- 1Skeletal Biology and Engineering Research Center, Department of Development and Regeneration, KU Leuven, Leuven, Belgium
- 2Prometheus, Division of Skeletal Tissue Engineering, KU Leuven, Leuven, Belgium
- 3Biomechanics Research Unit, GIGA In Silico Medicine, GIGA institute, University of Liège, Liège, Belgium
- 4Biomechanics Section, Department of Mechanical Engineering, Faculty of Engineering, KU Leuven, Leuven, Belgium
- 5Institute for Chemical Engineering Sciences, Foundation for Research and Technology–Hellas, Patras, Greece
The field of tissue engineering aspires to provide clinically relevant solutions for patients through the integration of developmental engineering principles with a bottom-up manufacturing approach. However, the manufacturing of cell-based advanced therapy medicinal products is hampered by protocol complexity, lack of non-invasive critical quality controls, and dependency on animal-derived components for tissue differentiation. We investigate a serum-free, chemically defined, xeno- and lipid-free chondrogenic differentiation medium to generate bone-forming callus organoids. Our results show an increase in microtissue homogeneity during prolonged differentiation and the high quality of in vivo bone-forming organoids. The low protein content of the culture medium potentially allows for the monitoring of relevant secreted biomarkers as (critical) quality attributes. Together, we envisage that this xeno- and lipid-free chondrogenic medium is compatible with industrial scale-up and automation while facilitating the implementation of non-invasive imaging and the use of quality control parameters based on secreted biomarkers.
Introduction
Over the last decade, the field of skeletal tissue engineering has been maturing and getting ready to bridge the translational gap. Despite extensive efforts and significant scientific progress, only a few products have reached the market (Negoro et al., 2018; Sallent et al., 2020). Typically, new tissue-engineered products belong to the class of advanced cell therapy medicinal products (ATMPs) and contain living cells combined with biomaterials or growth factors. As a result, regulatory bodies such as FDA and EMA implemented a stringent set of requirements before ATMPs are allowed into the market. One of these requirements is the use of a manufacturing process that is robust, scalable, provides ample opportunities for quality control measurements, is biologically defined and characterized, and ideally is predictive of the clinical outcome (Mendicino et al., 2014; Petricciani et al., 2017; Rousseau et al., 2018; Doulgkeroglou et al., 2020; Iglesias-Lopez et al., 2021). Yet, basic research–inspired laboratory-scale protocols are often complex, require careful handling, use media that are not chemically defined, contain historically included components, and most of the time rely on animal-derived constituents such as fetal bovine serum (FBS).
We have been focusing on new tissue engineering strategies for bone repair. Under normal conditions, between 5% and 10% of fractures do not heal properly and in particular non-unions constitute a challenging clinical problem. Recently, emerging types of ATMPs for large bone defects follow the use of microtissues as building blocks for the bottom-up engineering of larger living tissues (Burdis and Kelly, 2021) while closely mimicking developmental biology principles (Lenas et al., 2009; Lenas and Luyten, 2011). In short, skeletal progenitor cell aggregates proliferate and differentiate in vitro through the chondrogenic lineage into microtissue structures containing a cartilaginous extracellular matrix. These microtissues can be used to form larger cartilaginous constructs by spontaneous fusion (Nilson Hall et al, 2021) or bioprinting (De Moor et al., 2020). Cell aggregates allow cells to condensate and, with the appropriate chondrogenic medium in a 3D culture, the aggregates appear to recapitulate the process of endochondral ossification (Hall et al., 2018), which is the basis for both developmental bone growth and postnatal bone regeneration (Nilsson Hall et al., 2019; Salhotra et al., 2020). This maturation process leads chondrocytes to a terminal hypertrophic stage where cells enter an apoptosis program followed by cell death or transdifferentiation to osteoblasts (Yang et al., 2014; Park et al., 2015; Cervantes-Diaz et al., 2017). Finally, through vascular invasion and the appearance of osteoblasts, chondroclasts, and osteoclasts, the engineered cartilage template is remodelled into a bone. Following this biological pathway, implantation of in vitro created transient cartilage gives rise to a bone both in an ectopic mouse model as in an orthotopic long bone segmental defect model (Herberg et al., 2019; Mcdermott, 2019; Nilsson Hall et al., 2019). Hence, the next stage is to investigate and translate this approach further toward clinical application and ultimately larger-scale industrial production.
Much effort has been put in choosing the appropriate cell source (Roberts et al., 2015; Groeneveldt et al., 2020) and growth factor combination (Mendes et al., 2016) to generate cartilage-intermediate tissues which mature further post-implantation and eventually remodel into a bone by interaction with the host environment. However, the differentiation medium is often a combination of experimentally optimized and empirically defined components. Currently, cell expansion and differentiation medium formulations heavily rely on animal-derived supplements such as complete FBS or bovine serum-derived albumin (BSA) with variable composition and high batch-to-batch variability (Zheng et al., 2006; Francis, 2010; Gay et al., 2011). The variability associated with FBS results in an overall inconsistent reagent and thereby introduces variability between experimental results (Gstraunthaler et al., 2013). Although substantial work has been carried out for the use of serum-free media for the expansion of adult progenitor cells such as bone marrow-derived mesenchymal stromal cells (BM-MSCs) (Chase et al., 2010; Martin et al., 2015; Allen et al., 2019; Bhat et al., 2021), there is scarce literature on xeno-free media for chondrogenic differentiation (Fitzsimmons et al., 2004; Kawata et al., 2019; Lach et al., 2019; Horikoshi et al., 2021). Moreover, the presence of FBS has been shown to negatively affect chondrogenic differentiation (Fitzsimmons et al., 2004; Lee et al., 2009). Nonetheless, its main component, bovine serum–derived albumin, continues to be the most prominent protein component of chondrogenic media formulations, making up 99.9% of all total added proteins (Mendes et al., 2016). The albumin protein contains hydrophobic patches that are used to solubilize and increase the bioavailability of fatty acids such as linoleic acid (LA), yet albumin has many more functions that can influence cell and tissue behavior (Francis, 2010). Recently, the role of fatty acids was shown to be inhibiting chondrogenic differentiation (van Gastel et al., 2020). Not only is the composition of BSA inherently variable but also the regulatory aspect of implanting a tissue that contains both human and animal components becomes more complex as it brings a risk of xenoimmunization and xenotransmission (Dessels et al., 2016). In addition, the need for non-invasive quality controls to monitor the biological status of the cell-based ATMP is generally accepted, the maturation of living tissues coincides with it, and is directly controlled by the secretion of signaling proteins (Maes, 2017). Yet, the information from the secreted proteome is masked by the presence of high-abundant and large proteins (Shin et al., 2015; Shin et al., 2019; Das et al., 2020). While there are multiple methods available for serum replacements (Karnieli et al., 2017; Ylostalo et al., 2017), we instead opted for a general removal of BSA without replacement.
In this study, we perform an in-depth comparison of transcriptional changes during in vitro differentiation of human periosteum–derived cell (hPDCs) aggregates into cartilaginous microtissues cultured in a BSA- and LA-containing medium versus a xeno-free equivalent medium. Subsequently, the bone-forming potential of these microtissues assembled into larger meso-tissues structures was assessed ectopically in vivo.
Material and methods
Cell expansion
Human periosteum–derived cells (hPDCs) were isolated from periosteal biopsies of six different donors of which one cell pool was created (ages of 20–32) as previously described (Eyckmans et al., 2010). The hPDCs were expanded until passage 7 at 37°C, 5% CO2, and 95% humidity in Dulbecco’s modified Eagle medium (DMEM, Gibco, UK) with 10% fetal bovine serum (South Africa FBS, BioWest, France) and 1% antibiotic–antimycotic solution (Invitrogen, United Staes). The medium was changed every 3–4 days. At a confluence of 80%, cells were harvested with TrypLE Express (Life Technologies, UK). The Ethical Committee for Human Medical Research (Katholieke Universiteit Leuven) approved all procedures, and patients’ informed consent forms were obtained (ML7861).
