
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Chem. Biol. , 12 July 2024
Sec. Molecular Sciences
Volume 3 - 2024 | https://doi.org/10.3389/fchbi.2024.1428646
This article is part of the Research Topic Structure-function relationship of enzymes through a chemical lens View all 6 articles
 Nigel G. J. Richards1,2*
Nigel G. J. Richards1,2* James H. Naismith3
James H. Naismith3Remarkable progress has been made to elucidate the structural and mechanistic enzymology of the biosynthetic pathways that give rise to naturally occurring C-nucleosides. These compounds are generally cytotoxic and exhibit interesting antiviral, antibiotic and anti-parasitic activity. Here we review current knowledge concerning formycin biosynthesis and highlight deficiencies in our understanding of key chemical transformations in the pathway.
There is renewed interest in the synthesis and clinical application of modified nucleosides and nucleotides (Duffy et al., 2020; Schramm, 2018; Seley-Radtke and Yates, 2018; Yadav et al., 2019). These compounds represent lead structures for antiviral drug discovery (De Clercq, 2016), and are finding use as components of expanded genetic alphabets (Hoshika et al., 2019; Pfeiffer and Nidetzky, 2020), diagnostic reagents (Sefah et al., 2014), and drug delivery systems (Zhang L. et al., 2020). Notably, the effectiveness of remdesivir in treating SARS-CoV-2 infections (Mackman, 2022) has prompted a re-evaluation of the clinical utility of C-nucleosides; compounds in which the nucleobase is connected to the sugar by a C-C rather than the more labile C-N bond. In nature, C-nucleoside linkages are present in numerous microbial secondary metabolites that exhibit interesting antibiotic and anti-viral activities, and the past decade has seen considerable progress in understanding the biosynthetic origins of these natural products (Shiraishi and Kuzuyama, 2019; Sosio et al., 2018; Zhang et al., 2022). In this mini-review, we discuss current knowledge about the pathway leading to the formycins 1-3 (Figure 1A), which are pyrazole-containing bioactive compounds (Santos et al., 2020), and identify gaps in our understanding that remain to be closed.
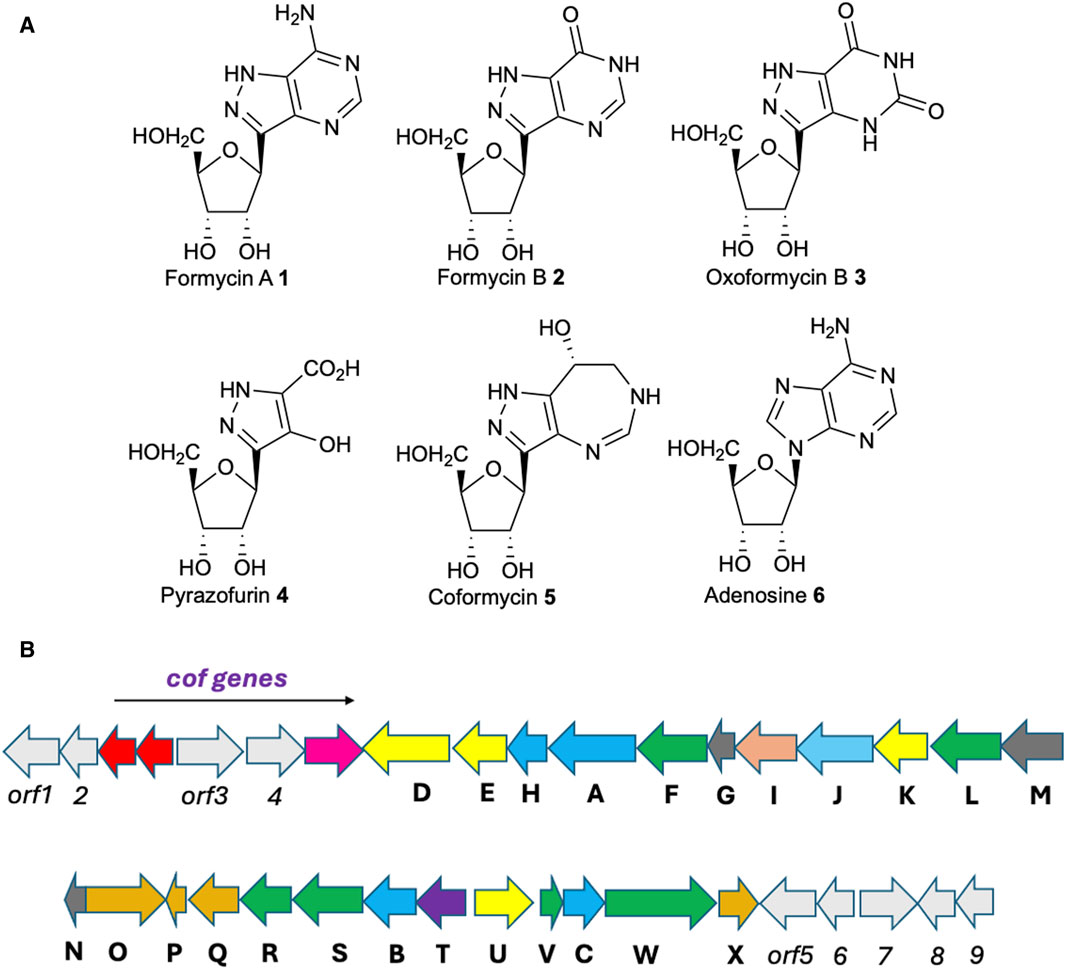
Figure 1. (A) Structures of compounds discussed in the text. (B) Gene cluster encoding coformycin (indicated by the arrow) and formycin biosynthesis. Color scheme: yellow, oxygenases; salmon, aminotransferase; orange, hydroxylases; purple, C-glycoside synthase; blue, homologs of enzymes involved in de novo purine biosynthesis; green, oxidoreductases; sky blue, MetRS homolog; grey, hypothetical enzymes. Functional annotations are provided in Table 1.
Formycins A 1 and B 2 (Figure 1A) were first isolated from Nocardia interforma approximately 60 years ago (Hori et al., 1964; Koyama and Umezawa, 1965), and subsequently in cultures of Streptomyces kaniharaensis SF-557 (Zhu et al., 2020), Streptomyces lavendulae (Aizawa et al., 1965) and Streptomyces resistomycificus NRLL 2290 (Zhang M. et al., 2020). There have been numerous studies into their structure, chemical properties, and biological activity (Robins et al., 1966; Ishizuka et al., 1968; Prusiner et al., 1973; Chenon et al., 1976). As well as being C-nucleosides, the pyrazolopyrimidine ring features an N-N bond, a functional group that is relatively uncommon in natural products (Blair and Sperry, 2013; Waldman et al., 2017). 1H NMR spectroscopy was used to establish the tautomeric preferences of the pyrazolopyrimidine in solution, with the major species being protonated at N7 (Krugh, 1973) consistent with quantum chemical calculations (Orozco and Luque, 1995).
