
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Chem. Biol. , 25 February 2025
Sec. Molecular Sciences
Volume 4 - 2025 | https://doi.org/10.3389/fchbi.2025.1441138
 Adam Štefek1,2
Adam Štefek1,2 Kamil Paruch1,2*
Kamil Paruch1,2*Homeodomain-interacting protein kinases (HIPKs) represent a relatively underexplored sub-family of serine/threonine protein kinases. However, the recently published studies point to the role of HIPKs in the developmental biology and etiology of pathological states, in particular cancer, and potential therapeutic applications of targeting this kinase family. This review summarizes the biology of HIPKs and their heretofore published small-molecule inhibitors.
Homeodomain-interacting protein kinases (HIPKs) are evolutionary conserved serine/threonine kinases (Kim et al., 1998; Manning et al., 2002). The four isoforms HIPK1-4 belong to the CMGC branch of the kinome, and form one of the three subunits of the DYRK kinase family (Figure 1). The isoforms HIPK1-3 were first described in 1998 (Kim et al., 1998), and HIPK4 in 2007 (Arai et al., 2007). HIPKs interact (acting as co-repressors) with homeobox proteins, which are prominent transcription factors. Unlike prototypical kinases, HIPKs act directly on transcription factors and other nuclear proteins. They play a role in terminal regulation rather than in activation of downstream signaling cascades (e.g., MAPK or PI3K/AKT) that consist of multiple sequential phosphorylation events (Pearson et al., 2001; Hemmings and Restuccia, 2012). In addition to their function as transcriptional repressors, HIPKs can also act as transcriptional activators influencing the expression of genes, depending on the status and requirements of the cell (Calzado et al., 2007). The highest concentrations of HIPK1-3 have been found in the nucleus, especially in interchromatin granules (nuclear speckles) (Kim et al., 1998; Rinaldo et al., 2008; Van Der Laden et al., 2015), while HIPK4 has been identified mainly in the cytoplasm (Arai et al., 2007). In contrast to HIPK1-3, which are present in all vertebrates, HIPK4 has been found only in mammalian cells (Schmitz et al., 2014).
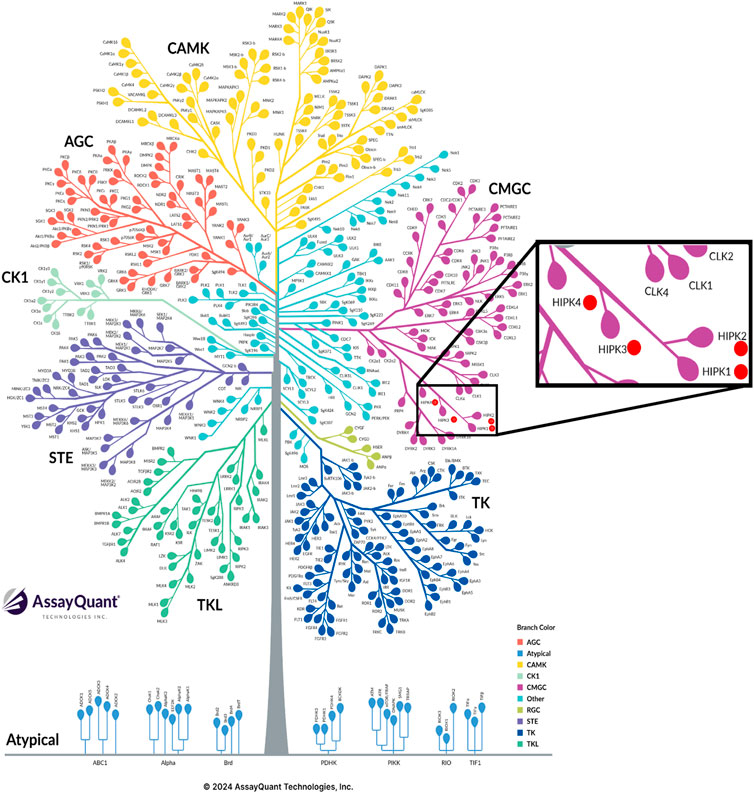
Figure 1. Graphical representation of the human kinome and the CMGC family. HIPKs are marked by red circles (Kinome Tree | Assayquant (29.10.2024)). https://www.assayquant.com/kinome-tree/#).
HIPK1-3 have a high degree of structural homology (Figure 2). The salient feature of HIPK4 is the absence of the homeobox-interacting domain, in contrast to the other three isoforms (Rinaldo et al., 2008; Agnew et al., 2019).
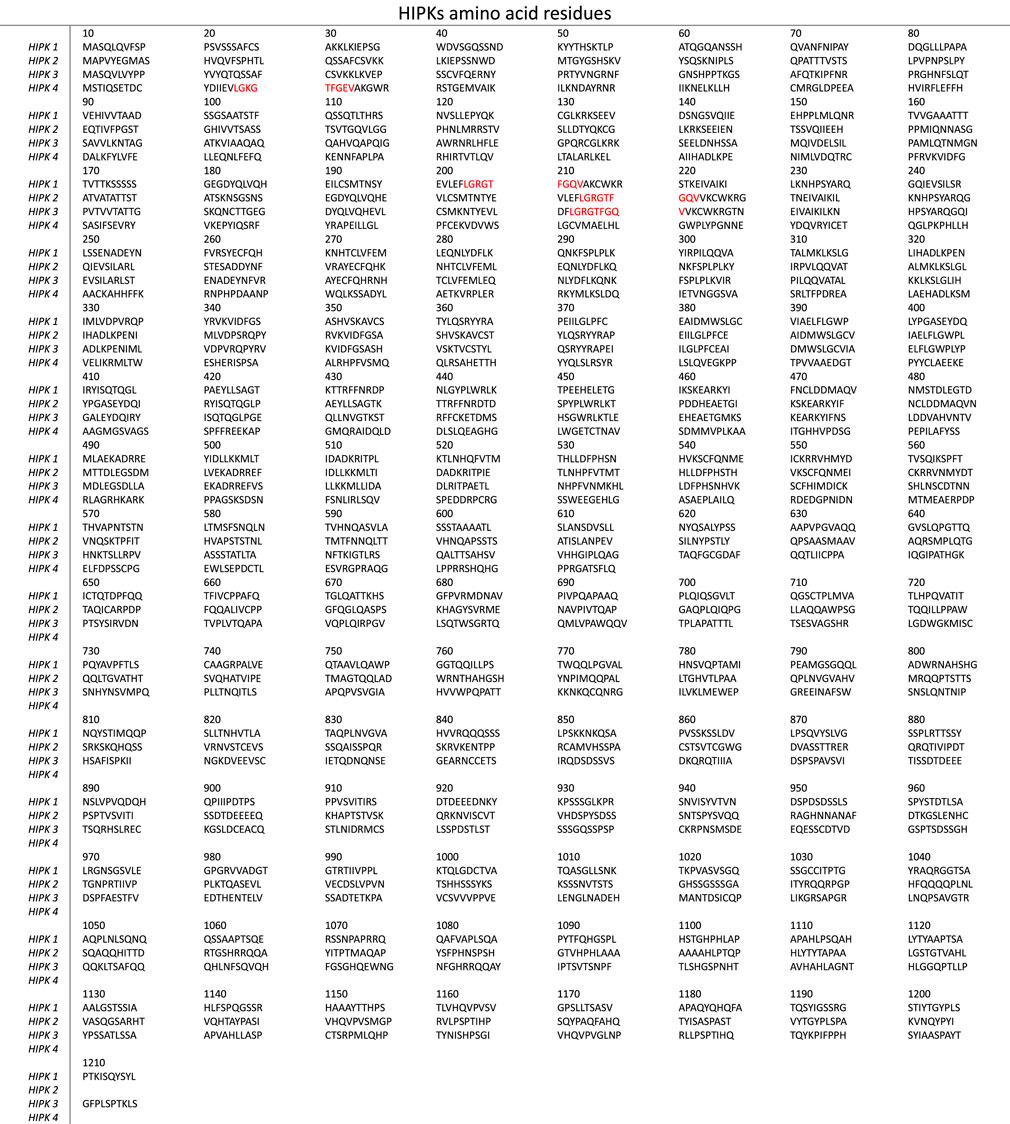
Figure 2. The amino acid sequences of human HIPK1-4 (obtained from UniProt database). The ATP-binding sites of HIPK1-3 sharing common amino acid sequences are highlighted in red.
The HIPK structure features a standard arrangement consisting of smaller N-terminal lobe and larger C-lobe, connected by the hinge region (Figure 3) (Schmitz et al., 2014; Van Der Laden et al., 2015).
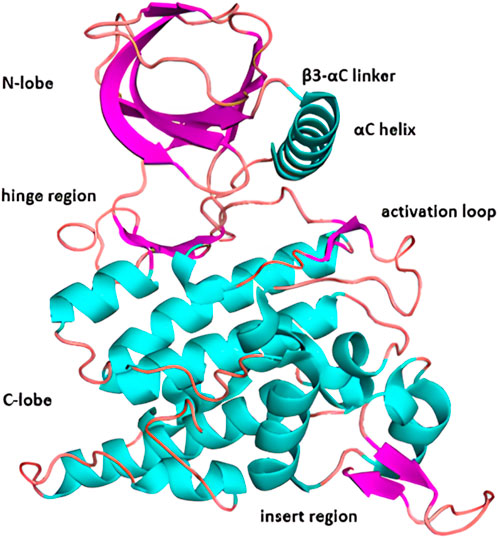
Figure 3. X-ray crystal structure of HIPK2 with highlighted major structural subunits (pdb code: 7NCF).
Of the HIPK isoforms, HIPK2 is the one most studied by X-ray crystallography; the published data include co-crystal structures of HIPK2 and several small-molecule inhibitors described below (Agnew et al., 2019; Němec et al., 2021).
As other kinases, HIPKs undergo various post-translational modifications (e.g., sumoylation, ubiquitinylation, and autophosphorylation) that regulate their activity, function and localization or mediate their degradation (Gresko et al., 2005; Hofmann et al., 2005; Winter et al., 2008; de la Vega et al., 2011; Saul et al., 2013).