Microtissue formation
A microwell platform (AggreWell™800, STEMCELL Technologies Inc, Canada) was coated with anti-adherence rinsing solution (STEMCELL Technologies Inc, Canada) to avoid cell attachment. Homogenous microwell coating was ensured by centrifugation, followed by a washing step with a basal medium before cell seeding. After eight passages, hPDCs were harvested with TrypLE Express (Life Technologies, UK) and seeded at 300,000 cells per well to contain 1,000 cells per microwell upon sedimentation. The cells self-aggregated and were differentiated for 21 days. In total, two serum-free, chemically defined chondrogenic media were compared. Both contain LG-DMEM (Gibco) supplemented with 1% antibiotic–antimycotic (Invitrogen, United States), 1 M × 10−3 M ascorbate-2 phosphate, 1 M × 10−7 M dexamethasone, 40 µg mL−1 L-proline, 20 M × 10−7 M of Rho- kinase inhibitor Y27632 (Axon Medchem), 100 ng ml−1 BMP2 (INDUCTOS), 100 ng ml−1 GDF5 (PeproTech), 10 ng ml−1 TGF-β1 (PeproTech), 1 ng ml−1 BMP-6 (PeproTech), and 0.2 ng ml−1 basic FGF-2 (R&D systems). xfCM was then supplemented with ITS Premix Universal Culture Supplement (containing 6.25 µg ml−1 insulin, 6.25 µg ml−1 transferrin, and 6.25 ng ml−1 selenious acid, Corning), while CM was supplemented with ITS + Premix Universal Culture Supplement (containing 6.25 µg ml−1 insulin, 6.25 µg ml−1 transferrin, 6.25 ng ml−1 selenious acid, 5.35 µg ml−1 linoleic acid, and 1,250 µg ml−1 bovine serum albumin to solubilize the linoleic acid, Corning). Half of the medium was changed on days 3, 7, 10, 14, and 17.
Image analysis
On days 7, 14, and 21 bright-field images were taken to assess microtissue movement and morphological appearance (n = 3 per time point). Fiji (ImageJ) CellCounter plugin was used to quantify microtissues that were in their microwells, floating outside their microwells, or fused into irregular shapes as well as the number of empty wells. The microtissue size was measured by fitting ovals over microtissues from four different wells through Fiji (ImageJ).
In vivo ectopic implantation
Custom round-bottom macrowells were created in 3% agarose (w/v) (Invitrogen) and sterilized using UV. Microtissues were gently flushed out from their microwells on day 21, concentrated, and added to the macrowells. CM or xfCM was added and the microtissues were incubated for 24 h at 37°C, 5% CO2, and 95% humidity to spontaneously fuse into one construct. Implants were removed from the agarose macrowell and washed in 1xDPBS. Then four implants per condition were implanted subcutaneously at the shoulder region of female immune-compromised mice (Rj:NMRInu/nu age 6–20 weeks). Each mouse had two implants, one for each condition and randomized left or right shoulder. Regarding the surgical procedure, after skin disinfection, a small incision was made on the back of the mouse under general anesthesia (ketamine/xylazine). In addition, two pockets were created at the shoulder region, each construct was placed in one pocket, and the skin was closed with an appropriate number of staples followed by postoperative administration of buprenorphine as painkiller. After 4 weeks, the mice were sacrificed by cervical dislocation and implants were extracted and fixed for 4 h in 4% PFA. All procedures on animal experiments were approved by the Local Ethical Committee for Animal Research, KU Leuven. The animals were housed according to the regulations of the Animalium Leuven (KU Leuven).
Nano-CT
3D quantification of mineralized tissue in PFA-fixed explants was performed through nano-CT (Phoenix Nanotom M, GE Measurement, and Control Solutions). Explants were scanned with a diamond target, mode 0, 500 ms exposure time, 1 frame average, 0 image skip, 2,400 images, and a 0.1 mm aluminum filter. Samples were scanned with 60 kV and 140 µA. CTAn (Bruker micro-CT, BE) was used for all image processing and quantification of mineralized tissue based on automatic Otsu segmentation, 3D space closing, and despeckle algorithm. The percentage of mineralized tissue was calculated with respect to the total explant volume. CTvox (Bruker micro-CT, BE) was used to create 3D visualization.
Histology and immunohistochemistry
Microtissues were flushed out, fixed in 2% PFA overnight, mixed in 3% agarose, dehydrated, and embedded in paraffin overnight. Ectopic explants were fixed in 4% PFA for 4 h, decalcified in ethylenediaminetetraacetic acid in PBS (pH 7.5) for 10 solution changes at 4°C, dehydrated, embedded in paraffin overnight, and sectioned at 5 µM thickness.
For safranin O (Sigma) staining, sections were deparaffinized and dehydrated, counterstained with hematoxylin (Merck, cat 6,525) for 1 min, briefly dipped in acid alcohol (1% HCL in 70% EtOH), rinsed in water, stained with 0.03% Fast Green (KLINIPATH, cat 80,051), and then dipped in 1% glacial acetic acid followed by 7-min staining in 0.25% safranin O (KLINIPATH, cat 640,780). Then the samples were washed in tap water, dehydrated with an ethanol series, cleared in Histoclear, and mounted on Pertex for microscopy imaging. Explant quantification of cartilage, bone marrow compartment, and bone regions was done through Ilastik pixel classification (Berg et al., 2019) on three different sections of each explant, with four explants per condition. The explant maturation score was calculated as follows:
For Collagen type 2 a1 immunostaining, sections were deparaffinized, followed by 15 min antigen retrieval using 1 mg/ml pepsin in 0.2 M HCL. Quenching was performed in 3% H2O2 for 10 min, followed by 30 min of blocking in 5% BSA in PBST. The primary anti-collagen 2 antibody (Merck Millipore, AB761, dilution 1:20) was incubated overnight. Samples were blocked for 40 min in 5% BSA in PBST and incubated for 30 min with a secondary HRP-conjugated antibody (Jackson ImmunoResearch, 111-035-003, dilution 1:500). Staining was performed using 3,3′-diaminobenzidine (DAB) (K3468, Dako, United Staes).
DNA and RNA analysis
In total, 300 pooled microtissues (n = 4) were gently flushed out from their microwells at different time points and centrifuged to separate microtissues from the conditioned medium. The conditioned medium was aliquoted, frozen in liquid nitrogen, and stored at −80°C. Microtissues were lysed in 350 µL RLT lysis buffer (Qiagen, Germany) supplemented with 3.5 µL β-mercaptoethanol (Sigma Aldrich, Germany), vortexed, and stored at −80°C. The Quant-iT dsDNA HS kit (Invitrogen) was used to quantify the DNA content from cell lysate, according to the manufacturer’s protocol. RNA was isolated from the lysate using the RNeasy Mini Kit (Qiagen), according to the manufacturer’s protocol, and quantified using NanoDrop 2000 (Thermo Fisher Scientific). cDNA libraries were generated using the Lexogen QuantSeq FWD Sample Preparation Kit. Libraries were sequenced on the Illumina HiSeq4000 sequencing system.
Bioinformatic analyses
Single-strand sequences were aligned to the Homo sapiens GRh37 reference genome using STAR v.2.7.9a (Dobin et al., 2013). Gene counts were obtained using the Subread package (Liao et al., 2013). DESeq2 v.1.26.0 (Love et al., 2014) was used to normalize the data and determine differentially expressed genes (based on log-fold change > 1 and adjusted p-value < 0.05). Differential expression was calculated using the likelihood ratio test as outlined in the DESeq2 documentation (Love et al., 2014).
Following the likelihood ratio test, the 500 most significant genes were selected and subdivided into up genes in CM or xfCM. For each set, an enrichment analysis of biological_processes_2021 and KEGG_pathways_2021 was performed through EnrichR (Kuleshov et al., 2016) to select relevant databases and significant gene ontology terms (adjusted p-value < 0.05). The time lapsed enrichment scores for gene ontology terms of interest were calculated as described by Tirosh et al. (2016). Here, the samples were scored based on the average normalized expression of the genes within a gene set of interest. From this, the enrichment score of randomly selected control genes with similar expression levels are subtracted. These control gene sets were defined by first binning all genes into 24 bins of aggregate expression level and then, for each gene in the gene set of interest, randomly selecting 100 genes from the same expression bin as that gene. In this way, the control gene sets have a comparable distribution to the gene set of interest and the control gene set is 100 times larger, such that the score for the gene set of interest is analogous to averaging over 100 randomly selected gene sets as the gene set of interest. The score was set to range from zero, meaning no enrichment compared to random sets of genes with similar expression, to one, reflecting the highest average expression of all genes within the gene set of interest. This is a repurposing of the AddModuleScore() function found in Seurat v3 (Stuart et al., 2019) toward bulk RNA-seq data processed with DESeq2.
Statistics
All statistical analyses were performed using standard functions in R (R core team). Statistical significance was defined at p < 0.05. Comparing two means was done through a two-sided, unpaired t-test. For non-normal data, a Wilcoxon test was performed. For multiple comparisons, a two-way ANOVA followed by Tukey’s post hoc test was used. Data is presented as mean and standard deviation from four samples. Symbols used are *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
Results
Xeno-free medium generates smaller but less motile and more homogenous microtissues
Here, we differentiated hPDCs as 3D microtissues in an established chondrogenic medium (Mendes et al., 2016; Nilsson Hall et al., 2019; Nilson Hall et al, 2021) (CM) versus a xeno- and lipid-free chondrogenic medium (xfCM) containing the same growth factors, but no bovine serum or fatty acids. We used the commercially available AggrewellTM800 24-well platform where we dropseeded 1,000 cells per microtissue at 150,000 cells/mL.