After cellular phosphorylation at C-5’ (Mehta and Gupta, 1985), formycins A 1 and B 2 become nonhydrolyzable analogues of AMP and GMP, respectively exhibiting anti-tumor, anti-viral (Dapp et al., 2014), antibiotic and anti-parasitic activity (Bzowska, 2008). The compounds likely exert their biological effects via a number of possible molecular mechanisms, including inhibition of the enzymes AMP nucleosidase (Ehrlich and Schramm, 1994) and purine nucleoside phosphorylase (PNP) (Kierdaszuk et al., 2000). Indeed, the biological properties of formycin A have inspired the synthesis of analogs that exhibit picomolar affinity for human PNP (Ho et al., 2010) and suppress T-cell activity by interfering with purine recycling (Cohen et al., 1978; Markert, 1991). The incorporation of formycin A into RNA, presumably via the triphosphate, is also thought to be the molecular mechanism underlying its ability to kill Leishmania parasites (Rainey and Santi, 1983). Given these findings, it is perhaps surprising that neither formycins nor their derivatives have found clinical use.
Although little was known about the molecular details of formycin biosynthesis until recently, early work established that formycin B 2 is a precursor of formycin A 1 in N. interforma (Sawa et al., 1968). Subsequent feeding experiments using 13C and 15N-labeled compounds then revealed that i) lysine provides (at least) one of the nitrogen atoms in the pyrazole moiety (Ochi et al., 1976), and ii) four carbon atoms in the pyrazolopyrimidine ring are obtained from glutamate (or a related metabolite such as 2-oxoglutarate) (Ochi et al., 1979). 14C-incorporation studies also demonstrated that 5′-phosphoribosyl-1′-pyrophosphate (PRPP) is the source of the ribose ring in the formycins (Kunimoto et al., 1971). Glutamate is a biosynthetic precursor of other C-nucleoside natural products (Elstner and Suhadolnik, 1972; Isono and Suhadolnik, 1977), including pyrazofurin A 4 (Figure 1A) (Buchanan et al., 1980; Suhadolnik and Reichenbach, 1981); the latter finding has subsequently proved essential for efforts to understand the chemistry of formycin biosynthesis (vide infra) (Ren et al., 2019; Zhang L. et al., 2020).
After approximately 35 years of little progress towards elucidating the metabolic pathway leading to the formycins, whole genome sequencing of S. kaniharaensis SF-557 allowed both our group (Zhu et al., 2020), and (independently) Liu and his co-workers (Ko et al., 2017; Wang et al., 2019), to identify the biosynthetic gene cluster (BGC) encoding the enzymes that mediate formycin synthesis (Figure 1B; Table 1). In addition to exploiting comparative bioinformatics analysis (Watam et al., 2018), identification of the BGC relied on the presence of genes encoding homologs to both lysine N-hydroxylating monooxygenase (Franceschini et al., 2012) and 4-(β-D-ribofuranosyl)aminobenzene 5′-phosphate (RFA-P) synthase, which forms a C-glycoside from PRPP and para-aminobenzoic acid (Rasche and White, 1998; Dumitru and Ragsdale, 2004; White, 2011; Bechard et al., 2019) during the biosynthesis of the modified folate methanopterin (White, 1996). Direct experimental evidence was provided, however, by Liu and co-workers (Wang et al., 2019). Thus, using the donor strain ET12567/pUSZ8002 (Paranthaman and Dharmalingam, 2003) to introduce Streptomyces-Escherichia coli shuttle vectors (Sun et al., 2009) into S. kaniharaensis SF-557, mutant strains were obtained containing in-frame deletions of the forC, forF, forH, forL, forT, and forU genes. Formycin A production in these deletion mutants was either abolished or significantly diminished, with the exception of the Δ forC and Δ forL mutants (Wang et al., 2019). Presumably, other enzymes in the organism can perform the transformations associated with ForC and ForL. Similar experiments to show that ForW and ForX are required in the biosynthetic pathway, however, have not yet been reported. Remarkably, the formycin BGC therefore consists of 24 ORFs, three of which encode two enzymes and a transporter involved in the biosynthesis of coformycin 5, an evolutionarily unrelated nucleoside (Figure 1A) (Ren et al., 2020). Formycin and coformycin (Nakamura et al., 1974), which is a potent inhibitor of adenosine deaminase (ADA) (Frieden et al., 1980), are therefore produced by S. kaniharaensis SF-557 at the same time. This co-production of adenosine-like, non-canonical nucleosides and ADA inhibitors is frequently observed, and may represent a general mechanism to prevent ADA-catalyzed deamination of nucleoside-containing natural products (Xu et al., 2018).
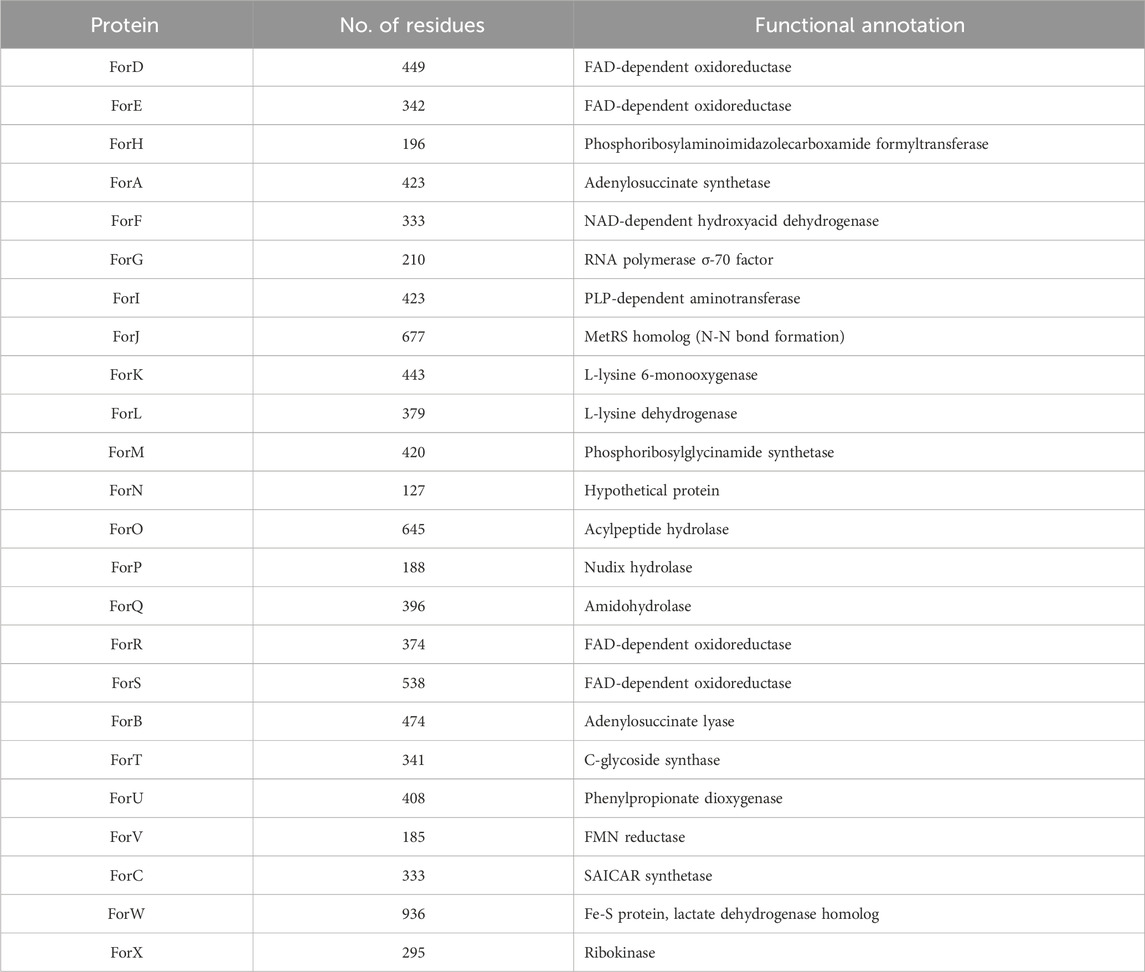
Table 1. Actual and putative (red or purple) functional annotations of proteins encoded by the 24 ORFs in the formycin BGC found in Streptomyces kaniharaensis SF-557.