Phosphorylation represents one of the most important post-translational modifications. Along this line, HIPK2 can be phosphorylated at multiple sites by Src kinase, and the resulting phosphorylated forms can be differentially re-distributed (Polonio-Vallon et al., 2014). HIPK autophosphorylation at tyrosine and serine residues also affects their activity (Van Der Laden et al., 2015), substrate specificity and the subcellular localization, and plays an important role in the regulation of various cellular processes, including apoptosis, DNA repair and responses to stressors (e.g., radiation or chemotherapeutics) (Saul et al., 2013; Siepi et al., 2013). In this regard, HIPKs are similar to their relatives in the CMGC kinase family, namely, the closely related DYRKs (Becker and Sippl, 2011; Van Der Laden et al., 2015).
SUMOylation affects the nuclear localization of HIPK2 and may be involved in both repression and activation of the kinase depending on the context of post-translational modifications. Reversible modification of HIPK2 at the Lys25 residue by SUMO-1 regulates the activation of c-Jun-NH2-terminal kinase (JNK), while deconjugation of SUMO-1 from HIPK2 mediated by the protease SuPr-1 increases the JNK activity (Hofmann et al., 2005). Binding of HIPKs to SUMO is mediated by the SUMO-interacting motif (SIM, Figure 4) - this interaction regulates the activity of HIPKs and their localization in nuclear speckles (de la Vega et al., 2011). Abnormal SUMOylation has been linked to the development of diverse pathological processes, including cancer (Han et al., 2018), and specifically, disrupted SUMOylation of mutated HIPK2 to acute myeloid leukemia (AML) and myelodysplastic syndrome (MDS) (Sung et al., 2019).

Figure 4. The schematic representation of the HIPK2 structure with the regions involved in the post-translational regulations and discussed in the text highlighted in yellow. (Homeobox-interacting domain: HID; speckle retention signal region: SRS; SUMO-interacting motive: SIM; auto-inhibitory domain: AID; serine, glutamine and alanine repeat region: SQA).
Another mechanism of positive regulation of HIPK2 is mediated by caspase 6, which in response to stress and DNA damage cleaves the auto-inhibitory domain of HIPK2 (Sombroek and Hofmann, 2009). This modification, occurring in HIPK2 at the residues D916 and D977 (Figure 4), results in increased kinase activity (Sombroek and Hofmann, 2009) and enhances apoptotic signaling via phosphorylation of p53 at Ser46. This process, crucial for promoting the transcription of pro-apoptotic genes, is discussed in greater detail below.
Negative regulation of HIPK2 proceeds via the ubiquitination pathway: ubiquitin ligase Siah-1 binds to HIPK2, which leads to polyubiquitination at the lysine residue K1182 (Figure 4), transfer to the proteasome and subsequent degradation (Winter et al., 2008).
Studies in Drosophila have demonstrated the importance of HIPK in the development of the eye as well as in neural and muscular development (Lee et al., 2009b; Blaquiere et al., 2014; Tettweiler et al., 2019; Wang et al., 2020).
Along this line, developmental defects were observed in mice with different degrees of HIPKs dysfunction; specifically, eye stunting was observed in 40% of Hipk1+/− Hipk2−/− mice (Inoue et al., 2010).
A degree of redundancy in HIPK1/2 functions has been observed in several in vivo studies. Absence of both HIPK1 and HIPK2 has been shown to cause early embryonic death due to insufficient development of the vascular and nervous systems (Aikawa et al., 2006; Isono et al., 2006). In this process, HIPK1/2-mediated phosphorylation of the p300 acetyltransferase appears to be essential (Aikawa et al., 2006). Another report revealed the role of HIPK1/2 downstream of the TGF-β-TAK1 signaling pathway regulating numerous angiogenic genes during early embryonic development (Shang et al., 2013).
HIPK1 was found to be important for proper spleen B cell function and homeostasis (Guerra et al., 2012). However, due to the dynamic cellular and tissue expression and high heterogeneity of the interacting molecules, the effects of HIPKs can be rather specific in a particular cellular context. In addition, HIPKs are involved in the regulation of several important cell signaling pathways in Drosophila–e.g., the Hippo pathway (SWH) responsible for the organ shape and size (Poon et al., 2012; Heidary Arash and Attisano, 2013). HIPK2 is involved in the regulation of the transcription factor IPF1/PDX1, which is critical for pancreatic development and proper β-cell function (Boucher et al., 2009). HIPKs are also involved in the regulation of the Wnt signaling pathway (through stabilization of the β-catenin arm) that is crucial in embryonic development and plays an important role in the etiology of cancer or type 2 diabetes (Lee et al., 2009a).
Biochemical studies demonstrated that HIPK3 participates in the regulation of the runt-related transcription factor 2 (Runx2) by the transcriptional regulators MINT+FGF2 that are critical for mammalian development (Sierra and Towler, 2010).
The role of the most recently discovered isoform HIPK4 in developmental biology is still poorly understood. High-throughput RNAi screening analysis revealed its function as a suppressor of skin epithelial cell differentiation (Larribère et al., 2017). Recent study revealed HIPK4 to be essential for spermiogenesis and fertility–mice lacking HIPK4 showed sperm head defects, deformation and shortening of the sperm flagella, and overall impaired sperm function associated with the inability to bind to the oocyte (Crapster et al., 2020). Subsequent report demonstrated an essential role of HIPK4 in the process of sperm head shaping, which is essential for male fertility (Liu et al., 2022).
Numerous studies link HIPKs to the mechanism of tumorigenesis (Conte et al., 2023). One of the key HIPK-interacting biomacromolecules is the protein p53 - one of the key DNA repair factors and tumor suppressor genes. Proper function of p53 is critical in cell aging and controlled cell death, and its mutation or deregulation is very common in a variety of malignancies (Kondo et al., 2003; Lavra et al., 2011). Mutations of p53 are associated with the genetic disorder Li-Fraumeni syndrome, characterized by the frequent occurrence of various malignancies early in life (Li and Fraumeni, 1969; Varley, 2003). HIPK2 has been reported to phosphorylate p53 at Ser46 (D’Orazi et al., 2002), and HIPK4 at Ser9 (Arai et al., 2007).
Phosphorylation of p53 is a crucial response of the cell to genotoxic stress (induced by, e.g., heat, ionizing radiation, xenobiotics or toxic metabolites) and the resulting DNA damage (Kuwano et al., 2016). Under normal conditions, the level of p53 is relatively low and the protein is gradually degraded through interaction with its negative regulator MDM2 (Ashcroft et al., 2000; Wade et al., 2010). Phosphorylation of p53 can lead to the protein stabilization and suppression of the MDM2-mediated degradation, and consequently increase the concentration of p53. This can halt the cell progression in the G1 phase, providing the cell sufficient time to repair DNA (Ashcroft et al., 2000; Oren, 2003). In case of severe DNA damage, apoptosis is induced via activated p53-mediated expression of pro-apoptotic genes such as Noxa, p53API1, Bax or PUMA (Oda et al., 2000a; Oda et al., 2000b; Fritsche et al., 2015). HIPK2 can control the mechanism of p53 degradation either directly through its phosphorylation, or indirectly by inactivating phosphorylation of MDM2 that promotes its export and degradation (Stefano et al., 2004; Di Stefano et al., 2005; Kuwano et al., 2016). This defines HIPK2 as an important regulatory factor, and further investigations will likely lead to better understanding of oncogenesis.
On the macroscopic level, HIPK1-deficient mice were found to be more susceptible to chemically induced skin cancer using DMBA initiator (Kondo et al., 2003).
However, HIPKs’ role in carcinogenesis can vary, depending on the tumor type–in particular the role of HIPK 2 (Torrente et al., 2017; Blaquiere et al., 2018). Faster skin tumor growth and disease progression were observed in HIPK2 −/− mice (Wei et al., 2007). In acute myeloid leukemia (AML), the level of HIPK1/3 mRNA was found to be significantly increased, while the level of HIPK2 mRNA was comparatively low (Gu et al., 2004). Recently, HIPK1 has been identified as one of 3 MSI-1-associated genes in group 3 medulloblastoma, and defined as an attractive therapeutic target (Kameda-Smith et al., 2022). Low levels of the HIPK2 protein expression have been associated with poor prognosis in pancreatic (Qin et al., 2019), colorectal, and thyroid cancers (Lavra et al., 2011; Soubeyran et al., 2011). However, rather opposite trends have been found in the cases of HPV-positive throat and cervical cancers (Al-Beiti and Lu, 2008; Kwon et al., 2017), and brain malignancies (Deshmukh et al., 2008; Schulten et al., 2016) where poor prognosis was associated with HIPK2 protein and mRNA overexpression. In addition, HIPK2 overexpression in squamous cell carcinoma has been linked to higher resistance of the tumors to radio- and chemotherapy (Kwon et al., 2017).
Low levels of HIPK3 mRNA has been found in renal tumors and in that context it may be considered as a negative prognostic factor (Xiao et al., 2021). In contrast, significantly elevated levels of HIPK3 non-coding RNA circHIPK3 were observed in esophageal squamous cell carcinoma, correlating with tumor progression and extent of metastasis (Ba et al., 2020). However, the cancer cell lines HCT116 and SW480 stably overexpressing HIPK3 were found to exhibit retarded cell growth, migration, and increased sensitivity to fluorouracil (Tao et al., 2022).
Of other diseases, chronic kidney diseases associated with fibrosis have been associated with HIPK2 over-activity (Jin et al., 2012; Xiao et al., 2020; Overstreet et al., 2022; Zhong et al., 2022). Mechanistically, HIPK2 regulates the TGF-β/Smad3 signaling pathway that is often associated with the development of fibrosis, which suggests that this kinase could be an attractive candidate for targeted therapy (Hu et al., 2024; Lee et al., 2024).
In the area of neurology, it has been experimentally shown that HIPK2 expression within the central nervous system (CNS) decreases with age, except in the cerebellum (Anzilotti et al., 2015). Genetic ablation of HIPK2 in mice led to the neurodegenerative process characterized by significant loss of Purkinje cells in the cerebellum, neuromotor impairment and ataxia (Anzilotti et al., 2015). Absence of HIPK2 negatively affected neural development, specifically the number and survival rate of dopaminergic neurons during early postnatal programmed cell death phases, where HIPK2−/− mutant mice developed numerous severe psychomotor abnormalities (Zhang et al., 2007). These observations define HIPK2 as a neuroprotective factor (Zhang et al., 2007). Conversely, overexpression of HIPK2 was found to be beneficial in a rat model of spinal cord injury where it reduced the inflammatory response, oxidative stress, and apoptosis (Li et al., 2018). HIPK2 may contribute to the pathogenesis of Alzheimer’s disease: soluble beta-amyloid peptides have been reported to be involved in HIPK2 degradation, thereby regulating the conformational state of p53 and the vulnerability to noxious stimuli, and triggering the amyloidogenic cascade (Lanni et al., 2010; Stanga et al., 2010). Finally, HIPK2 has been recently defined as a promising therapeutic target for the treatment of amyotrophic lateral sclerosis (ALS) – in the SOD1G93A mouse model, loss of HIPK2 was found to be associated with later disease onset, lesser extent of cell death of spinal motor neurons, and improved overall survival (Lee et al., 2016).
