and N. MacDowell (United Kingdom: Royal Society of Chemistry), 291–350.
Wong, J. M., and Hofmann, G. E. (2021). Gene expression patterns of red sea urchins (Mesocentrotus franciscanus) exposed to different combinations of temperature and pCO(2) during early development. BMC Genomics 22:32. doi: 10.1186/s12864-020-07327-x
Keywords: iron fertilisation, ocean afforestation, Ocean Alkalinity Enhancement, electrochemical approaches to CDR, meta barcoding, transcriptomics
Citation: Hook SE, Bodrossy L, Brewer EA and Willis A (2024) Genomics—based approaches may assist in the verification and accelerate responsible deployment of marine carbon dioxide removal. Front. Clim. 6:1471313. doi: 10.3389/fclim.2024.1471313
Edited by:
Wil Burns, American University, United StatesReviewed by:
Phillip Williamson, University of East Anglia, United KingdomCopyright © 2024 Hook, Bodrossy, Brewer and Willis. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sharon E. Hook, U2hhcm9uLkhvb2tAY3Npcm8uYXU=
 Sharon E. Hook
Sharon E. Hook Levente Bodrossy
Levente Bodrossy Elizabeth A. Brewer1
Elizabeth A. Brewer1 Anusuya Willis
Anusuya Willis