The large number of open reading frames (ORFs) in the formycin BGC was unexpected given the similarity of this natural product to adenosine 6 (Figure 1A), which is formed by microorganisms from α-D-ribose-5′-phosphate in only eleven enzyme-catalyzed steps (Buchanan and Hartman, 1959; Kappock et al., 2000). The enzymes ForA, ForB, ForC, and ForH are indeed homologs of those that build the six-membered ring in purine nucleobases (Ko et al., 2017). Moreover, some of the enzymes encoded in the formycin BGC are homologous to those that mediate pyrazofurin A biosynthesis, suggesting that the pathways to formycin and pyrazofurin A (Figure 1A) proceed via a series of common intermediates (Ren et al., 2019; M; Zhang et al., 2020b; Zhao et al., 2020). Due to some confusion in the literature, in this review we use the naming convention for enzymes in the pyrazofurin BGC devised by Ryan and Du (Zhao et al., 2020). Thus, genes encoding a lysine N6-hydroxylating monooxygenase are named ForK/PyrM and a methionyl-tRNA synthase homolog are denoted ForJ/PyrN (M. Zhang L. et al., 2020; Zhao et al., 2020). The function of these two enzymes, which are found in other microbial pathways leading to natural products possessing an N-N bond (He et al., 2022; Matsuda et al., 2022a; Twigg et al., 2019; Zheng et al., 2024), is to couple lysine to glutamate in the early stages of formycin and pyrazofurin 4 (Figure 1A) (Zhao et al., 2021). Similarly, the genes ForT and PyrE encode homologous enzymes that catalyse C-C bond formation (Gao et al., 2020; Ren et al., 2019; M; Zhang et al., 2020b).
The deduced primary structures of ForK, and the homologous enzyme PyrM, identify these enzymes as lysine N-hydroxylating monooxygenases, similar to the well-characterized enzyme used in kutzneride biosynthesis (Neumann et al., 2012; Setser et al., 2014). ForK seems likely to use dioxygen to hydroxylate the ε-amino group of lysine 7 (Supplementary Figure S1A) in an interesting reaction that likely proceeds via homolytic O-O cleavage in a flavin hydroperoxide intermediate (Badieyan et al., 2015). Work in our group has confirmed that ForK is an FAD-dependent enzyme for which NAD(P)H is the reducing agent. As observed for other microbial FAD-dependent N-hydroxylating monooxygenases (Mügge et al., 2020), ForK is a tetramer at high concentration but becomes dimeric as the protein concentration is decreased.
Given that the hydroxy substituent is a leaving group, especially if activated by acetylation or phosphorylation, the chemical logic of hydroxylating the side chain nitrogen is to activate it for nucleophilic attack by an amine to form an N-N bond. This transformation is likely accomplished by ForJ, based on recent studies of the catalytic activity of the homologous enzyme PyrN in pyrazofurin A biosynthesis, which mediates the coupling of N6-hydroxylysine 8 and glutamate 9 via an N-N bond (Supplementary Figure S1B) (Zhao et al., 2021). As is the case for ForK, PyrN is composed of two domains, one of which is homologous to methionyl-tRNA synthetase (MetRS) (Mechulam et al., 1999). The other, smaller domain binds one Zn2+ ion and is a member of the cupin superfamily of enzymes (Dunwell et al., 2000). Elegant experiments using truncated PyrN variants showed that the MetRS-like domain mediates formation of a reactive ester 10 (Supplementary Figure S1B), which undergoes acyl-transfer to form an N-hydroxyamide 11 in the absence of the cupin domain (Zhao et al., 2021). Full-length PyrN, however, yields the hydrazine-containing intermediate 12 when incubated with ATP, glutamic acid and N6-hydroxylysine (Supplementary Figure S1B) (Zhao et al., 2021). Thus, the cupin domain plays an essential role in promoting N-N bond formation. Although PyrN could not be crystallized, genome mining identified a cupin protein from Rhodococcus jostii RHA1 (also called RHS1) that catalyzes N-N bond formation (Zhao et al., 2021) and for which an X-ray crystal structure is available showing the presence of a Zn2+-binding site (PDB: 5UQP). QM/MM calculations support a mechanism in which N-N bond formation takes place by reaction of the ester formed by the MetRS-like domain and a second glutamate molecule, which are both coordinated to the Zn2+ center (Zhao et al., 2021). The N-N bond formation in the proposed mechanism, however, proceeds via a formal 6-endo-tet SN2 reaction, which is technically disfavored by Baldwin’s rules (Baldwin, 1976; Gilmore et al., 2016). The calculated barrier for this PyrN-catalyzed step, which is 13.3 kcal/mol, remains to be validated by kinetic experiments.
Based on their sequence homology to a monooxygenase encoded by the Spb39 gene (Matsuda et al., 2017), which forms hydrazinoacetic acid in the pathway leading to a dipeptide natural product containing an N-N bond (Matsuda et al., 2017), ForL and PyrL are probably FAD-dependent monooxygenases, which carry out C-H abstraction on 12 to give an intermediate that spontaneously breaks down to give the aldehyde 13 and hydrazinoglutamate 14 (Supplementary Figure S1B). The detailed kinetic and structural characterization of these enzymes, however, has not yet been reported. This machinery for making N-N bonds appears to be conserved in bacteria to make a wide variety of natural products in addition to pyrazofurin A and the formycins (Matsuda et al., 2022b; Matsuda and Wakimoto, 2024).