The recent report by Zhang et al. points to another role of HIPK2 in neurodegenerative processes, namely, through modulation of mitochondrial function and affecting the Parkin-mediated pathway (Zhang et al., 2020). Loss of HIPK2 has been found to provide increased resistance towards mitochondrial toxins such as MPP+, rotenone or paraquat. Mechanistically, depletion of HIPK2 disrupts HIPK2-promoted Parkin degradation via proteasome-mediated mechanism, which leads to elevation of the Parkin protein level and higher mitochondrial durability (Zhang et al., 2020). These observations suggest that HIPK2 may influence the progression of neurodegenerative processes via regulation of mitochondrial resistance, and make HIPK2 a potential target for the treatment of Parkinson´s disease (Zhang et al., 2020).
Recent studies have demonstrated that in Caenorhabditis elegans the HIPK2 homolog HPK-1 plays a role in the response to stress (Berber et al., 2016), and is involved in two different molecular mechanism of proteostasis maintenance (Das et al., 2017). In one of them, HPK-1 prevents SUMOylation of the heat shock transcription factor HSF-1, which induces molecular chaperones upon thermal stress and enhances longevity (Das et al., 2017). In the other mechanism, HPK-1 induces autophagosome formation and autophagy gene expression upon dietary restriction or inactivation of TORC1 (Das et al., 2017). Along a similar line, HPK-1 was recently discovered to be the most broadly upregulated kinase in C. elegans during normal aging, and essential for preservation of integrity of the nervous system (Lazaro-Pena et al., 2023). In the aging nervous system, HPK-1 induction overlaps with key longevity transcription factors, and restoration of the HPK-1 neuronal expression rescues the premature age-associated decline and reduces thermotolerance of hkp-1 null C. elegans (Lazaro-Pena et al., 2023). Specifically, the HPK-1 activity was found to be important for the heat shock response of serotonergic neurons, proper activity of GABAergic neurons, and proteome stability (Lazaro-Pena et al., 2023).
At the macroscopic level, HPK-1 overexpression in C. elegans increased the lifespan by up to 16% (Das et al., 2017). The recent study by Doering et al. identified HPK-1 as a central positive regulator of the nhr-49 dependent hypoxia response pathway, and revealed that null mutation of hpk-1 caused a significant reduction of the survival rate under hypoxia (Doering et al., 2022).
Additional HIPK2 targets are summarized in Table 1.
HIPK3 has been found to play a role in the metabolism of glucose and possibly in the pathogenesis of type 2 diabetes (Shojima et al., 2012). Specifically, mice lacking HIPK3 showed reduced beta cell proliferation and glucose-induced insulin secretion, associated with decreased phosphorylation of the kinase GSK3β and the transcription factor PDX1 (Shojima et al., 2012).
Collectively, the studies described above suggest that modulation of HIPK2 for therapeutic purposes may require finely balanced concentrations of HIPK inhibitors that would positively affect the maintenance of proteostasis and prevent age-induced neuronal cell death, but would not induce chronic stress response. Similarly delicately balanced scenario may be necessary for the development of potential anti-cancer drugs, where in some contexts inhibition of overexpressed HIPK2 (Al-Beiti and Lu, 2008; Kwon et al., 2017; Cao et al., 2021) could provide the desired anti-cancer effect, while in others it could negatively affect the p53 apoptotic function and promote tumorigenesis (D’Orazi et al., 2012). Similarly balanced regimes may be necessary for therapeutic targeting of the isoforms HIPK1, HIPK3 and HIPK4, whose biology (including post-translational modifications) is however comparatively less explored. In addition, modulation of the HIPK isoforms in specific compartments of the cell (Ritter and Schmitz, 2019) may require isoform-selective inhibitors.
The family of protein kinases contains >500 proteins that regulate numerous cellular signaling pathways in the cell. Targeting this class of enzymes represents one of the most dynamic areas of pharmaceutical research and >80 small-molecule kinase inhibitors approved for clinical use (Roskoski, 2024). Vast majority of them is represented by ATP-competitive inhibitors that are tightly anchored to the kinase hinge backbone. The ATP-binding site across the kinome is typically highly conserved, which makes identification of highly selective kinase inhibitors a non-trivial task (Breen and Soellner, 2015; Umezawa and Kii, 2021).
While sufficiently selective inhibitors have been identified for some kinases (and frequently served as the starting points for development of novel drugs), for many others that are only emerging as potential targets for pharmacological inhibition they are still to be discovered (Attwood et al., 2021).
To our best knowledge, there are no clinically profiled inhibitors having HIPKs as primary targets. However, several classes of recently discovered HIPK inhibitors summarized below can serve as starting points for the pre-clinical identification of suitable candidates for pharmacological inhibition of these kinases.
Kinase inhibitors are classified according to their mode of interaction with the target kinase. Most common ones are ATP-competitive inhibitors, i.e., compounds that occupy the binding site of the natural substrate ATP (Roskoski, 2021; Roskoski, 2024). Type I ATP-competitive inhibitors bind to active conformation of the kinase (Wu et al., 2015), whereas type II inhibitors bind to inactive conformation and can also affect regions adjacent to the ATP binding site (Wu et al., 2015).
Allosteric inhibitors, on the other hand, inactivate the kinase indirectly, most frequently by changing its conformation or disrupting an association to a partner that is essential for the kinase function (Thomson et al., 2024). Most inhibitors interact with active or inactive forms of kinases through non-covalent interactions - hydrogen bonds, pi-stacking, dipole or lipophilic interaction (Łukasik et al., 2021). Historically less common (but currently growing) group includes covalent kinase inhibitors, e.g., ibrutinib and afatinib (Abdeldayem et al., 2020; Cheke and Kharkar, 2024).
High-throughput profiling of a library consisting of 118 compounds yielded the compound A64 (Figure 5) (Miduturu et al., 2011). This commercially available compound (sold as the hydrochloride salt under the name Protein Kinase Inhibitor 1) inhibits HIPK1 and HIPK2 with the IC50 values of 136 nM and 74 nM, respectively (Miduturu et al., 2011). Recently, the compound A64 has been applied as a research tool to inhibit HIPK2 in cells (using 74 nM concentration of its dihydrochloride PKI1H) and in vivo (with the 100 mg/kg dose, Liang et al., 2020; Zhou et al., 2021).
A recently reported class of HIPK inhibitors is based on the non-routine furo[3,2-b]pyridine scaffold, which was previously used as the basis of potent and highly selective inhibitors of cdc-like kinases (CLKs) (Němec et al., 2019). Expansion of the SAR in the sub-series of 3,5-disubstituted furo[3,2-b]pyridines with the focus on HIPK inhibition yielded the compounds MU135 (HIPK1 IC50 = 248 nM; HIPK2 IC50 = 119 nM; HIPK3 IC50 = 476 nM) and MU1787 (HIPK1 IC50 = 285 nM; HIPK2 IC50 = 123 nM; HIPK3 IC50 = 283 nM) (Figure 6) with remarkable kinome-wide selectivity (Němec et al., 2021).
Crystallographic studies revealed that MU135 in HIPK2 adopts a rather unusual type-I binding mode: in contrast to standard type-I inhibitors, it features only a weak interaction of the furo[3,2-b]pyridine core to the hinge, combined with the hydrogen bond of the pyrazole with the salt bridge, and likely hydrophobic stacking with Phe 277 and Ile 345 in the back pocket (Figure 7).
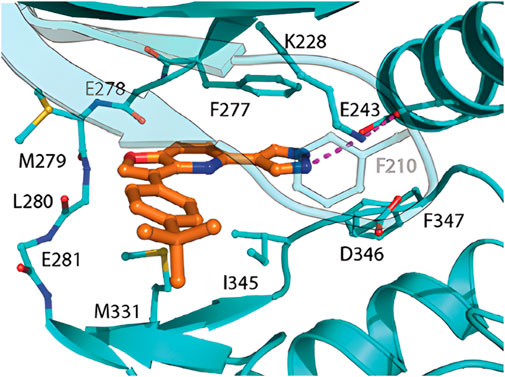
Figure 7. Crystal structure of MU135 in HIPK2; adopted from (Němec et al., 2021).
In contrast to the ATP-competitive inhibitors described above, the compound BT173 (Figure 8) binds to HIPK2 allosterically and does not directly inhibit its kinase activity. However, at 10 μM concentration, it blocks the ability of HIPK2 to associate with the protein Smad3 in 293T cells (Liu et al., 2017), thereby regulating the TGF-β1/Smad3 pathway, and attenuating renal fibrosis (Sato et al., 2003; Zhang et al., 2010; Chen et al., 2014; Liu et al., 2017; Lee et al., 2024). The compound has been found to be effective also in vivo–in the Tg26 mouse model, it ameliorated proteinuria and kidney fibrosis at the dose of 20 mg/kg (Liu et al., 2017).
First reported class of inhibitors with attractive off-target HIPK activity consists of polyhalogenated (benz)imidazoles, exemplified by the compounds TBI, TBiD and TMCB (Figure 9). The compound TBI (also referred to as TBBz) was firstly reported as a CK2 inhibitor (Zień et al., 2003). Subsequent studies identified this compound as an antiprotozoal agent effective against Acanthamoeba castellanii, and also as a HIPK inhibitor with HIPK2 IC50 = 0.7 μM (Kopańska et al., 2004; Pagano et al., 2008; Cozza et al., 2014). The compound TBID is more potent (HIPK2 IC50 = 0.33 μM), capable of inhibiting phosphorylation of p53 at serine 46 in the cell at 50 μM concentration (Cozza et al., 2014). Both compounds exhibit moderate activity towards the kinase CK2: TBI CK2 IC50 = 0.6 μM and TBID CK2 IC50 = 5.5 μM (Cozza et al., 2014). The commercially available compound TMCB is comparatively less potent (HIPK2 IC50 = 15.25 μM) and targets several other kinases (namely, CK2, ERK8, PIM1) with higher potency (Pagano et al., 2008; Cozza et al., 2010; Janeczko et al., 2012) – it has been used as a research tool in that context (Schneider et al., 2012; Bollacke et al., 2016; Wadey et al., 2017; Kim et al., 2023). TBI iodinated analogues were also prepared and studied (Gianoncelli et al., 2009).