As seen in Figure 1, microtissues cultured in both media have distinct morphological characteristics during long-term culture. Compared to CM, xeno-free microtissues are smaller, less variable in terms of size (Figure 1G) on day 7, and contain fewer cells as seen by both DNA and RNA content (Figures 1B,C). However, there is more DNA per area, indicating a lower amount of extracellular matrix per cell (CM: 0.0509 ± 0.008 ng/µm2 versus xfCM: 0.0748 ± 0.009 ng/µm2, p = 0.008) (Supplementary Figure S1).
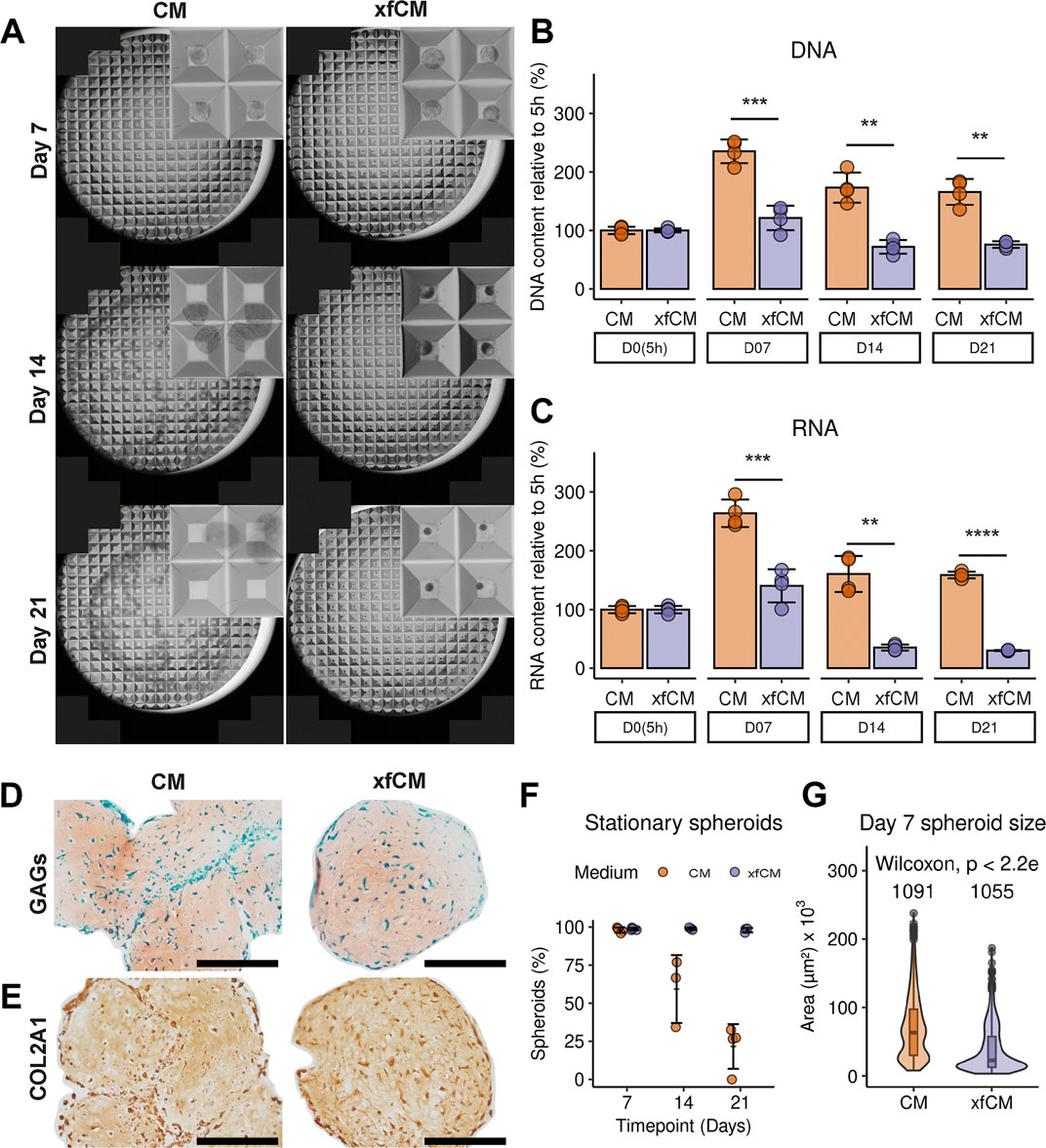
FIGURE 1. Microtissue culture in CM versus xfCM. (A) Bright-field microscopy full-well and detail images of microtissues on day 7, 14, and 21. Each microwell is 800 µM. (B,C) DNA and RNA content of one well, containing approximately 300 microwells, normalized to 5 h. (D) GAG deposition in day 21 microtissues determined by SafO/FG/H staining. (E) Collagen type 2 deposition in day 21 microtissues. (F) Percentage of microtissues that remained in their microwells over time. (G) Size distribution of microtissues on day 7. Data is presented as mean ± standard deviation. Scalebar = 200 µM.
During 21 days of culture, 98 ± 1.5% of the microtissues cultured in xfCM remained in their microwells, resulting in a homogenous microtissue population. In contrast, microtissues cultured with BSA and LA grew larger during the first week and by day 21 only 21 ± 15% remained in their microwells, whereas the remainder spontaneously fused into larger tissues (Figure 1A,D–F).
The histological analysis shows the production of an extracellular matrix (ECM) in both conditions with deposition of glycosaminoglycans (GAGs) (Figure 1D) as well as collagen type 2 (Figure 1E). Microtissues in xfCM are more homogenous in their GAG distribution compared to CM microtissues. However, microtissues in CM deposit more ECM per cell (Supplementary Figure S1).
Both chondrogenic media generate in vivo ectopic bone ossicles
The two media formulations are chondrogenic media containing the same experimentally defined growth factors. The combination of BMP2, BMP6, GDF5, bFGF2, and TGF-β1 has been validated on multiple occasions (Mendes et al., 2016; Nilsson Hall et al., 2019). Also, the addition of ROCK inhibitor (Woods et al., 2005; Mendes et al., 2016), ascorbic acid (Heng et al., 2004; Akiyama et al., 2006; Temu et al., 2010), dexamethasone (Heng et al., 2004; Langenbach and Handschel, 2013), L-proline (Patriarca et al., 2021), and ITS (Fitzsimmons et al., 2004; Liu et al., 2014) have proven beneficial effects on the differentiation potential and health of periosteum-derived cells. As we saw significant changes in microtissue morphology, we needed to investigate whether our adaptations to the chondrogenic medium formulations preserved the bone formation capacity upon implantation in vivo. Microcomputed tomography (µCT) showed explants from both conditions with a full or partially mineralized cortical shell, as well as some mineralized regions inside an unmineralized internal space (Figures 2A,B). There was no difference in the percentage of mineralization or the thickness of mineralized tissue, but explants from CM were more variable and larger compared to xfCM explants (Figures 2C–E). Through histochemical staining with safranin O/Fastgreen/Haematoxilin, we could see full bone marrow compartments, including some remains of mineralized cartilage and trabecular bone. Quantification of those regions showed no differences, which leads to identical ossicle maturation scores.