ForT is a C-glycoside synthase that couples PRPP 15 with 4-amino-1H-pyrazole-3,5-dicarboxylate (APDA) 16 (Supplementary Figure S2A) (Ren et al., 2019). Remarkably, the very closely related molecule 4-hydroxy-1H-pyrazole-3,5-dicarboxylate (HPDA) 17 is not a substrate, despite the homologous enzyme, PyrE, using both pyrazoles (M. Zhang L. et al., 2020). As a result, HPDA 17 is almost certainly converted to the APDA precursor 19 in the preceding step of the pathway by the PLP-dependent enzyme ForI using L-aspartate as a nitrogen source (Supplementary Figure S2B) (Gao et al., 2019). From an evolutionary perspective, ForT is structurally homologous to homoserine kinase, thereby placing it within the GHMP kinase superfamily of enzymes (Zhou et al., 2000). Using a combination of X-ray crystallography and kinetic studies of a series of ForT variants (Gao et al., 2020; Li et al., 2023), we have been able to obtain evidence that C-C bond formation proceeds via electrophilic substitution to form a tetrahedral intermediate. Subsequent decarboxylation then yields the C-nucleoside 18 (Supplementary Figure S2A), which can be elaborated to a pyrazolopyrimidine (Supplementary Figure S3). The crystal structure of the T138V ForT variant (in which Thr-138 is replaced by valine) bound to APDA 16 and MgATP reveals that the pyrazole substrate forms very few interactions with the enzyme, being held in place by hydrogen bonds to the carboxylate that is retained in the product, thereby correctly positioning APDA relative to the reactive C-1’ carbon of PRPP 15 (Li et al., 2023). The only other APDA/ForT interaction, which is mediated by a water molecule, involves a pyrazole nitrogen and the side chain carboxylate of Glu-221. It is therefore possible that Glu-221 acts as a general base to activate the pyrazole for nucleophilic attack, and replacing this residue with alanine does indeed yield an inactive ForT variant (Li et al., 2023). Given that ForT does not make hydrogen bonds with the amino substituent of the pyrazole, elucidating the mechanism by which it can discriminate between APDA 16 and HPDA 17 will likely require computational strategies (Zhang et al., 2023).
The lack of direct APDA/ForT interactions, however, raises the question of whether the enzyme can couple alternate substrates to PRPP (Gong et al., 2021; Pfeiffer and Nidetzky, 2023A; Pfeiffer and Nidetzky, 2023B). To date, we have shown that ForT can accept an isomeric pyrazole, but the product appears to be the N-nucleoside rather than a C-nucleoside (Li et al., 2023). It is possible that pyrroles may also be substrates for the coupling reaction, in the light of recent work on the biosynthesis of showdomycin (Ren et al., 2021), another C-nucleoside with interesting biological activity (De Clercq, 2016).
Elaboration of the pyrazole-containing C-nucleoside 18 to formycin B 5′-monophosphate 21 is accomplished by an identical series of steps to those used in the de novo formation of inosine 5′-monophosphate (IMP) (Supplementary Figure S3) (Ko et al., 2017). The enzymes ForC, ForB, ForH and ForA, which carry out these steps, are therefore homologs of those used in purine biosynthesis (Kappock et al., 2000). In a surprising observation, however, ForH is a truncated form of PurH, the enzyme that forms the pyrimidine ring during purine biosynthesis (Kappock et al., 2000). As ForH is composed only of a cyclohydrolase domain similar to that present in PurH (Wang et al., 2019), it does not exhibit any formylase activity and cannot convert the pyrazole amide 20 into the N-formylated intermediate 21 that must be cyclized to yield the pyrazolopyrimidine ring in formycin B 5′-monophosphate 22 (Supplementary Figure S3). As shown in a series of elegant experiments by Liu and co-workers, the conversion of 20 into 21 is catalyzed by PurH. On the other hand, although PurH catalyzes the synthesis of the formylated intermediate 21, it cannot use this compound as a substrate. ForH must therefore catalyze the cyclization of 21 to yield formycin B 5′-monophosphate 22 (Wang et al., 2019). Thus, formycin production depends on the presence of PurH, an enzyme required in purine biosynthesis. The enzymes ForA and ForB then convert formycin B 5′-monophosphate 22 into formycin A 5′-monophosphate 23 using identical amination chemistry used in the synthesis of AMP from IMP in purine biosynthesis (Kappock et al., 2000). Formycin B 5′-monophosphate 22 is also the precursor of oxoformycin B 3 (Figure 1A) (Sawa et al., 1968) although the enzyme that oxidizes the pyrazolopyrimidine ring has not yet been identified. Presumably, it is homologous to IMP dehydrogenase (Hedstrom, 2009). Formycin A, formycin B and oxoformycin B are isolated as the nucleosides, however, meaning the 5′-phosphates must be removed by phosphatases prior to release of the C-nucleosides into the environment. The apparent absence of a gene encoding a phosphatase in the formycin BGC implies that these dephosphorylation reactions are catalyzed by one, or more, phosphatases already present in the cell.
Despite the progress made in understanding the metabolic origins of the N-N and C-nucleoside bonds in the formycins, functions for only nine of the 24 proteins encoded in the formycin BGC have been elucidated. Even excluding the gene for a putative transporter that mediates excretion of these C-nucleoside natural products, this leaves a remarkable number of enzymes seemingly required for the conversion of hydrazinoglutamate 14 into HPDA 17. As a result, there is little consensus regarding the reactions needed to bridge these two intermediates in the biosynthetic pathway. The key chemical problems are to form a new C-N bond at C-4 and to hydroxylate C-3 of hydrazinoglutamate 14. In principle, both reactions can be accomplished by oxidizing 14 to obtain an intermediate containing a double bond between C-3 and C-4 but no such desaturase has been identified. The latest proposal therefore postulates a dehydropyridine intermediate, which is easily oxidized to the pyridine 24 (Supplementary Figure S4). Enzyme-catalyzed ring-opening followed by C-N then yields the target pyrazole skeleton (Ren et al., 2019). Although this “simple route” appears consistent with annotated oxygenases and hydroxylases encoded in the cluster (Table 1), C-N bond formation to give the pyrazole proceeds via an energetically disfavored five-endo-trig reaction (Baldwin, 1976; Gilmore et al., 2016). An approach based on in-frame deletion or modification of specific genes in the cluster (Alberti and Corre, 2019), as described for S. kaniharaensis by Liu and co-workers (Wang et al., 2019), coupled to identifying intermediates seems the most promising strategy to resolve the issue (Caesar et al., 2021). Whatever the outcome of such studies, the chemistry used by microorganisms to make and manipulate heterocyclic ring systems will continue to surprise and fascinate natural product chemists (McCarty et al., 2009; Palmu et al., 2017; Kong et al., 2019; Ren et al., 2022).
NR: Writing–original draft, Writing–review and editing. JN: Writing–review and editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. We gratefully acknowledge grants from the UKRI Biotechnology and Biological Sciences Research Council (grants BB/T006161/1 and BB/T006188/1).