Abemaciclib (Figure 10) is a small-molecule inhibitor developed by Eli Lilly, sold under the name Verzenio. The compound was approved by the FDA in 2015 for the treatment of metastatic breast cancer and later for other types of breast cancers (Finn et al., 2009; Coates et al., 2010; Lu, 2015; Palumbo et al., 2019; Johnston et al., 2021; Rugo et al., 2022; Wekking et al., 2023). The primary targets of abemaciclib are CDK4/6; however, the compound was found to possess notable activity towards HIPK2 and HIPK3 with the IC50 values of 668 nM and 467 nM, respectively (Poratti and Marzaro, 2019; Kaltheuner et al., 2021). The compound is significantly less active against HIPK1 and HIPK4 (HIPK1 IC50 = 4.53 μM; HIPK4 IC50 = 10.36 μM; Kaltheuner et al., 2021).
The compound STO-609 (Figure 11) is currently commercially available as a research tool for inhibition of calmodulin-dependent protein kinase kinase (CaM-KK) (Tokumitsu et al., 2002; Kukimoto-Niino et al., 2011; Hou et al., 2021; Wang et al., 2021; 2022). Profiling of STO-609 in a panel of 72 kinases revealed off-target inhibition of HIPK2 (65% inhibition at 1 μM) and HIPK3 (32% inhibition at 1 μM) (Bain et al., 2007).
Silmitasertib also known as CX-4945 (Figure 12) is a small-molecule inhibitor of casein kinase 2 (CK2), currently undergoing clinical trials focused on the treatment for bile duct cancer and medulloblastoma (Pierre et al., 2011; Purzner et al., 2018; Borad et al., 2021; 2023). It sparked interest during the COVID-19 pandemic due to its antiviral effect (Bouhaddou et al., 2020; Gordon et al., 2020; Naik et al., 2022), possibly caused by modulation of CK2-mediated extracellular matrix remodeling (Bouhaddou et al., 2020). The compound was found to be significantly active against HIPK3 isoform with the IC50 value of 45 nM (Pierre et al., 2011).
Off-target activity against HIPK2 was observed in case of the c-Jun NH2-terminal protein kinase (JNK) inhibitors SP600125 and AS601245 (Figure 13) (Slouka et al., 1982; Bain et al., 2003; Gaillard et al., 2005). The compound SP600125 is a broad-spectrum inhibitor of serine/threonine kinases and inhibits 65% of HIPK2 at 1 μM, while the more selective compound AS601245 71% at the same concentration (Bain et al., 2007).
HIPKs represent a relatively underexplored kinase family (Attwood et al., 2021). However, the biological studies published in the past decade (and summarized in this review) suggest that targeting HIPKs could bring therapeutic benefit. Development of sufficiently potent and selective small-molecule HIPK inhibitors therefore represents an attractive research area as it will likely afford quality chemical biology probes that would serve as valuable tools for further exploration of potential therapeutic relevance of targeting HIPKs. Those compounds will be likely useful also as the starting points for the development of clinical candidates. However, modulation of HIPKs for optimal therapeutic outcome may require finely balanced concentrations of HIPK inhibitors (in some cases those possessing sufficient isoform selectivity), especially in the areas of neurology and oncology. In addition, exploration and linking of the HIPK biology to new therapeutic applications could expand the medicinal use of the already approved drugs that inhibit HIPKs as off-targets.
AŠ: Data curation, Writing–original draft, Writing–review and editing. KP: Conceptualization, Funding acquisition, Supervision, Writing–original draft, Writing–review and editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. The authors gratefully acknowledge the support provided by the following sources of funding: the project CZ-OPENSCREEN: National Infrastructure for Chemical Biology (CZ-OPENSCREEN LM2023052), Ministry of Health of the Czech Republic - Agency for Healthcare Research of the Czech Republic (grant number NW24-08-00280), and Bader Philanthropies.
DeepL Pro and Grammarly (v.1.2.131.1585) were used to check grammar and search for more appropriate phrases or synonyms.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abdeldayem, A., Raouf, Y. S., Constantinescu, S. N., Moriggl, R., and Gunning, P. T. (2020). Advances in covalent kinase inhibitors. Chem. Soc. Rev. 49 (9), 2617–2687. doi:10.1039/C9CS00720B
Agnew, C., Liu, L., Liu, S., Xu, W., You, L., Yeung, W., et al. (2019). The crystal structure of the protein kinase HIPK2 reveals a unique architecture of its CMGC-insert region. J. Biol. Chem. 294 (37), 13545–13559. doi:10.1074/jbc.RA119.009725
Aikawa, Y., Nguyen, L. A., Isono, K., Takakura, N., Tagata, Y., Schmitz, M. L., et al. (2006). Roles of HIPK1 and HIPK2 in AML1- and p300-dependent transcription, hematopoiesis and blood vessel formation. EMBO J. 25 (17), 3955–3965. doi:10.1038/sj.emboj.7601273
Al-Beiti, M. A. M., and Lu, X. (2008). Expression of HIPK2 in cervical cancer: correlation with clinicopathology and prognosis. Aust. N. Z. J. Obstetrics Gynaecol. 48 (3), 329–336. doi:10.1111/j.1479-828X.2008.00874.x
Ann, E.-J., Kim, M. Y., Yoon, J. H., Ahn, J. S., Jo, E. H., Lee, H. J., et al. (2016). Tumor suppressor HIPK2 regulates malignant growth via phosphorylation of Notch1. Cancer Res. 76 (16), 4728–4740. doi:10.1158/0008-5472.CAN-15-3310
Anzilotti, S., Tornincasa, M., Gerlini, R., Conte, A., Brancaccio, P., Cuomo, O., et al. (2015). Genetic ablation of homeodomain-interacting protein kinase 2 selectively induces apoptosis of cerebellar Purkinje cells during adulthood and generates an ataxic-like phenotype. Cell Death and Dis. 6 (12), e2004. doi:10.1038/cddis.2015.298
Arai, S., Matsushita, A., Du, K., Yagi, K., Okazaki, Y., and Kurokawa, R. (2007). Novel homeodomain-interacting protein kinase family member, HIPK4, phosphorylates human p53 at serine 9. FEBS Lett. 581 (29), 5649–5657. doi:10.1016/j.febslet.2007.11.022
Ashcroft, M., Taya, Y., and Vousden, K. H. (2000). Stress signals utilize multiple pathways to stabilize p53. Mol. Cell. Biol. 20 (9), 3224–3233. doi:10.1128/MCB.20.9.3224-3233.2000
Attwood, M. M., Fabbro, D., Sokolov, A. V., Knapp, S., and Schiöth, H. B. (2021). Trends in kinase drug discovery: targets, indications and inhibitor design. Nat. Rev. Drug Discov. 20 (11), 839–861. doi:10.1038/s41573-021-00252-y
Ba, Y., Liu, Y., Li, C., Zhu, Y., and Xing, W. (2020). HIPK3 promotes growth and metastasis of esophageal squamous cell carcinoma via regulation of miR-599/c-MYC Axis. OncoTargets Ther., 13, pp. 1967–1978. doi:10.2147/OTT.S217087
Bain, J., McLAUCHLAN, H., Elliott, M., and Cohen, P. (2003). The specificities of protein kinase inhibitors: an update. Biochem. J. 371 (1), 199–204. doi:10.1042/bj20021535
Bain, J., Plater, L., Elliott, M., Shpiro, N., Hastie, C., Mclauchlan, H., et al. (2007). The selectivity of protein kinase inhibitors: a further update. Biochem. J. 408 (3), 297–315. doi:10.1042/BJ20070797
Becker, W., and Sippl, W. (2011). Activation, regulation, and inhibition of DYRK1A. FEBS J. 278 (2), 246–256. doi:10.1111/j.1742-4658.2010.07956.x
Berber, S., Wood, M., Llamosas, E., Thaivalappil, P., Lee, K., Liao, B. M., et al. (2016). Homeodomain-Interacting Protein Kinase (HPK-1) regulates stress responses and ageing in C. elegans. Sci. Rep. 6 (1), 19582. doi:10.1038/srep19582
Blaquiere, J. A., Lee, W., and Verheyen, E. M. (2014). Hipk promotes photoreceptor differentiation through the repression of Twin of eyeless and Eyeless expression. Dev. Biol. 390 (1), 14–25. doi:10.1016/j.ydbio.2014.02.024
Blaquiere, J. A., Wong, K. K. L., Kinsey, S. D., Wu, J., and Verheyen, E. M. (2018). Homeodomain-interacting protein kinase promotes tumorigenesis and metastatic cell behavior. Dis. Models and Mech. 11 (1), dmm031146. doi:10.1242/dmm.031146
Bollacke, A., Nienberg, C., Borgne, M. L., and Jose, J. (2016). Toward selective CK2alpha and CK2alpha’ inhibitors: development of a novel whole-cell kinase assay by Autodisplay of catalytic CK2alpha. J. Pharm. Biomed. Analysis 121, 253–260. doi:10.1016/j.jpba.2016.01.011