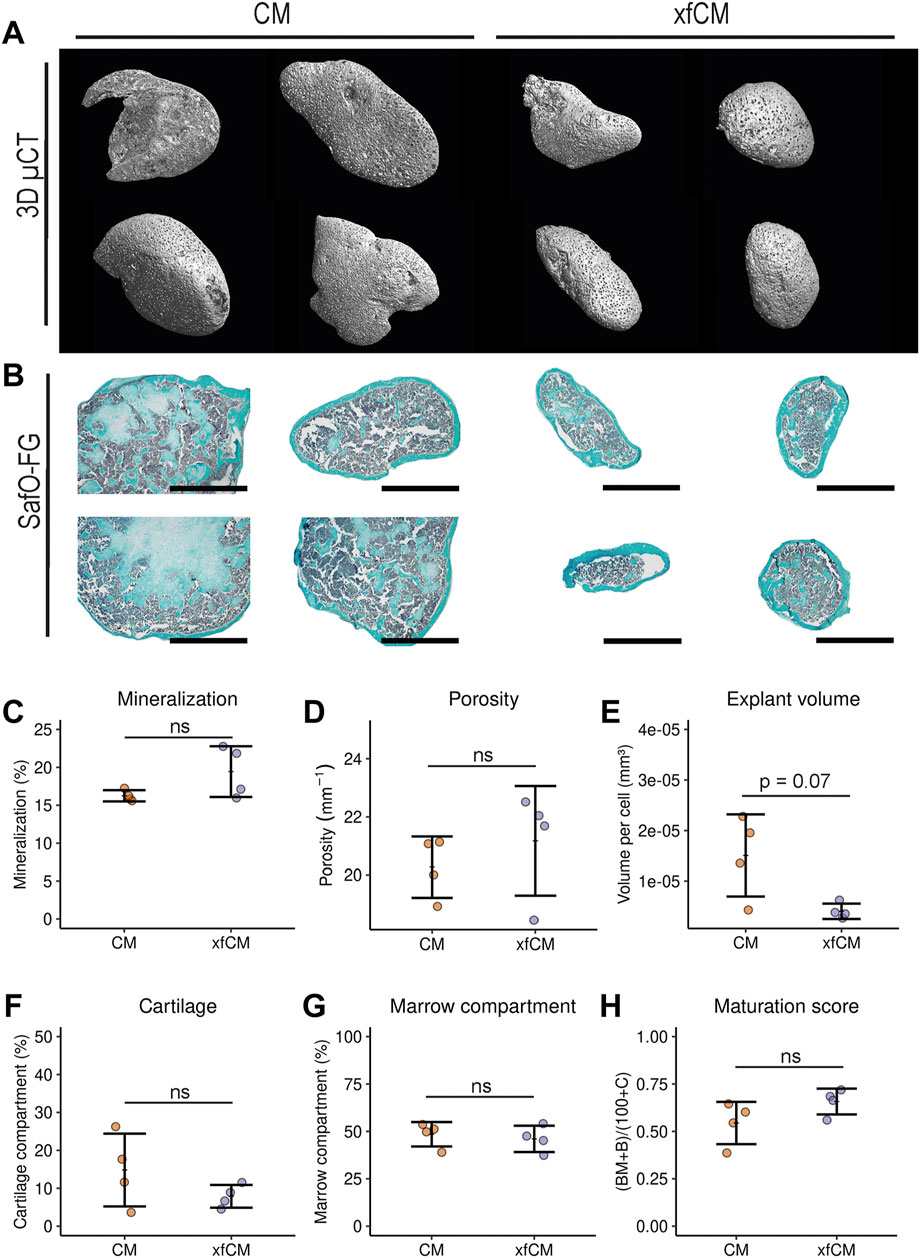
FIGURE 2. Ectopic implantations of bone-forming microtissues. (A) 3D renderings of 4-week ectopic explants by nano computed tomography (nano-CT). (B) Saf O/FG/H histology staining of 4-week ectopic explants after decalcification. (C–E) Percentage mineralization, porosity as 1/mineralized thickness and explant volume relative to DNA content derived from nano-CT quantification. (F,G) Percentage of cartilage tissue and bone marrow component per area as determined by SafO/FG/H histological staining. (H) Ossicle maturation score calculated from SafO/FG/H quantifications. Data are presented as mean ± standard deviation. Scalebar = 1 mm.
Bulk RNA sequencing reveals different ratios of endochondral and intramembranous ossification
While the quality of engineered bone seems similar for both conditions, it can be generated through a combination of direct and indirect ossification (Colnot, 2009). In fracture repair, a broad distinction is made between a non-displaced fracture and a full fracture with high displacement. Full fractures are more complex, less stable, and have a strong initial inflammatory phase leading to proliferation and the onset of endochondral ossification to heal the bone defect through a cartilage template. In contrast, non-displaced fractures are less complex, more stable, and can typically be healed to a higher degree through direct or intramembranous ossification. While the reality is often somewhere in between, the two extremes have distinct transcriptional signatures (Coates et al., 2019).
We performed bulk RNA sequencing of microtissues cultured in CM and xfCM at 5 h, 7, 14, and 21 days of differentiation. The 500 most differentially expressed genes (DEGs) between both conditions at any time point compared to the start of the treatment were divided in those upregulated in CM or xfCM. Gene ontology and KEGG pathway enrichment analysis in CM (Figures 3A,B) showed a high amount of glycolytic metabolism-related terms, terms related to hypoxia and angiogenesis, glycosaminoglycan synthesis, and cholesterol metabolism: all previously found in full fracture models (Tsiridis and Giannoudis, 2006; Kolar et al., 2011; Coates et al., 2019). Interestingly, CM was also enriched in steroid and estrogen pathways as well as protein digestion and absorption pathways.
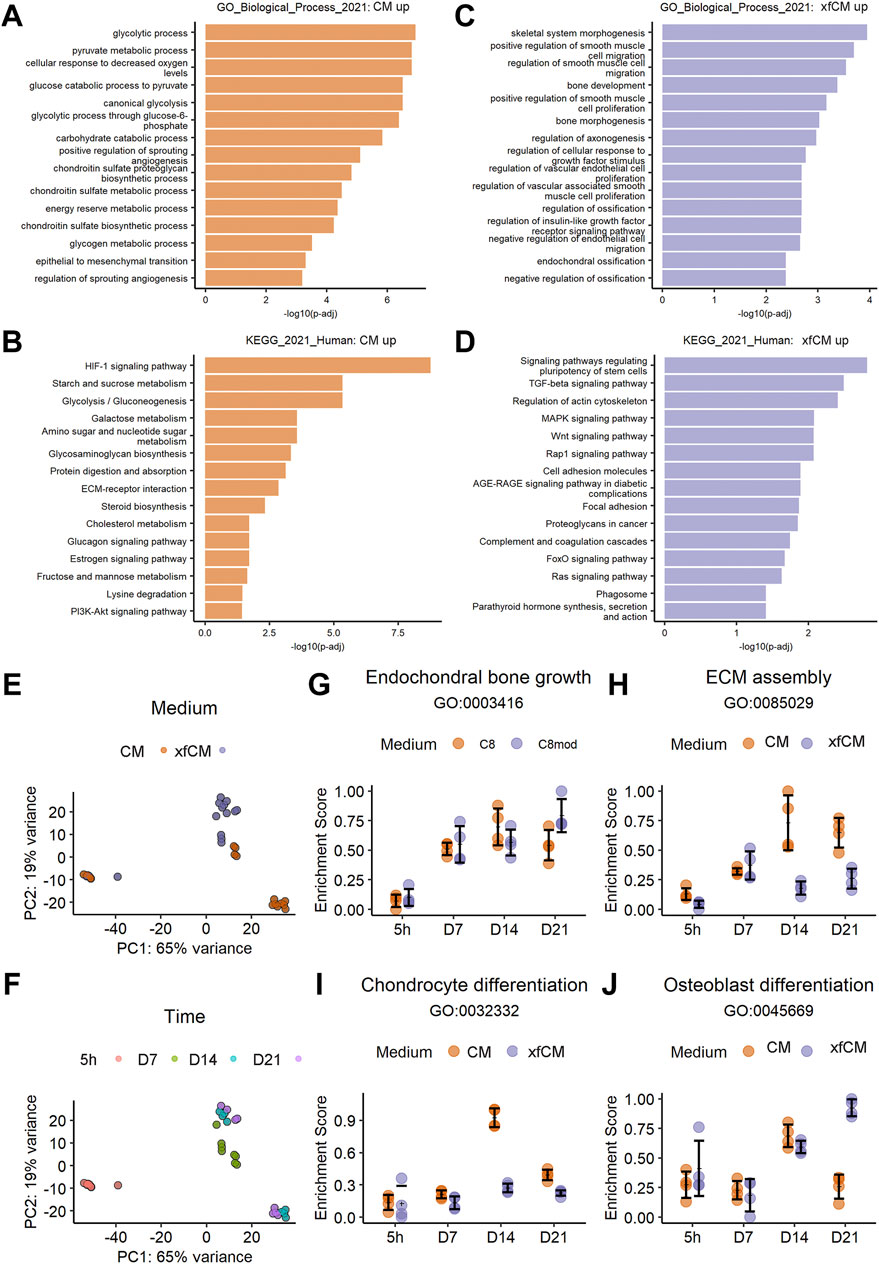
FIGURE 3. RNA sequencing of bone-forming microtissues during differentiation. (A,B) Gene ontology, biological process, and KEGG pathway terms upregulated in CM in the 500 most variable genes. (C,D) Gene ontology, biological process, and KEGG pathway terms upregulated in xfCM in the 500 most variable genes. (E,F) Principle component analysis performed on the full transcriptome of microtissues in CM and xfCM at 5 h, 7, 14, and 21 days. (G–J) Temporal gene set scoring on the full transcriptome over time for genes grouped in a given GO term. Data are presented as mean ± standard deviation.