We thank Sisi Gao, Georgina C. Girt, Wenbo Li, Ashish Radadiya, and Alan F. Scott for their work on the enzymes ForI and ForT. Valerie de Crécy-Lagard (University of Florida) and Wen Zhu played an essential part in identifying and annotating the biosynthetic gene cluster present in Streptomyces kaniharaensis Shomura and Niida SF-557. We also acknowledge helpful discussions with Hung-wen Liu concerning the chemical synthesis of 4-amino-1H-pyrazole-3,5-dicarboxylic acid.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fchbi.2024.1428646/full#supplementary-material
Aizawa, S., Hidaka, T., Otake, N., Yonehara, H., Isono, K., Igarashi, N., et al. (1965). Studies on a new antibiotic, laurusin. Agric. Biol. Chem. 29 (4), 375–376. doi:10.1080/00021369.1965.10858402
Alberti, F., and Corre, C. (2019). Editing streptomycete genomes in the CRISPR/Cas9 age. Nat. Prod. Rep. 36 (9), 1237–1248. doi:10.1039/c8np00081f
Badieyan, S., Bach, R. D., and Sobrado, P. (2015). Mechanism of N-hydroxylation catalyzed by flavin-dependent monooxygenases. J. Org. Chem. 80 (4), 2139–2147. doi:10.1021/jo502651v
Baldwin, J. E. (1976). Rules for ring closure. J. Chem. Soc. Chem. Commun. (18), 734–736. doi:10.1039/C39760000734
Bechard, M. E., Farahani, P., Greene, D., Pham, A., Orry, A., and Rasche, M. E. (2019). Purification, kinetic characterization, and site-directed mutagenesis of Methanothermobacter therautotrophicus RFAP synthase produced in Escherichia coli. AIMS Microbiol. 5 (3), 186–204. doi:10.3934/microbiol.2019.3.186
Blair, L. M., and Sperry, J. (2013). Natural products containing a nitrogen-nitrogen bond. J. Nat. Prod. 76 (4), 794–812. doi:10.1021/np400124n
Buchanan, J. G., Hamlin, M. R., Sood, G. R., and Wightman, R. H. (1980). The biosynthesis of pyrazofurin and formycin. J. Chem. Soc. Chem. Commun. (19), 917–918. doi:10.1039/c39800000917
Buchanan, J. M., and Hartman, S. C. (1959). Enzymic reactions in the synthesis of the purines. Adv. Enzym. - Relat. Areas Mol. Biol. 21, 199–261. doi:10.1002/9780470122662.ch5
Bzowska, A. (2008). “Formycins and their analogues: purine nucleoside phosphorylase inhibitors and their potential application in immunosuppression and cancer,” in Modified Nucleosides, ed. P. Herdewijn (Weinheim, Germany; Wiley-VCH Verlag GmbH & Co. KGaA), 473. doi:10.1022/9783527623112.ch19
Caesar, L. K., Montaser, R., Keller, N. P., and Kelleher, N. L. (2021). Metabolomics and genomics in natural products research: complementary tools for targeting new chemical entities. Nat. Prod. Rep. 38 (11), 2041–2065. doi:10.1039/d1np00036e
Chenon, M. T., Panzica, R. P., Smith, J. C., Pugmire, R. J., Grant, D. M., and Townsend, L. B. (1976). Carbon-13 magnetic resonance spectra of C-nucleosides. 3. Tautomerism in formycin and formycin B and certain pyrazolo[4,3-d]pyrimidines. J. Am. Chem. Soc. 98 (16), 4736–4745. doi:10.1021/ja00432a008
Cohen, A., Gudas, L. J., Ammann, A. J., Staal, G. E., and Martin, D. W. (1978). Deoxyguanosine triphosphate as a possible toxic metabolite in the immunodeficiency associated with purine nucleoside phosphorylase deficiency. J. Clin. Investig. 61 (5), 1405–1409. doi:10.1172/JCI109058
Dapp, M. J., Bonnac, L., Patterson, S. E., and Mansky, L. M. (2014). Discovery of novel ribonucleoside analogs with activity against human immunodeficiency virus type 1. J. Virol. 88 (1), 354–363. doi:10.1128/JVI.02444-13
De Clercq, E. (2016). C-Nucleosides to be revisited. J. Med. Chem. 59 (6), 2301–2311. doi:10.1021/acs.jmedchem.5b01157
Duffy, K., Arangundy-Franklin, S., and Holliger, P. (2020). Modified nucleic acids: replication, evolution, and next-generation therapeutics. BMC Biol. 18, 112. doi:10.1186/s12915-020-00803-6
Dumitru, R. V., and Ragsdale, S. W. (2004). Mechanism of 4-(β-D-Ribofuranosyl)aminobenzene 5′-phosphate synthase, a key enzyme in the methanopterin biosynthetic pathway. J. Biol. Chem. 279 (38), 39389–39395. doi:10.1074/jbc.M406442200
Dunwell, J. M., Khuri, S., and Gane, P. J. (2000). Microbial relatives of the seed storage proteins of higher plants: conservation of structure and diversification of function during evolution of the cupin superfamily. Microbiol. Mol. Biol. Rev. 64 (1), 153–179. doi:10.1128/MMBR.64.1.153-179.2000
Ehrlich, J. I., and Schramm, V. L. (1994). Electrostatic potential surface analysis of the transition state for AMP nucleosidase and for formycin 5’-phosphate, a transition-state inhibitor. Biochemistry 33 (30), 8890–8896. doi:10.1021/bi00196a005
Elstner, E. F., and Suhadolnik, R. J. (1972). Nucleoside antibiotics. Asymmetric incorporation of glutamic acid and acetate into the maleimide ring of showdomycin by Streptomyces showdoensis. Asymmetric incorporation glutamic acid acetate into maleimide ring showdomycin by Streptomyces showdoensis. Biochem. 11 (14), 2578–2584. doi:10.1021/bi00764a004
Franceschini, S., Fedkenheuer, M., Vogelaar, N. J., Robinson, H. H., Sobrado, P., and Mattevi, A. (2012). Structural insight into the mechanism of oxygen activation and substrate selectivity of flavin-dependent N-hydroxylating monooxygenases. Biochemistry 51 (36), 7043–7045. doi:10.1021/bi301072w
Frieden, C., Kurz, L. C., and Gilbert, H. C. (1980). Adenosine deaminase and adenylate deaminase: comparative kinetic studies with transition state and ground state analog inhibitors. Biochemistry 19 (23), 5303–5309. doi:10.1021/bi00564a024
Gao, S., Liu, H., de Crécy-Lagard, V., Zhu, W., Richards, N. G. J., and Naismith, J. H. (2019). PMP-diketopiperazine adducts form at the active site of a PLP-dependent enzyme involved in formycin biosynthesis. Chem. Commun. 55 (99), 14502–14505. doi:10.1039/c9cc06975e
Gao, S., Radadiya, A., Li, W., Liu, H., Zhu, W., de Crécy-Lagard, V., et al. (2020). Uncovering the chemistry of C-C bond formation in C-nucleoside biosynthesis: crystal structure of a C-glycoside synthase/PRPP complex. Chem. Commun. 56 (55), 7617–7620. doi:10.1039/d0cc02834g
Gilmore, K., Mohamed, R. K., and Alabugin, I. V. (2016). The Baldwin rules: revised and extended. WIREs Comput. Mol. Sci. 6 (5), 487–514. doi:10.1002/wcms.1261
Gong, R., Le, Y., Qin, Y., Price, N. P., He, X., Deng, Z., et al. (2021). Harnessing synthetic biology-based strategies for engineered biosynthesis of nucleoside natural products in actinobacteria. Biotechnol. Adv. 46, 107673. doi:10.1016/j.biotechadv.2020.107673
He, H.-Y., Niikura, H., Du, Y.-L., and Ryan, K. S. (2022). Synthetic and biosynthetic routes to N-N bonds. Chem. Soc. Rev. 51 (8), 2951–3046. doi:10.1039/c7cs0045c
Hedstrom, L. (2009). IMP dehydrogenase: structure, mechanism, and inhibition. Chem. Rev. 109 (7), 2903–2928. doi:10.1021/cr900021w
Ho, M. C., Shi, W., Rinaldo-Matthis, A., Tyler, P. C., Evans, G. B., Clinch, K., et al. (2010). Four generations of transition-state analogues for human purine nucleoside phosphorylase. Proc. Natl. Acad. Sci. 107 (11), 4805–4812. doi:10.1073/pnas.0913439107
Hori, M., Takita, T., Koyama, G., Tadeuchi, T., and Umezawa, H. (1964). A new antibiotic, formycin. J. antibiotics 17, 96–99.