Borad, M. J., Bai, L., Richards, D., Mody, K., Hubbard, J., Rha, S. Y., et al. (2023). Silmitasertib plus gemcitabine and cisplatin first-line therapy in locally advanced/metastatic cholangiocarcinoma: a Phase 1b/2 study. Hepatology 77 (3), 760–773. doi:10.1002/hep.32804
Borad, M. J., Bai, L. Y., Chen, M. H., Hubbard, J. M., Mody, K., Rha, S. Y., et al. (2021). Silmitasertib (CX-4945) in combination with gemcitabine and cisplatin as first-line treatment for patients with locally advanced or metastatic cholangiocarcinoma: a phase Ib/II study. J. Clin. Oncol. 39 (3_Suppl. l), 312. doi:10.1200/JCO.2021.39.3_suppl.312
Boucher, M.-J., Simoneau, M., and Edlund, H. (2009). The homeodomain-interacting protein kinase 2 regulates insulin promoter factor-1/pancreatic duodenal homeobox-1 transcriptional activity. Endocrinology 150 (1), 87–97. doi:10.1210/en.2007-0865
Bouhaddou, M., Memon, D., Meyer, B., White, K. M., Rezelj, V. V., Correa Marrero, M., et al. (2020). The global phosphorylation landscape of SARS-CoV-2 infection. Cell 182 (3), 685–712.e19. doi:10.1016/j.cell.2020.06.034
Breen, M. E., and Soellner, M. B. (2015). Small molecule substrate phosphorylation site inhibitors of protein kinases: approaches and challenges. ACS Chem. Biol. 10 (1), 175–189. doi:10.1021/cb5008376
Calzado, M. A., Renner, F., Roscic, A., and Schmitz, M. L. (2007). HIPK2, a versatile switchboard regulating the transcription machinery and cell death. Cell Cycle 6 (2), 139–143. doi:10.4161/cc.6.2.3788
Cao, Y., Xie, X., Li, M., and Gao, Y. (2021). CircHIPK2 contributes to DDP resistance and malignant behaviors of DDP-resistant ovarian cancer cells both in vitro and in vivo through circHIPK2/miR-338-3p/CHTOP ceRNA pathway. OncoTargets Ther. 14, 3151–3165. doi:10.2147/OTT.S291823
Cheke, R. S., and Kharkar, P. S. (2024). Covalent inhibitors: an ambitious approach for the discovery of newer oncotherapeutics. Drug Dev. Res. 85 (1), e22132. doi:10.1002/ddr.22132
Chen, J., Xia, Y., Lin, X., Feng, X. H., and Wang, Y. (2014). Smad3 signaling activates bone marrow-derived fibroblasts in renal fibrosis. Lab. Investig. 94 (5), 545–556. doi:10.1038/labinvest.2014.43
Conte, A., Valente, V., Paladino, S., and Pierantoni, G. M. (2023). HIPK2 in cancer biology and therapy: recent findings and future perspectives. Cell. Signal. 101, 110491. doi:10.1016/j.cellsig.2022.110491
Cozza, G., Bortolato, A., and Moro, S. (2010). How druggable is protein kinase CK2? How druggable is protein kinase CK2. Med. Res. Rev. 30 (3), 419–462. doi:10.1002/med.20164
Cozza, G., Zanin, S., Determann, R., Ruzzene, M., Kunick, C., and Pinna, L. A. (2014). Synthesis and properties of a selective inhibitor of homeodomain–interacting protein kinase 2 (HIPK2). PLOS ONE 9 (2), e89176. doi:10.1371/journal.pone.0089176
Crapster, J. A., Rack, P. G., Hellmann, Z. J., Le, A. D., Adams, C. M., Leib, R. D., et al. (2020). HIPK4 is essential for murine spermiogenesis, eLife. 9. doi:10.7554/eLife.50209
Das, R., Melo, J. A., Thondamal, M., Morton, E. A., Cornwell, A. B., Crick, B., et al. (2017). The homeodomain-interacting protein kinase HPK-1 preserves protein homeostasis and longevity through master regulatory control of the HSF-1 chaperone network and TORC1-restricted autophagy in Caenorhabditis elegans. PLOS Genet. 13 (10), e1007038. doi:10.1371/journal.pgen.1007038
de la Vega, L., Fröbius, K., Moreno, R., Calzado, M. A., Geng, H., and Schmitz, M. L. (2011). Control of nuclear HIPK2 localization and function by a SUMO interaction motif. Biochimica Biophysica Acta (BBA) - Mol. Cell Res. 1813 (2), 283–297. doi:10.1016/j.bbamcr.2010.11.022
Deshmukh, H., Yeh, T. H., Yu, J., Sharma, M. K., Perry, A., Leonard, J. R., et al. (2008). High-resolution, dual-platform aCGH analysis reveals frequent HIPK2 amplification and increased expression in pilocytic astrocytomas. Oncogene 27 (34), 4745–4751. doi:10.1038/onc.2008.110
Di Stefano, V., Mattiussi, M., Sacchi, A., and D'Orazi, G. (2005). HIPK2 inhibits both MDM2 gene and protein by, respectively, p53-dependent and independent regulations. FEBS Lett. 579 (25), 5473–5480. doi:10.1016/j.febslet.2005.09.008
Doering, K. R. S., Cheng, X., Milburn, L., Ratnappan, R., Ghazi, A., Miller, D. L., et al. (2022). Nuclear hormone receptor NHR-49 acts in parallel with HIF-1 to promote hypoxia adaptation in Caenorhabditis elegans. eLife 11, e67911. doi:10.7554/elife.67911
D’Orazi, G., Cecchinelli, B., Bruno, T., Manni, I., Higashimoto, Y., Saito, S., et al. (2002). Homeodomain-interacting protein kinase-2 phosphorylates p53 at Ser 46 and mediates apoptosis. Nat. Cell Biol. 4 (1), 11–19. doi:10.1038/ncb714
D’Orazi, G., Rinaldo, C., and Soddu, S. (2012). Updates on HIPK2: a resourceful oncosuppressor for clearing cancer. J. Exp. and Clin. Cancer Res. 31 (1), 63. doi:10.1186/1756-9966-31-63
Finn, R. S., Dering, J., Conklin, D., Kalous, O., Cohen, D. J., Desai, A. J., et al. (2009). PD 0332991, a selective cyclin D kinase 4/6 inhibitor, preferentially inhibits proliferation of luminal estrogen receptor-positive human breast cancer cell lines in vitro. Breast Cancer Res. 11 (5), R77. doi:10.1186/bcr2419
Fritsche, M. K., Metzler, V., Becker, K., Plettenberg, C., Heiser, C., Hofauer, B., et al. (2015). Cisplatin fails to induce puma mediated apoptosis in mucosal melanomas. Oncotarget 6 (12), 9887–9896. doi:10.18632/oncotarget.3195
Gaillard, P., Jeanclaude-Etter, I., Ardissone, V., Arkinstall, S., Cambet, Y., Camps, M., et al. (2005). Design and synthesis of the first generation of novel potent, selective, and in vivo active (Benzothiazol-2-yl)acetonitrile inhibitors of the c-jun N-terminal kinase. J. Med. Chem. 48 (14), 4596–4607. doi:10.1021/jm0310986
Gianoncelli, A., Cozza, G., Orzeszko, A., Meggio, F., Kazimierczuk, Z., and Pinna, L. A. (2009). Tetraiodobenzimidazoles are potent inhibitors of protein kinase CK2. Bioorg. and Med. Chem. 17 (20), 7281–7289. doi:10.1016/j.bmc.2009.08.047
Gordon, D. E., Jang, G. M., Bouhaddou, M., Xu, J., Obernier, K., White, K. M., et al. (2020). A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 583 (7816), 459–468. doi:10.1038/s41586-020-2286-9
Gresko, E., Möller, A., Roscic, A., and Schmitz, M. L. (2005). Covalent modification of human homeodomain interacting protein kinase 2 by SUMO-1 at lysine 25 affects its stability. Biochem. Biophysical Res. Commun. 329 (4), 1293–1299. doi:10.1016/j.bbrc.2005.02.113
Gu, C., Zheng, H. J., Chan, L. S., Brandwein, J., Kamel-Reid, S., Minden, M. D., et al. (2004). Disordered expression of HIPK family members in MDS and AML. Blood 104 (11), 4311. doi:10.1182/blood.V104.11.4311.4311
Guerra, F. M., Gommerman, J. L., Corfe, S. A., Paige, C. J., and Rottapel, R. (2012). Homeodomain-interacting protein kinase (HIPK)-1 is required for splenic B cell homeostasis and optimal T-independent type 2 humoral response. PLOS ONE 7 (4), e35533. doi:10.1371/journal.pone.0035533
Han, Z.-J., Feng, Y. H., Gu, B. H., Li, Y. M., and Chen, H. (2018). The post-translational modification, SUMOylation, and cancer (Review). Int. J. Oncol. 52 (4), 1081–1094. doi:10.3892/ijo.2018.4280
Heidary Arash, E., and Attisano, L. (2013). A role for Hipk in the Hippo pathway. Sci. Signal. 6 (275), pe18. doi:10.1126/scisignal.2004259
Hemmings, B. A., and Restuccia, D. F. (2012). PI3K-PKB/Akt pathway. Cold Spring Harb. Perspect. Biol. 4 (9), a011189. doi:10.1101/cshperspect.a011189
Hofmann, T. G., Jaffray, E., Stollberg, N., Hay, R. T., and Will, H. (2005). Regulation of homeodomain-interacting protein kinase 2 (HIPK2) effector function through dynamic small ubiquitin-related modifier-1 (SUMO-1) modification. J. Biol. Chem. 280 (32), 29224–29232. doi:10.1074/jbc.M503921200
Hofmann, T. G., Stollberg, N., Schmitz, M. L., and Will, H. (2003). HIPK2 regulates transforming growth factor-beta-induced c-Jun NH(2)-terminal kinase activation and apoptosis in human hepatoma cells. Cancer Res. 63 (23), 8271–8277.