In xfCM (Figures 3C,D), gene Ontology enrichment analysis showed more terms related to bone development and endochondral ossification, but also smooth muscle cell recruitment. KEGG pathway enrichment showed increased Wnt, PTH, and AGE-RAGE signaling, supporting osteoblast maturation, accelerated fracture healing, and osteoclast activation (Einhorn and Gerstenfeld, 2015; Wang et al., 2017a; Plotkin et al., 2019). Also, in xfCM, which is also deprived of linoleic acid, the FOXO signaling pathway is enriched (van Gastel et al., 2020).
The enrichment analysis is typically performed on a selection of variable genes for single timepoints or to directly compare conditions. However, microtissues created in CM or xfCM show a time-dependent phenotypical difference that increased over time (Figure 1). Therefore, we performed a principal component analysis (PCA) (Figures 3E,F) showing near-identical gene expression at the early 5 h timepoint. By day 7, the conditions start to differ and branch out, leading to high transcriptional differences on day 14 and day 21.
To evaluate the time dependence of gene ontology terms and KEGG pathways, we adjusted the geneset scoring system62. Here, the samples were scored based on the normalized expression of each gene in a gene set of interest. The score was set to range from zero, meaning no enrichment compared to random sets of genes with similar expression, to one, reflecting the highest average expression of all genes within the gene set of interest.
As shown in Figures 3G–J, time-series geneset scoring shows that indeed few differences are found before day 7, and both conditions show a stable increase of endochondral bone growth genes. Yet, on day 14, CM microtissues have a peak of chondrocyte differentiation, osteoblast differentiation, and ECM assembly genes that decreases again towards day 21. In contrast, microtissues in xfCM decrease their ECM assembly genes earlier from day 7 and have less chondrocyte differentiation at day 14, but a higher score of osteoblast differentiation genes.
Discussion
Developmental engineering strategies employ microtissues as building blocks for the engineering of larger living and functional tissue implants. They show promising advances, yet the manufacturing of cell-based tissue-engineered products is hampered by the complexity of the protocols including the use of animal components during cell differentiation. In this study, we compared an established chondrogenic differentiation medium (CM) to a xeno- and lipid-free chondrogenic medium (xfCM) containing the same growth factors, but no bovine serum albumin or fatty acids.
Culture in CM resulted in batches with a less homogenous size distribution of microtissues as compared to the condition cultured in xfCM. Microtissues cultured in CM were larger and tended to move out of their microwells and their containment when cultured longer than 7 days. As time progressed, the yield of single, non-fused, microtissues decreased. Instead, in many batches, single spheroids gave way to large, irregular-shaped tissues by spontaneous fusion. In contrast, xfCM microtissues were smaller, more homogenous, and resulted in an overall yield of 98 ± 1.5% microtissues that remained in their microwell for 21 days. This is an interesting observation that could have implications on avoiding batch failures especially when scale-up of this process is taken into account. Uncontrolled fusion of microtissues might affect the diffusion of oxygen, nutrients, and growth factors (Murphy et al., 2017; Nilson Hall et al, 2021) affecting the quality of the resulting tissue modules and the critical quality profile of the implant as an end tissue product (Mosaad et al., 2018).
The DNA and RNA increase during the first 7 days was significantly lower in xfCM, and the DNA to area ratio was higher. Our results indicate a lower stimulation of cell proliferation resulting in a lower cell number per batch. This can be explained by a difference in nutrient availability as linoleic acid and albumin can serve as a source of energy and amino acids. Alternatively, albumin depletion can decrease the bioavailability of cytokines that have a short half-life or are poorly soluble (Kumorek et al., 2015). In addition, we observed a higher amount of ECM per cell present in microtissues grown in the CM medium (Prein et al., 2016; Sarem et al., 2019). Yet, the extracellular matrix contains both GAGs and collagen type 2 in both conditions, thus confirming the creation of a cartilaginous structure. Chondrocytes do not depend on exogenous lipids, but osteoblasts and progenitor cells do have active fatty acid oxidation (van Gastel et al., 2020). Therefore, a high lipid availability in CM could induce an early activation of the progenitor cell population leading to more proliferation and larger microtissues, while xfCM retains this cell population that later contributes to bone formation in vivo. Despite the differences observed in terms of in vitro quality aspects of the cartilaginous organoids, both protocols generated implants that were able to form ossicles upon subcutaneous implantation. The quality of the formed bone is excellent in both cases, with the presence of critical components such as the cortical bone, trabecular bone, and large vascularized marrow compartments. In both cases, no contaminating fibrous tissue compartment was observed but low amounts of remaining cartilage tissue were observed. This indicates a high degree of cartilage to bone transition and remodeling, leading to high maturation scores (Nilson Hall et al, 2021; Pigeot et al., 2021).
The transcriptome analysis of microtissues during differentiation shows that both chondrogenic media induce “endochondral bone growth”, but as time progresses xfCM spheroids showed a higher degree of osteogenic differentiation while CM tissues show a longer chondrogenic differentiation and maturation period. A longer chondrogenic period would lead to more ECM production, and thus to larger implants. This trend was confirmed by the sequencing data, showing higher chondrocyte differentiation at day 14, and overall higher ECM assembly at day 14 and 21 in CM spheroids.
These differences could be also attributed to the cellular composition of the initial hPDC population, which is a cell population containing osteo- and chondroprogenitors as well as other cellular subtypes (Stich et al., 2017; Duchamp De Lageneste et al., 2018; Bolander et al., 2020). Potentially the lack of albumin and fatty acids in the differentiation media might affect the differentiation kinetics or composition of differentiated cells in the end tissue product. Both linoleic acid and bovine serum albumin can have a multitude of effects on cellular signaling, growth factor bioavailability, and nutrient availability that can lead to the observed effects (Pereira et al., 2008; Francis, 2010; Jung et al., 2012; Bastiaansen-Jenniskens et al., 2013; Villalvilla et al., 2013; van Gastel et al., 2020). We hypothesized that the different media might promote different subpopulations in the inherently variable starting population of autologous periosteum-derived cells, including osteo- and chondroprogenitors as well as other cellular subtypes (Stich et al., 2017; Duchamp De Lageneste et al., 2018). Small signaling proteins are notoriously instable (Ren et al., 2020), so BSA could increase the duration of bioavailability of added growth factors. CM could then promote a stronger component of proliferating chondroprogenitor cells that produce more ECM. A lower availability in xfCM would then lead to less proliferation in the chondroprogenitors and a relatively higher ratio of osteoprogenitors. As a result, xfCM promotes the production of a smaller quantity of similarly composed ECM. Therefore, the necessary requirements for ECM composition and cellular components to create ectopic bone through endochondral bone formation are met in both conditions. This difference could potentially be compensated by optimizing the use of growth factor availability through recombinant technologies (Ren et al., 2020) or alternative release technologies (Wang et al., 2017b).
The use of xfCM decreases the matrix volume output per cell, but it also increases protocol robustness and building block homogeneity. Furthermore, it allows the integration of on-line non-invasive quality controls for both the manufacturing process through bright-field imaging and the biological process through the investigation of the secreted proteome with a high signal to noise ratio (Power et al., 2020; Wang et al., 2022). While the creation of adequate engineered tissue volumes is a recognized challenge in tissue engineering (De Pieri et al., 2021), this robust bottom-up approach could be compatible in the future with a controlled automated manufacturing process (Heathman et al., 2015; Doulgkeroglou et al., 2020).
In conclusion, we envisage that this xeno- and lipid-free chondrogenic medium is compatible with industrial scale-out and automation, while facilitating the implementation of non-invasive quality control mechanisms related to imaging and protein quantification.
Data availability statement
The raw and processed RNA sequencing data are deposited at the Gene Expression Omnibus under accession code GSE198914. The code for our scoring function and notebooks demonstrating its application can be found on the GitHub page: https://github.com/HerpelinckT/geneset-modulescoring.
Ethics statement
The animal study was reviewed and approved by the Local Ethical Committee for Animal Research, KU Leuven. The ethical committee for Human Medical Research (Katholieke Universiteit Leuven) approved all procedures, and patients’ informed consent forms were obtained (ML7861).
Author contributions
ID contributed to the design, data acquisition, and analysis of the study and drafted and revised the manuscript. TH contributed to the design, data acquisition, and analysis of the study and revised the manuscript. LG and FL contributed to the manuscript revision. IP contributed to the conceptualization and design of the study and revision of the manuscript.