Hoshika, S., Leal, N. A., Kim, M.-Y., Kim, M.-S., Karalkar, N. B., Kim, H.-J., et al. (2019). Hachimoji DNA and RNA: a genetic system with eight building blocks. Science 363 (6429), 884–887. doi:10.1126/science.aat0971
Ishizuka, M., Sawa, T., Hori, S., Takayama, H., Takeuchi, T., and Umezawa, H. (1968). Biological studies on formycin and formycin B. J. Antibiot. (Tokyo) 21 (1), 5–12. doi:10.7164/antibiotics.21.5
Isono, K., and Suhadolnik, R. J. (1977). Biosynthesis of the C-nucleoside, minimycin: asymmetric incorporation of glutamate and acetate into the oxazine ring. J. Antibiot. 30 (3), 272–273. doi:10.7164/antibiotics.30.272
Kappock, T. J., Ealick, S. E., and Stubbe, J. (2000). Modular evolution of the purine biosynthetic pathway. Curr. Opin. Chem. Biol. 4 (5), 567–572. doi:10.1016/s1367-5931(00)00133-2
Kierdaszuk, B., Modrak-Wójcik, A., Wierzchowski, J., and Shugar, D. (2000). Formycin A and its N-methyl analogues, specific inhibitors of E. coli purine nucleoside phosphorylase (PNP): induced tautomeric shifts on binding to enzyme, and enzyme-->ligand fluorescence resonance energy transfer. Biochim. Biophys. Acta, Protein Struct. Mol. Biochimica Biophysica Acta (BBA) - Protein Struct. Mol. Enzym. 1476 (1), 109–128. doi:10.1016/s0167-4838(99)00225-3
Ko, Y., Wang, S.-A., Ogasawara, Y., Ruszczycky, M. W., and Liu, Hw (2017). Identification and characterization of enzymes catalyzing pyrazolopyrimidine formation in the biosynthesis of formycin A. Org. Lett. 19 (6), 11426–11429. doi:10.1021/acs.orglett.7b00355
Kong, L., Xu, G., Liu, X., Wang, J., Tang, Z., Cai, Y. S., et al. (2019). Divergent biosynthesis of C-nucleoside minimycin and indigoidine in bacteria. iScience 22, 430–440. doi:10.1016/j.isci.2019.11.037
Koyama, G., and Umezawa, H. (1965). Formycin B and its relation to formycin. J. antibiotics 18, 175–177.
Krugh, T. R. (1973). Tautomerism of the nucleoside antibiotic formycin, as studied by carbon-13 nuclear magnetic resonance. J. Am. Chem. Soc. 95 (14), 4761–4762. doi:10.1021/ja00795a053
Kunimoto, T., Sawa, T., Wakashiro, T., Hori, M., and Umezawa, H. J. (1971). Biosynthesis of formycin family. J. Antibiot. 24 (4), 253–258. doi:10.7164/antibiotics.24.253
Li, W., Girt, G. C., Radadiya, A., Stewart, J. J. P., Richards, N. G. J., and Naismith, J. H. (2023). Experimental and computational snapshots of C-C bond formation in a C-nucleoside synthase. Open Biol. 13 (1), 220287. doi:10.1098/rsob.220287
Mackman, R. L. (2022). Phosphoramidate prodrugs continue to deliver, the journey of remdesivir (GS-5734) from RSV to SARS-CoV-2. ACS Med. Chem. Lett. 13 (3), 338–347. doi:10.1021/acsmedchemlett.1c00624
Markert, M. L. (1991). Purine nucleoside phosphorylase deficiency. Immunodefic. Rev. 3 (3), 45–81. doi:10.1172/JCI108826
Matsuda, K., Arima, K., Akiyama, S., Yamada, Y., Abe, Y., Suenaga, H., et al. (2022a). A natural dihydropyridazone scaffold generated from a unique substrate for a hydrazine-forming enzyme. J. Am. Chem. Soc. 142 (29), 9083–9086. doi:10.1021/jacs.8b05354
Matsuda, K., Hasebe, F., Shiwa, Y., Kanesaki, Y., Tomita, T., Yoshikawa, H., et al. (2017). Genome mining of amino group carrier protein-mediated machinery: discovery and biosynthetic characterization of a natural product with unique hydrazone unit. ACS Chem. Biol. 12 (1), 124–131. doi:10.1021/acschembio.6b00818
Matsuda, K., Tomita, T., Shin-Ya, K., Wakimoto, T., Kuzuyama, T., Nishiyama, N., et al. (2022b). A natural dihydropyridazinone scaffold generated from a unique substrate for a hydrazine-forming enzyme. J. Am. Chem. Soc. 144 (28), 12954–12960. doi:10.1021/jacs.2c05269
Matsuda, K., and Wakimoto, T. (2024). Bacterial hydrazine biosynthetic pathways featuring cupin/methionyl tRNA synthetase-like enzymes. ChemBioChem e202300874. doi:10.1002/cbic.20230874
McCarty, R. M., Somogyi, A., Lin, G., Jacobsen, N. E., and Bandarian, V. (2009). The deazapurine biosynthetic pathway revealed: in vitro enzymatic synthesis of PreQ0 from guanosine 5’-triphosphate in four steps. Biochemistry 48 (18), 3847–3852. doi:10.1021/bi900400e
Mechulam, Y., Schmitt, E., Maveyraud, L., Zelwer, C., Nureki, O., Yokoyama, S., et al. (1999). Crystal structure of Escherichia coli methionyl-tRNA synthetase highlights species-specific features. J. Mol. Biol. 294 (5), 1287–1297. doi:10.1006/jmbi.1999.3339
Mehta, K. D., and Gupta, R. S. (1985). Involvement of adenosine kinase in the phosphorylation of formycin B in CHO cells. Biochem. Biophysical Res. Commun. 130 (2), 910–917. doi:10.1016/0006-291X(85)90503-0
Mügge, C., Heine, T., Baraibar, A. G., van Berkel, W. J. H., Paul, S. E., and Tischler, D. (2020). Flavin-dependent N-hydroxylating enzymes: distribution and application. Appl. Microbiol. Biotechnol. 104 (15), 6481–6499. doi:10.1007/s00253-020-10705-w
Nakamura, H., Koyama, G., Iitaka, Y., Ono, M., Yagiawa, N., Kondo, S., et al. (1974). Structure of coformycin, an unusual nucleoside of microbial origin. J. Am. Chem. Soc. 96 (13), 4327–4328. doi:10.1021/ja00820a049
Neumann, C. S., Jiang, W., Heemstra, J. R., Gontang, E. A., Kolter, R., and Walsh, C. T. (2012). Biosynthesis of piperazic acid via N5-hydroxy-ornithine in Kutzneria spp. 744. ChemBioChem 13 (7), 972–976. doi:10.1002/cbic.201200054