Hou, L., Jiang, F., Huang, B., Zheng, W., Jiang, Y., Cai, G., et al. (2021). Dihydromyricetin resists inflammation-induced muscle atrophy via ryanodine receptor-CaMKK-AMPK signal pathway. J. Cell. Mol. Med. 25 (21), 9953–9971. doi:10.1111/jcmm.16810
Hu, X., Wu, Y., Ouyang, H., Wu, J., Yao, M., Chen, Z., et al. (2024). Virtual screening, molecular dynamics, and mechanism study of homeodomain-interacting protein kinase 2 inhibitor in renal fibroblasts. Pharmaceuticals 17 (11), 1420. doi:10.3390/ph17111420
Inoue, T., Kagawa, T., Inoue-Mochita, M., Isono, K., Ohtsu, N., Nobuhisa, I., et al. (2010). Involvement of the Hipk family in regulation of eyeball size, lens formation and retinal morphogenesis. FEBS Lett. 584 (14), 3233–3238. doi:10.1016/j.febslet.2010.06.020
Isono, K., Nemoto, K., Li, Y., Takada, Y., Suzuki, R., Katsuki, M., et al. (2006). Overlapping roles for homeodomain-interacting protein kinases Hipk1 and Hipk2 in the mediation of cell growth in response to morphogenetic and genotoxic signals. Mol. Cell. Biol. 26 (7), 2758–2771. doi:10.1128/MCB.26.7.2758-2771.2006
Janeczko, M., Orzeszko, A., Kazimierczuk, Z., Szyszka, R., and Baier, A. (2012). CK2α and CK2α′ subunits differ in their sensitivity to 4,5,6,7-tetrabromo- and 4,5,6,7-tetraiodo-1H-benzimidazole derivatives. Eur. J. Med. Chem. 47, 345–350. doi:10.1016/j.ejmech.2011.11.002
Jin, Y., Ratnam, K., Chuang, P. Y., Fan, Y., Zhong, Y., Dai, Y., et al. (2012). A systems approach identifies HIPK2 as a key regulator of kidney fibrosis. Nat. Med. 18 (4), 580–588. doi:10.1038/nm.2685
Johnston, S., O’Shaughnessy, J., Martin, M., Huober, J., Toi, M., Sohn, J., et al. (2021). Abemaciclib as initial therapy for advanced breast cancer: MONARCH 3 updated results in prognostic subgroups. npj Breast Cancer 7 (1), 80–85. doi:10.1038/s41523-021-00289-7
Kaltheuner, I. H., Anand, K., Moecking, J., Düster, R., Wang, J., Gray, N. S., et al. (2021). Abemaciclib is a potent inhibitor of DYRK1A and HIP kinases involved in transcriptional regulation. Nat. Commun. 12 (1), 6607. doi:10.1038/s41467-021-26935-z
Kameda-Smith, M. M., Zhu, H., Luo, E. C., Suk, Y., Xella, A., Yee, B., et al. (2022). Characterization of an RNA binding protein interactome reveals a context-specific post-transcriptional landscape of MYC-amplified medulloblastoma. Nat. Commun. 13 (1), 7506. doi:10.1038/s41467-022-35118-3
Kanei-Ishii, C., Ninomiya-Tsuji, J., Tanikawa, J., Nomura, T., Ishitani, T., Kishida, S., et al. (2004). Wnt-1 signal induces phosphorylation and degradation of c-Myb protein via TAK1, HIPK2, and NLK. Genes and Dev. 18 (7), 816–829. doi:10.1101/gad.1170604
Kim, E.-A., Kim, J. E., Sung, K. S., Choi, D. W., Lee, B. J., and Choi, C. Y. (2010). Homeodomain-interacting protein kinase 2 (HIPK2) targets β-catenin for phosphorylation and proteasomal degradation. Biochem. Biophysical Res. Commun. 394 (4), 966–971. doi:10.1016/j.bbrc.2010.03.099
Kim, J.-E., Lee, D. S., Kim, T. H., Park, H., and Kang, T. C. (2023). Distinct roles of CK2- and AKT-mediated NF-κB phosphorylations in clasmatodendrosis (autophagic astroglial death) within the Hippocampus of chronic epilepsy rats. Antioxidants 12 (5), 1020. doi:10.3390/antiox12051020
Kim, Y. H., Choi, C. Y., Lee, S. J., Conti, M. A., and Kim, Y. (1998). Homeodomain-interacting protein kinases, a novel family of Co-repressors for homeodomain transcription factors. J. Biol. Chem. 273 (40), 25875–25879. doi:10.1074/jbc.273.40.25875
Kondo, S., Lu, Y., Debbas, M., Lin, A. W., Sarosi, I., Itie, A., et al. (2003). Characterization of cells and gene-targeted mice deficient for the p53-binding kinase homeodomain-interacting protein kinase 1 (HIPK1). Proc. Natl. Acad. Sci. 100 (9), 5431–5436. doi:10.1073/pnas.0530308100
Kopańska (née Zastąpiło), K., Najda, A., Żebrowska, J., Chomicz, L., Piekarczyk, J., Myjak, P., et al. (2004). Synthesis and activity of 1H-benzimidazole and 1H-benzotriazole derivatives as inhibitors of Acanthamoeba castellanii. Bioorg. and Med. Chem. 12 (10), 2617–2624. doi:10.1016/j.bmc.2004.03.022
Kukimoto-Niino, M., Yoshikawa, S., Takagi, T., Ohsawa, N., Tomabechi, Y., Terada, T., et al. (2011). Crystal structure of the Ca2+/Calmodulin-dependent protein kinase kinase in complex with the inhibitor STO-609. J. Biol. Chem. 286 (25), 22570–22579. doi:10.1074/jbc.M111.251710
Kuwano, Y., Nishida, K., Akaike, Y., Kurokawa, K., Nishikawa, T., Masuda, K., et al. (2016). Homeodomain-interacting protein kinase-2: a critical regulator of the DNA damage response and the epigenome. Int. J. Mol. Sci. 17 (10), 1638. doi:10.3390/ijms17101638
Kwon, M. J., Kang, S. Y., Nam, E. S., Cho, S. J., and Rho, Y. S. (2017). HIPK2 overexpression and its prognostic role in human papillomavirus-positive tonsillar squamous cell carcinoma. BioMed Res. Int. 2017, 1–10. doi:10.1155/2017/1056427
Lanni, C., Nardinocchi, L., Puca, R., Stanga, S., Uberti, D., Memo, M., et al. (2010). Homeodomain interacting protein kinase 2: a target for alzheimer’s beta amyloid leading to misfolded p53 and inappropriate cell survival. PLOS ONE 5 (4), e10171. doi:10.1371/journal.pone.0010171
Larribère, L., Galach, M., Novak, D., Arévalo, K., Volz, H. C., Stark, H. J., et al. (2017). An RNAi screen reveals an essential role for HIPK4 in human skin epithelial differentiation from iPSCs. Stem Cell Rep. 9 (4), 1234–1245. doi:10.1016/j.stemcr.2017.08.023
Lavra, L., Rinaldo, C., Ulivieri, A., Luciani, E., Fidanza, P., Giacomelli, L., et al. (2011). The loss of the p53 activator HIPK2 is responsible for galectin-3 overexpression in well differentiated thyroid carcinomas. PLOS ONE 6 (6), e20665. doi:10.1371/journal.pone.0020665
Lazaro-Pena, M. I., Cornwell, A. B., Diaz-Balzac, C. A., Das, R., Ward, Z. C., Macoretta, N., et al. (2023). “Homeodomain-interacting protein kinase maintains neuronal homeostasis during normal Caenorhabditis elegans aging and systemically regulates longevity from serotonergic and GABAergic neurons. eLife, 12. doi:10.7554/eLife.85792
Lazzari, C., Prodosmo, A., Siepi, F., Rinaldo, C., Galli, F., Gentileschi, M., et al. (2011). HIPK2 phosphorylates ΔNp63α and promotes its degradation in response to DNA damage. Oncogene 30 (48), 4802–4813. doi:10.1038/onc.2011.182
Lee, K., Drakas, R., and He, J. C. (2024). Small molecule allosteric inhibitor of HIPK2 as a novel therapy against kidney fibrosis. J. Am. Soc. Nephrol. 35, 809–811. doi:10.1681/ASN.0000000000000327
Lee, S., Shang, Y., Redmond, S., Urisman, A., Tang, A., Li, K., et al. (2016). Activation of HIPK2 promotes ER stress-mediated neurodegeneration in amyotrophic lateral sclerosis. Neuron 91 (1), 41–55. doi:10.1016/j.neuron.2016.05.021
Lee, W., Andrews, B. C., Faust, M., Walldorf, U., and Verheyen, E. M. (2009a). Hipk is an essential protein that promotes Notch signal transduction in the Drosophila eye by inhibition of the global co-repressor Groucho. Dev. Biol. 325 (1), 263–272. doi:10.1016/j.ydbio.2008.10.029
Lee, W., Swarup, S., Chen, J., Ishitani, T., and Verheyen, E. M. (2009b). Homeodomain-interacting protein kinases (Hipks) promote Wnt/Wg signaling through stabilization of β-catenin/Arm and stimulation of target gene expression. Development 136 (2), 241–251. doi:10.1242/dev.025460
Li, F. P., and Fraumeni, J. F. (1969). Soft-tissue sarcomas, breast cancer, and other neoplasms. Ann. Intern. Med. 71 (4), 747–752. doi:10.7326/0003-4819-71-4-747
Li, R., Shang, J., Zhou, W., Jiang, L., Xie, D., and Tu, G. (2018). Overexpression of HIPK2 attenuates spinal cord injury in rats by modulating apoptosis, oxidative stress, and inflammation. Biomed. and Pharmacother. 103, 127–134. doi:10.1016/j.biopha.2018.03.117
Liang, L., Xie, R., Lu, R., Ma, R., Wang, X., Wang, F., et al. (2020). Involvement of homodomain interacting protein kinase 2-c-Jun N-terminal kinase/c-Jun cascade in the long-term synaptic toxicity and cognition impairment induced by neonatal Sevoflurane exposure. J. Neurochem. 154 (4), 372–388. doi:10.1111/jnc.14910
Liu, R., Das, B., Xiao, W., Li, Z., Li, H., Lee, K., et al. (2017). A novel inhibitor of homeodomain interacting protein kinase 2 mitigates kidney fibrosis through inhibition of the TGF-β1/smad3 pathway. J. Am. Soc. Nephrol. 28 (7), 2133–2143. doi:10.1681/ASN.2016080841
Liu, X., Zang, C., Wu, Y., Meng, R., Chen, Y., Jiang, T., et al. (2022). Homeodomain-interacting protein kinase HIPK4 regulates phosphorylation of manchette protein RIMBP3 during spermiogenesis. J. Biol. Chem. 298 (9), 102327. doi:10.1016/j.jbc.2022.102327
Lu, J. (2015). Palbociclib: a first-in-class CDK4/CDK6 inhibitor for the treatment of hormone-receptor positive advanced breast cancer. J. Hematol. and Oncol. 8 (1), 98. doi:10.1186/s13045-015-0194-5
Łukasik, P., Baranowska-Bosiacka, I., Kulczycka, K., and Gutowska, I. (2021). Inhibitors of cyclin-dependent kinases: types and their mechanism of action. Int. J. Mol. Sci. 22 (6), 2806. doi:10.3390/ijms22062806
Manning, G., Whyte, D. B., Martinez, R., Hunter, T., and Sudarsanam, S. (2002). The protein kinase complement of the human genome. Science 298 (5600), 1912–1934. doi:10.1126/science.1075762