Funding
The authors gratefully acknowledge support from the KU Leuven R&D in the framework of the IMEC-KU Leuven dual core collaboration. The micro (or nano)-CT images were generated on the X-ray computed tomography facility of the Department of Development and Regeneration of the KU Leuven, financed by the Hercules Foundation (project AKUL/13/47). TH is supported by a fellowship of the Research Foundation Flanders (FWO Vlaanderen, project 1S80021N). The project leading to this publication has received funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement No 874837. This work was supported by the Flemish Government (department of Economy, Science and Innovation) through the Regenerative Medicine Crossing Borders (http://www.regmedxb.com) initiative.
Acknowledgments
We thank Kathleen Bosmans for performing the in vivo mice experiments. This work was performed in the context and with the support of members of Prometheus, the KU Leuven R&D division for skeletal tissue engineering (http://www.kuleuven.be/prometheus).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fceng.2022.892190/full#supplementary-material
References
Akiyama, M., Nonomura, H., Kamil, S. H., and Ignotz, R. A. (2006). Periosteal cell pellet culture system: a new technique for bone engineering. Cell. Transpl. 15, 521–532. doi:10.3727/000000006783981765
Allen, L. M., Matyas, J., Ungrin, M., Hart, D. A., and Sen, A. (2019). Serum-free culture of human mesenchymal stem cell aggregates in suspension bioreactors for tissue engineering applications. Stem Cells Int. 2019, 1–18. doi:10.1155/2019/4607461
Bastiaansen-Jenniskens, Y. M., Siawash, M., van de Lest, C., Verhaar, J., Kloppenburg, M., Zuurmond, A. M., et al. (2013). Monounsaturated and saturated, but not n-6 polyunsaturated fatty acids decrease cartilage destruction under inflammatory conditions: a preliminary study. Cartilage 4, 321–328. doi:10.1177/1947603513494401
Berg, S., Kutra, D., Kroeger, T., Straehle, C. N., Kausler, B. X., Haubold, C., et al. (2019). ilastik: interactive machine learning for (bio)image analysis. Nat. Methods 16, 1226–1232. doi:10.1038/s41592-019-0582-9
Bhat, S., Viswanathan, P., Chandanala, S., Prasanna, S. J., and Seetharam, R. N. (2021). Expansion and characterization of bone marrow derived human mesenchymal stromal cells in serum-free conditions. Sci. Rep. 11, 3403–3418. doi:10.1038/s41598-021-83088-1
Bolander, J., Herpelinck, T., Chaklader, M., Gklava, C., Geris, L., and Luyten, F. P. (2020). Single-cell characterization and metabolic profiling of in vitro cultured human skeletal progenitors with enhanced in vivo bone forming capacity. Stem Cells Transl. Med. 9, 389–402. doi:10.1002/sctm.19-0151
Burdis, R., and Kelly, D. J. (2021). Biofabrication and bioprinting using cellular aggregates, microtissues and organoids for the engineering of musculoskeletal tissues. Acta Biomater. 126, 1–14. doi:10.1016/j.actbio.2021.03.016
Nilson Hall, G., Rutten, L., and Lammertyn, J. (2021). Cartilaginous spheroid-assembly design considerations for endochondral ossification: towards robotic-driven biomanufacturing. Biofabrication. doi:10.1088/1758-5090/ac2208
Cervantes-Diaz, F., Contreras, P., and Marcellini, S. (2017). Evolutionary origin of endochondral ossification: the transdifferentiation hypothesis. Dev. Genes. Evol. 227, 121–127. doi:10.1007/s00427-016-0567-y
Chase, L. G., Lakshmipathy, U., Solchaga, L. A., Rao, M. S., and Vemuri, M. C. (2010). A novel serum-free medium for the expansion of human mesenchymal stem cells. Stem Cell. Res. Ther. 1, 8–11. doi:10.1186/scrt8
Coates, B. A., McKenzie, J. A., Buettmann, E. G., Liu, X., Gontarz, P. M., Zhang, B., et al. (2019). Transcriptional profiling of intramembranous and endochondral ossification after fracture in mice. Bone 127, 577–591. doi:10.1016/j.bone.2019.07.022
Colnot, C. (2009). Skeletal cell fate decisions within periosteum and bone marrow during bone regeneration. J. Bone Min. Res. 24, 274–282. doi:10.1359/jbmr.081003
Das, L., Murthy, V., and Varma, A. K. (2020). Comprehensive analysis of low molecular weight serum proteome enrichment for mass spectrometric studies. ACS Omega 5, 28877–28888. doi:10.1021/acsomega.0c04568
De Moor, L., Fernandez, S., Vercruysse, C., Tytgat, L., Asadian, M., De Geyter, N., et al. (2020). Hybrid bioprinting of chondrogenically induced human mesenchymal stem cell spheroids. Front. Bioeng. Biotechnol. 8, 484. doi:10.3389/fbioe.2020.00484
De Pieri, A., Rochev, Y., and Zeugolis, D. I. (2021). Scaffold-free cell-based tissue engineering therapies: advances, shortfalls and forecast. npj Regen. Med., 18–15. doi:10.1038/s41536-021-00133-3
Dessels, C., Potgieter, M., and Pepper, M. S. (2016). Making the switch: alternatives to fetal bovine serum for adipose-derived stromal cell expansion. Front. Cell. Dev. Biol. 4, 115. doi:10.3389/fcell.2016.00115
Dobin, A., Davis, C. A., Schlesinger, F., Drenkow, J., Zaleski, C., Jha, S., et al. (2013). Star: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. doi:10.1093/bioinformatics/bts635
Doulgkeroglou, M. N., Di Nubila, A., Niessing, B., Konig, N., Schmitt, R. H., Damen, J., et al. (2020). Automation, monitoring, and standardization of cell product manufacturing. Front. Bioeng. Biotechnol. 8 811. doi:10.3389/fbioe.2020.00811
Duchamp De Lageneste, O., Julien, A., Abou-Khalil, R., Frangi, G., Carvalho, C., Cagnard, N., et al. (2018). Periosteum contains skeletal stem cells with high bone regenerative potential controlled by Periostin. Nat. Commun. 919, 773–815. doi:10.1038/s41467-018-03124-z
Einhorn, T. A., and Gerstenfeld, L. C. (2015). Fracture healing: Mechanisms and interventions. Nat. Rev. Rheumatol. 11, 45–54. doi:10.1038/nrrheum.2014.164
Eyckmans, J., Roberts, S. J., Schrooten, J., and Luyten, F. P. (2010). A clinically relevant model of osteoinduction: a process requiring calcium phosphate and BMP/wnt signalling. J. Cell. Mol. Med. 14, 1845–1856. doi:10.1111/j.1582-4934.2009.00807.x
Fitzsimmons, J. S., Sanyal, A., Gonzalez, C., Fukumoto, T., Clemens, V. R., O'Driscoll, S. W., et al. (2004). Serum-free media for periosteal chondrogenesis in vitro. J. Orthop. Res. 22, 716–725. doi:10.1016/j.orthres.2003.10.020
Francis, G. L. (2010). Albumin and mammalian cell culture: implications for biotechnology applications. Cytotechnology 62, 1–16. doi:10.1007/s10616-010-9263-3
Gay, M., Pares, A., Carrascal, M., Bosch-i-Crespo, P., Gorga, M., Mas, A., et al. (2011). Proteomic analysis of polypeptides captured from blood during extracorporeal albumin dialysis in patients with cholestasis and resistant pruritus. PLoS One 6, e21850. doi:10.1371/journal.pone.0021850
Groeneveldt, L. C., Herpelinck, T., Marechal, M., Politis, C., van IJcken, W. F. J., Huylebroeck, D., et al. (2020). The bone-forming properties of periosteum-derived cells differ between harvest sites. Front. Cell. Dev. Biol. 8, 554984. doi:10.3389/fcell.2020.554984
Gstraunthaler, G., Lindl, T., and Van Der Valk, J. (2013). A plea to reduce or replace fetal bovine serum in cell culture media. Cytotechnology 65, 791–793. doi:10.1007/s10616-013-9633-8
Hall, G. N., Mendes, L., Geris, L., Papantoniou, I., and Luyten, F. (2018). Designing microtissue bioassemblies for skeletal regeneration: healing critical size long bone defects. Cytotherapy 20, S14. doi:10.1016/j.jcyt.2018.02.025