Ochi, K., Kikuchi, S., Yashima, S., and Eguchi, Y. (1976). Biosynthesis of formycin: incorporation and distribution of labeled compounds into formycin. J. Antibiot. 29 (6), 638–645. doi:10.7164/antibiotics.29.638
Ochi, K., Yashima, S., Eguchi, Y., and Matsushita, K. (1979). Biosynthesis of formycin. Incorporation and distribution of 13C-14C-and 15N-labeled compounds into formycin. J. Biol. Chem. 254 (18), 8819–8824. doi:10.1016/s0021-9258(19)86772-8
Orozco, M., and Luque, F. J. (1995). Theoretical study of the tautomerism and protonation of 7-aminopyrazolopyrimidine in the gas phase and in aqueous solution. J. Am. Chem. Soc. 117 (4), 1378–1386. doi:10.1021/ja00109a023
Palmu, K., Rosenqvist, P., Thapa, K., Ilina, Y., Siitonen, V., Baral, B., et al. (2017). Discovery of the showdomycin gene cluster from Streptomyces showdoensis ATCC 15227 yields insight in the biosynthetic logic of c-nucleoside antibiotics. ACS Chem. Biol. 12 (6), 1472–1477. doi:10.1021/acschembio.7b00078
Paranthaman, S., and Dharmalingam, K. (2003). Intergeneric conjugation in Streptomyces peucetius and Streptomyces sp. Strain C5. Chromosomal integration and expression of recombinant plasmids carrying the chiC gene. Appl. Environ. Microbiol. 69 (1), 84–91. doi:10.1128/AEM.69.1.84-91.2003
Pfeiffer, M., and Nidetzky, B. (2020). Reverse C-glycosidase reaction provides C-nucleotide building blocks of xenobiotic nucleic acids. Nat. Commun. 11 (1), 6270. doi:10.1038/s41467-020-20035-0
Pfeiffer, M., and Nidetzky, B. (2023). C-Ribosylating enzymes in the (bio)synthesis of C-nucleosides and C-glycosylated natural products. ACS Catal. 13 (24), 15910–15938. doi:10.1021/acscatal.3c04436
Pfeiffer, M., and Nidetzky, B. (2023b). Biocatalytic cascade transformations for the synthesis of C-nucleosides and N-nucleoside analogs. Curr. Opin. Biotechnol. 79, 102873. doi:10.1016/j.copbio.2022.102873
Prusiner, P., Brennan, T., and Sundaralingam, M. (1973). Crystal structure and molecular conformation of formycin monohydrates. Possible origin of the anomalous circular dichroic spectra in formycin mono- and polynucleotides. Biochemistry 12 (6), 1196–1202. doi:10.1021/bi00730a028
Rainey, P., and Santi, D. V. (1983). Metabolism and mechanism of action of formycin B in Leishmania. Proc. Natl. Acad. Sci. Proc. Natl. Acad. Sci. 80 (1), 288–292. doi:10.1073/pnas.80.1.288
Rasche, M. E., and White, R. H. (1998). Mechanism for the enzymatic formation of 4-(β-d-Ribofuranosyl)aminobenzene 5‘-phosphate during the biosynthesis of methanopterin. Biochemistry 37 (32), 11343–11351. doi:10.1021/bi973086q
Ren, D., Kim, M., Wang, S.-A., and Liu, H. (2021). Identification of a pyrrole intermediate which undergoes C-glycosidation and autoxidation to yield the final product in showdomycin biosynthesis. Angew. Chem. Int. Ed. 60 (31), 17148–17154. doi:10.1002/anie.202105667
Ren, D., Lee, Y.-H., Wang, S.-A., and Liu, Hw (2022). Characterization of the oxazinomycin biosynthetic pathway revealing the key role of a nonheme iron-dependent monooxygenase. J. Am. Chem. Soc. 144 (24), 10968–10977. doi:10.1021/jacs.2c04080
Ren, D., Ruzczycky, M., Ko, Y., Wang, S.-A., Ogasawara, Y., Kim, W., et al. (2020). Characterization of the coformycin biosynthetic gene cluster in Streptomyces kaniharaensis. Proc. Natl. Acad. Sci. Proc. Natl. Acad. Sci. 117 (19), 10265–10270. doi:10.1073/pnas.2000111117
Ren, D., Wang, S.-A., Ko, Y., Geng, Y., Ogasawara, Y., and Liu, H. (2019). Identification of the C-glycoside synthases during biosynthesis of the pyrazole C-nucleosides formycin and pyrazofurin. Angew. Chem. Int. Ed. 58 (46), 16512–16516. doi:10.1002/anie.201910356
Robins, R. K., Townsend, L. B., Cassidy, F., Gerster, J. F., Lewis, A. F., and Miller, R. L. (1966). Structure of the nucleoside antibiotics formycin, formyein B and laurusin. J. Heterocycl. Chem. 3 (3), 110–114. doi:10.1002/jhet.5570030130
Santos, N. E., Carreira, A. R. F., Silva, V. L. M., and Braga, S. S. (2020). Natural and biomimetic antitumor pyrazoles, a perspective. Molecules 25 (6), 1364. doi:10.3390/molecules25061364
Sawa, T., Fukagawa, Y., Homma, I., Wakashiro, T., Takeuchi, T., Hori, M., et al. (1968). Metabolic conversion of formycin B to formycin A and oxoformycin B in Nocardia interforma. J. Antibiot. 21 (5), 334–339. doi:10.7164/antibiotics.21.334
Schramm, V. L. (2018). Enzymatic transition states and drug design. Chem. Rev. 118 (22), 11194–11258. doi:10.1021/acs.chemrev.8b00369
Sefah, K., Yang, Z., Bradley, K. M., Hoshika, S., Jiménez, E., Zhang, L., et al. (2014). In vitro selection with artificial expanded genetic information systems. Proc. Natl. Acad. Sci. 111 (4), 1449–1454. doi:10.1073/pnas.1311778111
Seley-Radtke, K. L., and Yates, M. K. (2018). The evolution of nucleoside analogue antivirals: a review for chemists and non-chemists. Part 1: early structural modifications to the nucleoside scaffold. Antivir. Res. 154 (6), 66–86. doi:10.1016/j.antiviral.2018.04.004
Setser, J. W., Heemstra, J. R., Walsh, C. T., and Drennan, C. L. (2014). Crystallographic evidence of drastic conformational changes in the active site of a flavin-dependent N-hydroxylase. Biochemistry 53 (38), 6063–6077. doi:10.1021/bi500655q