McAllister-Williams, R. H., Bertrand, D., Rollema, H., Hurst, R. S., Spear, L. P., Kirkham, T. C., et al. (2010). Protein kinase inhibitors. Encycl. Psychopharmacol., 1075. doi:10.1007/978-3-540-68706-1_3498
Miduturu, C. V., Deng, X., Kwiatkowski, N., Yang, W., Brault, L., Filippakopoulos, P., et al. (2011). High-throughput kinase profiling: a more efficient approach toward the discovery of new kinase inhibitors. Chem. and Biol. 18 (7), 868–879. doi:10.1016/j.chembiol.2011.05.010
Naik, R. R., Shakya, A. K., Aladwan, S. M., and El-Tanani, M. (2022). Kinase inhibitors as potential therapeutic agents in the treatment of COVID-19. Front. Pharmacol. 13, 806568. doi:10.3389/fphar.2022.806568
Němec, V., Hylsová, M., Maier, L., Flegel, J., Sievers, S., Ziegler, S., et al. (2019). Furo[3,2-b]pyridine: a privileged scaffold for highly selective kinase inhibitors and effective modulators of the hedgehog pathway. Angew. Chem. Int. Ed. 58 (4), 1062–1066. doi:10.1002/anie.201810312
Němec, V., Maier, L., Berger, B. T., Chaikuad, A., Drápela, S., Souček, K., et al. (2021). Highly selective inhibitors of protein kinases CLK and HIPK with the furo[3,2-b]pyridine core. Eur. J. Med. Chem. 215, 113299. doi:10.1016/j.ejmech.2021.113299
Oda, E., Ohki, R., Murasawa, H., Nemoto, J., Shibue, T., Yamashita, T., et al. (2000a). Noxa, a BH3-only member of the bcl-2 family and candidate mediator of p53-induced apoptosis. Science 288 (5468), 1053–1058. doi:10.1126/science.288.5468.1053
Oda, K., Arakawa, H., Tanaka, T., Matsuda, K., Tanikawa, C., Mori, T., et al. (2000b). p53AIP1, a potential mediator of p53-dependent apoptosis, and its regulation by ser-46-phosphorylated p53. Cell 102 (6), 849–862. doi:10.1016/S0092-8674(00)00073-8
Oren, M. (2003). Decision making by p53: life, death and cancer. Cell Death and Differ. 10 (4), 431–442. doi:10.1038/sj.cdd.4401183
Overstreet, J. M., Gifford, C. C., Tang, J., Higgins, P. J., and Samarakoon, R. (2022). Emerging role of tumor suppressor p53 in acute and chronic kidney diseases. Cell. Mol. Life Sci. 79 (9), 474. doi:10.1007/s00018-022-04505-w
Pagano, M. A., Bain, J., Kazimierczuk, Z., Sarno, S., Ruzzene, M., Di Maira, G., et al. (2008). The selectivity of inhibitors of protein kinase CK2: an update. Biochem. J. 415 (3), 353–365. doi:10.1042/BJ20080309
Palumbo, A., Lau, G., and Saraceni, M. (2019). Abemaciclib: the newest CDK4/6 inhibitor for the treatment of breast cancer. Ann. Pharmacother. 53 (2), 178–185. doi:10.1177/1060028018795146
Pearson, G., Robinson, F., Beers Gibson, T., Xu, Be, Karandikar, M., Berman, K., et al. (2001). Mitogen-activated protein (MAP) kinase pathways: regulation and physiological functions. Endocr. Rev. 22 (2), 153–183. doi:10.1210/edrv.22.2.0428
Pierre, F., Chua, P. C., O’Brien, S. E., Siddiqui-Jain, A., Bourbon, P., Haddach, M., et al. (2011). Discovery and SAR of 5-(3-chlorophenylamino)benzo[ c ] [2,6]naphthyridine-8-carboxylic acid (CX-4945), the first clinical stage inhibitor of protein kinase CK2 for the treatment of cancer. J. Med. Chem. 54 (2), 635–654. doi:10.1021/jm101251q
Polonio-Vallon, T., Kirckpatrick, J., Krijgsveld, J., and Hofmann, T. (2014). Src kinase modulates the apoptotic p53 pathway by altering HIPK2 localization. Cell Cycle 13 (1), 115–125. doi:10.4161/cc.26857
Poon, C. L. C., Zhang, X., Lin, J., Manning, S., and Harvey, K. (2012). Homeodomain-interacting protein kinase regulates Hippo pathway-dependent tissue growth. Curr. Biol. 22 (17), 1587–1594. doi:10.1016/j.cub.2012.06.075
Poratti, M., and Marzaro, G. (2019). Third-generation CDK inhibitors: a review on the synthesis and binding modes of Palbociclib, Ribociclib and Abemaciclib. Eur. J. Med. Chem. 172, 143–153. doi:10.1016/j.ejmech.2019.03.064
Purzner, T., Purzner, J., Buckstaff, T., Cozza, G., Gholamin, S., Rusert, J. M., et al. (2018). Developmental phosphoproteomics identifies the kinase CK2 as a driver of Hedgehog signaling and a therapeutic target in medulloblastoma. Sci. Signal. 11 (547), eaau5147. doi:10.1126/scisignal.aau5147
Qin, Y., Hu, Q., Ji, S., Xu, J., Dai, W., Liu, W., et al. (2019). Homeodomain-interacting protein kinase 2 suppresses proliferation and aerobic glycolysis via ERK/cMyc axis in pancreatic cancer. Cell Prolif. 52 (3), e12603. doi:10.1111/cpr.12603
Rinaldo, C., Siepi, F., Prodosmo, A., and Soddu, S. (2008). HIPKs: jack of all trades in basic nuclear activities. Biochimica Biophysica Acta (BBA) - Mol. Cell Res. 1783 (11), 2124–2129. doi:10.1016/j.bbamcr.2008.06.006
Ritter, O., and Schmitz, M. L. (2019). Differential intracellular localization and dynamic nucleocytoplasmic shuttling of homeodomain-interacting protein kinase family members. Biochimica Biophysica Acta (BBA) - Mol. Cell Res. 1866 (10), 1676–1686. doi:10.1016/j.bbamcr.2019.04.009
Roskoski, R. (2021). Hydrophobic and polar interactions of FDA-approved small molecule protein kinase inhibitors with their target enzymes. Pharmacol. Res. 169, 105660. doi:10.1016/j.phrs.2021.105660
Roskoski, R. (2024). Properties of FDA-approved small molecule protein kinase inhibitors: a 2024 update. Pharmacol. Res. 200, 107059. doi:10.1016/j.phrs.2024.107059
Rugo, H. S., Kabos, P., Beck, J. T., Jerusalem, G., Wildiers, H., Sevillano, E., et al. (2022). Abemaciclib in combination with pembrolizumab for HR+, HER2− metastatic breast cancer: phase 1b study. npj Breast Cancer 8 (1), 118–8. doi:10.1038/s41523-022-00482-2
Rui, Y., Xu, Z., Lin, S., Li, Q., Rui, H., Luo, W., et al. (2004). Axin stimulates p53 functions by activation of HIPK2 kinase through multimeric complex formation. EMBO J. 23 (23), 4583–4594. doi:10.1038/sj.emboj.7600475
Sato, M., Muragaki, Y., Saika, S., Roberts, A. B., and Ooshima, A. (2003). Targeted disruption of TGF-β1/Smad3 signaling protects against renal tubulointerstitial fibrosis induced by unilateral ureteral obstruction. J. Clin. Investigation 112 (10), 1486–1494. doi:10.1172/JCI19270
Saul, V. V., de la Vega, L., Milanovic, M., Krüger, M., Braun, T., Fritz-Wolf, K., et al. (2013). HIPK2 kinase activity depends on cis-autophosphorylation of its activation loop. J. Mol. Cell Biol. 5 (1), 27–38. doi:10.1093/jmcb/mjs053
Schmitz, M. L., Rodriguez-Gil, A., and Hornung, J. (2014). Integration of stress signals by homeodomain interacting protein kinases. Biol. Chem. 395 (4), 375–386. doi:10.1515/hsz-2013-0264
Schneider, C. C., Kartarius, S., Montenarh, M., Orzeszko, A., and Kazimierczuk, Z. (2012). Modified tetrahalogenated benzimidazoles with CK2 inhibitory activity are active against human prostate cancer cells LNCaP in vitro. Bioorg. and Med. Chem. 20 (14), 4390–4396. doi:10.1016/j.bmc.2012.05.038
Schulten, H.-J., Hussein, D., Al-Adwani, F., Karim, S., Al-Maghrabi, J., Al-Sharif, M., et al. (2016). Microarray expression data identify DCC as a candidate gene for early meningioma progression. PLOS ONE 11 (4), e0153681. doi:10.1371/journal.pone.0153681
Shang, Y., Doan, C. N., Arnold, T. D., Lee, S., Tang, A. A., Reichardt, L. F., et al. (2013). Transcriptional corepressors HIPK1 and HIPK2 control angiogenesis via TGF-β–TAK1–dependent mechanism. PLOS Biol. 11 (4), e1001527. doi:10.1371/journal.pbio.1001527
Shojima, N., Hara, K., Fujita, H., Horikoshi, M., Takahashi, N., Takamoto, I., et al. (2012). Depletion of homeodomain-interacting protein kinase 3 impairs insulin secretion and glucose tolerance in mice. Diabetologia 55 (12), 3318–3330. doi:10.1007/s00125-012-2711-1
Siepi, F., Gatti, V., Camerini, S., Crescenzi, M., and Soddu, S. (2013). HIPK2 catalytic activity and subcellular localization are regulated by activation-loop Y354 autophosphorylation. Biochimica Biophysica Acta (BBA) - Mol. Cell Res. 1833 (6), 1443–1453. doi:10.1016/j.bbamcr.2013.02.018
Sierra, O. L., and Towler, D. A. (2010). Runx2 trans-activation mediated by the msx2-interacting nuclear target requires homeodomain interacting protein kinase-3. Mol. Endocrinol. 24 (7), 1478–1497. doi:10.1210/me.2010-0029
Slouka, J., Bekárek, V., and Lyčka, A. (1982). Cyclization reactions of some o-acylphenylhydrazones. Collect. Czechoslov. Chem. Commun. 47 (6), 1746–1756. doi:10.1135/cccc19821746
Sombroek, D., and Hofmann, T. G. (2009). How cells switch HIPK2 on and off. Cell Death and Differ. 16 (2), 187–194. doi:10.1038/cdd.2008.154
Soubeyran, I., Mahouche, I., Grigoletto, A., Leste-Lasserre, T., Drutel, G., Rey, C., et al. (2011). Tissue microarray cytometry reveals positive impact of homeodomain interacting protein kinase 2 in colon cancer survival irrespective of p53 function. Am. J. Pathology 178 (5), 1986–1998. doi:10.1016/j.ajpath.2011.01.021
Stanga, S., Lanni, C., Govoni, S., Uberti, D., D'Orazi, G., and Racchi, M. (2010). Unfolded p53 in the pathogenesis of Alzheimer’s disease: is HIPK2 the link? Aging 2 (9), 545–554. doi:10.18632/aging.100205
Stefano, V. D., Blandino, G., Sacchi, A., Soddu, S., and D'Orazi, G. (2004). HIPK2 neutralizes MDM2 inhibition rescuing p53 transcriptional activity and apoptotic function. Oncogene 23 (30), 5185–5192. doi:10.1038/sj.onc.1207656
Sung, K. S., Kim, S. J., Cho, S. W., Park, Y. J., Tae, K., and Choi, C. Y. (2019). Functional impairment of the HIPK2 small ubiquitin-like modifier (SUMO)-interacting motif in acute myeloid leukemia. Am. J. Cancer Res. 9 (1), 94–107.