Heathman, T. R., Nienow, A. W., McCall, M. J., Coopman, K., Kara, B., and Hewitt, C. J. (2015). The translation of cell-based therapies: clinical landscape and manufacturing challenges. Regen. Med. 10, 49–64. doi:10.2217/rme.14.73
Heng, B. C., Cao, T., and Lee, E. H. (2004). Directing stem cell differentiation into the chondrogenic lineage in vitro. Stem Cells 22, 1152–1167. doi:10.1634/stemcells.2004-0062
Herberg, S., McDermott, A. M., Dang, P. N., Alt, D. S., Tang, R., Dawahare, J. H., et al. (2019). Combinatorial morphogenetic and mechanical cues to mimic bone development for defect repair. Sci. Adv. 5, eaax2476. doi:10.1126/sciadv.aax2476
Horikoshi, S., Kajiya, M., Motoike, S., Yoshino, M., Morimoto, S., Yoshii, H., et al. (2021). Clumps of mesenchymal stem cells/extracellular matrix complexes generated with xeno-free chondro-inductive medium induce bone regeneration via endochondral ossification. Biomedicines 9, 1408. doi:10.3390/biomedicines9101408
Iglesias-Lopez, C., Obach, M., Vallano, A., and Agustí, A. (2021). Comparison of regulatory pathways for the approval of advanced therapies in the european union and the united States. Cytotherapy 23, 261–274. doi:10.1016/j.jcyt.2020.11.008
Jung, C. H., Lee, W. J., Hwang, J. Y., Seol, S. M., Kim, Y. M., La Lee, Y., et al. (2012). The role of Rho/Rho-kinase pathway in the expression of ICAM-1 by linoleic acid in human aortic endothelial cells. Inflammation 35, 1041–1048. doi:10.1007/s10753-011-9409-2
Karnieli, O., Friedner, O. M., Allickson, J. G., Zhang, N., Jung, S., Fiorentini, D., et al. (2017). A consensus introduction to serum replacements and serum-free media for cellular therapies. Cytotherapy, 19 155–169. doi:10.1016/j.jcyt.2016.11.011
Kawata, M., Mori, D., Kanke, K., Hojo, H., Ohba, S., Chung, U. i., et al. (2019). Simple and robust differentiation of human pluripotent stem cells toward chondrocytes by two small-molecule compounds. Stem Cell. Rep. 13, 530–544. doi:10.1016/j.stemcr.2019.07.012
Kolar, P., Timo, G., Carsten, P., and Georg, D. (2011). Human early fracture hematoma is characterized by inflammation and hypoxia. Clin. Orthop. Relat. Res. 469, 3118–3126. doi:10.1007/s11999-011-1865-3
Kuleshov, M. V., Jones, M. R., Rouillard, A. D., Fernandez, N. F., Duan, Q., Wang, Z., et al. (2016). Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44, W90–W97. doi:10.1093/nar/gkw377
Kumorek, M., Kubies, D., Filova, E., Houska, M., Kasoju, N., Mazl Chanova, E., et al. (2015). Cellular responses modulated by FGF-2 adsorbed on albumin/heparin layer-by-layer assemblies. PLoS One 10, e0125484. doi:10.1371/journal.pone.0125484
Lach, M. S., Wroblewska, J., Kulcenty, K., Richter, M., Trzeciak, T., and Suchorska, W. M. (2019). Chondrogenic differentiation of pluripotent stem cells under controllable serum-free conditions. Int. J. Mol. Sci. 20, 2711. doi:10.3390/ijms20112711
Langenbach, F., and Handschel, J. (2013). Effects of dexamethasone, ascorbic acid and β-glycerophosphate on the osteogenic differentiation of stem cells in vitro. Stem Cell. Res. Ther. 4, 117–7. doi:10.1186/scrt328
Lee, S., Kim, J. H., Jo, C. H., Seong, S. C., Lee, J. C., and Lee, M. C. (2009). Effect of serum and growth factors on chondrogenic differentiation of synovium-derived stromal cells. Tissue Eng. Part A 15, 3401–3415. doi:10.1089/ten.tea.2008.0466
Lenas, P., and Luyten, F. P. (2011). An emerging paradigm in tissue engineering : from chemical engineering to developmental engineering for bioartificial tissue formation through a series of unit operations that simulate the in vivo successive developmental stages. Ind. Eng. Chem. Res. 50, 482–522. doi:10.1021/ie100314b
Lenas, P., Moos, M., and Luyten, F. P. (2009). Developmental engineering : A new paradigm for the design and manufacturing of cell-based products . part I : from three-dimensional cell growth to biomimetics of in vivo development. Tissue Eng. Part B Rev. 15, 381–394. doi:10.1089/ten.teb.2008.0575
Liao, Y., Smyth, G. K., and Shi, W. (2013). The Subread aligner: Fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 41, e108. doi:10.1093/nar/gkt214
Liu, X., Liu, J., Kang, N., Yan, L., Wang, Q., Fu, X., et al. (2014). Role of insulin-transferrin-selenium in auricular chondrocyte proliferation and engineered cartilage formation in vitro. Int. J. Mol. Sci. 15, 1525–1537. doi:10.3390/ijms15011525
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 151215, 550–621. doi:10.1186/s13059-014-0550-8
Maes, C. (2017). Seminars in cell & developmental Biology signaling pathways effecting crosstalk between cartilage and adjacent tissues Seminars in cell and developmental biology : the biology and pathology of cartilage. Semin. Cell. Dev. Biol. 62, 16–33. doi:10.1016/j.semcdb.2016.05.007
Martin, C., Piccini, A., Chevalot, I., Olmos, E., Guedon, E., and Marc, A. (2015). Serum-free media for mesenchymal stem cells expansion on microcarriers. BMC Proc. 9, P70. doi:10.1186/1753-6561-9-s9-p70
Mcdermott, A. M. (2019). Recapitulating bone development through engineered mesenchymal condensations and mechanical cues for tissue regeneration. 7756, 1
Mendes, L. F., Tam, W. L., Chai, Y. C., Geris, L., Luyten, F. P., and Roberts, S. J. (2016). Combinatorial analysis of growth factors reveals the contribution of bone morphogenetic proteins to chondrogenic differentiation of human periosteal cells. Tissue Eng. Part C. Methods 22, 473–486. doi:10.1089/ten.tec.2015.0436
Mendicino, M., Bailey, A. M., Wonnacott, K., Puri, R. K., and Bauer, S. R. (2014). MSC-based product characterization for clinical trials: an FDA perspective. Cell. Stem Cell. 14, 141–145. doi:10.1016/j.stem.2014.01.013
Mosaad, E. O., Chambers, K. F., Futrega, K., Clements, J. A., and Doran, M. R. (2018). The microwell-mesh : a high- throughput 3D prostate cancer spheroid and drug-testing platform. Sci. Rep. 8, 253–312. doi:10.1038/s41598-017-18050-1
Murphy, K. C., Hung, B. P., Browne-Bourne, S., Zhou, D., Yeung, J., Genetos, D. C., et al. (2017). Measurement of oxygen tension within mesenchymal stem cell spheroids. J. R. Soc. Interface 14, 20160851. doi:10.1098/rsif.2016.0851
Negoro, T., Takagaki, Y., Okura, H., and Matsuyama, A. (2018). Trends in clinical trials for articular cartilage repair by cell therapy. npj Regen. Med. 313, 17–10. doi:10.1038/s41536-018-0055-2
Nilsson Hall, G., Mendes, L. F., Gklava, C., Geris, L., Luyten, F. P., and Papantoniou, I. (2019). Developmentally engineered callus organoid bioassemblies exhibit predictive in vivo long bone healing. Adv. Sci. 1–16. doi:10.1002/advs.201902295
Park, J., Gebhardt, M., Golovchenko, S., Perez-Branguli, F., Hattori, T., Hartmann, C., et al. (2015). Dual pathways to endochondral osteoblasts: a novel chondrocytederived osteoprogenitor cell identified in hypertrophic cartilage. Biol. Open 4, 608–621. doi:10.1242/bio.201411031
Patriarca, E. J., Cermola, F., D’Aniello, C., Fico, A., Guardiola, O., De Cesare, D., et al. (2021). The multifaceted roles of proline in cell behavior. Front. Cell. Dev. Biol. 9, 2236. doi:10.3389/fcell.2021.728576
Pereira, L. M., Hatanaka, E., Martins, E. F., Oliveira, F., Liberti, E. A., Farsky, S. H., et al. (2008). Effect of oleic and linoleic acids on the inflammatory phase of wound healing in rats. Cell. biochem. Funct. 26, 197–204. doi:10.1002/cbf.1432