Shiraishi, T., and Kuzuyama, T. (2019). Recent advances in the biosynthesis of nucleoside antibiotics. J. Antibiot. 72 (12), 913–923. doi:10.1038/s41429-019-0236-2
Sosio, M., Gaspari, E., Iorio, M., Pessina, S., Medema, M. H., Bernasconi, A., et al. (2018). Analysis of the pseudouridimycin biosynthetic pathway provides insights into the formation of C-nucleoside antibiotics. Cell. Chem. Biol. 25 (5), 540–549.e4. doi:10.1016/j.chembiol.2018.02.008
Suhadolnik, R. J., and Reichenbach, N. L. (1981). Glutamate as the common precursor for the aglycon of the naturally occurring C-nucleoside antibiotics. Biochemistry 20 (24), 7042–7046. doi:10.1021/bi00527a042
Sun, Y., He, X., Liang, J., Zhou, X., and Deng, Z. (2009). Analysis of functions in plasmid pHZ1358 influencing its genetic and structural stability in Streptomyces lividans 1326. Appl. Microbiol. Biotechnol. 82 (2), 303–310. doi:10.1007/s00253-008-1793-7
Twigg, F. F., Cai, W., Huang, W., Liu, J., Sato, M., Perez, T. J., et al. (2019). Identifying the biosynthetic gene cluster for triacsins with an N-hydroxytriazene moiety. ChemBioChem 20 (9), 1145–1149. doi:10.1002/cbic.201800762
Waldman, A. J., Ng, T. L., Wang, P., and Balskus, E. P. (2017). Heteroatom-heteroatom bond formation in natural product biosynthesis. Chem. Rev. 117 (8), 5784–5863. doi:10.1021/acs.chemrev.6b00621
Wang, S.-A., Ko, Y., Zeng, J., Geng, Y., Ren, D., Ogasawara, Y., et al. (2019). Identification of the formycin A biosynthetic gene cluster from Streptomyces kaniharaensis illustrates the interplay between biological pyrazolopyrimidine formation and de novo purine biosynthesis. J. Am. Chem. Soc. 141 (15), 6127–6131. doi:10.1021/jacs.9b00241
Watam, A. R., Brettin, T., Davis, J. J., Gerdes, S., Kenyon, R., Machi, D., et al. (2018). Assembly, annotation, and comparative genomics in PATRIC, the all bacterial bioinformatics resource center. Methods Mol. Biol. 1704, 79–101. doi:10.1007/978-1-4939-7463-4_4
White, R. H. (1996). Biosynthesis of methanopterin. Biochemistry 35 (11), 3447–3456. doi:10.1021/bi952308m
White, R. H. (2011). The conversion of a phenol to an aniline occurs in the biochemical formation of the 1-(4-aminophenyl)-1-deoxy-D-ribitol moiety in methanopterin. Biochemistry 50 (27), 6041–6052. doi:10.1021/bi200362w
Xu, G., Kong, L., Gong, R., Xu, L., Gao, Y., Jiang, M., et al. (2018). Coordinated biosynthesis of purine nucleoside antibiotics aristeromycin and coformycin in Actinomycetes. Appl. Environ. Microbiol. 84 (22), 018600–e1918. doi:10.1128/AEM.01860-18
Yadav, Y., Sharma, D., Kaushik, K., Kumar, V., Jha, A., Prasad, A. K., et al. (2019). Synthetic, structural, and anticancer activity evaluation studies on novel pyrazolylnucleosides. Molecules 24 (21), 3922. doi:10.3390/molecules24213922
Zhang, F., Zeng, T., and Wu, R. (2023). QM/MM modeling aided enzyme engineering in natural products biosynthesis. J. Chem. Inf. Model. 63 (16), 5018–5034. doi:10.1021/acs.jcim.3c00779
Zhang, L., Wang, S., Yang, Z., Hoshika, S., Xie, S., Lin, J., et al. (2020a). An aptamer-nanotrain assembled from six-letter DNA delivers doxorubicin selectively to liver cancer cells. Angew. Chem. Int. Ed. 59, 663–668. doi:10.1002/anie.201909691
Zhang, M., Kong, L., Gong, R., Iorio, M., Donadio, S., Deng, Z., et al. (2022). Biosynthesis of C-nucleoside antibiotics in actinobacteria: recent advances and future developments. Microb. Cell. Factories 21 (1), 2. doi:10.1186/s12934-021-01722-z
Zhang, M., Zhang, P., Xu, G., Zhou, W., Gao, Y., Gong, R., et al. (2020b). Comparative investigation into formycin A and pyrazofurin A biosynthesis reveals branch pathways for the construction of C-nucleoside scaffolds. Appl. Environ. Microbiol. 86, 019711. doi:10.1128/AEM.01971-19
Zhao, G., Peng, W., Song, K., Shi, J., Lu, X., Wang, B., et al. (2021). Molecular basis of enzymatic nitrogen-nitrogen formation by a family of zinc-binding cupin enzymes. Nat. Commun. 12, 7205. doi:10.1038/s41467-021-27523-x
Zhao, G., Yao, S., Rothchild, K. W., Liu, T., Liu, Y., Lian, J., et al. (2020). The biosynthetic gene cluster of pyrazofurin A – a C-nucleoside antibiotic with a rare pyrazole. ChemBioChem 21 (5), 645–649. doi:10.1002/cbic.201900449
Zhou, T., Daugherty, M., Grishin, N. V., Osterman, A., and Zhang, H. (2000). Structure and mechanism of homoserine kinase: prototype for the GHMP kinase superfamily. Structure 8 (12), 1247–1257. doi:10.1016/s0969-2126(00)00533-5
Keywords: formycin, C-nucleoside, biosynthesis, gene cluster, enzymology
Citation: Richards NGJ and Naismith JH (2024) The chemistry of Formycin biosynthesis. Front. Chem. Biol 3:1428646. doi: 10.3389/fchbi.2024.1428646
Received: 06 May 2024; Accepted: 24 June 2024;
Published: 12 July 2024.
Edited by:
Adam Offenbacher, East Carolina University, United StatesReviewed by:
Steven Gary Van Lanen, University of Kentucky, United StatesCopyright © 2024 Richards and Naismith. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nigel G. J. Richards, bnJpY2hhcmRzQGZmYW1lLm9yZw==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.