Tao, L., Xu, C., Shen, W., Tan, J., Li, L., Fan, M., et al. (2022). HIPK3 inhibition by exosomal hsa-miR-101-3p is related to metabolic reprogramming in colorectal cancer. Front. Oncol. 11. doi:10.3389/fonc.2021.758336
Tettweiler, G., Blaquiere, J. A., Wray, N. B., and Verheyen, E. M. (2019). Hipk is required for JAK/STAT activity during development and tumorigenesis. PLOS ONE 14 (12), e0226856. doi:10.1371/journal.pone.0226856
Thomson, R. J., Moshirfar, M., and Ronquillo, Y. (2024). “Tyrosine kinase inhibitors,” in StatPearls (Treasure Island (FL): StatPearls Publishing). Available at: http://www.ncbi.nlm.nih.gov/books/NBK563322/(Accessed: November 1, 2024).
Tokumitsu, H., Inuzuka, H., Ishikawa, Y., Ikeda, M., Saji, I., and Kobayashi, R. (2002). STO-609, a specific inhibitor of the Ca2+/Calmodulin-dependent protein kinase kinase. J. Biol. Chem. 277 (18), 15813–15818. doi:10.1074/jbc.M201075200
Torrente, L., Sanchez, C., Moreno, R., Chowdhry, S., Cabello, P., Isono, K., et al. (2017). Crosstalk between NRF2 and HIPK2 shapes cytoprotective responses. Oncogene 36 (44), 6204–6212. doi:10.1038/onc.2017.221
Umezawa, K., and Kii, I. (2021). Druggable transient pockets in protein kinases. Molecules 26 (3), 651. doi:10.3390/molecules26030651
Van Der Laden, J., Soppa, U., and Becker, W. (2015). Effect of tyrosine autophosphorylation on catalytic activity and subcellular localisation of homeodomain-interacting protein kinases (HIPK). Cell Commun. Signal. 13 (1), 3. doi:10.1186/s12964-014-0082-6
Varley, J. m. (2003). Germline TP53 mutations and Li-Fraumeni syndrome. Hum. Mutat. 21 (3), 313–320. doi:10.1002/humu.10185
Wade, M., Wang, Y. V., and Wahl, G. M. (2010). The p53 orchestra: mdm2 and Mdmx set the tone. Trends Cell Biol. 20 (5), 299–309. doi:10.1016/j.tcb.2010.01.009
Wadey, K. S., Brown, B., Sala-Newby, G., Jayaraman, P. S., Gaston, K., and George, S. (2017). Protein kinase CK2 inhibition suppresses neointima formation via a proline-rich homeodomain-dependent mechanism. Vasc. Pharmacol. 99, 34–44. doi:10.1016/j.vph.2017.09.004
Wang, L., Li, M., Sha, B., Hu, X., Sun, Y., Zhu, M., et al. (2021). Inhibition of deubiquitination by PR-619 induces apoptosis and autophagy via ubi-protein aggregation-activated ER stress in oesophageal squamous cell carcinoma. Cell Prolif. 54 (1), e12919. doi:10.1111/cpr.12919
Wang, S., Yi, X., Wu, Z., Guo, S., Dai, W., Wang, H., et al. (2022). CAMKK2 defines ferroptosis sensitivity of melanoma cells by regulating AMPK‒NRF2 pathway. J. Investigative Dermatology 142 (1), 189–200.e8. doi:10.1016/j.jid.2021.05.025
Wang, S. J. H., Sinclair, D. A. R., Kim, H. Y., Kinsey, S. D., Yoo, B., Shih, C. R. Y., et al. (2020). Homeodomain-interacting protein kinase (Hipk) plays roles in nervous system and muscle structure and function. PLOS ONE 15 (3), e0221006. doi:10.1371/journal.pone.0221006
Wei, G., Ku, S., Ma, G. K., Saito, S., Tang, A. A., Zhang, J., et al. (2007). HIPK2 represses β-catenin-mediated transcription, epidermal stem cell expansion, and skin tumorigenesis. Proc. Natl. Acad. Sci. 104 (32), 13040–13045. doi:10.1073/pnas.0703213104
Wekking, D., Lambertini, M., Dessì, M., Denaro, N., Bardanzellu, F., Garrone, O., et al. (2023). CDK4/6 inhibitors in the treatment of metastatic breast cancer: focus on toxicity and safety. Seminars Oncol. 50, 131–139. doi:10.1053/j.seminoncol.2024.01.002
Winter, M., Sombroek, D., Dauth, I., Moehlenbrink, J., Scheuermann, K., Crone, J., et al. (2008). Control of HIPK2 stability by ubiquitin ligase Siah-1 and checkpoint kinases ATM and ATR. Nat. Cell Biol. 10 (7), 812–824. doi:10.1038/ncb1743
Wu, P., Nielsen, T. E., and Clausen, M. H. (2015). FDA-approved small-molecule kinase inhibitors. Trends Pharmacol. Sci. 36 (7), 422–439. doi:10.1016/j.tips.2015.04.005
Xiao, W., Wang, T., Ye, Y., Wang, X., Chen, B., Xing, J., et al. (2021). Identification of HIPK3 as a potential biomarker and an inhibitor of clear cell renal cell carcinoma. Aging 13 (3), 3536–3553. doi:10.18632/aging.202294
Xiao, W., E, J., Bao, L., Fan, Y., Jin, Y., Wang, A., et al. (2020). Tubular HIPK2 is a key contributor to renal fibrosis. JCI Insight 5 (17), e136004. doi:10.1172/jci.insight.136004
Zhang, D., Sun, L., xian, W., Liu, F., Ling, G., Xiao, L., et al. (2010). Low-dose paclitaxel ameliorates renal fibrosis in rat UUO model by inhibition of TGF-β/Smad activity. Lab. Investig. 90 (3), 436–447. doi:10.1038/labinvest.2009.149
Zhang, F., Qi, L., Feng, Q., Zhang, B., Li, X., Liu, C., et al. (2021). HIPK2 phosphorylates HDAC3 for NF-κB acetylation to ameliorate colitis-associated colorectal carcinoma and sepsis. Proc. Natl. Acad. Sci. 118 (28), e2021798118. doi:10.1073/pnas.2021798118
Zhang, J., Pho, V., Bonasera, S. J., Holtzman, J., Tang, A. T., Hellmuth, J., et al. (2007). Essential function of HIPK2 in TGFβ-dependent survival of midbrain dopamine neurons. Nat. Neurosci. 10 (1), 77–86. doi:10.1038/nn1816
Zhang, J., Shang, Y., Kamiya, S., Kotowski, S. J., Nakamura, K., and Huang, E. J. (2020). Loss of HIPK2 protects neurons from mitochondrial toxins by regulating Parkin protein turnover. J. Neurosci. 40 (3), 557–568. doi:10.1523/JNEUROSCI.2017-19.2019
Zhang, Q., Nottke, A., and Goodman, R. H. (2005). Homeodomain-interacting protein kinase-2 mediates CtBP phosphorylation and degradation in UV-triggered apoptosis. Proc. Natl. Acad. Sci. 102 (8), 2802–2807. doi:10.1073/pnas.0409373102
Zhang, Q., and Wang, Y. (2007). Homeodomain-interacting protein kinase-2 (HIPK2) phosphorylates HMGA1a at ser-35, thr-52, and thr-77 and modulates its DNA binding affinity. J. Proteome Res. 6 (12), 4711–4719. doi:10.1021/pr700571d
Zhong, W., Hong, C., Dong, Y., Li, Y., Xiao, C., and Liu, X. (2022). ASH2L aggravates fibrosis and inflammation through HIPK2 in high glucose-induced glomerular mesangial cells. Genes 13 (12), 2244. doi:10.3390/genes13122244
Zhou, Q., Deng, J., Yao, J., Song, J., Meng, D., Zhu, Y., et al. (2021). Exercise downregulates HIPK2 and HIPK2 inhibition protects against myocardial infarction. eBioMedicine 74, 103713. doi:10.1016/j.ebiom.2021.103713
Keywords: HIPK, kinase, inhibitor, selectivity, neurology, cancer
Citation: Štefek A and Paruch K (2025) Biology and pharmacological inhibition of homeodomain-interacting protein kinases. Front. Chem. Biol. 4:1441138. doi: 10.3389/fchbi.2025.1441138
Received: 30 May 2024; Accepted: 24 January 2025;
Published: 25 February 2025.
Edited by:
Sandra Berndt, Leipzig University, GermanyReviewed by:
Andrew Samuelson, University of Rochester, United StatesCopyright © 2025 Štefek and Paruch. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kamil Paruch, cGFydWNoQGNoZW1pLm11bmkuY3o=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.