Petricciani, J., Hayakawa, T., Stacey, G., Trouvin, J. H., and Knezevic, I. (2017). Scientific considerations for the regulatory evaluation of cell therapy products. Biologicals 50, 20–26. doi:10.1016/j.biologicals.2017.08.011
Pigeot, S., Klein, T., Gullotta, F., Dupard, S. J., Garcia Garcia, A., Garcia-Garcia, A., et al. (2021). Manufacturing of human tissues as off-the-Shelf grafts programmed to induce regeneration. Adv. Mat. 33, 2103737. doi:10.1002/adma.202103737
Plotkin, L. I., Essex, A. L., and Davis, H. M. (2019). RAGE signaling in skeletal biology. Curr. Osteoporos. Rep. 17, 16–25. doi:10.1007/s11914-019-00499-w
Power, L. J., Fasolato, C., Barbero, A., Wendt, D. J., Wixmerten, A., Martin, I., et al. (2020). Sensing tissue engineered cartilage quality with Raman spectroscopy and statistical learning for the development of advanced characterization assays. Biosens. Bioelectron. X. 166, 112467. doi:10.1016/j.bios.2020.112467
Prein, C., Warmbold, N., Farkas, Z., Schieker, M., Aszodi, A., and Clausen-Schaumann, H. (2016). Structural and mechanical properties of the proliferative zone of the developing murine growth plate cartilage assessed by atomic force microscopy. Matrix Biol. 50, 1–15. doi:10.1016/j.matbio.2015.10.001
Ren, X., Zhao, M., Lash, B., Martino, M. M., and Julier, Z. (2020). Growth factor engineering strategies for regenerative medicine applications. Front. Bioeng. Biotechnol. 7, 469. doi:10.3389/fbioe.2019.00469
Roberts, S. J., van Gastel, N., Carmeliet, G., and Luyten, F. P. (2015). Uncovering the periosteum for skeletal regeneration: the stem cell that lies beneath. Bone 70, 10–18. doi:10.1016/j.bone.2014.08.007
Rousseau, C. F., Maciulaitis, R., Sladowski, D., and Narayanan, G. (2018). Cell and gene therapies: european view on challenges in translation and how to address them. Front. Med. 5, 158. doi:10.3389/fmed.2018.00158
Salhotra, A., Shah, H. N., Levi, B., and Longaker, M. T. (2020). Mechanisms of bone development and repair. Nat. Rev. Mol. Cell. Biol. 21, 696–711. doi:10.1038/s41580-020-00279-w
Sallent, I., Capella-Monsonis, H., Procter, P., Bozo, I. Y., Deev, R. V., Zubov, D., et al. (2020). The few who made it: commercially and clinically successful innovative bone grafts. Front. Bioeng. Biotechnol. 8 952. doi:10.3389/fbioe.2020.00952
Sarem, M., Otto, O., Tanaka, S., and Shastri, V. P. (2019). Cell number in mesenchymal stem cell aggregates dictates cell stiffness and chondrogenesis. Stem Cell. Res. Ther. 10, 10–18. doi:10.1186/s13287-018-1103-y
Shin, J., Kim, G., Kabir, M. H., Park, S. J., Lee, S. T., and Lee, C. (2015). Use of composite protein database including search result sequences for mass spectrometric analysis of cell secretome. PLoS One 10, e0121692. doi:10.1371/journal.pone.0121692
Shin, J., Rhim, J., Kwon, Y., Choi, S. Y., Shin, S., Ha, C. W., et al. (2019). Comparative analysis of differentially secreted proteins in serum-free and serum-containing media by using BONCAT and pulsed SILAC. Sci. Rep. 9, 3096–3112. doi:10.1038/s41598-019-39650-z
Stich, S., Loch, A., Park, S. J., Haupl, T., Ringe, J., and Sittinger, M. (2017). Characterization of single cell derived cultures of periosteal progenitor cells to ensure the cell quality for clinical application. PLoS One 12, e0178560. doi:10.1371/journal.pone.0178560
Stuart, T., Butler, A., Hoffman, P., Hafemeister, C., Papalexi, E., Mauck, W. M., et al. (2019). Comprehensive integration of single-cell data. Cell. 177, 1888e21–1902.e21. doi:10.1016/j.cell.2019.05.031
Temu, T. M., Wu, K. Y., Gruppuso, P. A., and Phornphutkul, C. (2010). The mechanism of ascorbic acid-induced differentiation of ATDC5 chondrogenic cells. Am. J. Physiology-Endocrinology Metabolism 299, 325–334. doi:10.1152/ajpendo.00145.2010
Tirosh, I., Izar, B., Prakadan, S. M., Wadsworth, M. H., Treacy, D., Trombetta, J. J., et al. (2016). Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Sci. (80-. ) 352, 189–196. doi:10.1126/science.aad0501
Tsiridis, E., and Giannoudis, P. V. (2006). Transcriptomics and proteomics: Advancing the understanding of genetic basis of fracture healing. Injury 37, S13–S19. doi:10.1016/j.injury.2006.02.036
van Gastel, N., Stegen, S., Eelen, G., Schoors, S., Carlier, A., Daniels, V. W., et al. (2020). Lipid availability determines fate of skeletal progenitor cells via SOX9. Nature 579, 111–117. doi:10.1038/s41586-020-2050-1
Villalvilla, A., Gómez, R., Largo, R., and Herrero-Beaumont, G. (2013). Lipid transport and metabolism in healthy and osteoarthritic cartilage. Int. J. Mol. Sci. 14 (14), 20793–20808. doi:10.3390/ijms141020793
Wang, B., Bowles‐Welch, A. C., Yeago, C., and Roy, K. (2022). Process analytical technologies in cell therapy manufacturing: State‐of‐the‐art and future directions. J. Adv. Manuf. Process. 4, e10106. doi:10.1002/amp2.10106
Wang, T., Zhang, X., and Bikle, D. D. (2017). Osteogenic differentiation of periosteal cells during fracture healing. J. Cell. Physiol. 232, 913–921. doi:10.1002/jcp.25641
Wang, Z., Lu, W. W., Zhen, W., Yang, D., and Peng, S. (2017). Novel biomaterial strategies for controlled growth factor delivery for biomedical applications. NPG Asia Mat., 9 e435–e435. doi:10.1038/am.2017.171
Woods, A., Wang, G., and Beier, F. (2005). RhoA/ROCK signaling regulates Sox9 expression and actin organization during chondrogenesis. J. Biol. Chem. 280, 11626–11634. doi:10.1074/jbc.m409158200
Yang, L., Tsang, K. Y., Tang, H. C., Chan, D., and Cheah, K. S. E. (2014). Hypertrophic chondrocytes can become osteoblasts and osteocytes in endochondral bone formation. Proc. Natl. Acad. Sci. U. S. A. 111, 12097–12102. doi:10.1073/pnas.1302703111
Ylostalo, J. H., Bazhanov, N., Mohammadipoor, A., and Bartosh, T. J. (2017). Production and administration of therapeutic mesenchymal stem/stromal cell (MSC) spheroids primed in 3-D cultures under xeno-free conditions. J. Vis. Exp. 2017, e55126. doi:10.3791/55126
Zheng, X., Baker, H., Hancock, W. S., Fawaz, F., McCaman, M., and Pungor, E. (2006). Proteomic analysis for the assessment of different lots of fetal bovine serum as a raw material for cell culture. Part IV. application of proteomics to the manufacture of biological drugs. Biotechnol. Prog. 22, 1294–1300. doi:10.1021/bp060121o
Keywords: advanced therapy medicinal products, skeletal tissue engineering, scalability, manufacturing, spheroid culture, cartilage microtissue, organoid
Citation: Decoene I, Herpelinck T, Geris L, Luyten FP and Papantoniou I (2022) Engineering bone-forming callus organoid implants in a xenogeneic-free differentiation medium. Front. Chem. Eng. 4:892190. doi: 10.3389/fceng.2022.892190
Received: 08 March 2022; Accepted: 18 July 2022;
Published: 25 August 2022.
Edited by:
Hesham Ali El Enshasy, University of Technology Malaysia, MalaysiaReviewed by:
Mark Allenby, The University of Queensland, AustraliaJames Cray, The Ohio State University, United States
Copyright © 2022 Decoene, Herpelinck, Geris, Luyten and Papantoniou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ioannis Papantoniou, aW9hbm5pcy5wYXBhbnRvbmlvdUBrdWxldXZlbi5iZQ==
†ORCID ID: Ioannis Papantoniou, orcid.org/0000-0002-4754-5492; Liesbet Geris, orcid.org/0000-0002-8180-1445; Tim Herpelinck, orcid.org/0000-0002-4500-4676
 Isaak Decoene
Isaak Decoene Tim Herpelinck
Tim Herpelinck Liesbet Geris
Liesbet Geris Frank P. Luyten1,2
Frank P. Luyten1,2 Ioannis Papantoniou
Ioannis Papantoniou